ABSTRACT
Marine sponges are a prolific source of novel enzymes with promising biotechnological potential. Especially halogenases, which are key enzymes in the biosynthesis of brominated and chlorinated secondary metabolites, possess interesting properties towards the production of pharmaceuticals that are often halogenated. In this study we used a polymerase chain reaction (PCR)-based screening to simultaneously examine and compare the richness and diversity of putative tryptophan halogenase protein sequences and bacterial community structures of six Aplysina species from the Mediterranean and Caribbean seas. At the phylum level, bacterial community composition was similar amongst all investigated species and predominated by Actinobacteria, Chloroflexi, Cyanobacteria, Gemmatimonadetes, and Proteobacteria. We detected four phylogenetically diverse clades of putative tryptophan halogenase protein sequences, which were only distantly related to previously reported halogenases. The Mediterranean species Aplysina aerophoba harbored unique halogenase sequences, of which the most predominant was related to a sponge-associated Psychrobacter-derived sequence. In contrast, the Caribbean species shared numerous novel halogenase sequence variants and exhibited a highly similar bacterial community composition at the operational taxonomic unit (OTU) level. Correlations of relative abundances of halogenases with those of bacterial taxa suggest that prominent sponge symbiotic bacteria, including Chloroflexi and Actinobacteria, are putative producers of the detected enzymes and may thus contribute to the chemical defense of their host.
Keywords: Halogenase, host-associated microbiome, phylogenetic diversity, marine sponges, bioactive compounds, environmental enzymes
A diverse range of novel halogenase genes was detected in six sponge species, indicating that microorganisms play a fundamental role in the production of sponge-derived bioactive compounds.
INTRODUCTION
Bioactive compounds and the role of halogenase enzymes
The need to discover novel drug candidates is high on the policy agenda due to the ongoing emergence of multi-drug resistant microbial strains (O'Neill 2016). The call for a better supply of new drugs against a large range of infectious diseases points towards novel natural products as a yet inexhaustible source of bioactivity. Especially the marine environment proved to be a particularly rich resource for novel bioactive compounds, and many of them are halogenated (Gribble 2015). Carbon–halogen bonds lead to an increase in thermal and oxidative stability as well as increased permeability through biological membranes compared to their non-halogenated counterparts (Smith et al. 2017). Halogenated molecules exhibit a wide range of biological activities and may have antifungal, antibacterial, antiviral, anticancer, and/or anti-inflammatory properties (Butler & Sandy 2009; Gribble 2015). Thus, many pharmaceutical and agrochemical compounds as well as polymers are halogenated (Bolton et al. 2011; Lu et al. 2012; Jeschke 2013), such as the commercially important antibiotics chloramphenicol, vancomycin and teicoplanin (Van Pée and Zehner 2003). In nature, halogens, including chloride, bromide, fluoride or iodide, get attached to organic molecular scaffolds by halogenase enzymes, which have been detected in fungi, bacteria and algae from terrestrial and marine environments (Atashgahi et al. 2018; Latham et al. 2018). While marine enzymes preferentially halogenate with bromide (Neubauer et al. 2018), chlorinated compounds are regularly detected in terrestrial sources (Latham et al. 2018). To date, more than 5000 naturally produced halogenated compounds (Gribble 2015) and six independently evolved halogenase enzyme families have been identified (Xu and Wang 2016). These enzyme families, namely cofactor-free haloperoxidase, vanadium-dependent haloperoxidase, heme iron-dependent haloperoxidase, non-heme iron-dependent halogenase, flavin-dependent halogenase, and S-adenosyl-l-methionine-dependent halogenase, differ in their cofactor dependence, sequence homology and mechanistic features (Xu and Wang 2016). The most extensively characterized halogenases are flavin (FADH2)-dependent halogenases (FDHs) (Van Pée and Patallo 2006; Zhao et al. 2015). In contrast to haloperoxidases, FDHs often halogenate regioselectively, and are often part of secondary metabolite gene clusters encoding for non-ribosomal peptide synthetases (NRPS) and polyketide synthetases (PKS)-producing halogenated molecules (Walsh et al. 2001; Van Pée and Unversucht 2003; Dorrestein et al. 2005; Van Pée and Patallo 2006). Regioselective halogenation under mild reaction conditions without harmful waste generation renders FDHs promising tools for biocatalysis of halogenated compounds (Smith, Grüschow and Goss 2013; Grüschow et al. 2015; Shepherd et al. 2015; Menon et al. 2016; Weichold, Milbredt and Van Pée 2016; Latham et al. 2018). The amino acid sequences of FDHs contain a highly conserved flavin binding site (GxGxxG) near the N-terminus, which binds the cofactor in order to create the reactive halogen, as well as a motif with two tryptophan residues (WxWxIP), which presumably prevents the binding of a substrate close to the flavin (Van Pée and Zehner 2003; Van Pée and Patallo 2006). These motifs can be detected in almost every FDH described today, and serve as identification marker for novel FDH genes (Bayer et al. 2013). FDHs can be distinguished into three classes depending on their preferred substrate moieties: phenols, pyrroles or tryptophan (Murphy 2006; Van Pée and Patallo 2006). In most cases, the exact substrates for halogenases encoded within biosynthetic gene clusters remain to be identified, however, many of the bacterial FDHs characterized to date halogenate a range of tryptophan derivatives and other aromatic substrates (Payne, Andorfer and Lewis 2013; Frese et al. 2014; Shepherd et al. 2015). These flavin-dependent tryptophan halogenases (THs) exhibit a broad substrate tolerance for other electron-rich organic scaffolds, although these are halogenated with lower reaction efficiencies in in vitro activity tests (Weichold, Milbredt and Van Pée 2016; Agarwal et al. 2017). Since halotryptophans occur fairly frequently in natural products (Smith et al. 2017), we focused our efforts on THs in this study. Most flavin-dependent THs described to date require the activity of an additional enzyme, a flavin reductase, to provide the reduced flavin cofactor, and are thus two-component systems (Agarwal et al. 2017). Single-component halogenating enzymes possess both domains for flavin-reduction and halogenation. To date, only two single-component halogenating enzymes have been reported from marine bacteria, but they remain poorly characterized (Agarwal and Moore 2014; Agarwal et al. 2014).
Sponges as source of novel halogenase sequence variants
Sponges are the most prolific marine invertebrates for the discovery of novel bioactive compounds (Blunt et al. 2009; Hu et al. 2011; Mehbub et al. 2014; Rocha-Martin et al. 2014; Sipkema 2017; Carroll et al. 2019) since many of them depend on a chemical arsenal to defend themselves against diseases, competitors and predators (Thoms, Ebel and Proksch 2006; Taylor et al. 2007; Pawlik 2011). These ancient, filter feeding animals harbour dense and diverse microbial communities including members of the bacterial phyla Actinobacteria, Acidobacteria, Bacteriodetes, Chloroflexi, Cyanobacteria, Planctomycetes, Proteobacteria, Nitrospira, Poribacteria, Tectomicrobia, Verrucomicrobia, as well as archaea and numerous microeukaryotes (Taylor et al. 2007; Webster and Taylor 2012; Thomas et al. 2016; Chaib De Mares et al. 2017). Some of these largely uncultured bacteria can produce highly potent bioactive natural products, many of which are halogenated (Gribble 2010; Smith et al. 2017). Amongst marine sponges, the Demosponge genus Aplysina represents a morphologically diverse group of species challenging to identify due to their lack of a mineral skeleton (Zea, Henkel and Pawlik 2014), as well as a high degree of sequence conservation in molecular marker genes (Erpenbeck et al. 2007; Cruz-Barraza et al. 2012). Chemotaxonomy was suggested as additional phylogenetic marker (Erpenbeck and van Soest 2007), since Aplysina species are strongly chemically defended and especially renowned for the production of more than 100 halogenated natural products (Lira et al. 2011; Loh and Pawlik 2014). Such metabolites can make up to 12% of the sponge dry weight (Turon, Becerro and Uriz 2000; Thoms, Ebel and Proksch 2006).
A survey of natural products from Caribbean Aplysina species suggested that the brominated alkaloids were sponge-derived rather than microbiome-derived, since metabolite profiles were highly correlated to sponge morphotype rather than location and depth (Puyana et al. 2015). It should be noted, however, that this study did not assess microbial composition, and hence, an essential or auxiliary role of the microbiome in metabolite production cannot be excluded. Halogenated compounds have been reported to be located in spherulous cells of Aplysina aerophoba, suggesting production by the sponge, or a complex symbiotic pattern with microorganisms involved at different levels of the biotransformation pathway (Turon, Becerro and Uriz 2000). Significant correlations of the relative abundances of a member of the Chloroflexi, a deltaproteobacterium and an unidentified bacterial OTU with the concentrations of three alkaloids (aerophobin-1, aplysinamisin-1 and isofistularin-3) in A. aerophoba indicated that bacteria were correlated to the production of brominated alkaloids (Sacristán-Soriano et al. 2011, Sacristán-Soriano, Banaigs and Becerro 2016). This finding corroborates observations that the majority of halogenating enzymes has been described from algae, fungi and bacteria (Xu and Wang 2016). Halogenated natural products or the corresponding biosynthetic gene clusters could previously be associated to specific bacterial symbionts in other marine sponges (Unson, Holland and Faulkner 1994; Flatt et al. 2005; Ridley et al. 2005; Hochmuth and Piel 2009; Della Sala et al. 2013; Öztürk et al. 2013; Smith et al. 2017), but to the best of our knowledge this remains unresolved for the metabolites of Aplysina species, from both the Mediterranean and Caribbean Sea (Puyana et al. 2015; Sacristán-Soriano, Banaigs and Becerro 2016). Currently, only dehalogenation mechanisms could be directly linked to the microbial community of A. aerophoba (Ahn et al. 2003). However, the conserved nature of key enzymes encoded in biosynthetic gene clusters allows the design of degenerate PCR primers, thus facilitating the screening and discovery of novel sequence variants of these enzymes from environmental DNA (Hornung et al. 2007; Kennedy, Marchesi and Dobson 2008; Borchert et al. 2016). Accordingly, PCR-based surveys have led to the identification of numerous putative flavin-dependent halogenase encoding genes from different environmental samples (Erol, Arends and Muyzer 2017), multiple marine sponges (Bayer et al. 2013; Öztürk et al. 2013) and cultivated Actinomycetes strains (Hornung et al. 2007; Gao and Huang 2009; Liao et al. 2016).
