Abstract
Rheumatoid arthritis (RA) is a systematic autoimmune disease, predominantly causing chronic polyarticular inflammation and joint injury of patients. For the treatment of RA, biologic disease-modifying antirheumatic drugs (bDMARDs) have been used to reduce inflammation and to interfere with disease progression through targeting and mediating the immune system. Although the therapeutic effects of bDMARDs in RA patients have been widely reported, whether these drugs also play important roles in T-cell repertoire status is still unclear. We therefore designed the study to identify the role of T-cell repertoire profiles in RA patients with different types of bDMARD treatments. A high-throughput sequencing approach was applied to profile the T-cell receptor beta chain (TCRB) repertoire of circulating T lymphocytes in eight patients given adalimumab (anti-TNF-α) with/without the following use of either rituximab (anti-CD20) or tocilizumab (anti-IL6R). We subsequently analyzed discrepancies in the clonal diversity and CDR3 length distribution as well as usages of the V and J genes of TCRB repertoire and interrogated the association between repertoire diversity and disease activities followed by the treatment of bDMARDs in these RA patients. All groups of patients showed well-controlled DAS28 scores (<2.6) after different treatment regimens of drugs and displayed no significant statistical differences in repertoire diversity, distribution of CDR3 lengths, and usage of V and J genes of TCRB. Nonetheless, a trend between overall TCRB repertoire diversity and disease activity scores in all bDMARD-treated RA patients was observed. Additionally, age was found to be associated with repertoire diversity in RA patients treated with bDMARDs. Through the profiling of the TCR repertoire in RA patients receiving different biologic medications, our study indicated an inverse tendency between TCR repertoire diversity and disease activity after biologic treatment in RA patients.
1. Introduction
Rheumatoid arthritis (RA) is a prevalent chronic systemic autoimmune disease that is characterized by severe synovial inflammation and pannus formation. These processes eventually cause irreversible damage to the normal architecture of bone, cartilage, tendon, and ligament tissue, affecting the structure and function of the entire joint. In many patients, RA may cause psychological disorders, permanent disabilities that impede daily activities, and considerable social and economic burdens. The pathogenesis and molecular etiology of RA are known to be multifactorial and highly complex. Several genetic markers have been identified as risk factors. HLA DRB1 alleles are critical markers that have been reported in several populations [1, 2]. Polymorphisms of chemokine (C-C motif) receptor 6 (CCR6) were reported to be significant in the susceptibility of RA [3, 4]. Additionally, dysregulation of T cells, B cells, antibodies, cytokines, osteoclasts, osteoblasts, amino acid citrullination, periodontal bacterial infection, and environmental factors, such as smoking and diet, are widely believed to be risk factors for rheumatoid arthritis [5, 6].
Conventional synthetic disease-modifying antirheumatic drugs (csDMARDs), such as methotrexate, leflunomide, sulfasalazine, and hydroxychloroquine, have been widely applied in clinical practice to slow disease progression. Moreover, advances in the understanding of disease mechanisms and novel molecular technologies have contributed to the development of new specific modalities for RA treatment, such as targeting of disease-relevant proinflammatory cytokines (e.g., antitumor necrosis factor-α (TNF-α) or anti-interleukin-6 (IL-6)), blocking the binding of antigen-presenting cells (APCs) to T-cell costimulating channels (cytotoxic T-lymphocyte-associated protein 4-immunoglobulin (CTLA4-Ig)), or the use of B-cell depleting agents (anti-CD20 antibody). Development of these biological DMARDs (bDMARDs) has led to dramatic improvements in clinical outcomes for RA patients.
The use of bDMARDs offers alternative and efficacious strategies for RA treatment via immunomodulation of cytokines and receptors. This mode of action subsequently affects the composition and function of the T-cell population. Because different bDMARDs (e.g., adalimumab, rituximab, and tocilizumab) target distinct immune-related molecules and pathways in RA, each therapy impacts T-cell populations in a unique way. For example, Nguyen et al. indicated that the treatment of peripheral blood mononuclear cells (PBMCs) from RA patients with adalimumab, an anti-TNF antibody, induced the IL-2/STAT5 signaling pathway and in vitro expansion of regulatory T cells (Treg), resulting in the decreased functionality and number of Th17 cells [7, 8]. When PBMCs from RA patients were treated with adalimumab, those samples with elevated Treg cells after in vitro stimulation corresponded to patients with better anti-TNF treatment outcomes, suggesting that Treg cell expansion may be useful to predict the outcomes of anti-TNF therapy [9]. By contrast, a substantial decrease of CD4+ T cells was observed after the treatment of RA patients with rituximab, an anti-CD20 antibody that is capable of inducing B-cell depletion. The extent of T-cell depletion was correlated with the response of patients to rituximab [10]. This phenomenon of rituximab-mediated T-cell depletion in RA patients might be explained by rituximab-dependent elimination of CD20+ Th17 cells affecting the overall T-cell population [11]. Subsequently, Lavielle et al. demonstrated that rituximab treatment led to B-cell depletion that would be predicted to accompany the reduction of T cells [12]. In another example, previous studies have reported that tocilizumab, an anti-IL6R antibody that blocks IL-6 signaling, induced fluctuations in the balance of Treg/Th17 cell subsets in PBMCs from RA patients [13, 14]. A further investigation performed by Kikuchi et al. identified an increased level of Treg cells and no significant alteration of Th17 cells in RA patients receiving long-term tocilizumab treatment [15]. The varied effects of bDMARDs on the numbers and function of T cells illustrate the high degree of complexity characterizing the immunomodulatory mechanisms by which bDMARDs control disease in RA.
Advances in next-generation sequencing technologies have enabled deeper profiling of the status and characteristics of the T-cell receptor (TCR) repertoire in patients with diseases such as cancer and autoimmune disorders [16–18]. Recent studies revealed somatic mutations in expended CD8+ T cells in circulating blood and enrichment of an inflammation-associated Treg cell population in both synovial fluid and bloodstream [19, 20]. However, the characteristics of TCR repertoires in the therapeutic outcomes of bDMARD for RA patients are still unclear. In this study, we profiled the TCR repertoire of RA patients to whom different courses of bDMARD therapies were given by high-throughput sequencing of TCR beta chain (TCRB) and interrogated the association of T-cell repertoire diversity with bDMARD therapeutic outcomes. Our results observed a tendency toward TCRB repertoire diversity and disease activities of bDMARD-treated RA patients.
