Abstract
Malus baccata is one of four wild apple species that can hybridize with the cultivated apple species (Malus domestica). It is widely used in high-latitude apple-producing areas as a rootstock and breeding resource because of its disease resistance, and cold tolerance. A lack of a reference genome has limited the application of M. baccata for apple breeding. We present a draft reference genome for M. baccata. The assembled sequence consisting of 665 Mb, with a scaffold N50 value of 452 kb, included transposable elements (413 Mb) and 46,114 high-quality protein-coding genes. According to a genetic map derived from 390 sibling lines, 72% of the assembly and 85% of the putative genes were anchored to 17 linkage groups. Many of the M. baccata genes under positive selection pressure were associated with plant–pathogen interaction pathways. We identified 2,345 Transcription factor-encoding genes in 58 families in the M. baccata genome. Genes related to disease defense and cold tolerance were also identified. A total of 462 putative nucleotide-binding site (NBS)-leucine-rich-repeat (LRR) genes, 177 Receptor-like kinase (RLK) and 51 receptor-like proteins (RLP) genes were identified in this genome assembly. The M. baccata genome contained 3978 cold-regulated genes, and 50% of these gene promoter containing DREB motif which can be induced by CBF gene. We herein present the first M. baccata genome assembly, which may be useful for exploring genetic variations in diverse apple germplasm, and for facilitating marker-assisted breeding of new apple cultivars exhibiting resistance to disease and cold stress.
Keywords: Siberian crab apple (Malus baccata), genome sequencing, resistance gene analogs (RGAs), cold-related genes
Apple is one of the most extensively cultivated temperate zone tree fruits, and is popular among consumers worldwide. Global apple production has rapidly increased in recent years, reaching about 89 million tons in 2016 (FAOSTAT: http://www.fao.org/faostat). Apple diseases and frost damage can cause significant decreases in yield. Developing apple varieties exhibiting cold tolerance and pathogen resistance is important for ensuring apple fruits can continue to be produced in countries that experience cold conditions and for overcoming the adverse effects of an increasing variety of diseases and climate change. The resistance to diseases and cold stress varies considerably among different apple species, with wild apple species exhibiting very good disease resistance and cold tolerance (Volk et al. 2015). Therefore, incorporating wild apple genetic resources into apple breeding programs to select disease- and cold-resistant varieties may be relevant for sustainable apple production.
Because apple fruits are a perennial crop, breeding new varieties is time consuming and labor intensive (Peace and Norelli 2009). Genome data and marker-assisted selection can greatly accelerate the breeding process (Badenes et al. 2016). Many perennial fruit tree genomes have recently been sequenced (Velasco et al. 2010; Zhang et al. 2012; Wu et al. 2013; International Peach Genome Initiative et al. 2013; Chagné et al. 2014; Li et al. 2016). Moreover, genome re-sequencing provides valuable information for facilitating marker-assisted breeding. However, the divergence between wild relatives and cultivated plant species is likely considerable. Consequently, genomic regions of interest in a wild relative may be absent in the corresponding domesticated crop. Additionally, the mapping of DNA sequences present only in wild relatives requires de novo assembly rather than resequencing. Many wild crop species genomes have recently been sequenced, and the molecular mechanisms regulating stress resistance have been analyzed and investigated (Bolger et al. 2014; Wang et al. 2014; Zhang et al. 2014; Aversano et al. 2015; Wu et al. 2016; Xu et al. 2017). However, the genomes of wild apple species have not been published, and this lack of genome data inhibits the progress of fruit tree breeding, especially regarding the introduction of unique characteristics from wild species to cultivated varieties (e.g., cold tolerance and disease resistance).
Malus baccata is one of four wild apple species that can be freely hybridized with cultivated apple varieties (Malus domestica) (Cornille et al. 2014). Malus baccata is widely used in high-latitude apple-producing areas as a rootstock because of its excellent environmental adaptability and resistance to cold stress (Volk et al. 2015). It is also widely distributed throughout China (Figure 1A) in habitats that overlap the apple-producing areas (Liguo and Tao 2003). Thus, M. baccata is an important germplasm resource for apple and apple rootstock breeding. We herein describe a high-quality draft genome sequence of the diploid wild apple species, Malus baccata (L.) Borkh ‘Shandingzi’. We compared the M. baccata genome architecture with that of cultivated apple to screen for genetic signatures of cold tolerance or pathogen resistance. This draft assembly revealed insights into the apple genes underlying cold adaptation and pathogen resistance. The presented data may benefit fundamental research involving the characterization of stress adaptations in fruit trees, but may also be relevant for targeting candidate genes for future breeding programs.
Figure 1.
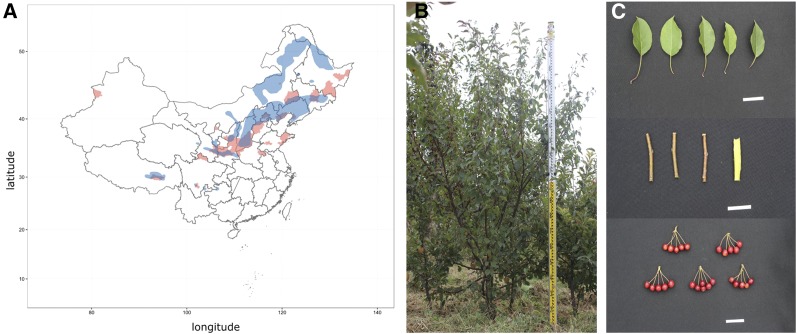
M. baccata plant and its distribution. (A) Distribution of apple-producing areas (red) and M. baccata natural habitats (blue) in China. (B) M. baccata plant. (B) Plant tissues of M. baccata. 1, leaf; 2; branch; 3, fruit.
Materials and Methods
Plant materials and sequencing
A Malus baccata tree from Taigu town, Shanxi province, China (accession No. Y-B094) was used for whole-genome shotgun sequencing. The tree was planted in the Apple Demonstration Nursery of Yangling Modern Agriculture Technology Park (Northwest Agriculture & Forestry University), Shaanxi province, China (34° 52′ N, 108° 7′ E) (Figure 1B and Figure 1C). The root, fruit, phloem, and leaf tissues underwent a transcriptome analysis to assess the annotated genes and evaluate genome quality. All harvested tissues were immediately frozen in liquid nitrogen and stored at −80° until used for DNA or RNA extraction.
