Abstract
Intrinsic and acquired drug resistance and induction of secondary malignancies limit successful chemotherapy. Because mutagenic translesion synthesis (TLS) contributes to chemoresistance as well as treatment-induced mutations, targeting TLS is an attractive avenue for improving chemotherapeutics. However, development of small molecules with high specificity and in vivo efficacy for mutagenic TLS has been challenging. Here, we report the discovery of a small molecule inhibitor, JH-RE-06, that disrupts mutagenic TLS by preventing recruitment of mutagenic POL ζ. Remarkably, JH-RE-06 targets a nearly featureless surface of REV1 that interacts with the REV7 subunit of POL ζ. Binding of JH-RE-06 induces REV1 dimerization, which blocks the REV1-REV7 interaction and POL ζ recruitment. JH-RE-06 inhibits mutagenic TLS and enhances cisplatin-induced-toxicity in cultured human and mouse cell lines. Co-administration of JH-RE-06 with cisplatin suppresses the growth of xenograft human melanomas in mice, establishing a framework for developing TLS inhibitors as a novel class of chemotherapy adjuvants.
Keywords: Translesion synthesis, REV1, REV7, POL ζ, cisplatin, chemotherapy, chemoresistance
Graphical Abstract

In Brief
A small molecule specifically targeting the mutagenic branch of translesion synthesis binds a featureless surface of REV1 to induce dimerization and block recruitment of POL ζ
INTRODUCTION
DNA-damaging chemotherapeutics, such as cisplatin, have been the mainstay of cancer treatment for decades. DNA lesions generated by these therapies cannot be utilized by high-fidelity replicative DNA polymerases as the template, thus blocking the progression of the replication fork, generating cytotoxicity, and ultimately causing cell death. To promote survival, cells employ specialized DNA polymerases to bypass the lesion site at the cost of replication fidelity in a process known as translesion synthesis (TLS) (Makarova and Burgers, 2015; Vaisman and Woodgate, 2017; Yang and Gao, 2018).
In mammalian cells, TLS occurs in a two-step process in which insertion TLS DNA polymerases such as POL κ, POL ι, POL η, or REV1 first introduce a nucleotide opposite the lesion. This is followed by elongation of the resulting 3’-terminus by an extension TLS DNA polymerase such as the B-family polymerase complex POL ζ (POL ζ4: REV3L/REV7/POLD2/POLD3 (Baranovskiy et al., 2012; Johnson et al., 2012; Makarova and Burgers, 2015; Makarova et al., 2012)). While TLS carried out by certain insertion DNA polymerases over their cognate lesions can be relatively accurate, for example POL η over a cyclobutane thymine-thymine dimer (McCulloch et al., 2004), the major mutagenic branch of TLS is characterized by its dependence on REV1 and POL ζ. The ca. 100 amino acid REV1 C-terminal domain (CTD) plays a major role in coordinating TLS, using one interface to recruit the insertion TLS polymerases POL κ, POL ι, POL η and a second interface to recruit POL ζ through an interaction with its REV7 component (Yamanaka et al., 2017). Genetic inhibition of TLS through RNA-mediated depletion of REV1 or REV3L, the catalytic subunit of mammalian POL ζ, strikingly sensitizes a variety of cancer cells to DNA-damaging chemotherapeutics (Doles et al., 2010; Xie et al., 2010; Xu et al., 2013) and suppresses the emergence of new tumor chemoresistance in vitro and in vivo (Xie et al., 2010), thereby highlighting the therapeutic potential of inhibiting the REV1-POL ζ mediated TLS in cancer therapy.
RESULTS
Discovery of a potent REV1-REV7 interface inhibitor, JH-RE-06
Although small molecule compounds interfering with aspects of TLS have been reported (Actis et al., 2016; Izuta, 2006; Mizushina et al., 2009; Sail et al., 2017; Vanarotti et al., 2018; Yamanaka et al., 2012), none has yet been shown to demonstrate in vivo efficacy. Obtaining a specific inhibitor of mutagenic TLS is inherently challenging since TLS and replicative polymerases share both common substrates and interaction partners (e.g. PCNA), and some components of TLS DNA polymerases, such as REV7, are additionally implicated in cellular functions beyond translesion synthesis (Bhat et al., 2015; Boersma et al., 2015; Xu et al., 2015). The evolutionarily conserved interaction between REV1 and POL ζ, mediated by a shallow pocket on the REV1 CTD and the REV7 subunit of POL ζ, plays a critical and specific role in mutagenic TLS, but not accurate lesion bypass (Hashimoto et al., 2012), rendering such a protein-protein interaction an ideal target for small molecule intervention. Therefore, we designed an ELISA assay to screen for small molecule inhibitors that specifically target the REV7-binding surface of the REV1 CTD to disrupt the REV1-REV7 interaction.
An initial obstacle to developing a robust assay for monitoring the REV1-REV7 interaction was the instability of the REV1 CTD in solution. However, by fusing the REV1 CTD C-terminally to the POL κ RIR (REV1-interacting region) peptide, which induces the folding of the disordered N-terminal loop of the REV1 CTD into a hairpin conformation (Wojtaszek et al., 2012b), we were able to dramatically improve the stability of the REV1 CTD. Our structural analysis of this chimeric REV1 CTD protein (referred to as cREV1 CTD below) at 2.03 Å resolution (Figure S1A, structure statistics shown in Table S1) revealed a REV1 CTD conformation nearly identical to that observed in our previously published structure of the REV1 CTD in complex with the free POL κ RIR peptide (backbone RMSD deviation of 0.45 Å), with the fused POL κ RIR peptide binding to the REV1-insertion polymerase interface (Wojtaszek et al., 2012b) and with the evolutionarily conserved REV7-binding surface of the REV1 CTD unoccupied (Wojtaszek et al., 2014) (Figure S1B). The linker loop connecting the POL κ RIR and the REV1 CTD and the C-terminal tail of the REV1 CTD were invisible in the crystal structure of the apo protein, reflecting their dynamic nature.
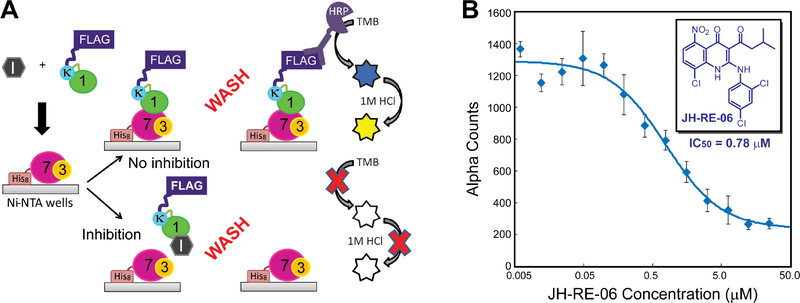
In the ELISA assay (Figure 1A), purified His8-REV7 co-expressed with a REV3 peptide (referred to as His8-REV7/3 below) was immobilized in Ni2+-NTA coated wells, and its ability to retain the FLAG-tagged cREV1 CTD in the presence of small molecule inhibitors was probed by the horseradish peroxidase (HRP) conjugated anti-FLAG antibody. Using this ELISA assay, we screened ~10,000 structurally diverse compounds from the LOPAC and PRESTWICK libraries, as well as representative compounds from the Korea Chemical Bank compound collection. We identified a 1,4-dihydroquinolin-4-one derivative, JH-RE-06, that potently inhibited the REV1-REV7 interaction at the 10 μM concentration. JH-RE-06 was re-synthesized in large scale (Figure S2A), and its ability to disrupt the REV1-REV7 interaction in a dose-dependent manner was determined in the quantitative AlphaScreen™ assay by using the anti-FLAG donor beads (to detect the FLAG-tagged cREV1 CTD) and anti-His acceptor beads (to detect His8-REV7/3), revealing an IC50 value of 0.78 μM (Figure 1B). Corroborating the AlphaScreen™ assay, a similar value of dissociation constant (Kd=0.42 μM) was determined by using the isothermal titration calorimetry (ITC) measurements (Figure S2B).
Figure 1. Discovery and characterization of JH-RE-06, a small molecule inhibitor that blocks the REV1 CTD-REV7 interaction.
(A) A schematic overview of the ELISA assay. (B) Dose-dependent inhibition of the REV1 CTD-REV7 interaction by JH-RE-06 in the AlphaScreen™ assay. Fitting of the inhibition curve yields an IC50 value of 0.78 ± 0.16 μM for JH-RE-06. Error bars represent standard error of the mean (n=3).
Structure of the REV1 CTD in complex with JH-RE-06
We next probed the atomic details of the interaction between the REV1 CTD and JH-RE-06 by determining the co-crystal structure of the cREV1 CTD/JH-RE-06 complex at 1.50 Å resolution (Figure 2A; structure statistics shown in Table S1). The location of the JH-RE-06 compound is very well defined by the omit electron density map (Figures 2A and 2B). Although the co-crystal structure indeed confirmed that the previously described evolutionarily conserved REV7-binding surface of the REV1 CTD (Wojtaszek et al., 2012a) is the target site of JH-RE-06, it also revealed an unexpected binding mode: JH-RE-06 binds to the REV1 CTD by inducing dimerization of the protein such that the compound is almost completely encapsulated inside the REV1 CTD dimer (Figures 2A and 2B).
