Abstract
DNA origami nanostructures are regarded as powerful and versatile vehicles for targeted drug delivery. So far, DNA origami-based drug delivery strategies mostly use intercalation of the therapeutic molecules between the base pairs of the DNA origami’s double helices for drug loading. The binding of nonintercalating drugs to DNA origami nanostructures, however, is less studied. Therefore, in this work, we investigate the interaction of the drug methylene blue (MB) with different DNA origami nanostructures under conditions that result in minor groove binding. We observe a noticeable effect of DNA origami superstructure on the binding affinity of MB. In particular, non-B topologies as for instance found in designs using the square lattice with 10.67 bp/turn may result in reduced binding affinity because groove binding efficiency depends on groove dimensions. Also, mechanically flexible DNA origami shapes that are prone to structural fluctuations may exhibit reduced groove binding, even though they are based on the honeycomb lattice with 10.5 bp/turn. This can be attributed to the induction of transient over- and underwound DNA topologies by thermal fluctuations. These issues should thus be considered when designing DNA origami nanostructures for drug delivery applications that employ groove-binding drugs.
Introduction
During the last three decades, the field of structural DNA nanotechnology has developed a variety of techniques to assemble DNA into increasingly complex two-dimensional (2D) and three-dimensional (3D) nanostructures.1−3 Currently, DNA nanostructures and especially DNA origami4,5 are widely investigated with regard to their applicability in fields as diverse as nanoelectronics,6−10 molecular sensing,11−14 and drug delivery.15−26 For the latter application, drug loading of the DNA origami delivery systems has been achieved mostly via intercalation between the base pairs of the DNA origami’s double helices.16−19,23,25,26 Intercalation induces unwinding of the double helices and may therefore lead to structural distortions of the DNA origami nanostructures.16,27 By employing DNA origami designs with deliberately underwound double helices, however, intercalator loading of the DNA origami can be enhanced.16,28
The interaction of DNA origami with other chemical species that undergo nonintercalative binding to DNA is less studied. Opherden et al. investigated Mg2+ and Eu3+ coordination of two different DNA origami nanostructures, that is, 2D triangles and 3D six-helix bundles (6HBs).29 By employing a variety of spectroscopic techniques, a superstructure-specific geometry of Eu3+-binding sites was revealed in the two DNA origami designs that furthermore deviates from that of genomic double-stranded DNA (dsDNA).
In this work, we investigate the binding of the drug methylene blue (MB) to different 2D and 3D DNA origami nanostructures under conditions favoring minor groove binding and observe a strong dependence on DNA origami superstructure. MB is a fluorescent azine dye that is widely used as an optical and electrochemical probe molecule in the study of DNA-based systems and reactions.30−33 In clinical practice, MB has been used extensively as a therapeutic agent to treat numerous diseases, including malaria and methemoglobinemia.34,35 More recently, MB has been rediscovered as a photosensitizer for the photodynamic therapy of various viral, bacterial, and fungal infections, as well as cancers.36−45 Consequently, also the delivery and controlled release of MB by various carrier systems has received significant attention in recent years.46−53
Results and Discussion
MB can interact with dsDNA via different binding modes. At low salt concentrations, intercalation into the G–C base pairs is favored, whereas at the high salt concentrations typically employed in DNA origami experiments, for example, 10 mM MgCl2 as used in the following experiments, a transition to nonintercalative binding occurs.54−57 Irrespective of the binding mode, however, interaction of MB with DNA is in general accompanied by a decrease in its absorption at 668 nm.57−59
In order to elucidate the nature of this nonintercalative binding mode, UV–vis absorbance spectra of MB with and without genomic dsDNA from salmon testes were recorded at different ionic strengths. This dsDNA has a GC content of 41% and is thus comparable to the fully hybridized M13mp18-based DNA origami scaffold with a GC content of 42%. As can be seen in the spectra presented in Figure 1a, the absorbance at 668 nm clearly decreases upon addition of dsDNA, both in water and in 1× TAE (Tris base, acetic acid, and ethylenediaminetetraacetic acid) buffer supplemented with 10 mM MgCl2. In water, this hypochromicity is accompanied by a significant red shift of the absorbance peak that results in a crossing of both spectral signatures at a wavelength of about 680 nm. In TAE/MgCl2 buffer, however, this shift is absent, indicating the nonintercalative binding of MB, which is in agreement with previous reports.58,59
Figure 1.
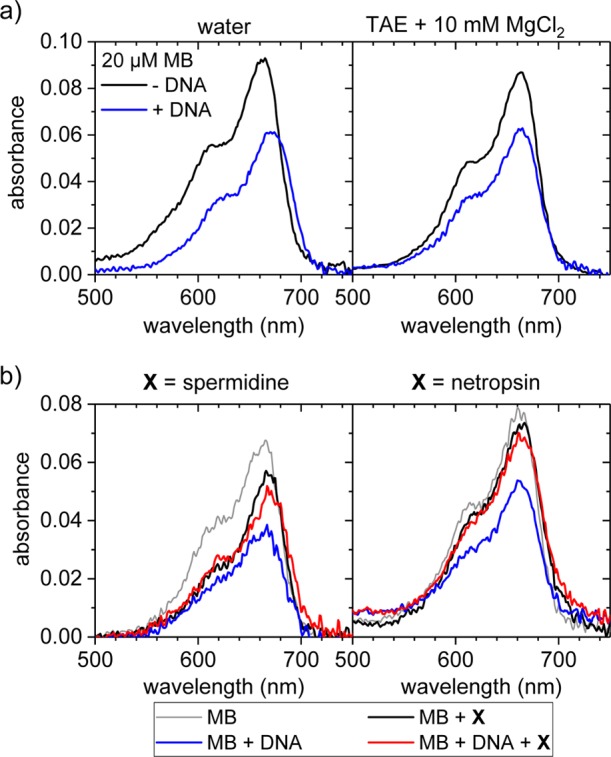
(a) UV–vis absorbance spectra of 20 μM MB with and without genomic dsDNA from salmon testes in water and MgCl2-containing TAE buffer. DNA concentrations were 13 μM (water) and 11 μM (TAE + 10 mM MgCl2) in phosphates. In order to enhance the signal-to-noise ratio and detect even small bathochromic shifts, the shown spectra have been averaged over 10 individual absorbance measurements. Individual spectra can be found in the Supporting Information. (b) UV–vis spectra of the competition between 20 μM MB and 500 μM spermidine (left) and netropsin (right), respectively, in 1× TAE buffer supplemented with 10 mM MgCl2. The spectra of the spermidine-containing samples exhibited a broad background which has been subtracted in the above plot. The concentration of the genomic dsDNA was 308 μM in phosphates.
