Abstract
Background.
Gastric adenocarcinoma is a heterogenous disease that results from complex interactions between environmental and genetic factors, which may contribute to the disparate outcomes observed between different patient populations. This study aimed to determine whether genomic differences exist in a diverse population of patients by evaluating tumor mutational profiles stratified by race.
Methods.
All patients with gastric adenocarcinoma between 2012 and 2016 who underwent targeted next-generation sequencing of cancer genes by the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets platform were identified. Patient race was categorized as Asian, African American, Hispanic, or Caucasian. Fisher’s exact test was used to examine differences in mutation rates between racial designations for the most common mutations identified. The p values in this study were adjusted using the false discovery rate method.
Results.
The study investigated 595 mutations in 119 patients. The DNA alterations identified included missense mutations (66%), frame-shift deletions (13%), and nonsense mutations (9%). Silent mutations were excluded. The most frequently mutated genes were ARID1A, CDH1, ERBB3, KRAS, PIK3CA, and TP53. Of these, TP53 was the most frequently mutated gene, affecting 50% of patients. The proportion of patients with TP53 mutations differed significantly between races (p = 0.012). The findings showed TP53 mutations for 89% (16/18) of the African American patients, 56% (10/18) of the Asian patients, 43% (9/21) of the Hispanic patients, and 40% (25/62) of the Caucasian patients.
Conclusions.
Significantly higher rates of TP53 mutations were identified among the African American patients with gastric adenocarcinoma. This is the first study to evaluate tumor genomic differences in a diverse population of patients with gastric adenocarcinoma.
Gastric adenocarcinoma is the third leading cause of cancer-related death worldwide, and the incidence and mortality rates differ greatly by geography and race/ethnicity.1 For instance, gastric adenocarcinoma in Western (Europe and the United States) versus Asian populations is associated with differences in both presenting stage and overall survival.2 The reported 5-year overall survival periods of patients in studies from Japan (54%), Korea (58%), and China (31%) are longer than those reported in studies from the United States (29%) and Canada (25%).3
The clinicopathologic presentation of gastric carcinoma also varies widely between Asian and Western countries. These variations include differences in histology (intestinal vs diffuse subtypes), location (distal vs proximal stomach), environmental exposures, dietary factors, and Helicobacter pylori status.4–6
Of the many factors associated with long-term survival of gastric cancer patients, techniques of gastrectomy and systemic therapy have been studied considerably. Although less studied, biologic variations in gastric tumors among races may in part drive some of the observed survival disparities. Recent molecular characterizations of gastric adenocarcinomas demonstrate a significant molecular heterogeneity in this cancer.7,8
Although gastric adenocarcinoma is less common in the United States than in the rest of the world, patterns of survival are confounded by the diverse racial and ethnic profile of the U.S. population.9 Data from the Surveillance, Epidemiology, and End Results (SEER) program of the National Cancer Institute showed a worse disease-specific survival for African American patients with gastric cancer than for their Asian and Caucasian counterparts.10 Although this discrepancy could derive from access to care and socioeconomic status, it could also be related to differences in tumor biology.
We hypothesized that differences in tumor genomics may account for some of the discrepency in outcomes between ethnic groups. Our primary aim was to evaluate tumor genomic differences in a diverse population of patients with gastric adenocarcinoma. Using single-institution data, we sought to identify alterations in the tumor genome among defined racial groups that may relate to differences in the biology of gastric cancer.
METHODS
All patients with gastric adenocarcinoma treated at the Memorial Sloan Kettering Cancer Center between 2012 and 2016 who had tumor evaluation by a targeted next-generation DNA sequencing assay were identified retrospectively. Patients were included in the study irrespective of race, stage of disease, prior treatment, or resection status. Tissue from primary tumors and metastasis, if no primary tumor sample was available, also were included in genomic evaluations.
Patients were excluded from the study if they had gastroesophageal (GE)-junction tumors or if the tumor could not be classified according to the Lauren classification (intestinal, diffuse, or mixed type). Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) analysis and clinicopathologic data [sex, age, race, tumor-node-metastasis (TNM), survival] were collected from a prospectively maintained institutional database.