Aim
With this study we aimed to investigate the phylogenetic diversity and distribution of flavin-dependent tryptophan halogenase protein sequence variants within six species of the marine sponge genus Aplysina. By using 16S rRNA and TH gene amplicon sequencing, we aimed to determine the resident sponge bacterial community of different Aplysina species and explore potential links between the microbial populations and the associated halogenase sequences. This will increase our understanding of the putative producers of halogenated secondary metabolites in sponges.
MATERIALS and METHODS
Sponge collection
Three individuals, each of the Caribbean sponge species Aplysina archeri (AAr), Aplysina cauliformis (ACa), Aplysina fistularis (AFi), Aplysina fulva (AFu) and Aplysina lacunosa (ALa), were collected around Bonaire by SCUBA diving at depths between 1 and 10 m on 8 and 9 October 2012 (Table 1). A sampling permit was given to Detmer Sipkema by the government of Bonaire. Sampled sponge individuals grew at least 1 m apart from each other. The sponge species were identified in the field by Dr. Shirley Pomponi. After sampling, the sponge fragments were rinsed three times with sterile artificial seawater (ASW, 33 g/L Reef Crystals, Blacksburg, VA, USA), cut into 1 cm3 pieces and stored in RNAlater (Sigma Aldrich) at −20°C. Additionally, three individuals of the Mediterranean sponge species Aplysina aerophoba (AAe) were collected by SCUBA diving at Cala Montgo, Spain (N 42.114, E 3.168), on 19 October 2010 and 15 January 2012. The collection of A. aerophoba samples was conducted in strict accordance with Spanish and European regulations within the rules of the Spanish National Research Council with the approval of the Directorate of Research of the Spanish Government. After sampling, the sponges were transported to the laboratory and rinsed three times with sterile artificial seawater (ASW) before grinding with a sterilized mortar and pestle. To obtain a homogenous cell suspension, two volumes of ASW were added. Cell suspensions were aliquoted and mixed with sterile glycerol in ASW for a final concentration of 17% glycerol. Samples were frozen at −20°C and stored at −80°C until DNA extraction (Sipkema et al. 2011).
Table 1.
Metadata, sample information and results of 16S rRNA gene as well as halogenase gene sequencing of the Aplysina species analyzed in this study. THs: tryptophan halogenases. (*) Samples did not pass quality criteria and were excluded from the analyses. Putative halogenase sequences were clustered at the amino acid sequence level.
| Sample ID | Species | Sea | Latitude | Longitude | Depth (m) | Temp. | Preservation | 16S rRNA genes | Halogenases | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reads | OTUs (97%) | Phyla | Reads | Putative THs (95%) | ||||||||
| AAe1 | Aplysina aerophoba | Mediterranean | 42.114 | 3.168 | 8 | NA | Cryopreservation | 8707 | 268 | 17 | 2813 | 10 |
| AAe2* | Aplysina aerophoba | Mediterranean | 42.114 | 3.168 | 8 | NA | Cryopreservation | NA | NA | NA | NA | NA |
| AAe3 | Aplysina aerophoba | Mediterranean | 42.115 | 3.168 | 12 | NA | Cryopreservation | 6032 | 241 | 19 | 1716 | 12 |
| AAr1 | Aplysina archeri | Caribbean | 12.160 | -68.283 | 10 | 28°C | RNALater | 9258 | 312 | 17 | 844 | 8 |
| AAr2 | Aplysina archeri | Caribbean | 12.160 | -68.283 | 10 | 28°C | RNALater | 9786 | 359 | 16 | 2927 | 18 |
| AAr3 | Aplysina archeri | Caribbean | 12.160 | -68.283 | 10 | 28°C | RNALater | 5397 | 361 | 16 | 1631 | 17 |
| ACa1 | Aplysina cauliformis | Caribbean | 12.026 | -68.251 | 16 | 29°C | RNALater | 11 664 | 429 | 18 | 907 | 15 |
| ACa2 | Aplysina cauliformis | Caribbean | 12.026 | -68.251 | 16 | 29°C | RNALater | 5523 | 341 | 18 | 1218 | 20 |
| ACa3 | Aplysina cauliformis | Caribbean | 12.026 | -68.251 | 16 | 29°C | RNALater | 3890 | 292 | 14 | 1210 | 17 |
| AFi1 | Aplysina fistularis | Caribbean | 12.094 | -68.232 | 1 | 30°C | RNALater | 4789 | 223 | 16 | 658 | 11 |
| AFi2 | Aplysina fistularis | Caribbean | 12.094 | -68.232 | 1 | 30°C | RNALater | 8429 | 181 | 17 | 688 | 11 |
| AFi3* | Aplysina fistularis | Caribbean | 12.094 | -68.232 | 1 | 30°C | RNALater | NA | NA | NA | NA | NA |
| AFu1 | Aplysina fulva | Caribbean | 12.160 | -68.283 | 10 | 28°C | RNALater | 9995 | 304 | 18 | 5324 | 12 |
| AFu2 | Aplysina fulva | Caribbean | 12.160 | -68.283 | 10 | 28°C | RNALater | 9988 | 295 | 17 | 2110 | 11 |
| AFu3 | Aplysina fulva | Caribbean | 12.160 | -68.283 | 10 | 28°C | RNALater | 10 140 | 424 | 17 | 4308 | 14 |
| ALa1 | Aplysina lacunosa | Caribbean | 12.160 | -68.283 | 10 | 28°C | RNALater | 4896 | 314 | 16 | 2360 | 15 |
| ALa2 | Aplysina lacunosa | Caribbean | 12.160 | -68.283 | 10 | 28°C | RNALater | 7198 | 459 | 18 | 6976 | 22 |
| ALa3 | Aplysina lacunosa | Caribbean | 12.160 | -68.283 | 10 | 28°C | RNALater | 6450 | 282 | 15 | 4497 | 18 |
DNA isolation
DNA was extracted using the FastDNA Spinkit for Soil (MP Biochemicals, Santa Ana, CA, USA) according to the manufacturer's instructions with the following modification for the first step: instead of using 500 mg of soil, 750 µl of cryopreserved Aplysina aerophoba cell suspension was centrifuged at 14 000 g for 10 min, and the pellet was used for the extraction. For the Caribbean sponge species, 500 mg of wet-weight sponge tissue was rinsed in sterile ASW, cut into small pieces and used for DNA extraction.
PCR amplification of tryptophan halogenase genes
A PCR-based method was used to screen the sponge samples for the presence and identity of potential flavin-dependent tryptophan halogenase genes. In a preliminary screening among previously reported primers, the degenerate halogenase gene targeted primers SZ002 and SZ005 (Zehner et al. 2005) were found the most suitable to amplify an approximately 500 bp DNA fragment. Barcoded PCR amplicons (1 specific barcode for each sample) were obtained through a two-step PCR reaction. For the first PCR reaction, the halogenase primers (SZ002 and SZ005) included linker sequences (341F and 806R): 341F-SZ002 5′-CMTAYGGGRBGCASCAG-TCGGYGTSGGCGARGCGACCRTCCC-3′ and 806R-SZ005 5′-GGACTACNNGGGTATCTAAT-GCCGGAGCAGTCGAYGAASAGGTC-3′. The linker sequences represent the binding regions for the barcoding primers in the second PCR. As we routinely use barcoded 16S rRNA gene targeted primers for prokaryotic community composition analysis, we employed those for barcoding of the less routinely used TH gene-targeted primers. Thus, the more widely used barcoding primers could be applied to barcode halogenase gene amplicons.
The first PCR amplification was performed in a volume of 50 µL using 10 µL 5x GoTaq buffer, 2 µL 10 mM dNTP mixture, 0.5 µL 5 U/µL GoTaq DNA polymerase (Promega, Madison, WI, USA), 3 µL 10 µM solution of both primer 341F-SZ002 and primer 806R-SZ005, 22.5 µL nuclease-free water and 1 µL template DNA (10–20 ng/µL) for each of the samples listed in Table 1. PCR conditions were initial denaturation (94°C for 5 min), followed by 35 cycles of denaturation (94°C for 30 s), annealing (60°C for 40 s), elongation (72°C for 50 s) and a final extension (72°C for 5 min). Amplification products were visualized on a 1.25% (w/v) agarose gel and purified using the Millipore DNA Gel Extraction Kit (Millipore, Billerica, MA, USA). A second barcoding PCR was performed as described earlier, except that a pyrosequencing adapter A (CCATCTCATCCCTGCGTGTCTCCGACTCAG) and 18 different barcodes of 10 nucleotides length connected to the 341F linker sequence were used as forward primer and pyrosequencing adapter B (CCTATCCCCTGTGTGCCTTGGCAGTCTCAG) connected to the 806R linker sequence as reverse primer. Furthermore, the number of amplification cycles was reduced to 15. PCR products were visualized on a 1% (w/v) agarose gel, and the bands of PCR products were excised from the gel and purified as described earlier. The amplified fragments with adapter and barcodes were quantified using a Qubit fluorometer (Invitrogen) and mixed in approximately equal concentrations (4 × 105 copies μL−1) to ensure equal representation of each sample in the pool. A 454-sequencing run was performed on a GS FLX Standard PicoTiterPlate (70 × 75) using a GS FLX pyrosequencing system according to the manufacturer's instructions (Roche, Mannheim, Germany) at the Technical University of Copenhagen. Pyrosequencing data of halogenase genes were deposited at the NCBI Sequence Read Archive under sample accession numbers SRR7853828–SRR7853845.
PCR amplification of 16S rRNA genes
Barcoded amplicons of bacterial 16S rRNA genes for all sponge samples were amplified from the extracted DNA. PCR reactions were performed in a volume of 100 μL containing 20 μL High Fidelity Buffer (ThermoFisher Scientific, Waltham, MA, USA), 2.5 μL 10 μM 338R-I, 2.5 μL 10 μM 338R-II reverse primer (Daims et al. 1999), 2 μL 10 mM dNTP mixture, 1 μL 2 U/μL Phusion Hot start II DNA polymerase, 65 μL nuclease free water. 5 µl 27F-DegS forward primer (van den Bogert et al. 2011) with Titanium adapter A and a sample-specific barcode (8nt) (Hamady et al. 2008) attached to the 5′- end as well as 2 µL template DNA (10–20 ng/µL) were added to each reaction. Amplification conditions were initial denaturation (98°C for 30 s), followed by 30 cycles of denaturation (98°C for 10 s), annealing (56°C for 20 s), elongation (72°C for 20 s) and a final extension (72°C for 10 min). Amplification products were visualized, purified, pooled and sequenced as described earlier. Pyrosequencing data of 16S rRNA genes were deposited at the NCBI Sequence Read Archive under sample accession numbers SRR7853935–SRR7853950.
