 This review provides a detailed examination and comparison of the diverse non-covalent albumin-binding ligands developed until now to extend the half-life of different small biotherapeutics.
This review provides a detailed examination and comparison of the diverse non-covalent albumin-binding ligands developed until now to extend the half-life of different small biotherapeutics.
Abstract
Peptides and small protein scaffolds are gaining increasing interest as therapeutics. Similarly to full-length antibodies, they can bind a target with a high binding affinity and specificity while remaining small enough to diffuse into tissues. However, despite their numerous advantages, small biotherapeutics often suffer from a relatively short circulating half-life, thus requiring frequent applications that ultimately restrict their ease of use and user compliance. To overcome this limitation, a large variety of half-life extension strategies have been developed in the last decades. Linkage to ligands that non-covalently bind to albumin, the most abundant serum protein with a circulating half-life of ∼19 days in humans, represents one of the most successful approaches for the generation of long-lasting biotherapeutics with improved pharmacokinetic properties and superior efficacy in the clinic.
1. Introduction
Peptides and small protein scaffolds have numerous potential benefits as biotherapeutics, including a high binding affinity, exquisite target specificity, low toxicity, and a relatively small size (<50 kDa) that enables them to diffuse into otherwise inaccessible spaces.1–3 However, their direct application as drugs is often hampered by their rapid renal clearance that results in a short systemic half-life. Though there are some peptide drugs that do not need half-life extenders as they exert their desired pharmacological effect at the administration site (e.g. toxin-derived peptide drugs),4 most peptides that have the potential to function as critical therapeutics are not available in the body long enough to be effective.1–3 Thus, maintaining effective concentrations requires high dosages and frequent injections, affecting patient compliance.5 In the last decades, a diverse set of strategies has been developed to prolong the half-lives of peptides and small proteins from minutes to several hours or even days. These approaches include conjugation to synthetic and natural polymers and direct linkage to genetically encoded, unstructured polypeptides and long-lived serum proteins.5,6 Additionally, non-covalent binding to these long-lived endogenous proteins, such as serum albumin,7–9 immunoglobulin (IgG),10–16 neonatal Fc receptor (FcRn),17–19 transthyretin,20 transferrin and its receptor,21–23 can enhance the pharmacokinetic properties of small biotherapeutics. In this approach, peptides and small proteins are directly connected to high-affinity binding moieties that non-covalently tether them to the serum proteins after injection, thus impairing their renal filtration. The affinity of the reversible ligands determines the percentage of free and bound bioactive molecule and balances the fraction available at the target site, which is irrelevant if the target is in the plasma where it can be acted upon by free or bound therapeutic. While most of these strategies have been thoroughly described elsewhere,5,6 this review focuses exclusively on ligands that bind non-covalently to serum albumin. The intrinsic capability of albumin to act as a non-covalent “taxi” for a plethora of exogenous and endogenous molecules has enabled the development of small biotherapeutics with an extended half-life and superior efficacy in the clinic.8,9
2. Albumin as carrier
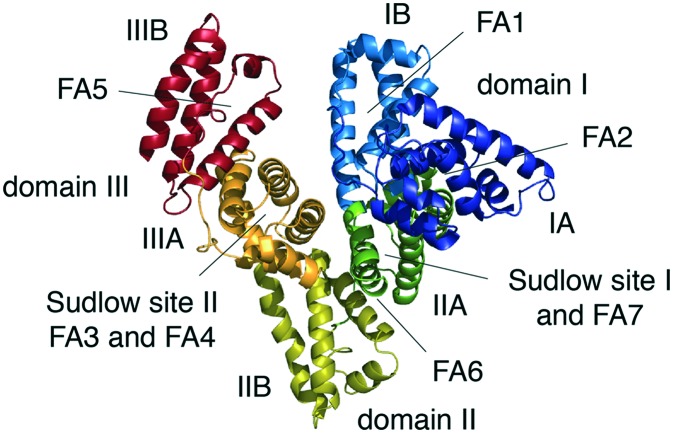
Albumin is the most abundant protein in plasma, with an average plasma concentration of ∼40 g L–1 (600 μM), and it possesses remarkable solubility and stability.8,24 It is a non-glycosylated, single-chain polypeptide comprising 585 amino acids in its mature form for a molecular weight of 66.5 kDa. The amino acid composition is characterized by a high percentage of ionic amino acids, with 83 and 98 positively and negatively charged residues, respectively.24 The resulting negative net charge highly increases the solubility of albumin in aqueous solution. The protein also contains 17 disulfide bridges that contribute to its stability and a single odd cysteine residue in position 34, which alone makes up 80% of the free thiols in plasma.7,24 The secondary structure of albumin is composed of 67% α-helix and no β-sheet elements, and the tertiary structure forms a monomer with a globular heart-shape for dimensions of 80 × 80 × 30 Å and a hydrodynamic diameter of approximately 5 nm.9,24 The protein has three homologous domains, named I, II and III, with similar amino acid sequences and structures (Fig. 1).25 Despite these similarities, each domain has its own unique interactions with the others, and the relative orientations create an asymmetrical module with several different ligand-binding sites distributed throughout its tertiary structure. Each domain contains two subdomains, denoted A and B, with four and six α-helices, respectively.25 The overall structure is extremely stable over a wide pH range from 4.0 to 9.0, though its fold and shape can undergo a number of structural changes in response to changes in pH.26
Fig. 1. Crystal structure of human serum albumin. The crystal structure of human serum albumin (PDB code 1AO6) representing the domains, subdomains, fatty acids and Sudlow's binding sites. The subdomains of albumin are shown in dark blue (IA), light blue (IB), dark green (IIA), olive (IIB), orange (IIIA) and dark red (IIIB). Legend: FA = fatty acid binding site.
The plasma concentration of albumin is the result of a sophisticated equilibrium among different synthesis, degradation, and distribution mechanisms in the body.7,24 About 10–15 grams of albumin are produced in the liver per day. A healthy person of 70 kilograms has approximately 360 grams of albumin, one third of which is located in the plasma, with the remaining amount found in the extravascular compartment.8,9 Several tissues catabolize an amount of albumin daily that is comparable to the produced amount, and this leaves a maximum circulatory half-life of 19 days in humans.8,24 This long half-life is mainly related to its structural properties and its ability to bind FcRn.27 Renal elimination is mostly limited by the molecular weight and net negative charge, while the pH-dependent recycling process mediated by the FcRn avoids intracellular lysosomal degradation. Mutagenesis and co-crystallization studies have revealed that the binding site for FcRn on albumin is localized on domain III and that it does not overlap with the receptor's IgG binding site.27 Notably, the FcRn is also involved in the rescue of albumin from glomerular filtration.27 In addition to FcRn, albumin interacts with other cell-surface receptors that are also responsible for its recycling, degradation, and intracellular transport.8,28 All of these receptors, along with its hydrodynamic radius, overall negative charge, and high concentration, ultimately contribute to albumin homeostasis and its long half-life in humans, allowing it to take part in numerous and important physiological activities. These include the maintenance of the oncotic pressure and plasma pH levels as well as the binding, transport, and distribution of a large variety of ligands.8,24 Both covalent and non-covalent binding to albumin are widely exploited, making it a suitable carrier for several different therapeutics. In particular, the capacity of albumin to extravasate from the bloodstream to reach the lymphatic system and accumulate in cancerous or inflamed areas has aroused a strong clinical interest in using it as a platform to extend the half-life of therapeutic peptides and small proteins.8,9
3. Albumin-binding small organic compounds
Serum albumin can intrinsically bind a large diversity of small endogenous and exogenous organic molecules, shielding their hydrophobic character and strongly increasing their solubility in plasma.8,24 In particular, albumin acts as the key lipid delivery vehicle for the tissues, binding up to seven molecules of long fatty acids simultaneously (Fig. 1).29 Two sites for binding medium-chain fatty acids have also been identified, yielding a total of nine distinct fatty acid binding locations distributed throughout its tertiary structure.30 Short- to medium-length fatty acids (6 to 12 carbons) bind albumin with affinities between 0.5 and 60 μM, while the longest ones (14 to 18 carbons) have 10-fold higher affinities (below 50 nM).31,32 The ability of serum albumin to bind long fatty acids with a high affinity inspired the use of post-translational acylation as a safe and natural platform for prolonging the half-life and the mode of action of peptides and small proteins.5,6 Currently, fatty acids and derivatives represent the only successful example of non-covalent albumin-binding ligands that has led to the development of four approved drugs from Novo Nordisk: insulin detemir (Levemir®), insulin degludec (Tresiba®), liraglutide (Victoza® and Saxenda®), and semaglutide (Ozempic®), used for the treatment of diabetes mellitus (type 1 and 2) and/or obesity.6,33 The properties and applications of lipid tails for extending the circulating half-life of small biotherapeutics has been extensively described elsewhere6,33 and it will not be further discussed here.
