Abstract
Pancreatic ductal adenocarcinoma (PDA) has a poor prognosis, and new strategies for prevention and treatment are urgently needed. We previously reported that histone H4 acetylation is elevated in pancreatic acinar cells harboring Kras mutations prior to the appearance of premalignant lesions. Since acetyl-CoA abundance regulates global histone acetylation, we hypothesized that altered acetyl-CoA metabolism might contribute to metabolic or epigenetic alterations that promote tumorigenesis. We found that acetyl-CoA abundance is elevated in KRAS mutant acinar cells and that its use in the mevalonate pathway supports acinar-to-ductal metaplasia (ADM). Pancreas-specific loss of the acetyl-CoA producing enzyme ATP-citrate lyase (ACLY) accordingly suppresses ADM and tumor formation. In PDA cells, growth factors promote AKT-ACLY signaling and histone acetylation, and both cell proliferation and tumor growth can be suppressed by concurrent BET inhibition and statin treatment. Thus, KRAS-driven metabolic alterations promote acinar cell plasticity and tumor development, and targeting acetyl-CoA-dependent processes exerts anti-cancer effects.
Keywords: ACLY, acetyl-CoA, pancreatic ductal adenocarcinoma, acinar-to-ductal metaplasia, KRAS
INTRODUCTION
Activating mutations of KRAS are found in >90% cases of pancreatic ductal adenocarcinoma (PDA), a disease that accounts for 50,000 new cases every year in the United States and is currently the third-leading cause of cancer-related deaths (1). Because pancreatic cancer metastasizes early in disease progression (2) and effective treatments for advanced disease are lacking, patients face an extremely poor prognosis (~9% 5-year survival rate) (3). Improved strategies to prevent PDA in at-risk patients, to detect the disease earlier when it is clinically more manageable, and to treat advanced disease are all urgently needed to reduce deaths from PDA (1).
Metabolism is extensively reprogrammed in pancreatic cancer cells to support proliferation and enable survival in an extremely nutrient- and oxygen-depleted microenvironment (4,5). Acetyl-CoA is a central metabolite with key roles in biosynthetic processes that are important for proliferation, including fatty acid and cholesterol biosynthesis, as well as signaling functions, through serving as the acetyl group donor for lysine acetylation. The two major enzymes that produce acetyl-CoA in the cytosol and nucleus are ATP-citrate lyase (ACLY), which generates acetyl-CoA from the cleavage of mitochondria-derived citrate, and acetyl-CoA synthetase 2 (ACSS2), which produces acetyl-CoA from acetate (6). How the dual metabolic and signaling roles of these enzymes are coordinated in cancer cells remain poorly understood.
Histone acetylation, a dynamic chromatin modification with key roles in gene regulation, is highly sensitive to the production and availability of acetyl-CoA (6–8). Acetyl-CoA fluctuates in response to a number of stimuli in mammalian cells, including nutrient availability (9), oxygen availability (10), circadian oscillations (11), diet (12), and PI3K-AKT signaling (9). In human PDA tumors, high levels of histone acetylation have been found to correlate with high stromal content (13) and poor prognosis (14), and co-culture of PDA cells with pancreatic stellate cells induces histone acetylation and gene expression changes (15). Elevated global levels of histone acetylation are acquired in human PDA metastatic clones, as compared to primary tumors or peritoneal metastatic clones, in a manner dependent on alterations in glucose metabolism (16). Moreover, targeting the reading of histone acetylation by BET inhibition, particularly in conjunction with histone deacetylase (HDAC) inhibition, has been shown to suppress pancreatic tumor formation and growth (17–19). Thus, histone acetylation is dynamically regulated in PDA cells, contributes to pancreatic tumor development and progression and may offer opportunities for therapeutic intervention in PDA.
ACLY is an AKT substrate, and in previous work, we reported that the AKT-ACLY signaling regulates histone acetylation in tumor cells (9). We also observed that global histone H4 acetylation was elevated in the acinar cells of young LSL-KrasG12D; p53L/+; Pdx1-Cre; RosaYFP (KPCY) versus wild-type (WT) mice, even prior to the appearance of premalignant lesions (9). Lineage-tracing studies in mutant KRAS-expressing animals have demonstrated that PDA can arise from cells that have undergone a metaplastic event termed Acinar-to-Ductal Metaplasia (ADM), which occurs as part of a normal response to pancreatic injury or inflammation (20,21). In WT cells, ADM is reversible and acini regenerate once the injury resolves. However, KRAS mutant cells that undergo ADM can progress to PanIN lesions. The metabolic and epigenetic mechanisms by which KRAS orchestrates this irreversible ADM remain poorly understood. Notably, PI3K signaling is required for pancreatic carcinogenesis (22–26), and AKT inhibition has been shown to suppress ADM (24). We thus wondered if ACLY as a substrate of AKT might contribute to the regulation of histone acetylation in acinar cells or play a role in facilitating ADM.
We hypothesized that elevated histone acetylation in KRAS mutant acinar cells might reflect early alterations in acetyl-CoA metabolism that may contribute to tumorigenesis or point towards metabolic and/or epigenetic vulnerabilities that could be exploited for PDA prevention or treatment. In this study, we identify a role for ACLY-dependent acetyl-CoA production in ADM and pancreatic tumor formation, and our data also point to the potential to target acetyl-CoA dependent processes in established tumors. Using mice in which Acly is deleted from the pancreas (Pdx1-Cre; Aclyf/f), we find that ACLY is required for mutant KRAS-mediated elevation of histone acetylation in acinar cells, as well as for efficient KRAS-driven ADM in vitro and pancreatic tumorigenesis in vivo. In human PDA cells, AKT-ACLY signaling is highly responsive to environmental conditions and regulates histone acetylation globally and H3K27ac at PDA enhancers. We find that targeting acetyl-CoA-dependent processes via BET inhibition and statin treatment suppresses PDA cell proliferation and can suppress tumor growth in vivo. These data establish a key role for ACLY in enabling KRAS-dependent tumor initiation and point to the potential for targeting acetyl-CoA-dependent processes in pancreatic cancer.
RESULTS
AKT inhibition suppresses histone acetylation and acinar-to-ductal metaplasia in KRAS mutant pancreatic acinar cells
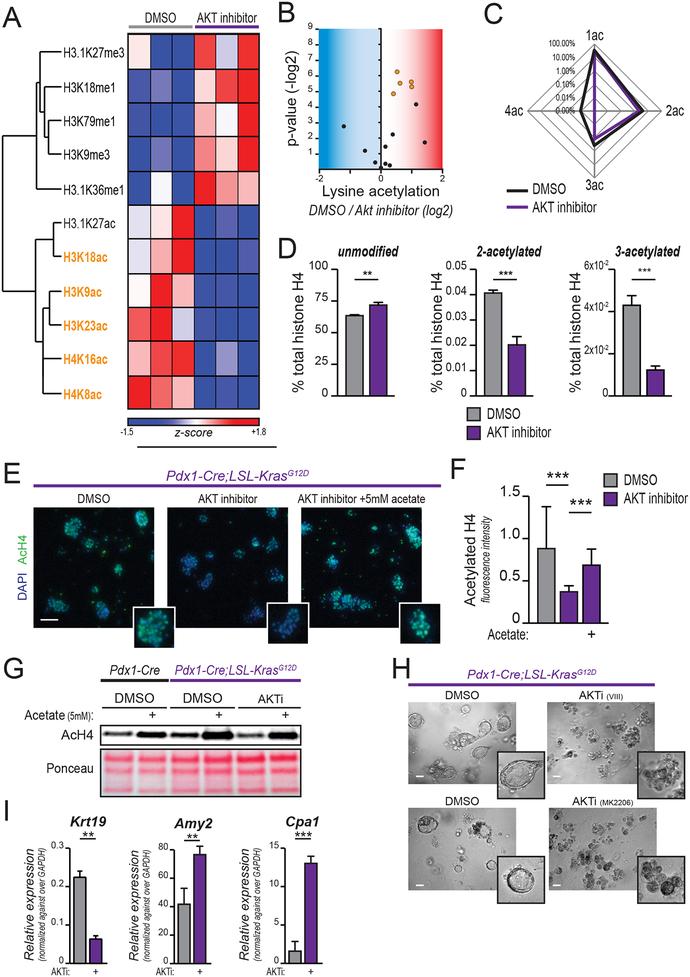
We previously reported that AKT-ACLY signaling promotes acetyl-CoA production and global increases in histone acetylation in cancer cells (9). To comprehensively define the AKT-dependent changes in chromatin modification in KRAS mutant PanIN cells, we analyzed histone modifications by mass spectrometry (MS) following a 24-hour treatment with a selective AKT1/2 inhibitor (AKTi). We found that AKT inhibition suppressed several histone acetyl marks, including H3K23ac, H3K18ac, H3K9ac, and H4K16ac, and H4K8ac (Figure 1A–B, Supplementary Figure S1A). AKT inhibition also reduced the presence of multiply acetylated histones (i.e., at lysines 5, 8, 12, and 16 of H4). AKT inhibition potently suppressed the abundance of H4 acetylated at 2, 3, or 4 sites, and reciprocally increased the abundance of the unmodified peptide (Figure 1C–D). Additionally, several methyl modifications increased with AKT inhibition (Figure 1A, Supplementary Figure S1A). Thus, AKT inhibition promotes extensive remodeling of histone modifications, including broadly suppressing histone acetylation, in PanIN cells.
Figure 1: AKT inhibition reduces histone acetylation in KRASG12D-expressing cells.
A-D, Mass spectrometry-based profiling of histone modifications in primary murine PanIN-derived cells treated with AKT inhibitor (CAS-612847-09-3, 10 μM) for 24 hours. A, Heatmap shows histone marks most strongly regulated (see Supplementary Figure S1A for heat map of full dataset). Columns show biological replicates (n=3, each treatment). Histone acetyl marks highlighted in orange are those reaching p<0.05 (H3.1K27ac p=0.057). Complete histone acetylome represented in B, volcano plot. Each dot represents an acetylated residue. Blue area contains downregulated marks with AKTi, red area represents upregulated marks. Orange dots represent those reaching p<0.05, as in part A. C, Spider graph shows percentage of 1-, 2-, 3-, 4-acetylated histone H4 (lysine residues 5, 8, 12, 16) over total histone H4. D, bar graphs depict abundance of indicated poly-acetylated H4 peptides, along with unmodified histone H4. E, AcH4 staining of acinar cells isolated from 6–8-week-old KC mice (n=3), embedded in Matrigel, and treated as indicated for 24 hours. Scale bar, 50 μm. F, quantification of E (25 optical fields acquired per experimental replicate). G, AcH4 western blot of acinar cells treated 24 hours in indicated conditions (AKT selective inhibitor VIII, 10 μM; acetate, 5 mM). H, morphology (scale bar, 50 μm) of Matrigel-embedded acinar cells after 48 hours culture in the presence or absence of indicated AKT inhibitors (VIII, upper panel; MK2206, lower panel). I, mRNA expression of indicated genes quantified by qPCR for cells in H. Bar graphs depict mean, +/− SD of triplicates (*, p<0.05; **, p<0.01; ***, p<0.001).
We next sought to determine the role of AKT in regulating histone acetylation in pancreatic acinar cells, since elevated histone H4 acetylation was observed in these cells in KPC mice even prior to appearance of PanIN lesions (9). We confirmed that pancreas-specific KrasG12D mutation alone was sufficient to promote elevated H4 and H3K27 acetylation in acinar cells (Supplementary Figure S1B). Primary pancreatic acinar cells were isolated from Pdx1-Cre;LSL-KrasG12D (KC, hereafter) mice and treated with AKTi for 24 hours. AKTi strongly reduced histone H4 acetylation as assessed by immunofluorescence (Figure 1E, quantified in F) and by western blotting (Figure 1G). Acetate supplementation boosted histone acetylation in all conditions tested (Figure 1E–G). Thus, Kras mutation promotes global histone acetylation in acinar cells in an AKT-dependent manner.
KRAS mutant acinar cells undergo ADM when cultured ex vivo in a 3D matrix (22). Time course analysis showed that histone acetylation further increases in primary KC acinar cells within 24 hours of plating in Matrigel (Supplementary Figure S1C–D), prior to changes in cell morphology (duct formation), which appear by 48 hours. Expression levels of lysine acetyl-transferases and deacetylases were minimally altered over this time course (Supplementary Figure SE–F). Consistent with prior work (24), AKT inhibition potently blocked the formation of ductal structures in vitro (Figure 1H). Treatment with AKTi also suppressed the activation of ductal-specific genes (e.g.: Krt19) and allowed expression of acinar-specific protease genes (Amy2, Cpa1) to be maintained (Figure 1I), indicating that AKT signaling is required for ADM in KRASG12D-expressing cells.
