Abstract
The Wnt signaling pathway, known for regulating genes critical to normal embryonic development and tissue homeostasis, is dysregulated in many types of cancer. Previously we identified that the anthelmintic drug niclosamide inhibited Wnt signaling by promoting internalization of Wnt receptor Frizzled 1 and degradation of Wnt signaling pathway proteins Dishevelled 2 and β-catenin, contributing to suppression of colorectal cancer growth in vitro and in vivo. Here we provide evidence that niclosamide-mediated inhibition of Wnt signaling is mediated through autophagosomes induced by niclosamide. Specifically, niclosamide promotes the co-localization of Frizzled 1 or β-catenin with LC3, an autophagosome marker. Niclosamide inhibition of Wnt signaling is attenuated in autophagosome-deficient ATG5 −/− MEF cells or cells expressing shRNA targeting Beclin1, a critical constituent of autophagosome. Treatment with the autophagosome inhibitor 3MA blocks niclosamide-mediated Frizzled 1 degradation. The sensitivity of colorectal cancer cells to growth inhibition by niclosamide is correlated with autophagosome formation induced by niclosamide. Niclosamide inhibits mTORC1 and ULK1 activities, and induces LC3B expression in niclosamide-sensitive cell lines, but not in the niclosamide-resistant cell lines tested. Interestingly, niclosamide is a less effective inhibitor of Wnt responsive genes (β-catenin, c-Myc and Survivin) in the niclosamide-resistant cells than in the niclosamide-sensitive cells, suggesting that deficient autophagy induction by niclosamide compromises the effect of niclosamide on Wnt signaling. Our findings provide a mechanistic understanding of the role of autophagosomes in the inhibition of Wnt signaling by niclosamide, and may provide biomarkers to assist selection of patients whose tumors are likely to respond to niclosamide.
INTRODUCTION
The Wnt signaling pathway plays key roles in tissue development and homeostasis. Wnt ligand binding to Frizzled/LRP co-receptors leads to the recruitment of Dishevelled proteins to prevent the phosphorylation, ubiquitination and destruction of β-catenin by the APC/Axin/GSK3β complex. As a result, β-catenin accumulates in the cytosol and then enters the nucleus to regulate Wnt-responsive genes (1). More than 80% of colorectal cancers have mutations that hyper-activate this pathway (2). As a consequence, drugs that modulate the Wnt pathway have been highly sought-after for 2–3 decades (1). Unfortunately, despite intensive efforts and large investments in research during this time, no drugs specifically targeting this pathway have been approved, in part due to historical difficulties in identifying targets druggable with small molecules (3). To overcome this barrier, we previously conducted a chemical-genetic high-throughput screen to identify modulators of the Wnt/β-catenin signaling pathway. We were the first to discover niclosamide, an FDA-approved anthelmintic drug, as a novel inhibitor of Wnt/β-catenin signaling (4). Subsequently, we found that niclosamide and its derivatives have anticancer effects in colorectal cancer cells both in vitro and in vivo, without observable toxicity (5–11).
Niclosamide was approved by the US FDA in 1982 for use in humans to treat tapeworm infection (9). Its mechanism of action has not been well-delineated, although it has been reported to involve uncoupling of oxidative phosphorylation (9). It is on the World Health Organization’s list of essential medicines and has been used safely to treat millions of patients (12). In the past several years, mounting evidence has accumulated that niclosamide is a multifunctional drug that regulates multiple signaling pathways and biological processes (9). One of its biological activities is the induction of autophagy (13). Autophagy is a normal physiological process by which cells degrade proteins and turn-over damaged cell organelles (14). By clearing away defective cell components, autophagy maintains homeostasis or normal functioning of cells. Autophagy is carried out by the formation of double-membrane vesicles called autophagosomes that are formed by the coordinated action of the ATG (autophagy-related) (15, 16) and Beclin1 (an essential mediator of autophagy) (17) proteins.
Here we used ATG5-deficient cells and Beclin1 knockdown cells to demonstrate that niclosamide-induced autophagosomes regulate the potency of Wnt signaling inhibition by niclosamide. Moreover, we show that the efficacy of niclosamide to inhibit proliferation of colorectal cancer cells varies between colorectal cancer cells that have blunted the induction of the autophagosome marker LC3 (Microtubule-associated protein 1A/1B-light chain 3) and cells without such deficiency in autophagosome formation. We further demonstrate that niclosamide affects the mammalian target of rapamycin complex 1 (mTORC1) activity and the mammalian autophagy-initiating kinase ULK1 in niclosamide sensitive colorectal cancer cells. Moreover, we demonstrate that niclosamide is more effective in inhibiting Wnt signaling in niclosamide-sensitive colorectal cancer cells that exhibit normal autophagosome formation, compared to niclosamide-resistant cells with deficient autophagy induction by niclosamide. Our results indicate a close association of niclosamide efficacy in colorectal cancer cell growth inhibition, autophagosome formation, and Wnt signaling.
MATERIALS AND METHODS
Reagents and antibodies
Niclosamide (N3510), chloroquine (C6628), MG132 (474790), 3-Methyladenine (3MA) (M9281), and DMSO (D2650) were purchased from Sigma-Aldrich. The following primary antibodies were used: GFP (A11122) from ThermoFisher Scientific; LC3B (3868), ULK1 (8054), phospho-ULK1 (14202), mTOR (2983), phospho-p70S6K1 (9205), and Survivin (2808) from Cell Signaling; Beclin1 (3495, Cell Signaling), β-catenin (sc-7963), c-Myc (sc-40), and β-actin (sc-47778) from Santa Cruz Biotechnology. Alexa Fluor™ 488 (A32723) and Alexa Fluor™ 647 (A32728) labeled secondary antibodies were from Invitrogen.
DNA constructs
The mCherry-LC3 plasmid was obtained from the Tsien Lab (University of California, San Diego). SNAP-Frizzled1 (SNAP-Fzd1) was obtained from the Clevers lab (University of Utrecht, Netherlands). The p8xTopFlash reporter plasmid was obtained from the Moon lab (University of Washington). The pRL-TK Renilla luciferase construct was purchased from Promega.
Cell culture
HEK293T cells were cultured in DMEM medium (D6429, Sigma). U2OS and WIDR cells were cultured in MEM medium (M4655, Sigma). DLD-1, CRC 240, COLO205, and CRC57 cells were cultured in RPMI-1640 medium (11875–093, Gibco). HCT116 cells were cultured in McCoy’s 5A medium (16600–082, Invitrogen). All media were supplemented with 10% fetal bovine serum (FBS, S11050, Atlanta Biologicals), 200 U/ml penicillin, and 50 ng/ml streptomycin (P/S, 15140122, Invitrogen). All cell lines, except CRC240 and CRC57, were from ATCC. CRC240 and CRC57 cells are colorectal cancer patient-derived xenograft cell lines obtained from the Hsu lab (Duke University). ATG5 +/+ and ATG5 −/− fibroblast cells were obtained from the Jin lab (Rutgers University), and cultured in DMEM plus 10% FBS, P/S and 2 mM L-glutamine (25030081, Invitrogen). TP6 cells used for the TopFlash luciferase assay were developed as described (4): these cells are HEK293 cells stably transfected with p8xTOPFlash, Renilla luciferase plasmid pRL-TK, and pLKO.1. TP6 cells were cultured in MEM medium plus 10% FBS, P/S and 1 μg/mL puromycin (P8833, Sigma). All cells were incubated at 37°C with 5% CO2 overnight to allow attachment to the plates before treatment.