2. Materials and Methods
2.1. Subject Enrollment
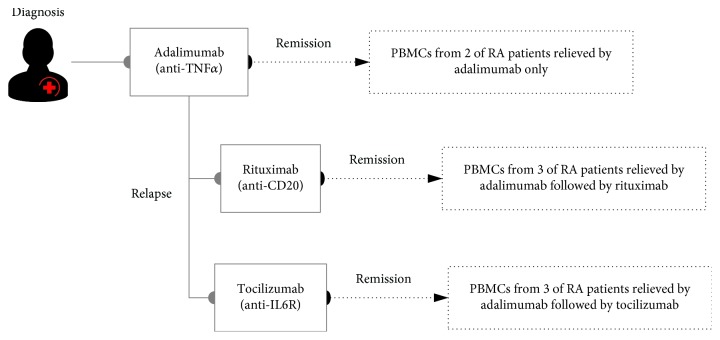
This study was approved by the Joint Institutional Review Board of Taipei Medical University and Taipei Municipal Wanfang Hospital. Informed consents were obtained from all participants. We enrolled eight rheumatoid arthritis (RA) patients who had inadequate response to previous conventional synthetic disease-modifying antirheumatic drug (csDMARD) combination therapy (including methotrexate 15 mg/week with hydroxychloroquine 400 mg/day or sulfasalazine 2000 mg/day) at least for 6 months and then shifted to receive long-term treatment of various biological DMARDs (bDMARDs) from the Outpatient Department of Taipei Municipal Wanfang Hospital. All patients fulfilled the 2010 ACR-EULAR classification criteria for diagnosis of RA [21]. All patients initially received a TNF-α inhibitor (adalimumab) as the first-line bDMARD. Six of the patients who experienced primary or secondary failure (relapse) to a TNF-α blocker were switched to either anti-CD20 (rituximab) or anti-IL6R (tocilizumab) as the second-line bDMARDs. Clinical and laboratory assessments were performed for all patients in the study.
2.2. Sample Preparation
Peripheral blood samples were harvested from RA patients and processed into peripheral blood mononuclear cells (PBMCs) using Histopaque separation as previously described [22]. Briefly, whole blood samples were mixed 1 : 1 with phosphate-buffered saline (PBS) and layered on top of Histopaque (Sigma-Aldrich, MO, USA). PBMCs were harvested from the interface between plasma and Histopaque after low-speed centrifugation and washed with PBS several times according to the manufacturer's instructions. Fresh PBMCs were then processed either to RNA extraction using an RNeasy Mini Kit (Qiagen, Hilden, Germany) or to cryopreservation in liquid nitrogen before proceeding to further procedures.
2.3. TCRB Sequencing
mRNAs from RA patients were synthesized into cDNA using a One-Step RT-PCR Kit (Qiagen, Hilden, Germany), followed by the amplification of the TCRB gene by the use of a Multiplex PCR Kit (Qiagen, Hilden, Germany) and a Human TCR Beta HTBI-M Primer Set (iRepertoire, AL, USA). DNA size selection and purification were carried out using a Pippin Prep System (Sage Science, MA, USA). The quality and quantity of the RNA and cDNA were confirmed using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, MA, USA) and a Qsep100 DNA Analyzer (BiOptic Inc., New Taipei City, Taiwan). The final products for the TCRB repertoire library were sequenced by a 2 × 250 bp MiSeq platform (Illumina Inc., CA, USA).
2.4. TCRB Repertoire Analysis
Raw reads of sequencing data were demultiplexed according to barcode sequences of the iRepertoire primer corresponding to TCRB libraries of different RA patients. For quality filtering, sequencing reads that were less than 150 bp after trimming 3′-adaptor contamination and removing 3′ nucleotides with a Phred quality score less than 30 were discarded. The filtered reads were mapped into V, D, and J gene segments of TCRB using the MiXCR software (version 3.0.3) with default parameters for both sequencing alignment and clonotype assembly [23]. The resulting nucleotide and amino acid sequences of CDR3 of TCRB were determined, and those with out-of-frame sequences were removed from the identified TCRB repertoire. The frequency of each TCRB clonotype was further defined by calculating the cumulative number of the TCRB clonotype with the same CDR3 nucleic/amino acid sequences.
To evaluate whether the sequencing depth was sufficient to determine the majority of the clonotypes of the TCR repertoire, a rarefaction analysis was performed on each sample. A rarefaction curve was plotted based on the numbers of TCR clonotypes at different sequencing depths, which were estimated by random subsampling of increasingly larger subsets of raw sequencing data.
To estimate TCRB repertoire diversity, the inverse Simpson diversity index (1/D) was calculated according to the following formula:
| (1) |
where N is the total number of TCRB clonotypes and p i is the proportion of clonotype i in the TCRB repertoire. To determine the usage of each V/J gene in the TCRB repertoire, the number of each V/J gene assigned to different TCRB clonotypes was summed.
To compare usages of TRBV and TRBJ genes as well as CDR3 lengths of the TCRB repertoire between patients, the repertoire dissimilarity index (RDI) was calculated for each pair of patients according to the methods of Rubelt et al. and Bolen et al. as described previously [24, 25].
2.5. Statistical Analysis
All statistical analyses of the study were carried by using R software and packages from Bioconductor as well as the Comprehensive R Archive Network (CRAN). The correlation between clinical characteristics and the diversity index was performed by linear regression and was evaluated using Pearson's correlation coefficient.
3. Results
3.1. Clinical Features and T-Cell Repertoire Sequencing of bDMARD-Treated RA Patients
We recruited eight RA patients who received different courses of bDMARD therapy—two receiving adalimumab (TNF-α inhibitor) only, three receiving adalimumab followed by rituximab (anti-CD20 antibody), and three receiving adalimumab followed by tocilizumab (anti-IL6R antibody)—and all patients achieved sustained remission after treatment, with the Disease Activity Score of 28-joints (DAS28) less than 2.6 at the time of sample collection (Table 1; Figure 1). Measurements of erythrocyte sedimentation rate (ESR) and C-reactive protein (CRP), which reflect the degree of inflammation and are basis of the DAS28 calculation, showed both indices being in normal ranges (ESR < 20, CRP < 1) in all patients after treatment. Clinical disease activity index (CDAI) and simple disease activity index (SDAI) of all patients after treatment were below 10, corresponding to low activity (CDAI < 10; SDIA < 11) or remission of the disease (CDAI < 2.8; SDAI < 3.3) [26]. We profiled T-cell repertoires of all biologic-treated RA patients using high-throughput sequencing of T-cell receptor β-chain (TCRB) genes. After quality filtering steps and VDJ gene alignment, 1,712,920 ± 695,199 (range: 1,017,720–2,408,119) sequencing reads were mapped into the CDR3 regions of the TCRB gene, while 14,423 ± 11,589 (2,834–26,012) unique TCRB clonotypes were identified among these RA patients (Table S1). The rarefaction curve displayed that all samples are saturated with sufficient sequencing depths (Figure S1).