Genomic DNA extracted from fresh leaves using a modified CTAB method (Dellaporta et al. 1983) was used to construct paired-end libraries. Nineteen paired-end libraries were prepared for sequencing the M. baccata genome. These included nine paired-end libraries with an insert of 200, 500, and 800 nt and 10 mate-pair libraries with insert sizes of 2, 5, 10, and 20 kb. All libraries were constructed following the instructions provided by Illumina. After filtering, 173.61 Gb high-quality clean data were retrieved (248-fold genome sequence coverage) (Table S2). Total RNA was isolated from each sample according to an SDS–phenol method (Hu et al. 2002). After an agarose gel electrophoresis step, suitable fragments were selected as templates for a polymerase chain reaction (PCR) amplification. During a quality control step, an Agilent 2100 Bioanalyzer and an ABI StepOnePlus Real-Time PCR system were used to quantify and assess the quality of the sample libraries. Finally, the constructed libraries were sequenced with an Illumina HiSeq 2000 system (BGI, Shenzhen, China).
k-mer analysis of the M. baccata genome
To estimate genome size and heterozygosity, 30.7 Gb high-quality short-insert reads underwent a k-mer analysis. The 17-mer frequency distribution derived from the sequencing reads was plotted (Figure S1). The peak for the 17-mer distribution was about 33, and the total k-mer count was 25,700,099,451. Thus, the genome size was estimated as 778 Mb (Table S1). A small peak was detected at 1/2 Peak-depth. Therefore, we used simulated heterozygosity rates for the wild apple genome, and conducted 17-mer analyses.
Genome assembly and evaluation of quality
Genomic DNA isolated from M. baccata leaf material was used to construct nine paired-end libraries and 10 mate-pair libraries. All libraries were sequenced using the Illumina HiSeq 2000 sequencing platform and assembled using the SOAPdenovo program (version 2.04.4) (http://soap.genomics.org.cn) (Luo et al. 2012). Mate-pair reads were used to construct super scaffolds with the SSPACE program (version 2.0) (Boetzer et al. 2011), with sequence gaps filled with GapCloser (version 1.10) (Luo et al. 2012).
A high-resolution genetic map with 3,137 SNP markers was created using a mapping population of 390 F1 progenies from a cross between M. baccata ‘Shandingzi’ and M. domestica ‘Danxia’ and restriction site-associated DNA sequencing technology. These markers were analyzed and filtered with Joinmap 4.1 (Ooijen 2006). Genome integrity was assessed using CEGMA (version 2.5) (Parra et al. 2007) and BUSCO (version 2.0) (Simão et al. 2015).
Gene and repeat annotations
We used de novo and homology-based approaches to analyze the repetitive elements in the M. baccata genome. The de novo method of annotating repetitive elements involved the RepeatModeler software (Smit and Hubley 2008). Tandem repeats in the genome were annotated with TRF software (version 4.04) (Benson 1999). Transposable elements were identified with a homology-based approach using RepeatMasker and RepeatProteinMask (version 4.0) (http://www.repeatmasker.org/) (Smit et al. 2013) as well as the RepBase database (version 16.10) (Jurka et al. 2005).
To annotate non-coding RNA, tRNA scan-SE software was used to detect tRNA sequences in the genome according to tRNA structural characteristics. The rRNAs were detected by aligning sequences against known plant rRNA sequences with the BLASTN tool. Using the Rfam family covariance model and Rfam’s INFERNAL software (Griffiths-Jones et al. 2005), we predicted the miRNA and snRNA sequence details for the M. baccata genome.
We used both homology-based and de novo methods to predict genes. Augustus (Stanke et al. 2006), GenScan (Burge and Karlin 1997), and glimmerHMM (Majoros et al. 2004) were applied for the de novo prediction of genes based on a repeat-masked genome. For the homology-based prediction, gene sets from M. domestica (Velasco et al. 2010), P. bretschneideri (Wu et al. 2013), P. persica (International Peach Genome Initiative et al. 2013), F. vesca (Shulaev et al. 2011), P. mume (Zhang et al. 2012) and A. thaliana (Arabidopsis Genome Initiative 2000), were mapped onto the assembled M. baccata genome using TBLASTN (Altschul et al. 1990). GeneWise 2.2.0 (Birney et al. 2004) was then used to predict gene structures and define gene models based on the complete regions. The complementary gene sets from homology-based and de novo predictions were merged to produce a non-redundant reference gene set using GLEAN (http://sourceforge.net/projects/glean-gene/). The RNA-seq data for the four analyzed tissues were also applied to improve the gene annotations. Moreover, the RNA-seq data were mapped to the assembled genome using TopHat (Trapnell et al. 2012), and the transcriptome-based gene structures were obtained using Cufflinks (http://cufflinks.cbcb.umd.edu/) (Trapnell et al. 2012). The predicted transcripts were used to complement the GLEAN gene set or were integrated as isoforms. We then used the Cuffcompare program (Trapnell et al. 2012) to compare the gene set with the previous GLEAN gene set and obtain the final non-redundant gene set.
The proteins encoded in the final non-redundant gene set were functionally annotated according to BLAST searches (E-value cutoff 1 × 10−5) of the InterproScan (Zdobnov and Apweiler 2001), SwissProt (Bairoch and Apweiler 2000), and TrEMBL (Bairoch and Apweiler 2000) databases. The pathways enriched among the genes were determined by identifying the best hit in the KEGG database (release 76) (Kanehisa and Goto 2000). We then obtained GO IDs from the corresponding InterPro entries.
Analysis of genome evolution
The Ka/Ks ratio was calculated with the KaKs_Calculator program (Zhang et al. 2006). Gene sets from M. domestica (NCBI version)(Velasco et al. 2010), P. bretschneideri (Wu et al. 2013), P. persica (International Peach Genome Initiative et al. 2013), F. vesca (Shulaev et al. 2011), P. mume (Zhang et al. 2012), A. thaliana (Arabidopsis Genome Initiative 2000), C. papaya (Ming et al. 2008), P. trichocarpa (Tuskan et al. 2006), and V. vinifera (Jaillon et al. 2007) were used for analyses of genome evolution. Gene clusters, phylogenetic relationships, estimated divergence time, and collinearity and gene family expansion/contraction were analyzed using MCScanX (Wang et al. 2012), MrBayes (Huelsenbeck and Ronquist 2001), the MCMCTree program of the PAML package (Yang 2007), and CAFÉ software (De Bie et al. 2006), respectively.
Identification of transcription factors, resistance gene analogs and cold-resistance genes
We searched for consensus transcription factors (TFs) in M. baccata using PlantTFDB (http://planttfdb.cbi.pku.edu.cn/) (Jin et al. 2014) in HMMER3.0 (http://hmmer.org/). The TFs were classified according to the consensus rules, including the required and prohibited protein domains for each TF gene family summarized on the PlantTFDB website. Accordingly, we predicted TFs for M. domestica (Velasco et al. 2010), P. bretschneideri (Wu et al. 2013), P. persica (International Peach Genome Initiative et al. 2013), and F. vesca (Shulaev et al. 2011).
We identified resistance gene analogs (RGAs) based on differences in their domains (2015). The TIR, NBS, LRR, and kinase domains were analyzed with hmmsearch in HMMER 3.0, with default thresholds for the Pfam database (http://www.prgdb.org) (Finn et al. 2014). The CC, SP, and TM motifs were identified with the paircoil2 program (http://www.cbs.dtu.dk/services/TMHMM/) (McDonnell et al. 2006), while SignalP motifs were identified with the SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/) (Nielsen 2017). We mapped the R gene markers from online resources (http://www.hidras.unimi.it/ and https://www.rosaceae.org/species/malus/all) against the M. baccata genome (e-value 1e-5; matched bases ≥ 50 bp; identity ≥ 80%).
