Abstract
How does novelty, a new, genetically based function, evolve? A compelling answer has been elusive because there are few model systems where both the genetic mechanisms generating novel functions and the ecological conditions that govern their origin and spread can be studied in detail. Here, I review what we have learned about the evolution of novelty from microbial selection experiments. This work reveals that the genetic routes to novelty can be more highly variable than standard models have led us to believe and underscores the importance of considering both genetics and ecology in this process.
Keywords: experimental evolution, epistasis, gene amplification, ecological opportunity
The paradox of novelty
The evolution of novelty – the origin of a new function – involves a paradox. How does something new come about if all that natural selection has to work with is something old? The answer, perhaps best articulated by Francois Jacob [1], is that new functions are not produced from scratch. Evolution, Jacob said, is more like a tinkerer that uses old materials in new ways. Appealing as this metaphor is, more precise statements about how tinkering happens – the genetic mechanisms that generate a novel function and the ecological conditions that promote its origin and spread – remain elusive.
The leading explanation, also known as the exaptation-amplification-diversification (EAD) model [1–4], attributes the origins of novelty to exaptation and amplification: some pre-existing function is co-opted for growth and reproduction under novel conditions, even if it only barely allows an organism to get by, and increases in fitness are caused by increases in production of a limiting enzyme, usually through gene amplification. Better to make more of what you already do, even if you do it poorly. The additional genetic material from gene amplification means that selection is free to modify one copy and not others, leading ultimately to functional divergence. While there are other means of acquiring novel gene function – horizontal gene transfer, reverse transcription of RNA back into DNA, exon shuffling, mobile element transposition, genome rearrangement, and even de novo selection from previously noncoding DNA [5–7] have all been suggested to play a role – the EAD model is very general, applying equally to both prokaryotes and eukaryotes, and among the most commonly cited explanations for the origin, as opposed to the transfer among lineages, of new genes and genetic functions [2,3,8].
Evaluating this model has proven challenging for three reasons. The first is disagreement and confusion over just what, precisely, a novelty – or its near synonym innovation – actually is (see Box 1). Does novelty refer to a trait, like wings for flight, or an ecological function, like the ability to occupy a new environment? The answers depend to a large extent on to whom one is talking: developmental biologists tend to focus on traits and their genetic basis, because that is the data they have available to them. Evolutionary ecologists tend to focus on, not surprisingly, ecology, as the ultimate driver of novel trait evolution and lineage diversification. Both are, obviously, important but few systems exist where the two processes can be studied in detail together.
Box 1. Novelty versus innovation.
Novelty is a familiar word but difficult to define, in part because it is often used synonymously with innovation. I think it is helpful to follow the Organisation for Economic Cooperation and Development (OECD), arguably the standard setters for global economic policy in research, who define innovation as a new or significantly improved product, service, or process [16] and interpret novelty as the extreme end of the innovation spectrum.
A more biological interpretation might be that a novel trait is one that confers a new (to a lineage) ecological function underlain by a qualitatively distinct (relative to the ancestral form) genetic architecture. A loss-of-function mutation that leads to constitutive expression of an otherwise inducible system, leading to the over-production of a pre-existing enzyme important for growth, would be counted as an innovation. The enzyme itself hasn’t changed, nor has the underlying genetic architecture governing how it is produced, though the ecological conditions have. The evolution of an enzyme capable of degrading a compound that the ancestral lineage could not otherwise use would be considered a novelty, especially if this new ability is underlain by genetic rearrangements and changes to enzyme activity [17]. Put another way, innovation is doing something better, novelty is doing something new.
Note that when an innovation or novelty also contributes to evolutionary diversification we say the trait is a key innovation [18,19]. Because ecological diversification requires a lineage first gain access to a range of ecological opportunities [4], key innovations must evolve before diversification. Interestingly, there is evidence that key innovations can evolve well before diversification happens [13], suggesting that the ecological conditions leading to the evolution of innovation and novelty could be distinct from the ecological opportunities that promote diversification.
The second, which is closely linked to the first, is that different disciplines have interpreted the problem of innovation and novelty through very different conceptual lenses. Developmental biology and protein biochemistry tend to see the trajectory of evolution being shaped by what genetic variation is available to selection [9–12]. Evolutionary ecology has assumed, by contrast, that genetic variation is unlimited in the long-run, with novelty being the result of conventional natural selection operating in unconventional ecological settings [13,14]. The central argument comes down to whether the rate at which novel traits and functions evolve is governed more by access to genetic variation or to novel ecological opportunities.
The third is that the model itself does not directly account for the striking variation in the time required for novelty to evolve. The evolution of aerobic citrate metabolism in Escherichia coli – a trait whose absence is actually diagnostic for the species – took ~31,500 generations, or approximately 15 years of daily sub-culturing the populations to evolve and, even then, it occurred in only one out of 12 replicate populations [15]. On the other hand, there are many examples (reviewed below) of more rapid adaptation, often on the order of tens to hundreds of generations, to novel environmental conditions such as the degradation of toxic compounds, the use of a novel substrate, or infection of a novel host. Why should one kind of novelty take so much longer than others?
