Abstract
Transcription pre-initiation complexes (PIC) are vital assemblies whose function underlies protein gene expression. Cryo-EM advances have begun to uncover their structural organization. Yet, functional analyses are hindered by incompletely modeled regions. Here we integrate all available cryo-EM data to build a practically complete human PIC structural model. This enables simulations that reveal the assembly’s global motions, define PIC partitioning into dynamic communities and delineate how structural modules function together to remodel DNA. We identify key TFIIE–p62 interactions linking core-PIC to TFIIH. P62 rigging interlaces p34, p44 and XPD while capping XPD DNA-binding and ATP-binding sites. PIC kinks and locks substrate DNA, creating negative supercoiling within the Pol II cleft to facilitate promoter opening. Mapping Xeroderma Pigmentosum, Trichothiodystrophy, and Cockayne syndrome disease mutations onto defined communities reveals clustering into three mechanistic classes, affecting TFIIH helicase functions, protein interactions and interface dynamics.
Keywords: Transcription initiation, Molecular dynamics, Gene regulation, Community network analysis, Global protein dynamics, RNA polymerase, ERCC2, ERCC3, NUPR1
Complexes of RNA Polymerase II (Pol II) are foundational for transcription since all mRNA in eukaryotic cells originates from Pol II synthesis.1–4 Additionally, Pol II transcribes most small regulatory non-coding RNAs controlling gene expression levels and acting in gene silencing. As transcription regulation governs all fundamental aspects of cell biology loss of transcriptional control is a hallmark of many autoimmune disorders, cancers, neurological, metabolic and cardiovascular diseases.5–9 To begin transcription, Pol II depends on key general transcription factors (GTFs) that recognize promoter DNA and assemble with the polymerase into a pre-initiation complex (PIC).4,9–12 Subsequently, the initial closed promoter complex (CC) transitions into an open complex (OC), wherein the melted single-stranded DNA template is inserted into the Pol II active site. This transient OC is then converted into an initial transcribing complex (ITC), competent to synthesize mRNA. When the nascent RNA chain grows to a critical length, Pol II clears the promoter and a stable elongation complex (EC) ensues.13,14 Formation of PIC and its conversion into a productive elongation complex are key for transcription regulation.15 Yet, PIC molecular architecture and its associated functional dynamics remain incompletely understood.
With the “resolution revolution” in cryo-electron microscopy, structures of these molecular machines have recently come into view.16,17 Two recent studies achieved near atomic visualization of Pol II core-PICs in multiple states (CC, OC and ITC) and enabled side-by-side comparison of the conformations leading to competent elongation complexes.16,18 Two subsequent studies showed TFIIH structure both in the absence (apo-TFIIH) and in the presence of core-PIC (holo-PIC).17,19 These breakthrough studies elucidated eukaryotic pre-initiation complex architectures; yet, the respective models were incomplete and, therefore, unsuitable as starting points for detailed molecular simulations and analysis.
Here we synthesized all available EM data to produce the most complete atomistic model of the human PIC to date. All previously omitted or unassigned regions have now been modelled into the corresponding EM densities (Figure 1 and Figure S1), including the ten-subunit TFIIH. The PIC assembly was fitted into the holo-PIC EM density. The quality of the new models makes them entirely suitable for molecular dynamics (MD) simulations on massively parallel computing platforms. Thus, we employed large-scale MD simulations to unveil the functional dynamics of Pol II holo- and core-PICs. Our analyses reveal the hierarchical organization of the PIC machinery into dynamic communities and unveil how its interwoven structural elements function together to remodel the DNA substrate and facilitate promoter opening. Strikingly, a mapping of patient-derived TFIIH mutations onto the newly discovered dynamic communities showed that mutations were clustered at critical junctures in the TFIIH dynamic network. Thus, our findings provide new understanding of PIC molecular mechanisms and the etiology of devastating autosomal recessive genetic disorders - Xeroderma pigmentosum (XP, cancer), trichothiodystrophy (TTD, aging) and Xeroderma pigmentosum/Cockayne syndrome (XP/CS, development, cancer). Importantly, our methods and models provide a roadmap for future structural, biochemical and mutational experiments to understand the interplay between TFIIH structural disruption and the complex XP, XP/CS and TTD disease phenotypes.20–25
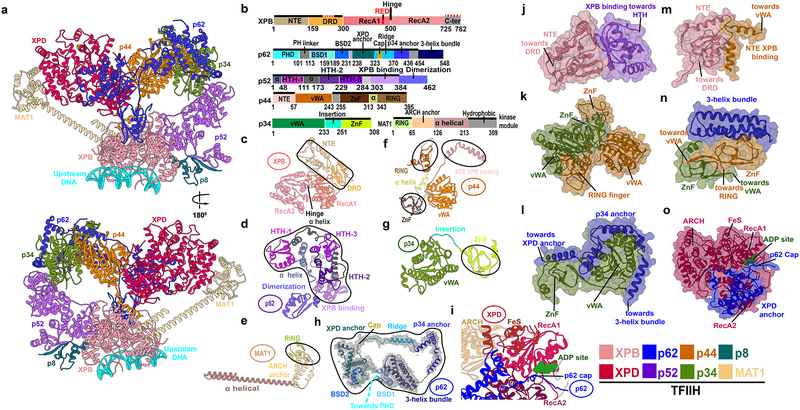
Figure 1. TFIIH integrative structure model based on comparative analysis of cryo-EM densities reveals “molecular rigging” formed by p62 and p44.
a, Anterior and posterior cartoon views of TFIIH GTF where missing regions or entire domains and proteins are built for XPB, p62, p52, p44, p34 and MAT1. Newly modeled p62 and p44 subunits act as molecular rigging, interlinking TFIIH. b, Motif schematic highlighting newly modeled regions (solid black lines). Two small regions in XPB, not present in the EM maps, were not modeled (red dashed lines). Abbreviations denote: DRD - damage recognition domain; NTE - N-terminal extension; HTH- helix-turn-helix. c-h, Cartoon of TFIIH subunits with newly modeled regions circled. h, Representative cryo-EM electron density from apo-TFIIH overlaid with p62 (PHD domain not shown). i, Zoom view of p62 cap region overlaying the XPD ATP binding cleft. Space filling views highlighting Interactions newly modeled in j, XPB NTE and p52; k, p34 and p44; l, p62 helices and p34; m, XPB N-terminus with p44; n, p62 3-helix bundle and p34 plus p44; and o, p62 and XPD.
Results
The molecular architecture of TFIIH underlies its role in the human PIC.
Promoter DNA opening by Pol II and the formation of the nascent transcription bubble critically depend on the transcription factor TFIIH.5,26–29 Specifically, a mechanism has been proposed wherein TFIIH-induced DNA translocation toward Pol II creates negative DNA supercoiling inside the polymerase cleft to facilitate promoter opening.16,18,30–32 To unravel the functional role of TFIIH, we first constructed a suitably complete model of human apo-TFIIH. Model building was based on comparative analysis of cryo-EM densities for apo-TFIIH (EMDB accession code: EMD-3802)19 and yeast core-PIC–TFIIH–DNA (EMDB accession code: EMD-3846).17 To guide our initial TFIIH model into the holo-PIC cryo-EM density, we employed the cascade MDFF method.33 TFIIH and core-PIC were separately flexibly fitted into the closed-state human holo-PIC density (EMDB accession code: EMD-3307)16 and then combined to assemble the full PIC–TFIIH–DNA complex.
