Abstract
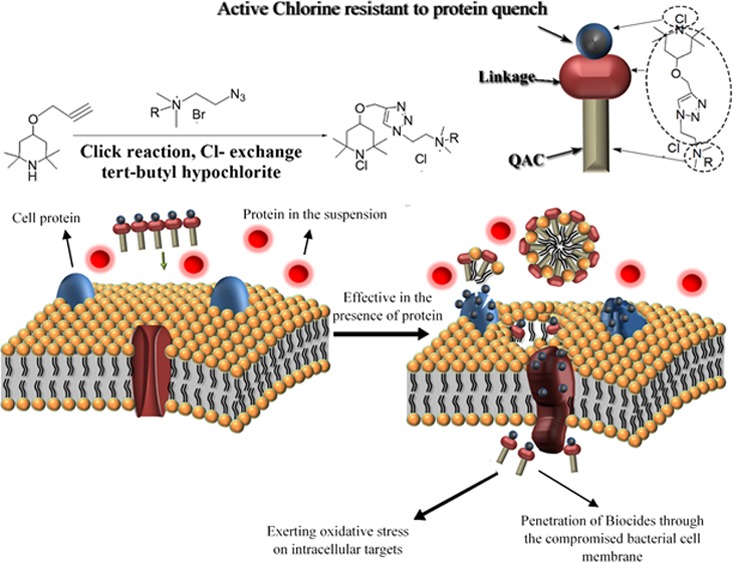
We previously reported that covalently joining an amide-based N-chloramine with a quaternary ammonium compound (QAC) can yield a new composite biocide with faster inactivation of various bacteria. Importantly, the composite biocide was found to reduce the risk for potential bacterial resistance associated with QAC. However, similar to other N-chloramines and QACs, this high-performance composite biocide becomes less potent against pathogenic bacteria in the presence of high protein fluids. In this study, we substituted the amide-based N-chloramine moiety in the previously reported composite biocide with a secondary amine-based N-chloramine to improve the biocidal efficacy in biological fluids. The N–Cl bond in the synthesized tetramethylpiperidine-based composite biocides is more stable in a high protein medium (HPM) than that in the hydantoin (amide)-based composite biocides. The composite biocide, 2-[4-(1-chloro-2,2,6,6-tetramethyl-piperidin-4-yloxymethyl)-[1,2,3]triazol-1-yl]-ethyl-dodecyl-dimethyl-ammonium chloride (6a), showed the best antibacterial activity in both phosphate-buffered saline and HPM among various composite biocides and benzyldodecyldimethylammonium chloride used in this study.
Introduction
Disinfection of hospital environment and food processing facilities has been an indispensable and valuable preventative measure for ensuring human health. In the context of rising antibiotic resistance and cross-resistance between antimicrobials and antibiotics,1−3 development of new potent non-resistance-inducing disinfectants becomes even more pivotal.
Membrane active compounds, such as quaternary ammonium compounds (QACs), account for a big portion of disinfectants. They have been widely used in hospitals for infection control and by public for better hygiene. Reports on bacterial resistance to QACs indicate a concerning trend.4−7 In 2012, up to 83% of methicillin-resistant Staphylococcus aureus (MRSA) isolates carry qac resistance genes in comparison to 10.2% in 1992.4,5 A recent study suggests that besides QacA efflux pump, membrane mutation also greatly contributes to bacterial resistance against QACs. Such strategies as creating multicationic centers have met with certain success in combating bacterial resistance.8,9 Similarly, covalently joining QACs with other disinfectants with different modes of action might also serve as an effective strategy in thwarting bacterial resistance against QACs.
Recently, we reported the synthesis of a series of composite biocides with both quaternary ammonium (QA) and N-chloramine moieties.10,11 The potential of one composite biocide, [3-(3-chloro-4,4-dimethyl-2,5-dioxo-imidazolidin-1-yl)-propyl]-dimethyl-tetradecyl-ammonium chloride (compound 2), and its corresponding precursor, [3-(4,4-dimethyl-2,5-dioxo-imidazolidin-1-yl)-propyl]-dimethyl-tetradecyl-ammonium chloride, (compound 1), in inducing bacterial resistance was studied.12 We were unsuccessful in isolating Pseudomonas aeruginosa mutants with decreased susceptibility to compound 2 despite arduous efforts of achieving the goal with two different strategies: recovering mutants from the zone of clearing and isolating mutants through exposure to gradually increasing concentrations of biocides. Interestingly, the presence of N-chloramine moiety in compound 2 seems potentially overcome the risk of selecting mutants of P. aeruginosa with reduced susceptibility against the quaternary ammonium part.
The antimicrobial action of N-chloramines occurs via different pathways, such as formation of chlorine cover and attack of the bacterial cell wall proteins, penetration into the bacterial cell followed by attacking the intracellular vital component. N-chloramines attack various targets in living cells, particularly, sulfur-containing amino acids, such as cysteine and methionine, interfere with hydrogen bonding in proteins and affect protein functions governed by their structures.13,14
On the other hand, the attraction between the positively charged nitrogen on QACs and negatively charged head groups of bacterial membrane phospholipids kicks off the antibacterial action of QACs. As soon as this bond forms, the long alkyl chain penetrates and integrates into the membrane core.4 When the concentration of QAC is high, the bacterial membrane destruction (or hole generation) occurs due to micelles formation and aggregation.15 Also, QACs can disrupt and denature the structure of proteins and enzymes.4 Covalently joining a QAC with an N-chloramine can give a composite biocide that could inactive bacteria through all of the above-mentioned modes of action. This can not only minimize the chance of bacterial resistance but also enhance the overall biocidal activity.
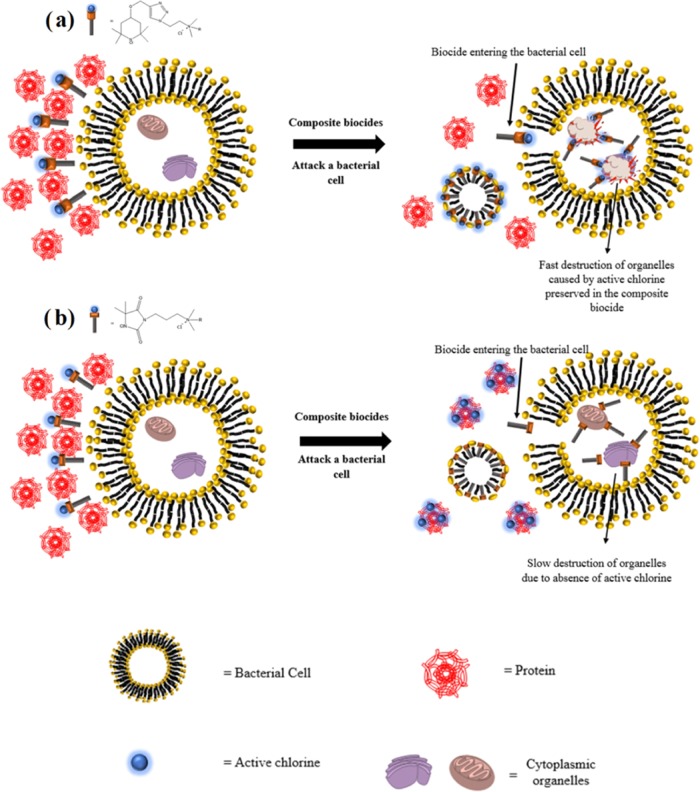
However, disinfectants often encounter interfering substances, such as proteins and carbohydrates, while fulfilling their tasks. Specifically, N-chloramines can be quenched by proteins through direct contact.16 This challenge presents a need to develop new antimicrobials, which are more resistant to interfering substances, such as proteins. Thiol-containing proteins can reduce N-chloramines and therefore quench their antimicrobial activity. Also, active chlorine (Cl+) in N-chloramines can transfer to amine, amide, and imide groups in proteins. In chlorine-transfer reaction (transchlorination), the oxidation capacity is a good measure for the ability of N-chloramines to exchange chlorine with another amine, amide, and imide groups. The reactivity of the active chlorine in N-chloramines is in the following order: amines < amides < imides, and therefore stability of N-chloramines follows the reverse order.17 The above-mentioned compound 2 contains an amide-based N-chloramine. Given the following stability sequence of N-chloramines: amines > amides > imides, an amine-based N-chloramine composite biocide was designed in this study to present improved antibacterial activity in a high protein medium (HPM) consisting of 5% fetal bovine serum (FBS). The proposed mode of action of a composite biocide with both N-chloramine and QA moieties is that the composite biocide punches holes in bacterial membrane due to the QA moiety (mode of action of QACs) and therefore facilitates the penetration of the whole composite molecule into the bacteria, allowing its N-chloramine component to exert oxidative stress inside cells causing fast inactivation of the bacterial cell. We hypothesized that an amine-based N-chloramine can better escape the quenching action of proteins and therefore be preserved to exert oxidative stress inside a bacterial cell to result a fast kill after the whole biocide gets into the cell through the holes in the bacterial membrane generated by the QA moiety (Scheme 1). Specifically, a secondary amine-based N-chloramine was linked to a QA moiety with an alkyl chain of 12 or 14 methylene units via a triazole ring linkage, and the antibacterial activity of these compounds was evaluated against Gram-negative and Gram-positive bacteria. This strategy led us to antimicrobial agents, which may be better suited for applications with high protein loadings, such as disinfection of wounds or food contact surfaces (Scheme 1).