Here I draw on the literature from experimental evolution with microbes (Table 1) to address these gaps in our understanding of the factors driving the evolution of novelty. Microbial selection experiments have the advantage of being performed under defined conditions where the genetic changes can be uncovered through whole genome sequencing and the impact of these changes on traits such as fitness, population size, and the degree of novelty in ecological function measured directly. It is thus possible to watch the evolution of novelty happening in real time and to dissect the genetic and ecological mechanisms responsible. Importantly, these experiments are performed in a region of evolutionary parameter space – large population sizes (on the order of 105 – 109 individuals in most cases), genetic variation introduced solely via mutation, often (though not exclusively) haploid asexuals – where natural selection can be very effective at generating adaptation. The generality of the inferences made must therefore be evaluated in other systems where conditions differ. My aim here is to point the way towards a theory of novelty that accounts more readily for the variety of genetic routes to novelty and the ecological conditions that lead to its evolution.
Table 1.
Microbial selection experiments on the evolution of innovation and novelty.
| Organism | Ecological novelty | Genetic mechanism | Generations | Comments | Citation |
|---|---|---|---|---|---|
| Salmonella typhymurium | Growth-limiting carbon sources | Amplification of genes associated with carbon source transport | 180 | Selection on each of arabinose, malate, and sorbitol leads to duplication of chromosomal regions containing permease genes | [36] |
| Cephalospor in resistance | Amplification of bla-TEM1 followed by second site point mutations | Not reported | Point mutations, which occurred only in strains with strains with amplified bla-TEM1, confer resistance by reducing porin expression | [40] | |
| Growth recovery from a costly mutation in hemC | Amplification of hemC followed by point mutations in amplified copies | Not reported | Non-mutated hemC copies were eventually lost, leaving only the mutated versions | [41] | |
| Salmonella enterica | Tryptophan synthesis in medium lacking tryptophan and histidine | Amplification and subsequent point mutations in hisA | 3000 | Ancestral strain lacks a key gene (trpF) for tryptophan biosynthesis | [42] |
| Lactose limitation | Amplificatio n via tandem duplication of lac | Not reported | lac is located on F′128 plasmid | [38] | |
| Escherichia coli | Limited glucose | Amplification via tandem duplication and subsequent deletion of orgK yegSR from one duplicated copy | 100 | Genetic target of selection unclear | [43] |
| Cefotaxime, a novel antibiotic | Amplification of bla-TEM1 | 80 | A designed experiment where duplications were present at the beginning and their evolution tracked | [44] | |
| Growth on L-1,2-propanediol | IS5 insertion leading to constitutive activation of fucAO operon | 700 | Glycerol added to growth medium as the ancestral strain cannot grow on L-1,2-propanediol alone | [51] | |
| Growth on glucose of a UargC strain | Structural and promoter mutations to proAB | Not reported | Mutations restore some arginine biosynthesis capacity at the cost of proline biosynthesis | [49] | |
| Limiting glucose in the presence of excess citrate | Duplication and rearrangement of citT downstream of aerobically active promoter rnk | 35,000 | Potentiation involved specialization on acetate, a by-product of glucose metabolism, in part via mutations in a gene (gltA) that codes for citrate synthase | [15,61,83,87] | |
| Rapid switching from glucose to acetate metabolism | Transposon-mediated mutation causing constitutive expression if aceB (malate synthase) allowing acetate metabolism | 1000 | Resource competition drives the evolution of the rapid switcher and helps support diversity | [56,68] | |
| Metabolism of propylene glycol (PG) and ethylene glycol (EG) | Overexpression of fucO allows growth on PG; fucO overexpression and amplification of aldA required for growth on EG | Not reported | Stewpise acquisition of metabolic activities, with metabolism of PG preceding that of EG. | [50] | |
| Growth on minimal glucose medium | 1–6 structural or regulatory mutations most common, amplifications were just 4% of all genomic changes | 145 | Starting strains each had one of 87 genes knocked out that prevented growth on minimal glucose; 22/87 strains showed evidence of recovery | [48] | |
| Pseudomon as sp. ADP | Atrazine as a sole source of nitrogen | Amplificatio n via tandem duplication of atzB | 320 | atzB, a gene involved in atrazine degradation, is located on a low-copy number, stable plasmid (pADP1) | [39] |
| Pseudomon as aeruginosa | Novel carbon sources | Mutations to regulatory genes associated with metabolism predominate; de novo gene duplication rare | 140 | Carbon sources are those found on commercially available Biolog plates | [47] |
| Fluorquinolone (ciprofloxac in) resistance | Exaptation resulting from single mutations | 80–100 | Mutations are often loss-of-function to efflux pump regulators (nfxB) or protein conformation changes to DNA gyrases (gyrA, gyrB) or topisomerases (parC, parE) | [27,75] | |
| Pseudomon as fluorescens | Biofilm formation at the air-broth interface | Loss-of-function mutation, usually in wspR, resulting in constitutive expression of wss operon | 50 | Resource competition, especially for oxygen, promotes the evolution of biofilm-forming genotypes | [52,54,64] |
| Growth on xylose | Unclear but mutations in transcriptional regulator (gntR) likely responsible | 100–200 | Ancestral strain grows very poorly on xylose because it lacks xylB | [55,65] | |
| Saccharomy ces cerevisiae | Glucose limitation | Amplification via tandem duplication leading to chimeric HTX7/6 | 450 | HTX7/6, a hexose transport chimera, is comprised of the upstream promoter of HTX7 and coding sequence of HTX6 presumed to result from unequal crossing over | [37] |
| Caenorhabd itis elegans | Growth recovery in a strain containing a costly mutation | Amplification via duplication | 200 | Genomic targets of selection unclear but duplications were often highly parallel across independently evolved lines, implying shared sites under strong selection | [45] |