In order to identify the nonintercalative binding mode of MB, competition assays were performed, employing spermidine and netropsin. While spermidine can bind to both the minor and major grooves,60,61 netropsin is a potent minor groove binder.62,63 Depending on the binding modes of MB and the competing groove binder, addition of a large excess of these groove binders will result in its displacement from the minor groove, the major groove, or both, and thereby reverse the hypochromicity observed upon MB binding to the dsDNA. Indeed, as can be seen in Figure 1b, addition of both groove binders results in a drastic increase in MB absorbance, which almost reaches the value of free MB. This verifies the binding of MB to the minor groove of dsDNA under the current buffer conditions, which also agrees with previous theoretical predictions.64
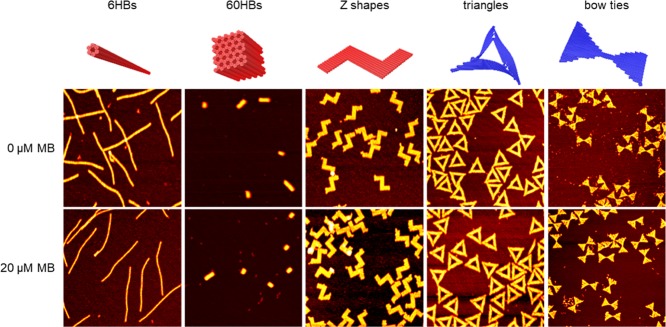
Next, we set out to investigate the interaction of MB with a selection of representative DNA origami nanostructures (see Figure 2). In particular, we chose two 3D DNA origami nanostructures, that is, 6HBs65 and 60-helix bundles (60HBs).66 The 6HBs have previously been found to have an extraordinarily high structural stability under physiological conditions, whereas more complex 3D structures such as the 60HB are typically less stable.67,68 Furthermore, we also selected three differently shaped 2D DNA origami. The Z shape69 is also designed on the honeycomb lattice and features a chiral shape that may serve as an indicator for visualizing MB-binding-induced structural distortions by atomic force microscopy (AFM).27 The Rothemund triangle,4 on the other hand, has previously been identified as a potent drug delivery vehicle in vivo.18 It is designed on the square lattice and appears highly strained (see the corresponding CanDo70,71 simulation in Figure 2). In order to evaluate the effect of such strain on MB binding, we have also evaluated a more relaxed DNA origami structure in the form of a bow tie that is designed on the same lattice but twist-corrected.69 These DNA origami nanostructures are characterized by AFM in Figure 2, both directly after assembly and after subsequent exposure to 20 μM MB. Obviously, all DNA origami nanostructures remain intact upon MB exposure. Furthermore, no major structural distortions induced by MB binding can be observed by AFM. This is in agreement with the minor groove binding of MB, which does not require any major structural alterations of the DNA helices.54,55,64,72 Intercalators, on the other hand, may induce unwinding of the helices and thereby twisting of the DNA origami.16,27,28 Such a twisting should be visible at least for the Z-shaped DNA origami.27
Figure 2.
CanDo70,71 simulations and AFM images (1 × 1 μm2) of 6HBs (height scales 4.5 nm), 60HBs (height scales 12 nm), Z-shaped DNA origami (height scales 3 nm), triangles (height scales 3 nm), and bow ties (height scales 2 nm) before and after exposure to 20 μM MB. The color coding in the CanDo simulations indicates the lattice type (red—honeycomb, blue—square).
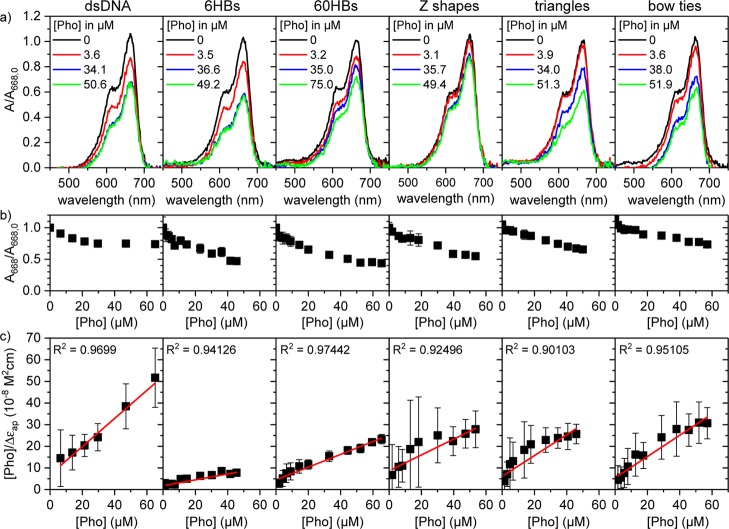
The binding of MB to these DNA origami nanostructures was investigated by UV–vis spectroscopy. In addition to the DNA origami, we also used synthetic dsDNA (15 bp) with a GC content close to that of the M13 scaffold (40%) as a reference. All the UV–vis spectra shown in Figure 3a were characterized by a decrease in the absorbance of MB at 668 nm with increasing DNA concentration. As in the case of genomic dsDNA, no bathochromic shift of the absorption peak is observed, indicating the binding of MB to the minor groove.
Figure 3.
(a) Representative UV–vis spectra of 20 μM MB in 1× TAE buffer containing 10 mM MgCl2 and different phosphate concentrations of dsDNA and different DNA origami nanostructures, respectively. The spectra are normalized to the absorbance at 668 nm in the absence of any DNA. (b) Representative normalized MB absorbance at 668 nm as a function of phosphate concentration. (c) Corresponding binding isotherms. The solid red lines in (c) are linear fits to the data that have been used for the determination of Kd values. R2 values of the fits are given in the plots. Error bars in (b,c) represent standard deviations obtained by averaging over five individual measurements. For Kd determination, these concentration-dependent measurements have been repeated at least once (see Methods).