Table 1.
Clinical characteristics of rheumatoid arthritis patients.
| ID | Gender | Age | Treatment | DAS1 | ESR2 | CRP3 | CDAI4 | SDAI5 |
|---|---|---|---|---|---|---|---|---|
| A1 | Female | 64 | Adalimumab | 2.27 | 5 | 0.1 | 6.5 | 6.6 |
| A2 | Female | 60 | Adalimumab | 1.46 | 6 | 0.1 | 3 | 3.1 |
| R1 | Female | 42 | Rituximab | 1.8 | 11 | 0.5 | 2 | 2.5 |
| R2 | Female | 72 | Rituximab | 2.13 | 14 | 0.1 | 3 | 3.1 |
| R3 | Female | 65 | Rituximab | 2.02 | 6 | 0.2 | 4.5 | 4.7 |
| T1 | Female | 61 | Tocilizumab | 1.96 | 2 | 0.1 | 8.5 | 8.6 |
| T2 | Female | 48 | Tocilizumab | 1.19 | 2 | 0.2 | 3 | 3.2 |
| T3 | Female | 47 | Tocilizumab | 2.2 | 5 | 0.1 | 6 | 6.2 |
1DAS: disease activity score; 2ESR: erythrocyte sedimentation rate; 3CRP: C-reactive protein; 4CDAI: clinical disease activity index; 5SDAI: simple disease activity index.
Figure 1.
A schematic diagram illustrating sample collection from bDMARD-treated RA patients.
3.2. TCR Repertoire Diversity in RA Patients after Treatment with bDMARDs
To evaluate the extent of the clonal expansion of T cells after bDMARD treatment, we first determined the proportions of clonotypes in the TCRB repertoire of RA patients. The cumulative proportions of clonotypes with a frequency of more than 0.001 were similar between therapeutic regimens (adalimumab only: 52.9 ± 4.28% (48.64–57.19%); adalimumab followed by rituximab: 33.37 ± 21.25% (12.12–54.62%); and adalimumab followed by tocilizumab: 32.67 ± 27.45% (5.22–60.12%)). The sums of the top 10 most abundant clonotypes were also similar (adalimumab only: 18.85 ± 13.08% (5.77–31.93%); adalimumab followed by rituximab: 21.42 ± 13.66% (7.75–35.08%); and adalimumab followed by tocilizumab: 10.04 ± 7.32% (2.72–17.36%)) (Figure S2). To further test whether patients treated with different courses of bDMARDs showed divergent degrees of clonal expansion, we compared both frequency criteria among the enrolled patients. However, no significant differences in either criterion were observed among patients with different therapeutic courses of bDMARDs (the Kruskal-Wallis test; p = 0.607 for a frequency of more than 0.001, and p = 0.236 for the top 10 most abundant) (Figure S2).
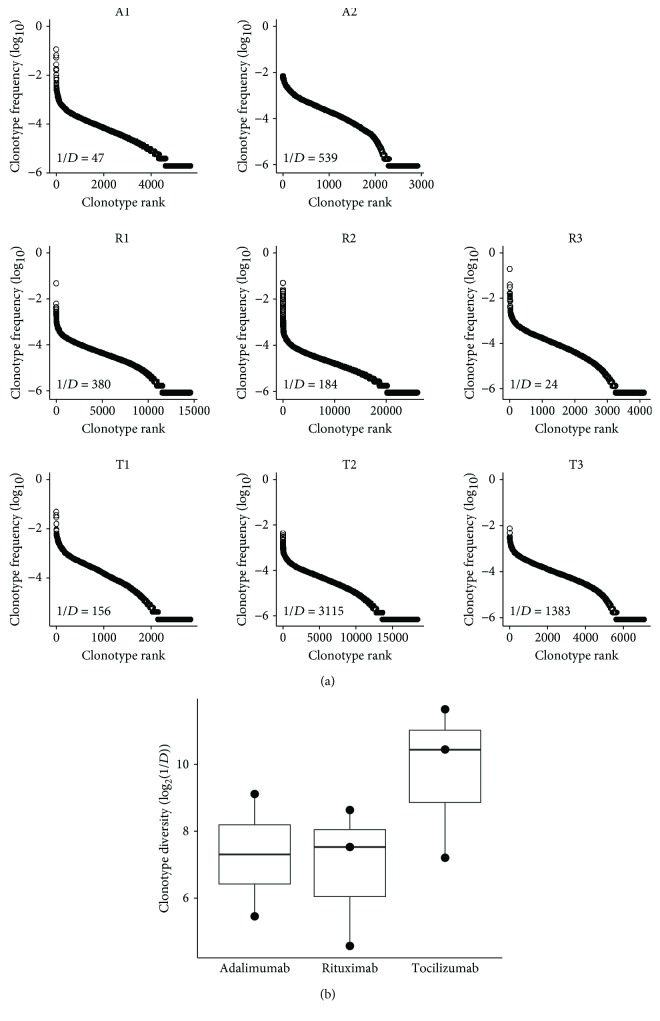
To further explore whether the diversity of the T-cell repertoire varied according to different bDMARD therapies, we next ranked all TCRB clonotypes according to their frequency and assessed the repertoire diversity based on the calculation of the inverse Simpson diversity index (1/D) (Figure 2(a)). The diversity indices appeared to be divergent (adalimumab only: 292.80 ± 246.26 (46.54–539.06); adalimumab followed by rituximab: 202.01 ± 178.32 (23.69–380.33); and adalimumab followed by tocilizumab: 1635.55 ± 1479.8 (155.74–3115.35)); however, statistical analysis showed that there was no significant difference in TCRB repertoire diversity among different bDMARD-treated patients (the Kruskal-Wallis test, p = 0.389) (Figure 2(b)).
