Abstract
Breast cancer is the most commonly diagnosed cancer and the leading cause of cancer death among women in the worldwide. Triple-negative breast cancer (TNBC) has a poor clinical outcome. The antitumor efficacy of Ilamycins, natural products with anti-tuberculosis activity isolated from deep sea-derived Streptomyces atratus, in TNBC has not been investigated, and the mechanisms remain elusive. Here, we demonstrated that Ilamycin-E, but not -F, decreases cell viability, inhibits G1/S cell cycle progression, and promotes apoptosis in the TNBC cell lines HCC1937 and MDA-MB-468. Ilamycin E promotes apoptosis via activation of endoplasmic reticulum (ER) stress, increasing the expression of CHOP, and down-regulating the expression of anti-apoptotic protein Bcl-2. Depletion of CHOP or overexpression of Bcl2 significantly rescued Ilamycin E-induced apoptosis. These findings indicate that Ilamycin E has anti-cancer activity in TNBC.
Keywords: Ilamycin E, triple-negative breast cancer, ER stress, CHOP, Bcl-2.
Introduction
Breast cancer is the leading cause of cancer-related deaths among women worldwide. It was reported that there were 626,679 estimated deaths and 2,088,849 estimated new cases of breast cancer in the worldwide in 2018 1. According to the expression status of estrogen receptor (ER) α, progesterone receptor (PR) and human epidermal growth factor 2 (HER-2), breast cancers are divided into the luminal (ER+ and/or PR+), HER-2 (HER-2+), and triple-negative (ER-/PR-/HER-2-) subtypes. Triple-negative breast cancer (TNBC), which accounts for 15-20 % of breast cancers, is the most malignant subtype with relatively poorer prognosis compared with other breast cancer subtypes. Because of the lack of molecular targets for therapy, surgery, radiation therapy and chemotherapy are still the main choices for TNBC patients 2-4. Thus, it is urgent to develop new effective therapies for TNBC.
The endoplasmic reticulum (ER) provides a suitable environment for secretory protein folding. Upon stress, the ER initiates a stress response via a well-coordinated and integrated signal transduction pathway, named the unfolded protein response (UPR) 5-7. The UPR is primarily mediated by three unique ER stress sensors: pancreatic ER kinase-like ER kinase (PERK), inositol requiring enzyme 1 (IRE1)α, and activating transcription factor 6 (ATF6) 8. In the presence of severe and irreparable ER stress, a switch in UPR from a pro-survival mode towards a pro-death response occurs through mitochondrial engagement, leading to the activation of the intrinsic apoptosis pathway. Apoptosis is mediated by transcriptional activation of pro-apoptotic transcription factor C/EBP homologous protein (CHOP) and the pro-apoptotic BH3-only protein Bim 9.
Natural compounds, which were extracted from medicinal plants and microorganisms, provide good resources for anti-cancer drug development 10-12. For example, Taxol and Vincristine are widely used anti-cancer drugs in the clinic 13. As the most abundant resource on the earth, ocean organisms are a very promising source for anti-cancer drug discovery 12, 14. Ilamycins are a series of cyclic peptides with anti-tuberculosis activity that are produced by Streptomyces atratus and Streptomyces islandicus 15-19. The anti-tumor activities of Ilamycins in MCF7 cells have been investigated 18. Whether Ilamycins have anti-cancer activities in triple-negative or other breast cancer cells has not been reported. Here, we demonstrated that Ilamycin E, a new analogue of Ilamycin, exhibits potent anti-cancer activity in TNBC cells in a time- and dosage-dependent manner. Ilamycin E significantly inhibited G1/S cell cycle progression and cell proliferation. Additionally, Ilamycin E effectively induced apoptosis via down-regulating Bcl-2 expression. We further demonstrated that Ilamycin E suppressed Bcl-2 expression via inducing ER stress, which activates CHOP in both HCC1937 and MDA-MB-468 breast cancer cell lines. These results suggest that Ilamycin E has anti-cancer activity in TNBC.
Materials and Methods
Compounds
The discovery, structural elucidation and biosynthesis of Ilamycin E and Ilamycin F have been described previously 18, 19. All compounds were dissolved in DMSO (Sigma, St Louis, MO, USA).
Cell lines and cell culture
All cell lines used in this study were obtained from the ATCC (American Type Culture Collection, Manassas, VA, USA) and were cultured at 37 °C with 5 % CO2. HCC1937 and HCC1806 cell lines were cultured in RPMI 1640 medium (Gibco, Carlsbad, CA, USA) supplemented with 10 % FBS (fetal bovine serum, Gibco). MDA-MB-468, MCF-7 and SKBR3 cell lines were maintained in DMEM medium (Gibco) with 10 % FBS. The MDA-MB-231 cell line was maintained in F12 (1:1) medium (Gibco) with 5 % FBS. T47D and BT474 cell lines were cultured in RPMI 1640 medium supplemented with 10 % FBS and 0.2 Units/ml insulin. MCF10A cell line was maintained in DMED F12 medium (Gibco) with 5 % horse serum (Gibco), 20 ng/ml epidermal growth factor (Peprotech, Rocky Hill, NJ, USA), 100 ng/ml cholera toxin (Sigma, St Louis, MO, USA), 0.16 Units/ml insulin and 500 ng/ml hydrocortisone (Sigma).
Cell viability and proliferation assays
Cell viability was measured by sulforhodamine B (SRB, Sigma, St. Louis, MO) assays 20. Briefly, cells were fixed with 10 % trichloroacetic acid (TCA, Sigma) at 4 °C for 60 minutes at the indicated time, followed by incubation with 0.4 % SRB (w/v) solution in 1 % acetic acid for 5 minutes at room temperature. Finally, the SRB was dissolved with 10 mM unbuffered Tris base, and the absorbance was measured at a single wavelength of 490-530 nm on plate reader (BioTek, VT, USA). Proliferation of HCC1937 and MDA-MB-468 cells was measured with Click-iT® EdU Flow Cytometry Assay Kits (Invitrogen, Carlsbad, CA, USA).
