DHA is an attractive triose molecule with a wide range of applications, notably in cosmetics and the food and pharmaceutical industries. DHA is found in many species, from microorganisms to humans, and can be used by Escherichia coli as a growth substrate. However, knowledge about the mechanisms and regulation of this process is currently lacking, motivating our investigation of DHA metabolism in E. coli. We show that under aerobic conditions, E. coli growth on DHA is far from optimal and is hindered by chemical, hierarchical, and possibly allosteric constraints. We show that optimal growth on DHA can be restored by releasing the hierarchical constraint. These results improve our understanding of DHA metabolism and are likely to help unlock biotechnological applications involving DHA as an intermediate, such as the bioconversion of glycerol or C1 substrates into value-added chemicals.
KEYWORDS: dihydroxyacetone, Escherichia coli, carbon metabolism, metabolic modeling
ABSTRACT
In this work, we shed light on the metabolism of dihydroxyacetone (DHA), a versatile, ubiquitous, and important intermediate for various chemicals in industry, by analyzing its metabolism at the system level in Escherichia coli. Using constraint-based modeling, we show that the growth of E. coli on DHA is suboptimal and identify the potential causes. Nuclear magnetic resonance analysis shows that DHA is degraded nonenzymatically into substrates known to be unfavorable to high growth rates. Transcriptomic analysis reveals that DHA promotes genes involved in biofilm formation, which may reduce the bacterial growth rate. Functional analysis of the genes involved in DHA metabolism proves that under the aerobic conditions used in this study, DHA is mainly assimilated via the dihydroxyacetone kinase pathway. In addition, these results show that the alternative routes of DHA assimilation (i.e., the glycerol and fructose-6-phosphate aldolase pathways) are not fully activated under our conditions because of anaerobically mediated hierarchical control. These pathways are therefore certainly unable to sustain fluxes as high as the ones predicted in silico for optimal aerobic growth on DHA. Overexpressing some of the genes in these pathways releases these constraints and restores the predicted optimal growth on DHA.
IMPORTANCE DHA is an attractive triose molecule with a wide range of applications, notably in cosmetics and the food and pharmaceutical industries. DHA is found in many species, from microorganisms to humans, and can be used by Escherichia coli as a growth substrate. However, knowledge about the mechanisms and regulation of this process is currently lacking, motivating our investigation of DHA metabolism in E. coli. We show that under aerobic conditions, E. coli growth on DHA is far from optimal and is hindered by chemical, hierarchical, and possibly allosteric constraints. We show that optimal growth on DHA can be restored by releasing the hierarchical constraint. These results improve our understanding of DHA metabolism and are likely to help unlock biotechnological applications involving DHA as an intermediate, such as the bioconversion of glycerol or C1 substrates into value-added chemicals.
INTRODUCTION
Dihydroxyacetone (DHA) is an attractive molecule that is used as a final product in a wide range of industries (i.e., the food, cosmetic, and pharmaceutical industries) (1–3) or as a growth substrate for various microorganisms (4–6). DHA is a ubiquitous molecule found in all domains. A physiological product of the body naturally present in human urine (7), it also plays a role in the osmoregulation of yeast and algae (8). DHA is produced as an intermediate of various metabolic pathways. In methylotrophic yeast, DHA is a key intermediate in methanol assimilation (9, 10), while in bacteria, DHA is produced by aldol cleavage of the glycolytic intermediary fructose 6-phosphate (11) or by oxidation of glycerol, the latter process being the basis for the current industrial production of DHA by Gluconobacter oxydans (12). DHA is also a highly reactive molecule, and its accumulation is presumed to be toxic. Indeed, DHA can react with DNA and proteins in Maillard-type reactions and thereby alter cell heredity (13, 14). In addition, DHA is unstable and can be converted nonenzymatically into several molecular species, among which is the highly toxic methylglyoxal (15–18).
Escherichia coli can metabolize DHA aerobically through at least three different metabolic pathways: (i) the dihydroxyacetone kinase (DAK) pathway, (ii) the glycerol (GLD) pathway, and (iii) the fructose-6-phosphate (FSA) pathway (17). The DAK pathway is named after dihydroxyacetone kinase, encoded by the dhaKLM operon. This operon is controlled by DhaR, a transcription factor activated by DHA (19). DhaKLM is composed of three subunits (DhaK, DhaL, and DhaM) and phosphorylates DHA to dihydroxyacetone phosphate (DHAP), an intermediate in the glycolytic pathway. DhaKLM has a high affinity for DHA (Km of <6 μM) and a catalytic constant (kcat) of 4.8 s−1 (20). This kinase resembles a phosphotransferase system (PTS) that uses phosphoenolpyruvate (PEP) as a phosphoryl donor (21). The DhaM subunit is first multiphosphorylated before the DhaL subunit uses the phosphate to convert ADP into ATP. Finally, this ATP is used directly by the DhaK subunit to phosphorylate DHA and form DHAP (22). The fact that dhaK transposon insertion prevents the growth of E. coli on DHA (23) indicates that DhaKLM is essential for this process (23).
The GLD pathway involves glycerol dehydrogenase (GldA) and glycerol kinase (GlpK). Hydroxyacetone induces gldA expression in the stationary phase (24) while the transcription of glpK is regulated by catabolite repression and by glycerol and glycerol-3-phosphate via the transcriptional repressor GlpR (25). During anaerobic glycerol fermentation, GldA enables the formation of DHA from glycerol (4), whereas under aerobic conditions, GldA enables the reverse process, i.e., the formation of glycerol from DHA (17, 26). GldA has a greater affinity for DHA (Km of 0.3 mM) than for glycerol (Km of 56 mM) (17), but the catalytic rate constants are close (kcat of 17.2 and 22.4 s−1, respectively). It has been proposed that the primary in vivo role of GldA is the removal of surplus dihydroxyacetone to limit its toxic effects (17). However, DhaKLM and GldA have been shown to constitute a fermentative route for the conversion of glycerol to glycolytic intermediates (27), demonstrating that GldA is also involved in fermentation of glycerol (4). Once formed from DHA, glycerol is phosphorylated by GlpK to form glycerol-3-phosphate, which is central for lipid biosynthesis. GlpK has been shown to play a crucial role in DHA assimilation in various microorganisms (28–31). In Haloferax volcanii, for instance, the deletion of glpK has a higher impact on growth than does dhaKLM deletion, and mutants with both genes deleted do not grow on DHA (29). It has been hypothesized that in these organisms, GlpK can either phosphorylate DHA into DHAP or control the transport of DHA into the cells (28, 29). In E. coli, in vitro results indicate that GlpK is able to use DHA as a substrate with a Km value of 0.5 mM (32); however, its in vivo implication in DHA metabolism has never been studied.
The central enzyme in the FSA pathway is fructose-6-aldolase, which condenses DHA with glyceraldehyde-3-phosphate (GAP) to form fructose-6-phosphate (F6P). Two different genes in E. coli (fsaA and fsaB or mipB and talC) code for FsaA and FsaB, respectively. These two enzymes share 70% identity and have similar affinities for DHA (Km of 32 mM for FsaA and 27 mM for FsaB), but FsaA has a higher catalytic constant (kcat) for DHA (116 s−1 versus 41 s−1) (11). The physiological role of FsaA in E. coli (33) and its regulation remain unknown. However, fsaB is in the same operon as gldA and ptsA, suggesting that fsaB is metabolically associated with DHA (17). The role of this operon and its regulation are not completely clear. However, PtsA has been identified as a component of a putative PTS system involved in anaerobic fructose catabolism (34). In addition, higher expression of gldA, fsaA, and fsaB has been observed in wild-type E. coli grown on glucose during a transition from aerobic to anaerobic cultivation (35).
While several pathways potentially can support DHA catabolism in E. coli, which ones are actually involved during growth on DHA remains unclear, and their regulation is poorly characterized. This missing information would help unravel the physiological significance of this versatile, ubiquitous, and high-potential metabolite. The objective of our study was to investigate DHA metabolism in E. coli using system-level analysis. We first predicted the metabolic fate of DHA using a genome-scale model of E. coli to evaluate its metabolic capabilities. We then used transcriptomic analysis to identify “genes that matter” for growth on DHA. Based on these results, we characterized the physiology effect of overexpression and deletion of genes involved in DHA metabolism, and we propose mechanisms that would explain the fate of DHA in E. coli.
RESULTS AND DISCUSSION
DHA metabolism in E. coli involves a complex network of chemical and biological processes.
