Abstract
Despite decades of efforts, non-small-cell lung cancer (NSCLC) remains the leading cause of cancer mortality globally primarily due to the challenge in early detection of the cancer. Being an important player in cancer development, the dysregulated miRNAs have been shown promising values as non-invasive diagnostic and prognostic biomarkers for NSCLC. The aim of our study is to access the efficacy and reliability of a potential circulating miRNA panel in early diagnosis of NSCLC. We first selected eight candidate miRNAs, miR-146b, miR-205, miR-29c, miR-31, miR-30b, miR-337, miR-411, and miR-708, which have been shown frequently aberrant in primary NSCLC patients based on our previous studies and other reports. The serum level of each of these miRNAs was evaluated by quantitative real-time PCR (qRT-PCR) in training and testing sets. We found that 5 out of 8 miRNAs (miR-146b, miR-205, miR-29c, miR-30b, and miR-337) were significantly up-regulated in NSCLCs patients compared to healthy or cancer-free controls in both training and testing sets. Based on the logistic regression model, a 4-miRNAs set (miR-146b, miR-205, miR-29c and miR-30b) was picked out of the 5 miRNAs owing to its excellent diagnostic power for NSCLC patients in the training set (AUC=0.99, accuracy=95.00%), the testing set (AUC=0.93, accuracy=89.69%), and the training-testing combined set ( AUC=0.96, accuracy=92.00%). When pathological subtypes of NSCLC are compared, this 4-miRNA panel carried a relatively higher prediction power and higher sensitivity for adenocarcinoma (AC) (AUC=0.98, sensitivity=99.10%) than for squamous cell carcinoma (SCC) (AUC=0.93, sensitivity=90.32%). Additionally, this panel demonstrated a comparable diagnostic capacity for stage I (AUC=0.96) and stage II-III (AUC=0.95) of NSCLC, suggesting its role in reflecting the tumor load. Importantly, the high levels of miR-146b and miR-29c in serum were significantly associated with poor 5-year overall survival (OS) (both p=0.04). Further survival analysis showed that high level of miR-146b in serum is specifically correlated with poor survival rate in SCC patients (p=0.0035) but not in AC patients (p=0.83), consistent with our previous finding that the high tissue expression of miR-146b in lung cancer specimen is indicative of a poor prognosis for SCC patients. Altogether, our study demonstrated that the 4-miRNA panel is a novel, sensitive and non-invasive serum marker for the early diagnosis of NSCLC.
Keywords: non-small-cell lung cancer (NSCLC), miRNA, serum biomarker, early diagnosis, prognosis
Introduction
Lung cancer is the most common cancer affecting both men and women, with approximately 80% cases being non-small-cell lung cancer (NSCLC) 1, 2. NSCLC has three major histological subtypes, adenocarcinoma (AC, 40%), squamous cell carcinoma (SCC, 25%), and large-cell lung cancer (LCC, 10%) 3. Despite decades of multidisciplinary efforts on improving the outcomes of lung cancer, it is still the leading cause of cancer-related mortality worldwide. One of the key reasons of high mortality rate is the low rate (<25%) of early diagnosis 4, which warrants the urgent need for new diagnostic strategies to screen individuals at risk. The low-dose computed tomography (LDCT) has been the workhorse in screening lung cancer, resulting in a reduction of the mortality rate by 20% 4, 5. Nevertheless, the high false positivity is the major drawback of LDCT on top of other disadvantages, such as the radiation exposure and poor cost-effectiveness 6. Therefore, we aim at developing non-invasive and sensitive biomarkers of NSCLC, which could lower the threshold age of screening and compensate the flaws of LDCT.
Liquid biopsy tests, which analyze different kinds of tumor material, such as DNA, RNA, proteins, and exosomes, have drawn increasing attention partly for its non-invasive nature in exploring biomarkers of tumors 7. MicroRNAs (miRNAs) are small non-coding, endogenous RNAs that regulate gene expression post-transcriptionally by interacting with the 3'UTR on mRNAs of targeted genes. They are pivotal modulators in multiple physiological and pathological processes with strong tissue specificities 8. Numerous reports have shown that altered expressions of miRNAs are associated with various cancers including lung cancer 9, 10. Those circulating miRNAs, present in plasma, serum, or whole-blood, are remarkable for their strong resistance to RNase activity and extreme stability in tough microenvironments such as high pH conditions. With all the advantages shown above, circulating miRNAs have evolved into potential non-invasive biomarkers to diagnose disease and monitor disease progression 11.
In the present study, we selected eight candidate miRNAs, miR-146b, miR-205, miR-29c, miR-31, miR-30b, miR-337, miR-411, and miR-708, which have been shown aberrantly expressed in NSCLC or other cancer patients based on our previous studies 12, 13 and other reports 14-17 (Supplementary Table S1). We assessed their capacity as non-invasive serum biomarkers for early diagnosis of NSCLC and prognostic evaluation in an independent cohort consisting of 128 NSCLC patients and 47 non-cancer controls.