The resulting structural model (Figure 1) reveals newly modelled TFIIH regions that are demarcated in Figure 1b and Table S1, indicating >95% completeness. TFIIH encompasses ten protein subunits.34 Seven subunits form the TFIIH core (Figure 1a and Video S1). Two helicase subunits, XPB and XPD, are adjacent while four intermediate subunits (p8, p52, p34, p44) lie in a characteristic horseshoe shape. The p62 subunit is the most extended: it traverses and interlaces the surfaces of p34, p44 and XPD. The MAT1 subunit connects the XPB DNA-damage recognition domain (DRD) to the XPD ARCH domain via an 86-Å long α-helix and a helical bundle (Figure 1a, 1b, 1e, and Figure S1a). The remaining three TFIIH subunits (CDK7, Cyclin-H and part of MAT1) form the flexible kinase (CAK) subcomplex, which is positioned away from the TFIIH core (Figure S2, Video S2 and Supplementary Notes) and is key for transcription regulation.35,36
Two subunits central to TFIIH’s function37,38, XPB and XPD, possess independent helicase and ATP-hydrolysis activities.39–42 Yet, in transcription XPD serves a structural role and its helicase activity is suppressed. In contrast, XPB engages promoter DNA downstream of the transcription start site (TSS) (Figure 1a, Figure 2 and Video S1), and its ATPase activity is obligatory for Pol II initiation. Human XPB features two RecA-like lobes (RecA1 and RecA2), the DRD, and an N-terminal extension domain (NTE) (Figure 1b,1c and Figure S1b). The DRD and NTE domains were built de novo after tracing the entire length of XPB in the apo-TFIIH density. The DRD domain recognizes distortions in DNA41 and may act in DNA damage detection. Not surprisingly, it has structural similarity to the mismatch recognition domain (MRD) of MutS–MSH43 and the SMARCAL1 HARP domain.44 The NTE domain, important for anchoring XPB within TFIIH34, consists of three α-helices and five β-strands (residues 1–159) that contact the XPB-interacting domain of p52 (Figure 1c,1d and Figure S1h). Human disease mutations map to the NTE, supporting TFIIH’s functional significance not only for transcription but also for nucleotide excision repair (NER).
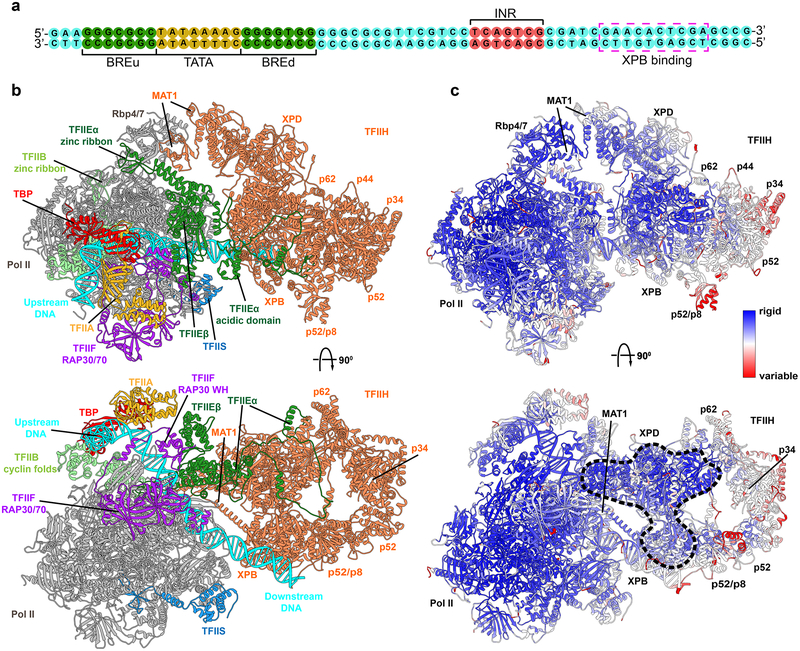
Figure 2. TFIIE links human PIC to TFIIH.
a, Sequence of the DNA substrate in PIC. b, Cartoon of human PIC structure including TFIIH highlighting non-TFIIH subunits. Most striking is TFIIE which crosses over a quarter of TFIIH. Model is based on integration and comparative analysis of cryo-EM densities for human apo-TFIIH, human closed-state holo-PIC density, and yeast core-PIC–TFIIH–DNA c, Computed B-factors mapped onto the PIC-TFIIH structural model with values colored from high (red) to low (blue) reveal a network of stable interactions. Black dashed outline highlights an unexpected rigid anchor region between TFIIH and PIC.
TFIIE, MAT1 and p62 are critical for the integrity of the core-PIC-TFIIH interface.
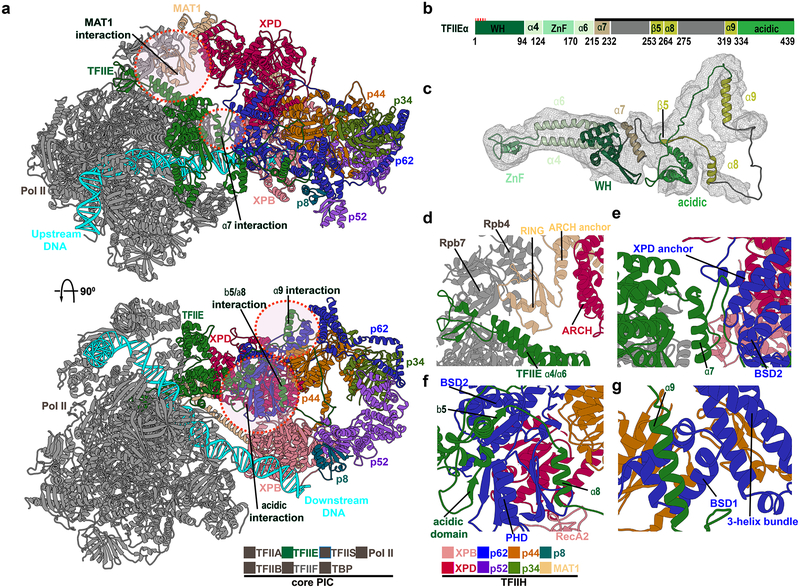
In Figure 2, TFIIH is revealed in the context of the complete PIC assembly. Notably, holo-PIC has a bipartite architecture with Pol II Rpb4/7, TFIIE, p62 and MAT1 principally responsible for the interface between core-PIC and TFIIH (Figure 3a, 3d and 3e; Table S2). Specifically, the MAT1 RING domain lodges in-between the Rpb4 and Rpb7 chains of Pol II while also contacting α4 and α6 helices of TFIIE. MAT1 ARCH anchor domain lies between the ARCH domain of XPD and Rpb4, stabilizing the TFIIH - core-PIC interface. Our model also highlights the critical role of p62. We built the entire length of p62 by comparing the human and yeast EM densities. We furthermore confirmed positional assignments of all p62 domains (N-terminal plekstrin homology domain (PHD), two BSD domains - BSD1 and BSD2, XPD–p34 anchor and C-terminal 3-helix bundle) by matching our model to existing chemical cross-linking data19,34 (Figure 1b, 1h, 1n, 1l, 1o, Figure S1d, S1k, S1l and S1m). Importantly, two domains from p62 (BSD2 and PHD) directly bind TFIIE through its α7 helix and adjacent loops (Figure 3e). Interfacial interactions include β-sheet formation (e.g. p62 PHD domain forms a beta sheet with the TFIIE acidic patch; Figure 3f) to strengthen the interface. Interestingly, we found that yeast and human PIC differ in the contacts at the TFIIH core-PIC juncture. In yeast, the PHD domain extends to make direct contact with the Pol II core.17 An analogous interaction in human PIC is impossible as the linker leading into the PHD domain is shortened by >50 residues (Figure S3). Instead, the PHD domain is unambiguously positioned between the XPB and XPD subunits in all existing holo-PIC EM reconstructions.16 Crosslinking data34 also supports this positioning, showing that PHD forms crosslinks to XPB in the human but not in the yeast PIC (Figure S3c and S3d). The linker deletion in human PIC has more subtle effects on the lower half of the TFIIH–core-PIC interface as compared to yeast PIC (Figure S4). Furthermore, our model supports p62 regulatory as well as structural roles: one p62 region (residues 274–293) tracks along the DNA path on XPD, based on a recent XPD ortholog structure45 and another (residues 333–342) caps XPD and could influence DNA binding and ATPase function, respectively (Figure 1i and Figure 1o). Notably, our results highlight a key role of TFIIE (TFIIEα and TFIIEβ) for PIC structural integrity. TFIIEβ is a crucial constituent of core-PIC, forming a cap over the Pol II cleft and making functionally important contacts with TFIIF. TFIIEα, on the other hand, consists primarily of three α-helices (α7, α8, α9) and a β5 strand, which are splayed on the surface of TFIIH and connected by long flexible linkers (Figure 3 and Figure S5). Specifically, α9 binds BSD1 and the 3-helix bundle of p62; α8 interacts with the p62 PHD domain and the α7-BSD2 interaction is critically important for the core-PIC–TFIIH interface. The unusual engagement mode between TFIIEα and TFIIH highlights the key architectural role of TFIIE for assembling the PIC. In essence, TFIIE serves as a structural adhesive to link TFIIH to the rest of the transcription initiation machinery – a finding that supports and extends current understanding of why TFIIE is required for TFIIH recruitment to the PIC.46,47
Figure 3. TFIIE, MAT1 and p62 are critical for the integrity of the core-PIC–TFIIH interface.