Halogenase gene amplicon data analysis
Halogenase gene pyrosequencing data were demultiplexed using QIIME version 1.9.0 (Caporaso et al. 2010). Sequences that (i) were shorter than 200 bp or longer than 1000 bp, (ii) contained more than one mismatch in the forward or reverse primer sequences, (iii) contained ambiguous bases or (iv) were represented with less than three reads were removed. Chimeric sequences were detected using the usearch61 algorithm (Edgar 2010) and removed. Two samples (AAe2 and AFi3) did not pass quality criteria of the 16S rRNA gene data analysis and were thus also removed from the halogenase gene analyses. The remaining sequences were translated into the three forward open reading frames using the transeq algorithm (Blankenberg et al. 2007) as implemented in Galaxy (Afgan et al. 2016), and ORFs containing stop codons were removed using customized Bash and R scripts (https://github.com/mibwurrepo/Gutleben_et.al_Halogenases_Aplysinas). Sequences were clustered at 95% amino acid sequence identity based on the average protein sequence identity in genomes of the same bacterial species (Rodriguez-R and Konstantinidis 2014; Chaib De Mares et al. 2018) using the uclust algorithm (Edgar 2010). The most abundant sequence per cluster was retained as representative sequence.
For identification of putative flavin-dependent TH sequences, a reference database was created, by subsetting the UniProt/SwissProt database (Bairoch 2002; Bateman et al. 2017) to 5427 ‘halogenase’ protein entries, since similarity searches to smaller databases return more sensitive results (Jagtap et al. 2013; Pearson 2014). Of these, 75 were manually annotated and reviewed entries (SwissProt). Representative amino acid sequences were aligned (blastp) against (i) the entire UniProt/SwissProt database (release 2018_02) and (ii) the halogenase database using the Diamond alignment tool (Buchfink et al. 2014). Protein families (Pfam) were assigned using the InterProScan pipeline 5.17 (Quevillon et al. 2005) based on an evalue cutoff of 10−6.
Amino acid sequences that aligned significantly (e-value < 0.001; Pearson 2014) against an entry in the halogenase database were retained for phylogenetic analyses. Sequences were aligned using the ClustalW algorithm (Larkin et al. 2007), together with the most closely related database entries, four reference sequences (tryptophan 5-halogenase PyrH (Streptomyces rugosporus, A4D0H5), flavin-dependent TH RebH (Lechevalieria aerocolonigenes, Q8KHZ8), flavin-dependent TH PrnA (Pseudomonas fluorescens, P95480), halogenase ClaH (Streptomyces uncialis, G3K6J6)), putative TH protein sequences previously found in the sponge C. crambe (Öztürk et al. 2013) and the outgroup sequences NADH-dependent flavin oxidoreductase BaiH (Clostridium scindens, P32370) and NADPH-flavin oxidoreductase Frp (Vibrio harveyi, Q56691). The halogenase sequences from A. aerophoba obtained by Bayer et al. (2013) could not be included in this analysis since they covered a different region of the gene.
The resulting alignment was manually refined and trimmed to the amplified regions excluding the primers using Jalview (Waterhouse et al. 2009). A maximum likelihood phylogenetic tree was calculated using RaXML HPC (Stamatakis 2006) under the PROTGAMMAWAG substitution model, and 100 bootstrap replicates were used to evaluate clusters. The best-scoring tree was visualized using iTol (Letunic and Bork 2016).
R (version 3.4.3) (Sasaki, Massaki and Kubo 2005) was used for diversity analyses of amino acid sequences identified as putative tryptophan halogenases. For visualization and interpretation, relative abundance information was used for interpretation and was visualized using the ggplot2 v.2.2.1 package (Wickham 2016). Weighted UniFrac dissimilarities (Lozupone et al. 2011) were calculated and ordinated using Principal Coordinates Analysis as implemented in the phyloseq package (McMurdie and Holmes 2013). Phyloseq and the microbiome package (Lahti et al. 2017) were used for calculating observed richness and Shannon index diversity. Faith's phylogenetic diversity was calculated using the package picante (Kembel et al. 2010). Venn diagrams were calculated and visualized using online tool jvenn (Bourtzis et al. 1996). Caribbean core halogenases were defined as being present in at least one sample from all the Caribbean species and identified using jvenn.
16S rRNA gene amplicon data analysis
Bacterial 16S rRNA gene pyrosequencing data were analyzed using mothur v.1.39.5 (P. D. Schloss et al. 2009) by following the 454 Standard operating procedure (https://www.mothur.org/wiki/454_SOP). In brief, sequences were demultiplexed, denoised, and sequences with (i) more than two mismatches in the primers, (ii) more than one mismatch in the barcode and (iii) more than 8 homopolymer were discarded (trim.flows and trim.seqs commands). Reads were reduced to unique sequences (unique.seqs) and aligned to the SILVA SSU 128 database (Quast et al. 2013) (align.seqs: flip = t). Aligned reads were kept (screen.seqs: optimize = start-end, criteria = 98, minlength = 250), and empty alignment columns were removed (filter.seqs: vertical = T, trump = .). Read counts for sequences that were within ≥99% sequence similarity to a more abundant sequence were merged (pre.cluster: diffs = 2). Chimeric sequences were detected with Vsearch (chimera.vsearch) (Rognes et al. 2016) and removed (remove.seqs). Taxonomy was assigned using the SILVA SSU 128 database (classify.seqs: cutoff = 80) (Wang et al. 2007). Sequences that were not classified at Domain level as well as chloroplast sequences were removed (remove.lineage). Uncorrected pairwise distances between aligned sequences were calculated (dist.seqs: cutoff = 0.15), OTUs were generated on the basis of 97% sequence identity (cluster: method = opti, cutoff = 0.03), and files were converted to .shared format (make.shared: list = , group = ,). Taxonomy was assigned to OTUs (classify.otu: list = , name = , taxonomy = , label = 0.03), and representative sequences for each OTU were picked (get.oturep: phylip = , list = , fasta = , label = 0.03, sorted = size). Further OTU table processing was done with Bash and R scripts (https://github.com/mibwurrepo/Gutleben_et.al_Halogenases_Aplysinas). Relative abundance information was used for visualization and interpretation. Calculation of community metrics and UniFrac dissimilarities were performed as described earlier.
Core taxa were defined as being present in at least one sample from all analyzed sponge species, or from all Caribbean sponge species (Caribbean core) and were identified using jvenn. Correlations between relative halogenase and bacterial abundances were expressed as Spearman coefficients for all taxa and all halogenase genes, as well as for the Caribbean core taxa and core halogenase genes. Only coefficients >±0.5 and with P < 0.05 were considered significant, and only taxa and halogenases shared by all Caribbean species were included in this analysis to maximize statistical power. Heatmaps were generated using the pheatmap v1.0.8 package (Kolde 2012). Analyses are available as R Markdown (https://github.com/mibwurrepo/Gutleben_et.al_Halogenases_Aplysinas).
RESULTS
Identification of putative tryptophan halogenases
To capture the diversity of PCR-amplified halogenase genes in Aplysina species, 454-pyrosequencing was performed. A total of 37 374 DNA reads, representing 3653 unique protein sequences, were obtained from TH gene-targeted amplicon sequencing. After clustering the unique sequences at 95% amino acid sequence identity, 1918 protein sequence clusters were retained with a maximum of 109 sequences per cluster. Detailed results per sample are given in Table 1.
Out of the representative (most abundant) sequences of these 1918 clusters, 1654 (86.24%) resulted in a hit against the entire UniProt protein database. However, only 40 sequences aligned significantly (e-value <0.001; Pearson 2014) against two flavin-dependent TH entries present in the UniProt database with low sequence identities (<45% amino acid identity) and low bitscores (mean = 56). Thus, the 1918 sequences were blasted against a manually curated ‘halogenase’ protein database containing the halogenase protein sequence subset (5427 entries) from UniProt. In total, 86 sequences (4.5%) resulted in a significant (e-value <0.001) hit against 1 of 16 entries from the ‘halogenase’ database with high amino acid sequence similarities (32.9–100%) and high bitscores (mean = 173; Table 2) and were thus identified as putative TH protein sequence fragments. Of these 86 sequences, 25 had 67–80% amino acid sequence identity to a TH from marine gammaproteobacterium HTCC2080, an abundant oligotrophic marine microorganism belonging to the NOR5/OM60 clade (Cho and Giovannoni 2004; Thrash et al. 2010). Another 19 sequences matched most closely (82–100%) to a TH fragment from Psychrobacter sp. D8, a gammaproteobacterium isolated from the sponge Crambe crambe (Öztürk et al. 2013). The closest database match of another 15 sequences, although with lower (36–51%) amino acid sequence identity, was a TH from the cyanobacterium Calothrix sp. NIES-2100 (Hirose et al. 2017).
Table 2.