Results
Expression profiles of eight candidate miRNAs in training set
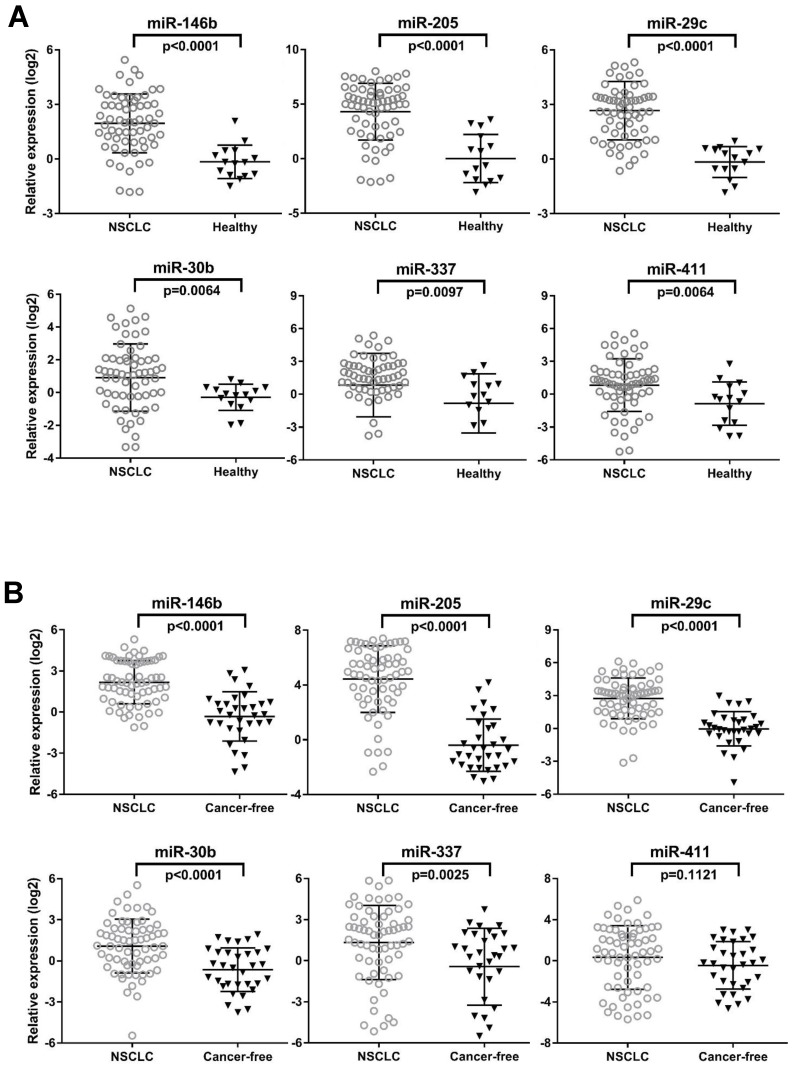
To determine whether aberrations at histological level in the specified miRNAs could be confirmed in independent serum samples, we first quantitated eight candidate miRNAs by qRT-PCR in individual serum samples in the training set (N=78) consisting of 63 NSCLC patients and 15 healthy controls. Notably in this training set, we intentionally enriched the enrollment of stage I and stage II lung cancer cases to investigate capacity of these miRNAs in detecting early staged patients. The clinical characteristics of the training set participants were summarized in Table 1. We found that 6 out of 8 miRNAs (miR-146b, miR-205, miR-29c, miR-30b, miR-337 and miR-411) were significantly up-regulated in NSCLC patients as compared to health subjects determined by the unpaired Mann-Whitney test (all p<0.01, Fig. 1A).
Table 1.
Characteristics of subjects in the training and testing set
| Training set (n=78) | Testing set (n=97) | ||||||
|---|---|---|---|---|---|---|---|
| NSCLC n=63 | Healthy control n=15 | p-value (NSCLC vs. Healthy control) | NSCLC n=65 | non-cancer control n=32 | p-value (NSCLC vs. non-cancer control) | ||
| COPD n=17 | Healthy control n=15 | ||||||
| Age (years) | |||||||
| Mean ± SD | 67.8 ± 9.0 | 64.7 ± 8.7 | 0.3016 | 67.4 ± 9.9 | 65.0 ±9.4 | 62.6 ±9.7 | 0.2468 |
| Gender | |||||||
| Male | 30 (48%) | 8 (53%) | 0.4795 | 38 (58%) | 11 ( 33%) | 7 (21%) | 0.4093 |
| Female | 33 (52%) | 7 (47%) | 27 (44%) | 7 (21%) | 8 (25%) | ||
| Smoking status | |||||||
| Nonsmoker | 8 (13%) | 6 (40%) | <0.0001 | 2 (3%) | 4 (13%) | 5 (16%) | <0.0001 |
| Smoker | 55 (87%) | 9 (60%) | 63 (97%) | 13 (40%) | 10(31%) | ||
| Histological type | |||||||
| Adenocarcinoma | 30 (48%) | NA | NA | 36 (55%) | NA | NA | NA |
| Squamous cell carcinoma | 33 (52%) | NA | 29 (45%) | NA | NA | ||
| TNM stage | |||||||
| I | 46 (73%) | NA | NA | 29 (45%) | NA | NA | NA |
| II | 17 (27%) | NA | 16 (25%) | NA | NA | ||
| III | 0 (0%) | NA | 20 (30%) | NA | NA | ||
| Histological grade | |||||||
| Well | 11(17%) | NA | NA | 5 (8%) | NA | NA | NA |
| Moderate | 25 (40%) | NA | 28 (43%) | NA | NA | ||
| Poor | 19 (30%) | NA | 25 (38%) | NA | NA | ||
| Unknown | 8 (13%) | NA | 7 (11%) | NA | NA | ||
Note: NA, not assessed
Figure 1.
The expression levels of 6 serum miRNAs (miR-146b, miR-205, miR-29c, miR-30b, miR-337 and miR-411), in the training set (A) and testing set (B). Mann-Whitney tests were performed to ascertain statistical significance between the expression levels across groups. Except miR-411, other five miRNAs were significantly overexpressed (p<0.05) in serum from NSCLC patients compared to controls in training and testing sets.
Validation of 6 differentially expressed miRNAs in an independent testing set
We further validated the six differentially expressed miRNAs from training set in an independent testing set by qRT-PCR. We asked whether this miRNA set can distinguish patients even with late stages of NSCLC from those healthy controls as well as from those with benign pulmonary diseases. To this end, we expanded the NSCLC group (N=65) with a relevant proportion of patients with more advanced cancer (stage IIIA-B, n=20). We also expanded the controls group (n=32) with 17 age-, gender- and smoking status-matched COPD patients and 15 healthy controls (Table 1). We first compared the expression of 6 miRNAs in COPD patients and healthy controls. No significant difference of each individual miRNA was observed between the two control groups (all p>0.05, data not shown). We subsequently combined them as cancer-free controls for the following comparison with NSCLC patients in the testing set. We found that, except miR-411(p=0.1121), all the other 5 miRNAs (miR-146b, miR-205, miR-29c, miR-30b, and miR-337) were significantly up-regulated in NSCLC patients compared to the cancer-free controls (all p<0.005, Fig. 1B). This is consistent with the results rendered from the training set. More importantly, the expression of these five miRNAs can distinguish NSCLC patients from a larger and tentatively more representative cancer-free population, which includes both healthy controls and COPD, a common benign pulmonary disease sharing the same risk factor with lung cancer. Therefore, these five miRNAs were subjected to the following steps, assessing their potential as diagnostic biomarkers for NSCLC.
A four-miRNA panel optimized as serum-based biomarkers for NSCLC diagnosis
To evaluate diagnostic reliability of these 5 miRNAs, we performed the univariate logistic regression analysis for each individual miRNA in the training set (N=78) and testing set (N=97). The regression coefficient (β) is a valid parameter to indicate an estimated increase in the odds of the outcome (i.e. NSCLC) with increased exposure (i.e. serum miRNA level). The 95% confidence interval (CI) was used as a measure of precision of the regression coefficient and to determine the presence of statistical significance by the p-value 18. Our analysis showed that all 5 miRNAs were found to be significantly associated with NSCLC in both sets (Table S2) with all p<0.05.