a, Human PIC structure in cartoon representation with colored TFIIH subunits. Circles demark zoomed regions in d.-g. b, Domain schematic of TFIIEα. c, TFIIEα cartoon. d, MAT1 - core PIC interaction. The MAT1 RING-finger docks into a groove between the Pol II stalk subunit Rpb7 and TFIIE α4-α7 helices. The RING-finger connects to the ARCH anchor which binds the XPD ARCH domain. e, TFIIEα helix α7 is wedged between TFIIE winged helix (eWH) domain and p62. f, TFIIE β5–α7 and acidic domain interacts with p62 PHD and BSD2. g, TFIIE helix α9 binds p62 BSD1 domain adjacent to the p62 3-helix bundle.
Promoter opening is linked to the global motions and dynamic networks within TFIIH.
Our holo-PIC model provided a starting point for MD simulations aimed at addressing the role of dynamics in driving the multifaceted functional responses of the transcription initiation machinery. We performed ~300-ns simulations of holo-PIC and core-PIC. To begin to dissect the staggering complexity of the simulations, we first tested if the relative rigidity or flexibility of the numerous PIC structural elements was linked to their putative functional roles. Thus, we mapped computed B-factors from the simulation onto the holo-PIC structure (Figure 2c). The core of Pol II (Rpb1 and Rpb2 chains) is the most rigid scaffold within the PIC, and also the best resolved region in cryo-EM (local resolution going down to ~3 Å).16 The low B-factors support the importance of this region, which establishes the path of the DNA substrate and confines it within the Pol II cleft. The DNA duplex upstream of the initiation region (INR; Figure 2a) is also structurally rigid, especially in the TBP-associated TATA box region. The downstream portion of DNA is more mobile, and its mobility is linked to the motion of XPB. Notably, there is a ridge of stability extending across the core-PIC–TFIIH interface starting with the Pol II core, continuing through TFIIE and encompassing core XPD, portion of p62 and lobe1 of XPB. In contrast, the middle domains (p8, p52, p34 and p44) are dynamic and appear to participate in concerted global motions.
To analyze and visualize such global motions, we relied on two methods - principal component analysis (PCA)48 and community network analysis.49 Briefly, PCA is a dimensionality reduction technique that involves three steps: 1) computing the matrix of residue-residue covariances from the MD trajectories; 2) diagonalizing the covariance matrix to yield eigenvectors (principal modes) and eigenvalues (mean square fluctuations); and 3) projecting the trajectory onto the principal modes to yield principal components. The first few principal modes are especially important as they capture the slow, large-amplitude motions that are also the most functionally significant. We focused on the second and third principal modes (denoted PC2 and PC3) (See supplementary material Video S3 and Video S4). PC2 reflects an out-of-plane twisting movement of TFIIH with respect to the Pol II core above the ringed plane of TFIIH defined by the p44, p34, p52 and XPD and XPB subunits. Interestingly, PC3 represents the in-plane swing motion of TFIIH that could push the DNA substrate toward the Pol II cleft, leading to DNA bending and deformation. Although differing in detail, both PC2 and PC3 imply the DNA substrate is rigidly held by Pol II in the TBP region while the downstream DNA duplex is held and pushed by XPB whose motion is directed by rotational movement of the TFIIH lever arm (comprised of p44, p34, p52 and p8).
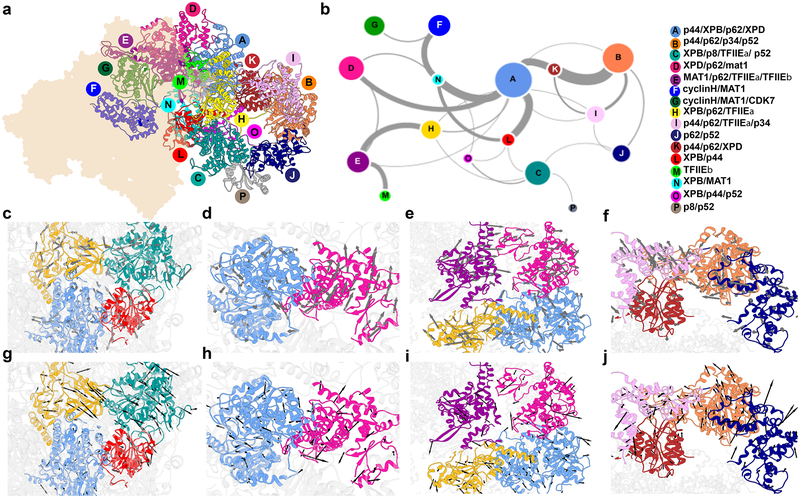
We employed community network analysis to partition PIC into dynamic communities (tightly connected clusters of residues that move together as modules). The PIC assembly was mapped onto a graph wherein each protein residue is a node and edges connect nodes that are in contact. All edges were assigned weights based on the covariance matrix data from the MD simulation. The Girvan-Newman algorithm was then used to subdivide the graph into strongly connected components. The magnitude of allosteric communication between communities was quantified by estimating the total betweenness for all edges that connect individual communities (betweenness is defined as the number of shortest paths that cross an edge). We identified sixteen dynamic communities in TFIIH, which were color-coded and mapped onto the holo-PIC structure (Figure 4a) and graphed the level of dynamic communication between communities (Figure 4b). An important observation from this analysis was that the motions of the two XPB lobes are largely decoupled. Lobe1 (community L) carries much stronger connection to community A (largely comprised of XPD). Lobe2 (community C) appears more closely associated with communities O and J that form the tip of the TFIIH lever arm (subunits p52, p34, p44) and community H that includes part of p62. In PC2 and PC3 lobe2 and p62 fragment from community H move concertedly in the same direction whereas lobe1 exhibits smaller magnitude motions and is most closely correlated to XPD (Figure 4c, 4g). Community network analysis also nicely captures the fact that the motion of XPB lobe1 is coupled to the motion of MAT1 through the long helix (strong connection between communities L and N). The XPD ARCH domain separates into its own community (Figure 4d,4h) while the TFIIE (community E) is in communication with p62 (community H) and the motions of these elements occur in the same direction (Figure 4e, 4i). Figure 4f and 4j capture the directionality and concerted rotational motion of the TFIIH lever arm. Notably, communities I and B and communities B and J are both separated by hinge regions.