The normalized absorbance values shown in Figure 3b are found to saturate at high DNA concentrations, indicating that at these concentrations, all MB in solution is DNA-bound. For all DNA origami nanostructures investigated here, saturation occurs at phosphate concentrations between 35 and 55 μM and with different saturation values of the absorbance. Also, the obtained loading efficiencies differ significantly for the different DNA origami nanostructures (see Table S1 for base pair contents) and range from about 5700 MB molecules per DNA origami for the 60HB to about 7400 for the bow tie (see Table S1).
The differences in the MB–DNA interaction observed in Figure 3a,b were assessed more quantitatively by converting the concentration-dependent absorbance values at 668 nm into binding isotherms (see Figure 3c) as described by Zhang and Tang.58 From the linear fits to the data shown in Figure 3c, dissociation constants Kd can be calculated as the ratio of intercept and slope.58 This approach is frequently employed for the quantitative investigation of the interaction of DNA with both intercalators58,73 and groove binders.74,75
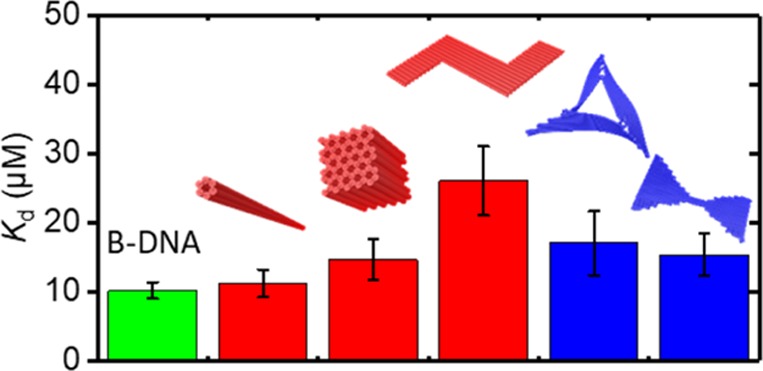
All the determined Kd values are compared in Figure 4. Obviously, DNA origami 6HBs present a Kd value virtually identical to that of synthetic dsDNA with the same GC content. This is to be expected because the 6HBs have been designed on a honeycomb lattice with 10.5 bp per helical turn and should thus exhibit a DNA topology close to the canonical B-form. The same argument of DNA topology also holds true for the 60HB, which indeed has a similar Kd as the 6HB and the dsDNA. Although in such compact 3D DNA origami nanostructures, access of MB to the inner helices may be restricted because of the shielding by the outer helices, this effect appears to be of minor importance in the 60HB.
Figure 4.
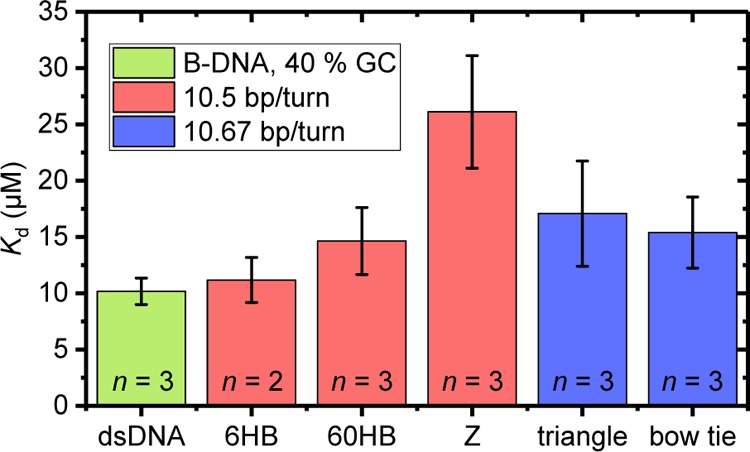
Determined Kd values for the investigated DNA structures. Values are presented as averages over n independent concentration series with the standard error of the mean as error bars (see Methods).
Such geometric effects, however, should be entirely absent in structures that consist just of a single honeycomb “unit cell” such as the 6HB or in objects that are purely two-dimensional. The latter is true for the Z-shaped 2D DNA origami that was constructed using the honeycomb lattice and is thus designed to exhibit 10.5 bp/turn just as the 6HB and the 60HB. Surprisingly, however, the Kd determined for the Z-shaped DNA origami is more than twice as large as those of the 6HB and the dsDNA (see Figure 4). On the basis of the overlap rule of standard error bars,76 the difference between the Z shape and the dsDNA is estimated to be statistically significant with p < 0.05.
In addition to the honeycomb lattice-based DNA origami, we have also evaluated MB binding to two 2D DNA origami nanostructures based on the square lattice.4,69 These structures are designed with 10.67 bp/turn and thus exhibit DNA topologies different from the canonical B-form of dsDNA.4 Efficient minor groove binding of MB64,72 and other molecules77,78 typically requires a tight fit. Therefore, underwinding of the helices will result not only in an increased spacing of base pairs but also in a widened minor groove79 and should thereby reduce the binding affinity of MB. This is in contrast to the case of intercalation, where underwinding results in lower Kd values because of easier access to the widened gaps between base pairs.16,28 In agreement with this hypothesis, both DNA origami triangles and twist-corrected bow-tie structures show Kd values slightly higher than that of dsDNA, although the effect is surprisingly small (see Figure 4).
Of all the DNA origami nanostructures studied in this work, the Z shape shows the strongest deviation from B-DNA, even though it is designed on the same honeycomb lattice as the 6HB and the 60HB, both of which show Kd values similar to dsDNA. We speculate that the high Kd of the Z-shaped DNA origami is caused by the presence of DNA topologies different from the canonical B-form. Despite being based on the honeycomb lattice, such non-B topologies may occur transiently, that is, resulting from thermal fluctuations. In the Z-shaped DNA origami, all helices are aligned parallel to its long axis, including those in the two arms (see Figure 2). These arms are therefore comparatively floppy and subject to strong fluctuations. These fluctuations travel from helix to helix via the crossovers, thereby inducing an oscillatory over- and underwinding of the helices in the vicinity of the crossovers (see CanDo70,71 simulations in the Supporting Information). Because groove binding requires a tight fit, both over- and underwinding will result in reduced binding and in sum to an increased Kd. The bow-tie design also features only parallel helices and could therefore show a similar behavior as the Z shape. However, because of the intrinsic underwinding of the helices in the square lattice, fluctuations in the bow tie may lead even to slightly improved groove binding because of the occurrence of transient B-form topologies. Note that in the case of intercalation, such fluctuations will not lead to different Kd values because here overwinding and underwinding result in lower and higher binding affinities, respectively, and thus compensate each other.
