Abstract
In recent years, large scale meta-analysis of genome-wide association studies (GWAS) have reliably identified genetic polymorphisms associated with neuropsychiatric disorders such as schizophrenia (SCZ), bipolar disorder (BPD) and major depressive disorder (MDD). However, the majority of disease-associated single nucleotide polymorphisms (SNPs) appear within functionally ambiguous non-coding genomic regions. Recently, increased emphasis has been placed on identifying the functional relevance of disease-associated variants via correlating risk polymorphisms with gene expression levels in etiologically relevant tissues. For neuropsychiatric disorders, the etiologically relevant tissue is brain, which requires robust postmortem sample sizes from varying genetic backgrounds. While small sample sizes are of decreasing concern, postmortem brain databases are composed almost exclusively of Caucasian samples, which significantly limits study design and result interpretation. In this review, we highlight the importance of gene expression and expression quantitative loci (eQTL) studies in clinically relevant postmortem tissue while addressing the current limitations of existing postmortem brain databases. Finally, we introduce future collaborations to develop postmortem brain databases for neuropsychiatric disorders from Chinese and Asian subpopulations.
Keywords: Postmortem brain, eQTL, GWAS, Neuropsychiatric disorders, Gene expression, Ethnic diversity
1. INTRODUCTION
In the past decade, a massive and concerted effort from multiple research groups has led to the generation of large ‘mega’ genome-wide association scans (GWAS) analyzing millions of genotyped and imputed single nucleotide polymorphisms (SNP) generated from thousands of subjects (MacArthur et al., 2017). This has resulted in the robust identification of replicable genetic variants associated with severe neuropsychiatric phenotypes such as schizophrenia (SZ), bipolar disorder (BP), and major depressive disorders (MDD) (Converge Consortium, 2015; Mühleisen et al., 2014; Ripke et al., 2013), with an increasing emphasis towards identifying risk associated genetic loci across different ethnic populations (Marigorta & Navarro, 2013). However, a major challenge of the postgenomic era is understanding how these genetic variants impact molecular function and disease etiopathology (Paul et al., 2013). Except for few common disorders, such as colon cancer or diabetes (Freedman et al., 2011; Morris et al., 2012), very little is known about the biological mechanism(s) by which genetic variants associated with neuropsychiatric disorder contribute to complex behavioral psychopathologies. One reason for increased success in identifying molecular pathways contributing to biomedical outcomes such as cancer or diabetes has been due to our ability to study these processes in etiologically relevant tissues.
The etiologically relevant tissue for studying molecular processes involved in neuropsychiatric disorders is, of course, the human brain. However, the collection of postmortem brain tissue is difficult due to extensive logistic requirements and substantial ethical and cultural hurdles. Regardless, efforts to collect postmortem brains from subjects with neuropsychiatric disorders and controls have been undergoing for more than three decades (Anderson et al., 2001; A. Schmitt et al., 2007). Research centers including (but not limited to) the Lieber Institute for Brain Development, the CommondMind Consortium, and Stanley Medical Research Institute have amassed a large number of brains with data on a wide variety of neuropsychiatric phenotypes. However, the majority of collected brains are mostly of European (EUR) descent. While, a growing number of studies, with steadily increasing sample sizes have been performed on brains from populations other than Caucasians i.e. African-American (AA) ancestry (Mighdoll & Hyde, 2018), to our knowledge there are few recent reports using brain tissues from Asian, (e.g. Chinese) populations. Understanding the biological mechanisms by which clinically relevant genetic variants, with varying allelic frequencies across different populations, contribute to the disease pathology, will require analysis of brain tissue from different ethnic groups.
Most studies using postmortem brain tissues have focused heavily on RNA expression at the expense of assessing other biomolecules such as proteins. One reason for this ‘bias’ is the significant technological advancements occurring within the last two decades underlying various genomic platforms used to assay DNA and RNA (van Dijk et al., 2014). This period serendipitously also coincided with the first attempts to utilize postmortem brain tissues to assess gene expression differences between a limited selection of cases and controls (Guidotti et al., 2000; Iwamoto et al., 2004). Thus, in this review our discussion centers on the importance of assessing transcriptomic differences between the brains of subjects diagnosed with neuropsychiatric disorders and neurotypical controls with the purpose of understanding underlying disease mechanisms and neuronal etiopathology. We further highlight the importance of collecting large and ethnically diverse postmortem brain samples to investigate disease mechanisms across cosmopolitan cohorts and inform how population specific risk variants affect the implicated molecular pathways for disease etiology. This will allow researchers to identify disease mechanisms confined to specific genetic backgrounds, facilitating the development of pharmacological treatments tailored to specific populations.
1.1. Postmortem Brain Databases:
The importance of establishing rigorously maintained postmortem brain banks designed to study neurobiological disorders cannot be overstated. Currently, 60 brain banks across Europe, North America, Australia, and Asia have been established with the goal of studying both neurodegenerative and neuropsychiatric traits (Palmer-Aronsten et al., 2016). Of those 60 brain banks, we have highlighted 19 of them as having neuropsychiatric specimens available for research (Table 1.0). As the number of postmortem brain banks expands, standardized protocols for brain tissue collection and maintenance is of increasing importance. This is especially relevant in the context of studying gene expression since preserving RNA integrity is critical for obtaining accurate results (Lewis, 2002; Barbara K. Lipska et al., 2006). For example, prolonged time between brain collection and storage, also known as postmortem interval (PMI), can confound result interpretation due to rapid RNA degradation (Blair et al., 2016). The agonal state of potential donors also impacts RNA quality independent from PMI due to changes in tissue pH (Harrison et al., 1995; Kingsbury et al., 1995). In addition to controlling the immediate effects of pre- and post-mortem factors on brain tissue quality, collecting information about lifestyle choices such as smoking, drinking, medication use and drug abuse from available hospital records or next-of-kin, can help further resolve confounds related to gene expression biases between different diagnostic groups (Lewis, 2002; Barbara K. Lipska et al., 2006).
Table 1.
International List of Active Postmortem Brain Banks
| Postmortem Brain Banks | Location |
|---|---|
| University of Miami Brain Endowment Bank a | Miami, FL |
| University of Maryland Brain and Tissue Bank a | Baltimore, MD |
| Stanley Brain Collection | Chevy Chase, MD |
| Harvard Brain Tissue Resource Center a | Belmont, MA |
| Pritzker Neuropsychiatric Consortium Brain Bank | Irvine, CA |
| dHuman Brain and spinal Fluid Resource Center a | Los Angeles, CA |
| Mt. Sinai Brain Banka | Bronx, NY |
| Brain Tissue Donation Program at the University of Pittsburg a | Pittsburgh, PA |
| Douglas - Bell Canada Brain Bank | Montreal, Quebec |
| Netherlands Brain Bank b | Amsterdam, Netherlands |
| Human Brain Tissue Bank Budapest b | Budapest, Hungary |
| Neurobiobank Munich b | Munich, Germany |
| The London Brain Bank for Neurodegenerative Diseases b | London, UK |
| New South Wales Tissue Resource Centre c | Sydney, Australia |
| South Australian Brain Bank c | Adelaide, Australia |
| Victorian Brain Bank c | Parkville, Australia |
| Neurological Foundation of New Zealand Human Brain Bank | Grafton, New Zealand |
| Human Brain Tissue Repository. NIMHM | Bangalore, India |
| Chinese Brain Bank Center | Wuhan City, China |
An active list of postmortem brain banks worldwide with available neuropsychiatric specimens. Some of these brain repositories are part of larger research consortiums and biobanks.
= NIH NeuroBiobank,
= BrainNet Europe,
= Australian Brain Bank Network
1.2. Demographic and Clinical Characterization:
Clinical characterization of the collected tissue is a different, yet equally important consideration in the postmortem brain expression studies. Prior to the collection of brain tissues, it is important that all potential donors are properly diagnosed. This insures that both subjects suffering from various neuropsychiatric disorders and neurotypical controls are properly identified. The latter is especially important to ensure no cryptic cases are misdiagnosed as controls, as this will affect downstream results and conclusions (Barbara K. Lipska et al., 2006). However, establishing diagnoses retrospectively is challenging and can introduce additional unwanted bias. One widely used approach is the collection of clinical and demographic information via psychiatric records review or structured interviews with immediate family members within one week of donation (T. M. Kelly & Mann, 1996). Psychiatric narratives are prepared for each case and then independently reviewed by two psychiatrists, who should arrive at a consensus for lifetime diagnoses status (American Psychiatric Association, 2013). Control status is defined by medical examiner documentation and a number of additional criteria. Individuals are only considered controls when subjects and their first-degree relatives have no history of psychological problems, psychiatric admissions, or any known history of psychiatric symptoms or substance abuse (Vonsattel et al., 2008).
In addition to obtaining accurate diagnoses, information regarding the potential use of licit and illicit drugs, including alcohol and tobacco, should also be thoroughly investigated and recorded due to their known neurobiological effects (Karege et al., 2005; Mighdoll & Hyde, 2018; Zahr et al., 2011). Generally, the history of nicotine and alcohol use at the time of death is collected and verified through toxicological analysis of cotinine and alcohol levels in blood and/or brain tissue. A certified forensic laboratory is necessary to perform toxicological testing on all subjects. Another important concern that may bias comparisons between neuropsychiatric cases and controls is controlling for the use of neuroleptic medications (Barbara K. Lipska et al., 2006). Over the years, several generations of neuroleptic medications for the treatment of different neuropsychiatric phenotypes have been employed in an attempt to minimize the medications harmful side effects (D. Cohen, 1997; Joukamaa et al., 2006; Lingjaerde et al., 1987; Tan et al., 2014). However, this poses an additional challenge: how should researchers control for different neuroleptics and prescribed dosage used in the treatment of potential donors? For clinical and research purposes, varying neuroleptic intake is converted into standardized doses that are widely accepted by the research community (Leucht et al., 2016); nowadays the common practice is dosage of different neuroleptic medications to be converted to chlorpromazine (CPZ) equivalent doses for median daily dose, total lifetime dose, and final dose before death (Lehman & Steinwachs, 1998; Woods, 2003). Finally, brain tissue integrity must also be examined by a board-certified pathologist to minimize the confounding effects of tissue degeneration (Mighdoll & Hyde, 2018; Vonsattel et al., 2008). Accurately assessing the clinical and demographic characteristics of individuals who have donated tissue is critical for controlling bias in postmortem experimental design.
2. IMPORTANCE OF THE POSTMORTEM BRAIN STUDIES
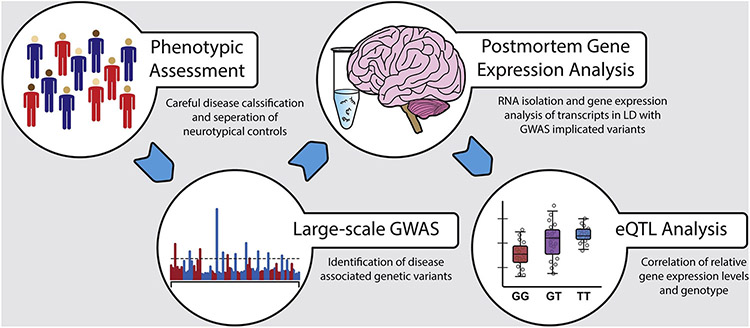
Large GWAS have successfully identified genetic associations for hundreds of complex disorders (Converge Consortium., 2015; MacArthur et al., 2017; Mühleisen et al., 2014; Ripke et al., 2013). However, a well-recognized limitation of GWAS data is the inability to elucidate potential biological mechanisms involving the identified associated variants. The first step in understanding the biological function of implicated SNPs involves gene expression (Figure 1.0); gene expression can be considered as the most intermediate phenotype between genetic polymorphisms and downstream biological processes. Although nearly all variants identified by GWAS occur outside of protein-coding genomic regions (Tak & Farnham, 2015), these variants can still regulate gene expression indirectly through various means. For example, polymorphisms can affect gene expression levels generally (Emilsson et al., 2008; Stranger et al., 2007) or result in the relative increases of different transcript isoform levels via a SNPs impact on alternative splicing mechanisms (Pickrell et al., 2010). GWAS polymorphisms can also affect transcript abundance by interfering with miRNA function, i.e. influencing the interactions between miRNAs and their gene targets. Such effects have already been observed in a limited number of studies investigating neuropsychiatric disorders such as MDD, SCZ and BPD (Jensen et al., 2014; Kandaswamy et al., 2014; Rahman et al., 2010). Additionally, sequence variants have been known to modify sequence-specific methylation patterning at gene regulatory elements, effectively altering gene expression (Gibbs et al., 2010). Determining the impact of genetic variants on gene regulation is crucial for elucidating the mechanisms by which variants increase the risk of disease (Emilsson et al., 2008). Therefore, to understand the biological implications of clinical variants on the brain pathophysiology, it is critical to assess their impact on gene expression within the target disease organ from varying ethnic populations and genetic backgrounds.
