The recent large influx of genome sequences from basidiomycetes, which are prolific producers of bioactive natural products, may provide opportunities to develop novel drug candidates. The development of a reliable expression system is essential for the genome mining of natural products because of the lack of a tractable host for heterologous expression of basidiomycete genes. For this purpose, we applied the ascomycete Aspergillus oryzae system for the direct expression of fungal natural product biosynthetic genes from genomic DNA. Using this system, 29 sesquiterpene synthase genes and diterpene biosynthetic genes for bioactive pleuromutilin were successfully expressed. Together with the use of computational tools for intron prediction, this Aspergillus oryzae system represents a practical method for the production of basidiomycete natural products.
KEYWORDS: Aspergillus oryzae, basidiomycete fungi, heterologous expression, terpene synthase, terpenes
ABSTRACT
Basidiomycete fungi are an attractive resource for biologically active natural products for use in pharmaceutically relevant compounds. Recently, genome projects on mushroom fungi have provided a great deal of biosynthetic gene cluster information. However, functional analyses of the gene clusters for natural products were largely unexplored because of the difficulty of cDNA preparation and lack of gene manipulation tools for basidiomycete fungi. To develop a versatile host for basidiomycete genes, we examined gene expression using genomic DNA sequences in the robust ascomycete host Aspergillus oryzae, which is frequently used for the production of metabolites from filamentous fungi. Exhaustive expression of 30 terpene synthase genes from the basidiomycetes Clitopilus pseudo-pinsitus and Stereum hirsutum showed two splicing patterns, i.e., completely spliced cDNAs giving terpenes (15 cases) and mostly spliced cDNAs, indicating that A. oryzae correctly spliced most introns at the predicted positions and lengths. The mostly spliced cDNAs were expressed after PCR-based removal of introns, resulting in the successful production of terpenes (14 cases). During this study, we observed relatively frequent mispredictions in the automated program. Hence, the complementary use of A. oryzae expression and automated prediction will be a powerful tool for genome mining.
IMPORTANCE The recent large influx of genome sequences from basidiomycetes, which are prolific producers of bioactive natural products, may provide opportunities to develop novel drug candidates. The development of a reliable expression system is essential for the genome mining of natural products because of the lack of a tractable host for heterologous expression of basidiomycete genes. For this purpose, we applied the ascomycete Aspergillus oryzae system for the direct expression of fungal natural product biosynthetic genes from genomic DNA. Using this system, 29 sesquiterpene synthase genes and diterpene biosynthetic genes for bioactive pleuromutilin were successfully expressed. Together with the use of computational tools for intron prediction, this Aspergillus oryzae system represents a practical method for the production of basidiomycete natural products.
INTRODUCTION
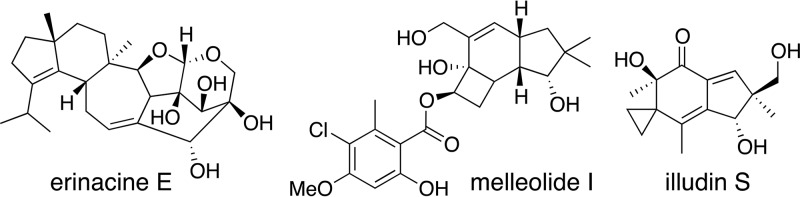
Mushroom-forming basidiomycete fungi are known to be prolific producers of structurally diverse, bioactive natural products (NPs) and have been used since ancient times in traditional medicine (1). This group of fungi is particularly known for their terpenoid NPs, which appear to be the major class of NPs produced by them (1, 2). Well-known examples of pharmaceutically relevant terpenoid-derived compounds include the widely used livestock antibiotic pleuromutilin (3), the anticancer compound illudin S (4), the melleolide antibiotics (5), and the nerve growth factor synthesis-promoting erinacines (6) (Fig. 1). The project involving 1,000 fungal genomes performed by the Joint Genome Institute (JGI) (7) has resulted in a large influx of basidiomycete genome sequences and genomic data from hundreds of basidiomycete taxa that can now be searched in silico for NP biosynthetic gene clusters.
FIG 1.
Chemical structures of biologically active terpenoids produced by basidiomycete fungi.
However, despite their incredible potential for NP discovery, basidiomycete fungi are largely unexplored territory for drug discovery compared to ascomycete fungi. While an abundance of genetic tools and techniques for transformation and genetic manipulation are available for yeast and filamentous fungi, such methods are mostly lacking for basidiomycete fungi (2, 8). In addition, many basidiomycete strains are difficult to grow under laboratory conditions, requiring the determination of suitable growth conditions and often long fermentation times. Characterization of their biosynthetic genes therefore typically requires heterologous expression in a more genetically tractable host. Unfortunately, basidiomycetes have very intron-rich genomes and genes that contain very small and unpredictable exons, especially in cytochrome P450 monooxygenase genes, that currently necessitate the amplification of genes from cDNA. Even if a gene is expressed, alternative splice variants may be produced that are not functional (2). All pioneering work to access the large diversity of terpenoid NPs made by basidiomycetes has therefore relied on the amplification of functional terpene synthase (TS) genes from cDNA for expression and characterization in Escherichia coli or yeasts, limiting the complete functional analysis of all computationally identified TSs in basidiomycete genomes (9–13). Being able to directly express basidiomycete biosynthetic genes from genomic DNA in a suitable fungal surrogate host would therefore greatly accelerate the functional characterization of NP pathways.
We reasoned that a genetically tractable ascomycete could potentially be used as a heterologous host for the functional expression of basidiomycete NP biosynthetic genes. Unlike yeasts, which have relatively few genes with introns, the splicing machinery of filamentous fungi such as Aspergillus spp. is more similar to that of basidiomycetes (14). Heterologous expression of biosynthetic genes from filamentous fungi directly from genomic DNA has become a powerful approach to elucidate biosynthetic pathways. We have used this strategy with the ascomycete Aspergillus oryzae for the heterologous production of several NPs and their pathway intermediates (15–18) and to access difficult-to-obtain pathway intermediates for the study of intriguing enzyme reactions (19–21). Recently, we successfully produced in A. oryzae the basidiomycete-derived diterpene pleuromutilin from cDNA-amplified biosynthetic genes. Like others, we found the cloning of functional genes from basidiomycete cDNA to be difficult and tedious. We and others also showed that the native A. oryzae NADPH cytochrome P450 reductase supports the activity of the basidiomycete P450 monooxygenase pathway (22, 23). Given our success with using A. oryzae for the direct expression of fungal NP biosynthetic genes from genomic DNA and the ability of A. oryzae to functionally express a basidiomycete terpenoid NP biosynthetic enzymes, including its P450 monooxygenase, we therefore hypothesized that this ascomycete would make an excellent platform strain for the characterization and production of basidiomycete NP pathways, in particular its terpenoid pathways.
