Abstract
The basic helix–loop–helix transcription factor Twist1 is a key regulator of craniofacial development. Twist1-null mouse embryos exhibit failure of cephalic neural tube closure and abnormal head development and die at E11.0. To dissect the function of Twist1 in the cranial mesoderm beyond mid-gestation, we used Mesp1-Cre to delete Twist1 in the anterior mesoderm, which includes the progenitors of the cranial mesoderm. Deletion of Twist1 in mesoderm cells resulted in loss and malformations of the cranial mesoderm-derived skeleton. Loss of Twist1 in the mesoderm also resulted in a failure to fully segregate the mesoderm and the neural crest cells, and the malformation of some cranial neural crest-derived tissues. The development of extraocular muscles was compromised whereas the differentiation of branchial arch muscles was not affected, indicating a differential requirement for Twist1 in these two types of craniofacial muscle. A striking effect of the loss of Twist1 was the inability of the mesodermal cells to maintain their mesenchymal characteristics, and the acquisition of an epithelial-like morphology. Our findings point to a role of Twist1 in maintaining the mesenchyme architecture and the progenitor state of the mesoderm, as well as mediating mesoderm–neural crest interactions in craniofacial development.
Keywords: Twist1, Cranial mesoderm, Mesp1-Cre, Conditional mutant, Epithelial, Mesenchymal, Tissue patterning, Craniofacial development, Mouse
Introduction
Craniofacial structures are derived from the cranial neural crest (CNC) and the cranial mesoderm (CM) (Bronner and Ledouarin, 2012; Couly et al., 1992; Kontges and Lumsden, 1996; Minoux and Rijli, 2010; Noden and Francis-West, 2006; Sauka-Spengler and Bronner-Fraser, 2006). The CNC cells, a population of migratory cells arising from the lateral region of the neural folds, contribute to all the elements of the viscerocranium (comprised of the lower jaw, upper jaw and snout), the frontal bones of the skull and the anterior skull base, as well as the connective tissues, cranial ganglia and the smooth muscle of the blood vessels in the head. The CM, which is part of the mesoderm layer formed during gastrulation, contributes to the muscles of the face and neck, endothelial cells of the blood vessels, and bones of the neurocranium and the posterior skull base (Noden and Francis-West, 2006; Saga et al., 1999; Sambasivan et al., 2011; Yoshida et al., 2008). The CM can be subdivided topographically into the prechordal, cranial paraxial and cranial lateral populations, all of which contribute to the craniofacial muscles. Cell transplantation and lineage tracing experiments performed on avian embryos show that the prechordal and rostral parts of the cranial paraxial mesoderm contain the precursors of the extraocular muscles (Couly et al., 1992) while the lateral part of the cranial mesoderm contributes to the muscles of the branchial arches. However, genetic lineage tracing studies in mouse embryos have revealed a clonal relationship between some cells of the extraocular muscles and the mandibular arch-derived muscles, suggesting that they are not derived from entirely separate lineages (Lescroart et al., 2010).
Twist1 encodes a basic helix–loop–helix transcription factor that is expressed in both the CNC and the CM during craniofacial development (Fuchtbauer, 1995; Stoetzel et al., 1995). In humans, haploin-sufficiency of TWIST1 is associated with the autosomal dominant Saethre–Chotzen syndrome (SCS) (El Ghouzzi et al., 1997; Qin et al., 2012; Rose and Malcolm, 1997), characterized by a varied pattern of craniofacial defects including craniosynostosis, ptosis and facial asymmetry. In Drosophila, loss of twist leads to abnormal gastrulation and failure of mesoderm formation (Thisse et al., 1987), pointing to a function in eliciting epithelial–mesenchymal transition (EMT). In the mouse, loss of Twist1 is associated with major craniofacial defects including a failure to close the cranial neural tube, branchial arch hypoplasia, poor vascular development and the demise of the embryo at around embryonic day (E) 11 (Chen and Behringer, 1995).
The embryonic lethality of the Twist1-null mutation does not allow analysis of Twist1 function in craniofacial development beyond mid-gestation. This obstacle can be overcome by conditional ablation of Twist1 in specific tissues, which minimizes the impact of loss of gene function on the viability of the embryo (Chen et al., 2007). Using this approach we have shown that loss of Twist1 in the CNC cells leads to reduced cell viability and impaired differentiation, loss or malformation of CNC-derived skeleton, poor frontonasal development and mandibular hypoplasia (Bildsoe et al., 2009). In addition, the formation the parietal and interparietal bones, which are derived from the CM, is also affected, suggesting that the CNC-derived tissues may play a supportive role in the development of some mesodermal bones.
To investigate the role of Twist1 in the cranial mesoderm, a similar conditional ablation strategy was employed to eliminate Twist1 activity by expressing Cre-recombinase in the nascent mesoderm during gastrulation, via a Cre transgene insertion into the Mesp1 locus (Loebel et al., 2012; Saga et al., 1999). In this study, Twist1 activity in the cranial mesoderm is shown to be essential for normal development of the mesoderm-derived components of the neurocranium and the formation of the extraocular muscles. We have also uncovered an unexpected effect of the loss of Twist1 on the ability of the CM cells to maintain their mesenchymal characteristics, resulting in the acquisition of an epithelial phenotype.
Material and methods
Mouse strains and genotyping
Twist13loxPneo/3loxPneo and Twist1del/+ mice were maintained and genotyped as previously described (Bildsoe et al., 2009; Chen et al., 2007). Mesp1-Cre mice (Saga et al., 1999) were maintained on a C57BL/6 background.
Crosses were performed as previously described (Loebel et al., 2012). We first crossed Mesp1-Cre mice (Saga et al., 1999) to Twist1del/+ mice to generate Twist1del/+; MespCre/+ mice. To generate embryos with a mesoderm-specific Twist1 deficiency, Twist1del/+;Mesp1Cre/+ mice were crossed with Twist13loxPneo/3loxPneo mice. Conditional knockout (CM–CKO) embryos of Twist13loxPneo/del;Mesp1Cre/+ genotype, wild-type (WT) Twist13loxPneo/+, Mesp1+/+, and heterozygous Twist13loxPneo/del; Mesp1+/+ or Twist13loxPneo/del; Mesp1+/Cre embryos were collected for phenotypic analysis.
To trace the distribution of the descendents of Mesp1-Cre expressing cells, mice carrying the Rosa26R allele (Soriano, 1999) the Twist13loxPneo/3loxPneo (Loebel et al., 2012) and Mesp1Cre/+ alleles were crossed to generate embryos in which Mesp1-Cre activity will activate the lacZ reporter in the wild type cells and also excise the floxed Twist1 allele such that Twist1-CKO cells can be tracked by lacZ expression.
Trp53 mutant mice (#002101), obtained from the Jackson Laboratory (Jacks et al., 1994), were crossed with Twist1-del mice (Bildsoe et al., 2009) to generate double heterozygous mice. These mice were crossed together and embryos with various combinations of Twist1 and Trp53 alleles collected at E9.5–E10.5. Mice and embryos were genotyped for Twist1 and Trp53 as described (Bildsoe et al., 2009; Chen et al., 2007; Jacks et al., 1994).
Bone and cartilage staining
Embryos were collected between E15.5 and E17.5 in PB1 and rinsed in cold PBS. Bone and cartilage was stained with alizarin red and alcian blue as previously described (Bildsoe et al., 2009; Hogan et al., 1994; Loebel et al., 2012). Stained specimens were washed, stored and photographed in aqueous 20% ethanol: 20% glycerol. Images were captured using a SPOT camera (SciTech) and Leica microscope.
β-galactosidase reporter staining
Embryos between E 7.5–11.5 were stained as whole mount specimens as described (Loebel et al., 2012; Watson et al., 2008). Briefly embryos were collected in PB1 media and rinsed in cold calcium- and magnesium-free PBS before fixing for a minimum of 2 h at 4 °C in glutaraldehyde fixative. After fixation the embryos were rinsed in X-gal washing buffer briefly and incubated in the X-gal staining solution at 37 °C for 2–6 h for color development. The embryos were washed twice in X-gal washing buffer, then in PBS and fixed in 4% PFA.
For staining of cryosections, heads of E13.5 and E15.5 embryos were dissected in PB1 medium, rinsed in calcium–magnesium free PBS and fixed on ice (Mani et al., 2010;Rivera-Perez et al., 1999). The heads were dehydrated in 10% sucrose/PBS overnight at 4 °C, followed by 25% sucrose/PBS overnight at 4 °C. Tissues were embedded in 25% sucrose/PBS:OCT (Tissue Tek) embedding medium (2:1). Sections were cut at 5–9 µm on a cryostat (M1900, Lexica), collected on Super frost Plus slides (Mensal-Glaser) and stored at −80 °C until needed. Sections were thawed at room temperature, rinsed in X-gal washing buffer three times and then incubated in the X-gal staining solution at 37 °C overnight for color development (Watson et al., 2008). Sections were washed in the X-gal washing buffer and water, and counterstained with nuclear fast red and mounted in Ultra-mount No.4 (Fronine).
In situ hybridization
Embryos from E7.5–11.5 were dissected in PB1, rinsed in cold PBS and fixed in 4% PFA. Automated whole mount in situ hybridization was carried out using an InsituPro machine (Intavis AG) (Bildsoe et al., 2009;Loebel et al., 2004). Stained embryos were washed in 0.1% Tween20 in H2O, fixed in 4% PFA and photographed. Probes to detect the following transcripts were used: Cre (Loebel et al., 2012), Myf5, Pitx2, Sox10, Tbx1 and Twist1. Embryos were imaged using a Leica dissecting microscope and SPOT digital camera (SciTech).
Cell death analysis
Apoptotic cells were detected by whole mount TUNEL staining with the ApopTag Plus Peroxidase In situ Apoptosis kit (Millipore) as previously described (Martinez-Barbera et al., 2002). Embryos were dissected in PB1, rinsed in cold PBS and fixed in 4% PFA overnight at 4 °C, and incubated in blocking buffer containing 2% blocking reagent (Roche) and 20% heat inactivated fetal calf serum in PBS/0.1% Tween. After antibody labeling, embryos were washed overnight at 4 °C in 2mg/ml BSA in PBS, 0.1% Tween with at least four changes of buffer. After treatment, embryos were fixed in 4% PFA and the embryos were imaged using a Leica microscope and SPOT digital camera (SciTech).
Histology
Embryos at E8.5–E15.5 were collected in PB1 and rinsed in cold PBS, followed by fixation in 4% PFA. For wax embedding, the embryos were dehydrated stepwise through an ethanol series (30%, 50%, 70%, 80%, 90% and 100%), cleared in xylene and embedded in paraffin wax. Serial sections cut at 5–10 µm were stained with haematoxylin and eosin.