Figure 2.
The TCRB repertoire diversity in bDMARD-treated RA patients. (a) TCRB clonotypes of RA patients receiving adalimumab only (A1 and A2), adalimumab followed by rituximab (R1, R2, and R3), and adalimumab followed by tocilizumab (T1, T2, and T3) are represented as dots and ranked according to clonal frequency from high to low. The diversity index of the TCRB repertoire for each patient was calculated as the inverse Simpson diversity index (1/D). (b) The boxplot shows the comparison of TCRB repertoire diversity between RA patients receiving different courses of bDMARD treatment. The upper, middle, and lower lines of the box corresponds to the third quartile, median, and first quartile of the diversity index of patients in each stratified group, respectively.
3.3. Distribution of CDR3 Length in RA Patients after Treatment with bDMARDs
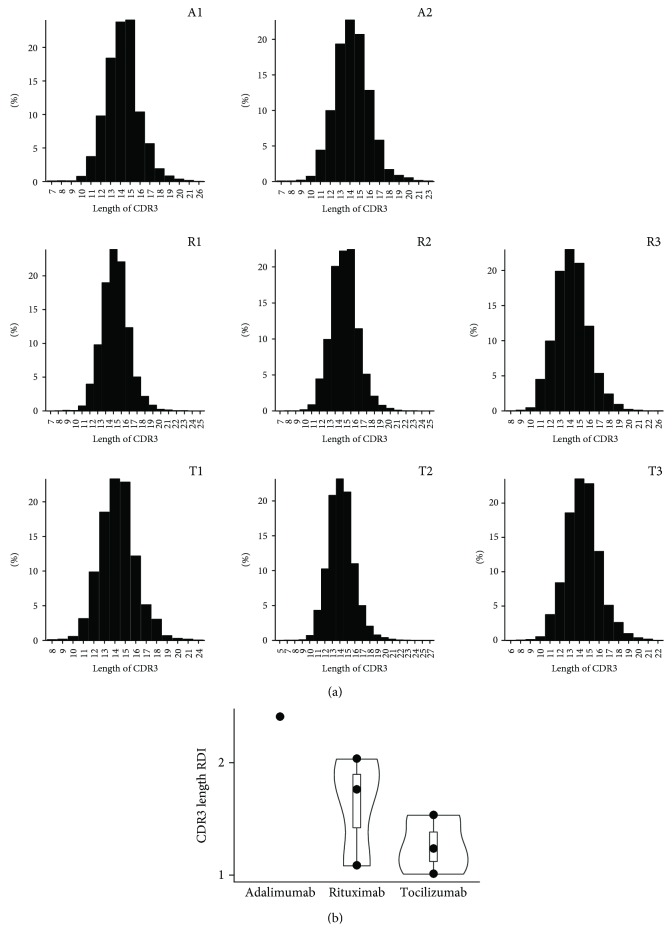
To assess the difference in junction diversity of the TCRB repertoire between different bDMARD regimens, we next depicted the distribution of the complementarity determining region (CDR) 3 lengths of TCR clonotypes in bDMARD-treated RA patients (Figure 3(a)). The length of CDR3 was predominantly distributed from 13 to 15 amino acids with the similar cumulative proportions of this range in all RA patients after bDMARD treatments (adalimumab only: 64.49 ± 1.77% (62.73–66.26%); adalimumab followed by rituximab: 64.3 ± 0.41% (63.88–64.71%); and adalimumab followed by tocilizumab: 64.9 ± 0.29% (64.61–65.18%)) (the Kruskal-Wallis test, p = 0.506). Furthermore, by calculating the repertoire dissimilarity index (RDI) for the CDR3 lengths of each pair of patients given the same regimen, we evaluated the difference in the distribution of the CDR3 lengths between RA patients with different bDMARD treatments. The RDI values between rituximab-treated patients and between tocilizumab-treated patients were relatively higher than between adalimumab-treated patients, but no significant difference in RDI was observed across patients with different bDMARD therapies (the Kruskal-Wallis test, p = 0.208) (Figure 3(b)).
Figure 3.
The CDR3 length of the TCRB repertoire in bDMARD-treated RA patients. (a) The distributions of the CDR3 amino acid lengths of the TCRB clonotypes in RA patients receiving adalimumab only (A1 and A2), adalimumab followed by rituximab (R1, R2, and R3), and adalimumab followed by tocilizumab (T1, T2, and T3) are illustrated. (b) The intragroup repertoire dissimilarity index (RDI) values of TCRB CDR3 lengths between each pair of RA patients are represented.
3.4. V- and J-Gene Usage in RA Patients after Treatment with bDMARDs
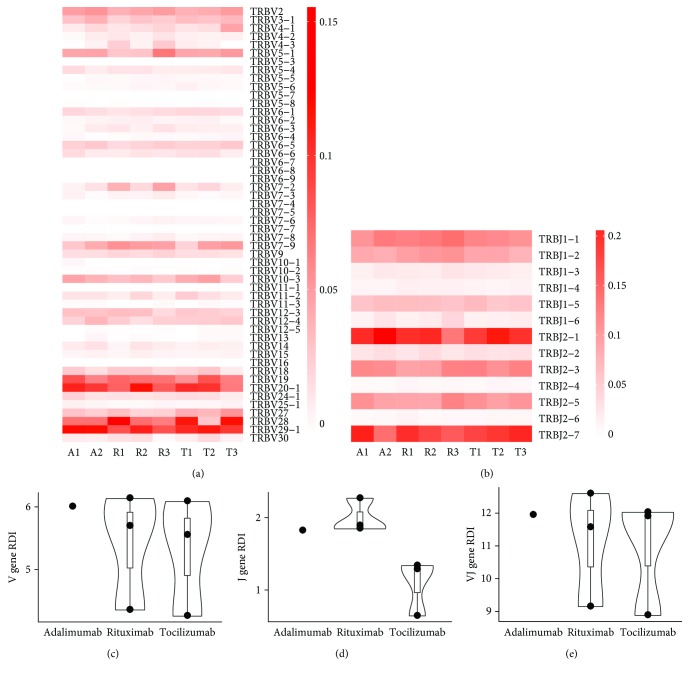
We next assessed the usage of TRBV and TRBJ genes in RA patients treated with different courses of bDMARDs. We observed that the usage rate of TRBV28 was relatively higher in patients treated with adalimumab plus rituximab (1/3) and adalimumab plus tocilizumab (2/3) than in those receiving adalimumab alone (0/2) (Figure 4(a)). However, all patients shared a similar profile of TRBV and TRBJ gene usage, regardless of their bDMARD treatment regimens (Figures 4(a) and 4(b)). To validate this observation, we further calculated the repertoire dissimilarity index (RDI) of TRBV and TRBJ genes for each pair of patients [24]. Our results showed no significant differences in the usage of either TRBV or TRBJ genes among patients with different courses of bDMARDs (the Kruskal-Wallis test, p = 0.751 and p = 0.076 for TRBV and TRBJ, respectively) (Figures 4(c) and 4(d)). In addition, the comparison of RDI for V-J combinations of the TCRB repertoire also indicated a nonsignificant difference across patients with different bDMARD therapies (the Kruskal-Wallis test, p = 0.867) (Figure 4(e)).