Here, we describe a new strategy using A. oryzae as an expression host for the heterologous production of basidiomycete NP biosynthetic genes. We selected two different terpenoid classes (pleuromutilin diterpene and sesquiterpenoids) from two different basidiomycetes and performed a systematic analysis of the predicted and experimentally obtained splicing patterns in A. oryzae. Our results indicated that a significant fraction of genes were correctly spliced by A. oryzae, while many partially spliced genes could be corrected based on gene model predictions and functionally expressed in A. oryzae.
RESULTS AND DISCUSSION
Expression of pleuromutilin biosynthetic genes from genomic DNA in Aspergillus oryzae.
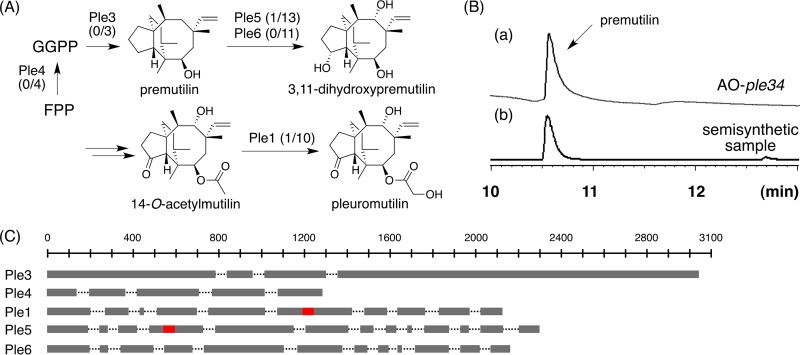
We previously achieved the heterologous production of the basidiomycete diterpene pleuromutilin using an A. oryzae expression system (22). Therefore, we initially examined the heterologous expression of pleuromutilin biosynthetic genes using genomic DNA sequences in this ascomycete host. When we tested the expression of the geranylgeranyl diphosphate (GGPP) synthase gene ple4 and the terpene synthase gene ple3 in A. oryzae, the resultant transformant (TF) successfully produced premutilin (compound 1) (Fig. 2). Sequencing of cDNA sequences recovered from the TF A. oryzae ple3 ple4 showed that all introns (the 3 in ple3 and the 4 in ple4) were correctly spliced (Fig. 2). We next examined the expression of intron-rich P450 genes (ple1, -5, and -6; 10/13/11). cDNA sequence analysis showed that all 11 introns of ple6 were successfully spliced, while ple1 and -5 each contained one unspliced intron (ple1, intron 6 out of 10; ple5, intron 4 out of 13), which can be readily removed by PCR-based methods. Based on these promising results, we speculated that gene splicing in basidiomycetes resembles that in ascomycete fungi (14, 24).
FIG 2.
(A) Biosynthetic pathway of pleuromutilin. The numbers in parentheses are the nonspliced introns in the cDNA recovered from the A. oryzae TF and the total number of introns of the corresponding gene. (B) GC-MS profiles of the metabolites from A. oryzae ple34 (a) and a synthetic sample (b). (C) Schematic view of the gene structures of ple3, ple4, ple1, ple5, and ple6. Exon regions are shown in shaded boxes. Nonspliced introns are shown in red boxes.
Functional gene expression analysis of STS genes.
Given that almost all introns of ple genes were successfully spliced in A. oryzae, we next performed a comprehensive analysis of basidiomycete gene expression from genomic DNA in this host. To establish the versatility of basidiomycete heterologous expression using A. oryzae, we chose to test the expression of sesquiterpene synthases (STSs) because their genes are prolific in basidiomycete genomes and they are small enzymes that convert abundant farnesyl pyrophosphate (FPP) in host cells into various cyclic, volatile products that can be readily identified by gas chromatography-mass spectrometry (GC-MS) analysis of cultures (25). In this work, we selected 31 STS genes for characterization from two basidiomycete fungi: our pleuromutilin producer Clitopilus pseudo-pinsitus and, for comparison, Stereum hirsutum, which was previously investigated for sesquiterpenoid production (11) (Fig. 3; see Table S1 in the supplemental material).
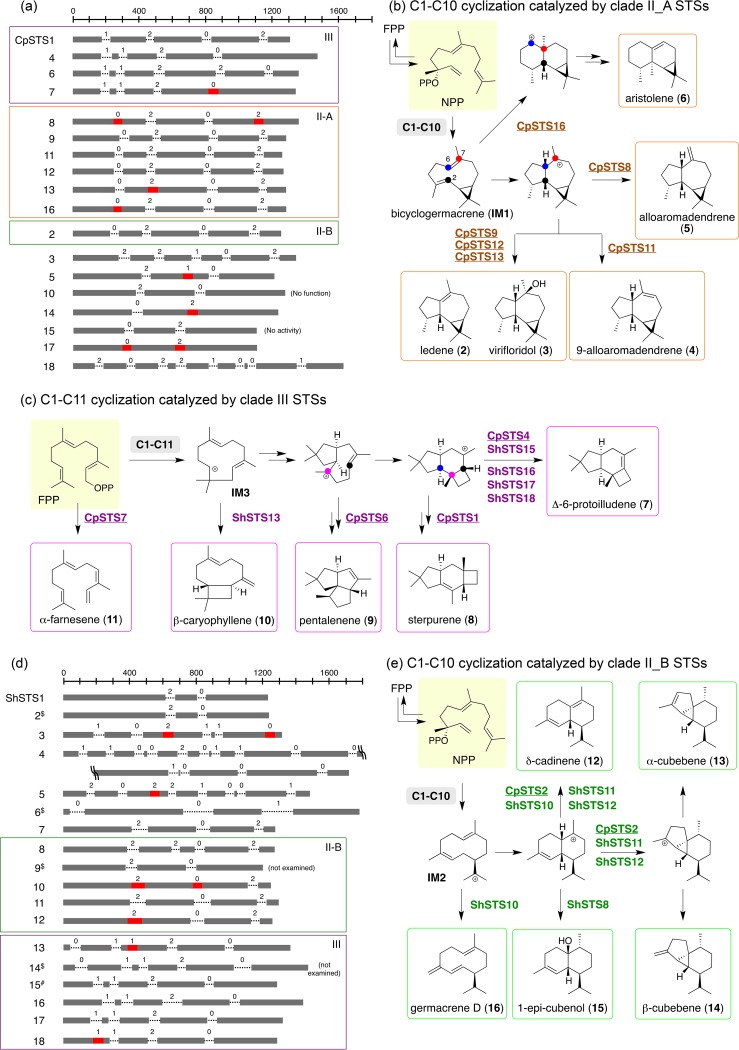
FIG 3.
