Small secreted proteins (SSPs) are common in fungal saprotrophs, but their roles remain elusive. As such, they comprise part of a gene pool which may be involved in governing fungal lifestyles not limited to symbiosis and pathogenicity, in which they are commonly referred to as “effectors.” We propose that Ssp1 in the white-rot fungus Pleurotus ostreatus regulates the transition from primary to secondary metabolism, development, aging, and fruiting body initiation. Our observations uncover a novel regulatory role of effector-like SSPs in a saprotroph, suggesting that they may act in fungal communication as well as in response to environmental cues. The presence of Ssp1 homologues in other fungal species supports a common potential role in environmental sensing and fungal development.
KEYWORDS: Pleurotus, idiophase, small secreted proteins, white-rot fungus
ABSTRACT
The function of small secreted proteins (SSPs) in saprotrophic fungi is, for the most part, unknown. The white-rot mushroom Pleurotus ostreatus produces considerable amounts of SSPs at the onset of secondary metabolism, during colony development, and in response to chemical compounds such as 5-hydroxymethylfurfural and aryl alcohols. Genetic manipulation of Ssp1, by knockdown (KDssp1) or overexpression (OEssp1), indicated that they are, in fact, involved in the regulation of the ligninolytic system. To elucidate their potential involvement in fungal development, quantitative secretome analysis was performed during the trophophase and the idiophase and at a transition point between the two growth phases. The mutations conferred a time shift in the secretion and expression patterns: OEssp1 preceded the entrance to idiophase and secondary metabolism, while KDssp1 was delayed. This was also correlated with expression patterns of selected genes. The KDssp1 colony aged at a slower pace, accompanied by a slower decline in biomass over time. In contrast, the OEssp1 strain exhibited severe lysis and aging of the colony at the same time point. These phenomena were accompanied by variations in yellow pigment production, characteristic of entrance of the wild type into idiophase. The pigment was produced earlier and in a larger amount in the OEssp1 strain and was absent from the KDssp1 strain. Furthermore, the dikaryon harboring OEssp1 exhibited a delay in the initiation of fruiting body formation as well as earlier aging. We propose that Ssp1 might function as a part of the fungal communication network and regulate the pattern of fungal development and metabolism in P. ostreatus.
IMPORTANCE Small secreted proteins (SSPs) are common in fungal saprotrophs, but their roles remain elusive. As such, they comprise part of a gene pool which may be involved in governing fungal lifestyles not limited to symbiosis and pathogenicity, in which they are commonly referred to as “effectors.” We propose that Ssp1 in the white-rot fungus Pleurotus ostreatus regulates the transition from primary to secondary metabolism, development, aging, and fruiting body initiation. Our observations uncover a novel regulatory role of effector-like SSPs in a saprotroph, suggesting that they may act in fungal communication as well as in response to environmental cues. The presence of Ssp1 homologues in other fungal species supports a common potential role in environmental sensing and fungal development.
INTRODUCTION
Regardless of their lifestyle, many members of the fungal kingdom harbor genes coding for small secreted proteins (SSPs). These proteins are typically less than 300 amino acids long and contain a signal peptide (1, 2). Many of them are encoded by orphan genes without known PFAM domains or similarities to known sequences in databases. The proportion of SSPs is 25% to 60% of the secretome across all lifestyles (2, 3). When present in pathogenic fungi, most of them have been designated “effectors,” namely, proteins that facilitate infection (4). Recent studies also highlight SSPs as molecular keys to promote symbiosis in mycorrhizal fungi. In Cenococcum geophilum, analysis of gene expression with interacting hosts was used to identify mycorrhiza-induced SSPs (MiSSPs), which can target the host plant (5). The MiSSP7 protein in Laccaria bicolor (6, 7) and the SP7 protein in Rhizophagus irregularis (8) have been shown to be targeted to the host nucleus and reshuffle plant defense pathways (2, 6). SSPs are also present in the genomes of saprotrophic fungi, where, for the most part, their role remains unclear (2, 3).
SSPs are part of the fungal secretome, which adapts to changes in the environment and allows fungi to proliferate under diverse conditions (9). Fungal secretomes are mostly comprised of proteins that participate in organic matter degradation, such as proteases, lipases, and carbohydrate-active enzymes (CAZymes) (1). The regulation of the fungal secretome involves sensing mechanisms and signaling to respond to external conditions and changes between different phases of the fungal life cycle (9).
The growth of fungi in batch culture is considered to be biphasic. The growing phase, or “trophophase,” is a period of active growth accompanied by the consumption of nutrients. Subsequently, the cultures undergo restricted growth, which usually controls the onset of mycelial differentiation and the biosynthesis of secondary metabolites, also known as the production phase or “idiophase” (10, 11). The shift from trophophase to idiophase is considered to be regulated by growth conditions and nutritional limitations (10–14). In the white-rot fungi (WRF), lignin-modifying enzymes are mainly expressed during idiophase under N or C starvation (12–15). In a previous study, we identified Pleurotus ostreatus SSPs (poSSPs), which are secreted during the idiophase and after exposure to aryl alcohol and the toxic compound 5-hydroxymethylfurfural. Genetic manipulations conferring changes in ssp1 expression levels resulted in an alteration of the fungal secretome and ligninolytic enzyme production, suggesting its role as a regulator of the ligninolytic system (16).
In the present study, the role of Ssp1 in P. ostreatus was further investigated, by identifying additional targets in the secretome that are affected by its genetic manipulation by either knockdown (KDssp1) or overexpression (OEssp1). A quantitative secretome analysis was performed, comparing the effects of Ssp1, during biphasic growth. This showed that Ssp1 influences fungal metabolism and development during idiophase.
RESULTS
Changes in Ssp1 affect the secretome of P. ostreatus.
Previously, it was reported that genetic manipulation of Ssp1, by either knockdown (KDssp1) or overexpression (OEssp1), resulted in a significant alteration of profiles of secreted proteins of P. ostreatus, mainly, but not limited to, lignin-modifying enzymes (16). To better elucidate the global effect and additional targets of Ssp1, quantitative secretome analysis was performed to compare the KDssp1 and OEssp1 mutants with the parental strain (PC9). The three strains were sampled at three different time points representing 8-, 10-, and 13-day-old cultures. The three sampling times were linked to distinct stages of fungal developmental (16, 17): trophophase (8 days), the transition from trophophase to idiophase (10 days), and idiophase (13 days). A total of 570 proteins were identified for the three different strains and sampling days (listed in Table S1 in the supplemental material).
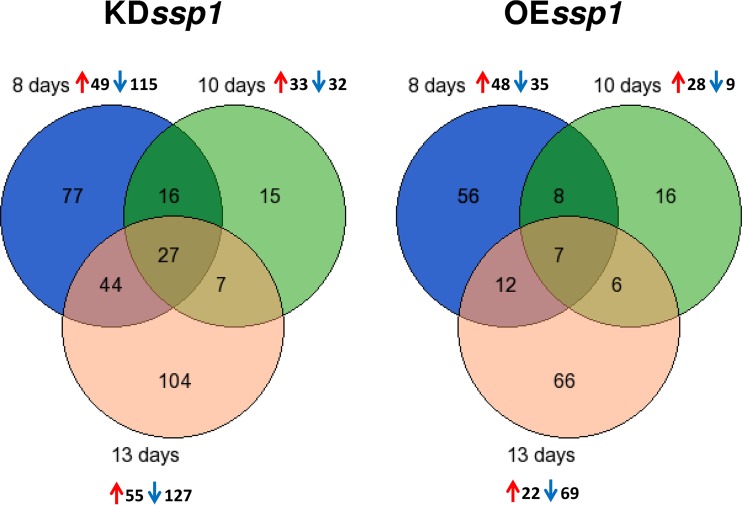
We focused on the proteins whose abundances were significantly changed (P value of <0.02) in the two mutant strains versus PC9 (Fig. 1). The combined change in the three tested time points showed that the KDssp1 and OEssp1 mutants altered a large portion of their secretome: 51% (290 proteins) and 30% (171 proteins), respectively. In both strains, the proteins whose abundances were most significantly changed for at least two different sampling days were comprised mainly of CAZymes and peptidases.
FIG 1.
Venn diagram of significantly changed proteins (P value of <0.02) in the secretomes of the KDssp1 and OEssp1 strains versus the PC9 strain, as determined with samples from three different time points representing 8-, 10-, and 13-day-old cultures. The significantly over- and underrepresented proteins at each time point are shown with red and blue arrows, respectively. See Table S1 in the supplemental material for the complete protein lists for each of the secretomes.