Immunofluorescence
Embryos at E8.0–E17.5 were fixed in 4% PFA for 1–24 h at 4 °C. Prior to staining the embryos were embedded in OTC solution, sectioned at 9–12 µm and stored at −80 °C. Slides were thawed at room temperature (RT) and permeablised using PBS, 0.1% BSA and 0.02% Triton followed by three washes with washing solution (PBS, 0.1% BSA, and 0.1% Tween) and blocked (PBS, 0.1% BSA, 0.1% Tween and 5–10% serum) for at least 1 h at RT. Slides were incubated with primary antibody in blocking solution overnight at 4 °C and washed 3×5 min in washing solution before adding secondary antibody in blocking solution and incubating for 1–2 h at RT. For F-actin staining, Alexa594 conjugated phalloidin (8 µl/ml, Molecular Probes) was added to blocking solution after secondary antibody incubation and the sections were incubated for 30 min, then washed in 2×5 min PBS, stained with DAPI and mounted in DABCO. Primary antibodies for specific factors used in this study were: myosin heavy chain (mf20, 1:20 dilution, Developmental Studies Hybridoma Bank, University of Iowa), Twist1 (1:50 dilution, Abcam), Ki67 (1/200, Abcam) E-cadherin (1/200 dilution, Life Technologies) and β-galactosidase (1/100, Abcam). Secondary antibodies used were goat anti-rat DyLight 488 (1/600 dilution, Thermo Scientific), donkey anti-mouse conjugated to Alexa594 or 488 (1:400 Jackson ImmunoResearch) and donkey anti-chicken conjugated to Alexa594 (1/500 Jackson ImmunoResearch).
qRT–PCR analysis
Total RNA was isolated from heads of E8.5, 9.5 and E10.5 embryos using an RNeasy Micro Kit (QIAGEN) according to the manufacturer’s instructions. The RNA samples were reverse transcribed into cDNA using Superscript III First-Strand Synthesis System (Invitrogen) according to manufacturer’s instructions. Quantitative RT–PCR was performed on Corbett Rotor-Gene Thermocyclers (QIAGEN) using Platinum Taq (Invitrogen) and SYBR Green I (Molecular Probes) with Polr2a or Tbp as the reference gene. The following primers were used:
Pitx2 forward; 5′-gctggaagccactttccagagaaa- 3′
Pitx2 reverse; 5′–tggacagagacgctgacgtgag-3′
Tbp forward: 5′-tctggaaaagttgtattaacag-3′
Tbp reverse: 5′-gctgcagggtgatttcagt-3′
Pdgfra forward: 5′-gaggataagctgaaggactgggaagg-3′
Pdgrfa reverse: 5′-tactggaacctgtctcgatggcact-3′
Snai2 forward: 5′-cacacattgccttgtgtctgcaagat
Snai2 reverse: 5′-atcagagtgggtctgcagatgtgcc
Vim forward:5′-tgtacgaggaggagatgcggga-3′
Vim reverse: 5′-tgtctgaatgactgcagggtgcttt-3′
Polr2a forward: 5′-gcaccacgtccaatgatat-3′
Polr2a reverse: 5′-gtgcggctgcttccataa-3′
Results
Conditional ablation of Twist1 in the cranial mesoderm
To investigate the impact of loss of Twist1 function in the cranial mesoderm tissues, Cre-recombinase expressed from the Mesp1 locus was used to ablate a floxed allele of Twist1 during mesoderm development (Loebel et al., 2012). Cre transcripts were detected by in situ hybridization in the nascent mesoderm localized next to the primitive streak of the gastrulating embryo [Fig. 1A, see also Loebel et al., (2012)]. In Mesp1cre/+; Rosa26R embryos, β-galactosidase positive cells populated the whole mesoderm layer of E7.5 embryo but not the other germ layers (Fig. 1B and Bi). In situ hybridization analysis of E7.5 embryos revealed that Twist1 mRNA was detected in the anterior-most mesoderm population (which contains the precursors of the cranial mesoderm) underlying the anterior ectoderm but were absent from the primitive streak and nascent mesoderm [Fig. 1C, see also Fuchtbauer, (1995)]. Taken together, the overlapping pattern of β-galactosidase reporter activity and Twist1 expression in the mesoderm suggests that Mesp1-Cre activity would have ablated Twist1 in the precursors of the cranial mesoderm.
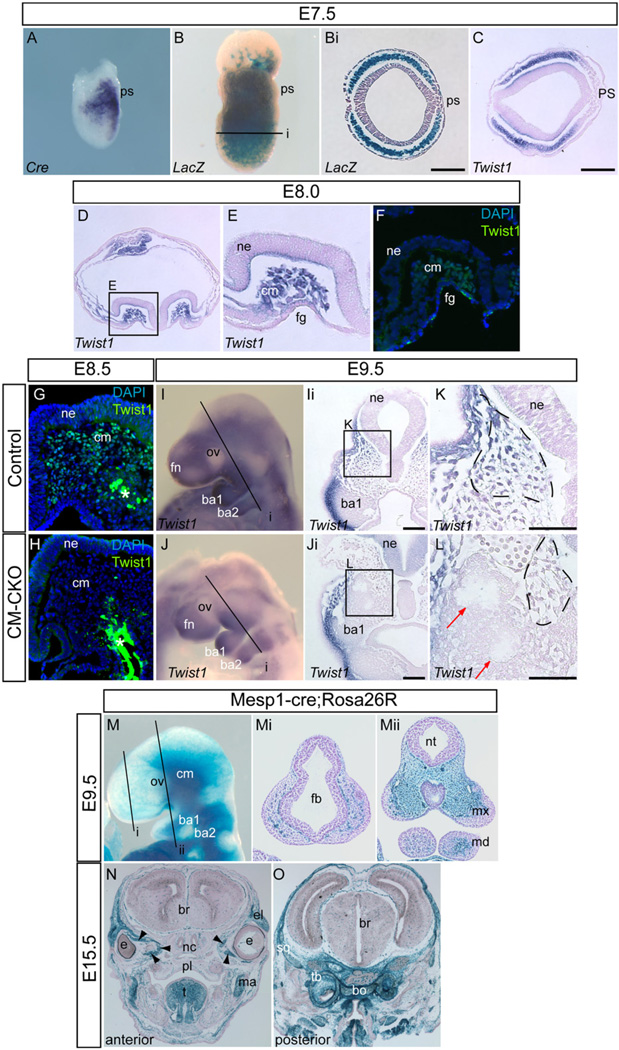
Fig. 1.
Expression of the Mesp1-Cre transgene and mesoderm-specific ablation of Twist1. (A) In situ hybridization showing Mesp1-Cre transcript expression in the nascent mesoderm. (B) Whole mount stained embryo showing distribution of Mesp1-Cre; Rosa26R positive cells in an E7.5 embryo. (Bi) Histology of age-matched E7.5 embryos showing Rosa26R-positive cells populating the entire mesoderm layer (plane of sectioning of Bi is shown in B). (C–E) Histological sections showing detection of Twist1 transcript by in situ hybridization. (C) At E7.5 Twist1 transcript expression overlaps with the distribution of Rosa26R-positive mesodermal cells shown in (B). (D) At headfold stage (E8.0) Twist1 expression is detected in the cranial mesoderm underlying the neural epithelium (E: magnification of boxed area in D). (F) Expression of Twist1 protein in the same region at headfold stage. The midline is to the right in E and F. (G,H) Twist1 protein is expressed in the cranial mesoderm of E8.5 control embryo (G) but absent in the CM–CKO (cranial mesoderm conditional Twist1 knockout) embryo (H), (White asterisk: nonspecific staining in the endoderm). (I–L) Twist1 expression in the cranial mesenchyme of control embryo at E9.5 (I, Ii and K) is absent in the CM–CKO (J, Ji and L). Corresponding regions in control and CM–CKO embryo are marked by the dashed line (K and L). The Twist1-deficient cranial mesenchyme displays early signs of acquiring a compacted morphology (L, red arrow). I and J: whole mount, Ii and Ji: sections of embryo at the plane (i) indicated in (I) and (J). Boxed areas K and L are magnified views of Ii and Ji. (M–O) Distribution of Mesp1-Cre; Rosa26R positive cells in the craniofacial region at E9.5 and E15.5. At E9.5 Mesp1-Cre; Rosa26R positive cells are observed in the paraxial regions of the cranial mesenchyme and in the core of the mandibular arch of the control embryo (M, whole mount, Mi, Mii: sections of the embryo at planes marked by i and ii in M), and in the craniofacial muscles in the peri-ocular, facial, jaw and tongue region and the skeletal elements of the chondrocranium of E15.5 embryo (N and O; arrowheads marks the extraocular muscles in N). Abbreviations: ba1, branchial arch 1; ba2, branchial arch 2; bo, basioccipital bone; br, brain; cm, cranial mesoderm; e, eye; el, eyelid; fb, forebrain; fg, foregut; ma, masseter muscle; md, mandible; mx, maxilla; nc, nasal cartilage; ne, neural epithelium; nt, neural tube; ov, optic vesicle; pl, palate; ps, primitive streak; sq, squamous part of the temporal bone (tb); t, tongue. Scale bar=100 µm.
In headfold stage embryos, Twist1 mRNAs were detected by in situ hybridization in the mesoderm beneath the cephalic neural folds (Fig. 1D and E). Nuclear localized Twist1 protein was detected in the cranial mesoderm at E8.0 (two somite stage, Fig. 1F), which is slightly earlier than E8.25 as previously reported (Gitelman, 1997). In contrast to the widespread presence of Twist1 protein in the cranial mesenchyme of E8.5 control (Twist13loxPneo/wt, Mesp1Cre/+) embryo (Fig. 1G), Twist1 was absent from the corresponding regions of age-matched Twist1 conditional mutant embryos (Twist13loxPneo/del, Mesp1Cre/+ = CM–CKO; Fig. 1H, n=3). Whole mount staining and histological sections of E9.5 CM–CKO embryos showed that Twist1 transcripts were not detectable in tissues that were populated by Rosa26R-positive cells (Fig. 1I–M), but were expressed by tissues in the Rosa26R-negative regions that were populated by the CNC cells, such as the lateral subectodermal mesenchyme, the branchial arches and the frontonasal tissues (data not shown). Mesp1-Cre was therefore effective for ablating Twist1 in the cranial mesoderm.