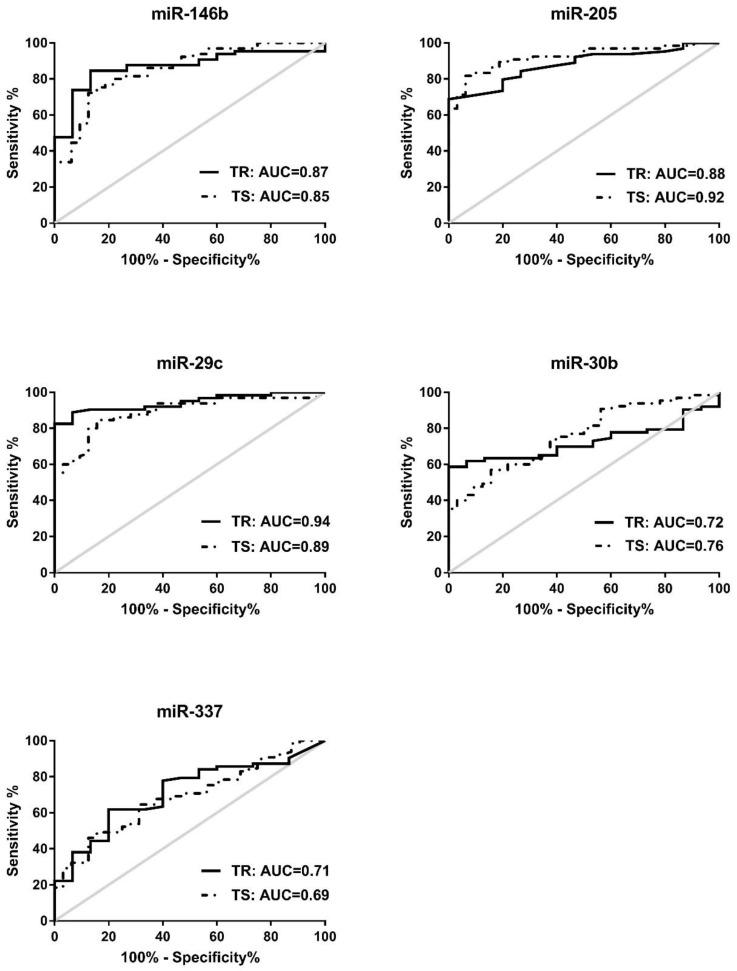
To assess the diagnostic capacity of the 5 miRNAs in the training set (n=78), ROC analysis of each miRNA was first conducted between NSCLC patients and healthy controls. The individual AUC value of the 5 miRNAs ranges between 0.71-0.94. When the best diagnostic cut-off point of each miRNA was determined by Youden's index (YI), these miRNAs yielded sensitivity of 61.9-88.89% and specificity of 80-93.33% (Fig. 2 and Table S3). Subsequently, ROC analyses were performed in the testing set (n=97), comparing NSCLC patients with cancer free controls. We found that these 5 miRNAs have similar diagnostic capacities in the testing set to the training set with the AUC values 0.69-0.92, sensitivities 63.08-84.62%, and specificity 62.5-93.75% (Fig. 2 and Table S3). Similar results were achieved when training set and testing set are combined (n=175). The AUCs, sensitivities, and specificities of these 5 miRNAs in the combined set 0.69-0.91, 63.28-83.59%, and 72.34-87.23% respectively when NSCLCs (n=128) are compared with cancer-free controls (N=47) (Table S3). Collectively, these data suggest that these individual 5 miRNAs are promising cancer-specific markers in serum for NSCLC patients.
Figure 2.
Receiver-operator characteristic (ROC) curve analyses of 5 serum miRNAs. MiR-146b, miR-205, miR-29c, miR-30b, and miR-337, is differentially expressed in NSCLC patients compared to either healthy controls in the training set (TR, shown in solid line) or cancer-free controls in the testing set (TS, shown in dotted line).
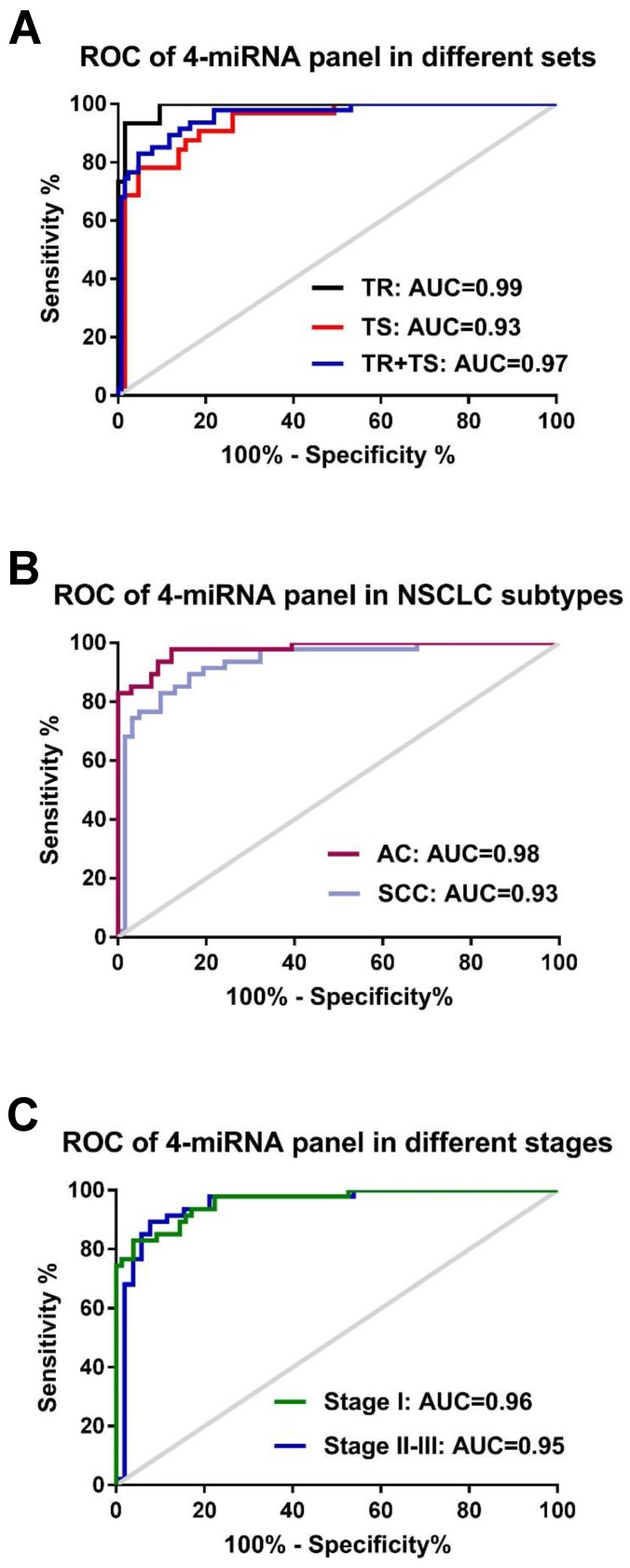
To assess if an optimal miRNA panel, instead of the single individual miRNA, could be chosen from these 5 candidate miRNAs for the early detection of NSCLC with higher sensitivity and specificity, logistic regression of all 5 miRNAs was performed using a backward elimination approach in the training set. One best prediction model was built by the composite of 4 miRNAs (miR-146b, miR-205, miR-29c, and miR-30b) following by the algorithm, In(p/1-p) = 0.311 + 0.06 × miR-146 + 0.023 × miR-205 + 0.180 × miR-29c - 0.1636 × miR-30b. ROC analysis based on this algorithm showed that the 4-miRNA panel produced an AUC value of 0.99 in the training set (Fig. 3A), which was considerably higher than AUC values of each individual miRNAs (0.71-0.94, in Table S3). When optimal cut-off (0.5606) was defined by Youden's index, the accuracy, sensitivity, and specificity were 95.00%, 93.65%, and 93.33% respectively in the training set (Table 2). By using the same logistic regression algorithm and cut-off value derived on the training set, we found that the 4-miRNA set performed equally well in differentiating NSCLC patients from cancer-free controls in testing set and the combined set. The corresponding AUC, accuracy, sensitivity, and specificity were 0.93, 89.69%, 95.38%, 78.13% in the testing set and 0.96, 92.00%, 95.31% and 82.98% in the combined set respectively (Fig. 3A and Table 2). All these results suggest that the 4-miRNA panel is a remarkable and reliable cancer-specific serum signature for the diagnosis of NSCLC patients.
Figure 3.