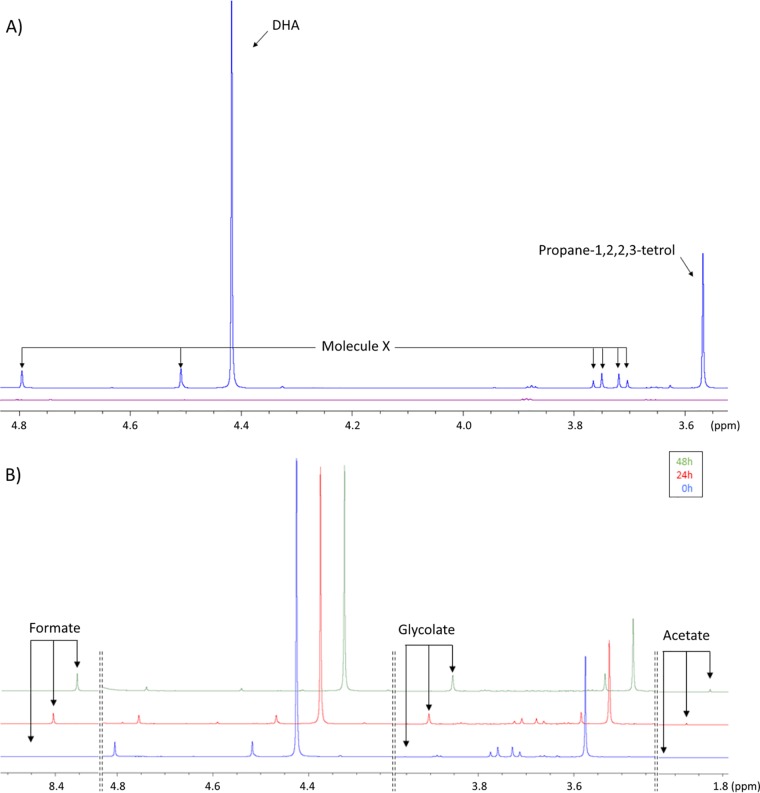
We carried out cultivation experiments to study the global behavior of E. coli during growth on DHA, monitoring the concentrations of biomass and extracellular metabolites as a function of time. DHA is an unstable compound that can be interconverted into different forms when dissolved in water (18) or autoxidized by Fenton’s reaction to form glycolate, other short-chained carbohydrates, and organic acids upon incubation (15, 16). We therefore determined beforehand whether nonenzymatic conversions of DHA could occur under our conditions. Noninoculated M9 minimal medium containing 15 mM DHA was incubated at 37°C for 48 h, and samples were collected after 0, 24, and 48 h of incubation and analyzed by 1H-nuclear magnetic resonance (NMR) (Fig. 1). Since DHA is a symmetrical molecule and all the detectable protons from the two CH2 moieties are equivalent, the 1H-NMR spectrum of pure DHA is expected to contain a singlet at 4.4 ppm. However, several signals were detected in the medium at time zero (Fig. 1A). None of these peaks were observed in a sample containing M9 medium without DHA (Fig. 1A), suggesting that they originated from the DHA solution. Two-dimensional (2D) NMR analysis (see Fig. S1 in the supplemental material) shows that the signal at 3.6 ppm corresponds to a hydrated form of DHA, propane-1,2,2,3-tetrol (or dihydroxyacetone monomer hydrate), as previously reported (18). Under our conditions, the proportions of nonhydrated and hydrated DHA were 65% ± 2% and 21% ± 2%, respectively. The remaining 14% ± 2% corresponds to the signals at 4.8, 4.7 (both singlets), and 3.7 (AB system) ppm, all originating from the same unidentified molecule X, which appeared after freezing/thawing the samples. Unfortunately, we were unable to identify the compound corresponding to these peaks.
FIG 1.
Nonenzymatic transformation of DHA in M9 medium with 5× diluted salts. (A) NMR spectrum overlay of nonincubated modified M9 medium with (blue) or without (purple) 15 mM DHA. (B) Kinetics of nonenzymatic transformation of DHA in modified M9 medium after 0 h (blue), 24 h (red), and 48 h (green) of incubation at 37°C. The three spectra are shown with a vertical step of 10% and a horizontal offstep of 0.05 ppm.
Upon incubation of the noninoculated medium, the peaks from DHA, propane-1,2,2,3-tetrol, and molecule X decreased in intensity, while additional signals appeared that were not seen with the initial spectrum (Fig. 1B). This phenomenon was influenced by the amount of salts (i.e., Na2HPO4, KH2PO4, NaCl, and NH4Cl) in the medium. At the salt concentration typically used for the M9 medium (36), 82% of DHA was degraded after 48 h, while only 34% was degraded when Na2HPO4, KH2PO4, NaCl, and NH4Cl were diluted five times (data not shown). Thus, a 5× reduced salt concentration was used for all the subsequent experiments. In this modified M9 medium, DHA was mainly converted into formate, glycolate, and acetate (Fig. 1B and Fig. S2). After 39 h of incubation, 27% of the DHA was converted into formate (36%), glycolate (26%), and acetate (1.3%). The nonlinear DHA concentration profile was fitted assuming first-order kinetics. The excellent agreement between experimental and fitted curves (Pearson’s r of >0.99) validates the proposed degradation kinetics; the estimated degradation rate constant is 0.0086 ± 0.0010 h−1, corresponding to a DHA half-life of 116 h (Fig. S2).
In the inoculated medium, the peaks from both forms of DHA decreased monotonously over time (data not shown). The proportions of both forms remained constant during the experiment, suggesting that chemical equilibrium is faster than DHA uptake. No accumulation of glycolate, formate, or acetate was observed, indicating that these products of nonenzymatic DHA degradation are coconsumed with DHA by wild-type E. coli. In order to infer quantitative flux information from these data, we developed an R program, PhysioFit, a mathematical model to estimate the growth rate and exchange (uptake or production) fluxes. This model, which takes into account nonenzymatic degradation of substrates, is described in detail in Materials and Methods. This program calculated a growth rate of 0.15 ± 0.03 h−1 with a DHA uptake rate of 5.2 ± 0.19 mmol gDW−1 h−1 (corresponding to 15.6 Cmmol gDW−1 h−1; gDW is grams of dry weight). Based on the nonenzymatic conversion yields of DHA, we estimated that DHA accounted for 74.3% of E. coli carbon uptake (expressed in Cmmol), with the remaining carbon taken up as formate (15.3%), glycolate (9.7%), and acetate (0.7%). This represents a specific uptake rate of 3.2, 1.0, and 0.1 mmol gDW−1 h−1, respectively (i.e., 3.2, 2.0, and 0.2 Cmmol gDW−1 h−1).
Overall, these results show that an additional set of reactions (i.e., nonenzymatic conversion of DHA) occur under these condition (Fig. 2) that have to be taken into account when studying the metabolism of DHA.
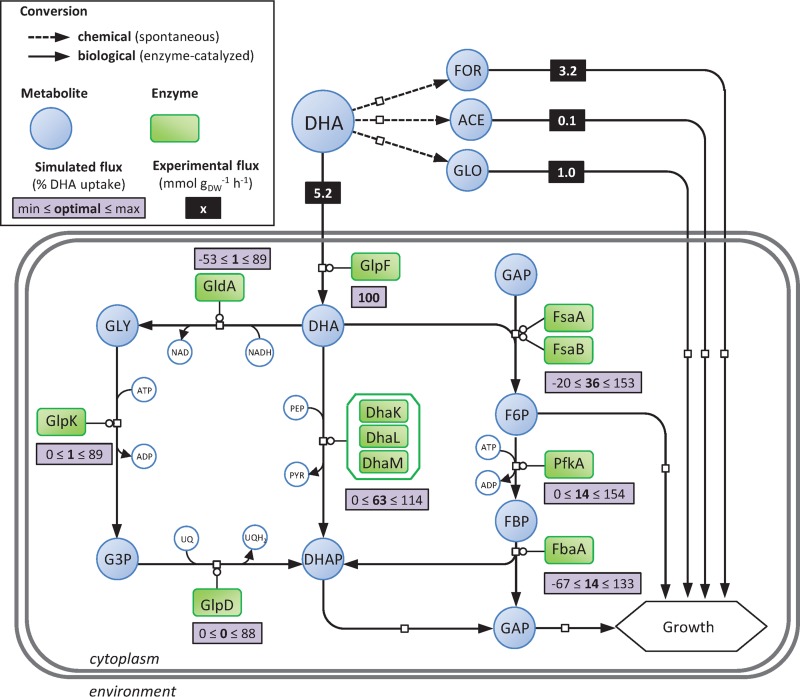
FIG 2.
Experimental and simulated fluxes through DHA metabolism of E. coli. Gray rectangles give the optimal and the ranges of simulated fluxes. Optimal simulated fluxes were obtained using flux balance analysis constrained with experimental uptake fluxes of the wild-type strain grown on modified M9 media containing 15 mM DHA (i.e., values given in black rectangle). Ranges of simulated fluxes were obtained using flux variability analysis when growth rate is constrained to 95% of the optimal value and with experimental uptake fluxes. Experimental flux values are given in mmol gDW−1 h−1, and simulated flux values are given in percent relative to DHA uptake rate. Shown are dihydroxyacetone kinase enzymes (DhaK, DhaL, and DhaM), fructose-6-phosphate aldolase enzymes (FsaA and FsaB), glycerol dehydrogenase enzyme (GldA), glycerol facilitator (GlpF), glycerol-3-phosphate dehydrogenase enzyme (GlpD), glycerol kinase enzyme (GlpK), fructose-bisphosphate aldolase enzyme (FbaA), 6-phosphofructokinase enzyme (PfkA), dihydroxyacetone (DHA), formate (FOR), acetate (ACE), glycolate (GLO), dihydroxyacetone phosphate (DHAP), fructose-6-phosphate (F6P), fructose-1,6-bisphosphate (FBP), glyceraldehyde-3-phosphate (GAP), glycerol (GLY), glycerol-3-phosphate (G3P), phosphoenolpyruvate (PEP), pyruvate (PYR), ubiquinone (UQ), ubiquinol (UQH2).