Immunostaining and images
U2OS cells that stably express Frizzled1-GFP (Fzd1-GFP-U2OS) (4), were transfected with mCherry-LC3. Fugene 6 (E269A, Promega) transfection was utilized throughout the study. The cells were then treated with DMSO or 10 μM niclosamide for 4h at 37°C, followed by fixation with 4% paraformaldehyde. Immunostaining of β-catenin was conducted in Fzd1-GFP-U2OS cells. These cells were treated with DMSO or 10 μM niclosamide for 4h at 37°C, followed by endogenous β-catenin immunostaining using β-catenin primary antibody and Alexa Fluor™ 547 secondary antibody. In addition, U2OS cells were transfected with mCherry-LC3, treated with DMSO or 10μM niclosamide for 4h at 37°C, followed by immunostaining using β-catenin antibody and Alexa Fluor™ 488 antibody.
HEK293T cells were transfected with SNAP-Fzd1 and mCherry-LC3. After 15 min room temperature labelling of SNAP-Fzd1 by SNAP-Surface 649 (S9159S, New England Biolabs), cells were treated with DMSO or 10μM niclosamide for 4h at 37°C. The cells were then fixed, followed by β-catenin immunostaining with β-catenin antibody and Alexa Fluor™ 488 antibody.
For immunostaining experiments, fixed cells were permeabilized in 0.2% Triton-X100 in PBS for 10 min and blocked in 5% bovine serum albumin (BSA) in PBS for 30 min at room temperature. The cells were then incubated with β-catenin antibody in 2% BSA in PBS overnight at 4°C, and subsequently incubated with the Alexa Fluor™ 488 antibody or Alexa-Flour Fluor™ 647 antibody in 2% BSA in PBS for 1h at room temperature.
Cells were imaged using a LSM 510-Meta confocal microscope (Carl Zeiss). Imaris 8.1 (Bitplane, an Oxford Instruments Company) was used to quantify co-localization in the images: mCherry-LC3 and Fzd1-GFP; β-catenin and Fzd1-GFP; mCherry-LC3 and β-catenin.
TopFlash Reporter Assay
TP6 cells were used to test the effect of niclosamide on chloroquine-treated cells following TopFlash Reporter Assay procedures as described (4). The cell lines to produce Wnt3A (ATCC® CRL-2647™) and Control (ATCC® CCL-1.3™) conditioned media were purchased from ATCC. The conditioned medium were prepared using published protocols (http://www.atcc.org/Products/All/CRL-2647.aspx#culturemethod). TP6 cells were seeded in 100 μl of growth medium in 96-well plates at 100% confluence. After allowing the cells to attach overnight, fifty microliters of Wnt-3A conditioned medium or control conditioned medium, containing 0.25 μM niclosamide or DMSO, and with or without 10 μM chloroquine, was added to each well. After 6h treatment, the cells were washed once with PBS and lysed with 55 μl of Passive Lysis Buffer. Luciferase activity was measured with 25 μl of cell lysate in a 96-well plate reader (FluoStar Optima, BMG Labtech) using the Dual-Luciferase Reporter Assay kit (E1960, Promega).
The shRNA targeting human Beclin1 in pLKO.1-puro vector was purchased from Sigma-Aldrich. Beclin1 knockdown cells were generated using lentivirus-mediated delivery of shRNA to TP6 cells, following the published protocol (http://www.addgene.org/tools/protocols/plko/). Scrambled shRNA virus was used to generate control cells. Cells were seeded in 100 μl of growth medium in 96-well plates at 100% confluence. After allowing the cells to attach overnight, fifty microliters of Wnt-3A conditioned medium or control conditioned medium, containing 0.25 μM niclosamide or DMSO, was added to each well, and after 6h the Dual Luciferase Reporter Assay was performed as described above.
ATG5 +/+ and ATG5 −/− cells were transiently co-transfected with p8xTopFlash and Renilla luciferase pRL-TK plasmids. Cells were seeded in 100 μl culture medium in 96-well plates at 100% confluence. After allowing the cells to attach overnight, fifty microliters of Wnt-3A conditioned medium or control conditioned medium, containing 0.25 μM niclosamide or DMSO, was added to each well, and after 6h, luciferase activity was measured using the Dual-Luciferase Reporter Assay.
Cell proliferation and Caspase activity assay
Colorectal cancer cell lines HCT116, DLD-1, CRC240, WIDR, COLO205, and CRC57 were used in the assays. For cell proliferation assay, the cells were plated at 5000 cells per well in 100 μl of growth medium in 96-well plates, and after allowing the cells to attach overnight, were treated with niclosamide from 0.04 to 10 μM for 72 h, after which time the cells were analyzed by colorimetric MTS assay (G358B, Promega). For Caspase activity assay, cells were plated at 4 × 105 cells per well in 100 μl of growth medium in 96-well plates. After overnight culture, the cells were treated with niclosamide at 2 μM for 48 h, after which time the cells were analyzed by Apo-One Homogenous Caspase 3/7 assay (G7790, Promega).
Western blotting
Cells were plated in 6 well plates for culture and treatment. After allowing the cells to grow to 70–80% confluency, Fzd1-GFP-U2OS cells were treated with DMSO, 10 μM niclosamide, 10 μM proteasome inhibitor MG132, 10 μM niclosamide plus MG132, 5 mM autophagy inhibitor 3MA, or 10 μM niclosamide plus 5 mM 3MA for 12h. Cell lysates were then collected directly using SDS sample buffer and used for anti-GFP antibody immunoblot. TP6 cells infected with Beclin1 or scrambled shRNA were blotted with anti-Beclin1 antibody. Colorectal cancer cell lines HCT116, DLD-1, CRC240, WIDR, COLO205, and CRC57 were treated with DMSO or niclosamide for 18h, followed by whole cell lysate preparation and immunoblots of LC3B, Beclin1, ULK1, phospho-ULK1, mTOR, phospho-p70S6K1, c-Myc, and Survivin, or cytosol lysate preparation for β-catenin immunoblot. Antibody to β-actin was used to measure levels of β-actin, which served as a loading control in all immunoblots.
Lysate samples were subjected to SDS-PAGE using 4–12% Novex™ Tris-Glycine Gels (XP04202BOX, Invitrogen), transferred to nitrocellulose membranes (1620115, Bio-Rad Laboratories), blocked with 5% nonfat milk powder in TBS-0.2% Tween-20 for 30 minutes, incubated with primary antibodies for 4h to overnight, and subsequently with horseradish peroxidase-conjugated secondary antibodies (Anti-Mouse IgG: 7076; Anti-Rabbit IgG: 7074, Cell Signaling). Peroxidase was detected using SuperSignal substrate (34577, Thermo Scientific), and bands detected and quantified using the ChemiDoc™ MP Imaging System (Bio-Rad Laboratories).