Receiver-operator characteristic (ROC) curve analyses of the 4-miRNA panel (miR-146b, miR-205, miR-29c and miR-30b). (A) when distinguishing NSCLC patients from either healthy controls in the training set (TR, black-colored line) or NSCLC patients from cancer-free controls in the testing set (TS, red-colored line) and in the two sets combined (TR+TS, blue-colored line); (B): distinguishing NSCLC patients with different histological subtypes: adenocarcinoma (AC, purpled-colored line) or squamous cell carcinoma (SCC, light blue-colored line) from all cancer-free controls; (C): distinguishing NSCLC patients with different stages (Stage I: green-colored line; stage II-III: blue-colored line) from all cancer-free controls.
Table 2.
Performance of 4-miRNA predictive panel in various sets
| Set | AUC | 95% CI | Accuracy (%) | Sensitivity (%) | Specificity (%) | |
|---|---|---|---|---|---|---|
| Training set (15 Healthy vs. 63 NSCLC) | 0.99 | 0.974-1.007 | 95.00 | 93.65 | 93.33 | |
| Testing set (32 Cancer-free vs. 65 NSCLC) | 0.93 | 0.885-0.985 | 89.69 | 95.38 | 78.13 | |
| Two sets combined: Cancer-free (n=47) vs. Cancer | NSCLC (n=128) | 0.96 | 0.925-0.988 | 92.00 | 95.31 | 82.98 |
| AC (n=66) | 0.98 | 0.957-1.000 | 92.92 | 99.10 | 82.98 | |
| SCC (n=62) | 0.93 | 0.884-0.982 | 87.16 | 90.32 | 82.98 | |
| Stage I (n=75) | 0.96 | 0.930-0.992 | 93.49 | 96.05 | 82.98 | |
| Stage II-III (n=53) | 0.95 | 0.904-0.996 | 89.00 | 94.23 | 82.98 | |
The correlation of the 4 miRNAs with NSCLC subtypes, TNM stages, tumor size and other clinical significance
Next, we asked whether the 4-miRNAs set has differential diagnostic capability in histological subtypes of NSCLC. To this end, we carried out pairwise group comparisons of these 4 individual miRNAs in 4 subgroups of all cases (n=175), adenocarcinoma (AC), squamous cell carcinoma (SCC), COPD, and healthy control groups. As shown in Fig. S1, we found that the serum level all 4 miRNAs were significantly higher in AC patients when compared to other 3 groups individually (SCC, COPD, and healthy controls) (all p<0.01). Similarly, all of the 4 miRNA levels were significantly higher in SCC group compared to COPD group (all p<0.01). Three individual miRNAs, miR-146b, miR-205, and miR-29c, but not miR-30b (p=0.7514), were significantly increased in SCC patients than in healthy controls (all p<0.01). When comparing COPD patients (n=17) to all healthy controls (n=30), only miR-30b was significantly decreased in COPD group than in healthy subjects (p=0.0011). No statistical significance of the other 3 miRNAs was identified between COPD and healthy individuals (all p>0.05) (Fig. S1).
We then performed ROC analysis of these 4 individual miRNAs in AC patients (n=66) and SCC patients (n=62) using cancer-free subjects as controls (n=47). We found that the AUC values in AC patients (AUC 0.87-0.98) are relative higher than those in SCC patients (AUC 0.60-0.88) (Fig. S2). Similarly, higher sensitivities and specificities were observed in AC patients (sensitivities 85.11-97.87% and specificities 80.3-92.42%) compared to SCC patients (sensitivities 78.72-87.23% and specificities of 40.32-80.65%) (Table S4). Further analyses of AUC and diagnostic accuracy were conducted in AC and SCC groups by the 4-miRNA panel. We found that this 4-miRNA panel has a relatively better diagnostic capacity in AC patients (AUC= 0.98, accuracy 92.92%) and in SCC patient (AUC=0.93, accuracy 87.16%) (Fig. 3B and Table 2). These data revealed that the 4 serum miRNAs displayed higher diagnostic power for lung AC patients than for SCC patients either individually or as a whole panel.