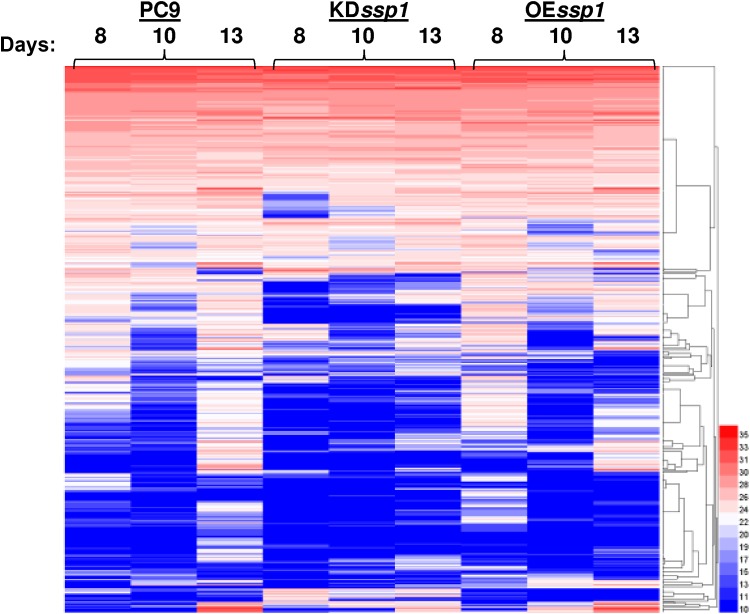
The most considerable changes in both strains were observed in the 8- and 13-day-old cultures, with minor differences in the 10-day-old cultures (Fig. 1 and 2). Furthermore, when focusing on the secretome of PC9, an interesting pattern was revealed: some of the proteins present in the trophophase (8-day-old cultures) were downregulated during the trophophase-idiophase transition (10-day-old cultures) and upregulated again in the idiophase (13-day-old cultures). Monitoring the expression levels of the genes encoding some of these proteins also revealed a two-peak pattern: a smaller peak in the 7- to 8-day-old cultures and a significantly higher peak in the 13-day-old cultures (Fig. S1). These results may explain the observed downregulation of proteins in the secretome of 10-day-old cultures, suggesting that this was a result of a transcriptionally regulated process.
FIG 2.
Heat maps of protein abundance in the secretomes of KDssp1, OEssp1, and PC9 strains, as determined in samples from three different time points representing 8-, 10-, and 13-day-old cultures. The values represent the averages of data from three biological replicates.
Overall, the manipulation of Ssp1 expression levels had major, age-dependent effects on the secretome composition of P. ostreatus, which was not limited to lignin-modifying enzyme production.
Manipulation of Ssp1 production affects the timing of the transition from trophophase to idiophase.
Based on monitoring the secretome composition of P. ostreatus during development, different patterns emerged, dividing it into 3 distinct groups: proteins whose abundance was increased at the onset of idiophase, proteins whose abundance was reduced at the onset of idiophase, and proteins whose abundance was not affected during fungal growth (Fig. 2).
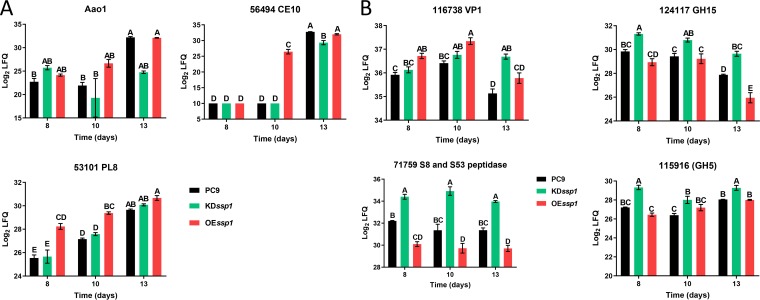
In order to determine whether Ssp1 might be involved in affecting these protein expression patterns, the effect of altering ssp1 expression on these patterns by genetic manipulation of the gene was characterized. The OEssp1 strain exhibited the same expression pattern as PC9, yet at earlier time points. Proteins whose levels were elevated at the transition from trophophase to idiophase were upregulated earlier in the OEssp1 strain, for example, proteins belonging to (i) the ligninolytic system (protein 69649, annotated as aryl alcohol oxidase 1 [Aao1]) and (ii) CAZymes (proteins 56494, annotated as carboxylesterase 10 [CE10], and 53101, annotated as polysaccharide lyase family 8 [PL8]) (Fig. 3A). On the other hand, proteins whose expression levels were reduced at the beginning of idiophase were downregulated earlier in the OEssp1 strain, for example, proteins belonging to (i) CAZymes (proteins 124117, annotated as a glycoside hydrolase family 15 protein [GH15], and 115916, annotated as a glycoside hydrolase family 5 protein [GH5]) and (ii) peptidases (protein 71759, annotated as a serine protease) (Fig. 3B).
FIG 3.
Individual proteins whose abundance was elevated (A) or reduced (B) at the transition from trophophase to idiophase, as measured by label-free quantification (LFQ), in PC9, KDssp1, and OEssp1 culture media. The specific proteins in panel A are 69649, annotated as aryl alcohol oxidase 1 (Aao1); 56494, annotated as carboxylesterase 10 (CE10); and 53101, annotated as polysaccharide lyase family 8 (PL8). The specific proteins in panel B are 116738, annotated as versatile peroxidase 1 (Vp1); 124117, annotated as a glycoside hydrolase family 15 protein (GH15); 115916, annotated as a glycoside hydrolase family 5 protein (GH5); and 71759, annotated as a serine protease. The values shown are the means of data from three replicates, and the error bars indicate standard errors. All means were compared with a Tukey-Kramer honestly significant difference (HSD) test (P < 0.05).
The KDssp1 strain also exhibited a time shift in the expression pattern, but in contrast to that observed in the secretome of the OEssp1 strain, it was delayed relative to PC9. For example, Aao1 was upregulated in a delayed manner. The proteins that had a delayed downregulation were 124117 (GH15), 115916 (GH5), Vp1, and 71759 (serine peptidase) (Fig. 3).
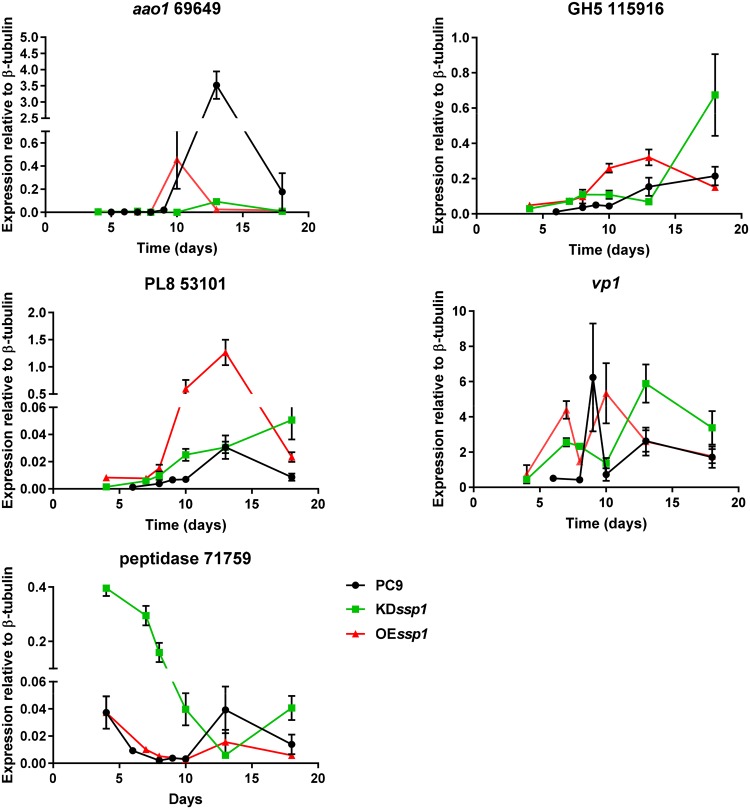
The effect of ssp1 manipulation was further examined, focusing on the expression pattern of selected genes over time. The expression of 53101 (PL8) and aao1 in PC9 peaked during the idiophase (13-day-old culture), while the expression of 115916 did not reach such a peak in PC9. In an OEssp1 background, the upregulation of these genes started at least 3 days earlier than in PC9, with an expression peak of 53101 at this time point (Fig. 4). In KDssp1, the expression of aao1 was almost abolished, and only a small peak was observed. In the case of proteins 53101 and 115916, the knockdown mutant did not reach a peak within the tested time frame. The expression pattern of vp1 in PC9 was composed of two peaks similar to those observed in the case of genes coding for Aao proteins (Fig. S1). The second peak, which was present in the idiophase, appeared earlier in the OEssp1 strain (Fig. 4). Peptidase 71759 had a significantly higher abundance in the secretome of the KDssp1 strain, and indeed, in PC9, the expression of the gene was downregulated in the trophophase and upregulated in the idiophase, peaking in the 13-day-old culture. In the KDssp1 strain, the downregulation slope was longer, and no peak was observed within the tested time frame (Fig. 4).
FIG 4.
Gene expression levels of individual genes (aao1, 115916, 53101, vp1, and 71759) at different developmental stages in the genetic background of the PC9, KDssp1, and OEssp1 strains. Expression levels shown are relative to those of β-tubulin.