Cell cycle analysis
HCC1937 and MDA-MB-468 cells were seeded in six-well plates at a density of 2.0×105 and 2.4×105 cells per well, respectively. One day after plating, the cells were treated with Ilamycin E at the indicated dosages. Twenty-four and thirty-six hours later, the cells were collected and fixed with 75 % cold ethanol at 4 °C overnight. After fixation, cells were stained with propidium iodide (PI, eBioscience, San Diego, CA, USA) buffer (0.3 % NP-40, 0.05 mg/ml PI, 0.5 mg/ml RNase A) at room temperature for 30 minutes followed by analysis on an Accuri C6 flow cytometer (BD Biosciences, San Diego, USA) within 4 h.
Apoptosis analysis
HCC1937 and MDA-MB-468 cells were seeded in six-well plates at densities of 2.0×105 and 2.4×105 cells per well, respectively. All cells and media were collected after being incubated with Ilamycin E at the indicated dosages for 24-36 hours. Cells were then collected and stained with anti-Annexin V antibody and PI (eBioscience, San Diego, CA, USA). Finally, all cells were analyzed on an Accuri C6 flow cytometer.
Western blot assay
HCC1937 and MDA-MB-468 cells were collected in lysis buffer containing protease inhibitors (P8340, Sigma) after being treated with Ilamycin E for 4-48 hrs. Cell lysate was subjected to SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes. The membranes were incubated with primary antibodies, horseradish peroxidase (HRP) conjugated secondary antibodies (Jackson ImmunoResearch Laboratory, West Grove, PA), and ImmobilonTM Western Chemiluminescent HRP Substrate (Millipore, MA, USA) and were finally visualized on an ImageQuant LAS4000 Biomolecular imager to determine the expression levels of specific proteins.
Anti-PARP, Caspase-3, 7, 8 and 9, Bcl-2, Mcl-1, CyclinD1, β-actin, PERK, IRE1α, CHOP, Bip, Bax, and Bim antibodies were purchased from Cell signaling (Cell Signaling Technology, Danvers, MA, USA). Anti-p21, GAPDH, and XBP1 antibodies were purchased from Santa Cruz (Santa Cruz Biotechnology, CA, USA). Anti-p27 and ATF6 antibodies were purchased from Abcam (Abcam, Cambridge, UK). Anti-DR5 antibody was purchased from Sigma (Sigma), and anti-Cyclin B1 antibody was purchased from Abnova (Abnova, Co. USA).
Transfection
We used Lipofectamine 2000 (Invitrogen) for siRNA and plasmid transfection according to the manufacturer's recommended protocols. Control siRNA was purchased from RiboBio Co., Ltd (Guangzhou, Guangdong, China). The sequences of CHOP siRNA are 5'-GGUCCUGUCCUCAGAUGAA-3' and 5'-CUGAUUGACCGAAUGGUGATT-3'. The siRNA target sequences for the human Bax gene are 5'-GTGCCGGAACTGATCAGAA-3' (Baxsi#1), 5'-GCTGGACATTGGACTTCCT-3' (Baxsi#2), 5'-AGTGGCAGCTGACATGTTT-3' (Baxsi#3). The siRNA target sequences for the human Bim gene are 5'-GCAACCTTCTGATGTAAGT-3' (Bimsi#1), 5'-CCTACAGACAGAGCCACAA-3' (Bimsi#2), and 5'-CCAGGCCTTCAACCACTAT-3' (Bimsi#3). Pooled siRNAs were used to knock down target genes.
Over-expression of Bcl-2
The Bcl-2 cDNA was amplified and cloned into the pCDH lentiviral vector using forward primer, 5'-CGGAATTCATGGCGCACGCTGGGAGAACA-3' and reverse primer 5'-TAAAGCGGCCGCTCACTTGTGGCCCAGATAGGCA-3'. The plasmid was confirmed by DNA sequencing. For lentivirus packaging, HEK293T cells were seeded in 10-cm plates at a density of 3×106 per dish. The next day, the cells were transfected with pCDH-Bcl2/pCDH and packaging plasmids using Lipofectamine 2000 according to the manufacturer's protocols. Lentiviruses were collected at both 48 and 72 hrs after transfection and used to infect HCC1937 cells. The infected cells were selected using 1 μg/ml puromycin to generate stable cell populations.
Real-time PCR
Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, USA) and cDNA was synthesized using the Reverse Transcription Kit (Applied Biosystems, Austin, TX, USA). For quantitative PCR, SYBR PCR Master Mix (Applied Biosystems) was used to quantify the expression level of target mRNA on a 7900HT FastReal-Time PCR System (Applied Biosystems). The Bcl2 gene primer sequences: Forward, 5'-CAACATCGCCCTGTGGATGA-3' and Reverse, 5'-GGCCAAACTGAGCAGAGTCT-3'. GAPDH was used as the loading control.
Statistical analysis
Data are shown as the means±s.d. Student's t-test was used for statistical analysis. GraphPad Prism 6 (GraphPad Software Inc. La Jolla, CA, USA) and SPSS 20.0 Software (IBM Inc., Armonk, NY, USA) were used for all statistical analyses. P-values<0.05 were considered significant.