Mesodermal contribution to the craniofacial tissues
At E9.5, β-galactosidase positive cells from Mesp1Cre/+; Rosa26R embryos were found widely in the craniofacial mesenchyme caudal to the optic vesicles (Fig. 1M, Mi, Mii). Mesenchymal cells in this region that were negative for β-galactosidase activity are likely to be descendants of CNC cells (Yoshida et al., 2008), while β-galactosidase positive cells that were scattered in the unstained anterior craniofacial mesenchyme have previously been identified as endothelial cell progenitors (Yoshida et al., 2008). Caudal to the optic vesicle, the β-galactosidase positive cells made up the majority of the mesenchymal tissues surrounding the neural tube but did not contribute to tissues in the lateral part of the maxillary eminences (Fig. 1Mii). In the mandibular arch, Mesp1Cre/+; Rosa26R-positive cells were localized in the core of the arch (Fig. 1Mii). At E15.5, β-galactosidase positive cells were found in mesoderm-derived tissues including the muscles of the tongue, jaws and neck, the extra ocular muscle (EOM), the endothelial cells and the skull vault and base (Fig. 1N and O).
Disrupted development of the skull vault and base in CM–CKO embryos
Loss of Twist1 in the mesoderm led to a severe disruption of skull development. The neural crest-derived frontonasal structures and the jaws were formed, but other parts of the skull were missing and the embryo displayed exencephaly at E13.5 (Fig. 2A and B, n=4). The CM–CKO embryos completely lacked the normal encasing of the brain as observed in the control embryo (Twist13loxPneo/+, Mesp1Cre/+, Rosa26R; Fig. 2A, B; Supplementary Fig. S1A, B). In control embryos, β-galactosidase positive cranial mesoderm-derived cells were located in the posterior part of the skull vault, whereas the β-galactosidase negative, presumed CNC-derived cells were found in the frontal bones of the skull vault, the upper and lower jaws and the frontonasal tissues (Fig. 2A). In CM–CKO embryos (Twist13loxPneo/del, Mesp1cre/+, Rosa26R) the β-galactosidase positive cells did not locate to the correct position. Instead, β-galactosidase-positive cells congregated ectopically at sites lateral to the CNC-derived presphenoid bone (Fig. 2Bi). The skulls of CM–CKO embryos were consistently smaller than those of control embryos (n=3 each) and most of the skull vault was missing (Fig. 2C, D). The mesoderm-derived parietal, interparietal and supraoccipital bones were absent and the CNC-derived frontal bones were reduced to small remnants located near the eyes (n=3, Fig. 2D). The nasal cartilage was foreshortened and the nasal bone was smaller in CM–CKO embryos compared to the controls (Twist13loxPneo/+). The maxilla and mandible were formed but reduced in size. The parietal plate, which intercalated between the squamous bone and the exoccipital bone was small and deformed (Fig. 2D). Formation of the skull base was also affected (Fig. 2E, F). The mesoderm-derived basioccipital bone made up a much smaller proportion of the skull base (Fig. 2E, F). The rostral part of this bone protruded dorsally and did not align with the skull base (Fig. 2D, black arrowhead; Supplementary Fig. S1A–D) and ectopic cartilaginous elements were found adjacent to the temporal bone (Supplementary Fig. S1Aii and Bii). In the peri-ocular region of the skull base the mesoderm-derived hypochiasmatic cartilage (McBratney-Owen et al., 2008), a skeletal element that is part of the presphenoid bone, was missing in the CM–CKO embryo (Fig. 2G, H). The CNC-derived elements in the skull base, including the presphenoid, basisphenoid and temporal bones were present but generally smaller (Fig. 2E, F).
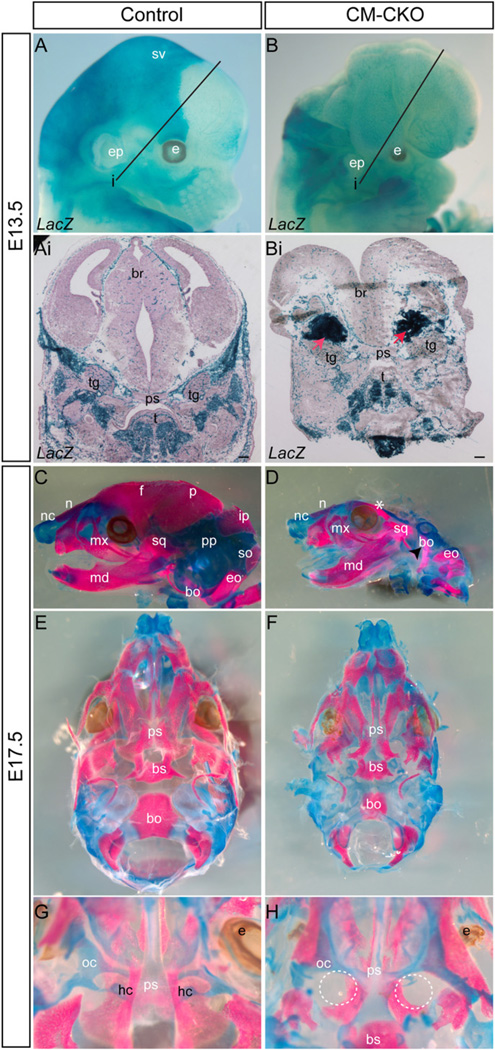
Fig. 2.
Formation of the craniofacial skeleton in the CM–CKO mutants. (A-Bi) Lineage tracing of the skeletogenic CM progenitors. The Mesp1-Cre; Rosa26R expressing mesodermal cells contribute to the skeletal elements of the base of neurocranium, the craniofacial and tongue muscles and the blood vessels in the E13.5 control embryos (Ai: section of embryo at the plane i indicated in A) but are found in ectopic compact masses (red arrows) in the lateral region underneath the open neural tube and sparsely in the craniofacial and tongue muscle and blood vessels in the E13.5 CM–CKO embryo (Bi: section of the embryo at plane i indicated in B). (C–H) Skeletal preparations of the skull vault and base. The E17.5 CM–CKO embryos (D, F, H) show loss of skeletal elements in the vault and the base of the skull when compared to control embryos (C, E and G). Dashed circles in H indicate the expected location of the missing hypochiasmatic cartilage. bo, basi-occipital; br, brain; bs, basisphenoid; e, eye; eo, exoccipital; ep, ear pinna; f, frontal; hc, hypochiasmatic cartilage; ip, interparietal; md, mandible; mx, maxilla; n, nasal bone; nc, nasal cartilage; oc, orbital cartilage; p, parietal; pp, parietal plate; ps, presphenoid; so, supra-occipital; sq, squamosal bone; sv, skull vault; t, tongue and tg, trigeminal ganglion; C and D: lateral view; E and F: ventral view with mandible removed, G and H: dorsal view with skull vault removed. Scale bar=100 µm.
Abnormal development of peri-ocular structures in CM–CKO embryos
In addition to the loss of the hypochiasmatic cartilage (Fig. 2H), the formation of the extraocular muscles (EOM) and upper eyelid was impaired (Fig. 3A–D). Anti-myosin heavy chain (MHC) immunofluorescence of E15.5 CM–CKO (n=2) and control (n=2) embryos and histology of E17.5 CM–CKO (n=2), and control embryos (n=1) revealed that EOM were missing or severely reduced in CM–CKO mutants (Fig. 3A D). All EOM groups appeared to be equally affected, as we did not detect any difference in the severity of the defect in the different EOMs. Quantitative RT–PCR of Pitx2, an early marker of the viability and myogenic property of the EOM progenitors (Zacharias et al., 2011), revealed that its expression was significantly reduced (P=0.03, n=3) in the CM–CKO head tissues compared to wild type and the heterozygous counterparts (Supplementary Fig. S2, n=3). Consistent with this, Pitx2 expression was not detected by in situ hybridization in the mesenchyme ventral to the eye that normally contains the EOM progenitors in E10.5 CM–CKO embryos (Fig. 3E, F, n=3). In contrast, Pitx2 expression in the myogenic tissues of the first and second branchial arches was maintained (Fig. 3G and H). At E11.5, a muscle condensation in the peri-ocular region that was visible in the control embryos (Twist13loxPneo/wt, Mesp1Cre/+, Rosa26R; Fig. 3I) was absent in the Mesp1Cre/+; Rosa26R positive CM–CKO embryos (Fig. 3J, n=2). The early loss of Pitx2 expression and the absence of muscle condensation were most probably due to the lack of EOM progenitors in the peri-ocular tissue of the CM–CKO embryos.
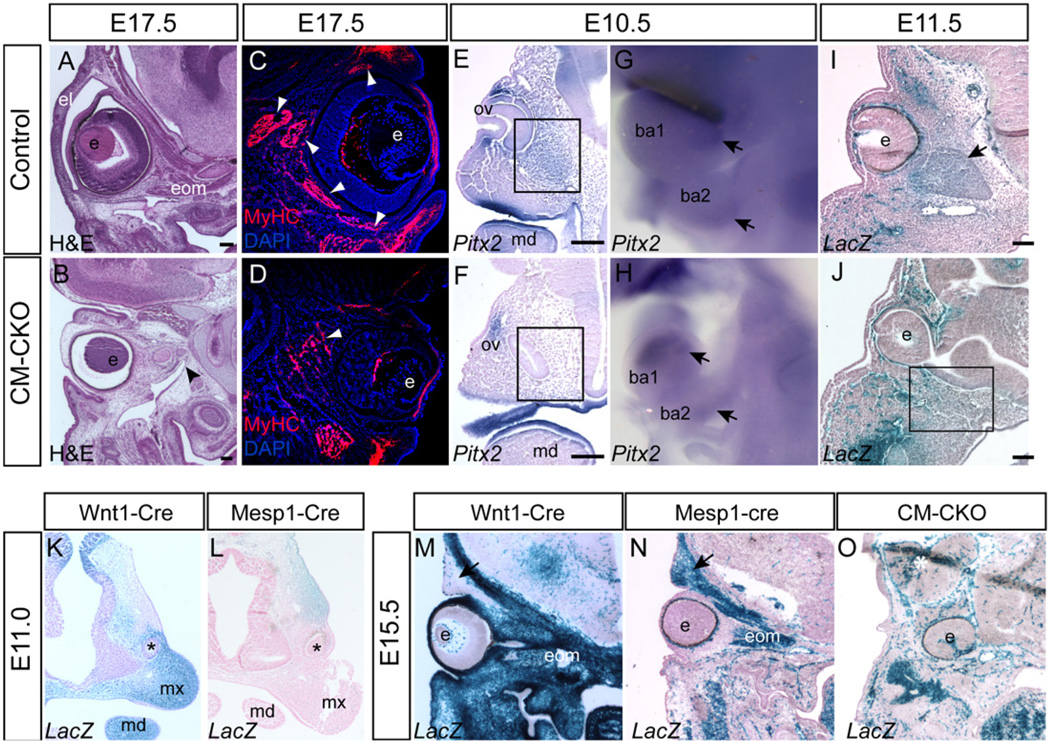
Fig. 3.