Targeting acetyllysine readers or the mevalonate pathway suppresses ADM
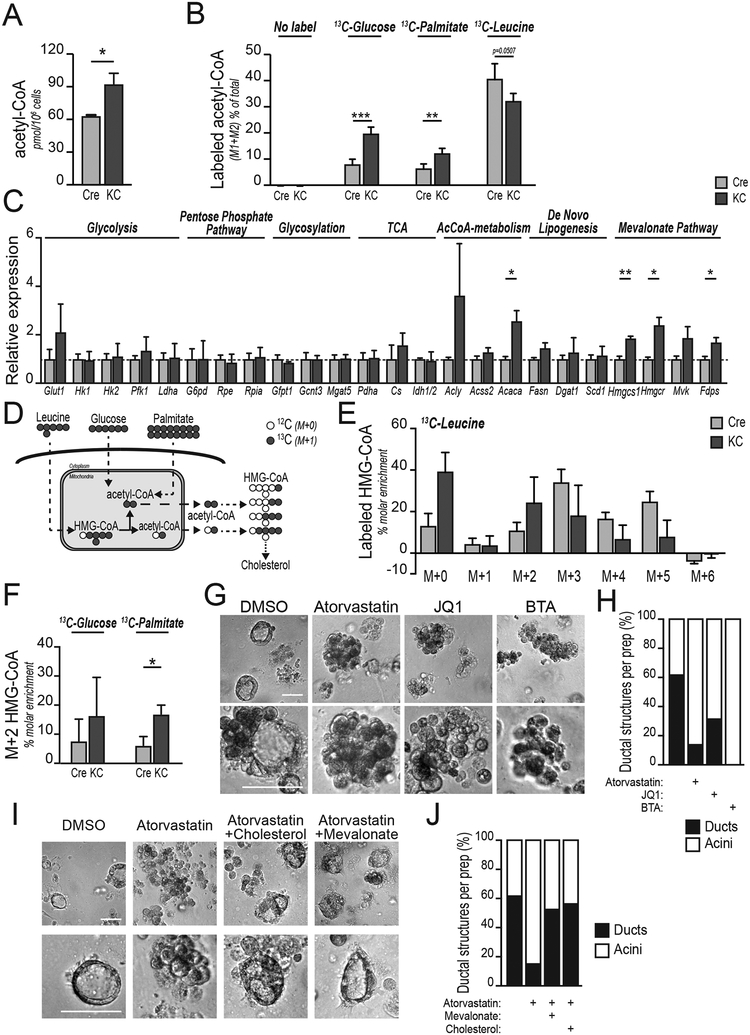
We hypothesized that KRAS may promote ADM in part through AKT-dependent regulation of acetyl-CoA production. We therefore investigated the effects of mutant KRAS on acetyl-CoA metabolism in primary pancreatic acinar cells. Consumption of glucose and glutamine, as well as lactate production were similar between WT and KC acinar cells (Supplementary Figure S2A). However, in a targeted analysis, we found that acetyl-CoA abundance was greater in KrasG12D-expressing acinar cells as compared to WT acinar cells (Figure 2A), consistent with elevated global histone acetylation in these cells. To define the carbon sources that supply acetyl-CoA in acinar cells, we traced [U-13C]- glucose, [U-13C]-palmitate and [U-13C]-leucine. In WT acinar cells, the branched-chain amino acid (BCAA) leucine was a major source of acetyl-CoA (Figure 2B), consistent with a recent in vivo isotope tracing study of BCAA metabolism, which reported high utilization of BCAAs by the pancreas (27). Glucose and palmitate were minor contributors (Figure 2B). In KC acinar cells, leucine was also a major source of acetyl-CoA, and in addition, labeling from both glucose and palmitate increased (Figure 2B). Together, these data indicate that KRASG12D expression impacts acetyl-CoA production in pancreatic acinar cells.
Figure 2: Acetyl-CoA abundance is elevated in KRAS mutant acinar cells and inhibition of acetyllysine reading or cholesterol synthesis impairs acinar-to-ductal metaplasia.
In all panels, pancreatic acinar cells were harvested from 6–8-week-old wild-type (Pdx1-Cre; Cre) or (Pdx1-Cre;KrasG12D; KC) mice. A, LC-MS quantification of acetyl-CoA in isolated acinar cells (n=3 mice, each group). B, isolated acinar cells (n=4 mice, each group) were cultured for 8 hours in the presence of the indicated 13C-labelled nutrient and acetyl-CoA labeling determined by LC-MS. C, mRNA expression of indicated genes in acinar cells after 48 hours culture in Matrigel, measured by qPCR (n=3 mice, each group). Mean value of each Cre column is equaled to 1, and data are normalized accordingly. Dashed blue line shows 1. D, schematic representation of acetyl-CoA and HMG-CoA labeling pattern from indicated, uniformly-labeled carbon sources. Compartmentalization also illustrated. E, HMG-CoA isotopologues after labeling of acinar cells (n=4 mice, each group) with 0.5 μM 13C-leucine for 8 hours. F, M+2 HMG-CoA in acinar cells labeled as in panel B. G, morphology of KC acinar cells embedded in collagen, treated with TGFα after 96 hours in the presence of the indicated inhibitors (n=3 mice, each group) Representative images shown. Scale bar, 50 μm. H, blinded quantification of ductal structures. Relative to Figure 2G. 35–50 images within 3 biological replicates were evaluated. I, collagen-embedded KC acinar cells after 96 hours treatment with atorvastatin, +/− mevalonate or cholesterol (n=3 mice, each group). Scale bar, 50 μm. J, blinded quantification of ductal structures. Relative to Figure 2E. 35–50 images within 3 biological replicates were evaluated. For all panels, columns show mean, +/− SD (*, p<0.05; **, p<0.01; ***, p<0.001).
To gain initial insight into how acetyl-CoA might be used by acinar cells, we surveyed metabolic gene expression in acinar cells undergoing ADM, finding that expression of mevalonate pathway gene expression was elevated in KRAS mutant acinar cells (Figure 2C). The mevalonate pathway, which is necessary for the synthesis of sterols and isoprenoids, is initiated through synthesis of HMG-CoA from three molecules of acetyl-CoA in the cytosol (28). HMG-CoA is also produced during leucine catabolism (Figure 2D). HMG-CoA M+5, produced in 13C-leucine catabolism in mitochondria, was clearly detectable in both WT and KC acinar cells, though this isotopologue was reduced in KC cells as a percent of the total HMG-CoA pool (Figure 2E). The other detected isotopologues (M+1, M+2, M+3, M+4) likely represent HMG-CoA synthesized from acetyl-CoA (Figure 2D–E). Consistent with acetyl-CoA labeling patterns, increased HMG-CoA synthesis from 13C-palmitate, and a trend towards increased synthesis from 13C-glucose, were observed in KC acinar cells (Figure 2F). Total HMG-CoA abundance was not different between the two genotypes (Supplementary Figure S2B). These data indicate that mevalonate pathway gene expression increases in KRAS mutant acinar cells and that HMG-CoA is dynamically synthesized from acetyl-CoA in acinar cells, with some differences in substrate utilization between genotypes.
Together, our data suggest that acetyl-CoA metabolism in acinar cells both regulates histone acetylation levels and supports the mevalonate pathway. We next asked whether acetyl-CoA utilization in these pathways is functionally important for ADM. Catabolism of leucine, glucose, or fatty acids results in acetyl-CoA production within mitochondria, and acetyl-CoA is transferred to the cytosol via citrate synthesis and export (6). Inhibition of the mitochondrial citrate carrier (CiC) potently suppressed TGFα-stimulated ADM in collagen-embedded primary KC acinar cells (Fig. 2G). We therefore next interrogated the roles in ADM of acetyllysine reading using the BET inhibitor JQ1 and the mevalonate pathway using the HMG-CoA reductase (HMGCR) inhibitor atorvastatin. Both JQ1 and atorvastatin suppressed duct formation compared to treatment with vehicle alone (Figure 2G–H). These findings were also validated in Matrigel-embedded primary KC acinar cells (Supplementary Figure S2C–S2E). Importantly, supplementation with either mevalonate-5-phosphate or cholesterol rescued duct formation in the presence of atorvastatin, suggesting that cholesterol is the key product of the mevalonate pathway that facilitates ADM in KRAS mutant acinar cells (Figure 2I–J; Supplementary Figure S2F–G). Cells can synthesize cholesterol or take it up from the circulation by receptor-mediated endocytosis. Publicly-available datasets (GTEx (29)), as well as quantitative PCR (qPCR) analysis showed that expression of LDLR [encoding the low density lipoprotein receptor (LDLR)] is low in normal pancreas in both human and mouse (Supplementary Figure S2H–I). Immunohistochemistry (IHC) staining confirmed that LDLR protein is lowly expressed in acinar cells, but is more highly expressed in areas of carcinoma (Supplementary Figure S2J). Consistently, isolated acinar cells expressed lower Ldlr than murine PDA cells (Supplementary Figure S2I). These data suggest that acinar cells may have limited capacity to uptake LDL-associated cholesterol, potentially underlying a need for de novo synthesis. In addition to cholesterol synthesis, the mevalonate pathway is also important for prenylation of membrane-targeted proteins such as KRAS. However, since cholesterol rescues duct formation (Figure 2I–J) and since minimal effects on MAPK phosphorylation are observed in acinar cells the presence of atorvastatin (Supplementary Figure S2K), it is unlikely that the statin treatment suppresses ADM through direct interference with KRAS-dependent signaling. Together, these data indicate that acetyl-CoA levels are elevated in KRASG12D acinar cells and that either BET inhibition or cholesterol synthesis blockade suppresses ADM.
Pancreas-specific deletion of Acly does not cause overt metabolic abnormalities
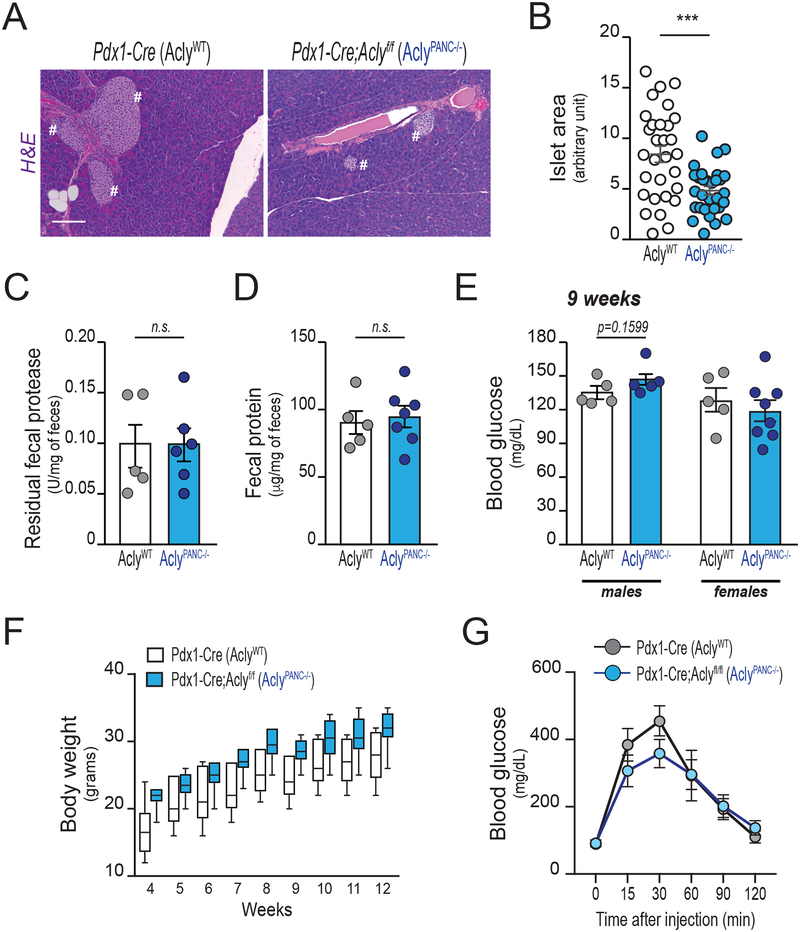
The generation of acetyl-CoA from citrate in the cytosol and nucleus is dependent on ACLY (6). To interrogate the role of ACLY in ADM and pancreatic tumorigenesis, we crossed Aclyf/f mice (30) with Pdx1-Cre mice, producing pancreas-specific ablation of Acly (AclyPANC−/− mice). These mice are viable and born at the expected Mendelian ratios. Lack of ACLY protein in whole pancreas lysates was confirmed by western blotting (Supplementary Figure S3A). Histological analysis of pancreata revealed no obvious abnormalities (Figure 3A), although islet size was significantly smaller (Figure 3B). Fecal content was not significantly different between genotypes, indicating that exocrine function is intact in the absence of ACLY (Figure 3C–D). In contrast to that observed in fibroblasts and in adipocytes upon Acly deletion (30), compensatory upregulation of ACSS2 was not observed in ACLY-deficient pancreas (Supplementary Figure S3A), suggesting that baseline levels of ACSS2 may be sufficient to supply nucleo-cytosolic acetyl-CoA in the healthy pancreas. Consistent with this interpretation, ACLY deficiency in vivo did not notably alter histone H4 acetylation in ductal or islet cells (Supplementary Figure 3kB). Histone H4 acetylation was low in acinar cells in WT mice, consistent with our prior findings (9). Since pancreatic islet size was reduced in the AclyPANC−/− mice, we further investigated whether these mice develop impairments in control of blood glucose levels. We observed minimal differences in fasting blood glucose levels (Figure 3E; Supplementary Figure S3C), or body weight in either male or female AclyPANC−/− mice (Figure 3F; Supplementary Figure S3D, respectively). Moreover, glucose tolerance was not significantly different between the two genotypes (Figure 3G), indicating that AclyPANC−/− mice can produce sufficient insulin to maintain glucose homeostasis. These data indicate that the lack of ACLY in mouse pancreas does not cause overt abnormalities in systemic glucose metabolism in unstressed mice fed a chow diet.
Figure 3: Acly deficiency in the murine pancreas does not cause overt metabolic abnormalities.
