Abstract
The preferred conformations of a dodecapeptide composed of l-valine (l-Val) and α-aminoisobutyric acid (Aib) residues, Boc-(l-Val-l-Val-Aib)4-OMe (3), were analyzed in solution and in the crystalline state. Peptide 3 predominantly folded into a mixture of α- and 310-(P) helical structures in solution and a (P) α helix in the crystalline state.
1. Introduction
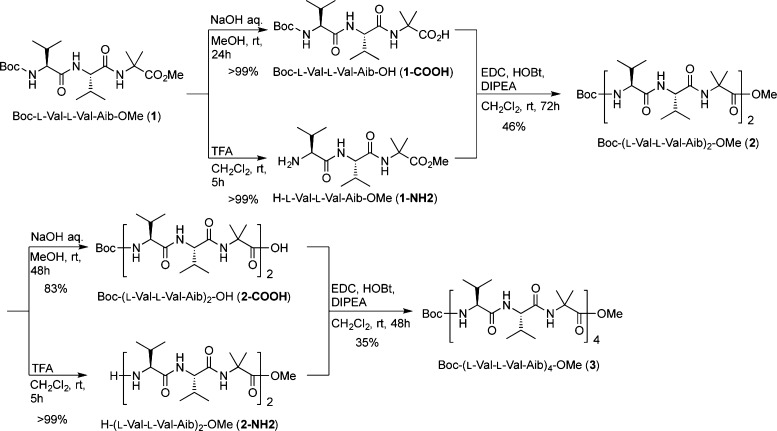
In proteins, helices are abundant and important secondary structures, which recognize macromolecules, such as other proteins and DNA. Helical peptides that mimic proteins are capable of inhibiting protein–protein interactions, and a variety of helix-stabilizing methods have been developed to aid the production of such peptides. As representative techniques, the introduction of α,α-disubstituted α-amino acids (dAA)1 or cyclic β-amino acids2 into short oligopeptides and side-chain stapling3 can all help to stabilize helical structures. In particular, α-aminoisobutyric acid (Aib) is the simplest dAA, and it is commonly used as a helical promoter.4 We have previously reported that the introduction of Aib residues into natural amino acid sequences stabilized helical structures. For example, the oligopeptides Boc-(l-Leu-l-Leu-Aib)n-OMe (n = 3 or 4) preferentially form stable right-handed (P) helical structures.5,6 These peptides are able to act as organocatalysts for asymmetric reaction, such as enantioselective epoxidation catalysts of α,β-unsaturated ketones6 and Michael addition of a malonate.7 Furthermore, the amphipathic peptides R-(l-Xaa-l-Xaa-Aib)3-NH2 (R = FAM-β-Ala and Xaa = Arg or R = H and Xaa = Lys) were also folded into stable helical structures and were used as antimicrobial peptides8 and cell-penetrating peptides,9 respectively. In addition, we have recently reported that the azidolysine (Azl)-based peptide Boc-(l-Azl-l-Azl-Aib)3-OMe formed a stable helical structure, and the azide groups could be replaced with several functional groups via click reactions without influencing the peptide’s helical structure.10 Thus, the insertion of Aib residues into α-amino acid-based oligopeptides is useful for stabilizing helical structures and providing a variety of functions. However, there have not been any reports about the secondary structural changes that occur when Aib residues are introduced into oligopeptides that form extended β-sheet structures. In general, oligopeptides composed of β-branched amino acids, such as valine (Val) and isoleucine (Ile), form β-sheet structures with extended conformations. In particular, oligovalines have a strong tendency to form β-sheet conformations.11 In this study, we designed a dodecapeptide composed of l-Val and Aib residues, Boc-(l-Val-l-Val-Aib)4-OMe (3), and analyzed its preferred conformations in solution and in the crystalline state.
2. Results and Discussion
The dodecapeptide Boc-(l-Val-l-Val-Aib)4-OMe (3) was synthesized using conventional solution-phase methods according to a fragment condensation strategy, in which 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide (EDC) hydrochloride and 1-hydroxybenzotriazole (HOBt) hydrate were used as coupling reagents. Briefly, alkaline hydrolysis of the tripeptide Boc-l-Val-l-Val-Aib-OMe (1) afforded the acid 1-COOH, whereas Boc deprotection by trifluoroacetic acid furnished the amine 1-NH2. The amine 1-NH2 was coupled with 1-COOH to give the hexapeptide Boc-(l-Val-l-Val-Aib)2-OMe (2). The dodecapeptide 3 was prepared in a manner similar to that used to prepare the hexapeptide (Scheme 1).
Scheme 1. Synthesis of Peptides 1–3.