Impact of loss of Twist1 in the cranial mesoderm on the development of peri-ocular structures. (A, B) The extra-ocular muscles (eom) are severely underdeveloped in the E17.5 CM–CKO embryo (B, black arrowhead), compared to the age-matched control (A). (C, D) Sparse and loosely organized myosin-heavy chain-positive fibers in the peri-ocular tissue of the E15.5 CM–CKO embryo (D), contrasting with the fiber bundles in the extraocular muscles of the control embryo (C, white arrowheads); the upper eyelids of the CM–CKO embryo (D) are rudimentary and lack muscle fibers. (E, F) In situ hybridization for Pitx2 expression at E10.5 showing reduced presence of Pitx2-expressing cells in the CM–CKO embryo (F) in the area ventral to the eye invagination compared to control embryo (E, boxed area), indicating that the extraocular muscle progenitor population is reduced. (G,H) Pitx2 expression is maintained (black arrow) in the first and second branchial arches of the CM–CKO (I,J) Staining of Mesp1-Cre; Rosa26R cells at E11.5. show a reduced population of Mesp1-Cre; Rosa26R positive mesoderm cells in the tissues surrounding the optic cup and the adjacent orbital area in the CM–CKO (J) compared to the control (I, black arrow, muscle anlagen). (K,L) CM and CNC contribution to the eyelids at E11.0. The Wnt1-Cre; Rosa26R positive CNC cells contributes to the tissues ventral to the eye primordium (asterisk, K) whereas the Mesp1-Cre; Rosa26R positive mesoderm cells to the tissues dorsal to the eye primordium (L). (M–O) Lack of eyelid contribution from the Twist1 deficient CM in the CM–CKO. The contribution of the Wnt1-Cre; Rosa26R positive neural crest cells to the lower eyelid (M), and tissues around the eye and in the neurocranium, compared to that of the Mesp1-Cre; Rosa26R positive mesoderm cells, which contribute to the upper eyelid (black arrow) and extraocular and facial muscles. At E13.5 (O) eyelids and extraocular muscles are absent in the CM–CKO embryo, and there is paucity of Mesp1-Cre; Rosa26R positive mesoderm cells around the eye that is displaced deep in the head (O). ba1, 1st branchial arch; ba2, 2nd branchial arch e, eye; el, eyelid; mx, maxilla; md, mandible; ov, optic vesicle. Scale bar=100 µm.
The formation of eyelids was affected by the loss of Twist1 in the CM (Fig. 3A, B). At E11.0 CM derived β-galactosidase positive mesenchyme, labeled by Mesp1-Cre was found dorsal to the developing eye whereas Wnt1-Cre labeled CNC-derived mesenchyme was located ventral to the eye primordia (Fig.3K and L) and they contributed to dorsal and ventral eyelid tissues at 13.5 respectively (Fig. 3M and N). However, in the E13.5 CM–CKO embryo, the contribution of the CM mesenchyme to the dorsal peri-ocular tissue was lacking when comparing to the control (Fig. 3N and O, n=3). This deficiency in mesoderm-derived tissue underpins the absence of the upper eyelid (Fig. 3O) and the failure of eyelid closure in the E17.5 CM–CKO embryos.
To test whether Twist1 expression was present only in the progenitors or whether it is also maintained in mesoderm-derived structures, we examined Twist1 and β-galactosidase expression by immunofluorescence at E13.5. By this stage Twist1 was not detectable in the lacZ-positive developing EOM or hypochiasmatic cartilage (Supplementary Fig. S3A, B, E–H), but was expressed in adjacent β-galactosidase-negative (neural crest-derived) tissues. In contrast, Twist1 expression was maintained in mesoderm-derived tissues that form the skull and upper eyelid (Supplementary Fig. 3A, D, E).
Whereas the extraocular muscles are derived from prechordal head and paraxial mesoderm (Couly et al., 1992; Lescroart et al., 2010), facial and jaw muscles are derived from the mesoderm of the first and second branchial arches [BA, (Depew et al., 2002)]. β-galactosidase positive mesoderm populated the core of the first (BA1) and second branchial arches (BA2) in both control and CM–CKO embryos (Fig. 4A and B, n=3). In addition to the expression of Pitx2, Tbx1, a pre-myogenic marker detected in the BA at E9.5, was expressed in the core of the BAs of the CM–CKO embryo [Fig. 4C (Rinon et al., 2007)] and expression was elevated in the tissues adjacent to the pharyngeal pouches in the CM–CKO embryo [Fig. 4D, n=3) (Rinon et al., 2007)]. At E11.5, Myf5 expression signified the onset of myogenic differentiation of the first branchial arch and periocular tissues in control embryos (Fig. 4E). In CM–CKO embryos, Myf5 was expressed properly in BA1 and BA2 but not in the periocular region, consistent with a disruption of extraocular muscle development (Fig. 4E and F, n=3).
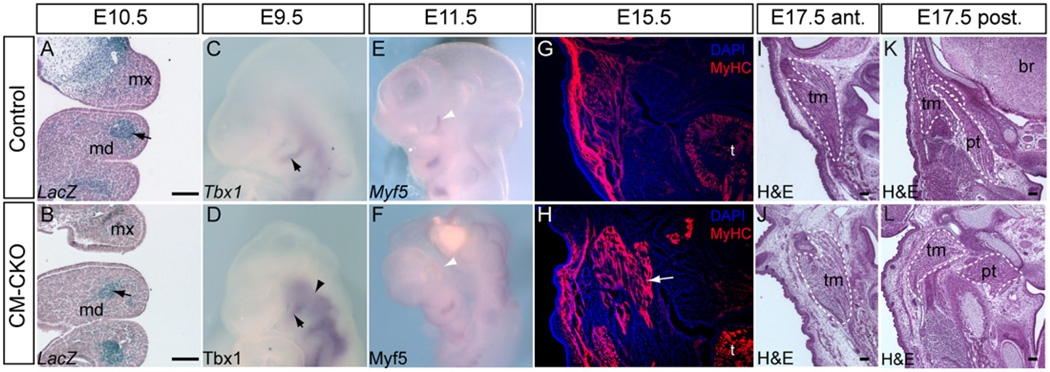
Fig. 4.
Myogenic differentiation of branchiomeric muscles is unaffected by the loss of Twist1 in the cranial mesoderm. (A-B) Mesp1-Cre; Rosa26R cells are located correctly in the first and second branchial arch of E10.5 CM–CKO embryos (black arrow, B), similar to the control (black arrow, A). (C–F) Correct initiation of myogenic differentiation. The mesodermal core in the branchial arches of control and CM–CKO embryos show comparable levels of Tbx1 (black arrow, D) and Myf5 expression (F) in the core tissues of the branchial arches of (C, D) E9.5 and (E, F) E11.5 embryos. The E9.5 CM–CKO embryo shows enhanced Tbx1 expression in the cranial paraxial mesoderm (arrowhead, D). Myf5 is expressed in the peri-ocular region of control but not CM–CKO embryos at E11.5 (white arrowheads, E, F). (G, H) Differentiation of muscle fibers is not affected in CM–CKO embryo at E15.5, revealed by the presence of myosin heavy chain (MyHC) protein in the facial muscles (H, white arrow: extra muscle masses are seen in this section of the mutant embryo, compared to the control (G). (I,J) In this example of the masticator muscles of the E17.5 CM–CKO embryo (J) shows more loosely packed fibers than (I) the control counterpart muscle (section plane: anterior to the otic capsule). Some muscles in the posterior head region are related inappropriately with the surrounding skeletal elements that are also disoriented. Dashed lines mark the corresponding muscles in the control (K) and the CM–CKO (L) embryos); br, brain; md, mandible; mx, maxilla; pt, pterygoid muscle; t, tongue; tm, temporal muscle. Scale bar=100 µm.
BA-derived muscles were formed in the CM–CKO embryo at E15.5 (n=2) and E17.5 (n=2; Fig. 4G and H, I and J). In CM–CKO embryos muscles in the posterior region, including the pterygoid (Fig. 4K and M) and the temporal muscles (Fig. 4K and M) displayed abnormal patterning, presumably due to either lack of or improper attachment to skeletal elements that were incorrectly positioned and shaped or completely absent.
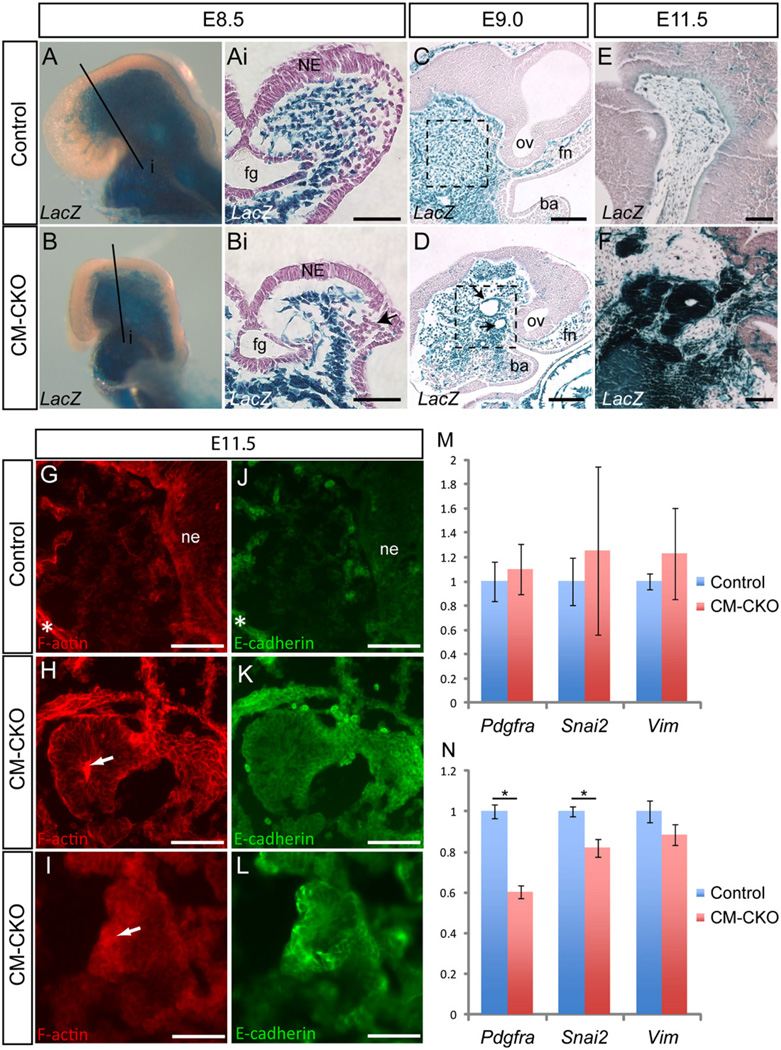
Abnormal distribution of Twist1-deficient mesoderm and acquisition of epithelial characteristics
In control (Twist13loxPneo/wt, Mesp1Cre/+, Rosa26R) and CM–CKO embryos at E8.5, the majority of cells in the cranial mesenchyme underneath the head folds were Mesp1-cre; Rosa26R positive (Fig. 5A and B). However the head folds of the CM–CKO embryo adopted a wide biconvex shape, instead of the elevated V-shaped configuration seen in control embryos (Fig. 5Ai and Bi). In control embryos, the mesoderm-derived cells ventral to the foregut region were densely packed, while those underneath the neural folds were loosely organized (Fig. 5Ai). In the CM–CKO embryos fewer β-galactosidase positive mesodermal cells were found underneath the neural plate while more cells were tightly clustered in the lateral region of the head folds (Fig. 5Bi, n=2). Ki67 immunostaining revealed decreased cell proliferation in the cranial mesoderm of E8.5 Twist1-deficient embryos (2.65%±s.e.m 0.002, n=4) compared to control (5.17%±0.019, n=3, p=0.045 by two-tailed t-test; Supplementary Fig. S4A, B). Whole mount TUNEL assay did not reveal elevated apoptosis in the cranial mesenchyme of E8.5 CM–CKO embryos (Supplementary Fig. S4C and D, n=3) or E9.5 (Supplementary Fig. S4E and F, n=3). To further test whether enhanced apoptosis may underpin the craniofacial phenotype, the effect of genetically ablating one or both functional Trp53 alleles (thus reducing apoptotic activity) on craniofacial development was examined in Twist1-null mutants, which display the same open neural tube phenotype as CM–CKO embryos. The craniofacial defects of Twist1-null embryos were not rescued by loss of one or both functional alleles of Trp53, suggesting that elevated cell death is unlikely to be a contributing factor of the null-mutant phenotype (Supplementary Fig. S4G–J).
