A. tumefaciens is used in the construction of genetically engineered plants, as it is able to transfer DNA to plant hosts. Knowledge of the mechanisms of DNA transfer and the genes required will aid in the understanding of this process. Manipulation of glycoside hydrolases may increase transformation and widen the host range of the bacterium. A. tumefaciens also causes disease (crown gall tumors) on a variety of plants, including stone fruit trees, grapes, and grafted ornamentals such as roses. It is possible that compounds that inhibit glycoside hydrolases could be used to control crown gall disease caused by A. tumefaciens.
KEYWORDS: Agrobacterium, B. daigremontiana, arabinosylfuranosidase, glycoside hydrolases, pectinases, tomato, xylanase
ABSTRACT
Agrobacterium tumefaciens is a rhizosphere bacterium that can infect wound sites on plants. The bacterium transfers a segment of DNA (T-DNA) from the Ti plasmid to the plant host cell via a type IV secretion system where the DNA becomes integrated into the host cell chromosomes. The expression of T-DNA in the plant results in tumor formation. Although the binding of the bacteria to plant surfaces has been studied previously, there is little work on possible interactions of the bacteria with the plant cell wall. Seven of the 48 genes encoding putative glycoside hydrolases (Atu2295, Atu2371, Atu3104, Atu3129, Atu4560, Atu4561, and Atu4665) in the genome of A. tumefaciens C58 were found to play a role in virulence on tomato and Bryophyllum daigremontiana. Two of these genes (pglA and pglB; Atu3129 and Atu4560) encode enzymes capable of digesting polygalacturonic acid and, thus, may play a role in the digestion of pectin. One gene (arfA; Atu3104) encodes an arabinosylfuranosidase, which could remove arabinose from the ends of polysaccharide chains. Two genes (bglA and bglB; Atu2295 and Atu4561) encode proteins with β-glycosidase activity and could digest a variety of plant cell wall oligosaccharides and polysaccharides. One gene (xynA; Atu2371) encodes a putative xylanase, which may play a role in the digestion of xylan. Another gene (melA; Atu4665) encodes a protein with α-galactosidase activity and may be involved in the breakdown of arabinogalactans. Limited digestion of the plant cell wall by A. tumefaciens may be involved in tumor formation on tomato and B. daigremontiana.
IMPORTANCE A. tumefaciens is used in the construction of genetically engineered plants, as it is able to transfer DNA to plant hosts. Knowledge of the mechanisms of DNA transfer and the genes required will aid in the understanding of this process. Manipulation of glycoside hydrolases may increase transformation and widen the host range of the bacterium. A. tumefaciens also causes disease (crown gall tumors) on a variety of plants, including stone fruit trees, grapes, and grafted ornamentals such as roses. It is possible that compounds that inhibit glycoside hydrolases could be used to control crown gall disease caused by A. tumefaciens.
INTRODUCTION
Agrobacterium tumefaciens is a rhizosphere bacterium that can infect wound sites on plants. These infections result in the development of crown gall tumors. The bacteria transfer a DNA fragment (transfer DNA [T-DNA]) from a plasmid (pTi) to the host cell using a type IV secretion system (T4SS) (1). The T-DNA is then integrated into the host chromosomes. T-DNA encodes enzymes for the biosynthesis of plant growth hormones behind constitutive eukaryotic promoters. Thus, the integration of the T-DNA results in plant cell overgrowth and tumor formation.
The initial interactions of A. tumefaciens with the plant surface have largely been studied by measuring binding of the bacteria to plant tissue culture cells and to root surfaces (2, 3). Root colonization has also been examined (4). These bacteria have several well-characterized mechanisms of binding to surfaces. The unipolar polysaccharide mediates polar bacterial binding to both animate and inanimate surfaces, including plant roots, soil particles, quartz sand, and nylon thread (3, 5). Planktonic cells generally do not make the unipolar polysaccharide. It is made immediately after surface contact and leads to reversible binding (6). Initial binding to surfaces may be mediated by the unipolar polysaccharide or by the Ctp pilus (7). The Ctp pilus is one of several types of pili encoded in the A. tumefaciens genome. It can mediate reversible binding to surfaces and may be involved in the initial stages of binding. However, neither the unipolar polysaccharide nor the Ctp pilus is required for tumor formation by A. tumefaciens (3).
After the bacteria have bound to plant surfaces, they begin to elaborate cellulose microfibrils (8). The production of these fibrils can be stimulated in planktonic cells by the addition of plant extracts to the medium. Their production results in irreversible binding to cellulose-containing surfaces, including plant cell walls and Whatman filter paper. However, cellulose synthesis, like the unipolar polysaccharide and the Ctp pilus, is not required for tumor formation by A. tumefaciens (9).
The T-pilus can also mediate binding to plant cell surfaces, although the level of binding is low compared to that mediated by the unipolar polysaccharide (2). It has been suggested that VirB2 and VirB5 may be involved in binding of the T-pilus to the plant plasma membrane (10). Whether T-pilus binding is required for tumor formation is unknown.
The mechanisms by which the T-DNA spans the bacterial cell membrane, cell wall, and outer membrane have been characterized (1, 11, 12). There is also some information about how the T-DNA crosses the plant cell membrane. However, very little is known about the interaction of the bacteria with the plant cell wall. Transfer of the T-DNA though the T4SS channel from the bacterium to the plant cytoplasm would presumably require the direct contact of the channel with the plant cell membrane (1). Thus, the plant cell wall in the region of DNA transfer would need to be loosened or removed to allow bacterial access. Plant cell walls contain a mixture of proteins and complex carbohydrates, including cellulose, xylan, arabinogalactan, arabinomannan, pectin, hemicellulose, and others (13, 14). The major constituents that hold the plant wall together tightly are polysaccharides. Thus, bacterial enzymes, which might loosen or remove a part of the wall, may play a role in T-DNA transfer. The genome of A. tumefaciens C58 contains 48 putative glycoside hydrolases and 1 lyase (15, 16). This study focuses on the possible roles of these genes in the interaction of the bacteria with the plant cell surface leading to tumor formation.
RESULTS
Seven putative glycoside hydrolase genes are required for virulence on tomato and Bryophyllum daigremontiana.