Scheme 1. (a) Proposed Mode of Action of the Secondary Amine-Based N-Chloramine and Its Resistance against Protein Quench, (b) Proposed Mode of Action of the Amide-Based N-Chloramine and Its Quench by Proteins.
Results
Chemistry Synthesis
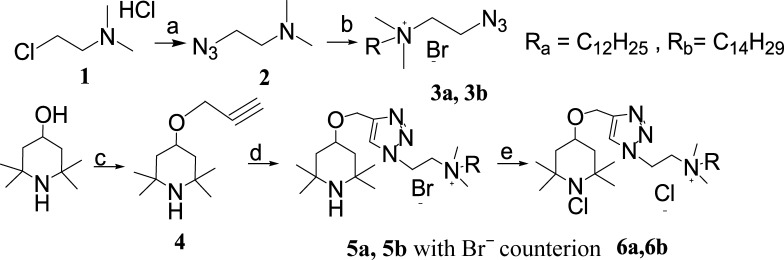
We previously demonstrated the antibacterial enhancement effect of covalently bonding long alkyl chain QAC moieties to an amide-based N-chloramine (compound 2 in Scheme 2).10 In this study, we joined an amine-based N-chloramine to QACs with an alkyl chain of 12 or 14 carbon atoms (6a, 6b in Scheme 2) with an intention to improve the antibacterial efficacy of the composite biocide in the presence of organic matters, such as proteins. [3-(4,4-Dimethyl-2,5-dioxo-imidazolidin-1-yl)-propyl]-dimethyl-tetradecyl-ammonium chloride (compound 1), dodecyl-dimethyl-{2-[4-(2,2,6,6-tetramethyl-piperidin-4-yloxymethyl)-[1,2,3]triazol-1-yl]-ethyl}-ammonium chloride (5a), dimethyl-tetradecyl-{2-[4-(2,2,6,6-tetramethyl-piperidin-4-yloxymethyl)-[1,2,3]triazol-1-yl]-ethyl}-ammonium chloride (5b) are the corresponding N-chloramine precursors (i.e., containing only a QA moiety) of the composite biocide, and benzyldodecyldimethylammonium chloride (BC) was used as a QAC control in antibacterial tests. The chemical reactions pathway is represented in Scheme 3. An overnight reaction between long chain alkyl bromides and 2-azido-N,N-dimethyl-ethanamine led us to azido-functionalized QACs 3a and 3b. 2,2,6,6-Tetramethyl-4-(prop-2-ynyloxy)piperidine 4 was prepared using the method reported by Barnes et al.18 from 2,2,6,6-tetramethylpiperidinol. Piperidinol derivative 4 and two QACs 3a/3b were then linked together via a triazole bridge to get 5a and 5b with Br− counterion. 5a, 5b and compound 1 with Br− counterion were passed through an ion-exchange resin to replace the counterion Br– with Cl– followed by performing the chlorination reaction using t-BuOCl to obtain {2-[4-(1-chloro-2,2,6,6-tetramethyl-piperidin-4-yloxymethyl)-[1,2,3]triazol-1-yl]-ethyl}-dodecyl-dimethyl-ammonium chloride (6a), {2-[4-(1-chloro-2,2,6-trimethyl-piperidin-4-yloxymethyl)-[1,2,3]triazol-1-yl]-ethyl}-dimethyl-tetradecyl-ammonium chloride (6b), and [3-(3-chloro-4,4-dimethyl-2,5-dioxo-imidazolidin-1-yl)-propyl]-dimethyl-tetradecyl-ammonium chloride (compound 2), respectively.
Scheme 2. Structures of Biocides Used in This Research.
Scheme 3. Chemical Reaction Pathways.
(a) NaN3, KOH, H2O, reflux, overnight, (b) R-Br, acetonitrile, reflux, overnight, (c) propargyl bromide, NaH, tetrahydrofuran (THF), N2, 60 °C, 5 h, (d) CuSO4, Cu, MeOH/H2O, room temperature, overnight, (e) anion-exchange resin (Amberlite R IRA-900, Cl–) tert-butyl hypochlorite, acetone/H2O, 0 °C, 1 h. Ra: Alkyl substitution on the compounds bearing the letter “a” in the name and Rb: alkyl substitution on the compounds bearing the letter “b” in the name.
Copper Analysis
Cu(II) was used as a catalyst in the click reaction to link 2,2,6,6-tetramethyl-piperidin with the long chain QA moiety. It is known that copper ion possesses antibacterial activity.19 To avoid unwanted interference in the antibacterial activity of the new composite biocides, copper ion was removed by using a chelating column made of Sorbtech CR20-01 beads. Cu327.395 in 5a and 5b was quantified to be 0.13 and 0.075 ppm, respectively. The amount of Cu324.754 also was measured and appeared to be 0.127 and 0.068 ppm for 5a and 5b. The amount of copper left in each compound found to be much less than the minimum concentration of copper required for antibacterial activity, reported to be 40 ppm by Gyawali et al.20
Redox Titration
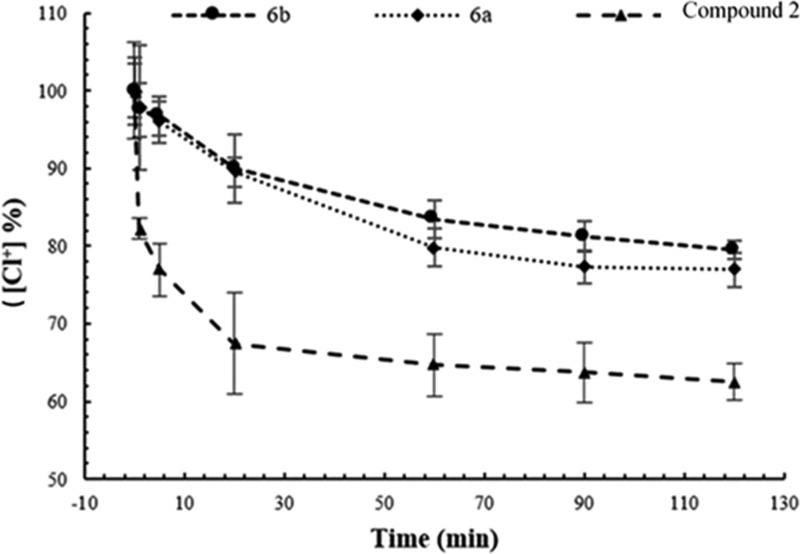
We proceeded to study the stability of N-chloramines in the HPM (5% FBS in phosphate-buffered saline (PBS)) using a redox titration assay, and the results are presented in Figure 1. The active chlorine content of compound 2 (amide-based N-chloramine) dropped to 80% within the first 5 min of contact, and the chlorine content reached a plateau at around 60% after 60 min. On the other hand, both 6a and 6b compounds (amine-based N-chloramines) were significantly more stable (p < 0.05), and their chlorine content was still maintained at >80% after 60 min. The loss rate of active chlorine in HPM was significantly lower (p < 0.05) for the two amine-based N-chloramines than the amide-based N-chloramine in the first 5 min. These results showed that the N–Cl bond of secondary amines (6a and 6b) was less reactive with proteins in FBS than that of amides.