Figure 2. Structural and biochemical characterization of the cREV1 CTD/JH-RE-06 complex.
(A) Structure of the cREV1 CTD/JH-RE-06 complex. Proteins are shown in the cartoon model, with protomer A colored in green and protomer B colored in cyan. JH-RE-06 is shown in the stick model, with the purple mesh representing the inhibitor omit map (2mFo-DFc) contoured at 1.0σ. (B) Surface representation of the cREV1 CTD/JH-RE-06 complex, illustrating the formation of a large ligand cavity at the dimeric REV1 CTD interface and the near encapsulation of JH-RE-06 within the cavity. (C) Interactions of JH-RE-06 with REV1 CTD residues. Denoted residue numbers correspond to the full-length REV1 protein. (D) Superimposition of the JH-RE-06-bound cREV1 CTD dimer (colored in green and cyan) with the POL κ RIR-REV1 CTD-REV7/3 translesionsome complex (colored in grey and pale cyan), illustrating the blockage of the REV1-REV7 interaction in the JH-RE-06-bound REV1 complex. (E) Binding of JH-RE-06 promotes the dimerization of the REV1 CTD in solution. Samples of the chimeric FLAG-tagged POL κ RIR-REV1 CTD in the presence of 0 μM or 100 μM JH-RE-06 were treated with increasing molar ratios of DSS and analyzed by SDS-PAGE followed by Western blotting with anti-FLAG antibody. Monomer and crosslinked dimer bands are labeled.
The REV1 CTD dimer is formed in an asymmetric, tail-to-tail fashion, with the REV7-binding pockets from the two protomers facing each other and merging into a deep cavity to accommodate the binding of JH-RE-06. The core four-helix bundle of the REV1 CTD and the POL κ RIR helix of the chimeric protein are largely unperturbed in comparison with those in the POL κ RIR-REV1 CTD-REV7/3 complex (Figure S3A), whereas the disordered loop connecting the POL κ RIR and the REV1 CTD in the chimeric protein displays different conformations in the two protomers of the inhibitor-bound complex. The C-terminal tail of the REV1 CTD, which is also dynamic and invisible in the apo structure, adopts distinct conformations in the two protomers: the C-terminal tail in protomer B of the REV1 CTD is nearly superimposable with that of the REV1 CTD in the POL κ RIR-REV1 CTD-REV7/3 complex (Figure S3A), whereas the C-terminal tail of protomer A is pushed out by the acyl chain and bulky 1,4-dihydroquinolinone group of JH-RE-06, with the C-terminal residue (T1249) swinging over a distance of 15.8 Å (Figure S3B). Remarkably, the relocated C-terminal tail of protomer A now pairs up with the unperturbed C-terminal tail of protomer B to form an antiparallel β-sheet as part of the ligand-binding pocket thereby stabilizing the REV1 CTD dimer (Figure 2A, inset). Thus, not only does asymmetric JH-RE-06 have the rare attribute of interacting simultaneously but differently with two REV1 CTDs, but it also has the second rare attribute that one of its binding modes induces a conformational change enabling dimerization.
Nearly all of the functional groups of JH-RE-06 are engaged in the interactions with one or the other of the two REV1 CTDs that form the dimer. A large number of hydrophobic interactions are observed between the acyl chain, the substituted aniline moiety, and the central 1,4-dihydroquinolinone group of JH-RE-06 with hydrophobic residues from both subunits of the REV1 CTD dimer (e.g., I1196, L1201, L1204, L1238, Y1242, L1246, and V1248; Figure 2C); direct polar interactions are also observed between the nitro, hydroxyl, and carbonyl groups of the 1,4-dihydroquinolinone moiety of JH-RE-06 and sidechains of Q1235 and S1244 of the REV1 CTD (Figure 2C). Of particular note, the formation of the REV1 CTD dimer not only creates a large binding pocket for JH-RE-06, but also conceals the REV7-binding surface of the REV1 CTD, thus blocking the REV1 CTD interaction with REV7 (Figure 2D).
In order to eliminate the possibility that the compound-binding-induced dimerization of the REV1 CTD reflects a crystal-packing artifact, we tested the ability of disuccinimidyl suberate (DSS) to crosslink the REV1 CTD in the absence and presence of JH-RE-06 in solution. We found that the level of the crosslinked, dimeric form of the cREV1 CTD was significantly enhanced in the presence of JH-RE-06, but not in the absence of the compound, confirming that JH-RE-06 binding indeed promotes the formation of the REV1 CTD dimer in solution (Figure 2E). Such a conclusion is further corroborated by our ITC measurements of the REV1 CTD interaction with JH-RE-06, which revealed a protein-to-inhibitor binding stoichiometry of 2.04 in solution (Figure S2B).
JH-RE-06 enhances cisplatin cytotoxicity and suppresses cisplatin-induced mutagenesis by interfering with REV1-dependent mutagenic TLS
After elucidating the atomic details of the JH-RE-06 binding mode to the REV1 CTD, we tested whether JH-RE-06 is active in living mammalian cells by evaluating its ability to enhance the cytotoxicity of cisplatin in a variety of different cell lines including HT1080 (human fibrosarcoma), A375 (human melanoma), KP (mouse KrasG12D;p53−/− lung adenocarcinoma), LNCap (human prostate adenocarcinoma), AG01522 (human primary cells), and as shown below MEFs (mouse embryonic fibroblasts). The addition of JH-RE-06 significantly decreased the ability of all the cisplatin-treated cancer cells to form colonies (Figures 3A–3D and 4A), except for AG01522 cells (Figure 3E), where JH-RE-06 does not further enhance cisplatin cytotoxicity in these non-cancerous cells. The enhanced cisplatin cytotoxicity caused by JH-RE-06 in cancer cells and not the primary cells was further confirmed using the CellTiter-Glo® assay (Figures S4A and S4B). We also used a variant of the previously published quantitative assay based on a gapped plasmid carrying an intrastrand cisplatin-1,2-GG adduct in the single-stranded region opposite to the gap (Avkin et al., 2004; Shachar et al., 2009; Ziv et al., 2012) to confirm that JH-RE-06 significantly suppresses TLS over a cisplatin adduct (Figure S5).
Figure 3. JH-RE-06 enhances cisplatin cytotoxicity and suppresses cisplatin-induced mutagenesis.
Cells were treated with DMSO or cisplatin (0.5 μM) for 24 hours, followed by JH-RE-06 (1.5 μM) treatment for additional 24 hours. Cells were washed and allowed to form colonies for 5–7 days and counted after staining with Coomassie brilliant blue R-250 stain. Shown in panels (A) HT1080 (human fibrosarcoma), (B) A375 (human melanoma), (C) KP (mouse KrasG12D;p53−/− lung adenocarcinoma), (D) LNCap (human prostate adenocarcinoma), and (E) AG01522 (human primary fibroblasts) are the relative colony forming ability of these cells in response to DMSO, JH-RE-06, cisplatin, and a combination dose of cisplatin and JH-RE-06. (F) The relative ability of HPRT+ HT1080 cells to mutate and form HPRT− colonies in 6-TG media in the presence of DMSO, 1.5 μM JH-RE-06, 0.5 μM cisplatin, and the combination dose of 0.5 μM cisplatin and 1.5 μM JH-RE-06. Error bars represent standard error of the mean (n=6 for panels A-E and n=12 for panel F). Statistical analysis: one-way ANOVA with Tukey HSD post-hoc test. **P<0.01; N.S., not significant.
Figure 4. JH-RE-06 sensitizes cells to cisplatin and reduces HPRT mutations in a REV1-dependent manner.
The combination treatment of JH-RE-06 (1.5 μM) and cisplatin (0.5 μM) significantly reduced the colony forming ability in Rev1+/+ MEF cells (A), but not in Rev1−/− MEF cells (B), in comparison with cisplatin treatment alone. (C) Complementation of Rev1−/− MEF cells with a plasmid encoding REV1 by nucleofection fully restored the JH-RE-06 (1.5 μM) mediated sensitization to cisplatin (1 μM). siRNA knock-down of REV1 abolished JH-RE-06 (1.5 μM) mediated sensitization to cisplatin treatment (1 μM) in HT1080 (D) and A375 (E) cells. Treatment with JH-RE-06 (1.5 μM) significantly reduced spontaneous or cisplatin-induced (0.5 μM) HPRT mutation rates in Rev1+/+ MEF cells (F), but not in Rev1−/− MEF cells (G). Relative cell survival reflects the normalized colony forming ability of treated cells to DMSO controls. Error bars represent standard error of the mean (n=12 for panels A-C and F-G; n=6 for panels D–E). Statistical analysis: one-way ANOVA with Tukey HSD post-hoc test. **P<0.01; N.S., not significant.