The carbohydrate active enzymes website (www.cazy.org) lists 48 putative glycoside hydrolases and 1 lyase identified by sequence homology in the genome of A. tumefaciens C58 (15, 16). Five of these genes have known functions as follows: Atu2730 chvB [β-(1,2)-glucan synthesis] (17), Atu3307 celC (cellulose synthesis) (18), Atu4055 exoK (succinoglycan synthesis) (19), Atu5162 avhB1 (a component of a type 4 secretion system), and Atu6167 virB1 (a component of a type 4 secretion system) (20). CelC, ExoK, and AvhB1 are not required for virulence (21). ChvB is required for virulence under most, but not all, conditions (21). VirB1 is one of the genes required for the elaboration on the T pilus and is required for virulence (1). Thirteen of the genes may be involved in cell wall metabolism according to their annotations in GenBank and BioCyc as follows: Atu0009 (mltA, membrane-bound lytic murein transglycosylase), Atu0092 (lytic murein transglycosylase), Atu0233 (lysozyme), Atu0572 (soluble lytic murein transglycosylase), Atu1022 (soluble lytic transglycosylase with homology to lysozyme), Atu2112 (soluble lytic transglycosylase with homology to lysozyme), Atu2117 (lytic murein transglycosylase), Atu2121 (lysozyme), Atu2122 (dolichol-phosphate mannosyltransferase), Atu2451 (membrane protein containing an N-acetyl-d-glucosamine binding site), Atu2489 (membrane-bound lytic murein transglycosylase B), Atu3779 (lytic murein transglycosylase), and Atu8140 (lysozyme-like) (22). Four of the genes are probably involved in starch and/or glycogen metabolism as follows: Atu4073 (glgX, glycogen debranching enzyme), Atu4077 (glgB, glycogen branching enzyme), Atu4833 (cga, glucoamylase), and Atu5284 (α-amylase). One of the genes appears to be involved in sucrose metabolism as follows: Atu6135 (splA, sucrose phosphorylase) (22). The effect of mutations in these genes on virulence has not been reported. Insertion or deletion mutations were created in each of the remaining 26 genes plus Atu1617. This gene has 78% identity to GenBank accession number EKJ97063, which is annotated as a xylanase from Bradyrhizobium lupini HPC(L) and was, therefore, included in this study (Atu1617 also has high identity [>85%] to alpha/beta hydrolases from the Agrobacterium/Rhizobium group).
Virulence of the wild-type C58 parent strain and each of the mutants was measured on tomato stems and B. daigremontiana leaves (see Table S1 in the supplemental material). In order to promote the growth of the inoculated bacteria, 10 μl of lysogeny (Luria) broth was placed in the wound site along with the bacteria. Most of the mutants were virulent or very slightly attenuated on both host plants. One mutant, Atu2596, was significantly reduced in tumor formation on B. daigremontiana (50% of inoculated sites formed tumors). This mutant was unable to grow on minimal AB medium with 0.2% glucose as a carbon source and was not studied further. Inoculations with seven of the mutants tested resulted in the development of tumors on less than 10% of the inoculated plants (Table 1; Fig. 1). These seven genes and mutants of these genes are the subject of the remainder of this study.
TABLE 1.
Activity predictions and virulence assays of mutant bacterial strains
| Gene mutated | CAZy and BLAST predictions of gene function | Virulence of mutant on (% tested sites forming tumors)a |
Virulence of the complemented mutant on (% tested sites forming tumors)a |
||
|---|---|---|---|---|---|
| B. daigremontiana | Tomato | B. daigremontiana | Tomato | ||
| bglA (Atu2295) | GH13, α-glucosidase | 0 (0/30) | 0 (0/20) | 30 (3/10) | 25 (2/8) |
| xynA (Atu2371) | GH10, endo-1,4-beta-xylanase | 3 (1/30) | 3 (1/31) | 33 (4/12) | 33 (2/6) |
| arfA (Atu3104) | GH51, arfA, arabinosylfuranosidase | 0 (0/20) | 0 (0/12) | 30 (6/20) | 37 (3/8) |
| pglA (Atu3129) | GH28, pgl, polygalacturonase | 0 (0/43) | 0 (0/19) | 62 (10/16) | 50 (5/10) |
| pglB (Atu4560) | GH28, polygalacturonase | 0 (0/24) | 0 (0/13) | 50 (8/16) | 30 (3/10) |
| bglB (Atu4561) | GH105, rhamnogalacturonyl hydrolase | 0 (0/16) | 0 (0/26) | 25 (4/16) | 25 (2/8) |
| melA (Atu4665) | GH4, melA, α-glucosidase | 0 (0/13) | 0 (0/14) | 30 (3/10) | 27 (4/14) |
Number of sites forming tumors/number of sites tested. Virulence of C58 parent was 87% (71/82) on B. daigremontiana and 100% (39/39) on tomato.
FIG 1.

Virulence of A. tumefaciens parent strain C58 and glycoside hydrolase mutants (A and C) and complemented mutants (B and D) on B. daigremontiana (A and B) and tomato (C and D). (A) B. daigremontiana leaf. C58 was inoculated at one site on the top side of the leaf, and C58 pglA::pBluescript KS− was inoculated at the other sites. On the bottom side of the leaf, all sites were inoculated with C58 pglB::pCDF-1b. (B) B. daigremontiana leaf. All sites were inoculated with C58 pglA::pBluescript KS− pBRpglA. (C) Tomato stem. The upper stem was inoculated with C58 pglB::pCDF-1b. The middle stem was inoculated with C58 pglA::pBluescript KS− and the lower stem with C58. (D) Tomato stem. The upper stem was inoculated with C58 pglA::pBluescript KS− and the lower stem with C58 pglA::pBluescript KS− pBRpglA.
xynA.
This gene is listed in CAZy as encoding a possible xylanase. It has homology to endo-1,4-beta-xylanase according to a Basic Local Alignment Search Tool (BLAST) (Table 1). It is located on the circular chromosome in a putative operon consisting of two genes. The second gene is annotated as a hypothetical protein (Fig. 2). The predicted substrate for the enzyme encoded by this gene (xylan) is a large molecule, which would not be able to enter the bacterial cytoplasm, suggesting that XynA might be secreted. The protein has a predicted signal sequence. It also has a predicted motif for twin arginine (TAT) secretion. None of the other six proteins studied here have either a signal sequence or a TAT sequence (23). Mutants in xynA were avirulent on tomato and B. daigremontiana (Table 1). C58 xynA::pBluescript KS− was partially complemented to virulence on tomato and B. daigremontiana by the introduction of pBBRxynA (Table 1).
FIG 2.
Maps of parts of the A. tumefaciens C58 genome showing the seven glycoside hydrolase genes required for virulence on B. daigremontiana and tomato (red striped) and the surrounding genes. Genes belonging to putative ABC transporters are indicated in blue. adeC, adenine deaminase; hypo, hypothetical protein; kdgK, 2-dehydro-3-deoxygluconokinase; exoU, UDP-hexose transferase; ABCm, ABC transporter transmembrane protein; ABCn, ABC transporter nucleotide-binding protein; ABCs, ABC transporter substrate binding protein; rspF, acetyltransferase; yteR, di-trans,poly-cis-decaprenylcistransferase; purR, transcriptional regulator; hlyD, HlyD family secretion protein. Maps are based on those in https://biocyc.org.
C58 xynA::pBluescript KS− showed normal growth in lysogeny broth and in liquid AB minimal medium with glucose, sucrose, cellobiose, or lactose as a carbon source (Table 2). It also showed normal growth on agar medium containing arabinose, xylose, melibiose, melezitose, raffinose, arabinomannan, arabinogalactan, polygalacturonic acid, or pectin as a carbon source. The mutant was unable to grow on AB agar medium containing xylan as a carbon source (Fig. 3A). The mutant strain carrying pBBRxynA was able to grow on xylan medium. These results suggest that xynA encodes a protein with xylanase activity.