Conclusions
In conclusion, we have investigated the interaction of the drug MB with different DNA origami nanostructures under conditions that result in binding to the minor groove. The DNA origami superstructure is found to have a noticeable influence on the binding efficiency of MB. In particular, non-B topologies as for instance found in designs using the square lattice with 10.67 bp/turn may result in reduced binding affinity. Furthermore, also flexible DNA origami shapes that are prone to structural fluctuations may exhibit reduced groove binding because of the induction of transient over- and underwound DNA topologies. These issues should be considered when rationally designing DNA origami nanostructures for drug delivery applications employing groove-binding drugs.
Methods
Preparation of dsDNA Samples
For the initial experiments addressing the binding mode of MB, lyophilized genomic dsDNA from salmon testes (Alfa Aesar) was dissolved at the desired concentration either in high-performance liquid chromatography (HPLC)-grade water (Carl Roth) or in 1× TAE (Calbiochem) with 10 mM MgCl2 (Sigma-Aldrich). MB, spermidine, and netropsin (all Sigma-Aldrich) were added to achieve the desired concentrations.
For comparison with the DNA origami nanostructures, synthetic dsDNA was prepared by hybridization of two complementary oligonucleotides (Metabion) with the sequences 5′-TTG GAA CAG CAT TGA-3′ and 5′-TCA ATG CTG TTC CAA-3′. To this end, the readily mixed sample (100 μM of each strand in 1× TAE with 10 mM MgCl2) was heated to 94 °C in a thermocycler Primus 25 advanced (PEQLAB), kept at this temperature for 5 min, and cooled down to 20 °C with a cooling rate of 1.5 °C per minute.
DNA Origami Synthesis and Purification
All DNA origami nanostructures were fabricated as previously reported (see below). All structures are based on the 7249 nt long M13mp18 scaffold (purchased from Tilibit Nanosystems).
Triangle: the staple sequences (purchased from Metabion) are taken from the article by Rothemund.4 DNA origami assembly was performed as previously described.80
6HB: the staple sequences (purchased from Metabion) are taken from the article by Bui et al.65 DNA origami assembly was performed as previously described.81
60HB: the staple sequences and the exact fabrication protocol (20 nM reactions) can be found in the article by Linko et al.66 Staple strands were purchased from IDT.
Z shape and bow tie: the staple sequences and the exact fabrication protocol (20 nM reactions) can be found in the article by Shen et al.69 Staple strands were purchased from IDT.
After assembly, the DNA origami nanostructures were purified as described previously80 by spin-filtering (Amicon Ultra filters from Millipore with 100 kDa molecular weight cutoff) and washing with 1× TAE supplemented with 10 mM MgCl2.
Determination of DNA Origami Concentrations
The concentrations of the DNA origami after purification were determined by UV–vis absorbance measurements using an Implen Nanophotometer P330 operated in dsDNA mode. Because different DNA origami nanostructures feature different numbers of base pairs and unpaired nucleotides, the so-determined concentration values had to be corrected.
The measured concentration cm is a result of the combined absorption the single-stranded DNA (ssDNA) and the dsDNA fractions in the DNA origami. Because the extinction of ssDNA is about 1.5 times higher than that of dsDNA,82 the real concentration creal is given by
Here, x is the fraction of paired bases relative to the total number of nucleotides (paired and unpaired) in the DNA origami. The numbers of base pairs and nucleotides of the DNA origami used in this work are given in Table S1.
UV–Vis Spectroscopy
UV–vis absorbance spectra with a spectral range from 200 to 800 nm were recorded with an Implen Nanophotometer P330 equipped with a sub-microliter cell. The sample volume for each measurement was 2 μL. After each measurement, the cell was washed at least twice with HPLC-grade water.
To record the UV–vis spectra of samples with different DNA concentrations, a 20 μM MB solution in 1× TAE with 10 mM MgCl2 was measured first. Then, a small aliquot of a DNA-containing solution (80–300 μM in phosphates depending on the DNA structure, 1× TAE, 10 mM MgCl2, 20 μM MB) was added to the initial MB solution and mixed well. After recording the UV–vis spectra of the resulting solution, the addition and measurement cycle was repeated several times to record a complete concentration series.
Determination of Kd Values
For the determination of the dissociation constants, the measured absorption at 668 nm was averaged over at least five individual measurements at each DNA concentration in a concentration series, and the resulting standard deviations were treated as error limits. After transforming the data according to the protocol of Zhang and Tang,58 the Kd of each concentration series was determined from a linear fit to the data with instrumental weighting of the errors (see below). For each DNA origami, this determination of Kd has been performed for two to three independent concentration series. The so-obtained Kd values for each DNA origami were averaged, and the error limits of the individual linear fits were used for determining the standard error of the mean for each Kd value given in Figure 4 via propagation of error (see Table S3).
The binding constant Kd follows from the relation
Here, [Pho] refers to the molar concentration of phosphates, which has been used instead of the concentration of base pairs in order to account for the different fractions of paired and unpaired nucleotides in the different DNA origami designs. Δεap is the difference between the measured extinction coefficient of the DNA–MB solution, εa, which features both DNA-bound and free MB and the extinction coefficient of free MB in a solution without DNA, εf, with
Here, εa and εf are determined by dividing the measured absorbance values by the molar MB concentration, which was kept constant at 20 μM in all experiments.
Δε represents the difference of the extinction coefficients of DNA-bound MB, εb, and free MB, εf, that is,
The dissociation constant Kd is
then
determined by plotting 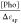 versus [Pho] and fitting the data with
a linear function. In this case, the linear fit has a slope of
versus [Pho] and fitting the data with
a linear function. In this case, the linear fit has a slope of 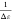 and intercepts the y-axis
at
and intercepts the y-axis
at 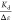 . Therefore, dividing
the intercept by the
slope yields the binding constant Kd.
. Therefore, dividing
the intercept by the
slope yields the binding constant Kd.
AFM Imaging
For AFM imaging, a 5 μL droplet of each DNA origami sample (10 nM in 1× TAE with 10 mM MgCl2) with or without 20 μM MB was deposited on a freshly cleaved mica substrate. After adding 100 μL of 1× TAE with 10 mM MgCl2 to spread the sample over the whole substrate surface, the sample was incubated for 5–15 min, subsequently dipped in HPLC-grade water, and dried in a stream of ultrapure air. AFM imaging was performed in air using Agilent 5100 AFM in intermittent contact mode and HQ:NSC18/Al BS cantilevers from MikroMasch.