Results
Ilamycin E inhibits breast cancer cell viability
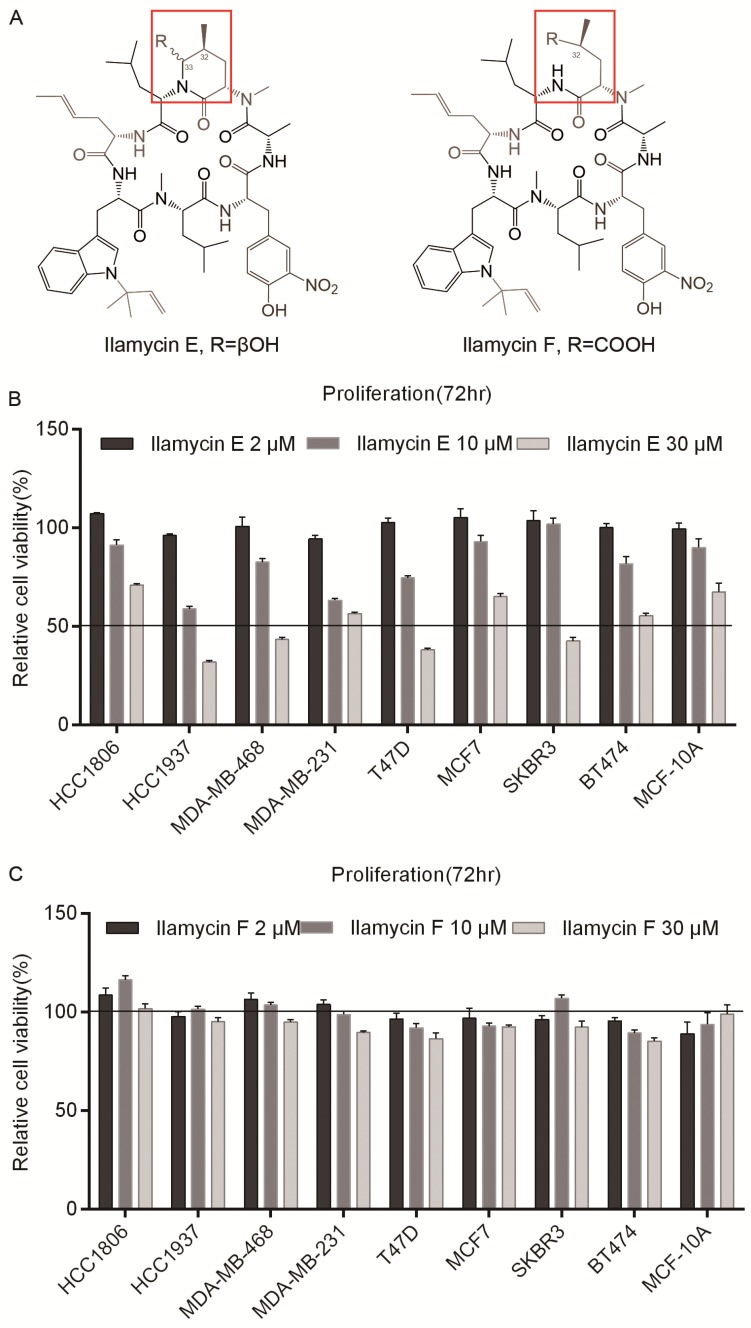
Ilamycin E and Ilamycin F, two Ilamycin congeners with known absolute stereochemistries, were isolated from the deep South China Sea-derived S. atratus SCSIO ZH16 and its genetically engineered mutants 18. To investigate whether these Ilamycin congeners (Fig. 1A) have anti-tumor activities in breast cancer, SRB assays were performed to test cell viabilities after Ilamycin E treatment in four TNBC cell lines (HCC1806, HCC1937, MDA-MB-468, and MDA-MB-231), two ERα positive breast cancer cell lines (T47D and MCF7), two Her-2 positive breast cancer cell lines (SKBR3 and BT474) and an immortalized breast cell line MCF-10A. Cell viabilities of all cell lines were significantly reduced by Ilamycin E (Fig. 1B), but not Ilamycin F (Fig. 1C), in a dosage-dependent manner.
Figure 1.
Ilamycin E showed cytotoxicity in breast cell lines (A) The chemical structures of Ilamycin E and Ilamycin F. (B) Ilamycin E decreased breast epithelial cell viability in a dose-dependent manner. The cell viability of 9 breast cell lines was determined by SRB assays after treating with Ilamycin E for 72 hrs. (C) Ilamycin F did not decrease breast epithelial cell viability.
We further determined the IC50 values of Ilamycin E in these cell lines. As the data shown in Table 1, the IC50 values of Ilamycin E varied from 14.24 to 49.19 μM. MCF-10A, an immortalized normal breast epithelial cell line showed the highest IC50.
Table 1.
IC50 of Ilamycin E in 9 different cell lines, including 4 TNBC cell lines, 2 ERα-positive breast cancer cell lines, 2 Her-2 positive breast cancer cell lines, and 1 control cell line.