In addition to fatty acids, serum albumin can bind numerous other small organic compounds by exploiting two major structurally dissimilar binding sites, known as Sudlow sites I and II (Fig. 1).34,35 Site I comprises a central zone from which three distinct compartments extend, while site II is smaller and consists of a single narrow cleft.35 Site I, also known as the warfarin–azapropazone binding site, usually accommodates dicarboxylic acids and/or bulky heterocycles carrying a central negative charge, whereas site II, also known as the benzodiazepine binding site, can discriminate ligands based on their size and stereoselectivity. Generally, aromatic carboxylic acids with a peripheric negative charge, that is distantly located from the hydrophobic center, lodge in this site.34,35 Because of their ability to non-covalently bind serum albumin, several organic moieties that are structurally similar to exogenous drugs (e.g. warfarin, ibuprofen and diazepam) and dye molecules (e.g. Evans blue) have also been used. For example, scientists at Genentech coupled several organic compounds, based on sulfonamide or phosphate ester moieties, to a peptide inhibitor of the coagulation factor VIIa (fVIIa).36,37 Koehler et al. identified novel albumin-binding affinity tags by screening a focused library of compounds appended to a model tetrapeptide and evaluated the resulting compounds' ability to bind to serum albumin immobilized on a column.36 The most promising hit, a naphthalene acyl sulfonamide tag (Fig. 2a), was coupled to a peptide with a potent affinity for fVIIa, and its activity was assessed in vitro and in vivo. The tagged peptide showed comparable affinities for fVIIa in the presence of human serum albumin, and when administered to rabbits, it exhibited a 4-fold increased plasma half-life compared to the peptide alone (Table 1).36 Similarly, Zoebel et al. envisioned identifying novel albumin-binding affinity tags by screening a focused library of phosphate esters,37 mainly due to the fact that the diphenylcyclohexanol phosphate ester moiety (Fig. 2b) of the FDA-approved contrast agent Gadofosveset trisodium (trade name Vasovist, Ablavar) bound to site II of albumin.38 The best conjugate retained its affinity for fVIIa and had a half-life 7-fold longer than the peptide tagged with naphthalene acyl sulfonamide moiety and 53-fold longer than the unmodified peptide in rabbits (Table 1).37
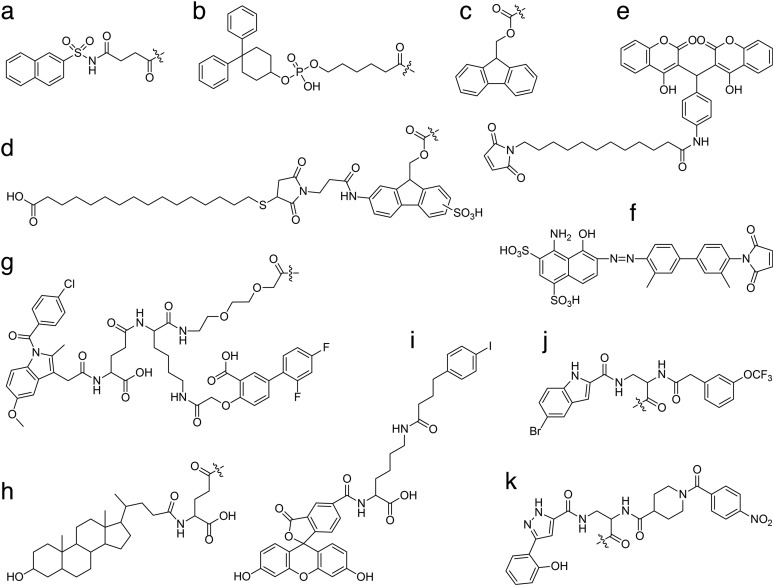
Fig. 2. Chemical structures of albumin-binding small organic compounds. a) naphthalene acyl sulfonamide moiety; b) diphenylcyclohexanol phosphate ester moiety; c) 9-fluorenylmethoxycarbonyl (Fmoc) moiety; d) Fmoc derivative linked to a 16-sulfanylhexadecanoic acid through a maleimide group; e) dicoumarol derivative with a maleimide group; f) Evans blue derivative with a maleimide group; g) divalent diflunisal-indomethacin moiety linked through a γGlu-Lys dipeptide coupled to a unit of 8-amino-3,6-dioxaoctanoic acid (O2Oc); h) lithocholic acid coupled to a γGlu linker; i) 6-(4-(4-iodophenyl)butanamido)hexanoate coupled to carboxyfluorescein through a d-Lys; j) A083/B134 and k) A099/B344.
Table 1. Albumin-binding small organic compounds.
| Cargo |
Albumin-binding molecule | Conjugate |
Factor | Ref. | ||
| Type | τ 1/2 | Affinity | τ 1/2 | |||
| fVIIa inhibitor† | 7.6 min (i.v.)a | Naphthalene acylsulfonamide | n.a. | 30 min (i.v.)a | 4a | 36 |
| 4.2 min (i.v.)a | Diphenylcyclohexanol phosphate ester | n.a. | 222 min (i.v.)a | 53a | 37 | |
| Insulin† | 3.3 h (i.v.)b | Fmoc derivative with a 16-sulfanyl hexadecanoic acid | 3.9c μM* | 17 h (i.v.)b | 5b | 40 |
| Exendin-4† | 2.8 h (s.c.)b | Dicoumarol derivative with maleimide | n.a. | 19–22 h (s.c.)b | 7–8b | 41 |
| 5.2 h (s.c.)d | Evans blue derivative with maleimide | n.a. | 36.3 h (s.c.)d | 7d | 43 | |
| GLP-1† | n.a. | Diflunisal-γGlu-Lys(±O2Oc)-indomethacin | n.a. | 5 h (s.c.)d | Liraglutide 4.4 h (s.c.)d | 46 |
| scFv§ | 20–30 min (i.v.)d | 6-(4-(p-Iodophenyl) butanamido) hexanoate | 3.2c, 3.6d μM | 16.6 h (i.v.)d | 33–50d | 52 |
| Fluorescein‡ | 4.6 min (i.v.)d | 108c, 118d nM# | 495 min (i.v.)d | 108d | 48 | |
| 330c nM$ | ||||||
| Gd-DTPA‡ | 8.6 min (i.v.)d | 3.3c μM | 408 min (i.v.)d | 47d | ||
The 9-fluorenylmethoxycarbonyl (Fmoc) organic moiety (Fig. 2c) associates with multiple albumin species with affinities of around 20 μM. Importantly, the speed of hydrolysis of Fmoc is significantly reduced (t1/2 ∼ 24 h) when incubated at 37 °C in aqueous buffer (pH 7.4) in the presence of a low concentration of serum albumin.39 Fridkin and co-workers exploited these properties to generate Fmoc derivatives (named FMS) that could react with the amino groups of therapeutic peptides and proteins, converting them into inactive pro-drugs with desirable pharmacokinetic properties that could reactivate in body fluids. An insulin derivative (FMS3-insulin) in which three FMS moieties were linked to three amine functional groups (Gly1 of chain A, Phe1 and Lys29 of chain B) recovered full biological potency only after four days of incubation in normal human serum at pH 7.4 and 37 °C. A single administration of this FMS3-insulin to streptozocin (STZ)-treated rats lowered circulating glucose levels for a prolonged period of 45 h.39 Afterwards, Sasson et al. developed an optimal reversible heterobifunctional agent that included a hydrolysable FMS derivative linked to a 16-sulfanylhexadecanoic acid with a higher affinity for albumin (Fig. 2d). An insulin derivative conjugated to this novel probe showed a half-life in rats exceeding more than five times that obtained for insulin alone (Table 1). When tested in mice, this derivative lowered glucose levels for a period of over 24 h.40
Han et al. has shown that conjugation of the small molecule dicoumarol (Fig. 2e), a former exogenous anticoagulant drug currently replaced by warfarin, to glucagon-like peptide-1 (GLP-1), an endogenous peptide hormone involved in the control of blood-glucose levels, resulted in an improved in vivo half-life in rats, which was approximately 2- and 7-fold greater than the long-acting GLP-1 analogs liraglutide and exendin-4, respectively (Table 1).41 Importantly, the dicoumarol-GLP-1 conjugate retained receptor activation efficacy, exhibited improved albumin affinity and plasma stability, and had prolonged in vivo antidiabetic effects.41
Evans blue (EB) is an azo dye that binds to site I of serum albumin with low micromolar affinity (2.5 μM) and has a long half-life in the blood.35 Because of its ability to bind albumin, EB has been used clinically for many years to estimate patient blood plasma volume and extravascular protein leakage. Over the last few years, a series of functionalized EB derivatives have been developed as diagnostic MRI and PET imaging tracers for detecting lymph nodes and identifying tumor lesions.42 Recently, novel chemical entities based on the structure of EB have been designed to extend the half-life of therapeutic peptides.43 Conjugation of a truncated EB moiety (Fig. 2f) to the peptide exendin-4 (denoted as Abextide) showed prolonged drug release and an enhanced hypoglycemic effect in a mouse model of type 2 diabetes.43 Abextide had a 7-fold extended half-life and sustained normal glycemia levels four times longer than the native exendin-4 (Table 1). An improved form, named Abextide II, displayed an even higher stability in both solution and powder form.44 Recently, Zhu et al. showed that co-delivery of CpG adjuvant and peptide antigens conjugated to an EB derivative (denoted as AlbiVax) to lymph nodes induced potent and durable T-cell responses in mice. AlbiVax in combination with an immune checkpoint inhibitor significantly reduced the progression of multiple established tumors.45
In an elegant study, Jensen and co-workers investigated divalent small-molecule albumin binders as an approach for improving peptide pharmacokinetics.46 Toward this goal, they performed a proof-of-principle study using GLP-1 analogues conjugated to either or both of the nonsteroidal anti-inflammatory drugs diflunisal and indomethacin (Fig. 2g).46 Diflunisal binds human albumin through both drug sites I and II with affinities of 3 μM, while indomethacin binds mainly to drug site I with an affinity of 0.7 μM.35 Studies on GLP-1 analogues functionalized with diflunisal, indomethacin, or both showed that albumin affinity was increased for the divalent analogue as a result of an avidity effect that arose from combining multiple ligands targeting different sites on serum albumin. In lean mice, the divalent GLP-1 analogue showed superior biological efficacy and a promising gain in circulatory half-life comparable to that reported for liraglutide (Table 1).46 Analogously to liraglutide, binding to albumin both prevented fast renal clearance and also protected the GLP-1 peptide from degradation, a process that occurs instantaneously in mouse plasma and is mediated by the dipeptidyl peptidase IV (DPP-IV).46
