Abstract
A point-of-care HIV-1 genotypic resistance assay that could be performed during a clinic visit would enable care providers to make informed treatment decisions for patients starting therapy or experiencing virologic failure on therapy. The main challenge for such an assay is the genetic variability at and surrounding each drug-resistance mutation (DRM). We analyzed a database of diverse global HIV sequences and used thermodynamic simulations to design an array of surface-bound oligonucleotide probe sets with each set sharing distinct 5′ and 3′ flanking sequences but having different centrally located nucleotides complementary to six codons at HIV-1 DRM reverse transcriptase position 103: AAA, AAC, AAG, AAT, AGA, and AGC. We then performed in vitro experiments using 80-mer oligonucleotides and PCR-amplified DNA from clinical plasma HIV-1 samples and culture supernatants that contained subtype A, B, C, D, CRF01_AE, and CRF02_AG viruses. Multiplexed solid-phase melt curve analysis discriminated perfectly among each of the six reported reverse transcriptase position 103 codons in both 80-mers and clinical samples. The sensitivity and specificity for detecting targets that contained AAC mixed with targets that contained AAA were >98% when AAC was present at a proportion of ≥10%. Multiplexed solid-phase melt curve analysis is a promising approach for developing point-of-care assays to distinguish between different codons in genetically variable regions such as those surrounding HIV-1 DRMs.
The increasing prevalence of HIV-1 drug resistance is a threat to HIV-1 treatment and prevention in the low- and middle-income countries (LMICs) hardest hit by the HIV-1 pandemic.1, 2 An inexpensive genotypic resistance test that could be performed during a clinic visit, heretofore called a point-of-care assay, would enable HIV care providers to rapidly make informed treatment decisions for antiretroviral-naive patients starting therapy and patients with virologic failure on therapy. Through the delivery of actionable information on detection of virologic failure, a point-of-care genotypic resistance test would increase the likelihood that treated patients achieve and maintain suppressed virus levels and would reduce treatment delivery costs, travel costs, and work absenteeism.
We previously reported that an assay that detected drug-resistance mutations (DRMs), including the nucleoside reverse transcriptase inhibitor (NRTI) DRMs K65R and M184V and the nonnucleoside RT inhibitor (NNRTI) DRMs K103N, V106M, Y181C, and G190A, would be highly sensitive for detecting intermediate/high-level acquired resistance in patients with virologic failure on a first-line NRTI/NNRTI regimen recommended by the World Health Organization and moderately sensitive for detecting intermediate/high-level transmitted drug resistance.3 Moreover, an assay that included each of the most common variants at these six DRM positions (K65R/N and M184V/I for NRTIs; K103N/S, V106A/M, Y181C/I/V, and G190A/S for NNRTIs) would have even greater sensitivity for both acquired and transmitted resistance.4 The main challenge to a successful point-of-care genotypic resistance test, however, is developing nucleic acid detection methods that have real-time or end-point PCR processes5, 6, 7 and offer DNA sequencing level sensitivity and specificity in the presence of genetic variability at and surrounding each DRM position.8
To address this challenge, we present here both in silico design methods and in vitro validation data from multiplexed experiments that demonstrated the accurate detection of DRMs in the HIV-1 RT gene in the presence of local sequence variability. A solid-phase nucleic acid melt curve analysis platform capable of measuring hybridization, including both capture and detachment (melt) of multiple nucleic acid targets to an array of multiple surface-bound oligonucleotide probes, was used. This study demonstrates how the multiplexing capability of this platform enables coverage of a large HIV sequence database without compromising specificity and sensitivity for six different codons at one key drug-resistance position. Finally, extensions that would be required to modify the multiplexed melt curve analysis platform used in this study to one suitable for point-of-care testing, including the use of a complementary metal oxide semiconductor (CMOS) biochip detection system, the HYDRA 1K (InSilixa, Inc., Sunnyvale, CA), have been discussed. This system, which has been used for the detection of a panel of upper respiratory viruses and of Mycobacterium tuberculosis DRMs, uses multiple integrated biosensors to perform PCR and hybridization detection in a closed-tube system without the need for an external light detector.9
Materials and Methods
Mutation Detection Platform
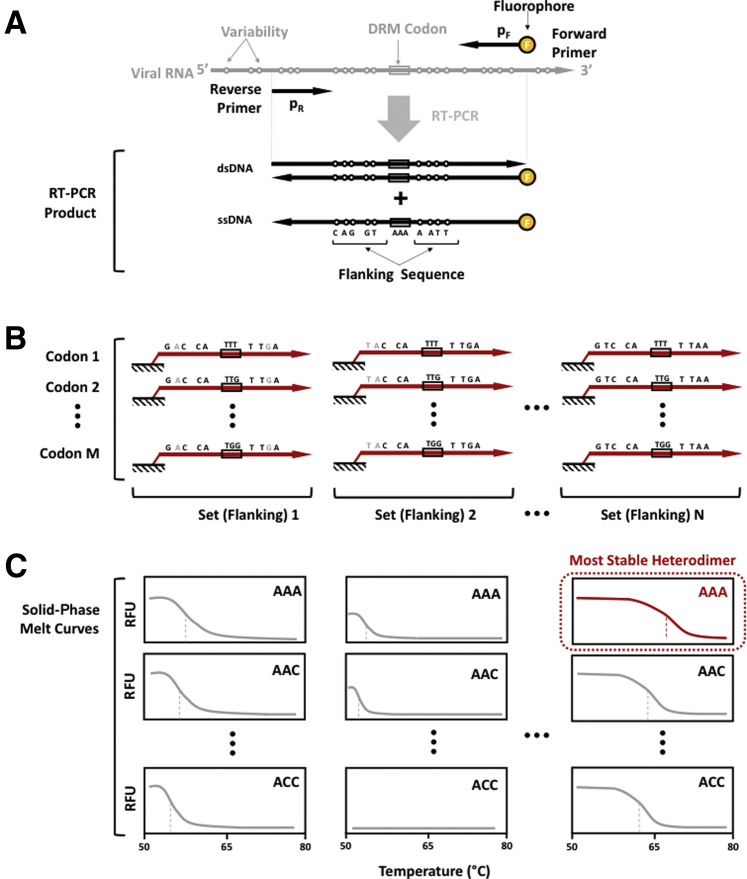
Mutation detection begins with the addition of fluorescently labelled DNA to an oligonucleotide microarray, which in this study, includes synthesized 80-mers and PCR-amplified cDNA from clinical plasma samples and culture supernatants (Figure 1A). Each probe in the array is designed to contain a distinct flanking sequence complementary to diverse global HIV-1 strains and a central nucleotide triplet complementary to each of the reported codons at a DRM position (Figure 1B). The probes are designed such that most published HIV-1 RT sequences that encompass the DRM position will form a stable heterodimer with one or more probes in the array. The concentrations of labeled DNA captured by the microarray's probes are measured in real time as the solution temperature gradually increases (Figure 1C). The time series data, which report the heterodimer duplex stability, are used to identify the correct codon at a DRM position.
Figure 1.
Nucleic acid detection methodology. A: Synthesized or amplified fluorescently labeled DNA was added to the oligonucleotide microarray. B: Labeled DNA hybridizes to microarray probe sets each sharing sequences with the same flanking region and a central nucleotide triplet complementary to each of the reported codons at a drug resistance mutation (DRM) position. C: Multiplexed melt curve analysis was used to identify the most stable heterodimer and thereby detect the correct codon at the DRM position. RFU, relative fluorescence unit.
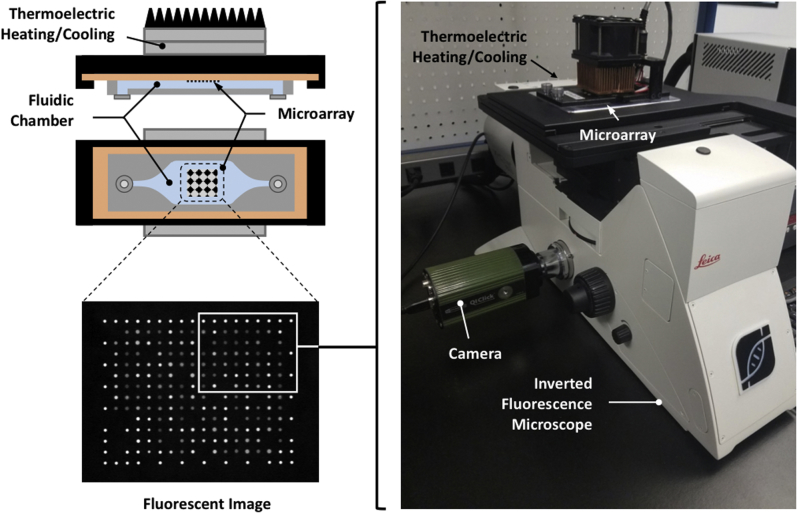
Figure 2 shows the custom real-time microarray slide structure and fluorescence microscope–based detection setup, including a Leica DMi8 microscope, a custom InSilixa slide stage with a Peltier heater, a light emitting diode source, and a charge-coupled device camera. This setup is the developer platform for the HYDRA-1K CMOS detection system that is used to test the performance of printed probe sets on silicon slides before printing them on CMOS chips.