(a) Schematic view of the gene structures of CpSTSs. The dark bars indicate exons and the dotted lines introns along with intron phases. The red bars represent the computationally predicted introns that were not spliced (skipped) in cDNA from A. oryzae TFs, removal of which resulted in functional STS expression in the E. coli TF. Genes with the colored boxes are proposed paralogs or orthologs in C. pseudo-pinsitus and S. hirsutum that belong to cyclization clade III (purple), II_A (orange), or II_B (green). Nonfunctional refers to CpSTS10 lacking conserved active-site motifs. (b) Proposed cyclization mechanism of terpene products catalyzed by clade II_A STSs. (c) Proposed cyclization mechanism of terpene products catalyzed by clade III STSs. (d) Schematic view of the gene structures of ShSTSs. #, gene structure of a functional ShSTS15 that was reported previously (35). $, computationally predicted gene structure of an STS not investigated in this work. (e) Proposed cyclization mechanism of terpene products catalyzed by clade II_B STSs. STSs from C. pseudo-pinsitus are underlined in panels b, c, and e. Stereochemistries of putative carbocation intermediates are predicted by those of cyclized products.
(i) STSs from Clitopilus pseudo-pinsitus.
Our local BLAST search using aristolochene synthase as a query sequence identified 18 STS candidate genes (encoding CpSTS1 to -18) in C. pseudo-pinsitus (Fig. 3; Table S1). Open reading frames (ORFs) were manually predicted by comparison with functionally characterized fungal STS genes. Except for CpSTS10, which lacked the conserved motifs (DDXXD and NSE) of STSs, the other 17 ORFs were selected for expression in A. oryzae. To ensure the expression of target genes, we used the recently developed fungal clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 system specifically optimized for A. oryzae (26). In the presence of the CRISPR-Cas9 plasmid with the protospacer sequence for the SC103 locus, which was found in a strain that produced betaenone in good yield (27), ligD-deficient A. oryzae NSPlD1 (28) was transformed using a donor DNA plasmid carrying a targeted STS gene. Usually, the knock-in rate ranged from 80% to 90%, much higher than in normal random integrations. GC-MS analysis of headspace volatile compounds from these TFs revealed that nine A. oryzae TFs expressed functional a STS that cyclized FPP into the cyclic products (Fig. 3; see Fig. S1 and S2 in the supplemental material). Sesquiterpenes were identified by retention indices (RI) and comparison of MS fragmentation patterns with those reported in databases or the literature (see Fig. S1 and Table S2 in the supplemental material).
To find out why the remaining eight TFs did not produce sesquiterpenoids, cDNAs of STSs were recovered and sequenced. The results showed that although A. oryzae correctly spliced the vast majority of introns that were computationally predicted by AUGUSTUS (29), some introns were (partially) skipped, thereby generating multiple splicing patterns leading to incomplete protein (e.g., one intron for CpSTS5, -7, -13, -14, and -16 and two introns for CpSTS8 and -17) (Fig. 3; see Fig. S3 and Table S3 in the supplemental material). Following the removal of nonspliced introns in amplified cDNA by PCR-based techniques, CpSTS5m -7, -8, -13, -14, -16, and -17 were expressed by E. coli TFs and yielded terpenes (Fig. 3, S1, and S2; Table S2). Only CpSTS15 did not give any product, although all introns were likely spliced correctly.
Sesquiterpenes produced by CpSTSs include (i) aromadendrene-type sesquiterpenes featuring the 5-7-3 tricyclic skeleton, such as ledene (compound 2) and virifloridol (compound 3) (C-1,C-10 cyclization products) (Fig. 3b), (ii) structurally unique sesquiterpenes such as 6-protoilludene (compound 7), sterpurene (compound 8), pentalenene (compound 9), and β-caryophyllene (compound 10) (C-1,C-11 cyclization products) (Fig. 3c), (iii) cadinene-type sesquiterpenes possessing the 6-6 bicyclic structure, such as δ-cadinene (compound 12) and γ-muurolene, and biosynthetically related ones such as α-cubebene (compound 13) (C-1,C-10 cyclization products) (Fig. 3e and S2), and (iv) linear and monocyclic sesquiterpenes such as α-farnesene (compound 11) and β-elemene (Fig. 3c and S2). Among them, compound 7 and cadinenes are frequently isolated from fungi, and the corresponding STSs have been functionally characterized (2). In contrast, compound 8 produced by CpSTS1 is known as a fungus-specific sesquiterpene, and a sterpurene synthase has not been identified. Successful functional characterization of CpSTS1 enabled us to search putative sterpurene synthases in public database and identify homologs in Psilocybe cyanescens (72% identity, accession no. PPQ78014.1), Gymnopilus dilepis (72%, PPQ64797.1), and Fibularhizoctonia sp. strain CBS 109695 (66%, KZP33092.1), although compound 8 and the related metabolites have not been isolated from them.
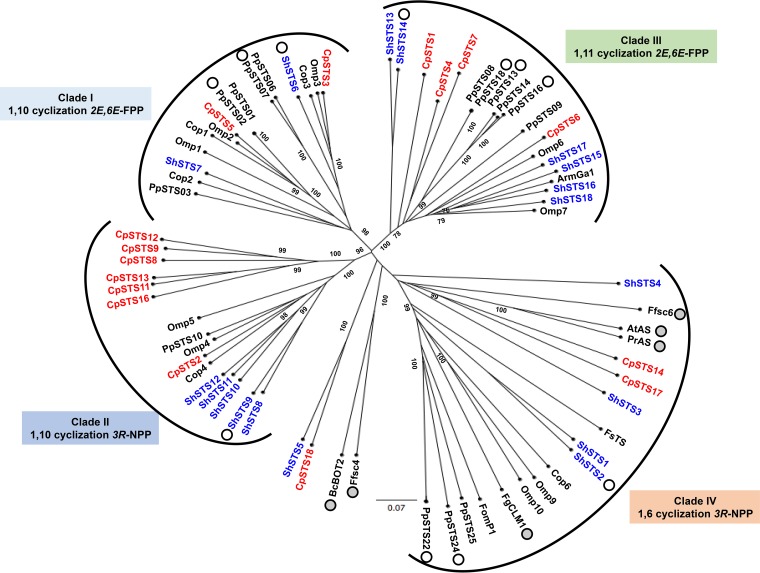
Based on a previous phylogenetic analysis (10), 69 basidiomycete STSs are divided into clades I to IV (Fig. 4). The majority of CpSTS (11/17) characterized in this work groups with fungal STS in clades II and III, which are proposed to catalyze the C-1,C-10 and C-1,C-11 cyclization of FPP, respectively (Fig. 3, 4, and S2). The remaining STS group with clade I (2 STSs), clade IV (2 STSs), and an unclassified clade (1 STS). We used this phylogenetic framework as a guide to investigate the relationship of homologous STSs in the context of sequence homology, gene structures, and chemical structures/reaction mechanisms. The species-specific STSs CpSTS8, -9, -11, -12, -13, and -16 (sequence identity, >40%) belonging to clade II have similar gene structures (Fig. 3a; Table S3) (30), possibly arising by gene duplication of ancestral STS, a well-known evolutionary mechanism in this class of fungi (31). These STSs produced the aromadendrene family members and aristolene (compound 6), which are frequently found as plant volatile compounds (Fig. 3b; see Fig. S4 in the supplemental material), but only one corresponding STS from Citrus (CsSesquiTPS5) has been characterized (32). All of them are most likely biosynthesized via bicyclogermacrene (compound IM1) (30, 32–34). These results suggested a strong relationship of the gene structure and the cyclization mechanism. It should be emphasized that comprehensive analysis of 17 STSs provides direct evidence regarding the gene structure and the enzyme function.