Bile acids and derivatives are examples of endogenous molecules used as albumin binders.6,47 The most promising analogue, a lithocholic acid (Fig. 2h) conjugated to insulin (NN344), showed a certain affinity for albumin, and upon injection it formed subcutaneous deposits that were gradually released in the blood. When administered to pigs, NN344 controlled glucose levels for more than 24 h.47 However, this molecule, as well as other cholic acid conjugates, appears not to have been further advanced, probably because of the low solubility of bile acids and their weak affinities for albumin.47
Finally, albumin ligands resembling the structures of known organic small molecules have been identified by Neri and co-workers by screening DNA-encoded chemical libraries.48–51 Dumelin et al. identified several albumin-binding affinity tags all featuring a 4-phenylbutanoic acid moiety with different hydrophobic substituents on the phenyl ring. The best 4-(p-iodophenyl)butyric acid derivative, named 428-d-Lys-FAM (Fig. 2i), displayed a stable non-covalent interaction with both mouse and human serum albumin and exhibited comparable binding affinities (Table 1). Competition experiments made using known ligands indicated that the derivative binds to site II of albumin.48 By using a larger DNA-encoded chemical library (>105 different compounds) composed of structurally diverse carboxylic acid building blocks, Franzini et al. identified several ligands capable of binding human serum albumin with sub-nanomolar binding affinities (A083/B134: 0.4 nM; A077/B286: 0.7 nM; A083/B321: 0.9 nM).50 Replacement of either of the building blocks by an acetamide group drastically decreased the affinity of the compounds for albumin. Particularly, the 5-bromoindole moieties were critical for albumin binding, and their substitution raised the binding affinities by more than 2000-fold. To the best of our knowledge, compound A083/B134 (Fig. 2j) is the tightest small-molecule binder for human albumin reported so far.50 By applying parallel affinity screening of a DNA-encoded chemical library against rat, bovine, and human serum albumin, Franzini et al. identified novel small-molecules with distinctive binding specificities for the individual albumins.51 Compounds with a binding preference to one of the proteins and ligands with an affinity to all three targets were isolated. For each serum albumin, the authors were able to identify a small molecule that bound with an affinity below 10 nM and exhibited at least 10-fold specificity over the other two proteins. Interestingly, the human selective A099/B344 (5.4 nM) is structurally distinct from the previously described small molecules binding human albumin (Fig. 2k).51
Even though numerous sub-nanomolar albumin binders have been recently isolated from these DNA-encoded chemical libraries, only a few 4-(p-iodophenyl)butyric acid derivatives have been characterized in vivo.48 Pharmacokinetic analyses of the best 4-(p-iodophenyl)butyric acid derivative, named Albutag, revealed a long half-life in the bloodstream of up to 24 h after injection.48 Attaching Albutag to an engineered therapeutic antibody fragment (scFv-F8) resulted into a >30-fold longer half-life and higher tumor uptake in a mouse model (Table 1).52 Although efficacious in prolonging the pharmacokinetic properties of small molecules or recombinant proteins,48,52 Albutag appears to have found its major success as portable albumin-binding moiety for the delivery of contrast agents for biomedical imaging. Conjugating Albutag to fluorescein derivatives used for angiographic analysis of the retina increased their half-life by approximately 100-fold in mice (Table 1).48 Similarly, linking Albutag to the magnetic resonance imaging (MRI) contrast agent gadolinium-diethylenetriaminepentaacetic acid (Gd-DTPA, Magnevist) enhanced its half-life by approximately 50-fold (Table 1).48 Analogue approaches have been applied to enhance the tumor-to-non-tumor ratio of multiple tumor-targeting contrast agents.53In vivo single-photon emission computed tomography (SPECT) imaging of an Albutag conjugated to a folic acid that was modified with a 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) chelator for coordination of a therapeutic radioisotope (cm09) showed 5- to 6-fold higher tumor-to-kidney ratios than those obtained with folate radioconjugated ligands that lacked an albumin-binding entity.53 Radionuclide therapy in tumor-bearing mice had positive results in terms of tumor remission and significantly prolonged survival times when compared with untreated controls.53 Babich and colleagues used Albutag to develop a dual-targeting radioiodinated ligand (RPS-027) that bound to both prostate-specific membrane antigen (PSMA) and albumin to enable longer blood circulation times.54 When tested in a preclinical model of prostate cancer, RPS-027 demonstrated a favorable tumor-to-kidney ratio and had a promising therapeutic profile that made it suitable for the targeted radiotherapy (TRT) of prostate cancer.54 The same research group has recently generated a novel class of trifunctional ligands, consisting of the high-affinity PSMA-binding domain, the Albutag, and the DOTA chelator for radiometals, to facilitate the modification of the three moieties independently and ultimately enable the generation of spatial optimized conjugates.55In vivo small-animal positron emission tomography (μPET) imaging demonstrated that the trifunctional ligands had high and persistent tumor uptake with absorbed doses that were four times greater than those observed for a similar compound lacking the albumin-binding moiety.55 A further optimized trifunctional ligand, named 68Ga-RPS-072, had an order of magnitude higher binding affinity for albumin, showed a significantly delayed blood clearance, and had a superior tumor-to-kidney ratio than those of the other radioligands tested in the cancer prostate xenograft model.56 Recently, Umbricht et al. used Albutag to develop novel long-circulating albumin-binding PSMA-targeting PET radioligands by substituting DOTA with 1,4,7-triazacyclononane,1-glutaric acid-4,7-acetic acid (NODAGA), a chelator that has been shown to increase the in vivo stability of longer-lived radionuclides.57 A PSMA-targeting PET radioligand bearing NODAGA (PSMA-ALB-89) had a more favorable in vivo stability and increased tumor accumulation when compared to the same ligand conjugated to DOTA (PSMA-ALB-56).57
4. Albumin-binding peptide ligands
Alternatively to small molecules, an increasing number of peptides have been used as albumin-binding molecules. Indeed, albumin is known to bind not only low molecular weight molecules, but also to peptides and proteins such as insulin, bradykinin, and interferons, and several hundreds of other binders have been identified or predicted.58,59 This observation suggested the application of in vitro techniques for screening peptides that can bind albumin with a high affinity and that could be used as carriers for half-life extension.60–63 In contrast to small chemical moieties, albumin-binding peptides can easily be fused to any protein or peptide, either recombinantly or chemically during solid-phase synthesis.64
Several linear peptides binding human albumin with nanomolar affinities have been identified by Revets and Boutton (WO2011095545).65 The best binder was an 18-amino acids peptide named 89D03 (Ac-WWEQDRDWDFDVFGGGTP-NH2) (Fig. 3a) that binds tightly to human albumin (15 nM).65 Fusing 89D03 to the C-terminus of an anti-HER2 nanobody developed by Ablynx (clone 5F7) prolonged its half-life by more than 7-fold in cynomolgus monkeys. The further addition of two 89D03 peptides in tandem to the C-terminus of the nanobody extended the half-life by more than 3 days (Table 2).65
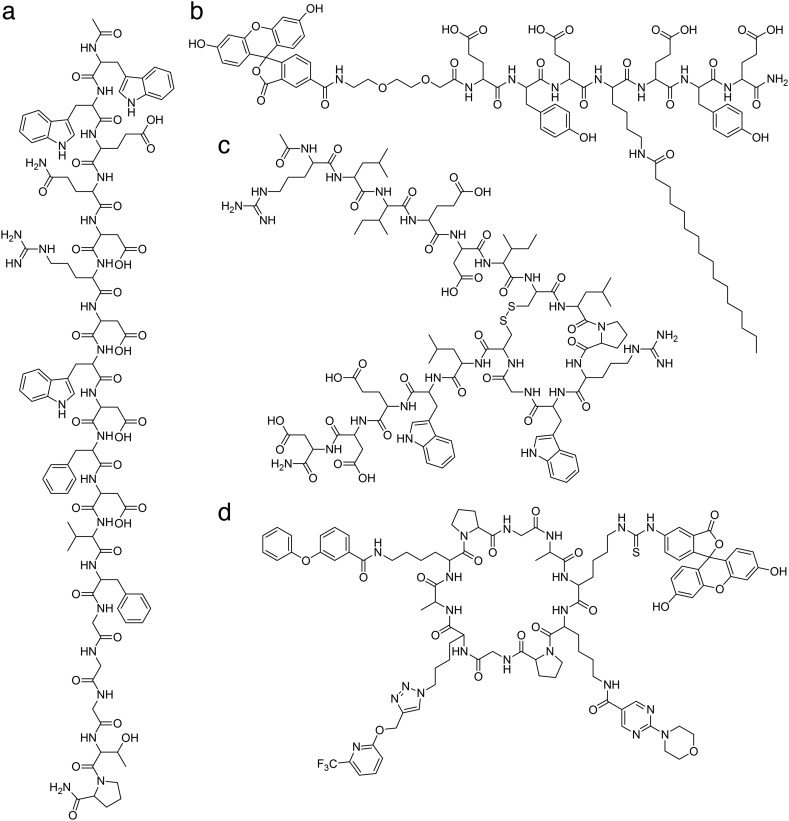
Fig. 3. Chemical structures of albumin-binding peptide ligands. a) linear peptide 89D03: Ac-WWEQDRDWDFDVFGGGTP-NH2; b) acylated heptapeptide F-tag: fluorescein-EYEK(palmitate)EYE-NH2; c) disulfide cyclized peptide SA21: Ac-RLIEDIC[combining low line]LPRWGC[combining low line]LWEDD-NH2; d) head-to-tail cyclized peptide HSA-1: A[combining low line]K*K*PGK*AK*PG[combining low line] with variable lysine (K*).
