 This review aims to provide an overview of the current small molecule strategies used for targeting S. mutans biofilms, and a perspective of the future for the field.
This review aims to provide an overview of the current small molecule strategies used for targeting S. mutans biofilms, and a perspective of the future for the field.
Abstract
The prevalence of biofilm diseases, and dental caries in particular, have encouraged extensive research on S. mutans biofilms, including methods of preventing its formation. Numerous small molecules with specific anti-biofilm activity against this pathogen have been isolated and synthesized. Generally, these molecules can be characterized into three categories: sucrose-dependent anti-adhesion, sucrose-independent anti-adhesion and cellular signaling interference. This review aims to provide an overview of the current small molecule strategies used for targeting S. mutans biofilms, and a perspective of the future for the field.
Introduction
The oral cavity is a complex, dynamic system, inhabited by over 700 different bacterial species. Under normal conditions, these bacterial communities live in symbiosis without causing harm to the host. However, a change in environmental or stress signal can tip the equilibrium toward pathogenic bacteria leading to oral diseases, such as dental caries, gingivitis, and periodontitis.1,2 In 2000, the US Surgeon General classified oral diseases as a “silent epidemic.”3 Almost 20 years later 3.5 billion people are still affected each year, with dental caries being a primary culprit.4 The role that dental caries plays in overall global health renders this an urgent matter. Many infections and diseases spanning the human body have been linked to oral pathogens. For example, bacteria residing in the oral cavity have been implicated in endocarditis and diabetes.5–8 Not only do these diseases pose a health-crisis, they contribute to a large portion of medical costs. According to the “Global Economic Impact of Dental Diseases,” the indirect and direct costs of dental diseases totaled $442 billion worldwide in 2010, providing global financial incentive to improve dental care.9
Dental caries is a biofilm-associated disease.10,11 According to the National Institutes of Health, 80% of bacterial infections are biofilm in nature.12 Bacteria exist in a dynamic state between planktonic and biofilm. Biofilms are polymicrobial, three-dimensional substances encapsulated by an exopolysaccharide (EPS) matrix. Mucosal surfaces, such as the gut, nasal, vaginal, and oral cavities, provide ideal surfaces for attachment, leading to high biofilm colonization.13
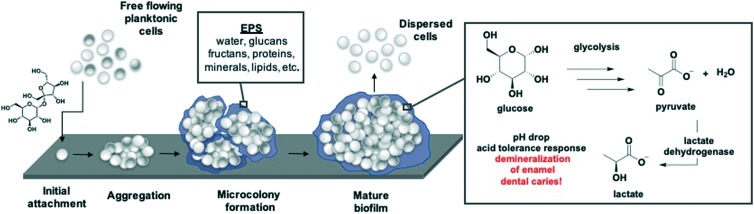
The formation of an oral biofilm begins with the attachment of a single planktonic cell to the tooth pellicle (Fig. 1). Following initial attachment, the primary colonizers, such as Streptococcus, Actinomyces, Haemophilus, Neisseria, Veillonella, Streptococcus mutans, either auto-aggregate (attachment between the same species) or co-aggregate (attachment between different species). The degree of biofilm formation depends on the similarity and attraction between bacterial species. During this attachment phase, metabolic activity is low. Secretion of an exopolymeric substance, containing polysaccharides, proteins, and DNA, envelops the bacterial cells forming the biofilm matrix. The microcolony phase, or rapid growth phase, is followed by the adherence of secondary colonizers, comprising of anaerobic Gram-negative bacteria, such as Porphyyromomonas gingivalis.14 Bacteria enclosed in the biofilm either slow their growth or become static, often times even showing signs of death. Concomitant with this steady state phase is the dispersal of biofilm back to their planktonic state. Dispersed cells either form a new biofilm at a different attachment site or enter the bloodstream. Alternatively, biofilms can develop into mature structures that are highly resistant to the innate host immune system and antibiotic treatment due to decreased metabolism.12,14,15
Fig. 1. S. mutans biofilm maturation process beginning with the attachment of a single cell, promoted by exposure to sucrose. Virulence traits including EPS formation, acid production, and acid tolerance are highlighted.
Many hypotheses have been developed to best characterize the etiology of dental caries, including the ecological plaque hypothesis and the specific-pathogen hypothesis.16 The ecological plaque hypothesis, created by Phillip D. Marsh, states that “disease is the result of an imbalance in the total microflora due to ecological stress, resulting in an enrichment of some “oral pathogens” or disease- micro-organisms.” The single pathogen hypothesis implicates a single organism, primarily S. mutans, as the cause for dental caries. While the former has recently garnered much interest, aspects of the latter remain critical in the prevention of dental caries.17 Focusing on the inhibition of pathogens enriched in plaque could minimize the severity of disease, and further colonization of the oral cavity. Furthermore, attempting to tackle all factors involved in caries progression runs the risk of excluding studies that could lessen the virulence of dental caries in many cases. For these reasons, this review explores S. mutans as a small molecule or natural product target for the reduction or prevention of dental caries.
S. mutans pathogenicity
Although dental caries is caused by microbial dysbiosis, S. mutans is the predominant pathogenic species. A reduction or elimination of S. mutans has been proven to prevent or lessen caries progression.18 Organic acid production, biofilm formation and acid tolerance are the main virulence traits associated with S. mutans.19,20 Before forming a biofilm, S. mutans exist as free-floating planktonic cells. The transition from planktonic cells to biofilm can proceed through a sucrose-independent or sucrose-dependent mechanism (Fig. 1). In the independent pathway, S. mutans binds to salivary pellicles on the teeth through cell surface adhesins (antigen I/II, SpaP, and Gbps).19,21 When exposed to sucrose, the bacterium begins synthesizing long polymer glycan chains via glucosyltransferases (Gtfs). Adherence to the tooth is mediated by the newly synthesized glucans, as well as glucan-binding proteins.22,23 GtfB synthesizes primarily insoluble glucans (α-1,3 glycosidic linkages), GtfC makes both insoluble and soluble glucans (α-1,6 glycosidic linkages), and GtfD produces soluble glucans. These glucans provide additional binding sites for planktonic cells and build the architecture of the growing biofilm. As the cells accumulate and excrete EPS, microcolonies form, eventually developing into mature biofilms (Fig. 1). Simultaneous increase in sugar uptake results in the production of organic acids. Continuous acid production plays a key role in S. mutans pathogenicity, resulting in demineralization of tooth enamel. S. mutans has evolved an acid tolerance response, due to the low pH environment they frequently reside.24 Mature biofilms exhibit increased aciduricity (ability to withstand low pH environments) due the evolutionary pressure to outcompete other bacteria that have also colonized the oral cavity. As a result, an acid-tolerant flora emerges which further promotes the formation of dental caries and other oral diseases because aciduricity trends with pathogenicity.24–26
A “sticky” pathogen to study
The diverse microflora in the oral cavity and the constantly changing environment (saliva, food intake, etc.) hinder our ability to study and fully understand the pathogenicity of this bacterium. For this reason, in vitro studies have not translated well in vivo. The variability in growth conditions for planktonic and biofilm assays, and the confusion between the two life states make data difficult to compare. The use of different acronyms (i.e. IC50, MBIC, MIC, MBEC) without clear definitions and the variance in experimental conditions leads to misrepresented data. Some small molecules have been described to have biofilm specific activity but under closer investigation varied experimental conditions led to inaccurate conclusions. The story of honokiol, a biphenyl natural product, demonstrates the importance of maintaining consistent experimental conditions. It was originally shown that honokiol exhibited biofilm inhibition and was later proven that the activity was due to lack of CO2 during bacterial growth.27,28 This is unsurprising since S. mutans has evolved to grow in the microaerophilic environment of the oral cavity.29,30 Despite these hurdles, S. mutans have been recognized as a model organism to study Gram-positive pathogenic bacteria because of the similarities in gene expression and metabolic pathways.20 For this reason, studying this bacterium and molecules that perturb it has far-reaching effects in Gram-positive biofilm diseases.