Statistics
One way ANOVA was used to compare the quantification of Fzd1-GFP immunoblots. Student’s t-test was used to analyze the data in TopFlash reporter assays and quantified colorectal cancer cell immunoblots. Differences at p <0.05 were considered statistically significant.
RESULTS
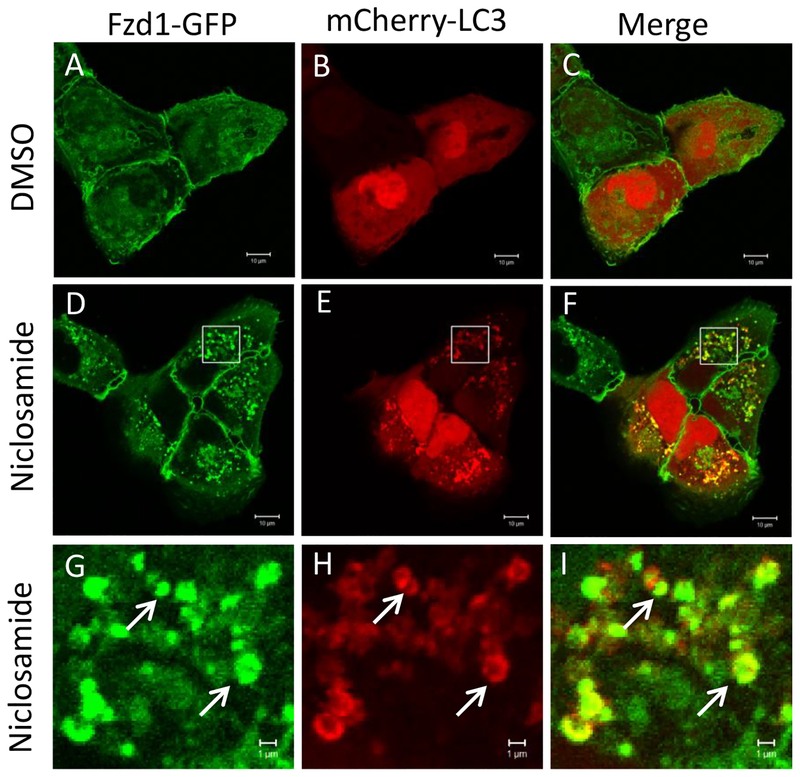
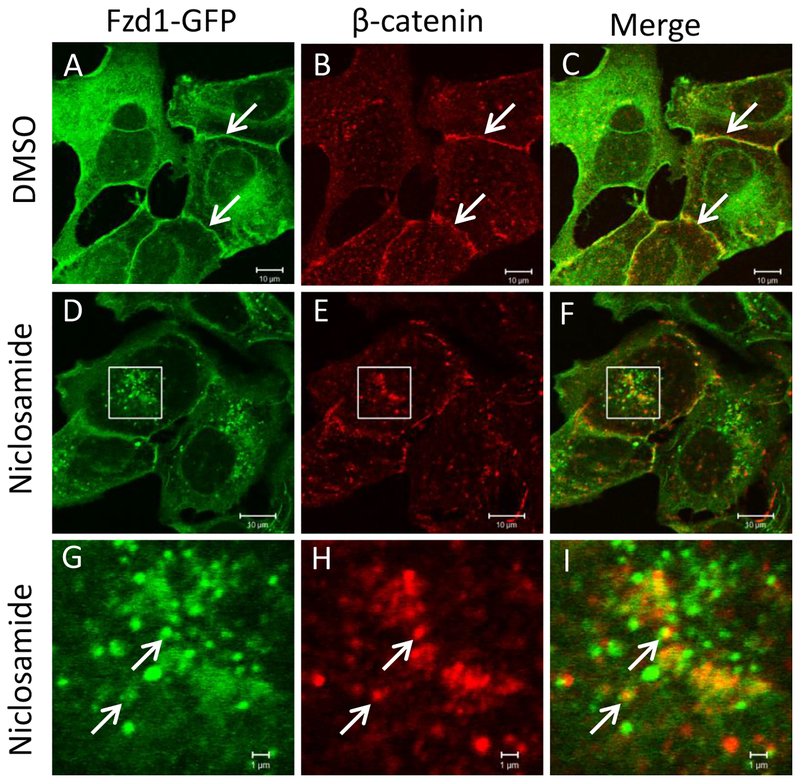
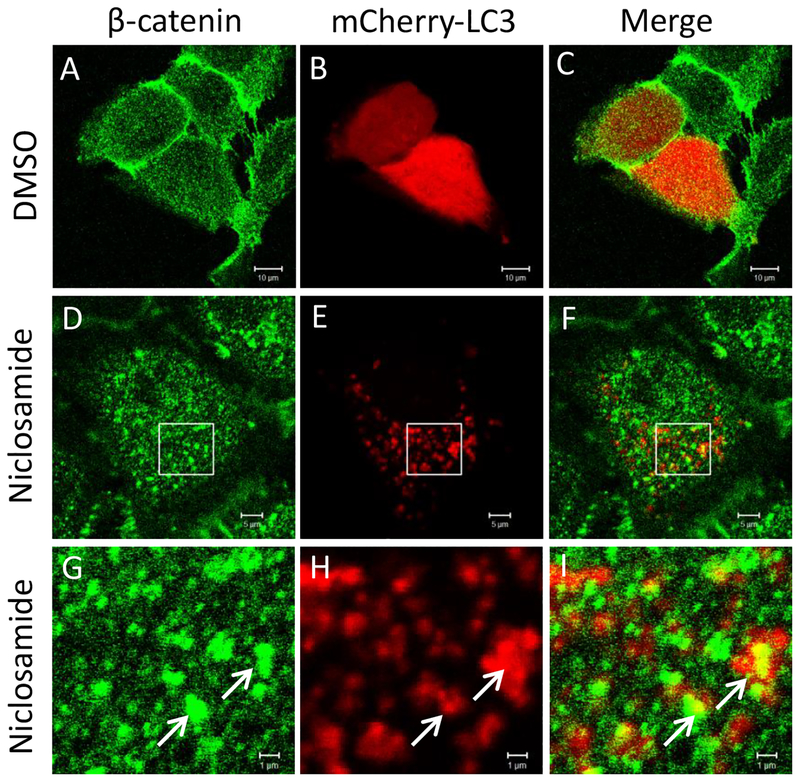
To determine the involvement of the autophagy pathway in niclosamide-mediated inhibition of Wnt signaling, we overexpressed the autophagy marker mCherry-LC3 in U2OS cells also expressing Fzd1-GFP. Niclosamide treatment induced the formation of numerous intracellular vesicles where Fzd1-GFP and mCherry-LC3 co-localized (Fig. 1). The percentage of mCherry-LC3 co-localizing to internalized Fzd1-GFP upon niclosamide treatment was significantly increased compared to that of DMSO treatment (Supplementary Fig. 1). Upon evaluation of co-localization of Fzd1-GFP and β-catenin, Fzd1-GFP was found to co-localize with endogenous β-catenin at the plasma membrane after DMSO treatment, but to co-localize in intracellular vesicular structures after niclosamide treatment (Fig. 2). The percentage of Fzd1-GFP co-localizing to internalized β-catenin upon niclosamide treatment was significantly increased compared to that of DMSO treatment (Supplementary Fig. 2). Similar to co-localization observed between Fzd1-GFP and mCherry-LC3, we also observed that endogenous β-catenin co-localized with mCherry-LC3 after niclosamide stimulation (Fig. 3). The percentage of mCherry-LC3 co-localizing to internalized β-catenin after niclosamide treatment was significantly increased compared to that of DMSO treatment (Supplementary Fig. 3). To determine if all three components indeed co-localize, we overexpressed SNAP-Fzd1 with mCherry-LC3 in HEK293T cells and labeled the cell surface Fzd1 prior to treating with niclosamide. We then stained for endogenous β-catenin, and found that niclosamide induced the co-localization of SNAP-Fzd1, mCherry-LC3, and β-catenin (Supplementary Fig. 4). These results suggest that niclosamide-induced autophagy could function as part of the mechanism of niclosamide-mediated Wnt signaling inhibition.