Figure 4.
Usages of the TRBV and TRBJ genes of the TCRB repertoire in bDMARD-treated RA patients. The heatmaps illustrate fractions of (a) TRBV and (b) TRBJ genes of RA patients receiving different courses of bDMARD treatment in a color spectrum between white (low) and red (high). The intragroup repertoire dissimilarity index (RDI) values of (c) TRBV, (d) TRBJ, and (e) V-J combinations between each pair of RA patients are illustrated.
3.5. Correlation between TCR Repertoire Diversity and Disease Activity and Age in RA Patients after Treatment of bDMARDs
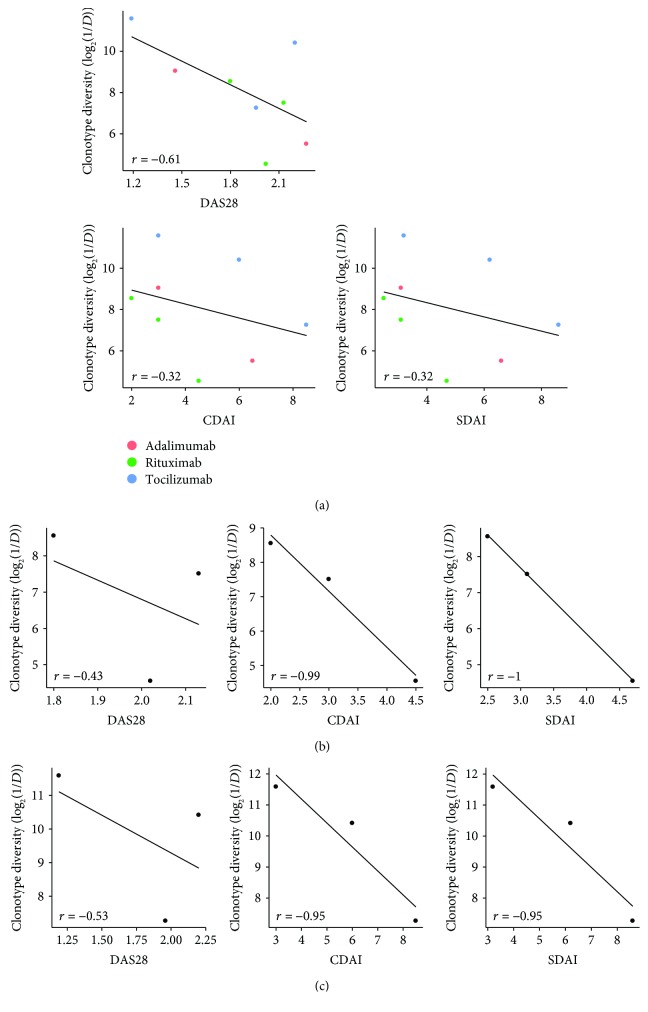
To investigate the potential association between the T-cell repertoire and clinical features in bDMARD-treated RA patients, we next analyzed the correlation between the log-transformed diversity index of the TCRB repertoire and disease activity score. Our results revealed that the reduction of TCRB repertoire diversity showed a trend toward increased DAS28 (r = −0.61, p = 0.11), CDAI (r = −0.32, p = 0.44), and SDAI (r = −0.32, p = 0.44) in all bDMARD-treated RA patients (Figure 5(a)). Based on these findings, we divided RA patients according to bDMARD regimens for further analysis and observed strong negative trends between TCRB repertoire diversity and CDAI and SDAI in patients treated with adalimumab followed by rituximab or tocilizumab (r = −0.99 for CDAI and -1 for SDAI of patients receiving adalimumab plus rituximab; r = −0.95 for both CDAI and SDAI of patients receiving adalimumab plus tocilizumab); however, a decrease of repertoire diversity showed only a moderate trend toward increased DAS28 in these patients (r = −0.43 for patients receiving adalimumab plus rituximab and -0.53 for patients receiving adalimumab plus tocilizumab) (Figures 5(b) and 5(c)).
Figure 5.
Correlation between TCRB repertoire diversity and disease activity in bDMARD-treated RA patients. The TCRB repertoire diversity showed a negative trend toward DAS28, CDAI, and SDAI scores (a) in all bDMARD-treated RA patients, (b) in patients receiving adalimumab followed by rituximab, and (c) in patients receiving adalimumab followed by tocilizumab. The statistical analysis was performed using the Pearson correlation.
We also classified all patients into elder (≥60 years old, median age 64) and younger (<60 years old, median age 47) groups by age at diagnosis and investigated whether age was associated with the diversity of TCRB clonotypes. Interestingly, a tendency toward decreased TCRB repertoire diversity in elder RA patients was observed, regardless of treatment group (Figure S3A). A significant difference in TCRB repertoire diversity was identified between elder and younger groups of bDMARD-treated RA patients (Student's t test, p = 0.036; suggestive significance in the Wilcoxon test, p = 0.071) (Figure S3B). In addition, a borderline significant moderate correlation between age and TCRB repertoire diversity was also found in all bDMARD-treated RA patients (r = −0.68, p = 0.065) (Figure S3C). When dividing patients according to their therapeutic regimens, we observed that an increase of age showed a moderate or a strong tendency toward decreased TCRB repertoire diversity in patients treated with adalimumab followed by rituximab (r = −0.53) or tocilizumab (r = −0.95), respectively (Figure S3D).