Genomic Evaluation
The MSK-IMPACT platform is a hybridization capture-based next-generation sequencing assay for targeted deep sequencing of all exons and selected introns of key cancer-associated genes. Initialy MSK-IMPACT included full exon coverage of 341 genes. In a second iteration, this was expanded to a total of 410 genes. Instead of sequencing a tumor’s whole genome or the entire part of the genome that codes for proteins, MSK-IMPACT analyzes the most important cancer genes, which are captured and sequenced on an illumina HiSeq platform. This targeted sequencing approach makes the analysis of tumor tissue more feasible and effective and increases the chance of finding clinically relevant genetic changes. The genes evaluated by MSK-IMPACT are those that have been shown to play a role in the development or phenotype of tumors.11
Statistical Analyses
Descriptive clinical, pathologic, and mutation characteristics were presented using medians and ranges for continuous variables, and using frequencies and percentages for categorical variables. Race was categorized as Caucasian, African American, Asian, or Hispanic. Three patients had both a primary sample and metastatic samples available, but we included only the primary tumor sample for the analysis. All silent mutations were excluded.
Heat maps were used to assess the overall mutation pattern and the patterns by race. Any gene with a mutation present in at least 10 patients, irrespective of race, was included in the analyses.
Fisher’s exact test was used to examine the differences in mutation rates between races. For adjustment of p values, the false discovery rate method was used. Adjusted p values lower than 0.05 and two-tailed were considered statistically significant. All analyses were performed using SAS 9.4 (The SAS Institute, Cary, NC, USA) and Cran R Version 3.3.0 (R Core Development Team, Vienna, Austria).
RESULTS
The analysis included 595 mutations in 119 patients. The study patients comprised 62 Caucasians (52.1%), 18 African Americans (15.1%), 18 Asians (15.1%), and 21 Hispanics (17.6%). Patient and pathologic characteristics as stratified by race are detailed in Tables 1 and 2. Of the 119 samples included, 87 (73%) were from the primary tumor, and the remaining samples were from the metastatic sites.
TABLE 1.
Patient characteristics (n = 119)
| n (%) | |
|---|---|
| Gender | |
| Male | 65 (55) |
| Female | 54 (45) |
| Age | |
| Median | 56 |
| Range | 23–84 |
| Race | |
| Caucasian | 62 (52) |
| African American | 18 (15) |
| Asian | 18 (15) |
| Hispanic | 21 (18) |
| Stage | |
| 1 | 11 (9) |
| 2 | 16 (13) |
| 3 | 18 (15) |
| 4 | 74 (62) |
Due to rounding, percentages may not add up exactly to 100%
TABLE 2.
Characteristics by race (n = 119)
| Caucasian (n = 62) | AA (n = 18) | Asian (n = 18) | Hispanic (n = 21) | All (n = 119) | |
|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | n (%) | |
| Lauren classifications | |||||
| Diffuse | 12 (19) | 3 (17) | 2 (11) | 7 (33) | 24 (20) |
| Intestinal | 11 (18) | 8 (44) | 6 (33) | 2 (10) | 27 (23) |
| Mixed | 9 (15) | 1 (6) | 4 (22) | 2 (10) | 16 (13) |
| Unknown | 30 (48) | 6 (33) | 6 (33) | 10 (48) | 52 (44) |
| Tumor location | |||||
| Distal | 19 (31) | 5 (28) | 7 (39) | 5 (24) | 36 (30) |
| Middle | 17 (27) | 7 (39) | 10 (56) | 7 (33) | 41 (34) |
| Proximal | 8 (13) | 0 (0) | 1 (6) | 1 (5) | 10 (8) |
| Whole | 0 (0) | 1 (6) | 0 (0) | 0 (0) | 1 (1) |
| Unknown | 18 (29) | 5 (28) | 0 (0) | 8 (38) | 31 (26) |
| Stage | |||||
| 1 | 5 (8) | 2 (11) | 4 (22) | 0 (0) | 11 (9) |
| 2 | 8 (13) | 3 (17) | 4 (22) | 1 (5) | 16 (13) |
| 3 | 8 (13) | 2 (11) | 1 (6) | 7 (33) | 18 (15) |
| 4 | 41 (66) | 11 (61) | 9 (50) | 13 (62) | 74 (62) |
| Grade | |||||
| Moderate | 11 (18) | 6 (33) | 3 (17) | 4 (19) | 24 (20) |
| Moderate to poor | 7 (11) | 3 (17) | 3 (17) | 3 (14) | 16 (13) |
| Poor | 42 (68) | 9 (50) | 11 (61) | 14 (67) | 76 (64) |
| Unknown | 2 (3) | 0 (0) | 1 (6) | 0 (0) | 3 (3) |
Due to rounding, percentages may not add up exactly to 100%
AA African American
The following six genes were mutated in 10 or more patients: ARID1A, CDH1, ERBB3, KRAS, PIK3CA, and TP53. The most prevalent mutated gene was TP53, with 60 patients (50%) expressing a nonsynonymous gene alteration.