Fig. 1.
Fzd1-GFP and mCherry-LC3 co-localize upon niclosamide treatment. U2OS cells stably expressing Fzd1-GFP were transfected with mCherry-LC3. The cells were then treated with DMSO (A, B, C) or 10 μM niclosamide (D, E, F) for 4h. Panels G, H, I show a higher magnification of the boxed regions in panels D, E, and F, respectively. Arrows show representative locations where co-localization of Fzd1-GFP and mCherry-LC3 were observed (G, H, I). Scale bar: 10 μm (A-F); 1 μm (G-I).
Fig. 2.
Fzd1-GFP and β-catenin co-localize upon niclosamide treatment. U2OS cells stably expressing Fzd1-GFP were treated with DMSO (A, B, C) or 10 μM niclosamide (D, E, F) for 4h. Panels G, H, I show a higher magnification of the boxed regions in panels D, E, and F, respectively. Endogenous β-catenin was visualized by immunostaining. Arrows in DMSO treated cells show overlap of Fzd1-GFP and β-catenin at membrane (A, B, C). Arrows in niclosamide treated cells indicate representative locations where co-localization of Fzd1-GFP and β-catenin were observed (G, H, I). Scale bar: 10 μm (A-F); 1 μm (G-I).
Fig. 3.
β-catenin and mCherry-LC3 co-localize upon niclosamide treatment. U2OS cells were transfected with mCherry-LC3 and treated with DMSO (A, B, C) or 10 μM niclosamide (D, E, F) for 4h. Panels G, H, I show a higher magnification of the boxed regions in panels D, E, and F, respectively. Endogenous β-catenin was visualized by immunostaining. Arrows in niclosamide treated cells indicate representative locations where co-localization of β-catenin and mCherry-LC3 were observed (G, H, I). Scale bar: 10 μm (A-C); 5 μm (D-E); 1 μm (G-I).
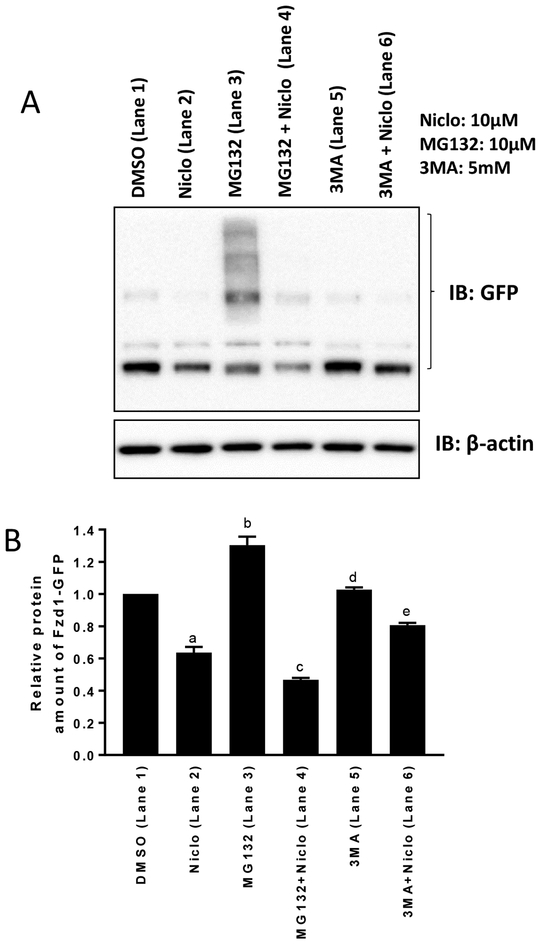
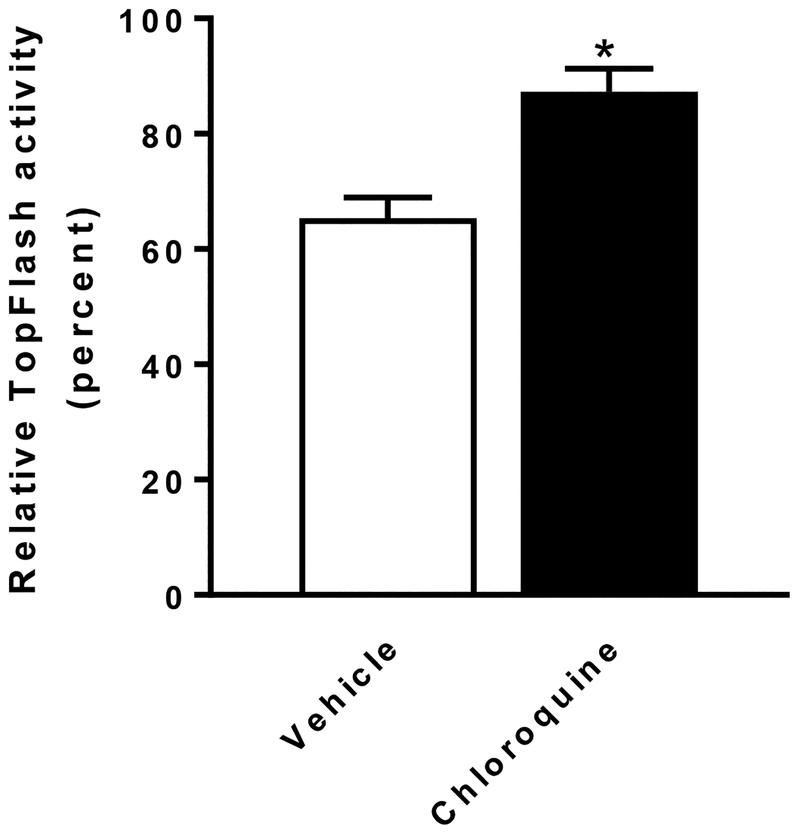
Based on the above observations, we set out to determine the effect of inhibiting autophagy on the degradation of the Fzd1 receptor and compared it to proteasomal degradation. U2OS cells expressing Fzd1-GFP were treated with DMSO, niclosamide, the autophagy inhibitor 3MA, 3MA plus niclosamide, the proteasome inhibitor MG132, or MG132 plus niclosamide, and cell lysates were immunoblotted using anti-GFP antibody (Fig. 4A). The immunoblots were then quantified (Fig. 4B). Niclosamide induced Fzd1-GFP degradation (Lane 2). The proteasome inhibitor MG132 increased the expression of Fzd1-GFP (Lane 3), indicating that spontaneous Fzd1-GFP degradation occurs mostly through the proteasome, since MG132 inhibited such degradation at a dose we have previously shown to inhibit proteasomal degradation of HER3 (18). However, MG132 failed to block the degradation of Fzd1-GFP upon niclosamide treatment (Lane 4), indicating that niclosamide induced degradation of Fzd1-GFP is not through the proteasome. The autophagy inhibitor 3MA had no effect on the expression of Fzd1-GFP (Lane 5). Interestingly, however, 3MA substantially reversed Fzd1-GFP degradation induced by niclosamide (Lane 6). This data indicates that niclosamide induced Fzd1-GFP degradation is occurring through autophagy. Chloroquine is also an inhibitor of autophagy (19). To confirm the results of autophagy inhibition by 3MA, we treated the TopFlash Wnt reporter cell line (TP6) with chloroquine. Chloroquine alone did not have a statistically significant effect on TopFlash luciferase activity in control conditioned media or in Wnt3A conditioned media (Supplementary Fig. 5), but chloroquine significantly reduced the ability of niclosamide to inhibit Wnt3A-induced TopFlash reporter activity (Fig. 5). Taken together, the data suggest the involvement of autophagy in niclosamide-mediated Wnt signaling inhibition.
