Abstract
Ferrochelatase (FeCh) is an essential enzyme catalyzing the synthesis of heme. Interestingly, in cyanobacteria, algae, and plants, FeCh possesses a conserved transmembrane chlorophyll a/b binding (CAB) domain that resembles the first and the third helix of light-harvesting complexes, including a chlorophyll-binding motif. Whether the FeCh CAB domain also binds chlorophyll is unknown. Here, using biochemical and radiolabeled precursor experiments, we found that partially inhibited activity of FeCh in the cyanobacterium Synechocystis PCC 6803 leads to overproduction of chlorophyll molecules that accumulate in the thylakoid membrane and, together with carotenoids, bind to FeCh. We observed that pigments bound to purified FeCh are organized in an energy-dissipative conformation and further show that FeCh can exist in vivo as a monomer or a dimer depending on its own activity. However, pigmented FeCh was purified exclusively as a dimer. Separately expressed and purified FeCH CAB domain contained a pigment composition similar to that of full-length FeCh and retained its quenching properties. Phylogenetic analysis suggested that the CAB domain was acquired by a fusion between FeCh and a single-helix, high light-inducible protein early in the evolution of cyanobacteria. Following this fusion, the FeCh CAB domain with a functional chlorophyll-binding motif was retained in all currently known cyanobacterial genomes except for a single lineage of endosymbiotic cyanobacteria. Our findings indicate that FeCh from Synechocystis exists mostly as a pigment-free monomer in cells but can dimerize, in which case its CAB domain creates a functional pigment-binding segment organized in an energy-dissipating configuration.
Keywords: plant biochemistry, photosynthetic pigment, photosynthesis, chloroplast, membrane protein, heme, carotenoid, chlorophyll, ferrochelatase, light harvesting complex (LHC)-like proteins, pigment binding, Synechocystis
Introduction
Heme is an essential cofactor for virtually all forms of life, and it is produced by a conserved tetrapyrrole pathway. The last step of heme biosynthesis is catalyzed by ferrochelatase (FeCh; protoheme ferro-lyase, EC 4.99.1.1),2 which inserts iron into protoporphyrin IX (PPIX), a metal-free tetrapyrrole ring. Apart from serving as a cofactor for a myriad of redox and sensor proteins and as a precursor for the synthesis of bilins, heme is also a crucial regulatory molecule (1). Among many other recognized regulatory functions, heme availability controls total metabolite flow through the tetrapyrrole pathway. This heme feedback loop, although not fully understood, seems to operate universally from unicellular bacteria to animals (mitochondria) and plants (plastids) (2–4).
In photosynthetic organisms, regulation of the tetrapyrrole pathway is more complicated, because PPIX is also a substrate for magnesium chelatase, which diverts PPIX toward the chlorophyll (Chl) branch (Fig. S1A) (4). Considering the general phototoxicity of tetrapyrroles, there must be regulatory mechanisms to prevent an overproduction of one of the tetrapyrrole products at the expense of the others or a harmful accumulation of intermediates. Thus, in Chl producing organisms, the activity of FeCh not only just controls a regulatory pool of heme but also competes with magnesium chelatase for PPIX, the last common substrate of both the heme and Chl branches. Indeed, the activity of FeCh has been shown to be crucial for the regulation of the tetrapyrrole pathway in both plants and cyanobacteria; an impairment of FeCh causes a massive accumulation of PPIX (5, 6). In contrast, low activity of magnesium chelatase resulted in a total down-regulation of the pathway but not in the accumulation of PPIX (7–9).
Interestingly, a unique feature of the FeCh enzymes of Chl-producing organisms is a conserved C-terminal transmembrane helix that resembles the first and third helix of light-harvesting complex (LHC) proteins including a Chl-binding motif (the so-called LHC or CAB domain; Fig. 1 and Fig. S1B). The FeCh in oxygenic phototrophs is thus the only known enzyme belonging to the LHC protein superfamily, a large and diverse group of proteins characterized by the presence of at least one LHC/CAB-domain. Apart from light-harvesting antennas, the superfamily includes LHC-like proteins that are apparently not involved in light harvesting but rather have regulatory or photoprotective functions (10–15). Cyanobacterial LHC-like proteins are limited to the single-helix high light-inducible proteins (Hlips) and FeCh, but in algae and plants, the spectrum of these proteins is much broader. Apart from FeCh and the homologs of Hlips called one-helix proteins (OHPs), eukaryotes possess various two- and three-helix LHC-like proteins such as Lil3 proteins, early light-inducible proteins (ELIPs), and Psb33 (16–21). ELIPs, Lil3s, and Hlips were already reported to bind Chl and carotenoids in vivo (22–25). Based on the known structure of various LHC proteins (26, 27), it is expected that two helices, each containing an LHC/CAB motif, must dimerize to create a functional Chl-binding site (25).
Figure 1.
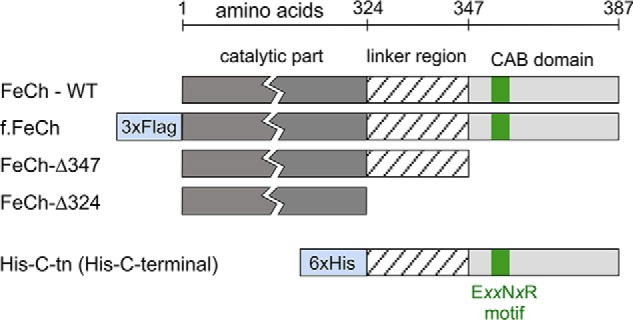
A schematic presentation of the FLAG-tagged and truncated Synechocystis FeCh proteins used in this study and the His-tagged C-terminal part of FeCh (His-C-tn protein) that possesses the FeCh linker and the CAB domain. The highly conserved Chl-binding motif (EXXNXR), which is characteristic for the whole LHC protein family, is also indicated.
The function of the FeCh CAB domain has been previously addressed by genetic and biochemical approaches (28–30); however, its role remains unclear. The CAB domain is connected to the catalytic part of FeCh via a variable linker (Fig. 1), and in the cyanobacterium Synechocystis PCC 6803 (hereafter Synechocystis), the FeCh enzyme lacking both the LHC domain and the linker still appeared to be fairly active but was unstable, and its accumulation in the cell was impaired (6). A truncation preserving the linker but eliminating the CAB domain yields an active and stable enzyme (29, 30). Such a truncated enzyme is, however, strictly monomeric, whereas the full-length enzyme can dimerize, at least in vitro (29, 30). In addition, a Synechocystis strain expressing FeCh without the CAB domain was sensitive to high light conditions (29).
In this work, we demonstrate that Synechocystis FeCh is present in the cell mostly as a pigment-less monomer but can conditionally dimerize, and in such cases, its CAB domain creates a functional pigment-binding segment. Interestingly, the Chl and carotenoids bound to the CAB domain are organized in an energy-dissipative configuration, as previously reported for Hlips (24). This is consistent with our phylogenetic analysis, suggesting that the FeCh CAB domain originates from a fusion between an ancient Hlip and FeCh enzyme early in cyanobacterial evolution.
Results
Partially inhibited FeCh binds Chl and carotenoids
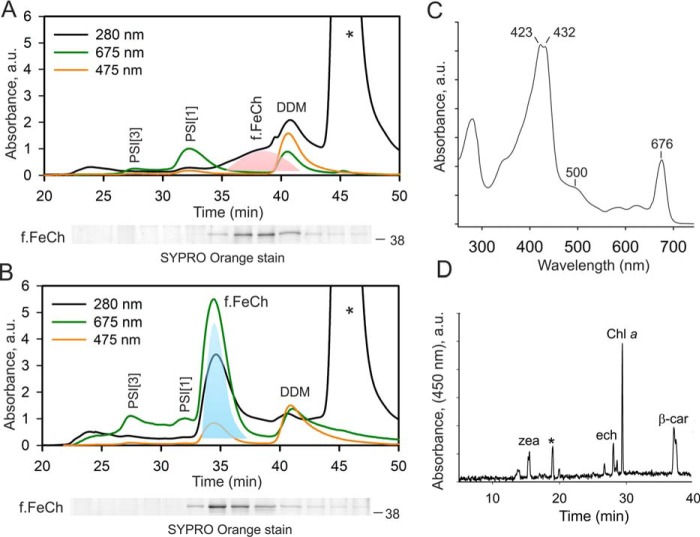
To address the longstanding question of whether the FeCh enzyme in oxygenic phototrophs can bind Chl, we affinity-purified a 3×FLAG-tagged version of FeCh (f.FeCh; Fig. 1) ectopically expressed in a Synechocystis strain with the original hemH gene deleted (29). The obtained eluate was further separated by size-exclusion chromatography (SEC), and the Chl absorbance was measured by a diode-array detector. However, the only detectable Chl in the eluate comigrated with traces of photosystem I complexes (PSI) and with detergent micelles (DDM), but not with the purified f.FeCh (Fig. 2A). The FeCh, migrating close to DDM micelles, was the only protein detectable on the stained gel after the separation of all fractions collected from the SEC column (Fig. S2). It should be noted that PSI complexes are extremely Chl-rich (90 Chl molecules per monomeric PSI), and thus their Chl absorbance is typically detectable even if the level of PSI protein subunits is below the detection limit of our protein stain (compare Fig. 1A and Fig. S2). We hypothesized that the FeCh could only bind (sense) Chl molecules if there was an abnormally large pool of unbound “free” Chl in the thylakoid membranes. Low FeCh activity in Synechocystis was previously shown to up-regulate the levels of Chl precursors (6, 31); however, whether such up-regulation leads to an accumulation of free Chl molecules in the membrane was not known.
Figure 2.