All panels depict characterization of age-matched Pdx1-Cre (AclyWT) and Pdx1-Cre;Aclyf/f (AclyPANC−/−) littermate mice (n=12, each group, unless otherwise reported). A, Hematoxylyn and eosin (H&E) staining of pancreata at 13 weeks of age (Representative images). Pound signs denote islets of Langerhans. Scale bars, 100 μm. B, Langerhans’ islets size, manually measured using ImageJ (6 slices per pancreas, each 50 μm spaced, were analyzed; n=5 mice per group). Each dot shows average islet size for each section analyzed (30 slides per group). Error bars show mean +/− SEM (***, p<0.001). C-D, fecal matter was harvested at 10 weeks of age from individually housed mice (n=6, each group). Fecal protease activity (C) and total residual protein content (D), normalized against fecal weight, mean +/− SEM. Each dot represents 1 mouse. E, blood glucose levels in male (left) and female (right) mice after overnight fast, mean +/− SEM. Each dot represents 1 mouse. F, body weight from age 4–12 weeks in male mice (n=5, each group). Boxes show 75% CI, lines show median, minimum, maximum. Difference between genotypes is not significant (ANOVA with Tukey-Kramer adjustment for multiple comparisons; p=0.9962). See Supplementary Figure 3 for data from female mice. G, Glucose tolerance test (GTT) on 10-week-old male mice (n=5, each group), mean +/− SEM.
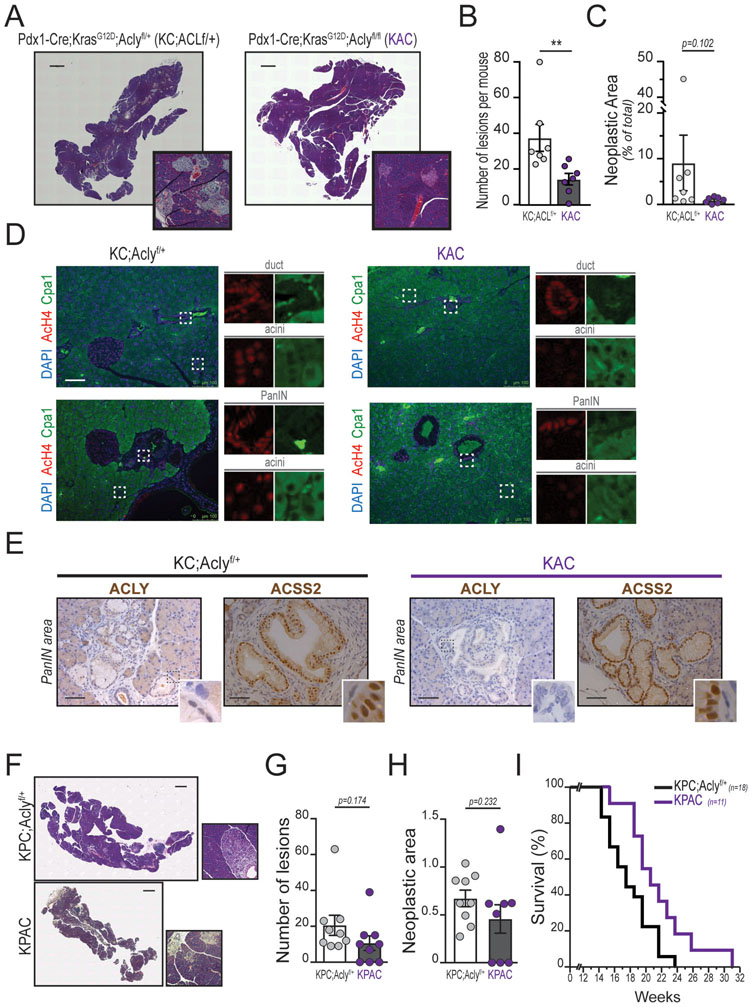
Genetic ablation of Acly inhibits acinar-to-ductal metaplasia and pancreatic tumorigenesis
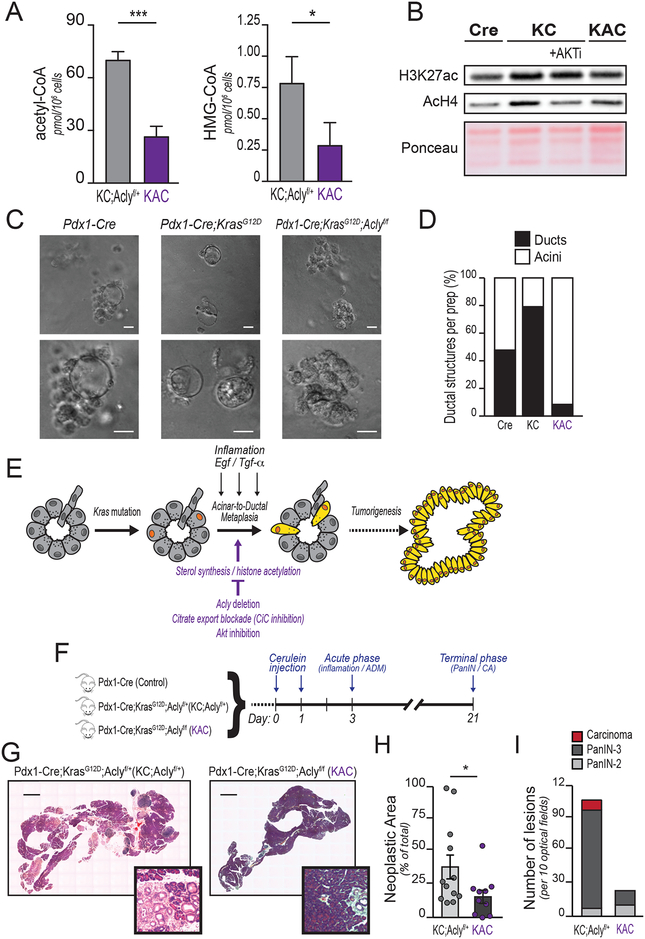
We next investigated if Acly deficiency impacts oncogenic KRAS-triggered ADM and pancreatic tumorigenesis. To test this, we crossed Pdx1-Cre;Aclyf/+ mice with LSL-KrasG12D;Aclyf/f mice to generate mice expressing mutant KRAS in the pancreatic epithelium that are either ACLY proficient (Pdx1-Cre;LSL-KrasG12D; Aclyf/+, hereafter referred to KC;Aclyf/+) or deficient (Pdx1-Cre;LSL-KrasG12D; Aclyf/f; KAC hereafter). Mice were born at the expected Mendelian ratios and developed normally. Deletion of Acly significantly reduced the abundance of acetyl-CoA and HMG-CoA in acinar cells (Figure 4A). Global histone acetylation (AcH4 and H3K27ac) levels were also suppressed in the absence of ACLY (Figure 4B). In ADM assays, ACLY deficiency strongly inhibited duct formation and preserved acinar morphology (Figure 4C–D), similar to that observed with AKT or CiC inhibition (Figure 1H and 2G, respectively). These data suggest that although ACLY is not required for normal pancreatic function, it participates in ADM, prompting us to further investigate its role in tumor formation (Figure 4E).
Figure 4: Acly deficiency impairs acinar-to-ductal metaplasia and tumor formation.
In vivo and ex vivo experiments were performed using Pdx1-Cre (Cre), Pdx1-Cre;KrasG12D (KC), Pdx1-Cre;KrasG12D;Aclyf/+ (KC;Aclyf/+) or Pdx1-Cre;KrasG12D;Aclyf/f (KAC) mice. A, LC-MS quantification of acetyl-CoA (left) and HMG-CoA (right) in isolated acinar cells (n=3 mice for KC;Aclyf/+, n=4 for KAC). Bars show mean +/− SD (***, p<0.001; *, p<0.05). B, Western blotting for acetylated histones of ex vivo acinar cells isolated from 8-week-old mice of the indicated genotypes, +/− AKTi (selective inhibitor VIII, 10 μM). Ponceau staining of acid-extracted histones is shown as a loading control. Representative of 3 independent biological repeats. C, acinar cell organoids were embedded in collagen and ADM induced with rTGFα (100 ng/mL). Images were acquired at day 5, representative images shown (n=3 independent repeat, each group). Scale bar, 20 μm. D, Day 5 organoids were scored as ductal-like (black) or acinar-like (white) in a blinded manner, according to morphology. 75 images over 3 independent experiments were evaluated. E, cartoon illustrating factors that promote ADM or restrict ADM. Kras mutant acinar cells (denoted by orange nuclei) undergo acinar-to-ductal metaplasia and become locked into a more undifferentiated morphology (cells highlighted in yellow), which eventually proliferate and evolve in carcinogenic lesions. F, Cerulein-injected mice were sacrificed at either Day 3 (inflammatory phase; n=3, each group) or Day 21 (terminal stage; n=12 KC;Aclyf/+, n=10 KAC). G, H&E staining of Day 21 pancreata. Representative images of whole tissue sections. Scale bar, 1 mm. PanIN-containing areas magnified in distinct panels. H, Total neoplastic area quantified. Each lesion’s area and whole organ surface were measured in ImageJ. Lesions areas were summed and denoted as “neoplastic area”. Percent of neoplastic area over total pancreas surface is shown. Each dot represents an animal. Error bars show mean +/− SD (*, p<0.05). I, Histopathological characterization. 10 high-power optical fields per mouse section were blindly analyzed by a veterinary pathologist. For all panels, columns show mean +/− SD (*, p<0.05; **, p<0.01; ***, p<0.001)
The cholecystokinin analog cerulein induces acute pancreatitis in mice, accompanied by widespread ADM. Whereas WT acini regenerate upon resolution of the inflammatory stimulus, in the context of Kras mutation, ADM is not resolved but instead progresses to PanIN (20). We assessed whether Acly deletion could impair tumorigenesis in the context of cerulein-induced pancreatitis (Figure 4F). After cerulein injection, mice showed a robust immune infiltration and disorganization of the pancreatic parenchyma, characterized by acinar cell loss, dilated interstitial space and presence of ADM foci, regardless of the genotype (Supplementary Figure S4A, H&E staining, upper panels). ERK phosphorylation, although less intense in the control group, was comparable in the inflammatory areas in KC;Aclyf/+ and KAC mice (Supplementary Figure S4A, pERK1/2 staining, middle panels). Staining for pACLY-S455 was observed in acinar cells of wild-type and KC;Aclyf/+ mice, but not in those of KAC mice (Supplementary Figure S4A, pACLY staining, bottom panels). Interestingly, strong pACLY positivity was also observed in infiltrating cells in all groups, serving as internal positive controls for the staining, and also suggesting that some infiltrating cells likely have high ACLY activity (Supplementary Figure S4A). By day 21, WT mice had recovered and exhibited normal pancreatic histology, as expected (data not shown). In contrast, KC;Aclyf/+ mice showed extensive neoplastic lesions and the occasional area of frank carcinoma. KAC mice had less acinar cell loss, reduced neoplastic area, and fewer high-grade lesions than KC;Aclyf/+ mice (Figure 4G–I). Western blotting of whole organ lysates confirmed efficient deletion of Acly, as well as ERK activation (Supplementary Figure S4B). ERK signaling was somewhat reduced in KAC animals (Supplementary Figure S4B), likely due to reduced neoplastic area in these animals. pERK1/2 positivity was more pronounced in neoplastic structures as compared to surrounding tissue (Supplementary Figure S4C). Lesions that formed in both genotypes were PanINs positive for ductal marker CK19 (Supplementary Figure S4D), and no differences in proliferation, as assessed by Ki67 positivity, were observed (Supplementary Figure S4E). Together, the data indicate that ACLY facilitates KRASG12D-dependent initiation of pancreatic tumorigenesis in the context of pancreatitis.
Genetic ablation of Acly extends PDA survival
We next investigated the role of ACLY in pancreatic tumorigenesis in the absence of injury. Kras mutation in murine pancreas results in the development of pre-neoplastic lesions (mostly PanIN), with PDA resulting after a long latency in 20–30% of mice (31). A lack of ACLY protein (residual signal ascribed either to infiltrating/stromal cells or to sub-absolute recombination efficiency) and robust ERK1/2 phosphorylation were confirmed (Supplementary Figure S5A). At 4 months of age, all KC;Aclyf/+ mice showed presence of numerous PanIN lesions. The number of neoplastic foci was significantly lower in age-matched KAC mice (Figure 5A, quantified in 5B), and total neoplastic area was also markedly reduced, though falling short of statistical significance due to variability in KC;Aclyf/+ mice (Figure 5C). Of those lesions that do form, both KC;Aclyf/+ and KAC mice develop similar lesions that are CK19- and Alcian Blue (mucin stain)-positive (Supplementary Figure S5B). Very few high-grade lesions (PanIN2 and PanIN3) were observed in KAC, in contrast to KC mice (Supplementary Figure S5C). Immunofluorescence staining for AcH4 and the acinar cell marker Carboxypeptidase A1 (Cpa1) showed that ACLY deletion strongly reduces histone H4 acetylation in KRAS mutant acinar cells (Figure 5D), consistent with our findings ex vivo (Figure 4B). Thus, ACLY is necessary for elevated histone acetylation in KRAS mutant acinar cells and facilitates, but is not absolutely required for, PanIN formation.
Figure 5: Pancreatic tumorigenesis is impaired in the absence of ACLY.
Aclyf/f mice were bred into a KC or KPC genotypes. A-E shows 4-month-old Pdx1-Cre;KrasG12D;Aclyf/+ (KC;Aclyf/+) or Pdx1-Cre;KrasG12D;Aclyflf (KAC) mice (n=7, each genotype). A, hematoxylin and eosin (H&E) staining transverse sections of whole pancreata. Scale bar, 1 mm. PanIN-containing areas magnified in distinct panels. Representative images shown. B, Neoplastic lesions were counted in each transverse section, and mean count per animal graphed. C, Each lesion’s area and whole organ surface were measured in ImageJ. Lesion areas were summed and denoted as “neoplastic area”. Percent of neoplastic area over total pancreas surface is shown. Each dot represents an animal, mean +/− SD (**, p<0.01). D, AcH4 immunofluorescence in pancreata of mice as in A. Whole organ sections were stained against Cpa1 (green), AcH4 (red); nuclei counterstained with DAPI. Split signals of relevant areas (denoted by white rectangles) are individually shown in distinct panels. Scale bar 100 μM. E, immunohistochemistry against ACLY and ACSS2. Nuclei counterstained with hematoxylin. Pictures show representative PanIN lesions. Scale bar 50 μM. F-I shows 9-week-old Pdx1-Cre;KrasG12D;p53L/+;Aclyf/+ (KPC;Aclyf/+) or Pdx1-Cre;KrasG12D; p53L/+;Aclyflf (KPAC) (n=9, each genotype). F, transverse sections (H&E) of whole pancreata. Magnifications of PanIN-containing areas are shown in distinct panels. Representative images shown. Scale bar 1 mm. G, Number of lesions and H, neoplastic area, assessed as in B, C. I, Kaplan-Meier survival curve. Logrank test was used to determine statistical significance (p=0.0246). Mice were sacrificed upon sudden weight loss (>15% body weight) or when showing signs of distress.