Figure 1.
Stability of N-chloramines 6a, 6b, compound 2 as reflected by the remaining percent of active chlorine ([Cl+]%) in the high protein medium (HPM). Studies were performed at 23 °C and pH 7.4. Values represent the mean ± standard deviation (SD). n = 3 for all three compounds.
Antibacterial Evaluation of the Synthesized Biocides
Table 1 lists the time needed (total kill (Tk)) for the antimicrobial agents to inactivate all of the chosen bacteria (total kill) in different solutions (PBS and HPM). The total kill is claimed when the bacteria level is equal to or below the 1.82 log detection limit and equivalent to 4.18 log bacterial reduction with the starting bacterial concentration of 106 CFU/mL.
Table 1. Time Required to Reach the Total Kill (Tk) (4.18 log Reduction) for Various Compounds in PBS and HPM against Four Strains of Bacteriaa.
| time
to total kill (Tk) (min) |
||||||||
|---|---|---|---|---|---|---|---|---|
| bacterial strain | medium | 5a | 6a | 5b | 6b | compound 1 | compound 2 | BC |
| methicillin-resistant Staphylococcus aureus (MRSA) (70065) | PBS | >60 | 3 | 60 | 5 | 10 | 3 | 10 |
| HPM | >60 | 10 | >60 | 30 | 5 | 10 | 20 | |
| Escherichia coli (25922) | PBS | >60 | 3 | >60 | 3 | 30 | 3 | 3 |
| HPM | >60 | 3 | >60 | 5 | 30 | 20 | 5 | |
| multidrug-resistant (MDR) P. aeruginosa (73104) | PBS | >60 | <1 | <1 | <1 | 5 | 5 | 5 |
| HPM | >60 | 3 | 60 | 20 | 60 | 30 | 10 | |
| wild-type P. aeruginosa (PA01) | PBS | >60 | 3 | 5 | 3 | 5 | 5 | 5 |
| HPM | >60 | 20 | 60 | >60 | >60 | >60 | 30 | |
Initial concentration of compounds in PBS and HPM are 141.0 and 423.1 μM, respectively. n = 3 for all compounds.
As can be found in Table 1, when the medium was changed from PBS to HPM, increase in Tk for 6a and 6b was 9 min on average, whereas Tk increase in the case of compound 2 was 16 min (excluding data of PA01). This means that the deterioration of antimicrobial activity in the case of piperidine (a secondary amine)-based N-chloramine compounds (6a, 6b) was less than compound 2. On the other hand, the performance of compound 2 in PBS was better than compound 1, but their activity was almost the same in HPM, which indicates that [Cl]+ on compound 2 is reduced in HPM, and compound 2 is partially converted back to compound 1, as revealed in Figure 1.
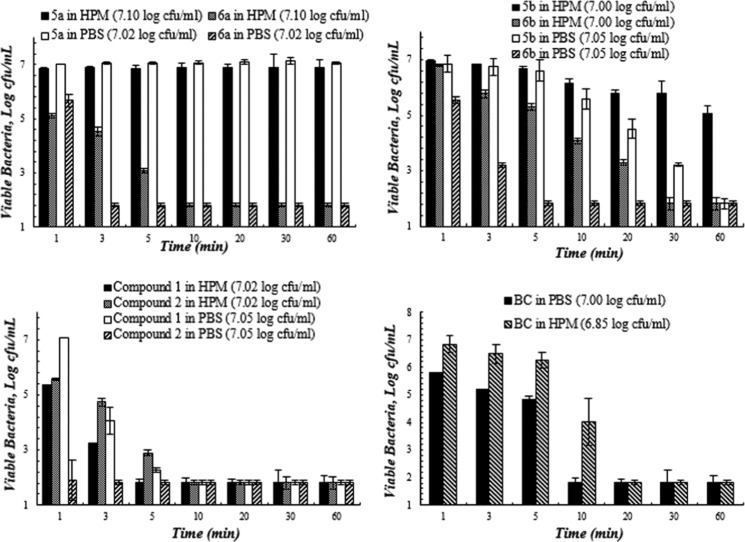
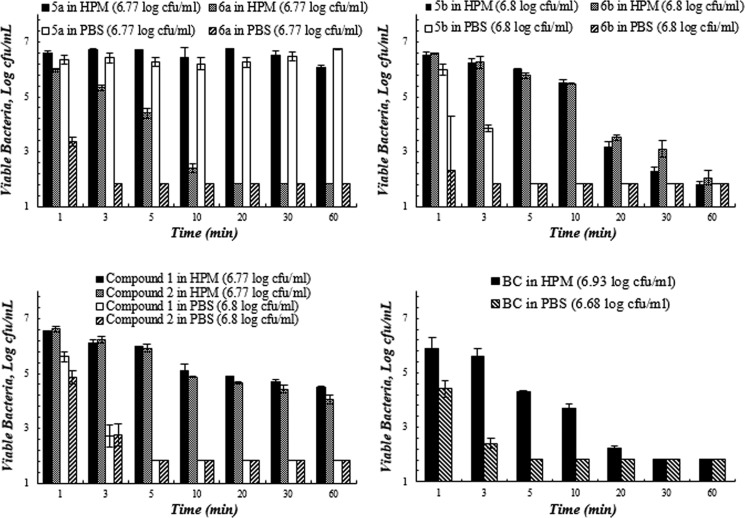
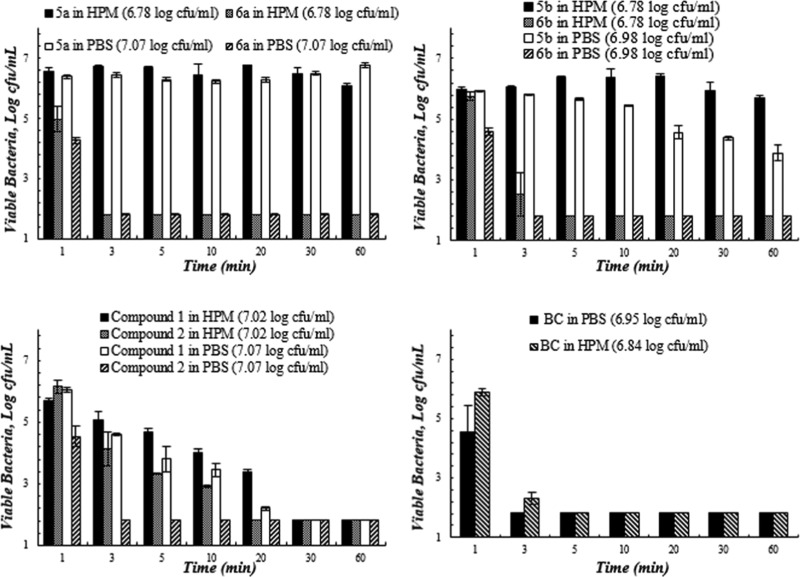
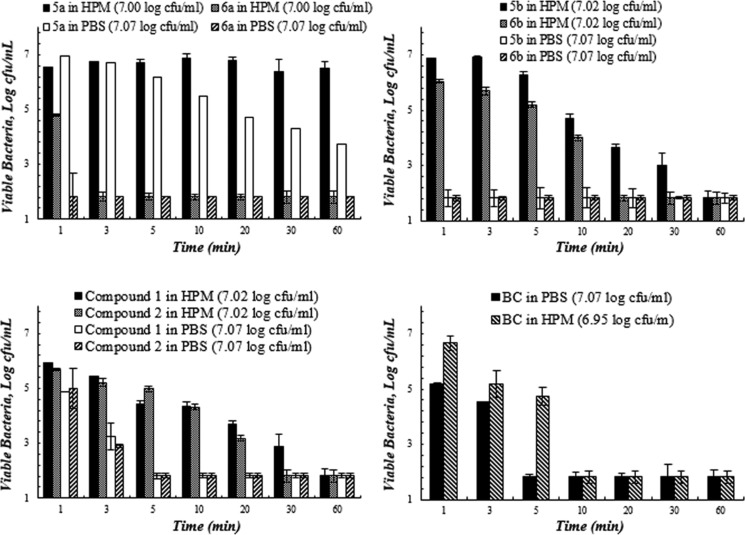
Figures 2–5 present the quantitative antibacterial results over the contact time against four bacteria in various media. The composite biocides appeared to be more effective against the Gram-negative strains (E. coli and P. aeruginosa) than Gram-positive one (MRSA).