Herein we try to compare activities while accounting for these differences. Minimum inhibitory concentration (MIC) is understood to be the lowest concentration at which bacteria does not grow.31 Some inhibition patterns are better suited to be represented by an IC50 value which denotes the concentration that bacterial growth is 50% inhibited. In this text, you will also see IC50 used to describe 50% enzyme inhibition. To complement those values, the minimum biofilm inhibitory concentration (MBIC) refers to the lowest concentration at which biofilm does not grow. In some cases, a MBIC50 will be used to signify 50% biofilm growth. If the researchers are testing the effect of a compound on preformed biofilm samples, they will measure eradication and dispersal. Minimum biofilm eradication concentration (MBEC) measures the ability for biofilm cells to regrow and refers to the lowest concentration in which the cells do not regrow past an OD650 of 0.1. Dispersal is usually represented with percent dispersal and commonly is accompanied with images (confocal, crystal violet, etc.) to demonstrate removal of biofilm mass. In some scenarios, researchers will test the viability of cells in the biofilm or in the supernatant and measure viability by counting colony forming units (CFUs). These definitions are nicely summarized in a review by Maciá et al.32
Current methods of prevention
Multiple methods of prevention have been used to limit carries formation. Mechanical methods such as brushing and flossing are used to remove the cariogenic bacteria that colonize the enamel and relies on human compliance to adequately control dental plaque buildup.33,34 Meanwhile fluoride treatments reinforce the enamel, protecting the teeth from dental plaque acidification.35 However, when sucrose intake is high and frequent, fluoride is unable to fully prevent demineralization.36 A newer method of prevention is to replace sugars in the diet with xylitol, a non-cariogenic and anti-cariogenic sugar substitute, to stop acidification in the oral cavity. Although useful, this method of replace cariogenic sugars with xylitol has not gained widespread.37
During mechanical removal, mouthwashes and toothpastes containing active antimicrobial agents are generally used. Common agents such as chlorohexidine or cetylpyridinium chloride are used to remove acidogenic bacteria from the oral cavity but they do not act in species specific or biofilm specific manners.38–40 For that reason, the broad spectrum activity disrupts the microbiome and causes many undesirable side effects, such as staining of the mouth and tongue.41 Alternate treatments include metal salts, enzymes, quaternary ammonium compounds and essential oils.42 The continuation of this health crisis and the adverse side-effects of current treatments proves the need for therapeutics that can selectively target S. mutans biofilm.
Small molecule strategies
As demonstrated above, there is a great need for therapeutics that selectively target S. mutans biofilm. Small molecules and natural products are a rich source for such compounds.43,44 Given the prominence of biofilms in infectious diseases, there has been an increased effort toward the development of small molecules that will modulate bacterial biofilm development and maintenance. In this review, we highlight the development of small molecules that inhibit and/or disperse bacterial biofilms through non-microbicidal mechanisms. Groups have taken various approaches to find these active molecules, such as screening large chemical libraries, screening natural products for biofilm activity or using synthesis to develop analogs of interesting lead structures.45–49
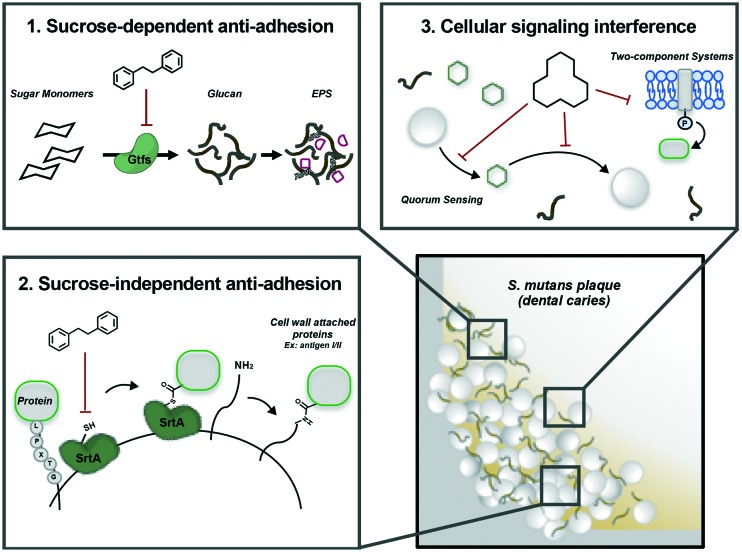
Herein we provide an overview of the current state of small molecules that to varying degrees selectively target S. mutans biofilm formation and growth, without disrupting planktonic cells, and in some cases the surrounding microbiome. Following a survey of the literature, it was found that these molecules generally act through two mechanisms: anti-adhesion and signal interference. Anti-adhesion mechanisms can be sucrose-dependent, such as blocking the formation of the biofilm polymer chains, or sucrose-independent, by blocking the surface protein attachment function of sortase A (Fig. 2, Box 1 and 2). The second mechanism, signaling interference, is directed toward quorum sensing and two-component systems (Fig. 2, Box 3). These mechanisms, along with the small molecules that target them, will be elaborated on in the subsequent sections. This arsenal of compounds is largely limited to inhibitory effects, primarily through targeting virulence traits, resulting in a weakened or less pathogenic biofilm.
Fig. 2. Three general mechanisms of perturbing S. mutans biofilms.
Sucrose-dependent anti-adhesion: attacking biofilm via glucosyltransferase inhibition
Molecules that affect S. mutans glucosyltransferases (gtfBCD) have been the focus of many studies.50–52 These enzymes are essential for attachment, biofilm formation, and virulence when sucrose is available in its growth conditions (Fig. 2, Box 1). Obstructing adhesion of planktonic cells will reduce biofilm formation or weaken the biofilm architecture enabling easier removal.
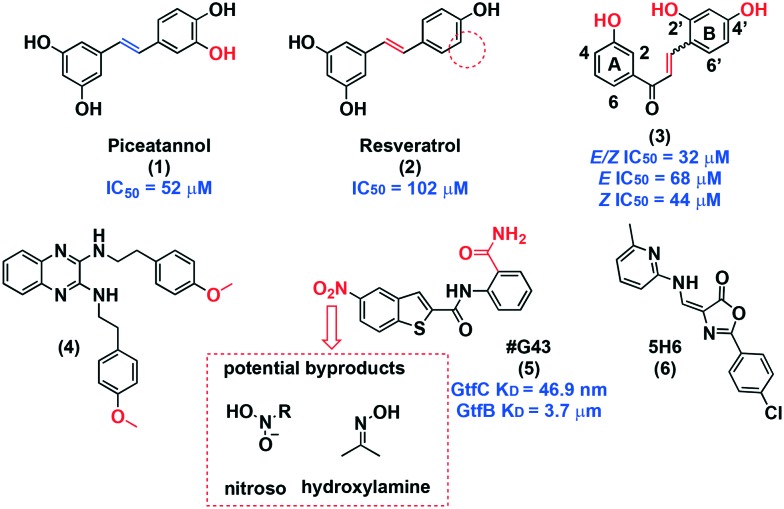
Piceatannol (1, Fig. 3), a polyphenol, was first discovered as a potential agent against S. mutans through in silico docking studies in the GtfC binding pocket.53 Nijampatnam and co-workers observed interactions were between the hydroxyl groups in piceatannol and Glu515, Trp517, and Asp477, all of which are in involved in the binding of acarbose, a weak GtfC inhibitor.54 To confirm the docking studies, the binding of 1 to GtfB and GtfC was quantitatively assessed using the Octet Red96 system, which provides binding kinetic characterization. Piceatannol 1 exhibited a KD of 1.58 and 14.6 μM for GtfB and GtfC, respectively. Moreover, piceatannol exhibits 10-fold selectivity for biofilm inhibition (IC50 = 52 μM) over planktonic inhibition. A sucrose-dependent Drosophila colonization model was used to determine potential effects on colonization in vivo. Visualization of the guts of flies infected with GFP-tagged bacteria and treated with 1 showed a drastic reduction in fluorescence at 50 μM, and the fluorescence closely resembled that of the gtfb mutant. This data further suggests GtfB as a target. Another small molecule identified in this screen was resveratrol (2). This natural product had been previously recognized as an anti-biofilm agent but all prior studies tested mixtures, rather than pure, isolated compounds.55 The decrease in activity of resveratrol is particularly interesting in comparison to piceatannol since the compounds are structurally similar. By removing one hydroxyl from piceatannol to reach resveratrol, biofilm potency is drastically reduced from 52 μM to 102 μM. Kinetic analysis provided KD values of 144 and 510 μM for GtfB and GtfC, respectively, showing a drastic decrease in binding affinity compared to 1. Interestingly, planktonic inhibition was similar for both 1 and 2. This subtle structural difference highlights the nuance for selectively disrupting biofilm when targeting Gtfs.