| Bacteriophage λ | Infection of E. coli host via novel receptor | Coevolution causes multiple mutations in phage protein J required for entry via novel receptor | Not reported | Potentiating mutations initially improved fitness via native receptor, LamB | [62,63] |
| Bacteriophage SBW25ϕ2 | Infection of novel P. fluorescens variants | Coevolution leading to multiple mutations in phage genes associated with infection | Not reported | Infection of novel hosts evolves only through coevolution with bacteria | [66] |
| Bacteriophage ϕ6 | Infection of novel Pseudomon as spp. | Not reported but previous work suggests single point mutations required for infection of each novel host | 168–254 | Strong competition among phage for access to hosts promotes emergence of novel host range mutants | [67] |
| Influenza A/H5N1 | Airborne transmission | 5–9 mutations required | 10 serial passages in ferrets | Ancestral strain for the serial passage experiments had three mutations introduced, two in HA and one in PB2; remaining mutations accumulated during serial passage | [78] |
| Poxvirus | Ability to infect human cells (HeLa) in the absence of host range gene E3L | Amplification of related host range gene, K3L | 10 passages | Amplifications faciltate rapid evolution in spite of low genome-wide mutation rates | [46] |
Genetics of innovation and novelty
The origins of innovation and novelty lie in exapted enzymes that perform both a native or canonical role but also possess a number of often fortuitous side functions that allow them to ‘moonlight’ in different roles if and when necessary. The nature and evolution of such enzyme promiscuity has been reviewed previously and interested readers can consult Copley [20,21], Bergthorsson et al. [3], and Kheronsky et al. [22] for further details. Microbial studies of adaptation to novel resources [17,23–25] or toxins such as antibiotics [26,27] provide many examples of the importance of exaptation as a first step in ecological innovation.
The second step involves population expansion, typically through gene amplification. There is good evidence that amplifications have contributed to the emergence of many different novel phenotypes, from proteins [9,28] to morphologies and body plans [6,29,30] in many taxonomic groups [31–33]. While they can be as small as a few base pairs or as large as entire chromosomes (aneuploidy) or genomes (polyploidy), the amplifications thought to most often underlie novel gene functions are of intermediate size and caused by homologous recombination between sites on sister chromatids resulting in tandem duplications of kilo- or mega-base regions [8,34]. Amplifications occur frequently, especially in microbes, but they are usually unstable and costly so can be lost quickly [35]. The EAD model solves this problem by invoking selection on the amplification itself through increased enzyme production leading to population expansion [8], and there is good evidence for this mechanism from microbial experiments [36–46] Yet, amplifications are not the only route to population expansion. Toll-Riera et al. [47], for example, found amplifications in only 4% of Pseudomonas aeruginosa lineages that had evolved the ability to metabolize a novel resource not previously used by the ancestral strain, with mutations in transcription factors leading to the de-regulation of alternative metabolic pathways being far more common. Similar results have been observed for the recovery of glucose metabolism [48,49] and growth on novel substrates [50,51] in E. coli as well as biofilm formation in P. fluorescens [52–54]. The environmental context within which selection happens can also play a role: growth on a readily-used, native substrate together with a novel resource, stressor, or toxin can support sufficiently large population sizes for long enough to access rare beneficial mutations allowing improved growth in the novel condition [15,51,55–57]. Indeed, this principle – ensuring population viability in the presence of a novel substrate by supplementing the growth medium with a native substrate – is common practice in microbiology, biochemistry, and bioengineering where the aim is to isolate novel metabolic or toxin-degrading variants [58,59].
The final step involves divergence of genes or genetic interactions involved in the novel function. Improvements to the novel function requiring multiple mutations can be built because each mutation on its own confers a fitness advantage at every step. It has also been suggested that multiple mutations accumulate through neutral processes for a time until some final mutation ‘discovers’ a new phenotype and the whole lot – driver and neutral mutations together – are driven to fixation by positive selection [60]. Microbial experiments, perhaps unsurprisingly given how effective selection can be in large populations like those usually studied in the laboratory, overwhelmingly come down on the side of selection as the driver of divergence, although the genetic and ecological routes taken can be variable. Three examples illustrate this point.
The first, by design, closely recapitulates the EAD model. Nasvall et al. [42] evolved populations of Salmonella enterica containing a modified hisA, which codes for an enzyme required for histidine synthesis as well as some rudimentary ability to synthesize tryptophan, on a plasmid prone to amplification in the absence of both histidine and tryptophan. Prolonged selection over ~3000 generations resulted in increased fitness driven by duplication to hisA and subsequent modification of one or both copies leading to either distinct enzymes specializing on either histidine and tryptophan synthesis, respectively, or generalist enzymes performing both functions. The other two examples involve more idiosyncratic pathways. Aerobic citrate metabolism (Cit+) in E. coli, for example, resulted from specialization on acetate (via a citrate synthase gene, gltA, also important for assimilating acetate), an overflow by-product of glucose metabolism, and then the fortuitous capture of a citrate transporter (citT) that is normally silent under aerobic conditions by an aerobically active promoter (rnk) following duplication and genomic rearrangement [15,61]. Meyer et al. [62] documented the role of coevolution between bacteriophage λ and its E. coli host leading to the fixation of at least four mutations all of which improve adsorption on the host [63], before access to a final key mutation allows the lineage to switch binding receptors from the ancestral LamB to the novel OmpF.