Hit table (blastp) of sequences aligned against the manually curated halogenase protein database. *TH: Tryptophan halogenase, THf: Tryptophan halogenase (Fragment). %ID is given as % amino acid sequence identity.
| No. of sequences | Closest match UniProt accession | Protein name | Organism | %ID min. | %ID max. | Bitscore avg. | Publication |
|---|---|---|---|---|---|---|---|
| 25 | A0Z0U8 | TH | marine Gammaproteobacterium HTCC2080 | 67.3 | 80.4 | 223 | (Thrash et al. 2010) |
| 19 | K7W8V2 | THf | Psychrobacter sp. D8 | 82.1 | 100 | 194 | (Öztürk et al. 2013) |
| 15 | A0A1Z4GVL5 | TH | Calothrix sp. NIES-2100 | 36.2 | 51.2 | 116 | (Hirose et al. 2017) |
| 7 | A0A2E9M8J4 | TH | Dehalococcoidales bacterium | 55.9 | 76.2 | 169 | (Tully, Graham and Heidelberg 2018) |
| 5 | A0A1Z4LA92 | TH | Nostoc linckia NIES-25 | 38.2 | 39.9 | 118 | (Tully, Graham and Heidelberg 2018) |
| 3 | A0A2D9NM13 | TH | Halieaceae bacterium | 71.2 | 83.9 | 172 | (Tully, Graham and Heidelberg 2018) |
| 3 | A0A2E6EA89 | TH | Woeseia sp. | 75.5 | 76.2 | 245 | (Bagnoud et al. 2015) |
| 1 | A0A0F2QFJ7 | TH | Hyphomonadaceae bacterium BRH_c29 | 67.7 | 67.7 | 84 | (Bagnoud et al. 2015) |
| 1 | A0A0F5Q235 | TH | Devosia psychrophila | 36.2 | 36.2 | 93 | (Lepp et al. 2015) |
| 1 | A0A0J7XXQ8 | TH | Novosphingobium barchaimii LL02 | 32.9 | 32.9 | 46 | (Pearce, Oakeshott and Pandey 2015) |
| 1 | A0A0M4LWL3 | TH | Altererythrobacter epoxidivorans | 42.6 | 42.6 | 48 | (Li et al. 2016) |
| 1 | A0A1M6AX62 | TH | Rubritalea squalenifaciens DSM 18 772 | 37.9 | 37.9 | 100 | (Varghese and Submissions 2016) |
| 1 | A0A2D9ICU6 | TH | Citromicrobium sp. | 45.5 | 45.5 | 41 | (Tully, Graham and Heidelberg 2018) |
| 1 | A0A2E1UEX3 | TH | Gammaproteobacteria bacterium | 80.7 | 80.7 | 200 | (Tully, Graham and Heidelberg 2018) |
| 1 | Q9RPF9 | TH | Myxococcus fulvus | 53.9 | 53.9 | 182 | (Hammer et al. 1999) |
| 1 | T1WAM0 | TH | Uncultured organism | 39.5 | 39.5 | 48 | (Nyyssönen et al. 2013) |
Additionally, the 86 protein sequences with significant ‘halogenase’ database hits were also investigated for protein domains using InterProScan (Finn et al. 2017). This confirmed for 84 sequences that they belong to the ‘Tryptophan halogenase’ protein family (PF04820) and all contained a flavin-TH domain (IPR006905). Only two short protein sequences (S1691 and S1792), which were identified as TH by blastp, did not contain a detectable domain.
Phylogenetic analyses of putative tryptophan halogenases
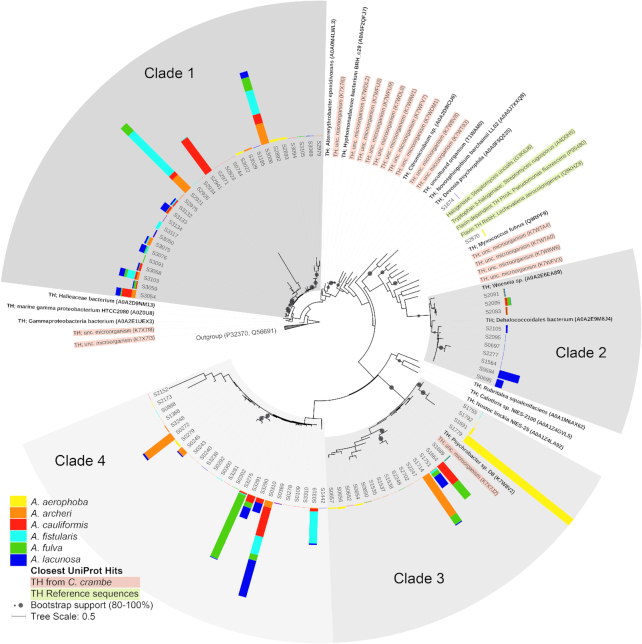
Phylogenetic analyses of the 86 identified putative TH protein sequences resulted in four distinct clades, and only two sequences (S1874 and S2870) did not cluster within these four clades (Fig. 1). Except for A. aerophoba, all analyzed sponge species contained halogenases from all four phylogenetic clades. The largest clade (Clade 1) contained 30 sequences, followed by Clade 4 (25 sequences), Clade 3 (19 sequences) and Clade 2 (10 sequences).
Figure 1.
Maximum likelihood phylogeny of putative tryptophan halogenase (TH) protein sequences from six Aplysina species. Sequences obtained in this study are labelled with S. The tree was constructed from 247 amino acid positions. Two NADH-dependent flavin oxidoreductases (P32370 from Clostridium scindens and Q56691 from Vibrio harveyi) were used as outgroup. Four well-studied TH obtained from UniProt (green) and putative TH from the sponge Crambe crambe (pink) are included as reference sequences. Bars indicate relative abundance of the sequences in the sponge species, with the outer rim of the grey shading indicating 88%. Bootstrap values >80% are indicated by grey circles at the branch points. Sequences in bold refer to the closest relatives from the ‘halogenase’ database. UniProt sequence accession numbers are given inside brackets. Tree scale corresponds to the mean expected number of amino acid substitutions per site.
Only one species-specific halogenase clade could be identified, namely an A. lacunosa-specific subclade within Clade 2 (7 out of 10 sequences; Fig. 1). The sequences within this subclade were all most closely related to a TH from a Dehalococcoidales bacterium (A0A2E9M8J4). Within Clade 3 (19 sequences), the halogenase most abundant (88%) in A. aerophoba (S1779) was closely related to a putative halogenase sequence (K7XJ2) previously detected in Mediterranean C. crambe as well as to a halogenase from Psychrobactersp. D8 isolated from the same sponge (Öztürk et al. 2013). Especially Clade 4 (25 sequences) was only distantly related to previously reported halogenases from Verrucomicrobia and Cyanobacteria and contained mainly sequences from Caribbean Aplysina spp., indicating a large number of novel halogenases within these sponges and their microbiota. The well-characterized flavin-dependent TH gene products RebH (L. aerocolonigenes, Q8KHZ8; Yeh et al. 2007) and PrnA (P. fluorescens, P95480; Dong et al. 2005) clustered outside the sponge-derived clades and were closely related to only one not abundant (<1%) sequence from A. aerophoba (S2870). While the Caribbean species shared many halogenases, it is noteworthy that sequences from the Mediterranean A. aerophoba were closely related to Caribbean halogenase sequences, albeit in no case identical. Furthermore it could be observed that some halogenases were highly abundant in only one sponge species such as S2941 in A. cauliformis or S3275 in A. fulva, while others such as S0310 or S3006 were detected in similar abundances in all Caribbean species.
Bacterial and halogenase diversity
A total of 122 142 high-quality bacterial 16S rRNA gene sequences were obtained from all sponge samples. These sequences clustered into 1993 OTUs at 97% sequence similarity. Bacterial composition at phylum level was similar for all Aplysina species, with Acidobacteria, Actinobacteria, Chloroflexi, Cyanobacteria, Gemmatimonadetes, SBR1093 and Proteobacteria representing the most predominant out of 20 detected bacterial phyla (Supplementary Fig. S1). The 50 overall most abundant OTUs cumulatively accounted for a relative abundance between 59% (ALa2) and 90% (AFi2) in individual samples (Supplementary Fig. S2). Each sponge species exhibited a large number of unique OTUs, while 39 OTUs were shared between all species. These core OTUs comprised between 6% and 45% relative abundance in the sponge samples and contained seawater-derived clades, such as Chloroflexi SAR202 (Morris et al. 2004), as well as putative sponge-symbiotic bacteria such as Rhodospirillales (Karimi et al. 2018), Acidobacteria and Nitrospira (Schmitt et al. 2012).
All Caribbean Aplysina species shared 85 OTUs, amongst which were a predominant sponge-associated member of the Cyanobacteria (Otu0001, 1–67% relative abundance in individual samples), an unclassified bacterial OTU (Otu0010, 0.6–8.2% relative abundance), a member of the actinobacterial OM1 clade (Otu0007, 0.6–8.2% relative abundance) and an SBR1093 OTU (Otu0019, 0.5–4.3% relative abundance). In total, 37 of the 50 overall most abundant OTUs were shared by all of the Caribbean species. Due to the large overlap in bacterial community structure of the Caribbean species, only A. fistularis clearly separated from the other Caribbean Aplysina spp. due to the high relative abundance of the cyanobacterial Otu0001 (>63% relative abundance). In addition, the Mediterranean A. aerophoba could be clearly separated from the other species in ordination plots (Fig. 2E).
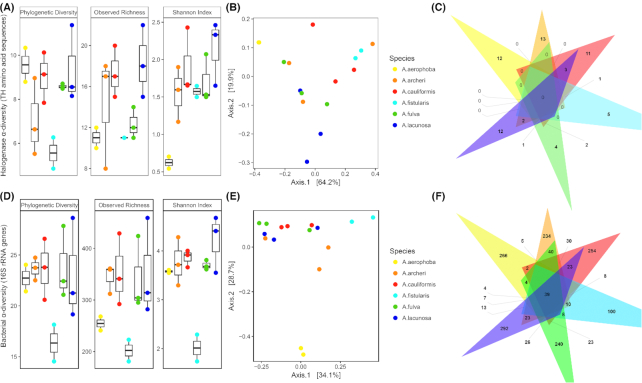
Figure 2.
Halogenase and bacterial 16S rRNA gene alpha diversity indices (A,D). PCoA analysis of weighted UniFrac distances on relative abundance information of tryptophan halogenase (TH) amino acid sequences (B) and 16S rRNA genes (E). A. aerophoba replicates are almost identical and thus overlap in plot B. Venn diagrams of shared and unique TH sequences (C) and 16S rRNA genes (F) per sponge species.