Fig. 3. Sucrose-dependent anti-adhesion small molecules.
In a separate in silico study using the GtfC crystal structure, Nijampatnam discovered 14 chalcones with potential activity against S. mutans. A preliminary biological screen conducted at 200 μM identified six compounds with activity against both S. mutans growth and biofilm with micromolar IC50 values.56 Compound 3, which was a mixture of E/Z isomers, stood out for its biofilm selective inhibition with an IC50 of 32 μM (>10-fold selectivity over planktonic growth). Following the synthesis of both possible isomers, the Z-isomer was most potent (IC50 = 44 μM), and the E-isomer exhibited decreased activity, indicating a synergistic effect between the isomers as evidenced by the mixture being most potent. Furthermore, at 200 μM only 10–18% of S. sanguinis and S. gordonii planktonic growth was inhibited and were completely unaffected at 44 μM. Similar binding interactions with Glu515 and Asp477 that were observed with piceatannol were seen. A dose-dependent zymogram assay that uses electrophoresis to measure proteolytic activity confirmed the inhibition of GtfC, as well as GtfB and GtfD. Again, using a Drosophila colonization model, they observed an impressive reduction in fluorescence, corroborating GtfB as a potential target. The presence of the enone moiety suggests a covalent interaction with an active site cysteine. With this in mind, the Z-isomer displaying more potent activity compared to the E-isomer can be explained sterically because the enone in the Z-isomer would be more accessible. Structure–activity relationships were derived from these biological studies. It was found that: (1) hydroxyl groups on either ring are necessary; (2) position 2′ on ring B contributes minimally to activity; (3) position 3 on ring A correlates to potent biofilm specific activity; and (4) the most active compound has hydroxyl groups on position 3 (ring A), 2′ and 4′ (ring B), suggesting that position 3 contributes most to bioactivity. These observations can help guide the development of compounds with similar scaffolds for use against S. mutans biofilm.
Following a similar screening method using the GtfC crystal structure, Ren et al. identified a potential S. mutans inhibitor, compound 4.57 The computational activity was confirmed in vitro by studying biofilm formation and EPS synthesis. Scanning electron microscopy confirmed a reduction in EPS and a deformed biofilm architecture. Bacterial viability was calculated by determining the number of CFU at 5 and 10 μg mL–1. This small molecule reduced cell viability by 46% and 79%, respectively, in a 24 hour-old biofilm. Compound 4 did not affect planktonic cells, as evidenced by OD595 measurements. Free GTF activity was analysed using zymography assays, and 4 was found to inhibit GtfC and GtfD activity, while interestingly enhancing GtfB activity. Using a rat model, treatment with compound 4 showed the reduction of incidence and severity of caries (both on the enamel and between the tissue) at 10 μg mL–1. Although the water solubility needs improvement, compound 4 is a promising scaffold for drug development. Based on the high presence of phenols with activity against glucosyltransferases, converting the ethers to alcohols could potentially enhance potency.
Compound #G43 (5) was also identified in a structure-based in silico study.58 At low micromolar concentration (12.5 μg mL–1) 85% of S. mutans biofilm was inhibited. Binding affinities for this compound were 3.7 μM and 46.9 nm for GtfB and GtfC, respectively, demonstrating a preference for GtfC. These data, in addition to the lack of activity against expression levels, suggest that (5) acts through direct enzyme binding. Planktonic cell viability and growth, and commensal strains, were undamaged up to 200 μg mL–1. In animal studies, cariogenicity was drastically reduced. Preliminary SAR analysis demonstrated the necessity of the primary ortho amide for activity. The presence of a nitro group poses potential problems through the production of unstable radical products, nitrosoamines, and hydroxylamines (Fig. 3).59 Therefore, follow-up investigation is needed to improve the activity of #G43 and remove the nitro group. The high selectivity for S. mutans biofilm and the preliminary in vivo results provide strong evidence for 5 as a useful inhibitor.
An oxazole derivative, designated 5H6 (6), was reported to decrease biofilm viability by 75% at 50 μg mL–1.60 This compound inhibited cell free GtfC and GtfD, but enhanced cell free GtfB activity. This is particularly surprising due to similar homology between these proteins. Further characterization of the binding interactions of each Gtf and 6 could increase our abilities to selectively inhibit different Gtfs. 5H6 exhibited dose-dependent EPS reduction and reduced both formation and maturation of biofilm at 50 μg mL–1. Planktonic growth was unaffected by 6. Most notably, rat caries were affected to the same extent when treated with 250 μg mL–1 fluoride or with 50 μg mL–1 of oxazole (6). The in vitro and in vivo data encourage further SAR studies on oxazole (6).
The GTF inhibitors discussed fall into two categories: 1) are hydroxychalcones, or 2) molecules with high heteroatom density, particularly nitrogen. Hydrogen bond donor and acceptor moieties were also common features of these molecules, demonstrating the importance of these interactions for enzyme binding. Future libraries of compounds that are targeting GTFs should use this SAR data to guide the discovery of new potent GTF inhibitors.
Sucrose-independent anti-adhesion: non-glucosyltransferase mechanisms
In sucrose-independent mediated attachment, sortase A, facilities the covalent attachment of surface proteins to the cell wall (Fig. 2, Box 2).61 Sortase recognizes the motif LPXTG in the substrate protein and cleaves between the threonine and glycine. The resulting free carboxy terminus on threonine is then attached to the cell wall. S. mutans encoded for a single sortase belonging to the SrtA subfamily, and six proteins (AgI/II, FruA, WapA, WapE, GbpC, and DexA) containing the LPXTG sequence.62,63 Sortase has been confirmed to facilitate the attachment of AgI/II,64 GbpC,65 and DexA66 to the cell wall. The inability of a sortase A mutant to adhere and colonize the oral cavity showcases the importance of this enzyme in S. mutans biofilm formation and cariogenicity. For this reason, small molecules targeting sortase A have been widely studied.
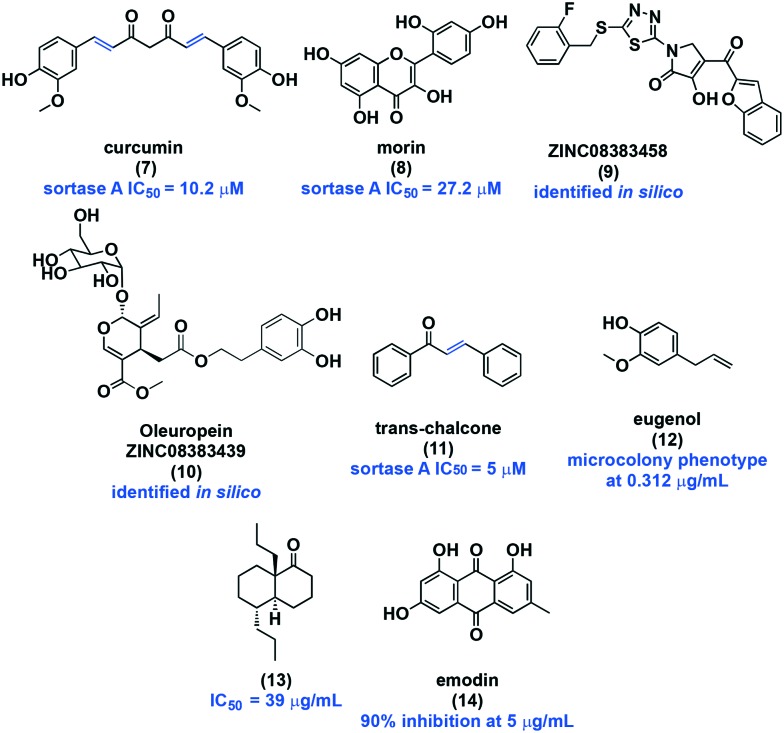
Curcumin (7, Fig. 4), a natural phenol, inhibits S. mutans sortase A with an IC50 of 10.2 μM.67,68 The surface protein AgI/II mRNA levels were not affected, but Western blot analysis showed decreased protein levels, suggesting inhibition via direct binding. Additionally, AgI/II was collected from the supernatant, further confirming inhibition of sortase A activity. The bacteria in compound-treated wells had reduced biofilm mass, indicated by OD600 measurements. Although 7 has an MIC of 125 μM, at a concentration of 15 μM, bacterial cells were less tightly bound to the well, suggesting potential use as an anti-caries treatment in combination with mechanical removal. The activity of curcumin against S. mutans is not surprising. Curcumin has been shown to act as a covalent inhibitor of sortase A in S. aureus through the addition of an active site Cys184 moiety to the enone functionality.69 This moiety is also present in S. mutans sortase A, suggesting a similar mode of action. Similar activity has been shown for the natural product morin (8), a flavonoid found in many Chinese herbs and fruits.70 Morin (8) was reported to inhibit sortase A activity (IC50 = 27.2 μM), and in a crystal violet assay was found to reduce biofilm mass. Western Blot analysis also showed reduced AgI/II levels at 30 μM. Morin exhibited biofilm specificity, as confirmed by similar CFU in the control versus compound treated planktonic cultures. The decrease in activity of 8 compared to curcumin (7) is possibly due to the branching structure which causes steric congestion and results in a less reactive Michael-acceptor.
