Abstract
Proteomic strategies provide a valuable tool kit to identify proteins involved in diseases. With recent progress in MS technology, high throughput proteomics has accelerated protein identification for potential biomarkers. Numerous biomarker candidates have been identified in several diseases, and many are common among pathologies. An overall strategy that could complement and strengthen the search for biomarkers is combining protein identity with biological outcomes. This review describes an emerging framework of bridging bioactivity to protein identity, exploring the possibility that some biomarkers will have a mechanistic role in the disease process. A review of pulmonary, cardiovascular, and CNS biomarkers will be discussed to demonstrate the utility of combining bioactivity with identification as a means to not only find meaningful biomarkers, but also to uncover functional mediators of disease.
Keywords: Acute respiratory distress syndrome, Asthma, Atherosclerosis, Bioassay, Inflammation, Multiple sclerosis, Proteoform, PTMs, Validation assay
1. Introduction
Scrutinizing relevant biological fluids and tissues to detect biomarkers pertinent to disease has become increasingly important for clinical research. New relevant biomarkers could be used to detect the risk of acquiring a disease, the presence or the stage of pathology, and responses to therapy. Genomic and transcriptional profiling studies have shown strong potential for biomarker discovery in a broad array of diseases while proteomic strategies are still emerging [1]. Considering all possible gene expression patterns, mutations, coding, and noncoding SNPs, RNA splicing, microRNA biology, and the immensity of protein PTMs, it was estimated in human that up to 1.8 million different protein forms, recently coined “proteoforms,” can be considered [2, 3]. A proteoform designates all of the different molecular forms in which the protein product of a single gene can be found, including changes due to genetic variations, alternatively spliced RNA transcripts and PTMs [2]. This enormous diversity, the dynamic turnover of proteins and their release in various biological fluids during pathology is the basis for identification of protein biomarkers. Proteomics of various diseases, especially using approaches that can identify proteoform diversity, offer new biomarker avenues.
Given the number of organ systems and their associated pathologies that can be explored using proteomics approaches, it becomes increasingly difficult to address them all. Proteomic analysis of brain, lung, and cardiovascular systems has shown that some of the same markers of disease are often identified across different tissues. For example, an abundant protein such as actin has been identified as a marker for hypersensitivity pneumonitis, multiple sclerosis, and aortic aneurysm [4–11]. Likewise, Cystatin C is simultaneously a putative biomarker for malignant pleural effusion, traumatic brain injury, and cardiac injury [12–14]. Other common protein biomarkers in various diseases, listed in Table 1, suggest that acute-phase proteins as well as proteins released during cellular injury are frequently detected and not necessarily specific to a disease. Interestingly, the same biomarker can be found in various biological fluids across different diseases. For example, clusterin has been detected in bronchoalveolar lavage fluid (BALF), plasma, and cerebrospinal fluid (CSF) in patients with lung cancer, multiple sclerosis, and Alzheimer disease [5, 15–17] (Table 1). Similarly, hemopexin is present in plasma, CSF, and the spinal cord of patients with lung injury, multiple sclerosis, and cardiopulmonary bypass [15, 18–21] (Table 1). Such markers, with often excellent sensitivity but poor specificity, are not necessarily the best candidates for disease detection and diagnosis. The issue of specificity will continue to be a problem unless the proteomic approach is combined with other diagnostics tests, including disease-related bioassays.
Table 1.
Biomarker proteins common to cardiac (C), neurological (N), and pulmonary (P) diseases
| Protein name | Condition | Tissue | Proteomic technology | Ref. | |
|---|---|---|---|---|---|
| Actin | C | Artherosclerotic aorta | Aortic tissues | LC-MS/MS | [10] |
| Aortic aneurysm | Aortic wall | 2D-DIGE and MS/MS | [11] | ||
| N | EAE | Spinal cord | 2D gel and MALDI-TOF MS | [7] | |
| Multiple sclerosis | CSF | 2D gel and LC/MS | [8] | ||
| Multiple sclerosis | CSF | 2D-DIGE, MALDI-TOF MS and UPLC/Q-TOF MS | [9] | ||
| P | Hypersensitivity pneumonitis | BALF | 2D gel and MS | [4] | |
| α-1 Antichymotrypsin | C | Valvular heart disease | Plasma | 2D gel and MS | [193] |
| N | Multiple sclerosis | CSF | 2D-DIGE, MALDI-TOF MS and UPLC/Q-TOF MS | [9] | |
| Multiple sclerosis | CSF | 2D gel and LC/MS | [8] | ||
| EAE | CSF | ChipLC-QTOF MS, Orbitrap XL | [194] | ||
| P | NSCLC | Plasma | Monoclonal antibody libraries | [195] | |
| β-2 Microglobulin | C | PAD | Plasma | SELDI-TOF MS | [196] |
| N | Multiple sclerosis | CSF | 2D gel and MALDI MS | [19] | |
| P | CBD | BALF | 2D gel and MS | [197] | |
| Calreticulin | C | AAA | Aortic tissue | 2D gel and MS | [198] |
| N | Parkinson’s disease | Striatum | 2D gel and MALDI TOF-MS | [199] | |
| P | Cigarette smoking | Lung tissue | 2D gel and MALDI TOF-MS | [200] | |
| Ceruloplasmin | C | Cardiopulmonary bypass | Plasma | 2-DIGE and MALDI-TOF/TOF | [21] |
| N | HAD | Serum | 2D gel and IB | [201] | |
| P | Lung injury | BALF | 1D gel and MS | [202] | |
| Clusterin | C | Alzheimer disease | Plasma | 2D gel and LC-MS/MS | [17] |
| N | Multiple sclerosis | Plasma | 2D gel and MS | [15] | |
| Multiple sclerosis | CSF | MALDI TOF-MS | [16] | ||
| P | Lung cancer | BALF | 2D gel and MS | [5] | |
| Complement C3 | C | Plasma clots | Plasma | 2G gel and MALDI-TOF-MS | [203] |
| N | EAE | CSF | QTOF LC-MS | [194] | |
| EAE | Spinal cord | iTRAQ MS/MS | [20] | ||
| P | NSCLC/RILT | Plasma | 2 ExacTag, LC/MS | [204] | |
| Cystatin C | C | Cardiac injury | Conditioned media of cardiomyocytes | LC-MS/MS | [14] |
| N | Traumatic brain injury | CSF | DIGE and MS | [13] | |
| P | Malignant pleural effusion | Adenocarcinoma cell lines | 1D gel and LC-MS/MS | [12] | |
| α-Enolase | C | CHF | Cardiomyocytes | 2D gel and MS | [205] |
| N | EAE | Spinal cord | 2DE and MALDI-TOF MS | [32] | |
| Multiple sclerosis | CSF | 2D-DIGE, MALDI-TOF MS and UPLC/Q-TOF MS | [9] | ||