In order to investigate connections between the halogenase diversity and the sponge-associated bacterial diversity, alpha and beta diversity indices were calculated for the 86 putative halogenase sequences and the trends were compared to the diversity indices calculated for the bacterial 16S rRNA gene sequences retrieved from the Aplysina samples (Fig. 2 and Supplementary Fig. S3). Phylogenetic Diversity, Observed Richness and Shannon Diversity indices for halogenases and bacterial 16S rRNA gene sequences followed a similar trend for all species (Fig. 2A and D), however, correlations of diversity indices were only significant for Observed Richness (Supplementary Fig. S3). Overall, a high phylogenetic diversity of both bacterial OTUs and halogenase sequences was detected for A. aerophoba, A. cauliformis and A. lacunosa, while A. fistularis exhibited the least phylogenetically diverse gene repertoire (Fig. 2A). The observed richness of A. aerophoba for both halogenases and bacterial 16S rRNA gene sequences was lower than that for the Caribbean species, however the phylogenetic diversity of both measured variables was high, indicating a large phylogenetic breadth. These differences were not statistically significant (anova Padj> 0.05). Beta diversity analyses showed no discernible species separation as well as a large spread in ordination space for the Caribbean species, while the A. aerophoba samples clustered apart from the other species and extremely close to each other, indicating an almost identical halogenase profile as well as bacterial 16S rRNA gene profile (Fig. 2B and E). Overall, the first two axes explained 84% of variation in the halogenase dataset, and 63% in the 16S rRNA gene dataset, where all Caribbean species spread along the first axis, separating only A. aerophoba along the second axis (Fig. 2E). This analysis revealed an overlap in the halogenase composition of the Caribbean sponges, who shared four halogenase sequences (S3006, S0310, S3103, and S3275). Additionally, each sponge species harbored between 4 (A. fulva) and 13 (A. archeri) species-specific halogenase sequences (Fig. 2C).
Co-correlation of bacterial 16S rRNA gene and halogenase relative abundances
Since diversity calculations indicated an interrelation between halogenase and bacterial 16S rRNA gene diversity, Spearman correlations on the relative abundances of halogenase amino acid sequences with bacterial 16S rRNA genes were calculated for the 50 most predominant bacterial OTUs. All 50 most predominant OTUs exhibited significant (P< 0.05, Spearman r > ±0.5) correlations with two or more of the 86 putative halogenases (Supplementary Fig. S4). It should be noted, however, that the large number of unique and distinct halogenases as well as bacterial OTUs derived from the Mediterranean species A. aerophoba caused a prominent block of strong correlations (Supplementary Fig. S4), and these results should thus be interpreted carefully. Therefore, we further evaluated only the relative abundance co-correlations of bacterial 16S rRNA genes and halogenases that were shared between all Caribbean species (Fig. 3).
Figure 3.
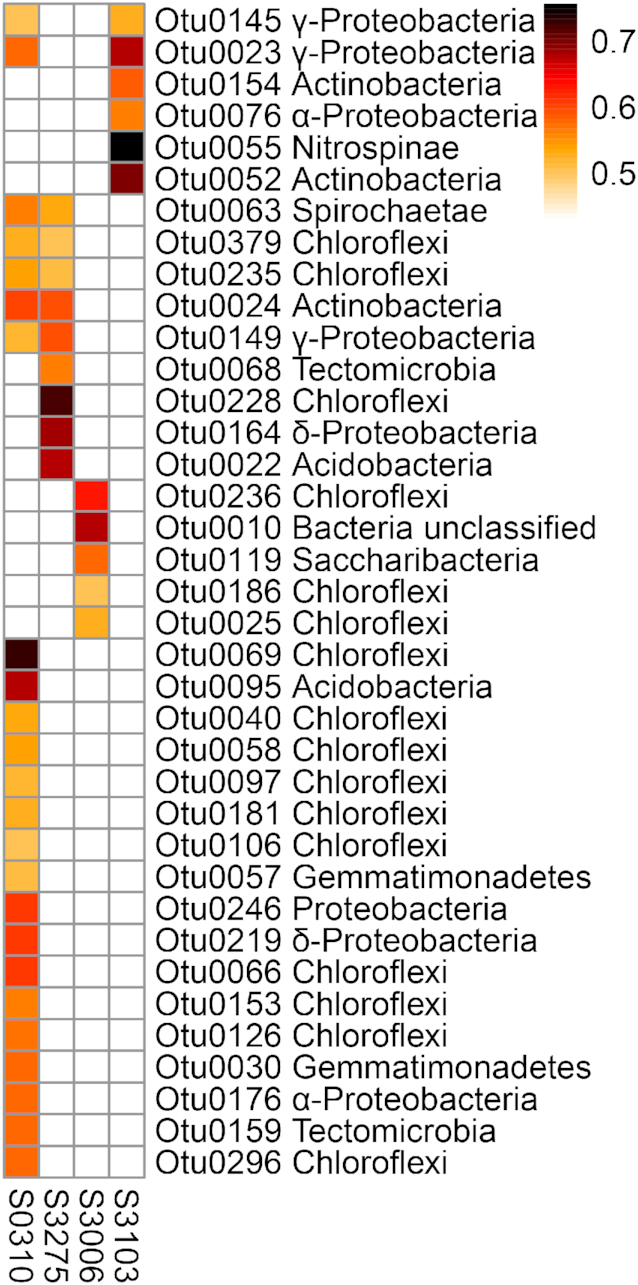
Heatmap displaying significant (Spearman r > 0.5, P< 0.05) co-occurance correlations of tryptophan halogenases (columns) and bacterial 16S rRNA gene OTUs (rows) shared between all Caribbean Aplysina species. OTU IDs and phyla are given to identify bacterial taxa. Columns and rows are clustered by Euclidian distance. Colors indicate correlation coefficients ranging from 0.5 (orange) to 1 (black).
Of the 85 shared bacterial OTUs, 37 were significantly (P < 0.05, Spearman r>0.5) positively correlated with the four shared putative halogenases. The shared bacterial OTUs constituted between 28% and 80% (mean = 46%) of the total relative abundance per sample, while the shared halogenases represent between 10% and 71% (mean = 30%) relative abundance. The majority of the correlated OTUs were affiliated to the phyla Chloroflexi (16 OTUs), Proteobacteria (8 OTUs, Alpha-, Delta- and Gamma-proteobacteria), Actinobacteria (3 OTUs) and Tectomicrobia (2 OTUs) (Fig. 3), all of which are renown for the production of halogenating enzymes (Bayer et al. 2013, 2018; Öztürk et al. 2013; Liao et al. 2016; Smith et al. 2017). For three halogenases, one specific bacterial OTU was found especially highly correlated (r> 0.72), suggesting this distinct bacterial taxon as potential halogenase producer: The highly predominant halogenases S0310 and S3275, both phylogenetically associated with clade 4, were suggested to be produced by the Chloroflexi Otu0069, member of the sponge-associated lineage TK10 (Schmitt et al. 2011; Burgsdorf et al. 2014), and an unclassified Chloroflexi Otu0228 with 98% 16S rRNA gene sequence identity to a sponge-associated member of the Chloroflexi (GenBank FJ481334, (Montalvo and Hill 2011)), respectively. The less abundant halogenase S3103, phylogenetically placed in clade 1, was most highly correlated to an unclassified Nitrospinae Otu0055, displaying 97% 16S rRNA gene sequence identity to an uncultured sponge-associated bacterium (GenBank FJ900348; Kamke et al. 2010). Despite this analysis being of only predictive nature, the co-occurrence of certain halogenases with specific bacterial taxa can narrow down the range for searching potential microbial producers of these proteins.
DISCUSSION
The marine environment is a prolific source of novel enzymes with potential industrial applications. To screen marine samples for such enzymes, metagenomics approaches are promising, whereby all extracted DNA is investigated with DNA sequence-based methods (Hugenholtz and Tyson 2008; Kennedy, Marchesi and Dobson 2008; Vakhlu et al. 2008; Wilson and Piel 2013; Barone et al. 2014; Loureiro et al. 2018). For genes where suitable PCR primers can be designed, a PCR-based screening approach is especially useful to rapidly explore diversity of the gene of interest in a larger number of environmental samples. Such approaches have previously been applied for the discovery of novel natural product biosynthesis genes (Zhao, Yang and Zeng 2008; Milshteyn, Schneider and Brady 2014; Amos et al. 2015; Müller et al. 2015; Borchert et al. 2016), lipases (Wang et al. 2010) or alcohol dehydrogenases (Itoh, Kariya and Kurokawa, 2014), to just name a few examples from a broad range of functional genes discovered in environmental samples (Kotik, 2009). In this study, the PCR-based screening approach resulted in the discovery of a large phylogenetic breadth of previously undescribed, putative halogenase protein fragments, which may be involved in the biosynthesis of numerous brominated natural products found in Mediterranean and Caribbean Aplysina species.
Sponges harbor numerous novel putative tryptophan halogenases
Out of all detected protein sequences clustered at 95% amino acid sequence identity, 86.24% had a match in the entire UniProt database, hinting at still a large number of unknown protein sequences in marine sponges. Additionally, only 86 out of 1918 sequences could successfully be identified as putative halogenases. This could indicate a low specificity of the primers applied, which is a regularly observed problem for primers targeting functional genes in microbial communities (Mohamed et al. 2010; Pereyra et al. 2010; Gaby and Buckley, 2012; Bonilla-Rosso et al. 2016) due to relatively low degrees of sequence conservation in comparison to the 16S rRNA gene.
On the other hand, the number (86) of putative THs is similar for sponges and other environmental samples such as freshwater, marine and soil environments or cultivated Actinomycetes strains (18 (Öztürk et al. 2013), 36 (Bayer et al. 2013), 38 (Erol, Arends and Muyzer, 2017), 103 (Hornung et al. 2007), 254 (Neubauer et al. 2018)). The low sequence identities (mean = 44%), and the match to only few (16) ‘halogenase’ entries from the UniProt database, further indicate a large, yet untapped resource of halogenases in marine sponges. Only one sequence (S2870 in A. aerophoba) with low abundance (<1%) was related with <44% amino acid sequence similarity to two well-charaterized flavin-dependent THs, PrnA and RebH (Fig. 1). PrnA is encoded in the biosynthetic gene cluster for the production of the antifungal compound pyrrolnitrin in Pseudomonas fluorescens (Harris et al. 1985), wheras RebH is part of the biosynthetic gene cluster for the anticancer compound rebeccamycin in Lechevalieria aerocolonigenes (Onaka et al. 2003).