Table 2. Albumin-binding peptide ligands.
| Cargo |
Albumin-binding molecule | Conjugate |
Factor | Ref. | ||
| Type | τ 1/2 | Affinity | τ 1/2 | |||
| Anti-HER2§ | 4.3 h (i.v.)a | 89D03 (15b nM) | n.a. | Single copy: 31.2 h (i.v.)a | 7a | 65 |
| Double copy: 84.5 h (i.v.)a | 20a | |||||
| huPA inhibitor† | 18 min (i.v.)c | F-tag (39b, 220c, 320d nM) | 168b, 780c nM | 7.4 h (i.v.)c | 25c | 62 |
| hFXIIa inhibitor† | 13 min (i.v.)d | 224a, 1600d nM | 5.2 h (i.v.)d | 24d | ||
| Fab§ | 53 min (i.v.)d | SA21 (467b, 266c, 320d, 7e nM) | 320d nM | 32.4 h (i.v.)d | 37d | 60 |
| 24 min (i.v.)e | 10.4 h (i.v.)e | 26e | ||||
| huPA inhibitor† | 30 min (i.v.)e | 354b, 14e nM | 24 h (i.v.)e | 48e | 67 | |
| Exendin-4† | 30 min (i.v.)c | 610a, 1560b, 210c nM | 2 d (i.v.)a | 22c | 83 | |
| 11 h (i.v.)c | ||||||
Recently, Heinis and co-workers engineered a novel chimeric albumin-binding molecule consisting of a fatty acid combined with a short linear peptide comprised of seven amino acids (Fig. 3b).62 By applying iterative rounds of amino acid substitution and affinity measurement by fluorescence polarization (FP), Zorzi et al. evolved an acylated heptapeptide tag, named F-tag, able to bind human, rat, and rabbit albumin with affinity values of 39, 220, and 320 nM, respectively (Table 2).62 Binding experiments revealed that the N-terminal fluorescein moiety used for FP analysis contributed, along with the palmitate moiety and the specific peptide sequence, to the affinity for albumin. Despite the presence of the long hydrophobic tail of the palmitic acid and the fluorescein, the lipidated F-tag peptide exhibits a high solubility due to the four negative charges in the peptide sequence. Importantly, the F-tag could easily be appended to cargo peptides by automated synthesis on standard peptide synthesizers. Pharmacokinetic experiments showed that the acylated heptapeptide extended the circulation time of two different bicyclic peptides in rats and rabbits by around 25-fold, reaching half-lives of 7 and 5 h, respectively (Table 2).62 Furthermore, the presence of the F-tag improved the proteolytic stability of the cargo peptides over the non-tagged peptides. Finally, conjugation of the F-tag to a picomolar bicyclic peptide inhibitor of the activated human coagulation factor XII (hFXIIa) led to an efficient blockage of the intrinsic coagulation pathway in rabbits for as long as 8 h, suggesting a potential application in humans as anti-thrombotic drug.62
Dennis et al. used phage display to isolate cyclic peptides bearing two fixed cysteine residues and varying loop sizes that bind albumin with high affinity.60 They selected several disulfide-cyclized binders for rabbit, rat, and human albumin, and a clone sharing high binding for all three species was further subjected to affinity maturation. The best albumin binder was an 18-amino acids peptide called SA21 (Ac-RLIEDIC[combining low line]LPRWGC[combining low line]LWEDD-NH2) (Fig. 3c) that bound albumin from different species with nanomolar affinities (Table 2). The pharmacokinetics of SA21 alone was tested in rabbits and showed an 18-fold prolonged half-life compared to an unrelated control peptide. The fusion of SA21 to the C-terminus of an antigen-biding antibody fragment (Fab) efficiently prolonged the half-life of the protein up to 37-fold in rabbits and 26-fold in mice (Table 2).60 The importance of developing high-affinity peptides was confirmed in a follow-up study where SA21 variants with 5- to 10-fold weaker binding to albumin showed two to three times shorter half-lives.66 The capacity of SA21 to improve the half-life of bicyclic peptides was recently investigated by Heinis and co-workers. Through the chemical conjugation of SA21 to UK18, a bicyclic peptide inhibitor of human urokinase-type plasminogen activator (huPA), Angelini et al. showed that non-covalent binding to albumin could improve both pharmacokinetic and metabolic characteristics of the peptide conjugates without interfering with the binding of the corresponding protein target.67 Experiments in mice revealed a ∼50-fold prolonged bicyclic peptide circulation time, and the conjugate remained fully intact and functional for more than 48 h (Table 2).67 In a second study, Pollaro et al. showed that the same conjugate diffused in high concentrations into tumor tissues and was not limited by the high affinity of SA21 for the endogenous murine albumin.68 The SA21 peptide was later applied to other small proteins and exhibited promising preclinical results.69,70
Recently, Neri and co-workers identified specific macrocyclic binders against human serum albumin by exploiting a novel DNA-encoded chemical library displaying multiple diversity elements into one side of a structurally defined β-sheet scaffold (Fig. 3d).63 The two enriched compounds featured the presence of diverse chemical element combinations and revealed binding affinities in the single-digit micromolar range (6.6 and 22 μM).63 Although not yet characterized in vivo, these macrocycles have high potential for the selective in vivo delivery of payloads to tumors.
5. Albumin-binding protein domains
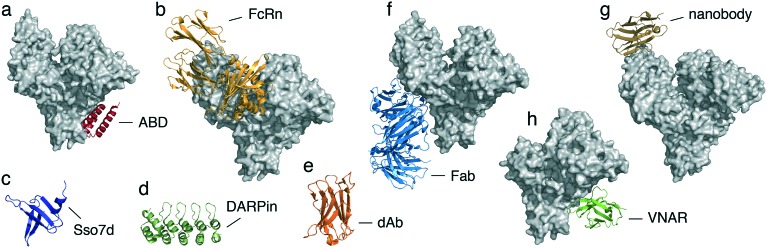
Protein domains usually have a large interaction interface with their target, leading to high binding affinities and specificities – key features for the development of albumin binders. Several albumin-binding domains (ABDs) were discovered in bacterial proteins, the best-characterized of which are derived from protein G of Streptococcus strain GI48 and from protein PAB of Finegoldia magna. These ABDs have 59% homologous amino acid sequence identities, are very stable despite their small size (∼5 kDa), and fold into a three-helix-bundle domain that interacts with serum albumin primarily along one of its faces.71 Importantly, both ABDs target the same binding site on domain II of human serum albumin (Fig. 4a) and do not overlap or interfere with binding to the FcRn-binding site on albumin, which is located in domain III (Fig. 4b). The affinity of ABDs for albumin was strongly improved by phage display and rational design, reaching a binding affinity for human albumin of 50–500 fM (variant ABD035).72 This engineered high-affinity variant, which forms the basis of Albumod, improved the pharmacokinetics of various recombinantly fused proteins, including soluble receptors,73 cytokines,74 bispecific single-chain diabodies (scDb),75,76 lipocalin-derived anticalin® proteins,77 targeted immunotoxins,78,79 and affibody® scaffold molecules.80–82 In a recent study, Levy et al. compared two exendin-4 conjugates obtained by appending either the optimized variant ABD035 or a shorter version of the cyclic peptide SA21 to its C-terminus. Very different affinities for albumin were reported between the two formats, ranging from 5 to 40 pM or 200 to 1500 nM, respectively.83 The SA21-exendin-4 conjugate showed a 10-fold higher potency in vitro compared to the ABD variant, most likely related to differences in steric hindrance. In contrast, the remarkable differences in albumin affinity strongly impacted the pharmacokinetics in monkeys, which resulted in a 7-fold longer half-life for the ABD-exendin-4 conjugate compared to the SA21 variant (14 days vs. 2 days) that potentially counterbalance its reduction in activity, while comparable values were obtained in rats (Tables 2 and 3).83 A de-immunized ABD variant, named ABD094, retained a high affinity for different serum albumin species and is currently being tested in the clinic (NCT02690142).82
Fig. 4. Three-dimensional structures of albumin-binding protein ligands. a) crystal structure of a bacterial ABD scaffold (GA, red) in complex with human serum albumin (grey, PDB code 1TF0); b) crystal structure of the human neonatal Fc receptor (FcRn, light orange) in complex with human serum albumin (grey, PDB code ; 4N0F); c) solution structure of the bacterial protein Sso7d (dark blue, PDB code ; 1SSO); d) crystal structure of a DARPin protein domain (dark green, PDB code 1 MJ0); e) crystal structure of a single-domain antibody (dAb, dark orange, PDB code ; 3BD3); f) crystal structure of the fab domain (CA645, light blue) in complex with human serum albumin (grey, PDB code ; 5FUO); g) crystal structure of a nanobody (Nb.b201, brown) in complex with human serum albumin (grey, PDB code ; 5VNW); h) crystal structure of a VNAR domain (E06, light green) in complex with human serum albumin (grey, PDB code ; 4HGM).
