Consumption of E+ tall fescue has an estimated annual $1 billion negative impact on the U.S. beef industry, with one driver of these costs being lowered weight gains. As global agricultural demand continues to grow, mitigating production losses resulting from grazing the predominant southeastern United States forage grass is of great value. Our investigation of the effects of E+ grazing on the fecal microbiota furthers our understanding of bovine fescue toxicosis in a real-world grazing production setting and provides a starting point for identifying easy-to-access fecal bacteria that could serve as potential biomarkers of animal productivity and/or FT severity for tall fescue-grazing livestock.
KEYWORDS: Epichloë coenophialum, fescue toxicosis, beef cattle, ergot alkaloids, microbiome, tall fescue
ABSTRACT
Tall fescue, the predominant southeastern United States cool-season forage grass, frequently becomes infected with an ergot alkaloid-producing toxic endophyte, Epichloë coenophialum. Consumption of endophyte-infected fescue results in fescue toxicosis (FT), a condition that lowers beef cow productivity. Limited data on the influence of ergot alkaloids on rumen fermentation profiles or ruminal bacteria that could degrade the ergot alkaloids are available, but how FT influences the grazing bovine fecal microbiota or what role fecal microbiota might play in FT etiology and associated production losses has yet to be investigated. Here, we used 16S rRNA gene sequencing of fecal samples from weaned Angus steers grazing toxic endophyte-infected (E+; n = 6) or nontoxic (Max-Q; n = 6) tall fescue before and 1, 2, 14, and 28 days after pasture assignment. Bacteria in the Firmicutes and Bacteroidetes phyla comprised 90% of the Max-Q and E+ steer fecal microbiota throughout the trial. Early decreases in the Erysipelotrichaceae family and delayed increases of the Ruminococcaceae and Lachnospiraceae families were among the major effects of E+ grazing. E+ also increased abundances within the Planctomycetes, Chloroflexi, and Proteobacteria phyla and the Clostridiaceae family. Multiple operational taxonomic units classified as Ruminococcaceae and Lachnospiraceae were correlated negatively with weight gains (lower in E+) and positively with respiration rates (increased by E+). These data provide insights into how E+ grazing alters the Angus steer microbiota and the relationship of fecal microbiota dynamics with FT.
IMPORTANCE Consumption of E+ tall fescue has an estimated annual $1 billion negative impact on the U.S. beef industry, with one driver of these costs being lowered weight gains. As global agricultural demand continues to grow, mitigating production losses resulting from grazing the predominant southeastern United States forage grass is of great value. Our investigation of the effects of E+ grazing on the fecal microbiota furthers our understanding of bovine fescue toxicosis in a real-world grazing production setting and provides a starting point for identifying easy-to-access fecal bacteria that could serve as potential biomarkers of animal productivity and/or FT severity for tall fescue-grazing livestock.
INTRODUCTION
Culture-independent next-generation sequencing (NGS)-based microbiota studies (e.g., 16S rRNA gene) in food-producing animals, like ruminants, are on the rise. The influence of the bovine microbiota on host energy status and metabolism has been studied (1–4), and recent NGS studies have linked enteric microbiota shifts to animal performance, e.g., weight gains, in multiple species (5–9). Notably, in beef cattle, the influence of feed additives and/or diet on the rumen and fecal microbial communities has been evaluated (10–13), and ubiquitous and foodborne pathogenic bacteria were identified in their fecal matter (14). Overall, the various dietary contributions to shifts in the bovine microbiome and subsequent changes in animal performance metrics are a major research focus; however, potential changes in the resident microbial community and the impact of these changes on animal performance in grazing beef, including tall fescue grazing, are not yet studied.
Tall fescue, Lolium arundinaceum, is the predominant southeastern United States forage grass, covering approximately 14 million hectares, and is commonly infected with the endophytic fungus Epichloë coenophialum (15). The wild-type fungus produces multiple metabolites. Some metabolites increase stand persistence through protection against extrinsic plant fitness factors. However, other fungal metabolites, namely, ergot alkaloids, are detrimental to grazing livestock and are key etiological agents of fescue toxicosis (FT) (16–19), a disease with an estimated $1 billion negative impact on the U.S. beef industry annually, when adjusted for inflation (20, 21). FT is a complex, multisystem disease manifesting as metabolic dysregulation (22), decreased volatile fatty acid (VFA) absorption (23, 24), and immune/inflammatory alterations (25–27). Other important signs of FT in grazing beef include increased respiration rates (RR) (28, 29), thermoregulatory impairments (i.e., decreased ability to regulate core body temperatures [25, 30]), and lowered weight gains (31).
Clavine alkaloids, a major alkaloid type produced by E. coenophialum, and other ergot alkaloid precursors have previously been shown to exhibit antibiotic-like properties (32, 33), indicating that they contribute to changes in the microbial populations of animals grazing toxic (E+) tall fescue. Although some previous studies report on how specific ruminal bacteria and/or fermentation profiles shift after exposure to E+ fescue or ergot alkaloids (34–36), understanding the relationship between toxic tall fescue grazing and the fecal microbiota is important, as beef cattle fecal microbiota is dynamic and shifts in response to dietary and/or management strategy changes (12, 37). This is particularly important for FT, as rotational grazing is a common management practice to minimize the impact of E+ grazing on animal productivity. Further, a previous study has found around 30% of operational taxonomic units (OTUs) from 16S rRNA gene sequencing were shared between the rumen and hindgut microbiota (38), indicating that some shifts in rumen populations are detectable in the lower gastrointestinal (GI) tract. Therefore, understanding how E+ grazing influences the fecal microbiota could provide an easier means of assessing global (rumen and hindgut) microbiota changes that could either contribute to or be associated with decreased animal productivity. Finally, these studies can be used in the future as a starting point to ascertain different management practices or therapeutic approaches that mitigate the effects of E+ fescue grazing on the fecal microbiota or to use the microbiota to diminish the adverse effects of the toxic fescue on the fescue-grazing beef.
Here, we sought to provide an initial characterization of the beef cattle fecal microbiota and to evaluate changes in the fecal microbiota community composition and dynamics that result from grazing toxic tall fescue. To accomplish this, we employed a next-generation sequencing-based analysis of the fecal microbiota of Angus steers by targeting the V4 region of the 16S rRNA gene. Fecal samples were collected from animals grazing tall fescue infected with either a toxic endophyte (E+) or a novel, nontoxic endophyte (Max-Q) across a 28-day grazing trial. Fecal microbiota changes due to E+ grazing were identified and correlated with animal performance and physiological responses. Taken together, these data describe the hindgut microbiota of beef cattle grazing toxic, ergot alkaloid-producing endophyte-infected tall fescue.
RESULTS
Environmental conditions, pathophysiological responses, and urinary ergot alkaloids.
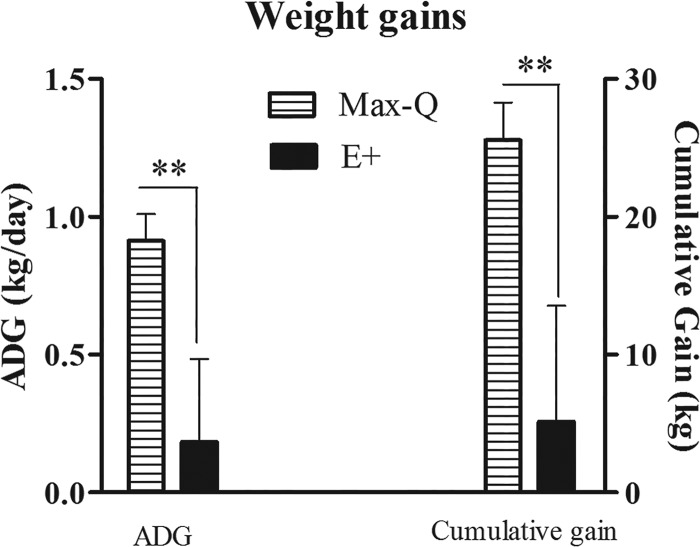
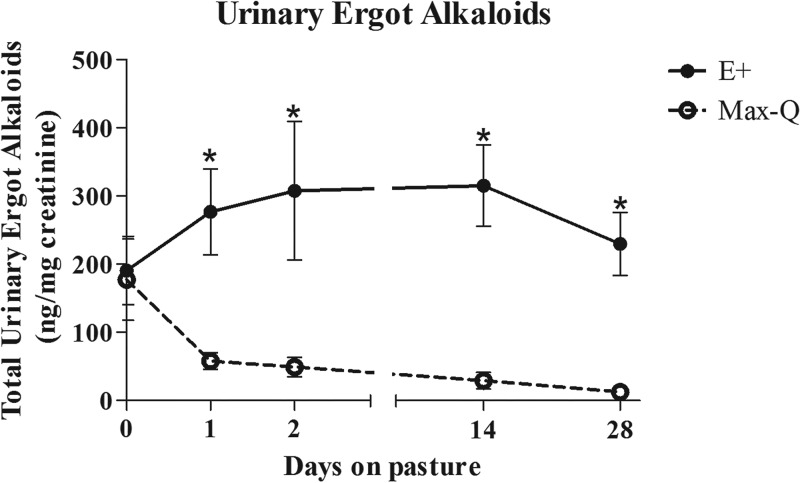
This study was conducted in spring 2016 (6 April to 4 May) at the J. Phil Campbell Natural Resources Conservation Center of the University of Georgia (Watkinsville, GA). The average 24-h temperature was 17.8°C (range, 8.3 to 23.3°C), and the average 24-h temperature-humidity index (THI) was 63.1 (range, 49.3 to 72.6). Pasture total ergot alkaloid levels on 11 April 2016 were 2,357.7 ± 19.70 ppb and 164.3 ± 11.30 ppb for E+ and Max-Q (i.e., nontoxic) pastures, respectively. Both cumulative and average daily weight gains were significantly lower (P < 0.01) in toxic fescue-grazing (E+) steers at the end of the grazing trial (Fig. 1). Respiration rates (RR) were significantly higher (P < 0.05) after 14 and 28 days of E+ grazing, and rectal temperatures (RT) tended (P = 0.096) to be elevated in E+ steers after 14 days of grazing (data not shown). Total urinary ergot alkaloids were significantly elevated (P < 0.05) in the E+ steers throughout the 28-day grazing period, reaching their maxima after 14 days of grazing (Fig. 2). Max-Q steers, while having measurable alkaloid levels prior to placement on the nontoxic Max-Q pastures (i.e., day 0), had markedly lower urinary alkaloids than the E+ steers throughout the 28-day grazing trial (Fig. 2).