Figure 2.
Detection platform. The instrument prototype included an inverted fluorescence microscope and a custom stage to place, heat, and image the oligonucleotide microarray. This platform was the developer platform for the HYDRA-1K system in that it used a silicon slide rather than a complementary metal oxide semiconductor chip and therefore required an external light detector.9
HIV Sequence Database and Target DRM Codon
To design probes for genotyping diverse viral isolates, a data set of >27,000 one-per-individual HIV-1 group M RT sequences belonging to multiple subtypes from six LMIC regions: southern, eastern, western, and central Africa and south and southeast Asia,4 were analyzed. This data set was used to enumerate the proportion of individuals with each RT 103 codon by subtype, and it was found that AAA (K), AAG (K), AAC (N), AAT (N), AGG (R), and AGC (S) accounted for 99.5% of all codons at this position (N and S reduce NNRTI susceptibility; K and R do not). This data set was also used to enumerate the proportions of all 5′ and 3′ flanking sequences in windows of various sizes. This study was approved by the Stanford University Institutional Review Board.
In Silico Probe Design
In silico thermodynamic simulations were performed to design isogenic probe sets that share the same 5′ and 3′ flanking sequences but have different centrally located nucleotides complementary to one of the six targeted DRM position codons (Figure 1). With the use of the Oligonucleotide Modeling Platform Developers Edition 1.0 software package (DNA Software Inc., Ann Arbor, MI), simulations were performed on 81,126 sequences: six codons × the 13,521 distinct 203-bp sequences (containing the 5′ and 3′ 100-bp flanking codon 103) in the LMIC data set. In each simulation, one probe per set was allowed to hybridize to one database-derived target over a range of temperatures from 10°C to 100°C. Because codon 103 variant AAA is the most commonly occurring, this analysis also assessed the discrimination by each isogenic probe of all non-AAA codon 103 variants from AAA.
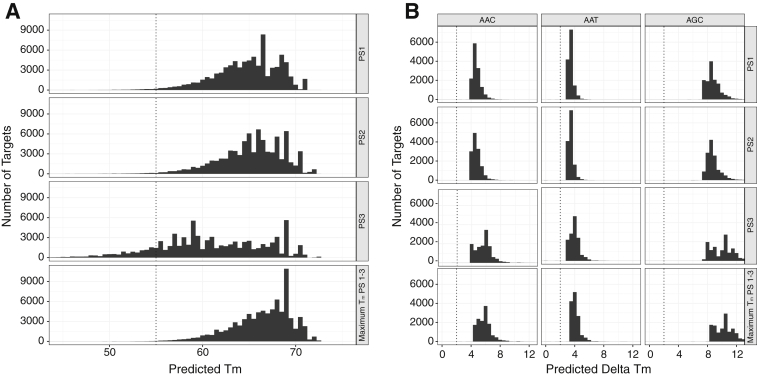
The simulations identified three sets of 34-bp probes, each containing the six RT position 103 codons but differing in their flanking regions (Table 1). One or more probe set was predicted to have a melting temperature (Tm) of ≥55°C for 99.7% of sequences (Figure 3A). One or more probe set was also predicted to have a ΔTm of ≥2°C for 99.8% of isogenic sequences compared with a sequence that contained the same flanking sequence but codon AAA at position 103 (Figure 3B). The simulations predicted that one or more of the probe sets would have a Tm ≥ 55°C for 98.5% of subtype G sequences, 99.0% of subtype A sequences, 99.5% of subtype B sequences, and ≥99.7% of sequences belonging to each of the remaining common subtypes, including subtypes C, D, CRF01_AE, and CRF02_AG and the pooled remaining subtypes and circulating recombinant forms (CRFs). One or more of the probe sets were predicted to have a Tm ≥ 55°C for 99.3% of sequences with AAT, 99.5% of sequences with AAA, and ≥99.8% of sequences with AAC, AAG, AGA, or AGC.
Table 1.
Probe Sets Used to Distinguish between Six Codons at HIV-1 RT Codon 103
| Probe set | Codon 103 | Probe sequence |
|---|---|---|
| 1 | AAA | linker-5′-GTACTGTTACTGATTTTTTCTTTTTTAAACCTGC-blocker |
| 1 | AAG | linker-5′-GTACTGTTACTGATTTCTTCTTTTTTAAACCTGC-blocker |
| 1 | AGA | linker-5′-GTACTGTTACTGATTTTCTCTTTTTTAAACCTGC-blocker |
| 1 | AAC | linker-5′-GTACTGTTACTGATTTGTTCTTTTTTAAACCTGC-blocker |
| 1 | AAT | linker-5′-GTACTGTTACTGATTTATTCTTTTTTAAACCTGC-blocker |
| 1 | AGC | linker-5′-GTACTGTTACTGATTTGCTCTTTTTTAAACCTGC-blocker |
| 2 | AAA | linker-5′-GTACTGTTACTGATTTTTTCTTTTTTAACCCTGC-blocker |
| 2 | AAG | linker-5′-GTACTGTTACTGATTTCTTCTTTTTTAACCCTGC-blocker |
| 2 | AGA | linker-5′-GTACTGTTACTGATTTTCTCTTTTTTAACCCTGC-blocker |
| 2 | AAC | linker-5′-GTACTGTTACTGATTTGTTCTTTTTTAACCCTGC-blocker |
| 2 | AAT | linker-5′-GTACTGTTACTGATTTATTCTTTTTTAACCCTGC-blocker |
| 2 | AGC | linker-5′-GTACTGTTACTGATTTGCTCTTTTTTAACCCTGC-blocker |
| 3 | AAA | linker-5′-GTACTGTCATTGATTTTTTCTTTTTTAACCCTGC-blocker |
| 3 | AAG | linker-5′-GTACTGTCATTGATTTCTTCTTTTTTAACCCTGC-blocker |
| 3 | AGA | linker-5′-GTACTGTCATTGATTTTCTCTTTTTTAACCCTGC-blocker |
| 3 | AAC | linker-5′-GTACTGTCATTGATTTGTTCTTTTTTAACCCTGC-blocker |
| 3 | AAT | linker-5′-GTACTGTCATTGATTTATTCTTTTTTAACCCTGC-blocker |
| 3 | AGC | linker-5′-GTACTGTCATTGATTTGCTCTTTTTTAACCCTGC-blocker |
Probe set 1 was complementary to the most commonly occurring flanking sequence in the low- and middle-income countries sequence data set. Probe sets 2 and 3 differed from probe set 1 by one and three nucleotides, respectively, shown in bold. The central codon position was complementary to the six most commonly reported position 103 codons.
Figure 3.
In silico simulations for probe set selection. A: Distribution of predicted melting temperature (Tm) for 81,126 sequences with three 34-bp probe sets (PS1, PS2, and PS3). Sequences included the 13,521 distinct low- and middle-income country 203-bp database sequences that encompassed codon 103 each with a different one of the six reported codons at position 103 (AAA, AAC, AAG, AAT, AGA, AGC). For each of the 81,126 (13,521 × 6) sequences, max_Tm_PS1-3 indicated the maximum Tm of PS1, PS2, and PS3. The temperature used for initial hybridization was 55°C. Dashed lines are at 55°C, the temperature at which binding is considered necessary for probe capture. B: Distribution of predicted ΔTm between probes complementary to AAC, AAT, and AGC versus probes complementary to AAA for each of the three probe sets (PS1, PS2, and PS3). Because the probes within a probe set shared the same flanking sequence, the ΔTm was caused entirely by the central mismatch at the codon 103 position. Dashed lines are at a ΔTm of 2°C, the minimum temperature considered necessary for distinguishing between the finding of two different targets to the same probe.
Array Fabrication and Probe Set Printing
Silicon substrates (25 × 76 × 0.625 mm with 3000 Å thermal SiO2) from Addison Engineering (San Jose, CA) were treated with i) oxygen plasma on a float plate at 600W, 1 Torr, 120°C for 3 minutes; ii) water vapor at 13 Torr, 120°C for 5 minutes; and iii) gaseous (3-glycidoxypropyl) trimethoxysilane (Gelest, Morrisville, PA) at 0.5 Torr, 120°C for 30 minutes. These process steps were serially performed, without breaking vacuum, on a model 1224P chemical vapor deposition instrument (Yield Engineering Systems, Livermore, CA). Coated substrates were cooled to room temperature and stored at 20% relative humidity under N2 until use.