The correlation of the 4-miRNA panel with clinical stages of NSCLC was also evaluated in our study. Similar diagnostic capacity of this panel were found between NSCLC stage I patients (n=75; AUC=0.96, sensitivity 96.05%, specificity 82.98%) and stage II-III patients (n=53; AUC=0.95, sensitivity 94.23%, specificity 82.98%), with cancer-free subjects (n=47) as controls (Fig. 3C and Table 2). However, the relevant accuracy in stage I patients is moderately higher than that in stage II-III patients (93.49% vs. 89.00%) (Table 2). All 4 individual miRNA expressions and the corresponding AUCs of NSCLC patients with different stages were shown in Fig. S3 and S4). These data suggest that the 4-miRNA panel has similar diagnostic power for stage I and stage II-III patients. The serum levels of these 4 miRNAs are able to reflect the presence of cancer load in NSCLC patients but not related with clinical stage. Nevertheless, a higher diagnostic accuracy is observed in early stage patients as compared to late stage patients.
We also analyzed the expression levels of 4 individual miRNAs with regard to different tumor size or T status (T1, T2, or T3) of TNM staging (Fig. S5). No significant correlation exists between the serum level of individual miRNA and tumor size or different T status (T1, T2, or T3) (all p > 0.05). However, significantly higher expressions of the 4 miRNAs were identified in serum from either T1, T2 or T3 NSCLC patients compared to cancer-free controls (all p < 0.001), reaffirming their diagnostic value as the cancer-specific biomarkers of NSCLC. No significant correlations were identified between the levels of each miRNA and other clinic-pathological features such as age, gender, history of smoking, or differentiation (all p > 0.05, data not shown).
The correlation of the 4 miRNAs with patient survival
We further investigated the association of these 4 serum miRNAs with survival rate of all NSCLC patients (n=128) in our current study. First of all, the serum levels of the miRNAs are categorized into high (n=42), moderate (n=44), and low (n=42) subgroups based on their serum levels using the two tripartite division values as cut-offs. The comparisons were subsequently performed only between high and low subgroups regarding their 5-year overall survival (OS). We found that high serum levels of miR-146b (p=0.0415) and miR-29c (p=0.0396) were associated with low 5-year OS of NSCLC patients (Fig. 4A-B). The other two miRNAs have no association with 5-year OS in NSCLC patients (all p>0.05, Fig. S6A-B).
Figure 4.
Kaplan-Meier analyses of 5-year overall survival curves for NSCLC patients stratified by high (dotted line) or low (solid line) serum expression levels of miR-146b (A) and miR-29c (B). The p-values were calculated using the log-rank test between patients with high- and low- (log2) fold changes.
Next, we asked whether these 4 miRNAs may have different association with prognosis with respect to histological subtypes of NSCLC. To achieve this, the serum level of each individual miRNA was categorized as high or low by using the median value as the cut-off. Then, we conducted survival analyses in AC (n=66) and SCC (n=62) patients respectively by comparing the high and the low miRNA groups. We found that high serum miR-146b level was significantly associated with poor OS in SCC patients (p=0.0035, Fig. S7B) but not in AC patients (p=0.8335, Fig. S7A). It is consistent with our previous finding that higher expression of miR-146 in lung cancer tissue is a robust predictor of poor overall survival in SCC patients 13. However, no significant correlation of miR-29c was discovered with survival either in AC group (p=0.2223, Fig. S7C) or SCC group (p=0.1138, Fig. S7D). MiR-205 and miR-30b fail to show any prognostic significance in AC or SCC subgroups by overall survival analysis as well (all p>0.05, data not shown). Additionally, no significant relevance was found between the levels of miRNAs and cancer recurrence (p>0.05, data not shown).
Analysis of miRNA target genes by DAVID Gene Ontology
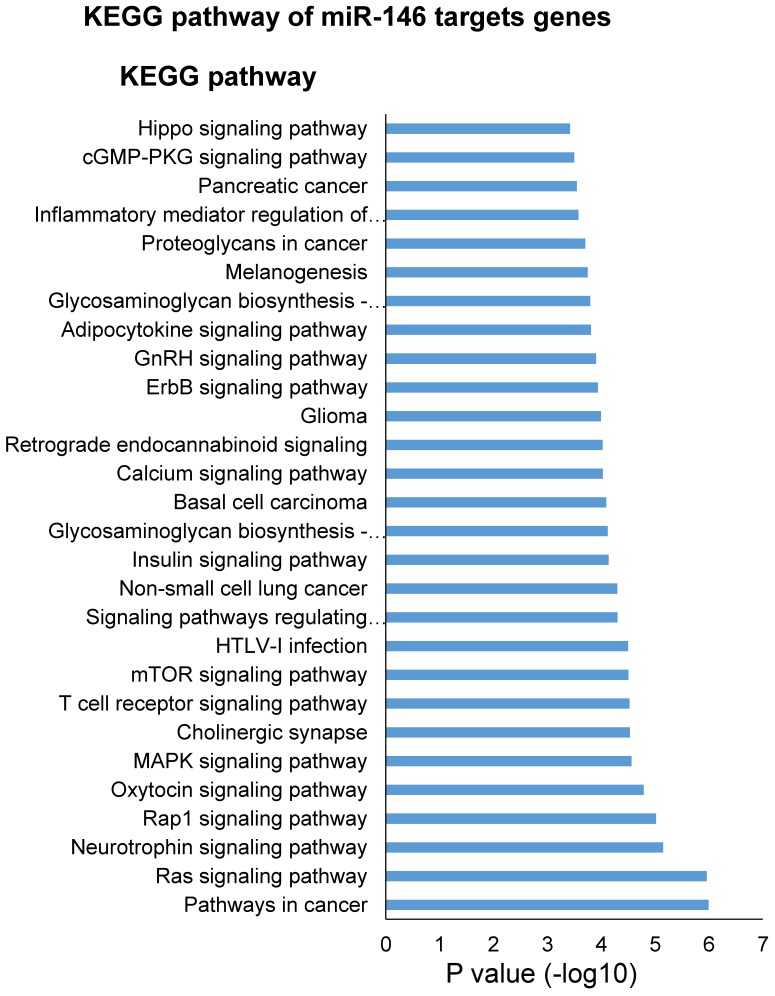
One miRNA could target multiple genes and one gene could be regulated by multiple miRNAs. In order to understand the potential biology process regulated by this 4-miRNA panel (miR-146b, miR-205, miR-29c and miR-30b), we first selected the miRNA target genes using Targetscan website (http://www.targetscan.org), then analyzed the biology process or pathway of these genes using DAVID pathway website (https://david.ncifcrf.gov). To our surprise, miR-146 target genes were involved in many oncogenic pathways of lung cancer such as EGFR, AKT, MAPK, mTOR, ErbB, Hippo signaling pathways, and T cell receptor signaling pathway (Fig. 5), which is congruent with its virtue of cancer-specific biomarker in NSCLC in our current study. In addition, we found that cellular metabolic processes were the most regulated biology processes by miR-205 and miR-30b (Fig. S9 and S10), while miR-205 and miR-29c are heavily involved in the nervous system development (Fig. S9 and S11).