Although TP53 mutations were not associated with histopathologic subtype, tumor grade, or tumor location, we did identify an association with stage 4 disease (compared with stages 1–3). Of the six genes, only TP53 mutation rates differed significantly between the races (p = 0.012). A TP53 mutation was present in 89% (16/17) of the African American patients, 56% (10/18) of the Asian patients, 43% (9/21) of the Hispanic patients, and 40% (25/of the Caucasian patients (Table 3).
TABLE 3.
Comparison of mutation rates between races (n = 119)
| Asian N (%) | AA n (%) | Hispanic n (%) | Caucasian n (%) | Adjusted p value | |
|---|---|---|---|---|---|
| ARID1A | |||||
| Yes | 3 (17) | 1 (6) | 5 (24) | 7 (11) | 0.54 |
| No | 15 (83) | 17 (94) | 16 (76) | 55 (89) | |
| CDH1 | |||||
| Yes | 3 (17) | 2 (11) | 1 (5) | 11 (18) | 0.61 |
| No | 15 (83) | 16 (89) | 20 (95) | 51 (82) | |
| ERBB3 | |||||
| Yes | 3 (17) | 2 (11) | 1 (5) | 5 (8) | 0.61 |
| No | 15 (83) | 16 (89) | 20 (95) | 57 (92) | |
| KRAS | |||||
| Yes | 0 (0) | 0 (0) | 2 (10) | 8 (13) | 0.54 |
| No | 18 (100) | 18 (100) | 19 (91) | 54 (87) | |
| PIK3CA | |||||
| Yes | 1 (6) | 1 (6) | 4 (19) | 4 (7) | 0.54 |
| No | 17 (94) | 17 (94) | 17 (81) | 58 (94) | |
| TP53 | |||||
| Yes | 10 (56) | 16 (89) | 9 (43) | 25 (40) | 0.012 |
| No | 8 (44) | 2 (11) | 12 (57) | 37 (60) | |
Due to rounding, percentages may not add up exactly to 100%
AA African American
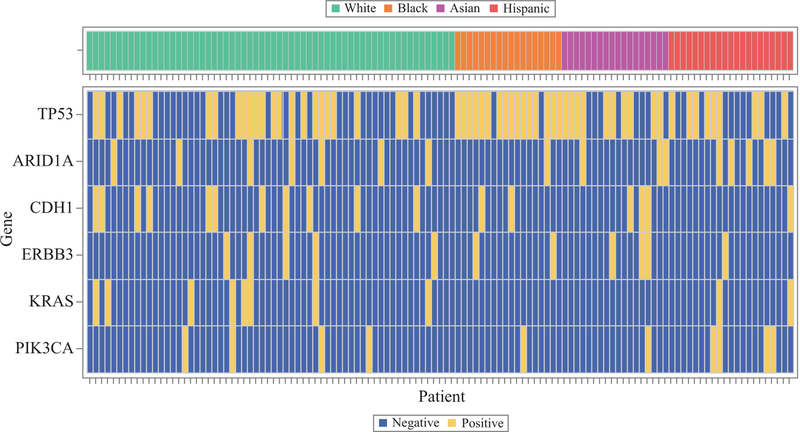
In the heat map for commonly mutated genes (Fig. 1), a discernable clustering for TP53 mutations was observed in the African American patients. This did not appear to be present among the other genes, either overall or by race. Only two of the African American patients analyzed had no TP53 mutation.
FIG. 1.
Heat map for the six genes
The majority of the gene mutations were missense mutations (66%, 390/595), followed by frame-shift deletions (13%, 77/595) and nonsense mutations (9%, 55/595) (data not shown). A breakdown of mutation classification by race for TP53 is outlined in Table 4, and the specific missense mutations are listed in Table 5.
TABLE 4.
TP53 variant classification by race
| Caucasian | AA | Asian | Hispanic | All | |
|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | n (%) | |
| Variant classification | |||||
| Missense mutation | 17 (65) | 7 (41) | 4 (40) | 4 (44) | 32 (52) |
| Frame shift deletion | 3 (12) | 2 (12) | 2 (20) | 0 (0) | 7 (11) |
| Nonsense mutation | 4 (15) | 6 (35) | 2 (20) | 1 (11) | 13 (21) |
| Frame shift Insertion | 1 (4) | 1 (6) | 0 (0) | 0 (0) | 2 (3) |
| Split site | 1 (4) | 1 (6) | 1 (10) | 3 (33) | 6 (10) |
| In frame deletion | 0 (0) | 0 (0) | 1 (10) | 0 (0) | 1 (2) |
| In frame insertion | 0 (0) | 0 (0) | 0 (0) | 1 (11) | 1 (2) |
Due to rounding, percentages may not add up exactly to 100%
AA African American
TABLE 5.