Fig. 4. Sucrose-independent anti-adhesion small molecules.
The similarities between S. aureus sortase A and S. mutans sortase A also encouraged Luo et al. to screen two pharmaceutical small molecule/natural product libraries in silico for sortase A inhibition.71 The most promising compounds featured benzofurans, thiadiazoles, and pyrroles that contributed to the hydrogen bond network that most likely engages with the cysteine active site of sortase A, shutting down enzymatic activity. Two compounds with the highest possibility for development are ZINC08383458 (9) and ZININC08383439 (10). Both compounds exhibit superior properties to curcumin, but in vitro activity has yet to be confirmed. Inspired by the structural similarities, such as the presence of an α,β-unsaturated ketone, between sortase A inhibitors, Wallock-Richards et al. identified trans-chalcone (11) in silico as a possible inhibitor.72 Preliminary in vitro studies showed a concentration dependent effect on biofilm formation and a sortase A IC50 of 5 μM, highlighting trans-chalcone as a potential anticariogenic therapeutic. Mass spectrometry confirmed an adduct between the enone of trans-chalcone (11) and the cysteine residue of sortase A. The anti-adhesion properties of this compound were tested using saliva-coated glass slides; and at 250 μM 90% of biofilm was inhibited. Despite these discoveries, further investigation is required to fully elucidate the molecular basis of sortase A inhibition in S. mutans to guide use in therapeutic application.
Another natural product, eugenol (12), that is commonly isolated from essential oils has been shown to suppress S. mutans attachment and virulence without affecting bacterial viability.73 Biofilm images of eugenol treated cells displayed a microcolony phenotype at sub-MIC levels (0.3125 μg mL–1). Reduction in AgI/II expression suggests that this phenylpropene acts through an anti-adhesion mechanism. However, many other biofilm-related genes were downregulated, such as gftB, ftf, vicKR, and glucan-binding proteins (Gbps). At much higher concentrations, Xu et al. found that eugenol disrupted acid production, adhesion, and glucan production. Although additional work is required to elucidate the mechanism of action of eugenol, it likely works via an anti-adhesion mechanism.
Another source of bioactive compounds is Trachyspermum ammi, an aromatic spice.74 To explore the potential arsenal of S. mutans inhibitors within this natural source, Kahn and co-workers isolated and screened compounds against S. mutans, leading to the discovery of compound 13. This bicyclic compound 13 was reported to have the most potent biofilm inhibition, with an MBIC50 of 39 μg mL–1. The MIC was 156.25 μg mL–1, demonstrating very moderate selectivity. At sub-MIC levels, adherence was reduced by 50%, and biofilm formation was completely inhibited. Glucan production by crude GTFase was found to be drastically suppressed at concentrations as low as 2.44 μg mL–1. However, GTF enzyme activity was not tested directly. Additionally, the cell wall was visibly disrupted as monitored with confocal imaging. Notably, compound treatment resulted in an increase in pH by 3 units compared to the control, indicating a disruption in S. mutans acidogenicity (ability to produce acid).
Another means of anti-adhesion is through the binding of surface proteins hindering the bacteria's ability to aggregate. Surface proteins are highly associated with cell hydrophobicity, which is speculated to mediate attachment to the tooth pellicle.19,75,76 The test agent 13 reduced the hydrophobicity by more than half, suggesting potential binding of these proteins, which could block aggregation. While compound 13 appears to work through a variety of mechanisms, adhesion appears to be the most dominant, but additional work is needed to identify a specific mode of action.74
Emodin (14), an anthraquinone, is a known biofilm inhibitor, but like resveratrol, was originally only tested in a mixture.55 As a pure compound, emodin 14 inhibited biofilm formation on a hydroxyapatite surface by 90% at 5 μg mL–1.77 Hydroxyapatite is a calcium phosphate mineral that closely resembles the human tooth. Concentrations >250 μg mL–1 were required for activity against planktonic cells. This small molecule has been reported to insert itself within the phospholipid bilayer.78 It is possible that the anti-biofilm activity of emodin is due to this disruption in membrane fluidity.
Inhibition of non-GTF mediated adhesion in S. mutans is quite under studied, but this section highlights the potential in this strategy. By weakening the attachment and undermining the overall stability of the biofilm, this inhibition strategy could potentiate current fluoride and mechanical removal strategies.
Non-attachment mechanisms of biofilm inhibition: interference of cellular signalling
Bacteria communicate via quorum sensing and two-component systems. These systems regulate when and how bacteria form biofilms based on certain environmental cues or stress signals. Quorum sensing signals are often modulated by two-component systems, making their inhibition tightly connected (Fig. 2, Box 3).79,80 These systems are typically comprised of a membrane histidine kinase sensor protein that senses the environmental cue, and a cytoplasmic response regulator that facilitates the cellular response through regulation of gene expression. Fig. 5 highlights a snapshot of small molecules and natural products targeting these systems. A benefit of this strategy is the minimal required concentration needed to impede signals initiating biofilm formation, without disrupting growth and survival. In theory, selective pressure will be bypassed and resistance development is reduced.81,82
Fig. 5. Small molecules that interfere with quorum sensing.
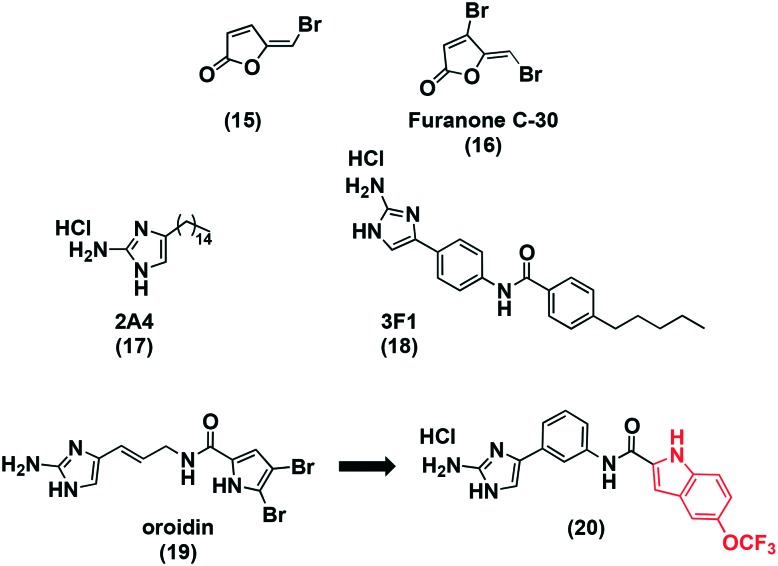
Brominated furanones from red algae were found to inhibit bacterial accumulation of algae, signalling to their potential use as an anti-biofilm agent.83 Brominated furanone (15, Fig. 5A) was inactive against planktonic growth below 60 μM as evidenced by OD600 measurements. Moderate effects on biofilm formation were observed at 6 μM when test compound was added to the growth media. The surface coating resulted in enhanced effects with a 63% reduction in biofilm at 0.06 μM, and no planktonic effects. The effect of 15 on quorum sensing was tested using a bioluminescence assay with a bacterial reporter strain that is activated by the presence of autoinducer-2 (AI-2), a signalling molecule produced and recognized by an array of bacterial species, including S. mutans.84,85 At 0.6 and 6 μM, bioluminescence was quenched suggesting an interefence in quorum sensing.