Genetics versus ecology in the evolution of novelty
Evolutionary developmental biologists have long argued that trait evolution cannot be understood independently of the developmental system that produces them; it is the spectrum of genetic variation that governs the evolution of novelty. Evolutionary ecologists, on the other hand, do not see genetic variation as a major constraint on adaptive evolution over very long time scales, and so view the range of ecological opportunities and interactions among species as the major driver of novelty and lineage diversification. Which view is more often correct?
A survey of the microbial evolution literature reveals there is merit to both. Ecological opportunity, or vacant niche space, is clearly a major driver of evolutionary innovation and novelty in these experiments. Ecological opportunity, or vacant niche space, creates the conditions for innovation and novelty to spread, once they have evolved. The citrate added to minimal glucose medium, for example, is an untapped ecological opportunity for E. coli that, eventually, a lineage evolved to exploit. When ecological opportunities themselves generate strong selection for novelty, the rate at which a novel trait evolves and spreads can be very rapid, provided genetic variation is not limiting (as it rarely is in microbial experiments). Selection for access to oxygen, which becomes rapidly limiting in liquid culture but is abundant at the air-broth interface, leads to the emergence of biofilm-forming genotyopes in static (unshaken) microcosms of Pseudomonas fluorescens within tens of generations, for example [52,54,64]. Moreover, we found that the same founder strain of Pseudomonas fluorescens, which lacks a key gene (xylB) for xylose metabolism, evolves the ability to grow rapidly on xylose within 100–200 generations when xylose is provided in abundance through mutations to gntR, a transcriptional regulator [55,65]. The literature is replete with similar examples [4,20]. Ecological interactions can also drive the rapid spread of novelty, as the co-evolution of bacteriophage λ with its E. coli host demonstrates [62,63,66]. Resource competition can also be important in acquiring novel bacteriophage hosts [67] or resources [68–70].
There is also growing evidence that the spectrum of genetic variation available to selection can be biased in ways that make it more likely that some genomic sites contribute to adaptation than others [71,72]. Local nucleotide context, repeats and homopolymer runs, and proximity to the replication terminus can be hotspots mutations in microbes [73,74] that could contribute disproportionately to adaptation associated with innovation and novelty. We have found, for example, that resistance to the fluoroquinolone antibiotic, ciprofloxacin in the opportunistic pathogen Pseudomonas aeruginosa occurs repeatedly through single base pair deletions in orfN in either poly-T or poly-G repeats, genomic regions that are prone to mutation [75]. More generally, Bailey et al. [76] have shown, using a modelling approach, that mutational heterogeneity could account for between 9–45% of the variation in parallelism in evolve-and-resequence studies in bacteria and yeast, depending on the study. Clearly, mutational heterogeneity along a genome biases the spectrum of genetic variation available to selection, at least in microbial selection experiments. It remains to be seen whether similar biases exist when selection for innovation and novelty occurs from standing genetic variation as well.
Variation in time to the emergence of new functions
The ability to aerobically grow on citrate in E. coli and the ability to grow rapidly on xylose in P. fluorescens are both examples of the evolution of novel substrate use. The examples are compelling because, in both cases, the absence of the ability to use each respective substrate was diagnostic for the strain. Why did the former take over 31,000 generations to evolve whereas the latter took only ~ 150?
One answer is ‘potentiation’, the evolution of a genetic background that affords a lineage access to genetic variation that would otherwise be inaccessible. The immediate ancestor to the E. coli lineage that evolved the ability to aerobically utilize citrate, for example, was far more likely to give rise to other Cit+ phenotypes than strain that founded the experiment [15]. By contrast, rapid adaptation to a novel resource, like in the case of xylose utilization in P. fluorescens, typically involves far fewer mutations, sometimes only one [47,52,77]. We have found, for example, that ciprofloxacin-resistance mutations resulting from knocking out the small molecule efflux pump regulator nfxB almost always evolve in under 100 generations in P. aeruginosa [27]. Similar results likely underlie many cases of rapid evolution of innovation. The ability of a strain to access relevant genetic variation can thus contribute to the time required for innovation or novelty to evolve.
Potentiation may be a common phenomenon that could explain what appears to be all-or-none epistasis, where multiple mutations that are neutral on their own appear to become beneficial in the presence of another, critical mutation. It has been seen in bacteriophage λ experiments by Meyer et al. [62,63] and may also be occurring in other gain-of-function experiments. The ability of avian influenza virus, for example, to be transmitted through the air to mammals requires multiple mutations, often on the order of at least 5 and possibly more [78]. It has been suggested that many proteins seem to be able to tolerate the introduction of mutations without severely compromising function [9], implying that potentiating mutations might fix through neutral processes that allow a gene to explore more mutational space before hitting on the ‘right’ combination of mutations that permit novelty to evolve under positive selection [60]. It is hard to see how this could happen in the experiments reviewed here. In bacteriophage λ, for example, the ability to infect via the novel OmpF receptor involved the fixation of at least four potentiating mutations within 9–17 days. Since neutral mutations fix at a rate that is equal to the mutation rate, which for most viruses is on the order of 10−6 per nucleotide per generation [79], this result that is hard to reconcile with the time required to fix the equivalent number of neutral mutations. However, selection, as explained earlier, is likely to be important in these kinds of experiments by design because population sizes are so large, so this result must be interpreted with caution.