| Cell types | Cell lines | IC50(μM) |
|---|---|---|
| TNBC | HCC1806 | 47.50 |
| HCC1937 | 14.24 | |
| MDA-MB-468 | 24.56 | |
| MDA-MB-231 | 33.72 | |
| ER+ breast cancer | T47D | 18.95 |
| MCF7 | 40.27 | |
| Her2+ breast cancer | SKBR3 | 26.18 |
| BT474 | 30.86 | |
| Immortalized | MCF-10A | 49.19 |
Ilamycin E inhibits TNBC cell proliferation and cell cycle progression
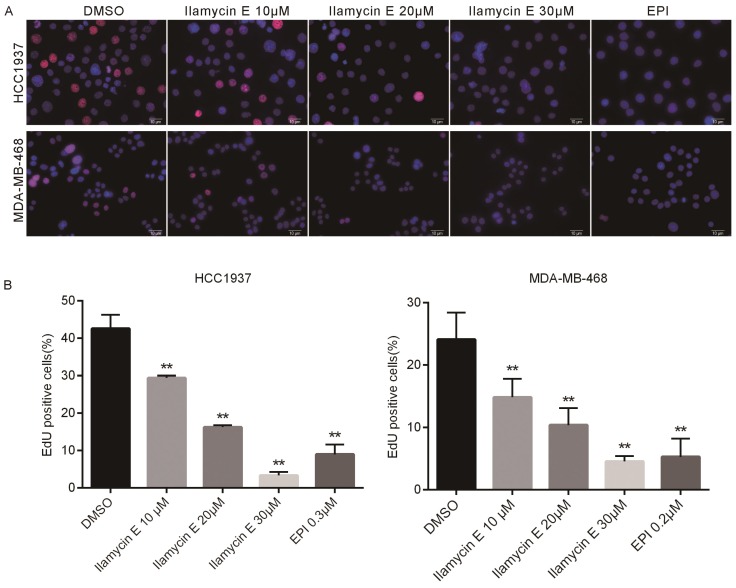
Considering Ilamycin E decreased TNBC cell viabilities and the effective therapies for TNBC are limited, we investigated whether Ilamycin E inhibits TNBC cell proliferation. EdU assays were performed to monitor the DNA synthesis of HCC1937 and MDA-MB-468, two TNBC cell lines. As shown in Fig. 2, the EdU positive HCC1937 and MDA-MB-468 cells were dramatically decreased by Ilamycin E treatment in a dose-dependent manner.
Figure 2.
Ilamycin E inhibits DNA synthesis of HCC1937 and MDA-MB-468 TNBC cell lines (A) Ilamycin E inhibits TNBC cell DNA synthesis, as measured by the EdU assay. HCC1937 and MDA-MB-468 cells were treated with Ilamycin E for 24 hrs. DMSO and Epirubicin (EPI) were used as negative and positive controls, respectively. (B) Quantitative data for the EdU assay in HCC1937 and MDA-MB-468 cells. Data represent the mean ± SD (n=3). *p < 0.05, **p < 0.01.
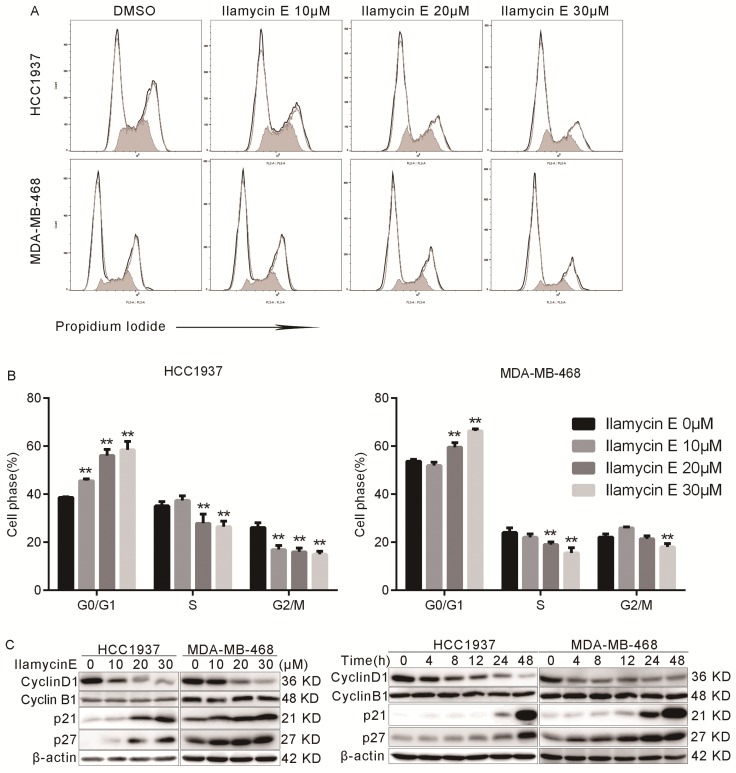
We further tested whether Ilamycin E inhibits cell cycle progression in HCC1937 and MDA-MB-468 cells. As shown in Fig. 3A-B, the proportion of cells in G0/G1 phase was significantly increased after Ilamycin E treatment, while those in S and G2 phases were dramatically decreased. These results indicated that Ilamycin E induced G1/S cell cycle arrest in these cell lines.
Figure 3.
Ilamycin E induces G1/S cell cycle arrest in HCC1937 and MDA-MB-468 (A) The cell cycle distributions of HCC1937 and MDA-MB-468 cells after Ilamycin E treatment. (B) Ilamycin E significantly increased the percentage of cells in the G1 phase and decreased the percentage of cells in the S phase in a dose dependent manner. Each experiment was conducted in triplicate and data are presented as the mean ± SD (n=3). *p < 0.05, **p < 0.01. (C) HCC1937 and MDA-MB-468 cells were treated with different concentrations of Ilamycin E for 48 hrs or treated with 30 μΜ Ilamycin E for 4 to 48 hrs. The expression levels of cell cycle related proteins, Cyclin D1, Cyclin B1, p21, and p27, were detected by WB.
Furthermore, we analyzed several cell cycle-associated proteins by Western blotting. As shown in Fig. 3C-D, Ilamycin E decreased Cyclin D1 and increased p21 and p27 protein levels in time and dose dependent manners in both HCC1937 and MDA-MB-468 cell lines.