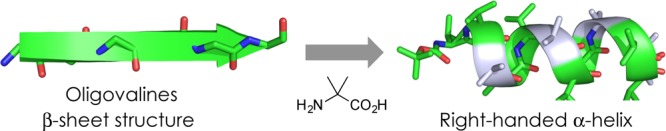
The dominant conformations of the synthesized peptides 1–3 in solution were analyzed based on their Fourier transform infrared (FT-IR), 1H nuclear magnetic resonance (NMR), and circular dichroism (CD) spectra. Figure 1 shows the IR spectra of the tri- (1), hexa- (2), and dodecapeptide (3) in the 3200–3500 cm–1 region (the amide A NH-stretching region) at a peptide concentration of 5.0 mM in CDCl3 solution. In the spectra, the weak bands in the 3425–3438 cm–1 region were assigned to free (solvated) peptide NH groups, and the strong bands in the 3325–3340 cm–1 region were assigned to peptide NH groups with N–H···O=C intramolecular hydrogen bonds. These IR spectra are similar to those of helical peptides containing Aib residues.12
Figure 1.
IR spectra of peptides 1 (green), 2 (blue), and 3 (red) in CDCl3 solution (peptide concentration: 5.0 mM).
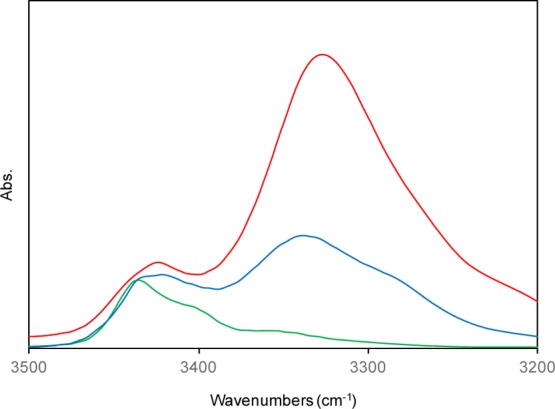
In the 1H NMR spectra of the dodecapeptide 3, the N-terminal urethane-type N(1)H proton signal was unambiguously determined by the high-field position but the remaining eleven peptide NH protons could not be assigned. Figure 2 shows a solvent perturbation experiment involving the addition of the strong H-bond acceptor solvent dimethyl sulfoxide (DMSO-d6) [0–10% (v/v)]. Two NH chemical shifts in the high-field positions were sensitive to the addition of DMSO-d6. These results are indicative of a 310- or α-helical structure in solution.13
Figure 2.
Plots of chemical shift values of the NH protons of peptide 3 as a function of the concentration of DMSO-d6 (v/v) in CDCl3 solution (peptide concentration: 5.0 mM).
The CD spectra of the dodecapeptide 3 in 2,2,2-trifluoroethanol (TFE) showed negative maxima at 207 and 222 nm indicating that 3 formed a right-handed (P) helical structure. Judging from the R([θ]222/[θ]208) value,14 the secondary structure of 3 (R = 0.64) was a mixture of α- and 310-helical structures (Figure 3). This spectrum is similar to that of Boc-(l-Leu-l-Leu-Aib)4-OMe (R = 0.51).15
Figure 3.
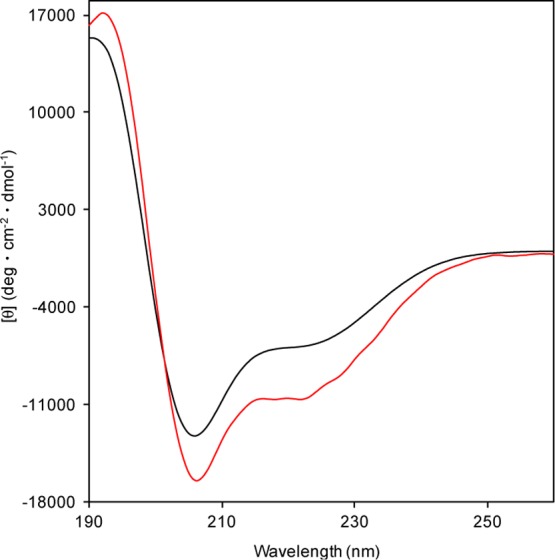
CD spectra of the dodecapeptide 3 (red) and Boc-(l-Leu-l-Leu-Aib)4-OMe (black) in TFE solution (peptide concentration: 0.1 mM).