Synechocystis FeCh binds Chl and carotenoids when its enzymatic activity is low. A, SEC separation of f.FeCh purified from Synechocystis cells. Chl, carotenoids, and UV absorptions were recorded by a diode-array detector; minute fractions were collected and separated by SDS-electrophoresis, and f.FeCh protein was stained by SYPRO Orange (below SEC chromatogram, see Fig. S2 for full-sized gel pictures). The peak of f.FeCh is highlighted by pink shading. The asterisk marks the absorption of the 3×FLAG peptide used to elute the f.FeCh protein from the anti-FLAG gel; DDM marks detergent micelles containing Chl molecules. PSI[3] and PSI[1] indicate Chl bound to traces of trimeric and monomeric photosystem I, respectively. B, SEC separation of f.FeCh purified from Synechocystis cells treated for 4 h with a specific FeCh inhibitor (MPP). The f.FeCh peak, highlighted by cyan shading, was collected for pigment analysis. C, absorbance spectrum of the pigmented form of f.FeCh; the unusually high absorbance in the region of 415–435 nm is due to the MPP bound to f.FeCh (see Fig. S4 for an HPLC analysis). D, pigments were extracted by methanol from the collected f.FeCh peak (B) and separated on a C18 column. Zea, ech, and β-car denote zeaxanthin, echinenone and, β-carotene, respectively. An asterisk indicates an unknown carotenoid species.
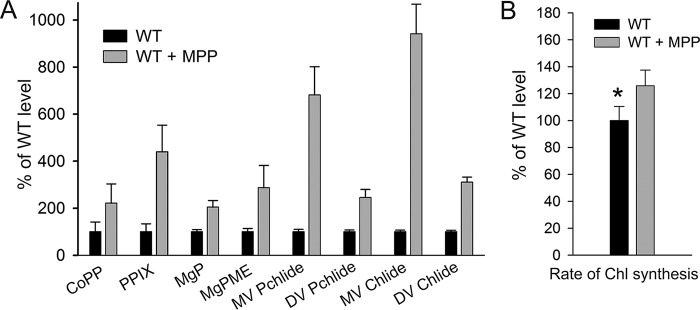
To clarify this point we treated the Synechocystis cells with a specific FeCh inhibitor, N-methyl mesoporphyrin IX (MPP) (32). After 4 h of the MPP treatment, the cells were fed with 14C-labeled glutamate to follow the rate of de novo Chl formation. The concentration of MPP (275 nm) was not significantly toxic to Synechocystis under our standard conditions, because the proliferation rate of the cells treated with this concentration of MPP for 24 h was only 6% slower than that of untreated cells (Fig. S3). However, the partial FeCh inhibition significantly accelerated Chl biosynthesis compared with untreated cells (Fig. 3, A and B). Further, to assess the amount of free (or loosely bound) Chl in the cell, the membrane fraction was isolated after 4 h of MPP-treatment and separated by clear-native electrophoresis. In the case of MPP-treated cells, the Chl fluorescence detected in the zone of free pigments (DDM micelles) was more than doubled than in untreated control (Fig. 4, A and B). We eluted and quantified pigments from this part of the gel, and the content of Chl correlated well with the fluorescence measurement (Fig. 4B). According to these results, ∼0.7% of the total Chl loaded on the clear-native gel comigrated with DDM micelles, and this amount is roughly doubled after treatment with MPP (Fig. 4B). Given these results, we concluded that decreased FeCh activity results in excess production of Chl in the cell.
Figure 3.
Low activity of FeCh accelerates the de novo synthesis of Chl molecules. A, tetrapyrrolic Chl precursors were extracted from WT and from WT cells treated for 4 h with MPP and separated using HPLC. Individual precursor species were detected and quantified using two fluorescence detectors essentially as described in Ref. 68. Error bars show the S.D., n = 3 independent experiments. CoPP, coproporphyrin III; MgP, magnesium protoporphyrin IX; MgPME, magnesium protoporphyrin IX monomethyl ester; MV/DV Pchlide, monovinyl/divinyl protochlorophyllide; MV/DV Chlide, monovinyl/divinyl chlorophyllide. B, WT control cells and cells treated with MPP for 3.5 h were supplemented with radiolabeled 14C[Glu]. After 60 min-pulse, pigments were extracted by methanol and separated on an HPLC equipped with a diode-array detector and a flow scintillation detector. The rate of Chl synthesis was quantified as the ratio of Chl absorbance and radioactivity peaks. Error bars show the S.D., n = 4 independent experiments. *, p = 0.029; Student's t test.
Figure 4.
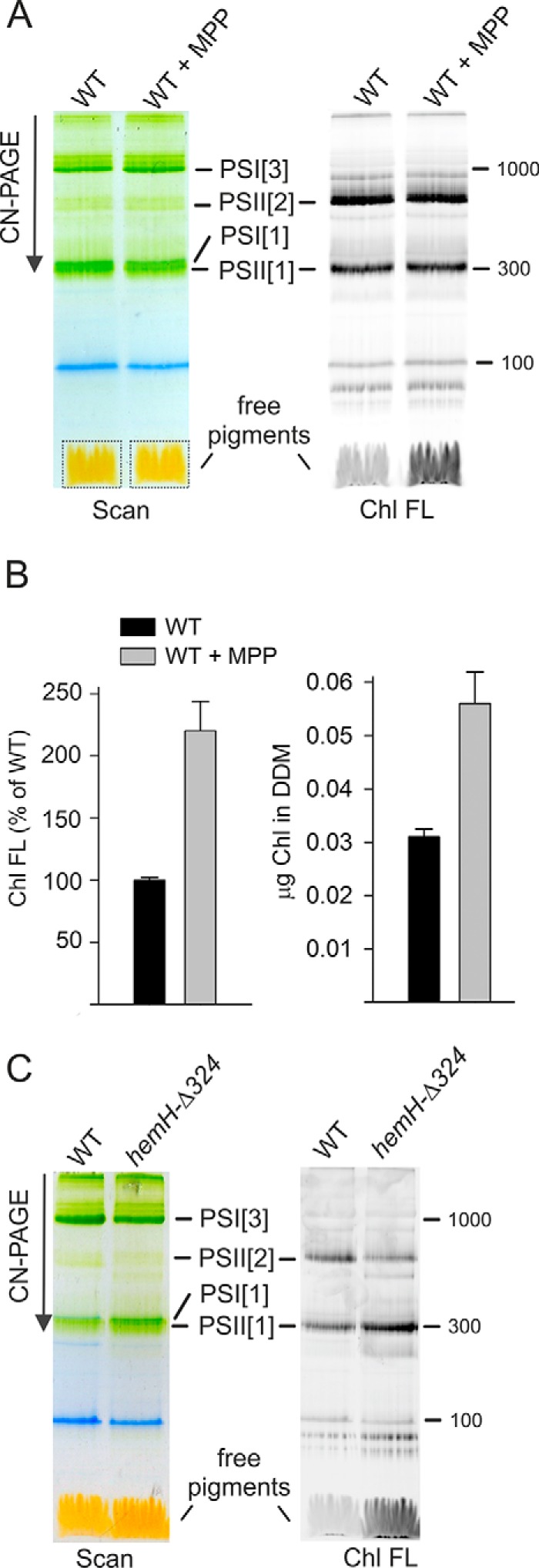
Oversynthesized Chl accumulates in thylakoid membranes. A, membrane complexes prepared from WT and from WT treated for 4 h with MPP were separated by clear native electrophoresis on 4–12% acrylamide gel and scanned (left panel); 4 μg of Chl was loaded per each sample. Chl fluorescence (right panel, Chl FL) in the gel was detected after excitation with blue light. PSI[1] and PSI[3], monomeric and trimeric photosystem I, respectively. PSII[1] and PSII[2], monomeric and dimeric photosystem II, respectively. Pigments migrating in gel with DDM micelles are indicated by dotted boxes. B, Chl fluorescence in the gel region marked by dotted boxes in A was quantified. This part of gel was further cut, the pigments were eluted by repeated incubation in an elution buffer containing DDM (see “Experimental procedures”), and the Chl content was quantified by HPLC. Error bars show S.D., n = 3 independent experiments. C, membrane complexes prepared from WT and hemH-Δ324 mutant were separated by clear native electrophoresis, and Chl fluorescence was detected as described for A.
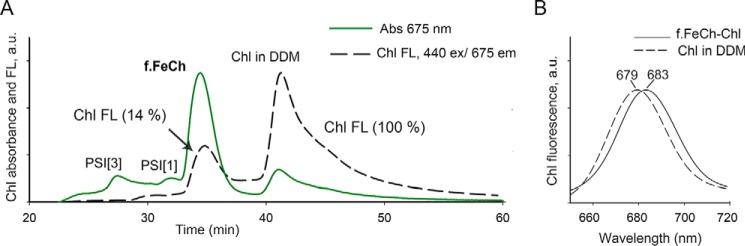
Notably, when we purified f.FeCh from the cells treated with MPP, it eluted from the SEC column as a higher-mass protein complex than when it was purified from cells without the inhibitor. Moreover, a peak of Chl absorbance (675 nm) clearly comigrated with the f.FeCh (Fig. 2B), and the absorbance spectrum of the f.FeCh peak (450–500 nm region; Fig. 2C) also signaled the presence of carotenoids. We extracted pigments from the collected f.FeCh peak and separated the extract by HPLC, which revealed a complex mixture of carotenoids in addition to Chl molecules. β-Carotene (β-car) was the most abundant carotenoid but zeaxanthin, echinenone, and an unknown carotenoid species were present as well (Fig. 2D). We also found MPP bound to f.FeCh, which explains the high absorbance of f.FeCh in the blue spectral region (Fig. 2C and Fig. S4). By recording Chl fluorescence during SEC, we discovered that the fluorescence of Chl molecules bound to f.FeCh is quenched: the fluorescence yield of Chl was only ∼14% when compared with the fluorescence of Chl molecules in detergent micelles (Fig. 5). From these observations, we conclude that the Synechocystis FeCh possesses a functional Chl and carotenoid-binding motif, which coordinates pigments in an energy-dissipative configuration.
Figure 5.
Quenching of Chl fluorescence in the FeCh–pigment complex. A, SEC separation of f.FeCh purified from Synechocystis cells treated for 4 h with MPP. Chl absorption and fluorescence were recorded by diode array and fluorescent detectors, respectively. PSI[3] and PSI[1] indicate Chl bound to traces of trimeric and monomeric PSI, respectively. Numbers in parentheses indicate Chl fluorescence yield. The efficiency of quenching was calculated from the ratio of fluorescence/absorbance at their respective maxima for Chls bound to FeCh and for Chl molecules in detergent micelles. The same extinction coefficient was assumed for both Chl populations. B, fluorescence maxima of the f.FeCh–pigment complex and free Chl molecules in DDM micelles recorded during SEC chromatography by a fluorescence detector after excitation at 440 nm.