In contrast to acinar cells, both ductal cells and PanIN cells showed high levels of AcH4 staining even in the absence of ACLY (Figure 5D). ACLY protein was not detected in KAC pancreatic epithelial cells (Supplementary Figure S5D), including in PanIN structures (Figure 5E), indicating that lesions likely did not form from cells in which recombination was incomplete. Since use of acetate is a major mechanism of compensation in the absence of ACLY (30), we next asked whether ACSS2 is upregulated in ACLY-deficient tumors, potentially supporting histone acetylation and PanIN growth. Surprisingly, ACSS2 levels actually tended to be slightly higher in ACLY proficient KC mice (Supplementary Figure S5A). ACSS2 is frequently upregulated in cancerous and hypoxic regions (10,32), and thus higher ACSS2 expression in KC mice is likely due to greater representation of premalignant cells. Indeed, by immunohistochemistry, we noted intense nuclear ACSS2 staining in PanIN lesions (Figure 5E). Neither proliferation nor apoptosis was significantly different in ACLY-deficient ADM or PanIN lesions (Supplementary Figure S5E–S5F), further suggesting that ACSS2 may take on a more prominent role once tumors develop. Together, these data indicate that ACLY plays a distinct role during tumor formation, but that ACSS2 is strongly expressed in PanIN lesions and may be a key source of extra-mitochondrial acetyl-CoA to support growth of neoplastic lesions once formed.
To investigate the role of ACLY in later stages of tumor development, we allowed a small cohort of KC;Aclyf/+ and KAC mice to age to 1 year of age. 2 out of 7 KC;Aclyf/+ mice developed PDA with large intraperitoneal tumor masses and had to be sacrificed prior to 1 year of age, while all KAC mice remained viable (Supplementary Figure S5G). Of those mice that remained alive at 1 year of age, there was a strong trend towards reduced neoplastic area in KAC as compared to KC mice (Supplementary Figure S5H–S5I). To further test the impact of ACLY deficiency on survival, we genetically deleted Acly in the aggressive KPC model of PDA tumorigenesis (33). We generated KPC mice that are either ACLY proficient (KPC;Aclyf/+) or deficient (KPAC). A cohort of mice was sacrificed at 9 weeks of age, an age at which PanIN lesions have developed in KPC mice (Figure 5F). Counting of lesions (Figure 5G) and calculation of neoplastic area (Figure 5H) revealed substantial heterogeneity in KPAC mice, with some animals lacking lesions altogether at 9 weeks of age and others with lesions comparable to those in KPC mice. Although all animals developed terminal PDA regardless of Acly status, survival was significantly extended in KPAC as compared to KPC;Aclyf/+ mice (Figure 5I; median survival 21 weeks vs 18 weeks for KPC;Aclyf/+; Log-rank test: p=0.0236 *). The cumulative data indicate that ACLY deficiency hampers tumor initiation in mice, thus reducing tumor burden and improving overall survival.
Environmental factors influence histone acetylation in human PDA cell lines
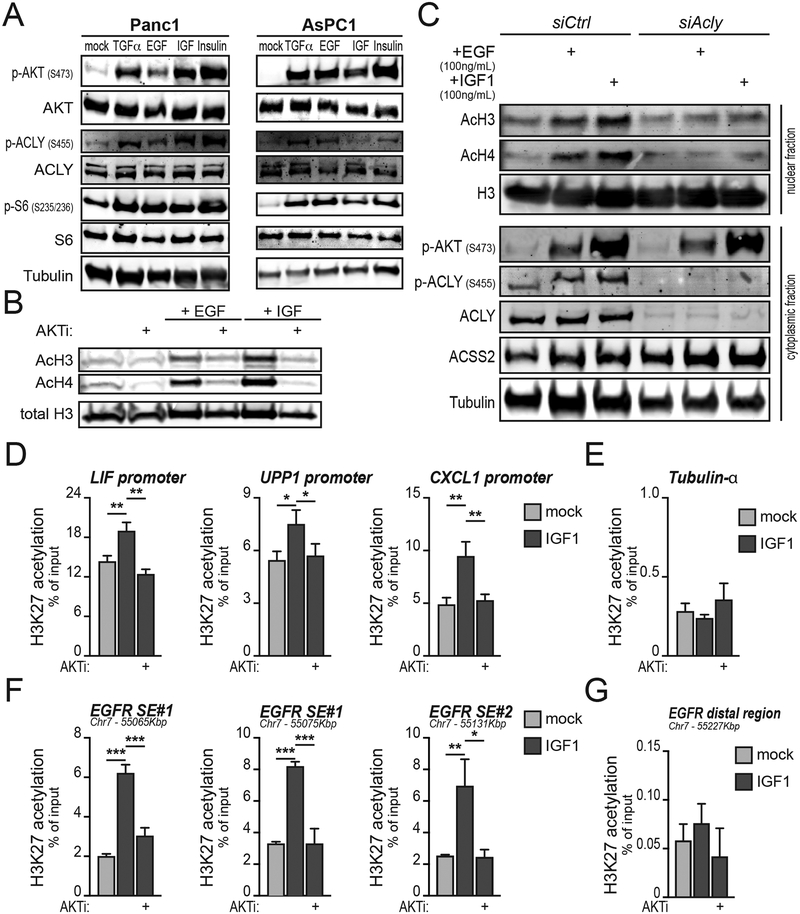
To probe the regulation of acetyl-CoA metabolism in established PDA cells, we examined the role of AKT signaling on ACLY phosphorylation and the regulation of histone acetylation in PDA cells. Surprisingly, AKTi treatment had minimal effect on histone acetylation in human PDA cells under standard culture conditions (denoted as DMEM in Supplementary Figure S6A–S6B). Although initially puzzling, this observation is consistent with evidence that in PDA, stromal elements promote AKT activation (34) and histone acetylation (15). Indeed, when cultured in an insulin-containing medium (denoted PDEC) Panc1 cells showed enhanced AKT signaling and increased histone acetylation that was suppressed by AKTi treatment (Supplementary Figure S6A). Similar findings were made in HPAC cells, but not in the KRAS WT cell line BxPC3 (Supplementary Figure S6B). PDEC medium did not alter glucose consumption rates, but did reduce lactate production in AsPC1 cells (Supplementary Figure S6C). We next specifically tested the role of exogenous growth factors in regulating AKT-ACLY signaling and histone acetylation in PDA cells. We found that EGF, TGFα, IGF, and insulin were all able to potently increase AKT and ACLY phosphorylation in PDA cell lines (Figure 6A; Supplementary Figure S6D–S6E). Growth factor stimulation increased histone acetylation, in a manner dependent on AKT and ACLY (Figure 6B–C). Thus, activation of the AKT-ACLY-histone acetylation axis in PDA cells is highly responsive to exogenous growth factor signals.
Figure 6: Environmental stimuli induce AKT-ACLY signaling and histone acetylation in PDA cells.
A, Panc1 or AsPC1 cells were serum-starved overnight and then treated with indicated growth factors for 1 hour and signaling analyzed by western blot. B, Western blotting of acetylated histones. AsPC1 cells were serum-starved overnight and then treated with either rEGF (100 ng/mL) or rIGF1 (100 ng/mL) for 24 hours, with or without AKTi VIII (10 μM), and histones acid-extracted. C, Western blotting of acetylated histones and selected cytoplasmic proteins. AsPC1 cells were transfected with siRNA targeting Acly or non-targeting control siRNA. After 56 hours, cells were treated with recombinant EGF or IGF as in B and histones acid-extracted. D-G, AsPC1 cells were treated as in B and H3K27ac ChIP was performed analyzing (D) stromal cell-regulated gene promoters (15), (E) a control gene promoter, (F) the EGFR superenhancer (SE) (35), or (G) a control (distal) region in the EFGR locus. Experiments are representative of 2 independent biological repeats. Columns show mean, +/− SD (*, p<0.05; **, p<0.01; ***, p<0.001).
To determine if growth factor stimulation regulates histone acetylation at functionally relevant loci, we investigated the regulation of previously defined PDA enhancers (35) and stromal-responsive loci (15). Chromatin immunoprecipitation (ChIP) experiments confirmed that IGF treatment significantly increased H3K27 acetylation at several stroma-regulated gene promoters in an AKT-dependent manner (Figure 6D). A control locus, TUBA1A (encoding tubulin-α) was not impacted under these conditions (Figure 6E). We also tested regulation of H3K27ac at the EGFR super-enhancer (35) using primers spanning the region (Supplementary Figure S6F). Treatment with IGF markedly increased H3K27 acetylation, and this was blocked by AKTi (Figure 6F). A distal region downstream of the enhancer was not affected (Figure 6G). Analogous results were obtained for the MYC SE locus (Supplementary Figure S6G–S6H). Thus, IGF-induced AKT signaling regulates H3K27ac at PDA enhancers and other stromal-responsive loci.
Targeting acetyl-CoA-dependent processes can inhibit PDA tumor growth
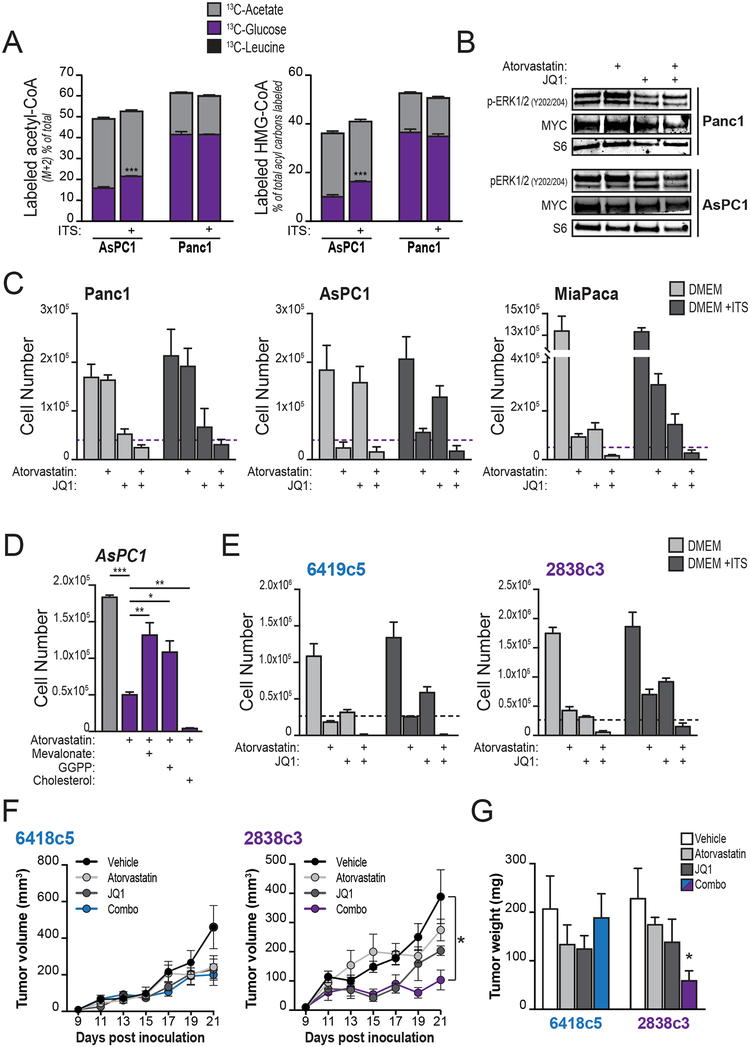
Although ACLY is important for efficient tumor formation and AKT-ACLY signaling is dynamically regulated in response to growth factor signals in PDA cells, ACSS2 may take on important roles in acetyl-CoA metabolism and allow tumors to grow independent of ACLY. To gain insights into the regulation of acetyl-CoA metabolic pathways in human PDA tumors, we surveyed gene expression using publicly available datasets (pipelined through GEPIA (36)). Expression of both ACLY and ACSS2 is elevated in human PDA (Supplementary Figure S7A). In addition, mevalonate pathway and sterol synthesis genes are upregulated in human PDA samples as compared to their expression in normal pancreata (Supplementary Figure S7A). In contrast, these genes are not upregulated in human lung adenocarcinoma (Supplementary Figure S7A). In cultured PDA cells, glucose and acetate, but not leucine, were found to be used for acetyl-CoA and HMG-CoA synthesis (Figure 7A). The insulin-containing supplement ITS also boosted the contribution of glucose to acetyl-CoA pools in AsPC-1 cells (Figure 7A). Thus, ACLY and ACSS2 likely both contribute to acetyl-CoA metabolism in PDA cells and may have at least partially overlapping roles in supplying acetyl-CoA-dependent pathways.
Figure 7: Targeting acetyl-CoA-dependent processes can suppress PDA growth.