To mimic the effect of OEssp1 on the expression of Ssp1-dependent genes, spent medium and the concentrated fraction of poSSPs from the 14-day-old OEssp1 culture were added to either the PC9 or KDssp1 strain. However, this did not alter the expression of either ssp genes, aao1, vp1, 53101, 115916, or 71759. In summary, the genetic manipulations of ssp1 conferred a time shift in the secretion pattern: overexpression of ssp1 resulted in expression and protein accumulation patterns similar to those of PC9, but the mutant entered the idiophase earlier, while the converse was observed when knockdown of ssp1 was imposed (i.e., the expression and patterns were similar but the mutant entered idiophase later).
Manipulation of Ssp1 production affects fungal growth patterns and physiology.
To further investigate the effect of Ssp1 manipulation on the transition from trophophase to idiophase, different aspects of fungal growth and physiology were monitored in the relevant strains.
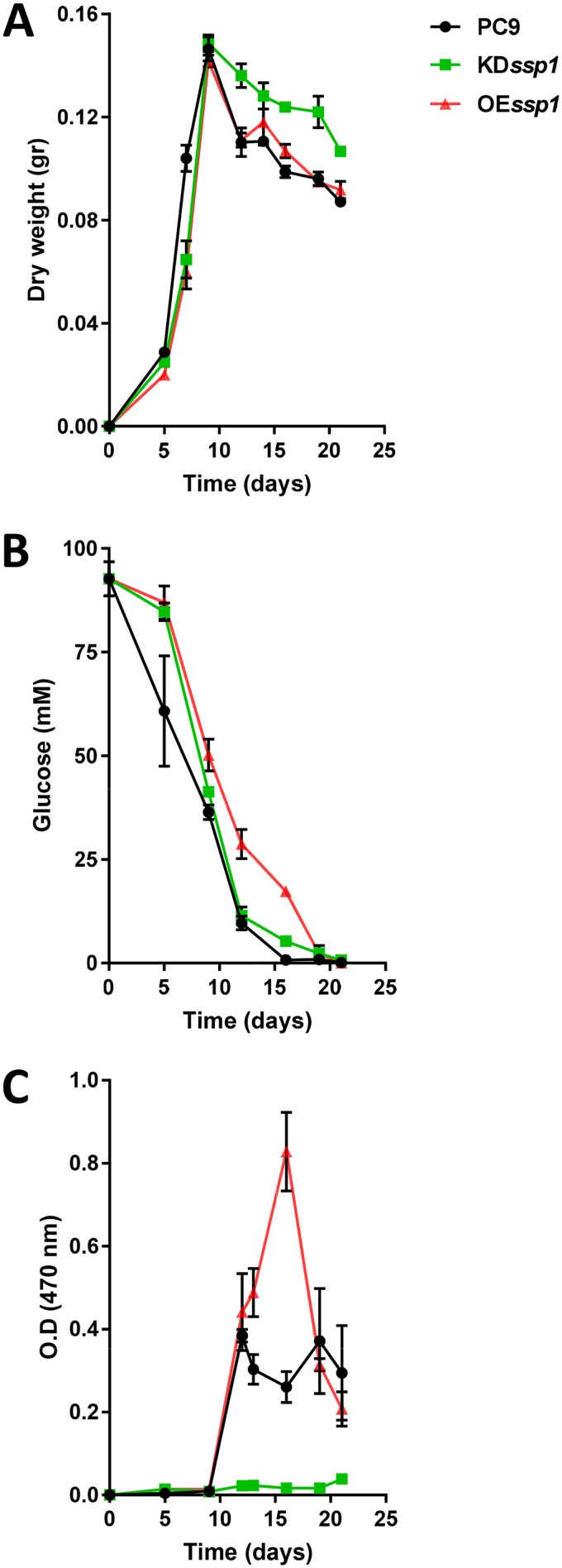
(i) Fungal growth pattern. Fungal growth was determined on the basis of biomass (dry weight) accumulation during a period of over 21 days. The two transformants OEssp1 and KDssp1 accumulated biomass more slowly than wild-type strain PC9 during the first 9 days of growth, which was explained by reduced growth, as previously reported (16). But all three strains reached a similar peak of ∼0.145 g/flask (Fig. 5). As expected, with the transition to idiophase, the biomass started to decline (11, 14). Unexpectedly, even though biomass reductions were similar in the case of the OEssp1 and PC9 strains, it was significantly slower (by 20 to 27%) in the KDssp1 strain. An example of the outcome of this slower biomass reduction can be seen by the fact that the biomass of the KDssp1 strain in the 21-day-old culture (0.107 g/flask) was similar to that of PC9 in a 16-day-old culture (0.099 g/flask) (Fig. 5A).
FIG 5.
Effect of Ssp1 manipulation on fungal growth pattern and physiology. (A) Fungal growth as measured by biomass accumulation (dry weight); (B) glucose utilization; (C) yellow pigment generation (absorbance at 470 nm). O.D., optical density.
(ii) Glucose utilization. To find out whether the slower biomass decline of the KDssp1 strain during the idiophase was a result of a difference in carbon utilization, the consumption of the main carbon source in the medium, glucose, was monitored. The residual glucose levels in the spent media of the KDssp1 and PC9 strains were similar during the idiophase (starting from 9-day-old cultures, as assumed from the growth curves of the fungi) (Fig. 5A). The OEssp1 strain utilized glucose at a significantly lower rate during the idiophase (9- to 19-old-day culture). At the measured time points, the level of glucose was 2.5 to 23 times higher in the OEssp1 culture medium than in the PC9 culture medium (Fig. 5B). In conclusion, the slower biomass decline of KDssp1 was not dependent on glucose utilization.
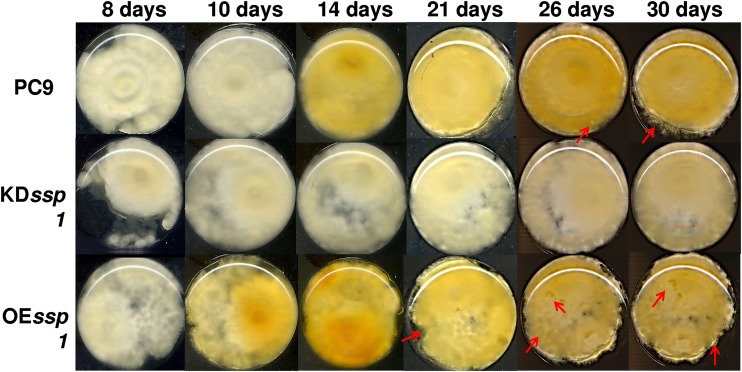
(iii) Colony morphology. In parallel to biomass accumulation and glucose consumption, the morphology of the fungal mat produced by the three strains was visually examined. Throughout all the experiments, the fungi were grown in stationary liquid medium, allowing the formation of a fungal mat. The morphology of the colony of PC9 changed during the idiophase. This was evident by the appearance of lysis in several areas of the mats in 21-day-old cultures, mainly at the periphery of the colony, suggesting that an aging process was occurring (Fig. 6). Furthermore, at an earlier stage, starting from a 14-day-old culture, a yellow pigment accumulated in the medium (Fig. 6). The morphology of the KDssp1 strain during idiophase was similar to that of a younger culture, lacking any signs of lysis, aging, or the generation of a yellow pigment. On the other hand, the morphology of the OEssp1 strain resembled an older and aged culture, as observed by a more severe phenotype of lysis that was not constrained to the periphery of the colony. The yellow pigmentation was more pronounced and appeared as early as after 10 days (Fig. 6).
FIG 6.
PC9, KDssp1, and OEssp1 strain colony morphologies at different developmental stages. The fungi were grown in stationary liquid medium, allowing the formation of a fungal mat. The bottoms of the Erlenmeyer flasks were scanned at different time points. Red arrows represent lysis areas formed with aging.