Figure 1: GWAS to Postmortem Gene Expression.
The workflow diagram outlines the steps involved in the integration of GWAS and gene expression data. The GWAS data are generated by genotyping and subsequent imputation of millions of SNPs in thousands of carefully selected cases and matched controls. Similarly, gene expression and SNP data are generated in brain tissues from various brain banks to identify brain specific eQTLs affecting gene expression. In the final steps, GWAS and eQTL data from these two sources are integrated to identify potentially causal GWAS variants.
2.1. Postmortem Brain Tissue vs. Proxy Tissues:
Proxy tissues such as blood, induced pluripotent stem cells (iPSC) or model organisms have been utilized to understand the etiology of neuropsychiatric disorders with unfortunately little success. None of these proxy tissues recapitulate the complex neurophysiological functions of the human brain nor encapsulate the long-term environmental interactions that underlie neuronal development. While these strategies enjoy the advantage of working with readily accessible tissues, it is unclear how variation in peripheral tissues or the brains of animal models accurately reflects changes associated with individual differences in the human brain. Many neuropsychiatric disorders such as BPD, SCZ or MDD are uniquely human illnesses and animal models have circumscribed utility for understanding their etiology. The regulatory landscape and transcriptional activities of the human brain are unique in many respects. The human brain is characterized by relatively high levels of expression when compared to other non-neuronal tissues (de la Grange et al., 2010; Ramsköld et al., 2009) or the brains of mammalian model organisms (Cáceres et al., 2003; Enard et al., 2002; Khaitovich et al., 2004; Lin et al., 2014). Human brain also has a greater transcriptome complexity, which is reflected by the higher levels of alternative isoforms and magnitude of alternative splice events showing qualitatively different splicing patterns relative to other tissue types (Mortazavi et al., 2008; E. T. Wang et al., 2008; Yeo et al., 2004). Additionally, the complex nature of diagnosing and assessing specific symptoms of neuropsychiatric disorders is not easily translatable to model organisms. For instance, animal studies rely on loose behavioral models to simulate desired phenotypes based on the disorder of interest (Nestler & Hyman, 2010). While some models have been validated though extensive research, e.g. stress-based tests for modeling anxiety like behaviors (Blanchard et al., 2001; Cryan & Holmes, 2005; Lister, 1990), there are no substantial paradigms for modeling psychosis at the behavioral level (Jones et al., 2011; B. K. Lipska & Weinberger, 2000). In fact, many animal studies of neuropsychiatric disorders rely on chemically or genetically induced dysregulation of specific neurochemical pathways already known to be associated with a specific disorder (Crawley, 2007; Gould & Gottesman, 2005). While this approach can be useful for conducting clinical trials or confirming electrophysiological abnormalities, it is far less useful for exploratory studies identifying how underlying genetic predispositions and associated aberrant gene regulation contribute to the behavioral variation we observe with psychiatric disorders (Nestler & Hyman, 2010). In contrast, a well-informed brain bank can provide the means to investigate psychiatric disorders under the guidelines of strict clinical criteria and without the influence of experimental manipulation. Therefore, due to such noteworthy differences in human brain expression levels and alternative splicing events compared to other proxy tissues, or the unique heterogeneous nature of psychiatric disorders and the challenges of translating this complexity to model organisms, there are distinct advantages to studying human brains directly.
3. CLINICAL RELEVANCE OF POSTMORTEM BRAIN TISSUE
The human brain is a complex organ composed of multiple distinct anatomical and functional regions, and it has been shown that gene expression patterns vary considerably across these different regions (Kang et al., 2011; Strand et al., 2007). Thus, it is important to study the transcriptome in those brain regions that are most relevant to the neuropsychiatric phenotype of interest. The following sections outline how current research has implicated various brain regions to different neuropsychiatric disorders.
3.1. Bipolar Disorder:
BPD is hallmarked by dysregulation of emotional states as afflicted individuals cycle through episodes of depression and mania (American Psychiatric Association, 2013). The brain correlates of these mood abnormalities remain an active area of investigation. Current evidence from multiple lines of research (postmortem, lesion and imaging studies) point to the crucial role of the limbic system for mood regulation. Other parts of the brain (i.e., dorsolateral prefrontal cortex (DLPFC, Brodmann area 45 and 46) and hippocampus) can also influence mood in conjunction with the structures in the limbic system (Rajkowska et al., 2016; Wise et al., 2017). Based on convergent evidence, two important structures within the limbic system, the amygdala and the subgenual Anterior Cingulate Cortex (sACC), were also demonstrated to govern mood regulation in the pathology of BPD (Drevets et al., 2008). For example, a meta-analysis of 98 studies found strong evidence for enlarged ventricles, left ACC, the amygdala and the globus pallidus in BPD (Kempton et al., 2008). A complementary meta-analysis found gray matter reductions in the anterior limbic regions and the left ACC (Bora et al., 2010). Larger meta-analyses that are currently being carried out by the ENIGMA consortium show significant volumetric differences in several limbic structures, especially the amygdala with respect to BPD (Selvaraj et al., 2012). A recent meta-analysis of brain imaging studies (Chen et al., 2011) found consistent evidence for over-activation throughout the limbic system (amygdala, striatum and the medial temporal lobe) in response to emotion eliciting tasks as well as under-activation in the prefrontal cortex, supporting the hypothesis that BPD may be associated with limbic overactivity and diminished prefrontal regulation (Strakowski et al., 2012). The sACC has also been prominently studied following an initial report where reduced PET activity was identified within the sACC of depressed patients with bipolar and unipolar disorder (Holtzheimer et al., 2012). Subsequent BPD studies have confirmed reduced fMRI activation in the sACC (Chen et al., 2011) with replicated evidence of glutamate/glutamine signaling abnormalities (Chitty et al., 2013). Targeting the sACC with deep brain stimulation has also shown initial promise in the treatment of the depression in both unipolar and bipolar disorders (Holtzheimer et al., 2012). The accumulation of evidence pointing to disruptions in connections within and between the limbic system and prefrontal brain regions highlight the importance of looking specifically at these brain structures to investigate the genomic processes at the root of observed neuropathological changes. These findings further illustrate the importance of studying the effects of ancestry specific genetic polymorphisms on gene regulation within disease relevant regions of interest.
3.2. Major Depressive Disorder:
In contrast to BPD, MDD is characterized by unipolar emotional dysregulation in which individuals experience recurring periods of severe depression (American Psychiatric Association, 2013). While many of the neuroanatomical correlates are common across mood disorders, investigators have identified additional structural, functional and connectivity abnormalities associated with MDD. As mentioned previously, the limbic system, beginning with the amygdala is the most heavily implicated neuroanatomical pathway associated with emotional regulation and affect (Mayberg, 1997; Whalen et al., 2002). Mood regulation and MDD pathology is not limited by limbic system functionality, but also extends to areas of the PFC and ACC (Davidson et al., 2002). More specifically, structural magnetic imaging (MRI) has identified reduced grey matter volume between MDD cases and controls among key regions of the limbic system (amygdala, hippocampus, insula), ventrolateral prefrontal cortex (VLPFC, Brodmann area 47), DLPFC (Brodmann area 45 and 46) and ACC (subgenual ACC and rostral anterior cingulate) (Koolschijn et al., 2009; Stratmann et al., 2014). In addition to structural abnormalities discovered via MRI, functional neuroimaging studies have identified MDD associated aberrant activation and functional anomalies within the limbic system and connecting brain regions. Studies involving cognitive and emotional tasks in conjunction with fMRI analysis identified hypoactivation of the amygdala and ACC, whereas hyperactivation of the DLPFC and striatum was associated with MDD related emotional processing (Diener et al., 2012). Findings from the ENIGMA meta-analysis showed that MDD cases have thinner cortical gray matter in the orbitofrontal cortex (OFC), anterior and posterior cingulate, insula and temporal lobes (Schmaal et al., 2017) as well as lower hippocampal volumes (Schmaal et al., 2016). Recent advances in resting state functional connectivity (RSFC) analyses, in which regional neuronal interactions are analyzed during task-negative or resting sate experimental conditions, have identified other neural networks associated with MDD (Kaiser et al., 2015). RSFC and resting-state networks have also implicated additional brain regions potentially associated with MDD such as the anterior insula, cerebellum, and thalamus (Greicius et al., 2007; Kaiser et al., 2015; Liu et al., 2010). In addition to MDD associated functional, structural and connectivity abnormalities, circadian rhythm disruption is a well-known symptom of MDD with brain region specific underpinnings (McCarthy & Welsh, 2012; Kronfeld-Schor & Einat, 2012). Circadian rhythm disruption is believed to be the product of dysregulated genes responsible for diurnal cycle i.e., the so called “clock genes” (Wu, et al., 2006; Ackermann, et al., 2007). The cyclical upregulation and downregulation of these select genes is weakened within the brains of patients with MDD (Li, et al., 2013). Interestingly, it was further shown that clock genes, such as brain and muscle Arnt-like protein-1 (BMAL1), period circadian regulator 1, 2, and 3, (PER1, PER2, PER3), and D-site binding protein (DBP), showed dysregulated expression within the ACC of MDD cases (Bunney et al., 2015). The substantial evidence pointing to MDD pathology as the potential product of neuroanatomical abnormalities only emphasizes the importance of postmortem brain studies to understand how genetic predispositions and gene regulation underlie dysfunction within MDD relevant tissues.
3.3. Schizophrenia:
The rationale for using postmortem tissue in the study of neuropsychiatric disorders is best highlighted when investigating the pathology of schizophrenia. SCZ is a debilitating psychiatric disorder characterized by neuroanatomical dysfunction resulting in cognitive impairment and psychosis (American Psychiatric Association, 2013). One main structural abnormality observed in SCZ patients is regional volume decreases in both grey and white matter. A meta-analysis of 15 voxel-base morphometry studies identified that the left superior temporal gyrus and the left medial temporal lobe showed the most significant deficits in grey matter volumes when comparing SCZ cases and controls (Honea et al., 2005). Additionally, longitudinal studies of SCZ patients have identified global and localized (left superior frontal area (Brodmann areas 9/10), left superior temporal gyrus (Brodmann area 42), right caudate nucleus, and right thalamus) decreases in grey matter density as the disease develops (van Haren et al., 2007). Structural brain abnormalities associated with SCZ are not limited to areas of the cortex and limbic systems but also include connecting white matter tracts (S. Kelly et al., 2018). More importantly, genetic risk variants associated with SCZ have been correlated with deficits in white matter and total brain volume (Terwisscha van Scheltinga et al., 2013). Contemporary dysconnectivity theories suggest that SCZ is the result of deficits in information integration between brain regions, specifically areas such as the PFC and ACC (Lynall et al., 2010; Shon et al., 2018), resulting in the progressive loss of synaptic plasticity (Stephan et al., 2006). There is additional evidence to suggest that functional connectivity deficits associated with SCZ might be under genetic control. For instance, the rs6039769 variant associated with both SCZ and BPD is in the promoter region of SNAP25, a gene involved in synaptic development and plasticity (Houenou et al., 2017). Based on familial history of SCZ diagnosis, genetically high-risk individuals show decreased neuronal connectivity within areas of the PFC and ACC (Jang et al., 2011). The convergent evidence suggesting that SCZ associated brain abnormalities have a potential genetic underpinning highlights the need for increased research on clinically relevant tissue types, specifically postmortem brain.
4. POSTMORTEM GENE EXPRESSION STUDIES
Availability of postmortem brain tissues is a must to investigate the impact of genetic variants on gene expression for a specific neuropsychiatric disorder. Transcriptomic studies have shown that the brain maintains the highest levels of gene expression and transcriptome complexity relative to other tissue types (GTEx Consortium, 2013; Su et al., 2004), including a substantial gene expression differences between brain regions and neuronal cell types (Hawrylycz et al., 2012). The unique transcriptome complexity in the brain is further augmented by the increased levels of alternative isoform expression resulting from neuronal tissue specific alternative splice events (Pan et al., 2008). Finally, several transcriptomic studies have reported that a significant proportion of the protein coding genes in brain show spatiotemporal expression patterns with greatest expression changes occurring prenatally (Kang et al., 2011; M. Li et al., 2018; D. Wang et al., 2018). Therefore, the significant distinctions between brain tissue and other tissue types further emphasizes the importance of studying human brains directly, especially in the context of neuropsychiatric disorders.