FIG 1.
Average daily gain (ADG; kg/day) and cumulative gain (kg) in the weight of Angus steers that grazed either nontoxic endophyte-infected (Max-Q; n = 6) or toxic endophyte-infected (E+; n = 6) tall fescue for a 28-day grazing trial. Two asterisks indicate significant difference between treatment groups (P < 0.01).
FIG 2.
Total ergot alkaloids in the urine of Angus steers before pasture placement (day 0) and after 1, 2, 14, and 28 days of grazing on either nontoxic (Max-Q; n = 6) or toxic (E+; n = 6) endophyte-infected tall fescue. Data are presented as ng/mg creatinine. An asterisk indicates significant difference between treatment groups on a sampling date (P < 0.05).
Fecal 16S rRNA gene sequencing results.
Following Illumina sequencing, the 60 fecal samples produced 3,199,179 raw reads. After sequence quality filtering using the mothur pipeline, which includes removal of short sequences, preclustering, and chimera removal, a total of 1,694,182 high-quality sequences were obtained; the mean number of sequences per sample was 28,235 ± 10,927 (x̄ ± standard deviations [SD]; range, 12,038 to 73,982). OTU determination resulted in 3,809 unique bacterial OTUs across all samples, with an average of 1,047 ± 200 OTUs per sample (x̄ ± SD; range, 491 to 1,462). Taxonomically, 95% of all sequences were classified into 21 different phyla, and 74% were further classified to at least the family level when queried against the Greengenes database.
Sequencing depth and coverage.
A rarefaction analysis for the bovine fecal microbiota by days on pasture is presented in Fig. S1 in the supplemental material. Although the collector’s curves did not appear, upon visual inspection, to reach an asymptote when rarefied at a minimum of 2,000 sequences per sample (Fig. S1), the calculated Good’s coverage indicated that the sampling depth captured most of the species diversity, with an average coverage of 99.00% ± 0.38%, irrespective of fescue cultivar, for the entirety of the grazing trial (i.e., before [termed Pre] and at days 1, 2, 14, and 28; data not shown).
Alpha-diversity metrics.
For alpha-diversity metrics, both diversity (Fig. S2A) and richness (Fig. S2B) were constant throughout the 28-day grazing period, with a trend for an increase in diversity and a slight decrease in richness after 14 days of grazing. A nonparametric permutational analysis of variance (PERMANOVA; 10,000 permutations) found a significant main effect of time (P < 0.001) without a significant fescue treatment (P = 0.156) or treatment and time interaction (P > 0.9) effect on the fecal bacterial communities using the Bray-Curtis (abundance and presence/absence) dissimilarity metric. Also, a significant main effect of time (P < 0.001) and a trend toward a fescue treatment effect (P = 0.095), with no interaction (P > 0.9), when using the Jaccard (presence/absence) dissimilarity metric was found. Further, there was a trend for a main effect of fescue treatment on the diversity of the fecal microbial community (P = 0.051 by inverse Simpson’s diversity), with no significant effect based on time (P = 0.21) or the treatment and time interaction (P = 0.89). There were no effects of fescue cultivar treatment (P > 0.5), time (P > 0.4), or treatment by time interaction (P > 0.8) on Chao1 richness. These data indicate that E+ grazing has a tendency to change the overall fecal microbial communities and that this effect is mainly on species evenness, not the total number of species.
Overall fecal microbial community composition of the Angus beef cattle.
Independent of fescue cultivar, Firmicutes and Bacteroidetes were the two predominant phyla in the Angus steers’ fecal samples before and throughout the grazing period, with combined sequences in these two phyla accounting for 90% (range, 53 to 61% and 27 to 37%, respectively) of the entire microbial community in both E+ and Max-Q steers (File S1). The remaining classified (e.g., Verrucomicrobia, Actinobacteria, Tenericutes, etc.) and unclassified bacterial phyla accounted for <4% of the total sequences (File S1). Overall, the Max-Q and E+ fecal microbiota at the phylum level was relatively stable, with no major differences between the two treatments. However, significant compositional differences between the Max-Q and E+ fecal microbiota were observed at the order, family, and genus levels (Fig. S3A, S3B, and S3C).
Throughout the grazing trial, bacteria in the Ruminococcaceae and Lachnospiraceae families accounted for 44% and 23%, respectively, of all Firmicutes sequences. An additional 16% of the sequences were unclassified at the family level, and all other families accounted for less than 3% of the total number of sequences within this phylum (File S1). For the Bacteroidetes phylum, 41% of the sequences were unclassified at the family level, with 14% of these being classified into the order Bacteroidales (File S1). The most abundant classified families included the Paraprevotellaceae (21%), Rikenellaceae (14%), Bacteroidaceae (10%), and the candidate family RF16 (4%; File S1). All other classified families made up less than 3% of the Bacteroidetes population.
The family Coriobacteriaceae accounted for 53% of all Actinobacteria sequences within the Max-Q steers and 84% in E+ steers; another major Actinobacteria family, the Corynebacteriaceae, constituted 45% and 9% of all Actinobacteria sequences within Max-Q and E+ steers, respectively, after 1 day of grazing (File S1). Max-Q steers had consistently higher Corynebacteriaceae and lower Coriobacteriaceae levels throughout the grazing period than the E+ steers (File S1). By the end of the study, the Coriobacteriaceae dominated the Actinobacteria phylum, with 93% (Max-Q) and 97% (E+) of all Actinobacteria sequences aligning to the Coriobacteriaceae family (File S1).
Prior to pasture (E+ or Max-Q) placement, the Streptococcaceae family made up >80% of all Bacilli within the Firmicutes; however, after placement on their respective pastures, steers from both Max-Q and E+ pastures had sharp decreases in the Streptococcaceae (down to 20%), accompanied by increases in the Planococcaceae and Turicibacteraceae families. These two families were more robustly represented in Max-Q (∼60% of Bacilli) than in E+ (∼30% of Bacilli) steers (File S1). Also, the Paraprevotellaceae family was greater than 10% of the total Bacteroidetes sequences and more abundant in E+ steers than Max-Q steers after 2, 14, and 28 days of grazing (File S1); most of the changes within the Paraprevotellaceae during the grazing period occurred for the candidate genera CF21 and YRC22 (File S1). There was also a sharp decrease in an OTU classified as belonging to Ruminococcus bromii in both E+ and Max-Q steers after tall fescue pasture placement, suggesting a fescue cultivar-independent effect (File S1).
Multivariate analyses to interrogate E+ grazing-related fecal microbiota shifts.
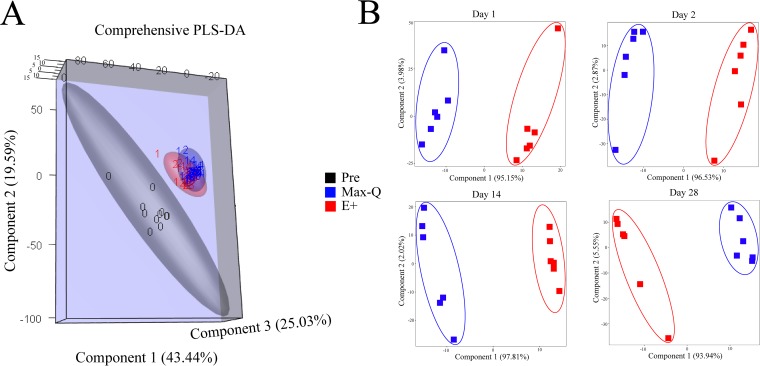
Principal component analysis (PCA) was conducted to interrogate sources of variability within the bovine fecal microbiota resulting from time spent grazing and from the E+ endophyte. The two principal components accounted for approximately 21% of the overall variance, with the first component being the major contributor (Fig. S4A). Although the 95% confidence intervals overlap between the Pre, Max-Q, and E+ data, PCA demonstrated distinct clustering and separation of Pre samples from Max-Q and E+, with this separation becoming greater as the grazing trial progressed (Fig. S4A). Importantly, while the Max-Q and E+ treatments clustered similarly along the first two components, they separated along the third principal component, most notably on days 14 and 28 of the grazing trial (Fig. S4B).
Partial least-squares discriminant analysis (PLS-DA) was also performed using the same normalized sequence count data set throughout the grazing trial (Fig. 3A) and individually for the 1-, 2-, 14-, and 28-day sampling dates (Fig. 3B). For the overall PLS-DA, three components were used to maximize the amount of cumulative variance explained (88.1%) by the analysis. The Pre, Max-Q, and E+ steers all formed distinctly separated clusters, with the Max-Q and E+ clusters being more similar yet remaining separated across the third component (Fig. 3A). Within sampling dates after pasture assignment, steers grazing Max-Q and E+ tall fescue formed two distinct clusters that separated primarily across the first principal component, with the second component contributing to clustering only in the 14-day E+ (Fig. 3B) and 28-day Max-Q (Fig. 3B) steers. The distinct PLS-DA clustering and separation between the Pre, Max-Q, and E+ steers is indicative of rapid changes of the Angus steers' fecal microbiota that occur after 1 day of tall fescue grazing, with the Max-Q and E+ steers developing and maintaining distinct fecal microbiota profiles.
FIG 3.