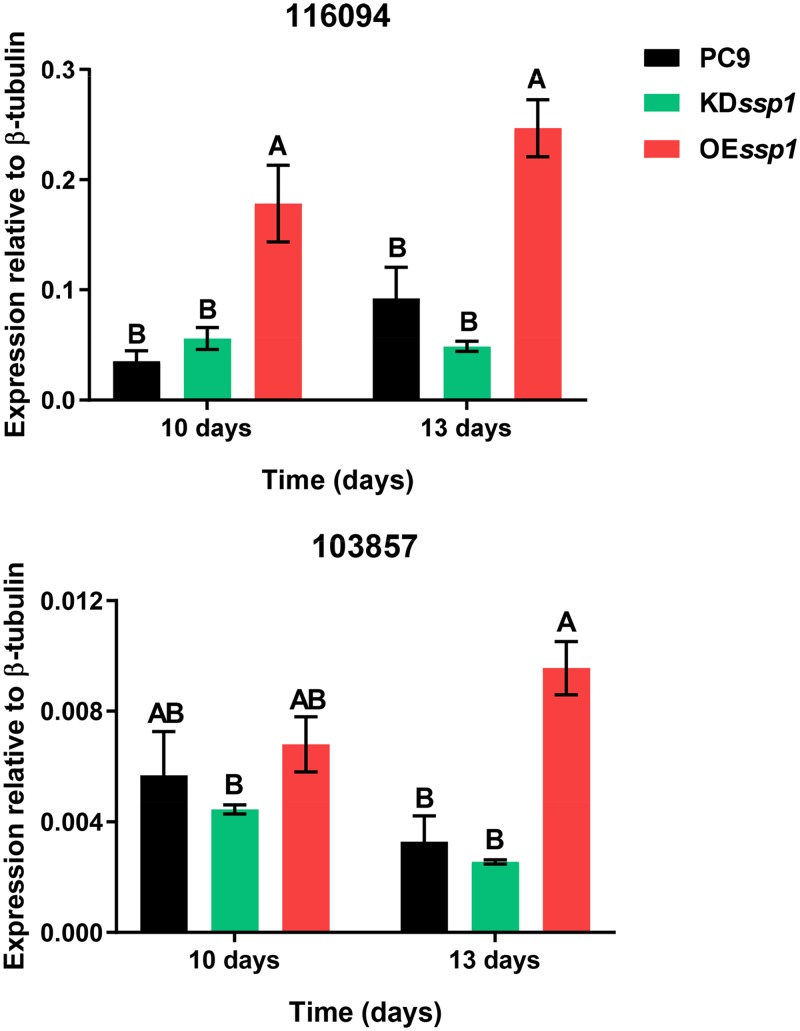
It was assumed that the colony lysis observed in PC9, and more profoundly in the OEssp1 strain, was a result of an aging process and a part of fungally regulated cell death (18, 19). To further test this possibility, the involvement of apoptosis-like pathways in this process was examined by monitoring the expression of metacaspases. There are seven genes harboring a KOG1546 domain, a hallmark of metacaspases known to be involved in the regulation of apoptosis, found in the P. ostreatus PC9 genome (20). The expression of two of these metacaspases was upregulated in 13-day-old-cultures of the OEssp1 strain. The metacaspases 103857 and 116094 were upregulated in the OEssp1 strain, relative to PC9, by 2.9- and 2.67-fold, respectively. The expression of 116094 was also upregulated in a 10-day-old culture, by 5-fold, relative to PC9. There was no significant difference in the expression of these metacaspases in the KDssp1 strain (Fig. 7).
FIG 7.
Gene expression of metacaspases in 10- and 13-day-old cultures of the PC9, KDssp1, and OEssp1 strains. Expression levels of genes encoding proteins 116094 and 103857, which have been annotated as putative metacaspases, were monitored, relative to the β-tubulin gene. All means were compared with a Tukey-Kramer HSD test (P < 0.05).
(iv) Generation of a yellow pigment. As the appearance of the yellow pigment paralleled the changes in colony morphology, its level was further characterized over time. The pigment absorbance spectrum (Fig. S2) was determined, and the absorbance at 470 nm was used to monitor its presence in the culture medium. In PC9, the pigment was produced in the 13-day-old culture, and the concentration remained the same until the end of the experiment (Fig. 5C). However, in the OEssp1 transformant, the absorbance peaked at 16 days, after which a decline occurred. In addition, the OEssp1 strain produced significantly more of the pigment than PC9 in the 16-day-old culture (Fig. 5C and Fig. 6). In contrast, the KDssp1 strain did not produce a 470-nm absorbing pigment at any developmental stage (Fig. 5C and Fig. 6).
Chemical analysis was performed in an attempt to reveal the nature of the yellow pigment. To do so, the media of the OEssp1 and KDssp1 strains were used, the latter in order to subtract the background of other chemicals. Both strains were grown for 16 days until production of a yellow pigment in the OEssp1 strain was evident. The separation performed on Sephadex LH-20 columns with a 1:1 mixture of methanol (MeOH)-water yielded seven fractions. The yellow substance was eluted from the column in the third fraction. The yellow fraction was loaded onto a medium-pressure liquid chromatography (MPLC) column and eluted with a 9:1 mixture of water-acetonitrile, and peaks were collected. Results of this analysis suggested that the pigment was associated with proteins. The yellow peaks were separated by SDS-PAGE, revealing ∼10-kDa bands (Fig. S2), and then identified by mass spectrometry (MS) as the proteins 49304 and 94118, which contain a cyanovirin-N homology (CNVH) domain; 114198, annotated as actin; and 89300, annotated as a hypothetical protein, with a lower score for Ssp1.
Hence, the main candidates to be associated with the pigment are the CNVH proteins, which have sugar-binding antiviral properties and can be found in filamentous fungi (21, 22). Both proteins 49304 and 94118 were produced during the idiophase in the PC9 and OEssp1 strains and in a reduced amount in the KDssp1 strain. 49304 also exhibited a pattern of Ssp1-regulated proteins and was upregulated in a 10-day-old culture of the OEssp1 strain (Table S1). Since CNVH proteins are not known to have an absorbance with yellow pigmentation, it is assumed that the association of the pigment may be dependent on an additional, as-yet-unknown interacting compound.
Overall, it seems that the manipulation of ssp1 by either knockdown or overexpression had a significant effect on fungal development during idiophase. The KDssp1 strain exhibited a slower reduction in biomass (Fig. 5A), which was accompanied by slower aging of the colony (Fig. 6), even though glucose utilization was similar to that of PC9 (Fig. 5B). Furthermore, the KDssp1 strain also did not generate the yellow pigment, which appeared to accompany the progression of fungal development (Fig. 5C). On the other hand, while the OEssp1 strain accumulated biomass similar to that of PC9 (Fig. 5A), the colony morphology exhibited more lysis and aging (Fig. 6), along with increased expression of metacaspases (Fig. 7). The OEssp1 strain also showed a reduction in glucose utilization (Fig. 5B) and a higher level of production of the yellow pigment (Fig. 5C).
OEssp1 delays fruiting body formation.
The two monokaryotic strains PC9 and PC15 were separated from the dikaryotic strain N001, a hybrid used for commercial cultivation (20). To determine the effect of ssp1 manipulation on the formation of fruiting bodies, redikaryotization of the PC9, KDssp1, or OEssp1 strain with the PC15 strain was induced. The transcription of ssp genes in dikaryotized 7-day-old cultures was examined using real-time PCR, and the OEssp1 strain with the PC15 strain (OEssp1+PC15) maintained elevated expression levels, ∼11-fold relative to PC9+PC15, but the ssp transcript abundance was not altered in KDssp1+PC15.
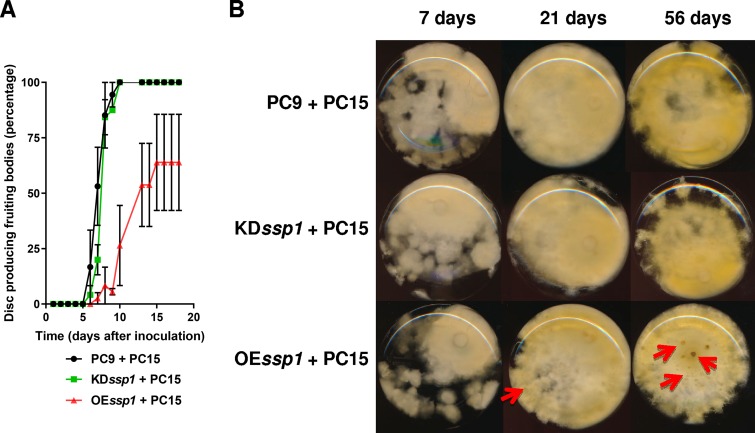
The growth and physiology of the hyphae formed by the dikaryons were further characterized. After the interaction between hyphae of PC9 or OEssp1 and those of PC15 occurred, pigmentation formed underneath the PC15 strain but was lacking in a KDssp1 background (Fig. S3). On solid medium, the hyphae of OEssp1+PC15 formed droplets that contained yellow liquid, unlike the other mono- or dikaryons studied (Fig. S3). Compared to the monokaryons, in the dikaryon KDssp1+PC15 and OEssp1+PC15 strains, there was no difference in biomass accumulation except for slower growth during the trophophase in OEssp1+PC15, and only a small delay in glucose utilization by OEssp1+PC15 was observed in the 12-day-old culture (Fig. S4). Similar to the OEssp1 monokaryon, OEssp1+PC15 aged faster, with appearance of extensive lysis in a 56-day-old culture (Fig. 8). The elusive 470-nm yellow pigment that was generated by the PC9 and OEssp1 strains was absent from all the dikaryons.
FIG 8.
The OEssp1+PC15 dikaryon has delayed fruiting body formation and faster aging. (A) Fruiting body formation was assessed in the PC9+PC15, KDssp1+PC15, and OEssp1+PC15 strains, during a period of 20 days. Initiation of fruiting bodies was quantified by calculating the percentage of discs showing initials. (B) Fungal mats at different developmental stages. The cultures were grown in stationary liquid medium, allowing the formation of a fungal mat. The bottoms of the Erlenmeyer flasks were scanned at different time points. Red arrows represent lysis areas formed with aging.