4.1. Alternative Splicing in the Brain:
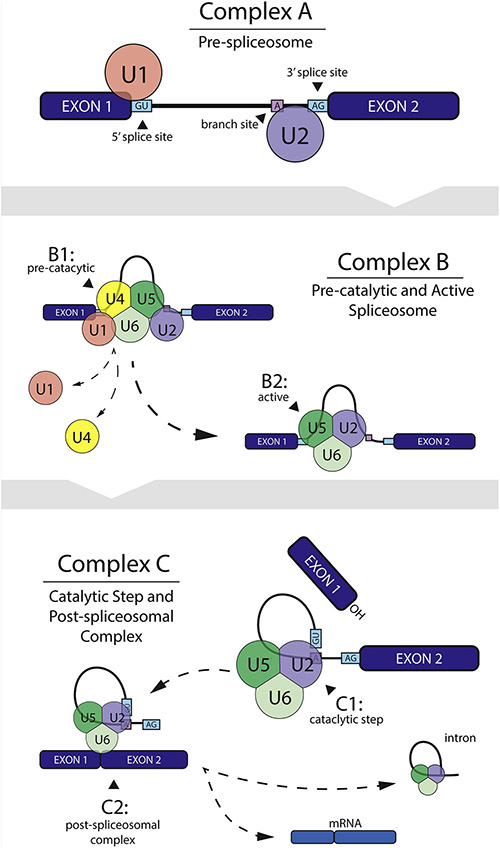
The splicing of precursor mRNA is important for the post-transcriptional regulation of gene expression. Alternative splicing allows for significant expansion of the human proteome relative to other species with a similar number of genes (Irimia & Blencowe, 2012; Will & Lührmann, 2011). This process is facilitated primarily by large ribonucleoprotein complexes called spliceosomes (Figure 2.0) (Will & Lührmann, 2011). As mentioned previously, the human transcriptome contains more alternative splice variants than any model organism with 88% of transcribed genes containing two or more isoforms (Barbosa-Morais et al., 2012; Merkin et al., 2012). Neuronal genes are especially susceptible to post-transcriptional regulation via increased alternative splicing events, in part due to brain-specific splicing factors (e.g., nPTB, NOVA1, NOVA2 and Hu/Elav proteins) (Ule et al., 2005). This is especially important for understanding disease etiology because differential expression of alternative transcripts within postmortem brains has been associated with the clinical diagnosis of neuropsychiatric disorders (Ernst et al., 2009; Lau & Zukin, 2007; Nakata et al., 2009a). For instance, dysregulated splicing events within exon11a of ENAH and the 3’ UTR of CPNE3 have been associated with SCZ (O. S. Cohen et al., 2012). These studies among others, highlight alternative splicing with disorder specific changes in alternative isoform expression as an underlying functional link between genetic variation and neuropsychiatric etiopathology.
Figure 2: U2-Type Spliceosome Facilitated pre-mRNA Splicing Mechanism.
Small nuclear ribonuceoprotein (snRNP) U2 facilitated spliceosome assembly begins with the U1 snRNP associating with the 5’ splice site and U2 at the branch point (complex A). The pre-assembled U4/U5/U6 tri-snRNP is recruited to form the pre-catalytic spliceosome (complex B1). U1 and U4 snRNPs are then destabilized by various protein interactions resulting in the catalytically active spliceosome (complex B2). The U2/U5/U6 spliceosome complex then facilitates intron excision and exon ligation via two step catalytic process (complex C).
4.2. Non-coding RNA species in the Brain:
Non-coding RNAs (ncRNA) are another class of molecule that have recently begun to be studied using postmortem brain tissue from cases with various neuropsychiatric disorders. These RNA molecules have very limited to no protein translation capabilities and include both short microRNA (miRNA), and long non-coding RNA (lncRNA) as well as certain transposable elements (TE) (Esteller, 2011).
4.2.1. MiRNA Studies in the Brain:
MiRNAs are small ncRNA (≈22 bp), the biogenesis of which is a three-step process starting in the cell nucleus and ending with the generation of the mature miRNA in the cytoplasm (Bartel, 2004). Most miRNAs regulate gene function mainly negatively through imperfect binding with the 3' untranslated region (3’UTR) of mRNA (John, et al., 2004; Ritchie, et al., 2009). Depending on homology, miRNA can affect either the degradation of an mRNA target or its translational inhibition (Lewis, et al., 2005). MiRNAs further contribute to the transcriptome complexity of the brain with two thirds of all miRNA being expressed in the brain and implicated in neurodevelopment and plasticity (Fineberg, et al., 2009; Xu, et al., 2010). Studies that have assessed miRNA expression in postmortem brain tissues have identified differentially expressed miRNA between cases with various neuropsychiatric disorders, such as schizophrenia, bipolar disorder and major depression (Perkins et al., 2007; Kim, et al., 2010; Moreau, et al., 2011; Smalheiser, et al., 2014; Azevedo et al., 2016). For example, one study profiling the expression of 256 different miRNAs identified 15 significantly upregulated miRNAs and one significantly downregulated within the prefrontal cortex of individuals with SCZ and schizoaffective disorder (Perkins, et al., 2007). Another study showed that downregulated miRNAs within the PFC of SCZ, BPD, and MDD cases are especially enriched at the synapse (Smalheiser, et al., 2014).
4.2.2. LncRNA expression in the Brain:
In contrast to miRNA, lncRNAs are defined as a larger non-coding RNA species (>200 bps), which interestingly share many features with the protein-coding genes, such as promoter regions, intron/exon junctions, and alternative splice variants (Quinn & Chang, 2016). Although, few isolated lncRNA (i.e. most notably the X-inactive specific transcript (XIST)) have been known and studied in great details (Yang, et al., 2018), it was not until recently that we have become aware of the sheer size of the lncRNA transcriptome and their importance for the development of many psychiatric and neurological disorders (Wu, et al., 2013; Zuo, et al., 2016). Not surprisingly, LncRNA have already been implicated in neuronal development (Qureshi & Mehler, 2012; Zhang, et al. 2017), higher order cognitive functioning (Barry & Mattick, 2012), and neuropsychiatric disorder etiopathology (Lin, et al., 2011; Liu et al., 2014). While the diverse functional properties of lncRNA in the brain are still poorly understood, the consensus is they participate in activity-dependent gene regulation through three-way binding interactions with DNA, RNA, and protein (Andersen & Lim, 2018). In respect to neuropsychiatric disorders, a limited number of studies have already identified differentially expressed lncRNAs in postmortem brains from subjects with SCZ and controls (Tian, et al., 2018; Liu, et al., 2018). One study in particular utilized a network based approach to determine the functional annotation of lncRNAs associated with SCZ disorder status (Tian, et al., 2018).
4.2.3. Transposable Elements (TE):
TEs, or transposons, are low-complexity genomic sequences that constitutes over half of the human genome, and primarily located in intergenic regions (Slotkin & Martienssen, 2007). Of the varied types of TEs present throughout the human genome, human endogenous retroviruses (HERVs) have been associated with multiple neuropsychiatric disorders. HERVS are a low-mobility TE that make up around 8% of the human genome and are believed to be the remnants of an ancient viral infection (Guffanti et al., 2014). A handful of studies have identified abnormal expression of HERVs, specifically the HERV(-K) and HERV(-W) families, within the postmortem brains of individuals with SCZ, BPD, and MDD (Yolken, et al., 2000 ;Frank et al., 2005; Weis et al., 2007).
4.3. Methods for Quantifying Gene Expression:
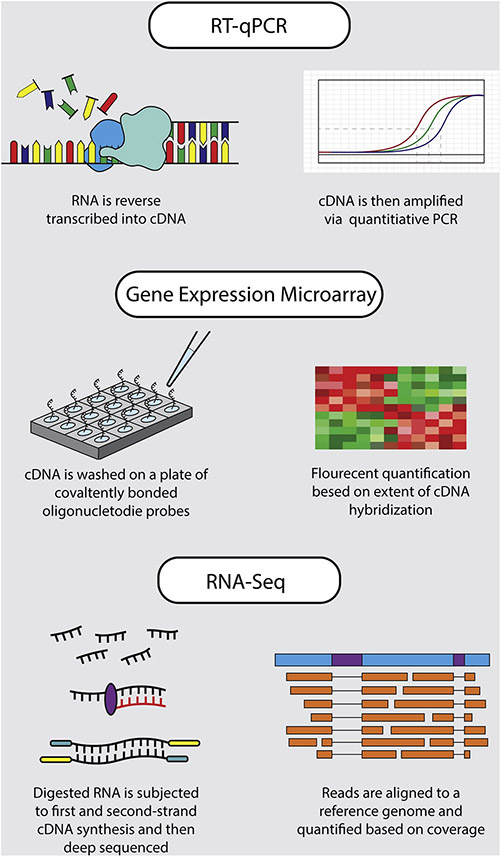
Over the years different approaches and platforms evolved to study gene expression in the brain (Figure 3.0). Initially, these were centering on low-throughput platforms such as polymerase chain reactions (PCR) that are ideal for detection of single gene expression differences. However, as the detection technologies were further developed, this shifted towards a high-throughput approaches capable of simultaneous assessment of the entire transcriptome either via the older hybridization (i.e. gene expression microarray) and/or the newer (RNA sequencing) platforms. As these platforms have their own advantages and limitations it is important to carefully consider, depending on the study design and sample size, which methodology is best suited to test the specific research hypothesis.
Figure 3: Gene Expression Analysis Methods.
A visual summary of the main steps for each of the three most commonly used methods for quantifying gene expression.
4.3.1. Reverse-Transcription Quantitative Polymerase Chain Reaction (RT-qPCR):
The reverse-transcription quantitative PCR (RT-qPCR) has become the “gold” standard for both targeted gene expressions analysis and experimental validation of microarray and RNA-seq (VanGuilder et al., 2008). Although the unparalleled specificity and sensitivity of RT-qPCR approaches is ideal for small scale study design and validation, it is not without disadvantages. More specifically, the widespread use of this method has resulted in the development of different protocols for each step of the experimental design. A concerted effort by various research groups attempted to solve this problem led to the creation of the “Minimum Information for Publication of Quantitative Real-time qPCR Experiments” (MIQE) (Bustin et al., 2009), which is a guideline to promote consistency between different protocols and laboratories. However, it is still not ubiquitously practiced, resulting in problems of reproducibility within and between research groups (Taylor et al., 2010). Additionally, the quality and purity of RNA samples has a significant impact on the PCR efficiency affecting transcript amplification and leading to confounding variability not associated with case/control status (Fleige & Pfaffl, 2006).
4.3.2. Gene Expression Microarray:
Based on the principals of complementary DNA (cDNA) synthesis and nucleic acid hybridization, gene expression microarrays investigate the expression of thousands of genes in parallel (Schena et al., 1995). Although parallel processing of multiple samples across thousands of genes has obvious utility, microarrays are not without limitation. The detection limit for determining significant differences in transcript abundance of both oligonucleotide and cDNA microarrays poses challenges for postmortem brain studies, specifically because many disease causing transcripts are expressed in low-abundance, below the detection limit of the microarray (Schulze & Downward, 2001). Additionally, the collection of probes available on any one specific array is usually limited to detecting main gene isoforms, thus eliminating the possibility of detecting differential expression of alternative transcripts. There are however, specialized microarrays that attempt to capture splice variants based on oligonucleotide probes designed to span unique exon junctions specific to alternative transcripts (Clark et al., 2002).
4.3.3. RNA-Sequencing:
RNA-seq provides the possibility for highly sensitive transcriptome profiling by deep-sequencing. This is incredibly useful for determining the gene expression levels for alternative splice variants in parallel and the potential for identifying novel RNA transcripts (Trapnell et al., 2012). Unlike microarrays, RNA sequencing allows for a greater dynamic range of expression levels based on the extent of coverage and depth in transcript detection (Z. Wang et al., 2009). While RNA-seq methodology has been significantly optimized since its first inception over a decade ago, it still has some technical issues and limitations (Marioni et al., 2008), such as 3’ sequence biases and gene length bias that occur during sample extraction and cDNA library preparation (Roberts et al., 2011). Additionally, the extent of sample preparation and data processing and storage involved in obtaining gene expression levels via RNA-seq is computationally expensive and laborious, requiring a team of experienced bioinformaticians and molecular geneticists (Z. Wang et al., 2009). While this method has become the new standard for large scale transcriptome profiling, it is still important to note that RNA-sequencing requires validation via independent platforms such as RT-qPCR.