Fig. 4.
Niclosamide (Niclo) induced degradation of Fzd1-GFP is reversed by autophagy inhibitor 3MA. (A) U2OS cells stably expressing Fzd1-GFP were treated with the indicated reagents for 12h and lysed with the SDS sample buffer for Western blot analysis. MG132 alone inhibited the Fzd1-GFP degradation through protein ubiquitination and resulted in multiple bands. (B) Quantification of the Western blots. Multiple bands resulted from each of the treatments shown in (A) were quantified by normalizing to β-actin and compared using one-way ANOVA. Results are mean ± SEM, n = 3, a: P<0.0001 of DMSO vs Niclo, b: P<0.0001 of DMSO vs MG132, c: P=0.009 of Niclo vs MG132 + Niclo, d: P=0.9801 of DMSO vs 3MA, e: P=0.009 of Niclo vs 3MA + Niclo.
Fig. 5.
Chloroquine antagonizes the inhibition of Wnt signaling by niclosamide in TP6 cells. Chloroquine (10 μM) treated cells are less sensitive to inhibition of Wnt signaling by 0.25 μM niclosamide treatment compared to vehicle (H2O) treated cells. The relative TopFlash activity upon niclosamide treatment was calculated by normalizing Wnt3A conditioned medium (plus DMSO) induced reporter activity as 100% in vehicle treated cells or in chloroquine treated cells, respectively. Results are means ± SEM. (n = 3; asterisk, P < 0.05 of student’s t test).
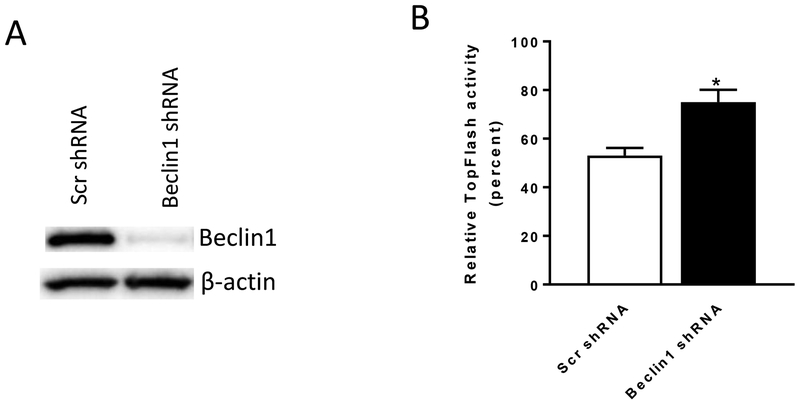
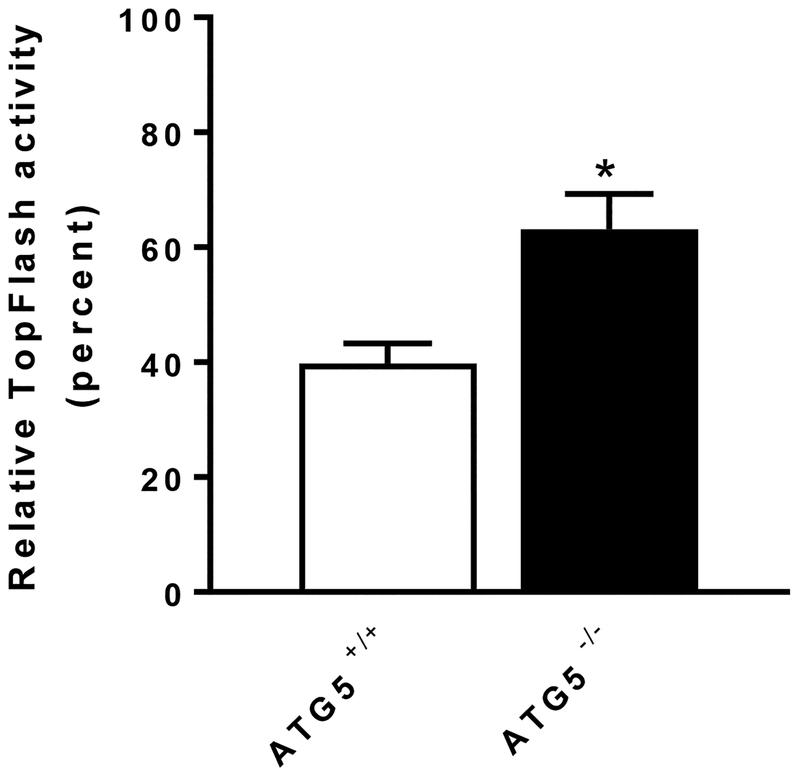
To further interrogate the role of autophagy in niclosamide-mediated Wnt signaling inhibition, we used two additional approaches to assess the involvement of autophagosomes. First, we used lentiviral shRNA to target the autophagosome component Beclin1. Upon treating cells with Beclin1-targeted shRNA, Beclin1 expression was decreased in the TopFlash Wnt reporter cells (Fig. 6A). In control conditioned media, TopFlash reporter activity in the cells infected with Beclin1 shRNA was significantly less than in cells infected with scramble shRNA (Supplementary Fig. 6). Stimulation of these cells with Wnt3A conditioned media resulted in increased reporter activity in cells infected with Beclin1 shRNA to a significantly higher level than in cells treated with scrambled shRNA (Supplementary Fig. 6). The results are consistent with the published data (20). Importantly, in cells infected with Beclin1 shRNA, the inhibitory effect of niclosamide was significantly reduced (Fig. 6B). Second, we used MEFs lacking the autophagosome component ATG5. Wild-type MEF (ATG +/+) cells and ATG5-deficient MEF cells (ATG −/−) were transfected with the TopFlash reporter plasmid and treated with control conditioned media or Wnt3A conditioned media. In control conditioned media, TopFlash reporter activity in ATG −/− cells was significantly less than in ATG +/+ cells (Supplementary Fig. 7). Upon Wnt3A conditioned media stimulation, TopFlash reporter activity in ATG −/− cells and in ATG +/+ cells was not significantly different (Supplementary Fig. 7). However, upon treatment with niclosamide, niclosamide was a significantly less effective inhibitor of Wnt-stimulated reporter activity in ATG5-knockout MEFs compared to wild type MEFs (Fig. 7). These data further support our findings that autophagy mediates Wnt signaling inhibition by niclosamide.
