Introduction
There has been substantial evidence showing the existence of cellular heterogeneity within an isogenic or clonal cell population. The cell-to-cell variations are caused by complex factors such as genetic drift, transcriptional events, differences in cell development and cell cycles, and the intrinsic stochasticity of cellular processes. The conventional bulk assays measure and analyze the population-average responses, and as a result, the unique characteristics of individual cells are concealed (Carlo & Lee, 2006). As single cells represent the fundamental functional units of life, inevitably, a great number of biological and medical questions can only be addressed by the approach of single-cell analysis. For example, clonal populations of progenitor cells exhibit diverse differentiation outcomes in response to the same stimuli, due to the stochastic effects in the expression level of basal signaling proteins, which are obscured in bulk assays but can be revealed via single-cell analysis on the cell population (Singh, Ku, Wichaidit, Steininger, Wu & Altschuler, 2010). Similarly, single-cellular level biomolecular analysis can isolate minority sub-populations that are resistant to chemotherapy or have a higher risk of metastasis from the heterogeneous tumor cells (Patel, Tirosh, Trombetta, Shalek, Gillespie, Wakimoto et al., 2014). Single-cell analysis also plays an essential role in rare cell-based studies, such as isolating the circulating-tumor-cells (CTCs) from peripheral blood cells for cancer diagnostics (Ramsköld, Luo, Wang, Li, Deng, Faridani et al., 2012), as rare CTCs would be masked by the abundant blood cells in bulk analysis, and each individual cell represents its unique source of tumor origin.
As it requires the data from a large number of individual cells to draw a statistically meaningful conclusion, high-throughput single-cell processing and analyzing technologies are of critical importance. In general, bench-top single-cell analyses are limited by their high cost, low throughput, and difficulties in analyzing low amount of starting materials. On the contrary, microfluidic technology manipulates samples in micrometer scale which is comparable to the single-cell diameter, requires low reagent volumes and cost, and attains high analysis efficiency. With parallelization, microfluidic processes can be high-throughput, automated, and multifunctional. Therefore, a variety of microfluidic platforms have been developed to isolate single cells and analyze them from genotype to phenotype.
In the past a few years, our group has developed microfluidic platforms with highly-packed microwell arrays for single-cell biomolecular and imaging analysis. We have built devices with serpentine-shape microfluidic trapping arrays capable of trapping 100, 1,600, and 76,800 single cells within 20 s, 3 min, and 6 min, respectively. This microwell array is conducive to single-cell mRNA physical probing when sealed by a 1-μm-thick PDMS membrane (Li, Tao, Lee, Wickramasinghe & Lee, 2017). The same microwell array can be designed to filter-out smaller cells and trap cells in the target size range (e.g., trap WBCs from RBCs) and is compatible with live-cell real-time imaging systems (Lee, Li, Ma, Digman & Lee, 2018). As the single-cell trapping efficiency is determined by the channel design instead of the flow rate, it can be coupled with other microfluidic sample processing units with various flow rates. In this chapter, we have described the detailed chip design and fabrication procedures, as well as its representative applications.
Chip Design and Rationale
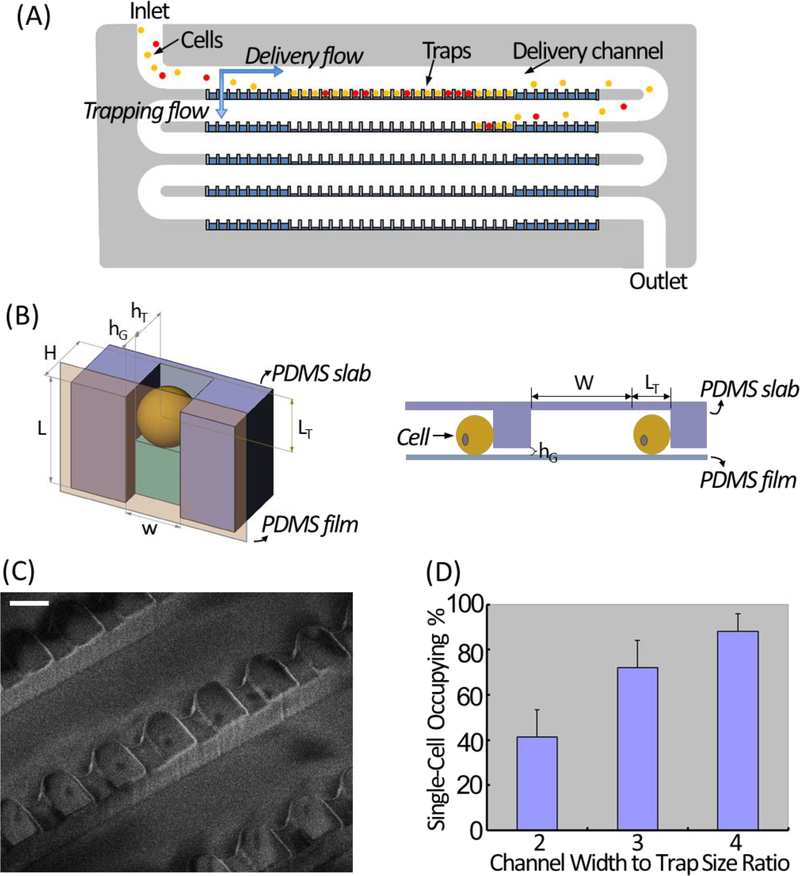
The design principle of our high-density single-cell trapping array was adapted from Kwanghun Chung et al (Chung, Rivet, Kemp & Lu, 2011). For a 100-trap single-cell array, it consists of a 5-row serpentine channel with 20 grooves arrayed along the channel edge of each row (Fig. 1A). As illustrated in Fig. 1B, for each trapping unit, the height of the trap (hT) is smaller than the height of the delivery channel (H), resulting in a gap area (hG = H − hT). The trapping principle relies on the two hydrodynamic flows – horizontal delivery flow and perpendicular trapping flow. While cells are delivered to the traps sequentially by the horizontal delivery flow, there is a perpendicular stream flowing through the gap area at each trapping unit, crossing each row of the delivery channel and pushing cells into traps. The width (w) and the length (LT) of each trap are the same as target cell diameter, so that once a cell occupies a trap, it physically excludes another cell from trapping at the same spot, which ensures that only one cell is trapped at each trapping unit. At the turning zone of each row, there are dummy traps with LT smaller than cell diameter, which do not trap cells but help generate perpendicular flow for cell focusing. The scanning electron microscopic (SEM) image illustrating the detailed structure of a finished single-cell trapping array is shown in Fig. 1C.
