Abstract
Transthyretin (TTR) is a major amyloidogenic protein associated with hereditary (ATTRm) and nonhereditary (ATTRwt) intractable systemic transthyretin amyloidosis. The pathological mechanisms of ATTR-associated amyloid fibril formation are incompletely understood, and there is a need for identifying compounds that target ATTR. C-terminal TTR fragments are often present in amyloid-laden tissues of most patients with ATTR amyloidosis, and on the basis of in vitro studies, these fragments have been proposed to play important roles in amyloid formation. Here, we found that experimentally-formed aggregates of full-length TTR are cleaved into C-terminal fragments, which were also identified in patients' amyloid-laden tissues and in SH-SY5Y neuronal and U87MG glial cells. We observed that a 5-kDa C-terminal fragment of TTR, TTR81–127, is highly amyloidogenic in vitro, even at neutral pH. This fragment formed amyloid deposits and induced apoptosis and inflammatory gene expression also in cultured cells. Using the highly amyloidogenic TTR81–127 fragment, we developed a cell-based high-throughput screening method to discover compounds that disrupt TTR amyloid fibrils. Screening a library of 1280 off-patent drugs, we identified two candidate repositioning drugs, pyrvinium pamoate and apomorphine hydrochloride. Both drugs disrupted patient-derived TTR amyloid fibrils ex vivo, and pyrvinium pamoate also stabilized the tetrameric structure of TTR ex vivo in patient plasma. We conclude that our TTR81–127–based screening method is very useful for discovering therapeutic drugs that directly disrupt amyloid fibrils. We propose that repositioning pyrvinium pamoate and apomorphine hydrochloride as TTR amyloid-disrupting agents may enable evaluation of their clinical utility for managing ATTR amyloidosis.
Keywords: amyloid, drug screening, drug discovery, protein aggregation, protein conformation, amyloid disrupters, apomorphine hydrochloride, pyrvinium pamoate, transthyretin (TTR)
Introduction
Amyloidosis comprises a group of hereditary or acquired intractable diseases that are characterized by extracellular deposits of insoluble amyloid fibrils derived from various kinds of proteins in singular or multiple organs such as heart, nerves, kidneys, gastrointestinal tract, and eyes (1–3). To date, 36 kinds of amyloidogenic proteins have been identified as disease-causing molecules in amyloid-related disorders, such as Alzheimer's disease, prion disease, systemic immunoglobulin light-chain amyloidosis, and systemic transthyretin (ATTR)2 amyloidosis (1–5). Aging, genetic mutations, inflammation, neoplastic disorders, obesity, and medical treatments reportedly affect development of these amyloidoses via overproduction, misfolding, and reduced clearance of disease-specific amyloid-related proteins, as well as proteolysis (1–5).
Transthyretin (TTR), mainly synthesized in the liver, forms a homotetramer that has a dimer–of–dimers configuration in the bloodstream, is a major amyloidogenic protein, and causes two types of intractable systemic amyloidosis (5–9). One is the rare hereditary TTR (ATTRm) amyloidosis, formerly known as familial amyloid polyneuropathy. More than 140 different point mutations, most of which are highly amyloidogenic, have been identified in the TTR gene (5, 6). Of these mutations, ATTR V30M is the most common genotype of ATTRm amyloidosis worldwide. Patients with ATTR V30M amyloidosis have sensorimotor polyneuropathy, autonomic dysfunction, cardiac failure, and other systemic symptoms and usually die within 10 years of disease onset if they are untreated. The other type of ATTR amyloidosis is wildtype TTR (ATTRwt) amyloidosis, formerly known as senile systemic amyloidosis, which has attracted increasing attention (7–9). ATTRwt amyloidosis is a nonhereditary aging-related systemic amyloidosis caused by WT TTR secreted by the liver and is often associated with cardiac failure and bilateral carpal tunnel syndrome in elderly patients (9, 10). Pathomechanisms of ATTRwt amyloidosis remain largely unclear, and specific disease-modifying therapy for ATTRwt amyloidosis is not available.
Liver transplantation has been utilized to treat ATTRm amyloidosis, by replacing unstable mutant TTR synthesized in the liver with a more stable WT TTR found in the bloodstream of patients (11). However, WT TTR synthesized by transplanted liver grafts reportedly continued to form amyloid deposits in certain patients even after liver transplantation (12). In vitro studies with acid-induced denaturation of TTR (13, 14) have indicated that dissociation of the tetrameric structure of TTR to monomers may be a crucial step in the initial phase of TTR amyloid formation (15), and several therapeutic compounds, such as diflunisal, tafamidis, AG10, and tolcapone, have been shown to stabilize the tetrameric TTR structure (16–20). In addition, gene-silencing therapies to reduce TTR expression by the liver have been developed (21, 22). Clinical trials of doxycycline plus tauroursodeoxycholic acid (23) and immunotherapies (24, 25), which aim to disrupt amyloid fibrils, are also being carried out. However, we do not fully understand the detailed mechanisms in later events directly associated with TTR amyloid formation after the dissociation of the TTR tetramer and have yet to develop amyloid-disruptors.
C-terminal fragments of TTR have been well-documented as often occurring in amyloid-laden tissues in ATTR amyloidosis (12, 26–33). In particular, patients with ATTRwt amyloidosis commonly have C-terminal fragments of WT TTR, in addition to full-length WT TTR, in amyloid-laden tissues (26–28). Several studies also indicated that late-onset ATTRm amyloidosis patients with the V30M mutation and ATTRm amyloidosis patients with various kinds of non-V30M mutations commonly had C-terminal fragments of TTR in addition to full-length TTR in amyloid-laden tissues, whereas only full-length TTR was usually found in amyloid deposits in early-onset ATTRm amyloidosis patients with the V30M mutation (12, 28–32). Swedish ATTR V30M Swedish ATTR V30M amyloidosis patients having of TTR in amyloid deposits reportedly showed thicker cardiac intraventricular septum and worse clinical outcome after liver transplantation than ATTR V30M amyloidosis patients without C-terminal fragments of TTR in amyloid deposits (32). In addition, TTR S52P and TTR E51_S52dup mutations that caused unusually aggressive systemic ATTRm amyloidosis were reported to be easily cleaved into C-terminal fragments by trypsin (34–36). In vitro studies of short peptides of TTR segments and tryptic TTR fragments have proposed that the C-terminal regions of TTR may play important roles in TTR amyloid formation (34–38). However, the detailed pathological mechanisms of TTR fragmentation and the clinicopathological impacts of the C-terminal fragments of TTR on ATTR amyloidosis remain to be clarified.
In this study, we first discovered that full-length TTR aggregates were cleaved into C-terminal fragments in cultured neuronal and glial cells and that a 5-kDa C-terminal fragment of TTR, TTR81–127, was highly amyloidogenic, even at neutral pH, in test tubes and in cultured cells. Second, with this highly amyloidogenic TTR81–127, we developed a novel cell-based high-throughput screening (HTS) method so as to detect a new class of drugs that would disrupt amyloid fibrils, and we succeeded in finding candidate repositioning drugs by using a screening library of 1280 off-patent drugs.
Results
Cleavage of full-length TTR aggregates into fragments in cultured neuronal and glial cells
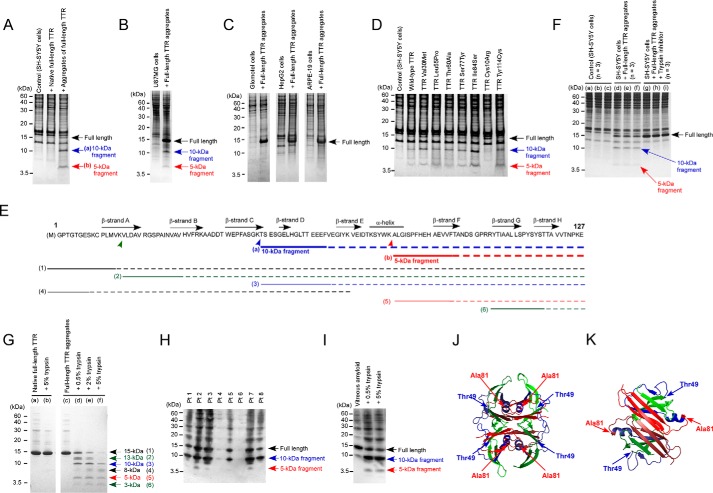
To investigate whether cultured neuronal cells cleave full-length TTR into fragments, we performed SDS-PAGE analyses with lysates of cultured SH-SY5Y neuronal cells treated with native and acid-induced aggregated full-length TTR V30M. We formed aggregates of full-length TTR V30M in acidic buffer at pH 4.0 for 24 h in test tubes, before adding it to the cells. The pre-formed aggregates of full-length TTR V30M were added to the extracellular culture medium. In cultured SH-SY5Y cells treated with acid-induced aggregates of full-length TTR V30M for 24 h, we found 5- and 10-kDa bands, which were smaller than the 15-kDa bands for full-length TTR V30M, and we suspected that they were TTR fragments (Fig. 1A). In contrast, we did not find the smaller bands in cells treated with native full-length TTR V30M. We did not find TTR fragments in TTR aggregates that were incubated with the cell culture medium of SH-SY5Y cells for 24 h (Fig. S1). In human U87MG glial cells, the acid-induced aggregates of full-length TTR V30M were also cleaved into shorter fragments (Fig. 1B), but we did not find TTR fragments in other kinds of cells, including human vascular smooth muscle-like glomotel cells, human hepatoma HepG2 cells, and human retinal pigment epithelium ARPE-19 cells (Fig. 1C). In SH-SY5Y neuronal cells, acid-induced aggregates of other full-length TTRs, such as WT, L55P, T60A, S77Y, I84S, and Y114C, except for C10R, were also cleaved into the smaller 5- and 10-kDa fragments (Fig. 1D). Amino acid sequence analyses revealed that the 5-kDa fragment started at position 81 of TTR, and the 10-kDa fragment of TTR started at position 49 (Fig. 1E). Mass spectrometric analyses of pieces of the gel revealed that the N-terminal fragments were hardly detected in cultured SH-SY5Y cells treated with pre-formed aggregates of full-length TTR V30M (Fig. S2). Both cleavage sites were at the carboxyl side of lysine, which suggests that trypsin or trypsin-like enzymes cleaved full-length TTR into 5- and 10-kDa fragments. As Fig. 1F shows, a trypsin inhibitor suppressed fragmentation of acid-induced aggregates of full-length TTR V30M in SH-SY5Y cells.
Figure 1.
Fragmentation of TTR in vitro. A, SDS-PAGE followed by CBB staining of SH-SY5Y cell lysates treated with native or acid-induced aggregates of full-length recombinant TTR V30M for 24 h. B, SDS-PAGE followed by CBB staining of U87MG cell lysates treated with acid-induced aggregates of full-length recombinant TTR V30M for 24 h. C, SDS-PAGE followed by CBB staining of glomotel cell lysates, HepG2 cell lysates, and ARPE-19 cell lysates treated with acid-induced aggregates of full-length recombinant TTR V30M for 24 h. D, SH-SY5Y cell lysates treated with acid-induced aggregates of full-length wildtype TTR, and various mutant TTRs for 24 h. E, amino acid sequences of TTR fragments detected in SH-SY5Y cells treated with acid-induced aggregates of full-length recombinant TTR V30M (a and b) and sequences of fragments detected in full-length TTR aggregates incubated with trypsin in test tubes (numbers 1–6). Solid lines indicate sequences detected via Edman degradation. Dotted lines indicate expected sequences of TTR fragments as based on SDS-PAGE analyses. F, SH-SY5Y cell lysates treated with acid-induced aggregates of full-length recombinant TTR V30M treated concomitantly with trypsin inhibitor (g–i) or not treated with trypsin inhibitor (d–f), for 24 h. a–c, control. G, SDS-PAGE followed by CBB staining of native or acid-induced aggregates of full-length recombinant TTR V30M incubated with trypsin in test tubes for 24 h. H, C-terminal fragments of TTR in vitreous amyloid fibrils isolated from two patients with ATTR V30M amyloidosis (Pt. 1, 60-year-old man; Pt. 2, 46-year-old woman; Pt. 3, 38-year-old woman; Pt. 4–8, age and sex were not available) detected by immunoblotting with anti-TTR81–127 antibodies. I, in vitro tryptic cleavage of vitreous amyloid fibrils isolated from a 60-year-old male patient with ATTR V30M amyloidosis; fibrils were incubated with 0.5 or 5% trypsin for 30 min. J and K, locations of the C-terminal regions in the tetrameric structure of TTR (H) and the dimeric structure of TTR (I). Red, TTR81–127 (C terminus); blue, TTR49–80 (middle part), and green, TTR1–48 (N terminus). Structural images were generated by using PyMOL.