Partial least-squares discriminant analysis (PLS-DA) plots of the fecal microbiota of Angus steers grazing on either nontoxic (Max-Q; n = 6) or toxic (E+; n = 6) endophyte-infected tall fescue over the 28-day grazing trial (A) or 1, 2, 14, and 28 days after pasture assignment (B).
Sampling date-specific PLS-DA loadings plots were generated to assess the top 50 loading weights (i.e., OTUs) (Fig. S5). The loadings of the first component were used, as the samples separated mainly across it. The families Ruminococcaceae and Lachnospiraceae (all dates), Bacteroidaceae and Mogibacteriaceae (1, 2, and 14 days), Erysipelotrichaceae and Coriobacteriaceae (1, 2, and 28 days), and Prevotellaceae and Clostridiaceae (1, 14, and 28 days) were the main contributors to the explained variance (Table S1). The candidate family S24-7 (1 and 2 days) and the Lactobacillaceae family (14 and 28 days) were also important to the loading weights (Table S1). Finally, several unclassified families were also drivers of the treatment separation seen in the PLS-DA (Table S1).
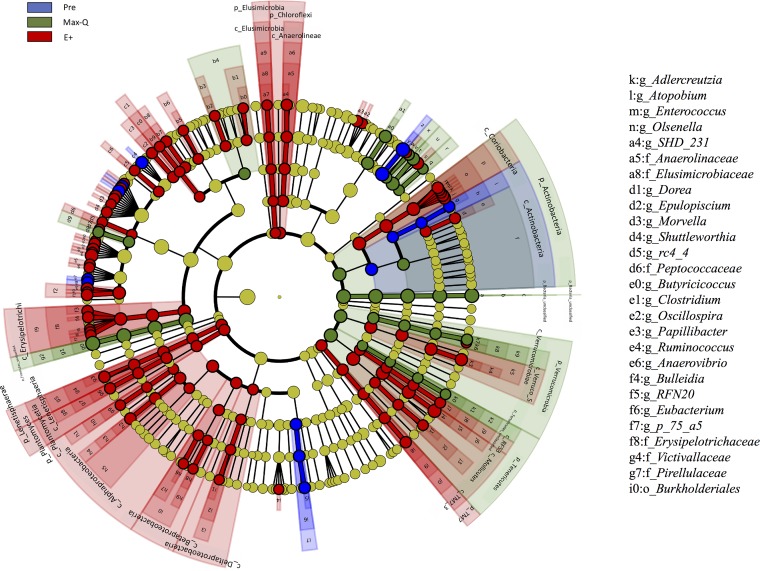
Linear discriminant analysis (LDA) of effect size (LEfSe) identified numerous bacteria with significantly different abundances between Pre, Max-Q, and E+ steers (Fig. 4). The abundances of the phyla Planctomycetes, Lentisphaerae, Elusimicrobia, Chloroflexi, and Proteobacteria were increased in E+ steers (Fig. 4). Further, a number of genera within the Lachnospiraceae, Ruminococcaceae, and Erysipelotrichaceae families were also increased in E+ steers (Fig. 4). Moreover, we found that overall abundance of bacteria in the Actinobacteria phylum was greater in Max-Q steers, but the Coriobacteriaceae family within this phylum was more abundant in E+ steers (Fig. 4). This suggests that suborder bacterial abundance was affected specifically, even if the cumulative abundance of all sequences within the phyla are affected differently. Although treatment differences became even more apparent at lower taxonomic levels (i.e., order, family, and genus) (Fig. S3A, S3B, and S3C), the LEfSe analysis also identified significant differences at the phylum level for some of the lower-abundance phyla (Fig. 4).
FIG 4.
LDA effect size (LEfSe; P < 0.05 by Kruskal-Wallis test; P < 0.05 by pairwise Wilcoxon test; logarithmic LDA score of >2.0) of the fecal microbiota of Angus steers before placement (Pre) or across a 28-day grazing trial after placement on either nontoxic (Max-Q; n = 6) or toxic (E+; n = 6) endophyte-infected tall fescue. Blue, green, and red shading indicate greater abundance in Pre, Max-Q, or E+ steers, respectively. Taxonomic rank labels are provided before bacterial names: p_, c_, o_, f_, and g_ indicate phylum, class, order, family, and genus, respectively. Letters and numbers within the cladogram refer to bacterial names located in the key to the right of the cladogram.
Specific microbiome (OTU) changes after E+ grazing.
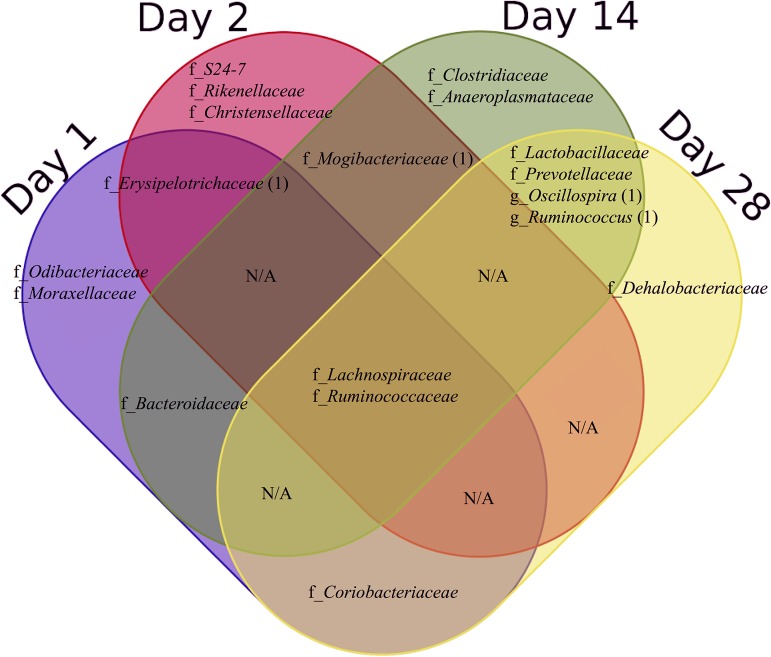
Totals of 25, 41, 55, and 37 OTUs were significantly (P < 0.05) different between Max-Q and E+ after 1, 2, 14, and 28 days of grazing, respectively. Of these, 1 OTU overlapped between days 1 and 2, 1 OTU overlapped between days 2 and 14, and 4 OTUs overlapped between days 14 and 28 (Fig. S5). The significantly different OTUs that overlapped between sampling dates include those classified to the families Erysipelotrichaceae (between 1 and 2 days), Bacteroidaceae (1 and 14 days), Coriobacteriaceae (1 and 28 days), Mogibacteriaceae (2 and 14 days), and Lactobacillaceae and Prevotellaceae (14 and 28 days). Lachnospiraceae, Ruminococcaceae, and another four unclassified families overlapped across all sampling dates (Fig. 5).
FIG 5.
Venn diagram demonstrating classified bacterial families and genera having OTUs that were significantly different between steers grazing on either nontoxic (Max-Q; n = 6) or toxic (E+; n = 6) endophyte-infected tall fescue throughout a 28-day grazing trial. f_, bacterial families; g_, bacterial genera. The number indicated in parentheses following the family or genus name represents specific OTUs within that family or genus that overlap between sampling dates.
To check for potential paddock effects within a fescue cultivar, heat maps were generated from the arcsine-normalized relative abundance data used for OTU comparisons by fescue treatment (Fig. S6). Overall, the heat maps for significantly (P < 0.05) different OTUs demonstrate OTU differences between Max-Q and E+ steers that are consistent across the three paddocks within treatment, indicating no apparent paddock effects (Fig. S6).
We then sought to determine if there were specific OTUs associated with the effects of E+ grazing on weight gain and obtain some preliminary information to that effect. We found that the majority of OTUs that differed between the most and least affected E+ steers belonged to the order Clostridiales (Fig. S7). For example, OTUs from the Lachnospiraceae genera Butyrivibrio and Clostridium had higher sequence counts in weight-gaining E+ steers, whereas the genus Blautia and two OTUs belonging to the genus Ruminococcus were increased in the most susceptible steers (Fig. S7). Clostridium genus OTUs were all increased in the susceptible steers. Of those OTUs not in the order Clostridiales, levels of the genus Akkermansia in the family Verrucomicrobiaceae were increased the most in susceptible steers, while the Coriobacteriaceae genus Atopobium had higher sequence counts in E+-resistant steers (Fig. S7).
Correlation analysis between OTU relative abundance and pathophysiological responses.
The relative abundance of several OTUs, referred to here simply as OTUs, correlated negatively with average daily gains (ADG) in E+ steers; the most frequently correlating families are presented in Table 1. Totals of 11, 7, 4, and 3 OTUs that correlated with ADG were classified to Lachnospiraceae, Ruminococcaceae, Coriobacteriaceae, and Erysipelotrichaceae families, respectively (Table 1). Ruminococcus was the only genus of the Ruminococcaceae family that had OTUs negatively correlated with ADG (4 OTUs) (Table 1). The Olsenella, Atopobium, and Adlercreutzia genera, all within the Coriobacteriaceae family, had OTUs negatively correlating with weight gain (Table 1). Finally, the Erysipelotrichaceae family had three negatively correlating OTUs classified at the genus level, with two OTUs and one OTU classified to the Bulleidia and the candidate p-75-a5 genera, respectively (Table 1).
TABLE 1.