A, AsPC1 or Panc1 cells were cultured overnight in DMEM with or without ITS+ (insulin-based supplement, BD Biosciences) and then labeled for 2 hours with indicated substrates (n=3, each condition). Percent labeling of acetyl-CoA and HMG-CoA were determined by LC-MS. Stars denote statistically different labeling from glucose. B, Western blotting shows levels of MYC and ERK1/2 phosphorylation in AsPC1 and Panc1 cells treated with atorvastatin (20 μM) and/or JQ1 (500 nM) for 4 days. C, indicated cell lines were cultured in DMEM with or without ITS+ and treated with either atorvastatin (20 μM), JQ1 (500 nM), or both for 4 days. Graphs show final cell counts. Dashed purple lines denote starting cell number (counted at day 0). Experiments are representative of 2 independent biological repeats. D, AsPC1 cells were treated with atorvastatin (20 μM) and counted after 4 days. Effect of supplementation with mevalonate (100 μM), geranylgeranyl pyrophosphate (GGPP, 100 μM) or cholesterol (12.5 μg/mL) is shown. Cholesterol was tested over a range of concentrations from 5–100 μg/mL and in all cases failed to rescue proliferation in the presence of atorvastatin (only 12.5 μg/mL data is shown). E, KPCY-derived mouse cell lines were cultured in DMEM with or without ITS+ and treated with either atorvastatin (20 μM), JQ1 (500 nM), or both for 4 days. Graphs show final cell counts. Dashed purple lines denote starting cell number (counted at day 0). Experiments are representative of 2 independent biological repeats. F, growth of two KPCY-derived tumor cell clones (6419c5, left; 2838c3, right) implanted subcutaneously into immune-competent C57Bl6/J mice and treated with either atorvastatin (10 mg/Kg), JQ1 (50 mg/Kg), or both, once a day after tumors became palpable (day 9 post-inoculation). G, tumor mass weight (same in F) after excision post-mortem. For panels A-E, bars show mean, +/− SD; for panels F-G, bars show mean +/− SEM (*, p<0.05; **, p<0.01; ***, p<0.001).
The expression of ACSS2 in tumors and major contribution of acetate to acetyl-CoA pools in PDA cells prompted us to consider that targeting acetyl-CoA-dependent processes, rather than acetyl-CoA production, might offer an opportunity to suppress tumor growth. Since mevalonate pathway gene expression is upregulated in PDA and histone acetylation is dynamically regulated in PDA cell lines (Figures 6B–F, Supplementary Figures S6A–S6B and S7A), we tested the effects of JQ1 and atorvastatin on PDA cell proliferation. At doses that only modestly impact p-ERK and MYC levels (Figure 7B), each drug individually had growth suppressive effects in some but not all of the cell lines tested (Figure 7C). Culture conditions impacted the effects of statin treatment; culturing cells in the presence of ITS partially alleviated atorvastatin-induced growth inhibition observed in DMEM (Figure 7C). The atorvastatin-mediated inhibition of proliferation was rescued by addition of either mevalonate or geranylgeranyl pyrophosphate (GGPP), but not with cholesterol (Figure 7D), suggesting that the mevalonate pathway plays a distinct role in supporting proliferation in PDA cells as compared to that in facilitating ADM. Combining JQ1 and atorvastatin profoundly inhibited cell proliferation in each of the PDA cell lines that we tested, in both the presence or absence of ITS (Figure 7C). Similar results were obtained in mouse PDA cell lines derived from Pdx1-Cre;KrasG12D;p53LSL-R172H/+;Aclyf/+;RosaYFP (KPCY) mice (37) (Figure 7E). Thus, at least in vitro, targeting the mevalonate pathway together with BET inhibition strongly suppresses PDA cell proliferation.
To test the impact of JQ1 and atorvastatin on tumor growth in vivo, we took advantage of the fact that the KPCY PDA cell lines were generated in C57Bl6/J mice, allowing us to conduct allograft experiments in immune competent animals. Growth of tumors from the 2838c3 clone was markedly suppressed upon combined administration of JQ1 and atorvastatin, demonstrating the proof-of-principle that targeting these acetyl-CoA-dependent pathways can suppress tumor growth (Figure 7F–G). Tumors generated from a second clone, 6419c5, however, were less responsive, suggesting that additional factors in vivo modify responses to these drugs (Figure 7F–G). Although determining the basis for this difference in responsiveness will require further investigation, these two clones elicit dramatic differences in the tumor microenvironment that impact therapeutic responses (37), raising the possibility that non-cell-autonomous effects influence the efficacy of this combination therapy. These data indicate that targeting acetyl-CoA-dependent pathways can suppress PDA tumor growth in vivo and that treatment responses may be impacted by factors such as the composition of the tumor microenvironment.
DISCUSSION
In this study we report that generation of nucleo-cytosolic acetyl-CoA by ACLY plays a key role in facilitating pancreatic tumor formation. We further find that once tumors form, ACSS2 is highly expressed and tumors can grow even in the absence of ACLY, but that targeting acetyl-CoA dependent pathways, using combined BET inhibition and statin treatment, can suppress proliferation and tumor growth. In regards to tumor formation, we show that KRAS mutant acinar cells exhibit increased acetyl-CoA abundance, correlating with elevated global histone acetylation. Inhibition of AKT, an upstream regulator of ACLY, inhibition of mitochondrial citrate export, or genetic deletion of Acly suppresses KRAS-induced histone acetylation and impairs ADM. Duct formation was also attenuated by either BET inhibition or mevalonate pathway inhibition, suggesting that acetyl-CoA is required for histone acetylation and cholesterol biosynthesis during ADM. We next specifically interrogated the role of ACLY in pancreatic tumorigenesis using genetic models. Importantly, mice lacking Acly in the pancreas developed normally and did not exhibit overt impairments in systemic glucose homeostasis. Strikingly, however, the absence of ACLY reduced acetyl-CoA levels in KRAS mutant acinar cells, impaired PanIN formation, and extended survival in the aggressive KPC model. Finally, we found that AKT-ACLY signaling potently regulates histone acetylation globally and H3K27ac at PDA enhancers. Concurrent BET inhibition and statin treatment potently suppressed proliferation in PDA cell lines and could also suppress tumor growth in vivo in immune-competent mice. Together the data point to a key role for ACLY in early pancreatic tumorigenesis and suggest the potential to target acetyl-CoA-dependent processes in PDA.
The findings of this study implicate ACLY-dependent production of acetyl-CoA as playing crucial roles in early stages of pancreatic tumorigenesis. Lineage tracing studies and genetic models in which mutant KRAS is expressed specifically in acinar cells have revealed that ADM can initiate a multi-step carcinogenic process in the mouse. Although its relevance for the human disease has not been definitively demonstrated, human acinar cells can undergo ADM in vitro (38), and putative precursors (AcTUB+/DLCK1+) are present both in mouse and human pre-neoplastic foci (39,40). In addition, ADM is observed in patients with chronic pancreatitis, who are at elevated risk for PDA, supporting the notion that human PDA can arise from ADM foci (20). Chromatin remodelers are known to participate in neoplastic lesion development (41), and dynamic changes in histone methylation and histone acetylation have been observed during ADM or tissue regeneration after pancreatic injury (42,43). Nevertheless, a comprehensive epigenomic analysis of ADM during tissue injury or in response to mutant KRAS signaling has not been reported. Our findings confirm and build on prior work that has shown that BET inhibition suppresses ADM and impairs pancreatic tumorigenesis (17). Further work is needed to evaluate the impact of ACLY on genome-wide histone modification, chromatin accessibility, and gene regulation during metaplasia. Of note, we found that supplementation with exogenous acetate does not rescue ADM upon AKT inhibition or in the absence of ACLY (data not shown), suggesting a unique role for ACLY-dependent generation of acetyl-CoA. Emerging evidence suggests that ACLY and ACSS2 have some ability to compensate for one another but also each have distinct roles, particularly in chromatin regulation, likely related to a need for spatial and temporal control of acetyl-CoA production for specific functions (44).
The roles of metabolism in ADM are also poorly understood, although there are hints in the literature that metabolic alterations might participate. In particular, mutant KRAS signaling in acinar cells has been shown to stimulate increased mitochondrial reactive oxygen species production, and that this is critical for ADM (45). Our data implicate for the first time a role for cholesterol synthesis in ADM. Although the mechanisms through which cholesterol acts remain to be determined, possibilities include regulation of hedgehog signaling (46), which has been implicated in acinar cell regeneration after injury and early carcinogenesis (47,48), or lipid rafts (49), which are cholesterol-rich and promote EGFR signaling (50–52). Cholesterol has also recently been shown to stimulate proliferation of intestinal stem cells and to promote tumor formation (53), suggesting that cholesterol metabolism might play similar roles in colon and pancreatic carcinogenesis. Our findings indicate that BCAAs are the primary source of acetyl-CoA used within the mevalonate pathway for cholesterol synthesis. This is consistent with a recent in vivo isotope tracing study that demonstrated that BCAAs contribute prominently to the TCA cycle in the pancreas (27). Although BCAA catabolism is suppressed in PDA as compared to normal pancreas and BCAT enzymes are dispensable for the growth of PDA tumors (54), the data suggest that the role of BCAAs in ADM and pancreatic carcinogenesis warrants further investigation. The data implicating cholesterol synthesis in pancreatic carcinogenesis thus open up a number of questions for future investigation and could have implications for disease prevention.
The findings that atorvastatin suppresses ADM and, when combined with BET inhibition, can reduce tumor growth aligns with an extensive epidemiological literature that suggest that statins, which are widely prescribed clinically as cholesterol-lowering agents, may also exert anti-cancer effects (55). Although analysis of two large prospective studies found no difference in pancreatic cancer risk between regular and non-regular statins users (56), a modest increase in median survival was observed for patients who had been regular statin users for the 2 years prior cancer diagnosis (57). In addition, numerous prior studies have found that statin use correlates with extended survival, particularly if precedent to pancreatic cancer diagnosis and in a high dose regimen (55). Notably, statins were found to decrease the risk of pancreatic cancer development in patients with chronic pancreatitis (58) and type-2 diabetes (59). Statins have been found to inhibit progression of PanIN to PDA and extend survival in KPC mice (60,61). Atorvastatin was also shown to delay PDA progression in a mouse model of acinar cell-derived PDA (62). Reciprocally, disruption of cilia in mouse pancreatic epithelial cells enhances KRAS-mediated tumorigenesis, via activation of the mevalonate pathway, and statin treatment reversed this effect (63). Sterol synthesis gene expression has also been linked to KRAS signaling in PDA (4,64), and inhibition of cholesterol uptake suppressed PDA cell proliferation and enhanced the effects of gemcitabine in suppressing tumor growth (64), further pointing to the importance of this pathway in PDA. Currently, a phase 3 clinical trial testing the use of atorvastatin combined with other treatments in patients with cancer is ongoing (). Our data support the hypothesis that use of statins in a combinatorial regimen has the potential to restrict tumor growth.
The data also suggest that it will be crucial to understand the role of the microenvironment and, in particular, the roles of immune cells in modulating the efficacy of statins and BET inhibitors. The two KPCY cell lines used to test the effects of these drugs on tumors growth have been shown to generate tumors with dramatically different immune compositions. Tumors grown from the 2838c3 clone, which are sensitive to the drug combination, are high in CD8+ T cells and low in myeloid derived suppressor cells (MDSCs). Tumors arising from the 6419c5 cell line, on the other hand, recruit few T cells and numerous MDSCs and are highly resistant to chemo- and immunotherapy (37). Notably, both statins and BET inhibitors can impact not only cancer cells but also immune cells, potentially contributing to their anti-cancer effects (65–68). Future studies aimed at elucidating how the tumor microenvironment impacts responses to these drugs will aid in identifying the most appropriate contexts and strategies for use of these inhibitors.
Further work is also needed to understand the distinct roles of ACLY and ACSS2 in established PDA. The data suggest that while tumor formation is impaired in the absence of ACLY, once PanINs form, it is possible for ACLY-deficient tumors to grow. Prominent nuclear ACSS2 expression and maintenance of global histone acetylation was observed in lesions, suggesting that ACSS2 may play important roles in chromatin regulation in pancreatic cancer. Although ACSS2 is known to promote tumor growth in several types of cancer including liver, breast, and brain (10,32,69), defining its role in PDA will require further study. We also noted that environmental factors potently activate AKT-ACLY signaling in PDA cells, driving elevated histone acetylation, globally and at PDA enhancers and stromal-responsive loci. Further elucidation of the impact of growth factor signals in the dynamic regulation of chromatin modifications in PDA and implications for gene regulation could also be important for understanding the impact of the tumor microenvironment on PDA tumor growth and therapeutic responses.
Collectively, these data provide evidence for involvement of acetyl-CoA metabolism in pancreatic cancer, and highlight the role of a metabolic enzyme in oncogene-driven acinar cell plasticity and tumorigenesis. These findings enhance our understanding of metabolic contribution to ADM and identify key questions for future investigation that could lead to improved strategies to prevent or potentially to treat pancreatic cancer.
METHODS
Animal studies.
All animal protocols were reviewed and approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania. Aclyf/f mice were previously described (30). To generate AclyPANC−/− mice, Aclyf/f mice were bred to Pdx1-Cre transgenic mice. To generate KAC mice, Aclyf/f mice were first bred either to Pdx1-Cre transgenic mice or LSL-KrasG12D transgenic mice. Pdx1-Cre;Aclyf/+ female mice were then crossed with LSL-KrasG12D;Aclyf/f male mice. Mice were largely born according to the Mendelian ratios, except for those concurrently carrying floxed Acly and p53 alleles, which were born at a much lower frequency, due to co-segregation (both encoded in Chromosome 11). To generate KPAC mice, LSL-KrasG12D;Aclyf/+ male mice were bred with Pdx1-Cre;p53L/+;Aclyf/f female mice. Genotyping was performed by PCR amplification of ear snips digested with Proteinase K, using primers listed in Supplementary Table 1. Effective recombination was tested by Western blotting for ACLY on whole pancreatic tissue lysates: pancreatic expression of ACLY was the sole exclusion parameter used in our studies. Unless otherwise stated, all experimental mice were a mix of male and female. The numbers of animals used per experiment are stated in the figure legends.