Peptide 3 formed good crystals for X-ray crystallographic analysis after the slow evaporation of methanol/water at room temperature. Its crystal and diffraction parameters, selected backbone and side-chain torsion angles, and intra- and intermolecular hydrogen-bond parameters are listed in the Supporting Information.16−19 The asymmetric unit in 3 contained two (P) α-helical structures with a flipped C-terminal Aib(12) residue (Figure 4a). The conformations of molecules A and B were well-matched, except for small differences in their side-chain conformations (Figure 4b). The mean ϕ and ψ torsion angles of the residues (2–11) were −63.1° and −39.9° for A and −62.6° and −40.7° for B, which are close to those of an ideal (P) α-helix (−60° and −45°, respectively). Regarding the intramolecular hydrogen bonds in molecules A and B, eight i ← i + 4 type hydrogen bonds were observed, respectively. In packing mode, molecules A and B were connected by intermolecular hydrogen bonds via methanol molecules, forming chains with head-to-tail alignments (···A···A···A··· and ···B···B···B···).
Figure 4.
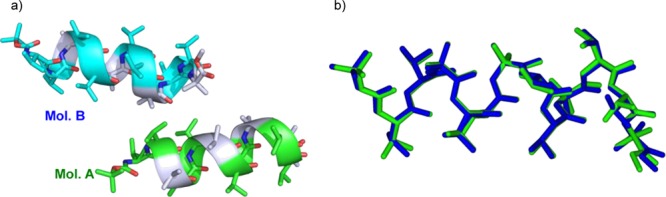
(a) X-ray diffraction structure of 3. The methanol molecules have been omitted. (b) Superimposed structures of molecules A (green) and B (blue).
3. Conclusions
We designed and synthesized a dodecapeptide-containing l-Val and Aib residues, Boc-(l-Val-l-Val-Aib)4-OMe (3), to investigate the influence of the helical promoter Aib on β-sheet structures. The conformation of 3 was analyzed based on its FT-IR, 1H NMR, and CD spectra in solution and X-ray diffraction analysis in the crystalline state. Peptide 3 predominantly folded into a mixture of α- and 310-(P) helical structures in solution and a (P) α helix in the crystalline state. Although oligopeptides composed of β-branched amino acids form β-sheet structures with extended conformations, the insertion of Aib residues into β-sheet-forming peptide sequences could change the conformations of helical structures. Thus, we revealed that the insertion of Aib residues into oligopeptides not only stabilized their helical structures but also markedly altered their secondary structures (from βsheets to helical structures). Not only helical but also unique secondary structures will be created by the combination of natural l- and/or d-amino acids and Aib residues,20 and these findings will be invaluable for the de novo design of peptide-based organic and bioorganic molecules.
4. Experimental Section
4.1. General
1H and 13C NMR spectra were recorded at 400 and 100 MHz in CDCl3 (tetramethylsilane as an internal standard). FT-IR spectra were recorded at 1 cm–1 resolution, with an average of 256 scans used for the CDCl3 solution method (0.1 mm path length for NaCl cell). High-resolution mass spectra were recorded with LCMS-IT-TOF spectrometer. CD spectra were recorded using a 1.0 mm path length cell in TFE.
4.2. Synthesis of Tripeptide 1
The tripeptide 1 was prepared by conventional solution-phase peptide synthesis strategy. Colorless crystals; mp 177–179 °C; [α]D24 = −95.7 (c 0.25, CHCl3); IR (CDCl3, cm–1): 3437, 2969, 2934, 2875, 1738, 1705, 1671; 1H NMR (400 MHz, CDCl3): δ 6.66 (s, 1H), 6.43 (d, J = 8.0 Hz, 1H), 4.99 (d, J = 7.2 Hz, 1H), 4.21–4.18 (m, 1H), 3.91 (dd, J = 6.8 Hz, 1H), 3.70 (s, 3H), 2.23–2.17 (m, 2H), 1.53 (s, 3H), 1.51 (s, 3H), 1.45 (s, 9H), 0.97 (d, J = 6.8 Hz, 3H), 0.94 (d, J = 6.8 Hz, 3H), 0.92 (d, J = 6.8 Hz, 3H), 0.91 (d, J = 6.8 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 174.6, 171.6, 167.0, 156.1, 80.4, 60.5, 58.3, 56.4, 52.5, 50.2, 30.3, 30.2, 28.3, 24.8, 24.7, 19.3, 19.2, 17.7, 17.5; [HR-ESI(+)-TOF] m/z: calcd for C20H37N3O6Na [M + Na]+, 438.2575; found, 438.2591.