Top classified bacterial families and genera that significantly negatively correlated with ADG and positively with RR
| Parameter and family or genus | No. of OTUs | Spearman’s avg correlation | P value |
|---|---|---|---|
| ADG | |||
| Lachnospiraceae | 11 | −0.50 | 0.036 |
| Ruminococcaceae | 7 | −0.55 | 0.020 |
| Ruminococcus | 4 | −0.54 | 0.024 |
| Coriobacteriaceae | 4 | −0.56 | 0.021 |
| Olsenella | 1 | −0.65 | 0.004 |
| Atopobium | 1 | −0.57 | 0.013 |
| Adlercreutzia | 1 | −0.48 | 0.045 |
| Erysipelotrichaceae | 3 | −0.53 | 0.025 |
| Bulleidia | 2 | −0.54 | 0.020 |
| p-75-a5 | 1 | −0.51 | 0.030 |
| RR | |||
| Ruminococcaceae | 11 | 0.56 | 0.022 |
| Clostridium | 2 | 0.58 | 0.024 |
| Oscillospira | 1 | 0.53 | 0.024 |
| Ruminococcus | 1 | 0.51 | 0.024 |
| Lachnospiraceae | 6 | 0.52 | 0.027 |
| Dorea | 1 | 0.58 | 0.011 |
| Coprococcus | 1 | 0.53 | 0.023 |
| Blautia | 1 | 0.52 | 0.028 |
| Victivallaceae | 4 | 0.52 | 0.026 |
| S24-7 | 3 | 0.58 | 0.018 |
Of the OTUs that positively correlated with RR, totals of 11, 6, 4, and 3 OTUs were classified into the families Ruminococcaceae, Lachnospiraceae, Victivallaceae, and candidate family S24-7, respectively (Table 1). The classified genera within the Ruminococcaceae included Clostridium (2 OTUs), Oscillospira (1 OTU), and Ruminococcus (1 OTU). Further, Dorea, Coprococcus, and Blautia were the classified Lachnospiraceae genera, all with 1 OTU positively correlating with RR (Table 1). Most OTU relative abundances that positively correlated with RR within the Ruminococcaceae and Lachnospiraceae were increased in E+ steers.
For rectal temperature, the top four most frequent correlates had totals of 18, 4, 2, and 2 OTUs classified to the families Lachnospiraceae, Ruminococcaceae, Coriobacteriaceae, and Paraprevotellaceae, respectively (Table S2). The genera within the family Lachnospiraceae included Butyrivibrio, Roseburia, Coprococcus, and Dorea, with Butyrivibrio having two OTUs while all other genera had one (Table S2). One Coriobacteriaceae OTU was classified as Enterococcus casseliflavus, and the two Paraprevotellaceae OTUs were classified into the candidate genera CF231 and YRC22 (Table S2).
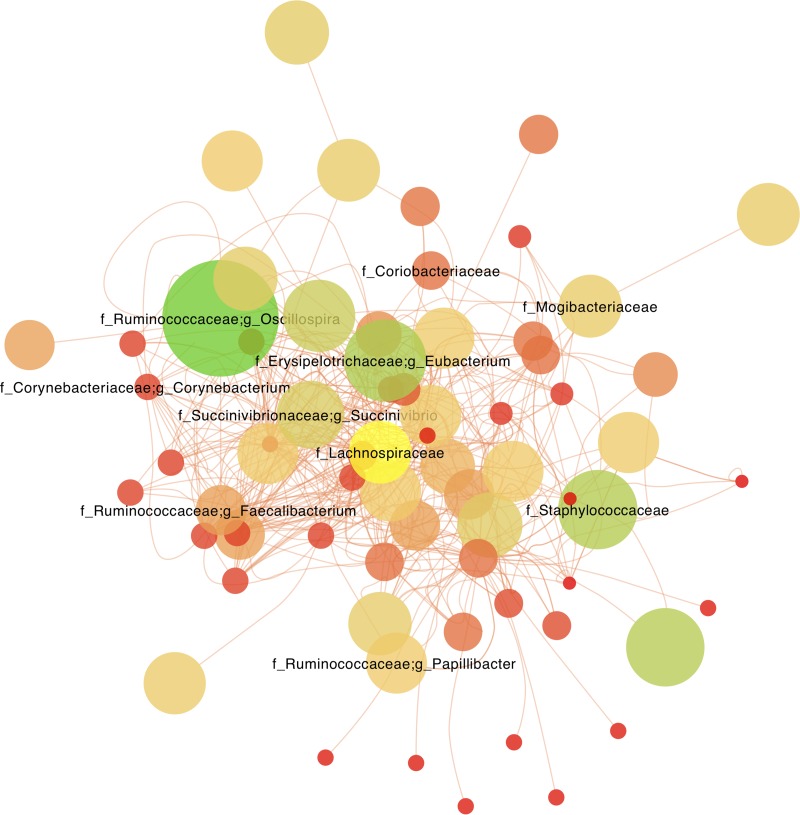
Cooccurrence network inference analysis.
Using the CoNet app within Cytoscape, we conducted a differential network analysis that resulted in one large, highly intraconnected and positively correlated cluster of OTUs within E+ steers. The anchors, defined here as the most highly connected nodes, were from OTUs within the phyla Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria (Fig. 6). Members of the families Lachnospiraceae, Ruminococcaceae, Prevotellaceae, and Paraprevotellaceae were the most prevalent families within the E+ network (Fig. 6). The other highly connected families in the E+ network included the Clostridiaceae, Mogibacteriaceae, Coriobacteriaceae, Succinovibrionaceae, and Staphylococcaceae. These data indicate that E+ grazing results in a distinct pattern of bacterial families and genera that are highly associated with one another.
FIG 6.
Cooccurrence network inference analysis of the fecal microbial communities of steers grazing toxic endophyte-infected tall fescue (E+; n = 6) after removal of nodes shared with the network of steers grazing a nontoxic endophyte-infected tall fescue (Max-Q). Node size is reflective of overall OTU abundance; node labels are the furthest phylogenetic classification of each node. Networks were generated using the CoNet app for Cytoscape.
DISCUSSION
Here, we describe the fecal microbiota of Angus steers grazing tall fescue, thereby increasing our current understanding of potential mechanisms underlying one of the costliest diseases to the United States beef industry, fescue toxicosis (FT) (31, 39). Lowered weight gains are an economic driver of production losses caused by ergot alkaloids found in E+ tall fescue and are a common finding in FT studies (31, 40, 41). Similarly, we found that after a 28-day grazing trial, E+ steers had significantly lower cumulative and average daily weight gains than steers grazing the nontoxic Max-Q cultivar. Urinary ergot alkaloid levels followed similar trends, as in previous studies (42–44), but excreted levels were substantially higher than those of a fall study of similar duration (22), supporting reported seasonal variability in urinary ergot alkaloid levels (45–47).
In our study, respiration rates (RR) were more sensitive to E+ grazing than rectal temperatures (RT) under spring thermoneutral conditions and were significantly higher after 14 days of E+ grazing, consistent with an acute ergot alkaloid challenge study (28). Effects of E+ on RT are dependent on E+ exposure duration and environmental conditions (25), which here were thermoneutral. Thus, based on urinary ergot alkaloid levels, decreased weight gains, and physiological responses (increased RR), steers in this study exhibited classic signs of FT.
Our analysis of the fecal microbiota throughout the grazing period revealed that, at the phylum level, most sequences aligned to Firmicutes (53 to 59%) and Bacteroidetes (33 to 37%). These data indicate that the grazing Angus beef cattle fecal microbiota, at this phylogenetic level, is in line with what is reported for other beef steers (48), poultry (49), swine (50), and dairy cattle (38). Therefore, it was not surprising that most compositional differences between the E+ and Max-Q grazing steers we found began at the suborder level.
By using dimension reduction analyses (PLS-DA and PLS-DA loadings), we were able to identify a number of OTUs that contributed to the differences in the fecal microbiota profiles of Max-Q and E+ steers at individual dates throughout the grazing trial. Interrogation of the data using these analyses also allowed identification of common bacterial families that had a significant response to E+ grazing. Further, the OTUs identified by these analyses, and the subsequent taxonomic identification of these OTUs, support the findings of the other analyses presented here. Many of the OTUs and bacterial families that were identified as contributing to the differences observed between Max-Q and E+ steers in our dimension reduction analyses were also identified as being significantly influenced by E+ grazing and significantly correlated to pathophysiological signs of interest within the context of FT (e.g., lowered weight gains). These data lend support to the results of the PLS-DA in selecting features that can differentiate fescue treatments. These analyses allowed us to identify the relationships between the data for each group (i.e., Pre, Max-Q, and E+), with both Max-Q and E+ fecal microbiota profiles being distinct from their fecal microbiota prior to fescue pasture placement yet responding to fescue treatment in an endophyte-dependent manner, as the plants used in the study are genetically identical. Further, the differential network analysis revealed a subnetwork of highly correlated OTUs, mostly belonging to families found to drive treatment separation in the PLS-DA analysis, that were specific to E+ grazing steers, indicating the toxic endophyte not only shifts the fecal microbiota profile but also that this shift results in a highly correlated structure of bacteria that have a mutualistic relationship.
Our findings indicate that, irrespective of the type of endophyte present, tall fescue grazing resulted in a marked shift in the fecal microbiota, reflecting the selective pressure imposed by fescue grazing. For example, we found one OTU was specifically affected by tall fescue grazing (discussed below), belonging to Ruminococcus bromii, which accounted for 35% of the Ruminococcus sequences prior to placement of the animals on the respective fescue pastures. Upon pasture placement and throughout the grazing trial, the sequence abundance of this OTU in the fecal microbiota of both E+ and Max-Q steers was negligible. Of note, this is a known dominant community member of cattle on a barley diet (51), and the steers prior to fescue pasture placement were fed mixed diets that contained some barley. The sharp decline of this OTU in the fescue-grazing steers is a likely indicator that the utilization of resistant starch from the fescue plant is not dependent on this bacterium, which is a well-known degrader of resistant starches (52), and that the animals likely relied on other bacteria instead.