FIG 4.
Phylogenic tree of functionally characterized STSs, except four ShSTSs (white circles) and seven PpSTSs (white circles), derived from basidiomycete fungi. STSs derived from C. pseudo-pinsitus and S. hirsutum are in red and blue, respectively. STSs derived from ascomycete fungi are indicated by gray circles.
(ii) STSs from S. hirsutum.
Previously, 18 STS genes were bioinformatically predicted in the mushroom S. hirsutum, and five STS genes were successfully amplified from cDNA for functional expression in E. coli (11, 35). The remaining STSs were not studied, partially because of cDNA availability limitations. We thought that this suite of STSs from S. hirsutum would be another good test system for our direct gene expression strategy. Of the 18 STS genes (ShSTS1 to -18), we excluded five genes from our study to avoid the expression of ShSTS2, -9, -14, -15, and -6, which showed >60% homology to ShSTS1, -8, -13, and -16 and CpSTS3 (see Table S4 in the supplemental material). GC-MS analysis of culture headspace showed that seven A. oryzae TFs (expressing ShSTS-1, -4, -7, -8, -11, -16, and -17) produced sesquiterpenes such as compounds 7, 12, and 15, β-barbatene, and hirsutene (Fig. 3, S1, and S2; Table S2). As in the case of CpSTSs, sequencing of cDNA showed intron skipping at one (ShSTS5, -12, -13, and -18) or two (ShSTS3 and -10) positions (Fig. 3d). Removal of predicted introns in ShSTS3, -5, -10, -12, -13, and -18 gave functional STSs in E. coli transformants that synthesized terpenes such as compounds 10, 11, 13, 14, and 16 and γ-cadinene (Fig. 3, S1, and S2; Table S2). The metabolite profiles revealed that clade III ShSTSs as well as those of CpSTSs produce structurally unique sesquiterpenes such as compounds 7 to 10 as a single product, while other STSs appear to give multiple biosynthetically related products.
A strong correlation between gene structure and function was also found in Stereum TSs: clade II ShSTS8 to -12 revealed a relatively high sequence identity (>48%), and the gene structures of ShSTS10 to -12 are nearly identical, whereas that of the ShSTS8 gene is likely modified by the insertion of an extra intron (Fig. 3d and 4). These STSs generated cadinene-type sesquiterpenes biosynthesized via the Z,E-germacrenedienyl cation (compound IM2) (Fig. 3e). In addition, a conservation of sequence identity (>54%) and gene structure is observed for clade III ShSTSs ShSTS15 to -18 (Fig. 3c and 4), which produce compound 7 as a characteristic and major sesquiterpene made by mushrooms. The gene structure of ShSTS13 resembles those of clade III ShSTSs except that an extra N-terminal intron is inserted (Fig. 3d). The structural difference between compounds 7 (ShSTS15 to -18) and 10 (ShSTS13) might reflect altered transition states derived from common intermediate compound IM3. When we searched for putative biosynthetic gene clusters surrounding clade II and III CpSTS and ShSTS paralogs/orthologs, we found quite divergent arrangements of putative biosynthetic genes for modifying the terpene scaffold (see Fig. S5 in the supplemental material).
Splicing of basidiomycete genes in A. oryzae.
To gain insights into the splicing patterns of basidiomycete genes in A. oryzae, we analyzed the sequences of spliced and nonspliced introns of the genes investigated in this study (Table S3). Overall, CpSTSs have a slightly smaller mean intron length (56 nucleotides [nt]; size range, 47 to 69 nt) and average number of introns (4 introns) than ShSTSs, with an intron length of 64 nt (size range, 48 to 122 nt) and 5 introns on average. The 5′and 3′splice site consensus sequences (GUxxGU and YAG, respectively) matched those from Cryptococcus neoformans (a basidiomycete fungus) and a Fusarium sp. (an ascomycete) (14, 24). Analysis of branch sites revealed that the majority of introns (82%, 108/132) have a CTNAN motif, while 13% (17/132) contain a TTNAN motif, and for the remaining (7/132), no motif could be determined. The branch site motif appears to be the most important predictor for splicing in A. oryzae, as 96% of introns with a CT.
NAN motifs are spliced in A. oryzae, while only 65% of introns with a TTNAN or 43% of discernible motifs are spliced. Only one spliced intron (CpSTS8 intron1) out of all introns spliced in A. oryzae (132 total) was misspliced in A. oryzae (Fig. S3), demonstrating that introns in basidiomycete genes that are recognized for splicing are faithfully and reliably spliced by A. oryzae. Intron skipping tends to occur at one or two positions in STS genes. A similar pattern was observed for the pleuromutilin genes investigated in this work (Fig. 2).
Estimation of reliability for computational prediction of introns and ORFs.
Comparison of the cDNA sequences of the 30 functional STS sequences characterized in this work with computationally predicted gene structures provides an excellent assessment of the reliability of bioinformatics tools such as AUGUSTUS (29) for basidiomycete gene prediction (see Tables S3 and S5 in the supplemental material). In AUGUSTUS, the accuracy of gene structure predictions depended on the gene model used; predictions with the mushroom Coprinus cinereus model instead of A. oryzae (the expression host) were much more accurate. Even with the mushroom gene model, AUGUSTUS mispredicted 11 out of 30 STS gene structures (Fig. 3 and S3; Table S5). Mispredictions included (i) insertion of an extra intron in CpSTS16, (ii) skipping of an essential intron in ShSTS5, (iii) prediction of shorter or longer introns in CpSTS18 and ShSTS12 and -16, and (iv) misprediction of introns located close to the 3′ ends of CpSTS4 and -13 and ShSTS17 (extra intron) and CpSTS9 (intron skipping) and the 5′ends of CpSTS8 and ShSTS11 (Fig. 5). Most errors concerning ORFs and introns located close to the 3′-terminal ends of the genes can be avoided by manual prediction by considering the amino acid sequences of the known STSs and the gene structures of paralogous STS genes (Fig. 3). Therefore, automated prediction may be useful when these measures are taken. We used AUGUSTUS prediction when we needed to remove a nonspliced intron(s) in A. oryzae expression. We successfully used this strategy to identify and subsequently remove nonspliced introns by A. oryzae in recovered cDNA for expression of active STSs by E. coli TFs (Fig. S1). Consequently, direct expression of basidiomycete genes from genomic DNA in A. oryzae in conjunction with bioinformatics proved to be an effective and reliable strategy to quickly obtain functional NP biosynthetic genes.
FIG 5.

Schematic view of the gene structures of active and mispredicted STSs. The blue bars indicate the mispredicted sites in computationally predicted (AUGUSTUS [26]) gene structures compared to the functional gene structure shown directly above.
Concluding remarks.