4.3. Synthesis of Hexapeptide 2
A solution of the tripeptide Boc-l-Val-l-Val-Aib-OMe (1) (415 mg, 1.0 mmol) and 1 M aqueous NaOH (2.0 mL, 2.0 mmol) in MeOH (10 mL) was stirred at room temperature for 24 h. The solution was neutralized with 1 M aqueous HCl and was extracted with AcOEt. Being dried over Na2SO4 and removing the solvent afforded the tripeptide-carboxylic acid 1-COOH, which was used for the next reaction without further purification. Trifluoroacetic acid (1 mL) was added to a solution of 1 (415 mg, 1.0 mmol) in CH2Cl2 (5 mL), and then the mixture was stirred at room temperature for 5 h. Removing the solvent afforded the crude N-terminal free tripeptide 1-NH2, which was used without further purification. A mixture of EDC (230 mg, 1.2 mmol), HOBt (162 mg, 1.2 mmol), N,N-diisopropylethylamine (418 μL, 2.4 mmol), the above 1-COOH (1.0 mmol), and the above 1-NH2 (1.0 mmol) in CH2Cl2 (10 mL) was stirred at room temperature for 3 days. The solution was washed with 3% aqueous HCl, saturated aqueous NaHCO3, and brine, before being dried over Na2SO4. After removing the solvent, the residue was purified by column chromatography on silica gel (n-hexane/AcOEt = 1:5) to give the hexapeptide 2 in 46% yield. Colorless crystals; mp 200–203 °C; [α]D24 = −27.4 (c 0.5, CHCl3); IR (CDCl3, cm–1): 3437, 3340, 2968, 2935, 2875, 1736, 1703, 1665; 1H NMR (400 MHz, CDCl3): δ 7.64 (s, 1H), 7.30 (d, J = 9.2 Hz, 1H), 7.17 (s, 1H), 6.93 (d, J = 6.8 Hz, 1H), 6.44 (d, J = 5.2 Hz, 1H), 5.01 (d, J = 2.6 Hz, 1H), 4.42 (dd, J = 8.8, 5.2 Hz, 1H), 4.18 (dd, J = 6.4, 4.4 Hz, 1H), 3.95 (dd, J = 4.4 Hz, 1H), 3.82 (dd, J = 4.4, 2.6 Hz, 1H), 3.68 (3H, s), 2.50–2.44 (m, 2H), 2.30–2.20 (m, 2H), 1.53 (3H, s), 1.52 (3H, s), 1.50 (9H, s), 1.50 (3H, s), 1.48 (3H, s), 1.06 (d, J = 6.8 Hz, 6H), 1.05–1.04 (m, 3H), 1.01 (d, J = 6.8 Hz, 3H), 1.00 (d, J = 6.8 Hz, 3H), 0.98 (d, J = 6.8 Hz, 3H), 0.95 (d, J = 6.8 Hz, 3H), 0.94 (d, J = 6.8 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 175.6, 175.2, 172.1, 171.9, 170.9, 170.5, 157.0, 81.8, 62.1, 60.8, 60.0, 58.6, 57.1, 55.8, 52.0, 29.4, 29.2, 28.9, 28.2, 27.5, 25.2, 24.7, 23.7, 19.6, 19.3, 19.2, 18.0, 17.5, 17.4, 17.2; [HR-ESI(+)-TOF] m/z: calcd for C34H62N6O9Na [M + Na]+, 721.4470; found, 721.4502.
4.4. Synthesis of Dodecapeptide 3
The dodecapeptide 3 was prepared using a method similar to that described for the preparation of 2. Yield 35%; colorless crystals; mp 302–304 °C; [α]D24 = −16.9 (c 0.5, CHCl3); IR (CDCl3, cm–1): 3425, 3325, 2967, 2936, 2876, 1734, 1703, 1656; 1H NMR (400 MHz, CDCl3): δ 7.80 (d, J = 4.8 Hz, 1H), 7.77 (s, 1H), 7.73 (s, 1H), 7.67 (d, J = 4.8 Hz, 1H), 7.53–7.51 (m, 3H), 7.21 (d, J = 5.6 Hz, 1H), 7.10 (d, J = 6.0 Hz, 1H), 7.03 (d, J = 7.6 Hz, 1H), 6.72 (br s, 1H), 5.39 (br s, 1H), 4.41 (dd, J = 9.0, 5.8 Hz, 1H), 4.25 (dd, J = 7.2, 5.6 Hz, 1H), 3.89–3.84 (m, 3H), 3.82–3.79 (m, 1H), 3.71–3.62 (m, 2H), 3.67 (s, 3H), 2.47–2.36 (m, 2H), 2.29–2.15 (m, 6H), 1.52–1.48 (m, 33H), 1.12–0.97 (m, 48H); 13C NMR (100 MHz, CDCl3): δ 175.9, 175.9, 175.5, 173.8, 173.8, 173.0, 172.7, 172.6, 172.3, 171.5, 171.0, 157.2, 81.7, 62.9, 62.7, 62.5, 62.3, 60.9, 60.7, 59.2, 57.0, 56.8, 56.6, 55.8, 51.9, 29.8, 29.6, 29.5, 29.2, 29.2, 28.9, 28.3, 27.5, 27.4, 25.2, 24.6, 23.4, 23.3, 23.0, 19.9, 19.7, 19.5, 19.4, 19.4, 19.3, 19.2, 19.1, 19.1, 19.0, 18.9, 18.5, 18.0, 18.0, 17.8; [HR-ESI(+)-TOF] m/z: calcd for C62H112N12O15Na [M + Na]+, 1287.8262; found, 1287.8333.