To annotate putative cold resistance genes in M. baccata, a set of reference proteins were selected from A. thaliana. In detail, 81 proteins annotated with the Gene Ontology term cold acclimation (CA), 40 proteins annotated as cellular response to cold (CRC), and 520 proteins as a response to cold (RC) were selected from TAIR10 GO annotation (https://www.arabidopsis.org/). We analyzed all M. baccata protein by BLASTP search using the threshold value as following: e-value < le-30, identity > 50%, query coverage > 80%.
Data Availability
The genome assembly have been deposited under CNGB Project ID CNA0002537 (https://db.cngb.org/search/project/CNP0000421/). The meta data for the genetic map can be also found in https://db.cngb.org/search/project/CNP0000421/. The genomic raw reads are available via NCBI SRR7248834, SRR7248835, SRR7248837, SRR7248847, SRR7248838 to SRR7248844, SRR7248849 to SRR7248858, SRR7248875 to SRR7248882, and the raw transcriptomic reads are available at NCBI SRR8156047 to SRR8156050. Supplemental material available at FigShare: https://doi.org/10.25387/g3.7523549.
Results and Discussion
Genome sequencing and de novo assembly
Our k-mer analysis indicated the M. baccata genome comprises nearly 779 Mb (Table S1), which was within the flow cytometry data range (709.05–792.18 Mb) (Korban et al. 2009). To estimate the heterozygosity of the genome, we approximated the k-mer distribution with simulated heterozygous genome sequences, which revealed that the best fit for the real k-mer distribution was a simulated k-mer distribution (k represents the chosen length of substrings) with 1.2% heterozygosity (Figure S1). The scaffolds totaling 719 Mb accounted for approximately 92.32% of the estimated M. baccata genome. The result statistics of our final assembly showed that the contig N50 and scaffold N50 values were 44.7 and 452.7 kb, respectively. Additionally, the longest contig and scaffold were 577.9 and 716.2 kb, respectively (Table 1). The M. baccata genome GC content is 38.06%, which is very close to that of the M. domestica genome (37.99%) (Velasco et al. 2010) (Table S3).
Table 1. Summary of wild apple (M. baccata) genome assembly features.
| Unit of assembly | Proportion/unit type | No. | Size | % assembly | Length of N50 (kb) | Longest (Mb) |
|---|---|---|---|---|---|---|
| Contigs | All | 320,531 | 665.80Mb | 92.6 | 44.7 | 0.6 |
| Scaffolds | All | 296,545 | 718.98Mb | 100 | 452.7 | 7.2 |
| Anchored | 1,561 | 528.25Mb | 73.5 | |||
| Repetitive sequences | Total | 421.05Mb (58.6%) | ||||
| Genes | Total | 41,113 | 126.46Mb (17.6%) | |||
| ncRNA | Total | 8,263 | 1,011.99Kb (0.14%) |
Construction of a genetic map and establishment of the pseudomolecules
To assemble pseudomolecules, we implemented the genotyping by restriction site-associated DNA sequencing method to construct a high-density M. baccata genetic map. We established a high-density genetic map with 3,065 markers using 390 F1 progenies from a cross between M. baccata ‘Shandingzi’ and M. domestica ‘Danxia’. A total of 1,480 scaffolds were anchored to the high-density genetic map by these markers, accounting for 72.57% of the assembly (521.75 of 718.98 Mb). We identified 17 chromosome pseudomolecules and determined the sequence orientation of 53.13% of the anchored scaffolds (382.02 Mb) based on genetic distances (Table S4, Table S5 and Figure S3). Moreover, 39,473 genes located in the anchored pseudochromosomes corresponded to 85.60% of all assembled scaffolds (Table S5). Genetic distance plotted against physical distance revealed that the genetic and physical positions were mostly consistent, except for chr11 (Figure S2).
Evaluation of assembled genome quality and sequence comparisons
We estimated the completeness of the M. baccata genome assembly by attempting to align 325,636 Malus species expressed sequence tag (EST) from the GenBank database with the assembly sequence. We observed that 95.22% of the EST were aligned. Because most ESTs were from M. domestica, only 84.84% of the EST had least 90% of their lengths covered in the alignments (Table S6). Meanwhile, RNA sequencing (RNA-seq) reads for root, fruit, phloem, and leaf tissues were aligned with the assembly sequence. An average of 95.00% of the read pairs were covered by the assembly sequence for four samples (Table S7). Additionally, we aligned 245 sequences from the nonredundant core eukaryotic genes (CEGs) with the genome assembly using the CEGMA (Core Eukaryotic Genes Mapping Approach) pipeline (Parra et al. 2007). A total of 222 (94%) CEG homologs were detected in the M. baccata genome (Table S8). We also used the BUSCO (Benchmarking Universal Single-Copy Orthologs) pipeline (Simão et al. 2015) to examine the coverage of highly conserved genes to validate the completeness of the M. baccata genome assembly. We observed that 93.20% of the plant BUSCO sequences searched were present in M. baccata scaffolds (Table S9). The percentage of BUSCOs for the M. baccata genome assembly was higher than that for the M. domestica (Velasco et al. 2010) and P. bretschneideri (Wu et al. 2013) genomes, but lower than that for the P. persica (International Peach Genome Initiative et al. 2013) and F. vesca (Shulaev et al. 2011) genomes (Figure S4). Thus, most of the conserved core gene set was present in the M. baccata genome assembly.
Heterozygosity of Malus Baccata genome
The M. baccata genome has high levels of heterozygosity because of self-incompatibility. We mapped the reads with 500-bp inserts onto the draft assembly. A total of 3,759,523 heterozygous single nucleotide polymorphisms (SNPs) were identified, corresponding to 5.2 SNPs per kb. Thus, the heterozygosity was about 0.5%, which was lower than our estimate (1.2%). M. baccata heterozygosity rate is similar to the corresponding rates for poplar (about 0.5%) (Tuskan et al. 2006), kiwifruit (0.536%) (Huang et al. 2013), and orchid (0.4%) (Cai et al. 2014), but is lower than the rate for pear (1.02%) (Wu et al. 2013), and tea (2.67%) (Xia et al. 2017).
We evaluated the structural and functional effects of heterozygous SNPs. The distribution profile revealed that 61.3% of SNPs were within 50 bp of each other, and nearly 25% were within < 10 bp of an adjacent SNP (Figure S5). Most of the identified SNPs (81.74%) were located in intergenic regions. Additionally, 32,895 genes included 18.26% of the SNPs, of which 11,608 genes had a SNP rate of < 1% (Table S10 and Figure S6). Genes with high SNP frequencies (> 3%) were associated with plant–pathogen interactions, protein processing in the endoplasmic reticulum, ascorbate and aldarate metabolism, isoquinoline alkaloid biosynthesis, and various pathways (Table S11).