Figure 2.
Bacterial viability (log) as a function of contact time between biocides and methicillin-resistant S. aureus (MRSA) in the high protein medium (HPM, 5% FBS in PBS) and PBS. Studies were performed at 23 °C and pH 7.4. Values represent the mean ± SD. n = 3 for all of the compounds (the starting inoculum size is indicated in the bracket after each compound in the legend).
Figure 5.
Bacterial viability (log) as a function of contact time between biocides and wild-type P. aeruginosa PA01 in the high protein medium (HPM, 5% FBS in PBS) and PBS. Studies were performed at 23 °C and pH 7.4. Values represent the mean ± SD. n = 3 for all of the compounds and results are significantly different (p < 0.05) (the starting inoculum size is indicated in the bracket after each compound in the legend).
Against MRSA (Figure 2), 6a and compound 2 showed the best performance in PBS. As expected, their activity deteriorated in HPM, but the extent of deterioration varied with antimicrobial agents. Among all biocides used in this study, compound 1 showed the best antibacterial activity in HPM.
Biocides demonstrated different antibacterial behavior against Gram-negative bacteria E. coli and MDR P. aeruginosa. The activity of compound 2 against E. coli significantly dropped (p < 0.05) in HPM in comparison to PBS (Figure 3). However, 6a and 6b showed insignificant change in their activity (p > 0.05) in two media and were significantly faster in inactivating E. coli in HPM than compound 2 (Tk = 3, 5 versus 20 min). In the case of MDR P. aeruginosa (Figure 4), 6a and 6b illustrated significantly superior killing than others in the first 5 min (p < 0.05) in PBS. In HPM, 6a displayed the least decline in their activity among the chlorinated biocides. compound 1 presented a slow killing kinetics and the most severe deterioration of antimicrobial activity in HPM against MDR P. aeruginosa.
Figure 3.
Bacterial viability (log) as a function of contact time between biocides and E. coli in the high protein medium (HPM, 5% FBS in PBS) and PBS. Studies were performed at 23 °C and pH 7.4. Values represent the mean ± SD. n = 3 for all of the compounds (the starting inoculum size is indicated in the bracket after each compound in the legend).
Figure 4.
Bacterial viability (log) as a function of contact time between biocides and MDR P. aeruginosa in the high protein medium (HPM, 5% FBS in PBS) and PBS. Studies were performed at 23 °C and pH 7.4. Values represent the mean ± SD. n = 3 for all of the compounds (the starting inoculum size is indicated in the bracket after each compound in the legend).
Wild-type P. aeruginosa was the least susceptible strain among the tested Gram-negative bacteria. All biocides, except 5a, were effective against this strain in PBS and reached total kill within 5 min of contact (Figure 5). However, only 6a (in 20 min), BC (30 min) and 5b (60 minutes) reached total kill of PA01 in HPM, and all other biocides failed to reach total kill in HPM in the tested time frame (60 min). 6a clearly stands out as the most potent biocide among all of the tested ones in both PBS and HPM when being challenged with the least susceptible bacterium.
MRSA, P. aeruginosa, E. coli, and wild-type P. aeruginosa were also challenged by sodium hypochlorite in PBS at 141.0 and 282.1 μM (equivalent to 5 and 10 ppm [Cl+]) and in HPM at 423.1 and 864.3 μM (equivalent to 15 and 30 ppm [Cl+]) for 10, 20, 30, and 60 min. The initial bacterial concentrations for the above bacteria were 4.2 × 106, 4.8 × 106, 5.4 × 106, and 3.6 × 106 CFU/mL, respectively. No significant bacterial reduction was observed at the tested concentrations within 1 h of contact of all of the bacteria (p > 0.05) except for one case: 23% reduction of MRSA achieved by 282.1 μM sodium hypochlorite (equivalent to 10 ppm [Cl+]) in PBS within 60 min of contact (p = 0.027 < 0.05).
Discussion
Antimicrobial efficacy of multiple QACs compounds was tested against different bacteria in PBS and HPM. It was observed from Table 1 that in PBS, chlorinated compounds (6a, 6b, and compound 2) performed better than nonchlorinated ones (5a, 5b, and compound 1). This indicates that the introduction of either amide or amine-based N-chloramine boosts the antibacterial activity of the whole composite biocides. However, in HPM, the antimicrobial efficacy of all biocides (chlorinated and nonchlorinated) (except compound 1 against MRSA) deteriorated, and Tks of all of the tested biocides (except compound 1 against MRSA) increased in comparison to those obtained in PBS. In the case of QACs, this deterioration in activity might be the result of ionic and hydrophobic interactions between QAC and negatively charged peptides and hydrophobic domains on proteins, which resulted in binding of QAC molecules to proteins and deactivation of QAC.21 On the other hand, N-chloramines could transfer their active chlorine to proteins through direct contact or be reduced by sulfur-containing amino acids, such as cysteine and methionine in HPM.22 The reaction kinetics of N-chloramine with proteins varies from one compound to another. Active chlorine loading on the composite biocides was monitored as a function of time in PBS and HPM (Figure 1). It is confirmed that secondary amine-based composite biocides (6a and 6b) are more resistant to protein quench than their amide-based counterpart (compound 2). This correlates well with their superior activity against E. coli in HPM as compared with compound 2 (Table 1). But the result that Tks of 6a against the other three bacteria in HPM were significantly shorter (p < 0.05) than those of both 6b and compound 2 reveals the existence of other important factors affecting their antibacterial activity in HPM. The proper balance of hydrophobicity and hydrophilicity in the composite biocide 6a with a C12 alkyl chain might contribute to more effective membrane damage (mode of action of QACs) and be one of the other factors. One more possible factor could be associated with less protein binding of a QAC with a C12 alkyl chain (6a) via hydrophobic interaction, as reported by Jono and co-workers.21 They found that C14-benzalkonium chloride was deactivated more than C12-benzalkonium chloride in the presence of proteins. This led to less quench of the biocide by proteins in HPM through the deactivation mechanism of QAC: electrostatic and hydrophobic interactions.