Fig. 5.
Altered tissue architecture of Twist1-deficient cranial mesenchyme. (A–F) Staining for Rosa26R activity in embryos at E8.5 (A-Bi), E9.0 (C, D) and E11.5 (E, F). At E8.5 the Mesp1-Cre; Rosa26R positive mesodermal cells are found, underlying the neural epithelium (A and Ai). In the CM–CKO these cells are more tightly packed in the lateral region and are especially sparse in the medial region of the head folds (B and 5Bi. Black arrow in this figure Bi indicates clustered migrating cranial neural crest cells). (C–F) At E9.0 the Twist1-deficient CM cells in the CM–CKO embryo exhibit multiple foci of cystic structure lined with a thin epithelium (arrows, D). This is in contrast to the mesenchymal appearance of the cranial mesoderm of the control embryo (C). Dashed boxes indicate equivalent regions in control and CM–CKO tissue. This phenotype is exacerbated at E11.5 with the Mesp1-Cre; Rosa26R positive cells in the CM–CKO forming compact tissue mass of epithelial-like structure with a central lumen (F). This is not observed in the control (E). (G–L) Phalloidin staining to detect F-actin distribution (G–I) and immunofluorescence for E-cadherin (J–L) reveals that mesoderm-derived cells in the CM–CKO adopt an epithelial morphology and is some cases display polarized apical-basal distribution of cytoskeletal F-actin (white arrow, H) and express E-cadherin (K, L); these epithelial characteristics are not observed in the cranial mesenchyme of the control embryo (G, J). White asterisk in G and J indicates nonspecific staining of cartilage elements. (M, N) Quantitative RT–PCR analysis of mesenchymal genes (Pdgfra, Snai2 and Vim) in control and CM–CKO embryo heads at E8.5 (M) and E9.5 (N). At E8.5, no difference is observed between control and CM–CKO embryos, whereas at E9.5, mesenchymal markers are down-regulated in CM–CKO tissues. *P < 0.05 by two-tailed t-test, error bar=s.e.m. n=3 for all groups. ba, branchial arch; fg, foregut; fn, frontonasal; ne, neuroepithelium; ov, optic vesicle Scale bar=100 µm.
In E9.0 CM–CKO embryo, Mesp1-Cre; Rosa26R positive mesoderm cells adjacent to the midbrain and upper hindbrain were condensed and some were organized as cystic structures that were not seen in the control embryos (Fig. 5C and D, black arrows). By E11.5, this aberrant tissue phenotype was exacerbated as the densely packed mesenchyme formed clusters of epithelial structures (Fig. 5E, F). The appearance of these epithelial cyst-like structures suggests that the Twist1-deficient mesenchyme has acquired an atypical epithelial morphology, which is reminiscent of mesenchymal-epithelial transition (MET). These Rosa26R-positive structures were unlikely to have been derived from the neuroepithelium or endoderm, which do not express Mesp1-Cre or Twist1. Phalloidin staining revealed F-actin was concentrated at the luminal side of the cells, highlighting an epithelium-like apical-basal polarity in the Twist1-deficient cells (Fig. 5G–I, n=3). E-cadherin, which is present in adherens junctions of epithelial cells, was also detected above background levels in these epithelial-like cells (Fig. 5K–L, n=3). A very low level of E-cadherin was found in the mesenchyme of the control embryo (Fig. 5J, n=2). The Twist1-deficient mesoderm also showed a loss of mesenchymal properties with significantly reduced expression of Pdgfra and Snai2 in the cranial tissues of E9.5 CM–CKO embryo coinciding with the appearance of the atypical epithelialisation of the mesenchyme (Fig. 5M and N). Together, the data indicate that Twist1-deficient mesoderm cells lose mesenchymal characteristics and gain epithelial properties.
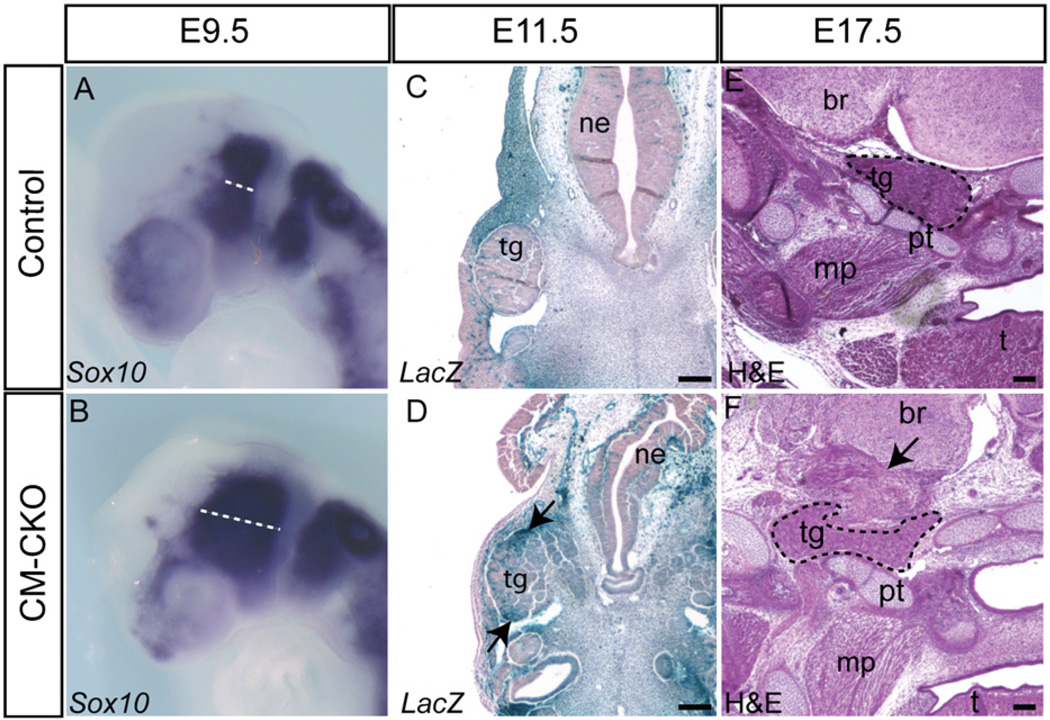
Altered neural crest cell distribution in CKO embryos
In Twist1-null embryos that lack Twist1 activity in both the CM and the CNC derivatives, migration of the CNC cells is affected and the patterning of the CNC-derived cranial ganglia is disrupted (Ota et al., 2004; Soo et al., 2002). In control embryos (Twist13loxPneo/wt, Mesp1Cre/+, Rosa26R), the unlabeled CNC cells were evenly distributed in the subectodermal region between the neural folds and the branchial arches (Fig. 5Ai). In contrast, the CNCs in the CM–CKO embryos clustered together in the space between the neural fold and dorsal surface ectoderm (Fig. 5Bi). In the CM–CKO embryos, Sox10-positive CNC cells of the anterior (pre-otic) and the posterior (post-otic) streams remained separate as in the control embryos, but both Sox10 positive streams appeared to spread to a wider domain (Fig. 6A and B, compare dashed lines n=3). At E17.5, the trigeminal ganglion was localized adjacent to part of the temporal bone in both the control and the CM–CKO embryos (Fig. 6E and F). However the ganglion of the CM–CKO embryos had an abnormal shape and the arrangement of skeletal elements around the ganglion was disorganized (Fig. 6E and F, black arrow). In E11.5 control embryos, CNC- derived trigeminal ganglion consisting mostly of unlabeled cells was clearly segregated from the surrounding β-galactosidase positive mesoderm-derived mesenchyme (Fig. 6C), whereas in CM–CKO embryos, Rosa26R positive cells intermingled with the Rosa26R negative cells (Fig. 6D). While no mesoderm-derived cells were localized inappropriately in the CNC-derived skeletal tissues, there was an increased contribution of β-galactosidase positive cells to the neuroepithelium (Fig. 3O, Supplementary Fig. S5). These invading cells could be endothelial, but the appearance and cellular arrangement of some clusters are reminiscent of the radial distribution of ES cell-derived neural progenitors that was observed in chimeric embryos (Tan et al., 1998). The intrusion of Twist1-deficient mesodermal cells to the CNC-derived tissues and the neurectoderm points to the possibility of a failure of the ability to segregate the mesoderm derivatives from other cranial tissues during morphogenesis.
Fig. 6.