4.4. Integrating GWAS Data and Differential Expression via eQTL:
There is a continued emphasis to provide functional explanations for disease associated genetic variants identified in GWAS. Expression quantitative trait loci (eQTL) mapping has become one of the most successful tools in assessing how genetic variants affects gene regulation. In eQTL studies, gene expression levels are treated as quantitative traits which are then interrogated via various genetic mapping techniques (Gilad et al., 2008). The advent of large GWAS studies for neuropsychiatric disorder and the identification of genome-wide significant polymorphisms (Wu & Pan, 2018) has highlighted the emerging need for eQTL studies in human brain tissue. To date, there have been several successful studies implementing this methodology to link genetic variants to differential gene expression in the study of neuropsychiatric phenotypes (Table 2.0). More specifically, one study implicates ZNF323 as a potential SCZ risk gene via eQTL analysis of the PGC GWAS identified SNP rs1150711 (Luo et al., 2015). Similarly, in another postmortem brain study a significant association was found between the differential expression of brain-specific AN3K isoforms and BP-associated risk variants (rs1938526) (Rueckert et al., 2013). Studying eQTLs is not just limited to the investigation of a single gene, but also to a whole gene networks in which the eQTL impact on the network functions is studied under a broader biological framework to better understand disease neuropathology (Mamdani et al., 2015). Thus, these studies highlight the clinical significance of integrating GWAS and eQTL data, specifically in postmortem brains, where careful consideration given to allelic frequency variation across different ethnic background will help to identify potential diagnostic or pharmacological targets.
Table 2.
Selection of Postmortem Gene Expression Studies
| Citation | Sample | Brain Regions | Gene Expression Method |
|---|---|---|---|
| Nakata et al., 2009 | 61 SCZ, 30 CON | Hippocampus | RT-qPCR |
| 29W, 62NW | |||
| Kim et al., 2010 | 35 SCZ, 35 BPD, 33 CON | DLPFC (BA46) | RT-qPCR (miRNA) |
| 102W, 1NW | |||
| Choi et al., 2011 | 40 BPD, 43 CON | PFC (BA46) | Microarray |
| 83W | |||
| Ramaker et al., 2017 | 24 SCZ, 24 BPD, 24 MDD, 24 CON | ACC, DLPFC (BA46), NAc | RNA-seq |
| 89W, 7NW | |||
| Darby et al., 2016 | 35 SCZ, 33 BPD, 32 CON | Hippocampus | RNA-seq |
| 98W, 2 NW | |||
| Pandey et al., 2018 | 31 SCZ, 24 CON | PFC (BA9) | RT-qPCR |
| 38W, 17NW | |||
| Pantazatos et al., 2017 | 30 MDD, 29 CON | PFC (BA9) | RNA-seq |
| 59 W | |||
| Hwang et al., 2013 | 14 SCZ, 15 CON | Hippocampus | RNA-seq |
| 29W | |||
| Schmitt et al., 2010 | 10 SCZ, 9 CON | Cerebellum | RT-qPCR |
| 19W | |||
| Fromer et al., 2016 | 258 SCZ, 279 CON | DLPFC (BA46, BA9) | RNA-seq |
| 433W, 104NW | |||
| Williamson et al., 2015 | 27 SCZ, 29 BPD, 22 CON | DLPFC (BA46) | RT-qPCR (miRNA) |
| 77W, 1NW | |||
| Kim et al., 2010 | 10 BPD, 10 CON | PFC (BA9) | RT-qPCR |
| 20W | |||
| Li et al., 2013 | 34 MDD, 55 CON | DLPFC, ACC, NAc, HIPP, CB, AMYG | Microarray |
| 89W | |||
| Kaalund et al., 2014 | 176 SCZ, 61 BPD, 138 MDD, 326 CON | DLPFC, HIPP, Caudate nucleus | RT-qPCR |
| 403W, 296NW | |||
| Gray et al., 2015 | 53 MDD, 32 CON | DLPFC | RT-qPCR |
| 58W, 27NW | |||
Postmortem gene expression studies with information about sample sizes and demographics, brain regions investigated, and method for gene expression quantification.
SCZ= schizophrenia, BPD= bipolar disorder, MDD= major depressive disorder CON= control
W= White, NW= non-white
4.5. Postmortem Gene Expression Studies and Neurodevelopment:
It is well documented that the biological underpinnings of neuropsychiatric etiopathology can begin well before the onset of symptoms, sometimes even prenatally (Birnbaum, et al., 2014; Cristino, et al., 2014). SCZ, in particular, has been extensively studied in respect to neurodevelopmental risk factors (Weinberger, 2017), with many of the GWAS of SCZ loci implicated in processes related to synaptic developmental (Fromer et al., 2014; Hall, et al., 2015). Furthermore, comparison of pre- and post-natal expression profiles have shown increased expression levels for genes within the SCZ GWAS implicated loci in fetal brains relative to postnatal brains (Birnbaum & Weinberger, 2017; Jaffe et al., 2015). However, in contrast to SCZ, the neurodevelopmental processes underlying BPD and MDD have been less studied and are much less understood (Bale et al., 2010). The lack of neurodevelopmental understanding in respect to neuropsychiatric disorders is a result of limited postmortem samples spanning different developmental periods and the often late onset of symptoms for BPD, MDD and SCZ making case/control identification difficult (Klein et al., 1999; Gogtay,et al., 2011; Perlis et al., 2009). Therefore, to improve our understanding of the neuropathology of psychiatric disorders, current and future postmortem brain banks will require, not only expansion of the ethnic diversity of their collection, but also an active effort towards collecting tissue across different neurodevelopmental stages.
5. FUTURE OF POSTMORTEM BRAIN STUDIES
GWAS of neuropsychiatric disorders continue to identify an ever increasing number of genetic variants associated with, as of now, still unknown functional significance. As previously stated, it is well supported that most of the pathological abnormalities underlying these disorders reside in localized regions of the brain. However, to properly use postmortem brain tissue to study neuropsychiatric disorders, we argue for implementation of larger postmortem brain banks from ethnically diverse populations. Below, we address two major limitations (i.e., sample size and ethnic diversity) that currently face the field and propose solutions to address these limitations in the postmortem brain research of neuropsychiatric disorders. While, due to a recent establishment of various genomic consortia (e.g. PsychEncode), the need for greater sample sizes has become less urgent, expanding the ethnic diversity of potential donors remains an important necessity for providing insight into the population specific disease associated genetic mechanisms.
5.1. Limited Power of Post Mortem Brain Studies:
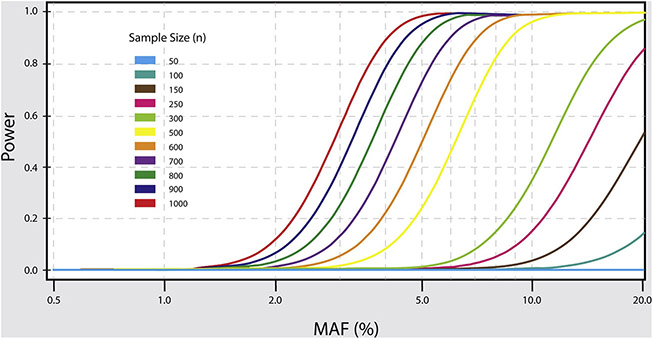
Over the past three decades there has been a concerted effort to collect postmortem brains from individuals afflicted with psychiatric disorder. In the last 10 years, postmortem brain databases worldwide have begun to increase substantially their postmortem brain repositories spanning multiple disease phenotypes and ethnic groups (Anderson et al., 2001; A. Schmitt et al., 2007). Unfortunately, with few exceptions until recently, the majority of the postmortem brain databases contain relatively small sample sizes due to logistic, ethical and cultural obstacles, effectively limiting the research community and inhibiting their ability to detect significant findings (Deep-Soboslay et al., 2011; Xiao et al., 2009). This is especially important in the context of current and future eQTL studies, in which the statistical power is a function of minor allele (MAF) frequency, effect size, and sample size (Schliekelman, 2008) (Figure 4.0). Majority of eQTL studies have suffered from limited statistical power that has had consistent and mostly negative impacts on genetic and eQTL studies of neuropsychiatric disorders (Huang et al., 2018). For example, the limited sample size has a substantial impact on identifying, the so-called trans-eQTLs (i.e. eQTL that are associated with expression of remote genes as opposed to cis-eQTL that reside close to the transcription start site of the target gene) (Yao et al., 2017), which even with the currently increasing sample sizes are still underpowered (Huang et al., 2018). Given that effect sizes recorded in neuropsychiatric disorders are notoriously low, study design requires large samples to identify reliable phenotypic differences between cases and controls (Bezeau & Graves, 2001). Additionally, the positive relationship between MAF and statistical power limits genetic studies to investigating only common variants (MAF >1%) (de Bakker et al., 2005). When working with small effect sizes and relatively low allele frequencies (MAF > 10%) studies have shown inflated false discovery rates (FDR) for eQTL associated genes (Huang et al., 2018). Although statistical methods attempt to mitigate issues of low MAF and small effect sizes (Zhu et al., 2016), increasing sample size remains an efficient way to increase statistical power and study design. This only further emphasizes the importance of expanding current postmortem brain banks to include greater samples sizes for genetic and transcriptomic studies of neuropsychiatric disorders.
Figure 4: Power Estimation for eQTL Studies.
The graph shows the relationship between statistical power, minor allele frequency (MAF) and sample size for detection of eQTLs. As shown by the graph successful detection of less common eQTLs is only possible with larger sample sizes, i.e. to detect eQTLs with MAF of 5% we need at least a sample size of N ≥800 brains.
5.2. Limited Diversity among Postmortem Brain Studies:
Aside from small sample sizes, the lack of ethnic diversity substantially impacts study design. Genetic studies have long identified population stratification as a major confounding factor when investigating disease associated polymorphisms due to the significant variation in allele frequencies between ethnic groups (T. G. P. Consortium, 2012; Rosenberg et al., 2002). Additionally, disregard for population specific genetic variation, can lead to substantial difficulties to interpret the eQTL data, especially for variants with significantly varying allele frequencies between populations or even not present in the analyzed sample. As an example, while the China Oxford and VCU Experimental Research on Genetic Epidemiology (CONVERGE) consortium has identified significantly associated genetic risk variants for MDD using a relatively large (N= 6000 cases/6000 controls) clinically and genetically homogenous sample of Han Chinese women (Converge Consortium, 2015), one immediate obstacle in translating these GWAS findings to clinically relevant neurobiological mechanisms is the overall lack of ethnically comparable brain repositories composed of Chinese postmortem samples that share the same allelic frequencies as the CONVERGE subjects. Moreover, all available postmortem brain banks worldwide are composed primarily from subjects with European or North American genetic backgrounds (Palmer-Aronsten et al., 2016). The limited tissue contribution to postmortem brain banks by donors from different ethnic groups may be a result of negative social, political and cultural ideations in respect to organ donation. It is well understood, at least in the North American medical community, that minority groups are less likely to participate in organ donation compared to Caucasians (Michie et al., 2011; Xiao et al., 2009). For these reasons, initiating international collaborations aimed at establishing postmortem brain repositories dedicated to expansion of the ethnic diversity of potential donors would greatly improve research outcomes for understanding how genetic predispositions impact neuropsychiatric etiopathology across different populations.
5.3. International Postmortem Brain Bank Collaborations:
One potential solution to diversify the ethnic composition of the current postmortem brain banks is through collaboration with research institutions from countries such as China. For example, the largest postmortem brain bank in China was established in 2012 by the Chinese Academy of Medical Sciences/Peking-Union Medical College (CAMS/PUMC) and currently contains samples from 224 donors (Zhang et al., 2018). To provide a shared platform and database for academic exchange, the China Human Brain Bank Consortium (CHBBC) was formally established in May 2016 through the joint efforts of 10 medical schools and universities across China (Qiu et al., 2018; Zhang et al., 2018). CHBBC goals for international sharing and integration of genomic data will undoubtedly contribute substantially to the clarification of differences among ethnic groups while paving the way for collaborations with other brain banks. Genetic and transcriptomic comparisons between different ethnic groups following postmortem brain tissue sharing between research institutions is fundamental to elucidating the molecular mechanisms underlying human diseases.