Figure 4. Community networks underlying TFIIH functional dynamics.
a, Communities identified from dynamic network analysis that transcend simple subunit divisions. b, Graph of allosteric communication among communities. Colored by community, nodes are sized by number of residues in each community. Thickness of edges between community pairs correlate to magnitude of dynamic communication (betweenness). c-j, Directional community motions (arrows) and magnitude (arrow size) of the corresponding component of the eigenvector for c, A, C, H and L along the second principal component; d. A and D along the second principal component; e, A, D, E and H along the second principal component; f, B, I, J and K along the second principal component; g, A, C, H and L along the third principal component; h, A and D along the third principal component; i, A, D, E and H along the third principal component; and j, B, I, J and K along the third principal component.
PIC global dynamics facilitate substrate DNA bending and deformation.
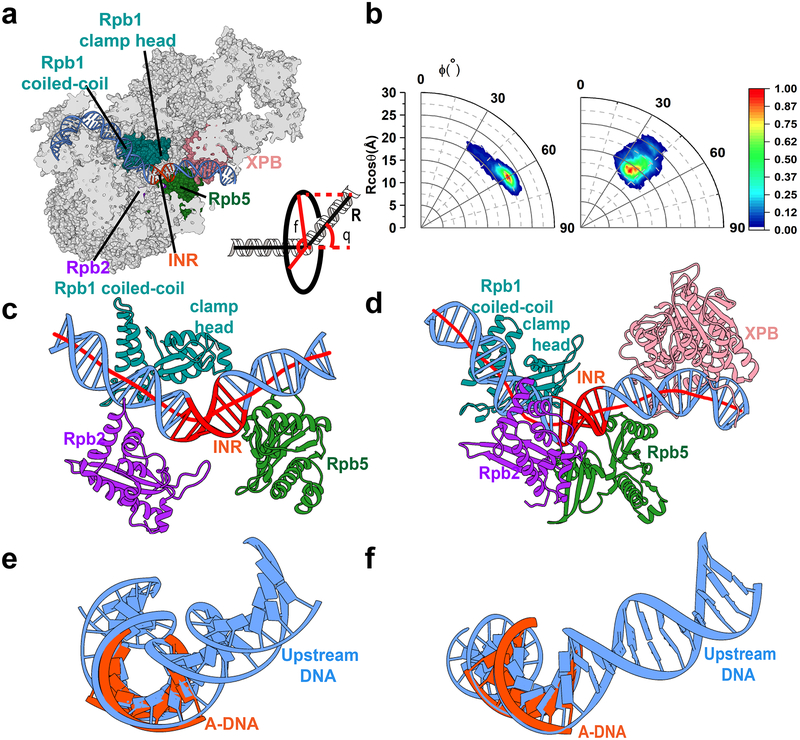
To examine the effect of global dynamics on the DNA substrate, we analyzed the DNA path through Pol II for our holo-PIC and core-PIC simulations (Figure 5). DNA traverses the Pol II cleft undergoing an ~ 90° bend at the position of TBP. The DNA path continues relatively straight between the Pol II Rpb2 and Rpb1 subunits. Interactions with Rpb5 lead to ~135° DNA kinking near the TSS. Surprisingly, the DNA duplex is kinked in this region both with and without TFIIH. Figure 5b shows a 2-D histogram of the MD data in terms of two angles ϕ and θ representing the bending and twisting of the DNA duplex around the axis defined by the straight region preceding the INR. While the bending angle θ appears to be approximately the same for holo- and core-PIC, the orientation angle ϕ is different, spanning a far greater range for core-PIC. Thus, besides kinking the DNA substrate, TFIIH also locks it into a fixed orientation.
Figure 5. Pol II induces DNA bending and distortion neighboring the initiation site.
a, DNA path within the PIC. The inset defines two geometric variables orientation angle (ϕ) and bending angle (θ) used in the analysis of the MD trajectories. b, Histogram of DNA bending and orientation angles in polar coordinates from the MD simulations of core PIC (left) and holo PIC (right). c, d, Pol II structural elements induce bending of downstream DNA from simulations of core PIC (c) and holo PIC (d). DNA axes (red lines) are computed by the CURVES+ code. e, f, Pol II induces structural distortion in the INR region besides bending. Comparison with canonical A DNA shows that in the INR region the DNA duplex is significantly underwound in core PIC and holo-PIC.
The kinked DNA conformation could be attributed entirely to interactions with structural elements from the Pol II cleft (Rpb2, Rpb1 coiled-coil and clamp head, Rpb5) (Figure 5c and 5d). XPB also induces a slight bend in the DNA as it passes between the two lobes but does not affect the INR region. Overlaying canonical A-DNA onto the INR region shows that the DNA substrate is not only bent but significantly under-wound (Figure 5e,5f). Negative DNA supercoiling should facilitate promoter opening. Thus, we propose that the role of TFIIH may be to further unwind the DNA until base flipping occurs leading to the formation of a nascent transcription bubble. Consistent with this proposition, negative DNA supercoiling relieves the requirement for TFIIH in basal transcription at multiple promoters.50,51
Disease mutations cluster at critical junctures of the TFIIH dynamic network.
XP, TTD and XP/CS are distinct autosomal recessive genetic disorders. Patients are often compound heterozygotes carrying two different mutations – one on each allele. The expressed phenotype results from contributions of both alleles.52 In general, TFIIH disease-causing point mutations relate specific sequence sites to distinct pathways and phenotypes: XP mutants are NER defective, TTD mutants have partial transcription defects, XP/TTD mutations exhibit both defects, and XP/CS mutations exhibit defective global genome repair (GGR) and transcription coupled repair (TCR).21–25,53
To link molecular features to disease phenotypes, we mapped 36 single-site patient mutations (Figure 6, Figure S6 and Table S3) onto our dynamic PIC model. These fall within XPD, XPB, p8 or the WH2 domain of TFIIEβ54 (Figure 6b), largely coinciding with the anchor region of reduced flexibility between TFIIH and core-PIC identified in our dynamics study (Figure 2c). Strikingly 80% of disease mutations localize to XPD helicase domain with none in transcription-essential XPB helicase domains. Of those not in the XPD helicase domains, two are in the XPB N-terminus; one in XPD Fe-S domain, two in XPD Arch domain, and one in p8. Notably, 20 XPD mutants localize to RecA2, pointing to RecA2’s pivotal role in TFIIH repair function. In our model, RecA2 is the central and most interactive helicase domain: it connects XPB, p62, and p44. The XPD helix (residues 712–725) at the three-community junction is a hot spot for patient mutations. Intriguingly, many XPD mutations lie along the path of p62 as it traverses across XPD, suggesting that p62 has regulatory as well as scaffolding functions: one p62 region (residues 274–293) tracks the XPD DNA binding groove and another (residues 333–342) caps the XPD ATP site. Most patient mutations map to secondary structure ends or loops, highlighting their significance (Table S3). Whereas half the TTD mutations fall within helices, this position is rare for XP and XP/CS mutations.