Ilamycin E induces apoptosis in HCC1937 and MDA-MB-468 cells
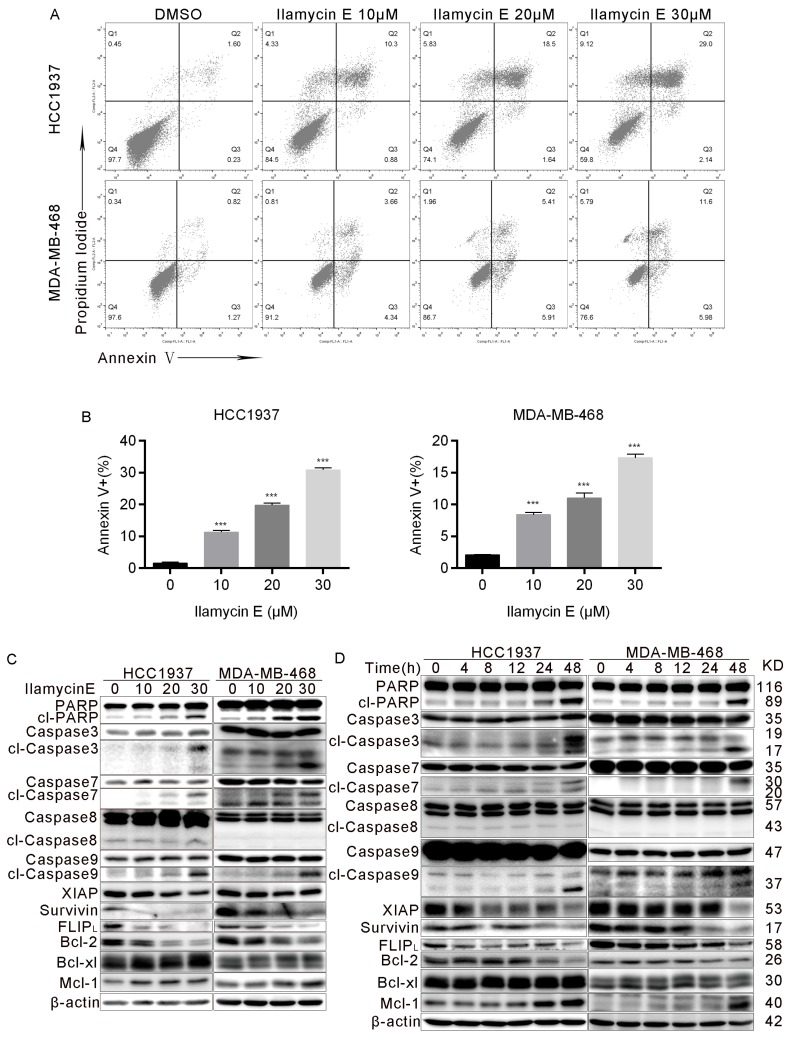
To investigate whether Ilamycin E induces apoptosis, we treated HCC1937 and MDA-MB-468 cells with Ilamycin E and measured apoptosis by AnnexinⅤstaining. As shown in Fig. 4A-B, Ilamycin E dramatically induced apoptosis in HCC1937 and MDA-MB-468 cells as indicated by the increase of Annexin V positive cells. We further examined the cleavage of PARP and Caspase-3 and -7 and found that Ilamycin E (20-30 μM) significantly increased the cleavage of these proteins (Fig. 4C-D). Meanwhile, Ilamycin E decreased the protein levels of Bcl-2, but not Bcl-xL and Mcl1, in dose and time dependent manners (Fig. 4C-D). We noticed that the cleavage of Caspase 9, but not Caspase 8, was significantly induced by Ilamycin E (Fig. 4C-D), suggesting that Ilamycin E induced TNBC apoptosis most likely via the intrinsic but not extrinsic pathway.
Figure 4.
Ilamycin E induces apoptosis in HCC1937 and MDA-MB-468 cells (A) Ilamycin E increased Annexin V positive apoptotic cells. HCC1937 and MDA-MB-468 cells were treated with Ilamycin E (30 μM) for 24 to 36 hrs, stained with Annexin V/PI, and analyzed by flow cytometry. (B) The quantitative data of panel A. The percentages of Annexin V-positive cells are shown. *p < 0.05, **p < 0.01. (C) HCC1937 and MDA-MB-468 cells were treated with different concentration of Ilamycin E for 48 hrs. The expression levels of apoptosis related proteins were detected by WB. (D) HCC1937 and MDA-MB-468 cells were treated with 30 μΜ Ilamycin E from 4 to 48 hrs. The expression levels of apoptosis related proteins were detected by WB.
Ilamycin E induces apoptosis partially through suppressing Bcl-2 expression
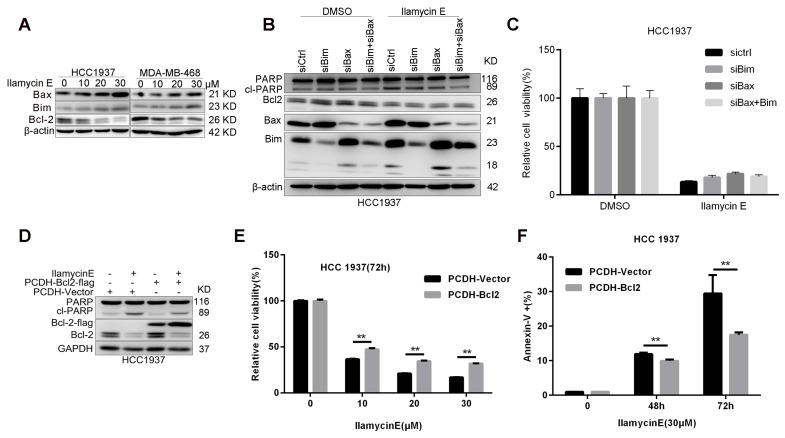
It is well known that, Caspase-9 is an initial caspase in the intrinsic apoptotic pathway, whereas Caspase-8 initializes the extrinsic apoptotic pathway. To understand the potential mechanism by which Ilamycin E induces apoptosis, we detected the expression of several anti-apoptotic proteins in the intrinsic apoptotic pathway and noticed that Bcl-2 was specifically and dramatically down-regulated in both HCC1937 and MDA-MB-468 cell lines 21 (Fig. 4C-D). Although Ilamycin E induces the expression of pro-apoptotic proteins Bim and Bax (Fig. 5A), knockdown of Bim and Bax did not block Ilamycin E induced apoptosis (Fig. 5B-C). To test whether Ilamycin E functions through down-regulating Bcl-2, we ectopically over-expressed Bcl-2 in HCC1937 cells (Fig. 5C). Bcl-2 overexpression indeed partially rescued Ilamycin E induced apoptosis (Fig. 5E) and loss of cell viability (Fig. 5F), indicating that Ilamycin E, at least partially, induced apoptosis through suppression of Bcl-2 expression.