Figure 5.
KEGG pathway analysis of miR-146 targets genes.
Discussion
NSCLC, as the main category of lung cancer, renders the highest cancer-related mortality in many countries of the world. The prognosis and survival rate for NSCLC patients are closely related to clinical stages, displaying a sharp decline from 68-92% for stage I/II patients to 1-13% for stage III/IV patients 19. Decades of endeavor to improve lung cancer outcomes only led to a little improvement with the global overall 5-year survival rate of NSCLC increased from 15% to 19% 2. The high mortality rate is mainly due to the lack of effective measures for early diagnosis with only 16% of lung cancer cases are diagnosed at an early stage 20. Therefore, multidisciplinary efforts are required to enhance the efficiency of early diagnosis to improve the overall survival. Ever since the implementation of low-dose computed tomography (LDCT) to screen high risk population, lung cancer mortality has been reduced for about 14-20% 5, 21. However, the high false-positive rates, high costs, and potential radiation harms of LDCT have curtailed its capacity as a screening tool, which provokes the need for discovering non-invasive biomarkers for early diagnosis 22.
Serum-based circulating miRNAs screening is a novel and crucial diagnostic tool partly owing to its non-invasive nature. It holds several other characteristics in clinical settings along with tremendous research advantages, such as: (i) serum specimens are much easier to obtain with less suffering compared to lung biopsy, (ii) it's considerably cheaper and more immediately implementable than LDCT especially for large-scale screening in population, (iii) circulating miRNAs showed high stability and tissue- or disease-specific expression patterns, (iv) the miRNA expression profiling pools from worldwide efforts make it possible to optimize the candidate miRNAs into a small set for tumor-specific origin and serve as a potential biomarkers for the detection of human malignancies 14, 22.
In line with this strategy, we selected 8 candidate miRNAs based on our previous studies 12, 13 and other relevant reports (detailed in Table S1) 14-17 and explored their potential as serum biomarkers for early diagnosis of NSCLC. We found that a 4-miRNA panel consisting miR-146b, miR-205, miR-29c and miR-30b can efficiently discriminated NSCLC patients from an expanded cancer-free group including COPD patients and healthy controls. We further revealed that the 4-miRNAs panel has relative better diagnostic power for AC (AUC=0.99, accuracy 95.00%, sensitivity 98.59%) than for SCC (AUC=0.93, accuracy 89.69%, sensitivity 90.32%). Meanwhile, we found that the 4-miRNA signature showed its remarkable diagnostic power in stage I NSCLC patients (AUC= 0.96, sensitivity 96.05%, specificity 82.98%) and stage II-III patients (AUC=0.95, sensitivity 94.23%, specificity 82.98%). Notably, this panel displayed relative higher diagnostic accuracy for stage I patients (93.49%) than for stage II-III patients (89.00%), supporting the 4-miRNA panel as biomarker in early diagnosis of NSCLC. Lastly, we assessed 4 individual miRNAs as potential prognostic biomarkers for lung cancer by one of the important clinic-pathological features, 5-year overall survival rate (OS). We found that high levels of serum miR-146b and miR-29c were associated with poor survival rate of NSCLC patients. It is especially interesting and important that high serum level of miR-146b was significantly correlated with poor overall survival in SCC patients but not in AC patients. This result corroborates our previous finding that higher tissue expression of miR-146b in lung cancer specimens is significantly related with worse overall survival in SCC patients 13.
It is of particular significance that the excellent diagnostic power of the 4-miRNA panel is established on a more representative cancer-free control by including not just healthy controls but also cancer-free COPD patients, evidenced by the AUC, accuracy, sensitivity, and specificity of 0.92, 91.72%, 94.53%, and 70.19% respectively (Fig. S8). Specifically speaking, COPD shares the same risk factor with lung cancer, cigarette smoking. On top of that, COPD is an independent risk factor for lung carcinoma 23. Therefore, the 4-miRNA panel revealed by our current study may provide an effective and powerful screening tool for the early detection of NSCLC in high-risk patients with COPD. It is worthwhile to note that the 4-miRNA panel has no discriminatory power to separate COPD from healthy subjects (AUC=0.51, data not shown). The optimized 4-miRNA panel out of the original 8 miRNA candidates may give a wider clinical application in differentiating NSCLC patients from COPD patients, especially in the presence of questionable lung densities by imaging studies.
It is also important to point out that we revealed a comparable diagnostic power of the 4-miRNA set for both early and advanced NSCLC patients. In other words, the serum level of the 4-miRNA set reflects the presence of the cancer load of NSCLC, which supports its application in screening high risk population, monitoring therapeutic response, and detecting NSCLC recurrence. A better diagnostic power for adenocarcinoma than for squamous cell carcinoma adds more clinical values of the 4-miRNA panel by aiding in subtype prediction.
Although these 4 miRNAs in our current study presented high serum levels in NSCLC patients than those in cancer-free subjects, we could not simply conclude that they are onco-miRNAs in NSCLC. Instead, they played diverse and even controversial roles (oncogenic or tumor suppressor miRNA) in NSCLC and other types of cancer, varied between different studies or different research groups. miR-146b has been demonstrated to be up-regulated in lung cancer tissues and related to patient survival in our previous study 13. Li et al. reported that increased tissue expression of miR-146b was related to short patient survival 24. DAVID pathway analysis indicated that miR-146 targeted genes were involved in key oncogenic signaling pathways of lung cancer such as EGFR, AKT, MAPK, mTOR, ErbB, and Hippo etc. (Fig. 5), indirectly supporting its virtue being a cancer-specific biomarker of NSCLC. miR-205 was reported to be increased in lung cancer 16, 25, 26, promoting the tumor growth, metastasis, and chemoresistence in lung cancer cells via targeting PTEN signaling pathways 27. The targets genes of miR-205 are also involved in the cellular metabolic processes and nervous system development (Fig. S9). However, miR-29c and miR-30b were shown as tumor suppressor miRNAs in lung cancer 17, 28-33. Oncogenes such as myc, gli, and NF-κb can bind to the promotor region of miR-29c to suppress its transcription in cholangiocarcinoma cells 34. Muller Fabbri has shown that miR-29 inhibits tumorigenicity both in vitro and in vivo by using NSCLC cell line A549 and H441 35. Zhong K. et al. demonstrated that miR-30b/c was down-regulated in NSCLC specimens compared with adjacent non-tumor tissues and miR-30b/c directly targeted and down-regulated Rab18 expression and inhibited NSCLC cells proliferation 28.