Tryptic cleavage of acid-induced aggregates of full-length TTR in vitro
In test tubes, trypsin directly cleaved acid-induced aggregates of full-length TTR V30M into several smaller fragments, including 5- and 10-kDa C-terminal fragments, which were similar to the TTR fragments found in cultured cells, in addition to a few other C- and N-terminal fragments (Fig. 1, E and G). In contrast, trypsin barely cleaved native full-length TTR V30M (Fig. 1G).
Fragmentation of TTR in amyloid fibrils from patients with ATTRm amyloidosis
We analyzed components of vitreous TTR amyloid fibrils from eight patients with ATTRm amyloidosis who had the TTR V30M mutation by means of immunoblotting with antibodies against the C-terminal fragment TTR81–127. We found 10- and 5-kDa TTR fragments in vitreous TTR amyloid in three (Pt. 2, 3, and 7) out of eight patients (Fig. 1H). In two patients (Pt. 1 and 5), we detected a clear 10-kDa fragment and a faintly positive 5-kDa fragment. In three patients (Pt. 4, 6, and 8), we found only the 10-kDa fragment, although the amount of vitreous TTR amyloid was small in two patients (Pt. 4 and 6). We also showed that trypsin cleaved vitreous TTR amyloid fibrils of Pt. 1 into the 5- and 10-kDa fragments (Fig. 1I).
To confirm the locations of the C-terminal regions in the tetrameric structure of full-length TTR, we generated structural images using PyMOL software. As Fig. 1, J and K, shows, the C-terminal regions TTR81–127 are located inside the tetrameric and dimeric structures of full-length TTR, but the N-terminal regions are exposed to the outside of the tetrameric and dimeric structures of full-length TTR.
Amyloid formation by TTR fragments in vitro
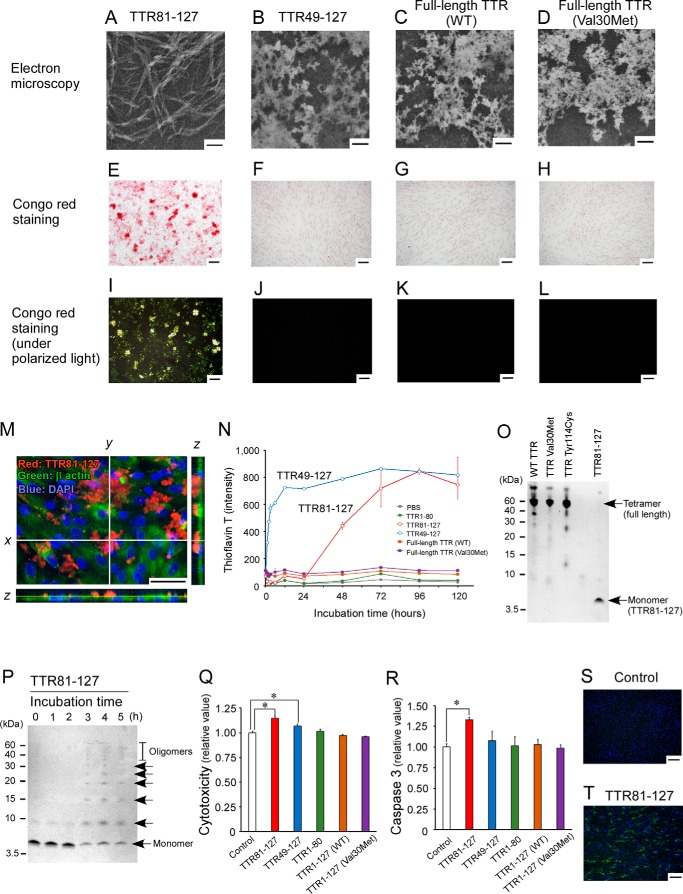
To investigate the in vitro amyloid-forming abilities of different regions of TTR, we analyzed in vitro amyloid-forming abilities of recombinant full-length and fragmented TTRs in PBS at neutral pH and at 37 °C. Electron microscopic analyses revealed that recombinant TTR81–127, which corresponded to the 5-kDa C-terminal fragment of TTR, formed amyloid fibrils in PBS at neutral pH (Fig. 2A). In contrast, recombinant full-length WT TTR, TTR V30M, and TTR49–127, the last of which corresponded to the 10-kDa C-terminal fragment of TTR, formed amorphous aggregates in PBS at neutral pH (Fig. 2, B–D); recombinant TTR1–80, which corresponded to the N terminus of TTR, did not form any aggregates or fibrils. As seen in Fig. 2, E–M, when we directly added the full-length TTR and fragments of TTR to the culture medium of glomotel cells, only TTR81–127 formed amyloid deposits in cultured cells (Fig. 2 E, I, and M); others did not (Fig. 2, F–H and J–L). TTR81–127 formed amyloid deposits mainly in the extracellular space of the cultured cells (Fig. 2M). Thioflavin T (ThT) intensities of TTR81–127, which formed amyloid fibrils in the electron microscopic analyses just mentioned (Fig. 2A), gradually increased over time, after a 24-h lag, in PBS at neutral pH (Fig. 2N), which is the typical kinetics of amyloid fibril formation. In contrast, ThT intensities of TTR49–127, which formed amorphous aggregates in the electron microscopic analyses (Fig. 2B), increased immediately after incubation in PBS at neutral pH (Fig. 2N). ThT intensities of other TTRs, including full-length WT TTR, full-length TTR V30M, and TTR1–80, did not increase in PBS (Fig. 2N). These results indicated that TTR81–27 was more amyloidogenic than the other regions of TTR, such as TTR1–80, TTR49–127, full-length WT TTR, and full-length TTR V30M.
Figure 2.
Amyloid formation of TTR81–127 at neutral pH. A–D, electronic microscopic findings for amyloid fibrils or amorphous aggregates derived from different regions of TTR, which had been incubated at neutral pH. Scale bars: 200 nm. E–L, Congo red staining of glomotel cells treated with 50 μm TTR81–127 (E and I); 50 μm TTR49–127 (F and J); 50 μm full-length WT TTR (G and K); and 50 μm full-length TTR V30M (H and L); for 24 h observed via light microscopy (E–H); and under polarized light (I–L). Scale bars, 200 μm. M, immunofluorescence analysis of TTR81–127 amyloid deposits in glomotel cells. Red fluorescence, TTR81–127; green fluorescence, β-actin; and blue fluorescence: DAPI. Scale bar, 50 μm. N, ThT intensities of different regions of TTR. Red line, TTR81–127; blue line, TTR49–127; green line, TTR1–80; brown line, full-length WT TTR; and purple line, full-length TTR V30M. O, tetramer formation assay of full-length TTR and TTR81–127. P, oligomer assay of TTR81–127 cross-linked with glutaraldehyde. Q, cytotoxicity of different regions of TTR. Glomotel cells were treated with different TTR fragments at 25 μm for 24 h. *, p < 0.05. R, caspase 3 activity in glomotel cells treated with different TTR fragments at 25 μm for 24 h. *, p < 0.05. S and T, annexin V staining in glomotel cells incubated with (S) and without (T) 25 μm TTR81–127 for 24 h. Scale bars, 200 μm.
To determine the intermolecular linking of TTR81–127, we performed SDS-PAGE analyses by using chemically cross-linked TTR81–127 with glutaraldehyde. Full-length TTR formed tetramers, whereas TTR81–127 existed as a monomer and did not form tetramers (Fig. 2O). TTR81–127 gradually polymerized and formed oligomers in PBS at neutral pH and at 37 °C (Fig. 2P).
Cytotoxicity of C-terminal fragments of TTR in cultured cells
We also studied whether different TTRs had cytotoxic effects on cultured cells. We added the recombinant TTRs to culture medium, followed by incubation. The C-terminal fragments of TTR, i.e. TTR81–127 and TTR49–127, showed cytotoxic effects (Fig. 2Q). Recombinant TTR81–127 increased caspase 3 activity and binding of annexin V in cultured cells (Fig. 2, R–T).
TTR81–127 amyloid deposits induced Fas-mediated apoptosis
TTR81–127 formed 1-fluoro-2,5-bis[(E)-3-carboxy-4-hydroxystyryl]benzene (FSB)–positive amyloid deposits and anti-TTR81–127 antibody–positive TTR deposits in cultured cells in a dose-dependent manner (Fig. S3, A and B), and 25 μm TTR81–127 increased caspase 8 and 9 activities (Fig. S3, C and D). In protein array analyses, Fas and BAX increased in glomotel cells treated with TTR81–127, and Bcl-2 decreased (Fig. S3E).
TTR81–127 increased expression of genes associated with inflammation in cultured cells
To investigate the effects of TTR81–127 amyloid deposits in cultured cells, we performed transcriptome analysis by means of RNA-Seq in glomotel cells treated with TTR81–127 for 24 h. Cellular TTR81–127 amyloid formation enhanced expression of inflammation-related genes in glomotel cells (Tables S1 and S2). By means of quantitative real-time PCR analyses, we also confirmed that TTR81–127 amyloid deposits increased the mRNA levels of those molecules, including ICAM1, VCAM1, RELB, IL32, CXCL1, CCL2, C3, and SOD2, and reduced KIT mRNA levels in glomotel cells (Fig. S4). We did not find increased LRWD1 mRNA levels in quantitative real-time PCR analyses, via the RNA-Seq analyses (Table S1). In SH-SY5Y cells derived from human neuroblastoma, we found similar changes in those mRNA levels except for three genes: CXCL1 and C3, which were not detected in SH-SY5Y cells, and SOD2, which was not changed in SH-SY5Y cells treated with TTR81–127 (Fig. S4). In another earlier experiment, gene expression of those molecules, in addition to that of the inflammatory cytokine genes TNF and IL6, increased significantly in glomotel cells treated with TTR81–127 (Fig. S5, A–D). FAS mRNA levels also increased significantly in glomotel cells treated with TTR81–127 (Fig. S5, E–G), which corresponded to the results of the protein array analyses described above. A selective inhibitor of the Fas ligand-dependent pathway, RKTS-33, suppressed increases in the mRNA of those inflammatory genes, including ICAM1, VCAM1, IL32, CCL2, and TNF, in glomotel cells treated with TTR81–127 (Fig. S6).
Novel screening method to detect antiamyloid drugs for drug repositioning
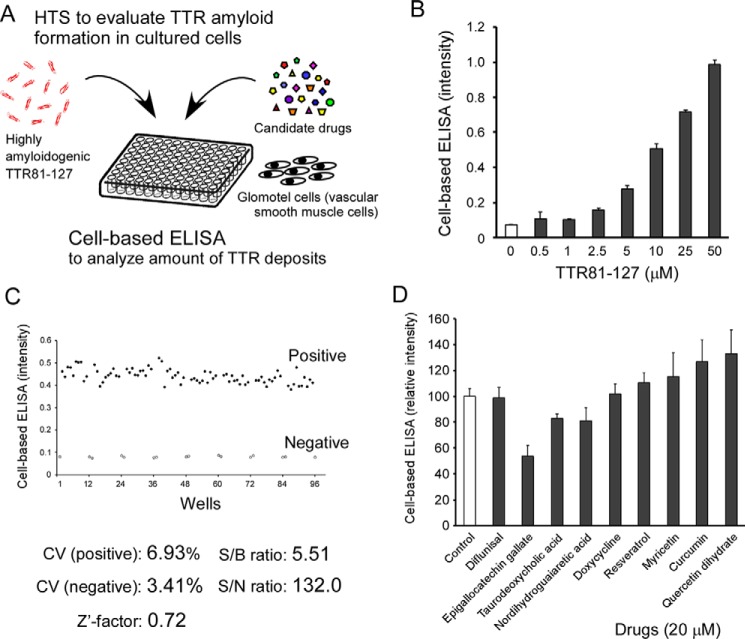
Using the highly amyloidogenic recombinant TTR81–127, we developed a novel cell-based HTS method with 96-well half-area microplates to evaluate TTR amyloid formation in cultured cells (Fig. 3A and Fig. S7). Through a cell-based ELISA with antiserum against TTR81–127, we quantitatively analyzed the amount of TTR81–127 deposits in cultured cells (Fig. 3B). The Z′-factor of our novel cell-based HTS method was 0.72, which we believed was sufficient for drug screening (Fig. 3C).