The pigment-binding form of FeCh is a homodimer
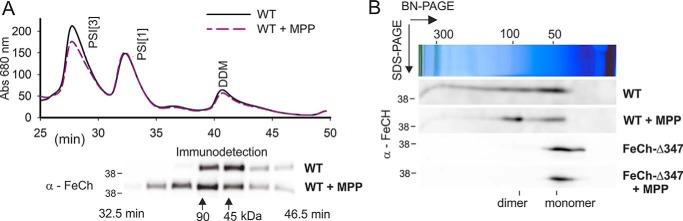
Based on the elution time from the SEC column, the apparent mass of f.FeCh was greater after incubation with MPP (Fig. 2, A and B). However, on the stained gel, we detected no other proteins except f.FeCh (Fig. S2), which implies the formation of a f.FeCh oligomer. According to the known structure of LHCII (33) and a structural model of Hlips (24, 25), two LHC/CAB domains should dimerize to create a canonical 4-Chls and 2-carotenoid binding motif. The observed shift in f.FeCh mobility suggests that the capability of the Synechocystis FeCh to bind Chl is related to its dimerization. To exclude the possibility that the f.FeCh oligomerization is an artifact of protein purification or its tagging, we separated Synechocystis membrane complexes by SEC and immunodetected FeCh in the collected minute fractions. A similar shift in the mass of FeCh was detected in membranes from WT cells: MPP treatment resulted in a higher-mass FeCh complex (Fig. 6A). According to the column calibration (Fig. S5), this shift corresponds to 50 kDa, which is very close to the shift expected for the presence of a FeCh dimer. Therefore, the discrepancy (∼2 min) in the elution times of FeCh and f.FeCh from the SEC column is most likely because of the presence of the highly charged 3×FLAG tag.
Figure 6.
MPP-treatment induces dimerization of Synechocystis FeCh via its CAB domain. A, membrane complexes prepared from WT cells and WT cells treated for 4 h with MPP were separated by SEC, and minute fractions were collected. Individual fractions were separated by SDS-electrophoresis, blotted, and probed with an anti-FeCh antibody. Calibration of the SEC column is indicated by arrows (see Fig. S5 for details). PSI[3] and PSI[1] show the position of trimeric and monomeric PSI complexes. B, solubilized membrane fractions were separated by 2D BN-PAGE and blotted, and the FeCh was immunodetected. Only slices of the blot with the signal of FeCh are shown (see Fig. S6 for full-sized gels). The position of monomeric and dimeric FeCh is indicated.
To verify the inducible FeCh dimerization by an independent approach, we separated WT membrane proteins by 2D blue-native/SDS electrophoresis (BN/SDS-PAGE), blotted, and immunodetected FeCh. Although the FeCh pattern was a bit smeared, after treatment with MPP, it was evident that a new spot corresponding to a size of a FeCh dimer appeared (Fig. 6B). We repeated the same experiment using membranes isolated from the Synechocystis hemH-Δ347 strain containing FeCh without the CAB domain (Fig. 1) (29). As expected, the FeCh-Δ347 enzyme was exclusively monomeric, and its mobility was not affected by MPP (Fig. 6B). These data demonstrate that the Synechocystis FeCh can exist in vivo as a monomer or as a homodimer, and the formation of FeCh dimer is stimulated by MPP.
To prove that the FeCh CAB domain is the pigment-binding site, the FeCh C terminus, including the linker, was expressed in Synechocystis as a His6-tagged protein (His-C-tn, 9 kDa; Fig. 1). We have previously shown that the His-C-tn protein interacts in vivo with the FeCh CAB domain and can be copurified with the full-length FeCh but not with a truncated, CAB-less derivative (29). To isolate the His-C-tn domain not “contaminated” by His-C-tn-FeCh heterodimers, we expressed this protein in the Synechocystis hemH-Δ324 strain containing FeCh lacking both the linker and the LHC domain (Fig. 1) (6). The FeCh-Δ324 truncation, apart from its inability to bind His-C-tn (29), lowers the cellular activity of FeCh because of impaired stability of the enzyme, thus mimicking the free Chl accumulation effect of MPP (Fig. 4C). We purified His-C-tn using a nickel affinity column, and after removing contaminating proteins and pigments by anion-exchange chromatography, we obtained pure His-C-tn protein (Fig. 7A). Fig. 7B shows the absorption spectrum of the isolated His-C-tn protein; as revealed by HPLC analysis of the extracted pigments, the protein binds Chl, β-car, and zeaxanthin at a molar ratio 6: 0.9: 1. A small amount of echinenone and the unknown carotenoid copurified with full-length FeCh was also detectable (Fig. 7C). Furthermore, the fluorescence of Chls bound to His-C-tn was also quenched (Fig. 7A) as in the case of purified FeCh. Therefore, we can conclude that the FeCh CAB domain is solely responsible for the binding of pigments; the catalytic domain is not required for its folding into a Chl-binding protein structure.
Figure 7.
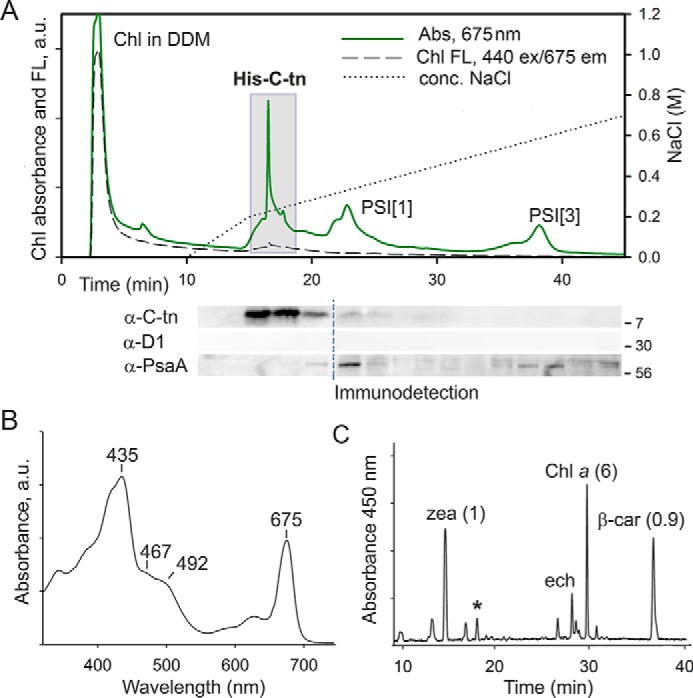
The C-terminal segment of FeCh lacking the catalytic domain (an artificial His-C-tn protein) binds Chl, β-car, and zeaxanthin in vivo. A, the His-C-tn protein was expressed in the Synechocystis hemH-Δ324 genetic background and purified on a nickel column. The obtained eluate was further separated by anion-exchange chromatography, and the Chl absorbance (675 nm) and Chl fluorescence were recorded (Chl FL; 440-nm excitation/675-nm emission). Note the strikingly different emission intensity of Chls in DDM micelles and Chls bound to His-C-tn. Minute fractions were collected and separated by SDS-electrophoresis, and the presence of His-C-tn protein was detected by antibodies raised against the FeCh C terminus. The core photosystem I and II subunits PsaA and D1 were also immunodetected as a control of purity; dashed line indicates blot splicing. Fractions highlighted by the gray box were pooled and used for the analysis of pigments associated with His-C-tn. B, the absorption spectrum of the His-C-tn protein purified by a combination of nickel-affinity and anion-exchange chromatography as described above. C, the extracted pigments from the purified His-C-tn protein were separated on a C18 column; the molar stoichiometry of the major constituents is indicated. zea, zeaxanthin; ech, echinenone. The unknown carotenoid (see Fig. 2D) is marked by an asterisk.
Occurrence and phylogeny of FeCh in cyanobacterial genomes
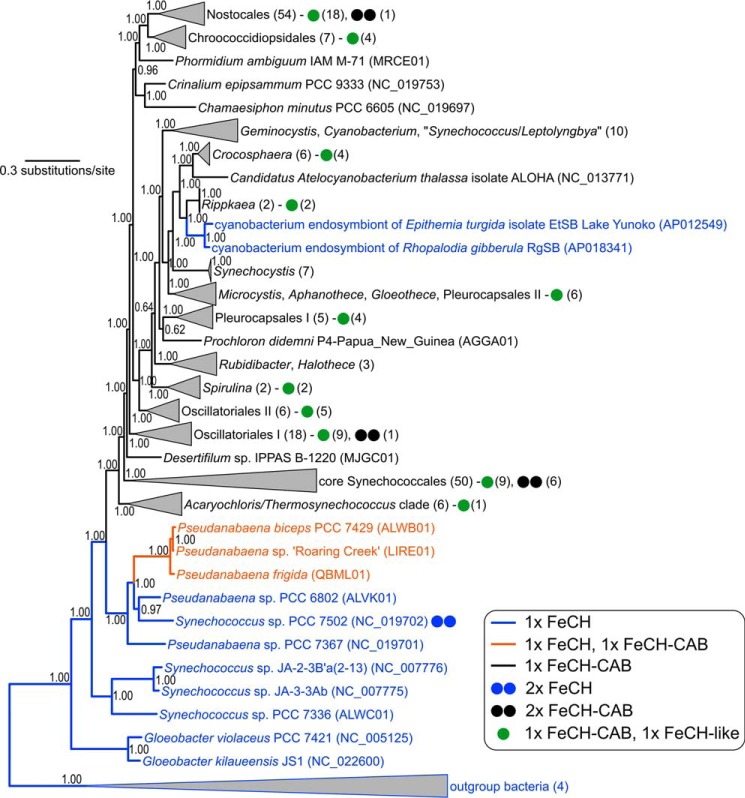
The FeCh enzyme found in distinct groups of oxygenic phototrophs, such as plants, green algae, diatoms, haptophytes, and glaucophytes, is of cyanobacterial origin and possesses a conserved CAB domain (Fig. S1) (34). However, the origin of the FeCh CAB domain, as well as its distribution in currently sequenced cyanobacterial species, remains unclear. We searched for hemH genes in all of the currently available cyanobacterial genomes (a total of 462 genomes) and found that the vast majority (453 genomes) code for FeCh with the CAB domain. The only exceptions are the most basal clades of the cyanobacterial evolutionary tree (primordial Gloeobacter, Synechococcus, and Pseudanabaena strains) and two members of a derived clade of unicellular endosymbionts of diatoms (Fig. 8 and Table 1). A complete list of the analyzed genomes is provided in Table S1. In all analyzed CAB domains (453 genomes), the EXXNXR Chl-binding motif is conserved.