Most bacterial OTUs that were significantly affected by E+ grazing were classified into the Ruminococcaceae and Lachnospiraceae families, which were more prominent later in the grazing trial. The relative abundances of the Lachnospiraceae OTUs increased up to 8-fold in E+ steers, whereas the number of significantly increased Ruminococcaceae OTUs in E+ steers increased with time. Both families include cellulose- and hemicellulose-degrading bacteria and members contributing to butyrate production (53–55). Butyrate, an important energy source for ruminants, has been shown to influence energy expenditure (56, 57), modulate nuclear receptor activity, and interact with free fatty acid receptors via the sympathetic nervous system to increase glucagon-like peptide 1 (GLP-1) and peptide YY (PYY) secretion from enteric L cells (58–60). However, the influence of butyrate on PYY production recently has been shown to be species dependent in vitro (61). Nonetheless, if excess butyrate reaches gastric receptors, the result would be reduced gastric emptying, suppressed gut motility, and increased satiety (62), potentially modulating host metabolism and feeding behavior in FT via the enteroendocrine system (63, 64). Although we did not directly measure butyrate levels here, elucidating the short-chain fatty acid profile and its interactions in the hindgut of tall fescue-grazing beef steers is important for future, more detailed studies.
We also found that certain E+ steers were relatively resistant to E+ grazing effects (i.e., lowering of weight gain), while others were more susceptible. There were also a select few OTUs that were differentially abundant between the “resistant” and “susceptible” steers, indicating a potentially epistatic microbiota profile as it relates to productivity. While these data are limited and preliminary, the role that gut bacteria play in susceptibility versus resistance to lowered weight gains caused by E+ grazing is worthy of future exploration, as it might lead to microbiota-directed treatments for FT.
Among the other notable effects was an increase in the sequence abundance for the genus Oscillospira in E+ steers after 14 and 28 days of grazing. One previous study has associated colonic Oscillospira presence with lowered ADG in crossbred steers (65). Oscillospira is a genus commonly found in both human and bovine gut microbiota (66, 67) and is known to degrade host glycans in multiple species (68), thereby altering glycoprotein homeostasis. For FT, the metabolic activity of Oscillospira may contribute to the lowered weight gains, as its increased presence in the human gut correlates with lower body weights (69). In addition to glycan breakdown, Oscillospira has also been inversely associated with plasma acetate levels (70), a VFA that is a main energy metabolite in ruminants.
The abundance of the family Erysipelotrichaceae significantly decreased after 1 and 2 days of grazing and was a major early driver of the E+ and Max-Q separation seen within PLS-DA. Notably, in humans, members of the Erysipelotrichaceae recently have been associated with host lipid metabolism (71, 72). Potential relationships between the Erysipelotrichaceae family and enteric leucine-rich repeat kinase 2 (LRRK-2) activity (73) and IgA production (72) have been suggested, indicating that this family of bacteria is involved in both the regulation of lipid metabolism and enteric nervous and immune system functioning. Disruption of lipid homeostasis is a feature of FT; dyslipidemia, as well as suppressed serum cholesterol and triglycerides, all have been associated with FT previously (22, 74). The early decreases in the Erysipelotrichaceae, along with the previously reported decreases in enzymes associated with lipid metabolism in FT (e.g., lipase) (75, 76), could contribute to the dyslipidemia.
Significant shifts in the families Clostridiaceae (increases after 14 days) and Prevotellaceae (decreases after 14 and 28 days), as well as increases in the Planctomycetes family Pirellulaceae, the Chloroflexi phylum, and the Betaproteobacteria order Burkholderiales, all could reflect ergot alkaloid-induced selection pressure. The Chloroflexi, Pirellulaceae, and Burkholderiales from the earthworm Eisenia fetida have members previously reported as being capable of degrading ergovaline, the main ergopeptine alkaloid in E+ tall fescue (77). Also, ex vivo ruminal sampling of tryptophan-utilizing bacteria demonstrated that Prevotellaceae are not heavily involved in ergovaline degradation, while Clostridiaceae strains degrade the majority of ergovaline (34). Although ergot alkaloids are quickly metabolized in the rumen (42), the changes identified here potentially reflect either ruminal ergot alkaloid selection pressure, as about 30% of OTUs are shared between the rumen, small intestine, and fecal samples (38), or, alternatively, ergot alkaloid bioavailability in the lower GI tract.
The genus Lactobacillus, which had OTUs with decreased abundances as the study progressed, was one of the main PLS-DA separation contributors and is known to produce indole metabolites (55). Recently, we reported that E+ grazing increased the level of plasma indole metabolites (22), and the results here indicate that these increases are not Lactobacillus derived; rather, we posit that breakdown of ingested ergot alkaloids and/or microbiota-related tryptophan metabolism by other bacteria, such as ruminal Clostridium sporogenes and/or Clostridium sticklandii (34), are likely partially responsible for the plasma indole metabolite increases we previously reported (22).
The bacterial families that positively correlated with RT were similar to the ones negatively correlated with ADG in E+ steers, with the Lachnospiraceae family OTUs being a major one. The enteric microbiota can influence thermogenesis by altering lipid availability and absorption (78), and thermoregulation is impaired in FT (25). Although these data are preliminary, the contribution of the enteric microbiota to increased RT is unclear yet worthy of more detailed investigation. Further, while E+ steers’ RT fluctuated concomitantly with environmental conditions, it was not affected by E+ grazing to a major extent, as our study was conducted under thermoneutral conditions. This suggests that the relationship between RT and E+ grazing should be investigated further under more extreme environmental conditions (e.g., high THI).
Finally, our study also determined correlations between the fecal microbiota and RR. To our knowledge, there are no current data establishing associations between fecal microbiota shifts and changes in pulmonary physiology in food-producing animals. However, there is increasing evidence of communication between the gut and lung mucosa, and respiratory microbiota influencing lung mucosa immunological homeostasis has been shown in both humans and rodents (79–82). The correlations between RR and fecal microbiota members were strong and, considering the effects of E+ grazing on RR and the potential for gut-lung mucosal cross talk, are worthy of future investigation.
Overall, our study contributes to an increased understanding of how the fecal microbiota functions in the overall health of fescue-grazing beef cattle. Our work also provides insights into the influence of E+ grazing on the Angus steer fecal microbiota and suggests that E+ grazing has a significant, rapid impact on the fecal microbiota, predominantly at the family/genus levels. Microbiota-derived metabolites likely will also be affected. These key changes potentially modulate energy expenditure, metabolic homeostasis, and feeding behaviors manifested in FT-related signs, such as dyslipidemia, shortened grazing periods, decreased feed intake, and lowered weight gains. However, additional targeted investigations are necessary to interrogate pathophysiological changes that result from microbiota dysbiosis induced by E+ grazing and determine if/which microbiota changes are the result of, or contribute to, the development of FT.
MATERIALS AND METHODS
Animals, pastures, and environmental conditions.
All animal handling and sampling procedures were preapproved by the University of Georgia’s Institutional Animal Care and Use Committee. Postweaning Angus steers (n = 12) were blocked by weight and randomly assigned to nontoxic (n = 6; weight, 332.8 ± 11.77 kg [x̅ ± SEM]; Max-Q; Jesup MaxQ with endophyte AR542; 3 paddocks, 2 steers per paddock) or toxic (n = 6; weight, 349.3 ± 19.34 kg [x̅ ± SEM]; E+; Jesup with wild-type endophyte; 3 paddocks, 2 steers per paddock) tall fescue treatments. Pastures, sown in fall 2004, have been described previously (22). Individual tillers were sampled on 11 April 2016 from 100 locations within the pastures, cutting the tiller at the soil surface, and transporting the samples to the laboratory on ice. Plant tissue was frozen, freeze-dried, ground to pass a 1-mm screen, and analyzed for total ergot alkaloid using a commercial enzyme-linked immunosorbent assay (ELISA) (END0899-96p; Agrinostics Ltd.). Steers were kept on the same pasture (0.8 ha) throughout the grazing trial. Temperature and humidity measurements were recorded for 24 h daily and across sampling times (8:00 to 11:00 a.m.) on sampling dates using an on-site weather station.
Statistical power calculations.
The power calculations performed utilized previously published average daily weight gain data, where the same two forage types (Max-Q and wild-type Jesup tall fescue) were used, and the overall average daily gains were used to estimate the potential treatment difference. The number of groups was set at two with six individuals per group, and the resultant power was 1.00. The power calculations for the correlational analysis (after pasture placement) with a set correlation coefficient (|r| = 0.4) and an alpha of 0.05 resulted in a calculated power of 0.493. Finally, for the t test comparison of arcsine-normalized relative abundances of OTUs, the difference in means and standard deviations was estimated based on the significantly different OTUs throughout this study; the power calculation was 0.347 based on an alpha of 0.05. These calculations ensured that the study was sufficiently powered.
Urinary ergot alkaloid analysis.
Total urinary ergot alkaloid concentrations were determined before and 1, 2, 14, and 28 days after pasture placement via ELISA (Agrinostics Ltd., Watkinsville, GA) as previously described (22, 43, 44). Lysergic acid was used as the standard, and serially diluted (1:2 to 1:16) urine samples were used for analysis. Urinary ergot alkaloid levels were creatinine normalized as described in reference 83 prior to statistical analysis.
Sample collection and processing.
Steer body weights were recorded before (Pre) and 14 and 28 days after pasture assignment with a digital scale. Fresh fecal samples were collected by hand using new gloves for every collection and stored on ice for transport before being stored at −80°C until DNA extraction. Respiration rates (RR) were monitored by counting full flank respiration movements for 60 s, twice per animal, and calculating the average, similar to previous procedures (28, 29, 84). Rectal temperatures (RT) were taken with a handheld DeltaTrak (Pleasanton, CA) digital thermometer once a stable reading for 15 s was acquired. Fecal samples, RR, and RT were taken before and 1, 2, 14, and 28 days after pasture assignment.
DNA extraction.
Genomic DNA was extracted from fecal samples using a mechanical disruption and phenol extraction protocol, established in reference 67, with the 25:24:1 phenol-chloroform-isoamyl alcohol modification used in reference 6. All DNA samples were resuspended in Tris-EDTA (TE) buffer and quantified using a Qubit fluorometer (Invitrogen, San Diego, CA). For each set of DNA extractions, a negative control using the TE extraction buffer was performed alongside each extraction and was taken through the amplification and sequencing protocol described below.