Acknowledgments
We thank K. Fahmy for many valuable discussions. Financial support from the Academy of Finland (grant nos. 286845 and 308578), the Sigrid Jusélius Foundation, and the Jane and Aatos Erkko Foundation is gratefully acknowledged.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b00934.
Individual UV–vis absorbance spectra of MB–DNA binding in water and buffer, paired and unpaired bases in the different DNA origami designs, netropsin competition assay for the different DNA origami nanostructures, additional independent concentration series and binding isotherms, loading efficiency of MB, and averaging of Kd values (PDF)
CanDo simulations of the Z-shaped DNA origami - top view (AVI)
(AVI) CanDo simulations of the Z-shaped DNA origami - side view 1 CanDo simulations of the Z-shaped DNA origami - side view 2
(AVI)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Jones M. R.; Seeman N. C.; Mirkin C. A. Programmable materials and the nature of the DNA bond. Science 2015, 347, 1260901. 10.1126/science.1260901. [DOI] [PubMed] [Google Scholar]
- Linko V.; Dietz H. The enabled state of DNA nanotechnology. Curr. Opin. Biotechnol. 2013, 24, 555–561. 10.1016/j.copbio.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Nummelin S.; Kommeri J.; Kostiainen M. A.; Linko V. Evolution of Structural DNA Nanotechnology. Adv. Mater. 2018, 30, 1703721. 10.1002/adma.201703721. [DOI] [PubMed] [Google Scholar]
- Rothemund P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Douglas S. M.; Dietz H.; Liedl T.; Högberg B.; Graf F.; Shih W. M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414–418. 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y.; Pearson A. C.; Gates E. P.; Uprety B.; Davis R. C.; Harb J. N.; Woolley A. T. Electrically conductive gold- and copper-metallized DNA origami nanostructures. Langmuir 2013, 29, 3482–3490. 10.1021/la305155u. [DOI] [PubMed] [Google Scholar]
- Pearson A. C.; Liu J.; Pound E.; Uprety B.; Woolley A. T.; Davis R. C.; Harb J. N. DNA origami metallized site specifically to form electrically conductive nanowires. J. Phys. Chem. B 2012, 116, 10551–10560. 10.1021/jp302316p. [DOI] [PubMed] [Google Scholar]
- Uprety B.; Westover T.; Stoddard M.; Brinkerhoff K.; Jensen J.; Davis R. C.; Woolley A. T.; Harb J. N. Anisotropic Electroless Deposition on DNA Origami Templates To Form Small Diameter Conductive Nanowires. Langmuir 2017, 33, 726–735. 10.1021/acs.langmuir.6b04097. [DOI] [PubMed] [Google Scholar]
- Teschome B.; Facsko S.; Schönherr T.; Kerbusch J.; Keller A.; Erbe A. Temperature-Dependent Charge Transport through Individually Contacted DNA Origami-Based Au Nanowires. Langmuir 2016, 32, 10159–10165. 10.1021/acs.langmuir.6b01961. [DOI] [PubMed] [Google Scholar]
- Shen B.; Linko V.; Dietz H.; Toppari J. J. Dielectrophoretic trapping of multilayer DNA origami nanostructures and DNA origami-induced local destruction of silicon dioxide. Electrophoresis 2015, 36, 255–262. 10.1002/elps.201400323. [DOI] [PubMed] [Google Scholar]
- Acuna G. P.; Moller F. M.; Holzmeister P.; Beater S.; Lalkens B.; Tinnefeld P. Fluorescence enhancement at docking sites of DNA-directed self-assembled nanoantennas. Science 2012, 338, 506–510. 10.1126/science.1228638. [DOI] [PubMed] [Google Scholar]
- Prinz J.; Schreiber B.; Olejko L.; Oertel J.; Rackwitz J.; Keller A.; Bald I. DNA Origami Substrates for Highly Sensitive Surface-Enhanced Raman Scattering. J. Phys. Chem. Lett. 2013, 4, 4140–4145. 10.1021/jz402076b. [DOI] [Google Scholar]
- Kuzuya A.; Sakai Y.; Yamazaki T.; Xu Y.; Komiyama M. Nanomechanical DNA origami “single-molecule beacons” directly imaged by atomic force microscopy. Nat. Commun. 2011, 2, 449. 10.1038/ncomms1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzyk A.; Urban M. J.; Idili A.; Ricci F.; Liu N. Selective control of reconfigurable chiral plasmonic metamolecules. Sci. Adv. 2017, 3, e1602803 10.1126/sciadv.1602803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrault S. D.; Shih W. M. Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano 2014, 8, 5132–5140. 10.1021/nn5011914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.-X.; Shaw A.; Zeng X.; Benson E.; Nyström A. M.; Högberg B. DNA origami delivery system for cancer therapy with tunable release properties. ACS Nano 2012, 6, 8684–8691. 10.1021/nn3022662. [DOI] [PubMed] [Google Scholar]
- Zhuang X.; Ma X.; Xue X.; Jiang Q.; Song L.; Dai L.; Zhang C.; Jin S.; Yang K.; Ding B.; et al. A Photosensitizer-Loaded DNA Origami Nanosystem for Photodynamic Therapy. ACS Nano 2016, 10, 3486–3495. 10.1021/acsnano.5b07671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.; Jiang Q.; Li N.; Dai L.; Liu Q.; Song L.; Wang J.; Li Y.; Tian J.; Ding B.; et al. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano 2014, 8, 6633–6643. 10.1021/nn502058j. [DOI] [PubMed] [Google Scholar]
- Jiang Q.; Song C.; Nangreave J.; Liu X.; Lin L.; Qiu D.; Wang Z.-G.; Zou G.; Liang X.; Yan H.; et al. DNA origami as a carrier for circumvention of drug resistance. J. Am. Chem. Soc. 2012, 134, 13396–13403. 10.1021/ja304263n. [DOI] [PubMed] [Google Scholar]