E. coli growth on DHA is robust but suboptimal.
Constraint-based metabolic models provide an attractive starting point for studying poorly characterized metabolisms, because they can predict growth rates and metabolite flows through a metabolic network with minimal a priori knowledge. We performed flux balance analysis (FBA) using a genome-scale E. coli model (iJO1366 [37]) to simulate the fate of DHA (Fig. 2). The only constraints used were the DHA, formate, glycolate, and acetate uptake rates of wild-type E. coli, calculated as detailed above. The model predicted a growth rate of 0.28 h−1, i.e., almost 2 times higher than the experimentally observed growth rate (0.15 h−1). Since FBA predictions assume optimal growth under stoichiometric and maximum uptake constraints, these results suggest that for some reason the system was not operating optimally in vivo. For optimal growth, the predicted model has 63% of the DHA converted into DHAP via the DAK-based pathway and 36% via the FSA pathway. The GLD-based pathway is only used to feed glycerol-3-phosphate for biomass synthesis (Fig. 2). Flux balance analysis computes an optimal objective value and a flux state that are consistent with that objective (and all the imposed constraints). While the objective value (i.e., the growth rate under our conditions) is unique, this can typically be supported by multiple flux states in genome-scale models. For this reason, we performed flux variability analysis (FVA) to find the minimum and maximum flux for each reaction in the network while maintaining growth at 95% of its optimal value (38). The results indicate that the fluxes through the GLD-, DAK-, and FSA-based pathways offer a wide range of possible values, including opposite fluxes (i.e., reverse reactions). These data suggest that, based solely on stoichiometric constraints, optimal growth does not in theory depend on any particular DHA metabolic pathway.
Overall, these data demonstrate flexible use of all the DHA catabolic pathways and suggest that this metabolism in E. coli is highly robust.
DHA-induced genes involved in a biofilm growth state.
In order to gain insight into the regulation of DHA metabolism, we compared the global transcriptional responses of wild-type E. coli grown on DHA (Fig. S3) and glucose medium. This revealed that several genes related to mobility, adherence, and biofilm formation and stress were expressed differently between DHA and glucose growth (Table 1 and Data File S1). For instance, the csg genes, encoding curli fibers, which promote cell adhesion during biofilm formation (39), were expressed at a higher level (by a factor of 10 on average) during DHA growth. Conversely, genes involved in motility and flagellum assembly were less expressed (e.g., by factors of 25- and 12-fold on average for the flg and fli genes, respectively) (Table 1 and Data File S1), indicating that motility is repressed on DHA. Genes related to lipopolysaccharide (LPS) synthesis were expressed by a factor of 4 on DHA (Table 1 and Data File S1). LPSs are a major component of the outer leaflet of the outer membrane of most Gram-negative bacteria, contributing to their structural integrity. LPSs govern many of the biological interactions between cells and their environment (40) and play a crucial role in the anchoring process (41). Taken together, these data suggest that E. coli switches from planktonic growth on glucose to biofilm growth on DHA. This might contribute to the low growth rate of E. coli on DHA, since cells in biofilms are known to grow more slowly than in the planktonic mode (42).
TABLE 1.
Functional classification of genes with statistically significant decreases and increases in mRNA level in E. coli strain BW25113a
| Category and expression status | GO concerned | P value | Gene examples |
|---|---|---|---|
| Overexpressed | |||
| Cell wall and adhesion | GO:0009103; lipopolysaccharide biosynthetic process | 2.66E−14 | waaO, wbbJ, arnC, wcaB, waaB, waaJ, waaY, waaU, rfbA, rfbC |
| GO:0022610; biological adhesion | 8.85E−12 | dgcZ, elfA, yadK, yehA, pgaC, yeeJ, csgB, csgF, csgE, csgG | |
| GO:0043711; pilus organization | 4.79E−10 | htrE, yehB, ybgQ, elfC, sfmD, ydeT, yqiG, fimD, sfmC, elfD | |
| Response to host | GO:0006952; defense response | 4.50E−07 | casE, casD, casC, casB, casA, abpA, abpB, rzpD, rrrQ, rzpR |
| GO:0009243; O antigen biosynthetic process | 4.86E−05 | rfbC, rfbD, rfbB, rfbA, wbbI, rfbX | |
| Response to acidic pH | GO:0010447; response to acidic pH | 6.07E−05 | cadB, yjaA, hdeA, iraM, oxc, frc, glsA, yagU, evgS, hdeB |
| Copper ion homeostasis | GO:0006878; cellular copper ion homeostasis | 7.59E−04 | cusA, cusB, cusC, cusF |
| Involved in glucarate catabolic process | GO:0019394; glucarate catabolic process | 1.67E−04 | garKLR, gudD |
| Underexpressed | |||
| Locomotion | GO:0040011; locomotion | 1.28E−24 | fliL, flgB, flgC, flgG, flgD, flgE, flgF, motA, fdrA, fliG |
| Cellular respiration | GO:0045333; cellular respiration | 6.09E−15 | frdB, hyaA, hyaB, pflD, fdoG, fdoH, frdA, frdC, frdD, hybB |
| Primary metabolism | GO:0019321; pentose metabolic process | 2.11E−10 | rbsD, rbsK, xylA, araD, araA, araB, xylF, xylG, xylH, rhaD |
| GO:0016052; carbohydrate catabolic process | 3.19E−9 | treC, lacZ, melA, malS, gldA, hybA, gatY, uidA, gntK, gntP, gntT | |
| GO:0008643; carbohydrate transport | 4.84E−09 | kdgT, gntP, gntT, ptsG, yiaO, melB, rbsA, srlB, frwC, treB | |
| GO:0071941; nitrogen cycle metabolic process | 2.45E−08 | argG, argI, argF, carA, ygeW, napC, nirB, narG, nirD, nirC | |
| GO:0006099; tricarboxylic acid cycle | 5.99E−07 | sdhD, sdhC, sdhB, acnA, sucA, fumB, mdh, sdhA, sucD, sucC |
mRNA levels were examined in E. coli strain BW25113 in modified M9-DHA medium and compared to values for M9 glucose medium (60). The Clusters of Orthologous Groups (COG) were used for grouping.
The DAK pathway is central but not essential for DHA metabolism.
The regulation of dhaKLM expression (19) and the properties of the enzyme (20) have led to the suggestion that the DAK pathway is important and even essential for growth on DHA in E. coli (23). To test this hypothesis, we investigated the expression levels of the genes involved in DHA metabolism in wild-type and ΔdhaKLM strains grown on DHA (Fig. 3 and Fig. S3) and studied the functional characteristics of DhaKLM (Fig. 4). Comparing the expression levels of wild-type strains grown on DHA versus glucose shows that the genes involved in glucarate metabolism (i.e., the gar and gud operons) are upregulated by a factor of about 5 on average (Fig. 3 and Data File S1), while those involved in glycolate metabolism (i.e., the glc operon) were not. As shown previously, DHA is spontaneously converted into glycolate, which is supposed to induce the glc genes (43); however, glycolate can also be recognized by the same transporter as glucarate (44) and catabolized using part of its metabolic pathway. The gene encoding the glycerol facilitator GlpF, a nonspecific channel protein capable of transporting straight-chain carbon compounds such as DHA, was upregulated about 3-fold, consistent with its putative role in DHA transport (23). As expected, both the dhaKLM operon and its transcription factor, dhaR, were strongly upregulated (Fig. 3 and Data File S1). However, the expression levels of glpK, fsaA, and fsaB were not affected, and gldA was downregulated by a factor of 2.5 (Fig. 3). Transcriptome analysis of the ΔdhaKLM and wild-type strains (Fig. 3) revealed that none of the genes of the FSA and GLD pathways were upregulated in the ΔdhaKLM strain, demonstrating that there is no compensatory activation of these pathways. Interestingly, the hyc and hyp operons and Fhl enzymes, involved in the production of dihydrogen and carbon dioxide in formate metabolism (45) and activated by formate, were upregulated by a factor of 10 (Fig. 3 and Data File S1). This points to the presence of formate in the medium, which is consistent with the nonenzymatic degradation of DHA into formate, and suggests that Fhl enzymes allow the ΔdhaKLM strain to convert formate from the medium and use the hydrogen as an electron donor for respiration (46). Deletion of dhaKLM resulted in a significant and strong reduction of the growth rate and a shift in the substrate profiles compared with those of the control strain (Fig. S3 and S4A), while complementation of the deleted genes restored normal growth (Fig. S4). Furthermore, in this strain the specific DHA uptake rate was strongly reduced. When dhaKLM was overexpressed (i.e., dhaKLM+++), the specific DHA uptake rate increased slightly, by 15% compared with that of the wild type, but the growth rate did not (Fig. 4). Taken together, these data support the hypothesis that DhaKLM plays a crucial role in DHA utilization, in agreement with the transcriptomics data. However, under our conditions, and contrary to previous observations in E. coli (23), inactivation of dhaKLM was not lethal. This is because the metabolism of DHA is shifted to formate, glycolate, and acetate metabolism (due to the instability of DHA) in this strain. Based on the nonenzymatic conversion yields of DHA into formate, glycolate, and acetate determined above, we estimate that formate accounted for 38.5% of the ΔdhaKLM strain’s carbon uptake (expressed in Cmmol), while DHA, glycolate, and acetate accounted for 34, 25.7, and 1.6%, respectively.
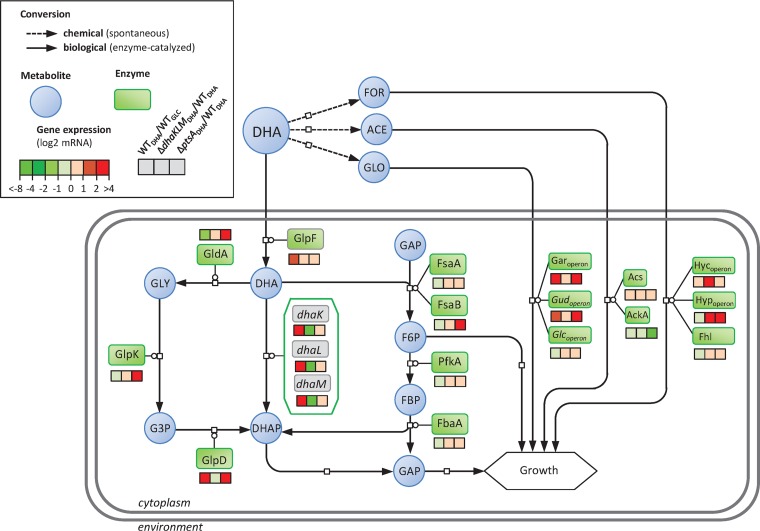
FIG 3.