Furanone C-30 (16) was synthesized due to its potential as a quorum sensing inhibitor, based on the activity of 15. An array of essential genes associated with biofilm formation were downregulated by this quorum sensing molecule, among these included gtfB, ftf, vicRK, brpA, smu630, comDE, and relA. Furanone C-30 had moderate activity against mature biofilms. At low microgram concentrations biofilm formation decreased by about 60% when 24 hour S. mutans biofilms were investigated. These biofilm effects were not a result of bacterial growth rate, nor were they due to antibacterial effects.86
The Wu group ran a HTS of 506 nitrogen-dense alkaloids that had previously displayed anti-biofilm activity against Gram-positive and Gram-negative bacteria, a 2-aminoimidazole small molecule was discovered to have specific inhibitory activity against S. mutans biofilms.87 The compound designated 2A4 (17) had a biofilm IC50 of 0.94 μM, with less than 13% of planktonic growth affected. AgI/II and GTF production was inhibited. In a planktonic culture, six biofilm-associated genes were down regulated suggesting a preference for biofilm-associated processes. S. sanguinis and S. gordonii, two oral commensal bacteria, were also unaffected by this small molecule inhibitor at concentrations much higher than the IC50. Biofilm treatment resulted in a decrease in expression in gtfb, pac, and relA. The former two genes are involved in surface attachment and the latter in acid tolerance. Commensal homologs of these genes were unaffected, but species selectivity needs further investigation. The structural resemblance to known quorum sensing inhibitors and the effect on expression level suggests interference with the signalling systems involved in biofilm formation. The same library was screened using a biofilm dispersion assay to discover a dispersion specific molecule. Compound 3F1 (18) was specific for biofilm dispersion at 5 μM, but lacked activity against planktonic cells and commensal strains.88 Additional work is required to determine the mechanism of action of 3F1, this small molecule is a promising scaffold for a biofilm specific drug, and as a probe to better understand the signalling system in S. mutans. Importantly, this molecule demonstrates not only inhibition affects, but also disperses biofilm which is more therapeutically relevant for S. mutans pathogenicity.
The 2-aminoimidazole-based alkaloid, oroidin (19), is a well-studied biofilm inhibitor and numerous analogs have been developed with similar activity.89,90 Hodnik et al. combined this compound with the idea that indoles are known to affect biofilm formation to develop novel S. mutans biofilm quorum sensing inhibitors.91,92 The MBIC50 of the most potent compound 20 was 20 μM, with others ranging from 50 to 120 μM. Planktonic studies need to be completed to determine the specificity and if it is a molecule worth pursuing further.
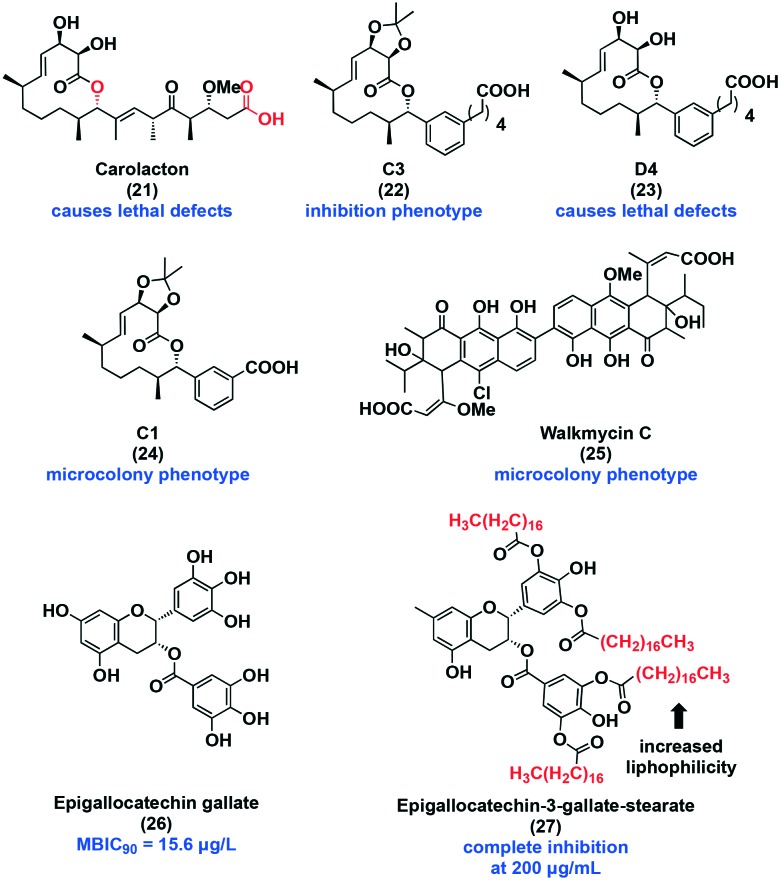
With respect to two-component systems, one potential inhibitor is carolacton (21, Fig. 6), a 12-membered macrolide that displays potent selectivity for biofilm over planktonic cells at low nanomolar concentrations, making it the target of many total synthesis campaigns.93–96 Between 0.053 μM and 53 μM, carolacton decreases biofilm viability by 55–65% as visualized with LIVE/DEAD™ staining. This macrocyclic natural product does not disperse biofilm cells. However, visible membrane damage and morphological changes have been observed. It is understood that carolacton specifically affects cells transitioning from the planktonic state to the biofilm state, but the exact target remains unknown. Potential targets include the serine/threonine protein kinase PknB,97,98 and the two-component systems, VicRX,99 CysR,100 and comX, all of which have been to be associated with its activity.101 Analog development by the Kirshning group has uncovered key structural features of carolacton. For instance, inversion of the stereocenter at C9 abolished activity, converting the acid to the methyl ester reduced activity, and the macrocylized lactone product also displayed lower activity. The reduced activity of the methyl ester and lactone are attributed to slow hydrolysis back to the free acid.96 To explore possible new bioactivity and to better understand the mechanism by which carolacton functions, our lab undertook the diverted total synthesis of carolacton analogs. Replacing the synthetically difficult trisubstituted alkene with an aryl isostere resulted in the discovery of three unique phenotypes. Representative examples include analog C3 (22), D4 (23), and C1 (24). Compound 22 inhibits 50% biofilm formation at 63 μM, and 23 acts similarly to carolacton in that it causes lethal defects in cells transitioning to biofilm. A new phenotype was discovered following treatment with 24. This compound arrested growth at the microcolony phase. This work highlighted the importance of the macrocycle for retaining activity. Target identification would be beneficial to advancing our understanding of carolacton's biofilm selectivity.
Fig. 6. Small molecules that interfere with two-component systems.
Walkmycin C (25) is a known histidine kinase inhibitor of the two-component system WalK/WalR in Bacillus Subtilis.102 WalK/WalR is a an ortholog of S. mutans VicK/VicR two-component system that plays a role in biofilm adhesion, formation, and stress tolerance. A microcolony phenotype was induced and biomass was reduced by about 60% at 0.63 μg mL–1. Planktonic growth was inhibited at 6.25 μg mL–1. Moreover, 25 inhibited the autophosphorylation activity of VicK, CiaH, and LiaS with IC50 values of 2.53, 4.29, and 4.96 μg mL–1, respectively. The authors also found that this natural product repressed the acid tolerance through inhibition of CiaH.103
Epigallocatechin gallate (26), a member of the catechin family, has an MBIC90 of 15.6 μg mL–1 and an MIC of 31.25 μg mL–1, displaying slight selectivity for biofilm formation over planktonic growth. Sucrose-dependent attachment, acid production, and acid tolerance were all affected at sub-MIC levels. F-ATPase activity was inhibited by 50%, which greatly affects the acid tolerance response, confirming the previous results. The genes, atpD, eno, ldh, and aguD were downregulated. Notably, aguD is an antiporter of AgDS. This system is regulated by the two-component systems: VicRK, ComDE, and CiaRH. When exposed to low pH or thermal stress, AgDS is induced, and is responsible for ammonia and ATP production, and thus plays an important role in regulating the cytoplasmic pH, extrusion of protons, and energy for growth. Therefore, inhibition of aguD can result in enhanced cellular stress via energy starvation and disruption of the pH gradient.104,105 The instability and poor bioavailability of this compound led to the synthesis of a lipophilic derivative, epigallocatechin-3-gallate-stearate (27).106 Congo red and crystal violet analysis shows that at 200 μg mL–1 biofilm formation was completely inhibited after 4 hours. Increasing the concentration to 250 μg mL–1 resulted in inhibition at 2 hours. Further studies are required to discern the mechanism of action. Although the bioactivity of many of the catechins is often attributed to the presence of hydroxyl groups in the 3, 4, and 5 positions on ring B, SAR studies are necessary to determine the important moieties for activity.107
Developing inhibitors of cellular signalling pathways has been successful for some compounds, but there are general drawbacks that limit the amount of success. One of the largest hurdles for discovering or developing these structures is untangling the intricate signalling networks that exist within S. mutans, and other microorganisms.