For the xenograft study, atorvastatin was diluted in water +5% glucose +0.1% DMSO; a total volume of 200 μL was orally administered through gavage. JQ1 was diluted in 10% (2-hydroxypropyl)-β-cyclodextrin (Cayman Chem): 1 part of JQ1 was diluted in 9 parts of solvent, as previously described (70).
C57BL/6J mice were injected subcutaneously (2.5×105 cells/mouse) with both 2838c3 and 6419c5 (37) in contralateral flanks. When tumors became palpable, mice were treated daily with atorvastatin (10 mg/Kg, gavage), JQ1 (50 mg/Kg, i.p.) and/or vehicle. Tumor volume was evaluated every other day using calipers and expressed in mm3 using the formula: V= π /6 × (Dmax2 × Dmin/2; (71)).
PanIN and PDA cell culture.
PanIN- and PDA-derived mouse primary cells were previously described (9,37). Unless otherwise noted in the text, they were cultured in a modified version of the PDEC medium as previously described (9). MiaPaca, Panc1 and BxPC3 were cultured in DMEM (GIBCO) supplemented with 10% Cosmic Calf Serum (CS). Cell lines were authenticated by short tandem repeat (STR) profiling using the Geneprint® 10 System (Promega). Data were matched against the ATCC reference database. All cell lines were matched to their identity with a score >80%. ATCC names and numbers for the cell lines used in this study are as follows; MiaPaca (ATCC# CRL-1420), Panc1 (ATCC# CRL-1469), HPAC (ATCC# CRL-2119), AsPC1 (ATCC# CRL-1682). All cell lines were routinely monitored for and confirmed to be free of mycoplasma. Our laboratories routinely perform testing for mycoplasma contamination and positive cells are immediately discarded (most recent testing: April, 13th, 2018 for AsPC1, Panc1, HPAC, BxPC3; January, 30th, 2018 for 2838c3, 6419c5. Cells are normally maintained in culture for no longer than 15 passages.
For histone extraction and ChIP experiments, cells were allowed to adhere overnight, and then medium was replaced with fresh medium containing indicated inhibitors and cultured for 24 hours, unless otherwise indicated. For gene silencing, siRNA smartpools targeting human ACLY and a non-silencing control (Horizon Discovery/ Dharmacon) were used. For proliferation assays, cells were plated in triplicate wells and allowed to adhere overnight. Cells were counted the following day (number indicated by a dashed blue line in the Figure) and culture medium was changed, and cells were allowed to proliferate for four days in the presence of indicated inhibitors. Medium was refreshed on the third day after plating.
Histone extraction.
Histones were purified using acid extraction, as previously described (9). Briefly, adherent cells were cultured in 6 well plates and nuclei were harvested in cold NIB buffer (15mM Tris-HCL pH7.5, 60mM KCl, 15mM NaCl, 5mM MgCl2, 1mM CaCl2, 250mM sucrose, freshly added: 1mM DTT, 1X protease inhibitors, 10mM sodium butyrate, 0.1% NP-40) and incubated in ice for 10 minutes with occasional vortexing. Suspension-growing acinar cells were resuspended in high-volume cold NIB buffer for 15 minutes. Nuclei were pelleted at 600 rcf for 5 min at 4 °C and washed twice using NIB buffer without NP-40. The nuclear pellets were immediately resuspended in 0.4N H2SO4 and rotated at 4 °C. After centrifugation, histones were precipitated from the supernatant by addition of 20% tricholoracetic acid (TCA) for at least 1 hour, followed by centrifugation. The pellet was washed once with acetone containing 0.1% HCl, and finally with 100% acetone. Histone proteins were dried at room temperature and resuspended in water.
Mass Spectrometry analysis of histones.
PanIN-derived cells were incubated for 24 hours in the presence of AKT inhibitor (see below) or vehicle control (DMSO). Histones were acid extracted, as described. Total histones were subjected to chemical derivatization using propionic anhydride (Sigma-Aldrich) and digested with sequencing-grade trypsin at a 10:1 substrate:enzyme ratio for 6 hr at 37°C. The digested peptides were processed as previously described (72). A resolution of 60,000 was used in the Orbitrap for the full mass spectrometry (MS), followed by MS/MS spectra collected in the ion trap. Data were subsequently analyzed with in-house software (72).
Chromatin Immunoprecipitation (ChIP).
ChIP was performed essentially as previously described (71). Briefly, were fixed on the dish with 1% formaldehyde for 10 minutes at room temperature. The reactions were quenched with 0.25 M glycine. The cells were then washed twice with 1X PBS and scraped in cell lysis buffer (10 mM Tris-HCl pH8.1, 10 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40), supplemented with protease inhibitors (Roche). The cell pellet was resuspended in 0.5 mL of nuclear lysis buffer (50 mM Tris-HCl pH 8.1, 5 mM EDTA, 1% SDS) supplemented with protease inhibitors. The chromatin was fragmented with a Diagenode Bioruptor Pico (12 cycles of 30 s on followed by 30 s off, at 4 °C). Samples were incubated with protein G magnetic beads (Millipore 16–662) and H3K27ac antibody (Abcam #ab4729) overnight at 4 °C. The next day, samples were washed 5 times with decreasingly stringent buffers. ChIP DNA was eluted off the beads by incubating beads in 125 μL elution buffer for 10 minutes at 65 °C. The combined supernatant was then incubated overnight at 65 °C to reverse crosslinks and proteinase K treated for 1 hour the next morning. Samples were purified using Macherey-Nagel DNA purification kit, with NTB binding buffer.
Quantitative PCR (qPCR).
Quantitative PCR for ChIP samples were diluted 1:20 and used as template in the Power Sybr Master Mix (ABI 4367659) and DNA was amplified using the ViiA-7 Real-Time PCR system. Primers are listed in Supplementary Table 1.
RNA was isolated from triplicate wells under each condition using TRIzol (Invitrogen) and cDNA synthesized using high-capacity RNA-to-cDNA master mix (Applied Biosystems), as per the kit instructions. cDNA was diluted 1:10 and used as template in the Power Sybr Master Mix (ABI 4367659) and DNA was amplified using the ViiA-7 Real-Time PCR system. Fold change in expression was calculated using ΔΔCt, with indicated reference gene (18S, Actin or GAPDH) as an endogenous control. All primers are listed in Supplementary Table 1.
Acinar cell isolation, culture and tracing experiment.
Acinar cells were isolated as previously described (22). Briefly, pancreata from 6–8-week-old mice were collected upon sacrifice, washed twice in cold Hank’s Balanced Salt Solution (HBSS) and subsequently minced. Tissue was then digested with 1 mg of Collagenase P (Roche #11215809103) in 5 mL HBSS at 37 °C for 30 minutes, occasionally inverting the tubes and interrupting the digestion every 10 minutes to mechanically disrupt the clogs by pipetting with progressively smaller pipette tubes. Reaction was stopped and tissue homogenate was washed twice with HBSS containing 5% CS and then filtered through a 500 μm mesh and a 100 μm cell strainer. The flow through was carefully laid onto a HBSS +30% CS solution and centrifuged. The pellet was resuspended in Waymouth’s medium containing 10% CS (supplemented with indicated inhibitors) for suspension culture. Cells were plated in low-adhesion petri dishes, incubated at 37°C for 24 hours and then harvested, unless otherwise reported.
For matrix-embedded culture, cells were resuspended in a 1:3 media:matrigel (growth-factor reduced, from Trevigen) or a 1:2 media:collagen solution (purified Rat Collagen I, from Trevigen). 250 μL of suspension were seeded onto a 48-well plate and incubated at 37 °C for solidification for at least 1 hour. Upon Matrigel solidification, 500 μL of Waymouth’s medium, containing 10% CS, 0.1 mg/mL Soybean Trypsin Inhibitor and indicated inhibitors/supplements, were added to each well. Cells were monitored daily and images acquired at Day 2 using a DMI6000B inverted light and fluorescent microscope. Upon Collagen solidification, 500 μL of Waymouth’s medium supplemented with 10% CS and 0.1 mg/mL Soybean Trypsin Inhibitor were added to each well. The following day, medium was replaced with Waymouth’s medium supplemented with 10% CS and 0.1 mg/mL Soybean Trypsin Inhibitor and Tgfα (100 ng/mL). Cells were monitored daily and images acquired at Day 5 (unless differently reported in the Figure Legend) using a DMI6000B inverted light and fluorescent microscope.
For nutrient tracing, suspension-plated cells were allowed to recover overnight in Waymouth’s medium supplemented with 10% CS and 0.1 mg/mL Soybean Trypsin Inhibitor. In the morning, cells were collected and rapidly spun down and resuspended in glucose- and leucine-free DMEM +10% dialyzed FBS (Gibco) supplemented with either 20 mM [U-13C]-glucose (Cambridge Isotope Laboratories #CLM-1396–1) or 500 μM [U-13C]-leucine (Isotec, MilliporeSigma #605239), plated in low-adhesion petri dishes and incubated at 37°C for 8 hours. 500 μM cold leucine and 20 mM of cold glucose were also respectively supplemented. For palmitate tracing, cells were spun down and resuspended in DMEM containing 120 μM of [13C]-palmitate and supplemented with 10% charcoal-stripped FBS (Gibco) and cultured as above for 8 hours.
Blood glucose measurement and glucose tolerance test.
Blood glucose levels were measured from tail in either 9-week-old or 13-week-old mice after overnight fast using a portable glucometer (Breeze2, Bayer Pharmaceuticals). For glucose tolerance test (GTT), mice were fasted overnight and weighed. Blood glucose levels were measured in the morning prior to glucose administration and values were used as t=0 reference points. Mice were injected with 2.0 g/kg body weight of glucose (resuspended in PBS, filtered) via intraperitoneal injection. Blood glucose was measured by glucometer at 15, 30, 60, 90, and 120 minutes after injection.
Immunofluorescence of acinar cell explants.
Acinar cells were harvested from mice, plated in matrigel, and treated as indicated. Cells were fixed with 4% paraformaldehyde for 15 minutes and permeabilized with 0.5% TritonX/PBS for 10 mins at 4 degrees. Cells were then blocked in 10% goat serum in IF Wash buffer for one hour at room temperature. Primary antibodies (AcH4; 1:250) were incubated overnight at 4 degrees in IF Wash buffer + 10% goat serum. Secondary antibody was incubated with DAPI in IF Wash (+ 10% goat serum) for 1 hour at RT. Cells were immediately imaged. IF Wash buffer: 130 mM NaCl2, 13 mM Na2HPO4, 3.5 mM NaH2PO4, 7.5 mM NaN3, 0.1% BSA, 0.2% Triton-X 100.
For quantification, fluorescence signals were quantified with ImageJ counting the number of pixels that exceeded a fixed background threshold, as previously described (73).
Antibodies and reagents.
Inhibitors used were as follows: Akt inhibitor VIII (Calbiochem, 10 μM), MK2206 (Selleckchem #S1078, 5 μM), PD325901 (LC Laboratories, 1 μM), FASN inhibitor (Orlistat: Cayman Chemical #CAS96829-58-2; 30 μM), HMGCR inhibitor (Atorvastatin, Cayman Chemicals #10493; 20 μM), JQ1 (AdooQ Biosciences #A12729 for in vivo experiments; in vitro quantities provided by James E Bradner laboratory, Harvard; 500nM unless specified in the text), 1,2,3-Benzenetricarboxylic acid (BTA, Sigma #B420–1).
Recombinant proteins and supplements used for cell treatments were: recombinant murine EGF (Peprotech #215–09; 100 ng/mL); recombinant TGFα (R&D Systems, 100 ng/mL); recombinant murine IGF-1 (Peprotech #250–19, 200 ng/mL); bovine-extracted insulin (Sigma #I5500; 50 ng/mL); Cholesterol (SyntheChol NSO supplement – synthetic cholesterol – Sigma #S5442; 12.5 μg/mL); Mevalonate ((±)-Mevalonic acid 5-phosphate trilithium salt hydrate; Sigma #S79849; 100 μM); ITS+ Premix (Fisher, #CB40352).
Antibodies used for Western blots include: AcH3, AcH4, H3, H4 (from Millipore), pAKT-Ser473, AKT, pACLY-Ser455, pERK, ERK, pS6, S6, H4K8ac, H3K23ac, ACSS2 (from Cell Signaling), tubulin (Sigma), H3K27ac, H3K9ac (from Abcam), ACLY (previously described (8)). Secondary antibodies were IRDye680RD Goat Anti-Mouse (LI-COR 926–68070) and IRDye800CW Goat Anti-Rabbit (LI-COR 926–32211). Blots were analyzed using a LICOR Odyssey CLx blot scanner.
Antibodies used for IHC on murine tissues as follows: AcH4 (1:4000; clone 06–759-MN, Millipore), Ki67 (1:250; #ab16667, AbCam) pACLY (1:100; #SAB4504020, Sigma), pERK (1:250; #9101, Cell Signaling), ACLY (1:100; #ARP42737_P050, AvivaBiosystems), cleaved Caspase-3 (1:200; #9661, Cell Signaling), ACSS2 (1:250; #3658S, Cell Signaling), H3K27ac (1:3000; #ab4729, Cell Signaling), LDLR (1:200; #AF2255, R&D Systems).
Antibodies used for IF on murine tissues as follows: AcH4 (1:2000; clone 06–759-MN, Millipore), CPA1 (1:200; #AF2765, R&D Systems).
Immunohistochemistry and analysis of murine pancreatic tissue.