4. Discussion
Rheumatoid arthritis is a systemic autoimmune disease characterized by chronic and destructive inflammatory synovitis and multiple organ manifestations that cause severe disability and mortality. Except for conventional synthetic disease-modifying antirheumatic drugs (csDMARDs), biologic DMARDs (bDMARDs) are an evolution of RA treatment. Clinical, laboratorial, or image remission is now a realistic target, achieved by a large proportion of RA patients who are rapidly and appropriately controlled by bDMARDs to halt joint damage and functional disabilities. Autoreactive T cells, B cells (action as professional antigen-presenting cells, antibody-dependent cell-mediated cytotoxicity, and proinflammatory cytokine-producing cells), and inflammatory cytokines such as TNF-α and IL-6 play a pivotal role in the pathological processes of RA through the accumulation of inflammatory cells, production of matrix metalloproteinase, and induction and/or activation of osteoclasts, leading to the destruction of cartilage and bone [27–29]. Many studies have shed light on how bDMARDs affect the size and function of different T-cell subtypes of RA patients at various extents [7–15], but the association between TCR repertoire status and bDMARD treatment and therapeutic outcomes is still unclear and not fully interrogated yet. Deep TCR sequencing is therefore important for characterizing the T-cell repertoire in bDMARD-treated RA patients and for investigating the correlation between repertoire and clinical features. Hence, we performed the first study using the high-throughput sequencing (HTS) method to identify and compare features of the TCR repertoire in RA patients with different bDMARD regimens.
In the study, we profiled the TCRB repertoire in three groups of RA patients given different courses of treatment with bDMARD (adalimumab only, adalimumab followed by rituximab, and adalimumab followed by tocilizumab) by the sequencing of the TCRB CDR3 region. We found that a reduction of the diversity index of the TCRB repertoire showed a visible trend toward increased disease activity scores, including DAS28, CDAI, and SDAI, in bDMARD-treated RA patients, although there was no statistically significant correlation possibly due to the limited sample size in our study. This finding can be reflected by a strong negative correlation between T-cell repertoire diversity and disease activity (as a measurement of DAS28) in RA patients as demonstrated previously [30]. When we further compared our data with the TCR profiling of healthy individuals in a Han Chinese cohort, we observed a nonsignificantly statistical difference in TCRB repertoire diversity (the inverse Simpson diversity index) between bDMARD-treated well-controlled RA patients and the healthy population. We therefore inferred a recovery of the T-cell repertoire in these patients after biologic therapy based on our observation and results from previous studies showing that CD4+ T-cell repertoire diversity in RA patients was reduced compared with healthy individuals and was increased after treatment [31–34]. Taken together, we suggested that the minor difference in disease activity between RA patients was linked to a divergence of their T-cell repertoire diversity, notwithstanding that these patients achieved remission/low disease activity and had TCRB repertoire diversities similar to those of healthy individuals after different courses of bDMARD treatment. Thus, the diversity of the T-cell repertoire might provide important clues for the effects of bDMARDs on the prognosis of RA patients.
We noticed that among RA patients treated with bDMARDs, those given adalimumab followed by tocilizumab had a higher diversity of the TCRB repertoire, implying better restoration of the T-cell repertoire in these patients [20, 35]. Additionally, tocilizumab-treated patients displayed a greater intragroup similarity of TRBJ gene usage compared to those receiving other therapeutic courses of bDMARDs. The difference in the V/J gene usage of the TCRB repertoire may suggest a distinct pattern of TCR rearrangement or selection under the effects of tocilizumab among bDMARD-treated patients. However, further investigation is needed for validation. Since age has been reported as a crucial factor shaping the constitution of the TCR repertoire across the lifespan [36, 37], we also evaluated the correlation between repertoire diversity and age of bDMARD-treated RA patients in our cohort. Our results showed that TCRB repertoire diversity was significantly lower in elder patients (≥60 years old) than in younger patients after bDMARD therapies, which was in line with previous findings indicating the association of the clonality index of the TCRB repertoire with age in healthy individuals and RA patients [20, 36].
Our study was constrained by a small sample size, which may limit the statistical power for detecting the variation of the T-cell repertoire in clinical assessments. Nevertheless, we still observed a trend toward a decrease of TCRB repertoire diversity and an increase of disease activity in biologic-treated RA patients; thus, we expected that the finding may be validated in a larger cohort study. Another limitation in this study was a lack of TCR repertoire profiles of RA patients before bDMARD therapies, which restricted the investigation of T-cell repertoire changes during the biologic treatment. Although our results and others' findings together suggested a potential recovery status of the T-cell repertoire in RA patients after therapy, comprehensively profiling the TCR repertoire of bDMARD-treated patients before and after treatment is recommended for a concrete conclusion regarding the effects of different bDMARDs on the constitution of the T-cell repertoire.
In summary, as the characterization of the T-cell repertoire is important for interrogating the pathogenesis and therapeutic actions of rheumatoid arthritis, we profiled the TCRB repertoire in bDMARD-treated RA patients in a comprehensive sequencing manner and analyzed the correlation between the repertoire diversity and clinical outcomes following the treatment of bDMARDs. We observed that a reduction of TCRB repertoire diversity had a tendency toward elevated disease activities after bDMARD therapies. Our findings provided a better understanding of the role of aging and the adaptive immune repertoire in the bDMARD treatment for RA patients.
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology, Taiwan (105-2628-B-038-001-MY4), Taipei Veterans General Hospital (V107D37-003-MY3-1), and Taipei Medical University (105TMU-CIT-02-3 and 106TMU-CIT-02-3).
Contributor Information
Hsien-Tzung Liao, Email: htliao@vghtpe.gov.tw.
Wei-Chiao Chang, Email: wcc@tmu.edu.tw.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Ethical Approval
The study was conducted with approval from the Taipei Medical University Joint Institutional Review Board (TMU-JIRB). The approval number is TMU-JIRB 201409020, registered on 19 December 2014.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Hsien-Tzung Liao and Wei-Chiao Chang contributed equally to this work.