Gilbert and Moore15 have shown that antibacterial activity of QACs is a function of their structure, alkyl chain length, and strain of bacteria. For example, to be effective against Gram-positive bacteria, the alkyl chain of QACs should consist of 12–14 carbon atoms, however, in the case of Gram-negative bacteria, the number of carbon atoms in the alkyl chain should be between 14 and 16. N-chloramines are more hydrophobic than their amine/amide precursors, and the added hydrophobicity in N-chloramine/QAC composite biocides might move the optimum alkyl chain length against Gram-negative bacteria from 14–16 to 12.
In antibacterial test against Gram-negative bacteria E. coli and P. aeruginosa, the antibacterial efficacy of 6a and 6b appeared to be less compromised by HPM than compound 2, whereas compound 2 and 6a showed similar changes in antibacterial kinetics in two media (PBS and HPM) against MRSA. These results still correlate with the stability of N-chloramine in HPM and can be explained using our hypothesized mechanism. Proteins in HPM neutralize the amine-based N-chloramine at a slower pace than the amide-based N-chloramine so that the amine-based N-chloramine is preserved to exert oxidative stress inside a bacterial cell after the whole molecule diffuses into the cell through the holes in the bacterial membrane due to the mode of action of the QA moiety. However, to what degree this advantage of better preserved amine-based N-chloramine can be manifested is also dependent on how effectively bacterial cells can adsorb the biocides, which is the first step of QACs’ action of disrupting bacterial membranes. E. coli has a much higher number density of negative charges (0.145 m–3 at pH 7) than S. aureus (0.025 m–3 at pH 7),23 which might be the reason why this advantage of better preserved amine-based N-chloramine is not clearly shown in the challenge test against MRSA in HPM (Figure 2). The thick cell wall of MRSA might also play a role by delaying the interaction of the positively charged biocides with bacterial cell membrane. As can be seen from Figures 2 and 3, BC showed a faster killing kinetics against E. coli than S. aureus.
5a and 5b were very ineffective against E. coli even in PBS (Figure 3). One possible reason is that the piperidine functional group is very basic (pKa of its conjugate acid is around 11)14 and become protonated in pH 7.4 PBS, greatly increasing the hydrophilicity of the whole molecules of 5a and 5b. QACs act on bacterial membranes by inserting their hydrophobic tails into the hydrophobic membrane core. The enhanced hydrophilicity due to the protonated piperidine functional group might impede this interaction, hence compromising the antibacterial potencies of both 5a and 5b. Chlorination of the synthesized QACs boosts their antibacterial activity via two different mechanisms. First, chlorine changes the hydrophilicity of QAC compounds by transforming the positively charged piperidine functional group in biological pH (7.4) to a neutral moiety (decreased basicity due to the presence of chlorine). Therefore, the hydrophilicity of the compounds drops, allowing QACs to better interdigitate into the bacterial membrane and negatively impact on its osmoregulatory and physiological functions.24 Secondly, active chlorine also attacks targets in bacterial cells. As reflected from the antibacterial results of sodium hypochlorite, active chlorine [Cl+] alone at the tested concentrations within the tested time frame (1 h) in either PBS or HPM is not effective in killing those bacteria. This indicates again that the N-chloramine moiety together with the QA moiety attacks bacterial cells in a potentially synergistic manner.
Conclusions
Nowadays, prevention of bacterial contaminations in food industries has become difficult due to long food production line, large production volume of food, and antimicrobial resistance.25 Also, the number of effective antimicrobial agents with a good performance in high protein containing environment is limited.26 In this study, two amine-based N-chloramine/QAC composite biocides were synthesized. These synthesized biocides were challenged with various bacterial strains, including Gram-positive and Gram-negative bacteria MRSA, E. coli, MDR P. aeruginosa, and wild-type P. aeruginosa (PA01) in both PBS and HPM. The results indicate that when a secondary amine as opposed of an amide-based N-chloramine is used, the loss of active chlorine in high protein-loaded fluids decreases, which results in less deterioration of antibacterial activity. The composite biocide 6a demonstrated the best antibacterial efficacy in both PBS and HPM among all biocides studied in this study. The relatively better antimicrobial activity of the amine-based N-chloramine (6a) in HPM is due to the relatively high stability of the active chlorine as compared to the amide-based N-chloramine. Transchlorination between the amine-based N-chloramines (6a, 6b) and proteins in the medium occurs at a slow rate as compared to the amide-based N-chloramine (compound 2). The amine-based N-chloramine then has a better chance to exert oxidative stress inside a bacterial cell after the whole molecule finds its way into the cell due to the membrane disruption action of the QA moiety. Interestingly, the amine-based composite biocide with a QAC unit of C12 alkyl chain (6a) is more potent in the protein medium than its counterpart with a QAC unit of C14 alkyl chain (6b). This is opposite to the general order of antibacterial activity of QACs with different alkyl chain lengths: C14 > C12 previously reported18,27 due to the presence of the piperidine-based N-chloramine moiety in the composite biocide. These interesting findings bring us one step closer to the design and synthesis of more potentially non-resistance-inducing biocides potent in real world where organic matters might be inevitable.
Experimental Section
Materials
All of the reagents and solvents, which were analytical grades and no further purification been used, were purchased from suppliers namely, Sigma, Fisher, and VWR. NMR spectra were performed using Bruker Avance 300 MHz NMR spectrometer at room temperature and in 5 mm NMR tubes. High-resolution mass measurements were recorded by AB SCIEX Triple TOF 5600+ (ESI-MS), Concord, ON with the direct infusion method. Multidrug-resistant (MDR) P. aeruginosa #73104 and community-associated-MRSA #70065 and wild-type P. aeruginosa (PA01), which were obtained from the CANWARD (Canadian Ward Surveillance) study assessing antimicrobial resistance in Canadian hospitals, http://www.canr.ca. Escherichia coli ATCC 25922 was sourced from ATCC.
Synthesis and Analysis
(2-Azido-ethyl)-dimethyl-amine (2) (Scheme 3)
2-Chloro-N,N-dimethylethylamine hydrochloride (1) (20 g, 139 mmol) was dissolved in 50 mL of water, and then 45.13 (0.69 mol, 5 equiv) of sodium azide was added to the solution and reaction continued overnight at reflux. Then, 7g of potassium hydroxide added to the mixture followed by extraction with dichloromethane (DCM) (3 × 100 mL). After solvent evaporation, 10.3 g (95 mmol, 65% yield) of a yellow liquid was achieved.
1H NMR (CDCl3, 300 Hz) δ [ppm]: 3.35 (t, J = 6.1 Hz, 2H: N–N=NCH2−), 2.5 (t, J = 6.2 Hz, 2H: −CH2N), 2.27 (s, 6H: −N(CH3)2).
13C NMR (CDCl3, 75 Hz) δ [ppm]: 46.6, 46.6, 48.7, 65.8; HRMS (ESI-TOF) m/z: [M + H]+ calculated for C4H10N4+, 114.0905; found: 114.0911.
3a, 3b (Scheme 3)
2-Azido-N,N-dimethylethanamine (2) (5.0 g, 44 mmol) was dissolved in 50 mL of acetonitrile, and then 48.4 mmol of 1-bromoalkane, 1-bromotetradecane (3a), or 1-bromotetradecane (3b) was added to the solutions and the reactions continued overnight at reflux. After solvent evaporation, compounds were dissolved in 5 mL of methanol followed by participation in 150 mL of 50:50 ethyl acetate/hexane 3a (11.20 g, 35.11 mmol, 79% yield) and 3b (11.6 g, 33.4 mmol, 76% yield) of white solids were achieved.
3a: 1H NMR (D2O, 300 Hz) δ [ppm]: 4.04 (t, J = 6.2 Hz, 2H: N–N=NCH2−), 3.62 (t, J = 6.2 Hz, 2H: −CH2N+), 3.43 (t, J = 7.8 Hz, 2H: N+CH2−), 3.22 (s, 6H: (CH3)2N+), 1.73–1.9 (m, 2H: −CH2CH2), 1.23–1.48 (m, 18H: −(CH2)9CH3), 0.91 (t, J = 6.3 Hz, 3H: −CH3).