Due to low sequence identies to well-studied halogenases, the putative THs could not be confidentially assigned to any of the known structural classes of halogenated compounds. These enzymes probably represent novel types, which halogenate different structural moieties compared to previously reported halogenases. Such a TH with an unusual substrate preference has been discovered in the metagenome of the sponge symbiont Candidatus Entotheonella serta (Smith et al. 2017). This halogenase displays only between 5% and 27% amino acid sequence identity to the halogenases from this study (data not shown) and represents another recent example of the large, yet untapped genetic resources for novel halogenases in marine sponges. Our phylogenetic analyses resulted in four distinct clades, potentially separating functionally divergent groups of halogenases. These findings expand on previously reported results for A. aerophoba (Bayer et al. 2013) and C. crambe (Öztürk et al. 2013), which showed three distinct sponge-specific clades of halogenases. Each analyzed sponge species harbored sequences from all four clades, hinting at a potentially highly diverse spectrum of halogenated molecules that can be synthesized within each sponge. Rua et al. (2018) hypothesized that larger microbiome diversity influences the potential of bioactive compound production in sponges. In our study we found that a high phylogenetic diversity and richness of microbial taxa corresponded to a high diversity of halogenases. The Mediterranean A. aerophoba harbored a bacterial community distinct from that of the Caribbean species, which is consistent with previous investigations (Thomas et al. 2016; Chaib De Mares et al. 2017), and while its halogenases were closely related to Caribbean ones, they were in no case identical.
All except for two of the detected novel putative halogenases as well as their closest relatives exhibited active binding sites for flavin and l-tryptophan. Since these enzymes are known to tolerate a wide range of organic scaffolds (Agarwal et al. 2017) and since all FDHs known to date can also function as brominases (Xu and Wang, 2016), they are potentially halogenating completely unknown organic substrates. Thus, these enzymes may contribute to the production of the more than 100 halogenated natural products reported from Aplysina species (Turon, Becerro and Uriz, 2000; Thoms, Ebel and Proksch, 2006; Lira et al. 2011; Loh and Pawlik, 2014; Puyana et al. 2015). However, further studies are necessary to unravel the exact functions of these enzymes as no closely related enzymes have been functionally characterized (Fig. 1).
Prominent sponge symbionts are potential halogenase producers
In an attempt to predict the potential bacterial producers of the detected halogenase genes, we correlated the relative abundances of the four 16S rRNA gene OTUs and the 85 putative halogenases shared by the Caribbean Aplysina species. We hypothesized that the shared halogenases are produced by bacteria that can be found amongst the shared bacterial taxa in the different sponge species. Furthermore, an increased relative abundance of the producers is expected to be reflected in an increased number of detected halogenase genes. Two halogenases (S0310 and S3275) were most highly correlated with sponge-associated Chloroflexi (Otu0069 and Otu0228, respectively; Fig. 3). These clades include predominant sponge symbionts with the genomic repertoire for chemical defense (Slaby et al. 2017; Bayer et al. 2018). Chloroflexi were previously found to contain halogenases (Bayer et al. 2013) and were linked to the production of brominated compounds in A. aerophoba (Sacristán-Soriano et al. 2011, Sacristán-Soriano, Banaigs and Becerro 2016). We furthermore detected strong correlation of a halogenase sequence to a Nitrospinae OTU, however little is known about this taxon in sponges to date. Nonetheless, Nitrospinae are closely related to the candidate phylum Tectomicrobia, which occurs in Aplysina species (Chaib De Mares et al. 2018) and of which some members are renowned for their large secondary metabolism gene repertoire (Wilson et al. 2014; Smith et al. 2017).
One highly predominant cyanobacterial OTU was present in all Caribbean species and was not significantly correlated to halogenases in this study, adding to the hypothesis that the role of Cyanobacteria in sponge-microbe symbioses of high microbial abundance sponges might be mainly related to nutrient production rather than chemical defense (Freeman et al. 2013; Burgsdorf et al. 2015). In low microbial abundance sponges, however, the production of brominated metabolites has been linked to cyanobacterial symbionts (Unson, Holland and Faulkner, 1994; Flatt et al. 2005). Thus, our results support previous findings and for the first time indicate potential microbial producers of brominated compounds found in Caribbean Aplysina species. However, further studies, including comparative and functinoal genomics of sponge symbionts, are necessary to reliably link halogenated compound production to their microbial producers.
CONCLUSION
The plethora of previously undescribed putative flavin-dependent THs from the metagenomic DNA of Mediterranean and Caribbean Aplysina species unraveled here indicates a large potential for the discovery of novel halogenating enzymes from these marine invertebrates and their associated microbiomes. The separation into four phylogenetically distinct clades of halogenase protein sequences indicates that multiple classes of organic scaffolds may be halogenated by sponge-associated microbes. High bacterial diversity was in most cases indicative of a high halogenase diversity, and while the Caribbean species shared many halogenases as well as bacterial OTUs, the Mediterranean A. aerophoba could be clearly distinguished. Based on co-occurrence, three prominent bacterial sponge symbionts belonging to the Chloroflexi and Nitrospinae were identified as potential sources of abundant halogenases. These results may thus contribute to explaining the origin of the numerous halogenated compounds discovered in Aplysina species.
Supplementary Material
ACKNOWLEDGEMENTS
We thank María J. Uriz from the Centre d'Estudis Avançats de Blanes (CEAB), CSIC, Blanes, Spain, for providing support for sample collection and access to the laboratories for prompt processing, Søren Sørensen and Waleed Abu Al-Soud with the sequencing of the halogenase amplicons and Sudarshan Shetty and Georg Steinert for help with the data analysis.
AUTHOR CONTRIBUTION
DS initiated this project and collected the samples with SP and RW. JG and KMP conducted the molecular lab bench work. JG, JK and DS planned and executed the analysis pipeline for the sequencing data. JG wrote the paper and all authors contributed to its improvement.
FUNDING
This work was supported by the People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme FP7/2007– 2013 [REA grant agreement no. 607786 (BluePharmTrain)] and the Dutch BE-Basic Foundation [project 7.1.5].
Conflicts of interest. None declared.
REFERENCES
- Afgan E, Baker D, van den Beek M et al.. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucl Acids Res. 2016;44:W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V, El Gamal AA, Yamanaka K et al.. Biosynthesis of polybrominated aromatic organic compounds by marine bacteria. Nat Chem Biol. 2014;10:640–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V, Miles ZD, Winter JM et al.. Enzymatic halogenation and dehalogenation reactions: pervasive and mechanistically diverse. Chem Rev. 2017;117:5619–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V, Moore BS.. Enzymatic synthesis of polybrominated dioxins from the marine environment. ACS Chem Biol. 2014;9:1980–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn Y-B, Rhee S-K, Fennell DE et al.. Reductive dehalogenation of brominated phenolic compounds by microorganisms associated with the marine sponge Aplysina aerophoba. Appl Environ Microbiol. 2003;69:4159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos GCA, Borsetto C, Laskaris P et al.. Designing and implementing an assay for the detection of rare and divergent NRPS and PKS clones in European, Antarctic and Cuban soils. PLoS One. 2015;10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atashgahi S, Liebensteiner MG, Janssen DB et al.. Microbial synthesis and transformation of inorganic and organic chlorine compounds. Front Microbiol. 2018;9:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnoud A, Chourey K, Hettich RL et al. Hyphomonadaceae bacterium BRH_c29, whole genome shotgun sequencing project. GenBank accession number LADW00000000.1, 2015.
- Bairoch A. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucl Acids Res. 2002;28:45–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone R, De Santi C, Palma Esposito F et al.. Marine metagenomics, a valuable tool for enzymes and bioactive compounds discovery. Front Mar Sci. 2014;1:1–6. [Google Scholar]
- Bateman A, Martin MJ, O'Donovan C et al.. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer K, Jahn MT, Slaby BM et al.. Marine sponges as chloroflexi hot spots: genomic insights and high-resolution visualization of an abundant and diverse symbiotic clade. mSystems. 2018;3:328013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer K, Scheuermayer M, Fieseler L et al.. Genomic mining for novel FADH2-dependent halogenases in marine sponge-associated microbial consortia. Mar Biotechnol. 2013;15:63–72. [DOI] [PubMed] [Google Scholar]
- Blankenberg D, Taylor J, Schenck I et al.. A framework for collaborative analysis of ENCODE data: making large-scale analyses biologist-friendly. Genome Res. 2007;17:960–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blunt JW, Copp BR, Hu WP et al.. Marine natural products. Nat Prod Rep. 2009;26:170–244. [DOI] [PubMed] [Google Scholar]
- Bolton O, Lee K, Kim HJ et al.. Activating efficient phosphorescence from purely organic materials by crystal design. Nat Chem. 2011;3:205–10. [DOI] [PubMed] [Google Scholar]
- Bonilla-Rosso G, Wittorf L, Jones CM et al.. Design and evaluation of primers targeting genes encoding NO-forming nitrite reductases: implications for ecological inference of denitrifying communities. Sci Rep. 2016;6:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert E, Jackson SA, O'Gara F. et al.. Diversity of natural product biosynthetic genes in the microbiome of the deep sea sponges Inflatella pellicula, Poecillastra compressa, and Stelletta normani. Front Microbiol. 2016;7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtzis K, Nirgianaki A, Markakis G et al.. Wolbachia infection and cytoplasmic incompatibility in Drosophila species. Genetics. 1996;144:1063–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2014;12:59–60. [DOI] [PubMed] [Google Scholar]
- Burgsdorf I, Erwin PM, López-Legentil S et al.. Biogeography rather than association with cyanobacteria structures symbiotic microbial communities in the marine sponge Petrosia ficiformis. Front Microbiol. 2014;5:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgsdorf I, Slaby BM, Handley KM et al.. Lifestyle Evolution in Cyanobacterial Symbionts of Sponges. MBio. 2015;6:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Sandy M. Mechanistic considerations of halogenating enzymes. Nature. 2009;460:848–54. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J et al.. QIIME allows analysis of high- throughput community sequencing data. Nat Publ Gr. 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll AR, Copp BR, Davis RA et al.. Marine natural products. Nat Prod Rep. 2019;36:122–73. [DOI] [PubMed] [Google Scholar]