Transposable elements and gene model annotation
Statistics regarding repetitive DNA:
Transposable elements (TEs) contributed 58.56% (413.09 Mb) of the M. baccata genome sequence (Table S12) which is similar to the annotation rate of M. domestica TEs. The long terminal repeat (LTR) retrotransposons were the most abundant transposable elements, representing 44.37% of the assembly. Among the LTR retrotransposons, Gypsy and Copia constituted 29.18% and 16.00% of the M. baccata genome sequence, respectively (Figure 2 and Table S13). Other transposable elements included CMC, DNA, hAT, and PIF (Table S14). Moreover, the abundance of LINE/RTE content (8.03%) in the M. baccata and M. domestica genomes is much greater than that of other Rosaceae plant species (Table S14), suggesting a unique evolutionary event occurred in the Malus genome.
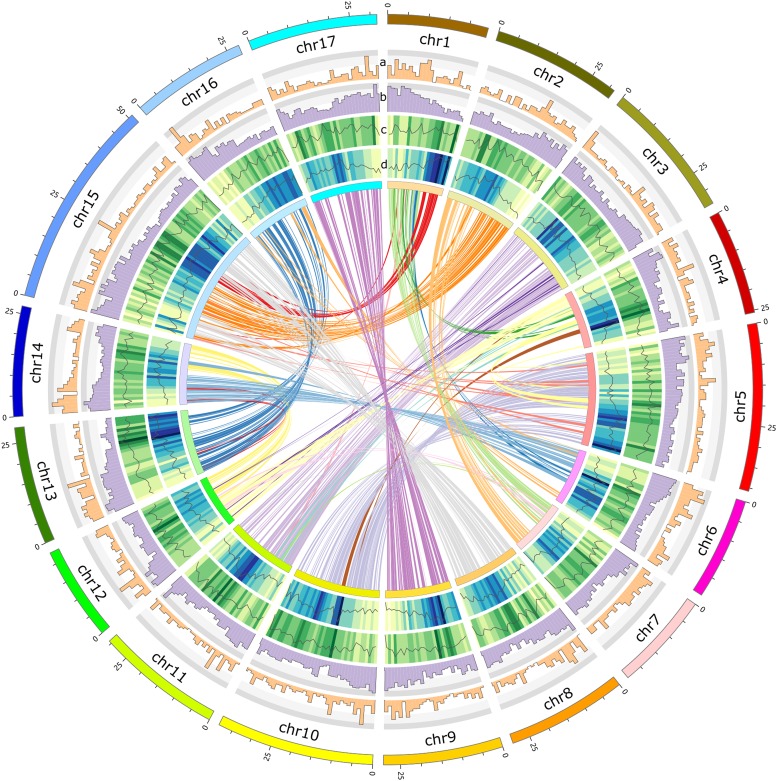
Figure 2.
Global view of the M. baccata genome. Transcription factor density (track a); gene density (track b); LTR-Gypsy number density (line) and size density (heat map) (track c); LTR-Copia number density (line) and size density (heat map) (track d). The innermost circle represents ideograms of 17 pseudochromosomes and the syntenic relationships of gene blocks from different pseudochromosomes.
Protein-coding gene annotation and evaluation:
We conducted RNA-seq experiments for root, phloem, leaf, and fruit libraries to identify genes, ultimately generating a 47.88-Mb transcriptome assembly. Additionally, publicly available Malus ESTs and homologs from the sequenced genomes of other Rosaceae species (i.e., Prunus persica (International Peach Genome Initiative et al. 2013), Prunus mume (Zhang et al. 2012), Pyrus bretschneideri (Wu et al. 2013), M. domestica (Velasco et al. 2010), and Fragaria vesca (Shulaev et al. 2011)) and from the Arabidopsis thaliana genome (Arabidopsis Genome Initiative 2000) were applied for predicting genes. Using evidence-based and de novo gene predictions, we identified 46,114 high-confidence protein-coding gene models (Table S15). A similar number of genes was predicted for M. baccata (46,114) and M. domestica (46,534), while fewer genes were predicted for P. bretschneideri (42,269).
The average gene size was 2,667 bp, with a mean of 4.4 exons per gene. The average gene length was similar to that other Rosaceae species such as P. persica (International Peach Genome Initiative et al. 2013), P. bretschneideri (Wu et al. 2013) and F. vesca (Shulaev et al. 2011) (Table S16). Among these genes, 82.01, 33.19, 66.26, and 68.89% were annotated using InterPro (Zdobnov and Apweiler 2001), Gene Ontology (GO) (http://www.geneontology.org/), Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto 2000) and Swiss-Prot (Bairoch and Apweiler 2000) databases, respectively, with 39,685 genes (86.06%) annotated by at least one database (Table S17 and Figure S7).
Non-coding RNAs:
We also identified and annotated various non-coding RNA sequences in the M. baccata genome, including 5,778 ribosomal RNA (rRNA), 1,553 transfer RNA (tRNA), 497 small nuclear RNA (snRNA), and 408 microRNA genes (Table S18).
Gene family evolution and comparisons
Functional annotation of specific genes:
A total of 21,930 (47.56%) M. baccata genes exhibited a one-to-one orthology with genes from M. domestica. The average Ka/Ks ratio (i.e., ratio of non-synonymous substitutions to synonymous substitutions) for these gene pairs was 0.4770, suggesting that most M. baccata genes evolved under purifying selection (Figure S8). A total of 1,574 genes had a Ka/Ks > 1, indicating they may be under positive selection pressure. Additionally, the KEGG pathways enriched among these genes were related to plant hormone signal transduction and plant–pathogen interactions. Furthermore, 25 of these genes were significant at the 0.05 p-value threshold (Table S19). The resulting gene list included four stress resistance genes with a complete structure and three genes with the TIR domain (Table S20).