13C NMR (D2O, 75 Hz) δ [ppm]: 13.8, 22.6, 26.2, 29.4, 29.5, 29.7, 29.8, 31.9, 44.9, 51.9, 61.6, 64.5; HRMS (ESI-TOF) m/z: [M – Br]+ calculated for C16H35N4+, 283.2856; found: 283.2864.
3b: 1H NMR (D2O, 300 Hz) δ [ppm]: 4.04 (t, J = 6.2 Hz, 2H: N–N=NCH2−), 3.62 (t, J = 6.2 Hz, 2H: −CH2N+), 3.43 (t, J = 7.5 Hz, 2H: N+CH2−), 3.22 (s, 6H: (CH3)2N+), 1.73–1.9 (m, 2H: −CH2CH2), 1.23–1.48 (m, 22H: −(CH2)11CH3), 0.91 (t, 3H: −CH3).
13C NMR (D2O, 75 Hz) δ [ppm]: 13.9, 22.7, 26.2, 29.2, 29.7, 29.9, 30.1, 32.0, 45.0, 52.1, 61.5, 64.3; HRMS (ESI-TOF) m/z: [M – Br]+ calculated for C18H39N4+, 311.3169; found: 311.3174.
2,2,6,6-tetramethyl-4-(prop-2-ynyloxy)piperidine (4) (Scheme 3)
2,2,6,6-tetramethylpiperidin-4-ol (9.0 g, 57 mmol) was added to 100 mL of anhydrous THF and kept under a nitrogen atmosphere for 30 min, stirring at room temperature, followed by addition of 2.26 g (57.0 mmol) of NaH (60%). After 30 min of mixing, the flask sealed with a rubber stopper and transferred into a 60 °C oil bath where 6.356 mL (57.00 mmol) of propargyl bromide (80%) was added to the mixture and the reaction continued overnight at 60 °C. Afterward, solid settlements were removed by filtration and THF evaporated using rotary evaporator. After solvent removal, the compound dissolved in 50 mL of 1N HCl solution and was washed with 3 × 50 mL of DCM. Then, sodium hydroxide was used to increase the pH to 10 (in an ice bath). Then, the aqueous solution was washed with DCM (3 × 50 mL). The organic layer was dried on sodium sulfate, and the solvent was evaporated. 5.10 g (26.2 mmol, 46% yield) of yellow solid was achieved.
1H NMR (CDCl3, 300 Hz) δ [ppm]: 4.17 (s, 2H: OCH2−), 3.83–3.96 (m, 1H: −CHO), 2.39 (s, 1H: −CH), 1.97–1.96 (d, 1H), 1.93–1.92 (d, J = 4 Hz, 2H: (CHCH)2), 1.17 (s, 6H: NH(C(CH3CH3))2), 1.12 (s, 6H: −(CH3CH3)2), 0.99 (t, J = 11.8 Hz, 2H: (CHCH)2−).
13C NMR (CDCl3, 75 Hz) δ [ppm]: 29.0, 35.0, 44.5, 51.6, 54.9, 71.9, 73.9; HRMS (ESI-TOF) m/z: [M + H]+ calculated for C12H22NO+, 196.1696; found: 196.1716.
Click Reaction, 5a, 5b (Scheme 3)
4: (2.91 g, 15.0 mmol) was dissolved in 15 mL of MeOH. Then, 12.5 mmol of azide 3a (4.62 g) or 3b (4.92 g) was added to the solution. Afterward, 0.312 g (1.95 mmol, 10%) of CuSO4 was dissolved in 2 mL of water and added to the solutions. Then, 2.38 g (37.4 mmol) of Cu was added to the solutions and reactions continued overnight at room temperature. After filtration to remove the copper particles, the solution passed through Sorbtech CR20-01 to remove Cu(I) and the anion-exchange resin to replace Br− with Cl−.
5a: 1H NMR (D2O, 300 Hz) δ [ppm]: 8.22 (s, 1H: −CHN–N=N), 5.057 (s, 2H: OCH2−), 4.72 (t, J = 6.1 Hz, 2H: −CH2), 4.01 (t, J = 6 Hz, 2H: −CH2), 4.09–4.24 (m, 1H: −CHO), 3.31 (t, J = 7.4 Hz, 2H: N+CH2−), 3.18 (s, 6H: N+(CH3)2), 2.30 (d, J = 3.5 Hz, 1H: −CHCH), 2.25 (d, J = 3.5 Hz, 1H: −CHCH), 1.61 (m, 2H: −CH2CH2), 1.50 (s, 6H: C(CH3CH3)2), 1.48 (s, 6H: C(CH3CH3)2), 1.23–1.38 (m, 18H: −(CH2)9CH3), 1.18 (t, J = 11.1 Hz, 2H, (CHCH)2−), 0.88 (t, J = 5.5, 3H: −CH3).
13C NMR (D2O, 75 Hz) δ [ppm]: 13.8, 22.5, 26.3, 28.7, 29.4, 29.5, 30.3, 31.8, 41.9, 44.1, 51.6, 54.5, 61.4, 64.4, 71.7, 125.4, 145.1; HRMS (ESI-TOF) m/z: [M – Cl]+ calculated for C28H56N5O+, 478.4479; found: 478.4482.
Purity based on QNMR result (internal standard: maleic acid) > 99%.
5b: 1H NMR (D2O, 300 Hz) δ [ppm]: 8.22 (s, 1H: −CHN–N=N), 5.057 (s, 2H: OCH2−), 4.72 (t, J = 6.1 Hz, 2H: −CH2), 4.01 (t, J = 6 Hz, 2H: −CH2), 4.09–4.24 (m, 1H: −CHO), 3.31 (t, J = 7.4 Hz, 2H: N+CH2−), 3.18 (s, 6H: N+(CH3)2), 2.30 (d, J = 3.5 Hz, 1H: −CHCH), 2.25 (d, J = 3.5 Hz, 1H: −CHCH), 1.61 (m, 2H: −CH2CH2), 1.50 (s, 6H: C(CH3CH3)2), 1.48 (s, 6H: C(CH3CH3)2), 1.23–1.38 (m, 22H, −(CH2)11CH3), 1.18 (t, 2H, J = 11.1 Hz, 2H, (CHCH)2−), 0.88 (t, J = 5.5, 3H: −CH3).
13C NMR (D2O, 75 Hz) δ [ppm]: 14.0, 22.7, 27.2, 29.1, 29.5, 29.7, 30.2, 32.0, 42.9, 43.1, 51.9, 52.8, 60.8, 64.13, 72.6, 125.4, 145.4; HRMS (ESI-TOF) m/z: [M – Cl]+ calculated for C30H60N5O+, 506.4792; found: 506.4788.
Purity based on QNMR result (internal standard: maleic acid) > 97%.
Chlorination of Compounds 5a and 5b (Scheme 3)
5a or 5b (500 mg) was dissolved in 10 mL of water/acetone (2:8 by volume), then 3 equiv of tert-butyl hypochlorite (450 μL) was added to vials, which completely wrapped with aluminum foil at 0 °C. Reactions continued for 60 min. Afterward, air flow was employed to remove acetone and unreacted tert-butyl hypochlorite, and water was removed using vacuum.
6a: 1H NMR (CDCl3, 300 Hz) δ [ppm]: 8.53 (s, 1H: −CHN–N=N), 5.28 (s, 2H: OCH2−), 4.55 (t, J = 6.3 Hz, 2H: −CH2), 4.36 (t, J = 6.4 Hz, 2H: −CH2), 3.68–3.81 (m, 1H: −CHO), 3.4 (t J = 7.1 Hz, 2H: N+CH2−), 3.34 (s, 6H: N+(CH3)2), 2.1 (d, J = 3.1 Hz, 1H: −CHCH), 2.05 (d, J = 3.1 Hz, 1H: CHCH), 1.52 (m, 2H: −CH2CH2), 1.23–1.38 (m, 32H: −(CH2)9CH3, C(CH3)2 and −CHCH), 0.88 (t, J = 6.5, 3H: −CH3).