High-performance liquid chromatography-purified capture probes that bear hexylamine and 6T at the 5′-terminus and a three-carbon blocker at the 3′-terminus (LGC Biosearch, Petaluma, CA) were reconstituted to 50 μmol/L in pH 8.5 phosphate buffer. Piezo type printing with a sciFLEXARRAYER SX (Scienion, Berlin, Germany) was used to deposit approximately 100-pL droplets at 200-μm pitch onto coated substrates in an 18 × 14 grid pattern (252 spots) with each probe represented at least five times. Covalent coupling of the probe to the substrate was accomplished with overnight incubation at 50% room humidity. Printed substrates were subsequently treated with excess Tris-buffered solution that contained ethanolamine to deactivate remaining epoxy groups and to remove non-immobilized probe. A flow-through laminated acrylic fluidic cap with an inlet and outlet (approximately 90 μL of total capacity) was bonded to the substrate with silicone-based pressure-sensitive adhesive to create the functional module. Finished arrays were stored in a desiccated N2 environment until use.
Development of an Asymmetric PCR Protocol
A 301-bp region that encompassed codon 103 was selected, and all corresponding database sequences from LMICs were analyzed to design primers that would produce amplicons of 100 to 200 bp. Oligonucleotide Modeling Platform Developers Edition was used to identify binding regions of relatively low-sequence entropy to ensure high complementarity to the 3′ extension terminus of candidate primers and to optimize primer design.10 Six combinations of three forward and two reverse primer candidates were selected for conventional solution-based real-time quantitative PCR standard curve experiments that used synthetic DNA gBlock templates. The primer pair with the highest efficiency and largest Cq value for the no-template control was selected for PCR: 5′-cyanine 3-TTYTGGGARGTYCAAYTAGGRATAC-3′ (forward, excess primer) and 5′-TCTAAAGGGACTGAGAAATATGCAT-3′ (reverse).
Synthesized 80-mer Targets
A set of 18 cyanine 3-labeled 80-bp HIV-1 targets (Integrated DNA Technologies, Redwood City, CA) that contained a 34-bp region complementary to each probe was synthesized. Although each target was perfectly complementary to one probe, it differed from the other probes by 1, 2, or 3 bp in its flanking sequence and by 1 or 2 bp at codon 103. Eighty-one experiments were performed with the 80-bp targets. Each of the 18 targets was tested in triplicate in 54 experiments, and each of the three targets with AAC was diluted to 50%, 25%, and 10% with targets that contained AAA and was tested in triplicate in 27 experiments (Table 2).
Table 2.
Complete List of the 80-bp Targets Used in 81 Experiments Designed to Determine the Correct Codon at RT Position 103
| Codon∗ | Flank∗ | Percentage, %† | Replicates, n | Total, n |
|---|---|---|---|---|
| AAA | 1, 2, 3 | 100 | 3 | 9 |
| AAC | 1, 2, 3 | 100 | 3 | 9 |
| AAG | 1, 2, 3 | 100 | 3 | 9 |
| AAT | 1, 2, 3 | 100 | 3 | 9 |
| AGA | 1, 2, 3 | 100 | 3 | 9 |
| AGC | 1, 2, 3 | 100 | 3 | 9 |
| AAC | 1, 2, 3 | 50 | 3 | 9 |
| AAC | 1, 2, 3 | 25 | 3 | 9 |
| AAC | 1, 2, 3 | 10 | 3 | 9 |
Targets are characterized by their codon 103 nucleotides, their flanking sequence, and percentage of purity. The results of these experiments are shown in Figures 5, 6, and 7, Supplemental Figures S1–S3, and Table 3.
Codon and 31-bp flanking sequence in the 80-mer targets.
In mixture experiments, AAC was diluted with AAA.
Clinical Samples
To evaluate the performance of the melt curve analysis on viruses with diverse sequences, plasma HIV-1 samples and culture supernatants that contained each of the six position 103 codons and that belonged to different subtypes and CRFs were identified. The plasma samples were previously sequenced at the Stanford HealthCare Diagnostic Virology Laboratory (Stanford, CA), and the culture supernatants were obtained from the Duke University External Quality Assurance Program Oversight Laboratory (Durham, NC) panel.11 RNA was extracted from 200 μL of plasma (or culture supernatant), using QIAamp MinElute Virus Spin Kit (Qiagen, Beverly, MA). Twenty microliters of eluted product was then reverse transcribed, using EnzScript (Qiagen), an RNaseH-free RT enzyme, at 50°C for 5 minutes and was performed with the reverse primer described in the next paragraph.
Asymmetric PCR of cDNA generated by RT of RNA extracted from clinical plasma samples and cultured supernatants was performed in four 25-μL reactions that used Phoenix Hot Start Taq Polymerase (Enzymatics, Beverly, MA), 300 nmol/L excess primer, 50 nmol/L limiting primer, and 5 μL of template. Amplification was performed on a CFX96 Touch (Bio-Rad, Hercules, CA) 94°C for 5 minutes, followed by 45 cycles of 95°C (15 seconds) and 60°C (30 seconds).
Hybridization/Melting
For each experiment, 100 nmol/L of the 80-mer targets or 90 μL of PCR-amplified DNA was injected onto a capped silicon substrate slide bearing the oligonucleotide capture probes. The injected DNA was then allowed to hybridize with the probes attached to the silicon slide surface for 1 hour at 55°C. Slides were then washed with buffer to remove unhybridized target. Hybridized target was then melted off the capture probes during a 20-minute period as the temperature was raised from 45°C to 90°C. Serial images were taken every 10 seconds. The fluorescent intensity of each feature over time was processed to generate the melt curve for each probe.
Melt Curve Analysis
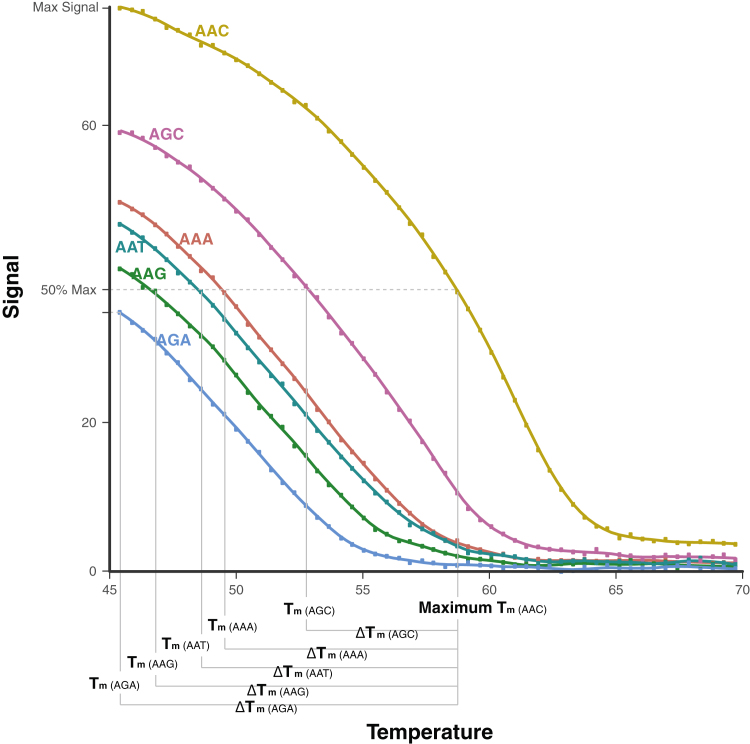
Fluorescent signal intensities recorded for each probe were the average of the quintuplicate signals obtained from each slide feature with that probe. One-hundred twenty signals were obtained per probe during the 20-minute melting period, yielding 2160 signals (18 probes × 120 signals) per experiment. For each probe, the signal intensity versus temperature was plotted to determine the probe's Tm, or temperature at which the probe's signal intensity was 50% of that of the probe with the highest signal intensity in the same probe set (Figure 4). Probes that were <50% hybridized at 45°C were assigned a Tm of 45°C.
Figure 4.
Melting temperature (Tm), maximum Tm, ΔTm. The Tm was defined as the temperature at which the probe's fluorescent signal intensity was 50% of that of the probe with the highest signal intensity in the same probe set. Probes that were <50% hybridized at 45°C were assigned a Tm of 45°C. For each probe set, the maximum Tm was defined as the highest Tm of the six probes (one for each K103 codon) with the same flanking sequence. The ΔTm was defined as the absolute value of the difference between the maximum Tm for the probe set to which the probe belonged and the Tm of the probe. By definition, the probe with the maximum Tm has a ΔTm of 0°C, whereas the remaining five probes in a set would have a ΔTm > 0°C. In each experiment, the mean and median ΔTm across each of the three probe sets were calculated for each of the six codon 103 variants.
For each probe set, the maximum Tm was defined as the highest Tm of the six probes (one for each K103 codon) that shared the same flanking sequence (Figure 4). Thus, for each experiment, three maximum Tms were calculated. A ΔTm for each of the 18 probes, defined as the absolute value of the difference between the maximum Tm in a probe set and the Tm of each probe in the set, was then calculated. By definition, the probe with the maximum Tm would have a ΔTm of 0°C, whereas the remaining five probes in a set would have a ΔTm >0°C. In each experiment, the mean and median ΔTm were then calculated across each of the three probe sets for each of the six codon 103 variants.