Table 3. Albumin-binding small protein scaffolds.
| Cargo |
Albumin-binding molecule | Conjugate |
Factor | Ref. | ||
| Type | τ 1/2 | Affinity | τ 1/2 | |||
| Exendin-4† | 30 min (i.v.)a | ABD035 (18a, 123b, 16c pM) | 8a, 40b, 5c pM | 16 h (i.v.)a | 32a | 83 |
| 14 d (i.v.)b | ||||||
| Fibronectin domain§ | n.a. | ABDCon (61b, 75c, 3200d pM) | 0.7c, 1.8d nM | 193 h (i.v.)b | n.a. | 84 |
| 60 h (i.v.)d | ||||||
| DARPins drug candidate§ | 0.008 d (i.v.)b | DARPins (91–266a, 63–110b, 11–21c, 56–142d, 77–185e nM) | n.a. | 2.7–20.7 d (i.v.)b | >330b | 89 |
| 21–82 h (i.v.)d | ||||||
| IL-1rα§ | 2 min (i.v.)d | AlbudAbs | 34–600 nMa,c,d | 4.3 h (i.v.)d | 130d | 91 |
| IFN-α2§ | 1.2 h (i.v.)a | 22.6 h (i.v.)a | 19a | 92 | ||
| 1.5 h (s.c.)a | 28.3 h (s.c.)a | |||||
| Anti-TNFR1§ | n.a. | 3.3–23.2 h (i.v.)d | n.a. | 93 | ||
| Exendin-4† | 2.5 h (s.c.)c | 6–10 d (s.c.)c | 58–96c | 94 | ||
| Fab§ | n.a. | dsFv CA645 (54a, 3.3b, 4.6c, 7.1d, 162f nM) | n.a. | 7.9 d (i.v.)b | n.a. | 96, 97 |
| 2.6 d (i.v.)d | ||||||
| Anti-TNF§ | 35–54 min (i.v.)d | Nanobody (22c nM) | n.a. | 2.2 d (i.v.)d | 91d | 99 |
| Anti-IL-6R§ | 4.3 h (i.v.)b | 1.7–6.6 d (i.v.)b | 9–37b | 100 | ||
| Naïve VNAR§ | 0.15 h (i.v.)d | VNAR (E06) (3a, 1c, 6d nM) | n.a. | 22–25 h (i.v.)a | 140–220d | 102 |
| 164–210 h (i.v.)b | ||||||
| 21–33 h (i.v.)d | ||||||
Recently, Jacobs et al. have engineered a novel three-helix-bundle albumin-binding domain, named ABDCon, that exhibits high binding affinity for different serum albumins (Table 3) while maintaining a high solubility and thermal stability (91 °C).84 The fusion of ABDCon to a fibronectin domain (Fn3) resulted in a long terminal half-life ranging from 3 to 8 days in mice and cynomolgus monkeys, respectively (Table 3).84 The capacity of ABDs for prolonging the half-life of conjugates does not correlate with further increased binding affinities for albumin, suggesting an intrinsic limit connected to either the FcRn-mediated recycling or albumin half-life.76,84
Other albumin-binding domains were recently engineered from the bacterial protein Sso7d from the hyperthermophilic archaeon Sulfolobus solfataricus. Sso7d is a small, cysteine-free DNA-binding protein (∼7 kDa, 63 amino acids) with a melting temperature of 98 °C. The three-dimensional structure consists of an oligonucleotide-binding (OB) fold with an incomplete five-stranded β-barrel closed off by a C-terminal α-helix (Fig. 4c).85 Traxlmayr et al. applied yeast display technology to engineer two high-affinity clones, M11.1.2 and M18.2.5, that bound mouse serum albumin with affinities of 7 and 26 nM, respectively.86 Sso7d binders against multiple albumin species have also been engineered by Fisk and colleagues.87 The tighter binder (13A8) had an affinity for albumin of 2.1 μM when displayed on the surface of phage and could discriminate among closely related human and bovine serum albumins.87 Even though numerous Sso7d albumin binders have recently been isolated, none have been characterized in vivo. Given their small size and high thermal stability, we expect the Sso7d-derived albumin-binding domains to have pharmacokinetic properties similar to other ABD domains.
Another strategy is the development of albumin-binding designed ankyrin repeat proteins (DARPins®).88 These scaffolds are derived from naturally occurring ankyrin proteins comprising up to 29 consecutive, tightly packed repeat modules. Each repeat is usually 33-amino acids long and forms a structural unit consisting of a β-turn followed by two antiparallel α-helices. DARPins comprise usually 4 to 5 repeat motifs with molecular masses of about 14 and 18 kDa, respectively (Fig. 4d). Similar to other albumin-binding domains, DARPins exhibit favorable biophysical properties, including a high thermal stability.88 Using ribosome display technology, Binz and co-workers selected several DARPin domains with mid-nanomolar affinities to different serum albumins (Table 3).89 Recombinant fusion of the best-engineered serum albumin-binding domain to a DARPin drug candidate led to a >300-fold prolonged half-life in cynomolgus monkey (Table 3).89 Interestingly, constructs including two DARPin albumin-binding domains with different affinities (2.5 and 11.2 nM) exhibited comparable serum half-lives in mice, indicating that a low nanomolar threshold affinity is sufficient for albumin-binding proteins, as half-lives are not extended when higher-affinity or higher-valency molecules are used.76,89
Similarly, antibody fragments have also been engineered as albumin binders with promising results. Tomlinson and co-workers applied phage display to isolate small (∼12 kDa), stable, and fully human domain antibodies (dAbs) against human, mouse, and rat albumins (Fig. 4e).90,91 Numerous dAbs, also known as AlbudAbs, were isolated with different degrees of species cross-reactivities and affinities (34–600 nM). Pharmacokinetic analyses with the high binding affinity AlbudAbs revealed half-lives similar to that of albumin itself.91 Fusion of AlbudAb to an interleukin-1 receptor antagonist (IL-1ra) exhibited similar in vitro potency, improved ex vivo stability, and a ∼130-fold longer in vivo half-life compared to IL-1ra alone (Table 3). The half-life improvement resulted in an improved efficacy of the fusion when tested in a mouse model of arthritis.91 The AlbudAb platform was recently applied to enhance the half-life of other small proteins, such as the interferon alpha-2 (IFN-α2)92 and an antibody domain antagonist for the mouse tumor necrosis factor receptor 1 (TNFR1),93 both of which have an increased half-life and a retained activity when compared to the small proteins alone (Table 3).92,93 The AlbudAb technology, now belonging to GlaxoSmithKline (GSK), has been recently tested in phase I in humans, wherein a fusion of AlbudAb to exendin-4 (GSK2374697) resulted in a 58- to 96-fold increase in the elimination half-lives in healthy patients (Table 3).94 While the AlbudAb has been mainly applied as genetic fusion, a recent study demonstrated that it can also be chemically linked representing a significant advance in the use of AlbudAb to extend the half-life of small biotherapeutics.95 The chemical conjugation of AlbudAb to an apelin-modified peptide agonist (MM202) resulted in an improved plasma half-life over both its parent compound and the endogenous agonist.95 Notably, the MM202-AlbudAb conjugate retained a high binding affinity to the human apelin receptor as well as functional activity in vitro and in vivo.95
Furthermore, Adams et al. identified a Fab, named CA645, with consistent cross-species reactivity and low nanomolar affinities for multiple serum albumins (Table 3).96 The crystal structure of CA645 in complex with human serum albumin revealed that the Fab binds to domain II (Fig. 4f). Importantly, the recognized epitope is distal from the main fatty acid and drug-binding sites and does not inhibit the binding of serum albumin to FcRn. When tested in mice, the CA645 exhibited a half-life of 3.5 days, 175-fold longer than a non-albumin-binding Fab variant. Interestingly, in line with what has been previously observed with other albumin-binding domains, a >140-fold reduction in the affinity for mouse serum albumin from 2.2 nM to 316 nM had no effect on the pharmacokinetic profile and half-life. Even with a reduction in affinity up to 62 μM, the half-life was still extended up to one day.96 Construction of a bispecific antibody format (Fab-dsFv), obtained by fusing an anti-albumin Fv domain (645Fv) derived from the humanized CA645 to another Fab with a different target specificity, led to serum half-lives of 2.6 days in mice and 7.9 days in cynomolgus monkeys (Table 3).97
A recent trend is the development of albumin binders based on the small (12–15 kDa) and stable variable domain of the heavy-chain-only (VHH) antibodies (Nanobodies®) naturally occurring in the Camelidae family.98 Structural analysis showed that nanobodies present a typical immunoglobulin variable domain (IgV) fold comprising nine β-strands and a conserved disulfide bond. As in traditional IgV domains, nanobodies contain three hypervariable loops connected by four conserved framework regions. Ablynx's proprietary half-life extension technology is based on a nanobody that binds to human serum albumin with an affinity of 22 nM. Fusion of this albumin-binding nanobody to two anti-TNF-targeting VHH domains led to a bispecific three-domain nanobody with a 90-fold prolonged half-life compared to a monomeric anti-TNF nanobody (Table 3) as well as superior therapeutic efficacy when tested in a mouse model of arthritis.99 Similarly, linkage of the albumin-binding nanobody to a VHH domain targeting the interleukin-6 receptor (IL-6R) resulted in a bispecific two-domains nanobody (ALX-0061) that exhibited high potency in vitro as well as promising in vivo efficacy when tested in non-human primate models of IL-6-induced inflammation and arthritis.100 The pharmacokinetic profile of ALX-0061 in cynomolgus monkeys showed half-lives ranging from 1.7 to 6.6 days, whereas the non-extended anti-IL-6R domain was cleared in few hours (Table 3).100 ALX-0061 is currently in clinical development for the treatment of autoimmune diseases such as rheumatoid arthritis. Moreover, a novel nanobody molecule (Nb.b201) with a modest affinity for human serum albumin (430 nM) and no binding to the related mouse serum albumin has recently been described by Kruse and co-workers.101 The crystal structure of the protein complex revealed that Nb.b201 recognized a convex protrusion on the surface of human albumin (Fig. 4g).101 However, no data have been reported on the pharmacokinetic profile of this molecule.