Conclusion and outlook
The prevention dental caries is not trivial. The complexity and nuance in the oral cavity and in the regulation of biofilm formation makes designing and testing small molecules with desirable activity quite challenging. Despite these challenges, many small molecules have been discovered from natural sources, or accessed synthetically with specific biofilm activity through inhibition and dispersal mechanisms. Often times, these mechanisms result in weakened, or less pathogenic biofilms that are more easily removed via mechanical measures, but this can have negative effects. Among these include further colonization on other mucosal surfaces and even sepsis.
Future efforts need to refocus on the important concepts of preventing S. mutans pathogenicity. First, drug design should focus on the eradication of pathogenic biofilms. By focusing on eradication, one can prevent the possibility of future infection in the oral cavity, greatly reducing the incidence of dental caries, and more serious diseases such as endocarditis. Compounds have been discovered that are capable of eradicating biofilm in other pathogenic microorganisms. One example comes from the Huigens Lab, where they found a potent class of halogenated phenazines that eradicates methicillin-resistant Staphylococcus aureus (MRSA) biofilm.108 It would be advantageous to study these active compounds, and others, against preformed S. mutans biofilms.
Secondly, it would be beneficial to incorporate more biologically relevant conditions while testing the efficacy of compounds against S. mutans biofilms because dental caries is a multifactorial disease. There are techniques currently available that should be considered when testing a new compound. Bjarnsholt et al. have outlined experimental biofilm systems in their 2013 review.109 Furthermore, commensal bacteria, other pathogenic bacteria, and nutrient flux are very influential on bacterial colonization in the oral cavity. When testing inhibition, it is important to take these factors into account. Incorporating co-culture testing into the array of experiments will increase the success of these compounds as they progress toward clinical trials.
Lastly, oral diseases are a prime example of the need for narrow spectrum antibiotics.110 With the overuse of antibiotics, we have seen increases in antibiotic resistance and have started to document the negative side effects that occur to our microbiome.111 As we have discussed, dental caries occurs through the persistence and reoccurrence of oral biofilms. We resort to constant exposure to therapeutics, such as mouthwashes, to prevent the growth of these biofilms. The constant exposure to antimicrobial agents has the potential to cause harm to our microbiome. Therefore, species- and biofilm-selective antimicrobial agents would be of great benefit. The optimal therapy would be one that could eradicate the pathogenic biofilm, specifically and completely without disrupting health-associated bacteria.
The molecules outlined in this review are viable starting points for the development of molecules that target S. mutans biofilms, which remains a prominent contributor to dental caries formation. However, new strategies need to be considered to identify more therapeutically relevant compounds.
Conflicts of interest
There are no conflicts to report.
Acknowledgments
The laboratory is supported by the National Institute of General Medical Sciences (GM119426), National Institute of Dental & Craniofacial Research (DE025837), and the National Science Foundation (CHE1755698). We thank Dr. William Shafer for the motivation in preparing this review.
References
- Kilian M., Chapple I. L. C., Hannig M., Marsh P. D., Meuric V., Pedersen A. M. L., Tonetti M. S., Wade W. G., Zaura E. Br. Dent. J. 2016;221:657. doi: 10.1038/sj.bdj.2016.865. [DOI] [PubMed] [Google Scholar]
- Avila M., Ojcius D. M., Yilmaz Ö. DNA Cell Biol. 2009;28:405–411. doi: 10.1089/dna.2009.0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services, Oral Health in America: A Report of the Surgeon General, 2000. [Google Scholar]
- GBD 2017 Disease and Injury Incidence and Prevalence Collaborators Lancet. 2018;392:1789–1858. doi: 10.1016/S0140-6736(18)32279-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Kolltveit K. M., Tronstad L., Olsen I. Clin. Microbiol. Rev. 2000;13:547–558. doi: 10.1128/cmr.13.4.547-558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K., Nomura R., Ooshima T. Jpn. Dent. Sci. Rev. 2008;44:29–37. [Google Scholar]
- McGhie D., Hutchison J. G., Nye F., Ball A. P. Br. Heart J. 1977;39:456–458. doi: 10.1136/hrt.39.4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J., Cai Q., Steinwandel M., Hargreaves M. K., Bordenstein S. R., Blot W. J., Zheng W., Shu X. O. J. Periodontal Res. 2017;52:636–643. doi: 10.1111/jre.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listl S., Galloway J., Mossey P. A., Marcenes W. J. Dent. Res. 2015;94:1355–1361. doi: 10.1177/0022034515602879. [DOI] [PubMed] [Google Scholar]
- Aas J. A., Griffen A. L., Dardis S. R., Lee A. M., Olsen I., Dewhirst F. E., Leys E. J., Paster B. J. J. Clin. Microbiol. 2008;46:1407–1417. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav K., Prakash S. J. Clin. Infect. Dis. Pract. 2017;2:1–15. [Google Scholar]
- Davies D. Nat. Rev. Drug Discovery. 2003;2:114. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- Donlan R. M. Emerging Infect. Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander P. E. Annu. Rev. Microbiol. 2000;54:437. doi: 10.1146/annurev.micro.54.1.413. [DOI] [PubMed] [Google Scholar]
- Huang R., Li M., Gregory R. L. Virulence. 2011;2:435–444. doi: 10.4161/viru.2.5.16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosier B. T., De Jager M., Zaura E., Krom B. P. Front. Cell. Infect. Microbiol. 2014;4:92. doi: 10.3389/fcimb.2014.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banas J. A., Drake D. R. BMC Oral Health. 2018;18:1–8. doi: 10.1186/s12903-018-0595-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L. Adv. Dent. Res. 2018;29:10–116. doi: 10.1177/0022034517736498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyściak W., Jurczak A., Kościelniak D., Bystrowska B., Skalniak A. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33:499–515. doi: 10.1007/s10096-013-1993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos J. A., Quivey R. G., Koo H., Abranches J. Microbiology. 2013;159:436–445. doi: 10.1099/mic.0.066134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto-Nakano M. Jpn. Dent. Sci. Rev. 2018;54:22–29. doi: 10.1016/j.jdsr.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banas J. A., Vickerman M. M. Crit. Rev. Oral Biol. Med. 2003;14:89–99. doi: 10.1177/154411130301400203. [DOI] [PubMed] [Google Scholar]
- Koga T., Asakawa H., Okahashi N., Hamada S. Microbiology. 1986;132:2873–2883. doi: 10.1099/00221287-132-10-2873. [DOI] [PubMed] [Google Scholar]
- Matsui R., Cvitkovitch D. Future Microbiol. 2010;5:403–417. doi: 10.2217/fmb.09.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welin-Neilands J., Svensäter G. Appl. Environ. Microbiol. 2007;73:5633–5638. doi: 10.1128/AEM.01049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton I. R., Buckley N. D. Oral Microbiol. Immunol. 1991;6:65–71. doi: 10.1111/j.1399-302x.1991.tb00453.x. [DOI] [PubMed] [Google Scholar]