For the analysis of experiments that contained mixtures of targets, receiver operating characteristic (ROC) curves were generated for the proportion of each target to determine the area under the ROC curve, optimal mean ΔTm cutoffs, and sensitivity and specificity associated with these cutoffs.12
Results
Single 80-mer Targets
Each of the 18 distinct 80-bp targets (six codons × three flanking sequences) were tested in triplicate, yielding 54 experiments (Table 2); each experiment yielded 18 melt curves (one per probe). Supplemental Figure S1 shows the results of the first of three replicate experiments for 9 of the 18 targets: targets that contained each of the three flanking sequences (1, 2, and 3) and three of the six codons: AAA (wild-type K), AAC (DRM N), and AGA (polymorphic variant R). Here, the x axis represented the temperature and the y axis represented the fluorescence signal. Supplemental Figure S2 shows the remaining 27 experiments performed on 80-mer targets that contained each of the three flanking sequences and codons AAG (wild-type K), AAT (DRM N), and AGC (DRM S).
Each row in these figures contained three experiments with different targets and each experiment contained three panels, one per probe set. In the first experiment in each row, the target was perfectly complementary to probe set 1 and contained one and three differences from probe sets 2 and 3, respectively (Table 1). In the second experiment, the target was perfectly complementary to probe set 2 and contained one and two differences from probe sets 1 and 3, respectively. In the third experiment, the target was perfectly complementary to probe set 3 and contained three and two differences from probe sets 1 and 2, respectively. Regardless of the number of flanking sequence mismatches between the probe and target, the maximum Tm occurred when the probe and target had the same codon.
The triplicate experiments for each target demonstrated high interexperiment reproducibility of the maximum Tm for each probe set (Table 3). The median coefficient of variation for the maximum Tm for a probe set across experiments was 1.68% (range, 0.5% to 5.3%). On average, the maximum Tm was inversely proportional to the number of mismatches between the target and probe flanking sequence. The mean maximum Tm was 62.1°C when there was an exact match between target and probe flanking sequence, 60.1°C when there was one mismatch (flanking sequences 1 versus 2), 59.1°C when there were two mismatches (flanking sequences 2 versus 3), and 55.0°C when there were three mismatches (flanking sequences 1 versus 3). The mean maximum Tm for probe sets in which the target and probes had the same flanking and codon sequences was lowest for AAA (60.5°C) and highest for AGC (63.6°C).
Table 3.
Influence of Target-Probe Flanking Sequence Mismatches on Hybridization for the Targets and Probes with Matching Codons: Reproducibility of Maximum Tm Across Triplicate Experiments
| Flanking sequence∗ |
Codon | Maximum Tm (replicate experiments), °C |
Mean, °C | SD, °C | CV, % | ||||
|---|---|---|---|---|---|---|---|---|---|
| Target | Probe | Target-probe mismatches, n | R1 | R2 | R3 | ||||
| 1 | 1 | 0 | AAA | 60.8 | 61.0 | 61.4 | 61.1 | 0.3 | 0.5 |
| 1 | 1 | 0 | AAC | 60.3 | 61.6 | 64.0 | 61.9 | 1.9 | 3.0 |
| 1 | 1 | 0 | AAG | 58.5 | 60.2 | 61.9 | 60.2 | 1.7 | 2.9 |
| 1 | 1 | 0 | AAT | 61.2 | 61.4 | 61.6 | 61.4 | 0.2 | 0.4 |
| 1 | 1 | 0 | AGA | 60.0 | 60.7 | 61.0 | 60.5 | 0.5 | 0.9 |
| 1 | 1 | 0 | AGC | 64.1 | 63.4 | 63.0 | 63.5 | 0.6 | 0.9 |
| 1 | 2 | 1 | AAA | 59.3 | 59.3 | 59.9 | 59.5 | 0.4 | 0.6 |
| 1 | 2 | 1 | AAC | 58.5 | 60.0 | 62.3 | 60.3 | 1.9 | 3.1 |
| 1 | 2 | 1 | AAG | 57.8 | 58.8 | 60.7 | 59.1 | 1.5 | 2.5 |
| 1 | 2 | 1 | AAT | 60.4 | 59.4 | 59.6 | 59.8 | 0.5 | 0.9 |
| 1 | 2 | 1 | AGA | 58.1 | 59.2 | 57.5 | 58.3 | 0.8 | 1.4 |
| 1 | 2 | 1 | AGC | 63.5 | 62.3 | 62.0 | 62.6 | 0.8 | 1.3 |
| 1 | 3 | 3 | AAA | 51.7 | 52.3 | 52.7 | 52.2 | 0.5 | 0.9 |
| 1 | 3 | 3 | AAC | 51.8 | 53.2 | 55.2 | 53.4 | 1.7 | 3.3 |
| 1 | 3 | 3 | AAG | 50.5 | 52.8 | 53.3 | 52.2 | 1.5 | 2.9 |
| 1 | 3 | 3 | AAT | 53.8 | 51.8 | 52.5 | 52.7 | 1.0 | 1.9 |
| 1 | 3 | 3 | AGA | 50.8 | 51.8 | 52.6 | 51.8 | 0.9 | 1.7 |
| 1 | 3 | 3 | AGC | 56.9 | 56.6 | 55.5 | 56.4 | 0.7 | 1.3 |
| 2 | 1 | 1 | AAA | 59.3 | 60.9 | 60.4 | 60.2 | 0.8 | 1.4 |
| 2 | 1 | 1 | AAC | 61.8 | 62.4 | 62.0 | 62.1 | 0.3 | 0.5 |
| 2 | 1 | 1 | AAG | 59.5 | 60.6 | 61.3 | 60.5 | 0.9 | 1.6 |
| 2 | 1 | 1 | AAT | 58.5 | 59.8 | 60.2 | 59.5 | 0.9 | 1.5 |
| 2 | 1 | 1 | AGA | 60.4 | 59.0 | 57.7 | 59.1 | 1.3 | 2.8 |
| 2 | 1 | 1 | AGC | 61.2 | 59.8 | 61.0 | 60.7 | 0.8 | 1.3 |
| 2 | 2 | 0 | AAA | 60.9 | 62.1 | 62.5 | 61.8 | 0.8 | 1.3 |
| 2 | 2 | 0 | AAC | 63.9 | 64.5 | 63.7 | 64.0 | 0.4 | 0.6 |
| 2 | 2 | 0 | AAG | 61.3 | 62.5 | 63.4 | 62.4 | 1.1 | 1.7 |
| 2 | 2 | 0 | AAT | 60.0 | 61.2 | 61.9 | 61.0 | 1.0 | 1.6 |
| 2 | 2 | 0 | AGA | 61.8 | 60.5 | 58.6 | 60.3 | 1.6 | 2.6 |
| 2 | 2 | 0 | AGC | 63.1 | 61.3 | 63.1 | 62.5 | 1.0 | 1.7 |
| 2 | 3 | 2 | AAA | 57.1 | 58.6 | 58.6 | 58.1 | 0.9 | 1.5 |
| 2 | 3 | 2 | AAC | 60.1 | 60.8 | 60.4 | 60.5 | 0.3 | 0.5 |
| 2 | 3 | 2 | AAG | 58.4 | 59.1 | 59.8 | 59.1 | 0.7 | 1.2 |
| 2 | 3 | 2 | AAT | 56.5 | 57.7 | 57.6 | 57.3 | 0.7 | 1.2 |
| 2 | 3 | 2 | AGA | 58.5 | 56.6 | 56.5 | 57.2 | 1.1 | 2.0 |
| 2 | 3 | 2 | AGC | 59.9 | 58.7 | 59.9 | 59.5 | 0.7 | 1.2 |
| 3 | 1 | 3 | AAA | 57.1 | 59.0 | 55.3 | 57.1 | 1.8 | 3.2 |
| 3 | 1 | 3 | AAC | 56.9 | 57.0 | 57.8 | 57.2 | 0.5 | 0.9 |
| 3 | 1 | 3 | AAG | 54.8 | 57.8 | 57.7 | 56.8 | 1.7 | 3.0 |
| 3 | 1 | 3 | AAT | 54.1 | 53.0 | 60.7 | 55.9 | 4.1 | 7.4 |
| 3 | 1 | 3 | AGA | 54.8 | 54.8 | 58.3 | 56.0 | 2.0 | 3.5 |
| 3 | 1 | 3 | AGC | 55.7 | 57.4 | 61.4 | 58.2 | 2.9 | 5.0 |
| 3 | 2 | 2 | AAA | 59.5 | 61.2 | 57.9 | 59.5 | 1.7 | 2.8 |
| 3 | 2 | 2 | AAC | 59.4 | 59.8 | 60.1 | 59.7 | 0.4 | 0.6 |
| 3 | 2 | 2 | AAG | 58.2 | 59.8 | 60.7 | 59.6 | 1.2 | 2.1 |
| 3 | 2 | 2 | AAT | 57.2 | 55.4 | 62.7 | 58.4 | 3.8 | 6.5 |
| 3 | 2 | 2 | AGA | 58.3 | 58.3 | 60.6 | 59.1 | 1.3 | 2.3 |
| 3 | 2 | 2 | AGC | 58.6 | 60.4 | 64.5 | 61.2 | 3.0 | 4.9 |
| 3 | 3 | 0 | AAA | 63.2 | 61.9 | 58.3 | 61.1 | 2.6 | 4.2 |
| 3 | 3 | 0 | AAC | 63.2 | 63.7 | 63.7 | 63.6 | 0.3 | 0.5 |
| 3 | 3 | 0 | AAG | 62.0 | 63.3 | 64.3 | 63.2 | 1.2 | 1.8 |
| 3 | 3 | 0 | AAT | 61.5 | 59.5 | 65.6 | 62.2 | 3.1 | 4.9 |
| 3 | 3 | 0 | AGA | 61.7 | 60.8 | 63.7 | 62.1 | 1.5 | 2.4 |
| 3 | 3 | 0 | AGC | 62.0 | 63.8 | 68.6 | 64.8 | 3.4 | 5.3 |
CV, coefficient of variation; Tm, melting temperature.