Fig. 6.
Inhibition of Wnt signaling by niclosamide is antagonized in Beclin1 knockout cells. TP6 cells infected with either a scrambled shRNA (Scr shRNA) or Beclin1 shRNA lentiviruses were used in the experiment. (A) Beclin1 expression is decreased in Beclin1 shRNA infected cells.
(B) Inhibition of Wnt signaling by 0.25 μM niclosamide is antagonized in Beclin1 shRNA infected cells compared to Scr shRNA control cells. The relative TopFlash activity upon niclosamide treatment was calculated by normalizing Wnt3A conditioned medium (plus DMSO) induced activity as 100% in Scr shRNA infected cells or in Beclin 1 shRNA infected cells, respectively. Results are means ± SEM. (n = 4; asterisk, P < 0.05 of student’s t test).
Fig. 7.
ATG5 knockout antagonizes inhibition of Wnt3A signaling by niclosamide. Inhibition of Wnt signaling by 0.25 μM niclosamide is antagonized in ATG5 −/− cells compared to ATG5 +/+ cells. The relative TopFlash activity upon niclosamide treatment was calculated by normalizing Wnt3A conditioned medium (plus DMSO) induced activity as 100% in ATG5 +/+ cells or in ATG5 −/− cells, respectively. Results are means ± SEM. (n = 4; asterisk, P < 0.05 of student’s t test).
Next we evaluated the ability of niclosamide to inhibit the proliferation of colorectal cancer cells using a panel of six cell lines and compared the results with the ability of niclosamide to induce autophagy. We found that niclosamide has distinct growth inhibitory effects on different cancer cells, with IC50 values ranging from 0.31 to 2000 μM (Table 1). The potency of inhibition of proliferation against these colorectal cancer cells enabled us to group the cells into two categories: niclosamide-sensitive cell lines (HCT116, DLD-1, and CRC240) and niclosamide-resistant cell lines (WIDR, CRC57, and COLO205). As expected, niclosamide induced significant apoptosis, as measured by Caspase 3/7 activity, in niclosamide-sensitive cell lines whereas apoptosis induction in niclosamide-resistant cells was minimal (Supplementary Fig. 8).
Table 1.
Inhibition of the cell proliferation by niclosamide in colorectal cancer cell lines. Cells were treated with niclosamide for 72 hours and cell proliferation evaluated by MTS assay. IC50 values were calculated using Graphpad Prism. APC and β-catenin mutation status were from literature reports (26, 27), wildtype (wt), mutation (mut), n.a (no report available).
| Colon cancer Cell line | IC50 (μM) | Niclosamide inhibition | APC mutation | β-Catenin mutation |
|---|---|---|---|---|
| HCT116 | 0.31 | Sensitive | wt | mut |
| CRC240 | 0.68 | Sensitive | n.a | n.a |
| DLD-1 | 1.78 | Sensitive | mut | wt |
| WIDR | 870 | Resistant | mut | n.a |
| COL0205 | 1655 | Resistant | mut | wt |
| CRC57 | 2000 | Resistant | n.a | n.a |
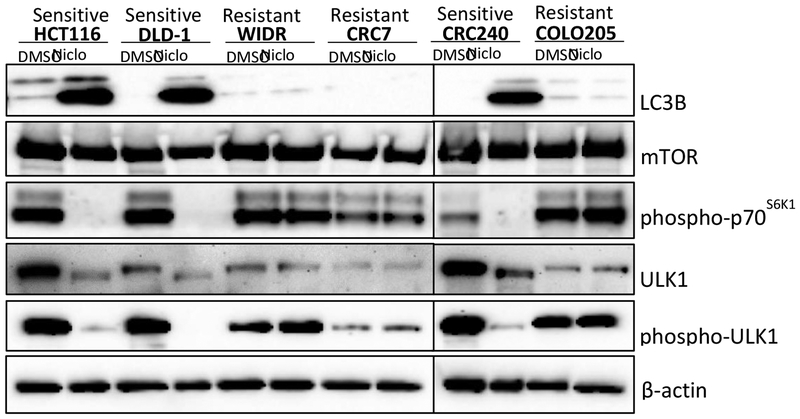
We then asked if autophagy differed in sensitive versus resistant cells. We found that upon niclosamide treatment, the niclosamide-sensitive cell lines induced LC3B expression normally, but that LC3B induction did not occur in the niclosamide-resistant cell lines (Fig. 8). It has been reported that niclosamide inhibits mTORC1 (13, 21), a key regulator of autophagy (22, 23). More specifically, mTORC1 inhibits autophagy through direct phosphorylation of ULK1, a homologue of yeast ATG1(24). We also investigated niclosamide’s effect on total mTOR and on mTORC1 activity by detecting p70S6K1 phosphorylation in the six cell lines. In addition, we assessed niclosamide’s effect on total ULK1 and ULK1 phosphorylation. Interestingly, while niclosamide had no effect on total mTOR (Fig. 8), we found that niclosamide inhibited p70S6K1 phosphorylation in niclosamide-sensitive cell lines but had no effect on p70S6K1 phosphorylation in the niclosamide-resistant cell lines (Fig. 8). Consistent with the mTORC1 activity data, we found that niclosamide decreased both total ULK1 and phosphorylation of ULK1 in niclosamide-sensitive cell lines but this reduction was not observed in the niclosamide-resistant cell lines (Fig. 8). Overall, our data suggest that autophagy is involved in mediating the sensitivity colorectal cancer cells to niclosamide.
Fig. 8.