Figure 1. Design and working principle of the microfluidic single-cell trapping array.
(A) Schematic illustration of the single-cell trapping array. (B) The trimetric view (top) and side view (bottom) of one microfluidic single-cell trapping unit. (C) SEM image showing the detailed structure of the single-cell trapping array. Scale bar: 20 μm. (D) Single-cell occupying efficiency at 3 tested W (delivery channel width) to w (trap width) ratios.
The single-cell occupying efficiency is not related to the flow rate, but related to the flow resistance ratio of the horizontal flow (Rhorizontal) and the perpendicular flow (Rvertical), which can be controlled by optimizing the ratio of the delivery channel width (W) to the trap width (w). A smaller W/w will lead to more empty traps, while a bigger W/w can cause cell clogging. Based on observations from our experiments, a W/w of 4 resulted in the best single-cell occupying efficiency (Fig. 1D), which was 94±4% calculated from 12 independent experiments tested using HeLa cells. We have also scaled up the 100-trap single-cell array by parallelization to accommodate 16×100 and 768×100 single-cell traps for high-throughput analyses.
Fabrication of the Microfluidic Single-Cell Trapping Array
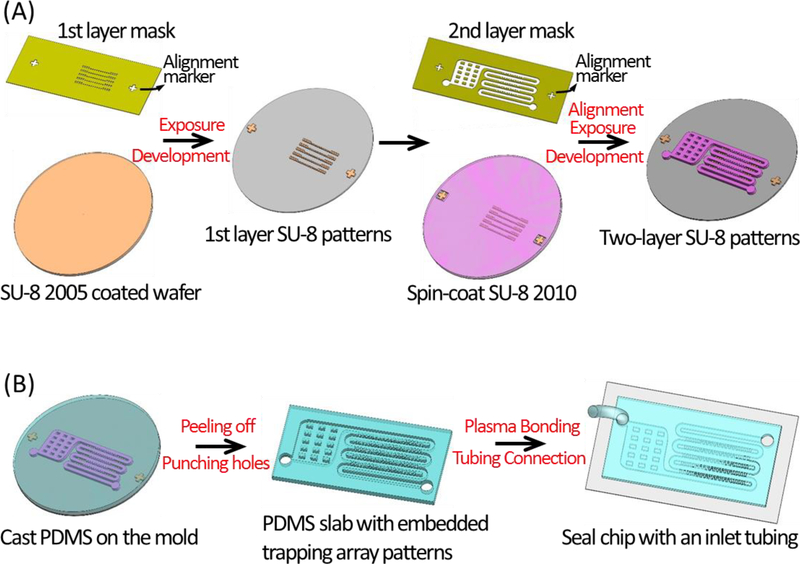
The single-cell trapping chip, consisting of a serpentine-shape single-cell trapping channel described above and a filter region, was fabricated with elastomer PDMS (Polydimethylsiloxane) using a master mold patterned on a silicon wafer. This master mold was fabricated by two-layer photolithography (Fig. 2A), and the PDMS slab with embedded patterns was cast from the mold by soft-lithography and sealed with a glass substrate by oxygen plasma bonding (Fig. 2B). The detailed fabrication process is described as below.
Figure 2. Fabrication procedure of the microfluidic single-cell trapping array.
(A) Fabricate the master mold by two-layer photolithography. The 1st layer (gap channels) is patterned using SU-8 2005. Thereafter SU-8 2010 is spin-coated on-top-of the 1st layer, and the alignment markers on the 2nd-layer mask and the 1st-layer patterns are aligned. After a 2nd time exposure and development, a double-layer SU-8 mold is patterned on the wafer. (B) Fabricate the PDMS slab with embedded trapping array patterns by casting the mold using soft-lithography, then punch the connection holes at the inlet and the outlet, and thereafter make a complete device by sealing the PDMS slab with a glass substrate using oxygen plasma bonding and connecting the Tygon tubing at the inlet.