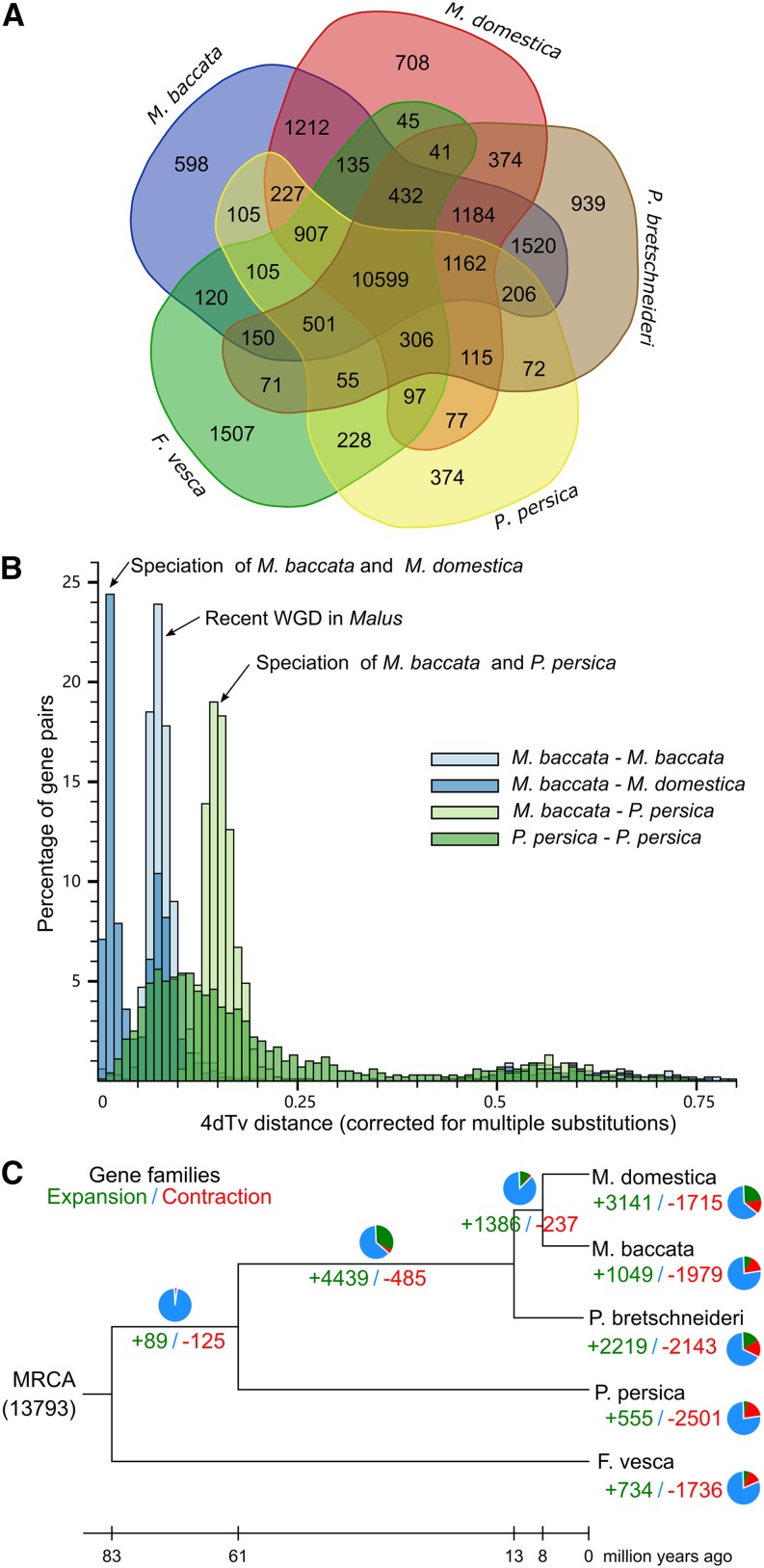
Comparative analyses involving the M. baccata genome and the M. domestica (Velasco et al. 2010), P. persica (International Peach Genome Initiative et al. 2013), P. bretschneideri (Wu et al. 2013) and F. vesca (Shulaev et al. 2011) genomes revealed that these five Rosaceae species contain a common core set of 10,599 gene families. However, 598 gene families were specific to M. baccata, while 708 gene families were specific to M. domestica (Figure 3A). The gene families specific to M. baccata were enriched in GO terms related to cell division and the cell cycle and KEGG pathways associated with purine metabolism, pyrimidine metabolism, the spliceosome, RNA polymerase, mRNA surveillance, RNA transport, the phagosome, and aminoacyl-tRNA biosynthesis. The identified genes may have contributed to the adaptation of M. baccata to environmental conditions. An analysis of the gene families specific to M. domestica revealed they were enriched in GO terms and KEGG pathways related to carbohydrate metabolism (e.g., fructose and mannose metabolism and starch and sucrose metabolism).
Figure 3.
Gene family evolution and comparisons with Rosaceae species, (A) Venn diagram of five Rosaceae species (M. baccata, M. domestica, P. bretschneideri, P. persica, and F. vesca). (B) Duplications in the M. baccata genome revealed via 4dTv analyses. (C) Gene family expansions and contractions in five Rosaceae species. Expansions and contractions are indicated in green and red, respectively. The corresponding proportions of the total changes are presented using the same colors in pie charts. The blue sections of the pie charts represent conserved gene families. MRCA, most recent common ancestor.
Contraction and expansion of the wild and cultivated apple gene families:
To study gene families that had expanded or contracted only in wild apple or cultivated apple species, we compared the M. baccata gene families with those of four other Rosaceae species and ancestral species. We determined that 1,049 and 1,715 gene families had expanded and contracted, respectively, in the M. baccata genome relative to its most recent common ancestor (Figure 3C). The results of a KEGG pathway enrichment analysis revealed that the expansion of these families involved genes related to cutin, suberin, and wax biosynthesis as well as fatty acid biosynthesis and degradation (Table S21).
We also examined the pathways associated with the expanded and contracted M. baccata and M. domestica gene families (Figure 3C). Most of the expansions and contractions were consistent between the two Malus species (Table S22). The only differences were that the gene families related to the isoflavonoid biosynthesis pathway and tyrosine metabolism pathway expanded in M. baccata, but contracted in M. domestica.
Phylogenetic analysis:
We estimated the divergence time of 10 sequenced plant species (i.e., M. baccata, M. domestica, P. bretschneideri, P. persica, P. mume, F. vesca, Carica papaya, A. thaliana, Populus trichocarpa, and Vitis vinifera) according to known ranges of divergence time as well as a phylogenetic tree. It is likely that M. baccata and M. domestica diverged from each other approximately 6.9–11.9 million years ago (Figure S9).
To further characterize the divergence between M. baccata and M. domestica, we measured the transversions at fourfold degenerate sites (4dTv) for orthologous gene pairs among M. baccata, M. domestica, and peach (Figure 3B). The 4dTv distribution indicated there are two significant groups of blocks, suggesting a recent whole-genome duplication (WGD) event occurred in M. baccata and M. domestica, but not in peach.
Synteny analysis:
An analysis of the synteny between M. baccata and other rosaceous species (Table S23) revealed that M. baccata and M. domestica share similar chromosome structures and organization. All 17 M. baccata chromosomes were similar to the corresponding M. domestica chromosomes (Figure S10). The self-collinearity of M. baccata (Figure 1 and Figure S10) enabled the identification of syntenic chromosome pairs, including LG3 and LG11, LG5 and LG10, LG9 and LG17, and LG13 and LG16, and revealed the chromosomal rearrangements in the M. domestica and P. bretschneideri genomes.
Identification of transcription factors and comparison between wild and domesticated apple
Transcription factors (TFs) are important for plant growth and development. We identified 2,345 TF-encoding genes in 58 families in the M. baccata genome. These genes represented 5.06% of the genes in the M. baccata genome (46,114). The proportion of the genome represented by TF-encoding genes was similar in M. baccata and P. bretschneideri, but was lower in M. domestica. In the M. baccata genome, the most abundant TF-encoding genes belonged to the following TF families: bHLH (208 genes), MYB (180 genes), NAC (175 genes), ERF (157 genes), and C2H2 (147 genes) (Table S24). The identification of these TFs may be useful for future functional verifications of M. baccata traits.