The ability of the dikaryons to form fruiting bodies was examined. Full emergence of fruiting body initials in the PC9+PC15 and KDssp1+PC15 strains was evident in 10-day-old cultures, within a similar time course (Fig. 8). However, the OEssp1+PC15 dikaryon had a significant delay, of about a week, in fruiting body initiation, and only 60% of the discs were able to produce fruiting body initials (Fig. 8 and Fig. S3).
DISCUSSION
White-rot fungi are saprotrophs that thrive on woody materials. They manipulate their secretome to acquire nutrients, by decomposing complex organic materials in the environment (23). SSPs are considered to be the least-characterized component of the secretome, and the understanding of their function is at an early stage (2, 3). In a previous study, a role for Ssp1 was demonstrated in the regulation of the ligninolytic system in P. ostreatus (16). In the present study, evidence is provided concerning the role of Ssp1 as a regulator of fungal developmental and growth of P. ostreatus.
The secretome of P. ostreatus was previously described, with a focus on the degradation of lignocellulosic biomass for the production of biofuel, such as woody substrates (24), wheat straw (24–26), or microcrystalline cellulose (26). In the present study, a total of 570 different proteins were identified in the secretomes of the PC9, KDssp1, and OEssp1 strains of P. ostreatus, grown in glucose-peptone (GP) medium, at three time points associated with different developmental stages: trophophase (8-day-old culture), the transition from trophophase to idiophase (10-day-old culture), and idiophase (13-day-old culture). The total number of identified proteins is slightly higher than those of the predicted bioinfosecretomes of P. ostreatus PC9, which include 538 proteins (20). Some of the proteins identified in the present study were annotated as cytoplasmic or lacked a classical signal peptide, similar to what was reported for other fungi and considered to be transported through the cell wall via vesicles (9).
The fungal secretome varies between the different phases of the fungal life cycle (9). By monitoring secreted protein production over time, at the onset of the fungal shift from trophophase to idiophase, an interesting phenomenon appeared. A group of proteins was downregulated in the trophophase-to-idiophase transition and upregulated again in the idiophase. The same pattern was also observed in the expression of the lignolytic enzymes encoded by aao genes and vp1, where expression was composed of two peaks. The first peak appeared during the trophophase, and the higher, second peak appeared during the idiophase. In the case of vp1, the heights of the two expression peaks were similar and could explain the results of a previous report concerning the presence of two peaks of Vp1 activity (17).
There are several lines of evidence linking Ssp1 to development. As seen from comparison of the secretomes, the KDssp1 strain entered idiophase earlier than PC9, while the converse was observed for the OEssp1 strain. This observation was further validated by monitoring the expression of selected genes over time. There was a shift in the production of ligninolytic enzymes and peptidases, which are considered markers for idiophase in WRF (12–15, 27). Many CAZymes were also strongly influenced by the transition to idiophase and Ssp1 manipulation. Although the biphasic growth of WRF is known and has been investigated for many years, most of the knowledge accumulated is based on growth conditions and nutrient limitations as triggers of the shift from trophophase to idiophase (10–14). Since the OEssp1 strain showed an expression pattern resembling the idiophase previous to glucose limitation, this suggests that Ssp1 may act as an additional regulator for the transition of P. ostreatus to the secondary growth phase.
Thus, it seems that the amount of Ssp1 produced and secreted into the medium influences the transition from trophophase to idiophase. During the trophophase, there is a small peak of upregulation of aao genes and vp1 (see Fig. S2 in the supplemental material). It is possible that the activity of these enzymes results in the modification or production of a specific chemical(s) in the medium, which in turn induces the production of Ssp1. It was previously shown that Ssp1 production is induced by 5-hydroxymethylfurfural (HMF) and aryl alcohols (16), which supports this assumption. The 7-day-old cultures already produced ssp genes, but the significant upregulation of idiophase-dependent genes occurred 2 to 3 days later (Fig. S2). Hence, the timeline supports the suggestion that the secretion of Ssp1 can act as a signal for the transition to idiophase and the subsequent production of phase-dependent enzymes.
Morphological observations and biomass accumulation provided additional evidence for developmental effects of Ssp1. Although the basic metabolism of glucose utilization remained similar to that of PC9, the KDssp1 colonies aged at a lower rate. The delayed aging morphology not only is a result of delayed entrance into idiophase but probably is also due to the lower gene expression levels and enzyme activities of idiophase-dependent proteins, as previously shown with the reduced aryl alcohol oxidase (AAO) activity in the KDssp1 background (16). The reciprocal effect was observed in the OEssp1 strain: the colony aged at a faster pace, accompanied by increased expression of metacaspases, previously linked to senescent cultures in Podospora anserina and Botrytis cinerea (19), and exhibited impaired glucose utilization. The production of a yellow pigment, which was generated during idiophase in PC9, was also observed and monitored; we suggest that the pigment is another marker of development. The pigment was produced earlier and more intensively in the OEssp1 strain and was absent from KDssp1 cultures. The pigment was associated with the protein fraction of the secretome, probably 2 CNVH proteins, which are known to bind high-mannose moieties (22). Polyketides are the most abundant fungal secondary metabolites that in some cases include yellow pigments (28), but no reports were found regarding their interaction with CNVH proteins. Hence, we propose that CNVH may bind to an unidentified chemical in the medium which contains sugar residues and has absorbance qualities at the yellow-orange spectrum.
Overall, the alterations in developmental timing and the magnitude of secondary metabolism in idiophase conferred by the genetic changes in Ssp1 have a direct effect on the aging of the colony. We assume that the slower aging of the KDssp1 strain is a result of a metabolic “trade-off”: less energy is required to produce enzymes and is utilized to keep the colony from lysing.
Additional evidence concerns the effect of Ssp1 on the redikaryotization process and the initiation of fruiting. Indeed, fruiting body secretomes contain a rich suite of small secreted proteins, many of which are developmentally regulated and conserved across species and may act as effectors (29). In this study, the formation of contact between KDssp1 and PC15 seems to be different morphologically compared to PC9 or even the OEssp1 strain. However, morphology and ssp expression in the KDssp1+PC15 dikaryon were similar to those seen when PC15 was paired with PC9. This fact can be explained by the sufficient supplementation of Ssp1 generated by the PC15 partner. On the other hand, the OEssp1+PC15 dikaryon was aging faster, similar to the monokaryon, which suggests that the effect of additional transcripts of ssp1 was not masked by the presence of the wild-type strain PC15. Nuclear ratios in heterokaryons of the basidiomycete Heterobasidion parviporum were found to be frequently imbalanced (30), which might suggest the high level of divergence in fruiting bodies emerging from the OEssp1+PC15 strain. Overall, the results suggest that Ssp1 is involved in morphogenesis during fruiting body initiation.
Studies by Zhong et al. and Luo et al. (31, 32) focused on analysis of differentially expressed genes and metabolites in P. ostreatus under an interspecific antagonistic interaction with the WRF Dichomitus squalens and Trametes versicolor. In their studies, they mentioned PLEOSDRAFT_156192, a hypothetical protein that is encoded by the most highly upregulated gene during the interaction between both fungi. PLEOSDRAFT_156192 is the closest homologue of Ssp1 in P. ostreatus PC15 (E value, 1.2E−104) and was upregulated by 660- and 707-fold with D. squalens and T. versicolor, respectively. Other homologues of Ssp1 (proteins 1080993, 1032581, and 1030599) were also upregulated by 2.5- to 490-fold. Based on the findings described here, it is tempting to speculate that the accumulated Ssp1 can act as a communication agent within the fungal colony or with other fungi. This was found in the plant kingdom, where increasing numbers of secreted peptides have been shown to influence a variety of developmental processes via cell-cell communication (33). In the fungal kingdom, plant-pathogenic and symbiotic fungi use SSP effectors to communicate with their hosts. However, there is growing evidence for the occurrence of facultative biotrophy in saprotrophic wood decay fungi, which can colonize roots of trees. It was found that P. ostreatus could colonize the surface, epidermis, cortex, and vascular of roots of Pinus sylvestris but not Picea abies (34). Based on the fungal effector prediction tool “EffectorP 2.0,” there is an average estimate of 11.7% effectors in the secretomes of saprophytes (4). Ssp1 is predicted to be an effector with a probability of 0.668. It was also suggested that SSPs might act as fruiting body effectors (29, 35). For Laccaria bicolor, it was suggested that MiSSP8 has a possible dual role, one in basic fungal biology regulating hyphal aggregation and another in mycorrhiza establishment; thus, it may be a potential self-acting regulatory protein (36). Here, we also suggest that Ssp1 may be considered part of a self-effecting mechanism and may function in a communication network analogous to quorum sensing (QS).