13C NMR (CDCl3, 75 Hz) δ [ppm]: 14.1, 22.5, 26.3, 29.1, 29.3, 29.5, 30.9, 31.9, 33.2, 44.2, 45.6, 51.5, 62.6, 70.4, 77.1, 125.1, 145.8; HRMS (ESI-TOF) m/z: [M – Cl]+ calculated for C28H55ClN5O+, 512.4090; found: 512.4080.
Purity based on QNMR result (internal standard: maleic acid) > 98%.
6b: 1H NMR (CDCl3, 300 Hz) δ [ppm]: 8.5 (s, 1H: −CHN–N=N), 5.24 (s, 2H: OCH2−), 4.58 (t, J = 6.4 Hz, 2H: −CH2), 4.32 (t, J = 6.4 Hz, 2H: −CH2), 3.7–3.83 (m, 1H: −CHO), 3.4 (t, J = 7.6 Hz, 2H: N+CH2−), 3.33 (s, 6H: N+(CH3)2), 2.10 (d, J = 3.3 Hz, 1H: −CHCH), 2.05 (d, J = 3.3 Hz, 1H: CHCH), 1.54 (m, 2H: −CH2CH2), 1.11–1.38 (m, 36H: −(CH2)11CH3, C(CH3)2 and −CHCH), 0.88 (t, J = 6.5, 3H: −CH3).
13C NMR (CDCl3, 75 Hz) δ [ppm]: 14.1, 22.5, 26.2, 29.2, 29.4, 29.6, 29.7, 31.9, 33.3, 44.4, 45.6, 51.6, 62.7, 70.4, 77.1, 125.1, 145.8; HRMS (ESI-TOF) m/z: [M – Cl]+ calculated for C30H59ClN5O+, 540.4403; found: 540.4386.
Purity based on QNMR result (internal standard: maleic acid) > 95%.
Copper Analysis
Antibacterial effect of copper has been reported in many studies.28 Copper has been used as the catalyst for click reaction, and this copper can interfere the antibacterial activity of compounds. Therefore, copper removal to the noneffective concentration was crucial. To this end, Sorbtech CR20-01 beads have been used. Copper concentration in compounds was measured using ICP optical emission spectrometer, Varian 725-ES.
Redox Titration
Redox titration is a titration technique in which a redox reaction takes place between an analyte and a titrant.19 A redox indicator and conductometer were used during the titration. In this study, redox titration was carried on, to track the chlorine lost in high protein-loaded fluid (5% FBS) for 6a, 6b, and compound 2.
To this end, 0.25 mmol of each compound dissolved in 10 mL of Milli-Q water to have an aqueous solution of each compound. Afterward, 50 μL of fetal bovine serum (FBS) was added to 950 μL of the prepared solution and mix them on an orbital shaker for different time frames (1, 5, 20, 60, 90, and 120 min). Mixture (100 μL) was added to a mixture of 30 mL of Milli-Q water, 2 mL of 5% acetic acid buffer solution, 1 g (6 mmol) potassium iodide, and five drops of freshly prepared 1% starch solution to get a solution with a dark purple color. Sodium thiosulfate standard solution (0.001 N) was used to titrate (dropwise addition) the mixture using buret. The titration stopped when the solution color turned clear, and the chlorine content was calculated as below
where Mw, m, and Vt stand for molecular weight (g/mol), mass of the compound (g), and buret reading (mL), respectively.
Quantitative Antimicrobial Assay (in PBS and High Protein Medium)
A bacterial suspension was prepared at a concentration of 108 CFU/mL in phosphate-buffered saline (PBS, 0.1 M, pH 7.4) using 0.5 McFarland standard. After 100 times dilution, 20 μL of diluted suspension was added to 60 mL of different broths based on the bacterial strains, Tryptone Soy Broth in the case of MRSA and Mueller Hinton Broth for P. aeruginosa and E. coli, followed by 18 h incubation at 37 °C. After incubation, 50 μL of bacterial suspension was added to a solution of 5 [Cl]+ ppm (141.0 μM) to 20 mL of PBS (0.1 M, pH 7.4) and 15 [Cl]+ ppm (423.1 μM) to a solution of 5% FBS in PBS and incubated on the orbital mixer at 37 °C. At each time point (1, 3, 5, 10, 20, 30, 60 min), 150 μL of mixture was added to 150 μL of neutralizer solution (PBS buffer consisting of 1.4% (w/v) l-α-phosphoatidylcholine, 10% (w/v) Tween 80, 1% peptone, and 0.316% sodium thiosulfate) to deactivate antimicrobial agent. Bacterial suspension (30 μL) and its serial dilutions were plated on Tryptone Soya Agar plates and incubated for 24 h, followed by colony counting. To eliminate the effect of bacterial growth in HPM and/or bacterial death due to malnutrition in PBS at each time point, there was a negative control. The bacterial log reduction was calculated as following
Lecithin, tween, and peptone have been proven to be effective in quenching QACs.11,29,30 We have also run adequate inactivation tests to confirm thorough quenching of our biocides at the tested concentrations (141.0 and 423.1 μM). Specially, we plated bacterial suspensions of one series of antibacterial experiments 30 and 90 min after mixing with the neutralizer, and no further decrease of bacterial concentration was observed from the enumeration results, indicating thorough quench of all biocides.
To compare the antibacterial efficacy of the synthesized biocides with free chlorine, an antibacterial test was also conducted against methicillin-resistant S. aureus (MRSA), P. aeruginosa, E. coli, and wild-type P. aeruginosa using sodium hypochlorite at two different concentrations (5 and 10 ppm [Cl+] (141.0 and 282.1 μM)) in PBS and two concentrations (15 and 30 ppm [Cl+] (423.1 and 864.3 μM)) in HPM.
Acknowledgments
The authors are grateful for the financial support from the Collaborative Health Research Project (CHRP) operating grant (Grant no.: CHRP 413713-2012) and the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery grant (Grant no.: RGPIN/04922-2014).
The authors declare no competing financial interest.