| Multiple sclerosis | Blood cells | [23] | |||
| P | NSCLC | Serum | 2D gel, IB, ELISA and MS | [206] | |
| Haptoglobin | C | Myocardial infarction | Plasma | 2D-DiGE and MALDi-TOF/TOF | [207] |
| N | Ischemic stroke | Plasma | 2D gel and MS | [208] | |
| P | Idiopathic pulmonary fibrosis | BALF | 2D gel, MS and WB | [6] | |
| Hemopexin | C | Cardiopulmonary bypass | Plasma | 2D gel and MS | [21] |
| N | Multiple sclerosis | CSF | 2D gel and MALDI-MS | [19] | |
| EAE | Spinal cord | iTRAQ, MALDI-MS | [20] | ||
| Multiple sclerosis | Plasma | 2D gel and MS | [15] | ||
| P | Chemical lung exposure | Plasma | 2D gel and MALDi-TOF | [18] | |
| Malate | C | Ischemia-reperfusion | Hearts mitochondria | 2DE and MALDI-TOF MS | [209] |
| Dehydrogenase | N | EAE | Spinal cord | 2D gel and MALDI-TOF MS | [7] |
| Multiple sclerosis | CSF | LC-ESI-MS/MS | [210] | ||
| P | ALI | Lung tissue | 2DE and MALDI-TOF MS | [94] | |
| MMP-9 | C | MACE | Plasma | ELISA | [211] |
| N | Brain stroke | Infarcted brains | Protein array | [212] | |
| P | Pulmonary tuberculosis | Serum | iTRAQ 2D LC-MS/MS | [213] | |
| Paraoxonase | C | ASCVD | Plasma | iTRAQ -MS | [214] |
| N | EAE | CSF | LC-MS | [194] | |
| P | SCLC | Serum | Affinity purification and iTRAQ | [215] | |
| T-kininogen | C | AMI | Serum | 2DE and MALDI-TOF/TOF MS | [216] |
| N | EAE | CSF | LC-MS and QTOF LC-MS | [194] | |
| P | Endotoxin challenge | BALF | 2D gel and MS | [217] | |
| Thrombospondin 1 | C | Myocardial infarction. | Left ventricle infarct tissue | 2D gel and MS | [218] |
| N | Ischemic stroke | Plasma | 1D gel and MS/MS | [219] | |
| P | Malignant mesothelioma | Pleural effusions | LC-MS/MS | [220] | |
| Vimentin | C | Degenerative aortic stenosis | Aortic valve | 2D-DIGE and MS | [221] |
| N | EAE | Spinal cord | iTRAQ, MALDI-MS | [20] | |
| MS | Blood/CNS | 1D, 2D gel IB, and MS | [24] | ||
| P | PM/DM | BALF | 2D gel and LC-MS/MS | [222] | |
| Malignant mesothelioma | Lung biopsies | 2D gel and MS | [223] |
Cardiac diseases. AAA, abdominal aortic aneurysms; AMI, acute myocardial infarction; ASCVD, atherosclerotic cardiovascular disease; CHF, congestive heart failure; MACE, major adverse cardiovascular event; PAD, peripheral arterial disease; RHD, rheumatic heart disease.
Neurological diseases. EAE, experimental autoimmune encephalomyelitis; HAD, HIV-1 associated dementia.
Pulmonary diseases. ALI, acute lung injury; IPF, idiopathic pulmonary fibrosis; PM/DM, polymyositis/ dermatomyositis; RILT, radiation-induced lung toxicity.
Tissues: BALF, bronchoalveolar lavage fluid; BCEC, brain capillary endothelial cells; CBD, chronic beryllium disease; CSF, cerebrospinal fluid; NSCLC, nonsmall cell lung cancer; SCLC, small cell lung cancer; TIF, tissue interstitial fluid.
Proteomic technologies: 2D-DIGE, 2dimensional fluorescence difference in-gel electrophoresis; iTRAQ, isobaric tag for relative and absolute quantification; LC/MS, liquid chromatography tandem mass spectrometry; MALDI-TOF, matrix-assisted laser desorption/ionization coupled to time of flight; MS, mass spectrometry, MS/MS, tandem mass spectrometry; SELDI, surface-enhanced laser desorption/ionization; UPLC-MSE, nano-ultra performance liquid chromatography coupled to electrospray ionisation mass spectrometry; UPLC/Q-TOF MS, ultra-performance liquid chromato-graph coupled with quadrupole time-of-flight mass spectrometry.
The ultimate goal of clinical proteomics is to find meaningful biomarkers associated, as specifically as possible, with the pathology of a particular tissue, preferably in a quantifiable manner. Clinically, the ideal biomarker should be easily obtained with minimum discomfort or risk to the patient, hence be present in a clinically accessible peripheral body tissue and/or fluid (e.g. blood, semen, saliva, urine, hair, or breath). Alternatively, biomarkers found in a tissue that cannot be sampled for routine examination may still be useful if they are detectable by noninvasive imaging. A reliable biomarker should be detected with high sensitivity and high reproducibility. To achieve this goal, rare proteins, expressed, or modified in pathological situations must first be identified. The search for novel protein biomarkers in various diseases such as multiple sclerosis, asthma, acute lung injury, and atherosclerosis are listed in this review. The broad utility of these systems will be addressed as well as the importance of validation using bioactivity-guided proteomics versus proteomics strategies that solely focus on identification. The PF2D platform will be used as an example of 2D-LC coupled to MS analysis to illustrate the need of validation and bridging bioactivity to protein identity.
2. Proteomics in clinical and basic research
2.1. Proteomics of a CNS disease, the case of multiple sclerosis
Multiple sclerosis is a chronic inflammatory disease of the CNS characterized by infiltrating auto-reactive immune cells that mediate demyelination and axonal degeneration of neurons [22]. Currently, its diagnosis is almost exclusively established by clinical examination and magnetic resonance imaging. Clinical biomarkers would help in differential diagnosis identifying disease subtypes (i.e. relapsing remitting, secondary progressive, primary progressive, progressive relapsing) with the possibility of earlier diagnosis, monitoring treatment effectiveness, and identifying new treatment targets. Human proteomic studies have used postmortem brain samples, CSF, and blood as a source to screen proteins [8, 9, 15, 19, 23, 24]. Since the tissue damage in multiple sclerosis is localized to the CNS, proteomics of postmortem brain are particularly relevant in revealing putative biomarkers and mechanisms linked to advanced pathology, although they do not qualify for longitudinal studies. Collecting CSF during disease progression is a more practical approach that can be applied to longitudinal studies [25]. However, the invasive procedure necessary to collect CSF is a major drawback for routine screening. Proteins from blood samples are easier to collect, however, the complexity of the blood proteome, the masking effect of highly abundant blood proteins, such as albumin, and the low concentration of CNS proteins in the blood, has not generated enthusiasm for using blood proteomics in the long run [26]. Hence, several methods have been developed to deplete albumin and other abundant serum proteins to gain sensitivity [27–29].