Although the external addition of Ssp1 did not trigger cellular and transcriptional responses similar to those observed after induction with HMF/aryl alcohols or overexpression by genetic modification, it does not disqualify our hypothesis regarding the role of Ssp1 in communication. It is not unlikely that, by supplementation, a cellular reaction that resulted in blocking of the signal may have been triggered. Another possibility is that other cellular components (such as a transporter) are needed for this function and that the recipient mechanisms were not expressed at the time of the experiment. Nevertheless, the data presented here support the notion that Ssp1 can mediate cell-to-cell communication within a fungal colony.
Even though SSPs are present in secretomes of many fungal saprophytes, little is known concerning their role. It is concluded that Ssp1 functions in P. ostreatus as an asexual and fruiting body developmental blueprint, perhaps by regulating the transition from trophophase to idiophase, in a self-affecting mode. Homologues of Ssp1 are present in at least 22 other fungi, many of which are wood decay or mycorrhizal fungi (16). The fact that some of them have already been reported to have a role in interspecies antagonistic interactions (31) indicates the importance of Ssp1 and Ssp1-like proteins for fungi in their natural environment.
MATERIALS AND METHODS
Fungal growth and experimental conditions.
(i) Strains. P. ostreatus monokaryon strain PC9 (Spanish Type Culture Collection [CECT] strain CECT20311), which is a protoclone derived by redikaryotization of the commercial dikaryon strain N001 (CECT20600) (37), was used throughout this study. The corresponding monokaryon strain PC15 (CECT20312) was used in mating trials. The KDssp1 and OEssp1 strains were generated during a previous study (16).
(ii) Gene and protein expression. Fungal strains were grown and maintained on GP medium (20 g liter−1 glucose, 5 g liter−1 peptone, 2 g liter−1 yeast extract, 1 g liter−1 K2HPO4, and 0.5 g liter−1 MgSO4·7H2O). When required, 15 g liter−1 agar was added.
The experiments were conducted on samples of fungal biomass or cell-free extracellular extracts prepared from cultures that were maintained in stationary 100-ml Erlenmeyer flasks containing 10 ml of liquid medium. The inoculum used for all experiments was one disc (5-mm diameter) of mycelium obtained from the edge of a young colony grown on solid medium and positioned at the center of the petri dish. Cultures were incubated at 28°C in the dark.
For analyzing the effect of the external addition of Ssp1, the spent medium of 14-day-old OEssp1 cultures was concentrated using a 10-kDa-cutoff PM-10 membrane and supplemented to 5-day-old PC9 and KDssp1 cultures. Gene expression was analyzed 4 and 24 h after supplementation.
(iii) Monitoring of a yellow pigment. The spectrum of the yellow pigment was subtracted from the spectral scan of the medium by using a Synergy HTX multimode microplate reader (BioTek, Winooski, VT, USA). The identified peak at 470 nm was further used to monitor the production of the pigment in the medium by using a Synergy HTX multimode microplate reader (BioTek, Winooski, VT, USA).
(iv) Glucose utilization. The estimation of glucose utilization was done with an Infinity glucose oxidase liquid stable reagent, according to the manufacturer’s procedures (Thermo Fisher Scientific Inc., VA, USA).
(v) Redikaryotization. Mating was performed as previously described (38). The tested strains (PC9, KDssp1, and OEssp1) were inoculated with PC15 on the same petri dish containing YMG medium (10 g liter−1 glucose, 10 g liter−1 malt extract, 0.4 g liter−1 yeast extract). The plate was then incubated at 28°C in the dark, until a contact zone was formed. A piece of mycelium was cut from the contact zone and placed on a new culture plate, and the morphology of the colony was examined under a microscope for the presence of clamp connections.
(vi) In vitro production of fruiting bodies. The cultures after dikaryotization were grown on GP medium. Discs were obtained from the edge of a young colony; each petri dish was inoculated with 3 to 4 discs (5-mm diameter). The medium used for fruiting body production was basidiomycete salt medium (BSM) [5 g liter−1 glucose, 1 g liter−1 K2HPO4, 0.6 g liter−1 asparagine, 0.1 g liter−1 yeast extract, 0.5 g liter−1 KCl, 0.5 g liter−1 MgSO4·7H2O, 3 mg liter−1 Zn(NO3)2·6H2O, 6 mg liter−1 Ca(NO3)2·4H2O, and 3 mg liter−1 CuSO4·5H2O] (39). The cultures were subjected to a photoperiod regime of 12 h of light/12 h of dark and incubated at room temperature. Fruiting body formation was examined on a daily basis. The experiment was performed in 3 biological triplicates; each contained 9 to 13 mycelium discs. The percentage of discs showing fruiting body initiation was calculated.
Mass spectrometry analysis of the secretome.
For the preparation of samples for the mass spectrometry analysis of the secretome, cultures of the PC9, KDssp1, and OEssp1 strains were grown as described above. The culture fluids were collected and concentrated at 3 time points representing 8-, 10-, and 13-day-old cultures. The experiment was performed in 3 biological repeats, each of which consisted of 3 pooled flasks. For each strain and time point, 3 different biological samples were used.
The culture fluids were filtered through Whatman no. 1 filter paper, followed by 0.45-μm mixed cellulose ester filter paper (Whatman, Buckinghamshire, UK). The samples were then concentrated using a 10-kDa-cutoff PM-10 membrane (Millipore, Amicon Division, Billerica, MA, USA) and treated with cOmplete (Roche Applied Science, Mannheim, Germany), after concentration.
Fifty microliters from each sample was added to a solution containing 8 M urea and 50 mM ammonium bicarbonate, reduced with 2.8 mM dithiothreitol (60°C for 30 min), and modified with 8.8 mM iodoacetamide (in the dark at room temperature for 30 min), and 10 μg was digested in a solution containing 2 M urea and 12.5 mM ammonium bicarbonate with modified trypsin (Promega) at a 1:50 enzyme-to-substrate ratio, overnight at 37°C. An additional second digestion was done for 4 h.
The tryptic peptides were desalted using C18 tips (homemade stage tips), dried, and resuspended in 0.1% formic acid. The peptides were resolved by reverse-phase chromatography on 0.075- by 180-mm fused silica capillaries (J&W) packed with Reprosil reversed-phase material (Dr. Maisch GmbH, Germany). The peptides were eluted with a linear 60-min gradient of 5 to 28% acetonitrile, a 15-min gradient of 28 to 95% acetonitrile, and 15 min with 95% acetonitrile with 0.1% formic acid in water at flow rates of 0.15 μl/min. Mass spectrometry was performed by using a Q Exactive plus mass spectrometer (Thermo) in a positive mode using a repetitively full MS scan followed by higher-energy collisional dissociation (HCD) of the 10 most dominant ions selected from the first MS scan.
The mass spectrometry data from the biological repeats were analyzed using MaxQuant software 1.5.2.8 (Mathias Mann’s group) versus the PleosPC9_1_GeneModels_Filteredmodels2_aa.fasta file and versus the Saccharomyces cerevisiae strain ATCC 204508 proteome from the UniProt database, with a 1% false discovery rate (FDR). The data were quantified by label-free analysis using the same software.
Statistical analysis of the identification and quantitation results was done using Perseus 1.5.0.31 software (Mathias Mann’s group). Statistical analysis was performed on the basis of t tests (P value of <0.02). Venn diagrams were generated with Venny 2.1 (40). The heat map illustrations were made with HemI (41).
Gene expression analyses.
Total RNA was extracted from culture biomass, first ground under liquid nitrogen with a mortar and pestle, and extracted using the RNeasy Plus minikit (Qiagen, Hilden, Germany). The RNA was treated with Turbo DNase (Invitrogen). cDNA was synthesized using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Carlsbad, CA, USA), with controls to ensure that DNA was not amplified. Gene expression analyses were performed with the ABI StepOne real-time PCR sequence detection system and software (Applied Biosystems, Foster City, CA, USA), using Power SYBR green PCR master mix (Applied Biosystems, Foster City, CA, USA). The PCR volume was 10 μl, using 20 ng of total cDNA and 300 nM oligonucleotide primers (Table 1). The thermal cycling conditions were as follows: an initial step at 95°C for 20 s and 40 cycles at 95°C for 5 s and 60°C for 30 s. To monitor the expression of ssp genes, 50 nM primers was used, and thermal cycling was done for 40 cycles at 95°C for 5 s and 64°C for 60 s. The β-tubulin gene (117235) was used as the endogenous control. The primer efficiency levels of the genes were within the range of 90% to 110%. Amplification data were compared on the basis of the of ΔCT method and presented as 2−ΔCT values or on the basis of the ΔΔCT method and presented as 2−ΔΔCT values.
TABLE 1.