Consistent with JH-RE-06 acting by inhibiting TLS, we found that JH-RE-06 sensitized KP cells to other DNA-damaging agents besides cisplatin, including the bulky DNA-damaging agent benzo[α]pyrene diol epoxide (BPDE), the UV-mimetic 4-nitroquinolone 1-oxide (4-NQO), and the alkylating agent methyl methanesulfonate (MMS) (Figures S4C). Even more importantly, our observations indicated that JH-RE-06 acts by inhibiting mutagenic TLS because it also decreased the frequency of both spontaneous and cisplatin-induced HPRT mutations in HT1080 cells (Figure 3F). In this assay, mutations that inactivate the HPRT gene prevent cells from incorporating the toxic guanine analog, 6-thioguanine (6-TG), into DNA and allow cells to survive in the 6-TG selection medium.
To test our prediction that the mutagenic TLS inhibited by JH-RE-06 is REV1-dependent, we employed an isogenic pair of wild-type (Rev1+/+) and knockout (Rev1−/−) MEF cell lines. We found that treatment of the Rev1+/+ and Rev1−/− MEF cells with JH-RE-06 alone had little effect on cell survival, indicating that JH-RE-06 has minimal toxicity on its own. However, JH-RE-06 strikingly increased the ability of cisplatin to kill Rev1+/+ MEF cells (Figure 4A) but not Rev1−/− MEF cells (Figure 4B). Furthermore, the lack of JH-RE-06 mediated sensitization to cisplatin in Rev1−/− MEF cells was completely reversed when these cells were complemented with plasmid-encoded Rev1 (Figure 4C). Similarly, treatment of wild-type (REV1+/+) HT1080 and A375 cells with siREV1 abolished JH-RE-06 induced sensitization to cisplatin in these cells (Figures 4D and 4E). These complementary genetic experiments in multiple cell lines mitigate any concerns of unpredictable compensatory changes in REV1−/− cells and unequivocally establish that JH-RE-06 enhances cisplatin cytotoxicity by specifically interfering with a REV1-dependent pathway and not by an off-target effect on some other aspect of the DNA repair or DNA damage response. Moreover, JH-RE-06 significantly reduced the frequency of spontaneous and cisplatin-induced HPRT mutations in Rev1+/+ MEF cells (Figure 4F) but not in Rev1−/− MEF cells (Figure 4G), again indicating that REV1 is the functional cellular target of JH-RE-06. Taken together, our results indicate that JH-RE-06 acts by specifically inhibiting REV1-dependent mutagenic TLS and thus has the potential to be a novel class of adjuvant that can sensitize cells to the cytotoxic effect of a DNA-damaging chemotherapeutic drug while simultaneously suppressing the attendant mutagenesis caused by the treatment.
JH-RE-06 improves tumor cell response to cisplatin in vivo
Encouraged by JH-RE-06’s ability to profoundly sensitize a variety of tumor cell lines to killing by DNA-damaging agents, we used the A375 xenograft mouse model of human melanoma to test whether JH-RE-06 could improve cisplatin chemotherapy in an in vivo model. A375 cells were injected into the NCRNU-F (nude) mice to grow xenograft tumors of approximately 100 mm3 size. The mice were randomly distributed into 4 groups to receive twice-weekly injections of saline, cisplatin alone, JH-RE-06 alone, and the JH-RE-06 and cisplatin combination for 5 weeks. The combination treatment resulted in virtually complete inhibition of tumor growth compared to the saline, JH-RE-06, or cisplatin alone treatments (Figure 5A), suggesting that suppression of the REV1-dependent mutagenic TLS by JH-RE-06-mediated specific inhibition of the REV1-REV7 interaction significantly improves chemotherapy. Strikingly, the mice treated with combination treatment of JH-RE-06 and cisplatin also survived longer than other groups (Figure 5B). These results validate REV1 inhibitors as viable adjuvants for DNA-damaging cancer therapy.
Figure 5. JH-RE-06 improves tumor cell response to cisplatin in a xenograft mouse model.
(A) Inhibition of A375 xenograft tumor growth with (i) saline, (ii) JH-RE-06, (iii) cisplatin, and (iv) cisplatin and JH-RE-06. Compounds were formulated in 10% DMSO, 10% ethanol, 40% PEG-400, and 50% saline. The doses of JH-RE-06 and cisplatin per injection were 1.6 mg/kg and 1.0 mg/kg, respectively. Error bars represent standard error of the mean (n=10–12 xenograft tumors from 6 mice per treatment group). p-values for tumor volumes between each treatment group were calculated by the Welch’s t-test (Graphpad Prism). *p<0.05; N.S., not significant. (B) Survival curves of tumor-bearing mice treated with the four formulations above. Day 10 represents the first day of the specified drug administration (n=6 mice per treatment group in one representative experiment shown here, from a total of three independent experiments). A p-value of 0.0017 for the JH-RE-06 and cisplatin combination treatment vs. cisplatin treatment for survival studies was determined by using the Mantel-Cox log-rank test.
DISCUSSION
The evolutionarily conserved scaffolding function between the REV1 CTD and the REV7 subunit of POL ζ plays an essential role in the mutagenic translesion synthesis that is responsible for the intrinsic and acquired resistance mechanisms of cancer cells to chemotherapy. Genetic suppression of REV1 and components of POL ζ profoundly sensitizes resistant tumors to chemotherapy and suppresses treatment-induced drug resistance in relapsed tumors (Doles et al., 2010; Xie et al., 2010; Xu et al., 2013). These observations render the REV1 CTD an outstanding novel target for the development of adjuvant cancer therapeutics for sensitizing recalcitrant cancers to chemotherapy. However, as the REV7-binding surface of the REV1 CTD is shallow and contains a conformationally dynamic C-terminal tail that is invisible in the crystal structure of the apo REV1 CTD, the REV1 CTD-REV7 interface might appear intractable for developing small molecule therapeutics. Contrary to the conventional wisdom, we have discovered a specific small molecule inhibitor, JH-RE-06 that unexpectedly induces dimerization of the REV1 CTD at its REV7-binding surface and blocks the REV1-REV7 interaction. Remarkably, such a dimerization interface is in part mediated by the pairing of the dynamic C-terminal tail of two REV1 proteins to forge a small antiparallel β-sheet. The formation of the JH-RE-06-induced REV1 CTD dimer creates a large cavity that nearly encapsulates the entire JH-RE-06 molecule, thus providing high affinity, unparalleled specificity, and excellent in vivo efficacy for this novel class of TLS inhibitor(s) in comparison with other in vitro active TLS inhibitors reported over the past decade (Actis et al., 2016; Izuta, 2006; Mizushina et al., 2009; Sail et al., 2017; Vanarotti et al., 2018; Yamanaka et al., 2012). Disrupting the buried hydrophobic interactions by replacing the buried dichloroaniline group in JH-RE-06 with a hydrophilic morpholine group yielded an inactive compound (JH-RE-25) that was unable to bind to the REV1 CTD or sensitize tumor cells to cisplatin in vitro (Figures S2C, S2D, and S2E). The formation of the dimeric REV1/JH-RE-06 complex in cells may be further facilitated by the enrichment of DNA repair proteins in biomolecular condensates in response to therapy-induced DNA damage, thus further enhancing the effectiveness and specificity of the compound in cells and in vivo. Co-administration of JH-RE-06 with cisplatin shows superior growth inhibition of drug-resistant tumors in mice relative to single agent chemotherapy, providing the first in vivo evidence of the therapeutic potential of a small molecule inhibitor that specifically disrupts the mutagenic TLS to overcome chemoresistance in tumors. In principle, such a strategy would also significantly decrease the frequency of secondary malignancies caused by treatment-induced mutagenesis, thereby mitigating an important undesired consequence of current chemotherapies. The remarkable observation of the JH-RE-06-induced asymmetric dimerization of the REV1 CTD at its conformationally flexible and shallow REV7-binding surface may furnish a new paradigm for developing high-affinity and high-specificity small molecule inhibitors targeting dynamic and featureless protein interfaces, which was once deemed unachievable.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Pei Zhou (peizhou@biochem.duke.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial Strains and Growth Media
BL21 Star™ (DE3) E. coli (ThermoFisher Scientific) and K12 JM109 E. coli (New England Biolabs) cells were used for protein expression and the gapped plasmid TLS assay, respectively. Selection and growth of E. coli was performed in the Lysogeny Broth (LB) medium supplemented with the appropriate antibiotics [100 μg/mL Ampicillin for the pET15b (Novagen/Sigma-Aldrich) or pUC19 (NEB) vectors or 50 μg/mL Streptomycin for the pCDFDuet-1 (Novagen/Sigma-Aldrich) vector] at 37 °C.