TABLE 2.
Growth of A. tumefaciens parent and mutant strains in liquid medium
| Bacterial strain | Doubling time (min) growing in: |
||||
|---|---|---|---|---|---|
| Lysogeny broth | AB glucosea | AB sucrose | AB cellobiose | AB lactose | |
| C58 | 120 ± 10b | 170 ± 15 | 195 ± 20 | 190 ± 20 | 190 ± 20 |
| C58 bglA::pBluescript KS− | 120 ± 10 | 170 ± 15 | 210 ± 30 | No growthc | No growth |
| C58 xynA::pBluescript KS− | 130 ± 10 | 165 ± 15 | 160 ± 30 | 200 ± 20 | 160 ± 30 |
| C58 arfA::pBluescript KS− | 120 ± 15 | 145 ± 20 | 160 ± 35 | 200 ± 20 | 180 ± 20 |
| C58 pglA::pBluescript KS− | 120 ± 10 | 160 ± 15 | 220 ± 30 | 120 ± 30 | 170 ± 25 |
| C58 pglB::pCDF-1b | 120 ± 10 | 145 ± 20 | 210 ± 20 | 190 ± 25 | 190 ± 35 |
| C58bglB | 110 ± 15 | 145 ± 15 | 190 ± 20 | No growth | No growth |
| C58 melA::pBluescript KS− | 120 ± 10 | 145 ± 15 | 120 ± 30 | 150 ± 30 | 230 ± 30 |
| C58 bglA::pBluescript KS− ΔbglB | 120 ± 15 | 160 ± 25 | 180 ± 30 | No growth | No growth |
AB minimal medium containing 0.2% of the indicated carbon source and 0.01% soytone as an inducer. A minimum of three replicates was carried out for each growth condition.
Average ± standard deviation of the average.
No growth was observed after 48 h of incubation.
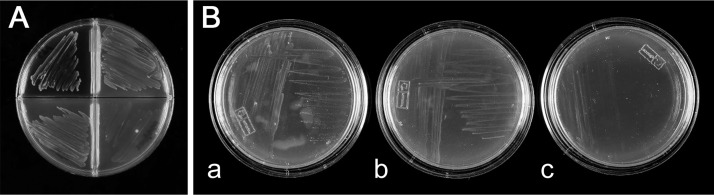
FIG 3.
(A) Growth of the parent C58 (a) and lack of growth of the mutant C58 xynA::pBluescript KS− (b) on AB minimal medium agar containing xylan as the carbon source and 0.02% soytone as plant extract inducer. (B) Growth of the parent C58 (a), C58 pglA::pBluescript KS− (b), and C58 pglB::pCDF-1b (c) and lack of growth of the C58 pglA::pBluescript KS− pglB::pCDF-1b (d) on AB minimal medium agar containing 0.2% polygalacturonic acid as the carbon source and 0.02% soytone as plant extract inducer. (C) Growth of the parent C58 (upper left), C58 bglA::pBluescript KS− (upper right), C58 xynA::pBluescript KS− (lower left), and C58 arfA::pBluescript KS− (lower right) on AB minimal medium agar containing 0.2% sucrose as the carbon source. (D) Growth of the parent C58 (upper left), C58 bglA::pBluescript KS− (upper right), and C58 ΔbglB (lower left) and lack of growth of the C58 bglA::pBluescript KS− ΔbglB (lower right) on AB minimal medium agar containing 0.2% cellobiose as the carbon source and 0.02% soytone as plant extract inducer. (E) Growth of C58 bglA::pBluescript KS− (right) and C58 ΔbglB (middle) and lack of growth of the C58 bglA::pBluescript KS− ΔbglB (left) on AB minimal medium agar containing 0.2% lactose as the carbon source and 0.02% soytone as plant extract inducer.
arfA.
A BLAST search and the CAZy database both identified this gene as encoding a possible arabinosylfuranosidase (Table 1). It is located on the linear chromosome in an operon containing genes for a putative ATP-binding cassette (ABC) transporter and one hypothetical protein (Fig. 2). Mutants in arfA were avirulent on tomato and B. daigremontiana (Table 1). C58 arfA::pBluescript KS− was partially complemented to virulence on tomato and B. daigremontiana by the introduction of pBBRarfA (Table 1).
C58 arfA::pBluescript KS− showed normal growth in lysogeny broth and in liquid AB minimal medium with glucose, sucrose, cellobiose, or lactose as a carbon source (Table 2). It also showed normal growth on agar medium containing xylose, melibiose, melezitose, raffinose, xylan, arabinomannan, arabinogalactan, polygalacturonic acid, or pectin as a carbon source. C58 arfA::pBluescript KS− grew poorly on arabinose. When a clone of Atu3100-3103 (pBR3100-3) carrying the ABC transporter genes from the operon containing arfA (Fig. 2) was introduced into C58 arfA::pBluescript KS−, the mutant bacteria were able to grow on arabinose, but they did not recover virulence.
C58, the parent strain, and C58 arfA::pBluescript KS− were assayed for arabinosylfuranosidase activity by measuring the hydrolysis of O-nitrophenyl-alpha-arabinose by bacteria grown in lysogeny broth with 0.01% soytone. C58 showed 50 ± 10 Miller units of activity while the mutant had no detectable activity. When a plasmid (pBBRarfA) carrying the intact arfA gene cloned behind the lac promoter (a weak constitutive promoter in A. tumefaciens) was introduced into the mutant bacteria, enzyme activity was restored to 30 ± 15 Miller units, supporting the sequence homology data which suggest that arfA encodes an arabinosylfuranosidase.
pglA and pglB.
Two of the seven genes were identified by both a BLAST search and the CAZy database as encoding possible polygalacturonases. One of these genes, Atu3129, was previously identified as a putative polygalacturonase and named pgl (24, 25). It is adjacent to picA (Atu3128), a gene whose expression is induced by carrot extracts. In some annotations, the two genes have been confused and Atu3129 is listed as picA. However, its sequence is identical with that reported previously for pgl and not picA. As there are two putative polygalacturonase genes in the C58 genome, we have renamed Atu3129 pglA and named the other putative polygalacturonase, Atu4560, pglB. The identification of a plant extract-inducible gene (picA) near the end of the operon containing pglA suggests that expression of the entire operon may be induced by plant extracts. Both pglA and pglB are located in operons on the linear chromosome (Fig. 2). pglA is downstream from genes encoding a putative ABC transporter and upstream from yteR (picA) and rspF, encoding di-trans,poly-cis-decaprenylcistransferase and an acetyltransferase, respectively. pglB is located in an operon downstream from a hypothetical protein and bglB. It is followed by genes encoding a putative ABC transporter (Fig. 2). Mutants in pglA and pglB were avirulent on tomato and B. daigremontiana (Table 1). C58 pglA::pBluescript KS− and C58 pglB::pCDF-1b were partially complemented to virulence on tomato and B. daigremontiana by the introduction of pBBRpglA or pBBRpglB, respectively (Table 1).