Supplementary Materials
Table S1: summary of sequencing results in bDMARD-treated patients with rheumatoid arthritis. Figure S1: rarefaction curves of a number of clonotypes in bDMARD-treated RA patients based on subsampled TCR-seq reads with different depths. A1 and A2: patient receiving adalimumab only; R1, R2, and R3: patient receiving adalimumab followed by rituximab; T1, T2, and T3: patient receiving adalimumab followed by tocilizumab. Figure S2: the clonality of the TCRB repertoire in bDMARD-treated RA patients. The fraction of TCRB clones with a clonal frequency of more than 0.001 in RA patients receiving adalimumab only (A1 and A2), adalimumab followed by rituximab (R1, R2, and R3), and adalimumab followed by tocilizumab (T1, T2, and T3) were illustrated with different colors. Figure S3: correlation between TCRB repertoire diversity and age in bDMARD-treated RA patients. (A) RA patients were classified into elder and younger groups and ranked by repertoire diversity. (B) The boxplot showed the comparison of repertoire diversity between elder and younger groups of RA patients. An increase of age displayed a tendency toward a decrease of repertoire diversity (C) in all bDMARD-treated RA patients and (D) in patients receiving adalimumab followed by rituximab/tocilizumab. The statistical analysis was performed using the Pearson correlation.
References
- 1.Wakitani S., Murata N., Toda Y., et al. The relationship between HLA-DRB1 alleles and disease subsets of rheumatoid arthritis in Japanese. British Journal of Rheumatology. 1997;36(6):630–636. doi: 10.1093/rheumatology/36.6.630. [DOI] [PubMed] [Google Scholar]
- 2.del Rincon I., Escalante A. HLA-DRB1 alleles associated with susceptibility or resistance to rheumatoid arthritis, articular deformities, and disability in Mexican Americans. Arthritis and Rheumatism. 1999;42(7):1329–1338. doi: 10.1002/1529-0131(199907)42:7<1329::AID-ANR5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 3.Kochi Y., Okada Y., Suzuki A., et al. A regulatory variant in CCR6 is associated with rheumatoid arthritis susceptibility. Nature Genetics. 2010;42(6):515–519. doi: 10.1038/ng.583. [DOI] [PubMed] [Google Scholar]
- 4.Chang W. C., Woon P. Y., Wei J. C., et al. A single-nucleotide polymorphism of CCR6 (rs3093024) is associated with susceptibility to rheumatoid arthritis but not ankylosing spondylitis, in a Taiwanese population. The Journal of Rheumatology. 2012;39(8):1765–1766. doi: 10.3899/jrheum.120040. [DOI] [PubMed] [Google Scholar]
- 5.Klareskog L., Gregersen P. K., Huizinga T. W. Prevention of autoimmune rheumatic disease: state of the art and future perspectives. Annals of the Rheumatic Diseases. 2010;69(12):2062–2066. doi: 10.1136/ard.2010.142109. [DOI] [PubMed] [Google Scholar]
- 6.Deane K. D., Demoruelle M. K., Kelmenson L. B., Kuhn K. A., Norris J. M., Holers V. M. Genetic and environmental risk factors for rheumatoid arthritis. Best Practice & Research Clinical Rheumatology. 2017;31(1):3–18. doi: 10.1016/j.berh.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGovern J. L., Nguyen D. X., Notley C. A., Mauri C., Isenberg D. A., Ehrenstein M. R. Th17 cells are restrained by Treg cells via the inhibition of interleukin-6 in patients with rheumatoid arthritis responding to anti-tumor necrosis factor antibody therapy. Arthritis and Rheumatism. 2012;64(10):3129–3138. doi: 10.1002/art.34565. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen D. X., Ehrenstein M. R. Anti-TNF drives regulatory T cell expansion by paradoxically promoting membrane TNF-TNF-RII binding in rheumatoid arthritis. The Journal of Experimental Medicine. 2016;213(7):1241–1253. doi: 10.1084/jem.20151255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen D. X., Cotton A., Attipoe L., Ciurtin C., Dore C. J., Ehrenstein M. R. Regulatory T cells as a biomarker for response to adalimumab in rheumatoid arthritis. The Journal of Allergy and Clinical Immunology. 2018;142(3):978–980.e9. doi: 10.1016/j.jaci.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melet J., Mulleman D., Goupille P., Ribourtout B., Watier H., Thibault G. Rituximab-induced T cell depletion in patients with rheumatoid arthritis: association with clinical response. Arthritis and Rheumatism. 2013;65(11):2783–2790. doi: 10.1002/art.38107. [DOI] [PubMed] [Google Scholar]
- 11.Eggleton P., Bremer E. Direct and indirect rituximab‐induced T cell depletion: comment on the article by Mélet et al. Arthritis & Rheumatology. 2014;66(4):p. 1053. doi: 10.1002/art.38347. [DOI] [PubMed] [Google Scholar]
- 12.Lavielle M., Mulleman D., Goupille P., et al. Repeated decrease of CD4+ T-cell counts in patients with rheumatoid arthritis over multiple cycles of rituximab treatment. Arthritis Research & Therapy. 2016;18(1):p. 253. doi: 10.1186/s13075-016-1152-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samson M., Audia S., Janikashvili N., et al. Brief report. Inhibition of interleukin-6 function corrects Th17/Treg cell imbalance in patients with rheumatoid arthritis. Arthritis and Rheumatism. 2012;64(8):2499–2503. doi: 10.1002/art.34477. [DOI] [PubMed] [Google Scholar]
- 14.Pesce B., Soto L., Sabugo F., et al. Effect of interleukin-6 receptor blockade on the balance between regulatory T cells and T helper type 17 cells in rheumatoid arthritis patients. Clinical and Experimental Immunology. 2013;171(3):237–242. doi: 10.1111/cei.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kikuchi J., Hashizume M., Kaneko Y., Yoshimoto K., Nishina N., Takeuchi T. Peripheral blood CD4(+)CD25(+)CD127(low) regulatory T cells are significantly increased by tocilizumab treatment in patients with rheumatoid arthritis: increase in regulatory T cells correlates with clinical response. Arthritis Research & Therapy. 2015;17(1):p. 10. doi: 10.1186/s13075-015-0526-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedensohn S., Khan T. A., Reddy S. T. Advanced methodologies in high-throughput sequencing of immune repertoires. Trends in Biotechnology. 2017;35(3):203–214. doi: 10.1016/j.tibtech.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 17.Rosati E., Dowds C. M., Liaskou E., Henriksen E. K. K., Karlsen T. H., Franke A. Overview of methodologies for T-cell receptor repertoire analysis. BMC Biotechnology. 2017;17(1):p. 61. doi: 10.1186/s12896-017-0379-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heather J. M., Ismail M., Oakes T., Chain B. High-throughput sequencing of the T-cell receptor repertoire: pitfalls and opportunities. Briefings in Bioinformatics. 