In all cases maximum Tm occurred when the target codon matched the probe codon. The flanking sequence used for each of the three probe sets is shown in Table 1. Flanking sequence 1 differs from flanking sequence 2 by one nucleotide; flanking sequence 2 differs from flanking sequence 3 by two nucleotides; and flanking sequence 1 differs from flanking sequence 3 by three nucleotides.
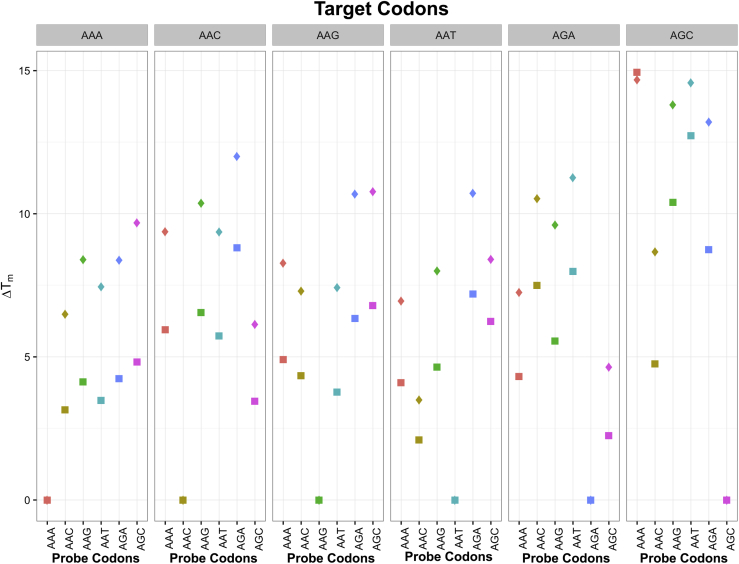
Figure 5 summarizes ΔTms by target codon, probe codon, and extent of mismatch between target and probe flanks. The strongest binding between target and probe occurred when the target and probe shared the same codon (ΔTm = 0). When the target and probe had different codons, the ΔTm varied from 2.3°C to 16.8°C. For the mismatched codons, the ΔTms were nearly always lower when the target and probe shared the same flanking sequence than when they differed by one to three nucleotides. Thus, the probes with flanking sequences that did not exactly match the target flanking sequence provided higher ΔTms and the most discriminatory information.
Figure 5.
Change in melting temperatures (ΔTms) according to target codon, probe codon, and the extent of mismatch between the target and probe flanking sequences in the 54 experiments with unmixed 80-mer targets. Each panel represents nine experiments with a target that contained a different codon. The target codon is indicated above each panel. The complementary codon for each probe is indicated vertically in plain text below each panel. The y axis shows the mean ΔTm for each of the three replicate experiments. For each combination of target and probe codon, data were obtained from a probe set in which the target and probe flanks were identical (squares) or differed by 1 to 3 nucleotides (diamonds). The strongest binding between target and probe occurred when the target and probe shared the same codon (ΔTm = 0), regardless of the extent of mismatch between the target and probe flanking sequences. Indeed, the squares and diamonds overlap whenever Tm was 0. However, in all but one case, the ΔTms for the mismatched codons were lower when the target and probe shared the same flanking sequence (squares), suggesting that the probes with flanking sequences that do not exactly match the target flanking sequence provided the most discriminatory information.
Mixtures of 80-mer Targets
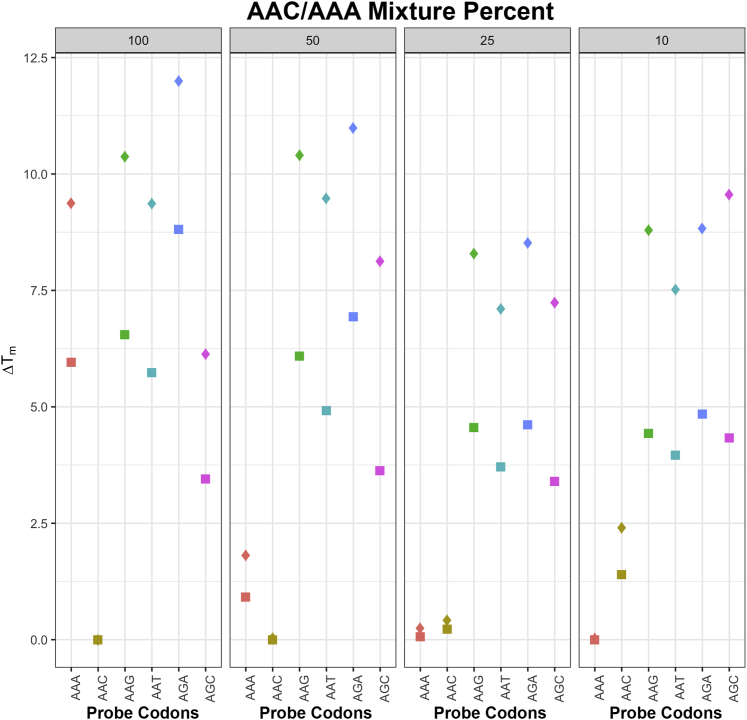
Targets that contained AAC and each of the three flanking sequences were diluted to 50%, 25%, and 10% with targets that contained AAA for a total of nine mixtures (three mixture proportions × three flanking sequences) (Table 2). Supplemental Figure S3 shows the first set of each replicate experiment in which AAC was mixed with AAA. For each probe set in each target mixture experiment, the probes complementary to AAA and AAC had the two highest Tms.
Figure 6 summarizes the ΔTms for each mixture experiment (50, 25, 10) displayed adjacent to the experiments with unmixed (100) AAC targets. Each point indicates the mean ΔTm of the three replicate experiments. For each combination of target and probe codon, data were obtained from a probe set in which the target and probe flanks were identical or differed by 1 to 3 nucleotides. For the nine distinct mixture experiments, the probes complementary to AAA and AAC consistently had the two lowest ΔTms, regardless of the extent of flanking sequence mismatch. Moreover, the probes with flanking sequences that did not exactly match the target flanking sequence provided higher ΔTms and the most discriminatory information in these experiments as well as those in with the unmixed targets.
Figure 6.
Change in melting temperatures (ΔTms) for each of the 27 mixture experiments according to the AAC dilution. The four panels each represent nine experiments. For comparison, the leftmost panel represents the experiments with the undiluted AAC target. The three rightmost panels each represent experiments with a different AAC/AAA dilution (50%, 25%, 10%). The AAC/AAA mixture percentage is indicated above each panel. The complementary codon for each probe is indicated below each panel vertically in plain text. The y axis shows the mean ΔTm for each of the three replicate experiments. For each combination of target codon and probe codon, data were obtained from a probe set in which the target and probe flanks were identical (squares) or differed by one to three nucleotides (diamonds). A 50% AAC/50% AAA mixture yielded lower ΔTms for probes complementary to AAC than those complementary to AAA, consistent with the stronger binding between C and G than between A and T at the codon's third position. At the 25% AAC/75% AAA mixture, the ΔTms for the probes complementary to AAC and AAA were both approximately 0°C and were >3°C lower than those of any other probes. At the 10% AAC/90% AAA mixture, the ΔTms for the probes complementary to AAA were 0°C, whereas those complementary to AAC were >0°C but lower than those of probes complementary to the other four codons.