Primers used for real-time PCR
| Transcript | Gene | Primer | Sequence (5′→3′) | Reference | Description |
|---|---|---|---|---|---|
| 117235 | β-Tubulin | tub_117235_F1374 | ACCAGTTCCACCACCAAGAG | 42 | Primers tub_117235_F1374 and tub_117235_R1506 were used for internal controls |
| tub_117235_R1506 | TGTTGTGCGTAAGGAAGCTG | ||||
| 69649 | aao1 | AAO_69649_F1495 | GATGCCTCGATCTTCCCATA | ||
| AAO_69649_R1688 | CTTAATGTCGTCGGCCATCT | ||||
| 93955 | aao3 | AAO_93955_F1013 | ATCCCCTCCTAGCCTGTGTT | ||
| AAO_93955_R1110 | GTCCGGTATGGTTTGCATTC | ||||
| 116309 | aao5 | AAO_116309_F1040 | TCTTCGTCAACAGCAACCAG | ||
| AAO_116309_R1149 | AGGCCAAGTGGTTAGCAATG | ||||
| 121882 | aao6 | AAO_121882_F941 | CTGGCATCGGTGATCCTACT | ||
| AAO_121882_R1041 | CACCGCAATGATATGGTCAG | ||||
| 71759 | 71759_294F | CACATTCGTCACCCAGACCA | This study | ||
| 71759_389R | GGAGGCGTCGTAGGTGTATG | ||||
| 53101 | 53101_238F | CAAACCCACTGGAGCCGTAT | |||
| 53101_325R | CGAGAGATAGCCGTGCGAAT | ||||
| 115916 | 115916_10F | TCATTCTTGAGGGCGGTGAC | |||
| 115916_104R | TCATTCTTGAGGGCGGTGAC | ||||
| 116094 | 116094_14F | ACAACCAAAACCAAGGCAGT | |||
| 116904_85R | TAGCCAGGATTGTAACCGCC | ||||
| 103857 | 103857_8F | CCTCAAACGCATTTGCCCTC | |||
| 103857_68R | GGAGACACAACCGCTGAGAT | ||||
| 117235 | β-Tubulin | tubF543 | GTGCGTAAGGAAGCTGAGGG | 43 | Primers tubF543 and tubR777 were used for internal controls |
| tubR777 | TGTGGCATTGTACGGCTCAAC | ||||
| 116738 | vp1 | MnP4F1269 | TTGTTGGCTAGAGACCCCCAGA | ||
| MnP4R1609 | CAAGTGGGCCGCTCCGAC | ||||
| 65712 | ssp1 | SSP_65712_F163 | TTTGGCTTCTCCGTGCAGTT | 16 | |
| SSP_65712_R488 | CGTAGCTGGCCAACTTGG | ||||
Chemical procedure.
Culture media of OEssp1 and KDssp1 strains were grown in stationary 250-ml Erlenmeyer flasks containing 50 ml of liquid GP medium, and after 16 days, the medium was collected. About 1 liter of the medium was applied to a preconditioned Amberlite XAD-2 column (80 by 5 cm). Following the elution of the medium, the column was washed with 1 liter of deionized water. The absorbed material was then discharged from the resin with 1 liter of methanol. After the methanol was evaporated, a dark yellow crude extract (1.35 g) was formed. The crude extract (1.35 g) was loaded onto a Sephadex LH-20 size exclusion column (25-cm height by 1.75-cm radius) eluted with a 1:1 mixture of MeOH-water to yield seven fractions (the yellow substance was eluted from the column in the third fraction [100 to 200 ml; 875 mg]): fraction I (0 to 40 ml), fraction II (40 to 100 ml; 32 mg), fraction IV (200 to 240 ml; 104 mg), fraction V (240 to 300 ml; 189 mg), fraction VI (300 to 320 ml; 6 mg), and fraction VII (320 to 380 ml; 185 mg). The yellow fraction (875 mg) was loaded (solid) onto a medium-pressure liquid chromatography (MPLC) column (Comby Flush EZ Prep RediSep, highly aqueous C18 bonded silica [C18Aq], 150 g gold) eluted with a 9:1 mixture of water-acetonitrile (20 ml/min) for 2 h, followed by washing with different solvents. The separation was monitored by UV at 214 nm and 254 nm. Nine peaks were collected: peak 1 (retention time [rt] 6.5 min; 657 mg), peak 2 (rt 42.2 min; 0.5 mg), peak 3 (rt 58.5 min; 3 mg), peak 4 (rt 110 min; 1 mg), peak 5 (methanol; 10 mg), peak 6 (0.1% aqueous trifluoroacetic acid [TFA]; 47 mg), peak 7 (acetonitrile; 32 mg), peak 8 (ethyl acetate; 53 mg), and peak 9 (waste; 9 mg).
Supplementary Material
ACKNOWLEDGMENTS
We thank the Smoler Proteomics Center at Technion, Israel, for their help in protein mass spectrometry and the statistical analysis of the secretome.
This study was supported by a grant from the Israel Science Foundation (ISF) to O.Y. and Y.H. D.F. was supported by a fellowship from the President of Israel fund granted by the Estates Committee.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00761-19.
REFERENCES
- 1.Alfaro M, Oguiza JA, Ramirez L, Pisabarro AG. 2014. Comparative analysis of secretomes in basidiomycete fungi. J Proteomics 102:28–43. doi: 10.1016/j.jprot.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Pellegrin C, Morin E, Martin FM, Veneault-Fourrey C. 2015. Comparative analysis of secretomes from ectomycorrhizal fungi with an emphasis on small-secreted proteins. Front Microbiol 6:1278. doi: 10.3389/fmicb.2015.01278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim K-T, Jeon J, Choi J, Cheong K, Song H, Choi G, Kang S, Lee Y-H. 2016. Kingdom-wide analysis of fungal small secreted proteins (SSPs) reveals their potential role in host association. Front Plant Sci 7:186. doi: 10.3389/fpls.2016.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sperschneider J, Dodds PN, Gardiner DM, Singh KB, Taylor JM. 2018. Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol Plant Pathol 19:2094–2110. doi: 10.1111/mpp.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Freitas Pereira M, Veneault-Fourrey C, Vion P, Guinet F, Morin E, Barry KW, Lipzen A, Singan V, Pfister S, Na H, Kennedy M, Egli S, Grigoriev I, Martin F, Kohler A, Peter M. 2018. Secretome analysis from the ectomycorrhizal ascomycete Cenococcum geophilum. Front Microbiol 9:141. doi: 10.3389/fmicb.2018.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plett JM, Kemppainen M, Kale SD, Kohler A, Legue V, Brun A, Tyler BM, Pardo AG, Martin F. 2011. A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr Biol 21:1197–1203. doi: 10.1016/j.cub.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 7.Plett JM, Daguerre Y, Wittulsky S, Vayssieres A, Deveau A, Melton SJ, Kohler A, Morrell-Falvey JL, Brun A, Veneault-Fourrey C, Martin F. 2014. Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc Natl Acad Sci U S A 111:8299–8304. doi: 10.1073/pnas.1322671111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kloppholz S, Kuhn H, Requena N. 2011. A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol 21:1204–1209. doi: 10.1016/j.cub.2011.06.044. [DOI] [PubMed] [Google Scholar]
- 9.McCotter SW, Horianopoulos LC, Kronstad JW. 2016. Regulation of the fungal secretome. Curr Genet 62:533–545. doi: 10.1007/s00294-016-0578-2. [DOI] [PubMed] [Google Scholar]
- 10.Betina V. 1995. Differentiation and secondary metabolism in some prokaryotes and fungi. Folia Microbiol (Praha) 40:51–67. doi: 10.1007/BF02816528. [DOI] [Google Scholar]
- 11.Demain AL. 1986. Regulation of secondary metabolism in fungi. Pure Appl Chem 58:219–226. doi: 10.1351/pac198658020219. [DOI] [Google Scholar]
- 12.Keyser P, Kirk TK, Zeikus JG. 1978. Ligninolytic enzyme system of Phanerochaete chrysosporium: synthesized in the absence of lignin in response to nitrogen starvation. J Bacteriol 135:790–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeffries TW, Choi S, Kirk TK. 1981. Nutritional regulation of lignin degradation by Phanerochaete chrysosporium. Appl Environ Microbiol 42:290–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Härtig C, Lorbeer H. 1993. Phenomenological principles of microbial lignin degradation. Acta Biotechnol 13:31–40. doi: 10.1002/abio.370130107. [DOI] [Google Scholar]
- 15.Knop D, Yarden O, Hadar Y. 2015. The ligninolytic peroxidases in the genus Pleurotus: divergence in activities, expression, and potential applications. Appl Microbiol Biotechnol 99:1025–1038. doi: 10.1007/s00253-014-6256-8. [DOI] [PubMed] [Google Scholar]
- 16.Feldman D, Kowbel DJ, Glass NL, Yarden O, Hadar Y. 2017. A role for small secreted proteins (SSPs) in a saprophytic fungal lifestyle: ligninolytic enzyme regulation in Pleurotus ostreatus. Sci Rep 7:14553. doi: 10.1038/s41598-017-15112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knop D, Ben-Ari J, Salame TM, Levinson D, Yarden O, Hadar Y. 2014. Mn2+-deficiency reveals a key role for the Pleurotus ostreatus versatile peroxidase (VP4) in oxidation of aromatic compounds. Appl Microbiol Biotechnol 98:6795–6804. doi: 10.1007/s00253-014-5689-4. [DOI] [PubMed] [Google Scholar]