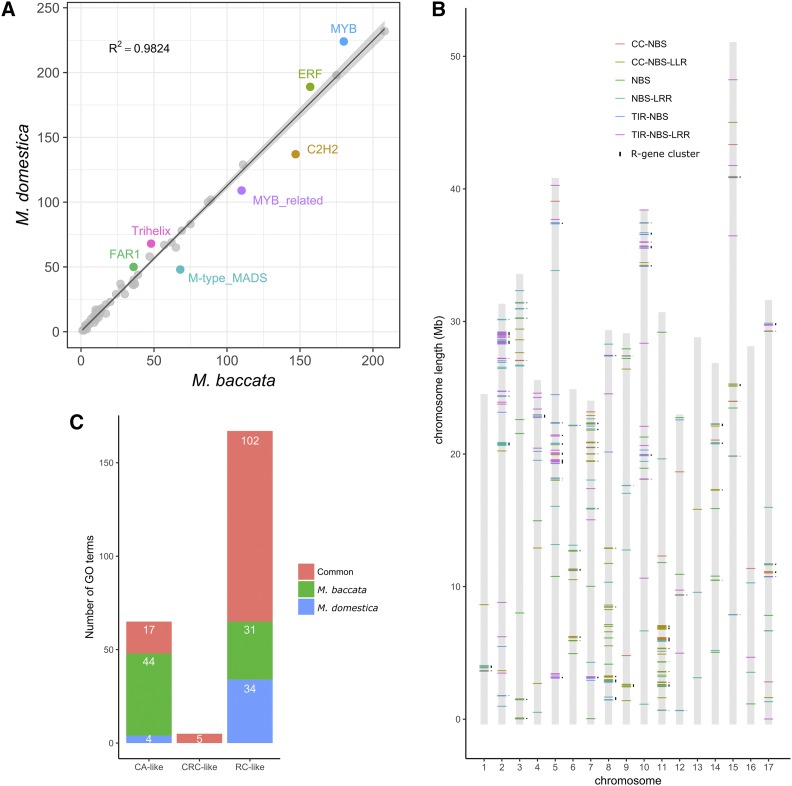
We compared the abundance of different TF types among Rosaceae genomes (Table S24). A comparison between M. baccata and M. domestica (Figure 4A) indicated there are fewer MYB and ERF TFs in the M. baccata genome than in the M. domestica genome. Some MYB family TFs induce fruit coloration, while the ERF family TFs promote fruit growth and ripening. The number of genes encoding these TFs increased in M. domestica, which may be related to the domestication of apple trees and their adaptation to the environmental conditions in central Asia. Meanwhile, the M. baccata genome was observed to carry more M-type MADS, MYB-related, and C2H2 TFs than the M. domestica genome.
Figure 4.
Transcription factor, R gene cluster and cold-responsive genes in M. baccata genome. (A) Transcription factor families with more than 50 members in M. baccata and M. domestica. The confidence interval (P = 0.01) of the regression curve is indicated (lines and shadow). Transcription factor families (with more than 50 members) that deviate significantly from the regression curve are indicated. (B) Distribution of R gene clusters along M. baccata chromosomes. Different colored horizontal bars represent different kinds of NBS R genes. The NBS R gene clusters are indicated next to the vertical bars representing chromosomes. (C) Results of GO analyses of the M. baccata and M. domestica COR (cold-responsive) genes. CA, cold acclimation; CRC, cellular response to cold; RC, response to cold.
Resistance gene analogs and cold-responsive genes
Disease resistance is the major focus of apple scion and rootstock breeding. The recognition of pathogen effectors is mainly mediated by plant disease-resistance genes, which can be grouped according to encoded motifs [i.e., nucleotide-binding site leucine-rich repeat (NBS-LRR) or transmembrane leucine-rich repeat (TM-LRR)] (Hammond-Kosack and Jones 1997). We compared the identified resistance gene analogs (RGAs) in the M. baccata and M. domestica genomes, and created a catalog comprising 462 and 800 NBS genes from M. baccata and M. domestica, respectively. We classified the corresponding genes into various structural categories based on the arrangement of the encoded domains (Table 2). In M. baccata, 119 coiled coil-NBS-LRR (CNL), 111 toll/interleukin receptor (TIR)-NBS-LRR (TNL), 126 NBS-LRR, 54 NBS, 23 CC-NBS, and 29 TIR-NBS genes were identified. In contrast, we detected 108 CNL, 153 TNL, 241 NBS-LRR, 136 NBS, 58 CC-NBS, and 104 TIR-NBS genes in M. domestica. The huge difference number of NBS R gene between the two genome is that M. domestica genome have a lot of single domains or incomplete structures NBS genes. Studies have reported that incomplete structures NBS genes can act as recruiters of or interact with other NBS-LRR proteins (Kohler et al. 2008; Lozano et al. 2012). Furthermore, the number and diversity of NBS genes containing the RPW8 domain were greater in M. baccata than in M. domestica; these genes are responsive to powdery mildew infection in many plant species (Table 2). We mapped the NBS genes to 17 pseudochromosomes and observed that they were nonrandomly distributed (Figure 4B). More NBS-LRR R genes were clustered in groups on M. baccata chromosomes (66%) than on M. domestica chromosomes (61%), and clusters were most abundant on chromosomes 2, 5, and 11 (Table S25). Receptor-like kinase (RLK) protein and receptor-like proteins (RLP) also act as positive regulators in plant innate immunity (Yang et al. 2012). And the M. baccata genome includes 177 RLK genes and 51 RLP genes, while the M. domestica genome carries 93 RLK genes and 74 RLP genes.
Table 2. R genes present in the M. baccata and M. domestica genomes.
| R gene type | M. baccata | M. domestica |
|---|---|---|
| Canonical R genes | ||
| CC-NBS-LLR | 119 | 108 |
| TIR-NBS-LRR | 111 | 153 |
| Single domains or incomplete structures | ||
| NBS-LRR | 126 | 241 |
| NBS | 54 | 136 |
| CC-NBS | 23 | 58 |
| TIR-NBS | 29 | 104 |
| Canonical transmembrane domains | ||
| RLK | 177 | 93 |
| RLP | 51 | 74 |
| NBS gene with RPW8 domain | ||
| RPW8-NBS | 17 | 19 |
| RPW8-SBP-NBS | 1 | 0 |
| RPW8-RPW8-RPW8-NBS | 1 | 0 |
| RPW8-NBS-LRR | 1 | 0 |
| RPW8-RPW8-SBP-NBS-HMA | 1 | 1 |
| RPW8-RPW8-SBP-NBS-Pkinase | 1 | 1 |
The CBF transcription factor along with other genes, can sensing low temperature, initiating the process of cold acclimation and inducing the expression of the cold regulated (COR) genes proteins. And the COR genes can reduce the damage of plant cells due to freeze-induced dryness and the presence of extracellular ice (Miura and Furumoto 2013). A total of 2,978 and 5,089 predicted protein sequences similar to A. thaliana cold-responsive proteins were identified in M. baccata and M. domestica, respectively (Table 3). In M. baccata, 408, 51, and 3,519 proteins were homologous to A. thaliana sequences annotated with the GO terms ‘cold acclimation’ (hereafter called CA-like), ‘cellular response to cold’ (hereafter called CRC-like), and ‘response to cold’ (hereafter called RC-like), respectively. In M. domestica, 466, 58, and 4,565 proteins were CA-like, CRC-like, and RC-like proteins, respectively (Table 3). Enriched GO term categories were detected in both species. For example, M. baccata and M. domestica CA (associated with cold acclimation) genes were annotated with 44 and 4 unique GO terms, respectively (Figure 4C). Among CBF dependent cold signaling pathways, CBF genes induce COR genes expression by binds to DREB motif in COR genes promoter (Sakuma et al. 2002). We identified the number of COR genes with DREB motif and observed that the proportion of COR genes that contained the DREB motif was higher for M. baccata (50.25%) than for M. domestica (42.84%) (Table 3), implying M. baccata can induce more cold-responsive biological processes.