Both C58 pglA::pBluescript KS− and C58 pglB::pCDF-1b showed normal growth in lysogeny broth and in liquid AB minimal medium with glucose, sucrose, cellobiose, or lactose as a carbon source (Table 2). They also showed normal growth on agar medium containing arabinose, xylose, melibiose, melezitose, raffinose, xylan, arabinomannan, arabinogalactan, polygalacturonic acid, or pectin as a carbon source (Table 3). Since both the pglA and pglB mutants were able to grow on polygalacturonic acid, a double pglA pglB mutant (C58 pglA::pBluescript KS− pglB::pCDF-1b) was constructed and its growth on polygalacturonic acid tested. C58 pglA::pBluescript KS− pglB::pCDF-1b grew on AB agar medium containing arabinose, xylose, melibiose, melezitose, raffinose, xylan, arabinomannan, arabinogalactan, or pectin as a carbon source. It was unable to grow on polygalacturonic acid, suggesting that either PglA or PglB must be active for the breakdown of polygalacturonic acid to occur (Fig. 3). Growth of the double mutant on pectin may be due to the complex structure of pectin, which contains many different sugar side chains as well as a polygalacturonic acid backbone; thus, the ability of C58 pglA::pBluescript KS− pglB::pCDF-1b to grow on pectin is not surprising (26). To confirm that PglA and PglB were able to break down polygalacturonic acid, the genes were cloned into pBluescript KS− and introduced into Escherichia coli ER2267. The presence of either pglA or pglB allowed growth of ER2267 on polygalacturonic acid and on pectin, which is consistent with their tentative identification as polygalacturonases (Table 4; Fig. 4).
TABLE 3.
Growth of A. tumefaciens strains on various carbon sources on agar plates
| Carbon source (0.2%) | Gene(s) mutated in the bacterial straina |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| None (C58 parent strain) | bglA | xynA | arfA | pglA | pglB | bglB | melA | pglA pglB | bglA bglB | |
| Glucose | + | + | + | + | + | + | + | + | + | + |
| Xylose | + | + | + | + | + | + | + | + | + | + |
| Galacturonic acid | + | + | + | + | + | + | + | + | + | + |
| Sucrose | + | + | + | + | + | + | + | + | + | + |
| Melibiose | + | + | + | + | + | + | + | + | + | + |
| Lactose | + | + | + | + | + | + | + | + | + | Nonec |
| Cellobiose | + | + | + | + | + | + | + | + | + | Nonec |
| Melezitose | + | + | + | + | + | + | + | + | + | NDb |
| Raffinose | + | + | + | + | + | + | + | + | + | ND |
| Polygalacturonic acid | + | + | + | + | + | + | + | + | Nonec | + |
| Pectin | + | + | + | + | + | + | + | + | + | + |
| Xylan | + | + | Nonec | + | + | + | + | + | + | ND |
| Arabinomannan | + | + | + | + | + | + | + | + | + | + |
| Arabinogalactan | + | + | + | + | + | + | + | + | + | + |
Growth was measured on modified AB minimal agar medium for a period of 14 days and is recorded as follows: +, growth; None, no growth.
ND, not determined.
Results which differ significantly from the parent strain. All results were replicated a minimum of 5 times.
TABLE 4.
Growth of E. coli carrying cloned genes on various carbon sources
| Carbon source (0.1%) | Growth of straina |
|||||||
|---|---|---|---|---|---|---|---|---|
|
ER2267 parent |
ER2267 pKSbglA |
ER2267 pKSxynA |
ER2267 pKSarfA |
ER2267 pKSpglA |
ER2267 pKSpglB |
ER2267 pKSbglB |
ER2267 pKSmelA |
|
| Lactose | None | + | None | None | None | None | + | None |
| Polygalacturonic acid | None | None | None | None | + | + | None | None |
| Pectin | None | None | None | None | + | + | None | None |
| Arabinogalactan | None | None | None | None | + | + | + | + |
None of the clones allowed growth on cellobiose or arabinomannan. E. coli ER2267 showed slow growth on xylan and raffinose. Therefore, these carbon sources could not be tested. Growth was measured on M9 minimal agar medium containing IPTG for a period of 4 days and is recorded as follows: +, growth; None, no growth. All results were replicated a minimum of 5 times.
FIG 4.
(A) Growth of E. coli ER2267 pBBRpglA (upper left) and ER2276 pBBRpglB (upper right) on 0.2% polygalacturonic acid as a carbon source and growth of ER2267 pBBRpglA (lower left) and lack of growth of ER2276 pBBR (lower right) on M9 minimal medium containing 0.2% pectin as a carbon source. (B) Growth of E. coli ER2267 pBBRmelA (a) and ER2267 pBBRbglB (b) and lack of growth of ER2267 pBBRbglA (c) on M9 minimal medium containing 0.2% arabinogalactan as a carbon source.
bglA and bglB.
bglA is listed in CAZy as encoding a possible α-glucosidase (Table 1) (16). It is the first gene in an operon on the circular chromosome (Fig. 2). The other two genes in the operon are hypothetical proteins. BglB is listed in CAZy as a possible rhamnogalacturonyl hydrolase. It is located in the same operon as pglB and is immediately upstream of pglB. The other genes in this operon are putative ABC transporter genes and a gene for a hypothetical protein. Since pglB and bglB are in the same operon, the mutation of bglB was an in-frame deletion of the gene so as not to affect the expression of the downstream gene, pglB. Mutants in bglA and bglB were avirulent on tomato and B. daigremontiana (Table 1). C58 bglA::pBluescript KS− and C58 ΔbglB were partially complemented to virulence on tomato and B. daigremontiana by the introduction of pBBRbglA or pBBRbglB, respectively (Table 1).
Both C58 bglA::pBluescript KS− and C58 ΔbglB showed normal growth in lysogeny broth and in liquid AB minimal medium with glucose or sucrose as a carbon source (Table 2). They were unable to grow in liquid AB medium with either lactose or cellobiose as a carbon source. They showed normal growth on agar medium containing arabinose, xylose, melibiose, melezitose, raffinose, xylan, arabinomannan, arabinogalactan, polygalacturonic acid, or pectin (Table 3). They also showed slow growth on AB agar medium containing either lactose or cellobiose as a carbon source (Fig. 3D). In order to explore the possibility that growth on solid medium containing lactose or cellobiose was due to a third gene not expressed (or for some reason ineffective) in liquid medium, a double mutant was constructed, C58 bglA::pBluescript KS−ΔbglB. This mutant was unable to grow on AB agar medium containing lactose or cellobiose as a carbon source, suggesting that these two genes are the only genes involved in growth on these substrates on AB minimal medium. To confirm that BglA and BglB were able to break down lactose, the genes were cloned into pBluescript KS− behind the lac promoter and introduced into E. coli ER2267. Expression of the cloned gene was induced with isopropyl-β-d-1-thiogalactopyranoside (IPTG) and the β-galactosidase activity of the bacteria determined. The presence of either bglA or bglB allowed growth of ER2267 on lactose, suggesting that the genes encoded proteins with β-galactosidase activity (Table 4). ER2267 carrying clones of bglA or bglB showed detectable β-galactosidase activity, 55 ± 5 and 131 ± 16 Miller units, respectively, suggesting that the proteins encoded by these genes are able to hydrolyze β-linked glycosides. This activity was specific to these two genes. Bacteria carrying clones of xynA, arfA, pglA, pglB, or melA showed no detectable enzyme activity (less than 5 Miller units). The substrate specificity of the two proteins appears to be different as the clone of bglB allowed growth of E. coli on arabinogalactan while the clone of bglA did not (arabinogalactan contains β-1-3 and β-1-6 linked galactose residues). Since these genes encode proteins with β-galactosidase activity, they were named bglA and bglB.
melA.