Ecological constraints that prevent the spread of novel genotypes is a second possibility. Patches containing novel substrates will, by definition, support fewer individuals than those containing preferred resources. For novelty to evolve the population must overcome drift and survive the swamping effect of immigrants arriving from more productive patches [80–83]. Competitors [69,70,83,84], parasites [85], and predators [86] can also reduce population size of a focal lineage, making it harder for it to access the relevant beneficial mutations leading to novelty, or by occupying ecological opportunities that effectively eliminate the opportunity for selection to do its work.
Rethinking the theory for the evolution of novelty
Because evolution is a process of descent with modification, novel phenotypes must originate from the re-tooling of existing genes and genetic sequences in new ways. The EAD model spells out more formally how, and in what order, this re-tooling is expected to happen. However, the model has remained for the most part untested simply because there are few systems where each step of the process can be rigorously and empirically evaluated.
Microbial selection experiments are especially valuable, then, because they provide an opportunity to confront the EAD model directly with data. The work reviewed here tells us that, while the EAD model can be an accurate description of how innovation and novelty evolve in some situations, reality can be more complex in at least two ways.
First, gene amplifications are not the only way for a lineage to increase fitness in a new environment. Other mechanisms including regulatory changes or the availability of alternative resources that can support growth can also be important in increasing population size and allowing a lineage to persist under novel conditions. If these regulatory changes also result in increased expression of downstream genetic regions, they could lead to the transcription and translation of non-coding sequences and so provide a substrate for the creation and selection of genes de novo [8]. Second, genotypes vary in their ability to access novel phenotypes through mutation, a feature that likely underlies both the distinction, as I have described it, between innovation and novelty and the time to the emergence of novel phenotypes. A genetic background that has ready access to novel phenotypic variation, for example through a loss-of-function mutation that results in deregulation of an otherwise inducible pathway, is an innovation that can evolve very quickly. On the other hand, the fixation of multiple mutations arising from adaptation to one function, like acetate metabolism, that fortuitously provides access to mutations that allow another, novel function to evolve, like the ability to aerobically utilize citrate, is more likely to take a much longer time and be counted as a genuine novelty.
Concluding Remarks
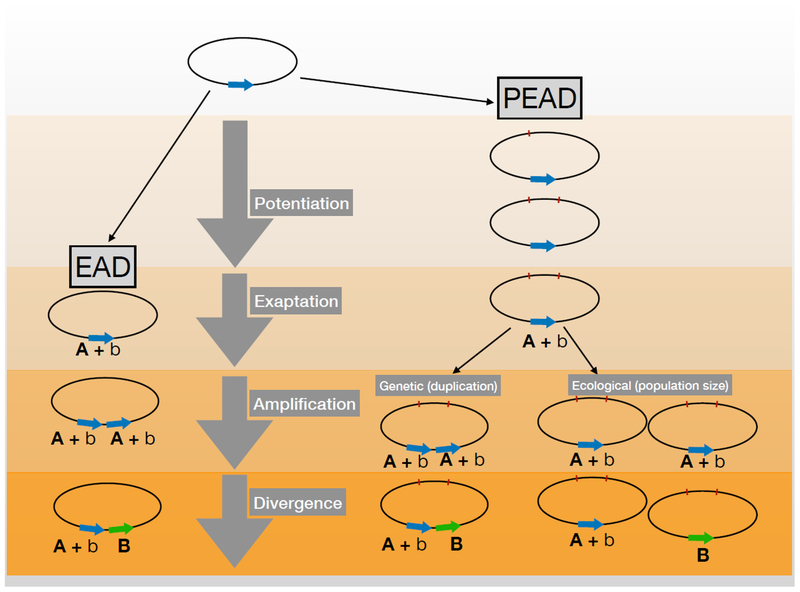
Taken together, it may be time to abandon the strict form of the EAD model. If so, it could be replaced, provisionally, with one that recognizes the importance of genetic factors like potentiation, alternative routes to increasing gene dosage beyond just gene amplification, and integrates key elements of ecology, like ecological opportunity and ecological interactions, as drivers of the evolution of novelty (see Fig. 1). A new acronym might help – call it the ‘PEAD’ model, where ‘P’ here stands for potentiation, ‘E’ stands for exaptation, and ‘A’ represents amplification of enzyme products either through conventional gene duplication or other mechanisms that increase population size of the exapted lineage. We can leave the last ‘D’ for divergence, but we need to be ready to expand on it dramatically and integrate ecology more directly into our thinking about how novelty evolves. We will need to first answer a number of key questions on the relative contribution of genetics and ecology in driving the emergence and spread of novel traits (see Outstanding Questions). We will also need to consider seriously how often, and under what conditions, novelty evolves through alternative mechanisms like de novo selection from noncoding sequences. Doing so, I suspect, will take us a long way towards understanding when and why novelty evolves, or not.
Figure 1.