Figure 3.
Novel cell-based HTS method targeting TTR amyloid deposits in cultured cells at neutral pH. A, schematic diagram of our novel cell-based HTS to determine the content of TTR amyloid deposits in cultured cells by using the highly amyloidogenic TTR81–127. B, dose-dependent TTR81–127 amyloid deposits in cultured cells. C, evaluation of the quality of the cell-based HTS method. We calculated the CV, S/B, S/N, and the Z′ factor in the cell-based ELISA and compared them with the acceptable values of those parameters (CV <20%, S/B >2, S/N >10, and Z′ factor >0.5) for HTS methods. D, amyloid-inhibitory effects of tested compounds in the cell-based HTS. We added these compounds, each at 20 μm, and 5 μm TTR81–127 to culture medium followed by incubation for 24 h and then analysis by using the cell-based ELISA.
By using this HTS, we analyzed the inhibitory effects of several compounds on TTR81–127 amyloid formation; these compounds reportedly inhibited TTR aggregation or stabilized the tetrameric structure of TTR by means of conventional methods with full-length TTR at acidic pH. Epigallocatechin gallate (EGCG) demonstrated the strongest inhibition of TTR81–127 amyloid formation, whereas diflunisal, which is well-known to stabilize the tetrameric structure of full-length TTR, did not show any inhibitory effect on TTR81–127 amyloid formation in cultured cells (Fig. 3D).
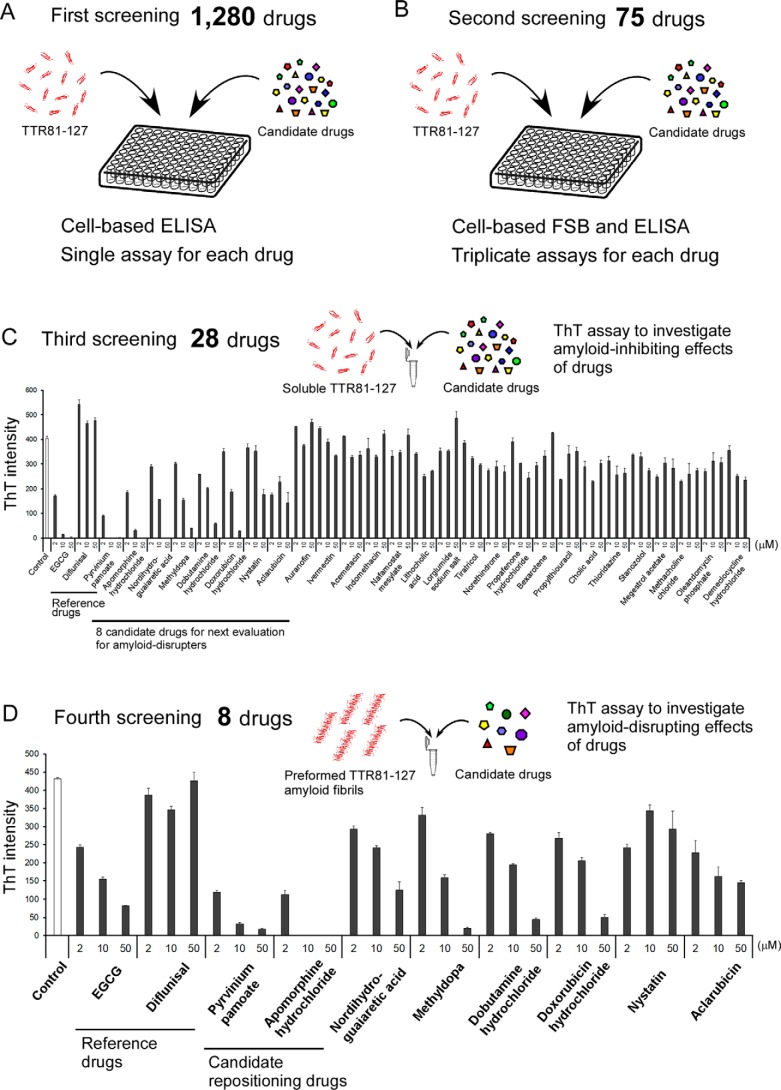
After using our novel HTS to directly evaluate TTR amyloid formation, we screened candidates for drug repositioning. We used a screening library of 1280 off-patent drugs (Drug Discovery Initiative, University of Tokyo, Tokyo, Japan) and tested EGCG and diflunisal as reference compounds for the HTS (Fig. 4). In the first screening, we found 75 candidate drugs that inhibited TTR amyloid deposits in the cell-based ELISA (Fig. 4A and Table S3). In the second step, we performed the cell-based FSB assay with the 75 drugs (triplicate assays) in addition to the cell-based ELISA with the 75 drugs (triplicate assays) and selected 28 candidate drugs for the next analysis (Fig. 4B and Table S3). Next, in the third screening, we determined whether those 28 candidate drugs directly inhibited TTR81–127 amyloid formation in PBS at neutral pH in test tubes (Fig. 4C). We found eight candidate drugs that strongly inhibited amyloid formation of TTR81–127 (Fig. 4C and Table S3) and used those eight candidate drugs in the next assay to detect amyloid disrupters. We incubated preformed TTR81–127 amyloid fibrils with those eight candidate drugs in PBS at neutral pH for 24 h and determined whether those drugs disrupted the preformed TTR81–127 amyloid fibrils. Pyrvinium pamoate and apomorphine hydrochloride significantly reduced the preformed TTR81–127 amyloid fibrils in a dose-dependent manner in test tubes (Fig. 4D and Table S3) and in the cell-based FSB assay (Fig. S8). Pamoate did not disrupt preformed TTR81–127 amyloid fibrils in vitro (Fig. S9).
Figure 4.
HTS of antiamyloid drugs by using a library containing 1280 off-patent drugs by means of the novel cell-based HTS at neutral pH. A, first screening of 1280 off-patent drugs by means of a cell-based ELISA. After concomitant incubation of candidate drugs at 10 and 5 μm TTR81–127 in cell culture medium for 24 h, cell-based ELISAs (one assay for each drug) were performed to analyze the inhibitory effects of candidate drugs on amyloid deposits in cultured cells. B, second screening of 75 off-patent drugs by means of the cell-based FSB assay and ELISA. After concomitant incubation of candidate drugs at 10 and 5 μm TTR81–127 in cell culture medium for 24 h, the cell-based FSB assay and ELISA (triplicate assays for each drug) were performed to determine the inhibitory effects of candidate drugs. C, third screening of 28 candidate off-patent drugs by means of the ThT assay to investigate the amyloid-inhibiting effects of the drugs. After concomitant incubation of 25 μm TTR81–127 and candidate drugs at 2–50 μm in PBS for 24 h at 37 °C in test tubes, ThT intensities were analyzed in triplicate assays. D, fourth screening of eight candidate off-patent drugs by means of the ThT assay to study the amyloid-disrupting effects of the drugs. After concomitant incubation of 25 μm preformed TTR81–127 amyloid fibrils and candidate drugs at 2–50 μm in PBS for 24 h at 37 °C, ThT intensities were analyzed in triplicate assays.
To investigate the effects of two repositioning compounds on tryptic cleavage of TTR aggregates, we performed SDS-PAGE of pre-formed aggregates of full-length TTR V30M incubated with 0.5% trypsin and 50 μm repositioning drugs for 24 h. We did not find inhibitory effects of the two repositioning drugs, pyrvinium pamoate and apomorphine hydrochloride, on tryptic cleavage of TTR aggregates (Fig. S10).
To investigate the effects of two repositioning compounds on amyloid formation of other amyloid proteins, we also examined the inhibitory effects on Aβ42 and Aβ40 amyloid fibril formation. Pyrvinium pamoate and apomorphine hydrochloride significantly inhibited Aβ42 and Aβ40 amyloid formation in a dose-dependent manner in vitro (Fig. S11).
Amyloid-disrupting effects of two candidate off-patent drugs on TTR amyloid fibrils
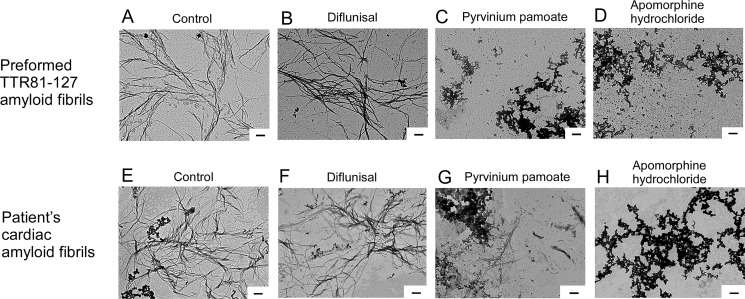
Electron microscopic analyses revealed that pyrvinium pamoate and apomorphine hydrochloride disrupted preformed TTR81–127 amyloid fibrils in vitro (Fig. 5, A–D).
Figure 5.
Effects of candidate off-patent drugs on preformed TTR81–127 amyloid fibrils and cardiac TTR amyloid fibrils isolated from a patient's cardiac tissue sample. A–D, electron microscopic images of preformed TTR81–127 amyloid fibrils incubated with candidate therapeutic drugs in PBS at 37 °C for 24 h. A, 0.2 μm preformed TTR81–127 amyloid fibrils. B–D, 0.2 μm preformed TTR81–127 amyloid fibrils treated with 50 μm diflunisal (B), pyrvinium pamoate (C), or apomorphine hydrochloride (D) in PBS at 37 °C for 24 h. E–H, electron microscopic findings of cardiac amyloid fibrils isolated from a cardiac sample from an autopsied 63-year-old male patient with TTR V30M. Fibrils were incubated with candidate therapeutic drugs in PBS at 37 °C for 24 h. E, cardiac amyloid fibrils isolated from the patient with ATTRm amyloidosis. F–H, cardiac amyloid fibrils treated with 50 μm diflunisal (F), pyrvinium pamoate (G), and apomorphine hydrochloride (H) in PBS at 37 °C for 24 h. Scale bars, 200 nm.
We also investigated the effects of two drugs with cardiac amyloid fibrils isolated from an amyloid-laden autopsied cardiac sample from a patient with ATTR V30M amyloidosis. Preformed TTR81–127 amyloid fibrils were ultrastructurally similar to the patient's amyloid fibrils (Fig. 5, A and E). We found that pyrvinium pamoate and apomorphine hydrochloride significantly disrupted the patient's cardiac amyloid fibrils ex vivo (Fig. 5, E–H). Diflunisal, a stabilizer of the tetrameric structure of TTR, did not disrupt those amyloid fibrils in vitro and ex vivo (Fig. 5, B and F).