2018;19(4):554–565. doi: 10.1093/bib/bbw138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossetti M., Spreafico R., Consolaro A., et al. TCR repertoire sequencing identifies synovial Treg cell clonotypes in the bloodstream during active inflammation in human arthritis. Annals of the Rheumatic Diseases. 2017;76(2):435–441. doi: 10.1136/annrheumdis-2015-208992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savola P., Kelkka T., Rajala H. L., et al. Somatic mutations in clonally expanded cytotoxic T lymphocytes in patients with newly diagnosed rheumatoid arthritis. Nature Communications. 2017;8(1, article 15869) doi: 10.1038/ncomms15869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aletaha D., Neogi T., Silman A. J., et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis & Rheumatism. 2010;62(9):2569–2581. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 22.Wong H. S., Chang C. M., Kao C. C., et al. V-J combinations of T-cell receptor predict responses to erythropoietin in end-stage renal disease patients. Journal of Biomedical Science. 2017;24(1):p. 43. doi: 10.1186/s12929-017-0349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolotin D. A., Poslavsky S., Mitrophanov I., et al. MiXCR: software for comprehensive adaptive immunity profiling. Nature Methods. 2015;12(5):380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 24.Rubelt F., Bolen C. R., McGuire H. M., et al. Individual heritable differences result in unique cell lymphocyte receptor repertoires of naïve and antigen-experienced cells. Nature Communications. 2016;7(1, article 11112) doi: 10.1038/ncomms11112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolen C. R., Rubelt F., Vander Heiden J. A., Davis M. M. The Repertoire Dissimilarity Index as a method to compare lymphocyte receptor repertoires. BMC Bioinformatics. 2017;18(1):p. 155. doi: 10.1186/s12859-017-1556-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aletaha D., Smolen J. S. The Simplified Disease Activity Index (SDAI) and Clinical Disease Activity Index (CDAI) to monitor patients in standard clinical care. Best Practice & Research Clinical Rheumatology. 2007;21(4):663–675. doi: 10.1016/j.berh.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 27.McInnes I. B., Schett G. The pathogenesis of rheumatoid arthritis. The New England Journal of Medicine. 2011;365(23):2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 28.Furst D. E., Emery P. Rheumatoid arthritis pathophysiology: update on emerging cytokine and cytokine-associated cell targets. Rheumatology. 2014;53(9):1560–1569. doi: 10.1093/rheumatology/ket414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burmester G. R., Kivitz A. J., Kupper H., et al. Efficacy and safety of ascending methotrexate dose in combination with adalimumab: the randomised CONCERTO trial. Annals of the Rheumatic Diseases. 2015;74(6):1037–1044. doi: 10.1136/annrheumdis-2013-204769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakurai K., Ishigaki K., Shoda H., et al. HLA-DRB1 shared epitope alleles and disease activity are correlated with reduced T cell receptor repertoire diversity in CD4+ T cells in rheumatoid arthritis. The Journal of Rheumatology. 2018;45(7):905–914. doi: 10.3899/jrheum.170909. [DOI] [PubMed] [Google Scholar]
- 31.Even J., Lim A., Puisieux I., et al. T-cell repertoires in healthy and diseased human tissues analysed by T-cell receptor beta-chain CDR3 size determination: evidence for oligoclonal expansions in tumours and inflammatory diseases. Research in Immunology. 1995;146(2):65–80. doi: 10.1016/0923-2494(96)80240-9. [DOI] [PubMed] [Google Scholar]
- 32.Wagner U. G., Koetz K., Weyand C. M., Goronzy J. J. Perturbation of the T cell repertoire in rheumatoid arthritis. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(24):14447–14452. doi: 10.1073/pnas.95.24.14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishigaki K., Shoda H., Kochi Y., et al. Quantitative and qualitative characterization of expanded CD4+ T cell clones in rheumatoid arthritis patients. Scientific Reports. 2015;5(1, article 12937) doi: 10.1038/srep12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imberti L., Scarsi M., Zanotti C., et al. Reduced T-cell repertoire restrictions in abatacept-treated rheumatoid arthritis patients. Journal of Translational Medicine. 2015;13(1):p. 12. doi: 10.1186/s12967-014-0363-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Britanova O. V., Putintseva E. V., Shugay M., et al. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. Journal of Immunology. 2014;192(6):2689–2698. doi: 10.4049/jimmunol.1302064. [DOI] [PubMed] [Google Scholar]
- 36.Qi Q., Liu Y., Cheng Y., et al. Diversity and clonal selection in the human T-cell repertoire. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(36):13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thome J. J., Grinshpun B., Kumar B. V., et al. Long-term maintenance of human naïve T cells through in situ homeostasis in lymphoid tissue sites. Science Immunology. 2016;1(6, article eaah6506) doi: 10.1126/sciimmunol.aah6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: summary of sequencing results in bDMARD-treated patients with rheumatoid arthritis. Figure S1: rarefaction curves of a number of clonotypes in bDMARD-treated RA patients based on subsampled TCR-seq reads with different depths. A1 and A2: patient receiving adalimumab only; R1, R2, and R3: patient receiving adalimumab followed by rituximab; T1, T2, and T3: patient receiving adalimumab followed by tocilizumab. Figure S2: the clonality of the TCRB repertoire in bDMARD-treated RA patients. The fraction of TCRB clones with a clonal frequency of more than 0.001 in RA patients receiving adalimumab only (A1 and A2), adalimumab followed by rituximab (R1, R2, and R3), and adalimumab followed by tocilizumab (T1, T2, and T3) were illustrated with different colors. Figure S3: correlation between TCRB repertoire diversity and age in bDMARD-treated RA patients. (A) RA patients were classified into elder and younger groups and ranked by repertoire diversity. (B) The boxplot showed the comparison of repertoire diversity between elder and younger groups of RA patients. An increase of age displayed a tendency toward a decrease of repertoire diversity (C) in all bDMARD-treated RA patients and (D) in patients receiving adalimumab followed by rituximab/tocilizumab. The statistical analysis was performed using the Pearson correlation.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.