- Chaib De Mares M, Jiménez DJ, Palladino G et al.. Expressed protein profile of a Tectomicrobium and other microbial symbionts in the marine sponge Aplysina aerophoba as evidenced by metaproteomics. Sci Rep. 2018;8:11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaib De Mares M, Sipkema D, Huang S et al.. Host Specificity for Bacterial, Archaeal and Fungal Communities Determined for High- and Low-Microbial Abundance Sponge Species in Two Genera. Front Microbiol. 2017;8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J-C, Giovannoni SJ.. Cultivation and Growth Characteristics of a Diverse Group of Oligotrophic Marine Gammaproteobacteria. Appl Environ Microbiol. 2004;70:432–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Barraza JA, Carballo JL, Rocha-Olivares A et al.. Integrative taxonomy and molecular phylogeny of genus Aplysina (Demospongiae: Verongida) from Mexican Pacific. PLoS One. 2012;7:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H, Brühl A, Amann R et al.. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: Development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–44. [DOI] [PubMed] [Google Scholar]
- Della Sala G, Hochmuth T, Costantino V et al.. Polyketide genes in the marine sponge Plakortis simplex: a new group of mono-modular type I polyketide synthases from sponge symbionts. Environ Microbiol Rep. 2013;5:809–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Flecks S, Unversucht S et al.. Structural biology: Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination. Science (80-). 2005;309:2216–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrestein PC, Yeh E, Garneau-Tsodikova S et al.. Dichlorination of a pyrrolyl-S-carrier protein by FADH2-dependent halogenase PltA during pyoluteorin biosynthesis. Proc Natl Acad Sci. 2005;102:13843–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. [DOI] [PubMed] [Google Scholar]
- Erol O, Arends I, Muyzer G.. Discovery of Putative Halogenases in Environmental Samples Using Metagenomics. Curr Biotechnol. 2017;6:17–25. [Google Scholar]
- Erpenbeck D, Duran S, Rützler K et al.. Towards a DNA taxonomy of Caribbean demosponges: A gene tree reconstructed from partial mitochondrial CO1 gene sequences supports previous rDNA phylogenies and provides a new perspective on the systematics of Demospongiae. J Mar Biol Assoc United Kingdom. 2007;87:1563–70. [Google Scholar]
- Erpenbeck D, van Soest RWM.. Status and perspective of sponge chemosystematics. Mar Biotechnol. 2007;9:2–19. [DOI] [PubMed] [Google Scholar]
- Finn RD, Attwood TK, Babbitt PC et al.. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017;45:D190–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt PM, Gautschi JT, Thacker RW et al.. Identification of the cellular site of polychlorinated peptide biosynthesis in the marine sponge Dysidea (Lamellodysidea) herbacea and symbiotic cyanobacterium Oscillatoria spongeliae by CARD-FISH analysis. Mar Biol. 2005;147:761–74. [Google Scholar]
- Freeman CJ, Thacker RW, Baker DM et al.. Quality or quantity: Is nutrient transfer driven more by symbiont identity and productivity than by symbiont abundance? ISME J. 2013;7:1116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese M, Guzowska PH, Voß H et al.. Regioselective enzymatic halogenation of substituted tryptophan derivatives using the FAD-dependent halogenase RebH. ChemCatChem. 2014;6:1270–6. [Google Scholar]
- Gaby JC, Buckley DH. A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PLoS One. 2012;7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Huang Y. Detection, distribution, and organohalogen compound discovery implications of the reduced flavin adenine dinucleotide-dependent halogenase gene in major filamentous actinomycete taxonomic groups. Appl Environ Microbiol. 2009;75:4813–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble GW. A recent survey of naturally occurring organohalogen compounds. Environ Chem. 2015;12:396–405. [Google Scholar]
- Gribble GW. ChemInform Abstract: Naturally Occurring Organohalogen Compounds - A Comprehensive Update 2010.
- Grüschow S, Smith DRM, Gkotsi DS et al.. Halogenases. In: Faber K, Fessner WD, Turner NJ(eds).Biocatalysis in Organic Synthesis. Stuttgart:Georg Thieme Verlag, 2015. [Google Scholar]
- Hamady M, Walker JJ, Harris JK et al.. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer PE, Burd W, Hill DS et al.. Conservation of the pyrrolnitrin biosynthetic gene cluster among six pyrollnitrin-producing strains. FEMS Microbiol Lett. 1999;180:39–44. [DOI] [PubMed] [Google Scholar]
- Harris CM, Kannan R, Kopecka H et al.. The Role of the Chlorine Substituents in the Antibiotic Vancomycin: Preparation and Characterization of Mono- and Didechlorovancomycin. J Am Chem Soc. 1985;107:6652–8. [Google Scholar]
- Hirose Y, Shimura Y, Fujisawa T et al.. Genome sequencing of cyanobacterial culture collection at National Institute for Environmental Studies (NIES). Unpublished.2017; https://www.ncbi.nlm.nih.gov/nuccore/AP018178.1. [Google Scholar]
- Hochmuth T, Piel J.. Polyketide synthases of bacterial symbionts in sponges - Evolution-based applications in natural products research. Phytochemistry. 2009;70:1841–9. [DOI] [PubMed] [Google Scholar]
- Hornung A, Bertazzo M, Dziarnowski A et al.. A genomic screening approach to the structure-guided identification of drug candidates from natural sources. ChemBioChem. 2007;8:757–66. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Tyson GW.. Metagenomics. Nature. 2008;455:481–3. [DOI] [PubMed] [Google Scholar]
- Hu GP, Yuan J, Sun L et al.. Statistical research on marine natural products based on data obtained between 1985 and 2008. Mar Drugs. 2011;9:514–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, Kariya S, Kurokawa J.. Efficient PCR-based amplification of diverse alcohol dehydrogenase genes from metagenomes for improving biocatalysis: screening of gene-specific amplicons from metagenomes. Appl Environ Microbiol. 2014;80:6280–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagtap P, Goslinga J, Kooren JA et al.. A two-step database search method improves sensitivity in peptide sequence matches for metaproteomics and proteogenomics studies. Proteomics. 2013;13:1352–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschke P. The Unique Role of Halogen Substituents in the Design of Modern Crop Protection Compounds. Modern Methods in Crop Protection Research, 2013. pp. 73–128. [Google Scholar]
- Kamke J, Taylor MW, Schmitt S.. Activity profiles for marine sponge-associated bacteria obtained by 16S rRNA vs 16S rRNA gene comparisons. ISME J. 2010;4:498–508. [DOI] [PubMed] [Google Scholar]
- Karimi E, Slaby BM, Soares AR et al.. Metagenomic binning reveals versatile nutrient cycling and distinct adaptive features in alphaproteobacterial symbionts of marine sponges. FEMS Microbiol Ecol. 2018;94:1–18. [DOI] [PubMed] [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR et al.. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–4. [DOI] [PubMed] [Google Scholar]
- Kennedy J, Marchesi JR, Dobson ADW.. Marine metagenomics: strategies for the discovery of novel enzymes with biotechnological applications from marine environments. Microb Cell Fact. 2008;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. pheatmap: {{Pretty Heatmaps}}. R Packag version. 2012;108:1–7.. https://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf. [Google Scholar]
- Kotik M. Novel genes retrieved from environmental DNA by polymerase chain reaction: Current genome-walking techniques for future metagenome applications. J Biotechnol. 2009;144:75–82. [DOI] [PubMed] [Google Scholar]
- Lahti L, Shetty S, Blake T et al.. Tools for microbiome analysis in R. Bioconductor. 2017; http://microbiome.github.io/microbiome. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP et al.. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. [DOI] [PubMed] [Google Scholar]
- Latham J, Brandenburger E, Shepherd SA et al.. Development of halogenase enzymes for use in synthesis. Chem Rev. 2018;118:232–69. [DOI] [PubMed] [Google Scholar]
- Lepp D, Hassan YI, Li XZ et al. Devosia psychrophila strain Cr7-05, whole genome shotgun sequencing project. GenBank accession number LAPV00000000.1, 2015.
- Letunic I, Bork P.. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucl. Acids Res. 2016;44:W242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L, Chen R, Jiang M et al.. Bioprospecting potential of halogenases from arctic marine actinomycetes. BMC Microbiol. 2016;16:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira NS, Montes RC, Tavares JF et al.. Brominated compounds from marine sponges of the genus Aplysina and a compilation of their 13C NMR spectral data. Mar Drugs. 2011;9:2316–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZY, Wu YH, Huo YY et al.. Complete genome sequence of a benzo[a]pyrene-degrading bacterium Altererythrobacter epoxidivorans CGMCC 1.7731. Mar Genomics. 2016;25:39–41. [DOI] [PubMed] [Google Scholar]
- Loh T-L, Pawlik JR.. Chemical defenses and resource trade-offs structure sponge communities on Caribbean coral reefs. Proc Natl Acad Sci. 2014;111:4151–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro C, Medema MH, van der Oost J et al.. Exploration and exploitation of the environment for novel specialized metabolites. Curr Opin Biotechnol. 2018;50:206–13. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Lladser ME, Knights D et al.. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011;5:169–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Liu Y, Xu Z et al.. Halogen bonding for rational drug design and new drug discovery. Expert Opin Drug Discov. 2012;7:375–83. [DOI] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S.. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehbub MF, Lei J, Franco C, Zhang W.. Marine sponge derived natural products between 2001 and 2010: trends and opportunities for discovery of bioactives. Mar Drugs. 2014;12:4539–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon BRK, Latham J, Dunstan MS et al.. Structure and biocatalytic scope of thermophilic flavin-dependent halogenase and flavin reductase enzymes. Org Biomol Chem. 2016;14:9354–61. [DOI] [PubMed] [Google Scholar]
- Milshteyn A, Schneider JS, Brady SF.. Mining the metabiome: identifying novel natural products from microbial communities. Chem Biol. 2014;21:1211–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed NM, Saito K, Tal Y et al.. Diversity of aerobic and anaerobic ammonia-oxidizing bacteria in marine sponges. ISME J. 2010;4:38–48. [DOI] [PubMed] [Google Scholar]
- Montalvo NF, Hill RT.. Sponge-associated bacteria are strictly maintained in two closely related but geographically distant sponge hosts. Appl Environ Microbiol. 2011;77: 7207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RM, Rappé MS, Urbach E et al.. Prevalence of the Chloroflexi-related SAR202 bacterioplankton cluster throughout the mesopelagic zone and deep ocean. Appl Environ Microbiol. 2004;70: 2836–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CD. Recent developments in enzymatic chlorination. Nat Prod Rep. 2006;23:147–52. [DOI] [PubMed] [Google Scholar]
- Müller CA, Oberauner-Wappis L, Peyman A et al.. Mining for nonribosomal peptide synthetase and polyketide synthase genes revealed a high level of diversity in the Sphagnumbog metagenome. Appl Environ Microbiol. 2015;81:5064–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer PR, Widmann C, Wibberg D et al.. A flavin-dependent halogenase from metagenomic analysis prefers bromination over chlorination. PLoS One. 2018;13:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyyssönen M, Tran HM, Karaoz U et al.. Coupled high-throughput functional screening and next generation sequencing for identification of plant polymer decomposing enzymes in metagenomic libraries. Front Microbiol. 2013;4:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill J.2016. https://amr-review.org/sites/default/files/160518_Final paper_with cover.pdf Tackling drug-resistant infections globally: Final report and recommendations.