Stabilizing effects of pyrvinium pamoate and apomorphine hydrochloride on the tetrameric structure of full-length TTR
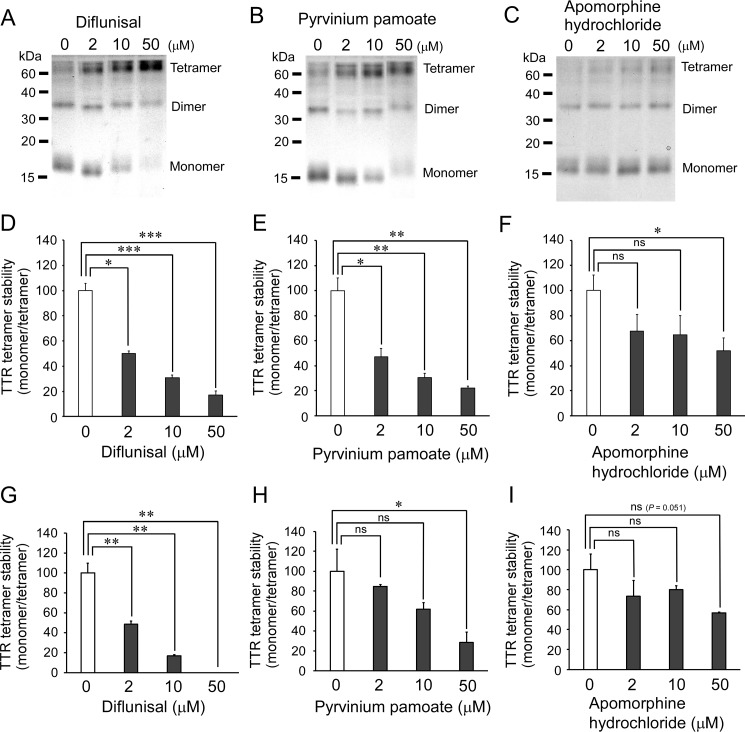
We also studied whether these two candidate repositioning drugs, pyrvinium pamoate and apomorphine hydrochloride, would stabilize the tetrameric structure of full-length TTR in addition to amyloid-disrupting effects. In vitro studies with recombinant full-length TTR V30M showed that pyrvinium pamoate had a stabilizing effect on TTR tetramers that was almost the same as that of diflunisal (Fig. 6, A–F).
Figure 6.
TTR tetramer-stabilizing effects of candidate off-patent drugs. A–C, SDS-PAGE analysis visualized by silver staining of cross-linked tetrameric full-length TTR V30M pretreated with diflunisal (A), pyrvinium pamoate (B), and apomorphine hydrochloride (C). D–F, quantitative analysis of tetramer stability of recombinant full-length TTR V30M treated with diflunisal (D), pyrvinium pamoate (E), and apomorphine hydrochloride (F). Incubation of 5 μm full-length TTR V30M and drugs at 2–50 μm in PBS for 30 min at 25 °C was followed by 4.5 m urea-induced denaturation of TTR tetramers for 24 h at 25 °C. Cross-linked samples were analyzed by means of SDS-PAGE. G–I, stability of plasma TTR tetramers against urea-induced denaturation. Plasma samples, which were obtained from an untreated 74-year-old man with ATTR V30M amyloidosis, were incubated with the drugs at 2–50 μm—diflunisal (G), pyrvinium pamoate (H), and apomorphine hydrochloride (I)— for 30 min at 25 °C, followed by 4.5 m urea-induced denaturation of TTR tetramers for 24 h at 25 °C. The plasma samples cross-linked via glutaraldehyde were analyzed by means of SDS-PAGE followed by immunoblotting with anti-TTR antibodies (Agilent Technologies). *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
We found that pyrvinium pamoate significantly stabilized the tetrameric structure of full-length TTR ex vivo in the plasma of a patient with ATTR V30M amyloidosis, although the stabilizing effect of pyrvinium pamoate on the tetrameric structure of plasma TTR was weaker than that of diflunisal (Fig. 6, G and H). In contrast, apomorphine hydrochloride did not stabilize the tetrameric structure of the patient's plasma TTR (Fig. 6I).
Discussion
In this study, we showed that trypsin cleaved aggregates of full-length TTR into C-terminal fragments. In addition, our in vitro studies revealed that the 5-kDa C terminus of TTR, TTR81–127, quite easily formed amyloid fibrils at neutral pH and induced apoptosis, probably via the Fas-dependent pathway in cultured cells. With this highly amyloidogenic TTR81–127 fragment, we developed a novel cell-based HTS method to find candidate drugs that would directly disrupt disease-causing TTR amyloid fibrils, and we successfully found candidate drugs by using a screening library of 1280 off-patent drugs.
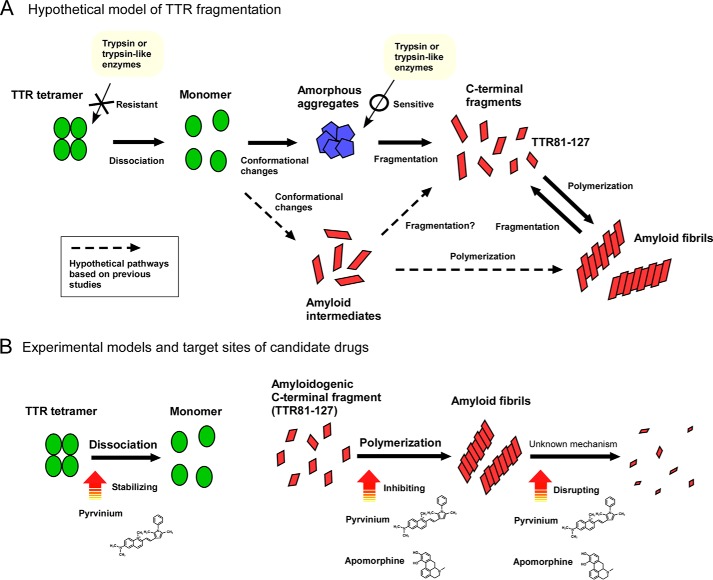
C-terminal fragments of TTR were reportedly found in tissue amyloid deposits in patients with ATTR amyloidosis (12, 26–33). Certain in vitro studies revealed that C-terminal fragments of TTR, especially the 10-kDa C terminus that corresponds to TTR49–127, promoted TTR amyloid formation (34–36). In this study, we found that trypsin cleaved acid-induced full-length TTR aggregates into the C-terminal fragments TTR81–127 and TTR49–127 in vitro, but it barely cleaved native soluble full-length TTR tetramers. These results indicated that fragmentation of TTR may require destruction of the native TTR tetrameric structure, which is generally believed to be caused by TTR mutations in ATTRm amyloidosis. Marcoux et al. (35) reported that the mechanical forces produced by magnetic bar agitation promoted fragmentation of full-length TTR by trypsin in vitro, which is consistent with our findings. We hypothesize that fragmentation of TTR occurs after conformational changes in TTR (Fig. 7A). To confirm our hypothesis, we must perform additional in vitro and in vivo investigations.
Figure 7.
Schematic illustration of a hypothetical model of TTR fragmentation and target sites of candidate repositioning drugs. A, hypothetical model of TTR fragmentation. Fragmentation of TTR occurs after conformational changes of TTR. Trypsin or trypsin-like enzymes barely cleave native soluble full-length TTR tetramers. B, experimental models and target sites of candidate repositioning drugs in TTR amyloid formation. Pyrvinium pamoate may have dual therapeutic effects—TTR-stabilizing and amyloid-disrupting effects—in ATTR amyloidosis. Apomorphine hydrochloride may disrupt TTR amyloid.
Unlike previous studies using a mixture of tryptic cleavage products of full-length TTR, in which the TTR49–127 fragment reportedly formed amyloid fibrils (34–36), our recombinant TTR49–127 protein only formed amorphous aggregates. There are two possibilities that could explain this discrepancy. First, the structural conformation of our recombinant TTR49–127 protein that was directly purified from Escherichia coli expressing human TTR49–127 may be different from the structural conformation of their TTR49–127 fragment, which was a tryptic cleavage product of full-length TTR. Second, their mixture of tryptic cleavage products of full-length TTR could contain TTR81–127 in addition to TTR49–127, as shown in their previous study (34). Further in vitro and in vivo studies are needed to determine amyloidogenesis of TTR49–129.
In previous in vitro studies of ATTR amyloidosis, acidic denaturation of full-length TTR was often used to form TTR aggregates (13, 14). We found, however, that a smaller C-terminal TTR81–127 fragment quite easily formed amyloid fibrils even at neutral pH in PBS and also in cell culture medium without any denaturing process. However, other forms of TTR, such as the 10-kDa C-terminal fragment TTR49–127, the N-terminal TTR1–80, and the full-length WT and mutant TTRs, never formed amyloid fibrils at neutral pH. Saelices et al. (37, 38) reported that two short peptides corresponding to the β-strands F and H, which are located within the TTR81–127 fragment, formed a “steric zipper” amyloid structure in vitro and could stimulate amyloid fibril formation of TTR. Our recent in vivo study also indicated that transgenic Caenorhabditis elegans models expressing a human TTR81–127 fragment formed aggregates of TTR81–127 and showed defective worm motility and a significantly shortened life span compared with other worm strains expressing full-length WT TTR, full-length TTR V30M, TTR1–80, and TTR49–127 (39). Those findings suggest that the C-terminal region of TTR, i.e. TTR81–127, plays important roles in TTR amyloid fibril formation.
TTR81–127 amyloid fibrils had features similar to those of our patient's amyloid fibrils. Ultrastructurally, morphological features of TT81–127 amyloid fibrils formed at neutral pH in vitro resembled cardiac amyloid fibrils isolated from an amyloid-laden tissue of our patient with ATTRm amyloidosis (Fig. 5, A and E). To investigate TTR81–127 amyloid formation and the effects of TTR81–127 amyloid fibrils on cultured cells, we mainly used glomotel cells, which were derived from vascular smooth muscle cells (8). There were two reasons why we used glomotel cells for the cell culture studies with TTR fragments. First, amyloid deposits were commonly found around vascular smooth muscle cells in patients with ATTR amyloidosis. Second, to avoid further cleavage of TTR fragments, we used glomotel cells, which did not cleave TTR as described earlier. Our cell culture studies revealed that TTR81–127 amyloid deposits induced Fas-mediated apoptosis and expression of inflammation-related genes such as ICAM1, VCAM1, RELB, IL32, CXCL1, CCL2, C3, and SOD2. In previous studies, full-length TTR aggregates also reportedly induced apoptosis and inflammation (40–42). As in our investigations with TTR81–127, a previous histopathological study found that Fas, caspase 8, and Bax increased in tissue samples obtained from patients with ATTRm amyloidosis and transgenic mice expressing human TTR V30M (42). Those findings suggest that TTR81–127 amyloid fibrils experimentally induce Fas-mediated apoptosis and inflammation-related changes in cultured cells, similar to the situation in patients with ATTRm amyloidosis. Further studies are needed to clarify the roles of these inflammation-related genes in ATTR amyloidosis.
On the basis of our results described above, we successfully developed a novel cell-based HTS method to directly target TTR amyloid fibril formation by using the highly amyloidogenic TTR81–127 fragment. The novel HTS method used TTR81–127 for discovering amyloid-disrupting drugs, the later process of TTR amyloid formation even at neutral pH. To investigate drug repositioning, we utilized a screening library of 1280 off-patent drugs and found two candidate repositioning drugs, pyrvinium pamoate and apomorphine hydrochloride. In the first to third steps of the screening, we analyzed amyloid-inhibiting effects of drugs and tested both amyloid-inhibiting and disrupting effects in the fourth step. We might have failed to screen some drugs with amyloid-disrupting effects in the first to third steps. We also used two reference drugs, diflunisal, which reportedly stabilized the tetrameric structure of TTR (14, 17), and EGCG, which inhibited amyloid formation (43, 44). In our assay, EGCG inhibited amyloid formation of TTR81–127, whereas diflunisal, which stabilized only tetrameric TTR (14, 17), did not show any effects. EGCG also inhibited TTR81–127 deposits in our in vivo transgenic C. elegans model that expressed TTR81–127 (39). We confirmed that two candidate repositioning drugs, pyrvinium pamoate and apomorphine hydrochloride, which we found by means of the novel HTS method using TTR81–127, disrupted the patient's cardiac amyloid fibrils ex vivo (Fig. 5, E–H). These data suggest that our novel HTS method using TTR81–127 is useful for discovering amyloid-disrupting drugs.