- Sakaue Y., Domon H., Oda M., Takenaka S., Kubo M., Fukuyama Y., Okiji T., Terao Y. Microbiol. Immunol. 2016;60:10–16. doi: 10.1111/1348-0421.12343. [DOI] [PubMed] [Google Scholar]
- Solinski A. E., Ochoa C., Lee Y. E., Paniak T., Kozlowski M. C., Wuest W. M. ACS Infect. Dis. 2018;4:118–122. doi: 10.1021/acsinfecdis.7b00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo O. E., Lefébure T., Pavinski Bitar P. D., Lang P., Richards V. P., Eilertson K., Do T., Beighton D., Zeng L., Ahn S. J., Burne R. A., Siepel A., Bustamante C. D., Stanhope M. J. Mol. Biol. Evol. 2013;30:881–893. doi: 10.1093/molbev/mss278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivey R. G., Kuhnert W. L., Hahn K. Crit. Rev. Oral Biol. Med. 2001;12:301–314. doi: 10.1177/10454411010120040201. [DOI] [PubMed] [Google Scholar]
- Cockerill III F. R., Wikler M. A., Alder J., Dudley M. N., Eliopoulos G. M., Ferraro M. J., Hardy D. J., Hecht D. W., Hindler J. A., Patel J. B., Powell M., Swenson J. M., Richard P., Thomson, Jr. B., Traczewski M. M., Turnidge J. D., Weinstein M. P. and Zimmer B. L., Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard — Ninth Edition M07-A9, 2012. [Google Scholar]
- Macià M. D., Rojo-Molinero E., Oliver A. Clin. Microbiol. Infect. 2014;20:981–990. doi: 10.1111/1469-0691.12651. [DOI] [PubMed] [Google Scholar]
- Zero D. T., Fontana M., Martinez-Mier E. A., Ferreira-Zandona A., Ando M., Gonzalez-Cabezas C., Bayne S. J. Am. Dent. Assoc., JADA. 2009;140:25S–34S. doi: 10.14219/jada.archive.2009.0355. [DOI] [PubMed] [Google Scholar]
- Lee Y. J. lifestyle Med. 2013;3:107–109. [PMC free article] [PubMed] [Google Scholar]
- Petersen P. E., Ogawa H. Community Dent. Health. 2016;33:66–68. [PubMed] [Google Scholar]
- Ccahuana-Vásquez R. A., Tabchoury C. P. M., Tenuta L. M. A., Del Bel Cury A. A., Vale G. C., Cury J. A. Caries Res. 2007;41:9–15. doi: 10.1159/000096100. [DOI] [PubMed] [Google Scholar]
- Milgrom P., Rothen M., Milgrom L. Suom. Hammaslaakarilehti. 2006;13:2–11. [PMC free article] [PubMed] [Google Scholar]
- Masadeh M. M., Gharaibeh S. F., Alzoubi K. H., Al-Azzam S. I., Obeidat W. M. J. Clin. Med. Res. 2013;5:389–394. doi: 10.4021/jocmr1535w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T., Oliveira-Neto J. M., Moore D. Cochrane Database Syst. Rev. 2015;4:1–57. doi: 10.1002/14651858.CD008457.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadoorian P. J., Williams K. B. J. Dent. Oral Hyg. 2008;82:1–5. [Google Scholar]
- Flötra L., Gjermo P., Rölla G., Waerhaug J. Eur. J. Oral Sci. 1971;79:119–125. doi: 10.1111/j.1600-0722.1971.tb02001.x. [DOI] [PubMed] [Google Scholar]
- Marsh P. D. J. Dent. 2010;38:S11–15. doi: 10.1016/S0300-5712(10)70005-1. [DOI] [PubMed] [Google Scholar]
- Worthington R. J., Richards J. J., Melander C. Org. Biomol. Chem. 2012;10:7457–7474. doi: 10.1039/c2ob25835h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H., Allan R. N., Howlin R. P., Stoodley P., Hall-Stoodley L. Nat. Rev. Microbiol. 2017;15:740–755. doi: 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paytubi S., de La Cruz M. L., Tormo J. R., Martín J., González I., González-Menendez V., Genilloud O., Reyes F., Vicente F., Madrid C., Balsalobre C. Front. Microbiol. 2017;8:1–13. doi: 10.3389/fmicb.2017.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker L. M., Clardy J. Antimicrob. Agents Chemother. 2007;51:3582–3590. doi: 10.1128/AAC.00506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley L., Shank E. A. Trends Microbiol. 2017;25:1016–1026. doi: 10.1016/j.tim.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. R., Tripathi A., Wu J., Schultz P. J., Yim I., McQuade T. J., Yu F., Arevang C. J., Mensah A. Y., Tamayo-Castillo G., Xi C., Sherman D. H. Nat. Commun. 2016;7:1–11. doi: 10.1038/ncomms10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin N., Zheng Y., Opoku-Temeng C., Du Y., Bonsu E., Sintim H. O. Future Med. Chem. 2015;7:647–671. doi: 10.4155/fmc.15.7. [DOI] [PubMed] [Google Scholar]
- Yoo S., Murata R. M., Duarte S. Caries Res. 2011;45:327–335. doi: 10.1159/000329181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrazzano G. F., Amato I., Ingenito A., Zarrelli A., Pinto G., Pollio A. Molecules. 2011;16:1486–1507. doi: 10.3390/molecules16021486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombo E. A. J. Evidence-Based Complementary Altern. Med. 2011;2011:1–15. doi: 10.1093/ecam/nep067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijampatnam B., Zhang H., Cai X., Michalek S. M., Wu H., Velu S. E. ACS Omega. 2018;3:8378–8385. doi: 10.1021/acsomega.8b00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbrun E., Hoover C. I., Walker G. J. Arch. Oral Biol. 1983;28:531–536. doi: 10.1016/0003-9969(83)90186-3. [DOI] [PubMed] [Google Scholar]
- Kwon Y. R., Son K. J., Pandit S., Kim J. E., Chang K. W., Jeon J. G. Oral Dis. 2010;16:204–209. doi: 10.1111/j.1601-0825.2009.01636.x. [DOI] [PubMed] [Google Scholar]
- Nijampatnam B., Casals L., Zheng R., Wu H., Velu S. E. Bioorg. Med. Chem. Lett. 2016;26:3508–3513. doi: 10.1016/j.bmcl.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z., Cui T., Zeng J., Chen L., Zhang W., Xu X., Cheng L., Li M., Li J., Zhou X., Li Y. Antimicrob. Agents Chemother. 2016;60:126–135. doi: 10.1128/AAC.00919-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Nijampatnam B., Hua Z., Nguyen T., Zou J., Cai X., Michalek S. M., Velu S. E., Wu H. Sci. Rep. 2017;7:5974. doi: 10.1038/s41598-017-06168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelsterli U., Ho H., Zhou S., Yeow Leow K. Curr. Drug Metab. 2006;7:715–727. doi: 10.2174/138920006778520606. [DOI] [PubMed] [Google Scholar]
- Chen L., Ren Z., Zhou X., Zeng J., Zou J., Li Y. Appl. Microbiol. Biotechnol. 2016;100:857–867. doi: 10.1007/s00253-015-7092-1. [DOI] [PubMed] [Google Scholar]
- Lévesque C. M., Voronejskaia E., Huang Y. C. C., Mair R. W., Ellen R. P., Cvitkovitch D. G. Infect. Immun. 2005;73:3773–3777. doi: 10.1128/IAI.73.6.3773-3777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre W. W., Schneewind O. Mol. Microbiol. 1994;14:115–121. doi: 10.1111/j.1365-2958.1994.tb01271.x. [DOI] [PubMed] [Google Scholar]
- Ajdic D., McShan W. M., McLaughlin R. E., Savic G., Chang J., Carson M. B., Primeaux C., Tian R., Kenton S., Jia H., Lin S., Qian Y., Li S., Zhu H., Najar F., Lai H., White J., Roe B. A., Ferretti J. J. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi T., Asaga E., Goto N. Oral Microbiol. Immunol. 2003;18:266–269. doi: 10.1034/j.1399-302x.2003.00076.x. [DOI] [PubMed] [Google Scholar]
- Igarashi T., Asaga E., Sato Y., Goto N. Oral Microbiol. Immunol. 2004;19:57–60. doi: 10.1046/j.0902-0055.2003.00104.x. [DOI] [PubMed] [Google Scholar]
- Igarashi T., Asaga E., Goto N. Oral Microbiol. Immunol. 2004;19:102–105. doi: 10.1046/j.0902-0055.2003.00123.x. [DOI] [PubMed] [Google Scholar]
- Hu P., Huang P., Chen M. W. Arch. Oral Biol. 2013;58:1343–1348. doi: 10.1016/j.archoralbio.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Hu P., Huang P., Chen W. M. Appl. Biochem. Biotechnol. 2013;171:396–402. doi: 10.1007/s12010-013-0378-9. [DOI] [PubMed] [Google Scholar]