Change in gene expression in DHA metabolism of the wild-type strain, ΔdhaKLM strain, and ΔptsA strain cultured on modified M9-DHA medium. For the wild-type strain (WTDHA), the fold changes in gene expression were calculated in reference to expression in an E. coli strain cultured in chemostat in M9-glucose medium at 0.1 h−1 (WTGLC). For the ΔdhaKLM (ΔdhaKLMDHA) and the ΔptsA (ΔptsADHA) strains, the fold changes in gene expression were calculated in reference to expression in an E. coli strain cultured in modified M9-DHA medium. The ptsA gene in the ΔptsA mutant was replaced by a kanamycin resistance cassette, leading to the overexpression of gldA and fsaB, which are part of the same operon. For all experiments, n = 2 biological replicates, and log2 values are given. Shown are dihydroxyacetone kinase genes (dhaK, dhaL, and dhaM), fructose-6-phosphate aldolase genes (fsaA and fsaB), glycerol dehydrogenase gene (gldA), glycerol facilitator (glpF), glycerol-3-phosphate dehydrogenase gene (glpD), glycerol kinase gene (glpK), fructose-bisphosphate aldolase gene (fbaA), 6-phosphofructokinase gene (pfkA), glucarate operon (garoperon and gudoperon), glycolate operon (glcoperon), acetyl-coenzyme A synthetase gene (acs), acetate kinase (ackA), formate hydrogenlyase system genes (hycoperon, hypoperon, and fhl), enzyme I gene (ptsI), dihydroxyacetone (DHA), formate (FOR), acetate (ACE), glycolate (GLO), dihydroxyacetone phosphate (DHAP), fructose-6-phosphate (F6P), fructose-1,6-bisphosphate (FBP), glyceraldehyde-3-phosphate (GAP), glycerol (GLY), and glycerol-3-phosphate (G3P).
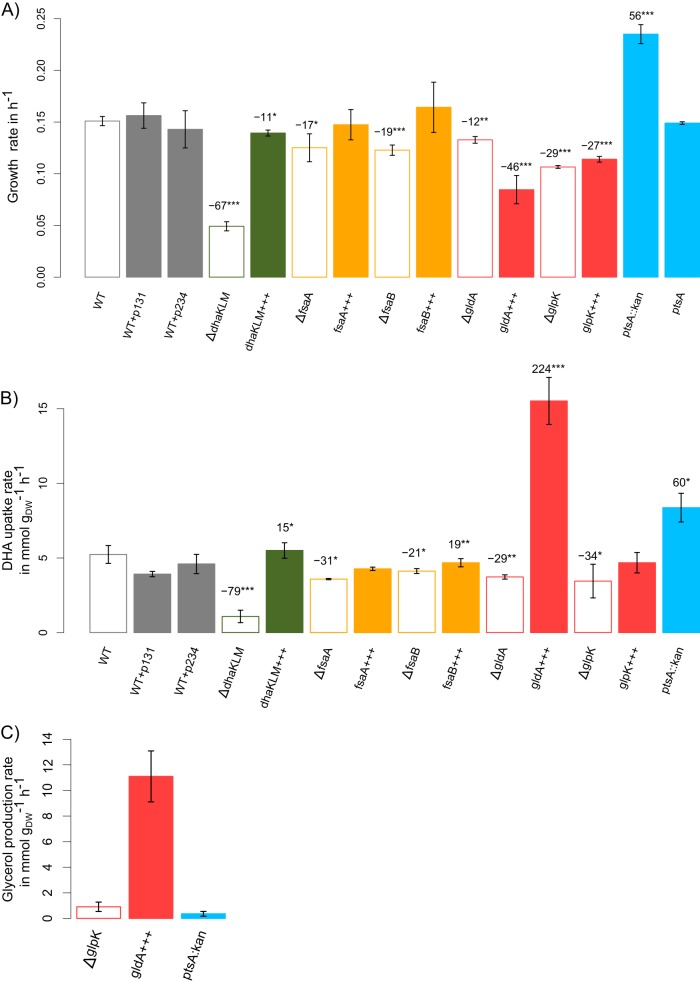
FIG 4.
Growth rate (A), specific DHA uptake rate (B), and specific glycerol production rate (C) of the strains with deletions (Δ) and overexpression (+++) of genes involved in DHA metabolism (Table 2). Cells were grown on modified M9 medium with 15 mM DHA at 37°C and shaking at 220 rpm. Growth rate is given in h−1. DHA uptake rate and glycerol production rate are given in mmol gDW−1 h−1. PhysioFit was used to estimate growth and exchange rates. Data shown represent means and standard deviations (n = 3). P values were obtained using a t test comparing mutant strains with the WT control (***, P < 0.001; **, P < 0.01; *, P < 0.05). Refer to Table 2 for details on each strain.
Overall these data show that the operon dhaKLM is predominantly expressed during growth on DHA, while the genes encoding the GLD and FSA pathways are not transcriptionally activated, even to rescue the functionality of a strain with dhaKLM deleted. Instead, this strain survives on DHA’s degradation products, leaving the alternative catabolic pathways dormant.
The GLD and FSA pathways are functionally involved in DHA metabolism.
To assess whether the GLD and FSA pathways are functionally involved in DHA metabolism even if their transcription is not activated on DHA, we performed functional analysis of the genes involved in these pathways. The results obtained for all the deletion or overexpressing strains (Table 2) grown on minimal medium containing DHA are shown in Fig. 4. Inactivation of gldA, glpK, fsaA, and fsaB resulted in significantly slower growth than that of the control strain (Fig. 4A), and complementation of the deleted genes restored the growth rate (Fig. S4). After ΔdhaKLM, the gene deletion that had the greatest impact on the growth rate and the specific DHA uptake rate was ΔglpK, reducing them by 29 and 34%, respectively (Fig. 4A and B). In H. volcanii (29), GlpK has a central role in DHA metabolism because it can directly phosphorylate DHA to DHAP using ATP. If GlpK could act as an ATP-dependent DHA kinase in E. coli, its overexpression would improve the fitness of the cells. Indeed, the conversion of DHAP to pyruvate generates two ATPs and only one PEP, and a doubling of the level of phosphate donors (ATP molecules) in the cytoplasm should support higher ATP-dependent DHA kinase activity than PEP-dependent DHA kinase and, thus, better growth. This has indeed been observed recently in an E. coli strain that overexpresses an ATP-dependent DHA kinase during anaerobic growth on DHA (47). However, this was not observed under our aerobic conditions, i.e., overexpressing glpK (i.e., glpK+++) decreased the growth rate (Fig. 4A), suggesting that GlpK does not act in vivo as an ATP-dependent DHA kinase enzyme. In addition, the production of glycerol observed in the ΔglpK strain (Fig. 4C) clearly indicates that GlpK is involved in glycerol phosphorylation rather than in DHA phosphorylation. Results obtained for the strain overexpressing gldA (i.e., gldA+++) also support this hypothesis. In this strain, glycerol was produced at almost the same rate that DHA was taken up (Fig. 4C) and started to be consumed only when DHA was exhausted (Fig. S5). This suggests that DHA has an effect in vivo on glycerol utilization. In E. coli, the catalytic activity of GlpK is inhibited allosterically by enzyme IIA from the glucose PTS system (48). It could also be inhibited allosterically by enzyme IIA from the DHA PTS system (i.e., DhaM) (22). Deletion of gldA (i.e, ΔgldA) reduced both the growth and the specific DHA uptake rate (Fig. 4A and B), most probably because the production of glycerol-3-phosphate, which is essential for biomass synthesis, is provided by just one pathway (i.e., from DHAP via glycerol-3-phosphate dehydrogenase, encoded by gpsA). Overexpressing gldA more than doubled the specific DHA uptake rate (Fig. 4B), which is consistent with its high affinity for DHA. However, the growth rate was not improved (Fig. 4A) because of a bottleneck at the GlpK level, possibly due to allosteric regulation by DHA.
TABLE 2.