Materials
Silicon wafers in test level (UniversityWafer)
Photoresists: SU-8 2005 and SU-8 2010 (MicroChem)
SU-8 developer (MicroChem, Y020100)
Teflon® AF solution (Dupont, 400S2–100-1)
Fluorinert™ FC-40 (Acros Organics)
PDMS (Dow Corning, Sylgard 184)
Glass microscope slides (Fisher Scientific)
Glass coverslips (Fisher Scientific)
2% (v/v) hydrofluoric acid (HF) solution (Fisher Scientific, 3819–232)
Isopropyl alcohol (IPA, Sigma Aldrich)
Deionized (DI) water
Equipment
Spin coater (Laurell Technologies)
Mask aligner (Suss Microtec, MA56)
Stylus-based profiler (Bruker Corporation, Dektak3)
Plasma cleaner (Harrick Plasma)
Hot plate (VWR)
Convection oven (VWR)
1.5 mm punch with plunger (Miltex)
Methods
Fabrication of the master mold by two-layer photolithography
Draw the single-cell trapping arrays in AutoCAD software, and print the designs into chrome-quartz photomasks (Front Range Photo Mask). Two layers of photomasks are required to fabricate the master mold, one for the gap channels and the other for the main channel, due to their height difference. Make sure to include alignment makers on both layers of the photomasks, and when printing photomasks, the background should be chrome, and the micro-channel patterns should be transparent.
Soak the silicon wafer in 2% HF solution for 2 min. Rinse the wafer with DI water and blow dry with N2. HF solution etches away the hydrophilic oxide layer on the surface of the silicon wafer, so that pure silicon, which is hydrophobic, is exposed. SU-8 photoresists adhere tighter to hydrophobic surfaces.
Spin coat SU-8 2005 (4000 RPM for 30 s) on the wafer, and bake the photoresist-coated wafer on the hot plate at 95 °C for 2 min. Then let the wafer cool down to room temperature.
Load the photomask with the gap channel patterns and the photoresist-coated wafer in the MA56 mask aligner. Expose the wafer to UV using the hard-contact mold at the exposure energy of 95 mJ/cm2.
Bake the exposed wafer on the hot plate at 95 °C for 3 min. Then let the wafer cool down to room temperature.
Develop the wafer in SU-8 developer for 1 min, and rinse the wafer with IPA to stop the developing. Rinse the wafer thoroughly in DI water and blow-dry with N2.
Spin coat SU-8 2010 (1200 RPM for 30 s) on the above wafer, and bake the photoresist-coated wafer on the hot plate at 95 °C for 3 min. Then let the wafer cool down to room temperature.
Dip the cotton swab with SU-8 developer, and use it to remove the portion of SU-8 2010 on top of the alignment marker that is already patterned in the first layer.
Load the photomask carrying the patterns of the main channel and the SU-8 2010 coated silicon wafer in the MA56 mask aligner. Align the alignment maker on the photomask with its complementary alignment marker that has already been patterned on the wafer in the first layer. Expose the wafer to UV using the hard-contact mold at the exposure energy of 140 mJ/cm2.
Bake the exposed wafer on the hot plate at 95 °C for 5 min. Then let the wafer cool down to room temperature.
Develop the wafer in SU-8 developer for 3 min, and rinse the wafer with IPA to stop the developing. Rinse the wafer thoroughly in DI water and blow-dry with N2.
Bake the wafer on the hot plate at 200 °C for 5 min. This step tightens the adherence of patterned SU-8 to the silicon wafer, and also makes the channel edges sharper. Then let the wafer cool down to room temperature.
Inspect the master mold using the Dektak3 surface profiler for the accuracy of the patterns.
Fabrication of the PDMS single-cell trapping array using the master mold
Dilute the Teflon® AF solution in FC-40 at a 1:5 (v/v) ratio. Spin coat the diluent (3000 RPM for 30 s) on the wafer with the patterned master mold, and followed by baking in a 120 °C oven for 10 min for curing the coating. Teflon coating avoids the adherence of PDMS to SU-8 photoresists, so that the PDMS slab can be easily peeled off after curing.
Prepare uncured PDMS by mixing the base pre-polymer and curing agent at a 10:1 (w/w) ratio.
Pour the uncured PDMS uniformly onto the master mold and degas it in a vacuum chamber for 15 min to remove air bubbles.
Cure the PDMS by baking in a 65 °C oven for 3 h. Then let it cool down to room temperature.
Gently peel off the cured PDMS from the wafer using a pair of tweezers. Cut the obtained PDMS slab to get individual single-cell trapping arrays.
Use a hole-puncher to punch two 1.5 mm diameter holes at the inlet and the outlet of each PDMS single-cell trapping array, respectively.
Clean the PDMS slab with the scotch tape to remove any debris and residues.
Sonicate the glass slides in IPA for 15 min, rinse them with DI water and blow-dry. Bake the cleaned glass slides in a 120 °C oven for 10 min to dehydrate. Then let the glass slides cool down to room temperature.
Put the cleaned PDMS slabs and glass slides in the plasma cleaner for a 60 s’ oxygen plasma treatment.
Take out the PDMS slabs and glass slides quickly from the plasma cleaner, and bond one PDMS slab with one glass slide. The two pieces are irreversibly annealed to each other immediately after getting into contact, due to the formation of covalent bonds.
Tighten the bonding by baking the device in a 120 °C oven for 5 min.