In this work, we developed an efficient approach for heterologous expression of basidiomycete NP biosynthetic genes directly from genomic DNA in the genetically tractable and well-studied ascomycete host A. oryzae. We show that direct expression of unspliced genes from genomic DNA from basidiomycetes in A. oryzae results in almost correctly spliced introns and that a combination of computational and manual gene structure prediction can be used to identify and correct nonspliced introns in recovered cDNA by a quick PCR step. Out of 30 STSs expressed in A. oryzae using this method, half of them were functionally expressed, and a simple PCR step yielded functional cDNA for the remaining STSs. Comprehensive analysis of gene structures indicated a strong correlation between gene structure preservation and evolution of enzyme function. Comparison of intron sequences revealed intron features such as branch sites that are less likely to be spliced by an ascomycete such as A. oryzae. This information will aid in the prediction of basidiomycete STS activities, provide insights into the diversification of NP clusters in this class of fungi, and improve computational gene prediction methods. Further, by knowing which introns are less likely to be processed, genomic sequences can be preemptively spot corrected prior to introduction into a versatile, heterologous ascomycete production host, thereby opening the door for the accessing the largely undiscovered NP diversity of basidiomycete mushrooms.
MATERIALS AND METHODS
General.
All reagents commercially supplied were used as received. Column chromatography was carried out on 60N silica gel (Kanto Chemicals). Optical rotations were recorded on Jasco P-2200 digital polarimeter. 1H nuclear magnetic resonance (NMR) spectra were recorded on a Bruker DRX-500 or Bruker AMX-500 spectrometer (500 MHz for 1H NMR). NMR spectra were recorded in CDCl3 (99.8 atom% enriched; Kanto). 1H chemical shifts were reported as δ values based on residual chloroform (7.26 ppm) as a reference. Data are reported as follows: chemical shift, multiplicity (s, singlet; d, doublet; t, triplet; q, quartet; m, multiplet; br, broad), coupling constant (Hz), and integration. GC-MS analyses were conducted with MS-2010 (Shimadzu). Mass spectra were obtained with a Waters Acquity QDa instrument (ESI mode).
Oligonucleotides for PCRs were purchased from Hokkaido System Science Co., Ltd. PCRs were performed with a Bio-Rad S1000 thermal cycler. All PCRs were performed with the KOD-Plus-Neo instrument (Toyobo). The assembly of DNA fragments was performed by using either the In-Fusion Advantage PCR cloning kit (Clontech Laboratories) or HiFi DNA Assembly Master Mix (New England Biolabs).
Strains and culture conditions.
Escherichia coli HST08 was used for cloning, following standard recombinant DNA techniques. E. coli BL21-Gold(DE3) was used for protein expression. Fungal host strains used in this study were A. oryzae NSAR1 (36), a quadruple auxotrophic mutant (niaD− sC− ΔargB adeA−), and A. oryzae NSPlD1 (28), a strain with a highly efficient gene-targeting background (niaD− sC− ΔpyrG ΔligD) for expression. A. oryzae bet123 (27), a transformant producing high levels of betaenone B, was used for genome sequencing.
Genome sequencing and analysis.
Genome sequencing of A. oryzae bet123 was performed by Hokkaido System Science Co., Ltd. (Hokkaido, Japan), with an Illumina HiSeq 2000 system. Read mapping was performed with the programs BWA, SAMtools, GATK, and Picard. The results showed a clear gap between AO090103000023 and AO090103000025 on chromosome 8. This site is tentatively defined as the SC103 region in this study.
Construction of Cas9 plasmid pC9SC103.
The Cas9 plasmid pC9SC103, harboring guide RNA (gRNA) sequences for SC103, was constructed as follows. The gRNA fragments (G-Fr1, U6P-SC103; G-Fr2, SC103-U6T) were amplified from ppAsACas9gwA (26) with the primer set shown in Table S6 in the supplemental material. These primary products were used as a template for the fusion PCR to afford guide RNA sequences flanked by the U6 promoter/terminator set. The PCR product (U6P-guide RNA sequence for SC103-U6T) was then inserted into SmaI-digested ppAsACas9 to construct pC9SC103.
Construction of donor vectors pDP1031 and pDP1032.
The donor vectors pDP1031 and pDP1032, harboring homologous arms of SC103 were constructed as follows. Two DNA fragments, SC103-up and SC103-down, were amplified from genomic DNA of Aspergillus oryzae NSAR1 with primer sets shown in Table S6. Each PCR product was inserted into HindIII/EcoRI-digested pUC19 to produce pUC19-SC103. The DNA fragment harboring primer/terminator sets for overexpression in A. oryzae were amplified from either pTAex3 or pUARA2 with primer sets shown in Table S6. Each PCR product was inserted into BamHI-digested pUC19-SC103 to produce pDP1031 (single primer/terminator set) and pDP1032 (tandem primer/terminator sets).
Bioinformatics analysis.
STS gene annotation, alignment, and gene structure prediction were performed as described previously (11). Briefly, gene predictions were made in AUGUSTUS (29) with different fungal gene models using the genomic region from 10 to 15 kb flanking identified STS ORFs. STS gene predictions were manually aligned with functionally characterized fungal STSs. All sequence alignments and phylogenetic analysis were performed in MEGA7 (37) using MUSCLE (38) for protein alignments. Phylogenetic analysis of fungal STSs (Fig. 2B) was done using the neighbor-joining method with the Poisson correction methods and 500 bootstrap replications (39, 40).
Accession numbers and reference protein sequences are as follows: for basidiomycete STSs, Coprinus cinereus (Cop1 to -4 and Cop6) (XP_001832573, XP_001836556, XP_001832925, XP_001836356, and XP_001832548), Omphalotus olearius (Omp1 to -10), Fomitopsis pinicola (FomPi84944), Stereum hirsutum (Stehi1|159379 [EIM83755.1], Stehi1|128017 [EIM91001.1], Stehi1|25180 [EIM88705.1], Stehi1|64702 [EIM82223.1], and Stehi1|73029 [EIM91236.1]), Armillaria gallica (ArmGa1) (P0DL13), and Postia placenta (PpSTS01 [BBD74517.1], PpSTS02 [BBD74518.1], PpSTS03 [BBD74519.1], PpSTS06 [BBD74520.1], PpSTS07 [BBD74521.1], PpSTS08 [BBD74522.1], PpSTS09 [BBD74523.1], PpSTS10 [BBD74524.1], PpSTS13 [BBD74525.1], PpSTS14 [BBD74526.1], PpSTS16 [BBD74527.1], PpSTS18 [BBD74528.1], PpSTS22 [BBD74529.1], PpSTS24 [BBD74530.1], PpSTS25 [BBD74531.1], and PpSTS29 [BBD74532.1]); and for ascomycete STSs, Fusarium fujikori (Ffsc4) (HF563560.1) and Ffsc6 (HF563561.1), Fusarium gramineareum (FgCLM1) (GU123140), Aspergillus terreus (atAS) (Q9UR08), Penicillium roqueforti (prAS) (W6Q4Q9), and Botrytis cinerea (BcBOT2) (AAQ16575.1).