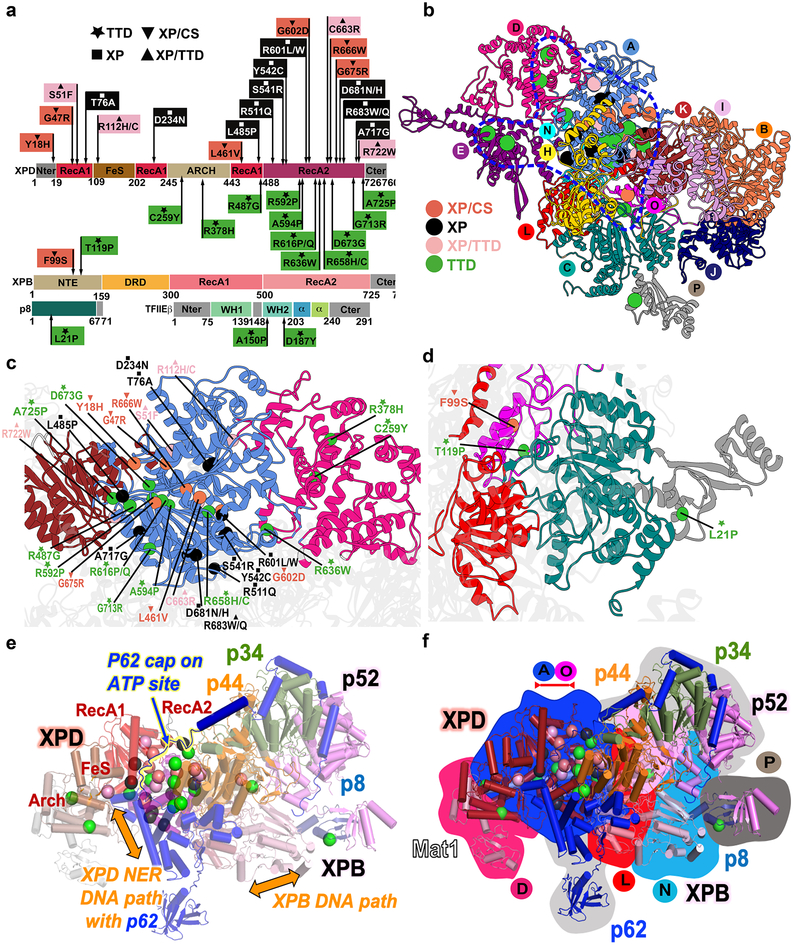
Figure 6. Human disease mutations mapped onto TFIIH and TFIIE show distinct patterns within protein-protein and community interfaces.
a, TTD, XP/TTD, XP and XP/CS point mutations mapped onto XPD, XPB, and TFIIE protein schematic do not co-localize by disease on primary sequence. b, Map of human disease mutations (spheres) onto TFIIH structure as cartoon colored by community show biased localization (blue outline). c, Zoom view of mutations on XPD. d, Zoom view of XPB and p8 mutations. e, Mutations and function mapped onto TFIIH cartoon colored by subunit. Regions of p62 and p44 are removed for clarity. f, Overlay of disease mutations, protein chain (cartoon view), and communities (background color). View matches e.
Single-site disease mutations22,39 exhibit an irregular spatial pattern (Figure 6). We find that patient mutations cluster primarily at protein and community interfaces (70%). The largest cluster at the intersection of communities A (XPD) and K (XPD, p44, p62), demarks the XPD–p44–p62 interaction as functionally important. Three mutations are directly at the interface: R592P (TTD), R722W (XP/TTD) and A725P (TTD, XP/CS/TTD), and 12 more are immediately adjacent: Y18H (XP/CS, TTD), G47R(XP/CS), S51F (XP/TTD); L485P (XP), R487G(TTD), R616P/Q (TTD), D673G (TTD), G675R(XP/CS), A594P(TTD), A596P(TTD), A717G (XP), and G713R (TTD). Switches to glycine and proline, which have the greatest impact on local backbone flexibility and dynamics, dominate in this region unveiling the critical functional role of dynamics at the XPD–p44 junction. The other key interfaces include communities A and D with mutations R636W (TTD), R112H/C(XP/TTD), A and L with mutations D681N (XP), R683W/Q (XP, XP/TTD), and R511Q (XP). Unlike XPD, neither of the XPB mutations (F99S (XP/CS) or T119P (TTD)) are in the helicase domains, but they center in four communities: C (XPB, p8, p52, TFIIEα), L (XPB, p44), N (XPB, Mat1) and O (XPB, p44, p52). Similarly, the sole p8 mutation, L21P (TTD), is at a community interface between C (XPB, p8, p52, TFIIEα) and P (p8, p52). Importantly, all these clusters correspond to critical junctures in the TFIIH dynamic network (Supplementary Video S5 and Video S6).
Considering mutations by disease, we find that 14 TTD or XP/TTD mutations mapped to protein-protein interfaces or interfacial helices in p8, XPB and XPD. The TTD mutations map to all helicase interfaces: XPB with XPD, p8, p44, or p52; or XPD with Mat1, p44, and p62. We therefore propose that TTD mutations disrupt protein-protein interfaces (directly or through breaks in helices at interfaces) to weaken assembly of TFIIH subunits while retaining residual XPB helicase activity for essential transcription function. Notably, TTD mutations were previously shown to result in lower levels of unstable TFIIH.20,25 Recently, two TTD patient-derived mutations, A150P and D187Y, were discovered in TFIIE.54 In our model, these positions map to the WH2 domain of TFIIEβ near the linchpin TFIIE α7 helix, which is key for the integrity of the TFIIE–p62 interface. These mutations are positioned to reduce WH2 stability by disrupting secondary structure packing (Table S3). In turn, this is expected to weaken interactions with TFIIEα and the interface between dynamic communities E (TFIIE) and H (p62).
All XP mutations map to XPD and fall into three categories. One set (R112H/C, R511H/C, S541R, Y542C, R601L/W, and R683W/Q) tracks the proposed DNA path on XPD, also traced by p62. A second set (residues S51F, T76A, D234N, and C663R) neighbors the Walker AB motifs and are expected to reduce ATPase activity. These two sets substitute charged or polar for bulky hydrophobic residues, potentially disrupting XPD-DNA binding and ATPase activities during NER. The third set (L485P, D681N, R683W/Q, A717G, and R722W) is on the opposite side where they appear to weaken interfaces with XPB, p44, and p62, suggesting that these XPD interactions with other TFIIH subunits are required for NER function. Mutation in residues 683 and 722 also have a TDD phenotype in some patients, suggesting that for these mutations both assembly and NER are defective.
All but one XPD XP/CS mutations lie close to p62, which is split between communities A and H. p62 wraps the XPD core (Figure S5, S6 and Supplementary Video S5) such that five XP/CS mutants (Y18H, G47R, G602D, R666W, G675W/Q) are near p62, which spans several dynamic communities, running along the XPD DNA binding path, capping ATPase site and linking TFIIH to TFIIE and core-PIC. The one XPB XP/CS mutation, F99S, is near p44, another rigging-like protein that links XPB to XPD, p62 and p34. XP/CS mutations are primarily at the center of communities and typically increase rigidity or distort conformation by removing glycines or changing to more rigid side chains (Table S3). Like a broken gear in a machine, these changes appear to break down community coordination. Therefore, we propose XP/CS mutations weaken TFIIH dynamic coordination explaining its hallmark TCR defects53.
Discussion
Transcription initiation complexes are amazingly dynamic macromolecular machines whose function and regulation underlie all of gene expression. By synthesizing cryo-EM data, we built and analyzed the functional dynamics of a practically complete atomic model of human PIC. Our results support a model for promoter opening in which the TFIIH XPB subunit serves as a DNA translocase to bend and unwind the DNA duplex in the cleft of Pol II. In this model, Pol II by itself can induce structural deformation in the DNA template. TFIIH’s role is to lock the downstream DNA duplex in a well-defined orientation and to use a ratcheting mechanism to induce negative supercoiling at the TSS. This action of TFIIH leads to strain-induced base flipping and the formation of a nascent transcription bubble. For this model to be operational the DNA duplex must be rigidly locked upstream of the transcription start site. The 90° bend in the DNA induced by TBP serves precisely this purpose. A second requirement is for the molecular motor twisting the downstream DNA duplex to be firmly attached to the rest of the initiation machinery. Correspondingly, we find a ridge of structural stability extending from the Pol II core through the TFIIE–p62 interface and into the central XPD subunit of TFIIH while also encompassing XPB lobe1. Disruption of this ridge by point mutations impairs TFIIH function as seen by the striking clustering of patient-derived mutants.