The experimental autoimmune encephalomyelitis (EAE) murine model stimulates T cells to trigger demyelination of the CNS in a way that simulates multiple sclerosis [30]. Proteomic analysis of human and animal tissues have revealed a large number of putative markers for multiple sclerosis and EAE, with some common to both [26, 31, 32]. The differences in induction and pathogenesis between EAE and multiple sclerosis argue for caution in interpreting proteomics changes in EAE alone. One example representing successful workflow from proteomic identification to in vivo model validation, is that of activated protein C (aPC) [33]. Global profiling of brain lesions from multiple sclerosis cases identified aPC within chronic active plaque. Administered in vivo, aPC ameliorated EAE through its anti-coagulant function and suppression of NFκB signaling. Among these common markers, those with altered PTM are particularly interesting. For example, occludin and neurofilament light-heavy chain have been found as phosphorylated proteins in both EAE [34–36] and multiple sclerosis [37–40]. Myelin basic protein and glial fibrillary acidic protein can be citrullinated during EAE [34, 41] and detected in human white matter of multiple sclerosis patients [42, 43]. Mitochondrial hsp70 and glyceraldehyde 3-phosphate dehydrogenase can be nitrated in EAE [44] and found in chronic inactive multiple sclerosis lesions [45, 46]. These proteins characterized by the presence of PTM on abundant proteins could be specific markers of disease. Such biomarkers can theoretically be followed by imaging of the brain by using specific ligand or antibody in a similar way as recently proposed for prostate cancer [47] and Alzheimer’s disease [48]. Proteomics-based studies have found many candidates for multiple sclerosis and have contributed to confirming the immunological and neurodegenerative process of the disease, yet there is no validated and consistent biomarker available for clinical use.
2.2. Proteomics of the cardiovascular system, the case of atherosclerosis
Atherosclerosis is one of the leading causes of death worldwide characterized by cholesterol deposition and chronic inflammation leading to progressive sclerosis of the arterial vessels that promote the pathophysiology of cardiovascular disease, stroke, and peripheral vascular disease [49]. The complex mechanism underlying plaque formation and disease progression involves matrix degradation, angiogenesis, oxidative stress lipid deposition and metabolism, apoptosis, and autophagy [50,51]. Advanced atherosclerotic disease can be diagnosed by clinical examination (e.g. vascular bruits) and by various cardiovascular imagining technologies [52]. Improving our knowledge about the molecular events involved at the onset of atherosclerosis and finding biomarkers of vascular atheroma would have considerable utility. Classical proteins commonly used in the clinic have been considered as biomarkers, but none of them have been implemented in the clinic due to their inconsistency [53,54]. More recent proteomics studies that used data mining techniques for biomarkers in plasma, serum, urine, atheroma, and secretome have revealed numerous potential candidates, but few have been submitted to functional validation [55]. The current proteomics approaches for atherosclerosis have produced a vast amount of experimental data [55–57]. Among them phospholipid-associated proteins: sod2, sod3, gst, hsp20, annexin A10, and fibrinogen fragment D are new candidate biomarkers. Many other proteins have been identified as putative biomarkers, but have not necessarily been confirmed by other investigators or fully validated [58–61]. On the contrary, several independent proteomics studies have shown that hsp27 plays an active role in atherosclerosis and appears to be a biomarker of myocardial ischemia [62–65]. Interestingly, 2DE and DIGE showed that three isoforms of phosphorylation-dependent hsp27 are modulated during atherosclerosis [62, 63]. The level of hsp27 circulating in the blood of patients with cardiovascular disease has been measured in patients and healthy volunteers, but its role as a biomarker has not always been consistent, perhaps because the different proteoforms of hsp27 have not been considered [65–68]. Moreover, serum levels of anti-hsp27 antibodies are significantly associated with acute coronary syndrome and stroke [69, 70]. Therefore, investigating the PTM of hsp27 might clarify its role as a biomarker for atherosclerosis. Soluble tumor necrosis factor-like weak inducer of apoptosis (sTWEAK) is another example of a potential candidate biomarker for atherosclerosis identified by proteomic analysis [71]. Isolation of an 18 kDa intact protein released in greater amounts by arteries of healthy patients than those with atherosclerotic plaques, was identified as sTWEAK [72]. ELISA and Western blot of secretome (culture of conditioned media from normal or pathological endarteries) validated that plaques release less than normal arteries [72]. As a functional validation, it was shown that mice treated with a TWEAK blocking monoclonal antibody and mice with genetic deletion of TWEAK are less prone to develop atherosclerosis [73]. In independent clinical studies, it was observed that circulating sTWEAK decreased in patients with coronary artery disease [74], chronic heart failure [75], and abdominal aortic aneurysms [76]. Additionally, sTWEAK has been shown to predict atherosclerosis in renal transplant patients [77]. Therefore, sTWEAK appears to match some of the requirements of a clinical biomarker, while also possessing a bioactivity role in disease.
2.3. Proteomics of the lung, the case of asthma, and acute respiratory distress syndrome
Asthma is a disease of the airways characterized by reversible airway obstruction, airway hyperresponsiveness, and chronic airway inflammation [78]. Acute respiratory distress syndrome (ARDS), and its experimental form, acute lung injury (ALI), are characterized by a widespread pulmonary inflammation with alveolar damage resulting in respiratory insufficiency [79]. Proteomics of asthma and ARDS/ALI have been used to pinpoint the pathophysiology of disease and to identify biomarkers. For both diseases, proteomes were analyzed using samples from BALF, sputum, lung tissue, plasma, and serum, and the findings have been reviewed in detail elsewhere [78–82]. Among all the putative biomarkers of asthma, S100A9 also called calgranulin b, is a potential candidate. Calgranulin b has been found in several proteomic studies to be upregulated in sputum of asthmatic patients [83, 84]. It was also upregulated in the sputum of patients with uncontrolled asthma analyzed by 2DE and MALDI-TOF MS, and was validated by Western blot and ELISA [85]. Interestingly, proteomics has identified four forms of calgranulin b with alternate start methionine codons and phosphorylation [86]. Moreover, calgranulin b is constitutively expressed in neutrophils [87], involved in lung inflammation as a damage-associated molecular pattern (DAMP) that binds receptor of advanced glycation end-products (RAGE) and Toll-like receptor-4 [88], and has also been shown to be oxidized, nitrosylated, and glutathionylated [89, 90]. Since calgranulin b is a DAMP, a bioassay could be designed to follow its bioactivity and validation of a specific proteoform of calgranulin b could be isolated using proteomics, not only as a specific biomarker, but perhaps also as a specific inflammatory agent in asthma.