Experimental Protocol for Single-Cell Trapping
Wash the chip with 70% ethanol for 30 min with a flow rate of 10 μL/min followed by 1% F-68 in PBS solution for 15 min with a flow rate of 10 μL/min. This step is to remove bubbles within the microfluidic trapping array, especially critical for high-throughput and complicated microfluidic structures.
Prepare the cell-in-media suspension at a concentration of 1× 106 cells/mL.
Load the cell suspension in a 1 mL syringe connected with a Tygon tubing.
Mount the syringe on a syringe pump, and set the flow rate at 2 μL/min.
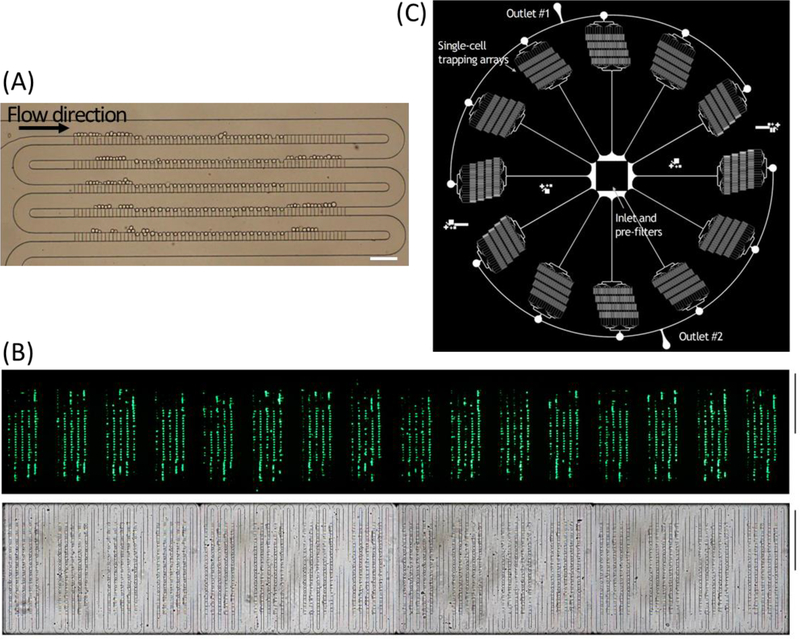
At this flow rate and cell concertation, a 100-trap single-cell-trapping array can be filled within 20 s as demonstrated using HeLa cells (Fig. 3A), and a real-time video recording the single-cell trapping process is included in the supplementary information (Movie. S1). The scaled-up trapping array with 1,600 traps and 76,800 traps are shown in Fig. 3B and Fig. 3C, respectively.
Figure 3. Microfluidic single-cell trapping arrays filled with cells.
(A) Bright-field image of trapping 100 single HeLa cells within the single-cell array. Scale bar: 100 μm. (B) Bright-field (top) and fluorescent (bottom) images of K562 cells trapped in the scaled-up microfluidic trapping array consisting of 16 identical arrays of highly packed 100 single-cell traps. Scale bar: 1 mm. (C) Schematic illustration of a paralleled device with 12 individual channels radially arrayed with a single inlet and two ring outlets consisting 76,800 single-cell traps in total.
Application 1: Single-Cell mRNA Probing
By sealing the single-cell trapping array with an ultra-thin PDMS membrane (≤1 μm-thick), external equipment such as an AFM nanoprobe or a micro-pipette, is able to penetrate into the microfluidic array and get access to a specific cell without cross-cell contamination and media evaporation. We have Au/Cr-coated an AFM probe into a dielectrophoretic nanotweezer (DENT, Fig.4A), wherein the application of an alternating-current (AC) field between the inner Si core and outer metal layer created a large gradient of the electric field square (∇E2), resulting in a dielectrophoretic (DEP) attractive force (FDEP = [(Vα)/2]∇E2; V, particle volume; α, polarizability) strong enough to attract mRNA molecules toward the probe-end (Fig. 4B) (Li et al., 2017; Nawarathna, Turan & Wickramasinghe, 2009). The DENT probe was accurately aligned with the target cell in the trapping array, penetrated through the PDMS membrane into the cell, and extracted mRNA molecules from the cytoplasm by DEP (Fig. 4C). It was then retracted from the chip, and mRNA molecules were released from the tip to perform RT-qPCR for quantitative gene expression analysis of the target cells. Because of the self-sealing capability of the ultrathin PDMS membrane, no punctured hole or leakage was observed under the microscope after retracting the probe from the cell-trapping array.