For histological evaluation, tissue samples were harvested as described (9). Importantly, pancreata were laid on a planar surface and fixed with formalin overnight. Sectioning of paraffin-embedded tissues exposed the transverse axis and revealed whole organ morphology (4 μm sections, Abramson Family Cancer Research Institute Histopathology Core). For tumor evaluation, images of whole pancreata were reconstructed through software-guided tile merging. ImageJ software was used for quantification of the area of entire pancreata as well as of individual neoplastic foci. Similarly, area of Langerhans’ islets was quantified. For each mouse, 3 consecutive tissue slides were analyzed. For each slide, area of Langerhans’ islets was summed. Data points represent islets’ total area in each individual slide. The number of animals evaluated in each experiment is reported in the Figure Legends.
Immunohistochemistry was performed on paraffin-embedded sections. Tissue sections were dewaxed and rehydrated. Antigen retrieval was performed by boiling samples in citrate buffer for 20 min and endogenous peroxidase was blunted by incubating samples with 3% H2O2 for 10 min. Primary antibody was incubated overnight at 4 °C. For Alcian blue staining, rehydrated paraffin sections were stained with 1% Alcian blue 8GX in 3% acetic acid (pH 2.5) for 30 minutes and counterstained with hematoxylin. Histopathological scoring was performed by a trained veterinary pathologist (E.L. Buza, UPenn) in a blinded manner. For each animal, 10 low-magnification images from different, non-overlapping microscopic fields were analyzed. Duct-like structures were counted and histologically evaluated according to the consensus criteria (74). At least 7 mice per genotype were analyzed, and data were represented as the average number of lesions per optical field.
Acyl-CoA quantification and isotopologue analysis.
Acyl-CoA analyses were performed by liquid chromatograpy-mass spectrometry/high-resolution mass spectrometry (LC-MS/HRMS) as previously described (75). Briefly, approximately 20–50 ×106 acinar cells were cultured in suspension in low adhesion 6 mm petri dishes. At harvest, cells were placed on ice, transferred to 15 ml falcon tubes and centrifuged at 600 xg for 2 minutes at 4 °C. Medium was aspirated and the cell pellet resuspended in 1 mL 10% (w/v) trichloroacetic acid (Sigma-Aldrich, catalog #T6399) for acyl-CoA extraction. For quantification, an equal amount (100 μL) of 13C315N1-labeled acyl-CoA internal standard (75) was added to each sample. Samples were pulse sonicated, centrifugated and the supernatant was purified by solid-phase extraction using Oasis HLB 1cc (30 mg) SPE columns (Waters). Eluate was evaporated to dryness under nitrogen gas and re-suspended in 50 μL of 5 % 5- sulfosalicylic acid (w/v) for injection. Samples were analyzed by an Ultimate 3000 autosampler coupled to a Thermo Q-Exactive Plus instrument in positive electrospray ionization (ESI) mode. For isotopic tracer analysis, isotopic enrichment from [U-13C]glucose or [U-13C]palmitate or [U-13C]leucine was calculated to compensate for the non-linearity of isotopic enrichment using the FluxFix calculator (76).
Cerulein treatment.
Acute pancreatitis was induced in 6–7-week-old mice of the indicated genotype. Mice were injected with caerulein (50 μg/kg diluted in saline; Sigma-Aldrich) or saline on two consecutive days once every hour for eight hours each day. First day of injection was considered Day 0. Number of animals are indicated in Figure legends.
Protein lysate preparation for western blotting.
The lysates were sonicated at 30% duty cycle and an output control setting of 3–4. The sonicated samples were pelleted for 5 minutes and 16,000 rcf and the supernatant was collected and quantified by BSA (Thermo Scientific).
YSI Metabolite measurements.
Glucose and glutamine consumption and lactate production were measured using a YSI 2950 Bioanalyzer. Measurements were conducted over a 24 hr time period and normalized to cell number area under the curve, as previously described (9).
Human gene expression profiling.
Gene expression was assessed though a web-based tool (GEPIA; (36)). Provisional datasets from TCGA and GTEx consortiums were used for the analysis.
Fecal analysis of exocrine activity.
To assess total fecal protein, 10 mg of feces was resuspended in lysis buffer (2% SDS, 150 mM NaCl, 0.5 M EDTA), sonicated and the protein concentration was assessed using a BCA assay. Total fecal protease activity was measured in 10 mg of fecal matter, resuspended in 1 ml of buffer A (0.1%Triton X-100, 0.5 M NaCl, 100 mM CaCl2), sonicated and centrifuged. The supernatant was then incubated with 3% Azo-Casein (Sigma-Aldrich, A2765) dissolved in 50 mM Tris·HCl buffer, pH 8.5 at 37 °C for 60 min. The reaction was stopped using 8% trichloroacetic acid and centrifuged. The absorbance of the supernatant (measured at 366 nm) was measured using a spectrophotometer.
Statistical analysis.
Data are presented as the means of experimental replicates with their respective standard deviations (SD), unless otherwise indicated. Student’s two-tailed t tests (two-sample equal variance, two-tailed distribution) were used for analyses, unless otherwise indicated. Repeated measures ANOVA with Tukey-Kramer adjustment for multiple comparisons was used to evaluate significant differences in body weight. For survival, logrank test was performed using GraphPad Prism. For tumor growth, two-way ANOVA was calculated using GraphPad Prism. Significance was defined as follows: *, p<0.05; **, p<0.01; ***, p<0.001.
Supplementary Material
SIGNIFICANCE.
Pancreatic cancer is amongst the deadliest of human malignancies. We identify a key role for the metabolic enzyme ACLY, which produces acetyl-CoA, in pancreatic carcinogenesis. The data suggest that acetyl-CoA use for histone acetylation and in the mevalonate pathway facilitates cell plasticity and proliferation, suggesting potential to target these pathways.
ACKNOWLEDGMENTS
The authors acknowledge Cynthia Clendenin and the staff at the Pancreatic Cancer Mouse Hospital for their support for the in vivo drug treatment experiments. The authors thank Jinyang Li (Stanger Lab) for providing C57Bl/6 KPCY PDA cell lines. The authors also thank the PennVet Core (Elizabeth Buza) for histological evaluation of murine pancreatic tumor samples. A.C. thanks Kyoung Lee for her help in establishing acinar cell culture.
FINANCIAL SUPPORT: This work was supported by the 2014 Pancreatic Cancer Action Network-AACR Career Development Award, Grant Number 14-20-25-WELL, and the NIH grants R01CA174761, and R01CA228339 to K.E.W. This project was also funded in part under a grant with the Pennsylvania Department of Health to K.E.W and B.Z.S. The Department specifically disclaims responsibility for any analyses, interpretations, or conclusions. N.W.S. is supported by the NIH grant R03HD092630. B.A.G. acknowledges NIH funding GM110174, AI118891 and CA196539. S.L.C. is supported by NIH pre-doctoral fellowship 1F31CA217070–01. S.T. is supported by the American Diabetes Association through post-doctoral fellowship #1–18-PDF-144. S.Z. received funding from the NIH transition award F99CA222741. J.L.D.P. was supported by the UPenn Baccalaureate Research Education fellowship R25-GM071745. H.C.A. is supported by NIH transition award 4K00CA212455–03.
Footnotes
CONFLICT OF INTEREST: The authors claim no conflicts with the presented work.
REFERENCES
- 1.Borazanci E, Dang CV, Robey RW, Bates SE, Chabot JA, Von Hoff DD. Pancreatic Cancer: “A Riddle Wrapped in a Mystery inside an Enigma”. Clin Cancer Res. 2017;23:1629–1637. Available from: http://clincancerres.aacrjournals.org/content/23/7/1629.abstract [DOI] [PubMed] [Google Scholar]
- 2.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, et al. EMT and dissemination precede pancreatic tumor formation. Cell. Elsevier Inc; 2012;148:349–61. Available from: 10.1016/j.cell.2011.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noone AM, Howlander N, Krapcho M, Miller D, Brest A, et al. , SEER Cancer Statistics Review, 1975–2015, National Cancer Institute. Bethesda, MD: Available from: https://seer.cancer.gov/csr/1975_2015/, based on November 2017 SEER data submission, posted to the SEER web site, April 2018. [Google Scholar]
- 4.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. Elsevier Inc; 2012;149:656–70. Available from: 10.1016/j.cell.2012.01.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halbrook CJ, Lyssiotis CA. Employing Metabolism to Improve the Diagnosis and Treatment of Pancreatic Cancer. Cancer Cell. Elsevier; 2017. page 5–19. [DOI] [PubMed] [Google Scholar]
- 6.Carrer A, Wellen KE. Metabolism and epigenetics: A link cancer cells exploit. Curr Opin Biotechnol. Elsevier Current Trends; 2015;34:23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi L, Tu BP. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr Opin Cell Biol. Elsevier Current Trends; 2015;33:125–31. Available from: https://www.sciencedirect.com/science/article/pii/S0955067415000125?via%3Dihub [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV., Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science (80-). American Association for the Advancement of Science; 2009;324:1076–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JV, Carrer A, Shah S, Snyder NW, Wei S, Venneti S, et al. Akt-dependent metabolic reprogramming regulates tumor cell Histone acetylation. Cell Metab. 2014;20(2):306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, Alam IS, et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. Elsevier; 2015=;27:57–71. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25584894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahar S, Masubuchi S, Eckel-Mahan K, Vollmer S, Galla L, Ceglia N, et al. Circadian control of fatty acid elongation by SIRT1 protein-mediated deacetylation of acetyl-coenzyme A synthetase 1. J Biol Chem. American Society for Biochemistry and Molecular Biology; 2014;289:6091–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24425865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carrer A, Parris JLD, Trefely S, Henry RA, Montgomery DC, Torres A, et al. Impact of a high-fat diet on tissue Acyl-CoA and histone acetylation levels. J Biol Chem. American Society for Biochemistry and Molecular Biology; 2017;292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dangi-Garimella S, Sahai V, Ebine K, Kumar K, Munshi HG. Three-Dimensional Collagen I Promotes Gemcitabine Resistance In Vitro in Pancreatic Cancer Cells through HMGA2-Dependent Histone Acetyltransferase Expression. Cukierman E, editor. PLoS One. Public Library of Science; 2013;8:e64566 Available from: http://dx.plos.org/10.1371/journal.pone.0064566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juliano CN, Izetti P, Pereira MP, dos Santos AP, Bravosi CP, Abujamra AL, et al. H4k12 and H3k18 Acetylation Associates With Poor Prognosis in Pancreatic Cancer. Applied Immunohistochemistry & Molecular Morphology; 2016;24:337–44. Available from: https://insights.ovid.com/pubmed?pmid=26067135 [DOI] [PubMed] [Google Scholar]
- 15.Sherman MH, Yu RT, Tseng TW, Sousa CM, Liu S, Truitt ML, et al. Stromal cues regulate the pancreatic cancer epigenome and metabolome. Proc Natl Acad Sci. National Academy of Sciences; 2017;114:1129–34. Available from: http://www.pnas.org/lookup/doi/10.1073/pnas.1620164114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald OG, Li X, Saunders T, Tryggvadottir R, Mentch SJ, Warmoes MO, et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet. Nature Publishing Group; 2017;49:367–76. Available from: http://www.nature.com/articles/ng.3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazur PK, Herner A, Mello SS, Wirth M, Hausmann S, Sánchez-Rivera FJ, et al. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat Med. Nature Publishing Group; 2015;21:1163–71. Available from: http://www.nature.com/articles/nm.3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy N, Malik S, Villanueva KE, Urano A, Lu X, Von Figura G, et al. Brg1 promotes both tumor-suppressive and oncogenic activities at distinct stages of pancreatic cancer formation. Genes Dev. Cold Spring Harbor Laboratory Press; 2015;29:658–71. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25792600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia PL, Miller AL, Kreitzburg KM, Council LN, Gamblin TL, Christein JD, et al. The BET bromodomain inhibitor JQ1 suppresses growth of pancreatic ductal adenocarcinoma in patient-derived xenograft models. Oncogene. Nature Publishing Group; 2016;35:833–45. Available from: http://www.nature.com/articles/onc2015126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storz P Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat Rev Gastroenterol Hepatol. Nature Publishing Group; 2017;14:296–304. Available from: http://www.nature.com/articles/nrgastro.2017.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey JM, DelGiorno KE, Crawford HC. The secret origins and surprising fates of pancreas tumors. Carcinogenesis. Oxford University Press; 2014;35:1436–40. Available from: https://academic.oup.com/carcin/article-lookup/doi/10.1093/carcin/bgu056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baer R, Cintas C, Dufresne M, Cassant-Sourdy S, Schönhuber N, Planque L, et al. Pancreatic cell plasticity and cancer initiation induced by oncogenic Kras is completely dependent on wild-type PI 3-kinase p110α. Genes Dev. Cold Spring Harbor Laboratory Press; 2014;28:2621–35. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25452273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanger BZ, Stiles B, Lauwers GY, Bardeesy N, Mendoza M, Wang Y, et al. Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer Cell. Elsevier; 2005;8:185–95. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16169464 [DOI] [PubMed] [Google Scholar]
- 24.Eser S, Reiff N, Messer M, Seidler B, Gottschalk K, Dobler M, et al. Selective requirement of PI3K/PDK1 signaling for kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell. Elsevier; 2013;23:406–20. [DOI] [PubMed] [Google Scholar]