Mammalian Cell Culturing
HT1080 cells (male, fibrosarcoma epithelial cells purchased from ATCC) were grown at 37 °C with 5% CO2 in RPMI 1640 (Gibco), 10% (v/v) FBS (HyClone), and 1% Penicillin-Streptomycin antibiotic (Corning). A375 cells (female, malignant melanoma; kindly gifted by Oliver Jonas, Koch Institute, MIT), KP cells (female, mouse KrasG12D;p53−/− lung adenocarcinoma; kindly gifted by Tyler Jackson Laboratory), and MEF (Mouse Embryonic Fibroblasts, sex unspecified in the original publication) wild-type (Rev1+/+) and Rev1 knockout (Rev1−/−) cells (Jansen et al., 2006) were grown at 37 °C with 5% CO2 in DMEM (Gibco), 10% (v/v) FBS (HyClone), and 1% Penicillin-Streptomycin antibiotic (Corning). LNCap cells (male, human prostate adenocarcinoma; kindly gifted by Michael Yaffe Lab, Koch Institute, MIT) were also grown at 37 °C with 5% CO2 in RPMI 1640 (-phenol) (Gibco), 10% (v/v) FBS (HyClone), and 1% Penicillin-Streptomycin antibiotic (Corning). AG01522 cells (male, human primary cells purchased from Coriell Institute) were grown at 37 °C with 5% CO2 in MEM (-Glutamine; +Earle’s Salts; +Non-Essential Amino Acids) (Gibco) and 20% (v/v) FBS (HyClone). All cells were trypsinized using 0.25% Trypsin-EDTA (ThermoFisher) for passaging.
Mice
Six- to eight-week-old, female NCRNU-F nude mice (immunodeficient; nomenclature: CrTac:NCr-Foxn1nu; genotype: homozygous sp/sp) were purchased from Taconic Biosciences for experimentation. All mice were drug and test naïve and were not involved in any previous procedure. All mice were housed in micro-isolator cages in the animal research facility of the MIT Division of Comparative Medicine (DCM), which is fully accredited by the AAALAC (Animal Welfare Assurance number A-3125) and meets NIH standards as set forth in the “Guide for Care and Use of Laboratory Animals” (DHHS). The MIT animal facility is maintained under specific pathogen free (SPF) conditions.
METHOD DETAILS
Molecular cloning and protein purification
The gene encoding the mouse POL κ RIR (K564-N577), a di-glycine linker, and the mouse REV1 CTD (F1150-T1249) was synthesized and cloned into a modified pET15b vector (Wojtaszek et al., 2014) as the C-terminal fusion protein to the solubility tag His10-GB1 (Zhou et al., 2001) separated by a TEV protease site. The FLAG-tagged chimeric POL κ RIR-REV1 CTD was generated by inserting the FLAG tag immediately after the TEV protease site. Both expression constructs were verified by DNA sequencing. The chimeric POL κ RIR-REV1 CTD and FLAG-tagged POL κ RIR-REV1 CTD were expressed in BL21 Star™ (DE3) E. coli cells (ThermoFisher Scientific). Cells were induced at O.D.600 of 0.5 with 0.1 mM isopropyl 1-thio-β-D-galactopyranoside (IPTG) at 18 °C overnight. Harvested cells were lysed in a buffer containing 50 mM sodium phosphate (pH 8.0), 300 mM sodium chloride, and 0.1% β-mercaptoethanol using a French Pressure cell at 1250 psi. His10-GB1-tagged REV1 proteins were purified using Ni-NTA affinity chromatography (HisPur Ni-NTA, Pierce Biotechnology) and eluted with the lysis buffer containing 300 mM imidazole. Elution fractions were combined and exchanged into the FPLC buffer containing 25 mM HEPES (pH 7.5), 100 mM KCl, and 2 mM tris(2-carboxyethyl)phosphine (TCEP). Following TEV protease cleavage (1:20 molar ratio, 4 hours at room temperature) and a second Ni-NTA column to remove the His10-GB1 tag, POL κ RIR-REV1 CTD and its FLAG-tagged counterpart were further purified to homogeneity by size-exclusion chromatography (Superdex 200, GE Healthcare Life Sciences) in the FPLC buffer.
Codon-optimized genes encoding mouse His8-REV7 containing a stabilizing R124A mutation and mouse REV3L (L1845-D1895) were synthesized, cloned into the pCDFDuet-1 vector, and verified by DNA sequencing (Wojtaszek et al., 2012a). His8-tagged REV7/3 was expressed in BL21 Star™ (DE3) cells (ThermoFisher Scientific). Cells were induced at O.D.600 of 0.5 with 1 mM IPTG at 37 °C for 6 hours. After lysing cells in a buffer containing 50 mM sodium phosphate (pH 8.0), 300 mM sodium chloride, and 0.1% β-mercaptoethanol using a French Pressure cell at 1250 psi, the His8-REV7/3 complex was purified by Ni-NTA chromatography (HisPur Ni-NTA, Pierce Biotechnology) and eluted with the lysis buffer containing 300 mM imidazole. The eluted His8-REV7/3 complex was further purified to homogeneity by size-exclusion chromatography (Superdex 200; GE Healthcare Life Sciences) in a buffer containing 25 mM HEPES (pH 7.5), 100 mM KCl and 2 mM TCEP.
Compound screening using the ELISA assay
The ELISA assay for probing the REV1 CTD-REV7 interaction was carried out by immobilizing 50 nM His8-tagged REV7/3 in 200 μL phosphate-buffered saline (PBS, Gibco) containing 0.2% BSA in a Ni-NTA coated 96-well plate (HisSorb, Qiagen) for 30 minutes. Unbound His8-tagged REV7/3 was removed by washing the wells four times with PBS containing 0.05% Tween-20. In parallel, 80 nM FLAG-tagged POL κ RIR-REV1 CTD was pre-incubated with 10 μM small molecules in 200 μL PBS containing 2% DMSO for 30 minutes before transferring to the His8-REV7/3 coated wells. After incubation for 30 minutes, the wells were washed four times with PBS containing 0.05% Tween-20 to remove the unbound FLAG-tagged POL κ RIR-REV1 CTD. A solution of the anti-FLAG horseradish peroxidase (HRP)-conjugated antibody (Sigma-Aldrich) in PBS containing 0.2% BSA was then added to the wells. After incubating for 1 hour, the antibody was washed off four times with PBS containing 0.05% Tween-20. The 3,3’,5,5’-tetramethylbenzedine (TMB) substrate (SureBlue TMB, Seracare) was added to the wells. After incubation of 20–30 minutes, the reaction was quenched with 1 M HCl. A SpectraMax plate reader (Molecular Devices) was used to measure absorbance at 450 nm.
The ELISA assay was used to screen several compound libraries, including the LOPAC library (Sigma-Aldrich) of 1,280 pharmacologically active compounds, the PRESTWICK library (Prestwick Chemical) of 1,200 FDA-approved drugs, and ~8,000 compounds from the Korea Chemical Bank that are structural representatives of ~430,000 diverse compounds. Among several hit compounds, JH-RE-06 was selected for further characterization due to its potency.
Chemical synthesis of JH-RE-06 (8-chloro-2-((2,4-dichlorophenyl)amino)-3-(3-methylbutanoyl)-5-nitroquinolin-4(1H)-one) and JH-RE-25 (8-chloro-3-(3-methylbutanoyl)-2-morpholino-5-nitroquinolin-4(1H)-one)
Upon identification of JH-RE-06 as a hit compound in the REV1 CTD-REV7/3 ELISA assay, we developed a modular synthetic route of the molecule (Figure S2A). Following literature reports on the synthesis of the 1,4-dihydroquinolinone scaffold (Choi et al., 2003; Pak et al., 1992), JH-RE-06 (9) was efficiently prepared from the commercially available Meldrum’s acid (1). The synthesis started with the preparation of the acyl Meldrum’s acid (3), which was reacted with 2-chloro-5-nitroaniline (4) under reflux. The reaction took place with the evolution of CO2 to provide the β-oxo amide (5). Subsequently, the β-oxo amide (5) was transformed into the acyl(arylcarbamoyl)-ketene dithioacetal (6) by using CS2 and Me2SO4 in the presence of K2CO3. Thermal cyclization in 1,2-dichlorobenzene followed by oxidation using H2O2 afforded the sulfoxide intermediate (7). Coupling of the sulfoxide intermediate (7) with 2,4-dichloroaniline (8) completed the synthesis of JH-RE-06 (9). In a similar manner, the sulfoxide intermediate (7) was coupled to morpholine to afford JH-RE-25 (10). The chemical identity and purity of the prepared compounds were verified by LC/MS and NMR
Dose-dependent inhibition of the REV1 CTD-REV7/3 interaction by the AlphaScreen assay
The FLAG-tagged mouse POL κ-REV1 CTD was diluted in PBS containing 1 mM Tris(2-carboxyethyl)phosphine (TCEP) and 0.005% Tween-20 at a final protein concentration of 1 nM and transferred to individual wells of a 96-well, half-area, white opaque plate (PerkinElmer). Serially diluted JH-RE-06 stock solutions in 50% DMSO were added to the wells to yield final inhibitor concentrations of 0–25 μM in 2% DMSO. After 30 min incubation, anti-FLAG Donor Beads (PerkinElmer) were added to a final concentration of 20 ng/μL to individual wells and incubated for an hour. His8-tagged mouse REV7/3 was subsequently added to the reaction mixture to a final concentration of 10 nM and incubated for 30 min. Anti-His Acceptor Beads (PerkinElmer) were added to a final concentration of 20 ng/μL and incubated for an hour. The chemiluminescent signals were observed with a PerkinElmer Enspire Reader at the excitation wavelength of 680 nm and detection wavelength of 615 nm.