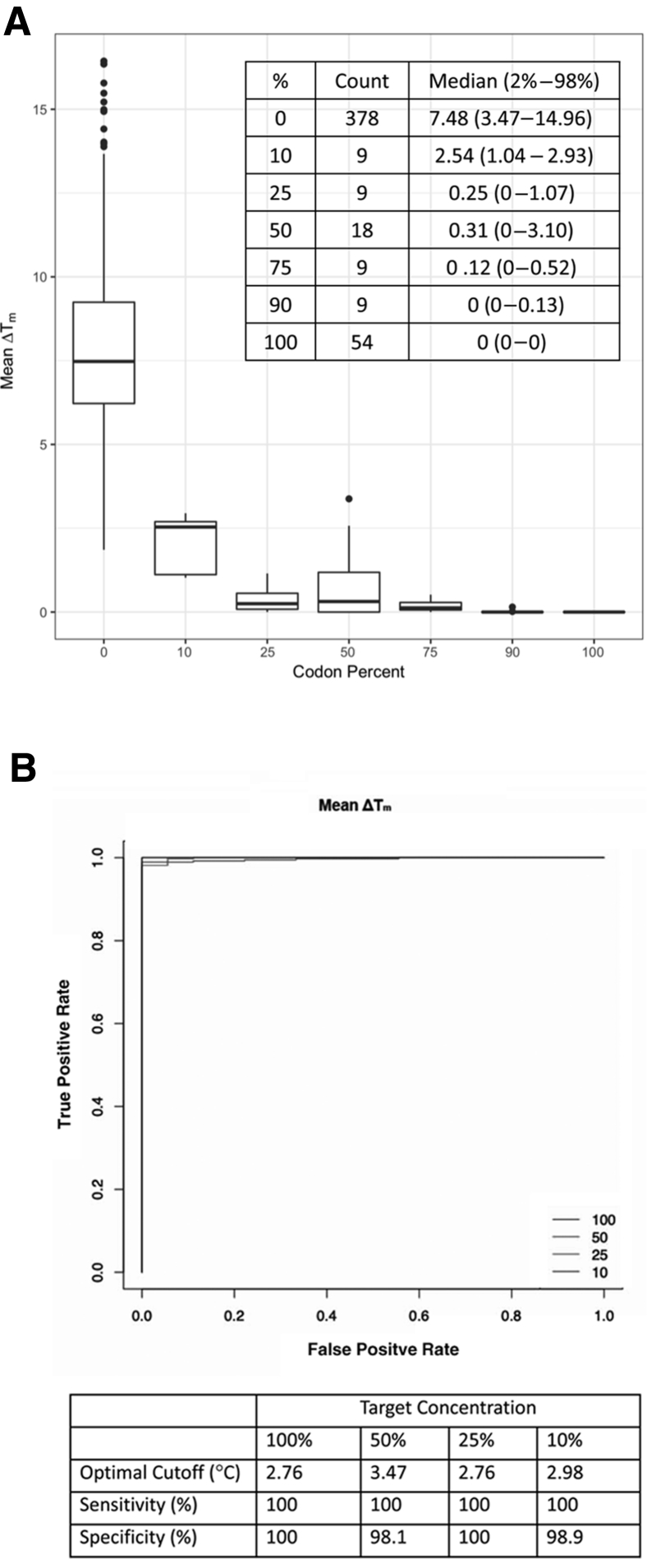
Figure 7A summarizes the distribution of the 486 mean ΔTms (81 experiments × six probes) defined as the mean ΔTm of each probe across the three probe sets in each experiment according to the percentage of a codon in experimental targets: 100% for targets that contained undiluted codons; 50%, 25%, and 10% for diluted AAC targets; 50%, 75%, and 90% for AAA targets used to dilute AAC; and 0% for absent codons. These experiments demonstrated little overlap in the mean ΔTm as determined by probes complementary to a target codon present at ≥10% compared with probes not complementary to a target's codon. The ROC curves showed that the area under the ROC curve was >0.98 for ΔTm at all target codon percentages (Figure 7B). The tabular insets indicate the sensitivity and specificity of the assay were >98% at the optimal cutoffs of 2.76°C to 3.47°C found to separate present and absent codons.
Figure 7.
Distribution of the mean change in melting temperatures (ΔTms) for each of the six probes in each of the 81 experiments (A) and receiver operating characteristic (ROC) curve analysis (B). A: The mean ΔTm for a probe in an experiment was the mean values for a probe in each of the three probe sets. The distribution of these values was plotted according to the percentage of a codon in the experimental targets (100% for undiluted targets; 50%, 25%, and 10% for diluted AAC targets; and 50%, 75%, and 90% for AAA targets used to dilute AAC). Median ΔTm for each codon dilution is shown. B: The ROC curves for targets at a concentration of 10%, 25%, 50%, and 100%. The sensitivity and specificity for detecting variant codons at these concentrations at optimized thresholds are shown.
Experiments Using Plasma Virus Samples and Culture Supernatants
Table 4 summarizes the melt curve analysis of 13 plasma virus samples and two culture supernatants that underwent RNA extraction, reverse transcription, and asymmetric PCR. There were six subtype B samples that contained five codons (AAA, AAC, AAT, AGA, and AGC), four subtype C samples that contained two codons (AAA and AAC), two CRF01_AE samples that contained two codons (AAC and AAG), and three samples that belonged to subtypes A, D, and CRF02_AG, each containing AAA.
Table 4.
Genotyping of RT Position 103 Using Plasma Virus Samples and Cultured Virus Supernatants: Melt Curve Analysis of SS-DNA Generated from Asymmetric PCR of Reverse-Transcribed Extracted RNA
| Sample∗ | Subtype | Codon | Mismatches PS1-PS2-PS3† | ΔTm‡ |
|||
|---|---|---|---|---|---|---|---|
| PS1 | PS2 | PS3 | Mean | ||||
| 7908 | B | AAA | 2-1-3 | 0 (16.3) | 0 (7.9) | 0 (14.3) | 0 (12.8) |
| 26339 | B | AAA | 4-3-4 | — | 0 (17.0) | — | 0 (17.0) |
| 112986 | B | AAC | 1-1-3 | 0 (16.1) | 0 (16.4) | — | 0 (16.2) |
| 175042 | B | AAT | 1-0-2 | 0 (3.9) | 0 (1.6) | 0 (16.2) | 0 (7.2) |
| 175881 | B | AGC | 4-3-5 | 0 (16.8) | 0 (17.1) | 0 (6.0) | 0 (13.3) |
| 220161 | B | AGA | 2-1-3 | 0 (15.2) | 0 (15.0) | 0 (11.2) | 0 (13.5) |
| 175872 | A | AAA | 2-3-5 | 0 (14.5) | 0 (5.3) | — | 0 (9.9) |
| 139002 | C | AAA | 3-2-2 | 0 (5.6) | 0 (8.3) | 0 (8.2) | 0 (7.3) |
| 139189 | C | AAA | 3-2-4 | 0 (13.4) | 0 (15.4) | 0 (7.1) | 0 (12.0) |
| 175134 | C | AAC | 2-1-1 | 0 (16.3) | 0 (18.8) | 0 (18.9) | 0 (18.0) |
| 57384 | C | AAC | 3-2-2 | 0 (13.4) | 0 (15.2) | 0 (15.2) | 0 (14.6) |
| JX140670 | D | AAA | 4-3-5 | 0 (13.4) | 0 (14.7) | — | 0 (14.0) |
| 175289 | CRF01 | AAC | 0-1-3 | 0 (8.9) | 0 (7.1) | 0 (6.3) | 0 (7.8) |
| 220168 | CRF01 | AAG | 3-3-5 | 0 (4.3) | 0 (4.4) | — | 0 (4.3) |
| JX140646 | CRF02 | AAA | 2-2-4 | 0 (7.0) | 0 (5.9) | 0 (6.8) | 0 (6.6) |
PS, probe set; —, probe sets not hybridizing with the PCR product; ΔTm, change in melting temperature.
JX140670 and JX140646 were cultured virus supernatants from the Duke University External Quality Assurance Program Oversight Laboratory sample collection.
Number of mismatches in the flanking sequence for each of the three PSs in the plasma virus: PS1-PS2-PS3.
ΔTm is the difference between the Tm of the probe complementary to the codon in the third column and the Tm of the probe with the highest Tm (maximum Tm) for PS1, PS2, PS3, and the mean of PS1, PS2, and PS3. In all cases, ΔTm was 0°C because the Tm of the probe with the complementary codon was the highest within the probe set. The parentheses contain the ΔTm for the probe with the second lowest ΔTm (ie, the second-best match).
Despite the marked variability of the flanking sequences surrounding position 103 (ie, having up to four or five mismatches from the flanking regions of one or more probes), the correct codon was identified in all assays with the probe with the second best having a mean ΔTm that ranged between 4.3°C and 18.8°C. Melt curve analysis yielded results for 21 of 21 probes that contained two or fewer flanking sequence differences from the clinical virus samples, 12 of 13 probes that contained three flanking sequence differences, four of six probes that contained four flanking sequence differences, and one of four probes that contained five flanking sequence differences.