Effects of niclosamide on cellular levels of LC3B, mTOR, phospho-p70S6K1, ULK1, and phospho-ULK1 in niclosamide sensitive (HCT116, DLD-1, and CRC240) versus resistant (WIDR, CRC57, and COLO205) colorectal cancer cell lines. Cells were treated with DMSO control or niclosamide (Niclo) at 5μM for 18h. Whole cell lysates were analyzed by Western blots to detect LC3B, mTOR, phosphor-p70S6K1, ULK1, and phosphor-ULK1. β-actin was used as a loading control.
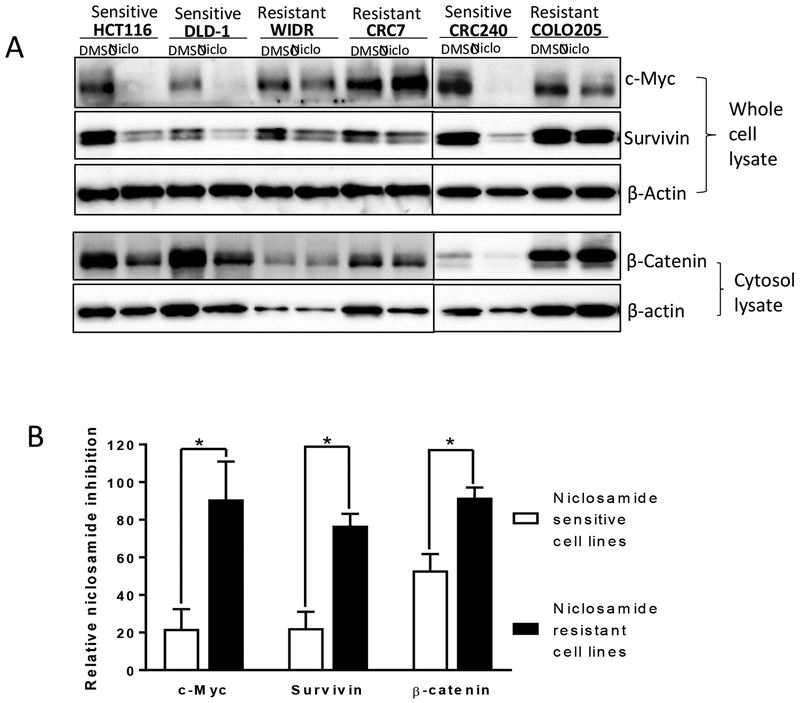
Autophagy is a negative regulator of Wnt signaling (20). Since niclosamide induces LC3 expression in cells sensitive to niclosamide, we anticipated that Wnt signaling would be inhibited by niclosamide in the niclosamide-sensitive HCT116, DLD-1, and CRC240 cells. Indeed, we found that niclosamide treatment reduced the expression of the Wnt responsive genes β-catenin, c-Myc and Survivin in these cells (Fig. 9A, B). Consistent with the role of autophagy in the response to niclosamide in the niclosamide-resistant WIDR, CRC57, and COLO205 cell lines, niclosamide was significantly less effective in inhibiting Wnt signaling in these cells (Fig. 9A, B), suggesting that deficient autophagy induction by niclosamide compromises the effect of niclosamide on Wnt signaling. Our data support the involvement of autophagosomes and Wnt signaling in niclosamide-mediated cancer cell growth inhibition.
Fig. 9.
Effect of niclosamide on Wnt signaling target genes (β-catenin, c-Myc, and Survivin) in niclosamide sensitive (HCT116, DLD-1, and CRC240) versus resistant (WIDR, CRC57, and COLO205) colorectal cancer cell lines. (A) Cells were treated with DMSO control or niclosamide (Niclo) at 5μM for 18 h. Whole cell lysates were analyzed by Western blot to detect c-Myc and Survivin. Cytosol lysates were analyzed to detect β-catenin. β-actin was used as a loading control. (B) Quantification showing niclosamide inhibits more Wnt signaling target genes in niclosamide sensitive cells than resistant cells. Western blots of c-Myc, Survivin, and β-catenin in the absence (DMSO) or presence of niclosamide shown in (A) were normalized to β-actin in each of the cell lines. The DMSO effect was set as 100% and niclosamide effect was normalized to DMSO effect to obtain the relative niclosamide inhibition data. The relative niclosamide inhibition data of the three sensitive cells as a group vs that of the three resistant cells as a group were compared using a two-tailed Student’s t test. Data represent mean ± SEM. (n = 3; asterisk, P <0.05).
DISCUSSION
Abnormal Wnt signaling is an important mechanism in colorectal cancer (2). Despite the clear rationale for inhibiting the Wnt/β-catenin pathway in colorectal cancer, developing effective strategies to intervene in this pathway clinically has proven challenging (1, 3). Through a high-content drug screen, we previously discovered the approved anthelmintic drug niclosamide as an inhibitor of the Wnt pathway (4) and have developed several niclosamide derivatives with improved pharmacological properties to overcome its low bioavailability and poor systemic exposure, including a nanoparticle formulation of a niclosamide conjugate (7, 11, 25). However, the clinical use of niclosamide and its derivatives to treat systemic diseases such as cancer, requires a more detailed understanding of the mechanism of its drug action. Here we provide multiple lines of evidence to support our conclusion that autophagy is critical to niclosamide’s effects on Wnt signaling and cancer cell growth inhibition. These include the co-localization of Fzd1, β-catenin, and LC3 upon niclosamide treatment, reduced Fzd1 degradation following inhibition of autophagy, reduced ability of niclosamide to inhibit Wnt signaling after inhibition of autophagy, reduced niclosamide mediated inhibition of Wnt signaling after knockdown of autophagy components Beclin1 and ATG5, and a strong correlation between autophagosome induction by niclosamide and sensitivity of colorectal cancer cell lines to niclosamide-mediated growth inhibition. Specifically, in cells resistant to the growth inhibitory effects of niclosamide, niclosamide was ineffective at inducing autophagosomes, indicating deficient autophagy induction by niclosamide in these cells. Since autophagy is a negative regulator of Wnt signaling, deficient autophagy may compromise Wnt signaling. Importantly, we found niclosamide is less potent in inhibiting Wnt signaling in niclosamide-resistant cells compared to sensitive cells.
The difference in autophagy induction by niclosamide between niclosamide-sensitive and niclosamide-resistant cells prompted us to investigate mTORC1, a key regulator of autophagy (22, 23). Our data showed inhibition of mTORC1 activity leads to autophagosome induction (Fig. 8). The results indicate that mTORC1 may play a major role in sensing niclosamide treatment and initiating autophagosome formation in niclosamide sensitive colorectal cancer cell lines. It is known that niclosamide disrupts cytoplasmic pH balance and inhibits mTORC1, perhaps by a protonophore mechanism (21). The failure of niclosamide to inhibit mTORC1 in resistant cell lines needs to be studied further. One possibility is that niclosamide may not affect cytoplasmic pH in the resistant cells. Another possibility is the mTORC1 signaling is impaired in niclosamide resistant cell lines, or that autophagosome induction by niclosamide is deficient. It is also possible efflux pumps remove niclosamide from the resistant cells, or that other mechanism are involved.