Bacterial strains, plasmids, and primers used in this study
| Plasmid, bacterial strain, or primer | Gene accession no. | Description (genotype and/or relevant characteristic[s]) or sequence of oligonucleotide primer | Reference or source |
|---|---|---|---|
| E. coli bacterial strains | |||
| WT | BW25113; rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | 49 | |
| ΔfsaA | b0825 | BW25113 ΔfsaA::frt | This study |
| ΔfsaB | b3946 | BW25113 ΔfsaB::frt | This study |
| ΔgldA | b3945 | BW25113 ΔgldA::frt | This study |
| ΔglpK | b3926 | BW25113 ΔglpK::frt | This study |
| ΔdhaKLM | b1200, b1199, b1198 | BW25113 ΔdhaKLM::frt | This study |
| ΔptsA::kan | b3947 | BW25113 ΔptsA::kan | 49 |
| ΔptsA | b3947 | BW25113 ΔptsA::frt | This study |
| WT+p131 | BW25113 + pSEVA131 | This study | |
| WT+p234 | BW25113 + pSEVA234 | This study | |
| fsaA+++ | b0825 | BW25113 + pSEVA234-fsaA | This study |
| fsaB+++ | b3946 | BW25113 + pSEVA131-fsaB | This study |
| gldA+++ | b3945 | BW25113 + pSEVA131-gldA | This study |
| glpK+++ | b3926 | BW25113 + pSEVA131-glpK | This study |
| dhaKLM+++ | b1200, b1199, b1198 | BW25113 + pSEVA131-dhaKLM | This study |
| ΔfsaA+++ | b0825 | ∆fsaA + pSEVA234-fsaA | This study |
| ΔfsaB+++ | b3946 | ∆fsaB + pSEVA131-fsaB | This study |
| ΔgldA+++ | b3945 | ∆gldA + pSEVA131-gldA | This study |
| ΔglpK+++ | b3926 | ∆glpK + pSEVA131-glpK | This study |
| ΔdhaKLM+++ | b1200, b1199, b1198 | ∆dhaKLM + pSEVA131-dhaKLM | This study |
| Plasmids | |||
| pSEVA131 | Medium copy number, lacIq/Ptrc promotor, pBBR1 ori, Ampr; original pSEVA131 plasmid does not contain a promotor | 66 | |
| pSEVA234 | Medium copy number, lacIq/Ptrc promotor, pBBR1 ori, Kmr | 67 | |
| pSEVA131-dhaKLM | Derivative of pSEVA-131 containing dhaKLM operon; used to overexpress dhaKLM in BW25113 | This study | |
| pSEVA131-fsaB | Derivative of pSEVA-131 containing fsaB gene; used to overexpress fsaB in BW25113 | This study | |
| pSEVA131-gldA | Derivative of pSEVA-131 containing gldA gene; used to overexpress gldA in BW25113 | This study | |
| pSEVA131-glpK | Derivative of pSEVA-131 containing glpK gene; used to overexpress glpK in BW25113 | This study | |
| pSEVA234-fsaA | Derivative of pSEVA-234 containing fsaA gene; used to overexpress fsaA in BW25113 | This study | |
| Primers | |||
| dhaKLM_knockout_F | CGTGTCGTTGAACATCATCCATGCCCTACCGTAATTGCTGGAGCAAAATAGTGTAGGCTGGAGCTGCTTC | ||
| dhaKLM_knockout_R | CATCAGAACGATGCCATCCGAACAGTGGCTTAACCCTGACGGTTGAAACGCATATGAATATCCTCCTTAG | ||
| glpK_knockout_F | TCCTTCAGAACAAAAAGCTTCGCTGTAATATGACTACGGGACAATTAA ACGTGTAGGCTGGAGCTGCTTC |
||
| glpK_knockout_R | ACGTTTCGGGACTACCGGATGCGGCATAAACGCTTCATTCGGCATTTACACATATGAATATCCTCCTTAG | ||
| Cm_F | AATCGTCGTGGTATTCACTCC |
Finally, the deletions of fsaA and fsaB (i.e., ΔfsaA and ΔfsaB) reduced both the growth rate and the specific DHA uptake rate (Fig. 4A and B). While their overexpression (i.e., fsaA+++ and fsaB+++) did not have a significant effect on the growth rate compared to that of the wild type, fsaB overexpression was associated with a significant increase in the DHA uptake rate. This demonstrates that FsaA and FsaB are involved in DHA utilization and suggests that the concentration of FsaB is limited in the wild-type strain.
Overall, these results show that GLD and FSA are both functionally involved in DHA metabolism but that their action is restricted. A number of strands of evidence suggest that fsaB and gldA are both anaerobic genes whose transcription is activated under anaerobic conditions (27, 34, 35). This would explain why these genes were not highly expressed under our aerobic conditions and, thus, why these pathways were only weakly used for DHA assimilation.
Overexpressing the GLD and FSA pathways leads to optimal growth on DHA.
We used a ptsA mutant from the KEIO collection (49) to test whether optimal DHA growth could be restored by overexpressing the FSA- and GLD-based pathways. The ptsA gene in this mutant is replaced by a kanamycin resistance cassette, leaving gldA and fsaB expression under the control of the kanamycin promoter. As a result, gldA and fsaB were overexpressed 28- and 123-fold, respectively, compared with expression of the wild-type strain (Fig. 2 and Fig. S3 and Data File S1). Interestingly, in the ΔptsA::kan mutant, glpK was also upregulated (by a factor of 10), making the GLD pathway fully activated. Furthermore, the ΔptsA::kan strain grew twice as fast as the wild type, at close to the in silico-predicted optimal rate on DHA (Fig. 4A). The specific DHA uptake rate was likewise increased by 60% (Fig. 4B). Removing the kanamycin resistance cassette of the ΔptsA::kan strain (i.e., ΔptsA) decreased the growth rate to a value similar to the one measured for the wild-type strain on DHA (Fig. 4A). This means that PtsA is not itself involved in the overexpression of genes encoding the GLD and FSA pathways. Overall, these data are consistent with our previous results that overexpressing fsaB increases the DHA uptake rate. However, glycerol was only slightly accumulated in the ΔptsA::kan strain (Fig. 4C), in contrast to what has been observed in the strain overexpressing gldA. This is consistent with the upregulation of glpK along with genes involved in sn-glycerol 3-phosphate (G3P) catabolism (i.e., glpABC and glpQ) and transport (glpT) (Table 3 and Data File S1) and, thus, with the use of the GLD pathway. The remaining question is why glpK is upregulated in the ΔptsA::kan strain. This cannot be due only to the overexpression of gldA, since glpK was not upregulated in the wild-type strain that only overexpressed gldA.
TABLE 3.
Functional classification of genes with statistically significant decreases and increases in mRNA level in E. coli BW25113 ΔptsA and WT strainsa
| Category and expression status | GO concerned | P value | Genes involved |
|---|---|---|---|
| Overexpressed | |||
| Secondary metabolism | GO:0019563; glycerol catabolic process | 3.82E−11 | gldA, hybA, glpK, glpC, glpB, glpD, glpA |
| GO:0045333; cellular respiration | 2.90E−19 | frdB, dmsA, dmsB, dmsC, hybB, fdnG, fdnH, frdA, frdC, frdD, hyaB, hybO, napB, napC, narG, narH, nirB, nirD, napA, glpD, glpA, glpB, glpC, ndh, hybA, hybC, hyaC, yjjI, cydA, cydB | |
| GO:0071941; nitrogen cycle metabolic process | 1.75E−09 | argF, napC, glnL, glnG, nirB, narG, nirD, nirC, narJ, narK, narH, napA, nrfA | |
| GO:0019394; glucarate catabolic process | 7.97E−06 | garL, gudD, garR, garK | |
| Response to stimuli | GO:0046688; response to copper ion | 1.14E−08 | cusS, copA, cusA, cusB, cusC, cusF, cusR, yobA, cueO |
| GO:0009432; SOS response | 1.74E−07 | recA, recN, sulA, umuC, umuD, yebG, recX, dinI, lexA, dinD, dinG | |
| Transport | GO:0015886; heme transport | 2.41E−08 | ccmA, ccmD, ccmB, ccmC, ccmF, dppF, ccmE, dppC |
| GO:0015675; nickel cation transport | 2.05E−05 | nikB, nikC, nikE, nikD, nikA | |
| GO:0006857; oligopeptide transport | 8.02E−04 | dppC, dppF, oppB, oppC, oppF, oppD, oppA | |
| Underexpressed | |||
| Iron | GO:0055072; iron ion homeostasis | 7.40E−15 | fepA, efeB, cirA, yqjH, fhuF, bfr, iscU, fhuA, fhuB, fhuC, fhuD, feoA, fepB, fes, fhuE, fecI, fecR, fepD, fepG, fepC, fiu |
| GO:0016226; iron-sulfur cluster assembly | 2.05E−06 | iscS, iscU, erpA, sufB, sufD, sufA, sufC, sufE, sufS | |
| Secondary metabolism | GO:0009447; putrescine catabolic process | 3.95E−10 | patA, patD, puuE, puuA, puuD, puuP, puuR, puuC, puuB |
| GO:0009065; glutamine family amino acid catabolic process | 1.41E−05 | putA, astE, astD, astC, astB, astA, gabD, aldA | |
| GO:0006790; sulfur compound metabolic process | 7.66E−08 | prpB, prpD, prpE, prpR, fadH, fadI, fadE, fadM, fadD, fadA, fadB, yqeF | |
| Tricarboxylic acid | GO:0006099; tricarboxylic acid cycle | 4.17E−09 | sdhC, sdhB, acnA, gltA, fumA, fumC, sdhA, aceB, aceA, aceK, sucA, prpC, prpD |
| Fatty acids | GO:0009062; fatty acid catabolic process | 4.62E−09 | prpB, prpD, prpE, prpR, fadH, fadI, fadE, fadM, fadD, fadA, fadB, yqeF |
| GO:0009712; catechol-containing compound metabolic process | 8.28E−08 | entA, entC, entE, entH, entF, entB, fes, paaJ | |
| Hydrogen sulfide | GO:0070814; hydrogen sulfide biosynthetic process | 1.25E−07 | astE, astD, astC, astB, astA, gabD |
| Enterobactin | GO:0042930; enterobactin transport | 3.10E−06 | fepA, fepD, fepG, fepC, fepB, entS |
Levels in E. coli BW25113 ΔptsA strain in M9-DHA medium were compared to those for E. coli BW25113 WT strain in M9-DHA medium. The Clusters of Orthologous Groups (COG) were used for grouping.