Construction of A. oryzae expression plasmids for ple and STS genes.
Pleuromutin biosynthetic genes (ple1, -3, -4, -5, and -6) were amplified from the genomic DNA of C. pseudo-pinsitus ATCC 20527 (22) with primer sets shown in Table S6. Each PCR product was inserted into the appropriate restriction sites (site 1 and/or site 2) of pDP1031 or pDP1032 to construct the expression plasmids pDP1031-ple5 (KpnI site), pDP1032-ple6 (NheI), pDP1032-ple3 (KpnI)-ple4 (SpeI), and pDP1032-ple7 (KpnI)-ple1 (SpeI).
Sesquiterpene synthase genes were amplified from the genomic DNA of C. pseudo-pinsitus ATCC 20527 and S. hirsutum FP-91666 S1 (11) with primer sets shown in Table S6. Each PCR product was then inserted into the appropriate restriction sites of expression vectors to construct the following expression plasmids: pTAex3-CpSTS1, pUARA2-CpSTS2, pUARA2-CpSTS3, pTAex3-CpSTS4, pDP1031-CpSTS5, pDP1031-CpSTS6, pDP1031-CpSTS7, pDP1031-CpSTS8, pDP1031-CpSTS9, pDP1031-CpSTS10, pDP1031-CpSTS11, pDP1031-CpSTS12, pUSA2-CpSTS13, pDP1031-CpSTS14, pDP1031-CpSTS15, pUSA2-CpSTS16, pDP1031-CpSTS17, pDP1031-CpSTS18, pDP1032-ShSTS1/18, pDP1031-ShSTS3, pDP1031-ShSTS4, pDP1031-ShSTS5, pDP1031-ShSTS7, pDP1031-ShSTS8, pDP1031-ShSTS10, pDP1032-ShSTS11/16, pDP1031-ShSTS12, pDP1031-ShSTS13, and pDP1031-ShSTS17.
Transformation of Aspergillus oryzae by the genome-editing method.
Most transformants used in this study was constructed by the genome-editing method. A spore suspension of A. oryzae NSPlD1 (1.0 × 108 cells) was inoculated into 100 ml of CD medium (0.3% NaNO3, 0.2% KCl, 0.1% K2HPO4, 0.05% MgSO4·7H2O, 2% dextrin, 0.002% FeSO4·7H2O, 0.15% methionine, 0.488% uracil, 0.2% uridine, pH 5.5) supplemented with appropriate nutrients. After 3 days of incubation at 30°C and 200 rpm, mycelia were collected by filtration and washed with water. Protoplasting was performed using Yatalase (TaKaRa; 5.0 mg ml−1) in solution 1 (0.8 mM NaCl, 10 mM NaH2PO4, pH 6.0) at 30°C for 2 h. Protoplasts were centrifuged at 2,000 rpm (Beckman JLA10.500 rotor) for 5 min and washed with 0.8 M NaCl solution. Protoplasts were then adjusted to 2.0 × 108 cells/ml by adding solution 2 (0.8 M NaCl, 10 mM CaCl2, 10 mM Tris-HCl, pH 8.0) and solution 3 (40% [wt/vol] polyethylene glycol [PEG] 4000, 50 mM CaCl2, 50 mM Tris-HCl, pH 8.0) in a 4/1 volume ratio. To the protoplast solution (200 μl) was added a Cas9 plasmid (2 μg) and a donor plasmid (5 μg). The aliquot was incubated on ice for 20 min and then solution 3 (1 ml) added to the aliquot. After 20 min of incubation at room temperature, solution 2 (10 ml) was added to the mixtures, and the mixture was centrifuged at 2,000 rpm (Beckman JLA10.500 rotor) for 5 min. After decantation, the residue was diluted with solution 2 (500 μl), and the mixture (100 μl) was poured onto a CD agar plate (1.5%) supplemented with 4.7% NaCl and then overlaid with the soft-top CD agar (0.6%) containing 21.8% sorbitol. The plates were incubated at 30°C for 3 to 7 days.
Analysis of the metabolites from A. oryzae TFs harboring an STS gene.
Mycelia of each transformant (TF) were inoculated into MPY medium (1 ml) containing 20 mM uridine and 0.2% uracil in a 10-ml test tube. Each culture was incubated at 30°C for 4 days. The volatile organic compounds were extracted with an SPME fiber (50/30 μm DVB/CAR/PDMS; Stableflex, 24Ga, manual holder), which was conditioned by inserting it into the GC injector to prevent contamination, for 30 min at room temperature. After extraction, the fiber was pulled into the needle sheath, and the SPME device was removed from the vial and then inserted into the injection port of a QP2010 GC-MS apparatus (Shimadzu, Kyoto, Japan) with an HP-5 capillary column (0.32 mm by 30 m, 0.25-μm film thickness; J&W Scientific, Folsom, CA). Each sample was injected onto the column at 60°C in the splitless mode. The column temperature was increased by 4°C min−1 to 180°C. The flow rate of the helium carrier gas was 0.66 ml min−1. This procedure is referred to as method A.
Isolation of sesquiterpenes.
(i) Sterpurene. Mycelia of A. oryzae-CpSTS1 were inoculated into a solid medium containing polished rice (100 g) and adenine (10 mg) in 500-ml Erlenmeyer flasks. Each culture was incubated at 30°C for 12 days. After extraction with ethyl acetate, the extract was concentrated in vacuo to produce crude extracts. The crude extracts were purified by silica gel column chromatography (hexane) to give sterpurene (20.0 mg from 1.3 kg of rice medium from A. oryzae-CpSTS1). The NMR data are in good agreement with the reported data (see Fig. S6 in the supplemental material) (41).
(ii) 9-Alloaromadendrene. Mycelia of A. oryzae-CpSTS11 were inoculated into MPY medium (30 ml) containing 20 mM uridine and 0.2% uracil in 200-ml Erlenmeyer flasks. Each culture was incubated at 30°C for 5 days. The crude extracts were purified by silica gel column chromatography (hexane) to yield 9-alloaromadendrene (1.0 mg from 150 ml of MPY medium containing uridine and uracil). The NMR data are in good agreement with the reported data (Fig. S6) (42).
cDNA preparation. Each transformant was grown on MPY medium containing 20 mM uridine and 0.2% uracil for 3 to 5 days at 30°C. Total RNA was extracted from each dried mycelium using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions and then treated with DNase I (Life Technologies) for reverse transcription. cDNA was synthesized with the PrimeScript II first-strand cDNA synthesis kit (TaKaRa) using the oligo(dT) primer according to the manufacturer’s instructions. The cDNA was used as a template for PCRs for direct sequencing and subcloning of each STS gene into the pColdI vector.
Estimation of sequences of functionally active STSs.
cDNA sequences derived from A. oryzae transformant were aligned with the predicted sequence by AUGUSTUS to identify the nonspliced introns in cDNA sequences. Alignments were computed by using ClustalW. The resultant transcripts of putative intronless STSs were then manually examined for the presence and proper alignment of the conserved motifs characteristic of sesquiterpene synthases.