6. CONCLUDING REMARKS
The importance of establishing robust postmortem brain banks with increased sample sizes from diverse ethnic backgrounds cannot be overstated, especially when investigating the biological mechanisms of disease risk eQTLs underlying neuropsychiatric etiopathology. Integrating findings from large GWAS and high-throughput gene expression studies will inform the functional relevance of disease-associated risk variants. While in recent years the statistical power has become less concerning, the lack of ethnic diversity among the current postmortem brain banks still significantly impedes the progress towards understanding the genetic mechanisms underlying neuropsychiatric disorders. Within the scientific community, a greater emphasis should be placed on the development of robust ethnically diverse postmortem brain databases through public education on the importance of organ donation and international scientific collaboration. The wealth of information provided from expanded postmortem brain studies will not only increase our understanding of individual differences role in disease susceptibility, but also yield important insights into ethnicity specific occurrence rates, progression and treatment of neurological diseases, and more specifically neuropsychiatric disorders.
Highlights.
Gene expression serves as a functional intermediate between genotype and phenotype.
The etiologically relevant tissue for studying neuropsychiatric disorders is brain.
Most current postmortem brain banks lack in ethnic diversity and sample sizes.
Acknowledgments
Funding
The authors are supported by grants from the National Institutes on Alcohol Abuse and Alcoholism [grant #1R21AA022749-01A1]; National Natural Science Foundation (#91546203); National Natural Science Foundation of China (NSFC #81771205, #91632113); the Natural Science Foundation and Major Basic Research Program of Shanghai (16JC1420500, 16JC1420502); and the CAMS Innovation Fund for Medical Sciences (CIFMS #2017-I2M-3-008).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ackermann K, Dehghani F, Bux R, Kauert G, & Stehle JH (2007). Day-night expression patterns of clock genes in the human pineal gland. Journal of Pineal Research, 43(2), 185–194. 10.1111/j.1600-079X.2007.00461.x [DOI] [PubMed] [Google Scholar]
- Anderson R, Balls M, Burke MD, Cummins M, Fehily D, Gray N, … Ylikomi T (2001). The establishment of human research tissue banking in the UK and several western European countries. The report and recommendations of ECVAM Workshop 44. Alternatives to laboratory animals: ATLA, 29(2), 125–134. [DOI] [PubMed] [Google Scholar]
- Andersen RE, & Lim DA (2018). Forging our understanding of lncRNAs in the brain. Cell and Tissue Research, 371(1), 55–71. 10.1007/S00441-017-2711-z [DOI] [PubMed] [Google Scholar]
- Association, A. P (2013). Diagnostic and Statistical Manual of Mental Disorders (DSM-5®): American Psychiatric Pub. [Google Scholar]
- Azevedo JA, Carter BS, Meng F, Turner DL, Dai M, Schatzberg AF, … Thompson RC (2016). The microRNA network is altered in anterior cingulate cortex of patients with unipolar and bipolar depression. Journal of Psychiatric Research, 82, 58–67. 10.1016/j.jpsychires.2016.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale TL, Baram TZ, Brown AS, Goldstein JM, Insel TR, McCarthy MM, … Nestler EJ (2010). Early Life Programming and Neurodevelopmental Disorders. Biological Psychiatry, 68(4), 314–319. 10.1016/j.biopsych.2010.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, … Blencowe BJ (2012). The evolutionary landscape of alternative splicing in vertebrate species. Science (New York, N.Y.), 338(6114), 1587–1593. doi: 10.1126/science.1230612 [DOI] [PubMed] [Google Scholar]
- Barry G, & Mattick JS (2012). The role of regulatory RNA in cognitive evolution. Trends in Cognitive Sciences, 16(10), 497–503. 10.1016/j.tics.2012.08.007 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116(2), 281–297. [DOI] [PubMed] [Google Scholar]
- Bezeau S, & Graves R (2001). Statistical power and effect sizes of clinical neuropsychology research. Journal of Clinical and Experimental Neuropsychology 23(3), 399–406. doi: 10.1076/jcen.23.3.399.1181 [DOI] [PubMed] [Google Scholar]
- Birnbaum R, Jaffe AE, Hyde TM, Kleinman JE, & Weinberger DR (2014). Prenatal expression patterns of genes associated with neuropsychiatric disorders. The American Journal of Psychiatry, 171(7), 758–767. 10.1176/appi.ajp.2014.13111452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JA, Wang C, Hernandez D, Siedlak SL, Rodgers MS, Achar RK, … Lee H.-g. (2016). Individual Case Analysis of Postmortem Interval Time on Brain Tissue Preservation. PLOS ONE, 11(3), e0151615. doi: 10.1371/journal.pone.0151615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard RJ, McKittrick CR, & Blanchard DC (2001). Animal models of social stress: effects on behavior and brain neurochemical systems. Physiology & Behavior, 73(3), 261–271. doi: 10.1016/S0031-9384(01)00449-8 [DOI] [PubMed] [Google Scholar]
- Bora E, Fornito A, Yücel M, & Pantelis C (2010). Voxelwise meta-analysis of gray matter abnormalities in bipolar disorder. Biological Psychiatry, 67(11), 1097–1105. doi: 10.1016/j.biopsych.2010.01.020 [DOI] [PubMed] [Google Scholar]
- Bunney BG, Li JZ, Walsh DM, Stein R, Vawter MP, Cartagena P, … Bunney WE (2015). Circadian dysregulation of clock genes: clues to rapid treatments in major depressive disorder. Molecular Psychiatry, 20(1), 48–55. 10.1038/mp.2014.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, … Wittwer CT (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry 55(4), 611–622. doi: 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- Cáceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, Geschwind DH, … Barlow C (2003). Elevated gene expression levels distinguish human from non-human primate brains. Proceedings of the National Academy of Sciences of the United States of America, 100(22), 13030–13035. doi: 10.1073/pnas.2135499100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-H, Suckling J, Lennox BR, Ooi C, & Bullmore ET (2011). A quantitative meta-analysis of fMRI studies in bipolar disorder. Bipolar Disorders, 13(1), 1–15. doi: 10.1111/j.1399-5618.2011.00893.x [DOI] [PubMed] [Google Scholar]
- Chitty KM, Lagopoulos J, Lee RSC, Hickie IB, & Hermens DF (2013). A systematic review and meta-analysis of proton magnetic resonance spectroscopy and mismatch negativity in bipolar disorder. European Neuropsychopharmacology: The Journal of the European College of Neuropsychopharmacology, 23(11), 1348–1363. doi: 10.1016/j.euroneuro.2013.07.007 [DOI] [PubMed] [Google Scholar]
- Choi KH, Higgs BW, Wendland JR, Song J, McMahon FJ, & Webster MJ (2011). Gene Expression and Genetic Variation Data Implicate PCLO in Bipolar Disorder. Biological Psychiatry, 69(4), 353–359. doi: 10.1016/j.biopsych.2010.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TA, Sugnet CW, & Ares M (2002). Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science (New York, N.Y.), 296(5569), 907–910. doi: 10.1126/science.1069415 [DOI] [PubMed] [Google Scholar]
- Cohen D (1997). A critique of the use of neuroleptic drugs in psychiatry In From placebo to panacea: Putting psychiatric drugs to the test (pp. 173–228). Hoboken, NJ, US: John Wiley & Sons Inc. [Google Scholar]
- Cohen OS, McCoy SY, Middleton FA, Bialosuknia S, Zhang-James Y, Liu L, … Glatt SJ (2012). Transcriptomic analysis of postmortem brain identifies dysregulated splicing events in novel candidate genes for schizophrenia. Schizophrenia Research, 142(1–3), 188–199. doi: 10.1016/j.schres.2012.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, G. T. (2013). The Genotype-Tissue Expression (GTEx) project. Nature Genetics, 45(6), 580–585. doi: 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, T. G. P. (2012). An integrated map of genetic variation from 1,092 human genomes. Nature, 491(7422), 56. doi: 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converge, C., Cai N, Bigdeli TB, Kretzschmar W, Li Y, Liang J, … Flint J (2015). Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature, 523(7562), 588–591. doi: 10.1038/nature14659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley JN (2007). What's wrong with my mouse? Behavioral phenotyping of transgenic and knockout mice, 2nd ed. Hoboken, NJ, US: John Wiley & Sons Inc. [Google Scholar]
- Cristino AS, Williams SM, Hawi Z, An J-Y, Bellgrove MA, Schwartz CE, … Claudianos C (2014). Neurodevelopmental and neuropsychiatric disorders represent an interconnected molecular system. Molecular Psychiatry, 19(3), 294 10.1038/mp.2013.16 [DOI] [PubMed] [Google Scholar]
- Cryan JF, & Holmes A (2005). Model organisms: The ascent of mouse: advances in modelling human depression and anxiety. Nature Reviews Drug Discovery 4(9), 775–790. doi: 10.1038/nrd1825 [DOI] [PubMed] [Google Scholar]
- Darby MM, Yolken RH, & Sabunciyan S (2016). Consistently altered expression of gene sets in postmortem brains of individuals with major psychiatric disorders. Translational Psychiatry 6(9), e890. doi: 10.1038/tp.2016.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RJ, Pizzagalli D, Nitschke JB, & Putnam K (2002). Depression: perspectives from affective neuroscience. Annual Review of Psychology 53, 545–574. doi: 10.1146/annurev.psych.53.100901.135148 [DOI] [PubMed] [Google Scholar]
- de Bakker PIW, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, & Altshuler D (2005). Efficiency and power in genetic association studies. Nature Genetics, 37(11), 1217–1223. doi: 10.1038/ng1669 [DOI] [PubMed] [Google Scholar]
- de la Grange P, Gratadou L, Delord M, Dutertre M, & Auboeuf D (2010). Splicing factor and exon profiling across human tissues. Nucleic Acids Research, 38(9), 2825–2838. doi: 10.1093/nar/gkq008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deep-Soboslay A, Benes FM, Haroutunian V, Ellis JK, Kleinman JE, & Hyde TM (2011). Psychiatric brain banking: three perspectives on current trends and future directions. Biological Psychiatry 69(2), 104–112. doi: 10.1016/j.biopsych.2010.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener C, Kuehner C, Brusniak W, Ubl B, Wessa M, & Flor H (2012). A meta-analysis of neurofunctional imaging studies of emotion and cognition in major depression. Neuroimage, 61(3), 677–685. doi: 10.1016/j.neuroimage.2012.04.005 [DOI] [PubMed] [Google Scholar]
- Drevets WC, Savitz J, & Trimble M (2008). The Subgenual Anterior Cingulate Cortex in Mood Disorders. CNS spectrums, 13(8), 663–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink E, Zhu J, … Stefansson K (2008). Genetics of gene expression and its effect on disease. Nature, 452(7186), 423–428. doi: 10.1038/nature06758 [DOI] [PubMed] [Google Scholar]
- Enard W, Khaitovich P, Klose J, Zöllner S, Heissig F, Giavalisco P, … Pääbo S (2002). Intra- and interspecific variation in primate gene expression patterns. Science (New York, N.Y.), 296(5566), 340–343. doi: 10.1126/science.1068996 [DOI] [PubMed] [Google Scholar]
- Ernst C, Deleva V, Deng X, Sequeira A, Pomarenski A, Klempan T, … Turecki G (2009). Alternative splicing, methylation state, and expression profile of tropomyosin-related kinase B in the frontal cortex of suicide completers. Archives of General Psychiatry, 66(1), 22–32. doi: 10.1001/archpsyc.66.1.22 [DOI] [PubMed] [Google Scholar]
- Esteller M (2011). Non-coding RNAs in human disease. Nature Reviews Genetics, 12(12), 861 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- Fineberg SK, Kosik KS, & Davidson BL (2009). MicroRNAs potentiate neural development. Neuron, 64(3), 303–309. 10.1016/j.neuron.2009.10.020 [DOI] [PubMed] [Google Scholar]
- Fleige S, & Pfaffl MW (2006). RNA integrity and the effect on the real-time qRT-PCR performance. Molecular Aspects of Medicine, 27(2–3), 126–139. doi: 10.1016/j.mam.2005.12.003 [DOI] [PubMed] [Google Scholar]