Figure 8.
Distribution of hemH genes coding for FeCh with and without the CAB domain in an evolutionary tree of cyanobacteria. FeCH lacking the CAB domain is plesiomorphic in cyanobacteria. The fusion of FeCH and CAB may have occurred twice (the orange clade and the black clade) or may have been lost once (sister clade to the orange one) after duplication of hemH. The tree inferred from 22 conserved proteins in 220 genomes was calculated using Bayesian inference posterior probabilities are shown near the nodes. Numbers in brackets indicate the number of genomes in a collapsed clade or possessing a particular organization of FeCH-coding genes as explained in the legend.
Table 1.
Numbers of cyanobacterial genomes coding for the Hlips, the FeCh with or without the CAB domain, and the FeCh-like proteins
A detailed list of all strains and identified hemH genes are provided in Table S1.
| Deduced content of genes coding for Hlips and FeChs | Number of genomes (462 in total) |
|---|---|
| Hlips | 462 |
| 1× FeCha | 9 |
| Gloeobacter violaceus PCC 7421 | |
| Gloeobacter kilaueensis JS1 | |
| Synechococcus sp. JA-3-3Ab | |
| Synechococcus sp. JA-2-3B′a(2-13) | |
| Pseudanabaena sp. PCC 6802 | |
| Synechococcus sp. PCC 7336 | |
| Pseudanabaena sp. PCC 7367 | |
| Cyanobacterium endosymbiont of Epithemia turgida | |
| Cyanobacterium endosymbiont of Rhopalodia gibberula | |
| 2× FeCha | 1 |
| Synechococcus sp. PCC 7502 | |
| 1× FeCh, 1× FeCh-CABb | 3 |
| Pseudanabaena biceps PCC 7429 | |
| Pseudanabaena sp. “Roaring Creek” | |
| Pseudanabaena frigida ULC066bin1 | |
| 1× FeCh-CAB | 359 |
| 2× FeCh-CABc | 10 |
| 1× FeCh, 1× FeCh-like | 0 |
| 1× FeCh-CAB, 1× FeCh-like | 90 |
a High similarity to cyanobacterial FeCh but lacking CAB domain.
b A cyanobacterial-type of FeCh containing CAB domain.
c Genes sharing high identity.
Three Pseudoanabena strains, forming a sister group to the basal, CAB domain-less clade, (Fig. 8) possess two similar hemH genes (identity ∼60%); one hemH copy contains the CAB domain, but this domain is absent in the second gene. This situation is unique, because no other cyanobacterial genomes contain a combination of hemH genes with and without the CAB domain (Table 1). Intriguingly, 90 cyanobacterial genomes code for a “FeCh-like” protein with very limited similarity to the cyanobacterial FeCh (20–30%; Table 1). This gene also seems to be cyanobacteria-specific, because we found no homologs in the NCBI database (by BLAST search) after excluding cyanobacteria from the search. Whereas the FeCh-like protein substantially differs from archeal, bacterial, or eukaryotic FeChs on a sequence level (not shown), its tertiary structure predicted by I-TASSER (35) aligned very tightly with the structure of canonical human FeCh (root-mean-square deviation between 256 pruned atom pairs = 0.37 Å; Fig. S7A). Moreover, His-263, Asp-340, and Glu-343 residues (if counted according to the human FeCh), which are essential for the extraction of protons from PPIX nitrogens (36), are conserved in all 90 sequences of the FeCh-like proteins. The same is true for the absolutely conserved Phe-337 residue placed onto the PPIX-binding pocked (Fig. S7B) (37). Strains containing the putative second FeCh are listed in Table S1.
Single-helix Hlips have already been proposed as ancestors of the whole LHC superfamily (16). Indeed, genes coding for Hlips are present in all of the analyzed genomes, including the strains lacking thylakoids and/or the FeCh CAB domain (Table 1). We therefore conclude that the FeCh CAB domain most likely originates from an ancient fusion between an Hlip protein and the FeCh enzyme in the ancestor of all modern cyanobacteria except for the three deepest-branching lineages (Fig. 8).
Discussion
The FeCh enzyme equipped with the pigment-binding CAB domain is apparently an old invention of cyanobacteria (Fig. 8). Comparing our phylogenetic tree to the recent calibrated molecular clock analyses, the evolution of this fused protein can be dated back to the early Proterozoic, roughly coinciding with the Great Oxygenation Event (38). Our data support a scenario in which the CAB domain originates from a fusion between an ancient cyanobacterial FeCh and an Hlip. Although we cannot completely exclude the possibility that the FeCh CAB domain is the progenitor of the whole LHC superfamily including Hlips, it is indeed very unlikely that the CAB segment of FeCh, in such a case already present in the last common cyanobacterial ancestor, would be selectively lost in basal cyanobacterial lineages (Fig. 8). Furthermore, in the literature there is an example of another fusion between an Hlip and a conserved cyanobacterial protein, a homolog of the Synechocystis Ssl2148 protein (39), supporting the plausibility of FeCh–Hlip fusion.
The function of Hlips and their plant homologs (OHPs) has been connected to the biogenesis of Chl-binding proteins, particularly the subunits of PSII (21, 40). All four Hlips present in Synechocystis (HliA–D) interact with either the D1 subunit (HliC/D) or the CP47 subunit of PSII (HliA/B/C) soon after these Chl proteins are synthesized (21). Hlips then remain associated with D1/CP47 to support certain stages of PSII assembly or, possibly, PSII repair (21). In addition, the HliC/D proteins associate with the Chl-synthase enzyme (41). The exact roles that Hlips play are not clear yet, but they most likely fulfill several different tasks: (i) photoprotection of PSII biogenesis (dissipation of absorbed energy), (ii) delivery of Chl molecules, and (iii) regulation of Chl metabolism (HliC/D) (10). We suggest that once the FeCh enzyme linked physically with an Hlip, the FeCh became targeted to the machinery responsible for the biogenesis and assembly of Chl-binding proteins. There is general agreement about the existence of specific membrane compartments in which the photosystems are assembled, in both cyanobacteria (42) and plastids (43). As a crucial regulatory enzyme, FeCh probably got close to a site where Chl molecules are produced (44) and distributed into Chl-binding apoproteins (45).
The ability of Hlips to bind Chl and carotenoids in an energy dissipative configuration (24) has been preserved in the FeCh CAB domain. The absorbance spectrum of the His-C-tn-pigment complex is similar to two Hlips that have been previously characterized (HliC and HliD) (24, 46). However, unlike HliC and HliD proteins that exclusively contain β-car, the CAB domain also binds zeaxanthin and echinenone (Fig. 7C). The calculated molar ratio of Chl:β-car:zeaxanthin (6:0.9:1) in the CAB domain is very similar to the ratio of Chl:β-car identified in HliC and HliD (6 or 4 Chl per 2 β-car, respectively) (24, 25). Two different β-car–binding sites have been unambiguously identified in Hlips (24, 47). However, in CAB domain, one carotenoid-binding site could be less specific and more polar than that in Hlips, preferentially accommodating xanthophylls or keto-carotenoids. A similar shift to polar xanthophylls (lutein, violaxanthin) also happened during the evolution of the eukaryotic major and minor antenna proteins (26, 48).
The original cyanobacterial FeCh was probably monomeric, because it is strictly monomeric if the CAB domain is eliminated (Fig. 6B) (29), and the recombinant full-length Synechocystis FeCh can be prepared in an active monomeric form (30). Although there is a small fraction of dimeric FeCh in Synechocystis grown under our standard laboratory conditions (29), the vast majority of FeCh appears to be monomeric in vivo (Fig. 6B). Because the pigment-binding motif in Hlips is, however, formed by two parallel helices (24), the monomeric CAB domain cannot serve a photoprotective or Chl carrier role. To be capable of pigment binding the FeCh needs to dimerize via CAB domain (29); most likely by mimicking the LHC-like pair of helices (26) clipped together by salt bridges between Glu and Arg residues on the opposite helices (see also Fig. S1B).
A mechanistic explanation or a function of the observed conditional FeCh dimerization is beyond the scope of this report and must be addressed by future studies; however, based on the present data, we speculate that the formation of FeCh dimers is triggered in response to disturbed tetrapyrrole metabolism. The rather mild dose of MPP used in our study allows treated cells to produce enough heme for close to normal proliferation (Fig. S3); yet the regulatory function of heme was apparently disturbed, resulting in a strongly up-regulated tetrapyrrole pathway. Under such conditions, a large pool of FeCh dimerizes. It should be noted that the His-C-tn protein (an artificial CAB domain) readily attaches to full-length FeCh via its CAB domain in vivo (29), suggesting that there is a control mechanism such that two FeChs form a stable dimer only under certain conditions. A possible explanation is that the monomeric FeCh is incorporated into a protein complex. A colocalization of cyanobacterial FeCh with protoporphyrinogen oxidase, the preceding enzyme in the pathway, has been reported (49), and it is worth noting that the mitochondrial FeCh is a component of large protein assembly dealing with heme metabolism (50). The proposed spatial separation of FeCh monomers in the cell is consistent with the fact that purified pigment-less Synechocystis FeCh spontaneously dimerizes (29). However, there is the possibility of in vitro artifacts caused by the high concentrations of the protein and/or handling and freezing the sample. In this work, the purified sample was immediately loaded into a SEC column. An alternative scenario is that the in vivo stability of the pigment-less FeCh dimer is weak, but after association with Chl and carotenoids, the dissociation constant of the pigmented dimer becomes much lower, resulting in the accumulation of FeCh dimer. This is indeed an attractive hypothesis, providing a mechanism for how the FeCh enzyme senses a risky accumulation of Chl molecules in the membrane, particularly if this occurs in the specialized compartment for Chl-protein biogenesis. The Synechocystis mutant lacking FeCh CAB domain was reported to be sensitive to high light and to accumulate chlorophyllide, the substrate of Chl synthase (29). We did not observe a large pool of FeCh dimer under high light conditions (not shown); however, it is possible that under physiological conditions, the level of FeCh dimer does not exceed a certain threshold and/or it is short-lived.