Construction of E. coli expression plasmids.
Intronless DNA clones were prepared from PCR-based removal of the nonspliced introns (Fig. 3 and 4) as follows. Two DNA fragments (upstream and downstream regions of the nonspliced intron) were separately amplified from cDNA with primers set shown in Table S6. Each PCR product was inserted into NdeI-, EcoRI-, or KpnI-digested pColdI to construct the following expression plasmids; pColdI-CpSTS5, pColdI-CpSTS7, pColdI-CpSTS8, pColdI-CpSTS13, pColdI-CpSTS14, pColdI-CpSTS16, pColdI-CpSTS17, pColdI-ShSTS3, pColdI-ShSTS5, pColdI-ShSTS10, pColdI-ShSTS12, pColdI-ShSTS13, and pColdI-ShSTS18.
Analysis of the metabolites from E. coli TFs.
The constructed plasmids were separately introduced into E. coli BL21-Gold(DE3) for overexpression. The transformant (TF) was grown at 37°C at an optical density at 600 nm (OD600) of ∼0.6 in a 500-ml flask. After cooling at 4°C, isopropyl β-d-thiogalactopyranoside (0.1 mM) was added to the culture. After incubation at 16°C for 17 h, the volatile organic compounds were extracted with an SPME fiber (50/30 μm DVB/CAR/PDMS; Stableflex, 24Ga, manual holder) for 30 min at room temperature. The extracts were then analyzed with a GC-MS apparatus by method A.
Accession number(s).
The sequences of the sesquiterpene synthase genes found in C. pseudo-pinsitus ATCC 20527 have been deposited in the DNA Data Bank of Japan (DDBJ) under accession numbers LC436345 to LC436362.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by Japan Society for the Promotion of Science grants-in-aid JP15H01835 to H.O., JP16H03277 and JP16H06446 to A.M., and JP18K14342 to C.L.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00409-19.
REFERENCES
- 1.Lorenzen K, Anke T. 1998. Basidiomycetes as a source for new bioactive natural products. Curr Org Chem 2:329–364. [Google Scholar]
- 2.Quin MB, Flynn CM, Schmidt-Dannert C. 2014. Traversing the fungal terpenome. Nat Prod Rep 31:1449–1473. doi: 10.1039/c4np00075g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kavanagh F, Hervey A, Robbins WJ. 1951. Antibiotic substances from Basidiomycetes. VIII. Pleurotus multilus (Fr.) Sacc. and Pleurotus passeckerianus Pilat. Proc Natl Acad Sci U S A 37:570–574. doi: 10.1073/pnas.37.9.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mcmorris TC, Anchel M. 1963. Structures of basidiomycete metabolites illudin S and illudin M. J Am Chem Soc 85:831–832. doi: 10.1021/ja00889a052. [DOI] [PubMed] [Google Scholar]
- 5.Midland SL, Izac RR, Wing RM, Zaki AI, Munnecke DE, Sims JJ. 1982. Melleolide, a new antibiotic from Armillaria mellea. Tetrahedron Lett 23:2515–2518. doi: 10.1016/S0040-4039(00)87383-9. [DOI] [Google Scholar]
- 6.Kawagishi H, Shimada A, Hosokawa S, Mori H, Sakamoto H, Ishiguro Y, Sakemi S, Bordner J, Kojima N, Furukawa S. 1996. Erinacines E, F, and G, stimulators of nerve growth factor (NGF)-synthesis, from the mycelia of Hericium erinaceum. Tetrahedron Lett 37:7399–7402. doi: 10.1016/0040-4039(96)01687-5. [DOI] [Google Scholar]
- 7.Martin F, Cullen D, Hibbett D, Pisabarro A, Spatafora JW, Baker SE, Grigoriev IV. 2011. Sequencing the fungal tree of life. New Phytol 190:818–821. doi: 10.1111/j.1469-8137.2011.03688.x. [DOI] [PubMed] [Google Scholar]
- 8.Stadler M, Hoffmeister D. 2015. Fungal natural products—the mushroom perspective. Front Microbiol 6:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agger S, Lopez-Gallego F, Schmidt-Dannert C. 2009. Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Mol Microbiol 72:1181–1195. doi: 10.1111/j.1365-2958.2009.06717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wawrzyn GT, Quin MB, Choudhary S, Lopez-Gallego F, Schmidt-Dannert C. 2012. Draft genome of Omphalotus olearius provides a predictive framework for sesquiterpenoid natural product biosynthesis in Basidiomycota. Chem Biol 19:772–783. doi: 10.1016/j.chembiol.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quin MB, Flynn CM, Wawrzyn GT, Choudhary S, Schmidt-Dannert C. 2013. Mushroom hunting by using bioinformatics: application of a predictive framework facilitates the selective identification of sesquiterpene synthases in Basidiomycota. Chembiochem 14:2480–2491. doi: 10.1002/cbic.201300349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ichinose H, Kitaoka T. 2018. Insight into metabolic diversity of the brown-rot basidiomycete Postia placenta responsible for sesquiterpene biosynthesis: semi-comprehensive screening of cytochrome P450 monooxygenase involved in protoilludene metabolism. Microb Biotechnol 11:952–965. doi: 10.1111/1751-7915.13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burkhardt I, Kreuzenbeck N, Beemelmanns C, Dickschat JS. 2019. Mechanistic characterization of three sesquiterpene synthases from the termite-associated fungus Termitomyces. Org Biomol Chem 17:3348–3355. doi: 10.1039/c8ob02744g. [DOI] [PubMed] [Google Scholar]
- 14.Kupfer DM, Drabenstot SD, Buchanan KL, Lai HS, Zhu H, Dyer DW, Roe BA, Murphy JW. 2004. Introns and splicing elements of five diverse fungi. Eukaryot Cell 3:1088–1100. doi: 10.1128/EC.3.5.1088-1100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujii R, Minami A, Tsukagoshi T, Sato N, Sahara T, Ohgiya S, Gomi K, Oikawa H. 2011. Total biosynthesis of diterpene aphidicolin, a specific inhibitor of DNA polymerase alpha: heterologous expression of four biosynthetic genes in Aspergillus oryzae. Biosci Biotechnol Biochem 75:1813–1817. doi: 10.1271/bbb.110366. [DOI] [PubMed] [Google Scholar]
- 16.Tagami K, Liu CW, Minami A, Noike M, Isaka T, Fueki S, Shichijo Y, Toshima H, Gomi K, Dairi T, Oikawa H. 2013. Reconstitution of biosynthetic machinery for indole-diterpene paxilline in Aspergillus oryzae. J Am Chem Soc 135:1260–1263. doi: 10.1021/ja3116636. [DOI] [PubMed] [Google Scholar]