- 18.Carmona-Gutierrez D, Bauer MA, Zimmermann A, Aguilera A, Austriaco N, Ayscough K, Balzan R, Bar-Nun S, Barrientos A, Belenky P, Blondel M, Braun RJ, Breitenbach M, Burhans WC, Buettner S, Cavalieri D, Chang M, Cooper KF, Côrte-Real M, Costa V, Cullin C, Dawes I, Dengjel J, Dickman MB, Eisenberg T, Fahrenkrog B, Fasel N, Froehlich K-U, Gargouri A, Giannattasio S, Goffrini P, Gourlay CW, Grant CM, Greenwood MT, Guaragnella N, Heger T, Heinisch J, Herker E, Herrmann JM, Hofer S, Jiménez-Ruiz A, Jungwirth H, Kainz K, Kontoyiannis DP, Ludovico P, Manon S, Martegani E, Mazzoni C, Megeney LA, Meisinger C, et al. 2018. Guidelines and recommendations on yeast cell death nomenclature. Microb Cell 5:4–31. doi: 10.15698/mic2018.01.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonçalves AP, Heller J, Daskalov A, Videira A, Glass NL. 2017. Regulated forms of cell death in fungi. Front Microbiol 8:1837. doi: 10.3389/fmicb.2017.01837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alfaro M, Castanera R, Lavín JL, Grigoriev IV, Oguiza JA, Ramírez L, Pisabarro AG. 2016. Comparative and transcriptional analysis of the predicted secretome in the lignocellulose-degrading basidiomycete fungus Pleurotus ostreatus. Environ Microbiol 18:4710–4726. doi: 10.1111/1462-2920.13360. [DOI] [PubMed] [Google Scholar]
- 21.Botos I, O’Keefe BR, Shenoy SR, Cartner LK, Ratner DM, Seeberger PH, Boyd MR, Wlodawer A. 2002. Structures of the complexes of a potent anti-HIV protein cyanovirin-N and high mannose oligosaccharides. J Biol Chem 277:34336–34342. doi: 10.1074/jbc.M205909200. [DOI] [PubMed] [Google Scholar]
- 22.Percudani R, Montanini B, Ottonello S. 2005. The anti-HIV cyanovirin-N domain is evolutionarily conserved and occurs as a protein module in eukaryotes. Proteins 60:670–678. doi: 10.1002/prot.20543. [DOI] [PubMed] [Google Scholar]
- 23.Singh AP, Singh T. 2014. Biotechnological applications of wood-rotting fungi: a review. Biomass Bioenerg 62:198–206. doi: 10.1016/j.biombioe.2013.12.013. [DOI] [Google Scholar]
- 24.Fernández-Fueyo E, Ruiz-Dueñas FJ, López-Lucendo MF, Pérez-Boada M, Rencoret J, Gutiérrez A, Pisabarro AG, Ramírez L, Martínez AT. 2016. A secretomic view of woody and nonwoody lignocellulose degradation by Pleurotus ostreatus. Biotechnol Biofuels 9:49. doi: 10.1186/s13068-016-0462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salvachúa D, Martínez AT, Tien M, López-Lucendo MF, García F, De Los Ríos V, Martínez MJ, Prieto A. 2013. Differential proteomic analysis of the secretome of Irpex lacteus and other white-rot fungi during wheat straw pretreatment. Biotechnol Biofuels 6:115. doi: 10.1186/1754-6834-6-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoav S, Salame TM, Feldman D, Levinson D, Ioelovich M, Morag E, Yarden O, Bayer EA, Hadar Y. 2018. Effects of cre1 modification in the white-rot fungus Pleurotus ostreatus PC9: altering substrate preference during biological pretreatment. Biotechnol Biofuels 11:212. doi: 10.1186/s13068-018-1209-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staszczak M, Nowak G, Grzywnowicz K, Leonowicz A. 1996. Proteolytic activities in cultures of selected white-rot fungi. J Basic Microbiol 36:193–203. doi: 10.1002/jobm.3620360306. [DOI] [Google Scholar]
- 28.Keller NP, Turner G, Bennett JW. 2005. Fungal secondary metabolism—from biochemistry to genomics. Nat Rev Microbiol 3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 29.Krizsán K, Almási É, Merényi Z, Sahu N, Virágh M, Kószó T, Mondo S, Kiss B, Bálint B, Kües U, Barry K, Cseklye J, Hegedüs B, Henrissat B, Johnson J, Lipzen A, Ohm RA, Nagy I, Pangilinan J, Yan J, Xiong Y, Grigoriev IV, Hibbett DS, Nagy LG. 2019. Transcriptomic atlas of mushroom development reveals conserved genes behind complex multicellularity in fungi. Proc Natl Acad Sci U S A 116:7409–7418. doi: 10.1073/pnas.1817822116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.James TY, Stenlid J, Olson Å, Johannesson H. 2008. Evolutionary significance of imbalanced nuclear ratios within heterokaryons of the basidiomycete fungus Heterobasidion parviporum. Evolution 62:2279–2296. doi: 10.1111/j.1558-5646.2008.00462.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhong Z, Li L, Chang P, Xie H, Zhang H, Igarashi Y, Li N, Luo F. 2017. Differential gene expression profiling analysis in Pleurotus ostreatus during interspecific antagonistic interactions with Dichomitus squalens and Trametes versicolor. Fungal Biol 121:1025–1036. doi: 10.1016/j.funbio.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 32.Luo F, Zhong Z, Liu L, Igarashi Y, Xie D, Li N. 2017. Metabolomic differential analysis of interspecific interactions among white rot fungi Trametes versicolor, Dichomitus squalens and Pleurotus ostreatus. Sci Rep 7:5265. doi: 10.1038/s41598-017-05669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tavormina P, De Coninck B, Nikonorova N, De Smet I, Cammue BPA. 2015. The plant peptidome: an expanding repertoire of structural features and biological functions. Plant Cell 27:2095–2118. doi: 10.1105/tpc.15.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith GR, Finlay RD, Stenlid J, Vasaitis R, Menkis A. 2017. Growing evidence for facultative biotrophy in saprotrophic fungi: data from microcosm tests with 201 species of wood-decay basidiomycetes. New Phytol 215:747–755. doi: 10.1111/nph.14551. [DOI] [PubMed] [Google Scholar]
- 35.Almási É, Sahu N, Krizsán K, Bálint B, Kovács GM. 2019. The genome of Auriculariopsis ampla sheds light on fruiting body development and wood-decay of bark-inhabiting fungi. bioRxiv doi: 10.1101/550103. [DOI]
- 36.Pellegrin C, Daguerre Y, Ruytinx J, Guinet F, Kemppainen M, Plourde MB, Hecker A, Morin E, Pardo AG, Germain H, Martin FM, Veneault-Fourrey C. 2017. Laccaria bicolor MiSSP8 is a small-secreted protein decisive for the establishment of the ectomycorrhizal symbiosis. bioRxiv doi: 10.1101/218131. [DOI] [PubMed]
- 37.Larraya LM, Perez G, Penas MM, Baars JJP, Mikosch TSP, Pisabarro AG, Ramirez L. 1999. Molecular karyotype of the white rot fungus Pleurotus ostreatus. Appl Environ Microbiol 65:3413–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salame TM, Knop D, Tal D, Levinson D, Yarden O, Hadar Y. 2012. Predominance of a versatile-peroxidase-encoding gene, mnp4, as demonstrated by gene replacement via a gene targeting system for Pleurotus ostreatus. Appl Environ Microbiol 78:5341–5352. doi: 10.1128/AEM.01234-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen R, Persky L, Hazan-Eitan Z, Yarden O, Hadar Y. 2002. Mn2+ alters peroxidase profiles and lignin degradation by the white-rot fungus Pleurotus ostreatus under different nutritional and growth conditions. Appl Biochem Biotechnol 102–103:415–430. doi: 10.1385/ABAB:102-103:1-6:415. [DOI] [PubMed] [Google Scholar]
- 40.Oliveros JC. 2016. Venny. An interactive tool for comparing lists with Venn’s diagrams, 2007–2015. http://bioinfogp.cnb.csic.es/tools/venny/.
- 41.Deng W, Wang Y, Liu Z, Cheng H, Xue Y. 2014. HemI: a toolkit for illustrating heatmaps. PLoS One 9:e111988. doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feldman D, Kowbel DJ, Glass NL, Yarden O, Hadar Y. 2015. Detoxification of 5-hydroxymethylfurfural by the Pleurotus ostreatus lignolytic enzymes aryl alcohol oxidase and dehydrogenase. Biotechnol Biofuels 8:63. doi: 10.1186/s13068-015-0244-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salame TM, Yarden O, Hadar Y. 2010. Pleurotus ostreatus manganese-dependent peroxidase silencing impairs decolourization of orange II. Microb Biotechnol 3:93–106. doi: 10.1111/j.1751-7915.2009.00154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.