Biomarkers for ARDS/ALI can be separated by factors induced during inflammation and those induced by cell injury [81]. Compared to other pathologies, a smaller number of proteomic analysis have been performed. Insulin-like growth factor has been identified in BALF of ARDS patients. Validation by Western blot and ELISA showed GFPB3 was overexpressed in BALF of ARDS patients compared to normal volunteers [91], but the opposite result was recently obtained using a much larger patient cohort [92]. Heparin-binding epidermal growth factor-like growth factor has also been identified in BALF of ARDS patients [91], and is a potential candidate because of its protective role in ALI [93]. Nevertheless, further studies are needed for its validation. Peroxiredoxin 1, cytochrome b5, and cytokeratin 17 have been identified and validated in animal models of ALI, but have not yet been investigated in human clinical studies [94, 95]. Proteomic studies of pulmonary edema fluid, plasma, and BALF from ARDS/ALI patients have detected many acute phase proteins and inflammatory proteins [96, 97]. Since none of the current biomarkers possess sufficient specificity and sensitivity, a new approach has highlighted a combination of biomarkers that has shown to be promising [98–100]. Specifically, due to the variety of causes of ARDS/ALI, a panel of biomarkers has a greater chance to pinpoint diagnosis compared to a single biomarker. Moreover, association of physiological markers can add accuracy to the diagnosis. For example, inflammatory cytokines like IL-6 and IL-8 are reliable predictors of oxygenation, ventilator-free days and 28-day mortality [99]. In cases of community-acquired pneumonia, levels of circulating soluble CD14 combined with a pathological scoring system enhanced the predictability of ARDS and mortality [101]. Also, there is excellent evidence suggesting metalloproteinases can fulfill a role in marking ARDS/ALI severity, especially MMP-1 and MMP-3 levels which correlated with increased mortality, disease severity, and multiorgan failure [102]. All the pathologies listed above, for different reasons, occur coincident with an inflammatory response. This facet could be exploited for biomarker mining. Multiple sclerosis is an inflammation-mediated disease characterized by an inflammatory environment in the CNS, activation of CD8 and CD4 T cells and the release of free radicals, proteolytic enzymes, and a variety of inflammatory mediators [103–105]. During ARDS/ALI, neutrophil infiltration and alveolar macrophage activation initiate the inflammatory response by producing high levels of proinflammatory cytokines, iNOS, free radicals, damage signals, and aberrant proteolysis [106–108]. Asthma is also characterized by inflammation, but classically considered a T-helper type 2 disease of the airways with eosinophil infiltration [109]. The inflammatory status in atherosclerosis is quite established as well, with continuous infiltration of leukocytes into the plaque, high levels of reactive oxygen species, and activation of metalloproteinases MMP2 and 9 [110, 111]. All these different types of inflammation lead to a large variety of factors able to modify proteins and creating additional proteoforms. As discussed below, the development of proteomics methods offers began to propose solutions for identifying these newly created proteoforms.
3. Evolution of proteomic approaches
3.1. Past and present proteomics
Proteomics in which intact proteins were first isolated and then identified, were the first to be used in proteomics research. One of the most popular methods was the 2DE separation of proteins, that is still used effectively today [112]. Proteins of interest, separated according to their pI and then mass, were excised from the gel and initially sequenced by Edman degradation [113]. More recently, trypsin-digest of excised spots are identified by mass spec and exact mass ions compared with databases to pinpoint protein identification. To eliminate gel-to-gel variability and allow higher sensitivity and more accurate quantification, DIGE was developed. In this newer version of 2D gel proteomics, proteins are labeled with fluorescent dyes before simultaneous separation in the same run allowing better sample comparison. Despite this advancement many limitations remain including the difficulty in detecting rare proteins, the masking effect of abundant proteins, the poor resolution of highly hydrophobic proteins, the limitation of pI range, and the lack of method automation. Recent advances in MS technologies, reviewed in greater detail elsewhere [55, 114], have bypassed some of the limitations of the gel-based systems and have advanced biomarker research. These technologies are called bottom-up proteomics or shotgun proteomics, which starts with peptide breakdown by enzymatic digestion of the entire proteome sample. Classical shotgun proteomics includes trypsin digestion of a complex mixture of proteins into a more complex mixture of peptides, followed by fractionation, fragmentation, sequencing of the peptides by MS, and identification of the proteins of origin via in silico digest protein databases. Shotgun proteomics have become broadly used and have provided immense quantities of protein biomarker potential candidates. This approach is unmatched for sensitivity and precise detection of known target peptides in a sample. However, there are certain drawbacks. Trypsin can miss proteins that are not properly denatured, proteolyze proteins into peptides too small for LC-mass spec detection, or skip a seemingly cleavable position [115]. This decreases the depth of the proteome to analyze and reduces the accuracy of the comparison with the theoretical in silico digests contained in databases used to identify the protein [116]. Also, shotgun proteomics can miss PTM that can lead to inaccurate protein quantification [117]. This limitation is unfortunate as protein PTM are frequently associated with a disease [118–121]. Furthermore, proteolytic digestion increases the complexity of the original protein sample, and prolongs the time of analysis, making high-throughput screening difficult. The vast quantity of peptides derived from the most abundant proteins, especially in the serum, limits the loading capacity of the LC-mass spec/mass spec columns and masks the presence of rare proteins. A large number of peptides increases considerably the chance of finding a significant number of false-positive matches when searched against a large database [122]. Another challenge common to many proteomics strategies, is the often underrepresentation of membrane proteins due to their low abundance and poor solubility [123]. Most importantly, and central to the concept developed in this review, trypsin digestion typically destroys protein biological function impeding validation in bioactivity assay.
One important aspect of proteomic strategies rely on accurate quantitative approaches based on stable isotope label, label-free statistical assessment methods and absolute quantification approaches [124]. Stable isotope labeling (e.g. iTRAQ, metabolic labeling, isotope-coded affinity tag reagents) uses stable isotopes or mass tag variants to label two different samples [125]. The proteins of a sample are tagged with a high molecular weight variant and the other one with low molecular weight variant or no tag. The samples are mixed, fractioned, and analyzed by MS. The peaks in the MS analysis indicate the relative ratio of the mass tag variants that indicate the relative abundance of the protein or peptide. Label-free statistical assessment or data-independent acquisition methods provide a sensitive approach for large-scale quantitative proteomic analyses [126]. With these methods, a sample is searched for the presence and quantity of a limited set of peptides that are specified prior to data acquisition. These kinds of targeted proteomics can precisely quantify specific proteins in many samples and are well suited for biomarker research. Finally, absolute quantification approaches, called AQUA strategy, or stable isotope dilution-MRM-MS permit absolute quantification of a given protein by spiking the sample with a peptide of interest synthesized using amino acids containing stable heavy isotopes, creating a slight increase of mass [127]. After protein digestion, the native and synthetic peptides are resolved together during the chromatography and ionized identically. By knowing the quantity of the labeled peptide, the precise abundance of the original peptide can be calculated. This approach is directed to the quantification of known biomarkers rather than the discovery of new ones [128].