Table 3. Cold-related (COR) genes and the proportion of COR genes with a DREB motif in the M. baccata and M. domestica genomes.
| M. baccata | M. domestica | |
|---|---|---|
| RC gene number | 3519 | 4565 |
| RC gene number with DREB motif | 1758 | 1940 |
| CRC gene number | 51 | 58 |
| CRC gene number with DREB motif | 29 | 29 |
| CA gene number | 408 | 466 |
| CA gene number with DREB motif | 212 | 211 |
| Total COR gene number | 3978 | 5089 |
| Total gene number with DREB motif | 1999(50.25%) | 2180(42.84%) |
CA: Cold Acclimation, CRC: Cellular Response to Cold, RC: Response to Cold.
Conclusions
We herein describe the first wild apple genome assembly, which was obtained by paired-end sequencing. The assembled genome sequence comprises 665 Mb, with a scaffold N50 value of 452 kb. Future genome-wide comparative studies will provide novel insights into the genomic evolution of Rosaceae species, especially Malus species. The annotation of the protein-coding genes and comparisons with the M. domestica genome provided insights into M. baccata-specific traits, particularly those involved in cold tolerance and pathogen resistance. The analyses of the M. baccata genome described herein may be relevant for future investigations of the genetic variations in wild apple germplasm, and may facilitate marker-assisted breeding for apple cultivars and stock exhibiting disease and cold resistance.
Acknowledgments
We thank Dr. Amandine Cornille (INRA, Univ. Paris-Sud, CNRS, AgroParisTech) for suggestions and comments on the project and manuscript. This work was supported by National Key Research and Development Program of China (2018YFD1000101), China Apple Research System (CARS-27), Shaanxi key research and development plan (2017ZDXM-NY-019) and Yangling Subsidiary Center Project of the National Apple Improvement Center.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25387/g3.7523549.
Communicating editor: D. J. de Koning
Literature Cited
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative , 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. 10.1038/35048692 [DOI] [PubMed] [Google Scholar]
- Aversano R., Contaldi F., Ercolano M. R., Grosso V., Iorizzo M., et al. , 2015. The Solanum commersonii Genome Sequence Provides Insights into Adaptation to Stress Conditions and Genome Evolution of Wild Potato Relatives. The Plant Cell Online 27: 954–968. 10.1105/tpc.114.135954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenes M. L., Fernández i Martí A., Ríos G., Rubio-Cabetas M. J., 2016. Application of Genomic Technologies to the Breeding of Trees. Front. Genet. 7: 198 10.3389/fgene.2016.00198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A., Apweiler R., 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28: 45–48. 10.1093/nar/28.1.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G., 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27: 573–580. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E., Clamp M., Durbin R., 2004. GeneWise and Genomewise. Genome Res. 14: 988–995. 10.1101/gr.1865504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M., Henkel C. V., Jansen H. J., Butler D., Pirovano W., 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27: 578–579. 10.1093/bioinformatics/btq683 [DOI] [PubMed] [Google Scholar]
- Bolger A., Scossa F., Bolger M. E., Lanz C., Maumus F., et al. , 2014. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 46: 1034–1038. 10.1038/ng.3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C., Karlin S., 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94. 10.1006/jmbi.1997.0951 [DOI] [PubMed] [Google Scholar]
- Cai J., Liu X., Vanneste K., Proost S., Tsai W.-C., et al. , 2014. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 47: 65–72. Erratum: 304. 10.1038/ng.3149 [DOI] [PubMed] [Google Scholar]
- Chagné D., Crowhurst R. N., Pindo M., Thrimawithana A., Deng C., et al. , 2014. The Draft Genome Sequence of European Pear (Pyrus communis L. ‘Bartlett’). PLoS One 9: e92644 10.1371/journal.pone.0092644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornille A., Giraud T., Smulders M. J., Roldán-Ruiz I., Gladieux P., 2014. The domestication and evolutionary ecology of apples. Trends Genet. 30: 57–65. 10.1016/j.tig.2013.10.002 [DOI] [PubMed] [Google Scholar]
- De Bie T., Cristianini N., Demuth J. P., Hahn M. W., 2006. CAFE: a computational tool for the study of gene family evolution. Bioinformatics 22: 1269–1271. 10.1093/bioinformatics/btl097 [DOI] [PubMed] [Google Scholar]
- Dellaporta S. L., Wood J., Hicks J. B., 1983. A plant DNA minipreparation: Version II. Plant Mol. Biol. Report. 1: 19–21. 10.1007/BF02712670 [DOI] [Google Scholar]
- Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., et al. , 2014. Pfam: the protein families database. Nucleic Acids Res. 42: D222–D230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S. R., et al. , 2005. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33: D121–D124. 10.1093/nar/gki081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack K. E., Jones J. D. G., 1997. PLANT DISEASE RESISTANCE GENES. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 575–607. 10.1146/annurev.arplant.48.1.575 [DOI] [PubMed] [Google Scholar]
- Hu C. G., Honda C., Kita M., Zhang Z., Tsuda T., et al. , 2002. A simple protocol for RNA isolation from fruit trees containing high levels of polysaccharides and polyphenol compounds. Plant Mol. Biol. Report. 20: 69 10.1007/BF02801935 [DOI] [Google Scholar]
- Huang S., Ding J., Deng D., Tang W., Sun H., et al. , 2013. Draft genome of the kiwifruit Actinidia chinensis. Nat. Commun. 4: 2640 10.1038/ncomms3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J. P., Ronquist F., 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Jaillon O., Aury J. M., Noel B., Policriti A., Clepet C., et al. , 2007. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467. 10.1038/nature06148 [DOI] [PubMed] [Google Scholar]
- Jin J., Zhang H., Kong L., Gao G., Luo J., 2014. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 42: D1182–D1187. 10.1093/nar/gkt1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V. V., Pavlicek A., Klonowski P., Kohany O., et al. , 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110: 462–467. 10.1159/000084979 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28: 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A., Rinaldi C., Duplessis S., Baucher M., Geelen D., et al. , 2008. Genome-wide identification of NBS resistance genes in Populus trichocarpa. Plant Mol. Biol. 66: 619–636. 10.1007/s11103-008-9293-9 [DOI] [PubMed] [Google Scholar]
- Korban S. S., Wannarat W., Rayburn C. M., Tatum T. C., Rayburn A. L., 2009. Genome size and nucleotypic variation in Malus germplasm. Genome 52: 148–155. 10.1139/G08-109 [DOI] [PubMed] [Google Scholar]