Acknowledgments
This study was supported, in part, by JSPS KAKENHI grant number 17k08385 (Y.D.), by a grant from the Takeda Science Foundation (Y.D.), a grant from the Terumo Life Science Foundation (Y.D.), a grant from the Suzuken Memorial Foundation (Y.D.), and a grant from the Naito Foundation (Y.D.).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b01030.
The authors declare no competing financial interest.
Supplementary Material
References
- a Crisma M.; Moretto A.; Peggion C.; Panella L.; Kaptein B.; Broxterman Q. B.; Formaggio F.; Toniolo C. Chiral, fully extended helical peptides. Amino Acids 2011, 41, 629–641. 10.1007/s00726-011-0839-9. [DOI] [PubMed] [Google Scholar]; b Tomsett M.; Maffucci I.; Le Bailly B. A. F.; Byrne L.; Bijvoets S. M.; Lizio M. G.; Raftery J.; Butts C. P.; Webb S. J.; Contini A.; Clayden J. A tendril perversion in a helical oligomer: trapping and characterizing a mobile screw-sense reversal. Chem. Sci. 2017, 8, 3007–3018. 10.1039/c6sc05474a. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Eto R.; Oba M.; Ueda A.; Uku T.; Doi M.; Matsuo Y.; Tanaka T.; Demizu Y.; Kurihara M.; Tanaka M. Diastereomeric Right- and Left-Handed Helical Structures with Fourteen (R)-Chiral Centers. Chem.—Eur. J. 2017, 23, 18120–18124. 10.1002/chem.201705306. [DOI] [PubMed] [Google Scholar]; d Yamashita H.; Oba M.; Misawa T.; Tanaka M.; Hattori T.; Naito M.; Kurihara M.; Demizu Y. A Helix-Stabilized Cell-Penetrating Peptide as an Intracellular Delivery Tool. ChemBioChem 2016, 17, 137–140. 10.1002/cbic.201500468. [DOI] [PubMed] [Google Scholar]; e Yamashita H.; Demizu Y.; Misawa T.; Shoda T.; Kurihara M. Synthesis of a bis-cationic α,α-disubstituted amino acid (9-amino-bispidine-9-carboxylic acid) and its effects on the conformational properties of peptides. Tetrahedron 2015, 71, 2241–2245. 10.1016/j.tet.2015.02.076. [DOI] [Google Scholar]; f Kobayashi H.; Misawa T.; Matsuno K.; Demizu Y. Preorganized Cyclic α,α-Disubstituted α-Amino Acids Bearing Functionalized Side Chains That Act as Peptide-Helix Inducers. J. Org. Chem. 2017, 82, 10722–10726. 10.1021/acs.joc.7b01946. [DOI] [PubMed] [Google Scholar]
- a Gellman S. H. Foldamers: A Manifesto. Acc. Chem. Res. 1998, 31, 173–180. 10.1021/ar960298r. [DOI] [Google Scholar]; b Shin Y.-H.; Gellman S. H. Impact of Backbone Pattern and Residue Substitution on Helicity in α/β/γ-Peptides. J. Am. Chem. Soc. 2018, 140, 1394–1400. 10.1021/jacs.7b10868. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Cheng R. P.; Gellman S. H.; DeGrado W. F. β-Peptides: From Structure to Function. Chem. Rev. 2001, 101, 3219–3232. 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]; d Seebach D.; Beck A. K.; Bierbaum D. J. The World of β- and γ-Peptides Comprised of Homologated Proteinogenic Amino Acids and Other Components. Chem. Biodiversity 2004, 1, 1111–1239. 10.1002/cbdv.200490087. [DOI] [PubMed] [Google Scholar]; e Goodman C. M.; Choi S.; Shandler S.; DeGrado W. F. Foldamers as versatile frameworks for the design and evolution of function. Nat. Chem. Biol. 2007, 3, 252–262. 10.1038/nchembio876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Kim Y.-W.; Grossmann T. N.; Verdine G. L. Synthesis of all-hydrocarbon stapled α-helical peptides by ring-closing olefin metathesis. Nat. Protoc. 2011, 6, 761–771. 