Discussion
The thermodynamic simulations described in this study predicted that three probe sets, each sharing the same flanking sequences but containing probes complementary to a different codon, would successfully genotype RT position 103 in >99% of a large set of diverse global HIV-1 sequences. The in vitro experiments demonstrated that a multiplexed solid-phase melt curve analysis platform that used these probe sets was capable of discriminating perfectly among the six codons at RT position 103 in undiluted oligonucleotide targets, despite surrounding sequence variability.
Experiments that used PCR-amplified DNA from 15 clinical plasma HIV-1 samples and culture supernatants that contained viruses belonging to diverse subtypes also demonstrated the ability of the platform to successfully genotype RT position 103. These experiments demonstrated the first major challenge to point-of-care testing in that probes containing four or more differences from a target sequence often did not capture target DNA. However, these experiments also demonstrated the usefulness of a platform that contained multiple probe sets per codon as in each of the 15 diverse samples, target DNA was captured by one or more probe sets.
The second major challenge to point-of-care testing is to determine the optimal ΔTm threshold for determining whether the second-best binding probe in a probe set is detecting a codon present at low abundance. The ability to reliably detect variants present at proportions of approximately 15% or higher is a worthwhile objective because this is the sensitivity of dideoxynucleotide Sanger sequencing and because DRMs detected at this level are highly reproducible and clinically interpretable.13, 14, 15 Although our experiments demonstrated a high sensitivity and specificity for detecting targets that contained AAC even at a proportion of 10% when mixed with targets that contained AAA, additional experiments are required to determine whether similarly high levels of sensitivity and specificity can be obtained regardless of which codon is present at a low frequency.
Previously described allele-specific methods for detecting HIV-1 DRMs include differential hybridization of PCR products,16, 17 allele-specific PCR,18, 19 and assays that incorporate a biochemical reaction such as ligation20, 21, 22 and sequence extension23, 24 to increase specificity. In addition, one study performed solution-based melt curve analysis to detect K103N, Y181C, and M184V in subtype C viruses from South Africa.25 Several of these assays, particularly the allele-specific PCR assays, were developed primarily for detecting low-abundance DRMs. Others were developed as low-cost options for LMICs. Each has had to contend with the sequence variability surrounding each DRM and the requirement for simultaneously detecting multiple DRMs.
The multiplexed solid-phase melt curve analysis approach used in this study contains several inherent advantages for point-of-care testing. First, it achieves sensitivity by capturing target at one temperature (eg, 55°C) and specificity by determining how well the probe–target duplex resists melting relative to isogenic probes complementary to different codons. Second, it has the potential for amplifying sample nucleic acid and performing melt curve analysis in the same reaction chamber. Third, it can be highly multiplexed by leveraging the many addressable positions on a solid surface to design an assay with multiple probe sets for each DRM position.
Additional work, however, is required to modify the multiplexed solid-phase melt curve analysis platform described here to one suitable for point-of-care testing. First, to perform the assay in a closed-tube format and to obviate the need for a wash step after hybridization, an inverse fluorescence approach is required in which the printed probes will be labeled with fluorophores and the PCR products with quenchers. Second, to detect the complete set of published NRTI- and NNRTI-associated DRMs at six RT positions, the PCR step will need to be multiplexed to create amplicons for the three RT regions that encompass position 65, positions 103 and 106, and positions 181, 184, and 190. Finally, the HYDRA-1K CMOS chip, which is the production version of the prototype platform used in this study, obviates the need for an external fluorescence detector because fluorescence is detected by the chip's integrated CMOS processes.9 Moreover, it has 1032 addressable positions rather than 252 on the system used in this study.
Conclusions
The described experiments represent critical proof-of-principle results, suggesting that multiplexed solid-phase melt curve analysis is a promising approach for the development of a point-of-care assay for detecting HIV-1 drug resistance. Even if four (rather than three) probe sets would be required to ensure hybridization to each drug resistance position, approximately 160 features would be required to detect the approximately 40 reported codons at RT positions 65, 103, 106, 181, 184, and 190.4 If these probes are printed in triplicate (n = 480 probes), approximately one-half of the 1032 addressable positions on the HYDRA-1K CMOS chip would still be available to detect DRMs in other HIV-1 genes, including protease and integrase.
Footnotes
Supported in part by NIH grant R21 AI131918 (R.W.S., B.A.P., and E.L.), and the Stanford University Center for Innovation in Global Health (D.S.C. and J.M.).
Disclosures: G.M., R.S., S.B., P.K., R.G.K., G.K.S., and A.H. were employees of InSilixa, Inc. at the time of this research. B.A.P. and R.W.S. have received research funding from InSilixa, Inc.
Portions of this work were presented at the Conference on Retroviruses and Opportunistic Infections 2018, held March 4–7, 2018, in Boston, MA.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2019.02.005.
Supplemental Data
Melt curve experiments with undiluted targets. One set of melt curves (the first of the three replicates) for each of the 18 undiluted 80-mer targets each containing a different codon and a different flanking sequence (1, 2, and 3). The results for targets that contained AAA (wild type K), AAC (drug-resistance mutation N), and AGA (polymorphic variant R) are shown. Each experiment contained three panels, one for each probe set. Probe sets 1, 2, and 3 contained the 31-bp flanking sequences 1, 2, and 3, respectively. The probes complement to red: AAA; tan: AAC; green: AAG; teal: AAT; blue: AGA; purple: AGC.
The results for the remaining 27 experiments performed on 80-mer targets for targets that contained AAG (wild-type K), AAT (drug-resistance mutation N), and AGC (drug-resistance mutation S) are shown. The x axis represents the temperature (°C) and the y axis represents the fluorescent signal. Each experiment contained three panels, one for each probe set. Probe sets 1, 2, and 3 contained the 31-bp flanking sequences 1, 2, and 3, respectively. The probes complement to red: AAA; tan: AAC; green: AAG; teal: AAT; blue: AGA; purple: AGC.
Melt curve experiments with target mixtures. One set of melt curves (the first of the three replicates) for targets that contained AAC diluted to 50%, 25%, and 10% with targets that contained the same flanking sequences and AAA. Each row contains targets with a different AAC dilution and each column contains targets with a different flanking sequence. The x axis represents the temperature (°C) and the y axis represents the fluorescent signal. Each of the six experiments contained three panels, one for each probe set. Probe sets 1, 2, and 3 contained the 31-bp flanking sequences 1, 2, and 3, respectively. The probes complement to red: AAA; tan: AAC; green: AAG; teal: AAT; blue: AGA; purple: AGC.