DNA amplification and sequencing.
Samples were diluted to 1 ng/μl for amplification, and universal bacterial primers for the 16S rRNA gene variable region V4, as previously described (85), were used in the amplification reactions. For each amplification, a negative PCR control containing the extraction buffer and PCR primers was used. A total of 5 μl of diluted DNA, 0.5 μl of 10 μM forward primer (5′-GTGCCAGCMGCCGCGGTAA-3′), 0.5 μl of 10 μM reverse primer (5′-GGACTACHVGGGTWTCTAAT-3′), 6.5 μl water, and 12.5 μl KAPA 2× HiFi master mix (Kapa Biosystems, Wilmington, MA) were used in each 25-μl reaction mixture with the following cycling conditions: initial denaturation at 95°C for 3 min; 25 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 30 s; and a final extension at 72°C for 5 min. Water was used for the PCR negative control. PCR products were purified with a 1% (wt/vol) low-melt agarose gel and recovered using a Zymoclean 96-well DNA recovery kit (Zymo Research, Irvine, CA). If bands were present in either the DNA extraction or PCR amplification negative controls, the sample were reextracted and amplified until no visible band was present in the gel. Samples were then quantified using a Qubit fluorometer and pooled in an equimolar fashion into the final library. The pooled library and 10% PhiX control DNA was sequenced on an Illumina MiSeq sequencing platform using the 2 × 250-bp paired-end MiSeq v2 sequencing kit (Illumina, San Diego, CA) using custom primers (85). The sequenced control and sample DNA were taken through quality-filtering and normalization procedures described below.
Fecal 16S rRNA gene sequence processing and bioinformatics analysis.
Raw sequence files were obtained in fastq format from the sequencer and processed using mothur v.1.38.1 (86) as described in reference 38. In brief, quality filters were applied to remove sequences with a quality score of less than 35, homopolymers longer than 8 bp, and greater than 2 bases different from those of the primer. Unique sequences next were aligned to the SILVA, version 119, reference alignment database (87), and chimeras were removed using chimera.uchime (http://drive5.com/uchime). Only bacterial sequences were retained and classified using the Greengenes database, v13.8 (http://greengenes.secondgenome.com) (88). Rarefaction curves and Good’s coverage were calculated in mothur. The OTU table was normalized for sequence depth in mothur using the normalize.shared function prior to statistical analysis. For all OTU or taxon-level statistical tests, the taxon abundance threshold was >0.1% and had to be present in >50% of samples in the respective analysis. Community diversity was estimated using Chao1 richness (89) and the inverse of Simpson’s diversity index (90); alpha-diversity metrics were tested for E+ grazing effects using the nonparametric Kruskal-Wallis test by ranks. PERMANOVA was used to test for E+ effects on the entire microbial community, with fescue treatment and time spent grazing set as the two factors. PCA, PLS-DA, and PLS-DA loadings were performed using the mixOmics R package (91, 92), and plots were recreated in Adobe Illustrator (Adobe Systems, San Jose, CA). PCA and PLS-DA were performed on the postsequencing depth-normalized OTU table, and the data were mean centered and total sum scale normalized prior to analysis. For the PLS-DA, the internal nearZeroVar function was used to flag and remove predictors (i.e., OTUs) that had low to zero variance, which is recommended for sparse data sets, such as 16S rRNA gene sequencing studies. LEfSe was performed within the Huttenhower laboratory’s galaxy instance using the relative abundance table as the input (http://huttenhower.sph.harvard.edu/galaxy/) (93), with Kruskal-Wallis test (P < 0.05), pairwise Wilcoxon test (P < 0.05), and logarithmic LDA score (>2.0). Comparison of differentially abundant OTUs was performed in R (92), with Bonferroni correction for multiple comparisons (α < 0.05).
All OTUs included in the analyses were determined at 97% sequence identity, allowing genus-level resolution (94), using the furthest neighbor (most conservative) clustering algorithm. Visualization of OTUs, at all levels, was completed using Krona Tools 2.7 (95) and is provided as an .html file in the supplemental material. Network analysis was performed using the CoNet app within Cytoscape, as in reference 96, with Pearson’s correlation, Spearman’s correlation, and the Bray-Curtis and Kullback-Leibler dissimilarity parameters with thresholds set so the 1,000 top- and bottom-scoring edges are kept in the network. Relationships between parent-child taxa were excluded within the algorithm to prevent overabundance of correlations based on similar lineages. Differential network analysis was performed within Cytoscape (97), and removal of all nodes and edges shared between Max-Q and E+ networks resulted in the final E+ steers’ network presented here.
Statistical analysis of nonmicrobiome data.
Statistical analyses of weight gains, RR, RT, and urinary ergot alkaloids were done with Sigma Plot, v12.5 (Systat Software, Inc., San Jose, CA), using two-way analysis of variance, with days of sampling and fescue treatment set as the two independent variables. If significant (P < 0.05) effects based on treatment or days spent grazing were observed, the Holm-Sidak post hoc analysis was applied to separate significant differences. Graphs were generated with GraphPad Prism 5 (La Jolla, CA).
Accession number(s).
All DNA sequences are publicly available in the NCBI Sequence Read Archive and are accessible under BioProject accession no. PRJNA540841.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded in part by a grant from the USDA, National Institute of Food and Agriculture (NIFA), Agriculture and Food Research Initiative (AFRI), grant 67030-25004 to N.M.F., and USDA NIFA AFRI grants 67015-21348 and 67015-23246 to G.S. Z.B.T.’s participation in this project was supported in part through the NIH Office of Research Infrastructure Programs (grant 4T35OD010433-10). We also thank the Interdisciplinary Toxicology Program, the Department of Physiology and Pharmacology, and the Graduate School of the University of Georgia for partial support to R.S.M.
Help with research, animal handling and care, and other assistance from the skillful personnel at the J. Phil Campbell Natural Resources Conservation Center of the University of Georgia (Watkinsville, GA) is greatly appreciated. We also thank Madison S. Cox for help and consulting with sequence analysis.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00032-19.
REFERENCES
- 1.Hungate RE. 1966. Chapter III. The rumen protozoa, p 91–147. In Hungate RE. (ed), The rumen and its microbes. Academic Press, San Diego, CA. [Google Scholar]
- 2.Hungate RE. 1966. Chapter IX. Host metabolism in relation to rumen processes, p 353–375. In Hungate RE. (ed), The rumen and its microbes. Academic Press, San Diego, CA. [Google Scholar]
- 3.Hungate RE. 1966. Chapter II. The rumen bacteria, p 8–90. In Hungate RE. (ed), The rumen and its microbes. Academic Press, San Diego, CA. [Google Scholar]
- 4.Mccann JC, Wickersham TA, Loor JJ. 2014. High-throughput methods redefine the rumen microbiome and its relationship with nutrition and metabolism. Bioinform Biol Insights 8:BBI.S15389. doi: 10.4137/BBI.S15389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan LL, Nkrumah JD, Basarab JA, Moore SS. 2008. Linkage of microbial ecology to phenotype: correlation of rumen microbial ecology to cattle's feed efficiency. FEMS Microbiol Lett 288:85–91. doi: 10.1111/j.1574-6968.2008.01343.x. [DOI] [PubMed] [Google Scholar]
- 6.Dill-McFarland KA, Breaker JD, Suen G. 2017. Microbial succession in the gastrointestinal tract of dairy cows from 2 weeks to first lactation. Sci Rep 7:40864. doi: 10.1038/srep40864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jewell KA, McCormick CA, Odt CL, Weimer PJ, Suen G. 2015. Ruminal bacterial community composition in dairy cows is dynamic over the course of two lactations and correlates with feed efficiency. Appl Environ Microbiol 81:4697–4710. doi: 10.1128/AEM.00720-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Y, Su Y, Zhu W. 2016. Microbiome-metabolome responses in the cecum and colon of pig to a high resistant starch diet. Front Microbiol 7:779. doi: 10.3389/fmicb.2016.00779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oikonomou G, Teixeira AG, Foditsch C, Bicalho ML, Machado VS, Bicalho RC. 2013. Fecal microbial diversity in pre-weaned dairy calves as described by pyrosequencing of metagenomic 16S rDNA. Associations of Faecalibacterium species with health and growth. PLoS One 8:e63157. doi: 10.1371/journal.pone.0063157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas M, Webb M, Ghimire S, Blair A, Olson K, Fenske GJ, Fonder AT, Christopher-Hennings J, Brake D, Scaria J. 2017. Metagenomic characterization of the effect of feed additives on the gut microbiome and antibiotic resistome of feedlot cattle. Sci Rep 7:12257. doi: 10.1038/s41598-017-12481-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petri RM, Schwaiger T, Penner GB, Beauchemin KA, Forster RJ, McKinnon JJ, McAllister TA. 2013. Changes in the rumen epimural bacterial diversity of beef cattle as affected by diet and induced ruminal acidosis. Appl Environ Microbiol 79:3744–3755. doi: 10.1128/aem.03983-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rice WC, Galyean ML, Cox SB, Dowd SE, Cole NA. 2012. Influence of wet distillers grains diets on beef cattle fecal bacterial community structure. BMC Microbiol 12:25. doi: 10.1186/1471-2180-12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carberry CA, Kenny DA, Han S, McCabe MS, Waters SM. 2012. The effect of phenotypic residual feed intake (RFI) and dietary forage content on the rumen microbial community of beef cattle. Appl Environ Microbiol 78:4949–4958. doi: 10.1128/aem.07759-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, Edrington TS. 2008. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol 8:125. doi: 10.1186/1471-2180-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young CA, Charlton ND, Takach JE, Swoboda GA, Trammell MA, Huhman DV, Hopkins AA. 2014. Characterization of Epichloe coenophiala within the US: are all tall fescue endophytes created equal?. Front Chem 2:95. doi: 10.3389/fchem.2014.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill NS, Stringer WC, Rottinghaus GE, Belesky DP, Parrott WA, Pope DD. 1990. Growth, morphological, and chemical component responses of tall fescue to Acremonium coenophialum. Crop Sci 30:156. doi: 10.2135/cropsci1990.0011183X003000010034x. [DOI] [Google Scholar]