This gene is listed in CAZy as encoding a possible α-glucosidase (Table 1). In some annotations of the C58 genome, it is identified as melA. It is located in the middle of an operon on the linear chromosome, which contains genes for a putative ABC transporter and a gene with homology to the HlyD family of secretion proteins, suggesting that this protein might be secreted (Fig. 2). A mutant in melA was avirulent on tomato and B. daigremontiana (Table 1). C58 melA::pBluescript KS− was partially complemented to virulence on tomato and B. daigremontiana by the introduction of pBBRmelA (Table 1).
C58 melA::pBluescript KS− showed normal growth in lysogeny broth and in liquid AB minimal medium with glucose, sucrose, cellobiose, or lactose as a carbon source (Table 2). It also showed normal growth on agar medium containing arabinose, xylose, melibiose, melezitose, raffinose, arabinomannan, arabinogalactan, xylan, polygalacturonic acid, and pectin. The melA gene was cloned into pBluescript KS− and expressed in E. coli ER2267. It did not allow ER2267 to grow on lactose, polygalacturonic acid, or pectin as a carbon source. It did allow ER2267 to grow on arabinogalactan. However, since arabinogalactan is a complex polysaccharide and commercial preparations contain 5 to 10% other plant cell wall polysaccharides, no conclusions can be drawn as to the sugar bonds hydrolyzed by MelA from this result. The cloned gene was also introduced into JW4080-3, which is an melA mutant of E. coli from the Keio collection (27). This E. coli strain was unable to grow on melibiose. When pKSmelA was introduced into the strain and its expression induced by the addition of IPTG, the bacteria were able to grow on melibiose. This result is consistent with the homology of the A. tumefaciens protein to E. coli MelA (68% identity) and suggests that the A. tumefaciens MelA has α-galactosidase activity.
DISCUSSION
Seven genes (xynA, arfA, pglA, pglB, bglA, bglB, and melA) predicted to encode glycoside hydrolases required for virulence on tomato and B. daigremontiana were identified. The probable enzymatic activity and possible substrates of these seven putative glycoside hydrolase genes were examined. A summary of the results is contained in Table 5.
TABLE 5.
Properties of glycoside hydrolase genes required for virulence on B. daigremontiana and tomato
| Gene | BLAST and/or CAZy identification | Results of tests of growth on carbon sources | Enzymatic activity | Predicted gene function |
|---|---|---|---|---|
| bglA (Atu2295) | Alpha-glucosidase | Either bglA or bglB is required for growth of A. tumefaciens on lactose or cellobiose, allows growth of E. coli lacZ mutant on lactose | β-Galactosidasea | β-Glycosidase |
| xynA (Atu2371) | Endo-1,4-beta-xylanase | Needed for growth of A. tumefaciens on xylan | NDb | Xylanase |
| arfA (Atu3104) | Arabinosylfuranosidase | Not required for growth on tested carbon sources | Arabinosylfuranosidasec | Arabinosylfuranosidase |
| pglA (Atu3129) | Polygalacturonase | Either pglA or pglB is required for growth of A. tumefaciens on PGA,d allows growth of E. coli on PGA | ND | Polygalacturonase |
| pglB (Atu4560) | Polygalacturonase | ND | Polygalacturonase | |
| bglB (Atu4561) | Rhamnogalacturonyl hydrolase | Either bglA or bglB is required for growth of A. tumefaciens on lactose or cellobiose, allows growth of E. coli lacZ mutant on lactose | β-Galactosidase | β-Glycosidase |
| melA (Atu4665) | MelA, α-galactosidase | Allows growth of E. coli melA mutant on melibiose and on arabinogalactan | ND | α-Galactosidase |
Enzyme activity of cloned gene expressed in E. coli.
ND, none detected. All cloned genes were tested for β-galactosidase activity.
Enzyme activity measured in A. tumefaciens.
PGA, polygalacturonic acid.
A mutation in one of these genes (xynA) resulted in the inability of A. tumefaciens to grow on xylan, suggesting that this gene is a xylanase as predicted by amino acid sequence homology. Xylans are common components of the plant cell wall, and this enzyme might aid in their digestion.
Wild-type C58 shows arabinosylfuranosidase activity. A mutant in another of these genes, arfA, lacked this activity. Arabinosylfuranosidase activity was restored in the mutant by the introduction of a clone carrying arfA, suggesting that ArfA has arabinosylfuranosidase activity. This result is consistent with the homology of this gene to known arabinosylfuranosidases. Polysaccharides carrying arabinose side chains are widely distributed in plant cell walls, including arabinogalactans, arabinomannans, and some pectins. ArfA may be able to remove the arabinose from these side chains.
Clones of either pglA or pglB introduced into E. coli allowed the bacteria to grow on polygalacturonic acid and on pectin (substances on which this E. coli strain does not otherwise grow). Single mutations in either of these genes in A. tumefaciens did not prevent the bacteria from growing on polygalacturonic acid, but the double mutant C58 pglA::pBluescript KS− pglB::pCDF-1b was unable to grow on this carbon source. These results are consistent with the amino acid sequence homology of these genes to polygalacturonases. Polygalacturonic acid makes up the backbone of pectin in plant cell walls.
Two other genes, bglA and bglB, appear to encode proteins with β-galactosidase activity. The expression of either bglA or bglB in a lacZ mutant E. coli strain resulted in β-galactosidase activity. Single mutations in either of these genes in A. tumefaciens prevented the bacteria from growing on lactose or cellobiose in liquid AB medium but not on agar. The reason for the lack of growth of the single mutants in liquid and their growth on solid medium is unknown. The double mutant C58 bglA::pBluescript KS− ΔbglB was unable to grow on lactose or cellobiose in liquid or on agar medium. Both of these sugars support the growth of the parent strain in liquid and on agar medium. The results suggest that BglA and BglB have β-glycosidase activity, but the substrate on which they act during plant infection remains unknown.
C58 melA::pBluescript KS− was able to grow on all carbon sources tested. A clone of this gene introduced into the E. coli JS4080-3 melA mutant allowed the growth of this E. coli strain on melibiose and on arabinogalactan, suggesting that MelA has α-galactosidase activity. This result is consistent with the amino acid homology of the protein to E. coli MelA. MelA may be involved in the breakdown of arabinogalactans or plant cell wall polysaccharides containing α-linked galactose residues.