The amount of FeCh in Synechocystis appears to be much higher than that needed for synthesizing a sufficient amount of heme: strains with ∼10% of normal FeCh activity do not exhibit any signs of phycobilisome deficiency, at least under low-stress conditions (6, 31). This situation resembles the reserve flux capacity that enables an immediate (<1 min) rerouting of glucose-6-phosphate into the oxidative pentose phosphate pathway in Escherichia coli (51). Synechocystis could have a similar capacity to promptly reroute PPIX into the heme pathway, away from magnesium chelatase. Whether FeCh dimerization together with pigment binding on the CAB domain influences its activity is unknown at this point. Unfortunately, we cannot assess the activity of pigmented FeCh because of the presence of MPP in the active site. However, in an in vitro assay, FeCh lacking the CAB domain shows increased catalytic turnover number (30), suggesting that there is an effect of this segment on the enzymatic parameters in vivo. In the same report, the addition of a methanol extract from Synechocystis membranes containing pigments lowered the FeCh activity irrespectively of the CAB domain (30). This observation might indicate that free Chl molecules have a direct effect on the enzyme catalytic site. However, the conditions in in vitro assay are very artificial, and also the methanol extract is a complex mixture of compounds; in the work of Storm et al. (30), a measurement with chemically pure Chl a is not provided. Potential modulation of FeCh activity by Chl, either via CAB domain or by interfering with catalytic site, thus remains an open question. Alternatively, instead of changing the catalytic properties, dimerization and binding of pigments on the CAB domain may alter the availability of PPIX for chelation by changing the FeCh interactome (as discussed above). In this scenario, FeCh would be regulated by substrate availability rather than by enzymatic activity.
Although FeCh activity is undoubtedly very important for the regulation of Chl biosynthesis (31, 52), the molecular mechanism of this regulation is not known. The classical model of labile free heme directly controlling the formation of ALA (53) appears to be oversimplified. In plants, it has been proposed that two spatially separated pools of PPIX are located in plastids, each used for the synthesis of heme, but only one pool of PPIX also serving as a substrate for magnesium chelatase (52, 54). In addition, there seems to be a dedicated regulatory heme pool produced by plastid FeCh1 isozyme (lacking the CAB domain) that is involved in plastid to nucleus signaling (55). Plant cells are much more complex than cyanobacterial cells; yet the CAB domain (FeCh2 isozyme) is conserved in plants, and there is no reason to expect that it has a fundamentally different function than in cyanobacteria. Predictably, FeCh2 is the key regulator of plant tetrapyrrole metabolism; FeCh1 activity does not affect the synthesis of tetrapyrroles in plastids (56). Because many cyanobacterial stains contain a putative second FeCh (Table 1), we speculate that the FeCh-controlled ALA synthesis and flow of PPIX into the Chl branch are limited to a specific compartment. This is in line with our proposed targeting of FeCh with the CAB domain into a site of Chl-protein biogenesis, where enzymes of the Chl pathway should be also localized (44, 57).
Different oligomeric states of the FeCh enzyme are most likely involved in the sophisticated regulatory machinery controlling Chl metabolism, which in cyanobacteria also includes Hlips (10) and other proteins (58, 59). Intriguingly, Synechocystis lycopene cyclase (CruA), an enzyme in carotenoid biosynthesis, was recently reported to bind Chl and carotenoids (60). This enzyme does not possess a CAB domain or helical transmembrane segments; the mechanism through which the lycopene cyclase binds Chl and carotenoids in a ratio and spectral characteristics resembling a typical Chl-complexes such as photosystem I thus remains enigmatic. In plants and algae, the conserved CAB/LHC motif is present within a broader spectrum of proteins that are otherwise diverse but also generally connected to Chl biosynthesis. For instance, the Msf1 protein recently identified in the alga Chlamydomonas reinhardtii is required for the accumulation of photosystem I and interacts with the cyclase enzyme in Chl biosynthesis (61). In Arabidopsis thaliana, Lil3.1/2 proteins are crucial for the synthesis of the Chl phytol chain (12, 13, 62–65), whereas OHPs assist during the synthesis of photosystem II as described above for the cyanobacterial Hlips (14, 15). Association with a biogenesis compartment and (regulatory) pigment-driven dimerization could be thus common themes for LHC-like proteins.
Experimental procedures
Synechocystis strains and growth conditions
The glucose-tolerant Synechocystis WT-P substrain is referred to here as WT (66). The Synechocystis strain expressing 3×FLAG-tagged FeCh and lacking the native hemH gene has been described previously (29), as well as the strains possessing truncated FeCh derivatives (hemH-Δ324, hemH-Δ347) (6, 29) or expressing the FeCh C-terminal domain as a His6-tagged protein (His-C-tn) (29). To express the His-C-tn protein in the hemH-Δ324 genetic background, the His-C-tn cells were transformed with chromosomal DNA isolated from the hemH-Δ324 cells, and transformants were selected on a BG-11 plate with Zeocin (3 mg/liter). Complete segregation of the mutant was verified by PCR. All strains were grown in BG-11 medium; solid media were further supplemented with 10 mm TES/NaOH (pH 8.2), 1.5% agar, and 0.3% sodium thiosulfate. Liquid cultures were grown either grown in 50 ml of BG-11 in 250-ml conical flasks on a rotary shaker or in 800 ml of BG-11 in 1-liter cylinders bubbled with air at 29 °C with a surface irradiance of 40 μmol photons m−2 s−1 of white light. The cultures were harvested in the exponential phase at a cell density of ∼108/ml (A750 in a range of 0.8–1.0) by centrifugation (5000 RCF, 5 min). Where applicable the FeCh inhibitor MPP (32) (Frontier Scientific) was added from a 10 mm stock in DMSO to a final concentration of 275 nm.
Affinity, size-exclusion, and anion-exchange chromatography
Pelleted cells were resuspended in isolation buffer A (20 mm MES, pH 6.5, 150 mm NaCl, complete protease inhibitor mixture; Roche) and mixed with glass beads of 100–200 μm diameter at a 1:1 ratio (1 volume of dense cell solution to 1 volume of glass beads). This suspension was vortexed six times for 10 s with 1 min on ice in between each cycle. After vortexing, the glass beads were washed four times with buffer A, and the obtained suspension of broken cells was pooled and centrifuged (40,000 RCF, 20 min). The pelleted membranes were resuspended in buffer A and frozen (−80 °C) for further use. Isolated membranes containing 500 μg Chl/ml were solubilized using 1% (w/v) DDM for 5 min on ice. His- and FLAG-tagged proteins were purified in buffer A at 10 °C according to the manufacturer's instructions using Protino nickel–nitrilotriacetic acid–agarose (Macherey–Nagel) and an anti-FLAG M2 affinity gel (Sigma–Aldrich), respectively. Prior to ion-exchange/size-exclusion chromatography, samples were concentrated approximately four times using 10-kDa cutoff microconcentrators (AmiconR Ultra 0.5 ml; Merck). The eluted and concentrated f.FeCh protein was immediately injected onto an Agilent-1200 HPLC system and separated on a Yarra SEC-3000 column (Phenomenex) using mobile phase (20 mm MES, pH 6.5, 100 mm NaCl, pH 6.5, containing 0.06% (w/v) DDM) at a flow rate of 0.25 ml min−1 at 15 °C. Eluted proteins and complexes were detected using a diode-array detector and a fluorescence detector set to 440/680 nm (excitation/emission wavelengths). The membrane fraction solubilized by 1% (w/v) DDM was separated with an identical procedure as that of the f.FeCh eluate.
For anion-exchange separation of the His-C-tn protein, the concentrated eluate from the nickel column was injected onto an Agilent-1200 HPLC system and separated on a MonoQ 4.6/100 PE column (GE Healthcare) using a gradient of NaCl in 20 mm HEPES, pH 8.1, 5 mm MgCl2, 0.06% (w/v) DDM. The high-salt (HS) buffer was identical to the low-salt buffer but contained 1 m NaCl. The gradient was run as follows: 10 min, 0% HS; 15 min, 20% HS; 45 min, 55% HS; and 47 min, 100% HS at a flow rate of 0.8 ml min−1 at 15 °C. Eluted proteins and complexes were detected using a diode-array detector and a fluorescence detector set to 440/675 nm (excitation/emission wavelengths).
Native and denaturing protein electrophoresis
Harvested cells were dissolved in isolation buffer B (25 mm MES, pH 6.5, 4 mm MgCl2, 4 mm CaCl2, 25% (w/v) glycerol, complete protease inhibitor mixture; Roche), mixed with glass beads of 100–200 μm diameter at a 1:1 ratio (1 volume of dense cell solution to 1 volume of glass beads), and broken (three times for 20 s) using a Mini-Beadbeater-16 (Biospec). The cells were kept on ice for 2 min between each cycle. The membranes were isolated as described above.
For the separation of membrane complexes on BN-PAGE, the isolated membranes containing 400 μg Chl/ml were solubilized using 1% (w/v) DDM for 5 min on ice. Nonsolubilized material was removed by centrifugation (20,000 RFC, 10 min). Extracted membrane proteins corresponding to 4 μg of Chl were mixed with 1/10 volume of sample buffer (750 mm aminocaproic acid, 5% (w/v) CBB-G) and separated at 4 °C on an 8–16% gradient polyacrylamide gel containing 500 mm aminocaproic acid and 50 mm Bis-Tris/HCl, pH 7.6, at room temperature according to Ref. 67. The starting blue cathode buffer (50 mm Tricine, 50 mm Bis-Tris/HCl, pH 7.0, 0.2% (w/v) CBB-G) was exchanged after one-third of the run for the clear cathode buffer (50 mm Tricine, 50 mm Bis-Tris/HCl, pH 7.0). As anode buffer, 50 mm Bis-Tris/HCl, pH 7.0, was used. In clear-native electrophoresis the cathode buffer contained 0.05% (w/v) sodium deoxycholate and 0.02% (w/v) DDM, and a 4–12% gradient polyacrylamide gel was used. After separation, Chl fluorescence in the gel was detected using LAS-4000 (Fuji) after illumination with blue light.