By now many sets of miRNAs were reported being able to distinguish NSCLC patients from those non-cancer controls 10. However, discrepancies in the performance of miRNA sets do exist among published reports, which can be explained by the variance in the source of miRNA (tissue or body fluid), the methods applied for miRNA analysis (microarray or qRT-PCR), the criteria for analyzed miRNAs of interest, the ethnicity, and/or clinicopathologic features of recruited patients. Even in our current study, among the eight candidate miRNAs screened out by us and other groups, only four out of them demonstrated the potential for early diagnosis of NSCLC. Therefore, it is necessary to further validate the 4-miRNA panel in larger cohorts of patients with reasonable stratifications of other relevant features, such as ethnicity.
In summary, our current study reported a 4-miRNA panel as a powerful, sensitive, and non-invasive serum biomarker in early diagnosis of NSCLC patients. Our findings are especially pertinent and beneficial to high risk populations (eg. smokers) who have not been considered to receive routine LDCT evaluations. By the same token, we will pursue the role of the 4-miRNA panel in monitoring NSCLC patients in monitoring therapeutic responses and cancer recurrence in the future.
Materials and Methods
Patients and Serum Samples Collection
In this study, we recruited 128 patients with stage IA-IIIB NSCLC, 17 age-, gender- and smoking status-matched chronic obstructive pulmonary disease (COPD) patients and 30 healthy individuals from University of Michigan Health System (UM) from 1991 to 2013. The COPD patients are free of lung cancer or any other malignancies. The diagnosis of NSCLC patients was based on the revised AJCC/UICC 7th edition TNM classification schema 36. Written informed consent was obtained from each participant after explaining the nature of the study, which was approved by the research ethics board of University of Michigan Health System and was performed according to the guidelines of the Declaration of Helsinki. Blood specimens of the cancer patients were collected at the time of lung tumor resection without preoperative adjuvant chemotherapy or radiotherapy. The main clinical and pathological data of patients as well as cancer-free controls including age, gender, smoking status were provided in Table S5. Peripheral blood (5~10 mL) from each participant was drawn and processed for serum extraction within 1 hour by centrifugation at 3000 g for 10 min at room temperature. Serum was transferred to new microfuge tubes (400 µL in each tube) and frozen instantly in liquid nitrogen, and then stored at -80°C.
RNA Isolation and miRNA quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR)
Total RNAs were isolated from a volume of 400 μL of serum using miRVana PARIS Kit (Ambion), following the manufacturer's protocol. RNA concentration was measured by Nanodrop 2000 spectrophotometer (Thermo Scientific, NY) and stored at -80°C. The cDNA was produced by reverse transcription of a total of 100ng RNA in a thermocycler using TaqMan MicroRNA Reverse Transcription Kit with specific TaqMan probes (Thermo Scientific, NY), following the procedure: incubation for 30 min at 16°C, 30 min at 42°C, and 5 min at 85°C and held at 4°C. qRT-PCR was carried out on an Applied BioSystems 7900HT thermocycler at 95°C for 10min, followed by 40 cycles of 95°C for 15sec and 60°C for 1min. The PCR reaction was repeated in duplicates for each sample. Since there is no consensus on the optimal housekeeping gene in serum, the relative miRNA expression was calculated using the equation 2-ΔCT, in which ΔCT = cycle threshold (CT) of NSCLC, healthy control or COPD individuals - mean of CT of all healthy controls, which was described by Hu et al. 37. The fold-change of miRNA value was log2 transformed.
Statistical analysis
The Pearson chi square test or Fisher's exact test was used to analyze categorical variables. Student's t-test was used to compare continuous variables. The values of miRNA expressions which were not normally distributed were analyzed by Mann-Whitney test. Each miRNA was categorized as high or low using the median value as the cut-off when assessing the correlation with clinicopathological features such as differentiation, recurrence of all NSLCL patients (n=128). To be more clearly assessing 5-year overall survival (OS) in NSLCL patients, candidate miRNAs were categorized as high (n=42), moderate (n=44) and low (n=42) subgroups based on their serum levels using the two tripartite division values as cut-offs. Kaplan-Meier survival analysis with Mantel-Cox statistics was subsequently performed only between high and low subgroups. Receiver-operating curve (ROC) analysis was conducted and the area under the ROC curve (AUC) was calculated to evaluate the diagnostic capability of each miRNA between NSCLC patients and their relative controls. Youden's index (YI) 38 was used to find the optimal cut-off point for both specificity and sensitivity of each miRNA and was defined by the equation YI=(sensitivity+specificity)-1. A logistic regression model was used to define the miRNA model for the best discriminating NSCLC from cancer-free controls. All values were expressed as mean ± standard deviation (SD). Values of P < 0.05 were considered statistically significant. All calculations were performed using the statistical software, GraphPad Prism8, (Graphpad Software Inc., CA) and Statistical Package for the Social Sciences software (SPSS Inc., IL).
Supplementary Material
Supplementary figures and tables.
Acknowledgments
This work was supported in part by the General Project of Key Research and Development Program of Shaanxi Province (Grant No. 2018SF-074) to X.Y. National Natural Science Foundation of China (NSFC) (81702270 to W.S.); Affiliated Hospital of Guangdong Medical University Doctoral Foundation (Grant No. 2018052638) to W.S.