10.1038/nprot.2011.324. [DOI] [PubMed] [Google Scholar]; b Moellering R. E.; Cornejo M.; Davis T. N.; Del Bianco C.; Aster J. C.; Blacklow S. C.; Kung A. L.; Gilliland D. G.; Verdine G. L.; Bradner J. E. Direct inhibition of the NOTCH transcription factor complex. Nature 2009, 462, 182–188. 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Oba M.; Kunitake M.; Kato T.; Ueda A.; Tanaka M. Enhanced and Prolonged Cell-Penetrating Abilities of Arginine-Rich Peptides by Introducing Cyclic α,α-Disubstituted α-Amino Acids with Stapling. Bioconjugate Chem. 2017, 28, 1801–1806. 10.1021/acs.bioconjchem.7b00190. [DOI] [PubMed] [Google Scholar]; d LaRochelle J. R.; Cobb G. B.; Steinauer A.; Rhoades E.; Schepartz A. Fluorescence Correlation Spectroscopy Reveals Highly Efficient Cytosolic Delivery of Certain Penta-Arg Proteins and Stapled Peptides. J. Am. Chem. Soc. 2015, 137, 2536–2541. 10.1021/ja510391n. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Demizu Y.; Yamagata N.; Nagoya S.; Sato Y.; Doi M.; Tanaka M.; Nagasawa K.; Okuda H.; Kurihara M. Enantioselective epoxidation of α,β-unsaturated ketones catalyzed by stapled helical L-Leu-based peptides. Tetrahedron 2011, 67, 6155–6165. 10.1016/j.tet.2011.06.075. [DOI] [Google Scholar]
- a Lister F. G. A.; Le Bailly B. A. F.; Webb S. J.; Clayden J. Ligand-modulated conformational switching in a fully synthetic membrane-bound receptor. Nat. Chem. 2017, 9, 420–425. 10.1038/nchem.2736. [DOI] [Google Scholar]; b Crisma M.; Formaggio F.; Moretto A.; Toniolo C. Peptide helices based on α-amino acids. Biopolymers 2006, 84, 3–12. 10.1002/bip.20357. [DOI] [PubMed] [Google Scholar]; c Tonlolo C.; Benedetti E. Trends Biochem Sci. 1991, 16, 350–353. 10.1016/0968-0004(91)90142-i. [DOI] [PubMed] [Google Scholar]; d Karle I. L.; Balaram P. Structural characteristics of α-helical peptide molecules containing Aib residues. Biochemistry 1990, 29, 6747–6756. 10.1021/bi00481a001. [DOI] [PubMed] [Google Scholar]; e Ousaka N.; Takeyama Y.; Iida H.; Yashima E. Chiral information harvesting in dendritic metallopeptides. Nat. Chem. 2011, 3, 856–861. 10.1038/nchem.1146. [DOI] [PubMed] [Google Scholar]; f Tsuchiya K.; Numata K. Chemoenzymatic synthesis of polypeptides containing the unnatural amino acid 2-aminoisobutyric acid. Chem. Commun. 2017, 53, 7318–7321. 10.1039/c7cc03095a. [DOI] [PubMed] [Google Scholar]; g Demizu Y.; Doi M.; Sato Y.; Tanaka M.; Okuda H.; Kurihara M. Screw-Sense Control of Helical Oligopeptides Containing Equal Amounts of L- and D-Amino Acids. Chem.—Eur. J. 2011, 17, 11107–11109. 10.1002/chem.201101809. [DOI] [PubMed] [Google Scholar]
- Demizu Y.; Doi M.; Kurihara M.; Okuda H.; Nagano M.; Suemune H.; Tanaka M. Conformational studies on peptides containing α,α-disubstituted α-amino acids: chiral cyclic α,α-disubstituted α-amino acid as an α-helical inducer. Org. Biomol. Chem. 2011, 9, 3303–3312. 10.1039/c0ob01146k. [DOI] [PubMed] [Google Scholar]
- Nagano M.; Doi M.; Kurihara M.; Suemune H.; Tanaka M. Stabilized α-Helix-Catalyzed Enantioselective Epoxidation of α,β-Unsaturated Ketones. Org. Lett. 2010, 12, 3564–3566. 10.1021/ol101435w. [DOI] [PubMed] [Google Scholar]
- Akagawa K.; Sakai N.; Kudo K. Histidine-Containing Peptide Catalysts Developed by a Facile Library Screening Method. Angew. Chem., Int. Ed. 2015, 54, 1822–1826. 10.1002/anie.201410268. [DOI] [PubMed] [Google Scholar]