- Frank O, Giehl M, Zheng C, Hehlmann R, Leib-Mösch C, & Seifarth W (2005). Human Endogenous Retrovirus Expression Profiles in Samples from Brains of Patients with Schizophrenia and Bipolar Disorders. Journal of Virology, 79(17), 10890–10901. 10.1128/JVI.79.17.10890-10901.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman ML, Monteiro ANA, Gayther SA, Coetzee GA, Risch A, Plass C, … Mills IG (2011). Principles for the post-GWAS functional characterization of cancer risk loci. Nature Genetics, 43(6), 513–518. doi: 10.1038/ng.840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, … O’Donovan MC (2014). De novo mutations in schizophrenia implicate synaptic networks. Nature, 506(7487), 179–184. 10.1038/nature12929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, … Sklar P (2016). Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nature Neuroscience, 19(11), 1442–1453. doi: 10.1038/nn.4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JR, Brug M. P. v. d., Hernandez DG, Traynor BJ, Nalls MA, Lai S-L, … Singleton AB (2010). Abundant Quantitative Trait Loci Exist for DNA Methylation and Gene Expression in Human Brain. PLOS Genetics, 6(5), e1000952. doi: 10.1371/journal.pgen.1000952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Rifkin SA, & Pritchard JK (2008). Revealing the architecture of gene regulation: the promise of eQTL studies. Trends in Genetics, 24(8), 408–415. doi: 10.1016/j.tig.2008.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogtay N, Vyas NS, Testa R, Wood SJ, & Pantelis C (2011). Age of Onset of Schizophrenia: Perspectives From Structural Neuroimaging Studies. Schizophrenia Bulletin, 37(3), 504–513. 10.1093/schbul/sbr030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould TD, & Gottesman II (2005). Psychiatric endophenotypes and the development of valid animal models. Genes, Brain and Behavior, 5(2), 113–119. doi: 10.1111/j.1601-183X.2005.00186.x [DOI] [PubMed] [Google Scholar]
- Gray AL, Hyde TM, Deep-Soboslay A, Kleinman JE, & Sodhi MS (2015). Sex differences in glutamate receptor gene expression in major depression and suicide. Molecular Psychiatry, 20(9), 1057. doi: 10.1038/mp.2015.91 [DOI] [PubMed] [Google Scholar]
- Greicius MD, Flores BH, Menon V, Glover GH, Solvason HB, Kenna H, … Schatzberg AF (2007). Resting-state functional connectivity in major depression: abnormally increased contributions from subgenual cingulate cortex and thalamus. Biological Psychiatry, 62(5), 429–437. doi: 10.1016/j.biopsych.2006.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Gerevini VD, Dwivedi Y, Grayson DR, … Costa E (2000). Decrease in Reelin and Glutamic Acid Decarboxylase67 (GAD67) Expression in Schizophrenia and Bipolar Disorder: A Postmortem Brain Study. Archives of General Psychiatry, 57(11), 1061–1069. doi: 10.1001/archpsyc.57.11.1061 [DOI] [PubMed] [Google Scholar]
- Guffanti G, Gaudi S, Fallon JH, Sobell J, Potkin SG, Pato C, & Macciardi F (2014). Transposable elements and psychiatric disorders. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 165(3), 201–216. 10.1002/ajmg.b.32225 [DOI] [PubMed] [Google Scholar]
- Hall J, Trent S, Thomas KL, O’Donovan MC, & Owen MJ (2015). Genetic risk for schizophrenia: convergence on synaptic pathways involved in plasticity. Biological Psychiatry, 77(1), 52–58. 10.1016/j.biopsych.2014.07.011 [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Heath PR, Eastwood SL, Burnet PWJ, McDonald B, & Pearson RCA (1995). The relative importance of premortem acidosis and postmortem interval for human brain gene expression studies: selective mRNA vulnerability and comparison with their encoded proteins. Neuroscience Letters, 200(3), 151–154. doi: 10.1016/0304-3940(95)12102-A [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, … Jones AR (2012). An anatomically comprehensive atlas of the adult human brain transcriptome. Nature, 489(7416), 391. doi: 10.1038/nature11405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzheimer PE, Kelley ME, Gross RE, Filkowski MM, Garlow SJ, Barrocas A, … Mayberg HS (2012). Subcallosal cingulate deep brain stimulation for treatment-resistant unipolar and bipolar depression. Archives of General Psychiatry, 69(2), 150–158. doi: 10.1001/archgenpsychiatry.2011.1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honea R, Crow TJ, Passingham D, & Mackay CE (2005). Regional deficits in brain volume in schizophrenia: a meta-analysis of voxel-based morphometry studies. The American Journal of Psychiatry, 162(12), 2233–2245. doi: 10.1176/appi.ajp.162.12.2233 [DOI] [PubMed] [Google Scholar]
- Houenou J, Boisgontier J, Henrion A, d'Albis M-A, Dumaine A, Linke J, … Jamain S (2017). A Multilevel Functional Study of a SNAP25 At-Risk Variant for Bipolar Disorder and Schizophrenia. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 37(43), 10389–10397. doi: 10.1523/JNEUROSCI.1040-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang QQ, Ritchie SC, Brozynska M, & Inouye M (2018). Power, false discovery rate and Winner’s Curse in eQTL studies. Nucleic Acids Research, 46(22), e133–e133. doi: 10.1093/nar/gky780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y, Kim J, Shin JY, Kim JI, Seo JS, Webster MJ, … Kim S (2013). Gene expression profiling by mRNA sequencing reveals increased expression of immune/inflammation-related genes in the hippocampus of individuals with schizophrenia. Translational Psychiatry, 3(10), e321. doi: 10.1038/tp.2013.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, & Blencowe BJ (2012). Alternative splicing: decoding an expansive regulatory layer. Current Opinion in Cell Biology, 24(3), 323–332. doi: 10.1016/j.ceb.2012.03.005 [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Kakiuchi C, Bundo M, Ikeda K, & Kato T (2004). Molecular characterization of bipolar disorder by comparing gene expression profiles of postmortem brains of major mental disorders. Molecular Psychiatry, 9(4), 406–416. doi: 10.1038/sj.mp.4001437 [DOI] [PubMed] [Google Scholar]
- Jaffe AE, Shin J, Collado-Torres L, Leek JT, Tao R, Li C, … Weinberger DR (2015). Developmental regulation of human cortex transcription and its clinical relevance at single base resolution. Nature Neuroscience, 18(1), 154–161. 10.1038/nn.3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JH, Jung WH, Choi J-S, Choi C-H, Kang D-H, Shin NY, … Kwon JS (2011). Reduced prefrontal functional connectivity in the default mode network is related to greater psychopathology in subjects with high genetic loading for schizophrenia. Schizophrenia Research, 127(1–3), 58–65. doi: 10.1016/j.schres.2010.12.022 [DOI] [PubMed] [Google Scholar]
- Jensen KP, Kranzler HR, Stein MB, & Gelernter J (2014). The effects of a MAP2K5 microRNA target site SNP on risk for anxiety and depressive disorders. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics, 165B(2), 175–183. doi: 10.1002/ajmg.b.32219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, & Marks DS (2004). Human MicroRNA targets. PLoS Biology, 2(11), e363 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CA, Watson DJG, & Fone KCF (2011). Animal models of schizophrenia. British Journal of Pharmacology 164(4), 1162–1194. doi: 10.1111/j.1476-5381.2011.01386.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukamaa M, Heliövaara M, Knekt P, Aromaa A, Raitasalo R, & Lehtinen V (2006). Schizophrenia, neuroleptic medication and mortality. The British Journal of Psychiatry, 188(2), 122–127. doi: 10.1192/bjp.188.2.122 [DOI] [PubMed] [Google Scholar]
- Kaalund SS, Newburn EN, Ye T, Tao R, Li C, Deep-Soboslay A, … Kleinman JE (2014). Contrasting changes in DRD1 and DRD2 splice variant expression in schizophrenia and affective disorders, and associations with SNPs in postmortem brain. Molecular Psychiatry, 19(12), 1258. doi: 10.1038/mp.2013.165 [DOI] [PubMed] [Google Scholar]
- Kaiser RH, Andrews-Hanna JR, Wager TD, & Pizzagalli DA (2015). Large-Scale Network Dysfunction in Major Depressive Disorder: A Meta-analysis of Resting-State Functional Connectivity. JAMA psychiatry, 72(6), 603–611. doi: 10.1001/jamapsychiatry.2015.0071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandaswamy R, McQuillin A, Curtis D, & Gurling H (2014). Allelic association, DNA resequencing and copy number variation at the metabotropic glutamate receptor GRM7 gene locus in bipolar disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 165(4), 365–372. doi: 10.1002/ajmg.b.32239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, … Šestan N (2011). Spatio-temporal transcriptome of the human brain. Nature, 478(7370), 483–489. doi: 10.1038/nature10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karege F, Vaudan G, Schwald M, Perroud N, & La Harpe R (2005). Neurotrophin levels in postmortem brains of suicide victims and the effects of antemortem diagnosis and psychotropic drugs. Molecular Brain Research, 136(1), 29–37. doi: 10.1016/j.molbrainres.2004.12.020 [DOI] [PubMed] [Google Scholar]
- Kelly S, Jahanshad N, Zalesky A, Kochunov P, Agartz I, Alloza C, … Donohoe G (2018). Widespread white matter microstructural differences in schizophrenia across 4322 individuals: results from the ENIGMA Schizophrenia DTI Working Group. Molecular Psychiatry, 23(5), 1261–1269. doi: 10.1038/mp.2017.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TM, & Mann JJ (1996). Validity of DSM-III-R diagnosis by psychological autopsy: a comparison with clinician ante-mortem diagnosis. Acta Psychiatrica Scandinavica, 94(5), 337–343. doi: 10.1111/j.1600-0447.1996.tb09869.x [DOI] [PubMed] [Google Scholar]
- Kempton MJ, Geddes JR, Ettinger U, Williams SCR, & Grasby PM (2008). Meta-analysis, database, and meta-regression of 98 structural imaging studies in bipolar disorder. Archives of General Psychiatry, 65(9), 1017–1032. doi: 10.1001/archpsyc.65.9.1017 [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, … Pääbo S (2004). Regional patterns of gene expression in human and chimpanzee brains. Genome Research, 14(8), 1462–1473. doi: 10.1101/gr.2538704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AH, Reimers M, Maher B, Williamson V, McMichael O, McClay JL, … Vladimirov VI (2010). MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophrenia Research, 124(1–3), 183–191. doi: 10.1016/j.schres.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H-W, Rapoport SI, & Rao JS (2010). Altered expression of apoptotic factors and synaptic markers in postmortem brain from bipolar disorder patients. Neurobiology of Disease, 37(3), 596–603. doi: 10.1016/j.nbd.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury AE, Foster OJF, Nisbet AP, Cairns N, Bray L, Eve DJ, … David Marsden C (1995). Tissue pH as an indicator of mRNA preservation in human post-mortem brain. Molecular Brain Research, 28(2), 311–318. doi: 10.1016/0169-328X(94)00219-5 [DOI] [PubMed] [Google Scholar]
- Klein DN, Schatzberg AF, McCullough JP, Dowling F, Goodman D, Howland RH, … Keller MB (1999). Age of onset in chronic major depression: relation to demographic and clinical variables, family history, and treatment response. Journal of Affective Disorders, 55(2), 149–157. 10.1016/S0165-0327(99)00020-8 [DOI] [PubMed] [Google Scholar]
- Koolschijn PCMP, van Haren NEM, Lensvelt-Mulders GJLM, Hulshoff Pol HE, & Kahn RS (2009). Brain volume abnormalities in major depressive disorder: a meta-analysis of magnetic resonance imaging studies. Human Brain Mapping, 30(11), 3719–3735. doi: 10.1002/hbm.20801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronfeld-Schor N, & Einat H. (2012). Circadian rhythms and depression: human psychopathology and animal models. Neuropharmacology, 62(1), 101–114. 10.1016/j.neuropharm.2011.08.020 [DOI] [PubMed] [Google Scholar]
- Lau CG, & Zukin RS (2007). NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nature Reviews. Neuroscience, 8(6), 413–426. doi: 10.1038/nrn2153 [DOI] [PubMed] [Google Scholar]
- Lehman AF, & Steinwachs DM (1998). Translating research into practice: the Schizophrenia Patient Outcomes Research Team (PORT) treatment recommendations. Schizophrenia Bulletin, 24(1), 1–10. [DOI] [PubMed] [Google Scholar]