References
- Neill J.Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. In Review on Antimicrobial Resistance; O’Neill J., Ed.; Wellcome Trust: London, 2014. [Google Scholar]
- Davin-Regli A.; Pagès J. Cross-resistance between biocides and antimicrobials: an emerging question. Rev. Sci. Tech. 2012, 31, 89–104. 10.20506/rst.31.1.2101. [DOI] [PubMed] [Google Scholar]
- Chuanchuen R.; Beinlich K.; Hoang T. T.; Becher A.; Karkhoff-Schweizer R. R.; Schweizer H. P. Cross-Resistance between Triclosan and Antibiotics in Pseudomonas aeruginosa Is Mediated by Multidrug Efflux Pumps: Exposure of a Susceptible Mutant Strain to Triclosan Selects nfxB Mutants Overexpressing MexCD-OprJ. Antimicrob. Agents Chemother. 2001, 45, 428–432. 10.1128/AAC.45.2.428-432.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffet-Bataillon S.; Tattevin P.; Bonnaure-Mallet M.; Jolivet-Gougeon A. Emergence of resistance to antibacterial agents: the role of quaternary ammonium compounds—a critical review. Int. J. Antimicrob. Agents 2012, 39, 381–389. 10.1016/j.ijantimicag.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Shamsudin M. N.; Alreshidi M.; Hamat R.; Alshrari A.; Atshan S.; Neela V. High prevalence of qacA/B carriage among clinical isolates of meticillin-resistant Staphylococcus aureus in Malaysia. J. Hosp. Infect. 2012, 81, 206–208. 10.1016/j.jhin.2012.04.015. [DOI] [PubMed] [Google Scholar]
- Müller A.; Rychli K.; Zaiser A.; Wieser C.; Wagner M.; Schmitz-Esser S. The Listeria monocytogenes transposon Tn 6188 provides increased tolerance to various quaternary ammonium compounds and ethidium bromide. FEMS Microbiol. Lett. 2014, 361, 166–173. 10.1111/1574-6968.12626. [DOI] [PubMed] [Google Scholar]
- Wassenaar T.; Ussery D.; Nielsen L.; Ingmer H. Review and phylogenetic analysis of qac genes that reduce susceptibility to quaternary ammonium compounds in Staphylococcus species. Eur. J. Microbiol. Immunol. 2015, 5, 44–61. 10.1556/EuJMI-D-14-00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings M. C.; Buttaro B. A.; Minbiole K. P.; Wuest W. M. Bio-organic investigation of multicationic antimicrobials to combat qac-resistant Staphylococcus aureus. ACS Infect. Dis. 2015, 1, 304–309. 10.1021/acsinfecdis.5b00032. [DOI] [PubMed] [Google Scholar]
- Schallenhammer S. A.; Duggan S. M.; Morrison K. R.; Bentley B. S.; Wuest W. M.; Minbiole K. P. Hybrid BisQACs: Potent Biscationic Quaternary Ammonium Compounds Merging the Structures of Two Commercial Antiseptics. ChemMedChem 2017, 12, 1931–1934. 10.1002/cmdc.201700597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning C.; Li L.; Logsetty S.; Ghanbar S.; Guo M.; Ens W.; Liu S. Enhanced antibacterial activity of new “composite” biocides with both N-chloramine and quaternary ammonium moieties. RSC Adv. 2015, 5, 93877–93887. 10.1039/C5RA15714E. [DOI] [Google Scholar]
- Li L.; Pu T.; Zhanel G.; Zhao N.; Ens W.; Liu S. New Biocide with Both N-Chloramine and Quaternary Ammonium Moieties Exerts Enhanced Bactericidal Activity. Adv. Healthcare Mater. 2012, 1, 609–620. 10.1002/adhm.201200018. [DOI] [PubMed] [Google Scholar]
- De Silva M.; Ning C.; Ghanbar S.; Zhanel G.; Logsetty S.; Liu S.; Kumar A. Evidence that a novel quaternary compound and its organic N-chloramine derivative do not select for resistant mutants of Pseudomonas aeruginosa. J. Hosp. Infect. 2015, 91, 53–58. 10.1016/j.jhin.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Arnitz R.; Sarg B.; Ott H. W.; Neher A.; Lindner H.; Nagl M. Protein sites of attack of N-chlorotaurine in Escherichia coli. Proteomics 2006, 6, 865–869. 10.1002/pmic.200500054. [DOI] [PubMed] [Google Scholar]
- Yoon J.; Jekle A.; Najafi R.; Ruado F.; Zuck M.; Khosrovi B.; Memarzadeh B.; Debabov D.; Wang L.; Anderson M. Virucidal mechanism of action of NVC-422, a novel antimicrobial drug for the treatment of adenoviral conjunctivitis. Antiviral Res. 2011, 92, 470–478. 10.1016/j.antiviral.2011.10.009. [DOI] [PubMed] [Google Scholar]
- Gilbert P.; Moore L. Cationic antiseptics: diversity of action under a common epithet. J. Appl. Microbiol. 2005, 99, 703–715. 10.1111/j.1365-2672.2005.02664.x. [DOI] [PubMed] [Google Scholar]
- Summers F. A.; Quigley A. F.; Hawkins C. L. Identification of proteins susceptible to thiol oxidation in endothelial cells exposed to hypochlorous acid and N-chloramines. Biochem. Biophys. Res. Commun. 2012, 425, 157–161. 10.1016/j.bbrc.2012.07.057. [DOI] [PubMed] [Google Scholar]
- Worley S. D.; Williams D.; Crawford R. A. Halamine water disinfectants. Crit. Rev. Environ. Control 1988, 18, 133–175. 10.1080/10643388809388345. [DOI] [Google Scholar]
- Barnes K.; Liang J.; Worley S.; Lee J.; Broughton R.; Huang T. Modification of silica gel, cellulose, and polyurethane with a sterically hindered N-halamine moiety to produce antimicrobial activity. J. Appl. Polym. Sci. 2007, 105, 2306–2313. 10.1002/app.26280. [DOI] [Google Scholar]
- Ruparelia J. P.; Chatterjee A. K.; Duttagupta S. P.; Mukherji S. Strain specificity in antimicrobial activity of silver and copper nanoparticles. Acta Biomater. 2008, 4, 707–716. 10.1016/j.actbio.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Gyawali R.; Ibrahim S. A.; Abu Hasfa S. H.; Smqadri S. Q.; Haik Y. Antimicrobial activity of copper alone and in combination with lactic acid against Escherichia coli O157: H7 in laboratory medium and on the surface of lettuce and tomatoes. J. Pathog. 2011, 2011, 650968 10.4061/2011/650968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jono K.; Takayama T.; Kuno M.; Higashide E. Effect of alkyl chain length of benzalkonium chloride on the bactericidal activity and binding to organic materials. Chem. Pharm. Bull. 1986, 34, 4215–4224. 10.1248/cpb.34.4215. [DOI] [PubMed] [Google Scholar]
- Dong A.; Wang Y.-J.; Gao Y.; Gao T.; Gao G. Chemical insights into antibacterial N-halamines. Chem. Rev. 2017, 117, 4806–4862. 10.1021/acs.chemrev.6b00687. [DOI] [PubMed] [Google Scholar]
- Sonohara R.; Muramatsu N.; Ohshima H.; Kondo T. Difference in surface properties between Escherichia coli and Staphylococcus aureus as revealed by electrophoretic mobility measurements. Biophys. Chem. 1995, 55, 273–277. 10.1016/0301-4622(95)00004-H. [DOI] [PubMed] [Google Scholar]
- Kaur R.; Liu S. Antibacterial surface design–Contact kill. Prog. Surf. Sci. 2016, 91, 136–153. 10.1016/j.progsurf.2016.09.001. [DOI] [Google Scholar]
- Langsrud S.; Sidhu M. S.; Heir E.; Holck A. L. Bacterial disinfectant resistance—a challenge for the food industry. Int. Biodeterior. Biodegrad. 2003, 51, 283–290. 10.1016/S0964-8305(03)00039-8. [DOI] [Google Scholar]
- Taylor J. H.; Rogers S.; Holah J. A comparison of the bactericidal efficacy of 18 disinfectants used in the food industry against Escherichia coli O157: H7 and Pseudomonas aeruginosa at 10 and 20 °C. J. Appl. Microbiol. 1999, 87, 718–725. 10.1046/j.1365-2672.1999.00916.x. [DOI] [PubMed] [Google Scholar]
- McDonnell G.; Russell A. D. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 1999, 12, 147–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copper C. L.; Koubek E. Analysis of an Oxygen Bleach: A Redox Titration Lab. J. Chem. Educ. 2001, 78, 652 10.1021/ed078p652. [DOI] [Google Scholar]
- Quisno R.; Gibby I. W.; Foter M. J. A neutralizing medium for evaluating the germicidal potency of the quaternary ammonium salts. Am. J. Pharm. 1946, 118, 320. [PubMed] [Google Scholar]
- Ioannou C. J.; Hanlon G. W.; Denyer S. P. Action of disinfectant quaternary ammonium compounds against Staphylococcus aureus. Antimicrob. Agents Chemother. 2007, 51, 296–306. 10.1128/AAC.00375-06. [DOI] [PMC free article] [PubMed] [Google Scholar]