Pyrvinium pamoate is a quinoline-derived cyanine dye that was originally used as an anthelmintic drug (45). Recent studies revealed that pyrvinium pamoate also had anticancer effects in various types of cancers via several mechanisms (46, 47). In this study, we determined that pyrvinium pamoate disrupted TTR amyloid fibrils and stabilized TTR tetramers in vitro and ex vivo. Because pyrvinium pamoate also inhibited Aβ42 and Aβ42 amyloid fibrils, amyloid-inhibiting effects of this drug may be not specific for TTR amyloid, similar to other anti-amyloid compounds such as polyphenols, doxycycline, and tauroursodeoxycholic acid (48, 49). The TTR-stabilizing effect of pyrvinium pamoate was almost the same as that of diflunisal, which is used as a disease-modifying therapy for patients with ATTRm amyloidosis (17). The present initial study to develop novel drugs indicated that pyrvinium pamoate may have dual therapeutic effects (TTR-stabilizing and amyloid-disrupting effects) in ATTR amyloidosis (Fig. 7B). We must perform additional in vitro studies to determine the detailed mechanisms of how pyrvinium pamoate binds to TTR, as well as in vivo studies.
When used as an Food & Drug administration-approved anthelmintic drug, pyrvinium pamoate is given orally to eliminate worms in the gastrointestinal tract. However, pyrvinium pamoate has limited absorption into the bloodstream when orally administered (50). For ATTR amyloidosis, we must modify pyrvinium pamoate so that it has improved intestinal absorption and is soluble in blood. We must also identify other ways to administer the drug, which have been attempted in the repositioning of this drug for anticancer therapy (46, 47).
The other candidate drug, apomorphine hydrochloride, is a morphine derivative that is a nonergoline dopamine agonist and was originally used to treat Parkinson disease (51). In this study, we found that apomorphine inhibited and disrupted TTR amyloid fibril formation (Fig. 7B). This drug also inhibited Aβ-amyloid formation as reported previously (52). Although its oral bioavailability was thought to be low, bioavailability after subcutaneous injection was reportedly 100% (53). Because apomorphine hydrochloride is said to efficiently cross the blood–brain barrier after subcutaneous injection, the drug will likely have potential therapeutic effects not only for systemic ATTR amyloid deposits but also for leptomeningeal amyloid deposits of ATTR amyloidosis, for which the present treatments, including liver transplantation, diflunisal, tafamidis, and gene-silencing therapies, are believed to be ineffective because delivery of those drugs is insufficient. We must conduct additional investigations of the effects of this drug on TTR amyloid formation in vivo.
There were several limitations to this study. Although we have shown that TTR fragmentation occurred after forming aggregates of TTR in vitro, detailed mechanism of TTR fragmentation remains to be elucidated in vivo. It also remains to be determined whether fragmentation of TTR is a primary or secondary process in TTR amyloid fibril formation. In addition, the pathological roles of TTR fragments, such as TTR81–127 and TTR49–127, are still subject to debate. Because the number of patients having TTR81–127 in amyloid deposits was limited, we should analyze TTR fragments in a larger number of patients. Because the HTS method using TTR81–127 is targeted to the later process of TTR amyloid fibrils' formation for discovering amyloid-disrupting drugs, we should note that this method is not appropriate for assessing the early process of TTR amyloid formation, such as dissociation of TTR tetramer to monomer.
In conclusion, the highly amyloidogenic TTR81–127 fragment is valuable in in vitro studies at neutral pH and in cell culture studies. Our novel cell-based HTS method is quite useful for discovering therapeutic drugs that disrupt amyloid fibrils directly. Repositioning pyrvinium pamoate and apomorphine hydrochloride may have potential therapeutic effects on ATTR amyloidosis.
Experimental procedures
Recombinant TTR expression and purification
Human WT TTR and its variants were obtained as described previously (54). The DNA sequences of TTR1–48 and TTR1–80 fragments were inserted into an NdeI/BamHI-predigested pOPTH vector (55, 56). TTR1–48 and TTR1–80 fragments with an N-terminal His6 tag were expressed in E. coli C41 (DE3) RIPL cells and purified with Ni-affinity chromatography (57). TTR49–127 and TTR81–127 fragments were expressed as a fusion protein with an N-terminal His6-tagged full-length WT TTR in E. coli M15 cells and purified with Ni-affinity chromatography (57). The His6 tag and TTR tag were cleaved by factor Xa or tobacco etch virus protease, and the TTR fragments were purified by reversed- phase HPLC. Those recombinant proteins were dissolved in 0.02% NH3 solution, and aliquots were stored at −80 °C.
Acid-induced formation of amorphous aggregates of full-length TTR
To make acid-induced amorphous aggregates of full-length TTR, we incubated 50 μm recombinant full-length TTR at pH 4.0 in 50 mm sodium acetate buffer containing 100 mm KCl for 24 h at 37 °C, according to methods used in our previous study (58).
Cells and cell culture studies
We used several cell lines, including the human vascular smooth muscle cell line glomotel (58), human neuroblastoma cell line SH-SY5Y, human glioblastoma cell line U87MG, human hepatocellular carcinoma cell line HepG2, and human retinal pigment epithelium cell line ARPE-19. Those cells were purchased from the American Type Culture Collection (Manassas, VA) and were maintained in culture medium containing Gibco Dulbecco's modified Eagle's medium (Thermo Fisher Scientific, Waltham, MA), 10% fetal calf serum (Sigma), and 1% penicillin/streptomycin solution (Thermo Fisher Scientific) in 5% CO2 at 37 °C. Cell culture medium was replaced every 2–3 days.
To investigate the mechanisms of TTR fragmentation in cultured cells, we seeded cells in 12-well culture plates. We induced formation of aggregates of full-length TTR V30M at pH 4.0 in 50 mm sodium acetate buffer containing 100 mm KCl for 24 h in test tubes. The pre-formed aggregates of full-length TTR V30M were added to the extracellular culture medium. Cells were treated with native full-length TTR or acid-induced aggregates of full-length TTR and were incubated for 48 h with or without 10 μm trypsin inhibitor (Sigma). Cells were washed once in ice-cold PBS and then lysed by adding Laemmli sample buffer (Bio-Rad), followed by SDS-PAGE analyses with the Tricine system. We stained gels with Coomassie Brilliant Blue R-250 (CBB).
Amino acid sequences of TTR fragments
To identify amino acid sequences of bands derived from TTR fragments, samples were electrophoretically transferred to polyvinylidene difluoride membranes (Bio-Rad). After staining with CBB, polyvinylidene difluoride membrane bands were digested with trypsin (Promega, Fitchburg, WI) in 0.1 m ammonium bicarbonate, and the resulting peptides were fractionated by reversed-phase HPLC on a Beckman Ultrasphere ODS column (Beckman Coulter, Brea, CA). Edman degradation analysis of samples was performed on an Applied Biosystems Model 491 cLC protein sequencer (Thermo Fisher Scientific) according to the manufacturer's standard cycles and methods.
Mass spectrometric analysis of components of cultured SH-SY5Y cells treated with pre-formed aggregates of full-length TTR V30M
To identify the components of cultured SH-SY5Y cells treated with pre-formed aggregates of full-length TTR V30M, we performed in-gel digestion of the 5-kDa band and the 10-kDa band according to the methods used in our previous study (59). Gel bands were cut into small pieces and were destained, dried, reduced with 10 mm DTT at 56 °C for 60 min, then alkylated with 55 mm iodoacetamide in the dark at room temperature for 45 min. Samples were washed with ammonium bicarbonate, dehydrated with acetonitrile, lyophilized, and then digested with 50 μg/ml trypsin (Promega, Fitchburg, WI) at 37 °C overnight. The supernatant was collected, lyophilized, and dissolved in 2% acetonitrile and 0.1% TFA. These peptide solutions were analyzed by using a nanoflow reversed-phase LC-MS/MS system (AMR Inc., Tokyo, Japan). To identify variable peptide modifications, carbamidomethylation of cysteine residues, methionine oxidation, and N-formylation, including formyl (Arg), formyl (Lys), and formyl (N terminus), were included in the search criteria.
Detection of TTR fragments in vitreous amyloid deposits
To detect TTR fragments in vitreous amyloid deposits in eight patients with the TTR V30M mutation of ATTRm amyloidosis, we performed immunoblotting analyses with rabbit polyclonal antiserum against TTR81–127 as the primary antibody, which was diluted 1:1000. We used a goat anti-rabbit antibody conjugated with horseradish peroxidase (Agilent Technologies, Santa Clara, CA), diluted 1:1,000, as the secondary antibody. The reaction was visualized by using the LAS-4000 Mini (GE Healthcare).
Fragmentation of full-length TTR by trypsin in vitro
Recombinant full-length TTR was incubated with trypsin (Promega) at 37 °C for 24 h. To detect smaller TTR fragments, we performed SDS-PAGE followed by CBB staining. Vitreous amyloid fibrils isolated from patients with ATTRm amyloidosis were incubated with trypsin (Promega) at 37 °C for 24 h. To detect smaller TTR fragments, we performed SDS-PAGE and immunoblotting as described above.
In vitro formation of amyloid fibrils at neutral pH
To induce amyloid fibril formation in vitro at neutral pH, 25 or 50 μm full-length WT TTR, full-length TTRV30M, TTR1–80, TTR49–127, TTR81–127, Aβ42 (Peptide Institute, Inc., Osaka, Japan), and Aβ40 (Peptide Institute) were incubated in PBS at 37 °C for 24 h.
Electron microscopic analyses of the morphology of amyloid fibrils
We investigated the morphology of amyloid fibrils or amorphous aggregates of TTR by using an electron microscope (HT7700; Hitachi High Technologies, Tokyo, Japan) at an accelerating voltage of 100 kV. In brief, 1-μl samples were placed on carbon/collodion-coated grids, after which samples were stained with 1 μl of 0.2% uranyl acetate for 1 min before examination with the electron microscope.
ThT fluorescence assay
During in vitro incubation of TTR, Aβ42, and Aβ40 in each condition, we analyzed ThT fluorescence with a spectrofluorometer (F-2700; Hitachi High Technologies) using a quartz cuvette having 10-mm path length under excitation and emission wavelengths of 442 and 489 nm, respectively. The 3-μl samples were mixed with 600 μl of 5 μm ThT in 50 mm glycine/NaOH buffer, pH 9.5, and were used for the measurements. We obtained three scans per sample. Measurements were performed at room temperature.
Evaluation of TTR tetramers and oligomers via chemical cross-linking
To evaluate tetramers and oligomers of TTR81–127, we utilized chemical cross-linking with glutaraldehyde according to a previous study (60). Briefly, we added 2.5 μl of 25% glutaraldehyde to 25 μl of 50 μm TTR in PBS for 4 min at room temperature followed by quenching via addition of 2.5 μl of 7% NaBH4 in 0.1 m NaOH. Cross-linked samples were analyzed by means of SDS-PAGE and were visualized by using CBB staining.
Amyloid formation in cultured cells and detection of TTR amyloid deposits
To induce amyloid formation in cultured cells, cells were seeded in half-area 96-well culture plates (Greiner Bio-One, Kremsmünster, Austria) or Lab-Tek chamber slides (Thermo Fisher Scientific). Cells were treated with various TTRs in Opti-MEM (Thermo Fisher Scientific) containing 1% penicillin/streptomycin solution (Thermo Fisher Scientific) in 5% CO2 at 37 °C.
We detected amyloid deposits in cultured cells by means of staining with Congo red (CR) or FSB (Dojindo Laboratories, Kumamoto, Japan) (59). We performed alkaline CR staining as described previously and confirmed CR reactivity under polarized light. For FSB staining, sections were immersed in 0.0001% FSB in 50% ethanol for 30 min and washed with 50% ethanol before fluorescence microscopy.
Immunofluorescence staining of amyloid formation in cultured cells and detection of TTR amyloid deposits
To detect the TTR81–127 amyloid deposits in glomotel cells, cells were seeded in Lab-Tek chamber slides (Thermo Fisher Scientific). Cells were treated with 50 μm TTR81–127 in Opti-MEM (Thermo Fisher Scientific) containing 1% penicillin/streptomycin solution (Thermo Fisher Scientific) in 5% CO2 at 37 °C. After 24 h of incubation, cells were fixed in 4% paraformaldehyde at room temperature for 30 min. After washing with PBS, the fixed cells were incubated with a sheep anti-TTR antibody (Abcam, Cambridge, UK) and a rabbit anti-β-actin antibody (Proteintech, Rosemont, IL) as the primary antibody and diluted 1:100 at room temperature for 1 h. After washing with PBS, we used a donkey anti-sheep IgG antibody conjugated with Alexa Fluor 647 (Abcam) and a swine anti-rabbit immunoglobulin antibody conjugated with FITC (CiteAb, Bath, UK), diluted 1:100, as the secondary antibodies. To mount cells, we used VECTASHIELD antifade mounting medium with 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA). Images of stained cells were obtained using a fluorescence microscope BZ-X800 (Keyence, Osaka, Japan).