To separate proteins using denaturing SDS-PAGE, a 16–20% linear gradient polyacrylamide gel containing 7 m urea, 650 mm Tris/HCl, pH 8.6, was prepared according to (68). For the second dimension, gel strips from the native gel were excised and incubated for 30 min in 25 mm Tris/HCl, pH 7.5, containing 1% (w/v) SDS and 2% (w/v) DTT, and placed on top of the denaturing gel; two strips were analyzed on a single denaturing gel. Cathode buffer (25 mm Tris, 192 mm glycine, 0.2% (w/v) SDS) and anode buffer (25 mm Tris/HCl, pH 8.3) were used. A ratio of 1:60 for acrylamide to bis-acrylamide was used for all gels.
Protein immunodetection
Proteins separated by SDS-PAGE were stained for 50 min with SYPRO Orange (Thermo Fisher) 5000× diluted in 10 mm Tris/HCl, pH 7.6, 150 mm NaCl, 10% (v/v) methanol and subsequently transferred (3 mA/cm2, 3h) onto an Immobilon-P membrane (0.45 μm; Millipore). After electrotransfer, the membrane was blocked by 0.05% (w/v) Tween 20 in 10 mm Tris/HCl, pH 7.6, 150 mm NaCl for 30 min and subsequently incubated with a specific primary antibody and then with a secondary antibody–horseradish peroxidase conjugate (Sigma). The peroxidase activity was visualized by incubating the membrane for 20 s in Luminata Crescendo Western HRP substrate (Sigma) and detected using LAS-4000 (Fuji).
The primary antibodies used in this study were as follows: anti-FeCh antibody (1:20,000 dilution) raised in rabbit against the Synechocystis recombinant FeCh (29); a rabbit polyclonal antiserum raised against residues 325–353 of the D1 protein from Pisum sativum (67) at a dilution of 1:5000; a polyclonal purified anti-PsaA antibody (Agrisera; dilution 1:10,000) raised against residues 2–18 of the Synechocystis PsaA; and a polyclonal purified antibody raised against the synthetic peptide VPHPKKNMKMYPQER, which corresponds to the linker region of Synechocystis FeCh (29) (dilution 1:10,000).
Chl radiolabeling and measurement of Chl precursors and pigments
To assess the rate of de novo Chl synthesis, cells corresponding to 50 mg of Chl in an exponential growth phase at density ∼5 × 107/ml (OD750 = 0.5) were harvested by centrifugation (2700 RCF, 3 min), washed, and resuspended in fresh BG-11 medium containing 20 mm TES to a final volume of 540 μl. The cell suspension was shaken in 10-ml glass tubes for 30 min at 29 °C at 40 μmol photons m−2 s−1. Then 60 μl of [14C]Glu with specific activity 248 mCi/mmol (Moravek Biochemicals) was added. After 60 min, the cells were harvested (10,000 RCF, 2 min) and frozen in liquid nitrogen. MPP (275 nm) was added to the cells 3.5 h before the addition of [14C]Glu and also kept at the 275 nm concentration during radiolabeling. Pelleted cells were washed and resuspended in 50 μl of water; Chl was extracted with 1 ml of methanol, and insoluble material was discarded by centrifugation (10,000 RCF, 4 min). The extract was injected into an Agilent-1260 HPLC system equipped with a Radiomatic 150 TR scintillation detector (PerkinElmer Life Sciences) and a diode-array detector. Pigment separation was carried out on a Zorbax Eclipse Plus C18 column (4.6-μm particle size, 3.9 × 100 mm; Agilent) with 35% methanol, 15% acetonitrile in 0.25 m pyridine as solvent A and 20% methanol, 20% acetone in acetonitrile as solvent B. Pigments were eluted with a linear gradient of solvent B (30–95% in 25 min) followed by 95% solvent B at a flow rate of 0.8 ml min−1 at 40 °C. The relative rate of Chl synthesis was estimated from the ratio of Chl radio emission to Chl absorbance between MPP-treated and untreated samples.
To quantify free Chl in the native gel, a piece of gel (∼4 × 5 mm) corresponding to the region of DDM micelles was cut and further chopped up into pieces (1 × 1 mm). The obtained gel pieces were incubated for 2 h in 125 μl of buffer A containing 0.04% DDM, and then the buffer was collected after centrifugation (10,000 RCF, 2 min). The incubation of gel pieces in the buffer A was repeated, the buffer fractions were pooled, and the eluted pigments were separated by HPLC as described above. The absolute Chl concentration was calculated from peak area using a Chl-a standard.
Chl precursors were quantified as described in Ref. 69. To identify pigments bound to the isolated f.FeCh and His-C-tn proteins, fractions collected during SEC and anion-exchange chromatography were concentrated on 10-kDa cutoff microconcentrators (AmiconR Ultra 0.5 ml; Merck). This concentrated solution was extracted with 90% methanol, and the extract was analyzed using the Agilent-1260 HPLC system, essentially as described above for the radiolabeled samples. Pigment stoichiometry was calculated from calibration curves prepared using authentic standards.
Nonphotochemical quenching of Chl bound to f.FeCh
Quenching of Chl fluorescence was estimated from the ratio of fluorescence/absorbance of Chl molecules bound to f.FeCh compared with unquenched Chl in DDM micelles. Identical fluorescence constants and extinction coefficients were assumed for both Chl populations.
Phylogenetic and bioinformatic analysis
A representative set of 550 publicly available cyanobacterial genome assemblies was downloaded from NCBI and utilized to create a custom BLAST database (70). FeCh protein from Synechocystis (slr0839 gene) was used as the query for a tBLASTn search with a cutoff value of 1.e−10 against this database, allowing multiple hits for each genome. All hits were automatically harvested and aligned using MAFFT v. 7 (71). Sequence analysis in Geneious Prime v. 2019 allowed us to remove short hits, reducing the number of genomes in the final data set to 462 and to assess the presence of a terminal CAB domain, specific conserved residues (Chl-binding motif), and pairwise amino acid sequence identity between individual deduced proteins in the matrix.
The multilocus phylogenetic tree was generated using a set of 22 conserved single-copy orthologous protein-coding genes selected from loci previously tested for congruent cyanobacterial phylogenies (72, 73). The selection was made based on the presence of a full unambiguous sequence of the genes in a representative subset of genomes, and comprised these loci: dnaG, frr, infC, nusA, pgk, pyrG, rplB, rplC, rplD, rplE, rplF, rplM, rplP, rplT, rpoB, rpsC, rpsE, rpsI, rpsK, rpsM, rpsS, and smpB. First, BLAST queries were derived from respective proteins mined from Synechocystis (BA000022). Each of the 22 queries was then used in tBLASTn (cutoff value 1.e−15) searches against the custom database. Hits for each protein were aligned using MAFFT v. 7, and the alignments were manually reviewed to remove ambiguous sites and gap regions. A maximum likelihood tree was calculated using each of the 22 alignments with 1000 bootstrap pseudoreplications in RaxML v. 8 (74). The resulting phylogenies were inspected manually for topological incongruence. Because individual gene trees were generally consistent, all alignments were concatenated into a 5117–amino acid–long matrix containing 220 sequence lines. A Bayesian inference (BI) tree was obtained using MrBayes v. 3.2.6 (75). Two independent runs of eight Markov chains were performed for 1 million generations, sampling every 100th tree, until the likelihood values were stable, and the divergence criterion was lower than 0.01. The BI calculation employed a common LG + I + G evolutionary model because of the excessive computational demands of a calculation using partitioned models. Posterior probabilities were estimated from branch frequencies in the sampled trees, discarding the first 25% of the harvested data as burn-in. The maximum likelihood and BI calculations were run via the CIPRES supercomputing facility (76).
Author contributions
M. P. and R. S. conceptualization; M. P., J. M., J. P., and R. S. data curation; M.P., J. M., J. P., and R. S. formal analysis; M. P. and R. S. funding acquisition; M. P. and R. S. investigation; M. P. and R. S. visualization; M.P., J. M., J. P., and R. S. methodology; M. P. and R. S. writing-original draft; M. P. and R. S. project administration; M.P., J. M., J. P., and R. S. writing-review and editing; J. M. software; R. S. resources; R. S. supervision; R. S. validation.
Supplementary Material
Acknowledgments
We thank Eva Kiss and Josef Komenda for reading the manuscript.
This work was supported by Czech Science Foundation Grant 17-08755S and Czech Ministry of Education Project LO1416. This work was also supported by University of South Bohemia Grant 04-026/2017 (to M. P.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S7.
- used are: FeCh
- ferrochelatase
- Chl
- chlorophyll
- MPP
- N-methyl mesoporphyrin
- CAB
- chlorophyll a/b–binding
- DDM
- n-dodecyl β-d-maltoside
- LHC
- light-harvesting complex
- Hlip
- high light–inducible protein
- ELIP
- early light–inducible protein
- PPIX
- protoporphyrin IX
- MV/DV Pchlide
- monovinyl/divinyl protochlorophyllide
- OHP
- one-helix protein
- SEC
- size-exclusion chromatography
- PSI
- photosystem I
- β-car
- β-carotene
- BN/SDS-PAGE
- blue-native/SDS polyacrylamide gel electrophoresis
- HS
- high-salt
- BI
- Bayesian inference
- RCF
- relative centrifugal force.