Finally, DNA remodeling to produce the transcription bubble appears to be a global conformational transition that critically depends on TFIIH global dynamics. Our MD simulation powerfully elucidates concerted motions of this complex machinery. We show that the XPB translocase lobes move independently. Lobe1’s motion is correlated with XPD while lobe2 tracks the large-scale collective motion of the TFIIH lever arm (subunits p44, p34, p52). In the absence of ATP such motion is bidirectional. However, during cycles of XPB ATP binding and hydrolysis the backward motion could be disallowed, leading to the simultaneous unwinding and pushing of the DNA toward the TSS. Interestingly, mapping of patient-derived mutations onto the TFIIH community structure further informs the above model for promoter opening. Specifically, this initiation model and disease phenotypes argue that mutants affecting XPD stability and/or its community integrity are functionally significant. Conversely, interfaces between the p44, p34 and p52 subunits lack disease causing mutations, indicating that either mutations at these interfaces are so disruptive as to be invariably lethal or, more probably, TFIIH lever arm mutations are too distal from XPB to disrupt the global motions, thus, resulting in mild or no disease phenotype. Also, XPD is well positioned in the TFIIH center for potential Fe-S-based charge transport signaling between transcription, replication, and repair.
During the review of this manuscript, Greber and colleagues published a higher resolution cryo-EM structure of apo-TFIIH55, which is consistent with our hybrid structure (overall RMSD of 2.3 Å). Notably, our model is more complete, allowing for dynamic analyses. Where overlapping, the two models follow a similar topological path. There are no significant divergences except in regions of the XPD chain, which was partly retraced in the higher resolution structure without impacting any of our conclusions.
Collectively, our results elucidate the functional dynamics of the human transcription initiation machinery, providing a framework for future experiments aimed at unraveling the intricate molecular choreography of TFIIH in nucleotide excision repair and transcription initiation.
Methods
Building the apo-TFIIH model.
To create a model of apo-TFIIH, we used two existing cryo-EM densities: apo human TFIIH (EMDB accession code: EMD-3802)19 and yeast PIC (EMDB accession code: EMD-3846)17. The corresponding structure19 (PDB accession codes: 5OF4) served as a starting point for model building. The following TFIIH elements had no known structural homologues and were, therefore, built de novo: the XPB DNA-damage recognition domain (DRD) (residues 159–300), the XPB N-terminal extension domain (NTE) (residues 1–158), the p52 XPB binding domain (residues 284–384), the p34 insertion (residues 233–251), the p44 N-terminus and α-helix insertion (residues 1–57 and 313–343), the MAT1 ARCH anchor and helices (residues 65–309), the p62 subunit except the BSD1 and PHD domain (residues 101–173 and 159–548). We used the GeneSilico metaserver56 for consensus secondary structure prediction and applied the results to establish the sequence register in the EM density. Individual secondary-structure fragments were constructed using COOT57 to generate a backbone only model by tracing the EM density. The resulting polypeptide chain segments were connected by extending the main-chain trace. Side chain orientations were built and manually inspected/corrected based on the electron density. Bulky residues such as phenylalanine, tyrosine, tryptophan, and arginine were instrumental in validating model construction and sequence registration.
To model the p62 BSD1 domain, the NMR structure of the p62 BSD1 domain (PDB ID: 2DII) was rigid-body docked into the EM density. The human p52 subunit resembles the yeast Tfb2 and shares 40% sequence identity (64% similarity). Therefore, the p52 helix-turn-helix (HTH) domain (residues 1–282) was constructed by homology modeling using MODELER 9V15 software58 and alignment to yeast Tfb2 (PDB ID: 5OQJ).17 To model the p44 subunit and the p34 ZINC finger domain (ZnF), the structures of the yeast Ssl1 and Tfb4 subunit (PDB ID: 5OQJ)17 were used as templates to construct the human p34–p44 ZnF homology structure. The RING domain of p44 was taken from the X-ray p34–p44 structure (PDB accession code: 5O85)59 and docked into the density after overlaying the p34 vWA domain. To model MAT1, the solution structure of the human MAT1 RING domain (PDB ID:1G25)60 was docked into the density ascribed to MAT1 RING by superposing the yeast Tfb1 density onto the human MAT1 density. We then built the entire apo TFIIH structure by docking the newly constructed XPB, p62, p52, p34, p44 and MAT1 subunits into the apo-TFIIH EM density.
Building the holo-PIC model.
To model TFIIH holo-PIC (core-PIC–TFIIH–DNA), we first docked the human apo-TFIIH structure into the yeast PIC density (accession code: EMD-3846).17 We then used the cascade molecular dynamics flexible fitting (cMDFF) method33 to fit apo-TFIIH into the density allowing the model to be fit sequentially to a series of maps (computationally blurred derivatives of the original map with lower-resolution). Thus, larger-scale features of the Gaussian-smoothed EM densities guided the initial stages of flexible fitting. Smaller-scale refinements were then introduced when fitting to the higher-resolution maps. The p62 PHD domain was excluded from fitting to the yeast PIC density as its orientation in the human PIC density was clearly different. Gaussian-smoothed maps were generated using Chimera61 starting with a half-width of σ = 3 Å and decreasing by 1 Å for each subsequent map. In total, 4 maps were used in cMDFF runs, including the original EM density. The structure was independently fitted using direct MDFF62 to each individual map obtained by Gaussian blurs. 4-ns MDFF simulations were performed at each of the 4 resolutions to achieve convergence during the cMDFF protocol. The final structure from cMDFF was further refined with direct MDFF to the human PIC density (accession code: EMD-3307).19 The MDFF bias was applied in each stage with a scaling factor ξ of 0.2.
Building the interface of core-PIC with TFIIH.
To model the C-terminus of TFIIEα, we employed the human closed-complex PIC EM density and the yeast PIC EM density (EMDB accession codes: EMD-330716 and EMD-384617, respectively). The C-terminal region of TFIIEα (residues 215–439) comprises three helices and one beta-strand (α7–β5–α8–α9) connected by loop regions. We inspected all holo-PIC densities (EMD-3307, EMD-8132 and EMD-8133)16 and positioned α7 between the TFIIHα WH domain and the p62 BSD2 domain. The predicted β5–α8–α9 elements were modeled based on the corresponding positions in the yeast PIC density (EMD-3846). The NMR structure for a short TFIIEα C-terminal segment bound to PHD (PDB accession code: 2RNR)46 was positioned between XPD and XPB and subsequently validated by crossing-linking.34 We then built the linker connecting the p62 BSD1 and PHD domains. TFIIH, TFIIEα C-terminus, the p62 PHD domain, core-PIC (PDB accession code: 5IYA)16 and duplex DNA were then fitted to the human closed-complex PIC density (EMD-3307) to produce the complete holo-PIC assembly. The apo-TFIIH and holo-PIC models were refined in real space with the PHENIX package63,64. MolProbity results for the apo-TFIIH and holo-PIC models are presented in Table S4. Map-to-model cross correlation values of 0.75 and 0.72 were computed for apo-TFIIH and holo-PIC, respectively. Table S5 summarizes map-to-model validation statistics for TFIIH fitted against the EMD-3802 and EMD-3846 cryo-EM maps.
Molecular dynamics simulations of core-PIC and holo-PIC.