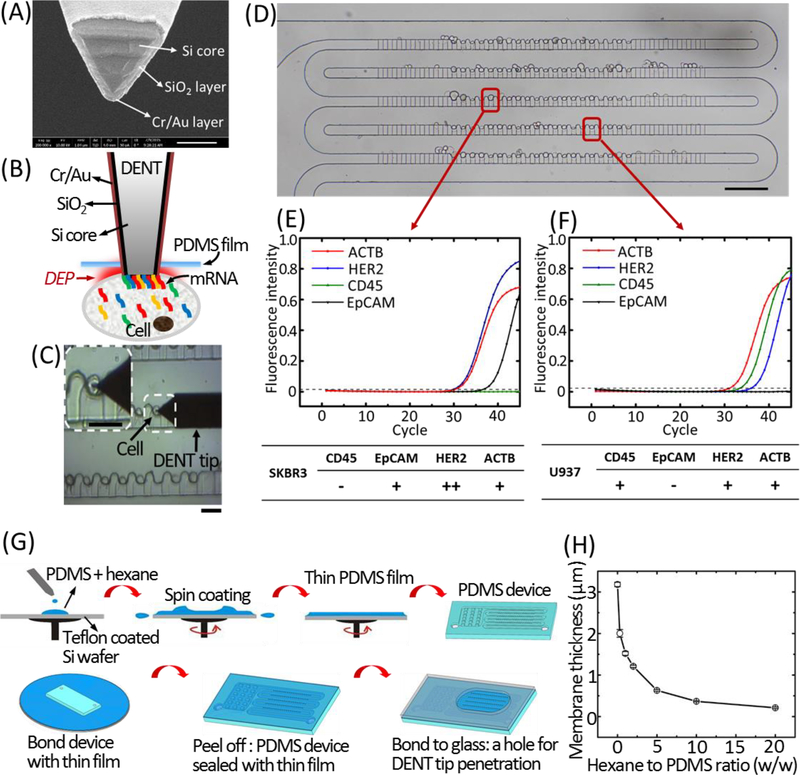
Figure 4. Single-cell mRNA probing from the ultra-thin PDMS membrane-sealed microfluidic trapping array.
(A) SEM image of the DENT probe (scale bar: 200 nm). (B) Schematic illustration of single-cell mRNA extraction using DENT after it penetrates through the PDMS film and enters into the cytoplasm. Application of AC field between the inner Si core and the outer metal layer creates a DEP attractive force to attract mRNA molecules toward the probe-end. (C) Bright-field microscopic image capturing the single-cell probing process. White dashed box indicates the cell of interest. The probe was moved downward toward a target cell, penetrated through the PDMS membrane and inserted into the target cell to extract mRNAs by DEP. Scale bars: 30 μm. (D) Bright-field image of trapping single cells of SK-BR-3 and U937 in the microwell array. Scale bar: 100 μm. The RT-qPCR fingerprints of the 4 target mRNAs (CD45, EpCAM, HER2 and ACTB) extracted by DENT from a trapped SK-BR-3 cell and a trapped U937 cell are shown in (E) and (F), respectively. (G) Fabrication process for the ultra-thin PDMS membrane-sealed microfluidic trapping array. (H)The thickness of the PDMS membrane according to the different ratios of hexane to PDMS pre-polymer at the spin coating condition of 5000 RPM, 5 min.
As a proof of concept, a mixture of SK-BR-3 (human adenocarcinoma cell line) and U937 (monocyte cell line, representing the dominant type of WBCs in human blood) cells, mimicking a blood sample after primary circulating-tumor-cell (CTC) enrichment, were trapped in our microwell array (Fig. 4D), and in situ selective mRNA probing with an applied field of 1.5 Vpp, 10 MHz was performed on arbitrary cells of interest. Although the two types of cells were in a similar size range and could not be differentiated easily from the optical images, the RT-qPCR fingerprints of a specific single cell’s mRNAs extracted by DENT revealed its specific gene expression levels and cell identity. If a cell expressed EpCAM (epithelial cellular adhesion molecule) as a CTC marker (Punnoose, Atwal, Spoerke, Savage, Pandita, Yeh et al., 2010), and overexpressed HER2 (human epidermal growth factor receptor 2) as a breast cancer cell marker (Fig.4E) (Roussidis, Theocharis, Tzanakakis & Karamanos, 2007), it was an SK-BR-3 cell. On the contrary, if a cell expressed CD45 as a leucocyte marker (Stonehouse, Woodhead, Herridge, Ashrafian, George, Chain et al., 1999), did not express EpCAM (Lorenzi, Turi, Lorenzi, Paolinelli, Mancioli, La Sala et al., 2012), and had a much lower HER2 expression level (Karagiannis, Singer, Hunt, Gan, Rudman, Mechtcheriakova et al., 2009) compared to breast cancer cells (Fig.4F), it was a U937 cell. There existed obvious differences in gene expression between SK-BR-3 and U937 cells, and also gene-expression heterogeneity within the same population can be further revealed.
DEP-based mRNA extraction using DENT is a nondestructive method that does not require cell lysing or mRNA purification, and is sensitive enough to detect the expression level of low-copy-number genes (e.g. HPRT) within a single living cell among the cell population. As it works at the frequency specific for mRNAs and avoids the removal of cytosol, cells are protected from losing essential molecules. The probed cells in the microfluidic trapping array remain viable and can be retrieved and cultured for further analysis. The detailed fabrication protocol of this ultra-thin PDMS membrane-sealed single-cell trapping array is illustrated in Fig. 2G and described as below.
Fabrication of the ultra-thin PDMS membrane-sealed single-cell array
Spin coat the 1:5 (v/v) Teflon® AF-FC 40 solution on a cleaned silicon wafer at 3000 RPM for 30 s. Bake the wafer in a 120 °C oven for 10 min. Teflon coating efficiently reduces the adhesion of PDMS to the silicon wafer to facilitate an easy peeling off of the sealed device.
Prepare uncured PDMS by mixing the base pre-polymer and curing agent at a 10:1 (w/w) ratio.