- Schüller V. J.; Heidegger S.; Sandholzer N.; Nickels P. C.; Suhartha N. A.; Endres S.; Bourquin C.; Liedl T. Cellular immunostimulation by CpG-sequence-coated DNA origami structures. ACS Nano 2011, 5, 9696–9702. 10.1021/nn203161y. [DOI] [PubMed] [Google Scholar]
- Linko V.; Ora A.; Kostiainen M. A. DNA Nanostructures as Smart Drug-Delivery Vehicles and Molecular Devices. Trends Biotechnol. 2015, 33, 586–594. 10.1016/j.tibtech.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Li S.; Jiang Q.; Liu S.; Zhang Y.; Tian Y.; Song C.; Wang J.; Zou Y.; Anderson G. J.; Han J.-Y.; et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 2018, 36, 258–264. 10.1038/nbt.4071. [DOI] [PubMed] [Google Scholar]
- Liu J.; Song L.; Liu S.; Jiang Q.; Liu Q.; Li N.; Wang Z.-G.; Ding B. A DNA-Based Nanocarrier for Efficient Gene Delivery and Combined Cancer Therapy. Nano Lett. 2018, 18, 3328–3334. 10.1021/acs.nanolett.7b04812. [DOI] [PubMed] [Google Scholar]
- Douglas S. M.; Bachelet I.; Church G. M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012, 335, 831–834. 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- Zeng Y.; Liu J.; Yang S.; Liu W.; Xu L.; Wang R. Time-lapse live cell imaging to monitor doxorubicin release from DNA origami nanostructures. J. Mater. Chem. B 2018, 6, 1605–1612. 10.1039/c7tb03223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L.; Jiang Q.; Liu J.; Li N.; Liu Q.; Dai L.; Gao Y.; Liu W.; Liu D.; Ding B. DNA origami/gold nanorod hybrid nanostructures for the circumvention of drug resistance. Nanoscale 2017, 9, 7750–7754. 10.1039/c7nr02222k. [DOI] [PubMed] [Google Scholar]
- Chen H.; Zhang H.; Pan J.; Cha T.-G.; Li S.; Andréasson J.; Choi J. H. Dynamic and Progressive Control of DNA Origami Conformation by Modulating DNA Helicity with Chemical Adducts. ACS Nano 2016, 10, 4989–4996. 10.1021/acsnano.6b01339. [DOI] [PubMed] [Google Scholar]
- Ke Y.; Bellot G.; Voigt N. V.; Fradkov E.; Shih W. M. Two design strategies for enhancement of multilayer-DNA-origami folding: underwinding for specific intercalator rescue and staple-break positioning. Chem. Sci. 2012, 3, 2587–2597. 10.1039/c2sc20446k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opherden L.; Oertel J.; Barkleit A.; Fahmy K.; Keller A. Paramagnetic Decoration of DNA Origami Nanostructures by Eu3+ Coordination. Langmuir 2014, 30, 8152–8159. 10.1021/la501112a. [DOI] [PubMed] [Google Scholar]
- Zhang F.-T.; Nie J.; Zhang D.-W.; Chen J.-T.; Zhou Y.-L.; Zhang X.-X. Methylene blue as a G-quadruplex binding probe for label-free homogeneous electrochemical biosensing. Anal. Chem. 2014, 86, 9489–9495. 10.1021/ac502540m. [DOI] [PubMed] [Google Scholar]
- Gorodetsky A. A.; Barton J. K. Electrochemistry using self-assembled DNA monolayers on highly oriented pyrolytic graphite. Langmuir 2006, 22, 7917–7922. 10.1021/la0611054. [DOI] [PubMed] [Google Scholar]
- Hosseini M.; Khaki F.; Dadmehr M.; Ganjali M. R. Spectroscopic Study of CpG Alternating DNA-Methylene Blue Interaction for Methylation Detection. J. Fluoresc. 2016, 26, 1123–1129. 10.1007/s10895-016-1804-5. [DOI] [PubMed] [Google Scholar]
- Zhang L. Z.; Cheng P. Study of Ni(II) ion-DNA interactions with methylene blue as fluorescent probe. J. Inorg. Biochem. 2004, 98, 569–574. 10.1016/j.jinorgbio.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Wainwright M.; Crossley K. B. Methylene Blue - a Therapeutic Dye for All Seasons?. J. Chemother. 2002, 14, 431–443. 10.1179/joc.2002.14.5.431. [DOI] [PubMed] [Google Scholar]
- Ginimuge P. R.; Jyothi S. D. Methylene blue: revisited. J. Anaesthesiol., Clin. Pharmacol. 2010, 26, 517–520. PMID: 21547182.. [PMC free article] [PubMed] [Google Scholar]
- Fadel M. A.; Tawfik A. A. New topical photodynamic therapy for treatment of hidradenitis suppurativa using methylene blue niosomal gel: A single-blind, randomized, comparative study. Clin. Exp. Dermatol. 2015, 40, 116–122. 10.1111/ced.12459. [DOI] [PubMed] [Google Scholar]
- Guffey J. S.; Payne W.; Roegge W. In vitro fungicidal effects of methylene blue at 625-nm. Mycoses 2017, 60, 723–727. 10.1111/myc.12652. [DOI] [PubMed] [Google Scholar]
- Kofler B.; Romani A.; Pritz C.; Steinbichler T. B.; Schartinger V. H.; Riechelmann H.; Dudas J. Photodynamic Effect of Methylene Blue and Low Level Laser Radiation in Head and Neck Squamous Cell Carcinoma Cell Lines. Int. J. Mol. Sci. 2018, 19, 1107. 10.3390/ijms19041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E. J.; Oak C.-H.; Heo J.; Kim Y.-H. Methylene blue-mediated photodynamic therapy enhances apoptosis in lung cancer cells. Oncol. Rep. 2013, 30, 856–862. 10.3892/or.2013.2494. [DOI] [PubMed] [Google Scholar]
- Pinto J. G.; Martins J. F. d. S.; Pereira A. H. C.; Mittmann J.; Raniero L. J.; Ferreira-Strixino J. Evaluation of methylene blue as photosensitizer in promastigotes of Leishmania major and Leishmania braziliensis. Photodiagn. Photodyn. Ther. 2017, 18, 325–330. 10.1016/j.pdpdt.2017.04.009. [DOI] [PubMed] [Google Scholar]