To address the functional dynamics of the holo-PIC and core-PIC assemblies, we performed extensive molecular dynamics simulations. holo-PIC is a ~1M Dalton complex, encompassing substrate DNA and some 31 individual protein chains. Modeling the systems (comprised of >1,000,000 atoms) used resources of the Texas Advanced Computing Center and the Oak Ridge Leadership Computing Facility. The systems were set up with the TLeap module of AMBER 1465 and solvated with TIP3P water molecules66. The minimum distance from the surface atoms of the complex to the edge of the periodic simulation box was 12.0 Å. In addition to Na+ counterions to neutralize the total charge in the simulation box, we introduced 150-mM NaCl concentration to mimic physiological conditions. Energy minimization was conducted for 3000 steps with fixed protein backbone atoms and for an additional 1500 steps with harmonic restraints on the backbone atoms (k = 10 kcal mol−1 Å−2). The temperature of the simulated systems was then gradually increased to 300 K over 500 ps of dynamics in the NVT ensemble. The equilibration was continued for another 4 ns in the NPT ensemble, and the harmonic restraints were gradually released. Production runs were performed in the NPT ensemble (1 atm and 300 K) for 300 ns for each of the two models of the core PIC and holo-PIC. The particle mesh Ewald (PME) method was used to treat long‐range electrostatic interactions. The r-RESPA multiple time step method67 was employed with a 2-fs time step for bonded, 2-fs time step for short-range nonbonded interactions, and 4-fs for long-range electrostatic interactions. The short-range nonbonded interactions were evaluated by using a cutoff distance of 10 Å and a switching function at 8.5 Å. All covalent bonds to hydrogen atoms were constrained using the SHAKE algorithm. The simulations were carried out with the NAMD 2.12 code68,69 and the AMBER Parm14SB force field70. Snapshots from the MD trajectories were collected at 2.0 ps intervals. We then selected and sampled 50,000 conformations from the last 280 ns of the MD trajectories for principal component analysis (PCA) and community network analysis. DNA structural parameters were analyzed with the program CURVES+71. All figures were generated using UCSF Chimera.60
Principal component analysis.
Principal component analysis (PCA)72 was performed based on the covariance matrix whose elements are defined as:
where xi is a Cartesian coordinate of atom i, and 〈xi〉 represents the time average over all the configurations obtained in the simulation. In PCA, diagonalization of the covariance matrix yields a complete set of orthogonal eigenvectors with corresponding eigenvalues. Eigenvectors with the larger eigenvalues contribute more to the total variance in the data and, therefore, to the overall motion seen in the MD trajectories. In this way, PCA helped to separate out the slower global motions, essential for PIC dynamics. Prior to construction of the covariance matrix the MD trajectory was aligned on a reference configuration to remove all translational and rotational motion. The covariance matrix C was computed using all protein Cα atoms and P atoms in the DNA backbone and then diagonalized to produce the PCA eigenvectors and eigenvalues. PCA analysis was performed using the CPPTRAJ module in AmberTools1773.
Community network analysis
The dynamic community network of TFIIH was constructed using the NetworkView plugin in VMD49,74. In community network analysis, the protein topology was represented as a collection of nodes connected by edges whose weights were derived from the MD simulation. Nodes were associated with the protein Cα atoms. Edges were added to the network connecting pairs of in-contact nodes. Two non-adjacent nodes were connected by an edge if the nodes are within 4.5 Å of each other for at least 75% of the simulation trajectory. The edge weights, wi,j, were computed from the correlation coefficients, ci,j, of the i-j node pair:
Here, ci,j is the residue-residue correlation calculated between the i-j residue pair in the MD trajectory. Residue-residue correlations were calculated using the program CARMA75. The contact map was generated within the NetworkView plugin. After constructing the TFIIH network the Girvan-Newman algorithm was employed to determine the underlying community structure using the betweenness centrality measure. The betweenness centrality measure (betweenness) of an edge is measured by calculating the number of shortest paths that cross that edge and is indicative of the probability of information transfer between nodes (protein residues). In Girvan-Newman, the betweenness is calculated for all edges and the edge with the largest betweenness value (most central edge) is subsequently removed. This process was repeated and a modularity score tracked to identify the division that resulted in an optimal community structure. The algorithm was run iteratively resulting in a modularity score of 0.871 and a network partitioning of 16 distinct communities. We then computed the summation of the betweenness for all edges between communities to determine the strength of communication between dynamically correlated sets of residues within TFIIH.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. The structural models of apo-TFIIH and holo-PIC have been deposited in the Protein Data Bank (PDB) with accession codes 6O9M and 6O9L, respectively.
Supplementary Material
Acknowledgments
We thank S. Schilbach and P. Cramer for sharing the EMD-3846 cryo-EM density map and model before these became available from the EMDB. We thank E. Nogales for insightful discussions. This work was supported by National Institutes of Health (NIH) grant GM110387 to I.I. Work on TFIIH by Y.H., S.E.T. and J.A.T. is supported by NIH P01 CA092584. J.A.T. is also supported for structural analyses by NIH R35 CA220430, a Robert A. Welch Chemistry Chair, and the Cancer Prevention and Research Institute of Texas (RR140052 and RP180813). Computational resources were provided in part by an allocation from the National Science Foundation XSEDE program CHE110042 to I.I. An award of computer time to I.I. was provided by the INCITE program. This research also used resources of the Oak Ridge Leadership Computing Facility, which is a DOE Office of Science User Facility supported under Contract DE-AC05–00OR22725
Footnotes
Competing Interests
The authors declare no competing financial interests.
References
- 1.Moore MJ & Proudfoot NJ Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136, 688–700 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Proudfoot NJ, Furger A & Dye MJ Integrating rnRNA processing with transcription. Cell 108, 501–512 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Bentley DL Coupling mRNA processing with transcription in time and space. Nat Rev Genet 15, 163–175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roeder RG The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci 21, 327–35 (1996). [PubMed] [Google Scholar]
- 5.Zurita M & Cruz-Becerra G TFIIH: New discoveries regarding its mechanisms and impact on cancer treatment. J Cancer 7, 2258–2265 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hishikawa A, Hayashi K & Itoh H Transcription factors as therapeutic targets in chronic kidney disease. Molecules 23, 1123–1136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villard J Transcription regulation and human diseases. Swiss Med Wkly 134, 571–579 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Chen XF, Zhang YW, Xu HX & Bu GJ Transcriptional regulation and its misregulation in Alzheimer’s disease. Molecular Brain 6, 44–53(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee TI & Young RA Transcriptional regulation and its misregulation in disease. Cell 152, 1237–1251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodrich JA, Cutler G & Tjian R Contacts in context: Promoter specificity and macromolecular interactions in transcription. Cell 84, 825–830 (1996). [DOI] [PubMed] [Google Scholar]
- 11.Boeger H et al. Structural basis of eukaryotic gene transcription. Febs Lett 579, 899–903 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Buratowski S, Hahn S, Guarente L & Sharp PA Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56, 549–61 (1989). [DOI] [PubMed] [Google Scholar]
- 13.Liu X, Bushnell DA, Silva DA, Huang XH & Kornberg RD Initiation complex structure and promoter proofreading. Science 333, 633–637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung ACM, Sainsbury S & Cramer P Structural basis of initial RNA polymerase II transcription. Embo J 30, 4755–4763 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sim RJ, Belotserkovskaya R & Reinberg D Elongation by RNA polymerase II: the short and long of it. Genes Dev 18, 2437–2468 (2004). [DOI] [PubMed] [Google Scholar]