Colorectal cancer is the second leading cause of cancer death in the US. More than 80% of all sporadic and hereditary colorectal cancers show hyperactivation of the Wnt/β-catenin pathway, mainly due to mutations in APC or β-catenin genes. The anti-proliferative effect of niclosamide did not correlate with any specific mutation in the Wnt/β-catenin pathway (26, 27) (Table 1). The correlation of niclosamide’s efficacy to inhibit cell growth and niclosamide’s efficacy to induce the autophagy marker LC3B suggest that autophagosome induction may be a potential biomarker to select patients for enrollment in clinical trials designed to evaluate the clinical utility of niclosamide in colorectal cancer. In addition, niclosamide is a multifunctional drug that is also reported to modulate STAT3, NF-κB and Notch signaling pathways in multiple cancers (9). Whether niclosamide-induced autophagy is involved in mediating these additional biological pathways or whether there are other distinct mechanisms is an interesting question for further investigation.
In summary, we provide evidence demonstrating that niclosamide-induced autophagosomes play a role in modulating the potency of niclosamide in inhibiting Wnt signaling. As a consequence, niclosamide is a less potent inhibitor of proliferation of colorectal cancer cells that exhibit deficient autophagy induction by niclosamide. These findings improve our understanding of the mechanism of niclosamide’s action and provide potential biomarkers for clinical studies of niclosamide.
Supplementary Material
Acknowledgements
WC is a V Foundation Scholar and an American Cancer Society Research Scholar. The assistance of Genevieve Jing in conducting the Western blot studies in colorectal cancer cell lines is also gratefully acknowledged. We also thank Dr. Peter Nicholls for help in quantifying protein co-localization in the images.
Funding
This work was funded in part by 5 R01 CA172570 (WC) and Clinical Oncology Research Center Development Grant 5K12-CA100639–08 (RAM).
Abbreviations
- APC
Adenomatous Polyposis Coli
- Fzd1
Frizzled 1
- GSK3β
Glycogen synthase kinase 3 beta
- LC3
Microtubule-associated protein 1A/1B-light chain 3
- LC3B
LC3-phosphatidylethanolamine conjugate
- LRP
Lipoprotein receptor-related protein
- mTORC1
mammalian target of rapamycin complex 1
- 3MA
3-Methyladenine
Footnotes
Competing interests
The authors declare that they have no conflict of interests.
REFERENCES
- 1.Nusse R, Clevers H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169(6):985–99. [DOI] [PubMed] [Google Scholar]
- 2.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kahn M Can we safely target the WNT pathway? Nat Rev Drug Discov. 2014;13(7):513–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen MY, Wang JB, Lu JY, Bond MC, Ren XR, Lyerly HK, et al. The Anti-Helminthic Niclosamide Inhibits Wnt/Frizzled1 Signaling. Biochemistry. 2009;48(43):10267–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osada T, Chen MY, Yang XY, Spasojevic I, Vandeusen JB, Hsu D, et al. Antihelminth Compound Niclosamide Downregulates Wnt Signaling and Elicits Antitumor Responses in Tumors with Activating APC Mutations. Cancer Research. 2011;71(12):4172–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mook RA Jr., Chen M, Lu J, Barak LS, Lyerly HK, Chen W. Small molecule modulators of Wnt/beta-catenin signaling. Bioorganic & medicinal chemistry letters. 2013;23(7):2187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mook RA Jr., Wang J, Ren XR, Chen M, Spasojevic I, Barak LS, et al. Structure-activity studies of Wnt/beta-catenin inhibition in the Niclosamide chemotype: Identification of derivatives with improved drug exposure. Bioorganic & medicinal chemistry. 2015;23(17):5829–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mook RA Jr., Ren XR, Wang J, Piao H, Barak LS, Kim Lyerly H, et al. Benzimidazole inhibitors from the Niclosamide chemotype inhibit Wnt/beta-catenin signaling with selectivity over effects on ATP homeostasis. Bioorganic & medicinal chemistry. 2017;25(6):1804–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen W, Mook RA Jr., Premont RT, Wang J. Niclosamide: Beyond an antihelminthic drug. Cellular signalling. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen W, Chen M, Barak LS. Development of small molecules targeting the Wnt pathway for the treatment of colon cancer: a high-throughput screening approach. American journal of physiology Gastrointestinal and liver physiology. 2010;299(2):G293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Mook RA Jr., Ren XR, Zhang Q, Jing G, Lu M, et al. Identification of DK419, a potent inhibitor of Wnt/beta-catenin signaling and colorectal cancer growth. Bioorganic & medicinal chemistry. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.WHO, editor. The Selection and Use of Essential Medicines Geneva: World Health Organization 2007.
- 13.Balgi AD, Fonseca BD, Donohue E, Tsang TC, Lajoie P, Proud CG, et al. Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PloS one. 2009;4(9):e7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong Y, Undyala VV, Gottlieb RA, Mentzer RM Jr., Przyklenk K Autophagy: definition, molecular machinery, and potential role in myocardial ischemia-reperfusion injury. Journal of cardiovascular pharmacology and therapeutics. 2010;15(3):220–30. [DOI] [PubMed] [Google Scholar]
- 15.Shibutani ST, Yoshimori T. A current perspective of autophagosome biogenesis. Cell research. 2014;24(1):58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nature reviews Molecular cell biology. 2013;14(12):759–74. [DOI] [PubMed] [Google Scholar]
- 17.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402(6762):672–6. [DOI] [PubMed] [Google Scholar]
- 18.Ren XR, Wang J, Osada T, Mook RA Jr., Morse MA, Barak LS, et al. Perhexiline promotes HER3 ablation through receptor internalization and inhibits tumor growth. Breast Cancer Res. 2015;17:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nature reviews Cancer. 2017;17(9):528–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T, et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12(8):781–90. [DOI] [PubMed] [Google Scholar]
- 21.Fonseca BD, Diering GH, Bidinosti MA, Dalal K, Alain T, Balgi AD, et al. Structure-activity analysis of niclosamide reveals potential role for cytoplasmic pH in control of mammalian target of rapamycin complex 1 (mTORC1) signaling. J Biol Chem. 2012;287(21):17530–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584(7):1287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noda T Regulation of Autophagy through TORC1 and mTORC1. Biomolecules. 2017;7(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhattacharyya J, Ren XR, Mook RA, Wang J, Spasojevic I, Premont RT, et al. Niclosamide-conjugated polypeptide nanoparticles inhibit Wnt signaling and colon cancer growth. Nanoscale. 2017;9(34):12709–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mouradov D, Sloggett C, Jorissen RN, Love CG, Li S, Burgess AW, et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014;74(12):3238–47. [DOI] [PubMed] [Google Scholar]
- 27.Berg KCG, Eide PW, Eilertsen IA, Johannessen B, Bruun J, Danielsen SA, et al. Multi-omics of 34 colorectal cancer cell lines - a resource for biomedical studies. Mol Cancer. 2017;16(1):116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.