Isothermal titration calorimetry
Isothermal titration calorimetry measurements were carried out using a MicroCal VP-ITC instrument at 25 °C, with the chimeric REV1 CTD protein (300 μM) in the syringe and compound (15 μM) in the cell. Compounds were initially dissolved in 50% MPD. Protein and compound samples were diluted in a buffer containing 50 mM HEPES pH 7.5, 50 mM KCl, 2 mM TCEP, 2% DMSO and 0.1% MPD. Microcal Origin 7 software was used to analyze the data.
X-ray crystallography
Apo POL κ RIR-REV1 CTD. A sample of 0.6 mM chimeric mouse POL κ-REV1 CTD in 25 mM HEPES pH 7.2, 100 mM KCl, 30 mM CHAPS, and 2 mM TCEP was mixed with the mother liquor containing 0.1 M sodium acetate, 25% w/v PEG 4000, 8% w/v isopropanol at a 1:1 drop ratio and crystallized upon incubation at 20°C. Crystals were flash frozen in liquid nitrogen without additional cryoprotectants.
The POL κ RIR-REV1 CTD/JH-RE-06 complex. A random micro-seed matrix screen was performed using a sample solution containing 0.6 mM of the chimeric POL κ RIR-REV1 CTD and 4 mM JH-RE-06 NaOH salt in a crystallization buffer of 25 mM HEPES pH 7.0, 100 mM KCl, 16.7% MPD and 0.1% β-mercaptoethanol and crystal seeds derived from apo protein crystals, yielding diffracting crystals in a mother liquor containing 20% PEG 3350, and 0.2 M magnesium formate. High-quality crystals were obtained through repeated seeding, and the final crystallization conditions contain 12.5 mM HEPES (pH 7.5), 50 mM KCl, 8.35% MPD, 0.05% β-mercaptoethanol, 10% PEG3350 and 0.1 M magnesium formate. The crystals were harvested and cryo-protected with the mother liquor containing 15% MPD and 1.88 mM JH-RE-06 NaOH salt.
X-ray diffraction datasets were collected on the SERCAT 22-ID beamline at Argonne National Laboratory and processed with XDS (Kabsch, 2010). The structures of the apo POL κ RIR-REV1 CTD and the POL κ RIR-REV1 CTD/JH-RE-06 complex were determined by molecular replacement using the coordinate of the mouse REV1 CTD and POL κ RIR components of our previously determined quaternary complex crystal structure (PDB 4FJO) as the search model. The final coordinates were constructed by iterative cycles of model building with COOT (Emsley and Cowtan, 2004)) and refinement with PHENIX (Adams et al., 2002) and were deposited to the Protein Data Bank with accession numbers of 6C59 and 6C8C for the apo protein and the inhibitor-bound complex, respectively.
In vitro DSS-crosslinking experiment
Chimeric FLAG-tagged POL κ RIR-REV1 CTD in a buffer containing 25 mM HEPES (pH 7.0), 100 mM KCl, and 4 mM TCEP was mixed with either MPD (control) or JH-RE-06 NaOH salt in MPD to yield a reaction solution containing 1 μM protein, 5% MPD, and either 0 or 100 μM compound. Appropriate dilutions of DSS in DMSO were added to the reaction mixture to yield DSS-to-protein molar ratios of 0:1, 0.5:1, 1:1, 5:1, 10:1, and 50:1 and a final DMSO concentration of 5% (v/v). The reaction mixture was incubated for 30 min at room temperature and then quenched by addition of 1 M Tris (pH 8.5). The SDS-loading dye containing 4 mM TCEP and 10.8 mM iodoacetamide (to block free cysteines) was added to each reaction mixture, and the samples were loaded onto a Pre-cast 4–20% gradient SDS-PAGE gel (Bio-Rad). The gel samples were transferred to a 0.45 μm nitrocellulose membrane (Bio-Rad) for Western blotting with the anti-FLAG primary antibody M2 (Sigma-Aldrich) and the HRP-conjugated secondary antibody (LI-COR) and imaging with the LI-COR Odyssey system.
Clonogenic survival assay
300 cells were plated in triplicate in 6-well plates for 24 hours. Cisplatin (cis-diammineplatinum(II) dichloride, Sigma-Aldrich) was added to relevant wells for 24 hours. All plates were incubated at 37 °C for 24 hours. Media were changed the next day and in fresh media JH-RE-06 (at 1.5 μM concentration) was added to untreated or cisplatin-treated cells for another 24 hours. Media were changed at the end of these combination treatments, and cells were allowed to recover for 7 days. To stain the resulting colonies, media were aspirated and the fixative (50% methanol and 10% glacial acetic acid) was added for 10 minutes, followed by the addition of 0.02% Coomassie brilliant blue R-250 stain (ThermoFisher) in methanol: acetic acid: water in a ratio of 46.5:7:46.5 (v/v/v) (Mochizuki and Furukawa, 1987). Colonies that stained blue and contained at least 40 cells were counted. Relative cell survival or colony formation was calculated by dividing colony counts from treated samples by the DMSO or untreated controls.
Viability assay
Relative viability of cells in response to JH-RE-06 and DNA-damaging agents was assessed by the CellTiter-Glo Luminescence cell viability assay (Promega) that determines the number of viable cells based on the relative amount of ATP in the culture, which is directly proportional to the number of metabolically active cells. Briefly, 10,000 cells were plated in each well of a 96-well, white, clear flat bottom plate (Corning). Increasing doses of drugs in various combinations—JH-RE-06 alone or in combination with DNA-damaging agents—were added into the plates after 24 hours. The JH-RE-06 compound was dissolved in 0.1% DMSO and other drugs were dissolved in solvents ascribed by the manufacturer. In all cases, DMSO controls were run in parallel to the drug treatments. The relative viability of cells was monitored after 24 hours of drug treatment by adding CellTiter-Glo Luminescence stain to an equilibrated plate per the manufacturer’s instructions. Luminescence was measured on the plate reader (Tecan Spark 10M). Relative luminescence, which is indicative of relative survival of metabolically active cells, was calculated by dividing the luminescence of treated samples with DMSO controls.
HPRT mutagenesis assay
For the hypoxanthine-guanine phosphoribosyl transferase (HPRT) mutagenesis assay, cells were first grown in HAT (complete media with 100 μM Hypoxanthine, 0.4 μM Aminopterin and 16 μM Thymidine) media (ThermoFisher) for 14 days to weed out any spontaneous HPRT mutants. After HAT selection, cells were exposed to cisplatin at the 0.5 μM concentration for 24 hours. Then, in fresh media, JH-RE-06 at a concentration of 1.5 μM was added to cells. After 24 hours of drug treatment, cells were trypsinized and washed with PBS. While 200–600 cells were plated in complete media in triplicates in 6-well plates to determine clonal efficiency, the rest of the cells were plated in complete media to allow the expression of the phenotype for 8 days. Then, 500,000 cells per treatment were plated in sextuplicate in 10 cm dishes in 6-TG media to allow the proliferation of mutated HPRT− cells. Colonies were fixed (50% methanol and 10% glacial acetic acid), stained (0.02% Coomassie brilliant blue R-250 stain in methanol: acetic acid: water in a ratio of 46.5:7:46.5 (v/v/v)), and counted after 14–20 days. The HPRT mutation frequency was calculated as the ratio of the number of HPRT− colonies in 6-TG media to the number of surviving colonies plated in complete media to determine clonal efficiency (Silva et al., 2005).
Nucleofection
REV1 was knocked down by transiently transfecting SMARTpool: ON-TARGETplus REV1 siRNA by nucleofection. The siRNA was mixed with the nucleofection buffer Mouse/Rat Hepatocyte Nucleofector™ Kit (Lonza) and electroporated using the Nucleofector™ 2b device. Full-length mouse Rev1 on the pC3 plasmid (Clontech) was nucleofected using the same buffers and device into Rev1−/− cells to complement the REV1 function.
Synthesis of the 16mer oligonucleotide containing a cisplatin 1,2-GG lesion and construction of the gapped plasmid
The 16mer oligonucleotide containing a cisplatin 1,2-GG lesion was synthesized as described below. The platination reaction was carried out with aquated derivatives of the platinum complexes to facilitate their reaction with a 16mer oligonucleotide containing a 1,2-GG sequence (5’-CTCTCTCGGCCTTCTA-3’). The aquated complexes were obtained by overnight stirring in the dark at room temperature of a solution containing cisplatin and 1.98 equivalent of silver nitrate. The precipitated silver chloride was removed by a 0.2 μm syringe filter. DNA was mixed with aquated platinum complex in a 1:2 ratio at 37 °C for 2 hours. The target 16mer oligonucleotide containing the cisplatin 1,2-GG lesion was purified by reverse-phase HPLC with a C18 column (5um, 100Å, 150×4.6 mm, Phenomenex). The molecular weight and lesion location were characterized by LC-MS (AB Sciex).