- Park B.-S., Kim J.-G., Kim M.-R., Lee S.-E., Takeoka G. R., Oh K.-B., Kim J.-H. J. Agric. Food Chem. 2005;53:9005–9009. doi: 10.1021/jf051765z. [DOI] [PubMed] [Google Scholar]
- Huang P., Hu P., Zhou S. Y., Li Q., Chen W. M. Curr. Microbiol. 2014;68:47–52. doi: 10.1007/s00284-013-0439-x. [DOI] [PubMed] [Google Scholar]
- Luo H., Liang D. F., Bao M. Y., Sun R., Li Y. Y., Li J. Z., Wang X., Lu K. M., Bao J. K. Int. J. Oral Sci. 2017;9:53–62. doi: 10.1038/ijos.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallock-Richards D. J., Marles-Wright J., Clarke D. J., Maitra A., Dodds M., Hanley B., Campopiano D. J. Chem. Commun. 2015;51:10483–10485. doi: 10.1039/c5cc01816a. [DOI] [PubMed] [Google Scholar]
- Adil M., Singh K., Verma P. K., Khan A. U. J. Glob. Antimicrob. Resist. 2014;2:286–292. doi: 10.1016/j.jgar.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Khan R., Zakir M., Khanam Z., Shakil S., Khan A. U. J. Appl. Microbiol. 2010;109:2151–2159. doi: 10.1111/j.1365-2672.2010.04847.x. [DOI] [PubMed] [Google Scholar]
- Weiss E., Rosenberg M., Judes H., Rosenberg E. Curr. Microbiol. 1982;7:125–128. [Google Scholar]
- Krasowska A., Sigler K. Front. Cell. Infect. Microbiol. 2014;4:112. doi: 10.3389/fcimb.2014.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenye T., Honraet K., Rigole P., Jimenez P. N., Nelis H. J. Antimicrob. Agents Chemother. 2007;51:1541–1544. doi: 10.1128/AAC.00999-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves D. S., Pérez-Fons L., Estepa A., Micol V. Biochem. Pharmacol. 2004;68:549–561. doi: 10.1016/j.bcp.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Beier D., Gross R. Curr. Opin. Microbiol. 2006;9:143–152. doi: 10.1016/j.mib.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Cvitkovitch D. G., Li Y. H., Ellen R. P. J. Clin. Invest. 2003;112:1626–1632. doi: 10.1172/JCI20430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj A. K., Vinothkumar K., Rajpara N. Recent Pat. Anti-Infect. Drug Discovery. 2013;8:68–83. doi: 10.2174/1574891x11308010012. [DOI] [PubMed] [Google Scholar]
- Gerdt J. P., Blackwell H. E. ACS Chem. Biol. 2014;9:2291–2299. doi: 10.1021/cb5004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lönn-Stensrud J., Petersen F. C., Benneche T., Scheie A. A. Oral Microbiol. Immunol. 2007;22:340–346. doi: 10.1111/j.1399-302X.2007.00367.x. [DOI] [PubMed] [Google Scholar]
- Federle M. J. Contrib. Microbiol. 2009;16:18–32. doi: 10.1159/000219371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztajer H., Lemme A., Vilchez R., Schulz S., Geffers R., Yip C. Y. Y., Levesque C. M., Cvitkovitch D. G., Wagner-Döbler I. J. Bacteriol. 2008;190:401–415. doi: 10.1128/JB.01086-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Wang Q., Hu Y., Liang J., Jiang Y., Ma R., Tang Z., Huang Z. Int. J. Antimicrob. Agents. 2012;40:30–35. doi: 10.1016/j.ijantimicag.2012.03.016. [DOI] [PubMed] [Google Scholar]
- Liu C., Worthington R. J., Melander C., Wu H. Antimicrob. Agents Chemother. 2011;55:2679–2687. doi: 10.1128/AAC.01496-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia S. S., Blackledge M. S., Michalek S., Su L., Ptacek T., Eipers P., Morrow C., Lefkowitz E. J., Melander C., Wu H. J. Dent. Res. 2017;96:807–814. doi: 10.1177/0022034517698096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard T. E., Richards J. J., Wolfe A. L., Melander C. Chem. – Eur. J. 2008;14:10745–10761. doi: 10.1002/chem.200801419. [DOI] [PubMed] [Google Scholar]
- AleŠ žula J. I., Kikelj D. Mini-Rev. Med. Chem. 2013;13:1921–1943. doi: 10.2174/1389557511313130007. [DOI] [PubMed] [Google Scholar]
- Hu M., Zhang C., Mu Y., Shen Q., Feng Y. Indian J. Microbiol. 2011;50:362–868. doi: 10.1007/s12088-011-0142-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodnik ž., Łoś J. M., žula A., Zidar N., Jakopin ž., Łoś M., Sollner Dolenc M., Ilaš J., WWȩgrzyngrzyn G., Peterlin Mašič L., Kikelj D. Bioorg. Med. Chem. Lett. 2014;24:2530–2534. doi: 10.1016/j.bmcl.2014.03.094. [DOI] [PubMed] [Google Scholar]
- Kunze B., Reck M., Dötsch A., Lemme A., Schummer D., Irschik H., Steinmetz H., Wagner-Döbler I. BMC Microbiol. 2010;10:1–13. doi: 10.1186/1471-2180-10-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R., Irschik H., Huch V., Schummer D., Steinmetz H., Bock M., Schmidt T., Kirschning A., Müller R. Eur. J. Org. Chem. 2010;7:1284–1289. [Google Scholar]
- Schmidt T., Kirschning A. Angew. Chem., Int. Ed. 2012;51:1063–1066. doi: 10.1002/anie.201106762. [DOI] [PubMed] [Google Scholar]
- Stumpp N., Premnath P., Schmidt T., Ammermann J., Dräger G., Reck M., Jansen R., Stiesch M., Wagner-Döbler I., Kirschning A. Org. Biomol. Chem. 2015;13:5765–5774. doi: 10.1039/c5ob00460h. [DOI] [PubMed] [Google Scholar]
- Reck M., Rutz K., Kunze B., Tomasch J., Surapaneni S. K., Schulz S., Wagner-Döbler I. J. Bacteriol. 2011;193:5692–5706. doi: 10.1128/JB.05424-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banu L. D., Conrads G., Rehrauer H., Hussain H., Allan E., van der Ploeg J. R. Infect. Immun. 2010;78:2209–2220. doi: 10.1128/IAI.01167-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senadheera M. D., Guggenheim B., Spatafora G. A., Huang Y. C. C., Choi J., Hung D. C. I., Treglown J. S., Goodman S. D., Ellen R. P., Cvitkovitch D. G. J. Bacteriol. 2005;187:4064–4076. doi: 10.1128/JB.187.12.4064-4076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio B., Gautier C., Pons N., Ehrlich D. S., Renault P., Guédon E. J. Bacteriol. 2010;192:3464–3473. doi: 10.1128/JB.00119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspiras M. B., Ellen R. P., Cvitkovitch D. G. FEMS Microbiol. Lett. 2004;238:167–174. doi: 10.1016/j.femsle.2004.07.032. [DOI] [PubMed] [Google Scholar]
- Okada A., Igarashi M., Okajima T., Kinoshita N., Umekita M., Sawa R., Inoue K., Watanabe T., Doi A., Martin A., Quinn J., Nishimura Y., Utsumi R. J. Antibiot. 2010;63:89. doi: 10.1038/ja.2009.128. [DOI] [PubMed] [Google Scholar]
- Eguchi Y., Kubo N., Matsunaga H., Igarashi M., Utsumi R. Antimicrob. Agents Chemother. 2011;55:1475–1484. doi: 10.1128/AAC.01646-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhou X. D., Wu C. D. Antimicrob. Agents Chemother. 2011;55:1229–1236. doi: 10.1128/AAC.01016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhou X. D., Wu C. D. Arch. Oral Biol. 2012;57:678–683. doi: 10.1016/j.archoralbio.2011.10.021. [DOI] [PubMed] [Google Scholar]
- Melok A., Lee L., Yussof S. Mohamed, Chu T. Dent. J. 2018;6:1–8. [Google Scholar]
- Sakanaka S., Kim M., Taniguchi M., Yamamoto T. Agric. Biol. Chem. 1989;53:2307–2311. [Google Scholar]
- Yang H., Abouelhassan Y., Burch G. M., Kallifidas D., Huang G., Yousaf H., Jin S., Luesch H., Huigens R. W. Sci. Rep. 2017;7:2003. doi: 10.1038/s41598-017-01045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnsholt T., Ciofu O., Molin S., Givskov M., Høiby N. Nat. Rev. Drug Discovery. 2013;12:791–808. doi: 10.1038/nrd4000. [DOI] [PubMed] [Google Scholar]
- Brown E. D., Wright G. D. Nature. 2016;529:336. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- Gilbert J. A., Blaser M. J., Caporaso J. G., Jansson J. K., Lynch S. V., Knight R. Nat. Med. 2018;24:392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]