Finally, the variable domain (VNAR) derived from the immunoglobulin new antigen receptor (IgNAR) of cartilaginous fish (IgNAR) represents another example of a single antibody scaffold that has been successfully engineered to bind albumin with a high affinity.102 Similarly to VHHs, VNARs are small (∼12 kDa) and stable proteins with a β-sandwich fold.103 By screening an immunized VNAR phage library, Müller et al. have selected multiple clones with high affinities against different albumin species.102 The lead VNAR clone (E06) exhibited single digit nanomolar affinities for different serum albumins (Table 3) and binds a non-overlapping epitope on domain II of serum albumin (Fig. 4h), enabling it to retain binding to FcRn. When linked to a random naïve VNAR domain in different orientations, clone E06 prolonged the half-life of the fusions up to one day in rats and mice as well as more than seven days in cynomolgus macaques (Table 3).102
6. Conclusions
The use of non-covalent albumin-binding ligands has proven to be an effective strategy to prolong the in vivo plasma residence time of numerous small biotherapeutics. When bound to albumin, the peptides and small proteins are sterically shielded from proteolytic degradation and protected from rapid renal filtration due to the relatively larger hydrodynamic volume of albumin and its ability to bind the recycling FcRn. Moreover, the choice of a reversible, non-covalent association with serum albumin allows for the detachment of the biotherapeutic, facilitating its interaction with its target as well as its penetration and diffusion into small regions that are not otherwise accessible to larger molecules.
The ability of albumin to bind molecules of such different natures through multiple sites distributed throughout its tertiary structure has led to the development of numerous extension tags, each with its own advantages and disadvantages. Many have been tested in preclinical models, while others are already in clinical use. The choice of one albumin-binding ligand at the expense of another depends primarily on the intrinsic properties of the bioactive molecule and its clinical application. Therefore, an albumin-binding ligand must be carefully evaluated for each biotherapeutic and the consideration of other parameters besides half-life, such as biodistribution, penetration and accumulation in tissue, and immunogenicity, are important factors.
Conflicts of interest
There are no conflicts to declare.
Acknowledgments
We thank Dr. Giulia Pasqual, Dr. Laura Cendron, Dr. Monique Jacqueline Kauke, and Dr. Kaycie Butler for the critical reading of this review. The financial contribution from the Swiss National Science Foundation (research grant BSCGI0_157842) to A. Z. is gratefully acknowledged.
References
- Vazquez-Lombardi R., Phan T. G., Zimmermann C., Lowe D., Jermutus L., Christ D. Drug Discovery Today. 2015;20(10):1271–1283. doi: 10.1016/j.drudis.2015.09.004. [DOI] [PubMed] [Google Scholar]
- Kintzing J. R., Filsinger Interrante M. V., Cochran J. R. Trends Pharmacol. Sci. 2016;37(12):993–1008. doi: 10.1016/j.tips.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henninot A., Collins J. C., Nuss J. M. J. Med. Chem. 2018;61(4):1382–1414. doi: 10.1021/acs.jmedchem.7b00318. [DOI] [PubMed] [Google Scholar]
- Stepensky D. Toxins. 2018;10(11):483. doi: 10.3390/toxins10110483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontermann R. E. Expert Opin. Biol. Ther. 2016;16(7):903–915. doi: 10.1517/14712598.2016.1165661. [DOI] [PubMed] [Google Scholar]
- van Witteloostuijn S. B., Pedersen S. L., Jensen K. J. ChemMedChem. 2016;11(22):2474–2495. doi: 10.1002/cmdc.201600374. [DOI] [PubMed] [Google Scholar]
- Fanali G., di Masi A., Trezza V., Marino M., Fasano M., Ascenzi P. Mol. Aspects Med. 2012;33(3):209–290. doi: 10.1016/j.mam.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Sleep D. Expert Opin. Drug Delivery. 2015;12(5):793–812. doi: 10.1517/17425247.2015.993313. [DOI] [PubMed] [Google Scholar]
- Larsen M. T., Kuhlmann M., Hvam M. L., Howard K. A. Mol. Cell. Ther. 2016;4:3. doi: 10.1186/s40591-016-0048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. L., Ultsch M. H., de Vos A. M., Wells J. A. Science. 2000;287(5456):1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- Mersich C., Billes W., Jungbauer A. Biotechnol. J. 2007;2(6):672–677. doi: 10.1002/biot.200700049. [DOI] [PubMed] [Google Scholar]
- Menegatti S., Hussain M., Naik A. D., Carbonell R. G., Rao B. M. Biotechnol. Bioeng. 2013;110(3):857–870. doi: 10.1002/bit.24760. [DOI] [PubMed] [Google Scholar]
- Sockolosky J. T., Kivimae S., Szoka F. C. PLoS One. 2014;9(7):e102566. doi: 10.1371/journal.pone.0102566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutt M., Farber-Schwarz A., Unverdorben F., Richter F., Kontermann R. E. J. Biol. Chem. 2012;287(7):4462–4469. doi: 10.1074/jbc.M111.311522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unverdorben F., Hutt M., Seifert O., Kontermann R. E. PLoS One. 2015;10(10):e0139838. doi: 10.1371/journal.pone.0139838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J. M., Zer C., Avery K. N., Bzymek K. P., Horne D. A., Williams J. C. Proc. Natl. Acad. Sci. U. S. A. 2013;110(43):17456–17461. doi: 10.1073/pnas.1307309110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seijsing J., Lindborg M., Hoiden-Guthenberg I., Bonisch H., Guneriusson E., Frejd F. Y. Proc. Natl. Acad. Sci. U. S. A. 2014;111(48):17110–17115. doi: 10.1073/pnas.1417717111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezo A. R., McDonnell K. A., Hehir C. A., Low S. C., Palombella V. J., Stattel J. M. Proc. Natl. Acad. Sci. U. S. A. 2008;105(7):2337–2342. doi: 10.1073/pnas.0708960105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sockolosky J. T., Tiffany M. R., Szoka F. C. Proc. Natl. Acad. Sci. U. S. A. 2012;109(40):16095–16100. doi: 10.1073/pnas.1208857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penchala S. C., Miller M. R., Pal A., Dong J., Madadi N. R., Xie J. Nat. Chem. Biol. 2015;11(10):793–798. doi: 10.1038/nchembio.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronwall C., Sjoberg A., Ramstrom M., Hoiden-Guthenberg I., Hober S., Jonasson P. Biotechnol. J. 2007;2(11):1389–1398. doi: 10.1002/biot.200700053. [DOI] [PubMed] [Google Scholar]
- Zhang J., Williams B. A., Nilsson M. T., Chaput J. C. Chem. Commun. 2010;46(41):7778–7780. doi: 10.1039/c0cc01475c. [DOI] [PubMed] [Google Scholar]
- Lee J. H., Engler J. A., Collawn J. F., Moore B. A. Eur. J. Biochem. 2001;268(7):2004–2012. doi: 10.1046/j.1432-1327.2001.02073.x. [DOI] [PubMed] [Google Scholar]
- Peters T., All About Albumin: Biochemistry, Genetics, and Medical Applications, Academic Press, San Diego, CA, 1996, p. 432. [Google Scholar]