The double stranded plasmid pUC19 with ampicillin resistance was modified to include the oligonucleotide containing cisplatin 1,2-GG lesion on one strand and a gapped region across it. Briefly, the 16mer cisplatin-containing oligonucleotide was flanked by two 21mer regular DNA strands (5’-GCCCGTCGTAGCGCGCATGCA-3’ on the 5’ end and 5’-TCTCGAGTGTTCCGTCAGCAC-3’ on the 3’ end) and elongated to a 58mer lesion-containing single strand DNA. After linearizing the plasmid by restriction endonucleases BstAPI and BspQI (New England Biolabs), the 58mer oligonucleotide was mixed with two scaffolds (5’-TGCATGCGCGCTACGACG-3’ and 5’-AGCGTGCTGACGGAACACTCGAGA-3’) and ligated with the linear pUC19 plasmid to build up a circular plasmid containing a site-specific cisplatin 1,2-GG lesion and a 16-nucleotide gap on the opposite strand.
The quantitative assay of the gapped plasmid containing a cisplatin 1,2-GG lesion
A competitor gapped plasmid that was three bases longer (started from a 19mer oligonucleotide 5’- CTCTCTAGGCTCACTTCTA -3’) than the lesion-containing plasmid was used as the internal control. The cells were pre-treated with either DMSO or JH-RE-06 (1.5, 3.0 and 15.0 μM) for 24 hours. Gapped-lesion plasmid (200 ng) and competitor plasmid (50 ng) were transfected in a 4:1 ratio into 300,000 HT1080 cells using Lipofectamine 3000 (ThermoFisher). Transfected cells were incubated at 37 °C for 4 hours. Next, the cells were trypsinized (0.25% trypsin-EDTA), and DNA was extracted using the Qiagen DNA isolation kit. The isolated DNA was then transformed into the recA- E. coli strain, JM109, to propagate fully closed plasmids obtained from the mammalian cells. After 16 hours, total plasmid DNA was isolated from the E. coli cells and the region encompassing the cisplatin lesion from both the cisplatin and competitor plasmids was amplified by PCR (forward primer: 5’-TTGTACTGAGAGTGCACCATGCCCGT-3’, reverse primer: 5’-GAGTCAGTGAGCGAGGAAGCGTGCTG-3’). Two restriction endonucleases XhoI and SphI (New England Biolabs, Ipswich, MA) were used to digest the PCR products into short DNA pieces, 20mer for the cisplatin plasmid and 23mer for the competitor plasmid. The digestion products were chromatographed on a PolarAdvantage C18 column (250 × 2.1 mm, 3μm, 120Å, ThermoFisher) eluted at 0.1 mL/min with a methanol gradient (15% - 50%, 400 mM hexafluoro-2-propanol), followed by the ESI triple quadrupole time-of-flight mass spectrometry (AB Sciex 4600) to detect the final nucleoside signal in the negative ion mode. The input ratio is the initial 4:1 ratio of the cisplatin 1,2-GG lesion and the competitor plasmid that were used to transfect mammalian cells. The gap-filling efficiency by TLS was calculated by dividing the output ratio obtained from HPLC-MS with the input ratio (4:1) and the results were normalized to 100%.
Murine xenograft tumor model
Prior to in vivo experiments, we verified that the DNA damage response pathways were intact in A375 cells by detecting elevated levels of the p21 biomarker via qRT-PCR in response to cisplatin or cisplatin/JH-RE-06 treatment (Figure S4D). Total RNA was isolated by using the RNeasy Mini kit (Qiagen) from cells treated with JH-RE-06 (1.5 μM), cisplatin (1 μM) and a combination of both JH-RE-06 (1.5 μM) and cisplatin (1 μM). 5 ng of RNA from each sample was mixed with 10 μl of Applied Biosystems™ PowerUp™ SYBR™ Green Master Mix (ThermoFisher), 1 μl of MultiScribe™ Reverse Transcriptase (ThermoFisher), 0.1 μl of RNaseOUT™ Recombinant Ribonuclease Inhibitor and 5.9 μl of RNase-free water, and run in a one-step qRT-PCR reaction. Each reaction was run in triplicate. Primers used were: p21 F 5’ GTC ACT GTC TTG TAC CCT TGT G 3’, p21 R 5’ CGG CGT TTG GAG TGG TAG AAA 3’; GAPDH F 5’ GGA GCG AGA TCC CTC CAA AAT 3’, GAPDH R 5’ GGC TGT TGT CAT ACT TCT CAT GG 3’.
NCRNU-F (nude) female, 6–8-week-old mice were divided into 4 groups (with 6 animals per group) for saline, cisplatin alone, JH-RE-06 alone, and cisplatin and JH-RE-06 combination treatments. Three million A375 cells mixed in matrigel (Corning) were injected into each flank of the 6 mice to generate 10–12 xenograft tumors per treatment group. After the tumors grew to a total tumor volume of at least 100 mm3, the drugs (saline, cisplatin alone, JH-RE-06 alone, and cisplatin and JH-RE-06 combination) with a total volume of 100 μL per injection were injected directly into the tumor. Treatments were carried out twice per week for 5 weeks. On the dosing day, tumors were first measured with calipers, weights were recorded, and then the drugs would be injected directly into the tumors. The mice were sedated with isofluorane prior to measurements and treatments.
The drugs were formulated in 10% EtOH, 40% PEG400, and 50% saline for all the four types of treatments. Cisplatin was injected at a dose of 1 mg/kg per animal and JH-RE-06 was administered at a 1.6 mg/kg per animal. In the combination treatment of cisplatin and JH-RE-06, the same doses of 1 mg/kg and 1.6 mg/kg respectively per animal were administered. Tumor volumes were calculated by the formula (W2 × L)/2 as described previously (Faustino-Rocha et al., 2013).
QUANTIFICATION AND STATISTICAL ANALYSIS
Cell culture results were statistically analyzed using one-way analysis of variance (ANOVA) followed by Tukey’s post hoc tests or with the Student’s t-test. For murine xenograft tumor studies, differences of tumor volumes and survival curves of tumor-bearing mice between treatment groups were analyzed by the Welch’s t-test and the Mantel-Cox log-rank test, respectively. Multiple biological replicates (n ≥ 3) were performed in all cases, unless otherwise noted. Variation is indicated using standard error of the mean (SEM) and presented as mean ± SEM unless otherwise noted. Significance was defined as *p<0.05 or **p<0.01. Statistical details of individual experiments can be found in the corresponding Figure Legends.
DATA AND SOFTWARE AVAILABILITY
The coordinates of the chimeric POL κ RIR-REV1 CTD and its complex with JH-RE-06 have been deposited to the Protein Data Bank with accession numbers of 6C59 and 6C8C respectively.
Supplementary Material
Table S1: X-ray data collection and refinement statistics of the chimeric POL κ RIR-REV1 CTD and its complex with JH-RE-06, Related to Figures 2 and S1.
Figure S1. Crystal structure of the chimeric POL κ RIR-REV1 CTD (cREV1 CTD) in the apo state, Related to Figure 1. (A) Ribbon diagram of the cREV1 CTD, with the N-terminus in blue and C-terminus in red. The POL κ RIR-connecting loop and the C-terminal tail of the REV1 CTD, which are invisible in the electron density, are indicated by dashed grey lines. (B) Superimposition of the apo cREV1 CTD with the corresponding components in the POL κ RIR-REV1 CTD-REV7/3 translesionsome complex (PDB 4FJO) reveals similar conformations of the REV1 CTD and POL κ RIR and verifies that the REV7-binding surface of the REV1 CTD (indicated by blue arrows) is unoccupied. The C-terminal tail, which is dynamic and invisible in the apo protein but structured in the translesionsome complex, is labeled in red.
Figure S2. Synthesis and characterization of JH-RE-06 and its analog JH-RE-25, Related to Figure 2. (A) The synthetic routes of JH-RE-06 and JH-RE-25. (B) Isothermal titration calorimetry measurements of the REV1 CTD/JH-RE-06 interaction yielded a dissociation constant (Kd) of 0.42 ± 0.11 μM and a protein-to-ligand stoichiometry of 2.04 ± 0.03. (C) Substituting the hydrophobic dicholoraniline group in JH-RE-06 with a hydrophilic morpholine group in JH-RE-25 abolished the REV1 CTD binding as measured by isothermal titration calorimetry. Relative cell viabilities of HT1080 and A375 cells treated with DMSO, cisplatin (1 μM), JH-RE-25 (1.5 μM), or the combination of cisplatin (1 μM) and JH-RE-25 (1.5 μM) are shown in panels (D) and (E) respectively. Cell viability was assessed by the Promega CellTiter-Glo Luminescence cell viability assay and normalized to DMSO control samples. Error bars represent standard error of the mean (n=6 for all measurements). Statistical analysis: one-way ANOVA with Tukey HSD post-hoc test. N.S., not significant.