- 17.Ye Y, Minami A, Mandi A, Liu C, Taniguchi T, Kuzuyama T, Monde K, Gomi K, Oikawa H. 2015. Genome mining for sesterterpenes using bifunctional terpene synthases reveals a unified intermediate of di/sesterterpenes. J Am Chem Soc 137:11846–11853. doi: 10.1021/jacs.5b08319. [DOI] [PubMed] [Google Scholar]
- 18.Heneghan MN, Yakasai AA, Halo LM, Song Z, Bailey AM, Simpson TJ, Cox RJ, Lazarus CM. 2010. First heterologous reconstruction of a complete functional fungal biosynthetic multigene cluster. Chembiochem 11:1508–1512. doi: 10.1002/cbic.201000259. [DOI] [PubMed] [Google Scholar]
- 19.Liu C, Tagami K, Minami A, Matsumoto T, Frisvad JC, Suzuki H, Ishikawa J, Gomi K, Oikawa H. 2015. Reconstitution of biosynthetic machinery for the synthesis of the highly elaborated indole diterpene penitrem. Angew Chem Int ed Engl 54:5748–5752. doi: 10.1002/anie.201501072. [DOI] [PubMed] [Google Scholar]
- 20.Ye Y, Minami A, Igarashi Y, Izumikawa M, Umemura M, Nagano N, Machida M, Kawahara T, Shin-Ya K, Gomi K, Oikawa H. 2016. Unveiling the biosynthetic pathway of the ribosomally synthesized and post-translationally modified peptide ustiloxin B in filamentous fungi. Angew Chem Int Ed Engl 55:8072–8075. doi: 10.1002/anie.201602611. [DOI] [PubMed] [Google Scholar]
- 21.Matsuda Y, Iwabuchi T, Wakimoto T, Awakawa T, Abe I. 2015. Uncovering the unusual D-ring construction in terretonin biosynthesis by collaboration of a multifunctional cytochrome P450 and a unique isomerase. J Am Chem Soc 137:3393–3401. doi: 10.1021/jacs.5b00570. [DOI] [PubMed] [Google Scholar]
- 22.Yamane M, Minami A, Liu C, Ozaki T, Takeuchi I, Tsukagoshi T, Tokiwano T, Gomi K, Oikawa H. 2017. Biosynthetic machinery of diterpene pleuromutilin isolated from basidiomycete fungi. Chembiochem 18:2317–2322. doi: 10.1002/cbic.201700434. [DOI] [PubMed] [Google Scholar]
- 23.Bailey AM, Alberti F, Kilaru S, Collins CM, de Mattos-Shipley K, Hartley AJ, Hayes P, Griffin A, Lazarus CM, Cox RJ, Willis CL, O'Dwyer K, Spence DW, Foster GD. 2016. Identification and manipulation of the pleuromutilin gene cluster from Clitopilus passeckerianus for increased rapid antibiotic production. Sci Rep 6:25202. doi: 10.1038/srep25202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phasha MM, Wingfield BD, Coetzee MPA, Santana QC, Fourie G, Steenkamp ET. 2017. Architecture and distribution of introns in core genes of four Fusarium species. G3 (Bethesda) 7:3809–3820. doi: 10.1534/g3.117.300344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dickschat JS. 2014. Capturing volatile natural products by mass spectrometry. Nat Prod Rep 31:838–861. doi: 10.1039/c3np70080a. [DOI] [PubMed] [Google Scholar]
- 26.Katayama T, Tanaka Y, Okabe T, Nakamura H, Fujii W, Kitamoto K, Maruyama J. 2016. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol Lett 38:637–642. doi: 10.1007/s10529-015-2015-x. [DOI] [PubMed] [Google Scholar]
- 27.Ugai T, Minami A, Fujii R, Tanaka M, Oguri H, Gomi K, Oikawa H. 2015. Heterologous expression of highly reducing polyketide synthase involved in betaenone biosynthesis. Chem Commun (Camb) 51:1878–1881. doi: 10.1039/c4cc09512j. [DOI] [PubMed] [Google Scholar]
- 28.Maruyama J, Kitamoto K. 2008. Multiple gene disruptions by marker recycling with highly efficient gene-targeting background (ΔligD) in Aspergillus oryzae. Biotechnol Lett 30:1811–1817. doi: 10.1007/s10529-008-9763-9. [DOI] [PubMed] [Google Scholar]
- 29.Hoff KJ, Stanke M. 2013. WebAUGUSTUS—a web service for training AUGUSTUS and predicting genes in eukaryotes. Nucleic Acids Res 41:W123–W128. doi: 10.1093/nar/gkt418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller DJ, Allemann RK. 2012. Sesquiterpene synthases: passive catalysts or active players?. Nat Prod Rep 29:60–71. doi: 10.1039/c1np00060h. [DOI] [PubMed] [Google Scholar]
- 31.Syed K, Shale K, Pagadala NS, Tuszynski J. 2014. Systematic identification and evolutionary analysis of catalytically versatile cytochrome P450 monooxygenase families enriched in model basidiomycete fungi. PLoS One 9:e86683. doi: 10.1371/journal.pone.0086683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alquezar B, Rodriguez A, de la Pena M, Pena L. 2017. Genomic analysis of terpene synthase family and functional characterization of seven sesquiterpene synthases from Citrus sinensis. Front Plant Sci 8:1481. doi: 10.3389/fpls.2017.01481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chou WKW, Fanizza I, Uchiyama T, Komatsu M, Ikeda H, Cane DE. 2010. Genome mining in Streptomyces avermitilis: cloning and characterization of SAV_76, the synthase for a new sesquiterpene, avermitilol. J Am Chem Soc 132:8850–8851. doi: 10.1021/ja103087w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dickschat JS, Martens T, Brinkhoff T, Simon M, Schulz S. 2005. Volatiles released by a Streptomyces species isolated from the North Sea. Chem Biodiv 2:837–865. doi: 10.1002/cbdv.200590062. [DOI] [PubMed] [Google Scholar]
- 35.Flynn CM, Schmidt-Dannert C. 2018. Sesquiterpene synthase—3-hydroxy-3-methylglutaryl coenzyme A synthase fusion protein responsible for hirsutene biosynthesis in Stereum hirsutum. Appl Environ Microbiol 84:e00036-18. doi: 10.1128/AEM.00036-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin FJ, Maruyama J, Juvvadi PR, Arioka M, Kitamoto K. 2004. Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae. FEMS Microbiol Lett 239:79–85. doi: 10.1016/j.femsle.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 37.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. [DOI] [PubMed] [Google Scholar]
- 40.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 41.Ayer WA, Saeedi-Ghomi MH. 1981. 1-Sterpurene-3,12,14-triol and 1-sterpurene, metabolites of silver-leaf disease fungus Stereum purpureum. Can J Chem 59:2536–2538. doi: 10.1139/v81-364. [DOI] [Google Scholar]
- 42.Friedel HD, Matusch R. 1987. Neue aromadendran-derivate aus tolu-balsam. Helv Chim Acta 70:1753–1759. doi: 10.1002/hlca.19870700711. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.