- Curry S. Drug Metab. Pharmacokinet. 2009;24(4):342–357. doi: 10.2133/dmpk.24.342. [DOI] [PubMed] [Google Scholar]
- Dockal M., Carter D. C., Ruker F. J. Biol. Chem. 2000;275(5):3042–3050. doi: 10.1074/jbc.275.5.3042. [DOI] [PubMed] [Google Scholar]
- Sand K. M., Bern M., Nilsen J., Noordzij H. T., Sandlie I., Andersen J. T. Front. Immunol. 2014;5:682. doi: 10.3389/fimmu.2014.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot A. M., Kalinowski D. S., Richardson D. R. Front. Physiol. 2014;5:299. doi: 10.3389/fphys.2014.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vusse G. J. Drug Metab. Pharmacokinet. 2009;24(4):300–307. doi: 10.2133/dmpk.24.300. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A. A., Grune T., Curry S. J. Mol. Biol. 2000;303(5):721–732. doi: 10.1006/jmbi.2000.4158. [DOI] [PubMed] [Google Scholar]
- Spector A. A. J. Lipid Res. 1975;16(3):165–179. [PubMed] [Google Scholar]
- Kragh-Hansen U., Watanabe H., Nakajou K., Iwao Y., Otagiri M. J. Mol. Biol. 2006;363(3):702–712. doi: 10.1016/j.jmb.2006.08.056. [DOI] [PubMed] [Google Scholar]
- Bech E. M., Pedersen S. L., Jensen K. J. ACS Med. Chem. Lett. 2018;9(7):577–580. doi: 10.1021/acsmedchemlett.8b00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudlow G., Birkett D. J., Wade D. N. Mol. Pharmacol. 1975;11(6):824–832. [PubMed] [Google Scholar]
- Ghuman J., Zunszain P. A., Petitpas I., Bhattacharya A. A., Otagiri M., Curry S. J. Mol. Biol. 2005;353(1):38–52. doi: 10.1016/j.jmb.2005.07.075. [DOI] [PubMed] [Google Scholar]
- Koehler M. F., Zobel K., Beresini M. H., Caris L. D., Combs D., Paasch B. D. Bioorg. Med. Chem. Lett. 2002;12(20):2883–2886. doi: 10.1016/s0960-894x(02)00610-8. [DOI] [PubMed] [Google Scholar]
- Zobel K., Koehler M. F., Beresini M. H., Caris L. D., Combs D. Bioorg. Med. Chem. Lett. 2003;13(9):1513–1515. doi: 10.1016/s0960-894x(03)00209-9. [DOI] [PubMed] [Google Scholar]
- Zhang H., Trisodium-[(2-(R)-[(4,4-diphenylcyclohexyl)phosphono-oxymethyl]-diethylenetriamin epentaacetato)(aquo)gadolinium(III)Gadofosveset, Molecular Imaging and Contrast Agent Database (MICAD), Bethesda (MD), 2004. [Google Scholar]
- Shechter Y., Mironchik M., Saul A., Gershonov E., Precido-Patt L., Sasson K. Int. J. Pept. Res. Ther. 2007;13(1–2):12. [Google Scholar]
- Sasson K., Marcus Y., Lev-Goldman V., Rubinraut S., Fridkin M., Shechter Y. J. Controlled Release. 2010;142(2):214–220. doi: 10.1016/j.jconrel.2009.10.028. [DOI] [PubMed] [Google Scholar]
- Han J., Sun L., Chu Y., Li Z., Huang D., Zhu X. J. Med. Chem. 2013;56(24):9955–9968. doi: 10.1021/jm4017448. [DOI] [PubMed] [Google Scholar]
- Jacobson O., Kiesewetter D. O., Chen X. Bioconjugate Chem. 2016;27(10):2239–2247. doi: 10.1021/acs.bioconjchem.6b00487. [DOI] [PubMed] [Google Scholar]
- Chen H., Wang G., Lang L., Jacobson O., Kiesewetter D. O., Liu Y. Theranostics. 2016;6(2):243–253. doi: 10.7150/thno.14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wang G., Zhang H., Ma Y., Lang L., Jacobson O. Bioconjugate Chem. 2016;27(1):54–58. doi: 10.1021/acs.bioconjchem.5b00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Lynn G. M., Jacobson O., Chen K., Liu Y., Zhang H. Nat. Commun. 2017;8(1):1954. doi: 10.1038/s41467-017-02191-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bech E. M., Martos-Maldonado M. C., Wismann P., Sorensen K. K., van Witteloostuijn S. B., Thygesen M. B. J. Med. Chem. 2017;60(17):7434–7446. doi: 10.1021/acs.jmedchem.7b00787. [DOI] [PubMed] [Google Scholar]
- Jonassen I., Havelund S., Ribel U., Plum A., Loftager M., Hoeg-Jensen T. Pharm. Res. 2006;23(1):49–55. doi: 10.1007/s11095-005-9047-1. [DOI] [PubMed] [Google Scholar]
- Dumelin C. E., Trussel S., Buller F., Trachsel E., Bootz F., Zhang Y. Angew. Chem., Int. Ed. 2008;47(17):3196–3201. doi: 10.1002/anie.200704936. [DOI] [PubMed] [Google Scholar]
- Trüssel S. S., Scheuermann J. and Neri D., Half-Life Extension by Binding to Albumin through Small Molecules, in: Therapeutic Proteins: Strategies to Modulate Their Plasma Half-Lives, ed. R. Kontermann, Wiley-VCH Verlag GmbH & Co., 2012, ch. 15. [Google Scholar]
- Franzini R. M., Ekblad T., Zhong N., Wichert M., Decurtins W., Nauer A. Angew. Chem., Int. Ed. 2015;54(13):3927–3931. doi: 10.1002/anie.201410736. [DOI] [PubMed] [Google Scholar]
- Franzini R. M., Nauer A., Scheuermann J., Neri D. Chem. Commun. 2015;51(38):8014–8016. doi: 10.1039/c5cc01230a. [DOI] [PubMed] [Google Scholar]
- Trussel S., Dumelin C., Frey K., Villa A., Buller F., Neri D. Bioconjugate Chem. 2009;20(12):2286–2292. doi: 10.1021/bc9002772. [DOI] [PubMed] [Google Scholar]
- Muller C., Struthers H., Winiger C., Zhernosekov K., Schibli R. J. Nucl. Med. 2013;54(1):124–131. doi: 10.2967/jnumed.112.107235. [DOI] [PubMed] [Google Scholar]
- Kelly J. M., Amor-Coarasa A., Nikolopoulou A., Wustemann T., Barelli P., Kim D. J. Nucl. Med. 2017;58(9):1442–1449. doi: 10.2967/jnumed.116.188722. [DOI] [PubMed] [Google Scholar]
- Kelly J., Amor-Coarasa A., Ponnala S., Nikolopoulou A., Williams Jr. C., Schlyer D. Eur. J. Nucl. Med. Mol. Imaging. 2018;45(11):1841–1851. doi: 10.1007/s00259-018-4004-5. [DOI] [PubMed] [Google Scholar]
- Kelly J. M., Amor-Coarasa A., Ponnala S., Nikolopoulou A., Williams Jr. C., DiMagno S. G. J. Nucl. Med. 2018;60(5):656–663. doi: 10.2967/jnumed.118.221150. [DOI] [PubMed] [Google Scholar]
- Umbricht C. A., Benesova M., Hasler R., Schibli R., van der Meulen N. P., Muller C. Mol. Pharmaceutics. 2018;15(12):5556–5564. doi: 10.1021/acs.molpharmaceut.8b00712. [DOI] [PubMed] [Google Scholar]
- Lowenthal M. S., Mehta A. I., Frogale K., Bandle R. W., Araujo R. P., Hood B. L. Clin. Chem. 2005;51(10):1933–1945. doi: 10.1373/clinchem.2005.052944. [DOI] [PubMed] [Google Scholar]
- Gundry R. L., Fu Q., Jelinek C. A., Van Eyk J. E., Cotter R. J. Proteomics: Clin. Appl. 2007;1(1):73–88. doi: 10.1002/prca.200600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis M. S., Zhang M., Meng Y. G., Kadkhodayan M., Kirchhofer D., Combs D. J. Biol. Chem. 2002;277(38):35035–35043. doi: 10.1074/jbc.M205854200. [DOI] [PubMed] [Google Scholar]
- Sato A. K., Sexton D. J., Morganelli L. A., Cohen E. H., Wu Q. L., Conley G. P. Biotechnol. Prog. 2002;18(2):182–192. doi: 10.1021/bp010181o. [DOI] [PubMed] [Google Scholar]
- Zorzi A., Middendorp S. J., Wilbs J., Deyle K., Heinis C. Nat. Commun. 2017;8:16092. doi: 10.1038/ncomms16092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., De Luca R., Cazzamalli S., Pretto F., Bajic D., Scheuermann J. Nat. Chem. 2018;10(4):441–448. doi: 10.1038/s41557-018-0017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollaro L. H. Med. Chem. Commun. 2010;1:5. [Google Scholar]
- Revets H. A. P. and Boutton C., Peptides capable of binding to serum albumin and compounds, constructs and polypeptides comprising the same, WO2011095545, 2011.