- Rineh A.; Dolla N. K.; Ball A. R.; Magana M.; Bremner J. B.; Hamblin M. R.; Tegos G. P.; Kelso M. J. Attaching the NorA Efflux Pump Inhibitor INF55 to Methylene Blue Enhances Antimicrobial Photodynamic Inactivation of Methicillin-Resistant Staphylococcus aureus in Vitro and in Vivo. ACS Infect. Dis. 2017, 3, 756–766. 10.1021/acsinfecdis.7b00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaksharam J.; Nayaki K. P. T.; Panneer Selvam N. Treatment of oral lichen planus with methylene blue mediated photodynamic therapy - a clinical study. Photodermatol., Photoimmunol. Photomed. 2012, 28, 97–101. 10.1111/j.1600-0781.2012.00647.x. [DOI] [PubMed] [Google Scholar]
- Samy N. A.; Salah M. M.; Ali M. F.; Sadek A. M. Effect of methylene blue-mediated photodynamic therapy for treatment of basal cell carcinoma. Lasers Med. Sci. 2015, 30, 109–115. 10.1007/s10103-014-1609-1. [DOI] [PubMed] [Google Scholar]
- Yang K.; Wen J.; Chao S.; Liu J.; Yang K.; Pei Y.; Pei Z. A supramolecular photosensitizer system based on the host-guest complexation between water-soluble pillar6arene and methylene blue for durable photodynamic therapy. Chem. Commun. 2018, 54, 5911. 10.1039/c8cc02739k. [DOI] [PubMed] [Google Scholar]
- Dos Santos A. F.; Terra L. F.; Wailemann R. A. M.; Oliveira T. C.; Gomes V. d. M.; Mineiro M. F.; Meotti F. C.; Bruni-Cardoso A.; Baptista M. S.; Labriola L. Methylene blue photodynamic therapy induces selective and massive cell death in human breast cancer cells. BMC Cancer 2017, 17, 194. 10.1186/s12885-017-3179-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S.; Sethi K.; Roy I. Magnetic nanoscale metal-organic frameworks for magnetically aided drug delivery and photodynamic therapy. New J. Chem. 2017, 41, 11860–11866. 10.1039/c7nj02032e. [DOI] [Google Scholar]
- Tada D. B.; Vono L. L. R.; Duarte E. L.; Itri R.; Kiyohara P. K.; Baptista M. S.; Rossi L. M. Methylene blue-containing silica-coated magnetic particles: A potential magnetic carrier for photodynamic therapy. Langmuir 2007, 23, 8194–8199. 10.1021/la700883y. [DOI] [PubMed] [Google Scholar]
- He X.; Wu X.; Wang K.; Shi B.; Hai L. Methylene blue-encapsulated phosphonate-terminated silica nanoparticles for simultaneous in vivo imaging and photodynamic therapy. Biomaterials 2009, 30, 5601–5609. 10.1016/j.biomaterials.2009.06.030. [DOI] [PubMed] [Google Scholar]
- Junqueira M. V.; Borghi-Pangoni F. B.; Ferreira S. B. S.; Rabello B. R.; Hioka N.; Bruschi M. L. Functional Polymeric Systems as Delivery Vehicles for Methylene Blue in Photodynamic Therapy. Langmuir 2016, 32, 19–27. 10.1021/acs.langmuir.5b02039. [DOI] [PubMed] [Google Scholar]
- Boccalini G.; Conti L.; Montis C.; Bani D.; Bencini A.; Berti D.; Giorgi C.; Mengoni A.; Valtancoli B. Methylene blue-containing liposomes as new photodynamic anti-bacterial agents. J. Mater. Chem. B 2017, 5, 2788–2797. 10.1039/c6tb03367a. [DOI] [PubMed] [Google Scholar]
- Hosseinzadeh R.; Khorsandi K.; Hosseinzadeh G. Graphene oxide-methylene blue nanocomposite in photodynamic therapy of human breast cancer. J. Biomol. Struct. Dyn. 2017, 36, 2216. 10.1080/07391102.2017.1345698. [DOI] [PubMed] [Google Scholar]
- Darabpour E.; Kashef N.; Amini S. M.; Kharrazi S.; Djavid G. E. Fast and effective photodynamic inactivation of 4-day-old biofilm of methicillin-resistant Staphylococcus aureus using methylene blue-conjugated gold nanoparticles. J. Drug Delivery Sci. Technol. 2017, 37, 134–140. 10.1016/j.jddst.2016.12.007. [DOI] [Google Scholar]
- Sahu A.; Choi W. I.; Lee J. H.; Tae G. Graphene oxide mediated delivery of methylene blue for combined photodynamic and photothermal therapy. Biomaterials 2013, 34, 6239–6248. 10.1016/j.biomaterials.2013.04.066. [DOI] [PubMed] [Google Scholar]
- Fujimoto B. S.; Clendenning J. B.; Delrow J. J.; Heath P. J.; Schurr M. Fluorescence and Photobleaching Studies of Methylene Blue Binding to DNA. J. Phys. Chem. 1994, 98, 6633–6643. 10.1021/j100077a033. [DOI] [Google Scholar]
- OhUigin C.; McConnell D. J.; Kelly J. M.; van der Putten W. J. M. Methylene blue photosensitised strand cleavage of DNA: Effects of dye binding and oxygen. Nucleic Acids Res. 1987, 15, 7411–7427. 10.1093/nar/15.18.7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordén B.; Tjerneld F. Structure of methylene blue-DNA complexes studied by linear and circular dichroism spectroscopy. Biopolymers 1982, 21, 1713–1734. 10.1002/bip.360210904. [DOI] [PubMed] [Google Scholar]
- Tuite E.; Norden B. Sequence-Specific Interactions of Methylene Blue with Polynucleotides and DNA: A Spectroscopic Study. J. Am. Chem. Soc. 1994, 116, 7548–7556. 10.1021/ja00096a011. [DOI] [Google Scholar]
- Zhang L.; Tang G.-Q. The binding properties of photosensitizer methylene blue to herring sperm DNA: a spectroscopic study. J. Photochem. Photobiol., B 2004, 74, 119–125. 10.1016/j.jphotobiol.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Tong C.; Hu Z.; Wu J. Interaction Between Methylene Blue and CalfThymus Deoxyribonucleic Acid by Spectroscopic Technologies. J. Fluoresc. 2010, 20, 261–267. 10.1007/s10895-009-0549-9. [DOI] [PubMed] [Google Scholar]
- Ruiz-Chica J.; Medina M. A.; Sánchez-Jiménez F.; Ramírez F. J. Fourier Transform Raman Study of the Structural Specificities on the Interaction between DNA and Biogenic Polyamines. Biophys. J. 2001, 80, 443–454. 10.1016/s0006-3495(01)76027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouameur A. A.; Tajmir-Riahi H.-A. Structural Analysis of DNA Interactions with Biogenic Polyamines and Cobalt(III)hexamine Studied by Fourier Transform Infrared and Capillary Electrophoresis. J. Biol. Chem. 2004, 279, 42041–42054. 10.1074/jbc.m406053200. [DOI] [PubMed] [Google Scholar]