- 16.He Y et al. Near-atomic resolution visualization of human transcription promoter opening. Nature 533, 359–365(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schilbach S et al. Structures of transcription pre-initiation complex with TFIIH and Mediator. Nature 551, 204–209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Y, Fang J, Taatjes DJ & Nogales E Structural visualization of key steps in human transcription initiation. Nature 495, 481–486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greber BJ et al. The cryo-electron microscopy structure of human transcription factor IIH. Nature 549, 414–417 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubaele S et al. Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol Cell 11, 1635–1646 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Coin F & Egly JM Ten years of TFIIH. Cold Spring Harb Symp Quant Biol 63, 105–110 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Lehmann AR The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev 15, 15–23 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Berneburg M & Lehmann AR Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription. Adv Genet 43, 71–102 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Fassihi H et al. Deep phenotyping of 89 xeroderma pigmentosum patients reveals unexpected heterogeneity dependent on the precise molecular defect. Proc Natl Acad Sci U S A 113, E1236–E1245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyle J et al. Persistence of repair proteins at unrepaired DNA damage distinguishes diseases with ERCC2 (XPD) mutations: cancer-prone xeroderma pigmentosum vs. non-cancer-prone trichothiodystrophy. Hum Mutat 29, 1194–208 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rimel JK & Taatjes DJ The essential and multifunctional TFIIH complex. Protein Sci 27, 1018–1037 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Compe E & Egly JM Nucleotide excision repair and transcriptional regulation: TFIIH and Beyond. Annu Rev Biochem, 85, 265–290 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Singh A, Compe E, Le May N & Egly JM TFIIH subunit alterations causing Xeroderma Pigmentosum and Trichothiodystrophy specifically disturb several steps during transcription. Am J Hum Genet 96, 194–207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Compe E & Egly JM TFIIH: when transcription met DNA repair. Nat Rev Mol Cell Biol 13, 343–354 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Grunberg S & Hahn S Structural insights into transcription initiation by RNA polymerase II. Trends Biochem Sci 38, 603–611 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grunberg S, Warfield L & Hahn S Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat Struct Mol Biol 19, 788–96 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fishburn J, Tomko E, Galburt E & Hahn S Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proc Natl Acad Sci U S A 112, 3961–3966 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singharoy A et al. Molecular dynamics-based model refinement and validation for sub-5 angstrom cryo-electron microscopy maps. Elife 5, e16105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo J et al. Architecture of the human and yeast general transcription and DNA repair factor TFIIH. Mol Cell 59, 794–806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu QZ, Wani G, Sharma N & Wani A Lack of CAK complex accumulation at DNA damage sites in XP-B and XP-B/CS fibroblasts reveals differential regulation of CAK anchoring to core TFIIH by XPB and XPD helicases during nucleotide excision repair. DNA Repair 11, 942–950 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drapkin R, LeRoy G, Cho H, Akoulitchev S & Reinberg D Human cyclin-dependent kinase-activating kinase exists in three distinct complexes. Proc Natl Acad Sci USA 93, 6488–6493 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuss JO & Tainer JA XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair 10, 697–713 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coin F, Oksenych V & Egly JM Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol Cell 26, 245–256 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Fan L et al. XPD helicase structures and activities: Insights into the cancer and aging phenotypes from XPD mutations. Cell 133, 789–800 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan L & DuPrez KT XPB: An unconventional SF2 DNA helicase. Prog Biophys Mol Biol 117, 174–181 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Fan L et al. Conserved XPB core structure and motifs for DNA unwinding: Implications for pathway selection of transcription or excision repair. Mol Cell 22, 27–37 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Abdulrahman W et al. ARCH domain of XPD, an anchoring platform for CAK that conditions TFIIH DNA repair and transcription activities. Proc Natl Acad Sci USA 110, E633–E642 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Obmolova G, Ban C, Hsieh P & Yang W Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature 407, 703–710 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Mason AC et al. A structure-specific nucleic acid-binding domain conserved among DNA repair proteins. Proc Natl Acad Sci USA 111, 7618–7623 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng K & Wigley DB DNA translocation mechanism of an XPD family helicase. Elife 7, e42400 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okuda M et al. Structural insight into the TFIIE-TFIIH interaction: TFIIE and p53 share the binding region on TFIIH. Embo J 27, 1161–1171 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohkuma Y, Hashimoto S, Wang CK, Horikoshi M & Roeder RG Analysis of the role of TFIIE in basal transcription and TFIIH-mediated carboxy-terminal domain phosphorylation through structure-function studies of TFIIE-alpha. Mol Cell Biol 15, 4856–4866 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.David CC & Jacobs DJ Principal component analysis: A method for determining the essential dynamics of proteins. Methods Mol Biol 1084, 193–226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eargle J & Luthey-Schulten Z NetworkView: 3D display and analysis of protein center dot RNA interaction networks. Bioinformatics 28, 3000–3001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parvin JD, Shykind BM, Meyers RE, Kim JS & Sharp PA Multiple sets of basal factors initiate transcription by RNA-Polymerase-II. J Biol Chem 269, 18414–18421 (1994). [PubMed] [Google Scholar]
- 51.Parvin JD & Sharp PA DNA Topology and a minimal set of basal factors for transcription by RNA Polymerase-II. Cell 73, 533–540 (1993). [DOI] [PubMed] [Google Scholar]
- 52.Ueda T, Compe E, Catez P, Kraemer KH & Egly JM Both XPD alleles contribute to the phenotype of compound heterozygote xeroderma pigmentosum patients. J Exp Med 206, 3031–3046 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DiGiovanna JJ & Kraemer KH Shining a light on xeroderma pigmentosum. J Invest Dermatol 132, 785–796 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuschal C et al. GTF2E2 mutations destabilize the general transcription factor complex TFIIE in individuals with DNA repair-proficient Trichothiodystrophy. Am J Hum Genet 98, 627–42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greber BJ, Toso DB, Fang J & Nogales E The complete structure of the human TFIIH core complex eLife 8, e44771, doi: 10.7554/eLife.44771, (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kurowski MA & Bujnicki JM GeneSilico protein structure prediction meta-server. Nucleic Acids Res 31, 3305–3307 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Sali A & Blundell TL Comparative protein modeling by satisfaction of spatial restraints. J Mol Biol 234, 779–815 (1993). [DOI] [PubMed] [Google Scholar]
- 59.Radu L et al. The intricate network between the p34 and p44 subunits is central to the activity of the transcription/DNA repair factor TFIIH. Nucleic Acids Res 45, 10872–10883 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gervais V et al. Solution structure of the N-terminal domain of the human TFIIH MAT1 subunit - New insights into the RING finger family. J Biol Chem 276, 7457–7464 (2001). [DOI] [PubMed] [Google Scholar]
- 61.Pettersen EF et al. UCSF chimera - A visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Trabuco LG, Villa E, Mitra K, Frank J & Schulten K Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Afonine PV et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Case DA et al. The Amber biomolecular simulation programs. J Comput Chem 26, 1668–1688 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW & Klein ML Comparison of simple potential functions for simulating liquid water. J Chem Phys 79, 926–935 (1983). [Google Scholar]
- 67.Tuckerman M, Berne BJ & Martyna GJ Reversible multiple time scale molecular-dynamics. J Chem Phys 97, 1990–2001 (1992). [Google Scholar]
- 68.Kale L et al. NAMD2: Greater scalability for parallel molecular dynamics. J Comput Phys 151, 283–312 (1999). [Google Scholar]
- 69.Phillips JC et al. Scalable molecular dynamics with NAMD. J Comput Chem 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maier JA et al. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput 11, 3696–3713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blanchet C, Pasi M, Zakrzewska K & Lavery R CURVES plus web server for analyzing and visualizing the helical, backbone and groove parameters of nucleic acid structures. Nucleic Acids Res 39, W68–W73 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amadei A, Linssen AB & Berendsen HJ Essential dynamics of proteins. Proteins 17, 412–25 (1993). [DOI] [PubMed] [Google Scholar]
- 73.Roe DR & Cheatham TE 3rd. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J Chem Theory Comput 9, 3084–95 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Humphrey W, Dalke A & Schulten K VMD: visual molecular dynamics. J Mol Graph 14, 33–8, 27–8 (1996). [DOI] [PubMed] [Google Scholar]
- 75.Glykos NM Software news and updates. Carma: a molecular dynamics analysis program. J Comput Chem 27, 1765–8 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. The structural models of apo-TFIIH and holo-PIC have been deposited in the Protein Data Bank (PDB) with accession codes 6O9M and 6O9L, respectively.