Figure 5.
Ilamycin E induces HCC1937 apoptosis partially through down-regulate of Bcl-2, but not upregulation of Bax and Bim (A) Ilamycin E inhibited anti-apoptotic protein Bcl2 expression and induced pro-apoptotic protein Bax and Bim expression in HCC1937 and MDA-MB-468 cells in a dose dependent manner, as measured by WB. Both HCC1937 and MDA-MB-468 cells were treated with Ilamycin E for 48 hrs. (B) Knockdown of Bax and Bim in HCC1937 was measured by WB. (C) Knockdown of Bax and Bim only slightly blocked Ilamycin E-induced apoptosis in HCC1937 cells, as measured by Annexin V staining and flow cytometry. (D) Ectopic overexpression of Bcl-2-Flag partially blocked Ilamycin E-induced PARP cleavage, as measured by WB. (E) Ectopic overexpression of Bcl-2-Flag partially blocked Ilamycin E-induced apoptosis, as measured by Annexin V staining and flow cytometry. **p < 0.01. (F) Ectopic overexpression of Bcl-2-Flag partially blocked Ilamycin E-induced loss of cell viability at 48 and 72 hrs. **p < 0.01.
Ilamycin E down-regulates Bcl-2 mRNA expression by inducing ER stress and CHOP
It has been reported that the CHOP transcription factor can directly inhibit Bcl-2 gene transcription and reduce Bcl-2 protein expression 22, 23 and that CHOP is activated by ER stress 24. We then checked whether Ilamycin E suppresses Bcl-2 expression via inducing ER stress. As shown in Fig. 6A-B, Ilamycin E induced the expression of PERK, IRE1α, Bip, XBP1, and CHOP in time- and dose-dependent manners in both HCC1937 and MDA-MB-468 cell lines. As expected, the Bcl2 mRNA levels were downregulated by Ilamycin E in both HCC1937 and MDA-MB-468 cell lines (Fig. 6C). Interestingly, the inhibition of CHOP by short-interfering RNAs blocked Ilamycin E induced Bcl-2 down regulation (Fig. 6D). Furthermore, CHOP knockdown also significantly inhibited Ilamycin E induced apoptosis in HCC1937 (Fig. 6E), indicating that Ilamycin E down-regulates Bcl-2 expression at least partially through inducing ER stress and CHOP.
Figure 6.
Ilamycin E induced Bcl-2 expression down-regulation is mediated by the activation of ER Stress and the induction of CHOP (A) Ilamycin E induced ER stress related protein expression in HCC1937 and MDA-MB-468 cells in a dose dependent manner, as measured by WB. Both HCC1937 and MDA-MB-468 cells were treated with Ilamycin E for 48 hrs. (B) Ilamycin E induced ER stress related protein expression in HCC1937 and MDA-MB-468 cells in a time dependent manner, as measured by WB. Both HCC1937 and MDA-MB-468 cells were treated with 30 μM of Ilamycin E. (C) Ilamycin E significantly decreased Bcl2 mRNA expression levels in both HCC1937 and MDA-MB-468 cells, as measured by RT-qPCR. **p < 0.01. (D) Knockdown of CHOP decreased Ilamycin E induced Bcl-2 expression down-regulation in HCC1937, as measured by WB. (E) Knockdown of CHOP significantly blocked Ilamycin E-induced apoptosis in HCC1937 cells, as measured by Annexin V staining and flow cytometry. *p < 0.05.
Discussion and Conclusion
Ilamycin E is a new natural product with anti-tuberculosis activities and anti-tumor activities isolated from the deep sea-derived Streptomyces atratus 18; however, it is unclear whether this compound has anti-tumor activities to TNBC cancer cells or other breast cancer cell lines. In this study, we demonstrated that Ilamycin E exhibits potent anti-tumor activities in eight breast cancer cell lines, including TNBCs, with IC50 values varying from 14 to 48 µM. Ilamycin E not only strongly inhibited TNBC cell proliferation and cell cycle progression, but also potently induced apoptosis. Ilamycin E up-regulated the expression of cell cycle inhibitors p21 and p27, down-regulated the expression of CyclinD1 and anti-apoptotic protein Bcl-2. Furthermore, we demonstrated that Ilamycin E decreased the transcription of Bcl-2 via inducing ER stress downstream transcription factor CHOP 25(Fig. 4). Knockdown of CHOP or over-expression of Bcl-2 significantly reduced Ilamycin E-induced apoptosis. These findings suggest that Ilamycin E inhibits TNBC partially through the ER stress-CHOP-Bcl-2 axis. It is worth pointing out that the anti-cancer activities of Ilamycin E are not limited to TNBC.
ER is a central eukaryotic cellular organelle that provides crucial biosynthetic, stress-sensing, and signaling functions 26, 27. Pathophysiological conditions, including hypoxia, Ca2+ depletion, oxidative injury, hypoglycemia, and viral infections, disrupt ER homeostasis, hinder the protein folding process and activate the ER stress 28. Once ER stress is triggered, sustained activation of PERK signaling induces phosphorylation of EIF2α, which in turn increases the translation of ATF4 29. ATF4 is a transcription factor for C/EBP homologous protein (CHOP) transcription 30. CHOP is also known as growth arrest and DNA damage-inducible gene 153 (GADD153) and plays an important role in ER stress-induced cellular death 31. CHOP suppresses Bcl-2 transcription, linking the ER stress response to mitochondrial apoptosis 23, 25, 32.