Degas the PDMS mixture in a vacuum chamber for 15 min to remove air bubbles.
Mix the degassed PDMS mixture with hexane at a 1:2 (w/w) ratio. Diluting PDMS pre-polymer mixture in hexane reduces its viscosity, so that a much thinner membrane is generated at similar spin coating parameters (Thangawng, Ruoff, Swartz & Glucksberg, 2007).
Spin coat the PDMS-hexane mixture on the baked silicon wafer at 5000 RPM for 5 min. Fig. 2H summarizes the resulting membrane thickness according to different ratios of hexane to PDMS pre-polymer at the spin coating condition of 5000 RPM, 5 min.
Leave the spin-coated PDMS membrane in a 120 °C oven for 45 min to evaporate the hexane and then in a 65 °C oven for overnight to ensure complete crosslinking. Let the wafer cool down to room temperature, thereafter.
Clean the PDMS slab (with punched holes at the inlet and outlet of the trapping array) using scotch tape to remove any debris and residues.
Put the cleaned PDMS slab and the silicon wafer with cured PDMS membrane in the plasma cleaner for a 120 s’ oxygen plasma treatment.
Remove the PDMS slab and the silicon wafer with cured PDMS membrane quickly from the plasma cleaner, and bond the PDMS slab with the membrane. The two pieces are irreversibly annealed to each other immediately after getting into contact, due to the formation of covalent bonds.
Tighten the bonding by baking the device in a 120 °C oven for 5 min.
Gently peel off the sealed device from the silicon wafer.
Optional: bond the PDMS membrane-sealed device to a glass slide with a through-hole upon 120 s’ oxygen plasma treatment. The glass slide works as a supporting substrate for the device, and the through-hole enables DENT probe or other external tools to penetrate through the membrane.
Application 2: Single-Cell Array for Live-Cell Imaging
When bonded with an imaging slide, the highly-packed single-cell trapping array is compatible with various microscopic techniques. Due to the limited working distance of lenses when imaging cells at high magnifications, the PDMS slab with embedded single-cell trapping arrays is sealed with an imaging slide (glass coverslip) with a thickness of ~0.17 mm, instead of a normal glass slide with a thickness of ~1mm. We have used our chip in fluorescence lifetime imaging microscopy (FLIM) to rapidly distinguish leukemia cells from normal white blood cells (WBCs) in a label-free and non-invasive manner based on their difference in metabolism (Lee et al., 2018).
As leukemia cells and normal WBCs both have a diameter in the range of 8~20 μm, they cannot be separated easily by size. However, they are both bigger than red blood cells (RBCs), which have a disk shape in a diameter of ~6.2–8.2 μm and a thickness at the thickest point of 2–2.5 μm (Nguyen, Wei, Zheng, Wang & Sun, 2015). Therefore, RBCs can be successfully filtered out using our microfluidic trapping array: the perpendicular flow through the gap area (hG) of each trapping unit deforms and migrates RBCs while WBCs/leukemia cells flow into traps, and the combination of the perpendicular deformation flow and the horizontal delivery flow enables continuous WBC/leukemia cell trapping and simultaneous RBC filtration (Movie. S2). The height of hG is critical in determining the WBC/leukemia cell capturing efficiency, as an hG that is too large allows WBCs/leukemia cells to squeeze through the traps during RBC filtration, while hG that is too small would prohibit both RBC passing through and WBC/leukemia cell trapping. Based on our experiments with various hG values, an optimal height of ~3.3 μm enables complete RBC filtration and a single WBC/leukemia cell trapping efficiency of ~73.48%. When the 20× diluted blood sample was introduced into the chip at a flow rate of 300 μL/hr, 1,600 highly-packed single-cell traps were filled in 3 min.
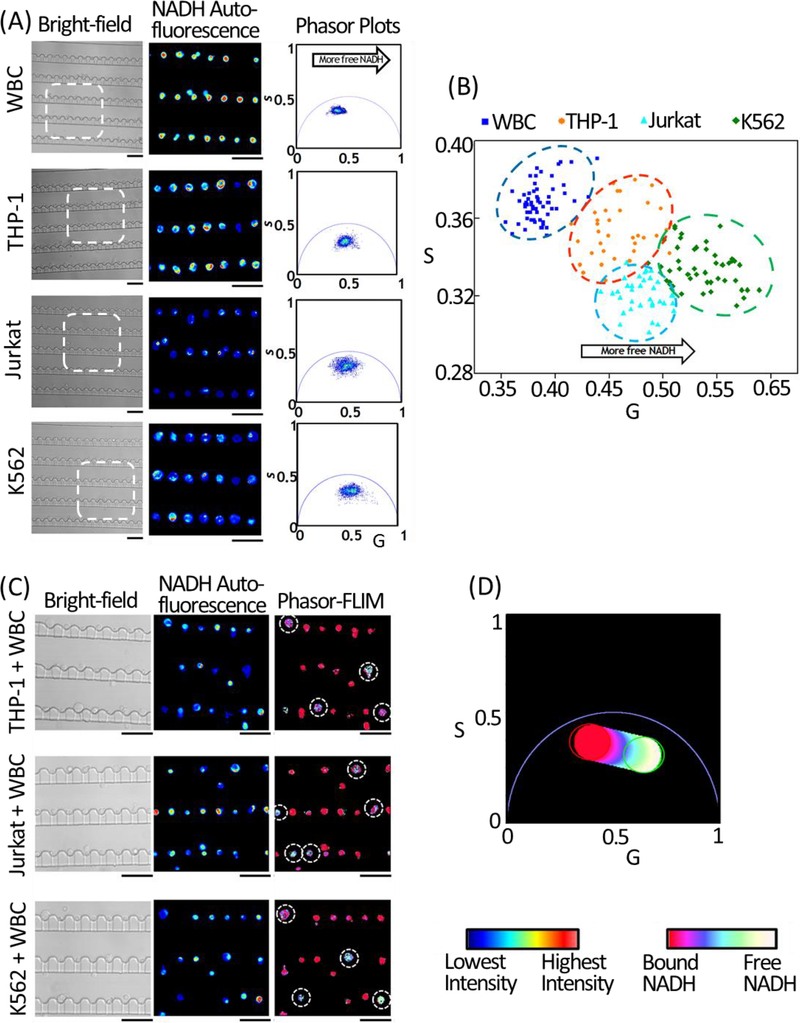