Impact of the loss of Twist1 in the cranial mesoderm on cranial neural crest cells. (A, B) Sox10-expressing neural crest cells remain segregated into pre- and post-otic streams but are more widely spread in the CM–CKO embryo at E9.5 (B) compared to the control (A, dashed line). (C, D) Staining for Rosa26R activity showing that, while Rosa26R -positive mesodermal cells are clearly segregated from the trigeminal ganglion in the control embryo (C), those in the CM–CKO embryos (D) breach the tissue border and intermingle with cells in the ganglion (black arrow). (E, F) H and E staining showing that the trigeminal ganglion of E17.5 CM–CKO (F) adopts an abnormal shape compared to the control (E) and is packed amongst an ectopic tissue mass (arrow). Abbreviations: br, brain; mp, medial pterygoid muscle; ne, neurepithelium; pt, petrous part of temporal bone; t, tongue; tg, trigeminal ganglion. Scale bar=100 µm.
Discussion
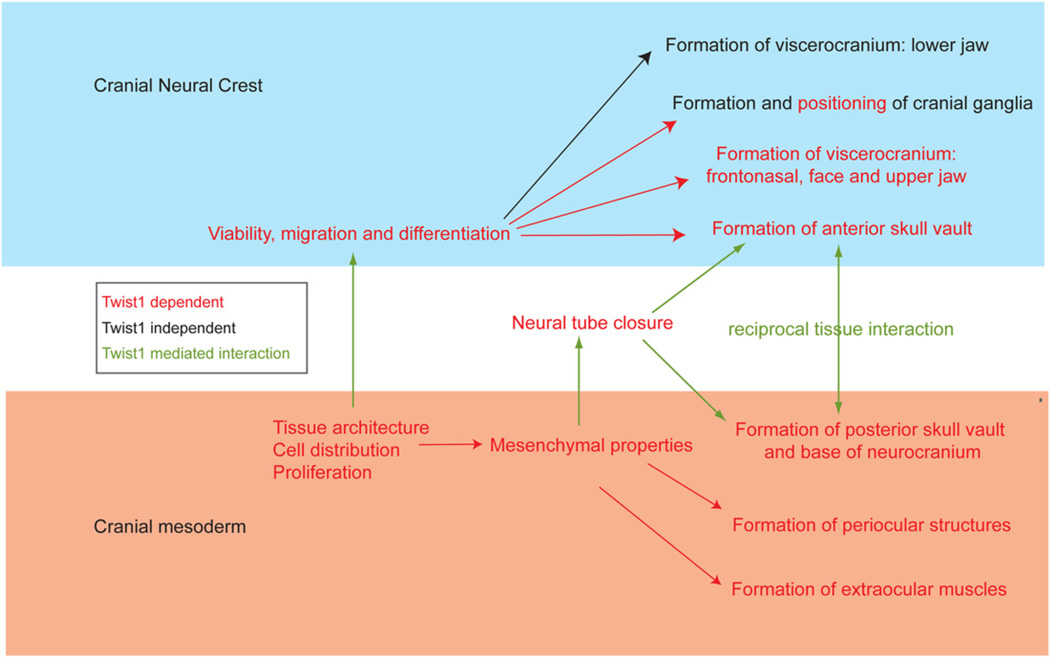
Twist1 is required for maintaining mesenchymal tissue architecture
In the present study, we have uncovered a mesoderm-specific requirement for Twist1, which differs from its role in the CNC (Fig. 7). Although the Twist1 transcript can be detected in the mesoderm during gastrulation (this study; Fuchtbauer, 1995; Stoetzel et al., 1995), Twist1 protein was not detectable until the 2-somite stage. At this stage, the mesenchymal cells are evenly distributed in the cephalic region in wild-type embryos. In CM–CKO embryos, the earliest defect that could be detected was a loss of the normal mesenchymal properties, initially revealed by changes in cell density and later a tendency to undergo a transition to an epithelial architecture. This indicates a failure to properly maintain the cells in a mesenchymal progenitor state. At E9.0, localized clumps of Mesp1-Cre; Rosa26R positive cells were found in the cranial mesoderm, and some compacted clusters acquired an epithelial cystic morphology. This phenomenon is reminiscent of a reversal of the transition from an epithelial to mesenchymal cell morphology. Activation of Twist1 expression is known to play a role in inducing epithelial–mesenchymal transition (EMT), and overexpression of Twist1 has been associated with the metastatic behavior of tumor cells (Karreth and Tuveson, 2004; Qin et al., 2012; Wu, 2011). This may be mediated either directly by the interaction of Twist1 with the regulatory element of E-cadherin to repress its expression (Yang et al., 2004; Yang et al., 2007) (Fu et al., 2011) or through activation of Snail2 gene expression, which is also known to repress E-cadherin (Casas et al., 2011). Reversion of EMT has been demonstrated in Twist1-depleted gastric cancer cells (Feng et al., 2009). To the best of our knowledge, the present study provides the first demonstration that Twist1 is required for the maintenance of mesenchymal properties of mesoderm cells during embryonic development. Consistent with the concept that loss of Twist1 activity can lead to the acquisition of epithelial characteristics, expression of E-cadherin was up-regulated in these de novo epithelial structures in the CM–CKO embryo, whilst Pdgrfa and Snai2 were down-regulated. The inability to maintain the expression of these mesenchymal specific genes and the repression of E-cadherin in the absence of Twist1 expression might account for this aberrant behavior of the Twist1-deficient mesenchyme.
Fig. 7.
The functional roles of Twist1 in craniofacial development. Twist1 is required to sustain the viability and the regionalization of the cranial neural crest cells destined to form the skull vault, base and frontonasal region including the upper jaw and the cranial ganglia. In the cranial mesoderm, Twist1 is required to maintain the mesenchymal architecture and proliferative capacity, which secondarily influences the migration of the cranial neural crest cells and the closure of the neural tube. Normal neural tube morphogenesis offers the morphogenetic options for the cranial neural crest cells and the mesoderm to localize to the sites of the skull vault. The formation of the bones in the skull vault is also dependent on the reciprocal interactions of these two cell types which is dependent on Twist1 activity in both cell populations.
Tissue-specific effect of loss of Twist1 function on neural tube closure
Previously we have shown that ablation of Twist1 in the CNC results in increased apoptosis and a reduced capacity for the remaining cells to initiate osteogenic differentiation [Fig. 7, upper part (Bildsoe et al., 2009; Ota et al., 2004)]. The combination of these effects may account for the loss of CNC-derived skeletal structures in CNC conditional mutant embryos (Bildsoe et al., 2009). In contrast, loss of Twist1 in the early cranial mesoderm has no effect on apoptosis but leads to less cell proliferation, suggesting that the population of mesoderm progenitors may be reduced (Fig. 7 lower part). In addition to a paucity of mesenchymal cells underneath the neural plate, the bulk of them are sequestered away from to the surface ectoderm and they are densely packed with very little extracellular space, suggesting that the ECM may be deficient. This change in the mesenchyme characteristics may have a secondary impact on the closure of the neural tube. The cranial mesenchyme is known to be essential for supporting neural tube formation. The morphogenetic forces generated by the increasing rigidity and the volume of the mesenchyme due to expansion of the hyaluronate-enriched extracellular matrix (ECM) can facilitate the elevation and folding of the neural plate (Morris-Wiman and Brinkley, 1990). Since the neural tube closure is unaffected by the loss of Twist1 in the cranial neural crest cells, the lack of growth or ECM expansion of the Twist1-deficient cranial mesenchyme is therefore likely to directly impact on the closure of the neural tube (Fig. 7, and see Zohn and Sarkar, 2012).
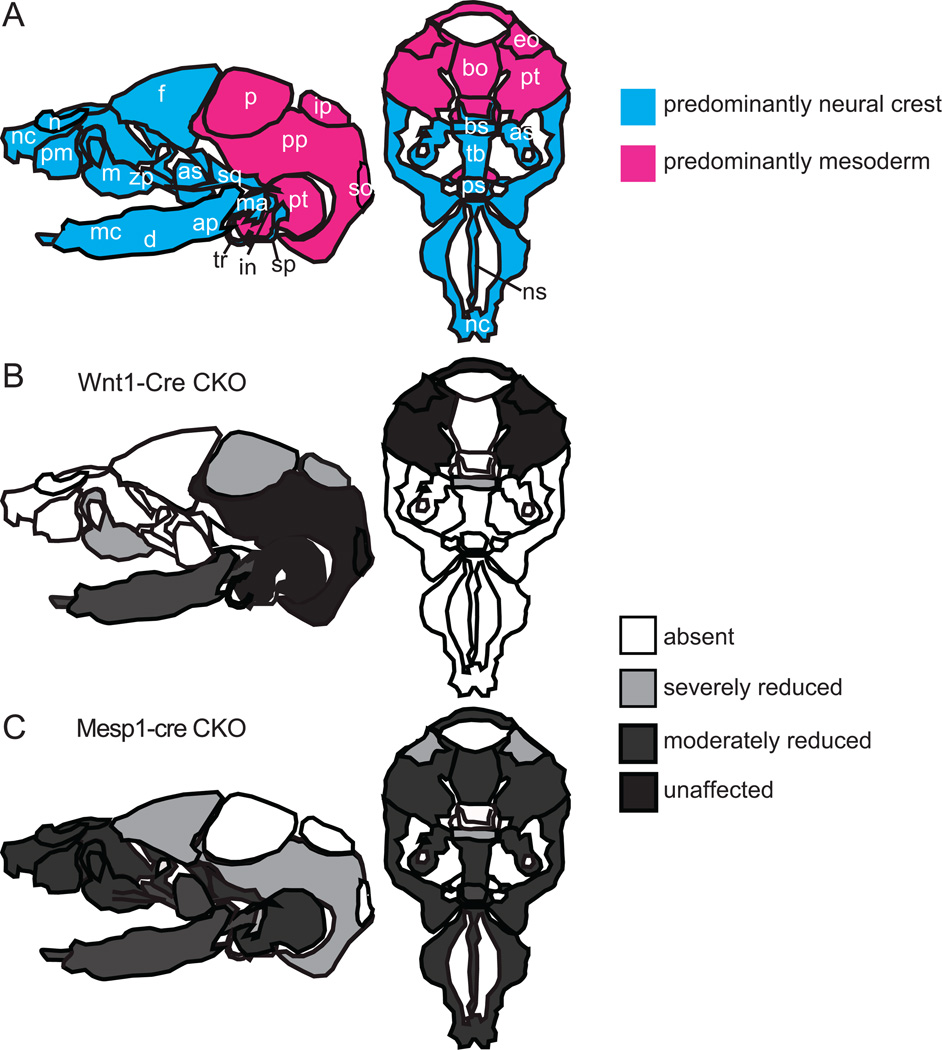
Loss of Twist1 in the mesoderm results in defects of both mesodermal and neural crest-derived bones
In this study, we have shown that tissue specific deletion of Twist1 leads to a loss or reduction of mesoderm-derived bones as well as some CNC-derived skull vault elements (Fig. 8A, C). It is unclear whether the loss of Twist1 function affects the differentiation of membranous bones resulting in malformation of the CM derived skeletal elements, or this is due to a secondary affect caused by the failure of neural tube closure creating a physical hindrance that disrupts the correct localization of the skull progenitor tissues in the head. The brain lacks a covering of mesoderm-derived cells. Instead, large aggregates of Mesp1-Cre; Rosa26R-positive cells was observed in the regions lateral to the occipital part of the head. The precise origin and fate of these Twist1-deficient mesoderm cells are not known, but it is possible that they are bone precursor cells that failed to localize to the appropriate sites and did not receive the proper inductive interactions for bone formation. Loss of Twist1 also affects mesoderm-derived bones in the skull base, leading to reduced size and abnormal shape of the bones and, in the case of the basioccipital, its mis-alignment with the skull base. These abnormalities are unlikely to be due to any physical hindrance to cell localization. Twist1 is known to negatively modulate the progression of chondrogenesis and osteogenesis and to maintain the mesenchymal cells in an immature state in vitro (Dong et al., 2007; Miraoui et al., 2010; Miraoui and Marie, 2010). Twist1 also interacts with Sox9 and Runx2 to inhibit their action in chondrogenesis and osteoblast differentiation respectively (Bialek et al., 2004; Gu et al., 2012; Rice et al., 2000). The maintenance of the immature state is likely to be a prerequisite for maintaining the capacity to differentiate, since differentiation of Twist1-deficient CNC cells is impaired in conditional knockout embryos (Bildsoe et al., 2009).