In this study, we found that Ilamycin E induced prolonged ER stress, the activation of PERK and IRE1α, the enhancement of CHOP expression, and the down-regulation of Bcl-2 expression. It is well-known that Bcl-2 inhibits apoptosis by maintaining the integrity of the mitochondrial out membrane. The cell viability of HCC1937 was significantly increased by restoring expression of Bcl-2. We noticed that Ilamycin E also decreased the expression levels of anti-apoptotic XIAP, Survivin, and c-FLIP proteins; therefore, we could not exclude the possibility that Ilamycin E functions through these proteins. In agreement with this, Bcl2 overexpression could only rescued Ilamycin E-induced apoptosis in part. Interestingly, long time treatment of Ilamycin E increased the Mcl1 protein expression. We demonstrated that knockdown of CHOP also blocked Ilamycin E-induced apoptosis. Consistently, targeting Bcl-2 with the BH3 mimetic ABT-199 is promising for breast cancer therapy 33. Previously, we found that several small molecular compounds with anti-breast cancer activities, including mifepristone, CYD-6-28, and YD-277, induced apoptosis through triggering ER stress 34-36. XBP1/miR-153, DR5, and IRE1α mediate the pro-apoptotic functions of these compounds 37, respectively. We found that Ilamycin E indeed induced IRE1α, DR5, Bim and Bax. Interestingly, knockdown of Bim and Bax did not block Ilamycin E induced apoptosis. Nevertheless, it's reasonable to believe that the CHOP-Bcl-2 axis plays a key role in Ilamycin E induced apoptosis. The direct target of Ilamycin E needs further investigation.
In summary, we showed that Ilamycin E inhibits TNBC cell growth and induces apoptosis through activating the intrinsic apoptotic pathway. Ilamycin E effectively suppressed cell survival at least partially via the ER stress/CHOP/Bcl-2 axis, providing a potential therapeutic agent for breast cancer treatment.
Acknowledgments
We sincerely appreciate the kind help from Prof. Qinghua Cui at Yunnan University for providing the Bcl-2 plasmid. This work was supported by grants from National Natural Science Foundation of China (81672624 to Feng, J., 81830087, U1602221, and 31771516 to Chen, C., 81672639 to Zhou, Z., 81772847 and U1502222 to Liu, R.), the Seed Fund Program of Shanghai University of Medicine & Health Sciences (SFP-17-22-15-030 to Wang, L and SFP-18-21-15-004 to Zhao, W) and the Scientific Research Foundation for Talented Scholars of Hubei University of Medicine (2017QDJZR15 to Zhou, W).
Abbreviations
- ATF6
Activating Transcription Factor 6
- CHOP
C/EBP Homologous Protein
- ER
Endoplasmic Reticulum
- ERα
Estrogen Receptor α
- HER-2
Human Epidermal growth factor 2
- IRE1α
Inositol Requiring Enzyme 1α
- PERK
Pancreatic ER kinase-like ER kinase
- PR
Progesterone Receptor
- TNBC
Triple-Negative Breast Cancer
- UPR
Unfolded Protein Response
- WB
Western blotting
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians; 2018. [DOI] [PubMed] [Google Scholar]
- 2.Collignon J, Lousberg L, Schroeder H, Jerusalem G. Triple-negative breast cancer: treatment challenges and solutions. Breast cancer. 2016;8:93–107. doi: 10.2147/BCTT.S69488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cetin I, Topcul M. Triple negative breast cancer. Asian Pacific journal of cancer prevention: APJCP. 2014;15:2427–31. doi: 10.7314/apjcp.2014.15.6.2427. [DOI] [PubMed] [Google Scholar]
- 4.Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nature reviews Clinical oncology. 2010;7:683–92. doi: 10.1038/nrclinonc.2010.154. [DOI] [PubMed] [Google Scholar]
- 5.Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nature reviews Cancer. 2014;14:581–97. doi: 10.1038/nrc3800. [DOI] [PubMed] [Google Scholar]
- 6.Clarke HJ, Chambers JE, Liniker E, Marciniak SJ. Endoplasmic reticulum stress in malignancy. Cancer cell. 2014;25:563–73. doi: 10.1016/j.ccr.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 7.Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene. 2013;32:805–18. doi: 10.1038/onc.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maas NL, Diehl JA. Molecular pathways: the PERKs and pitfalls of targeting the unfolded protein response in cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2015;21:675–9. doi: 10.1158/1078-0432.CCR-13-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verfaillie T, Garg AD, Agostinis P. Targeting ER stress induced apoptosis and inflammation in cancer. Cancer letters. 2013;332:249–64. doi: 10.1016/j.canlet.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 10.Gammone MA, Riccioni G, Galvano F, D'Orazio N. Novel Therapeutic Strategies Against Cancer: Marine-derived Drugs May Be the Answer? Anti-cancer agents in medicinal chemistry. 2016;16:1549–57. doi: 10.2174/1871520616666160211123841. [DOI] [PubMed] [Google Scholar]
- 11.Russo P, Del Bufalo A, Fini M. Deep sea as a source of novel-anticancer drugs: update on discovery and preclinical/clinical evaluation in a systems medicine perspective. EXCLI journal. 2015;14:228–36. doi: 10.17179/excli2014-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russo P, Nastrucci C, Cesario A. From the sea to anticancer therapy. Current medicinal chemistry. 2011;18:3551–62. doi: 10.2174/092986711796642652. [DOI] [PubMed] [Google Scholar]
- 13.Hawkins MJ. New anticancer agents: taxol, camptothecin analogs, and anthrapyrazoles. Oncology. 1992;6:17–23. discussion 7-30. [PubMed] [Google Scholar]
- 14.Skropeta D, Wei L. Recent advances in deep-sea natural products. Natural product reports. 2014;31:999–1025. doi: 10.1039/c3np70118b. [DOI] [PubMed] [Google Scholar]