Although A. tumefaciens C58 could grow on monosaccharides and disaccharides in liquid medium, it was only able to grow on the polysaccharides tested when they were added to solid agar medium. Little or no growth was obtained in liquid medium. A small amount of plant extract (0.01% soytone) was also required for growth; presumably, this substance acts as an inducer since it was present in amounts too small to support growth. One of the genes (pglA) is found in a putative operon containing a gene whose expression has been shown to be induced by carrot extracts (Atu3128; picA) (24, 25). Thus, it is not surprising that the expression of some of these glycoside hydrolase genes may be regulated by substances coming from the plant. The mechanism(s) of the regulatory interactions between these genes, their products, and plant root exudates is currently under study.
Proteins involved in the breakdown of polysaccharides need to be present on the bacterial surface or secreted to reach their substrate. Of these seven proteins, only XynA has a putative signal sequence. It also has a motif for twin arginine secretion (TAT). None of the other proteins have either a signal sequence or a TAT motif (23). One gene (melA) is located next to a gene (Atu4666) whose protein product has homology to HlyD, a membrane-fusion protein of the type 1 secretion system (T1SS) (Fig. 2), and thus MelA may be secreted using this system (28). All seven proteins lack the basic region identified as involved in protein secretion via the T4SS (1).
All of these mutants could be partially complemented to virulence by the introduction of the intact gene cloned in a plasmid (pBBR1mcs-5) behind the lac promoter. The reasons for the failure of the cloned gene to fully complement the mutations are unclear. Possible reasons include difference in timing and level of expression of the gene when placed behind the lac promoter on the plasmid and decreased fitness of the bacteria due to the lack of expression of genes downstream of the insertion.
The function of these genes in tumor formation by A. tumefaciens is unclear. There are several possible roles for these genes. The products of the digestion of polysaccharides by the enzymes encoded by one or more of these genes could serve as signal molecules alerting the bacteria to their proximity to plant cell walls. Alternatively, the enzymes encoded by these genes may loosen the plant cell wall and permit the translocation channel to reach the plant cell membrane for T-DNA transfer. There are additional possible roles for these genes. Some of the enzymes encoded by the genes could be involved in the alteration of surface polysaccharides of the bacteria. Such alterations are known to be involved in the interaction of some bacteria with plant hosts. For example, in Sinorhizobium meliloti, the extracellular digestion of high-molecular-weight succinoglycan into low-molecular-weight succinoglycan plays a role in the interaction of the bacteria with the plant (29–31). It is also possible that some of the proteins encoded by these genes play a role in modifying the plant responses to bacteria. For example, PglA and PglB could be involved in the degradation of pectin-derived elicitors. Some of these genes may play a role in the fitness of the bacteria in the plant environment, and the decrease in fitness may be responsible for the loss of virulence.
Five of these seven glycoside hydrolases are located in operons that also contain genes for ABC transporters, suggesting that the sugars produced by their action are transported into the cell where they could be used as carbon sources and/or serve as signal molecules to indicate the presence of a plant cell adjacent to the bacteria. When the mutant bacteria were introduced into wound sites with the addition of lysogeny broth and 0.1% sucrose or xylose, they still failed to induce tumors. Thus, although the sugars generated by these enzymes may serve as a carbon source for the bacteria, it is unlikely that reduced access to carbon sources is responsible for the lack of virulence of these mutants.
These seven genes were identified as required for virulence on tomato and B. daigremontiana by screening mutants of genes encoding glycoside hydrolases. There is no reason to think that they all function in the same pathway in tumor formation. The individual genes may have different roles. However, the functions of pglB and bglB, which are located in the same operon, may be related. The other genes are all located in separate operons and may function in unrelated processes. In addition, it is possible that a single protein may play more than one role in the interaction of the bacteria with the plant.
In conclusion, 7 of the 48 putative glycoside hydrolases (bglA, xynA, arfA, pglA, pglB, bglB, and melA) in the genome of A. tumefaciens were found to play a role in virulence on tomato stems and B. daigremontiana leaves. Two of these genes (pglA and pglB) encode enzymes capable of digesting polygalacturonic acid and, thus, may play a role in the digestion of pectin. One gene (arfA) encodes an arabinosylfuranosidase, which could remove arabinose from the ends of polysaccharide chains. Two genes (bglA and bglB) encode proteins with β-glycosidase activity and could digest a variety of plant cell wall oligosaccharides and polysaccharides. One gene (xynA) encodes a putative xylanase, which may play a role in the digestion of xylan. One gene (melA) encodes a protein with α-galactosidase activity and could play a role in the degradation of arabinogalactans. It appears that limited digestion of plant cell wall polysaccharides by A. tumefaciens may be involved in tumor formation on tomato stems and B. daigremontiana leaves.
MATERIALS AND METHODS
Bacterial growth.
E. coli and A. tumefaciens strains were grown in lysogeny broth and on lysogeny agar plates at 37°C and 25°C, respectively. M9 was used as a minimal medium for E. coli, and AB medium with the concentration of phosphate increased from 0.025 M to 0.05 M was used as the minimal medium for A. tumefaciens (32). Carbon sources were added at 0.2%. Soytone used as an inducer rather than a carbon source was added at 0.01%. Antibiotics were added to the medium as needed at the following concentrations: 3 mg/liter tetracycline; 25 mg/liter carbenicillin, spectinomycin, and gentamicin; and 20 mg/liter kanamycin. In some experiments, 20 μg/ml IPTG was added to M9 to induce expression of cloned genes in E. coli. Growth curves of A. tumefaciens strains in liquid medium were carried out in side-arm flasks at 25°C and 150 rpm. Flasks were inoculated with a 1 to 50 dilution of a fresh 24-h culture of the bacteria grown in lysogeny broth. Measurements of optical density at 600 nm were made over a 48-h period.
Construction of mutants.
The bacterial strains used in these experiments are listed in Table 6. Mutants in A. tumefaciens putative glycoside hydrolase genes were constructed as follows. A segment approximately 300 bp long from the middle of the gene was amplified by PCR using primers that carried restriction sites not found in the sequence being amplified. The primers used for PCR in these experiments are listed in Table 7. The resulting DNA fragment and the plasmid pBluescript KS− were digested with the appropriate restriction enzymes and the DNA purified by gel electrophoresis. The plasmid DNA and the DNA fragment produced by PCR were ligated together and transformed into competent E. coli ER2267 using calcium-mediated transformation. Transformants carrying the desired insert were selected on carbenicillin and checked for the proper insert using gel electrophoresis. The resulting plasmid was transformed into competent A. tumefaciens C58 using a freeze-thaw method, and transformants were selected on carbenicillin (33). The pBluescript plasmid cannot replicate in A. tumefaciens, so the resulting transformants should have the plasmid integrated into the host cell DNA. That the insertion was in the expected gene was confirmed by PCR using primers for the ends of the gene (Table 7) followed by gel electrophoresis. Deletions of bglB were made using the plasmid pK18 and the technique described by Schafer et al. (34). The PCR primers used to generate the upstream and downstream fragments required for this procedure are listed in Table 7.