Figure S3. Distinct conformations of the C-terminal tails of the cREV1 CTD subunits in the JH-RE-06-bound complex, Related to Figure 2. (A) Superimposition of the subunits of the cREV1 CTD to the corresponding fragments in the POL κ RIR-REV1 CTD-REV7/3 translesionsome complex (PDB 4FJO). Protomers A and B are colored in green and cyan respectively; and the translesionsome complex is colored in grey, except for the REV1 CTD, which is colored in pale cyan. (B) Superimposition of the two protomers in the cREV1 CTD/JH-RE-06 complex, with protomer A colored in green and protomer B colored in cyan. The distance of the C-terminal residue movement is labeled. Proteins are shown in Cα traces whereas JH-RE-06 is shown in the stick model.
Figure S4. Viability and cellular response of cells treated with JH-RE-06 and DNA-damaging agents, Related to Figures 3 and 4. (A) Relative survivability with increasing doses of JH-RE-06 in cisplatin-treated cells. (B) Relative survivability with increasing doses of cisplatin in JH-RE-06 treated cells. (C) JH-RE-06 sensitizes cells to DNA-damaging agents beyond cisplatin. (D) qRT-PCR measurements of p21 mRNA induction in A375 cells treated with DMSO, JH-RE-06, cisplatin and the cisplatin/JH-RE-06 combination. Viabilities of HT1080 (human fibrosarcoma), A375 (human melanoma), LNCap (human prostate adenocarcinoma), KP (mouse KrasG12D;p53−/− lung adenocarcinoma), and AG01522 (human primary fibroblasts) cells were assessed by the Promega CellTiter-Glo Luminescence cell viability assay. Relative luminescence intensities to samples treated with 0 μM JH-RE-06 (panel A) or with 0 μM DNA-damaging agents (panels B and C) are plotted. Blue lines represent single agent treatments, and orange lines represent JH-RE-06 combination treatments. JH-RE-06 at a concentration of 1.5 μM was used in all experiments except for the JH-RE-06 dose response tests in panel A. Combination treatments in panel A contained cisplatin at concentrations of 0.5 μM for HT1080, A375, LNCap, and AG01522 cells, and 10 μM for KP cells. Error bars represent standard error of the mean (n=6 for panels A–C and n=3 for panel D).
Figure S5. JH-RE-06 suppresses bypass efficiency of 1,2-GG cisplatin DNA adduct in HT1080 cells, Related to Figures 3 and 4. (A) Experimental design for the gapped plasmid TLS assay. (B) JH-RE-06 inhibits bypass efficiency in cells. (C) Dose-dependent inhibition of bypass efficiency with different concentrations of JH-RE-06. Error bars represent standard error of the mean (n=3 for all measurements). Statistical analysis: Student’s t-test. *P< 0.05; **P<0.01.
Highlights.
Discovery of JH-RE-06, a compound disrupting Rev1-Pol ζ mediated mutagenic TLS
JH-RE-06 induces Rev1 dimerization to block the Rev1-Rev7 interaction
JH-RE-06 sensitizes tumors to cisplatin and reduces mutagenesis in vitro
JH-RE-06 suppresses tumor progression in mice and prolongs animal survival
ACKNOWLEDGEMENTS
This work was supported in part by grants from the National Cancer Institute (CA191448 to P.Z. and J.H.; CA213042 to D.L.), the Alexander and Margaret Stewart Trust (to P.Z.), the National Institute of Environmental Health Sciences (ES028303 to G.C.W.; ES028865 to D.L.), Duke University (to J.H.), and Center for Precision Cancer Medicine at MIT (to M.T.H). G.C.W. is an American Cancer Society Professor. The chemical library used in this study was kindly provided by Korea Chemical Bank (http://www.chembank.org/) of Korea Research Institute of Chemical Technology. X-ray diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory, supported by the US Department of Energy, Office of Science and the Office of Basic Energy Sciences under Contract number W-31-109-Eng-38. NMR spectra were acquired at the Duke University NMR Center funded by the NSF, NIH, NC Biotechnology Center, and Duke University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures and one table and can be found with this article online.
DECLARATION OF INTERESTS
P.Z. and J.H. are inventors of a patent on JH-RE-06. The remaining authors declare no competing interests.
REFERENCES
- Actis ML, Ambaye ND, Evison BJ, Shao Y, Vanarotti M, Inoue A, McDonald ET, Kikuchi S, Heath R, Hara K, et al. (2016). Identification of the first small-molecule inhibitor of the REV7 DNA repair protein interaction. Bioorganic & medicinal chemistry 24, 4339–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, and Terwilliger TC (2002). PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58, 1948–1954. [DOI] [PubMed] [Google Scholar]
- Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, and Livneh Z (2004). Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: the role of DNA polymerase kappa. The Journal of biological chemistry 279, 53298–53305. [DOI] [PubMed] [Google Scholar]
- Baranovskiy AG, Lada AG, Siebler HM, Zhang Y, Pavlov YI, and Tahirov TH (2012). DNA polymerase delta and zeta switch by sharing accessory subunits of DNA polymerase delta. The Journal of biological chemistry 287, 17281–17287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat A, Wu Z, Maher VM, McCormick JJ, and Xiao W (2015). Rev7/Mad2B plays a critical role in the assembly of a functional mitotic spindle. Cell Cycle 14, 3929–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersma V, Moatti N, Segura-Bayona S, Peuscher MH, van der Torre J, Wevers BA, Orthwein A, Durocher D, and Jacobs JJL (2015). MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5’ end resection. Nature 521, 537–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi EB, Yon GH, Lee HK, Yang HC, Yoo CY, and Pak CS (2003). Synthesis of ß-Lactam from Acyl(Arylcarbamoyl)-S,S-bis(alkylketene) Dithioacetal: Revised Structure of the Product from Thermal Cyclization of Acyl(Arylcarbamoyl)-S,S-bis(alkylketene) Dithioacetal. Synthesis 2003, 2771–2776. [Google Scholar]
- Doles J, Oliver TG, Cameron ER, Hsu G, Jacks T, Walker GC, and Hemann MT (2010). Suppression of Rev3, the catalytic subunit of Pol{zeta}, sensitizes drug-resistant lung tumors to chemotherapy. Proceedings of the National Academy of Sciences of the United States of America 107, 20786–20791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Faustino-Rocha A, Oliveira PA, Pinho-Oliveira J, Teixeira-Guedes C, Soares-Maia R, da Costa RG, Colaco B, Pires MJ, Colaco J, Ferreira R, et al. (2013). Estimation of rat mammary tumor volume using caliper and ultrasonography measurements. Lab Anim (NY) 42, 217–224. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Cho Y, Yang IY, Akagi J, Ohashi E, Tateishi S, de Wind N, Hanaoka F, Ohmori H, and Moriya M (2012). The vital role of polymerase zeta and REV1 in mutagenic, but not correct, DNA synthesis across benzo[a]pyrene-dG and recruitment of polymerase zeta by REV1 to replication-stalled site. The Journal of biological chemistry 287, 9613–9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izuta S (2006). Inhibition of DNA polymerase eta by oxetanocin derivatives. Nucleic Acids Symp Ser (Oxf), 269–270. [DOI] [PubMed] [Google Scholar]
- Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, and de Wind N (2006). Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med 203, 319–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Prakash L, and Prakash S (2012). Pol31 and Pol32 subunits of yeast DNA polymerase delta are also essential subunits of DNA polymerase zeta. Proceedings of the National Academy of Sciences of the United States of America 109, 12455–12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010). XDS. Acta Crystallogr D Biol Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova AV, and Burgers PM (2015). Eukaryotic DNA polymerase zeta. DNA Repair (Amst) 29, 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova AV, Stodola JL, and Burgers PM (2012). A four-subunit DNA polymerase zeta complex containing Pol delta accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res 40, 11618–11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch SD, Kokoska RJ, Chilkova O, Welch CM, Johansson E, Burgers PM, and Kunkel TA (2004). Enzymatic switching for efficient and accurate translesion DNA replication. Nucleic Acids Res 32, 4665–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushina Y, Motoshima H, Yamaguchi Y, Takeuchi T, Hirano K, Sugawara F, and Yoshida H (2009). 3-O-methylfunicone, a selective inhibitor of mammalian Y-family DNA polymerases from an Australian sea salt fungal strain. Mar Drugs 7, 624–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki Y, and Furukawa K (1987). Application of coomassie brilliant blue staining to cultured hepatocytes. Cell Biol Int Rep 11, 367–371. [DOI] [PubMed] [Google Scholar]
- Pak CS, Yang HC, and Choi EB (1992). Aminolysis of 5-Acyl-2,2-dimethyl-1,3-dioxane-4,6-diones (Acyl Meldrum’s Acids) as a Versatile Method for the Synthesis of ß-Oxo Carboxamides. Synthesis 1992, 1213–1214. [Google Scholar]