- Leucht S, Samara M, Heres S, & Davis JM (2016). Dose Equivalents for Antipsychotic Drugs: The DDD Method. Schizophrenia Bulletin, 42(Suppl 1), S90–S94. doi: 10.1093/schbul/sbv167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA (2002). The Human Brain Revisited: Opportunities and Challenges in Postmortem Studies of Psychiatric Disorders. Neuropsychopharmacology, 26(2), 143–154. doi: 10.1016/S0893-133X(01)00393-1 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, & Bartel DP (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120(1), 15–20. 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, Vawter MP, … Bunney WE (2013). Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proceedings of the National Academy of Sciences, 201305814. doi: 10.1073/pnas.1305814110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Santpere G, Kawasawa YI, Evgrafov OV, Gulden FO, Pochareddy S, … Sestan N (2018). Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science, 362(6420), eaat7615. doi: 10.1126/science.aat7615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Lin Y, Nery JR, Urich MA, Breschi A, Davis CA, … Snyder MP (2014). Comparison of the transcriptional landscapes between human and mouse tissues. Proceedings of the National Academy of Sciences, 111(48), 17224–17229. doi: 10.1073/pnas.1413624111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng D, & Lachman HM (2011). RNA-Seq of Human Neurons Derived from iPS Cells Reveals Candidate Long Non-Coding RNAs Involved in Neurogenesis and Neuropsychiatric Disorders. PLOS ONE, 6(9), e23356 10.1371/journal.pone.0023356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingjaerde O, Ahlfors UG, Bech P, Dencker SJ, & Elgen K (1987). The UKU side effect rating scale. A new comprehensive rating scale for psychotropic drugs and a cross-sectional study of side effects in neuroleptic-treated patients. Acta Psychiatrica Scandinavica. Supplementum, 334, 1–100. [DOI] [PubMed] [Google Scholar]
- Lipska BK, Deep-Soboslay A, Weickert CS, Hyde TM, Martin CE, Herman MM, & Kleinman JE (2006). Critical Factors in Gene Expression in Postmortem Human Brain: Focus on Studies in Schizophrenia. Biological Psychiatry, 60(6), 650–658. doi: 10.1016/j.biopsych.2006.06.019 [DOI] [PubMed] [Google Scholar]
- Lipska BK, & Weinberger DR (2000). To model a psychiatric disorder in animals: schizophrenia as a reality test. Neuropsychopharmacology: Official Publication of the American College of Neuropsychopharmacology, 23(3), 223–239. doi: 10.1016/S0893-133X(00)00137-8 [DOI] [PubMed] [Google Scholar]
- Lister RG (1990). Ethologically-based animal models of anxiety disorders. Pharmacology & Therapeutics, 46(3), 321–340. doi: 10.1016/0163-7258(90)90021-S [DOI] [PubMed] [Google Scholar]
- Liu Z, Xu C, Xu Y, Wang Y, Zhao B, Lv Y, … Du C (2010). Decreased regional homogeneity in insula and cerebellum: a resting-state fMRI study in patients with major depression and subjects at high risk for major depression. Psychiatry Research, 182(3), 211–215. doi: 10.1016/j.pscychresns.2010.03.004 [DOI] [PubMed] [Google Scholar]
- Liu Z, Li X, Sun N, Xu Y, Meng Y, Yang C, … Zhang K (2014). Microarray Profiling and Co-Expression Network Analysis of Circulating lncRNAs and mRNAs Associated with Major Depressive Disorder. PLOS ONE, 9(3), e93388 10.1371/journal.pone.0093388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chang X, Hahn C-G, Gur RE, Sleiman PAM, & Hakonarson H (2018). Non-coding RNA dysregulation in the amygdala region of schizophrenia patients contributes to the pathogenesis of the disease. Translational Psychiatry, 8(1), 44 10.1038/s41398-017-0030-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X-J, Mattheisen M, Li M, Huang L, Rietschel M, Børglum AD, … Yao Y-G (2015). Systematic Integration of Brain eQTL and GWAS Identifies ZNF323 as a Novel Schizophrenia Risk Gene and Suggests Recent Positive Selection Based on Compensatory Advantage on Pulmonary Function. Schizophrenia Bulletin, 41(6), 1294–1308. doi: 10.1093/schbul/sbv017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynall M-E, Bassett DS, Kerwin R, McKenna PJ, Kitzbichler M, Muller U, & Bullmore E (2010). Functional Connectivity and Brain Networks in Schizophrenia. Journal of Neuroscience, 30(28), 9477–9487. doi: 10.1523/JNEUROSCI.0333-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, … Parkinson H (2017). The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Research, 45(D1), D896–D901. doi: 10.1093/nar/gkw1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamdani M, Williamson V, McMichael GO, Blevins T, Aliev F, Adkins A, … Vladimirov VI (2015). Integrating mRNA and miRNA Weighted Gene Co-Expression Networks with eQTLs in the Nucleus Accumbens of Subjects with Alcohol Dependence. PLOS ONE, 10(9), e0137671. doi: 10.1371/journal.pone.0137671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marigorta UM, & Navarro A (2013). High Trans-ethnic Replicability of GWAS Results Implies Common Causal Variants. PLOS Genetics, 9(6), e1003566. doi: 10.1371/journal.pgen.1003566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, & Gilad Y (2008). RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Research, 18(9), 1509–1517. doi: 10.1101/gr.079558.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayberg HS (1997). Limbic-cortical dysregulation: a proposed model of depression. The Journal of Neuropsychiatry and Clinical Neurosciences, 9(3), 471–481. doi: 10.1176/jnp.9.3.471 [DOI] [PubMed] [Google Scholar]
- McCarthy MJ, & Welsh DK (2012). Cellular circadian clocks in mood disorders. Journal of Biological Rhythms, 27(5), 339–352. 10.1177/0748730412456367 [DOI] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen R, & Burge CB (2012). Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science (New York, N.Y.), 338(6114), 1593–1599. doi: 10.1126/science.1228186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michie M, Henderson G, Garrett J, & Corbie-Smith G (2011). If I Could in a Small Way Help”: Motivations for and Beliefs about Sample Donation for Genetic Research. Journal of Empirical Research on Human Research Ethics, 6(2), 57–70. doi: 10.1525/jer.2011.6.2.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mighdoll MI, & Hyde TM (2018). Brain donation at autopsy: clinical characterization and toxicologic analyses. Handbook of Clinical Neurology, 150, 143–154. doi: 10.1016/B978-0-444-63639-3.00011-6 [DOI] [PubMed] [Google Scholar]
- Moreau MP, Bruse SE, David-Rus R, Buyske S, & Brzustowicz LM (2011). Altered MicroRNA Expression Profiles in Postmortem Brain Samples from Individuals with Schizophrenia and Bipolar Disorder. Biological Psychiatry, 69(2), 188–193. 10.1016/j.biopsych.2010.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, Steinthorsdottir V, … McCarthy MI (2012). Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nature Genetics, 44(9), 981–990. doi: 10.1038/ng.2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, & Wold B (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods, 5(7), 621–628. doi: 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Mühleisen TW, Leber M, Schulze TG, Strohmaier J, Degenhardt F, Treutlein J, … Cichon S (2014). Genome-wide association study reveals two new risk loci for bipolar disorder. Nature Communications, 5, 3339. doi: 10.1038/ncomms4339 [DOI] [PubMed] [Google Scholar]
- Nakata K, Lipska BK, Hyde TM, Ye T, Newburn EN, Morita Y, … Kleinman JE (2009a). DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proceedings of the National Academy of Sciences of the United States of America, 106(37), 15873–15878. doi: 10.1073/pnas.0903413106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K, Lipska BK, Hyde TM, Ye T, Newburn EN, Morita Y, … Kleinman JE (2009b). DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proceedings of the National Academy of Sciences, pnas.0903413106. doi: 10.1073/pnas.0903413106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, & Hyman SE (2010). Animal models of neuropsychiatric disorders. Nature Neuroscience, 13(10), 1161–1169. doi: 10.1038/nn.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer-Aronsten B, Sheedy D, McCrossin T, & Kril J (2016). An International Survey of Brain Banking Operation and Characterization Practices. Biopreservation and Biobanking, 14(6), 464–469. doi: 10.1089/bio.2016.0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, & Blencowe BJ (2008). Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genetics, 40(12), 1413. doi: 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- Pandey GN, Rizavi HS, Zhang H, & Ren X (2018). Abnormal gene and protein expression of inflammatory cytokines in the postmortem brain of schizophrenia patients. Schizophrenia Research, 192, 247–254. doi: 10.1016/j.schres.2017.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazatos SP, Huang Y.-y., Rosoklija GB, Dwork AJ, Arango V, & Mann JJ (2017). Whole-transcriptome brain expression and exon-usage profiling in major depression and suicide: evidence for altered glial, endothelial and ATPase activity. Molecular Psychiatry, 22(5), 760. doi: 10.1038/mp.2016.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul DS, Soranzo N, & Beck S (2013). Functional interpretation of non-coding sequence variation: Concepts and challenges. BioEssays, 36(2), 191–199. doi: 10.1002/bies.201300126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, … Pritchard JK (2010). Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature, 464(7289), 768–772. doi: 10.1038/nature08872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, … Hammond SM (2007). microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biology, 8(2), R27 10.1186/gb-2007-8-2-r27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlis RH, Dennehy EB, Miklowitz DJ, DelBello MP, Ostacher M, Calabrese JR, … Sachs G (2009). Retrospective age-at-onset of bipolar disorder and outcome during two-year follow-up: results from the STEP-BD study. Bipolar Disorders, 11(4), 391–400. 10.1111/j.1399-5618.2009.00686.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Zhang H, Bao A, Zhu K, Huang Y, Yan X, … Ma C (2018). Standardized Operational Protocol for Human Brain Banking in China. Neuroscience Bulletin. doi: 10.1007/s12264-018-0306-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, & Mehler MF (2012). Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nature Reviews. Neuroscience, 13(8), 528–541. 10.1038/nrn3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman OA, Sasvari-Szekely M, Szekely A, Faludi G, Guttman A, & Nemoda Z (2010). Analysis of a polymorphic microRNA target site in the purinergic receptor P2RX7 gene. Electrophoresis, 31(11), 1790–1795. doi: 10.1002/elps.200900664 [DOI] [PubMed] [Google Scholar]
- Rajkowska G, Clarke G, Mahajan G, Licht CMM, van de Werd HJJM, Yuan P, … Uylings HBM (2016). Differential effect of lithium on cell number in the hippocampus and prefrontal cortex in adult mice: a stereological study. Bipolar Disorders, 18(1), 41–51. doi: 10.1111/bdi.12364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaker RC, Bowling KM, Lasseigne BN, Hagenauer MH, Hardigan AA, Davis NS, … Myers RM (2017). Post-mortem molecular profiling of three psychiatric disorders. Genome Medicine, 9. doi: 10.1186/s13073-017-0458-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsköld D, Wang ET, Burge CB, & Sandberg R (2009). An Abundance of Ubiquitously Expressed Genes Revealed by Tissue Transcriptome Sequence Data. PLOS Computational Biology, 5(12), e1000598. doi: 10.1371/journal.pcbi.1000598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kähler AK, Akterin S, … Sullivan PF (2013). Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nature Genetics, 45(10), 1150. doi: 10.1038/ng.2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie W, Flamant S, & Rasko JEJ (2009). Predicting microRNA targets and functions: traps for the unwary. Nature Methods, 6(6), 397–398. 10.1038/nmeth0609-397 [DOI] [PubMed] [Google Scholar]
- Roberts A, Trapnell C, Donaghey J, Rinn JL, & Pachter L (2011). Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biology, 12(3), R22. doi: 10.1186/gb-2011-12-3-r22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, & Feldman MW (2002). Genetic Structure of Human Populations. Science, 298(5602), 2381–2385. doi: 10.1126/science.1078311 [DOI] [PubMed] [Google Scholar]
- Rueckert EH, Barker D, Ruderfer D, Bergen SE, O’Dushlaine C, Luce CJ, … Sklar P (2013). Cis-Acting Regulation of Brain-Specific ANK3 Gene Expression by a Genetic Variant Associated with Bipolar Disorder. Molecular Psychiatry, 18(8), 922–929. doi: 10.1038/mp.2012.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, & Brown P O. (1995). Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science (New York, N.Y.), 270(5235), 467–470. [DOI] [PubMed] [Google Scholar]