Analyses of cytotoxicity and apoptosis in cultured cells
Cytotoxicity was analyzed by using the MultiTox-Fluor multiplex cytotoxicity assay (Promega), according to the manufacturer's instructions. This cytotoxicity assay measured dead-cell and live-cell protease activities using a cell-impermeant fluorogenic peptide substrate (bis-alanyl-alanyl-phenylalanyl-rhodamine 110) and a cell-permeant fluorogenic peptide substrate (glycyl-phenylalanyl-amino fluorocoumarin). Caspase 3/7 activity was determined by using the Apo-ONE homogeneous caspase-3/7 assay (Promega), according to the manufacturer's instructions. Caspase 8 and 9 activities were also analyzed by using Caspase-Glo 8 assay (Promega) and Caspase-Glo 9 assay (Promega) systems, respectively. Fluorescence and luminescence were measured with the FilterMax F5 microplate reader (Molecular Devices, Sunnyvale, CA). Annexin V staining was performed by using apoptotic, necrotic, and healthy cells quantitation kit plus (Biotium, Fremont, CA) with Biozero BZ-8000 fluorescence microscopy (Keyence, Osaka, Japan).
Analysis of apoptosis-related proteins in cultured cells treated with the highly amyloidogenic TTR81–127
We analyzed the expression of apoptosis-related proteins in cultured glomotel cells, which were treated with 25 μm TTR81–127 in Opti-MEM for 6 h, by using the Proteome Profiler human apoptosis array kit (R&D Systems, Minneapolis, MN), according to the manufacturer's instructions. The reaction was visualized by using LAS-4000 Mini (GE Healthcare). The relative ratio of signal intensity was determined in cultured cells treated with TTR81–127 compared with nontreated cells.
RNA-seq analysis in cultured cells treated with the highly amyloidogenic TTR81–127
We extracted total RNA from cultured glomotel cells treated with the highly amyloidogenic TTR81–127 at 25 μm or not so treated in Opti-MEM for 24 h by using the TRIzol RNA isolation reagents according to the manufacturer's instructions. The RNA obtained was analyzed with the Bioanalyzer 2100 (Agilent Technologies).
With 1 μg of RNA extracted from cultured cells, we constructed the RNA-seq library with the mRNA-seq sample preparation kit by following the manufacturer's instructions (Illumina, San Diego). We generated 36-bp single-end-read RNA-seq tags via an Illumina GA sequencer according to a standard protocol. We used RNA-seq tags that mapped to human reference genome sequences (hg18) without mismatches. RNA-seq tags corresponded to RefSeq transcripts or TSCs when their genomic coordinates overlapped. We calculated fragments per kilobase million (FPKM) values for cultured cells treated with TTR81–127 and compared this result with FPKM values for nontreated cultured cells in each gene (Table S1).
Real-time quantitative reverse transcription-PCR
We extracted total RNA from cultured cells by using the RNeasy mini kit (Qiagen, Venlo, Netherlands). Total RNA (0.5 μg) was reverse-transcribed to cDNA via the PrimeScript RT Master Mix reagent (Takara Bio, Shiga, Japan) according to the manufacturer's instructions. Each PCR assay was performed with the LightCycler 480 system (Roche Applied Science, Penzberg, Germany) with SYBR Premix DimerEraser (Takara Bio) under the following conditions: initialization for 10 s at 95 °C, then 45 cycles of amplification, with 5 s at 95 °C for denaturation and 20 s at 60 °C for annealing and elongation. The primers (Table S4) were purchased from Takara Bio. GAPDH was used as the internal reference gene. All standards and samples were analyzed in triplicate.
Inhibition of the Fas ligand-dependent pathway in cultured cells
To inhibit the Fas ligand-dependent signal pathway, we used RKTS-33 (FUJIFILM Wako Pure Chemical Corp., Osaka, Japan), an epoxycyclohexenone derivative that specifically inhibits this Fas ligand-dependent pathway (61), in cultured cells simultaneously treated with TTR81–127.
HTS assays with the highly amyloidogenic TTR81–127
We developed the cell-based FSB assay by using FSB, which is the fluorescent amyloid-specific probe, to determine the amount of amyloid in deposits in cultured cells treated with TTR81–127; we also used the cell-based enzyme-linked immunosorbent assay (ELISA) with rabbit polyclonal antiserum against TTR81–127 to analyze the amount of TTR81–127 in deposits in cultured cells (Fig. S6).
In the cell-based FSB assay, we seeded glomotel cells in half-area 96-well culture plates (Greiner Bio-One). After 24 h, we added TTR81–127 to the cell culture medium and incubated the samples at 37 °C for 24 h. We fixed cultured cells with 10% formaldehyde neutral buffer solution (Nacalai Tesque, Kyoto, Japan) for 30 min. After washing the fixed cells with PBS, we stained cultured cells with 0.0001% FSB in 50% EtOH for 30 min. We washed FSB-stained cells with 50% EtOH once, followed by washing with PBS three times. We analyzed the amount of amyloid in deposits by using IN Cell Analyzer 2200 (GE Healthcare).
To quantify the amount of TTR81–127 in deposits, we performed a cell-based ELISA with the rabbit polyclonal anti-TTR81–127 antiserum. Briefly, we seeded glomotel cells in half-area 96-well culture plates (Greiner Bio-One). After 24 h, we added TTR81–127 to the cell culture medium and incubation proceeded at 37 °C for 24 h. We fixed the cultured cells with 10% formaldehyde neutral buffer solution (Nacalai Tesque) for 30 min. After washing the fixed cells with PBS, we added Blocking One solution (Nacalai Tesque) and incubated the cells for 30 min. After washing the samples with PBS three times, we added rabbit polyclonal antiserum against TTR81–127 diluted 1:2000 in 5% BSA dissolved in PBS, and incubation was continued for 1 h. After we washed the samples with PBS three times, we added a goat anti-rabbit antibody conjugated with horseradish peroxidase (Agilent Technologies) diluted 1:5000 in 5% BSA dissolved in PBS, and incubation continued for 1 h. After we washed the samples with PBS five times, we added KPL SureBlue TMB Microwell Peroxidase Substrate (1-Component) (SeraCare, Milford, MA), followed by incubation for 15 min, after which we added 1 n HCl to stop the reaction. We read wells at 450 nm by using the xMark Microplate Spectrophotometer (Bio-Rad). To evaluate the quality of this screening method, we calculated the Z′ factor, signal-to-background ratio (S/B), signal-to-noise ratio (S/N), and coefficient of variation (CV) in the cell-based ELISA and compared the results with the acceptable values of those parameters (Z′ factor >0.5, S/B >2, S/N >10, and CV <20%) for HTS methods.
Drug library consisting of off-patent drugs
A library consisting of 1280 off-patent drugs was provided by the Drug Discovery Initiative, University of Tokyo (Tokyo, Japan). The concentrations of all compounds were 20 mm, dissolved in DMSO.
High-throughput screening for drug repositioning
For the first screening, we performed the cell-based ELISA with 5 μm TTR81–127 with concomitant administration of 1280 off-patent drugs at 10 μm (Fig. 4 and Table S3). We analyzed each drug in a single assay and calculated values relative to the control, in which we did not use drugs. We obtained the names of the compounds that showed relative values of <88% and used these drugs for the next (second) screening.
For the second screening, we performed the cell-based FSB assay in triplicate in addition to the cell-based ELISA (triplicate assays) with 75 off-patent drugs (Fig. 4 and Table S3). We used compounds that had relative values of <85% in the cell-based FSB assay, for the third screening.
In the third screening, we investigated the amyloid-inhibiting effects of drugs by means of the ThT assay with 25 μm TTR81–127 with concomitant administration of 28 off-patent drugs at 2–50 μm in test tubes (Fig. 4C and Table S3). We used candidate drugs that had relative values of <50% in the ThT assay for the next screening.
In the fourth screening, we evaluated the amyloid-disrupting effects of drugs by means of the ThT assay with 25 μm preformed TTR81–127 amyloid fibrils with eight off-patent drugs at 2–50 μm in test tubes (Fig. 4D and Table S3).
Isolation of amyloid fibrils from a patient's cardiac tissue sample
Cardiac amyloid fibrils were isolated from a patient's amyloid-laden cardiac tissue sample as described previously (62). Briefly, 150 mg of a nonfixed frozen cardiac sample from an autopsied patient with ATTR V30M amyloidosis, who died at 63 years old, was diced with a scalpel and washed 10 times with 1 ml of Tris-calcium buffer (20 mm Tris, 138 mm NaCl, 2 mm CaCl2, 0.1% (w/v) NaN3, pH 8.0). The pellet was resuspended in 1 ml of a freshly prepared solution of 5 mg/ml Clostridium histolyticum collagenase (Sigma) in Tris-calcium buffer. After overnight incubation at 37 °C, the tissue sample was centrifuged at 3100 × g for 30 min at 4 °C, and the retained pellet was resuspended in 1 ml of Tris-EDTA (EDTA) buffer (20 mm Tris, 140 mm NaCl, 10 mm EDTA, 0.1% (w/v) NaN3, pH 8.0) and homogenized with a pellet pestle using 10 cycles consisting of 1 s on and 1 s off. The homogenate was centrifuged for 5 min at 3100 × g at 4 °C, and the supernatant was removed carefully. This step was repeated 10 times. After the homogenization step in Tris-EDTA buffer, the remaining pellet was homogenized with a pellet pestle in 0.25 ml of ice-cold water. The homogenate was centrifuged for 5 min at 3100 × g at 4 °C, and the supernatant was removed and stored as water extract 1. This step was repeated 10 times.
Evaluation of the stability of TTR tetramers
To evaluate the effects of drugs on the stability of TTR tetramers to urea denaturation, recombinant TTR or plasma samples were incubated at 25 °C for 2 h with 2–50 μm drugs in test tubes followed by denaturation with 4.5 m urea for 24 h at 25 °C, as described previously (60). After SDS-PAGE, TTR was detected by silver staining with the ProteoSilver silver stain kit (Sigma) or immunoblotting with a polyclonal rabbit anti-TTR antibody (Agilent Technologies). Band intensities were quantified by using the NIH ImageJ program 0 (http://rsbweb.nih.gov/ij/).
Statistical analysis
Data were evaluated with Student's t test. All analyses were performed with JMP Version 5.1 (SAS Institute Japan, Tokyo, Japan). p values less than 0.05 were considered to be statistically significant.
Ethics
The study protocol was approved by the Human Ethics Review Committee of Kumamoto University and abided by the Declaration of Helsinki principles. Signed consent forms were obtained from all patients or family members.
Author contributions
M. U., H. M., M. D. B., and Y. A. conceptualization; M. U., M. O., M. M., B. K.-B., K. K., A. I., Y. M., M. T., A. U., A. K., R. S., T. M., Y. I., T. N., S. S., T. S., H. K., T. Y., H. M., M. D. B., and Y. A. data curation; M. U., M. D. B., and Y. A. funding acquisition; M. U., M. O., M. M., B. K.-B., K. K., A. I., Y. M., M. T., A. U., A. K., R. S., T. N., S. S., T. S., H. K., T. Y., H. M., M. D. B., and Y. A. investigation; M. U., M. M., and Y. A. methodology; M. U. writing-original draft; M. U., M. O., M. M., B. K.-B., K. K., A. I., Y. M., M. T., A. U., A. K., R. S., T. M., Y. I., T. N., S. S., T. S., H. K., T. Y., H. M., M. D. B., and Y. A. writing-review and editing; M. M., B. K.-B., K. K., H. M., M. D. B., and Y. A. supervision.
Supplementary Material
Acknowledgments
We are indebted to Hiroko Katsura and Mika Oka for excellent technical assistance and to Judith B. Gandy, Precision Editing, for providing professional English editing of the manuscript.