3.2. Proteomics approach for intact proteins
New proteomics during which proteins are first fractionated, and sometimes totally purified and identification by MS, provides complementary approaches to shotgun proteomics that have been reviewed in detail [112, 129, 130]. Processing of intact proteins into the mass spectrometer permits advanced characterization of PTM and bypasses the difficulties associated with peptide-based proteomics [112]. In these strategies, separation of intact proteins based on mass, charge, and/or hydrophobicity increases the separation of proteoforms. Samples containing simple protein mixtures or fully purified protein can allow for 100% sequence coverage and full characterization. The most popular ways to separate intact proteins rely on classical LC methods like ion exchange, IEF, or reverse phase. More recently, capillary electrophoresis has allowed separation of intact proteins and peptides in small capillaries under high voltage, using low sample and reagent consumption, and with high efficiency [131]. Some of these approaches, called top-down proteomics, although less employed than bottom-up proteomics, have shown their usefulness in investigating disease biomarkers [132–139]. They have been described as a complementary cost-effective approach in clinical research [140], and recently promoted by a new consortium for the comprehensive analysis of intact proteins [www.topdownproteomics.org.].
3.3. PF 2D fractionation platform
Among the proteomics platforms with integrated proteome analysis capability, the PF2D offers a way to fractionate any kind of proteome by 2D LC. Quantitative and qualitative proteomic maps of fractions containing proteins are easily generated, which can be used to identify proteins by MS sequencing. Like other platforms [141], the PF2D isolates intact proteoforms that can be tested using bioassays before their identification by MS. Similar to other proteomic strategies, some investigators have added additional dimensions before or after 2D LC fractionation to increase separation power. Immunoaffinity depletion of abundant proteins in plasma samples has been performed before PF2D separation [142], and 1D SDS-PAGE analysis of the PF2D reverse-phase fractions has been used as a way to select protein bands before MS [142–144].
3.3.1. Sample preparation
To achieve maximum proteome depth, a sample must include proteins in a buffer that allow their separation while keeping them in solution. Sample preparation for PF2D proteomics uses solubilization buffers containing 6 M urea and a nonionic detergent detergent, n-octyl-β-D-glucopyranoside, that can be largely removed by processing samples through the reverse phase chromatography. High urea concentration with detergent has the advantage of being chemically stable and denaturing, hence inhibiting most enzyme activities including proteases. Membrane proteins, known to be difficult to analyze, have been successfully identified on PF2D platforms, suggesting this technology could be used for routine analysis of membrane proteins [145, 146], and for large extracellular matrix proteins that are usually not resolved by 2DE [147]. The protease inhibitors contained in the buffer and high urea concentration prevent protein degradation and loss of PTM. Samples as diverse as eukaryote and prokaryote cell lysate, serum, plasma, mitochondria, extracellular matrix, urine, and BALF have been successfully analyzed by PF2D proteomics [95, 144, 146–154]. Conveniently, the sample can be prepared with or without reducing agents, allowing for isolation of proteins linked by disulfide bonds. This allows the study of proteins which structure and enzymatic activity is modulated by thioredoxin and other disulfide modifying proteins [155].
3.3.2. First dimension IEF
Proteins are separated in a first dimension by IEF. Most investigators have used buffers in the range of pH 4 to 8.5 that cover the pI of mostproteins [144–147, 150, 152, 154, 156–161], but a few studies have tried additional pH ranges. For example, proteins from the same samples have been separated using two different pH ranges (4.0–8.5 and 7.0–10.0) [154], and basic proteins from macrophages infected with Candida albicans have been successfully identified using a pH range from 8.3 to 11.3 [162]. The elution of proteins from the IEF column is performed by a decreasing pH gradient, fractions are collected in a 96-deep well plate refrigerated at 12°C as they are automatically reinjected for a second fractionation. The first dimension collects fractions, not at a constant interval of time, but every 0.3 unit of pH. This aids in comparison of pH fractionation from run to run, and increases reproducibility. In fact, the reproducibility of the PF2D platform has actually been evaluated by running 10 consecutive aliquots of urine from the same patient [153], and improved protocols extend the life of the column to maximize cost-effectiveness [150, 163].
3.3.3. Second dimension, reverse phase chromatography
After the first dimension, fractions are reinjected through an automated autoloader onto a C-18 column heated at 50°C for reverse phase chromatography. Bound material is eluted through a concentration gradient of acetonitrile and peaks are detected at 214 nm. Typically, 12–20 first dimension fractions, selected from the linear part of first dimension gradient are fractionated by reverse phase chromatography and separated by hydrophobicity. Thus each first dimension run is fractionated into 36 fractions, generating a total of 432–720 fractions. Two-dimensional proteomic maps displaying p! versus hydrophobicity are constructed using the Proteoview/Deltaview software built in the PF2D platform.
3.3.4. Analysis of proteome maps
Analysis usually reveals a large majority of identical chromatographic patterns that we refer to as signatures and differential ones as fingerprints [143]. For example, Raf kinase inhibitor protein (RKIP), which was similarly expressed in resting and restimulated primed lymphocytes, was detected in a fingerprint suggesting the presence of an alternative proteoform [143]. Shotgun proteomics strategies that start with a trypsin digestion of the proteome are not well suited to find proteins that are physiologically cleaved, or proteins slightly modified and would have likely missed the differential presence of RKIP. After analysis of the proteomic maps, proteins contained in the selected fingerprint fractions can be directly identified by MS or subjected to further analysis for either purification refinement (e.g. SDS-PAGE), and validation using biological assays that will confirm the relevance of the protein.
3.3.5. Limitations
One difficulty in using the PF2D is its great sensitivity to pH variation. The first IEF dimension relies on precise fractionation every 0.3 unit of pH. Another related difficulty is the 16–18 h required to run a sample through the 2D LC. This can affect the reproducibility when a large cohort of samples are analyzed because the pH of the buffers need to be maintained at a precise value for several days. Moroever, considering the large number of fractions produced at each run there is a need for substantial −80°C storage area. A possible area of improvement is the loading capacity of the first dimension. Currently, no more than 4 mg of total protein can be analyzed per run without lose in resolution. Loading a larger quantity of protein would facilitate the detection of rare proteins.