- 25.Wu C-YC, Carpenter ES, Takeuchi KK, Halbrook CJ, Peverley LV, Bien H, et al. PI3K regulation of RAC1 is required for KRAS-induced pancreatic tumorigenesis in mice. Gastroenterology. Elsevier; 2014;147:1405–16.e7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25311989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ying H, Elpek KG, Vinjamoori A, Zimmerman SM, Chu GC, Yan H, et al. PTEN is a major tumor suppressor in pancreatic ductal adenocarcinoma and regulates an NF-κB-cytokine network. Cancer Discov. American Association for Cancer Research; 2011;1:158–69. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21984975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neinast MD, Jang C, Hui S, Anthony TG, Rabinowitz JD, Arany Z, et al. Quantitative Analysis of the Whole-Body Metabolic Fate of Branched-Chain Amino Acids Article Quantitative Analysis of the Whole-Body Metabolic Fate of Branched-Chain Amino Acids. Cell Metabol. Elsevier; 2019;1–13. [Google Scholar]
- 28.Mullen PJ, Yu R, Longo J, Archer MC. The interplay between cell signalling and the mevalonate pathway in cancer. Nat Rev Cancer. Nature Publishing Group; 2016;16:718–31. Available from: 10.1038/nrc.2016.76 [DOI] [PubMed] [Google Scholar]
- 29.Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. Nature Publishing Group; 2013;45:580 Available from: 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao S, Torres AM, Henry RA, Trefely S, Wallace M, Lee JVJV, et al. ATP-Citrate Lyase Controls a Glucose-to-Acetate Metabolic Switch. Cell Rep. Elsevier; 2016;17:1037–52. Available from: 10.1016/j.celrep.2016.09.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. Elsevier; 2003;4:437–50. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14706336 [DOI] [PubMed] [Google Scholar]
- 32.Comerford SA, Huang Z, Du X, Wang Y, Cai L, Witkiewicz AK, et al. Acetate dependence of tumors. Cell. Elsevier; 2014;159:1591–602. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25525877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. Elsevier; 2005;7:469–83. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15894267 [DOI] [PubMed] [Google Scholar]
- 34.Tape CJ, Ling S, Dimitriadi M, McMahon KM, Worboys JD, Leong HS, et al. Oncogenic KRAS Regulates Tumor Cell Signaling via Stromal Reciprocation. Cell. Elsevier; 2016;165:910–20. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27087446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. Elsevier; 2013;155:934–47. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24119843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Byrne KT, Yan F, Yamazoe T, Chen Z, Baslan T, et al. Tumor Cell-Intrinsic Factors Underlie Heterogeneity of Immune Cell Infiltration and Response to Immunotherapy. Immunity. Elsevier; 2018;1–16. Available from: 10.1016/j.immuni.2018.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houbracken I, de Waele E, Lardon J, Ling Z, Heimberg H, Rooman I, et al. Lineage tracing evidence for transdifferentiation of acinar to duct cells and plasticity of human pancreas. Gastroenterology. Elsevier; 2011;141:731–41, 741.e1–4. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21703267 [DOI] [PubMed] [Google Scholar]
- 39.Delgiorno KE, Hall JC, Takeuchi KK, Pan FC, Halbrook CJ, Washington MK, et al. Identification and manipulation of biliary metaplasia in pancreatic tumors. Gastroenterology. Elsevier; 2014;146:233–244.e5. Available from: 10.1053/j.gastro.2013.08.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey JM, Alsina J, Rasheed ZA, McAllister FM, Fu YY, Plentz R, et al. DCLK1 marks a morphologically distinct subpopulation of cells with stem cell properties in preinvasive pancreatic cancer. Gastroenterology. Elsevier, Inc; 2014;146:245–56. Available from: 10.1053/j.gastro.2013.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Von Figura G, Fukuda A, Roy N, Liku ME, Morris Iv JP, Kim GE, et al. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol. Nature Publishing Group; 2014;16:255–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, Iacobuzio-Donahue CA, et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. Elsevier; 2003;3:565–76. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12842085 [DOI] [PubMed] [Google Scholar]
- 43.Chen N-M, Singh G, Koenig A, Liou G-Y, Storz P, Zhang J-S, et al. NFATc1 Links EGFR Signaling to Induction of Sox9 Transcription and Acinar-Ductal Transdifferentiation in the Pancreas. Gastroenterology. Elsevier; 2015;148:1024–1034.e9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25623042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sivanand S, Viney I, Wellen KE. Spatiotemporal Control of Acetyl-CoA Metabolism in Chromatin Regulation. Trends Biochem Sci. Elsevier Current Trends; 2018;43:61–74. Available from: 10.1016/j.tibs.2017.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liou GY, Döppler H, DelGiorno KE, Zhang L, Leitges M, Crawford HC, et al. Mutant KRas-Induced Mitochondrial Oxidative Stress in Acinar Cells Upregulates EGFR Signaling to Drive Formation of Pancreatic Precancerous Lesions. Cell Reports. Elsevier; 2016;14:2325–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.di Magliano MP, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer. Nature Publishing Group; 2003;3:903–11. Available from: http://www.nature.com/doifinder/10.1038/nrc1229 [DOI] [PubMed] [Google Scholar]
- 47.Fendrich V, Esni F, Garay MVR, Feldmann G, Habbe N, Jensen JN, et al. Hedgehog Signaling Is Required for Effective Regeneration of Exocrine Pancreas. Gastroenterology. Elsevier; 2008;135:621–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pasca M, Sekine S, Ermilov A, Ferris J, Dlugosz AA, Hebrok M. Hedgehog / Ras interactions regulate early stages of pancreatic cancer. Genes Dev. Cold Spring Harbor Laboratory Press; 2006;3161–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sezgin E, Levental I, Mayor S, Eggeling C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat Rev Mol Cell Biol. Nature Publishing Group; 2017;18:361–74. Available from: http://www.nature.com/doifinder/10.1038/nrm.2017.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lubeseder-Martellato C, Alexandrow K, Hidalgo-Sastre A, Heid I, Boos SL, Briel T, et al. Oncogenic KRas-induced Increase in Fluid-phase Endocytosis is Dependent on N-WASP and is Required for the Formation of Pancreatic Preneoplastic Lesions. EBioMedicine. Elsevier; 2017;15:90–9. Available from: 10.1016/j.ebiom.2016.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kishi S, Fujiwara-Tani R, Luo Y, Kawahara I, Goto K, Fujii K, et al. Pro-metastatic signaling of the trans fatty acid elaidic acid is associated with lipid rafts. Oncol Lett. Spandidos Publishing Group; 2018;15:4423–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sukhanova A, Gorin A, Serebriiskii IG, Gabitova L, Zheng H, Restifo D, et al. Targeting C4-demethylating genes in the cholesterol pathway sensitizes cancer cells to EGF receptor inhibitors via increased EGF receptor degradation. Cancer Discov. American Association for Cancer Research; 2013;3:96–111. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23125191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang B, Rong X, Palladino END, Wang J, Fogelman AM, Martín MG, et al. Phospholipid Remodeling and Cholesterol Availability Regulate Intestinal Stemness and Tumorigenesis. Cell Stem Cell. Elsevier; 2018;22:206–220.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayers JR, Torrence ME, Danai LV, Papagiannakopoulos T, Davidson SM, Bauer MR, et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science. American Association for the Advancement of Science; 2016;353:1161–5. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27609895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clendening JW, Penn LZ. Targeting tumor cell metabolism with statins. Oncogene. Macmillan Publishers Limited; 2012;31:4967 Available from: 10.1038/onc.2012.6 [DOI] [PubMed] [Google Scholar]
- 56.Hamada T, Khalaf N, Yuan C, Babic A, Morales-Oyarvide V, Qian ZR, et al. Statin use and pancreatic cancer risk in two prospective cohort studies. J Gastroenterol. Springer Japan; 2018;53(8):959–966. Available from: 10.1007/s00535-018-1430-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hamada T, Khalaf N, Yuan C, Morales-Oyarvide V, Babic A, Nowak JA, et al. Pre-diagnosis Use of Statins Associates With Increased Survival Times of Patients With Pancreatic Cancer. Clin Gastroenterol Hepatol. Elsevier; 2018;16(8):1300–1306.e3. Available from: http://linkinghub.elsevier.com/retrieve/pii/S1542356518301678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bang UC, Watanabe T, Bendtsen F. The relationship between the use of statins and mortality, severity, and pancreatic cancer in Danish patients with chronic pancreatitis. Eur J Gastroenterol Hepatol. Wolters Kluwer; 2018;30(3):346–351. Available from: http://insights.ovid.com/crossref?an=00042737-900000000-98139 [DOI] [PubMed] [Google Scholar]
- 59.Chen M-J, Tsan Y-T, Liou J-M, Lee Y-C, Wu M-S, Chiu H-M, et al. Statins and the risk of pancreatic cancer in Type 2 diabetic patients-A population-based cohort study. Int J Cancer. Wiley-Blackwell; 2016;138:594–603. Available from: http://doi.wiley.com/10.1002/ijc.29813 [DOI] [PubMed] [Google Scholar]
- 60.Liao J, Chung YT, Yang AL, Zhang M, Li H, Zhang W, et al. Atorvastatin inhibits pancreatic carcinogenesis and increases survival in LSL-Kras G12D -LSL-Trp53 R172H -Pdx1-Cre mice. Mol Carcinog. Wiley-Blackwell; 2013;52:739–50. Available from: http://doi.wiley.com/10.1002/mc.21916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fendrich V, Sparn M, Lauth M, Knoop R, Plassmeier L, Bartsch DK, et al. Simvastatin delay progression of pancreatic intraepithelial neoplasia and cancer formation in a genetically engineered mouse model of pancreatic cancer. Pancreatology. Elsevier; 2013;13:502–7. Available from: http://linkinghub.elsevier.com/retrieve/pii/S1424390313007734 [DOI] [PubMed] [Google Scholar]
- 62.Mohammed A, Qian L, Janakiram NB, Lightfoot S, Steele VE, Rao CV. Atorvastatin delays progression of pancreatic lesions to carcinoma by regulating PI3/AKT signaling in p48 Cre/+ LSL-Kras G12D/+ mice. Int J Cancer. Wiley-Blackwell; 2012;131:1951–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deng Y-Z, Cai Z, Shi S, Jiang H, Shang Y-R, Ma N, et al. Cilia loss sensitizes cells to transformation by activating the mevalonate pathway. J Exp Med. Rockefeller University Press; 2018;215:177–95. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29237705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guillaumond F, Leca J, Olivares O, Lavaut M-N, Vidal N, Berthezene P, et al. Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc Natl Acad Sci. 2013;110:3919–24. Available from: http://www.pnas.org/cgi/doi/10.1073/pnas.1219555110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gong J, Sachdev E, Robbins LA, Lin E, Hendifar AE, Mita MM. Statins and pancreatic cancer. Oncol Lett. Spandidos Publications; 2017;13:1035–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xia Y, Xie Y, Yu Z, Xiao H, Jiang G, Zhou X, et al. The Mevalonate Pathway Is a Druggable Target for Vaccine Adjuvant Discovery Article The Mevalonate Pathway Is a Druggable Target for Vaccine Adjuvant Discovery. Cell. Elsevier Inc; 2018;175:1059–1073.e21. Available from: 10.1016/j.cell.2018.08.070 [DOI] [PubMed] [Google Scholar]
- 67.Zhu H, Bengsch F, Svoronos N, Rutkowski MR, Bitler BG, Allegrezza MJ, et al. BET Bromodomain Inhibition Promotes Anti-tumor Immunity by Suppressing PD-L1 Expression. Cell Reports. Elsevier; 2016;16:2829–37. Available from: https://www.ncbi.nlm.nih.gov/pubmed/27626654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kagoya Y, Nakatsugawa M, Yamashita Y, Ochi T, Guo T, Anczurowski M, et al. BET bromodomain inhibition enhances T cell persistence and function in adoptive immunotherapy models. J Clin Invest. The American Society for Clinical Investigation; 2016;126:3479–94. Available from: 10.1172/JCI86437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mashimo T, Pichumani K, Vemireddy V, Hatanpaa KJ, Singh DK, Sirasanagandla S, et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. Elsevier; 2014;159:1603–14. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25525878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mazur PK, Herner A, Mello SS, Wirth M, Hausmann S, Sánchez-Rivera FJ, et al. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat Med. Nature Publishing Group; 2015;21:1163–71. Available from: http://www.nature.com/articles/nm.3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee JV, Berry CT, Kim K, Sen P, Kim T, Carrer A, et al. Acetyl-CoA promotes glioblastoma cell adhesion and migration through Ca2+-NFAT signaling. Genes Dev. Cold Spring Harbor Laboratory Press; 2018;32(7–8):497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuan Z, Lin S, Molden RC, Cao X, Bhanu NV, Wang X, et al. Modifications by Extracting Retention Time and Intensity in High-resolution Mass Spectra. Mol Cell Proteomics. 2015;1696–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carrer A, Moimas S, Zacchigna S, Pattarini L, Zentilin L, Ruozi G, et al. Neuropilin-1 identifies a subset of bone marrow Gr1-monocytes that can induce tumor vessel normalization and inhibit tumor growth. Cancer Res. American Association for Cancer Research; 2012;72(24):6371–81. [DOI] [PubMed] [Google Scholar]
- 74.Hruban RH, Adsay NV, Albores-Saavedra J, Anver MR, Biankin AV., Boivin GP, et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: Consensus report and recommendations. Cancer Res. American Association for Cancer Research; 2006;66:95–106. [DOI] [PubMed] [Google Scholar]
- 75.Frey AJ, Feldman DR, Trefely S, Worth AJ, Basu SS, Snyder NW. LC-quadrupole/Orbitrap high-resolution mass spectrometry enables stable isotope-resolved simultaneous quantification and 13C-isotopic labeling of acyl-coenzyme A thioesters. Analytical and Bioanalytical Chemistry; 2016;408:3651–8. Available from: 10.1007/s00216-016-9448-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Trefely S, Ashwell P, Snyder NW. FluxFix: Automatic isotopologue normalization for metabolic tracer analysis. BMC Bioinformatics [Internet]. BMC Bioinformatics; 2016;17:1–8. Available from: 10.1186/s12859-016-1360-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.