- 17.Clay K. 1990. Fungal endophytes of grasses. Annu Rev Ecol Syst 21:275–297. doi: 10.1146/annurev.ecolsys.21.1.275. [DOI] [Google Scholar]
- 18.Clay K. 1990. Comparative demography of three graminoids infected by systemic, clavicipitaceous fungi. Ecology 71:558–570. doi: 10.2307/1940309. [DOI] [Google Scholar]
- 19.Clay K. 1993. The ecology and evolution of endophytes. Agric Ecosyst Environ 44:39–64. doi: 10.1016/0167-8809(93)90038-Q. [DOI] [Google Scholar]
- 20.Craig AM, Klotz JL, Duringer JM. 2015. Cases of ergotism in livestock and associated ergot alkaloid concentrations in feed. Front Chem 3:8. doi: 10.3389/fchem.2015.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthews K, Haley M. 2015. Livestock, dairy, and poultry outlook. USDA Economic Research Service, Washington, DC. [Google Scholar]
- 22.Mote RS, Hill NS, Uppal K, Tran VT, Jones DP, Filipov NM. 2017. Metabolomics of fescue toxicosis in grazing beef steers. Food Chem Toxicol 105:285–299. doi: 10.1016/j.fct.2017.04.020. [DOI] [PubMed] [Google Scholar]
- 23.Foote AP, Kristensen NB, Klotz JL, Kim DH, Koontz AF, McLeod KR, Bush LP, Schrick FN, Harmon DL. 2013. Ergot alkaloids from endophyte-infected tall fescue decrease reticuloruminal epithelial blood flow and volatile fatty acid absorption from the washed reticulorumen. J Anim Sci 91:5366–5378. doi: 10.2527/jas.2013-6517. [DOI] [PubMed] [Google Scholar]
- 24.Foote AP, Penner GB, Walpole ME, Klotz JL, Brown KR, Bush LP, Harmon DL. 2014. Acute exposure to ergot alkaloids from endophyte-infected tall fescue does not alter absorptive or barrier function of the isolated bovine ruminal epithelium. Animal 8:1106–1112. doi: 10.1017/S1751731114001141. [DOI] [PubMed] [Google Scholar]
- 25.Strickland JR, Aiken GE, Spiers DE, Fletcher LR, Oliver JW, Fribourg HA, Hannaway DB, West CP. 2009. Physiological basis of fescue toxicosis. Agron Monogr 53:203–227. [Google Scholar]
- 26.Oliver JW, Al-Tamimi H, Waller JC, Fribourg HA, Gwinn KD, Abney LK, Linnabary RD. 2004. Effect of chronic exposure of beef steers to the endophytic fungus of tall fescue; comparative effects on nitric oxide synthase activity and nitrate/nitrite levels in lateral saphenous veins, p 55–56. In Lang DJ. (ed), SERAIEG-8. Tall Fescue Toxicosis/Endophyte Workshop, Chapel Hill, TN. [Google Scholar]
- 27.Oliver JW, Cox SB, Waller JC, Fribourg HA, Gwinn KD, Rohrbach BW, Linnabary RD. 2004. Effect of chronic exposure of beef steers to the endophytic fungus of tall fescue; comparative effects on serum arginine levels, p 56–57. In Lang DJ. (ed), SERAIEG-8. Tall Fescue Toxicosis/Endophyte Workshop, Chapel Hill, TN. [Google Scholar]
- 28.Browning R Jr, Leite-Browning ML. 1997. Effect of ergotamine and ergonovine on thermal regulation and cardiovascular function in cattle. J Anim Sci 75:176–181. doi: 10.2527/1997.751176x. [DOI] [PubMed] [Google Scholar]
- 29.Al-Haidary A, Spiers DE, Rottinghaus GE, Garner GB, Ellersieck MR. 2001. Thermoregulatory ability of beef heifers following intake of endophyte-infected tall fescue during controlled heat challenge. J Anim Sci 79:1780–1788. doi: 10.2527/2001.7971780x. [DOI] [PubMed] [Google Scholar]
- 30.Aldrich CG, Paterson JA, Tate JL, Kerley MS. 1993. The effects of endophyte-infected tall fescue consumption on diet utilization and thermal regulation in cattle. J Anim Sci 71:164–170. doi: 10.2527/1993.711164x. [DOI] [PubMed] [Google Scholar]
- 31.Klotz JL. 2015. Activities and effects of ergot alkaloids on livestock physiology and production. Toxins (Basel) 7:2801–2821. doi: 10.3390/toxins7082801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eich E, Eichberg D, Müller W. 1984. Clavines: new antibiotics with cytostatic activity. Biochem Pharmacol 33:523–526. doi: 10.1016/0006-2952(84)90301-0. [DOI] [PubMed] [Google Scholar]
- 33.Eich E, Eichberg D, Schwarz G, Clas F, Loos M. 1985. Antimicrobial activity of clavines. Arzneimittelforschung 35:1760–1762. [PubMed] [Google Scholar]
- 34.Harlow BE, Goodman JP, Lynn BC, Flythe MD, Ji H, Aiken GE. 2017. Ruminal tryptophan-utilizing bacteria degrade ergovaline from tall fescue seed extract. J Anim Sci 95:980–988. doi: 10.2527/jas.2016.1128. [DOI] [PubMed] [Google Scholar]
- 35.Schumann B, Lebzien P, Ueberschär KH, Spilke J, Höltershinken M, Dänicke S. 2008. Effects of the level of feed intake and ergot contaminated concentrate on ruminal fermentation and on physiological parameters in cows. Mycotoxin Res 24:57–72. doi: 10.1007/BF02985283. [DOI] [PubMed] [Google Scholar]
- 36.De Lorme MJM, Lodge-Ivey SL, Craig AM. 2007. Physiological and digestive effects of Neotyphodium coenophialum-infected tall fescue fed to lambs. J Anim Sci 85:1199–1206. doi: 10.2527/jas.2005-430. [DOI] [PubMed] [Google Scholar]
- 37.Bessegatto JA, Paulino LR, Lisbôa JAN, Alfieri AA, Montemor CH, Medeiros LP, Kobayashi RKT, Weese JS, Costa MC. 2017. Changes in the fecal microbiota of beef cattle caused by change in management and the use of virginiamycin as a growth promoter. Res Vet Sci 114:355–362. doi: 10.1016/j.rvsc.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 38.de Oliveira MN, Jewell KA, Freitas FS, Benjamin LA, Totola MR, Borges AC, Moraes CA, Suen G. 2013. Characterizing the microbiota across the gastrointestinal tract of a Brazilian Nelore steer. Vet Microbiol 164:307–314. doi: 10.1016/j.vetmic.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 39.Klotz JL, Smith DL. 2015. Recent investigations of ergot alkaloids incorporated into plant and/or animal systems. Front Chem 3:23. doi: 10.3389/fchem.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Filipov NM, Thompson FN, Stuedemann JA, Elsasser TH, Kahl S, Sharma RP, Young CR, Stanker LH, Smith CK. 1999. Increased responsiveness to intravenous lipopolysaccharide challenge in steers grazing endophyte-infected tall fescue compared with steers grazing endophyte-free tall fescue. J Endocrinol 163:213–220. doi: 10.1677/joe.0.1630213. [DOI] [PubMed] [Google Scholar]
- 41.Hoveland CS. 1993. Importance and economic-significance of the acremonium endophytes to performance of animals and grass plant. Agric Ecosyst Environ 44:3–12. doi: 10.1016/0167-8809(93)90036-O. [DOI] [Google Scholar]
- 42.Ayers AW, Hill NS, Rottinghaus GE, Stuedemann JA, Thompson FN, Purinton PT, Seman DH, Dawe DL, Parks AH, Ensley D. 2009. Ruminal metabolism and transport of tall fescue ergot alkaloids. Crop Science 49:2309–2316. doi: 10.2135/cropsci2009.01.0018. [DOI] [Google Scholar]
- 43.Hill NS, Thompson FN, Stuedemann JA, Dawe DL, Hiatt EE III.. 2000. Urinary alkaloid excretion as a diagnostic tool for fescue toxicosis in cattle. J Vet Diagn Investig 12:210–217. doi: 10.1177/104063870001200303. [DOI] [PubMed] [Google Scholar]
- 44.Stuedemann JA, Hill NS, Thompson FN, Fayrer-Hosken RA, Hay WP, Dawe DL, Seman DH, Martin SA. 1998. Urinary and biliary excretion of ergot alkaloids from steers that grazed endophyte-infected tall fescue. J Anim Sci 76:2146–2154. doi: 10.2527/1998.7682146x. [DOI] [PubMed] [Google Scholar]
- 45.Rogers WM, Roberts CA, Andrae JG, Davis DK, Rottinghaus GE, Hill NS, Kallenbach RL, Spiers DE. 2011. Seasonal fluctuation of ergovaline and total ergot alkaloid concentrations in tall fescue regrowth. Crop Sci 51:1291–1296. doi: 10.2135/cropsci2010.07.0402. [DOI] [Google Scholar]
- 46.Rottinghaus GE, Garner GB, Cornell CN, Ellis JL. 1991. Hplc method for quantitating ergovaline in endophyte-infested tall fescue–seasonal-variation of ergovaline levels in stems with leaf sheaths, leaf blades, and seed heads. J Agric Food Chem 39:112–115. doi: 10.1021/jf00001a022. [DOI] [Google Scholar]
- 47.McCulley RL, Bush LP, Carlisle AE, Ji H, Nelson JA. 2014. Warming reduces tall fescue abundance but stimulates toxic alkaloid concentrations in transition zone pastures of the U.S. Front Chem 2:88. doi: 10.3389/fchem.2014.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shanks OC, Kelty CA, Archibeque S, Jenkins M, Newton RJ, McLellan SL, Huse SM, Sogin ML. 2011. Community structures of fecal bacteria in cattle from different animal feeding operations. Appl Environ Microbiol 77:2992–3001. doi: 10.1128/AEM.02988-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan D, Yu Z. 2014. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes 5:108–119. doi: 10.4161/gmic.26945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Isaacson R, Kim HB. 2012. The intestinal microbiome of the pig. Anim Health Res Rev 13:100–109. doi: 10.1017/S1466252312000084. [DOI] [PubMed] [Google Scholar]