Fig. 8.
The impact of loss of Twist1 function on the formation of skull bones. (A) Components of the neurocranium and viscerocranium that are derived predominantly from cranial neural crest (CNC) and cranial mesoderm (CM) respectively. CNC cells give rise to the entire viscerocranium (upper and lower jaw and the nasal region) and to the anterior part of the skull vault and base (blue elements), whereas the CM make up the posterior elements of the skull vault and base (pink elements). (B) Skull bones that are affected by the loss of Twist1 in the CNC cells. Most severely affected are the elements of the CNC-derived viscerocranium and the frontal bones, which are either absent or reduced in size. In addition, CM-derived bone such as the parietal and interparietal bones are also affected. (C) Loss Twist1 in the CM leads to absence of parietal, interparietal, supraoccipital and suprachiasmatic bones and deformation of skull base bones, as well as the drastically reduced or lost CNC-derived frontal bones. Abbreviations: ap, angular process; as, alisphenoid; bo, basioccipital; bs, basisphenoid; d, dentary; eo, exoccipital; f, frontal; in, incus; ip, interparietal; m, maxilla; ma, malleus; mc, Meckel’s cartilage; n, nasal; nc, nasal capsule; ns, nasal septum; p, parietal; pm, premaxilla; pp, parietal plate; ps, prespenoid; pt, petrous part of temporal bone; so, supraoccipital; sp, styloid process; sq, squamous; tb, trabecular basal plate; tr, tympanic ring; zp, zygomatic process. Data are collated from Bildsoe et al., 2009 and this study.
The impact of loss of Twist1 in the mesoderm also affects skull bones derived from the CNC cells such as the frontal bone, which was greatly reduced in size (Fig. 8C). This is consistent with an indirect effect due to the physical barrier created by the failure to close the cranial neural tube, preventing the bone precursors from forming proper frontal bones. However, previous studies have shown that Twist1 is required for interactions between the mesoderm and the CNC cells. In embryos in which Twist1 was ablated in the CNC (Bildsoe et al., 2009), loss of skull elements also encompasses those of mesoderm origin (such as the parietal, interparietal and supraoccipital bones; Fig. 8B). This is most likely due to the lack of inductive interactions between CNC-derived meninges, which are lost in the absence of Twist1, and the mesodermal bone progenitors (Gagan et al., 2007). Twist1 may also mediate the influence of the mesoderm on migratory CNC cells. Cell transplantation experiments (Soo et al., 2002) have shown that wild-type CNC cells were less efficient in homing to the branchial arches in the Twist1-null host embryos. In the present study, we observed that migrating CNC cells occupied a wider domain in CKO embryos and that the mesoderm cells breached the boundary between the mesenchyme and the CNC-derived ganglia and intermingled with the cells of the ganglion. Collectively, these findings suggest Twist1 may be required for proper osteogenic differentiation of both CM and CNC cells, and that the loss of gene function in one tissue may affect the other due to the disruption of the reciprocal tissue interactions.
Development of peri-ocular structures is affected by loss of Twist1 in the mesoderm
A negative correlation between Twist1 expression and myoblast differentiation has been observed in muscle development (Koutsoulidou et al., 2011). Twist1 can dimerize competitively with other basic helix–loop–helix proteins such as MyoD preventing them from binding with E-proteins and inhibiting the trans-activation of downstream targets, suppressing myogenesis (Franco et al., 2011; Spicer et al., 1996). In the absence of Twist1 activity in the mesoderm, there was no negative effect on myogenic differentiation. Branchial arch (BA)-derived muscles of appropriate histology and molecular characteristics were formed in the CM–CKO embryos. However, the arrangement of the muscles differs from that in the wild type embryo. This may be related to the absence and misplacement of the skull elements to which the muscles are attached. In contrast to the BA-derived muscles, the formation of the EOM was severely impaired in CM–CKO mutant embryos.
EOM are derived from prechordal mesoderm with additional contributions from the cranial paraxial mesoderm, whereas the BA-derived muscle mainly originate from lateral parts of the cranial mesoderm (Lescroart et al., 2010; Noden and Francis-West, 2006; Sambasivan et al., 2011b). One of the elements in the peri-orbital skeleton is the hypochiasmatic cartilage, which develops as an isolated mesoderm-derived structure embedded in the CNC-derived anterior skull base. This cartilage forms a bridge between the orbital cartilage and the skull base, to which the EOM attaches. It has been proposed, based on their spatial association, that the EOM and the hypochiasmatic cartilage share a common origin in the anterior cranial mesoderm (McBratney-Owen et al., 2008). At E9.5, Twist1 is no longer expressed in the mesoderm cells in the core of the branchial arches, which have initiated myogenic differentiation (Rinon et al., 2007) but Twist1 expression is sustained in the tissues in other parts of the CM at this stage, including the region that is likely to contain the progenitors of the EOMs. Twist1 expression is also maintained in a cell line derived from EOM, but not in a skeletal muscle-derived cell line (Porter et al., 2006). Our Rosa26R reporter results show that the poor development or absence of EOM may be caused by a reduction of the progenitor population. Absence of Twist1 activity in the peri-ocular tissue may lead to the failure to maintain the progenitors prior to the formation of the EOMs and the hypochiasmatic cartilage. This scenario is consistent with the concept that the function of Twist1 is to maintain the progenitor state of the mesoderm cells and thereby control the initiation of lineage differentiation.
Supplementary Material
Acknowledgments
We thank Atsushi Yoshiki, Yumiko Saga and Richard Harvey for providing the Mesp1-Cre mice, Phillipe Soriano for Rosa26R mice and the following scientists for providing the molecular reagents: S. Tajbakhsh (Myf5) J.C. Izpisúa Belmonte (Pitx2), M. Hargrave (Sox10), E. Tzahor (Tbx1) and W. Shawlot (Twist1). We thank the staff from the CMRI Bioservices Unit for animal husbandry. This project is supported by a grant from the Australian Research Council (DP 1094008) and by Mr James Fairfax. HB is a National Health and Medical Research Council (NHMRC) Postgraduate Scholar, DAFL is a CMRI Kimberly-Clark Research Fellow and PPLT is a NHMRC Senior Principal Research Fellow (Grant 1003100).
Footnotes
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2012.12.004.