References
- 1.Gupta R.K., Gregson J., Parkin N., Haile-Selassie H., Tanuri A., Andrade Forero L., Kaleebu P., Watera C., Aghokeng A., Mutenda N., Dzangare J., Hone S., Hang Z.Z., Garcia J., Garcia Z., Marchorro P., Beteta E., Giron A., Hamers R., Inzaule S., Frenkel L.M., Chung M.H., de Oliveira T., Pillay D., Naidoo K., Kharsany A., Kugathasan R., Cutino T., Hunt G., Avila Rios S., Doherty M., Jordan M.R., Bertagnolio S. HIV-1 drug resistance before initiation or re-initiation of first-line antiretroviral therapy in low-income and middle-income countries: a systematic review and meta-regression analysis. Lancet Infect Dis. 2018;18:346–355. doi: 10.1016/S1473-3099(17)30702-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phillips A.N., Stover J., Cambiano V., Nakagawa F., Jordan M.R., Pillay D., Doherty M., Revill P., Bertagnolio S. Impact of HIV drug resistance on HIV/AIDS-associated mortality, new infections, and antiretroviral therapy program costs in Sub-Saharan Africa. J Infect Dis. 2017;215:1362–1365. doi: 10.1093/infdis/jix089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhee S.Y., Jordan M.R., Raizes E., Chua A., Parkin N., Kantor R., Van Zyl G.U., Mukui I., Hosseinipour M.C., Frenkel L.M., Ndembi N., Hamers R.L., Rinke de Wit T.F., Wallis C.L., Gupta R.K., Fokam J., Zeh C., Schapiro J.M., Carmona S., Katzenstein D., Tang M., Aghokeng A.F., De Oliveira T., Wensing A.M., Gallant J.E., Wainberg M.A., Richman D.D., Fitzgibbon J.E., Schito M., Bertagnolio S., Yang C., Shafer R.W. HIV-1 drug resistance mutations: potential applications for point-of-care genotypic resistance testing. PLoS One. 2015;10:e0145772. doi: 10.1371/journal.pone.0145772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clutter D.S., Rojas Sanchez P., Rhee S.Y., Shafer R.W. Genetic variability of HIV-1 for drug resistance assay development. Viruses. 2016;8 doi: 10.3390/v8020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niemz A., Ferguson T.M., Boyle D.S. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 2011;29:240–250. doi: 10.1016/j.tibtech.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gubala V., Harris L.F., Ricco A.J., Tan M.X., Williams D.E. Point of care diagnostics: status and future. Anal Chem. 2012;84:487–515. doi: 10.1021/ac2030199. [DOI] [PubMed] [Google Scholar]
- 7.Ioannidis P., Papaventsis D., Karabela S., Nikolaou S., Panagi M., Raftopoulou E., Konstantinidou E., Marinou I., Kanavaki S. Cepheid GeneXpert MTB/RIF assay for Mycobacterium tuberculosis detection and rifampin resistance identification in patients with substantial clinical indications of tuberculosis and smear-negative microscopy results. J Clin Microbiol. 2011;49:3068–3070. doi: 10.1128/JCM.00718-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larder B.A., Kohli A., Kellam P., Kemp S.D., Kronick M., Henfrey R.D. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature. 1993;365:671–673. doi: 10.1038/365671a0. [DOI] [PubMed] [Google Scholar]
- 9.Hassibi A., Manickam A., Singh R., Bolouki S., Sinha R., Jirage K.B., McDermott M.W., Hassibi B., Vikalo H., Mazarei G., Pei L., Bousse L., Miller M., Heshami M., Savage M.P., Taylor M.T., Gamini N., Wood N., Mantina P., Grogan P., Kuimelis P., Savalia P., Conradson S., Li Y., Meyer R.B., Ku E., Ebert J., Pinsky B.A., Dolganov G., Van T., Johnson K.A., Naraghi-Arani P., Kuimelis R.G., Schoolnik G. Multiplexed identification, quantification and genotyping of infectious agents using a semiconductor biochip. Nat Biotechnol. 2018;36:738–745. doi: 10.1038/nbt.4179. [DOI] [PubMed] [Google Scholar]
- 10.SantaLucia J., Jr. Physical principles and visual-OMP software for optimal PCR design. Methods Mol Biol. 2007;402:3–34. doi: 10.1007/978-1-59745-528-2_1. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez A.M., DeMarco C.T., Hora B., Keinonen S., Chen Y., Brinkley C., Stone M., Tobler L., Keating S., Schito M., Busch M.P., Gao F., Denny T.N. Development of a contemporary globally diverse HIV viral panel by the EQAPOL program. J Immunol Methods. 2014;409:117–130. doi: 10.1016/j.jim.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sing T., Sander O., Beerenwinkel N., Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 13.Woods C.K., Brumme C.J., Liu T.F., Chui C.K., Chu A.L., Wynhoven B., Hall T.A., Trevino C., Shafer R.W., Harrigan P.R. Automating HIV drug resistance genotyping with RECall, a freely accessible sequence analysis tool. J Clin Microbiol. 2012;50:1936–1942. doi: 10.1128/JCM.06689-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohamed S., Penaranda G., Gonzalez D., Camus C., Khiri H., Boulme R., Sayada C., Philibert P., Olive D., Halfon P. Comparison of ultra-deep versus Sanger sequencing detection of minority mutations on the HIV-1 drug resistance interpretations after virological failure. AIDS. 2014;28:1315–1324. doi: 10.1097/QAD.0000000000000267. [DOI] [PubMed] [Google Scholar]
- 15.Church J.D., Jones D., Flys T., Hoover D., Marlowe N., Chen S., Shi C., Eshleman J.R., Guay L.A., Jackson J.B., Kumwenda N., Taha T.E., Eshleman S.H. Sensitivity of the ViroSeq HIV-1 genotyping system for detection of the K103N resistance mutation in HIV-1 subtypes A, C, and D. J Mol Diagn. 2006;8:430–432. doi: 10.2353/jmoldx.2006.050148. quiz 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozal M.J., Shah N., Shen N., Yang R., Fucini R., Merigan T.C., Richman D.D., Morris D., Hubbell E., Chee M., Gingeras T.R. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 17.Stuyver L., Wyseur A., Rombout A., Louwagie J., Scarcez T., Verhofstede C., Rimland D., Schinazi R.F., Rossau R. Line probe assay for rapid detection of drug-selected mutations in the human immunodeficiency virus type 1 reverse transcriptase gene. Antimicrob Agents Chemother. 1997;41:284–291. doi: 10.1128/aac.41.2.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson J.A., Li J.F., Wei X., Lipscomb J., Bennett D., Brant A., Cong M.E., Spira T., Shafer R.W., Heneine W. Simple PCR assays improve the sensitivity of HIV-1 subtype B drug resistance testing and allow linking of resistance mutations. PLoS One. 2007;2:e638. doi: 10.1371/journal.pone.0000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metzner K.J., Giulieri S.G., Knoepfel S.A., Rauch P., Burgisser P., Yerly S., Gunthard H.F., Cavassini M. Minority quasispecies of drug-resistant HIV-1 that lead to early therapy failure in treatment-naive and -adherent patients. Clin Infect Dis. 2009;48:239–247. doi: 10.1086/595703. [DOI] [PubMed] [Google Scholar]
- 20.Shi C., Eshleman S.H., Jones D., Fukushima N., Hua L., Parker A.R., Yeo C.J., Hruban R.H., Goggins M.G., Eshleman J.R. LigAmp for sensitive detection of single-nucleotide differences. Nat Methods. 2004;1:141–147. doi: 10.1038/nmeth713. [DOI] [PubMed] [Google Scholar]
- 21.Ellis G.M., Vlaskin T.A., Koth A., Vaz L.E., Dross S.E., Beck I.A., Frenkel L.M. Simultaneous and sensitive detection of human immunodeficiency virus type 1 (HIV) drug resistant genotypes by multiplex oligonucleotide ligation assay. J Virol Methods. 2013;192:39–43. doi: 10.1016/j.jviromet.2011.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panpradist N., Beck I.A., Chung M.H., Kiarie J.N., Frenkel L.M., Lutz B.R. Simplified paper format for detecting HIV drug resistance in clinical specimens by oligonucleotide ligation. PLoS One. 2016;11:e0145962. doi: 10.1371/journal.pone.0145962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang G., Cai F., de Rivera I.L., Zhou Z., Zhang J., Nkengasong J., Gao F., Yang C. Simultaneous detection of major drug resistance mutations of HIV-1 subtype B viruses from dried blood spot specimens by multiplex allele-specific assay. J Clin Microbiol. 2016;54:220–222. doi: 10.1128/JCM.02833-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang G., Cai F., Zhou Z., DeVos J., Wagar N., Diallo K., Zulu I., Wadonda-Kabondo N., Stringer J.S., Weidle P.J., Ndongmo C.B., Sikazwe I., Sarr A., Kagoli M., Nkengasong J., Gao F., Yang C. Simultaneous detection of major drug resistance mutations in the protease and reverse transcriptase genes for HIV-1 subtype C by use of a multiplex allele-specific assay. J Clin Microbiol. 2013;51:3666–3674. doi: 10.1128/JCM.01669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sacks D., Ledwaba J., Morris L., Hunt G.M. Rapid detection of common HIV-1 drug resistance mutations by use of high-resolution melting analysis and unlabeled probes. J Clin Microbiol. 2017;55:122–133. doi: 10.1128/JCM.01291-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Melt curve experiments with undiluted targets. One set of melt curves (the first of the three replicates) for each of the 18 undiluted 80-mer targets each containing a different codon and a different flanking sequence (1, 2, and 3). The results for targets that contained AAA (wild type K), AAC (drug-resistance mutation N), and AGA (polymorphic variant R) are shown. Each experiment contained three panels, one for each probe set. Probe sets 1, 2, and 3 contained the 31-bp flanking sequences 1, 2, and 3, respectively. The probes complement to red: AAA; tan: AAC; green: AAG; teal: AAT; blue: AGA; purple: AGC.
The results for the remaining 27 experiments performed on 80-mer targets for targets that contained AAG (wild-type K), AAT (drug-resistance mutation N), and AGC (drug-resistance mutation S) are shown. The x axis represents the temperature (°C) and the y axis represents the fluorescent signal. Each experiment contained three panels, one for each probe set. Probe sets 1, 2, and 3 contained the 31-bp flanking sequences 1, 2, and 3, respectively. The probes complement to red: AAA; tan: AAC; green: AAG; teal: AAT; blue: AGA; purple: AGC.
Melt curve experiments with target mixtures. One set of melt curves (the first of the three replicates) for targets that contained AAC diluted to 50%, 25%, and 10% with targets that contained the same flanking sequences and AAA. Each row contains targets with a different AAC dilution and each column contains targets with a different flanking sequence. The x axis represents the temperature (°C) and the y axis represents the fluorescent signal. Each of the six experiments contained three panels, one for each probe set. Probe sets 1, 2, and 3 contained the 31-bp flanking sequences 1, 2, and 3, respectively. The probes complement to red: AAA; tan: AAC; green: AAG; teal: AAT; blue: AGA; purple: AGC.