- Schliekelman P (2008). Statistical power of expression quantitative trait loci for mapping of complex trait loci in natural populations. Genetics, 178(4), 2201–2216. doi: 10.1534/genetics.107.076687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaal L, Hibar DP, Sämann PG, Hall GB, Baune BT, Jahanshad N, … Veltman DJ (2017). Cortical abnormalities in adults and adolescents with major depression based on brain scans from 20 cohorts worldwide in the ENIGMA Major Depressive Disorder Working Group. Molecular Psychiatry, 22(6), 900. doi: 10.1038/mp.2016.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaal L, Veltman DJ, van Erp TGM, Sämann PG, Frodl T, Jahanshad N, … Hibar DP (2016). Subcortical brain alterations in major depressive disorder: findings from the ENIGMA Major Depressive Disorder working group. Molecular Psychiatry, 21(6), 806–812. doi: 10.1038/mp.2015.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A, Bauer M, Heinsen H, Feiden W, li T. C. o. B. E., Falkai P, … Kretzschmar H (2007). How a neuropsychiatric brain bank should be run: a consensus paper of Brainnet Europe II. Journal of Neural Transmission, 114(5), 527–537. doi: 10.1007/s00702-006-0601-8 [DOI] [PubMed] [Google Scholar]
- Schmitt A, Koschel J, Zink M, Bauer M, Sommer C, Frank J, … Henn FA (2010). Gene expression of NMDA receptor subunits in the cerebellum of elderly patients with schizophrenia. European Archives of Psychiatry and Clinical Neuroscience, 260(2), 101–111. doi: 10.1007/s00406-009-0017-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A, & Downward J (2001). Navigating gene expression using microarrays--a technology review. Nature Cell Biology 3(8), E190–195. doi: 10.1038/35087138 [DOI] [PubMed] [Google Scholar]
- Selvaraj S, Arnone D, Job D, Stanfield A, Farrow TF, Nugent AC, … McIntosh AM (2012). Grey matter differences in bipolar disorder: a meta-analysis of voxel-based morphometry studies. Bipolar Disorders, 14(2), 135–145. doi: 10.1111/j.1399-5618.2012.01000.x [DOI] [PubMed] [Google Scholar]
- Shon S-H, Yoon W, Kim H, Joo SW, Kim Y, & Lee J (2018). Deterioration in Global Organization of Structural Brain Networks in Schizophrenia: A Diffusion MRI Tractography Study. Frontiers in Psychiatry, 9, 272. doi: 10.3389/fpsyt.2018.00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G, Zhang H, Rizavi H, Cook EH, & Dwivedi Y (2014). Expression of microRNAs and Other Small RNAs in Prefrontal Cortex in Schizophrenia, Bipolar Disorder and Depressed Subjects. PLoS ONE, 9(1). 10.1371/journal.pone.0086469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan KE, Baldeweg T, & Friston KJ (2006). Synaptic plasticity and dysconnection in schizophrenia. Biological Psychiatry, 59(10), 929–939. doi: 10.1016/j.biopsych.2005.10.005 [DOI] [PubMed] [Google Scholar]
- Slotkin RK, & Martienssen R (2007). Transposable elements and the epigenetic regulation of the genome. Nature Reviews Genetics, 8(4), 272 10.1038/nrg2072 [DOI] [PubMed] [Google Scholar]
- Strakowski SM, Adler CM, Almeida J, Altshuler LL, Blumberg HP, Chang KD, … Townsend JD (2012). The functional neuroanatomy of bipolar disorder: a consensus model. Bipolar Disorders, 14(4), 313–325. doi: 10.1111/j.1399-5618.2012.01022.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand AD, Aragaki AK, Baquet ZC, Hodges A, Cunningham P, Holmans P, … Olson JM (2007). Conservation of Regional Gene Expression in Mouse and Human Brain. PLOS Genetics, 3(4), e59. doi: 10.1371/journal.pgen.0030059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, … Dermitzakis ET (2007). Population genomics of human gene expression. Nature Genetics, 39(10), 1217. doi: 10.1038/ng2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann M, Konrad C, Kugel H, Krug A, Schöning S, Ohrmann P, … Dannlowski U (2014). Insular and Hippocampal Gray Matter Volume Reductions in Patients with Major Depressive Disorder. PLOS ONE, 9(7), e102692. doi: 10.1371/journal.pone.0102692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, … Hogenesch JB (2004). A gene atlas of the mouse and human protein-encoding transcriptomes. Proceedings of the National Academy of Sciences of the United States of America, 101(16), 6062–6067. doi: 10.1073/pnas.0400782101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak YG, & Farnham PJ (2015). Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics & Chromatin, 8. doi: 10.1186/s13072-015-0050-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan ML, Dyck BA, Gabriele J, Daya RP, Thomas N, Sookram C, … Mishra RK (2014). Synapsin II gene expression in the dorsolateral prefrontal cortex of brain specimens from patients with schizophrenia and bipolar disorder: effect of lifetime intake of antipsychotic drugs. The Pharmacogenomics Journal, 14(1), 63–69. doi: 10.1038/tpj.2013.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S, Wakem M, Dijkman G, Alsarraj M, & Nguyen M (2010). A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods (San Diego, Calif.), 50(4), S1–5. doi: 10.1016/j.ymeth.2010.01.005 [DOI] [PubMed] [Google Scholar]
- Terwisscha van Scheltinga AF, Bakker SC, van Haren NEM, Derks EM, Buizer-Voskamp JE, Boos HBM, … Psychiatric Genome-wide Association Study, C. (2013). Genetic schizophrenia risk variants jointly modulate total brain and white matter volume. Biological Psychiatry, 73(6), 525–531. doi: 10.1016/j.biopsych.2012.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Wei Z, Chang X, Liu Y, Gur RE, Sleiman PMA, & Hakonarson H (2018). The Long Noncoding RNA Landscape in Amygdala Tissues from Schizophrenia Patients. EBioMedicine, 34, 171–181. 10.1016/j.ebiom.2018.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, … Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols, 7(3), 562–578. doi: 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Ule A, Spencer J, Williams A, Hu J-S, Cline M, … Darnell RB (2005). Nova regulates brain-specific splicing to shape the synapse. Nature Genetics, 37(8), 844. doi: 10.1038/ng1610 [DOI] [PubMed] [Google Scholar]
- van Dijk EL, Auger H, Jaszczyszyn Y, & Thermes C (2014). Ten years of next-generation sequencing technology. Trends in genetics: TIG, 30(9), 418–426. doi: 10.1016/j.tig.2014.07.001 [DOI] [PubMed] [Google Scholar]
- van Haren NEM, Hulshoff Pol HE, Schnack HG, Cahn W, Mandl RCW, Collins DL, … Kahn RS (2007). Focal gray matter changes in schizophrenia across the course of the illness: a 5-year follow-up study. Neuropsychopharmacology: Official Publication of the American College of Neuropsychopharmacology, 32(10), 2057–2066. doi: 10.1038/sj.npp.1301347 [DOI] [PubMed] [Google Scholar]
- VanGuilder HD, Vrana KE, & Freeman WM (2008). Twenty-five years of quantitative PCR for gene expression analysis. BioTechniques, 44(5), 619–626. doi: 10.2144/000112776 [DOI] [PubMed] [Google Scholar]
- Vonsattel JPG, Amaya M. P. d., & Keller CE (2008). Twenty-first century brain banking. Processing brains for research: the Columbia University methods. Acta Neuropathologica, 115(5), 509–532. doi: 10.1007/s00401-007-0311-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, … Gerstein MB (2018). Comprehensive functional genomic resource and integrative model for the human brain. Science, 362(6420), eaat8464. doi: 10.1126/science.aat8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, … Burge CB (2008). Alternative isoform regulation in human tissue transcriptomes. Nature, 456(7221), 470–476. doi: 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, & Snyder M (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews. Genetics, 10(1), 57–63. doi: 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger DR (2017). Future of Days Past: Neurodevelopment and Schizophrenia. Schizophrenia Bulletin, 43(6), 1164–1168. 10.1093/schbul/sbx118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis S, Llenos IC, Sabunciyan S, Dulay JR, Isler L, Yolken R, & Perron H (2007). Reduced expression of human endogenous retrovirus (HERV)-W GAG protein in the cingulate gyrus and hippocampus in schizophrenia, bipolar disorder, and depression. Journal of Neural Transmission, 114(5), 645–655. 10.1007/s00702-006-0599-y [DOI] [PubMed] [Google Scholar]
- Whalen PJ, Shin LM, Somerville LH, McLean AA, & Kim H (2002). Functional neuroimaging studies of the amygdala in depression. Seminars in Clinical Neuropsychiatry, 7(4), 234–242. [DOI] [PubMed] [Google Scholar]
- Will CL, & Lührmann R (2011). Spliceosome structure and function. Cold Spring Harbor Perspectives in Biology 3(7). doi: 10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson VS, Mamdani M, McMichael GO, Kim AH, Lee D, Bacanu S, & Vladimirov VI (2015). Expression quantitative trait loci (eQTLs) in microRNA genes are enriched for schizophrenia and bipolar disorder association signals. Psychological Medicine, 45(12), 2557–2569. doi: 10.1017/S0033291715000483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise T, Radua J, Via E, Cardoner N, Abe O, Adams TM, … Arnone D (2017). Common and distinct patterns of grey-matter volume alteration in major depression and bipolar disorder: evidence from voxel-based meta-analysis. Molecular Psychiatry, 22(10), 1455. doi: 10.1038/mp.2016.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods SW (2003). Chlorpromazine equivalent doses for the newer atypical antipsychotics. The Journal of Clinical Psychiatry, 64(6), 663–667. [DOI] [PubMed] [Google Scholar]
- Wu Y-H, Fischer DF, Kalsbeek A, Garidou-Boof M-L, van der Vliet J, van Heijningen C, … Swaab DF (2006). Pineal clock gene oscillation is disturbed in Alzheimer’s disease, due to functional disconnection from the “master clock.” FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology, 20(11), 1874–1876. 10.1096/fj.05-4446fje [DOI] [PubMed] [Google Scholar]
- Wu P, Zuo X, Deng H, Liu X, Liu L, & Ji A (2013). Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Research Bulletin, 97, 69–80. 10.1016/j.brainresbull.2013.06.001 [DOI] [PubMed] [Google Scholar]
- Wu C, & Pan W (2018). Integrating eQTL data with GWAS summary statistics in pathway-based analysis with application to schizophrenia. Genetic Epidemiology, 42(3), 303–316. doi: 10.1002/gepi.22110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Krueger GRF, Buja LM, & Covinsky M (2009). The Impact of Declining Clinical Autopsy: Need for Revised Healthcare Policy. The American Journal of the Medical Sciences, 337(1), 41–46. doi: 10.1097/MAJ.0b013e318184ce2b [DOI] [PubMed] [Google Scholar]
- Yao C, Joehanes R, Johnson AD, Huan T, Liu C, Freedman JE, … Levy D (2017). Dynamic Role of trans Regulation of Gene Expression in Relation to Complex Traits. American Journal of Human Genetics, 100(4), 571–580. doi: 10.1016/j.ajhg.2017.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Karayiorgou M, & Gogos JA (2010). MicroRNAs in psychiatric and neurodevelopmental disorders. Brain Research, 1338, 78–88. 10.1016/j.brainres.2010.03.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Jiang X, Jiang X, & Zhao H (2018). X-inactive-specific transcript: A long noncoding RNA with complex roles in human cancers. Gene, 679, 28–35. 10.1016/j.gene.2018.08.071 [DOI] [PubMed] [Google Scholar]
- Yeo G, Holste D, Kreiman G, & Burge CB (2004). Variation in alternative splicing across human tissues. Genome Biology, 5(10), R74. doi: 10.1186/gb-2004-5-10-r74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yolken RH, Karlsson H, Yee F, Johnston-Wilson NL, & Torrey EF (2000). Endogenous retroviruses and schizophrenia. Brain Research. Brain Research Reviews, 31(2–3), 193–199. [DOI] [PubMed] [Google Scholar]
- Zahr NM, Kaufman KL, & Harper CG (2011). Clinical and pathological features of alcohol-related brain damage. Nature Reviews. Neurology, 7(5), 284–294. doi: 10.1038/nrneurol.2011.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X-Q, Wang Z-L, Poon M-W, & Yang J-H (2017). Spatial-temporal transcriptional dynamics of long non-coding RNAs in human brain. Human Molecular Genetics, 26(16), 3202–3211. 10.1093/hmg/ddx203 [DOI] [PubMed] [Google Scholar]
- Zhang H, Chen K, Wang N, Zhang D, Yang Q, Zhang Q, … Ma C (2018). Analysis of Brain Donors' Demographic and Medical Characteristics to Facilitate the Construction of a Human Brain Bank in China. Journal of Alzheimer's disease: JAD, 66(3), 1245–1254. doi: 10.3233/JAD-180779 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, … Yang J (2016). Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nature Genetics, 48(5), 481. doi: 10.1038/ng.3538 [DOI] [PubMed] [Google Scholar]
- Zuo L, Tan Y, Wang Z, Wang K-S, Zhang X, Chen X, … Luo X (2016). Long non-coding RNAs in psychiatric disorders. Psychiatric Genetics, 26(3), 109–116. 10.1097/YPG.0000000000000129 [DOI] [PMC free article] [PubMed] [Google Scholar]