- Li X., Kui L., Zhang J., Xie Y., Wang L., et al. , 2016. Improved hybrid de novo genome assembly of domesticated apple (Malus x domestica). Gigascience 5: 35 10.1186/s13742-016-0139-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liguo F., Tao H., 2003. HIGHER PLANTS OF CHINA. Qingdao Publishing House, Qingdao, Shandong. [Google Scholar]
- Lozano R., Ponce O., Ramirez M., Mostajo N., Orjeda G., 2012. Genome-Wide Identification and Mapping of NBS-Encoding Resistance Genes in Solanum tuberosum Group Phureja. PLoS One 7: e34775 10.1371/journal.pone.0034775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R., Liu B., Xie Y., Li Z., Huang W., et al. , 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1: 18 Erratum: 30. 10.1186/2047-217X-1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoros W. H., Pertea M., Salzberg S. L., 2004. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20: 2878–2879. 10.1093/bioinformatics/bth315 [DOI] [PubMed] [Google Scholar]
- McDonnell A. V., Jiang T., Keating A. E., Berger B., 2006. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics 22: 356–358. 10.1093/bioinformatics/bti797 [DOI] [PubMed] [Google Scholar]
- Ming R., Hou S., Feng Y., Yu Q., Dionne-Laporte A., et al. , 2008. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452: 991–996. 10.1038/nature06856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Furumoto T., 2013. Cold Signaling and Cold Response in Plants. Int. J. Mol. Sci. 14: 5312–5337. 10.3390/ijms14035312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H., 2017. Predicting Secretory Proteins with SignalP, pp. 59–73 in Protein Function Prediction, Methods in Molecular Biology, Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- Ooijen, V., 2006 JoinMap4 Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Kyazma BV, Wageningen 10–1371. [Google Scholar]
- Parra G., Bradnam K., Korf I., 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23: 1061–1067. 10.1093/bioinformatics/btm071 [DOI] [PubMed] [Google Scholar]
- Peace C., Norelli J., 2009. Genomics Approaches to Crop Improvement in the Rosaceae, pp. 19–53 in Genetics and Genomics of Rosaceae, edited by Folta K. M., Gardiner S. E. Plant Genetics and Genomics: Crops and Models 6, Springer, New York. [Google Scholar]
- Sakuma Y., Liu Q., Dubouzet J. G., Abe H., Shinozaki K., et al. , 2002. DNA-Binding Specificity of the ERF/AP2 Domain of Arabidopsis DREBs, Transcription Factors Involved in Dehydration- and Cold-Inducible Gene Expression. Biochem. Biophys. Res. Commun. 290: 998–1009. 10.1006/bbrc.2001.6299 [DOI] [PubMed] [Google Scholar]
- Shulaev V., Sargent D. J., Crowhurst R. N., Mockler T. C., Folkerts O., et al. , 2011. The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 43: 109–116. 10.1038/ng.740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Smit, AFA, and Hubley, 2008 RepeatModeler Open-1.0.
- Smit, AFA, Hubley, and R & Green, 2013 RepeatMasker Open-4.0.
- Stanke M., Keller O., Gunduz I., Hayes A., Waack S., et al. , 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34: W435–W439. 10.1093/nar/gkl200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Peach Genome Initiative , I. Verde, A. G. Abbott, S. Scalabrin, S. Junget al. , 2013. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 45: 487–494. 10.1038/ng.2586 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., et al. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. Erratum: 2513. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G. A., DiFazio S., Jansson S., Bohlmann J., Grigoriev I., et al. , 2006. The Genome of Black Cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604. 10.1126/science.1128691 [DOI] [PubMed] [Google Scholar]
- Velasco R., Zharkikh A., Affourtit J., Dhingra A., Cestaro A., et al. , 2010. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 42: 833–839. 10.1038/ng.654 [DOI] [PubMed] [Google Scholar]
- Volk G. M., Chao C. T., Norelli J., Brown S. K., Fazio G., et al. , 2015. The vulnerability of US apple (Malus) genetic resources. Genet. Resour. Crop Evol. 62: 765–794. 10.1007/s10722-014-0194-2 [DOI] [Google Scholar]
- Wang Y., Tang H., DeBarry J. D., Tan X., Li J., et al. , 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40: e49 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Yu Y., Haberer G., Marri P. R., Fan C., et al. , 2014. The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat. Genet. 46: 982–988. 10.1038/ng.3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Wang Z., Shi Z., Zhang S., Ming R., et al. , 2013. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 23: 396–408. 10.1101/gr.144311.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, W., Y.-L. Yang, W.-M. He, M. Rouard, W.-M. Li et al., 2016 Whole genome sequencing of a banana wild relative Musa itinerans provides insights into lineage-specific diversification of the Musa genus. Scientific Reports 6: srep31586. [DOI] [PMC free article] [PubMed]
- Xia E.-H., Zhang H.-B., Sheng J., Li K., Zhang Q.-J., et al. , 2017. The Tea Tree Genome Provides Insights into Tea Flavor and Independent Evolution of Caffeine Biosynthesis. Mol. Plant 10: 866–877. 10.1016/j.molp.2017.04.002 [DOI] [PubMed] [Google Scholar]
- Xu S., Brockmöller T., Navarro-Quezada A., Kuhl H., Gase K., et al. , 2017. Wild tobacco genomes reveal the evolution of nicotine biosynthesis. Proc. Natl. Acad. Sci. USA 114: 6133–6138. 10.1073/pnas.1700073114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Yang X., Deng F., Ramonell K. M., 2012. Receptor-like kinases and receptor-like proteins: keys to pathogen recognition and defense signaling in plant innate immunity. Frontiers in Biology 7: 155–166. 10.1007/s11515-011-1185-8 [DOI] [Google Scholar]
- Zdobnov E. M., Apweiler R., 2001. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848. 10.1093/bioinformatics/17.9.847 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Chen W., Sun L., Zhao F., Huang B., et al. , 2012. The genome of Prunus mume. Nat. Commun. 3: 1318 10.1038/ncomms2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Li J., Zhao X.-Q., Wang J., Wong G. K.-S., et al. , 2006. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4: 259–263. 10.1016/S1672-0229(07)60007-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.-J., Zhu T., Xia E.-H., Shi C., Liu Y.-L., et al. , 2014. Rapid diversification of five Oryza AA genomes associated with rice adaptation. Proc. Natl. Acad. Sci. USA 111: E4954–E4962. 10.1073/pnas.1418307111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome assembly have been deposited under CNGB Project ID CNA0002537 (https://db.cngb.org/search/project/CNP0000421/). The meta data for the genetic map can be also found in https://db.cngb.org/search/project/CNP0000421/. The genomic raw reads are available via NCBI SRR7248834, SRR7248835, SRR7248837, SRR7248847, SRR7248838 to SRR7248844, SRR7248849 to SRR7248858, SRR7248875 to SRR7248882, and the raw transcriptomic reads are available at NCBI SRR8156047 to SRR8156050. Supplemental material available at FigShare: https://doi.org/10.25387/g3.7523549.