In the ΔptsA::kan strain, genes coding for the DAK pathway were not upregulated, while genes encoding the glucarate pathway (used for glycolate assimilation) were highly expressed (7-fold on average) (Fig. 2 and Data File S1). Surprisingly, several genes associated with anaerobic or O2-limited conditions were upregulated (Table 3 and Data File S1). Several operons encoding enzymes in anaerobic respiratory chains (46, 50), i.e., (i) anaerobic dehydrogenases (GlpABC, FdnGHI, HyaABC, and HybABC), (ii) anaerobic terminal reductases (NarG, NapABCGH, NirBD, FrdABCD, and DmsABC), and (iii) microaerobic terminal oxidase (CydAB), were upregulated. All of these dehydrogenases are theoretically able to transfer electrons to each of the terminal reductases or oxidases, provided the enzymes react with the same type of quinone. This may provide the ΔptsA::kan strain with a large variety of respiratory chains (46) to fulfill its energy requirements. Second, genes encoding enzymes in the tricarboxylic acid cycle and involved in fatty acid degradation were downregulated along with genes involved in putrescine catabolism. These genes are known to be repressed under anaerobic conditions (51–53). However, no change in the expression of Fnr, the major regulator governing the physiological switch between aerobic and anaerobic growth conditions and controlling the expression of all these genes (54, 55), was observed. Altogether, these data suggest that the ΔptsA::kan strain adopts anaerobic growth behavior in a fully aerated medium, taking advantage of all available sources of carbon to grow on DHA while meeting its energetic needs.
In sum, these results demonstrate that optimal growth on DHA can be achieved by releasing hierarchical constraints on DHA metabolism, opening additional routes for its assimilation. However, the regulation mechanisms involved in the activation of anaerobic genes in the ΔptsA::kan strain remain to be elucidated.
Concluding remarks.
This study shows that aerobic DHA metabolism in E. coli is far from optimal because of chemical, hierarchical, and possibly allosteric constraints. However, our results show that removing hierarchical constraints optimizes growth on DHA. Beyond contributing to a better system-level understanding of DHA metabolism, these results are likely to accelerate the development of microbial biocatalysts. Enabling the rational design of strains that use DHA efficiently should improve biotechnological applications involving DHA as an intermediate, a well-known example being the bioconversion of glycerol into value-added chemicals (56). This should also facilitate research aiming at constructing synthetic methylotrophs (i.e., engineering of nonnative methylotrophs for methane and methanol-based production of chemicals) for which DHA can be an intermediate (47, 57).
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli K-12 BW25113 was selected as the model wild-type strain. Single-deletion mutants were taken from the Keio collection (49), and the ΔdhaKLM strain was constructed using the gene deletion method described previously (58). The dhaKLM operon was replaced by a Cmr cassette using the primers listed in Table 2. All of the antibiotic resistance cassettes were removed by FLP recombination. The plasmids listed in Table 2 were purchased from BaseClear (Leiden, Netherlands). Cells were transformed according to the rubidium chloride protocol (59). All of the strains are listed in Table 2, and genetic modifications were checked by PCR.
Growth conditions.
E. coli was cultivated overnight at 37°C with agitation at 220 rpm in LB broth (10 g tryptone, 5 g yeast extract, 10 g NaCl per liter) with appropriate antibiotics (100 μg/ml ampicillin or 50 μg/ml kanamycin) and 0.25 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) if needed. The optical density at 600 nm (OD600) was measured by spectrophotometry, and 5-ml samples of preculture were centrifuged at 8,000 × g for 5 min. The pellets were resuspended in modified M9 medium (3.48 g/liter Na2HPO4·12H2O, 0.606 g/liter KH2PO4, 0.102 g/liter NaCl, 0.408 g/liter NH4Cl, 0.49 g/liter MgSO4, 4.38 mg/liter CaCl2, 15 mg/liter Na2EDTA·2H2O, 4.5 mg/liter ZnSO4·7H2O, 0.3 mg/liter CoCl2·6H2O, 1 mg/liter MnCl2·4H2O, 1 mg/liter H3BO3, 0.4 mg/liter Na2MoO4·2H2O, 3 mg/liter FeSO4·7H2O, 0.3 mg/liter CuSO4·5H2O, 0.1 g/liter thiamine) with no carbon source.
The cultures were incubated at 37°C and 220 rpm in 250-ml baffled shake flasks containing 50 ml of M9 medium supplemented with DHA (99.9% purity; Merck) at a final concentration of 15 mM. Cells were inoculated at an initial OD600 of 0.05, and growth was analyzed every 2 h for 48 h using a Thermo Genesys6 spectrophotometer (Thermo Scientific). Seven hundred fifty microliters of medium was collected and centrifuged at 8,000 × g for 3 min. Two hundred fifty microliters of supernatant was stored at −20°C before NMR or high-performance liquid chromatography (HPLC) analysis.
Sampling, RNA extraction, and microarray procedures.
Cells were grown in duplicate under the same conditions as those described above. When an OD600 around 0.5 was reached (see Fig. S3 in the supplemental material), the cells were harvested and immediately frozen in liquid nitrogen. At such OD cells are in exponential growth phase, and DHA and formate are present in the medium for all of them (Fig. S3). For the WT strain, as the sampling time occurs during the night, the cultivation was diluted the next day in a fresh modified M9 medium at an OD of 0.2, and sampling was done at an OD600 of around 0.5. Total RNA was extracted by following the Qiagen RNeasy minikit procedure and quantified using a NanoDrop spectrophotometer. Double-stranded complementary DNA (cDNA) synthesis and array processing were performed using the Agilent Technologies one-color microarray-based gene expression analysis protocol. The images were analyzed with the software DEVA (v1.2.1). All array procedures were performed using the GeT-Biopuces platform (http://get.genotoul.fr/). For the wild-type strain, gene expression of batch cultures with DHA was expressed relative to the ratio of gene expression of chemostatic growth with glucose at low (μ = 0.1 h−1) and high (μ = 0.6 h−1) growth rates (data from reference [60]). This allowed us to focus only on genes whose level of expression changes in relation to the nature of the substrate while avoiding highlighting of genes whose expression changes because of the very different growth rate. For the ΔdhaKLM and ΔptsA::kan mutants, gene expression on DHA was compared to the wild type’s gene expression on DHA. Gene ontology analyses were performed using EcoCyc (61).
NMR and HPLC analysis.
NMR analyses were performed on an Avance III 800-MHz spectrometer (Bruker, Rheinstetten, Germany) equipped with a 5-mm QPCI cryogenic probe head at 280K. Supernatants were analyzed by quantitative 1H one-dimensional NMR at 280K using a zgpr30 sequence with water presaturation. A total of 32 scans were accumulated after 8 dummy scans. The time domain function (the FID) was converted to the frequency domain function (the spectrum) by Fourier transform. The phase of the spectra was adjusted manually, the baseline was corrected automatically, and the spectra were aligned using the signal from 3-trimethylsilylpropionic-2,2,3,3-d4 acid sodium salt (TSP-d4), an internal standard, with the Bruker software TopSpin (v3.5). Propane-1,2,2,3-tetrol was identified using 2D heteronuclear single quantum correlation spectroscopy (HSQC) and heteronuclear multiple-bond correlation spectroscopy (HMBC) spectra. The HSQC experiment was acquired with 2,000 by 1,024 points for a spectral width of 13.95 by 140 ppm in the 1H and 13C dimensions, respectively. It was processed with 4,000 by 1,024 points. The HMBC experiment was acquired with 2,000 by 1,024 points, for a spectral width of 13.95 by 220 ppm in the 1H and 13C dimensions, respectively. It was processed with 8,000 by 1,024 points. The HSQC and HMBC data were acquired with 16 scans per increment. Both 2D spectra were calibrated in the frequency domain by setting the peak from TPS-d4 to 0 ppm in both dimensions. The spectra were processed using TopSpin 3.5.
For transcriptomic analysis, HPLC analyses were performed using a column made of an H+ chromatography resin (Zorbax-C18; Agilent Technologies). A solution of sulfuric acid (5%, vol/vol) was used as the eluent at a flow rate of 0.5 ml min−1 and a volume of injection of 20 μl. The oven temperature was set to 48°C. Ranges of standards of glycerol, formate, and DHA were realized in order to quantify extracellular metabolites. Retention times in minutes for each compound were the following: glycerol, 16.6 (refractometry detection); DHA, 16.3 (UV detection); and formate, 16.9 (UV detection). Calibration curves of these three compounds were established and used to calculate their concentrations in the culture supernatants.
Calculation of growth, substrate uptake, and degradation rates.
We developed a mathematical model to infer quantitative growth and exchange flux information from the measured time-dependent concentrations of biomass and extracellular metabolites. The general model, which accounts for the nonenzymatic degradation of substrates or products, relates changes of concentrations to fluxes using the following system of ordinary differential equations:
| (1) |
| (2) |
where X is the biomass concentration (gDW liter−1), μ is the growth rate (h−1), and Mi is the concentration of exchanged metabolite i (mmol liter−1) with a degradation constant, k (h−1), and exchange flux, qMi (mmol gDW −1 h−1). Integrating equations 1 and 2 provides the following analytical functions:
| (3) |
| (4) |
This formalism has been implemented in PhysioFit, a flexible R program that allows growth rates and exchange fluxes to be quantified by fitting time variations of extracellular metabolite and biomass concentrations using the nlsic algorithm (62). PhysioFit includes options to account (or not) for the degradation of extracellular compounds and a lag phase before cells start to grow, and it implements sensitivity analyses to evaluate the precision of the estimated fluxes. PhysioFit is provided open source at https://github.com/MetaSys-LISBP/PhysioFit.