Schematic illustration of the differences between the standard (exaptation-amplification-diversification; EAD) and revised (potentiation-exaptation-amplification-diversification; PEAD) models for the evolution of innovation and novelty. Each oval represents the genome of an individual. Under the EAD model (left), the exaptation is afforded because a gene (blue arrow) that produces a primary product with function A can also perform side activity b that has become important in a new environment. Amplification occurs through gene duplication, leading to increased production of b, and divergence occurs due to positive selection for improved B. The PEAD model (right) differs because of an additional potentiation step preceding exaptation, where mutations accumulate elsewhere in the genome (red marks) that allow the side function of the focal gene (b) to become important. Amplification can then proceed as it does in the EAD model, through gene duplication, or through other mechanisms allowing population expansion without gene duplication. Divergence proceeds as before, through positive selection for improved B.
Box 2 – Outstanding questions.
To what extent is the evolution of novelty limited primarily by access to genetic variation versus access to novel environmental conditions?
What is the relative importance of internal genetic changes like gene amplifications and rearrangements versus those coming externally through horizontal gene transfer in driving the emergence of novel traits?
What causes potentiation and how does it allow a lineage to gain access to novel variation?
How does the distribution of fitness effects among mutations – and especially the ability to access novel phenotypes – change with genetic background?
How often, and under what conditions, does novelty evolve through alternative mechanisms involving, for example, the de novo origins of genes from non-coding sequence?
Highlights.
Microbial systems provide a unique opportunity to dissect both the genetic mechanisms and ecological conditions that lead to the evolution of novel traits and functions.
All novel functions are derived from pre-existing ones but the major obstacle to their evolution is accessing novel kinds of genetic variation under ecological conditions that allow this variation to spread.
A range of genetic mechanisms can promote the generation of novel genetic variation and bias the kind of variation produced.
Ecological factors that influence population size impact the likelihood that novelty will spread through a population.
Selection – sometimes driven by adaptation to conditions not obviously connected to the novel function that eventually evolves – can move lineages into regions of mutational space that allow novel variation to be accessed.
Acknowledgements
This work benefited from conversations at the KITP EcoEvo 2017 program and from comments provided by A. Wong and R. Sargent. My research is supported by an NSERC Discovery Grant and the KITP programs are supported in part by the National Science Foundation under Grant No. NSF PHY-1748958, NSF Grant No. PHY-1748958, NIH Grant No. R25GM067110, and the Gordon and Betty Moore Foundation Grant No. 2919.01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Refrences
- 1.Jacob F (1977) Evolution and tinkering. Science 196, 1161–1166 [DOI] [PubMed] [Google Scholar]
- 2.Francino MP (2005) An adaptive radiation model for the origin of new gene functions. Nature Genetics 37, 573–578 [DOI] [PubMed] [Google Scholar]
- 3.Bergthorsson U et al. (2007) Ohno’s dilemma: Evolution of new genes under continuous selection. Proceedings of the National Academy of Sciences 104, 17004–17009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kassen R (2014) Experimental evolution and the nature of biodiversity, Roberts and Company. [Google Scholar]
- 5.Ochman H et al. (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304 [DOI] [PubMed] [Google Scholar]
- 6.Chen S et al. (2013) New genes as drivers of phenotypic evolution. Nature Reviews Genetics 14, 645–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall JPJ et al. (2017) Sampling the mobile gene pool: innovation via horizontal gene transfer in bacteria. Philosophical Transactions of the Royal Society B: Biological Sciences 372, 20160424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersson DI et al. (2015) Evolution of new functions de novo and from preexisting genes. Cold Spring Harbor Perspectives in Biology 7, a017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soskine M and Tawfik DS (2010) Mutational effects and the evolution of new protein functions. Nature Reviews Genetics 11, 572–582 [DOI] [PubMed] [Google Scholar]
- 10.Wagner GP and Lynch VJ (2010) Evolutionary novelties. Current Biology 20, R48–R52 [DOI] [PubMed] [Google Scholar]
- 11.Hallgrímsson B et al. (2012) The generation of variation and the developmental basis for evolutionary novelty: variation and developmental basis for evolutionary novelty. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution 318, 501–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harms MJ and Thornton JW (2013) Evolutionary biochemistry: revealing the historical and physical causes of protein properties. Nature Reviews Genetics 14, 559–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erwin DH (2015) Novelty and innovation in the history of life. Current Biology 25, R930–R940 [DOI] [PubMed] [Google Scholar]
- 14.Schluter D (2000) The ecology of adaptive radiation, Oxford University Press. [Google Scholar]
- 15.Blount ZD et al. (2008) Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proceedings of the National Academy of Sciences 105, 7899–7906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Organisation for Economic Co-operation and Development and Statistical Office of the European Communities, eds. (2005) Oslo manual: guidelines for collecting and interpreting innovation data, 3rd edOrganisation for Economic Co-operation and Development : Statistical Office of the European Communities. [Google Scholar]
- 17.Clarke PH and Drew R (1988) An experiment in enzyme evolution studies with Pseudomonas aeruginosa amidase. Bioscience Reports 8, 103–120 [DOI] [PubMed] [Google Scholar]