TABLE 6.
Bacterial strains and plasmids used in these experiments
| Bacterial strain or plasmid | Relevant characteristic(s) | Source |
|---|---|---|
| E. coli strains | ||
| ER2267 | K12 F′ proA+B+ lacIq Δ(lacZ)M15 zzf::mini-Tn10 (KanR)/Δ(argF-lacZ)U169 glnV44 e14−(McrA−) rfbD1? recA1 relA1? endA1 spoT1? thi-1 Δ(mcrC-mrr)114::IS10, Kmr | New England Biolabs |
| JW4080-3 | Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ−, rph-1, Δ(rhaD-rhaB)568, ΔmelA745::kan, hsdR514 | CGSC, the Coli Genetic Stock Center, Yale University |
| A. tumefaciens strains | ||
| C58 | Wild type, virulent | Laboratory collection |
| C580594 | C58 with an insertion of pBluescript KS− in Atu0594 | This study |
| C580944 | C58 with an insertion of pBluescript KS− in Atu0944 | This study |
| C581523 | C58 with an insertion of pBluescript KS− in Atu1523 | This study |
| C581617 | C58 with an insertion of pBluescript KS− in Atu1617 | This study |
| C581709 | C58 with an insertion of pBluescript KS− in Atu1709 | This study |
| C58 bglA::pBluescript KS− | C58 with an insertion of pBluescript KS− in bglA | This study |
| C58 xynA::pBluescript KS− | C58 with an insertion of pBluescript KS− in xynA | This study |
| C582575 | C58 with an insertion of pBluescript KS− in Atu2575 | This study |
| C582596 | C58 with an insertion of pBluescript KS− in Atu2596 | This study |
| C583025 | C58 with an insertion of pBluescript KS− in Atu3025 | This study |
| C583093 | C58 with an insertion of pBluescript KS− in Atu3093 | This study |
| C583 arfA::pBluescript KS− | C58 with an insertion of pBluescript KS− in arfA | This study |
| C583128 | C58 with an insertion of pBluescript KS− in Atu3128 | This study |
| C58 pglA::pBluescript KS− | C58 with an insertion of pBluescript KS− in pglA | This study |
| C583285 | C58 with an insertion of pBluescript KS− in Atu3285 | This study |
| C583312 | C58 with an insertion of pBluescript KS− in Atu3312 | This study |
| C583553 | C58 with an insertion of pBluescript KS− in Atu3553 | This study |
| C583564 | C58 with an insertion of pBluescript KS− in Atu3564 | This study |
| C583914 | C58 with an insertion of pBluescript KS− in Atu3419 | This study |
| C584485 | C58 with an insertion of pBluescript KS− in Atu4485 | This study |
| C58 pglB::pCDF-1b | C58 with an insertion of pCDF-1b in pglB | This study |
| C58 ΔbglB | C58 with an in-frame deletion of bglB | This study |
| C58 melA::pBluescript KS− | C58 with an insertion of pBluescript KS− in melA | This study |
| C585072 | C58 with an insertion of pBluescript KS− in Atu5072 | This study |
| C585082 | C58 with an insertion of pBluescript KS− in Atu5082 | This study |
| C585284 | C58 with an insertion of pBluescript KS− in Atu5284 | This study |
| C588097 | C58 with an insertion of pBluescript KS− in Atu8079 | This study |
| Plasmids | ||
| pBluescript KS− | Cloning vector Carbr | Stratagene |
| pBBR1mcs-5 | Cloning vector Gentr | Kovach et al. (37) |
| pCDF-1b | Cloning vector Specr | Novagen |
| pBRbglA | bglA cloned into pBBR1mcs-5 | This study |
| pBRxynA | xynA cloned into pBBR1mcs-5 | This study |
| pBR3100-3 | Atu3100-3103 inclusive cloned into pBR1mcs-5 | This study |
| pBRarfA | arfA cloned into pBBR1mcs-5 | This study |
| pBRpglA | pglA cloned into pBBR1mcs-5 | This study |
| pBRpglB | pglB cloned into pBBR1mcs-5 | This study |
| pBRbglB | bglB cloned into pBBR1mcs-5 | This study |
| pBRmelA | melA cloned into pBBR1mcs-5 | This study |
| pK18 | Kanr, sacB, pRP4 oriV and oriT | Schafer et al. (34) |
| pKSbglA | bglA cloned into pBluescript KS− | This study |
| pKSxynA | xynA cloned into pBluescript KS− | This study |
| pKSarfA | arfA cloned into pBluescript KS− | This study |
| pKSpglA | pglA cloned into pBluescript KS− | This study |
| pKSpglB | pglB cloned into pBluescript KS− | This study |
| pKSbglB | bglB cloned into pBluescript KS− | This study |
| pKSmelA | melA cloned into pBluescript KS− | This study |
TABLE 7.
PCR primers used in these experiments
Construction of plasmids.
For enzyme assays and complementation of mutants, plasmids carrying intact genes were constructed. The wild-type gene from the start codon to the stop codon was amplified using PCR with sites for the appropriate restriction enzymes on the ends of the primers. The resulting DNA fragment was purified. It and the plasmid pBluescript KS− or pBBR1-mcs5 were digested with restriction enzymes and the products purified by gel electrophoresis. The plasmid and the PCR DNA fragment were ligated and transformed into competent E. coli ER2267 or JW4080-3 using calcium-mediated transformation. pBBR1-mcs5 plasmids were transformed into A. tumefaciens strains using a freeze-thaw method (33).
Virulence assays.
Bacterial virulence was determined by inoculating wounds on leaves of Bryophyllum daigremontiana and stems of tomato (Lycopersicum esculentum) with approximately 107 bacteria from a fresh plate of lysogeny agar with appropriate antibiotics using previously described techniques (9, 35). Lysogeny broth (10 μl) was added to the wound site immediately after bacterial inoculation. The plants were kept at room temperature and scored for tumor formation weekly for 6 weeks.
Enzyme activity assays.
β-Galactosidase assays were carried out using the method described by Miller (36). For assays of E. coli strains carrying plasmids, expression of the plasmid-borne gene was induced by the addition of 20 μg/ml IPTG 1 h prior to the assay. Arabinosylfuranosidase assays were carried out using the same protocol but with the substrate O-nitrophenyl-alpha-arabinose.
Supplementary Material
ACKNOWLEDGMENTS
We thank Susan Whitfield for assistance with the illustrations. Dane Meredith originally had the idea to look for polygalacturonases in the A. tumefaciens genome when he was an undergraduate at UNC. We thank Bruce Stone and Linda Spermulli for helpful discussions and Janine Corley and Lauren Burke for assistance with some of the virulence assays.
This research was supported in part by a summer undergraduate research fellowship from the University of North Carolina to S.L.M.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00603-19.