FLIM identifies leukemia cells from normal WBCs based on the Warburg effect: differentiated cells rely on oxidative phosphorylation (OXPHOS) in ATP production, and have more enzyme-bound nicotinamide adenine dinucleotide (NADH); whereas tumor cells exhibit increased rate of glycolysis, so that a large portion of NADH is in free state (non-enzyme-bound state) (Vander Heiden, Cantley & Thompson, 2009). Thus, the ratio of free/bound NADH is used to distinguish tumor cells from normal cells. As NADH has different auto-fluorescence lifetimes in its free and enzyme-bound states, this measurement can be done via FLIM in a non-disruptive and label-free manner (Stringari, Cinquin, Cinquin, Digman, Donovan & Gratton, 2011). And a phasor algorithm (phasor-FLIM) was established for fluorescence lifetime data analysis allowing straightforward interpretation of intrinsic fluorescence signals from live tissues directly regarding physiological relevant fluorophores (Digman, Caiolfa, Zamai & Gratton, 2008). In our experiment, the single-cell array was imaged using a 40 × 1.2 NA oil-immersion objective (Carl Zeiss), and was excited via two-photon excitation at a wavelength of 740 nm with a laser power of ~5 mW. The fluorescence emission in the band-pass of 420–500 nm, which covered the emission wavelength of free and protein-bound NADH, was collected and the lifetime signatures were calculated. We first collected the FLIM signatures from trapping arrays of normal WBCs, and three leukemia cell lines, i.e., THP-1 (human acute monocytic leukemia cell line), Jurkat (human acute T cell leukemia cell line) and K562 (human chronic myelogenous leukemia cell line), respectively (Fig.5A). As shown in the scatter plot of single-cell average phasor-FLIM values in Fig. 5B, while intra-population heterogeneity was observed through the cell-cell variation of the phasor-FLIM values, different types of cells had specific phasor-FLIM signatures, and there was a clear trend that leukemia cells were shifted toward the lower right direction in the phasor plot compared to WBCs, demonstrating a shorter lifetime, and therefore a higher free/bound NADH ratio. Based on the above population-averaged phasor-FLIM patterns, we then used the microfluidic trapping array to process leukemia cell-spiked human blood samples, and identified single leukemic cells from normal WBCs in the trapping array according to their phasor-FLIM signatures (Fig.5C and D).
Figure 5. Live-cell FLIM imaging using the single-cell trapping array.
(A) Bright-field images (left), NADH auto-fluorescence emission intensity images of the selected regions of interest (middle) highlighted by the white-dashed squares in the bright-field images, and the corresponding lifetime phasor plots (right) of the single-cell arrays of WBCs, THP-1, Jurkat, and K562 cells. Scale bars: 50 μm. (B) Scatter plot of the average G and S phasor-FLIM values of trapped single cells based on their NADH auto-fluorescence phasor-FLIM signatures. A total number of 65 WBCs (blue), 35 THP-1 cells (cyan), 35 Jurkat cells (orange), and 46 K562 cells (green) were measured and plotted. While the heterogeneity between individual cells among the sample population was observed, the FLIM signatures of all the leukemia cells shifted toward the bottom-right compared to WBCs, indicating a higher free-to-bound NADH ratio and a more glycolytic state. (C) Bright-field images (left) and NADH auto-fluorescence emission intensity images (middle) of the THP-1, Jurkat, and K562 cells-spiked blood samples, respectively. In the phasor plot (D), the color scale from red/pink to white/yellow represents a linear increase of free to protein-bound NADH ratio. Based on the above color-code, the NADH lifetime maps of the leukemia cell-spiked blood samples were plotted in the right panel in (C). Leukemia cells demonstrated a significant shift toward a shorter lifetime and a higher free/bound NADH ratio indicating a higher glycolytic state. Scale bars: 50 μm.
In comparison to conventional biomarker-based screenings, phasor-FLIM does not require any labeling procedures and is cell-safe (the 740-nm excitation wavelength is in the infra-red range which is nondestructive to cells). By integrating phasor-FLIM with the microfluidic trapping array which is capable of high-density single-cell trapping with simultaneous RBC filtration, the integrated platform has demonstrated rapid and label-free individual-leukemia-cell screening through non-invasive metabolic imaging, and has the potential to screen blood in clinical volumes through array parallelization.