Missense TP53 mutation variants by racea
| Caucasian (n = 17) | AA (n = 7) | Asian (n = 4) | Hispanic (n = 4) | All (n = 32) | |||
|---|---|---|---|---|---|---|---|
| Codon | WT AA | Mutant AA | n (%) | n (%) | n (%) | n (%) | n (%) |
| 175 | Arg | His | 3 (18) | 0 (0) | 0 (0) | 2 (50) | 5 (16) |
| 266 | Gly | Arg | 1 (6) | 2 (29) | 0 (0) | 0 (0) | 3 (9) |
| 273 | Arg | His | 2 (12) | 0 (0) | 1 (25) | 0 (0) | 3 (9) |
| 237 | Met | Ile | 1 (6) | 0 (0) | 0 (0) | 1 (25) | 2 (6) |
| 273 | Arg | Cys | 2 (12) | 0 (0) | 0 (0) | 0 (0) | 2 (6) |
| 277 | Cys | Phe | 1 (6) | 1 (14) | 0 (0) | 0 (0) | 2 (6) |
| 282 | Arg | Trp | 0 (0) | 0 (0) | 2 (50) | 0 (0) | 2 (6) |
| 37 | Ser | Cys | 0 (0) | 1 (14) | 0 (0) | 0 (0) | 1 (3) |
| 135 | Cys | Phe | 1 (6) | 0 (0) | 0 (0) | 0 (0) | 1 (3) |
| 141 | Cys | Tyr | 1 (6) | 0 (0) | 0 (0) | 0 (0) | 1 (3) |
| 145 | Leu | Gln | 1 (6) | 0 (0) | 0 (0) | 0 (0) | 1 (3) |
| 173 | Val | Leu | 1 (6) | 0 (0) | 0 (0) | 0 (0) | 1 (3) |
| 195 | Ile | Thr | 1 (6) | 0 (0) | 0 (0) | 0 (0) | 1 (3) |
| 205 | Tyr | Cys | 0 (0) | 1 (14) | 0 (0) | 0 (0) | 1 (3) |
| 244 | Gly | Cys | 0 (0) | 1 (14) | 0 (0) | 0 (0) | 1 (3) |
| 245 | Gly | Ser | 0 (0) | 0 (0) | 0 (0) | 1 (25) | 1 (3) |
| 248 | Arg | Trp | 1 (6) | 0 (0) | 0 (0) | 0 (0) | 1 (3) |
| 248 | Arg | Gln | 0 (0) | 1 (14) | 0 (0) | 0 (0) | 1 (3) |
| 274 | Val | Gly | 0 (0) | 0 (0) | 1 (25) | 0 (0) | 1 (3) |
| 278 | Pro | Ser | 1 (6) | 0 (0) | 0 (0) | 0 (0) | 1 (3) |
Due to rounding, percentages may not add up exactly to 100%
AA African American
DISCUSSION
We sought to evaluate genomic differences in gastric cancers among patients of different races. In doing so, we identified a significantly higher frequency of TP53 mutations in African American patients. This is the first study to suggest a difference in tumor genetics between African American patients with gastric cancer.
African Americans have the highest cancer-related death rate of any racial group in the United States for most cancers, with a lower 5-year overall survival rate than Caucasian patients (60 vs. 69%).12 Recent data on racial disparities in cancer statistics demonstrated that gastric cancer had the largest discrepancy in survival rates between African Americans and Caucasians in the United States. For both male and female patients with gastric cancer, African Americans were 2.5 times more likely to die of their disease than Caucasians.13 These inequalities are thought to reflect social and economic disparities that may limit access to medical care, delay diagnosis, and postpone definitive treatment.12,13 However, some studies suggest that racial disparities persist even after accounting for differences in access to care and other socioeconomic factors.14,15 Our data suggest that the poor outcomes reported for African American patients with gastric cancer also may be related to tumor biologic differences, and more specifically to frequency of TP53 mutation.