This work was supported in part by the Japan Society for the Promotion of Science KAKENHI Grants 18K07502, 15K09318, 15H04841, and 25870541 and by the Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology, Japan. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S11 and Tables S1–S4.
- ATTR
- amyloid transthyretin
- TTR
- transthyretin
- HTS
- high-throughput screening
- CBB
- Coomassie Brilliant Blue R-250
- FSB
- 1-fluoro-2,5-bis[(E)-3-carboxy-4-hydroxystyryl]benzene
- ThT
- thioflavin T
- DAPI
- 4′,6-diamidino-2-phenylindole
- CV
- coefficient of variation
- S/B
- signal-to-background ratio
- S/N
- signal-to-noise ratio.
- FPKM
- fragments per kilobase million
- Pt.
- patient
- EGCG
- epigallocatechin gallate
- CR
- Congo red
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- RNA-seq
- RNA sequencing.
References
- 1. Benson M. D., Buxbaum J. N., Eisenberg D. S., Merlini G., Saraiva M. J. M., Sekijima Y., Sipe J. D., and Westermark P. (2018) Amyloid nomenclature 2018: recommendations by the International Society of Amyloidosis (ISA) nomenclature committee. Amyloid 25, 215–219 10.1080/13506129.2018.1549825 [DOI] [PubMed] [Google Scholar]
- 2. Chiti F., and Dobson C. M. (2017) Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68 10.1146/annurev-biochem-061516-045115 [DOI] [PubMed] [Google Scholar]
- 3. Iadanza M. G., Jackson M. P., Hewitt E. W., Ranson N. A., and Radford S. E. (2018) A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 19, 755–773 10.1038/s41580-018-0060-8 [DOI] [PubMed] [Google Scholar]
- 4. Merlini G., Dispenzieri A., Sanchorawala V., Schönland S. O., Palladini G., Hawkins P. N., and Gertz M. A. (2018) Systemic immunoglobulin light chain amyloidosis. Nat. Rev. Dis. Primers 4, 38 10.1038/s41572-018-0034-3 [DOI] [PubMed] [Google Scholar]
- 5. Ando Y., Coelho T., Berk J. L., Cruz M. W., Ericzon B.-G., Ikeda S., Lewis W. D., Obici L., Planté-Bordeneuve V., Rapezzi C., Said G., and Salvi F. (2013) Guideline of transthyretin-related hereditary amyloidosis for clinicians. Orphanet. J. Rare Dis. 8, 31 10.1186/1750-1172-8-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benson M. D., and Kincaid J. C. (2007) The molecular biology and clinical features of amyloid neuropathy. Muscle Nerve 36, 411–423 10.1002/mus.20821 [DOI] [PubMed] [Google Scholar]
- 7. van den Berg M. P., Mulder B. A., Klaassen S. H. C., Maass A. H., van Veldhuisen D. J., van der Meer P., Nienhuis H. L. A., Hazenberg B. P. C., and Rienstra M. (2019) Heart failure with preserved ejection fraction, atrial fibrillation, and the role of senile amyloidosis. Eur. Heart J. 40, 1287–1293 10.1093/eurheartj/ehz057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ueda M., Horibata Y., Shono M., Misumi Y., Oshima T., Su Y., Tasaki M., Shinriki S., Kawahara S., Jono H., Obayashi K., Ogawa H., and Ando Y. (2011) Clinicopathological features of senile systemic amyloidosis: an ante- and post-mortem study. Mod. Pathol. 24, 1533–1544 10.1038/modpathol.2011.117 [DOI] [PubMed] [Google Scholar]
- 9. Sperry B. W., Reyes B. A., Ikram A., Donnelly J. P., Phelan D., Jaber W. A., Shapiro D., Evans P. J., Maschke S., Kilpatrick S. E., Tan C. D. W., Rodriguez E. R., Monteiro C., Tang W. H., Kelly J. W., et al. (2018) Tenosynovial and cardiac amyloidosis in patients undergoing carpal tunnel release. J. Am. Coll. Cardiol. 72, 2040–2050 10.1016/j.jacc.2018.07.092 [DOI] [PubMed] [Google Scholar]
- 10. Sueyoshi T., Ueda M., Jono H., Irie H., Sei A., Ide J., Ando Y., and Mizuta H. (2011) Wild-type transthyretin-derived amyloidosis in various ligaments and tendons. Hum. Pathol. 42, 1259–1264 10.1016/j.humpath.2010.11.017 [DOI] [PubMed] [Google Scholar]
- 11. Yamashita T., Ando Y., Okamoto S., Misumi Y., Hirahara T., Ueda M., Obayashi K., Nakamura M., Jono H., Shono M., Asonuma K., Inomata Y., and Uchino M. (2012) Long-term survival after liver transplantation in patients with familial amyloid polyneuropathy. Neurology 78, 637–643 10.1212/WNL.0b013e318248df18 [DOI] [PubMed] [Google Scholar]
- 12. Oshima T., Kawahara S., Ueda M., Kawakami Y., Tanaka R., Okazaki T., Misumi Y., Obayashi K., Yamashita T., Ohya Y., Ihse E., Shinriki S., Tasaki M., Jono H., Asonuma K., et al. (2014) Changes in pathological and biochemical findings of systemic tissue sites in familial amyloid polyneuropathy more than 10 years after liver transplantation. J. Neurol. Neurosurg. Psychiatry 85, 740–746 10.1136/jnnp-2013-305973 [DOI] [PubMed] [Google Scholar]
- 13. Lai Z., Colón W., and Kelly J. W. (1996) The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry 35, 6470–6482 10.1021/bi952501g [DOI] [PubMed] [Google Scholar]
- 14. Baures P. W., Peterson S. A., and Kelly J. W. (1998) Discovering transthyretin amyloid fibril inhibitors by limited screening. Bioorg. Med. Chem. 6, 1389–1401 10.1016/S0968-0896(98)00130-8 [DOI] [PubMed] [Google Scholar]
- 15. Miroy G. J., Lai Z., Lashuel H. A., Peterson S. A., Strang C., and Kelly J. W. (1996) Inhibiting transthyretin amyloid fibril formation via protein stabilization. Proc. Natl. Acad. Sci. U.S.A. 93, 15051–15056 10.1073/pnas.93.26.15051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ueda M., and Ando Y. (2014) Recent advances in transthyretin amyloidosis therapy. Transl. Neurodegener. 3, 19 10.1186/2047-9158-3-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Berk J. L., Suhr O. B., Obici L., Sekijima Y., Zeldenrust S. R., Yamashita T., Heneghan M. A., Gorevic P. D., Litchy W. J., Wiesman J. F., Nordh E., Corato M., Lozza A., Cortese A., Robinson-Papp J., et al. (2013) Repurposing diflunisal for familial amyloid polyneuropathy. JAMA 310, 2658–2667 10.1001/jama.2013.283815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Penchala S. C., Connelly S., Wang Y., Park M. S., Zhao L., Baranczak A., Rappley I., Vogel H., Liedtke M., Witteles R. M., Powers E. T., Reixach N., Chan W. K., Wilson I. A., Kelly J. W., et al. (2013) AG10 inhibits amyloidogenesis and cellular toxicity of the familial amyloid cardiomyopathy-associated V122I transthyretin. Proc. Natl. Acad. Sci. U.S.A. 110, 9992–9997 10.1073/pnas.1300761110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sant'Anna R., Gallego P., Robinson L. Z., Pereira-Henriques A., Ferreira N., Pinheiro F., Esperante S., Pallares I., Huertas O., Almeida M. R., Reixach N., Insa R., Velazquez-Campoy A., Reverter D., Reig N., and Ventura S. (2016) Repositioning tolcapone as a potent inhibitor of transthyretin amyloidogenesis and associated cellular toxicity. Nat. Commun. 7, 10787 10.1038/ncomms10787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maurer M. S., Schwartz J. H., Gundapaneni B., Elliott P. M., Merlini G., Waddington-Cruz M., Kristen A. V., Grogan M., Witteles R., Damy T., Drachman B. M., Shah S. J., Hanna M., Judge D. P., Barsdorf A. I., et al. (2018) Tafamidis treatment for patients with transthyretin amyloid cardiomyopathy. N. Engl. J. Med. 379, 1007–1016 10.1056/NEJMoa1805689 [DOI] [PubMed] [Google Scholar]
- 21. Adams D., Gonzalez-Duarte A., O'Riordan W. D., Yang C.-C., Ueda M., Kristen A. V., Tournev I., Schmidt H. H., Coelho T., Berk J. L., Lin K.-P., Vita G., Attarian S., Planté-Bordeneuve V., Mezei M. M., et al. (2018) Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 379, 11–21 10.1056/NEJMoa1716153 [DOI] [PubMed] [Google Scholar]
- 22. Benson M. D., Waddington-Cruz M., Berk J. L., Polydefkis M., Dyck P. J., Wang A. K., Planté-Bordeneuve V., Barroso F. A., Merlini G., Obici L., Scheinberg M., Brannagan T. H. 3rd., Litchy W. J., Whelan C., Drachman B. M., et al. (2018) Inotersen treatment for patients with hereditary transthyretin amyloidosis. N. Engl. J. Med. 379, 22–31 10.1056/NEJMoa1716793 [DOI] [PubMed] [Google Scholar]
- 23. Obici L., Cortese A., Lozza A., Lucchetti J., Gobbi M., Palladini G., Perlini S., Saraiva M. J., and Merlini G. (2012) Doxycycline plus tauroursodeoxycholic acid for transthyretin amyloidosis: a phase II study. Amyloid 19, 34–36 10.3109/13506129.2012.678508 [DOI] [PubMed] [Google Scholar]
- 24. Higaki J. N., Chakrabartty A., Galant N. J., Hadley K. C., Hammerson B., Nijjar T., Torres R., Tapia J. R., Salmans J., Barbour R., Tam S. J., Flanagan K., Zago W., and Kinney G. G. (2016) Novel conformation-specific monoclonal antibodies against amyloidogenic forms of transthyretin. Amyloid 23, 86–97 10.3109/13506129.2016.1148025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Richards D. B., Cookson L. M., Barton S. V., Liefaard L., Lane T., Hutt D. F., Ritter J. M., Fontana M., Moon J. C., Gillmore J. D., Wechalekar A., Hawkins P. N., and Pepys M. B. (2018) Repeat doses of antibody to serum amyloid P component clear amyloid deposits in patients with systemic amyloidosis. Sci. Transl. Med. 10, eaan3128 10.1126/scitranslmed.aan3128 [DOI] [PubMed] [Google Scholar]
- 26. Westermark P., Sletten K., Johansson B., Cornwell G. G. 3rd. (1990) Fibril in senile systemic amyloidosis is derived from normal transthyretin. Proc. Natl. Acad. Sci. U.S.A. 87, 2843–2845 10.1073/pnas.87.7.2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yanagisawa A., Ueda M., Sueyoshi T., Okada T., Fujimoto T., Ogi Y., Kitagawa K., Tasaki M., Misumi Y., Oshima T., Jono H., Obayashi K., Hirakawa K., Uchida H., Westermark P., et al. (2015) Amyloid deposits derived from transthyretin in the ligamentum flavum as related to lumbar spinal canal stenosis. Mod. Pathol. 28, 201–207 10.1038/modpathol.2014.102 [DOI] [PubMed] [Google Scholar]
- 28. Bergström J., Gustavsson A., Hellman U., Sletten K., Murphy C. L., Weiss D. T., Solomon A., Olofsson B.-O., and Westermark P. (2005) Amyloid deposits in transthyretin-derived amyloidosis: cleaved transthyretin is associated with distinct amyloid morphology. J. Pathol. 206, 224–232 10.1002/path.1759 [DOI] [PubMed] [Google Scholar]
- 29. Ihse E., Rapezzi C., Merlini G., Benson M. D., Ando Y., Suhr O. B., Ikeda S., Lavatelli F., Obici L., Quarta C. C., Leone O., Jono H., Ueda M., Lorenzini M., Liepnieks J., et al. (2013) Amyloid fibrils containing fragmented ATTR may be the standard fibril composition in ATTR amyloidosis. Amyloid 20, 142–150 10.3109/13506129.2013.797890 [DOI] [PubMed] [Google Scholar]