The procedure of bonding PDMS slab with imaging slide is described as below.
Clean the PDMS slab (with punched holes at the inlet and outlet of the trapping array) using scotch tape to remove any debris and residues.
Sonicate the coverslips in IPA for 15 min, rinse them with DI water and blow-dry.
Put the cleaned PDMS slabs and coverslips in the plasma cleaner for a 100 s oxygen plasma treatment.
Remove the PDMS slabs and coverslips quickly from the plasma cleaner, and bond one PDMS slab with one piece of coverslip. The two pieces are irreversibly annealed to each other immediately after contact, due to the formation of covalent bonds.
Tighten the bonding by baking the device in a 120 °C oven for 5 min.
Conclusion
In conclusion, we have described the design rationale and detailed fabrication protocol of a highly-packed serpentine-shape microfluidic array for single-cell isolation and trapping. This platform forms an essential link from an upstream sample preparation device and a downstream single-cell analysis approach. We demonstrated both a non-invasive single cell array imaging platform for phenotyping and a minimally-invasive nanoprobe method for genotyping. These are simply two examples but there could be many other techniques that could leverage such a platform. In this chapter, when sealed by a thin PDMS membrane, the microfluidic array enables external equipment to penetrate through and access trapped individual cells for applications such as live-cell mRNA extraction using the DENT probe. When sealed by an imaging slide, the microfluidic array can be adapted in various live-cell real-time imaging systems, such as fluorescence lifetime imaging microscopy (FLIM). As the single-cell trapping efficiency is determined by the channel design instead of the flow rate, it can be integrated with other upper-stream functional units with various flow rates, and is expected to have broad applications in single-cell analysis.
Supplementary Material
References
- Carlo DD, & Lee LP, 2006. Dynamic single-cell analysis for quantitative biology. ACS Publications. [DOI] [PubMed] [Google Scholar]
- Chung K, Rivet CA, Kemp ML, & Lu H (2011). Imaging single-cell signaling dynamics with a deterministic high-density single-cell trap array. Analytical chemistry, 83, 7044–7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman MA, Caiolfa VR, Zamai M, & Gratton E (2008). The phasor approach to fluorescence lifetime imaging analysis. Biophysical journal, 94, L14–L16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagiannis P, Singer J, Hunt J, Gan SK, Rudman SM, Mechtcheriakova D, et al. (2009). Characterisation of an engineered trastuzumab IgE antibody and effector cell mechanisms targeting HER2/neu-positive tumour cells. Cancer immunology, immunotherapy, 58, 915–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D-H, Li X, Ma N, Digman MA, & Lee AP (2018). Rapid and label-free identification of single leukemia cells from blood in a high-density microfluidic trapping array by fluorescence lifetime imaging microscopy. Lab on a Chip, 18, 1349–1358. [DOI] [PubMed] [Google Scholar]
- Li X, Tao Y, Lee D-H, Wickramasinghe HK, & Lee AP (2017). In situ mRNA isolation from a microfluidic single-cell array using an external AFM nanoprobe. Lab on a Chip, 17, 1635–1644. [DOI] [PubMed] [Google Scholar]
- Lorenzi T, Turi A, Lorenzi M, Paolinelli F, Mancioli F, La Sala L, et al. (2012). Placental expression of CD100, CD72 and CD45 is dysregulated in human miscarriage. PloS one, 7, e35232. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nawarathna D, Turan T, & Wickramasinghe HK (2009). Selective probing of mRNA expression levels within a living cell. Applied physics letters, 95, 083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen J, Wei Y, Zheng Y, Wang C, & Sun Y (2015). On-chip sample preparation for complete blood count from raw blood. Lab on a Chip, 15, 1533–1544. [DOI] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. (2014). Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science, 1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punnoose EA, Atwal SK, Spoerke JM, Savage H, Pandita A, Yeh R-F, et al. (2010). Molecular biomarker analyses using circulating tumor cells. PloS one, 5, e12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsköld D, Luo S, Wang Y-C, Li R, Deng Q, Faridani OR, et al. (2012). Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nature biotechnology, 30, 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussidis A, Theocharis A, Tzanakakis G, & Karamanos NK (2007). The importance of c-Kit and PDGF receptors as potential targets for molecular therapy in breast cancer. Current medicinal chemistry, 14, 735–743. [DOI] [PubMed] [Google Scholar]
- Singh DK, Ku CJ, Wichaidit C, Steininger RJ, Wu LF, & Altschuler SJ (2010). Patterns of basal signaling heterogeneity can distinguish cellular populations with different drug sensitivities. Molecular systems biology, 6, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonehouse T, Woodhead V, Herridge P, Ashrafian H, George M, Chain B, et al. (1999). Molecular characterization of U937-dependent T-cell co-stimulation. Immunology, 96, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringari C, Cinquin A, Cinquin O, Digman MA, Donovan PJ, & Gratton E (2011). Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proceedings of the National Academy of Sciences, 108, 13582–13587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangawng AL, Ruoff RS, Swartz MA, & Glucksberg MR (2007). An ultra-thin PDMS membrane as a bio/micro–nano interface: fabrication and characterization. Biomedical microdevices, 9, 587–595. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, & Thompson CB (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. science, 324, 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.