A study by The Cancer Genome Atlas (TCGA) defined the following four molecular subgroups of gastric cancer by profiling 295 primary gastric adenocarcinomas: Epstein-Bar virus-infected tumors, microsatellite instable tumors, genomically stable tumours, and chromosomally unstable tumors. In the latter, a significantly higher TP53 mutation was observed (71% of tumors). No difference was found in overall survival rates among these molecular subtypes, likely due to the small number of samples and relatively short follow-up period. Although not significant, an higher recurrence rate for the chromosomally unstable tumors was observed. It must be noted that African American patients were largely not included in these data.7
In a separate study, Cristescu et al.16 reported gene expression data of 300 primary gastric tumors to establish the following four molecular subtypes associated with distinct disease progression and prognosis: microsatellite instable (MSI) subtype, microsatellite stable/epithelial-to-mesenchymal transition (MSS/EMT) subtype, MSS/TP53? (intact P53 activity) subtype, and MSS/TP53− (functional loss of P53 gene) subtype. Interestingly, the subtype of MSS/TP53? showed better prognosis than the MSS/TP53− group.
In more than half of all human cancers, TP53 gene alterations and dysfunction have been identified,17,18 and the presence of a mutation has been correlated with a shorter survival.19 The predictive value of TP53 mutations in breast cancer have been analyzed in multiple studies. The TP53 gene alterations in breast cancer have been associated with response to specific treatment regimens, and findings have shown these alterations to be useful in identifying patients at higher risk of disease recurrence and death.20–23
Mutations in TP53, found in the vast majority of triple-negative breast cancers, is associated with one of the worst prognoses among breast cancers and shown to be pre-dominant among African American patients.24–27 It should be noted that universal clinical use of TP53 mutation analysis has been difficult because the predictive significance is extremely variable according to tumor type, treatment, or both.28
Missense TP53 mutations are the most common and represent about 72% of all mutations based on the UMD TP53 Mutations Database (http://p53.fr). Interestingly, we identified missense mutations in 41% of African American patients, and 65% of Caucasion patients. Missense mutations, while potentially preserving some wild-type function, generally act as dominant-negative and/or gain-of-function alterations occurring most commonly at corresponding amino acid residues 248, 273, and 175.17–20 Intriguingly, these three “hotspot” mutations occured in 47% of Caucasian patients and 14% of African American patients. Although the numbers are too small for a conclusion to be drawn, our results suggest that not only do the rates of TP53 mutations differ across racial groups in the United States, but the type of mutation may differ as well.
Several important limitations need to be considered when the results of this study are interpreted. First, the sample for all the patients was small, particularly for the non-Caucasian patients included in the study. Because our study involved a single tertiary cancer center in the Northeast, our African American patients may not be representative of all African American patients in the United States. Second, the disease characteristics of these patients were heterogeneous and could not be controlled for in this small sample. The mutational status may have been related to an underlying disease factor more than to race. Additionally, we also included both primary and metastatic samples in our study, which may present different mutational profiles. Finally, race is a complex factor, and our study relied on patient-reported race designation, which can be complex in a diverse population such as that seen in the United States, and categorical delineations may be an oversimplification.
In conclusion, this is the first study to identify more mutations in the tumor-supressor gene TP53 among African American patients with gastric adenocarcinoma than among other racial groups in the United States. In doing so, we have provided a potential biologic explanation for the poor survival associated with gastric cancer in African American patients. This study is particularly timely because drugs targeting TP53 mutations have entered early-phase clinical trials. Moreover, African American patients as a population have been difficult to accrue to clinical trials and may be ideal candidates for inclusion. Future larger studies are warranted to validate the results reported in this study.
Footnotes
DISCLOSURE No disclosures.
REFERENCES
- 1.World Health Organization. GLOBOCAN 2012: Stomach cancer estimated cancer incidence, mortality, and prevalence worldwide in 2012 http://globocan.iarc.fr. Accessed 13 July 2017.