- 15.Kaku R. [Studies on the antitubercular activity of a new antibiotic, ilamycin. 1. Antitubercular activity of water-insoluble ilamycin] The Journal of antibiotics Ser B. 1963;16:93–8. [PubMed] [Google Scholar]
- 16.Takita T. L-Gamma-Formyl-N-Methyl-Norvaline, a New Amino Acid in Ilamycin. The Journal of antibiotics. 1963;16:175–7. [PubMed] [Google Scholar]
- 17.Cary LW, Takita T, Ohnishi M. A study of the secondary structure of ilamycin B(1) by 300 MHz proton magnetic resonance. FEBS letters. 1971;17:145–8. doi: 10.1016/0014-5793(71)80584-7. [DOI] [PubMed] [Google Scholar]
- 18.Ma J, Huang H, Xie Y, Liu Z, Zhao J, Zhang C. et al. Biosynthesis of ilamycins featuring unusual building blocks and engineered production of enhanced anti-tuberculosis agents. Nature communications. 2017;8:391. doi: 10.1038/s41467-017-00419-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Q, Chen Q, Song Y, Huang H, Li J, Ma J. et al. Deciphering the sugar biosynthetic pathway and tailoring steps of nucleoside antibiotic A201A unveils a GDP-l-galactose mutase. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:4948–53. doi: 10.1073/pnas.1620191114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou W, Song F, Wu Q, Liu R, Wang L, Liu C. et al. miR-217 inhibits triple-negative breast cancer cell growth, migration, and invasion through targeting KLF5. PloS one. 2017;12:e0176395. doi: 10.1371/journal.pone.0176395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siddiqui WA, Ahad A, Ahsan H. The mystery of BCL2 family: Bcl-2 proteins and apoptosis: an update. Archives of toxicology. 2015;89:289–317. doi: 10.1007/s00204-014-1448-7. [DOI] [PubMed] [Google Scholar]
- 22.Wu J, Kaufman RJ. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell death and differentiation. 2006;13:374–84. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- 23.McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Molecular and cellular biology. 2001;21:1249–59. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu TH, Tseng TJ, Su CC, Tang FC, Yen CC, Liu YY. et al. Arsenic induces reactive oxygen species-caused neuronal cell apoptosis through JNK/ERK-mediated mitochondria-dependent and GRP 78/CHOP-regulated pathways. Toxicology letters. 2014;224:130–40. doi: 10.1016/j.toxlet.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 25.Puthalakath H, O'Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND. et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129:1337–49. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 26.Sasaki K, Yoshida H. Organelle autoregulation-stress responses in the ER, Golgi, mitochondria and lysosome. Journal of biochemistry. 2015;157:185–95. doi: 10.1093/jb/mvv010. [DOI] [PubMed] [Google Scholar]
- 27.Wang S, Kaufman RJ. The impact of the unfolded protein response on human disease. The Journal of cell biology. 2012;197:857–67. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhat TA, Chaudhary AK, Kumar S, O'Malley J, Inigo JR, Kumar R. et al. Endoplasmic reticulum-mediated unfolded protein response and mitochondrial apoptosis in cancer. Biochimica et biophysica acta. 2017;1867:58–66. doi: 10.1016/j.bbcan.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J. et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nature cell biology. 2013;15:481–90. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Averous J, Bruhat A, Jousse C, Carraro V, Thiel G, Fafournoux P. Induction of CHOP expression by amino acid limitation requires both ATF4 expression and ATF2 phosphorylation. The Journal of biological chemistry. 2004;279:5288–97. doi: 10.1074/jbc.M311862200. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Guo Y, Tang J, Jiang J, Chen Z. New insights into the roles of CHOP-induced apoptosis in ER stress. Acta Biochim Biophys Sin (Shanghai) 2014;46:629–40. doi: 10.1093/abbs/gmu048. [DOI] [PubMed] [Google Scholar]
- 32.Gurlo T, Rivera JF, Butler AE, Cory M, Hoang J, Costes S. et al. CHOP Contributes to, But Is Not the Only Mediator of, IAPP Induced beta-Cell Apoptosis. Molecular endocrinology. 2016;30:446–54. doi: 10.1210/me.2015-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaillant F, Merino D, Lee L, Breslin K, Pal B, Ritchie ME. et al. Targeting BCL-2 with the BH3 mimetic ABT-199 in estrogen receptor-positive breast cancer. Cancer cell. 2013;24:120–9. doi: 10.1016/j.ccr.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 34.Chen Z, Wu Q, Ding Y, Zhou W, Liu R, Chen H. et al. YD277 Suppresses Triple-Negative Breast Cancer Partially Through Activating the Endoplasmic Reticulum Stress Pathway. Theranostics. 2017;7:2339–49. doi: 10.7150/thno.17555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu J, Ding Y, Chen CH, Zhou Z, Ding C, Chen H. et al. A new oridonin analog suppresses triple-negative breast cancer cells and tumor growth via the induction of death receptor 5. Cancer letters. 2016;380:393–402. doi: 10.1016/j.canlet.2016.06.024. [DOI] [PubMed] [Google Scholar]
- 36.Liu R, Shi P, Nie Z, Liang H, Zhou Z, Chen W. et al. Mifepristone Suppresses Basal Triple-Negative Breast Cancer Stem Cells by Down-regulating KLF5 Expression. Theranostics. 2016;6:533–44. doi: 10.7150/thno.14315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang H, Xiao J, Zhou Z, Wu J, Ge F, Li Z, Hypoxia induces miR-153 through the IRE1alpha-XBP1 pathway to fine tune the HIF1alpha/VEGFA axis in breast cancer angiogenesis. Oncogene; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]