REFERENCES
- 1.Li YG, Christie PJ. 2018. The Agrobacterium VirB/VirD4 T4SS: mechanism and architecture defined through in vivo mutagenesis and chimeric systems. Curr Top Microbiol Immunol 418:233–260. doi: 10.1007/82_2018_94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matthysse AG. 2014. Attachment of Agrobacterium to plant surfaces. Front Plant Sci 5:252. doi: 10.3389/fpls.2014.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson MA, Onyeziri MC, Fuqua C. 2018. Function and regulation of Agrobacterium tumefaciens cell surface structures that promote attachment. Curr Top Microbiol Immunol 418:143–184. doi: 10.1007/82_2018_96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matthysse AG, McMahan S. 1998. Root colonization by Agrobacterium tumefaciens is reduced in cel, attB, attD, and attR mutants. Appl Environ Microbiol 64:2341–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomlinson AD, Fuqua C. 2009. Mechanisms and regulation of polar surface attachment in Agrobacterium tumefaciens. Curr Opin Microbiol 12:708–714. doi: 10.1016/j.mib.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu J, Kim J, Koestler BJ, Choi JH, Waters CM, Fuqua C. 2013. Genetic analysis of Agrobacterium tumefaciens unipolar polysaccharide production reveals complex integrated control of the motile-to-sessile switch. Mol Microbiol 89:929–948. doi: 10.1111/mmi.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Haitjema CH, Fuqua C. 2014. The Ctp type IVb pilus locus of Agrobacterium tumefaciens directs formation of the common pili and contributes to reversible surface attachment. J Bacteriol 196:2979–2988. doi: 10.1128/JB.01670-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matthysse AG. 1983. Role of bacterial cellulose fibrils in Agrobacterium tumefaciens infection. J Bacteriol 154:906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sykes LC, Matthysse AG. 1986. Time required for tumor induction by Agrobacterium tumefaciens. Appl Environ Microbiol 52:597–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Backert S, Fronzes R, Waksman G. 2008. VirB2 and VirB5 proteins: specialized adhesins in bacterial type-IV secretion systems? Trends Microbiol 16:409–413. doi: 10.1016/j.tim.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Lacroix B, Citovsky V. 2013. The roles of bacterial and host plant factors in Agrobacterium-mediated genetic transformation. Int J Dev Biol 57:467–481. doi: 10.1387/ijdb.130199bl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gelvin SB. 2012. Traversing the cell: Agrobacterium T-DNA's journey to the host genome. Front Plant Sci 3:52. doi: 10.3389/fpls.2012.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houston K, Tucker MR, Chowdhury J, Shirley N, Little A. 2016. The plant cell wall: a complex and dynamic structure as revealed by the responses of genes under stress conditions. Front Plant Sci 7:984. doi: 10.3389/fpls.2016.00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert HJ. 2010. The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol 153:444–455. doi: 10.1104/pp.110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantarel BL, Coutinho PM, Rancurel C, Ernard T, Lombard V, Enrissat B. 2009. The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puvanesarajah V, Schell FM, Stacey G, Douglas CJ, Nester EW. 1985. Role for 2-linked-beta-d-glucan in the virulence of Agrobacterium tumefaciens. J Bacteriol 164:102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthysse AG, White S, Lightfoot R. 1995. Genes required for cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol 177:1069–1075. doi: 10.1128/jb.177.4.1069-1075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cangelosi GA, Hung L, Puvanesarajah V, Stacey G, Ozga DA, Leigh JA, Nester EW. 1987. Common loci for Agrobacterium tumefaciens and Rhizobium meliloti exopolysaccharide synthesis and their roles in plant interactions. J Bacteriol 169:2086–2091. doi: 10.1128/jb.169.5.2086-2091.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baron C, Llosa M, Zhou S, Zambryski PC. 1997. VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens, is processed to a C-terminal secreted product, VirB1. J Bacteriol 179:1203–1210. doi: 10.1128/jb.179.4.1203-1210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthysse AG. 2018. Exopolysaccharides of Agrobacterium tumefaciens. Curr Top Microbiol Immunol 418:111–141. doi: 10.1007/82_2018_100. [DOI] [PubMed] [Google Scholar]
- 22.Caspi R, Billington R, Ferrer L, Foerster H, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Karp PD. 2016. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 44:D471–D480. doi: 10.1093/nar/gkv1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding Z, Christie PJ. 2003. Agrobacterium tumefaciens twin-arginine-dependent translocation is important for virulence, flagellation, and chemotaxis but not type IV secretion. J Bacteriol 185:760–771. doi: 10.1128/JB.185.3.760-771.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rong L, Karcher SJ, O'Neal K, Hawes MC, Yerkes CD, Jayaswal RK, Hallberg CA, Gelvin SB. 1990. picA, a novel plant-inducible locus on the Agrobacterium tumefaciens chromosome. J Bacteriol 172:5828–5836. doi: 10.1128/jb.172.10.5828-5836.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rong LJ, Karcher SJ, Gelvin SB. 1991. Genetic and molecular analyses of picA, a plant-inducible locus on the Agrobacterium tumefaciens chromosome. J Bacteriol 173:5110–5120. doi: 10.1128/jb.173.16.5110-5120.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cosgrove DW. 1997. Assembly and enlargement of the primary cell wall in plants. Annu Rev Cell Dev Biol 13:171–201. doi: 10.1146/annurev.cellbio.13.1.171. [DOI] [PubMed] [Google Scholar]
- 27.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holland I, Peherstorfer S, Kanonenberg K, Lenders M, Reimann S, Schmitt L. 13 December 2016. Type I protein secretion—deceptively simple yet with a wide range of mechanistic variability across the family. EcoSal Plus doi: 10.1128/ecosalplus.ESP-0019-2015. [DOI] [PubMed] [Google Scholar]
- 29.Cheng H, Walker G. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J Bacteriol 180:5183–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Battisti L, Lara JC, Leigh JA. 1992. Specific oligosaccharide form of the Rhizobium meliloti exopolysaccharide promotes nodule invasion in alfalfa. Proc Natl Acad Sci U S A 89:5625–5629. doi: 10.1073/pnas.89.12.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marczak M, Mazur A, Koper P, Zebracki K, Skorupska A. 2017. Synthesis of rhizobial exopolysaccharides and their importance for symbiosis with legume plants. Genes (Basel) 8:360. doi: 10.3390/genes8120360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morton ER, Fuqua C. 2012. Laboratory maintenance of Agrobacterium. Curr Protoc Microbiol Chapter 1:Unit3D. doi: 10.1002/9780471729259.mc03d01s24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holsters M, de Waele D, Depicker A, Messens E, van Montagu M, Schell J. 1978. Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet 163:181–187. doi: 10.1007/BF00267408. [DOI] [PubMed] [Google Scholar]
- 34.Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 35.Matthysse AG, Jaeckel P, Jeter C. 2008. attG and attC mutations of Agrobacterium tumefaciens are dominant negative mutations that block attachment and virulence. Can J Microbiol 54:241–247. doi: 10.1139/w08-005. [DOI] [PubMed] [Google Scholar]
- 36.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 37.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.