- 51.Klieve AV, O’Leary MN, McMillen L, Ouwerkerk D. 2007. Ruminococcus bromii, identification and isolation as a dominant community member in the rumen of cattle fed a barley diet. J Appl Microbiol 103:2065–2073. doi: 10.1111/j.1365-2672.2007.03492.x. [DOI] [PubMed] [Google Scholar]
- 52.Ze X, Duncan SH, Louis P, Flint HJ. 2012. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J 6:1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.La Reau AJ, Meier-Kolthoff JP, Suen G. 2016. Sequence-based analysis of the genus Ruminococcus resolves its phylogeny and reveals strong host association. Microb Genomics 2:e000099. doi: 10.1099/mgen.0.000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.La Reau AJ, Suen G. 2018. The Ruminococci: key symbionts of the gut ecosystem. J Microbiol 56:199–208. doi: 10.1007/s12275-018-8024-4. [DOI] [PubMed] [Google Scholar]
- 55.Zhang LS, Davies SS. 2016. Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions. Genome Med 8:46. doi: 10.1186/s13073-016-0296-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.den Besten G, Bleeker A, Gerding A, van Eunen K, Havinga R, van Dijk TH, Oosterveer MH, Jonker JW, Groen AK, Reijngoud DJ, Bakker BM. 2015. Short-chain fatty acids protect against high-fat diet-induced obesity via a PPARgamma-dependent switch from lipogenesis to fat oxidation. Diabetes 64:2398–2408. doi: 10.2337/db14-1213. [DOI] [PubMed] [Google Scholar]
- 57.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. 2013. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin HV, Frassetto A, Kowalik EJ Jr, Nawrocki AR, Lu MM, Kosinski JR, Hubert JA, Szeto D, Yao X, Forrest G, Marsh DJ. 2012. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS One 7:e35240. doi: 10.1371/journal.pone.0035240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, Cameron J, Grosse J, Reimann F, Gribble FM. 2012. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes 61:364–371. doi: 10.2337/db11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Larraufie P, Dore J, Lapaque N, Blottiere HM. 2017. TLR ligands and butyrate increase Pyy expression through two distinct but inter-regulated pathways. Cell Microbiol 19:e12648. doi: 10.1111/cmi.12648. [DOI] [PubMed] [Google Scholar]
- 61.Larraufie P, Martin-Gallausiaux C, Lapaque N, Dore J, Gribble FM, Reimann F, Blottiere HM. 2018. SCFAs strongly stimulate PYY production in human enteroendocrine cells. Sci Rep 8:74. doi: 10.1038/s41598-017-18259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Conterno L, Fava F, Viola R, Tuohy KM. 2011. Obesity and the gut microbiota: does up-regulating colonic fermentation protect against obesity and metabolic disease? Genes Nutr 6:241–260. doi: 10.1007/s12263-011-0230-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Holzer P, Farzi A. 2014. Neuropeptides and the microbiota-gut-brain axis. Adv Exp Med Biol 817:195–219. doi: 10.1007/978-1-4939-0897-4_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sternini C, Anselmi L, Rozengurt E. 2008. Enteroendocrine cells: a site of “taste” in gastrointestinal chemosensing. Curr Opin Endocrinol Diabetes Obes 15:73–78. doi: 10.1097/MED.0b013e3282f43a73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Myer PR, Wells JE, Smith TP, Kuehn LA, Freetly HC. 2015. Microbial community profiles of the colon from steers differing in feed efficiency. Springerplus 4:454. doi: 10.1186/s40064-015-1201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE. 2014. Human genetics shape the gut microbiome. Cell 159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stevenson DM, Weimer PJ. 2007. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biotechnol 75:165–174. doi: 10.1007/s00253-006-0802-y. [DOI] [PubMed] [Google Scholar]
- 68.Kohl KD, Amaya J, Passement CA, Dearing MD, McCue MD. 2014. Unique and shared responses of the gut microbiota to prolonged fasting: a comparative study across five classes of vertebrate hosts. FEMS Microbiol Ecol 90:883–894. doi: 10.1111/1574-6941.12442. [DOI] [PubMed] [Google Scholar]
- 69.Konikoff T, Gophna U. 2016. Oscillospira: a central, enigmatic component of the human gut microbiota. Trends Microbiol 24:523–524. doi: 10.1016/j.tim.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 70.Org E, Blum Y, Kasela S, Mehrabian M, Kuusisto J, Kangas AJ, Soininen P, Wang Z, Ala-Korpela M, Hazen SL, Laakso M, Lusis AJ. 2017. Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol 18:70. doi: 10.1186/s13059-017-1194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martinez I, Perdicaro DJ, Brown AW, Hammons S, Carden TJ, Carr TP, Eskridge KM, Walter J. 2013. Diet-induced alterations of host cholesterol metabolism are likely to affect the gut microbiota composition in hamsters. Appl Environ Microbiol 79:516–524. doi: 10.1128/AEM.03046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaakoush NO. 2015. Insights into the role of Erysipelotrichaceae in the human host. Front Cell Infect Microbiol 5:84. doi: 10.3389/fcimb.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maekawa T, Shimayama H, Tsushima H, Kawakami F, Kawashima R, Kubo M, Ichikawa T. 2017. LRRK2: an emerging new molecule in the enteric neuronal system that quantitatively regulates neuronal peptides and IgA in the gut. Dig Dis Sci 62:903–912. doi: 10.1007/s10620-017-4476-3. [DOI] [PubMed] [Google Scholar]
- 74.Nihsen ME, Piper EL, West CP, Crawford RJ Jr, Denard TM, Johnson ZB, Roberts CA, Spiers DA, Rosenkrans CF Jr.. 2004. Growth rate and physiology of steers grazing tall fescue inoculated with novel endophytes. J Anim Sci 82:878–883. doi: 10.2527/2004.823878x. [DOI] [PubMed] [Google Scholar]
- 75.Brown KR, Anderson GA, Son K, Rentfrow G, Bush LP, Klotz JL, Strickland JR, Boling JA, Matthews JC. 2009. Growing steers grazing high versus low endophyte (Neotyphodium coenophialum)-infected tall fescue have reduced serum enzymes, increased hepatic glucogenic enzymes, and reduced liver and carcass mass. J Anim Sci 87:748–760. doi: 10.2527/jas.2008-1108. [DOI] [PubMed] [Google Scholar]
- 76.Oliver JW. 1997. Physiological manifestations of endophyte toxicosis in ruminant and laboratory species, p 311–346. In Bacon CW, Hill NS (ed), Neotyphodium/grass interactions. Plenum Publishing, New York, NY. [Google Scholar]
- 77.Perumbakkam S, Rattray RM, Delorme MJM, Duringer JM, Craig AM. 2007. Discovery of novel microorganisms involved in ergot alkaloid detoxification: an approach, p 395–398. Proc 6th Int Symp Fungal Endophytes Grasses. New Zealand Grassland Association, Palmerston North, New Zealand. [Google Scholar]
- 78.Nicholls HT, Krisko TI, LeClair KB, Banks AS, Cohen DE. 2016. Regulation of adaptive thermogenesis by the gut microbiome. FASEB J 30:854.2. [Google Scholar]
- 79.Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Gao Z, Chen H, Berger KI, Goldring RM, Rom WN, Blaser MJ, Weiden MD. 2013. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 1:19. doi: 10.1186/2049-2618-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Segal LN, Rom WN, Weiden MD. 2014. Lung microbiome for clinicians. New discoveries about bugs in healthy and diseased lungs. Ann Am Thorac Soc 11:108–116. doi: 10.1513/AnnalsATS.201310-339FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Noverr MC, Falkowski NR, McDonald RA, McKenzie AN, Huffnagle GB. 2005. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect Immun 73:30–38. doi: 10.1128/IAI.73.1.30-38.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Noverr MC, Noggle RM, Toews GB, Huffnagle GB. 2004. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun 72:4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murray RL. 1989. Creatinine, p 1015–1021. In Kaplan LA, Pesce AJ (ed), Clinical chemistry, 2nd ed. C.V. Mosby, St. Louis, MO. [Google Scholar]
- 84.Koontz AF, Bush LP, Klotz JL, McLeod KR, Schrick FN, Harmon DL. 2012. Evaluation of a ruminally dosed tall fescue seed extract as a model for fescue toxicosis in steers. J Anim Sci 90:914–921. doi: 10.2527/jas.2011-4292. [DOI] [PubMed] [Google Scholar]
- 85.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chao A. 1984. Nonparametric-estimation of the number of classes in a population. Scand J Stat 11:265–270. [Google Scholar]
- 90.Simpson EH. 1949. Measurement of diversity. Nature 163:688–688. doi: 10.1038/163688a0. [DOI] [Google Scholar]
- 91.Le Cao K, Gonzalez I, Dejean S, Rahart F, Gautier B, Monget P, Coquery J, Yao F, Liquet B. 2015. mixOmics: Omics data integration project. http://CRAN.R-project.org/package=mixOmics.
- 92.R Development Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org. [Google Scholar]
- 93.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AK, Kent AD, Daroub SH, Camargo FA, Farmerie WG, Triplett EW. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ondov BD, Bergman NH, Phillippy AM. 2011. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics 12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Faust K, Raes J. 2016. CoNet app: inference of biological association networks using Cytoscape. F1000Res 5:1519. doi: 10.12688/f1000research.9050.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.