References
- 1. Yuan X., Rietzschel N., Kwon H., Walter Nuno A. B., Hanna D. A., Phillips J. D., Raven E. L., Reddi A. R., and Hamza I. (2016) Regulation of intracellular heme trafficking revealed by subcellular reporters. Proc. Natl. Acad. Sci. U.S.A. 113, E5144–E5152 10.1073/pnas.1609865113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ikushiro H., Nagami A., Takai T., Sawai T., Shimeno Y., Hori H., Miyahara I., Kamiya N., and Yano T. (2018) Heme-dependent inactivation of 5-aminolevulinate synthase from Caulobacter crescentus. Sci. Rep. 8, 14228 10.1038/s41598-018-32591-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Armas-Ricard M., Levicán G., Katz A., Moser J., Jahn D., and Orellana O. (2011) Cellular levels of heme affect the activity of dimeric glutamyl-tRNA reductase. Biochem. Biophys. Res. Commun. 405, 134–139 10.1016/j.bbrc.2011.01.013 [DOI] [PubMed] [Google Scholar]
- 4. Brzezowski P., Richter A. S., and Grimm B. (2015) Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochim. Biophys. Acta 1847, 968–985 10.1016/j.bbabio.2015.05.007 [DOI] [PubMed] [Google Scholar]
- 5. Papenbrock J., Mishra S., Mock H. P., Kruse E., Schmidt E. K., Petersmann A., Braun H. P., and Grimm B. (2001) Impaired expression of the plastidic ferrochelatase by antisense RNA synthesis leads to a necrotic phenotype of transformed tobacco plants. Plant J. 28, 41–50 10.1046/j.1365-313X.2001.01126.x [DOI] [PubMed] [Google Scholar]
- 6. Sobotka R., McLean S., Žuberová M., Hunter C. N., and Tichý M. (2008) The C-terminal extension of ferrochelatase is critical for enzyme activity and for functioning of the tetrapyrrole pathway in Synechocystis strain PCC 6803. J. Bacteriol. 190, 2086–2095 10.1128/JB.01678-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Papenbrock J., Mock H. P., Tanaka R., Kruse E., and Grimm B. (2000) Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway. Plant Physiol. 122, 1161–1169 10.1104/pp.122.4.1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Papenbrock J., Pfündel E., Mock H. P., and Grimm B. (2000) Decreased and increased expression of the subunit CHL I diminishes Mg chelatase activity and reduces chlorophyll synthesis in transgenic tobacco plants. Plant J. 22, 155–164 10.1046/j.1365-313x.2000.00724.x [DOI] [PubMed] [Google Scholar]
- 9. Crawford T. S., Eaton-Rye J. J., and Summerfield T. C. (2016) Mutation of Gly195 of the ChlH subunit of Mg-chelatase reduces chlorophyll and further disrupts PS II assembly in a Ycf48-deficient strain of Synechocystis sp. PCC 6803. Front. Plant Sci. 7, 1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu H., Vavilin D., Funk C., and Vermaas W. (2002) Small Cab-like proteins regulating tetrapyrrole biosynthesis in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 49, 149–160 10.1023/A:1014900806905 [DOI] [PubMed] [Google Scholar]
- 11. Sinha R. K., Komenda J., Knoppová J., Sedlářová M., and Pospíšil P. (2012) Small CAB-like proteins prevent formation of singlet oxygen in the damaged photosystem II complex of the cyanobacterium Synechocystis sp PCC 6803. Plant Cell Environ. 35, 806–818 10.1111/j.1365-3040.2011.02454.x [DOI] [PubMed] [Google Scholar]
- 12. Hey D., Rothbart M., Herbst J., Wang P., Müller J., Wittmann D., Gruhl K., and Grimm B. (2017) LIL3, a light-harvesting complex protein, links terpenoid and tetrapyrrole biosynthesis in Arabidopsis thaliana. Plant Physiol. 174, 1037–1050 10.1104/pp.17.00505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou F., Wang C. Y., Gutensohn M., Jiang L., Zhang P., Zhang D., Dudareva N., and Lu S. (2017) A recruiting protein of geranylgeranyl diphosphate synthase controls metabolic flux toward chlorophyll biosynthesis in rice. Proc. Natl. Acad. Sci. U.S.A. 114, 6866–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hey D., and Grimm B. (2018) One-helix protein 2 (OHP2) is required for the stability of OHP1 and assembly factor HCF244 and is functionally linked to PSII biogenesis. Plant Physiol. 177, 1453–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Myouga F., Takahashi K., Tanaka R., Nagata N., Kiss A. Z., Funk C., Nomura Y., Nakagami H., Jansson S., and Shinozaki K. (2018) Stable accumulation of photosystem II requires one-helix protein 1 (OHP1) of the light harvesting-like family. Plant Physiol. 176, 2277–2291 10.1104/pp.17.01782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Engelken J., Brinkmann H., and Adamska I. (2010) Taxonomic distribution and origins of the extended LHC (light-harvesting complex) antenna protein superfamily. BMC Evol. Biol. 10, 233 10.1186/1471-2148-10-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Engelken J., Funk C., and Adamska I. (2012) The extended light-harvesting complex (LHC) protein superfamily: classification and evolutionary dynamics. In Functional Genomics and Evolution of Photosynthetic Systems (Burnap R. L., and Vermaas W. F. J., eds) pp. 265–284, Springer, New York [Google Scholar]
- 18. Heddad M., Engelken J., and Adamska I. (2012) Light stress proteins in viruses, cyanobacteria and photosynthetic eukaryota. In Photosynthesis: Plastid Biology, Energy Conversion and Carbon Assimilation (Eaton-Rye J. J., Tripathy B. C., and Sharkey T. D., eds) pp. 299–317, Springer, New York [Google Scholar]
- 19. Neilson J. A., and Durnford D. G. (2010) Evolutionary distribution of light-harvesting complex-like proteins in photosynthetic eukaryotes. Genome 53, 68–78 10.1139/G09-081 [DOI] [PubMed] [Google Scholar]
- 20. Jansson S. (1999) A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci 4, 236–240 10.1016/S1360-1385(99)01419-3 [DOI] [PubMed] [Google Scholar]
- 21. Komenda J., and Sobotka R. (2016) Cyanobacterial high-light-inducible proteins: Protectors of chlorophyll-protein synthesis and assembly. Biochim. Biophys. Acta 1857, 288–295 10.1016/j.bbabio.2015.08.011 [DOI] [PubMed] [Google Scholar]
- 22. Adamska I., Roobol-Bóza M., Lindahl M., and Andersson B. (1999) Isolation of pigment-binding early light-inducible proteins from pea. Eur. J. Biochem. 260, 453–460 10.1046/j.1432-1327.1999.00178.x [DOI] [PubMed] [Google Scholar]
- 23. Reisinger V., Plöscher M., and Eichacker L. A. (2008) Lil3 assembles as chlorophyll-binding protein complex during deetiolation. FEBS Lett. 582, 1547–1551 10.1016/j.febslet.2008.03.042 [DOI] [PubMed] [Google Scholar]
- 24. Staleva H., Komenda J., Shukla M. K., Šlouf V., Kaňa R., Polívka T., and Sobotka R. (2015) Mechanism of photoprotection in the cyanobacterial ancestor of plant antenna proteins. Nat. Chem. Biol. 11, 287–291 10.1038/nchembio.1755 [DOI] [PubMed] [Google Scholar]
- 25. Shukla M. K., Llansola-Portoles M. J., Tichý M., Pascal A. A., Robert B., and Sobotka R. (2018) Binding of pigments to the cyanobacterial high-light-inducible protein HliC. Photosyn. Res. 137, 29–39 10.1007/s11120-017-0475-7 [DOI] [PubMed] [Google Scholar]
- 26. Standfuss J., Terwisschavan Scheltinga A. C., Lamborghini M., and Kühlbrandt W. (2005) Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5A resolution. EMBO J. 24, 919–928 10.1038/sj.emboj.7600585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wei X. P., Su X., Cao P., Liu X., Chang W., Li M., Zhang X., and Liu Z. (2016) Structure of spinach photosystem II-LHCII supercomplex at 3.2 angstrom resolution. Nature 534, 69–74 10.1038/nature18020 [DOI] [PubMed] [Google Scholar]
- 28. Funk C., and Vermaas W. (1999) A cyanobacterial gene family coding for single-helix proteins resembling part of the light-harvesting proteins from higher plants. Biochemistry 38, 9397–9404 10.1021/bi990545+ [DOI] [PubMed] [Google Scholar]
- 29. Sobotka R., Tichý M., Wilde A., and Hunter C. N. (2011) Functional assignments for the carboxyl-terminal domains of the ferrochelatase from Synechocystis PCC 6803: the CAB domain plays a regulatory role, and region II is essential for catalysis. Plant Physiol. 155, 1735–1747 10.1104/pp.110.167528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Storm P., Tibiletti T., Hall M., and Funk C. (2013) Refolding and enzyme kinetic studies on the ferrochelatase of the cyanobacterium Synechocystis sp. PCC 6803. PLoS One 8, e55569 10.1371/journal.pone.0055569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sobotka R., Komenda J., Bumba L., and Tichý M. (2005) Photosystem II assembly in CP47 mutant of Synechocystis sp PCC 6803 is dependent on the level of chlorophyll precursors regulated by ferrochelatase. J. Biol. Chem. 280, 31595–31602 10.1074/jbc.M505976200 [DOI] [PubMed] [Google Scholar]
- 32. Shipovskov S., Karlberg T., Fodje M., Hansson M. D., Ferreira G. C., Hansson M., Reimann C. T., and Al-Karadaghi S. (2005) Metallation of the transition-state inhibitor N-methyl mesoporphyrin by ferrochelatase: Implications for the catalytic reaction mechanism. J. Mol. Biol. 352, 1081–1090 10.1016/j.jmb.2005.08.002 [DOI] [PubMed] [Google Scholar]
- 33. Liu Z., Yan H., Wang K., Kuang T., Zhang J., Gui L., An X., and Chang W. (2004) Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 428, 287–292 10.1038/nature02373 [DOI] [PubMed] [Google Scholar]
- 34. Kořený L., Sobotka R., Janouškovec J., Keeling P. J., and Oborník M. (2011) Tetrapyrrole synthesis of photosynthetic chromerids is likely homologous to the unusual pathway of apicomplexan parasites. Plant Cell 23, 3454–3462 10.1105/tpc.111.089102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang J., Yan R., Roy A., Xu D., Poisson J., and Zhang Y. (2015) The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Medlock A., Swartz L., Dailey T. A., Dailey H. A., and Lanzilotta W. N. (2007) Substrate interactions with human ferrochelatase. Proc. Natl. Acad. Sci. U.S.A. 104, 1789–1793 10.1073/pnas.0606144104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dailey H. A., Wu C. K., Horanyi P., Medlock A. E., Najahi-Missaoui W., Burden A. E., Dailey T. A., and Rose J. (2007) Altered orientation of active site residues in variants of human ferrochelatase: evidence for a hydrogen bond network involved in catalysis. Biochemistry 46, 7973–7979 10.1021/bi700151f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sánchez-Baracaldo P., Raven J. A., Pisani D., and Knoll A. H. (2017) Early photosynthetic eukaryotes inhabited low-salinity habitats. Proc. Natl. Acad. Sci. U.S.A. 114, E7737–E7745 10.1073/pnas.1620089114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kilian O., Steunou A. S., Grossman A. R., and Bhaya D. (2008) A novel two domain-fusion protein in cyanobacteria with similarity to the CAB/ELIP/HLIP superfamily: evolutionary implications and regulation. Mol. Plant 1, 155–166 10.1093/mp/ssm019 [DOI] [PubMed] [Google Scholar]