- 18.Heard SB and Hauser DL (1995) Key evolutionary innovations and their ecological mechanisms. Historical Biology 10, 151–173 [Google Scholar]
- 19.Hunter JP (1998) Key innovations and the ecology of macroevolution. Trends in Ecology & Evolution 13, 31–36 [DOI] [PubMed] [Google Scholar]
- 20.Copley SD (2009) Evolution of efficient pathways for degradation of anthropogenic chemicals. Nature Chemical Biology 5, 559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Copley SD (2014) An evolutionary perspective on protein moonlighting. Biochemical Society Transactions 42, 1684–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khersonsky O et al. (2006) Enzyme promiscuity: evolutionary and mechanistic aspects. Current Opinion in Chemical Biology 10, 498–508 [DOI] [PubMed] [Google Scholar]
- 23.Lin ECC et al. (1976) Experimental models of acquisitive evolution. BioScience 26, 548–555 [Google Scholar]
- 24.Hall BG (1989) Selection, adaptation, and bacterial operons. Genome 31, 265–271 [DOI] [PubMed] [Google Scholar]
- 25.Mortlock RP (1983) Experiments in evolution using microorganisms. BioScience 33, 308–313 [Google Scholar]
- 26.Weinreich DM (2006) Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312, 111–114 [DOI] [PubMed] [Google Scholar]
- 27.Melnyk AH et al. (2017) Evolution of cost-free resistance under fluctuating drug selection in Pseudomonas aeruginosa. mSphere 2, e00158–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conant GC and Wolfe KH (2008) Turning a hobby into a job: How duplicated genes find new functions. Nature Reviews Genetics 9, 938–950 [DOI] [PubMed] [Google Scholar]
- 29.Zhang J (2003) Evolution by gene duplication: an update. Trends in Ecology & Evolution 18, 292–298 [Google Scholar]
- 30.Carroll SB (2008) Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134, 25–36 [DOI] [PubMed] [Google Scholar]
- 31.Lynch M and Conery JS The evolutionary demography of duplicate genes. J. Struct. Funct. Genomics 3, 35–44 [PubMed] [Google Scholar]
- 32.Andersson DI and Hughes D (2009) Gene amplification and adaptive evolution in bacteria. Annual Review of Genetics 43, 167–195 [DOI] [PubMed] [Google Scholar]
- 33.Katju V and Bergthorsson U (2013) Copy-number changes in evolution: rates, fitness effects and adaptive significance. Frontiers in Genetics 4, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reams AB and Roth JR (2015) Mechanisms of gene duplication and amplification. Cold Spring Harbor Perspectives in Biology 7, a016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor JS and Raes J (2004) Duplication and divergence: the evolution of new genes and old ideas. Annual Review of Genetics 38, 615–643 [DOI] [PubMed] [Google Scholar]
- 36.Sonti RV and Roth JR (1989) Role of gene duplications in the adaptation of Salmonella typhimurium to growth on limiting carbon sources. Genetics 123, 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown CJ et al. (1998) Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Molecular Biology and Evolution 15, 931–942 [DOI] [PubMed] [Google Scholar]
- 38.Kugelberg E et al. (2006) Multiple pathways of selected gene amplification during adaptive mutation. Proceedings of the National Academy of Sciences 103, 17319–17324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Devers M et al. (2008) Fitness drift of an atrazine-degrading population under atrazine selection pressure. Environmental Microbiology 10, 676–684 [DOI] [PubMed] [Google Scholar]
- 40.Sun S et al. (2009) Contribution of gene amplification to evolution of increased antibiotic resistance in Salmonella typhimurium. Genetics 182, 1183–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pränting M and Andersson DI (2011) Escape from growth restriction in small colony variants of Salmonella typhimurium by gene amplification and mutation. Molecular Microbiology 79, 305–315 [DOI] [PubMed] [Google Scholar]
- 42.Nasvall J et al. (2012) Real-time evolution of new genes by innovation, amplification, and divergence. Science 338, 384–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maharjan RP et al. (2013) A case of adaptation through a mutation in a tandem duplication during experimental evolution in Escherichia coli. BMC Genomics 14, 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dhar R et al. (2014) Increased gene dosage plays a predominant role in the initial stages of evolution of duplicate tem-1 beta lactamase genes: experimental evolution of duplicate genes. Evolution 68, 1775–1791 [DOI] [PubMed] [Google Scholar]
- 45.Farslow JC et al. (2015) Rapid Increase in frequency of gene copy-number variants during experimental evolution in Caenorhabditis elegans. BMC Genomics 16, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elde NC et al. (2012) Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell 150, 831–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toll-Riera M et al. (2016) The genomic basis of evolutionary innovation in pseudomonas aeruginosa. PLOS Genetics 12, e1006005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blank D et al. (2014) The predictability of molecular evolution during functional innovation. PNAS 111, 3044–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McLoughlin SY and Copley SD (2008) A compromise required by gene sharing enables survival: Implications for evolution of new enzyme activities. Proceedings of the National Academy of Sciences 105, 13497–13502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szappanos B et al. (2016) Adaptive evolution of complex innovations through stepwise metabolic niche expansion. Nature Communications 7, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee D-H and Palsson BO (2010) Adaptive evolution of escherichia coli k-12 mg1655 during growth on a nonnative carbon source, l-1,2-propanediol. Applied and Environmental Microbiology 76, 4158–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bantinaki E et al. (2007) Adaptive divergence in experimental populations of pseudomonas fluorescens. Iii. Mutational origins of wrinkly spreader diversity. Genetics 176, 441–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McDonald MJ et al. (2009) Adaptive Divergence in Experimental Populations of Pseudomonas fluorescens. IV. Genetic Constraints Guide Evolutionary Trajectories in a Parallel Adaptive Radiation. Genetics 183, 1041–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lind PA et al. (2015) Experimental evolution reveals hidden diversity in evolutionary pathways. eLife 4, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bailey SF and Kassen R (2012) Spatial structure of ecological opportunity drives adaptation in a bacterium. The American Naturalist 180, 270–283 [DOI] [PubMed] [Google Scholar]