References
- 1.Bray F, Ferlay J, Soerjomataram I. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Wood SL, Pernemalm M, Crosbie PA. et al. Molecular histology of lung cancer: from targets to treatments. Cancer Treat Rev. 2015;41:361–375. doi: 10.1016/j.ctrv.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Wood DE, Kazerooni EA, Baum SL. et al. Lung Cancer Screening, Version 3.2018, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2018;16:412–441. doi: 10.6004/jnccn.2018.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aberle DR, Berg CD, Black WC. et al. The National Lung Screening Trial: overview and study design. Radiology. 2011;258:243–253. doi: 10.1148/radiol.10091808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snowsill T, Yang H, Griffin E. et al. Low-dose computed tomography for lung cancer screening in high-risk populations: a systematic review and economic evaluation. Health Technol Assess. 2018;22:1–276. doi: 10.3310/hta22690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung A, Kirchner T. Liquid Biopsy in Tumor Genetic Diagnosis. Dtsch Arztebl Int. 2018;115:169–174. doi: 10.3238/arztebl.2018.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu S, Kong H, Hou Y. et al. Two plasma microRNA panels for diagnosis and subtype discrimination of lung cancer. Lung Cancer. 2018;123:44–51. doi: 10.1016/j.lungcan.2018.06.027. [DOI] [PubMed] [Google Scholar]
- 10.Moretti F, D'Antona P, Finardi E. et al. Systematic review and critique of circulating miRNAs as biomarkers of stage I-II non-small cell lung cancer. Oncotarget. 2017;8:94980–94996. doi: 10.18632/oncotarget.21739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inamura K, Ishikawa Y. MicroRNA In Lung Cancer: Novel Biomarkers and Potential Tools for Treatment. J Clin Med; 2016. p. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nadal E, Zhong J, Lin J. et al. A MicroRNA cluster at 14q32 drives aggressive lung adenocarcinoma. Clin Cancer Res. 2014;20:3107–3117. doi: 10.1158/1078-0432.CCR-13-3348. [DOI] [PubMed] [Google Scholar]
- 13.Raponi M, Dossey L, Jatkoe T. et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 14.Bianchi F, Nicassio F, Marzi M. et al. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med. 2011;3:495–503. doi: 10.1002/emmm.201100154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Zhang H, Dong Y. et al. MiR-146b-5p functions as a suppressor miRNA and prognosis predictor in non-small cell lung cancer. J Cancer. 2017;8:1704–1716. doi: 10.7150/jca.16961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sromek M, Glogowski M, Chechlinska M. et al. Changes in plasma miR-9, miR-16, miR-205 and miR-486 levels after non-small cell lung cancer resection. Cell Oncol (Dordr) 2017;40:529–536. doi: 10.1007/s13402-017-0334-8. [DOI] [PubMed] [Google Scholar]
- 17.Zhu W, He J, Chen D. et al. Expression of miR-29c, miR-93, and miR-429 as potential biomarkers for detection of early stage non-small lung cancer. PLoS One. 2014;9:e87780. doi: 10.1371/journal.pone.0087780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matin F, Jeet V, Moya L. et al. A Plasma Biomarker Panel of Four MicroRNAs for the Diagnosis of Prostate Cancer. Sci Rep. 2018;8:6653. doi: 10.1038/s41598-018-24424-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Detterbeck FC. The eighth edition TNM stage classification for lung cancer: What does it mean on main street? J Thorac Cardiovasc Surg. 2018;155:356–359. doi: 10.1016/j.jtcvs.2017.08.138. [DOI] [PubMed] [Google Scholar]
- 20.U.S. National Institute Of Health, National Cancer Institute. SEER Cancer Statistics Review, 1975-2015. Available from: https://seer.cancer.gov/csr/1975_2015/sections.html.
- 21.de Koning HJ, Meza R, Plevritis SK. et al. Benefits and harms of computed tomography lung cancer screening strategies: a comparative modeling study for the U.S. Preventive Services Task Force. Ann Intern Med. 2014;160:311–320. doi: 10.7326/M13-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu GCW, Lazare K, Sullivan F. Serum and blood based biomarkers for lung cancer screening: a systematic review. BMC Cancer. 2018;18:181. doi: 10.1186/s12885-018-4024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durham AL, Adcock IM. The relationship between COPD and lung cancer. Lung Cancer. 2015;90:121–127. doi: 10.1016/j.lungcan.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Yang H, Li Y. et al. microRNA-146 up-regulation predicts the prognosis of non-small cell lung cancer by miRNA in situ hybridization. Exp Mol Pathol. 2014;96:195–199. doi: 10.1016/j.yexmp.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Halvorsen AR, Bjaanaes M, LeBlanc M. et al. A unique set of 6 circulating microRNAs for early detection of non-small cell lung cancer. Oncotarget. 2016;7:37250–37259. doi: 10.18632/oncotarget.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li JH, Sun SS, Li N. et al. MiR-205 as a promising biomarker in the diagnosis and prognosis of lung cancer. Oncotarget. 2017;8:91938–91949. doi: 10.18632/oncotarget.20262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lei L, Huang Y, Gong W. miR-205 promotes the growth, metastasis and chemoresistance of NSCLC cells by targeting PTEN. Oncol Rep. 2013;30:2897–2902. doi: 10.3892/or.2013.2755. [DOI] [PubMed] [Google Scholar]
- 28.Zhong K, Chen K, Han L. et al. MicroRNA-30b/c inhibits non-small cell lung cancer cell proliferation by targeting Rab18. BMC Cancer. 2014;14:703. doi: 10.1186/1471-2407-14-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun DM, Tang BF, Li ZX. et al. MiR-29c reduces the cisplatin resistance of non-small cell lung cancer cells by negatively regulating the PI3K/Akt pathway. Sci Rep. 2018;8:8007. doi: 10.1038/s41598-018-26381-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arechaga-Ocampo E, Lopez-Camarillo C, Villegas-Sepulveda N. et al. Tumor suppressor miR-29c regulates radioresistance in lung cancer cells. Tumour Biol. 2017;39:1010428317695010. doi: 10.1177/1010428317695010. [DOI] [PubMed] [Google Scholar]
- 31.Qi Z, Zhang B, Zhang J. et al. MicroRNA-30b inhibits non-small cell lung cancer cell growth by targeting the epidermal growth factor receptor. Neoplasma. 2018;65:192–200. doi: 10.4149/neo_2018_170217N118. [DOI] [PubMed] [Google Scholar]
- 32.Liu L, Bi N, Wu L. et al. MicroRNA-29c functions as a tumor suppressor by targeting VEGFA in lung adenocarcinoma. Mol Cancer. 2017;16:50. doi: 10.1186/s12943-017-0620-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhan S, Wang C, Yin F. MicroRNA-29c inhibits proliferation and promotes apoptosis in non-small cell lung cancer cells by targeting VEGFA. Mol Med Rep. 2018;17:6705–6710. doi: 10.3892/mmr.2018.8678. [DOI] [PubMed] [Google Scholar]
- 34.Mott JL, Kurita S, Cazanave SC. et al. Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-kappaB. J Cell Biochem. 2010;110:1155–1164. doi: 10.1002/jcb.22630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fabbri M, Iliopoulos D, Trapasso F. et al. WWOX gene restoration prevents lung cancer growth in vitro and in vivo. Proc Natl Acad Sci USA. 2005;102:15611–15616. doi: 10.1073/pnas.0505485102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Travis WD, Brambilla E, Noguchi M. et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Z, Chen X, Zhao Y. et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 38.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.