- Misawa T.; Imamura M.; Ozawa Y.; Haishima K.; Kurihara M.; Kikuchi Y.; Demizu Y. Development of helix-stabilized antimicrobial peptides composed of lysine and hydrophobic α,α-disubstituted α-amino acid residues. Bioorg. Med. Chem. Lett. 2017, 27, 3950–3953. 10.1016/j.bmcl.2017.07.074. [DOI] [PubMed] [Google Scholar]
- Yamashita H.; Demizu Y.; Shoda T.; Sato Y.; Oba M.; Tanaka M.; Kurihara M. Amphipathic short helix-stabilized peptides with cell-membrane penetrating ability. Bioorg. Med. Chem. 2014, 22, 2403–2408. 10.1016/j.bmc.2014.03.005. [DOI] [PubMed] [Google Scholar]
- Misawa T.; Kanda Y.; Demizu Y. Rational Design and Synthesis of Post-Functionalizable Peptide Foldamers as Helical Templates. Bioconjugate Chem. 2017, 28, 3029–3035. 10.1021/acs.bioconjchem.7b00621. [DOI] [PubMed] [Google Scholar]
- a Balcerski J. S.; Pysh E. S.; Bonora G. M.; Toniolo C. Vacuum ultraviolet circular dichroism of β-forming alkyl oligopeptides. J. Am. Chem. Soc. 1976, 98, 3470–3473. 10.1021/ja00428a013. [DOI] [PubMed] [Google Scholar]; b Baron M. H.; De Loze C.; Toniolo C.; Fasman G. D. Infrared and Raman study in the solid state of fully protected, monodispersed homooligopeptides of L-valine, L-isoleucine, and L-phenylalanine. Biopolymers 1979, 18, 411–424. 10.1002/bip.1979.360180216. [DOI] [Google Scholar]; c Toniolo C. Structural Role of Valine and Isoleucine Residues in Proteins. A Proposal. Macromolecules 1978, 11, 437–438. 10.1021/ma60062a033. [DOI] [Google Scholar]; d Toniolo C.; Bonora G. M.; Mutter M. Conformations of poly(ethylene glycol) bound homooligo-L-alanines and -L-valines in aqueous solution. J. Am. Chem. Soc. 1979, 101, 450–454. 10.1021/ja00496a030. [DOI] [Google Scholar]; e Toniolo C.; Bonora G. M.; Salardi S. Study of the relationship between conformation and nature of side chains in linear oligopeptides: homologous series derived from β-branched amino acid residues. Int. J. Biol. Macromol. 1981, 3, 377–383. 10.1016/0141-8130(81)90093-3. [DOI] [Google Scholar]; f Narayanan C.; Dias C. L. Hydrophobic interactions and hydrogen bonds in β-sheet formation. J. Chem. Phys. 2013, 139, 115103. 10.1063/1.4821596. [DOI] [PubMed] [Google Scholar]
- a Crisma M.; Bonora G. M.; Toniolo C.; Benedetti E.; Bavoso A.; Di Blasio B.; Pavone V.; Pedone C. Structural versatility of peptides from Cα,α-dialkylated glycines: an infrared absorption and 1H n.m.r. study of homopeptides from 1-aminocyclopentane-1-carboxylic acid. Int. J. Biol. Macromol. 1988, 10, 300–304. 10.1016/0141-8130(88)90008-6. [DOI] [Google Scholar]; b Benedetti E.; Barone V.; Bavoso A.; Di Blasio B.; Lelj F.; Pavone V.; Pedone C.; Bonora G. M.; Toniolo C.; Leplawy M. T.; Kaczmarek K.; Redlinski A. Structural versatility of peptides from Cαα-dialkylated glycines. I. A conformational energy computation and x-ray diffraction study of homo-peptides from Cα,α-diethylglycine. Biopolymers 1988, 27, 357–371. 10.1002/bip.360270302. [DOI] [Google Scholar]; c Toniolo C.; Bonora G. M.; Bavoso A.; Benedetti E.; Di Blasio B.; Pavone V.; Pedone C.; Barone V.; Lelj F.; Leplawy M. T.; Kaczmarek K.; Redlinski A. Structural versatility of peptides from C?,?-dialkylated glycines. II. An IR absorption and1H-nmr study of homo-oligopeptides from Cα,α-diethylglycine. Biopolymers 1988, 27, 373–379. 10.1002/bip.360270303. [DOI] [Google Scholar]