- Sail V, Rizzo AA, Chatterjee N, Dash RC, Ozen Z, Walker GC, Korzhnev DM, and Hadden MK (2017). Identification of Small Molecule Translesion Synthesis Inhibitors That Target the Rev1-CT/RIR Protein-Protein Interaction. ACS chemical biology 12, 1903–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, et al. (2009). Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J 28, 383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MJ, Costa P, Dias A, Valente M, Louro H, and Boavida MG (2005). Comparative analysis of the mutagenic activity of oxaliplatin and cisplatin in the Hprt gene of CHO cells. Environmental and molecular mutagenesis 46, 104–115. [DOI] [PubMed] [Google Scholar]
- Vaisman A, and Woodgate R (2017). Translesion DNA polymerases in eukaryotes: what makes them tick? Crit Rev Biochem Mol Biol 52, 274–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanarotti M, Evison BJ, Actis ML, Inoue A, McDonald ET, Shao Y, Heath RJ, and Fujii N (2018). Small-molecules that bind to the ubiquitin-binding motif of REV1 inhibit REV1 interaction with K164-monoubiquitinated PCNA and suppress DNA damage tolerance. Bioorganic & medicinal chemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek J, Lee CJ, D’Souza S, Minesinger B, Kim H, D’Andrea AD, Walker GC, and Zhou P (2012a). Structural Basis of Rev1-mediated Assembly of a Quaternary Vertebrate Translesion Polymerase Complex Consisting of Rev1, Heterodimeric Polymerase (Pol) zeta, and Pol kappa. The Journal of biological chemistry 287, 33836–33846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek J, Liu J, D’Souza S, Wang S, Xue Y, Walker GC, and Zhou P (2012b). Multifaceted Recognition of Vertebrate Rev1 by Translesion Polymerases zeta and kappa. The Journal of biological chemistry 287, 26400–26408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek JL, Wang S, Kim H, Wu Q, D’Andrea AD, and Zhou P (2014). Ubiquitin recognition by FAAP20 expands the complex interface beyond the canonical UBZ domain. Nucleic Acids Res 42, 13997–14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Doles J, Hemann MT, and Walker GC (2010). Error-prone translesion synthesis mediates acquired chemoresistance. Proceedings of the National Academy of Sciences of the United States of America 107, 20792–20797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Chapman JR, Brandsma I, Yuan J, Mistrik M, Bouwman P, Bartkova J, Gogola E, Warmerdam D, Barazas M, et al. (2015). REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 521, 541–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Xie K, Zhang XQ, Pridgen EM, Park GY, Cui DS, Shi J, Wu J, Kantoff PW, Lippard SJ, et al. (2013). Enhancing tumor cell response to chemotherapy through nanoparticle-mediated codelivery of siRNA and cisplatin prodrug. Proceedings of the National Academy of Sciences of the United States of America 110, 18638–18643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Chatterjee N, Hemann MT, and Walker GC (2017). Inhibition of mutagenic translesion synthesis: A possible strategy for improving chemotherapy? PLoS genetics 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Dorjsuren D, Eoff RL, Egli M, Maloney DJ, Jadhav A, Simeonov A, and Lloyd RS (2012). A comprehensive strategy to discover inhibitors of the translesion synthesis DNA polymerase kappa. PLoS One 7, e45032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, and Gao Y (2018). Translesion and Repair DNA Polymerases: Diverse Structure and Mechanism. Annu Rev Biochem 87, 239–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Lugovskoy AA, and Wagner G (2001). A solubility-enhancement tag (SET) for NMR studies of poorly behaving proteins. J Biomol NMR 20, 11–14. [DOI] [PubMed] [Google Scholar]
- Ziv O, Diamant N, Shachar S, Hendel A, and Livneh Z (2012). Quantitative measurement of translesion DNA synthesis in mammalian cells. Methods Mol Biol 920, 529–542. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: X-ray data collection and refinement statistics of the chimeric POL κ RIR-REV1 CTD and its complex with JH-RE-06, Related to Figures 2 and S1.
Figure S1. Crystal structure of the chimeric POL κ RIR-REV1 CTD (cREV1 CTD) in the apo state, Related to Figure 1. (A) Ribbon diagram of the cREV1 CTD, with the N-terminus in blue and C-terminus in red. The POL κ RIR-connecting loop and the C-terminal tail of the REV1 CTD, which are invisible in the electron density, are indicated by dashed grey lines. (B) Superimposition of the apo cREV1 CTD with the corresponding components in the POL κ RIR-REV1 CTD-REV7/3 translesionsome complex (PDB 4FJO) reveals similar conformations of the REV1 CTD and POL κ RIR and verifies that the REV7-binding surface of the REV1 CTD (indicated by blue arrows) is unoccupied. The C-terminal tail, which is dynamic and invisible in the apo protein but structured in the translesionsome complex, is labeled in red.
Figure S2. Synthesis and characterization of JH-RE-06 and its analog JH-RE-25, Related to Figure 2. (A) The synthetic routes of JH-RE-06 and JH-RE-25. (B) Isothermal titration calorimetry measurements of the REV1 CTD/JH-RE-06 interaction yielded a dissociation constant (Kd) of 0.42 ± 0.11 μM and a protein-to-ligand stoichiometry of 2.04 ± 0.03. (C) Substituting the hydrophobic dicholoraniline group in JH-RE-06 with a hydrophilic morpholine group in JH-RE-25 abolished the REV1 CTD binding as measured by isothermal titration calorimetry. Relative cell viabilities of HT1080 and A375 cells treated with DMSO, cisplatin (1 μM), JH-RE-25 (1.5 μM), or the combination of cisplatin (1 μM) and JH-RE-25 (1.5 μM) are shown in panels (D) and (E) respectively. Cell viability was assessed by the Promega CellTiter-Glo Luminescence cell viability assay and normalized to DMSO control samples. Error bars represent standard error of the mean (n=6 for all measurements). Statistical analysis: one-way ANOVA with Tukey HSD post-hoc test. N.S., not significant.
Figure S3. Distinct conformations of the C-terminal tails of the cREV1 CTD subunits in the JH-RE-06-bound complex, Related to Figure 2. (A) Superimposition of the subunits of the cREV1 CTD to the corresponding fragments in the POL κ RIR-REV1 CTD-REV7/3 translesionsome complex (PDB 4FJO). Protomers A and B are colored in green and cyan respectively; and the translesionsome complex is colored in grey, except for the REV1 CTD, which is colored in pale cyan. (B) Superimposition of the two protomers in the cREV1 CTD/JH-RE-06 complex, with protomer A colored in green and protomer B colored in cyan. The distance of the C-terminal residue movement is labeled. Proteins are shown in Cα traces whereas JH-RE-06 is shown in the stick model.
Figure S4. Viability and cellular response of cells treated with JH-RE-06 and DNA-damaging agents, Related to Figures 3 and 4. (A) Relative survivability with increasing doses of JH-RE-06 in cisplatin-treated cells. (B) Relative survivability with increasing doses of cisplatin in JH-RE-06 treated cells. (C) JH-RE-06 sensitizes cells to DNA-damaging agents beyond cisplatin. (D) qRT-PCR measurements of p21 mRNA induction in A375 cells treated with DMSO, JH-RE-06, cisplatin and the cisplatin/JH-RE-06 combination. Viabilities of HT1080 (human fibrosarcoma), A375 (human melanoma), LNCap (human prostate adenocarcinoma), KP (mouse KrasG12D;p53−/− lung adenocarcinoma), and AG01522 (human primary fibroblasts) cells were assessed by the Promega CellTiter-Glo Luminescence cell viability assay. Relative luminescence intensities to samples treated with 0 μM JH-RE-06 (panel A) or with 0 μM DNA-damaging agents (panels B and C) are plotted. Blue lines represent single agent treatments, and orange lines represent JH-RE-06 combination treatments. JH-RE-06 at a concentration of 1.5 μM was used in all experiments except for the JH-RE-06 dose response tests in panel A. Combination treatments in panel A contained cisplatin at concentrations of 0.5 μM for HT1080, A375, LNCap, and AG01522 cells, and 10 μM for KP cells. Error bars represent standard error of the mean (n=6 for panels A–C and n=3 for panel D).
Figure S5. JH-RE-06 suppresses bypass efficiency of 1,2-GG cisplatin DNA adduct in HT1080 cells, Related to Figures 3 and 4. (A) Experimental design for the gapped plasmid TLS assay. (B) JH-RE-06 inhibits bypass efficiency in cells. (C) Dose-dependent inhibition of bypass efficiency with different concentrations of JH-RE-06. Error bars represent standard error of the mean (n=3 for all measurements). Statistical analysis: Student’s t-test. *P< 0.05; **P<0.01.
Data Availability Statement
The coordinates of the chimeric POL κ RIR-REV1 CTD and its complex with JH-RE-06 have been deposited to the Protein Data Bank with accession numbers of 6C59 and 6C8C respectively.