A conversion factor was used to obtain cellular dry weights (DWs) from OD600 (0.37 gDW/OD600) and calculate specific DHA uptake rates (mmol gDW−1 h−1), formate production yields (mmol gDW−1 h−1), and biomass yields (g mol−1). Experiments were performed in triplicate to calculate averages and standard deviations.
In silico analysis of DHA metabolism.
In silico analyses of DHA metabolism were carried out with the E. coli genome-scale metabolic model iJO1366 (37) (Biomodels identifier MODEL1108160000) constrained with experimental uptake fluxes of the wild-type strain (DHA = 5.2 mmol gDW−1 h−1, formate = 3.2 mmol gDW−1 h−1, glycolate = 1.0 mmol gDW−1 h−1, and acetate = 0.1 mmol gDW−1 h−1), using Sybil package (v2.1.2) (63) of the R environment (v3.2.4) (64, 65). The model and all the scripts used to run calculations are available at https://github.com/MetaSys-LISBP/Peiro_2019 under the GPLv3 open source license to ensure reproducibility and reusability.
Data availability.
Gene expression data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-7666.
Supplementary Material
ACKNOWLEDGMENTS
We thank MetaToul-MetaboHUB (Metabolomics & Fluxomics Facitilies, Toulouse, France, www.metatoul.fr, ANR-11-INBS-0010, http://www.metabohub.fr/) and its staff members for technical support and access to NMR facilities. We also thank students from INSA Toulouse, Ophélie Besse, Johan Benestad, Anaïs Bonnin, Maéva Brunet, Tanguy Soulié, Marion Urvoy, and Maxant Vivier, for their help in producing transcriptomic data and initiating the data analysis.
This work was funded by the ANR (ANR-16-CE20-0018-01).
C.P. performed the physiological analysis, P.M. performed the modeling experiments, A.D.S. built the strains, E.C. and L.P. performed the NMR analysis, B.E. performed the transcriptomic analysis, and S.H. designed the study and wrote the paper with the help of all the coauthors. We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00768-19.
REFERENCES
- 1.Ciriminna R, Fidalgo A, Ilharco LM, Pagliaro M. 2018. Dihydroxyacetone: an updated insight into an important bioproduct. ChemistryOpen 7:233–236. doi: 10.1002/open.201700201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hekmat D, Bauer R, Fricke J. 2003. Optimization of the microbial synthesis of dihydroxyacetone from glycerol with Gluconobacter oxydans. Bioprocess Biosyst Eng 26:109–116. doi: 10.1007/s00449-003-0338-9. [DOI] [PubMed] [Google Scholar]
- 3.Stanko RT, Robertson RJ, Spina RJ, Reilly JJ Jr, Greenawalt KD, Goss FL. 1990. Enhancement of arm exercise endurance capacity with dihydroxyacetone and pyruvate. J Appl Physiol 68:119–124. doi: 10.1152/jappl.1990.68.1.119. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez R, Murarka A, Dharmadi Y, Yazdani SS. 2008. A new model for the anaerobic fermentation of glycerol in enteric bacteria: Trunk and auxiliary pathways in Escherichia coli. Metab Eng 10:234–245. doi: 10.1016/j.ymben.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Molin M, Norbeck J, Blomberg A. 2003. Dihydroxyacetone kinases in Saccharomyces cerevisiae are involved in detoxification of dihydroxyacetone. J Biol Chem 278:1415–1423. doi: 10.1074/jbc.M203030200. [DOI] [PubMed] [Google Scholar]
- 6.Streekstra H, Teixeira de Mattos MJ, Neijssel OM, Tempest DW. 1987. Overflow metabolism during anaerobic growth of Klebsiella aerogenes NCTC 418 on glycerol and dihydroxyacetone in chemostat culture. Arch Microbiol 147:268–275. doi: 10.1007/BF00463487. [DOI] [Google Scholar]
- 7.Bales JR, Higham DP, Howe I, Nicholson JK, Sadler PJ. 1984. Use of high-resolution proton nuclear magnetic resonance spectroscopy for rapid multi-component analysis of urine. Clin Chem 30:426–432. [PubMed] [Google Scholar]
- 8.Akhtar N, Blomberg A, Adler L. 1997. Osmoregulation and protein expression in a pbs2Δ mutant of Saccharomyces cerevisiae during adaptation to hypersaline stress. FEBS Lett 403:173–180. doi: 10.1016/S0014-5793(97)00048-3. [DOI] [PubMed] [Google Scholar]
- 9.Ahmad M, Hirz M, Pichler H, Schwab H. 2014. Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl Microbiol Biotechnol 98:5301–5317. doi: 10.1007/s00253-014-5732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Connor MB, Quayle JR. 1980. Pentose phosphate dependent fixation of formaldehyde by methanol-grown Hansenula polymorpha and Candida boidinii. Microbiology 120:219–255. doi: 10.1099/00221287-120-1-219. [DOI] [Google Scholar]
- 11.Sánchez-Moreno I, Nauton L, Théry V, Pinet A, Petit J-L, de Berardinis V, Samland AK, Guérard-Hélaine C, Lemaire M. 2012. FSAB: a new fructose-6-phosphate aldolase from Escherichia coli. Cloning, over-expression and comparative kinetic characterization with FSAA. J Mol Catal B Enzym 84:9–14. doi: 10.1016/j.molcatb.2012.02.010. [DOI] [Google Scholar]
- 12.Deppenmeier U, Hoffmeister M, Prust C. 2002. Biochemistry and biotechnological applications of Gluconobacter strains. Appl Microbiol Biotechnol 60:233–242. doi: 10.1007/s00253-002-1114-5. [DOI] [PubMed] [Google Scholar]
- 13.Benov L, Beema AF. 2003. Superoxide-dependence of the short chain sugars-induced mutagenesis. Free Radic Biol Med 34:429–433. doi: 10.1016/S0891-5849(02)01331-X. [DOI] [PubMed] [Google Scholar]
- 14.Tessier FJ, Monnier VM, Sayre LM, Kornfield JA. 2003. Triosidines: novel Maillard reaction products and cross-links from the reaction of triose sugars with lysine and arginine residues. Biochem J 369:705–719. doi: 10.1042/BJ20020668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lip H, Yang K, MacAllister SL, O’Brien PJ. 2013. Glyoxal and methylglyoxal: autoxidation from dihydroxyacetone and polyphenol cytoprotective antioxidant mechanisms. Chem Biol Interact 202:267–274. doi: 10.1016/j.cbi.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 16.Maksimović V, Mojović M, Vučinić Ž. 2006. Monosaccharide–H2O2 reactions as a source of glycolate and their stimulation by hydroxyl radicals. Carbohydr Res 341:2360–2369. doi: 10.1016/j.carres.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 17.Subedi KP, Kim I, Kim J, Min B, Park C. 2008. Role of GldA in dihydroxyacetone and methylglyoxal metabolism of Escherichia coli K12. FEMS Microbiol Lett 279:180–187. doi: 10.1111/j.1574-6968.2007.01032.x. [DOI] [PubMed] [Google Scholar]
- 18.Yaylayan VA, Harty-Majors S, Ismail AA. 1999. Investigation of dl-glyceraldehyde–dihydroxyacetone interconversion by FTIR spectroscopy. Carbohydr Res 318:20–25. doi: 10.1016/S0008-6215(99)00077-4. [DOI] [Google Scholar]
- 19.Bächler C, Schneider P, Bähler P, Lustig A, Erni B. 2005. Escherichia coli dihydroxyacetone kinase controls gene expression by binding to transcription factor DhaR. EMBO J 24:283–293. doi: 10.1038/sj.emboj.7600517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Alles LF, Siebold C, Nyffeler TL, Flükiger-Brühwiler K, Schneider P, Bürgi H-B, Baumann U, Erni B. 2004. Phosphoenolpyruvate- and ATP-dependent dihydroxyacetone kinases: covalent substrate-binding and kinetic mechanism. Biochemistry 43:13037–13045. doi: 10.1021/bi048575m. [DOI] [PubMed] [Google Scholar]
- 21.Jin RZ, Lin E. 1984. An inducible phosphoenolpyruvate: dihydroxyacetone phosphotransferase system in Escherichia coli. Microbiology 130:83–88. doi: 10.1099/00221287-130-1-83. [DOI] [PubMed] [Google Scholar]
- 22.Gutknecht R, Beutler R, Garcia-Alles LF, Baumann U, Erni B. 2001. The dihydroxyacetone kinase of Escherichia coli utilizes a phosphoprotein instead of ATP as phosphoryl donor. EMBO J 20:2480–2486. doi: 10.1093/emboj/20.10.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulsen IT, Reizer J, Jin RZ, Lin ECC, Saier MH Jr.. 2000. Functional genomic studies of dihydroxyacetone utilization in Escherichia coli. Microbiology 146:2343–2344. doi: 10.1099/00221287-146-10-2343. [DOI] [PubMed] [Google Scholar]
- 24.Truniger V, Boos W. 1994. Mapping and cloning of gldA, the structural gene of the Escherichia coli glycerol dehydrogenase. J Bacteriol 176:1796–1800. doi: 10.1128/jb.176.6.1796-1800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freedberg WB, Lin E. 1973. Three kinds of controls affecting the expression of the glp regulon in Escherichia coli. J Bacteriol 115:816–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang CT, Ruch FE, Lin CC. 1979. Purification and properties of a nicotinamide adenine dinucleotide-linked dehydrogenase that serves an Escherichia coli mutant for glycerol catabolism. J Bacteriol 140:182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durnin G, Clomburg J, Yeates Z, Alvarez PJ, Zygourakis K, Campbell P, Gonzalez R. 2009. Understanding and harnessing the microaerobic metabolism of glycerol in Escherichia coli. Biotechnol Bioeng 103:148–161. doi: 10.1002/bit.22246. [DOI] [PubMed] [Google Scholar]