- Bailly C.; Chaires J. B. Sequence-specific DNA minor groove binders. Design and synthesis of netropsin and distamycin analogues. Bioconjugate Chem. 1998, 9, 513–538. 10.1021/bc980008m. [DOI] [PubMed] [Google Scholar]
- Kopka M. L.; Yoon C.; Goodsell D.; Pjura P.; Dickerson R. E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc. Natl. Acad. Sci. U.S.A. 1985, 82, 1376–1380. 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R.; Sklenar H. Methylene blue binding to DNA with alternating AT base sequence: minor groove binding is favored over intercalation. J. Biomol. Struct. Dyn. 2004, 21, 699–711. 10.1080/07391102.2004.10506960. [DOI] [PubMed] [Google Scholar]
- Bui H.; Onodera C.; Kidwell C.; Tan Y.; Graugnard E.; Kuang W.; Lee J.; Knowlton W. B.; Yurke B.; Hughes W. L. Programmable periodicity of quantum dot arrays with DNA origami nanotubes. Nano Lett. 2010, 10, 3367–3372. 10.1021/nl101079u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linko V.; Shen B.; Tapio K.; Toppari J. J.; Kostiainen M. A.; Tuukkanen S. One-step large-scale deposition of salt-free DNA origami nanostructures. Sci. Rep. 2015, 5, 15634. 10.1038/srep15634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J.; Wickham S. F. J.; Shih W. M.; Perrault S. D. Addressing the instability of DNA nanostructures in tissue culture. ACS Nano 2014, 8, 8765–8775. 10.1021/nn503513p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielar C.; Xin Y.; Shen B.; Kostiainen M. A.; Grundmeier G.; Linko V.; Keller A. On the Stability of DNA Origami Nanostructures in Low-Magnesium Buffers. Angew. Chem., Int. Ed. 2018, 57, 9470–9474. 10.1002/anie.201802890. [DOI] [PubMed] [Google Scholar]
- Shen B.; Linko V.; Tapio K.; Pikker S.; Lemma T.; Gopinath A.; Gothelf K. V.; Kostiainen M. A.; Toppari J. J. Plasmonic nanostructures through DNA-assisted lithography. Sci. Adv. 2018, 4, eaap8978 10.1126/sciadv.aap8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-N.; Kilchherr F.; Dietz H.; Bathe M. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 2012, 40, 2862–2868. 10.1093/nar/gkr1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro C. E.; Kilchherr F.; Kim D.-N.; Shiao E. L.; Wauer T.; Wortmann P.; Bathe M.; Dietz H. A primer to scaffolded DNA origami. Nat. Methods 2011, 8, 221–229. 10.1038/nmeth.1570. [DOI] [PubMed] [Google Scholar]
- Rohs R.; Bloch I.; Sklenar H.; Shakked Z. Molecular flexibility in ab initio drug docking to DNA: binding-site and binding-mode transitions in all-atom Monte Carlo simulations. Nucleic Acids Res. 2005, 33, 7048–7057. 10.1093/nar/gki1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brglez J.; Nikolov P.; Angelin A.; Niemeyer C. M. Designed Intercalators for Modification of DNA Origami Surface Properties. Chem.—Eur. J. 2015, 21, 9440–9446. 10.1002/chem.201500086. [DOI] [PubMed] [Google Scholar]
- Sarwar T.; Ishqi H. M.; Rehman S. U.; Husain M. A.; Rahman Y.; Tabish M. Caffeic acid binds to the minor groove of calf thymus DNA: A multi-spectroscopic, thermodynamics and molecular modelling study. Int. J. Biol. Macromol. 2017, 98, 319–328. 10.1016/j.ijbiomac.2017.02.014. [DOI] [PubMed] [Google Scholar]
- Moghadam N. H.; Salehzadeh S.; Shahabadi N. Spectroscopic and molecular docking studies on the interaction of antiviral drug nevirapine with calf thymus DNA. Nucleosides, Nucleotides Nucleic Acids 2017, 36, 553–570. 10.1080/15257770.2017.1287379. [DOI] [PubMed] [Google Scholar]
- Cumming G.; Fidler F.; Vaux D. L. Error bars in experimental biology. J. Cell Biol. 2007, 177, 7–11. 10.1083/jcb.200611141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.; Krah A.; Wien F.; Garman E.; Mckenna R.; Sanderson M.; Neidle S. A DNA-porphyrin minor-groove complex at atomic resolution: the structural consequences of porphyrin ruffling. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 9476–9481. 10.1073/pnas.160271897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins C. A. Structural analysis of the binding modes of minor groove ligands comprised of disubstituted benzenes. Nucleic Acids Res. 2001, 29, 936–942. 10.1093/nar/29.4.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall G. L.; Zechiedrich L.; Pettitt B. M. In the absence of writhe, DNA relieves torsional stress with localized, sequence-dependent structural failure to preserve B-form. Nucleic Acids Res. 2009, 37, 5568–5577. 10.1093/nar/gkp556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan S.; Krainer G.; Grundmeier G.; Schlierf M.; Keller A. Structural stability of DNA origami nanostructures in the presence of chaotropic agents. Nanoscale 2016, 8, 10398–10405. 10.1039/c6nr00835f. [DOI] [PubMed] [Google Scholar]
- Teshome B.; Facsko S.; Keller A. Topography-controlled alignment of DNA origami nanotubes on nanopatterned surfaces. Nanoscale 2014, 6, 1790–1796. 10.1039/c3nr04627c. [DOI] [PubMed] [Google Scholar]
- Tinoco I. Hypochromism in Polynucleotides. J. Am. Chem. Soc. 1960, 82, 4785–4790. 10.1021/ja01503a007. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.