- 30. Suhr O. B., Wixner J., Anan I., Lundgren H.-E., Wijayatunga P., Westermark P., and Ihse E. (2019) Amyloid fibril composition within hereditary V30M (p. Val50Met) transthyretin amyloidosis families. PLoS ONE 14, e0211983 10.1371/journal.pone.0211983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ihse E., Ybo A., Suhr O., Lindqvist P., Backman C., and Westermark P. (2008) Amyloid fibril composition is related to the phenotype of hereditary transthyretin V30M amyloidosis. J. Pathol. 216, 253–261 10.1002/path.2411 [DOI] [PubMed] [Google Scholar]
- 32. Gustafsson S., Ihse E., Henein M. Y., Westermark P., Lindqvist P., and Suhr O. B. (2012) Amyloid fibril composition as a predictor of development of cardiomyopathy after liver transplantation for hereditary transthyretin amyloidosis. Transplantation 93, 1017–1023 10.1097/TP.0b013e31824b3749 [DOI] [PubMed] [Google Scholar]
- 33. Thylén C., Wahlqvist J., Haettner E., Sandgren O., Holmgren G., and Lundgren E. (1993) Modifications of transthyretin in amyloid fibrils: analysis of amyloid from homozygous and heterozygous individuals with the Met30 mutation. EMBO J. 12, 743–748 10.1002/j.1460-2075.1993.tb05708.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mangione P. P., Porcari R., Gillmore J. D., Pucci P., Monti M., Porcari M., Giorgetti S., Marchese L., Raimondi S., Serpell L. C., Chen W., Relini A., Marcoux J., Clatworthy I. R., Taylor G. W., et al. (2014) Proteolytic cleavage of S52P variant transthyretin triggers its amyloid fibrillogenesis. Proc. Natl. Acad. Sci. U.S.A. 111, 1539–1544 10.1073/pnas.1317488111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marcoux J., Mangione P. P., Porcari R., Degiacomi M. T., Verona G., Taylor G. W., Giorgetti S., Raimondi S., Sanglier-Cianférani S., Benesch J. L., Cecconi C., Naqvi M. M., Gillmore J. D., Hawkins P. N., Stoppini M., et al. (2015) A novel mechano-enzymatic cleavage mechanism underlies transthyretin amyloidogenesis. EMBO Mol. Med. 7, 1337–1349 10.15252/emmm.201505357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klimtchuk E. S., Prokaeva T., Frame N. M., Abdullahi H. A., Spencer B., Dasari S., Cui H., Berk J. L., Kurtin P. J., Connors L. H., and Gursky O. (2018) Unusual duplication mutation in a surface loop of human transthyretin leads to an aggressive drug-resistant amyloid disease. Proc. Natl. Acad. Sci. U.S.A. 115, E6428–E6436 10.1073/pnas.1802977115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saelices L., Johnson L. M., Liang W. Y., Sawaya M. R., Cascio D., Ruchala P., Whitelegge J., Jiang L., Riek R., and Eisenberg D. S. (2015) Uncovering the mechanism of aggregation of human transthyretin. J. Biol. Chem. 290, 28932–28943 10.1074/jbc.M115.659912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saelices L., Sievers S. A., Sawaya M. R., and Eisenberg D. S. (2018) Crystal structures of amyloidogenic segments of human transthyretin. Protein Sci. 27, 1295–1303 10.1002/pro.3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsuda Y., Yamanaka K., Toyoshima R., Ueda M., Masuda T., Misumi Y., Ogura T., and Ando Y. (2018) Development of transgenic Caenorhabditis elegans expressing human transthyretin as a model for drug screening. Sci. Rep. 8, 17884 10.1038/s41598-018-36357-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Suenaga G., Ikeda T., Masuda T., Motokawa H., Yamashita T., Takamatsu K., Misumi Y., Ueda M., Matsui H., Senju S., and Ando Y. (2017) Inflammatory state exists in familial amyloid polyneuropathy that may be triggered by mutated transthyretin. Sci. Rep. 7, 1579 10.1038/s41598-017-01775-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gonçalves N. P., Teixeira-Coelho M., and Saraiva M. J. (2014) The inflammatory response to sciatic nerve injury in a familial amyloidotic polyneuropathy mouse model. Exp. Neurol. 257, 76–87 10.1016/j.expneurol.2014.04.030 [DOI] [PubMed] [Google Scholar]
- 42. Macedo B., Batista A. R., do Amaral J. B., and Saraiva M. J. (2007) Biomarkers in the assessment of therapies for familial amyloidotic polyneuropathy. Mol. Med. 13, 584–591 10.2119/2007-00068.Macedo [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ferreira N., Cardoso I., Domingues M. R., Vitorino R., Bastos M., Bai G., Saraiva M. J., and Almeida M. R. (2009) Binding of epigallocatechin-3-gallate to transthyretin modulates its amyloidogenicity. FEBS Lett. 583, 3569–3576 10.1016/j.febslet.2009.10.062 [DOI] [PubMed] [Google Scholar]
- 44. Miyata M., Sato T., Kugimiya M., Sho M., Nakamura T., Ikemizu S., Chirifu M., Mizuguchi M., Nabeshima Y., Suwa Y., Morioka H., Arimori T., Suico M. A., Shuto T., Sako Y., Momohara M., et al. (2010) The crystal structure of the green tea polyphenol (−)-epigallocatechin gallate–transthyretin complex reveals a novel binding site distinct from the thyroxine binding site. Biochemistry 49, 6104–6114 10.1021/bi1004409 [DOI] [PubMed] [Google Scholar]
- 45. Beck J. W., Saavedra D., Antell G. J., and Tejeiro B. (1959) The treatment of pinworm infections in humans (enterobiasis) with pyrvinium chloride and pyrvinium pamoate. Am. J. Trop. Med. Hyg. 8, 349–352 10.4269/ajtmh.1959.8.349 [DOI] [PubMed] [Google Scholar]
- 46. Li B., Fei D. L., Flaveny C. A., Dahmane N., Baubet V., Wang Z., Bai F., Pei X.-H., Rodriguez-Blanco J., Hang B., Orton D., Han L., Wang B., Capobianco A. J., Lee E., and Robbins D. J. (2014) Pyrvinium attenuates hedgehog signaling downstream of smoothened. Cancer Res. 74, 4811–4821 10.1158/0008-5472.CAN-14-0317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barbarino M., Cesari D., Intruglio R., Indovina P., Namagerdi A., Bertolino F. M., Bottaro M., Rahmani D., Bellan C., and Giordano A. (2018) Possible repurposing of pyrvinium pamoate for the treatment of mesothelioma: a pre-clinical assessment. J. Cell. Physiol. 233, 7391–7401 10.1002/jcp.26579 [DOI] [PubMed] [Google Scholar]
- 48. Dionísio P. A., Amaral J. D., Ribeiro M. F., Lo A. C., D'Hooge R., and Rodrigues C. M. (2015) Amyloid-β pathology is attenuated by tauroursodeoxycholic acid treatment in APP/PS1 mice after disease onset. Neurobiol. Aging 36, 228–240 10.1016/j.neurobiolaging.2014.08.034 [DOI] [PubMed] [Google Scholar]
- 49. Giorgetti S., Greco C., Tortora P., and Aprile F. A. (2018) Targeting amyloid aggregation: an overview of strategies and mechanisms. Int. J. Mol. Sci. 19, E2677 10.3390/ijms19092677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Smith T. C., Kinkel A. W., Gryczko C. M., and Goulet J. R. (1976) Absorption of pyrvinium pamoate. Clin. Pharmacol. Ther. 19, 802–806 10.1002/cpt1976196802 [DOI] [PubMed] [Google Scholar]
- 51. Katzenschlager R., Poewe W., Rascol O., Trenkwalder C., Deuschl G., Chaudhuri K. R., Henriksen T., van Laar T., Spivey K., Vel S., Staines H., and Lees A. (2018) Apomorphine subcutaneous infusion in patients with Parkinson's disease with persistent motor fluctuations (TOLEDO): a multicentre, double-blind, randomised, placebo-controlled trial. Lancet Neurol. 17, 749–759 10.1016/S1474-4422(18)30239-4 [DOI] [PubMed] [Google Scholar]
- 52. Lashuel H. A., Hartley D. M., Balakhaneh D., Aggarwal A., Teichberg S., and Callaway D. J. (2002) New class of inhibitors of amyloid-β fibril formation. Implications for the mechanism of pathogenesis in Alzheimer's disease. J. Biol. Chem. 277, 42881–42890 10.1074/jbc.M206593200 [DOI] [PubMed] [Google Scholar]
- 53. Smith R. V., Wilcox R. E., Soine W. H., Riffee W. H., Baldessarini R. J., and Kula N. S. (1979) Plasma levels of apomorphine following intravenous, intraperitoneal and oral administration to mice and rats. Res. Commun. Chem. Pathol. Pharmacol. 24, 483–499 [PubMed] [Google Scholar]
- 54. Mizuguchi M., Hayashi A., Takeuchi M., Dobashi M., Mori Y., Shinoda H., Aizawa T., Demura M., and Kawano K. (2008) Unfolding and aggregation of transthyretin by the truncation of 50 N-terminal amino acids. Proteins 72, 261–269 10.1002/prot.21919 [DOI] [PubMed] [Google Scholar]
- 55. Samson R. Y., Obita T., Freund S. M., Williams R. L., and Bell S. D. (2008) A role for the ESCRT system in cell division in archaea. Science 322, 1710–1713 10.1126/science.1165322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mizuguchi M., Obita T., Serita T., Kojima R., Nabeshima Y., and Okazawa H. (2014) Mutations in the PQBP1 gene prevent its interaction with the spliceosomal protein U5–15 kD. Nat. Commun. 5, 3822 10.1038/ncomms4822 [DOI] [PubMed] [Google Scholar]
- 57. Matsubara K., Mizuguchi M., and Kawano K. (2003) Expression of a synthetic gene encoding human transthyretin in Escherichia coli. Protein Expr. Purif. 30, 55–61 10.1016/S1046-5928(03)00069-X [DOI] [PubMed] [Google Scholar]
- 58. Misumi Y., Ando Y., Ueda M., Obayashi K., Jono H., Su Y., Yamashita T., and Uchino M. (2009) Chain reaction of amyloid fibril formation with induction of basement membrane in familial amyloidotic polyneuropathy. J. Pathol. 219, 481–490 10.1002/path.2618 [DOI] [PubMed] [Google Scholar]
- 59. Tasaki M., Ueda M., Hoshii Y., Mizukami M., Matsumoto S., Nakamura M., Yamashita T., Ueda A., Misumi Y., Masuda T., Inoue Y., Torikai T., Nomura T., Tsuda Y., Kanenawa K., et al. (2019) A novel age-related venous amyloidosis derived from EGF-containing fibulin-like extracellular matrix protein 1. J. Pathol. 247, 444–455 10.1002/path.5203 [DOI] [PubMed] [Google Scholar]
- 60. Sekijima Y., Dendle M. A., and Kelly J. W. (2006) Orally administered diflunisal stabilizes transthyretin against dissociation required for amyloidogenesis. Amyloid 13, 236–249 10.1080/13506120600960882 [DOI] [PubMed] [Google Scholar]
- 61. Mitsui T., Miyake Y., Kakeya H., Hayashi Y., Osada H., and Kataoka T. (2005) RKTS-33, an epoxycyclohexenone derivative that specifically inhibits Fas ligand-dependent apoptosis in CTL-mediated cytotoxicity. Biosci. Biotechnol. Biochem. 69, 1923–1928 10.1271/bbb.69.1923 [DOI] [PubMed] [Google Scholar]
- 62. Annamalai K., Gührs K. H., Koehler R., Schmidt M., Michel H., Loos C., Gaffney P. M., Sigurdson C. J., Hegenbart U., Schönland S., and Fändrich M. (2016) Polymorphism of amyloid fibrils in vivo. Angew. Chem. Int. Ed. Engl. 55, 4822–4825 10.1002/anie.201511524 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.