- Onaka H, Taniguchi S, Igarashi Y et al.. Characterization of the biosynthetic gene cluster of Rebeccamycinfrom Lechevalieriaaerocolonigenes ATCC 39243. Biosci Biotechnol Biochem. 2003;67:127–38. [DOI] [PubMed] [Google Scholar]
- Öztürk B, de Jaeger L, Smidt H et al.. Culture-dependent and independent approaches for identifying novel halogenases encoded by Crambe crambe (marine sponge) microbiota. Sci Rep. 2013;3:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlik JR. The chemical ecology of sponges on caribbean reefs: natural products shape natural systems. Bioscience. 2011;61:888–98. [Google Scholar]
- Payne JT, Andorfer MC, Lewis JC.. Regioselective arene halogenation using the FAD-dependent halogenase RebH. Angew Chemie - Int Ed. 2013;52:5271–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce SL, Oakeshott JG, Pandey G. Insights into ongoing evolution of the hexachlorocyclohexane catabolic pathway from comparative genomics of then Sphingomonadaceae strains. G3. 2015;5:1081–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR. An introduction to sequence similarity (“Homology”) searching. Curr Protoc Bioinforma. 2014;1:1286–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereyra LP, Hiibel SR, Prieto Riquelme M V et al.. Detection and quantification of functional genes of cellulosedegrading, fermentative, and sulfate-reducing bacteria and methanogenic archaea. Appl Environ Microbiol. 2010;76:2192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puyana M, Pawlik J, Blum J et al.. Metabolite variability in caribbean sponges of the genus aplysina. Brazilian J Pharmacogn. 2015;25:592–9. [Google Scholar]
- Quast C, Pruesse E, Yilmaz P et al.. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl Acids Res. 2013;41:590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S et al.. InterProScan: protein domains identifier. Nucl Acids Res. 2005;33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley CP, Bergquist PR, Harper MK et al.. Speciation and biosynthetic variation in four dictyoceratid sponges and their cyanobacterial symbiont, Oscillatoria spongeliae. Chem Biol. 2005;12:397–406. [DOI] [PubMed] [Google Scholar]
- Rocha-Martin J, Harrington C, Dobson ADW et al.. Emerging strategies and integrated systems microbiology technologies for biodiscovery of marine bioactive compounds. Mar Drugs. 2014;12:3516–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-R LM, Konstantinidis KT.. Bypassing cultivation to identify bacterial species. Microbe. 2014;9:111–8. [Google Scholar]
- Rognes T, Flouri T, Nichols B et al.. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rua CPJ, de Oliveira LS, Froes A et al.. Microbial and functional biodiversity patterns in sponges that accumulate bromopyrrole alkaloids suggest horizontal gene transfer of halogenase genes. Microb Ecol. 2018;76:825–38. [DOI] [PubMed] [Google Scholar]
- Sacristán-Soriano O, Banaigs B, Becerro MA. Can light intensity cause shifts in natural product and bacterial profiles of the sponge Aplysina aerophoba? Mar Ecol. 2016;37:88–105. [Google Scholar]
- Sacristán-Soriano O, Banaigs B, Casamayor EO et al.. Exploring the links between natural products and bacterial assemblages in the sponge aplysina aerophoba. Appl Environ Microbiol. 2011;77:862–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Massaki N, Kubo T.. Wolbachia variant that induces two distinct reproductive phenotypes in different hosts. Heredity (Edinb). 2005;95:389–93. [DOI] [PubMed] [Google Scholar]
- Schloss P. D., Westcott S. L., Ryabin T. et al.. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S, Deines P, Behnam F et al.. Chloroflexi bacteria are more diverse, abundant, and similar in high than in low microbial abundance sponges. FEMS Microbiol Ecol. 2011;78:497–510. [DOI] [PubMed] [Google Scholar]
- Schmitt S, Tsai P, Bell J et al.. Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges. ISME J. 2012;6:564–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd SA, Karthikeyan C, Latham J et al.. Extending the biocatalytic scope of regiocomplementary flavin-dependent halogenase enzymes. Chem Sci. 2015;6:3454–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipkema D, Schippers K, Maalcke WJ et al.. Multiple approaches to enhance the cultivability of bacteria associated with the marine sponge Haliclona(gellius) sp. Appl Environ Microbiol. 2011;77:2130–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipkema D. Marine biotechnology: diving deeper for drugs. Microb Biotechnol. 2017;10:7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaby BM, Hackl T, Horn H et al.. Metagenomic binning of a marine sponge microbiome reveals unity in defense but metabolic specialization. ISME J. 2017;11:2465–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DRM, Grüschow S, Goss RJM.. Scope and potential of halogenases in biosynthetic applications. Curr Opin Chem Biol. 2013;17:276–83. [DOI] [PubMed] [Google Scholar]
- Smith DRM, Uria AR, Helfrich EJN et al.. An unusual flavin-dependent halogenase from the metagenome of the marine sponge Theonella swinhoei WA. ACS Chem Biol. 2017;12:1281–7. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. [DOI] [PubMed] [Google Scholar]
- Taylor MW, Radax R, Steger D et al.. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev. 2007;71:295–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Moitinho-Silva L, Lurgi M et al.. Diversity, structure and convergent evolution of the global sponge microbiome. Nat Commun. 2016;7:11870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms C, Ebel R, Proksch P.. Activated chemical defense in Aplysina sponges revisited. J Chem Ecol. 2006;32:97–123. [DOI] [PubMed] [Google Scholar]
- Thrash JC, Cho JC, Ferriera S et al.. Genome sequences of strains HTCC2148 and HTCC2080, belonging to the OM60/NOR5 clade of the Gammaproteobacteria. J Bacteriol. 2006;192:3842–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully BJ, Graham ED, Heidelberg. JF. The reconstruction of 2,631 draft genomes from the global oceans. Sci Data. 2018;5:170203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turon X, Becerro MA, Uriz MJ.. Distribution of brominated compounds within the sponge Aplysina aerophoba: coupling of X-ray microanalysis with cryofixation techniques. Cell Tissue Res. 2006;301:311–22. [DOI] [PubMed] [Google Scholar]
- Unson MD, Holland ND, Faulkner DJ.. A brominated secondary metabolite synthesized by the cyanobacterial symbiont of a marine sponge and accumulation of the crystalline metabolite in the sponge tissue. Mar Biol. 1994;119:1–11. [Google Scholar]
- Vakhlu J, Sudan AK, Johri BN.. Metagenomics: future of microbial gene mining. Indian J Microbiol. 2008;48:202–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bogert B, de Vos WM, Zoetendal EG et al.. Microarray analysis and barcoded pyrosequencing provide consistent microbial profiles depending on the source of human intestinal samples. Appl Environ Microbiol. 2011;77:2071–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Pée KH, Patallo EP.. Flavin-dependent halogenases involved in secondary metabolism in bacteria. Appl Microbiol Biotechnol. 2006;70:631–41. [DOI] [PubMed] [Google Scholar]
- Van Pée KH, Unversucht S.. Biological dehalogenation and halogenation reactions. Chemosphere. 2003;52:299–312. [DOI] [PubMed] [Google Scholar]
- Van Pée KH, Zehner S.. Enzymology and molecular genetics of biological halogenation. Handbook Environ Chem, 2003;3:171–99. [Google Scholar]
- Varghese N, Submissions S. Rubritalea squalenifaciens DSM 18772, whole genome shotgun sequencing project. GenBank accession number FQYR00000000.1, 2016.
- Walsh CT, Chen H, Keating TA et al.. Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Curr Opin Chem Biol. 2001;5:525–34. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM et al.. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Wu H, Wang A et al.. Prospecting metagenomic enzyme subfamily genes for DNA family shuffling by a novel PCR-based approach. J Biol Chem. 2010;285:41509–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA et al.. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster NS, Taylor MW.. Minireview Marine sponges and their microbial symbionts : love. Environ Microbiol. 2012;14:335–46. [DOI] [PubMed] [Google Scholar]
- Weichold V, Milbredt D, Van Pée KH.. Specific enzymatic halogenation – from the discovery of halogenated enzymes to their applications in vitro and in vivo. Angew Chemie - Int Ed. 2016;55:6374–89. [DOI] [PubMed] [Google Scholar]
- Wickham H. Ggplot2. New York: Springer-Verlag, 2016; http://ggplot2.org. [Google Scholar]
- Wilson MC, Mori T, Rückert C et al.. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506:58–62. [DOI] [PubMed] [Google Scholar]
- Wilson MC, Piel J.. Metagenomic approaches for exploiting uncultivated bacteria as a resource for novel biosynthetic enzymology. Chem Biol. 2013;20:636–47. [DOI] [PubMed] [Google Scholar]
- Xu G, Wang BG.. Independent evolution of six families of halogenating enzymes. PLoS One. 2014;11:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Blasiak LC, Koglin A et al.. Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases. Biochemistry. 2014;46:1284–92. [DOI] [PubMed] [Google Scholar]
- Zea S, Henkel TP, Pawlik JR. The Sponge Guide: A Picture Guide to Caribbean Sponges. 3rd ed 2014.; www.spongeguide.org. [Google Scholar]
- Zehner S, Kotzsch A, Bister B et al.. A regioselective tryptophan 5-halogenase is involved in pyrroindomycin biosynthesis in Streptomyces rugosporus LL-42D005. Chem Biol. 2005;12:445–52. [DOI] [PubMed] [Google Scholar]
- Zhao J, Yang N, Zeng R.. Phylogenetic analysis of type I polyketide synthase and nonribosomal peptide synthetase genes in Antarctic sediment. Extremophiles. 2005;12:97–105. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yan B, Yang T et al.. Purification and crystallographic analysis of a FAD-dependent halogenase from Streptomyces sp. JCM9888. Acta Crystallogr Sect Struct Biol Commun. 2015;71:972–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.