- 2.Ohtsu A, Yoshida S, Saijo N. Disparities in gastric cancer chemotherapy between the East and West. J Clin Oncol 2006;24:2188–96. [DOI] [PubMed] [Google Scholar]
- 3.Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet 2015;385:977–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strong VE, Song KY, Park CH, et al. Comparison of gastric cancer survival following R0 resection in the United States and Korea using an internationally validated nomogram. Ann Surg 2010;251:640–6. [DOI] [PubMed] [Google Scholar]
- 5.Noguchi Y, Yoshikawa T, Tsuburaya A, Motohashi H, Karpeh MS, Brennan MF. Is gastric carcinoma different between Japan and the United States? Cancer 2000;89:2237–46. [PubMed] [Google Scholar]
- 6.You WC, Li JY, Zhang L, Jin ML, Chang YS, Ma JL, Pan KF. Etiology and prevention of gastric cancer: a population study in a high-risk area of China. Chin J Dig Dis 2005;6:149–54. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cristescu R, Lee J, Nebozhyn M, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21:449–56. [DOI] [PubMed] [Google Scholar]
- 9.American Cancer Society. Cancer Facts & Figures 2015. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2015/cancer-facts-and-figures-2015.pdf. Accessed 13 June 2017.
- 10.Howard JH, Hiles JM, Leung AM, Stern SL, Bilchik AJ. Race influences stage-specific survival in gastric cancer. Am Surg 2015;81:259–67. [PubMed] [Google Scholar]
- 11.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015;17:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.American Cancer Society. Cancer Facts and Figures 2012–2014 https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/cancer-facts-and-figures-for-Africanamericans/cancer-facts-and-figures-for-Africanamericans-2013-2014.pdf. Accessed 13 June 2017.
- 13.DeSantis CE, Siegel RL, Sauer AG, Miller KD, Fedewa SA, Alcaraz KI, Jemal A. Cancer Statistics for African Americans, 2016: progress and opportunities in reducing racial disparities. CA Cancer J Clin 2016;66:290–308. [DOI] [PubMed] [Google Scholar]
- 14.Du XL, Lin CC, Johnson NJ, Altekruse S. Effects of individual-level socioeconomic factors on racial disparities in cancer treatment and survival: findings from the National Longitudinal Mortality Study, 1979–2003. Cancer 2011;117:3242–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albain KS, Unger JM, Crowley JJ, Coltman CA Jr, Hershman DL. Racial disparities in cancer survival among randomized clinical trials patients of the Southwest Oncology Group. J Natl Cancer Inst 2009;101:984–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cristescu R, Lee J, Nebozhyn M, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21:449–56. [DOI] [PubMed] [Google Scholar]
- 17.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science 1991;253:49–53. [DOI] [PubMed] [Google Scholar]
- 18.Hainaut P, Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv Cancer Res 2000;77:81–137. [DOI] [PubMed] [Google Scholar]
- 19.Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene 2007;26:2157–65. [DOI] [PubMed] [Google Scholar]
- 20.Olivier M, Langerod A, Carrieri P, et al. The clinical value of somatic TP53 gene mutations in 1794 patients with breast cancer. Clin Cancer Res 2006;12:157–1167. [DOI] [PubMed] [Google Scholar]
- 21.Bergh J, Norberg T, Sjogren S, Lindgren A, Holmberg L. Complete sequencing of the p53 gene provides prognostic information in breast cancer patients, particularly in relation to adjuvant systemic therapy and radiotherapy. Nat Med 1995;1:1029–34. [DOI] [PubMed] [Google Scholar]
- 22.Aas T, Borresen AL, Geisler S, et al. Specific P53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nat Med 1996;2:811–4. [DOI] [PubMed] [Google Scholar]
- 23.Bull SB, Ozcelik H, Pinnaduwage D, et al. The combination of p53 mutation and neu/erbB-2 amplification is associated with poor survival in node-negative breast cancer. J Clin Oncol 2004;22:86–96. [DOI] [PubMed] [Google Scholar]
- 24.Kumar P, Aggarwal R. An overview of triple-negative breast cancer. Arch Gynecol Obstetr 2016;293:247–69. [DOI] [PubMed] [Google Scholar]
- 25.Papa A, Caruso D, Tomao S, Rossi L, Zaccarelli E, Tomao F. Triple-negative breast cancer: investigating potential molecular therapeutic target. Expert Opin Ther Targets 2015;19:55–75. [DOI] [PubMed] [Google Scholar]
- 26.Mathe A, Scott RJ, Avery-Kiejda KA. MiRNAs and other epigenetic changes as biomarkers in triple-negative breast cancer. Int J Mol Sci 2015;16:28347–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner N, Moretti E, Siclari O, et al. Targeting triple-negative breast cancer: is p53 the answer? Cancer Treat Rev 2013;39:541–50. [DOI] [PubMed] [Google Scholar]
- 28.Bertheau P, Espié M, Turpin E, et al. TP53 status and response to chemotherapy in breast cancer. Pathobiology 2008;75:132–9. [DOI] [PubMed] [Google Scholar]