References
- Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, Justice MJ, Karsenty G. A twist code determines the onset of osteoblast differentiation. Dev. Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- Bildsoe H, Loebel DA, Jones VJ, Chen YT, Behringer RR, Tam PP. Requirement for Twist1 in frontonasal and skull vault development in the mouse embryo. Dev. Biol. 2009;331:176–188. doi: 10.1016/j.ydbio.2009.04.034. [DOI] [PubMed] [Google Scholar]
- Bronner ME, Ledouarin NM. Development and evolution of the neural crest: an overview. Dev. Biol. 2012;366:2–9. doi: 10.1016/j.ydbio.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas E, Kim J, Bendesky A, Ohno-Machado L, Wolfe CJ, Yang J. Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer Res. 2011;71:245–254. doi: 10.1158/0008-5472.CAN-10-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YT, Akinwunmi PO, Deng JM, Tam OH, Behringer RR. Generation of a Twist1 conditional null allele in the mouse. Genesis. 2007;45:588–592. doi: 10.1002/dvg.20332. [DOI] [PubMed] [Google Scholar]
- Chen ZF, Behringer RR. Twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 1995;9:686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- Couly GF, Coltey PM, Le Douarin NM. The developmental fate of the cephalic mesoderm in quail–chick chimeras. Development. 1992;114:1–15. doi: 10.1242/dev.114.1.1. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Tucker AS, Sharpe PT. Craniofacial Development. In: Rossant J, Tam PPL, editors. Mouse Development. San Diego: Academic Press; 2002. pp. 421–498. [Google Scholar]
- Dong YF, Soung Y, Chang Y, Enomoto-Iwamoto M, Paris M, O’Keefe RJ, Schwarz EM, Drissi H. Transforming growth factor-beta and Wnt signals regulate chondrocyte differentiation through Twist1 in a stage-specific manner. Mol. Endocrinol. 2007;21:2805–2820. doi: 10.1210/me.2007-0199. [DOI] [PubMed] [Google Scholar]
- El Ghouzzi V, Le Merrer M, Perrin-Schmitt F, Lajeunie E, Benit P, Renier D, Bourgeois P, Bolcato-Bellemin AL, Munnich A, Bonaventure J. Mutations of the TWIST gene in the Saethre–Chotzen syndrome. Nat. Genet. 1997;15:42–46. doi: 10.1038/ng0197-42. [DOI] [PubMed] [Google Scholar]
- Feng MY, Wang K, Shi QT, Yu XW, Geng JS. Gene expression profiling in TWIST-depleted gastric cancer cells. Anat. Rec. (Hoboken.) 2009;292:262–270. doi: 10.1002/ar.20802. [DOI] [PubMed] [Google Scholar]
- Franco HL, Casasnovas J, Rodriguez-Medina JR, Cadilla CL. Redundant or separate entities?—Roles of Twist1 and Twist2 as molecular switches during gene transcription. Nucl. Acids Res. 2011;39:1177–1186. doi: 10.1093/nar/gkq890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Qin L, He T, Qin J, Hong J, Wong J, Liao L, Xu J. The Twist/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res. 2011;21:275–289. doi: 10.1038/cr.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchtbauer EM. Expression of M-Twist during postimplantation development of the mouse. Dev Dyn. 1995;204:316–322. doi: 10.1002/aja.1002040309. [DOI] [PubMed] [Google Scholar]
- Gagan JR, Tholpady SS, Ogle RC. Cellular dynamics and tissue interactions of the dura mater during head development. Birth Defects Res. C Embryo. Today. 2007;81:297–304. doi: 10.1002/bdrc.20104. [DOI] [PubMed] [Google Scholar]
- Gitelman I. Twist protein in mouse embryogenesis. Dev. Biol. 1997;189:205–214. doi: 10.1006/dbio.1997.8614. [DOI] [PubMed] [Google Scholar]
- Gu S, Boyer TG, Naski MC. Basic helix–loop–-helix transcription factor twist1 inhibits the transacivator function of the master chondrogenic regulator Sox9. J. Biol. Chem. 2012;287(25):21082–21092. doi: 10.1074/jbc.M111.328567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Second. Plainview, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Karreth F, Tuveson DA. Twist induces an epithelial–mesenchymal transition to facilitate tumor metastasis. Cancer Biol. Ther. 2004;3:1058–1059. doi: 10.4161/cbt.3.11.1302. [DOI] [PubMed] [Google Scholar]
- Kontges G, Lumsden A. Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development. 1996;122:3229–3242. doi: 10.1242/dev.122.10.3229. [DOI] [PubMed] [Google Scholar]
- Koutsoulidou A, Mastroyiannopoulos NP, Furling D, Uney JB, Phylactou LA. Endogenous TWIST expression and differentiation are opposite during human muscle development. Muscle Nerve. 2011;44:984–986. doi: 10.1002/mus.22241. [DOI] [PubMed] [Google Scholar]
- Lescroart F, Kelly RG, Le Garrec JF, Nicolas JF, Meilhac SM, Buckingham M. Clonal analysis reveals common lineage relationships between head muscles and second heart field derivatives in the mouse embryo. Development. 2010;137:3269–3279. doi: 10.1242/dev.050674. [DOI] [PubMed] [Google Scholar]
- Loebel DA, Hor AC, Bildsoe H, Jones V, Chen YT, Behringer RR, Tam PP. Regionalized Twist1 activity in the forelimb bud drives the morphogenesis of the proximal and preaxial skeleton. Dev. Biol. 2012;362:132–140. doi: 10.1016/j.ydbio.2011.11.020. [DOI] [PubMed] [Google Scholar]
- Loebel DA, Tsoi B, Wong N, O’Rourke MP, Tam PP. Restricted expression of ETn-related sequences during post-implantation mouse development. Gene Expr. Patterns. 2004;4:467–471. doi: 10.1016/j.modgep.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Mani P, Jarrell A, Myers J, Atit R. Visualizing canonical Wnt signaling during mouse craniofacial development. Dev. Dyn. 2010;239:354–363. doi: 10.1002/dvdy.22072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Barbera JP, Rodriguez TA, Greene NDE, Weninger WJ, Simeone A, Copp AJ, Beddington RSP, Dunwoodie S. Folic acid prevents exencephaly in Cited2 deficient mice. Human Mol. Genet. 2002;11:283–293. doi: 10.1093/hmg/11.3.283. [DOI] [PubMed] [Google Scholar]
- McBratney-Owen B, Iseki S, Bamforth SD, Olsen BR, Morriss-Kay GM. Development and tissue origins of the mammalian cranial base. Dev. Biol. 2008;322:121–132. doi: 10.1016/j.ydbio.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoux M, Rijli FM. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development. 2010;137:2605–2621. doi: 10.1242/dev.040048. [DOI] [PubMed] [Google Scholar]
- Miraoui H, Marie PJ. Pivotal role of Twist in skeletal biology and pathology. Gene. 2010;468:1–7. doi: 10.1016/j.gene.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Miraoui H, Severe N, Vaudin P, Pages JC, Marie PJ. Molecular silencing of Twist1 enhances osteogenic differentiation of murine mesenchymal stem cells: implication of FGFR2 signaling. J. Cell Biochem. 2010;110:1147–1154. doi: 10.1002/jcb.22628. [DOI] [PubMed] [Google Scholar]
- Morris-Wiman J, Brinkley LL. Changes in mesenchymal cell and hyaluronate distribution correlate with in vivo elevation of the mouse mesencephalic neural folds. Anat. Rec. 1990;226:383–395. doi: 10.1002/ar.1092260316. [DOI] [PubMed] [Google Scholar]
- Noden DM, Francis-West P. The differentiation and morphogenesis of craniofacial muscles. Dev. Dyn. 2006;235:1194–1218. doi: 10.1002/dvdy.20697. [DOI] [PubMed] [Google Scholar]
- Ota MS, Loebel DA, O’Rourke MP, Wong N, Tsoi B, Tam PP. Twist is required for patterning the cranial nerves and maintaining the viability of mesodermal cells. Dev. Dyn. 2004;230:216–228. doi: 10.1002/dvdy.20047. [DOI] [PubMed] [Google Scholar]
- Porter JD, Israel S, Gong B, Merriam AP, Feuerman J, Khanna S, Kaminski HJ. Distinctive morphological and gene/protein expression signatures during myogenesis in novel cell lines from extraocular and hindlimb muscle. Physiol. Genomics. 2006;24:264–275. doi: 10.1152/physiolgenomics.00234.2004. [DOI] [PubMed] [Google Scholar]
- Qin Q, Xu Y, He T, Qin C, Xu J. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res. 2012;22:90–106. doi: 10.1038/cr.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice DP, Aberg T, Chan Y, Tang Z, Kettunen PJ, Pakarinen L, Maxson RE, Thesleff I. Integration of FGF and TWIST in calvarial bone and suture development. Development. 2000;127:1845–1855. doi: 10.1242/dev.127.9.1845. [DOI] [PubMed] [Google Scholar]
- Rinon A, Lazar S, Marshall H, Buchmann-Moller S, Neufeld A, Elhanany-Tamir H, Taketo MM, Sommer L, Krumlauf R, Tzahor E. Cranial neural crest cells regulate head muscle patterning and differentiation during vertebrate embryogenesis. Development. 2007;134:3065–3075. doi: 10.1242/dev.002501. [DOI] [PubMed] [Google Scholar]
- Rivera-Perez JA, Wakamiya M, Behringer RR. Goosecoid acts cell autonomously in mesenchyme-derived tissues during craniofacial development. Development. 1999;126:3811–3821. doi: 10.1242/dev.126.17.3811. [DOI] [PubMed] [Google Scholar]
- Rose CS, Malcolm S. A TWIST in development. Trends Genet. 1997;13:384–387. doi: 10.1016/s0168-9525(97)01296-1. [DOI] [PubMed] [Google Scholar]
- Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- Sambasivan R, Kuratani S, Tajbakhsh S. An eye on the head: the development and evolution of craniofacial muscles. Development. 2011;138:2401–2415. doi: 10.1242/dev.040972. [DOI] [PubMed] [Google Scholar]
- Sambasivan R, Kuratani S, Tajbakhsh S. An eye on the head: the development and evolution of craniofacial muscles. Development. 2011b;138:2401–2415. doi: 10.1242/dev.040972. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. Development and evolution of the migratory neural crest: a gene regulatory perspective. Curr. Opin. Genet. Dev. 2006;16:360–366. doi: 10.1016/j.gde.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Soo K, O’Rourke MP, Khoo PL, Steiner KA, Wong N, Behringer RR, Tam PP. Twist function is required for the morphogenesis of the cephalic neural tube and the differentiation of the cranial neural crest cells in the mouse embryo. Dev. Biol. 2002;247:251–270. doi: 10.1006/dbio.2002.0699. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Spicer DB, Rhee J, Cheung WL, Lassar AB. Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science. 1996;272:1476–1480. doi: 10.1126/science.272.5267.1476. [DOI] [PubMed] [Google Scholar]
- Stoetzel C, Weber B, Bourgeois P, Bolcato-Bellemin AL, Perrin-Schmitt F. Dorso-ventral and rostro-caudal sequential expression of M-twist in the postimplantation murine embryo. Mech. Dev. 1995;51:251–263. doi: 10.1016/0925-4773(95)00369-x. [DOI] [PubMed] [Google Scholar]
- Tan SS, Kalloniatis M, Sturm K, Tam PP, Reese BE, Faulkner-Jones B. Separate progenitors for radial and tangential cell dispersion during development of the cerebral neocortex. Neuron. 1998;21:295–304. doi: 10.1016/s0896-6273(00)80539-5. [DOI] [PubMed] [Google Scholar]
- Thisse B, el MM, Perrin-Schmitt F. The twist gene: isolation of a Drosophila zygotic gene necessary for the establishment of dorsoventral pattern. Nucleic Acids Res. 1987;15:3439–3453. doi: 10.1093/nar/15.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CM, Trainor PA, Radziewic T, Pelka GJ, Zhou SX, Parameswaran M, Quinlan GA, Gordon M, Sturm K, Tam PP. Application of lacZ transgenic mice to cell lineage studies. Meth. Mol. Biol. 2008;461:149–164. doi: 10.1007/978-1-60327-483-8_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KJ. Direct activation of Bmi1 by Twist1: implications in cancer stemness, epithelial–mesenchymal transition, and clinical significance. Chang Gung. Med. J. 2011;34:229–238. [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhang X, Gang H, Li X, Li Z, Wang T, Han J, Luo T, Wen F, Wu X. Up-regulation of gastric cancer cell invasion by Twist is accompanied by N-cadherin and fibronectin expression. Biochem. Biophys. Res. Commun. 2007;358:925–930. doi: 10.1016/j.bbrc.2007.05.023. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Vivatbutsiri P, Morriss-Kay G, Saga Y, Iseki S. Cell lineage in mammalian craniofacial mesenchyme. Mech. Dev. 2008;125:797–808. doi: 10.1016/j.mod.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Zacharias AL, Lewandoski M, Rudnicki MA, Gage PJ. Pitx2 is an upstream activator of extraocular myogenesis and survival. Dev. Biol. 2011;349:395–405. doi: 10.1016/j.ydbio.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohn IE, Sarkar AA. Does the cranial mesenchyme contribute to neural fold elevation during neurulation? Birth Defects Res. A Clin. Mol. Teratol. 2012;94:841–848. doi: 10.1002/bdra.23073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.