- Nguyen A., Reyes 2nd A. E., Zhang M., McDonald P., Wong W. L., Damico L. A. Protein Eng., Des. Sel. 2006;19(7):291–297. doi: 10.1093/protein/gzl011. [DOI] [PubMed] [Google Scholar]
- Angelini A., Morales-Sanfrutos J., Diderich P., Chen S., Heinis C. J. Med. Chem. 2012;55(22):10187–10197. doi: 10.1021/jm301276e. [DOI] [PubMed] [Google Scholar]
- Pollaro L., Raghunathan S., Morales-Sanfrutos J., Angelini A., Kontos S., Heinis C. Mol. Cancer Ther. 2015;14(1):151–161. doi: 10.1158/1535-7163.MCT-14-0534. [DOI] [PubMed] [Google Scholar]
- Dennis M. S., Jin H., Dugger D., Yang R., McFarland L., Ogasawara A. Cancer Res. 2007;67(1):254–261. doi: 10.1158/0008-5472.CAN-06-2531. [DOI] [PubMed] [Google Scholar]
- Langenheim J. F., Chen W. Y. J. Endocrinol. 2009;203(3):375–387. doi: 10.1677/JOE-09-0211. [DOI] [PubMed] [Google Scholar]
- Nilvebrant J., Hober S. Comput. Struct. Biotechnol. J. 2013;6:e201303009. doi: 10.5936/csbj.201303009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson A., Dogan J., Herne N., Abrahmsen L., Nygren P. A. Protein Eng., Des. Sel. 2008;21(8):515–527. doi: 10.1093/protein/gzn028. [DOI] [PubMed] [Google Scholar]
- Makrides S. C., Nygren P. A., Andrews B., Ford P. J., Evans K. S., Hayman E. G. J. Pharmacol. Exp. Ther. 1996;277(1):534–542. [PubMed] [Google Scholar]
- Li R., Yang H., Jia D., Nie Q., Cai H., Fan Q. J. Controlled Release. 2016;228:96–106. doi: 10.1016/j.jconrel.2016.03.004. [DOI] [PubMed] [Google Scholar]
- Stork R., Muller D., Kontermann R. E. Protein Eng., Des. Sel. 2007;20(11):569–576. doi: 10.1093/protein/gzm061. [DOI] [PubMed] [Google Scholar]
- Hopp J., Hornig N., Zettlitz K. A., Schwarz A., Fuss N., Muller D. Protein Eng., Des. Sel. 2010;23(11):827–834. doi: 10.1093/protein/gzq058. [DOI] [PubMed] [Google Scholar]
- Masuda Y., Yamaguchi S., Suzuki C., Aburatani T., Nagano Y., Miyauchi R. J. Pharmacol. Exp. Ther. 2018;365(2):368–378. doi: 10.1124/jpet.117.246652. [DOI] [PubMed] [Google Scholar]
- Guo R., Guo W., Cao L., Liu H., Liu J., Xu H. Int. J. Pharm. 2016;511(1):538–549. doi: 10.1016/j.ijpharm.2016.07.046. [DOI] [PubMed] [Google Scholar]
- Wei J., Bera T. K., Liu X. F., Zhou Q., Onda M., Ho M. Proc. Natl. Acad. Sci. U. S. A. 2018;115(15):E3501–E3508. doi: 10.1073/pnas.1721780115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J. T., Pehrson R., Tolmachev V., Daba M. B., Abrahmsen L., Ekblad C. J. Biol. Chem. 2011;286(7):5234–5241. doi: 10.1074/jbc.M110.164848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova A., Jonsson A., Rosik D., Lundqvist H., Lindborg M., Abrahmsen L. J. Nucl. Med. 2013;54(6):961–968. doi: 10.2967/jnumed.112.110700. [DOI] [PubMed] [Google Scholar]
- Frejd F. Y., Kim K. T. Exp. Mol. Med. 2017;49(3):e306. doi: 10.1038/emm.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy O. E., Jodka C. M., Ren S. S., Mamedova L., Sharma A., Samant M. PLoS One. 2014;9(2):e87704. doi: 10.1371/journal.pone.0087704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S. A., Gibbs A. C., Conk M., Yi F., Maguire D., Kane C. Protein Eng., Des. Sel. 2015;28(10):385–393. doi: 10.1093/protein/gzv040. [DOI] [PubMed] [Google Scholar]
- Gao Y. G., Su S. Y., Robinson H., Padmanabhan S., Lim L., McCrary B. S. Nat. Struct. Biol. 1998;5(9):782–786. doi: 10.1038/1822. [DOI] [PubMed] [Google Scholar]
- Traxlmayr M. W., Kiefer J. D., Srinivas R. R., Lobner E., Tisdale A. W., Mehta N. K. J. Biol. Chem. 2016;291(43):22496–22508. doi: 10.1074/jbc.M116.741314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N., Schmitt M. A., Fisk J. D. Rev. Geophys. 2016;283(7):1351–1367. doi: 10.1111/febs.13674. [DOI] [PubMed] [Google Scholar]
- Pluckthun A. Annu. Rev. Pharmacol. Toxicol. 2015;55:489–511. doi: 10.1146/annurev-pharmtox-010611-134654. [DOI] [PubMed] [Google Scholar]
- Steiner D., Merz F. W., Sonderegger I., Gulotti-Georgieva M., Villemagne D., Phillips D. J. Protein Eng., Des. Sel. 2017;30(9):583–591. doi: 10.1093/protein/gzx022. [DOI] [PubMed] [Google Scholar]
- Winter G., Griffiths A. D., Hawkins R. E., Hoogenboom H. R. Annu. Rev. Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- Holt L. J., Basran A., Jones K., Chorlton J., Jespers L. S., Brewis N. D. Protein Eng., Des. Sel. 2008;21(5):283–288. doi: 10.1093/protein/gzm067. [DOI] [PubMed] [Google Scholar]
- Walker A., Dunlevy G., Rycroft D., Topley P., Holt L. J., Herbert T. Protein Eng., Des. Sel. 2010;23(4):271–278. doi: 10.1093/protein/gzp091. [DOI] [PubMed] [Google Scholar]
- Goodall L. J., Ovecka M., Rycroft D., Friel S. L., Sanderson A., Mistry P. PLoS One. 2015;10(9):e0137065. doi: 10.1371/journal.pone.0137065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor-Semmes R. L., Lin J., Hodge R. J., Andrews S., Chism J., Choudhury A. Clin. Pharmacol. Ther. 2014;96(6):704–712. doi: 10.1038/clpt.2014.187. [DOI] [PubMed] [Google Scholar]
- Read C., Yang P., Kuc R. E., Nyimanu D., Williams T. L., Glen R. C. Basic Clin. Pharmacol. Toxicol. 2019:1–8. doi: 10.1111/bcpt.13227. [DOI] [PubMed] [Google Scholar]
- Adams R., Griffin L., Compson J. E., Jairaj M., Baker T., Ceska T. mAbs. 2016;8(7):1336–1346. doi: 10.1080/19420862.2016.1185581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave E., Adams R., Zaccheo O., Carrington B., Compson J. E., Dugdale S. mAbs. 2016;8(7):1319–1335. doi: 10.1080/19420862.2016.1210747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeland S., Vandenbroucke R. E., Libert C. Drug Discovery Today. 2016;21(7):1076–1113. doi: 10.1016/j.drudis.2016.04.003. [DOI] [PubMed] [Google Scholar]
- Coppieters K., Dreier T., Silence K., de Haard H., Lauwereys M., Casteels P. Arthritis Rheum. 2006;54(6):1856–1866. doi: 10.1002/art.21827. [DOI] [PubMed] [Google Scholar]
- Van Roy M., Ververken C., Beirnaert E., Hoefman S., Kolkman J., Vierboom M. Arthritis Res. Ther. 2015;17:135. doi: 10.1186/s13075-015-0651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon C., Baier A. S., Pascolutti R., Wegrecki M., Zheng S., Ong J. X. Nat. Struct. Mol. Biol. 2018;25(3):289–296. doi: 10.1038/s41594-018-0028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M. R., Saunders K., Grace C., Jin M., Piche-Nicholas N., Steven J. mAbs. 2012;4(6):673–685. doi: 10.4161/mabs.22242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielonka S., Empting M., Grzeschik J., Konning D., Barelle C. J., Kolmar H. mAbs. 2015;7(1):15–25. doi: 10.4161/19420862.2015.989032. [DOI] [PMC free article] [PubMed] [Google Scholar]