3.4. Proteomics require validation assays
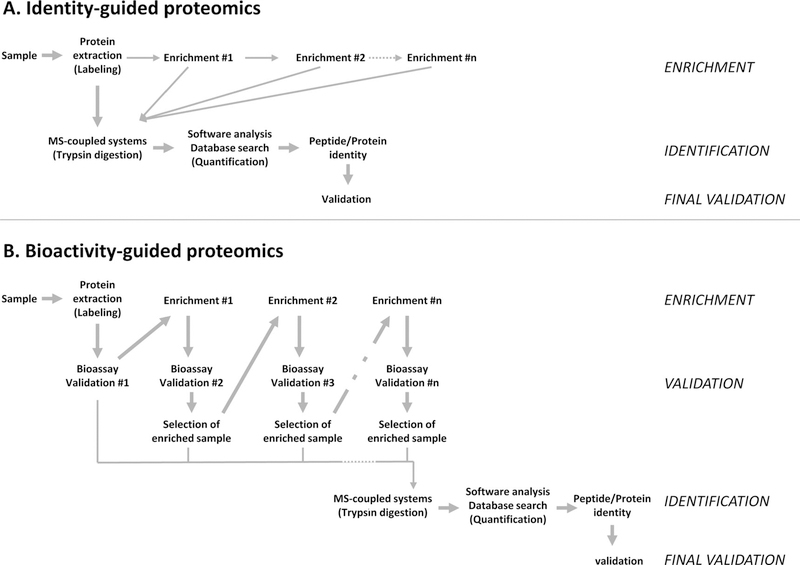
Validation has been recently described as one of the key bottlenecks contributing to the high failure rate of candidate biomarkers [164, 165]. First, validation should identify protein biomarker in the sample of interest and not in the control, or at least to lower levels in the control. This task is often performed using ELISA, Western blotting, protein microarrays as well as MS. The second goal is significantly linking the identified protein to a biological, pathological, or clinical outcome. Without stringent validation, overly simplistic selection of biomarkers results in a large number of potential candidates that fail after initial promise. One possibility to improve the success rate in biomarker research would be to develop new strategies driven more by bioactivity of the protein than by its identity. A general view of the difference and complementarity between identity-guided proteomics and bioactivity-guided proteomics is presented in Fig. 1. Identity-guided proteomics focuses first on efficient protein extraction and processing through MS-coupled system for identification, whereas bioactivity-guided proteomics is driven by the biological activity of protein(s). Consequently, validation of the protein(s) of interest occurs at the final stage of identity-guided proteomics but is built in bioactivity-guided proteomics. Hence, sequential and thorough validation performed during bioactivity-guided proteomics could improve the success rate in identifying new biomarkers.
Figure 1.
From identity to bioactivity-guided proteomics. (A) Identity-guided proteomics focuses on the most efficient and reliable way to identify proteins and peptides from a complex sample. Priority is given to efficient protein extraction, sometimes after several enrichment steps, and processing through a MS-coupled system for identification. Validation of the protein(s) of interest occurs at the final stage of the entire strategy. Identity-guided proteomics can theoretically identify and quantify intact or fragmented proteins from any kind of sample, but often suffers from low success rate in identifying new biomarkers due to incomplete validation. (B) Bioactivity-guided proteomics focuses on the most biologicaly relevant protein(s) from a complex sample. Emphasis is given to sequential validation using bioassay(s), before processing through MS-coupled system and identification. It requires intact proteins or at least biologically active ones and the development of reliable and sensitive bioassay(s). Additional validation of the protein of interest can be performed at the end of the strategy. The sequential validation performed during bioactivity-guided proteomics should improve the success rate of biomarkers usable in the clinic.
Since PF2D fractionates intact proteins, it can also integrate different kinds of validation strategies and begin to test the feasibility of bioactivity-guided proteomics. As mentioned earlier, RKIP was targeted because of the PF2D data but was also validated using locostatin, a specific small molecule RKIP-inhibitor [143]. Cytokine inhibition was observed in locostatin-treated mouse and human T cells, and in vivo treatment with locostatin inhibited TNF-α production upon triggering antigen specific T cells. A third line of validation was obtained using a model of T cell induced systemic inflammatory response syndrome (SIRS) where RKIP deletion inhibited production of IFN-γ due to intrinsic altered T-cell receptor activity [166]. Thus, the bioactivity validation of RKIP qualifies this protein as a potential marker and druggable protein for the treatment of SIRS.
Another example of validation linked to PF2D proteomics is the identification of T-cell specific human tumor antigens [167]. Specifically, tumor lysate was fractionated by chromato-focusing, then 20 fractions were tested by ELISPOT in a cross-presentation assay using autologous T cells, and seven reactive fractions were selected for the second dimension. Subfractions displaying protein peaks were tested using the same assay and proteins were identified by MS. Tumor expression of the proteins was first validated by both RT-PCR and immunofluorescence. Further validation was obtained for Transthyretin and Calgranulin that were recognized as autologous tumor antigens by T cells from patients with brain tumors. By integrating 2D LC proteomics with an assay that tracks biological activity of proteins through the purification scheme, two new tumor antigens were identified. A similar strategy could theoretically be applied to identify antigens in autoimmune or asthma-related illnesses not only for T cells but also B cells.
Similar in principle to the T-cell screening of tumor antigens present in the PF2D fractions, antibodies have been used in technologies called PF2D-SERPA (serological proteome analysis) andimmuno-PF2D-MS/MS [144, 168]. PF2D-SERPA can test the efficiency of a novel vaccine by screening sera reactivity of vaccinated individuals. Proteins from complex targets, like inactivated pathogens or opportunistic micro-organisms, separated by 2D LC and incubated with the sera revealed the specific antigens. The resulting liquid fractions can be validated further by Western blot for subsequent peptide sequencing by MS/MS. Such analyses can uncover the most immunogenic antigens expressed by the pathogen and be tested in vaccine development [169, 170]. This method can theoretically detect proteoforms (isoforms and PTM) that are valuable variations of an antigen for vaccine design [171]. Immuno-PF2D-MS/MS is a similar approach in which fractions from the first and second PF2D dimensions were successively selected by dot-blot and Western blot until final identification by MS. This strategy has been proposed for identification of immunogens of poorly characterized bacteria [144].
Finally, an example of identification and validation of factors associated with ALI was obtained using a combination of proteomics and in situ analysis. In a model of ALI induced by Staphylococcal Enterotoxin A inhalation, mouse BALF was separated by PF2D. Cytochrome b5 and Cytokeratin 17 were identified as putative biomarkers of ALI [95]. Validation was performed by immunohistochemistry in lung tissue sections that showed staining in the epithelial cells of the bronchioles. Although Cytochrome b5 and Cytokeratin 17 are potentially valuable biomarkers for ALI, their presence did not explain the exacerbation of lung inflammation. Nevertheless, it is likely that these biomarkers are an indicator of cell death since they are not typically secreted like cytokines or acute phase proteins. Thus, there are clear advantages to automated 2D LC fractionation but validation is still the centerpiece of proteomics.