- 40. Rochaix J. D., and Bassi R. (2019) LHC-like proteins involved in stress responses and biogenesis/repair of the photosynthetic apparatus. Biochem. J. 476, 581–593 10.1042/BCJ20180718 [DOI] [PubMed] [Google Scholar]
- 41. Niedzwiedzki D. M., Tronina T., Liu H., Staleva H., Komenda J., Sobotka R., Blankenship R. E., and Polívka T. (2016) Carotenoid-induced non-photochemical quenching in the cyanobacterial chlorophyll synthase–HliC/D complex. Biochim. Biophys. Acta 1857, 1430–1439 10.1016/j.bbabio.2016.04.280 [DOI] [PubMed] [Google Scholar]
- 42. Rast A., Heinz S., and Nickelsen J. (2015) Biogenesis of thylakoid membranes. Biochim. Biophys. Acta 1847, 821–830 10.1016/j.bbabio.2015.01.007 [DOI] [PubMed] [Google Scholar]
- 43. Uniacke J., and Zerges W. (2007) Photosystem II assembly and repair are differentially localized in Chlamydomonas. Plant Cell 19, 3640–3654 10.1105/tpc.107.054882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chidgey J. W., Linhartová M., Komenda J., Jackson P. J., Dickman M. J., Canniffe D. P., Koník P., Pilný J., Hunter C. N., and Sobotka R. (2014) A cyanobacterial chlorophyll synthase-HliD complex associates with the Ycf39 protein and the YidC/Alb3 insertase. Plant Cell 26, 1267–1279 10.1105/tpc.114.124495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bučinská L., Kiss É., Koník P., Knoppová J., Komenda J., and Sobotka R. (2018) The ribosome-bound protein Pam68 promotes insertion of chlorophyll into the CP47 subunit of photosystem II. Plant Physiol. 176, 2931–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Llansola-Portoles M. J., Sobotka R., Kish E., Shukla M. K., Pascal A. A., Polívka T., and Robert B. (2017) Twisting a β-carotene, an adaptive trick from nature for dissipating energy during photoprotection. J. Biol. Chem. 292, 1396–1403 10.1074/jbc.M116.753723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hontani Y., Kloz M., Polívka T., Shukla M. K., Sobotka R., and Kennis J. T. M. (2018) Molecular origin of photoprotection in cyanobacteria probed by watermarked femtosecond stimulated Raman spectroscopy. J. Phys. Chem. Lett. 9, 1788–1792 10.1021/acs.jpclett.8b00663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pan X., Li M., Wan T., Wang L., Jia C., Hou Z., Zhao X., Zhang J., and Chang W. (2011) Structural insights into energy regulation of light-harvesting complex CP29 from spinach. Nat. Struct. Mol. Biol. 18, 309–315 10.1038/nsmb.2008 [DOI] [PubMed] [Google Scholar]
- 49. Masoumi A., Heinemann I. U., Rohde M., Koch M., Jahn M., and Jahn D. (2008) Complex formation between protoporphyrinogen IX oxidase and ferrochelatase during haem biosynthesis in Thermosynechococcus elongatus. Microbiology 154, 3707–3714 10.1099/mic.0.2008/018705-0 [DOI] [PubMed] [Google Scholar]
- 50. Medlock A. E., Shiferaw M. T., Marcero J. R., Vashisht A. A., Wohlschlegel J. A., Phillips J. D., and Dailey H. A. (2015) Identification of the mitochondrial heme metabolism complex. PLoS One 10, e0135896 10.1371/journal.pone.0135896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Christodoulou D., Link H., Fuhrer T., Kochanowski K., Gerosa L., and Sauer U. (2018) Reserve flux capacity in the pentose phosphate pathway enables Escherichia coli's rapid response to oxidative stress. Cell Syst. 6, 569–578.e7 10.1016/j.cels.2018.04.009 [DOI] [PubMed] [Google Scholar]
- 52. Czarnecki O., and Grimm B. (2012) Post-translational control of tetrapyrrole biosynthesis in plants, algae, and cyanobacteria. J. Exp. Bot. 63, 1675–1687 10.1093/jxb/err437 [DOI] [PubMed] [Google Scholar]
- 53. Cornah J. E., Terry M. J., and Smith A. G. (2003) Green or red: what stops the traffic in the tetrapyrrole pathway? Trends Plant Sci. 8, 224–230 10.1016/S1360-1385(03)00064-5 [DOI] [PubMed] [Google Scholar]
- 54. Huang L., and Castelfranco P. A. (1990) Regulation of 5-aminolevulinic acid (ALA) synthesis in developing chloroplasts: evidence for functional-heterogeneity of the ALA pool. Plant Physiol. 92, 172–178 10.1104/pp.92.1.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Terry M. J., and Smith A. G. (2013) A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front. Plant Sci. 4 10.3389/fpls.2013.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Scharfenberg M., Mittermayr L., von Roepenack-Lahaye E., Schlicke H., Grimm B., Leister D., and Kleine T. (2015) Functional characterization of the two ferrochelatases in Arabidopsis thaliana. Plant Cell Environ. 38, 280–298 10.1111/pce.12248 [DOI] [PubMed] [Google Scholar]
- 57. Sobotka R. (2014) Making proteins green; biosynthesis of chlorophyll-binding proteins in cyanobacteria. Photosyn. Res. 119, 223–232 10.1007/s11120-013-9797-2 [DOI] [PubMed] [Google Scholar]
- 58. Kopečná J., Cabeza de Vaca I., Adams N. B., Davison P. A., Brindley A. A., Hunter C. N., Guallar V., and Sobotka R. (2015) Porphyrin binding to Gun4 protein, facilitated by a flexible loop, controls metabolite flow through the chlorophyll biosynthetic pathway. J. Biol. Chem. 290, 28477–28488 10.1074/jbc.M115.664987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schottkowski M., Ratke J., Oster U., Nowaczyk M., and Nickelsen J. (2009) Pitt, a novel tetratricopeptide repeat protein involved in light-dependent chlorophyll biosynthesis and thylakoid membrane biogenesis in Synechocystis sp. PCC 6803. Mol. Plant 2, 1289–1297 10.1093/mp/ssp075 [DOI] [PubMed] [Google Scholar]
- 60. Xiong W., Shen G., and Bryant D. A. (2017) Synechocystis sp. PCC 6803 CruA (sll0147) encodes lycopene cyclase and requires bound chlorophyll a for activity. Photosyn. Res. 131, 267–280 10.1007/s11120-016-0316-0 [DOI] [PubMed] [Google Scholar]
- 61. Zhao L., Cheng D., Huang X., Chen M., Dall'Osto L., Xing J., Gao L., Li L., Wang Y., Bassi R., Peng L., Wang Y., Rochaix J. D., and Huang F. (2017) A light harvesting complex-like protein in maintenance of photosynthetic components in Chlamydomonas. Plant Physiol. 174, 2419–2433 10.1104/pp.16.01465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tanaka R., Rothbart M., Oka S., Takabayashi A., Takahashi K., Shibata M., Myouga F., Motohashi R., Shinozaki K., Grimm B., and Tanaka A. (2010) LIL3, a light-harvesting-like protein, plays an essential role in chlorophyll and tocopherol biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 16721–16725 10.1073/pnas.1004699107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Takahashi K., Takabayashi A., Tanaka A., and Tanaka R. (2014) Functional analysis of light-harvesting-like protein 3 (LIL3) and its light-harvesting chlorophyll-binding motif in Arabidopsis. J. Biol. Chem. 289, 987–999 10.1074/jbc.M113.525428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lohscheider J. N., Rojas-Stütz M. C., Rothbart M., Andersson U., Funck D., Mendgen K., Grimm B., and Adamska I. (2015) Altered levels of LIL3 isoforms in Arabidopsis lead to disturbed pigment-protein assembly and chlorophyll synthesis, chlorotic phenotype and impaired photosynthetic performance. Plant Cell Environ 38, 2115–2127 10.1111/pce.12540 [DOI] [PubMed] [Google Scholar]
- 65. Mork-Jansson A., Bue A. K., Gargano D., Furnes C., Reisinger V., Arnold J., Kmiec K., and Eichacker L. A. (2015) Lil3 assembles with proteins regulating chlorophyll synthesis in barley. PLoS One 10, e0133145 10.1371/journal.pone.0133145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tichý M., Bečková M., Kopečná J., Noda J., Sobotka R., and Komenda J. (2016) Strain of Synechocystis PCC 6803 with aberrant assembly of photosystem II contains tandem duplication of a large chromosomal region. Front. Plant Sci. 7, 648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schägger H., and von Jagow G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 10.1016/0003-2697(91)90094-A [DOI] [PubMed] [Google Scholar]
- 68. Komenda J., Lupínková L., and Kopecký J. (2002) Absence of the psbH gene product destabilizes photosystem II complex and bicarbonate binding on its acceptor side in Synechocystis PCC 6803. Eur. J. Biochem. 269, 610–619 10.1046/j.0014-2956.2001.02693.x [DOI] [PubMed] [Google Scholar]
- 69. Pilný J., Kopečná J., Noda J., and Sobotka R. (2015) Detection and quantification of heme and chlorophyll precursors using a high performance liquid chromatography (HPLC) system equipped with two fluorescence detectors. Bio-protocol 5, e1390 [Google Scholar]
- 70. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., and Madden T. L. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10, 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Katoh K., and Standley D. M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shih P. M., Wu D., Latifi A., Axen S. D., Fewer D. P., Talla E., Calteau A., Cai F., Tandeau de Marsac N., Rippka R., Herdman M., Sivonen K., Coursin T., Laurent T., Goodwin L., et al. (2013) Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 110, 1053–1058 10.1073/pnas.1217107110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mareš J. (2018) Multilocus and SSU rRNA gene phylogenetic analyses of available cyanobacterial genomes, and their relation to the current taxonomic system. Hydrobiologia 811, 19–34 10.1007/s10750-017-3373-2 [DOI] [Google Scholar]
- 74. Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., Larget B., Liu L., Suchard M. A., and Huelsenbeck J. P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Miller M. A., Schwartz T., Pickett B. E., He S., Klem E. B., Scheuermann R. H., Passarotti M., Kaufman S., and O'Leary M. A. (2015) A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evol. Bioinform. Online 11, 43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.