- 28.Jin RZ, Forage RG, Lin EC. 1982. Glycerol kinase as a substitute for dihydroxyacetone kinase in a mutant of Klebsiella pneumoniae. J Bacteriol 152:1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouellette M, Makkay AM, Papke RT. 2013. Dihydroxyacetone metabolism in Haloferax volcanii. Front Microbiol 4:376. doi: 10.3389/fmicb.2013.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinhouse H, Benziman M. 1976. Phosphorylation of glycerol and dihydroxyacetone in Acetobacter xylinum and its possible regulatory role. J Bacteriol 127:747–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kremer DR, Hansen TA. 1987. Glycerol and dihydroxyacetone dissimilation in Desulfovibrio strains. Arch Microbiol 147:249–256. doi: 10.1007/BF00463484. [DOI] [Google Scholar]
- 32.Hayashi S-I, Lin E. 1967. Purification and properties of glycerol kinase from Escherichia coli. J Biol Chem 242:1030–1035. [PubMed] [Google Scholar]
- 33.Schurmann M. 2001. Fructose-6-phosphate aldolase is a novel class I aldolase from Escherichia coli and is related to a novel group of bacterial transaldolases. J Biol Chem 276:11055–11061. doi: 10.1074/jbc.M008061200. [DOI] [PubMed] [Google Scholar]
- 34.Reizer J, Reizer A, Saier MH Jr.. 1995. Novel phosphotransferase system genes revealed by bacterial genome analysis—a gene cluster encoding a unique enzyme I and the proteins of a fructose-like permease system. Microbiology 141:961–971. doi: 10.1099/13500872-141-4-961. [DOI] [PubMed] [Google Scholar]
- 35.Rolfe MD, Ocone A, Stapleton MR, Hall S, Trotter EW, Poole RK, Sanguinetti G, Green J, SysMO-SUMO Consortium . 2012. Systems analysis of transcription factor activities in environments with stable and dynamic oxygen concentrations. Open Biol 2:120091. doi: 10.1098/rsob.120091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heux S, Philippe B, Portais JC. 2011. High-throughput workflow for monitoring and mining bioprocess data and its application to inferring the physiological response of Escherichia coli to perturbations. Appl Environ Microbiol 77:7040–7049. doi: 10.1128/AEM.05838-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orth JD, Conrad TM, Na J, Lerman JA, Nam H, Feist AM, Palsson BØ. 2011. A comprehensive genome-scale reconstruction of Escherichia coli metabolism—2011. Mol Syst Biol 7:535. doi: 10.1038/msb.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahadevan R, Schilling CH. 2003. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng 5:264–276. doi: 10.1016/j.ymben.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 39.Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu Rev Microbiol 60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang G, Meredith TC, Kahne D. 2013. On the essentiality of lipopolysaccharide to Gram-negative bacteria. Curr Opin Microbiol 16:779–785. doi: 10.1016/j.mib.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacques M. 1996. Role of lipo-oligosaccharides and lipopolysaccharides in bacterial adherence. Trends Microbiol 4:408–409. doi: 10.1016/0966-842X(96)10054-8. [DOI] [PubMed] [Google Scholar]
- 42.Jefferson KK. 2004. What drives bacteria to produce a biofilm? FEMS Microbiol Lett 236:163–173. doi: 10.1016/j.femsle.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Pellicer MT, Fernandez C, Badía J, Aguilar J, Lin ECC, Baldomà L. 1999. Cross-induction of glc and ace operons of Escherichia coli attributable to pathway intersection—characterization of the glc promoter. J Biol Chem 274:1745–1752. doi: 10.1074/jbc.274.3.1745. [DOI] [PubMed] [Google Scholar]
- 44.Roberton AM, Sullivan PA, Jones-Mortimer MC, Kornberg HL. 1980. Two genes affecting glucarate utilization in Escherichia coli K12. J Gen Microbiol 117:377–382. doi: 10.1099/00221287-117-2-377. [DOI] [PubMed] [Google Scholar]
- 45.McDowall JS, Murphy BJ, Haumann M, Palmer T, Armstrong FA, Sargent F. 2014. Bacterial formate hydrogenlyase complex. Proc Natl Acad Sci U S A 111:E3948–E3956. doi: 10.1073/pnas.1407927111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unden G, Steinmetz PA, Degreif-Dunnwald P. 14 May 2014. The aerobic and anaerobic respiratory chain of Escherichia coli and Salmonella enterica: enzymes and energetics. EcoSal Plus 2014. doi: 10.1128/ecosalplus.ESP-0005-2013. [DOI] [PubMed] [Google Scholar]
- 47.Wang L, Chauliac D, Rhee MS, Panneerselvam A, Ingram LO, Shanmugam KT. 2018. Fermentation of dihydroxyacetone by engineered Escherichia coli and Klebsiella variicola to products. Proc Natl Acad Sci U S A 115:4381–4386. doi: 10.1073/pnas.1801002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Boer M, Broekhuizen CP, Postma PW. 1986. Regulation of glycerol kinase by enzyme IIIGlc of the phosphoenolpyruvate:carbohydrate phosphotransferase system. J Bacteriol 167:393–395. doi: 10.1128/jb.167.1.393-395.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tseng CP, Albrecht J, Gunsalus RP. 1996. Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J Bacteriol 178:1094–1098. doi: 10.1128/jb.178.4.1094-1098.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iuchi S, Lin EC. 1991. Adaptation of Escherichia coli to respiratory conditions: regulation of gene expression. Cell 66:5–7. doi: 10.1016/0092-8674(91)90130-Q. [DOI] [PubMed] [Google Scholar]
- 52.Gunsalus RP, Park SJ. 1994. Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res Microbiol 145:437–450. doi: 10.1016/0923-2508(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 53.Partridge JD, Scott C, Tang Y, Poole RK, Green J. 2006. Escherichia coli transcriptome dynamics during the transition from anaerobic to aerobic conditions. J Biol Chem 281:27806–27815. doi: 10.1074/jbc.M603450200. [DOI] [PubMed] [Google Scholar]
- 54.Constantinidou C, Hobman JL, Griffiths L, Patel MD, Penn CW, Cole JA, Overton TW. 2006. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J Biol Chem 281:4802–4815. doi: 10.1074/jbc.M512312200. [DOI] [PubMed] [Google Scholar]
- 55.Kang Y, Weber KD, Qiu Y, Kiley PJ, Blattner FR. 2005. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J Bacteriol 187:1135–1160. doi: 10.1128/JB.187.3.1135-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wendisch VF, Lindner SN, Meiswinkel TM. 2011. Use of glycerol in biotechnological applications, biodiesel—quality, emissions, and by-products. IntechOpen doi: 10.5772/25338. [DOI] [Google Scholar]
- 57.Heux S, Brautaset T, Vorholt JA, Wendisch VF, Portais JC. 2018. Synthetic methylotrophy: past, present, and future, p 133–151. In Kalyuzhnaya MG, Xing X-H (ed), Methane biocatalysis: paving the way to sustainability. Springer International Publishing, Cham, Switzerland. doi: 10.1007/978-3-319-74866-5_9. [DOI] [Google Scholar]
- 58.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Green R, Rogers EJ. 2013. Transformation of chemically competent E. coli. Methods Enzymol 529:329–336. doi: 10.1016/B978-0-12-418687-3.00028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Esquerre T, Moisan A, Chiapello H, Arike L, Vilu R, Gaspin C, Cocaign-Bousquet M, Girbal L. 2015. Genome-wide investigation of mRNA lifetime determinants in Escherichia coli cells cultured at different growth rates. BMC Genomics 16:275. doi: 10.1186/s12864-015-1482-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keseler IM, Mackie A, Santos-Zavaleta A, Billington R, Bonavides-Martínez C, Caspi R, Fulcher C, Gama-Castro S, Kothari A, Krummenacker M, Latendresse M, Muñiz-Rascado L, Ong Q, Paley S, Peralta-Gil M, Subhraveti P, Velázquez-Ramírez DA, Weaver D, Collado-Vides J, Paulsen I, Karp PD. 2017. The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res 45:D543–D550. doi: 10.1093/nar/gkw1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sokol S, Millard P, Portais JC. 2012. influx_s: increasing numerical stability and precision for metabolic flux analysis in isotope labeling experiments. Bioinformatics 28:687–693. doi: 10.1093/bioinformatics/btr716. [DOI] [PubMed] [Google Scholar]
- 63.Gelius-Dietrich G, Desouki AA, Fritzemeier CJ, Lercher MJ. 2013. Sybil—efficient constraint-based modeling in R. BMC Syst Biol 7:125. doi: 10.1186/1752-0509-7-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.R Core Development Team. 2009. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 65.R Team. 2015. RStudio: integrated development for R. RStudio, Inc, Boston, MA. [Google Scholar]
- 66.Muller JEN, Meyer F, Litsanov B, Kiefer P, Potthoff E, Heux S, Quax WJ, Wendisch VF, Brautaset T, Portais JC, Vorholt JA. 2015. Engineering Escherichia coli for methanol conversion. Metab Eng 28:190–201. doi: 10.1016/j.ymben.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 67.Silva-Rocha R, Martínez-García E, Calles B, Chavarría M, Arce-Rodríguez A, de Las Heras A, Páez-Espino AD, Durante-Rodríguez G, Kim J, Nikel PI, Platero R, de Lorenzo V. 2013. The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res 41:D666–D675. doi: 10.1093/nar/gks1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-7666.