4. Concluding remarks and perspectives
One reason proteomics of intact proteins is used less than bottom-up approaches is due to the slow development of high-throughput technology. Recent improvements have changed that perception with large-scale experiments where over 5000 proteoforms were observed [172, 173]. In general, proteomics have also recently benefited from advances in instrumentation and LC separation [172, 174, 175]. The PF2D proteomics presented in this review could also be improved with slip-flow technology. This new chromatographic technique, which uses capillaries packed with 0.47 μm silica colloidal crystal particles, have shown higher efficiency in reversed-phase separation of intact proteins [176, 177]. This could replace the second dimension of the PF2D, currently a C-18 reverse phase chromatography step, and provide a higher resolution for the final protein fractionation. Proteomics have already demonstrated, in a diverse range of diseases, that analysis of proteoforms of intact proteins are useful and relevant for translational studies [132, 143, 178, 179]. One area particularly well suited for this approach is bioassays, where this might advance biomarker discovery in diseases associated with acute inflammation. The pathways activated during inflammation induce phosphorylation of signaling proteins, active proteases, and protein ubiquitination, nitration, sumoylation, and numerous other protein modifications. The functional significance of these new proteoforms can be tested by immunological assays and technologies currently used to study the immune system. Single-cell mass cytometry, also known as cytometry by TOF or CyTOF, is a recent example of merging flow cytometry, a technology classically used to study immune cells, with MS [180]. CyTOF uses antibodies tagged to transition metals, which potentially allow up to 100 independent measurements on a single cell. This technology can be applied to the screening of a patient’s blood sample to track, for example, a specific phospho-protein biomarker in a rare cell population [181]. Multiplexed ion beam imaging is a related technology for advanced immunohistochemistry studies [182, 183]. Antibodies tagged with transition metals have been used to stain tissue sections following routine immunohistochemistry protocols. The samples were then ablated spot by spot in a laser ablation chamber and processed through a CyTOF mass cytometer. This approach avoides sample autofluorescence, does not require an amplification step and has been used to simultaneously image 32 proteins and protein modifications at subcellular resolution [183].
Another emerging area that complements proteomics, is detection and bioactivity of enzymes in complex proteomes using activity-based probes (ABPs) [184, 185]. Click chemistry can engineer these synthetic ABPs with both a reactive and a reporting group [186]. The reactive group is designed to covalently bind a specific class of enzyme while the detection group enables its analysis and this strategy has been used for activity-based protein profiling (ABPP) [187]. ABPP has been successful in the detection and analysis of more than 80% of mammalian Serine Hydrolases [188]. ABPP has also been applied to the screening of anti-inflammatory small molecules using an in vitro macrophage bioassay [189]. In a viral infection model, Caspase, Hydrolase, and Tyrosine Phosphatase enzyme activities have been analyzed at the protein isoform level by using proteomics followed by ABPP [190]. Besides the detection of specific enzymatic activity, ABPP has also enhanced identification and characterization of unknown protein functions [191] and the targets of natural products [192].
In conclusion, there are at least two methods to enhance proteomic biomarker discovery. The first being classical validation using relevant biological samples and the second using bioactivity assays (Fig. 1). The latter is the newest challenge, but advances in automated protein fractionation and cell culture strategies should drive the field past protein identification alone and could contribute further in understanding disease mechanism.
Abbreviations
- ABP
activity-based probe
- ABPP
activity-based protein profiling
- ALI
acute lung injury
- ARDS
acute respiratory distress syndrome
- BALF
bronchoalveolar lavage fluid
- CSF
cerebrospinal fluid
- DAMP
damage-associated molecular pattern
- EAE
experimental autoimmune encephalomyelitis
- RAGE
receptor of advanced glycation end-products
- RKIP
raf kinase inhibitor protein
- SIRS
systemic inflammatory response syndrome
Footnotes
The authors have declared no conflict of interest.
5 References
- [1].Altelaar AF, Munoz J, Heck AJ, Next-generation proteomics: towards an integrative view of proteome dynamics. Nat. Rev. Genet 2013, 14, 35–48. [DOI] [PubMed] [Google Scholar]
- [2].Smith LM, Kelleher NL, Proteoform: a single term describing protein complexity. Nat. Methods 2013, 10, 186187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jensen ON, Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr. Opin. Chem. Biol 2004, 8, 33–41. [DOI] [PubMed] [Google Scholar]
- [4].Okamoto T, Miyazaki Y, Shirahama R, Tamaoka M, Inase N, Proteome analysis of bronchoalveolar lavage fluid in chronic hypersensitivity pneumonitis. Allergol. Int 2011, 61, 83–92. [DOI] [PubMed] [Google Scholar]
- [5].Okano T, Kondo T, Kakisaka T, Fujii K et al. , Plasma proteomics of lung cancer by a linkage of multi-dimensional liquid chromatography and two-dimensional difference gel electrophoresis. Proteomics 2006, 6, 3938–3948. [DOI] [PubMed] [Google Scholar]
- [6].Lilja-Maula LI, Palviainen MJ, Heikkila HP, Raekallio MR, Rajamaki MM, Proteomic analysis of bronchoalveolar lavage fluid samples obtained from West Highland White Terriers with idiopathic pulmonary fibrosis, dogs with chronic bronchitis, and healthy dogs. Am. J. Vet. Res 2012, 74, 148–154. [DOI] [PubMed] [Google Scholar]
- [7].Farias AS, Martins-de-Souza D, Guimaraes L, Pradella F et al. , Proteome analysis of spinal cord during the clinical course of monophasic experimental autoimmune encephalomyelitis. Proteomics 2012, 12,2656–2662. [DOI] [PubMed] [Google Scholar]
- [8].Dumont D, Noben JP, Raus J, Stinissen P, Robben J, Proteomic analysis of cerebrospinal fluid from multiple sclerosis patients. Proteomics 2004, 4, 2117–2124. [DOI] [PubMed] [Google Scholar]