- 56.Spencer CC et al. (2007) Adaptive diversification in genes that regulate resource use in Escherichia coli. PLoS Genetics 3, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Q et al. (2011) Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science 333, 1764–1767 [DOI] [PubMed] [Google Scholar]
- 58.Hegeman GD and Rosenberg SL (1970) The evolution of bacterial Eenzyme systems. Annual Review of Microbiology 24, 429–462 [DOI] [PubMed] [Google Scholar]
- 59.Winkler JD and Kao KC (2014) Recent advances in the evolutionary engineering of industrial biocatalysts. Genomics 104, 406–411 [DOI] [PubMed] [Google Scholar]
- 60.Wagner A (2008) Neutralism and selectionism: a network-based reconciliation. Nature Reviews Genetics 9, 965–974 [DOI] [PubMed] [Google Scholar]
- 61.Quandt EM et al. (2015) Fine-tuning citrate synthase flux potentiates and refines metabolic innovation in the Lenski evolution experiment. eLife 4, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meyer JR et al. (2012) Repeatability and contingency in the evolution of a key innovation in phage lambda. Science 335, 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burmeister AR et al. (2016) Host coevolution alters the adaptive landscape of a virus. Proceedings of the Royal Society B: Biological Sciences 283, 20161528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rainey PB and Travisano M (1998) Adaptive radiation in a heterogeneous environment. Nature 394, 69. [DOI] [PubMed] [Google Scholar]
- 65.Bailey SF et al. (2015) The Effect of Selection Environment on the Probability of Parallel Evolution. Molecular Biology and Evolution 32, 1436–1448 [DOI] [PubMed] [Google Scholar]
- 66.Hall AR et al. (2011) Bacteria‐phage coevolution and the emergence of generalist pathogens. The American Naturalist 177, 44–53 [DOI] [PubMed] [Google Scholar]
- 67.Bono LM et al. (2012) Competition and the origins of novelty: experimental evolution of niche-width expansion in a virus. Biology Letters 9, 20120616–20120616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tyerman JG et al. (2008) Experimental demonstration of ecological character displacement. BMC Evolutionary Biology 8, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Q-G et al. (2012) The effect of a competitor on a model adaptive radiation. Evolution 66, 1985–1990 [DOI] [PubMed] [Google Scholar]
- 70.Bailey SF et al. (2013) Competition both drives and impedes diversification in a model adaptive radiation. Proceedings of the Royal Society B: Biological Sciences 280, 20131253–20131253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stoltzfus A and Yampolsky LY (2009) Climbing mount probable: mutation as a cause of nonrandomness in evolution. Journal of Heredity 100, 637–647 [DOI] [PubMed] [Google Scholar]
- 72.Blount ZD (2016) A case study in evolutionary contingency. Studies in History and Philosophy of Science Part C: Studies in History and Philosophy of Biological and Biomedical Sciences 58, 82–92 [DOI] [PubMed] [Google Scholar]
- 73.Lee H et al. (2012) Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proceedings of the National Academy of Sciences 109, E2774–E2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dettman JR et al. (2016) The properties of spontaneous mutations in the opportunistic pathogen Pseudomonas aeruginosa. BMC Genomics 17, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wong A et al. (2012) Genomics of adaptation during experimental evolution of the opportunistic pathogen Pseudomonas aeruginosa. PLoS Genetics 8, e1002928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bailey SF et al. (2017) What drives parallel evolution?: How population size and mutational variation contribute to repeated evolution. BioEssays 39, e201600176. [DOI] [PubMed] [Google Scholar]
- 77.Schick A et al. (2015) Evolution of fitness trade-offs in locally adapted populations of pseudomonas fluorescens. The American Naturalist 186, S48–S59 [DOI] [PubMed] [Google Scholar]
- 78.Herfst S et al. (2012) Airborne transmission of Influenza A/H5N1 virus between ferrets. Science 336, 1534–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sanjuán R et al. (2010) Viral mutation rates. Journal of Virology 84, 9733–9748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holt RD and Gomulkiewicz R (1997) How does immigration influence local adaptation? A reexamination of a familiar paradigm. American Naturalist 149, 563–572 [Google Scholar]
- 81.Jasmin J-N and Kassen R (2007) On the experimental evolution of specialization and diversity in heterogeneous environments. Ecology Letters 10, 272–281 [DOI] [PubMed] [Google Scholar]
- 82.Hall AR and Colegrave N (2007) How does resource supply affect evolutionary diversification? Proceedings of the Royal Society B: Biological Sciences 274, 73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leon D et al. (2018) Innovation in an E. coli evolution experiment is contingent on maintaining adaptive potential until competition subsides. PLOS Genetics 14, e1007348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fukami T et al. (2007) Immigration history controls diversification in experimental adaptive radiation. Nature 446, 436–439 [DOI] [PubMed] [Google Scholar]
- 85.Buckling A and Rainey PB (2002) The role of parasites in sympatric and allopatric host diversification. Nature 420, 496–499 [DOI] [PubMed] [Google Scholar]
- 86.Meyer JR and Kassen R (2007) The effects of competition and predation on diversification in a model adaptive radiation. Nature 446, 432–435 [DOI] [PubMed] [Google Scholar]
- 87.Blount ZD et al. (2012) Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature 489, 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]