- a Bonora G. M.; Toniolo C.; Di Blasio B.; Pavone V.; Pedone C.; Benedetti E.; Lingham I.; Hardy P. Folded and extended structures of homooligopeptides from α,α-dialkylated α-amino acids. An infrared absorption and proton nuclear magnetic resonance study. J. Am. Chem. Soc. 1984, 106, 8152–8156. 10.1021/ja00338a025. [DOI] [Google Scholar]; b Toniolo C.; Bonora G. M.; Barone V.; Bavoso A.; Benedetti E.; Di Blasio B.; Grimaldi P.; Lelj F.; Pavone V.; Pedone C. Conformation of pleionomers of α-aminoisobutyric acid. Macromolecules 1985, 18, 895–902. 10.1021/ma00147a013. [DOI] [Google Scholar]; c Demizu Y.; Doi M.; Kurihara M.; Maruyama T.; Suemune H.; Tanaka M. One-Handed Helical Screw Direction of Homopeptide Foldamer Exclusively Induced by Cyclic α-Amino Acid Side-Chain Chiral Centers. Chem.—Eur. J. 2012, 18, 2430–2439. 10.1002/chem.201102902. [DOI] [PubMed] [Google Scholar]
- a Toniolo C.; Polese A.; Formaggio F.; Crisma M.; Kamphuis J. Circular Dichroism Spectrum of a Peptide 310-Helix. J. Am. Chem. Soc. 1996, 118, 2744–2745. 10.1021/ja9537383. [DOI] [Google Scholar]; b Yoder G.; Polese A.; Silva R. A. G. D.; Formaggio F.; Crisma M.; Broxterman Q. B.; Kamphuis J.; Toniolo C.; Keiderling T. A. Conformational Characterization of Terminally Blockedl-(αMe)Val Homopeptides Using Vibrational and Electronic Circular Dichroism. 310-Helical Stabilization by Peptide–Peptide Interaction. J. Am. Chem. Soc. 1997, 119, 10278–10285. 10.1021/ja971392l. [DOI] [Google Scholar]; c Mammi S.; Rainaldi M.; Bellanda M.; Schievano E.; Peggion E.; Broxterman Q. B.; Formaggio F.; Crisma M.; Toniolo C. Concomitant Occurrence of Peptide 310- and α-Helices Probed by NMR. J. Am. Chem. Soc. 2000, 122, 11735–11736. 10.1021/ja002710a. [DOI] [Google Scholar]
- Demizu Y.; Okitsu K.; Yamashita H.; Doi M.; Misawa T.; Oba M.; Tanaka M.; Kurihara M. α-Helical Structures of Oligopeptides with an Alternating l-Leu-Aib Segment. Eur. J. Org. Chem. 2016, 2815–2820. 10.1002/ejoc.201600327. [DOI] [Google Scholar]
- See Supporting Information.
- Sheldrick G. M.Program for Crystal Structure Refinement (SHELXL 97); University of Göttingen: Germany, 1997. [Google Scholar]
- Beurskens P. T.; Admiraal G.; Beurskens G.; Bosman W. P.; Gelder R. D.; Israel R.; Smits J. M. M.. The DIRDIF-99 Program System, Technical Report of the Crystallography Laboratory; University of Nijmegen: The Netherlands, 1994. [Google Scholar]
- CCDC-1837130 contains the supplementary crystallographic data for this paper. These data can be obtained free of charge at www.ccdc.cam.ac.uk/conts/retrieving.html (or from the Cambridge Crystallographic Data Centre, 12, Union Road, Cambridge CB2 1EZ, UK; Fax: (+44) 1223-336-033; or deposit@ccdc.cam.ac.uk).
- a Demizu Y.; Nagoya S.; Doi M.; Sato Y.; Tanaka M.; Kurihara M. Twisted Structure of a Cyclic Hexapeptide Containing a Combination of Alternating l-Leu-d-Leu-Aib Segments. J. Org. Chem. 2012, 77, 9361–9365. 10.1021/jo301509c. [DOI] [PubMed] [Google Scholar]; b Demizu Y.; Yamashita H.; Yamazaki N.; Sato Y.; Doi M.; Tanaka M.; Kurihara M. Oligopeptides with Equal Amounts of l- and d-Amino Acids May Prefer a Helix Screw Sense. J. Org. Chem. 2013, 78, 12106–12113. 10.1021/jo402133e. [DOI] [PubMed] [Google Scholar]; c Demizu Y.; Yamashita H.; Doi M.; Misawa T.; Oba M.; Tanaka M.; Kurihara M. Topological Study of the Structures of Heterochiral Peptides Containing Equal Amounts of l-Leu and d-Leu. J. Org. Chem. 2015, 80, 8597–8603. 10.1021/acs.joc.5b01541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.