- [9].Liu S, Bai S, Qin Z, Yang Y et al. , Quantitative proteomic analysis of the cerebrospinal fluid of patients with multiple sclerosis. J. Cell Mol. Med 2009, 13, 1586–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kaga E, Karademir B, Baykal AT, Ozer NK, Identification of differentially expressed proteins in atherosclerotic aorta and effect of vitamin E. J. Proteomics 2013, 92, 260–273. [DOI] [PubMed] [Google Scholar]
- [11].Ando T, Nagai K, Chikada M, Okamoto K et al. , Proteomic analyses of aortic wall in patients with abdominal aortic aneurysm. J. Cardiovasc. Surg 2011, 52, 545–555. [PubMed] [Google Scholar]
- [12].Yu CJ, Wang CL, Wang CI, Chen CD et al. , Comprehensive proteome analysis of malignant pleural effusion for lung cancer biomarker discovery by using multidimensional protein identification technology. J. Proteome Res 2011, 10,4671–4682. [DOI] [PubMed] [Google Scholar]
- [13].Gao WM, Chadha MS, Berger RP, Omenn GS et al. , A gel-based proteomic comparison of human cerebrospinal fluid between inflicted and non-inflicted pediatric traumatic brain injury. J. Neurotrauma 2007, 24, 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Xie L, Terrand J, Xu B, Tsaprailis G et al. , Cystatin C increases in cardiac injury: a role in extracellular matrix protein modulation. Cardiovasc. Res 2010, 87, 628–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rithidech KN, Honikel L, Milazzo M, Madigan D et al. , Protein expression profiles in pediatric multiple sclerosis: potential biomarkers. Mult. Scler 2009, 15, 455–464. [DOI] [PubMed] [Google Scholar]
- [16].Stoop MP, Dekker LJ, Titulaer MK, Burgers P C. et al. , Multiple sclerosis-related proteins identified in cerebrospinal fluid by advanced mass spectrometry. Proteomics 2008, 8, 1576–1585. [DOI] [PubMed] [Google Scholar]
- [17].Yu HR, Kuo HC, Sheen JM, Wang L et al. , A unique plasma proteomic profiling with imbalanced fibrinogen cascade in patients with Kawasaki disease. Pediatr. Allergy Immunol 2009, 20, 699–707. [DOI] [PubMed] [Google Scholar]
- [18].Kap-Soon N, Do-Youn L, Hak CJ, Joo WA et al. , Protein biomarkers in the plasma of workers occupationally exposed to polycyclic aromatic hydrocarbons. Proteomics 2004, 4,3505–3513. [DOI] [PubMed] [Google Scholar]
- [19].Hammack BN, Fung KY, Hunsucker SW, Duncan MW et al. , Proteomic analysis of multiple sclerosis cerebrospinal fluid. Mult. Scler 2004, 10, 245–260. [DOI] [PubMed] [Google Scholar]
- [20].Liu T, Donahue KC, Hu J, Kurnellas MP et al. , Identification of differentially expressed proteins in experimental autoimmune encephalomyelitis (EAE) by proteomic analysis of the spinal cord. J. Proteome Res 2007, 6, 2565–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lull ME, Carkaci-Salli N, Freeman WM, Myers JL et al. , Plasma biomarkers in pediatric patients undergoing cardiopulmonary bypass. Pediatr. Res 2008, 63, 638–644. [DOI] [PubMed] [Google Scholar]
- [22].Iwanowski P, Losy J, Immunological differences between classical phenothypes of multiple sclerosis. J. Neurol. Sci 2015, 349, 10–14. [DOI] [PubMed] [Google Scholar]
- [23].De Masi R, Vergara D, Pasca S, Acierno R et al. , PBMCs protein expression profile in relapsing IFN-treated multiple sclerosis: a pilot study on relation to clinical findings and brain atrophy. J. Neuroimmunol 2009, 210, 80–86. [DOI] [PubMed] [Google Scholar]
- [24].Almeras L, Lefranc D, Drobecq H, de Seze J et al. , New antigenic candidates in multiple sclerosis: identification by serological proteome analysis. Proteomics 2004, 4, 2184–2194. [DOI] [PubMed] [Google Scholar]
- [25].Wilson ME, Boumaza I, Lacomis D, Bowser R, Cystatin C: a candidate biomarker for amyotrophic lateral sclerosis. PLoS One 2010, 5, e15133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rosenling T, Attali A, Luider T M., Bischoff, R., The experimental autoimmune encephalomyelitis model for proteomic biomarker studies: from rat to human. Clin. Chim. Acta 2011,412,812–822. [DOI] [PubMed] [Google Scholar]
- [27].Uzun L, Armutcu C, Bicen O, Ersoz A et al. , Simultaneous depletion of immunoglobulin G and albumin from human plasma using novel monolithic cryogel columns. Colloids Surf. B Biointerfaces 2013, 112, 1–8. [DOI] [PubMed] [Google Scholar]
- [28].Haudenschild DR, Eldridge A, Lein PJ, Chromy BA, High abundant protein removal from rodent blood for biomarker discovery. Biochem. Biophys. Res. Commun 2014, 455, 84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Henning AK, Albrecht D, Riedel K, Mettenleiter TC, Karger A, An alternative method for serum protein depletion/enrichment by precipitation at mildly acidic pH values and low ionic strength. Proteomics 2015, 11, 1935–1940. [DOI] [PubMed] [Google Scholar]
- [30].Steinman L, Zamvil SS, How to successfully apply animal studies in experimental allergic encephalomyelitis to research on multiple sclerosis. Ann. Neurol 2006, 60, 12–21. [DOI] [PubMed] [Google Scholar]
- [31].Dagley LF, Emili A, Purcell AW, Application of quantitative proteomics technologies to the biomarker discovery pipeline for multiple sclerosis. Proteomics Clin. Appl 2012, 7, 91–108. [DOI] [PubMed] [Google Scholar]
- [32].Farias AS, Pradella F, Schmitt A, Santos LM, Martins-de-Souza D, Ten years of proteomics in multiple sclerosis. Proteomics 2013, 14,467–480. [DOI] [PubMed] [Google Scholar]
- [33].Han MH, Hwang SI, Roy DB, Lundgren DH et al. , Proteomic analysis of active multiple sclerosis lesions reveals therapeutic targets. Nature 2008, 451, 1076–1081. [DOI] [PubMed] [Google Scholar]
- [34].Grant JE, Hu J, Liu T, Jain MR et al. , Post-translational modifications in the rat lumbar spinal cord in experimental autoimmune encephalomyelitis. J. Proteome Res 2007, 6, 2786–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Morgan L, Shah B, Rivers LE, Barden L et al. , Inflammation and dephosphorylation of the tight junction protein occludin in an experimental model of multiple sclerosis. Neuroscience 2007, 147, 664–673. [DOI] [PubMed] [Google Scholar]
- [36].Gresle MM, Shaw G, Jarrott B, Alexandrou EN et al. , Validation of a novel biomarker for acute axonal injury in experimental autoimmune encephalomyelitis. J. Neurosci. Res 2008, 86, 3548–3555. [DOI] [PubMed] [Google Scholar]
- [37].Norgren N, Sundstrom P, Svenningsson A, Rosengren L et al. , Neurofilament and glial fibrillary acidic protein in multiple sclerosis. Neurology 2004, 63, 1586–1590. [DOI] [PubMed] [Google Scholar]
- [38].Petzold A, Gveric D, Groves M, Schmierer K et al. , Phosphorylation and compactness of neurofilaments in multiple sclerosis: indicators of axonal pathology. Exp. Neurol 2008, 213, 326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Proia P, Schiera G, Salemi G, Ragonese P et al. , Neuronal and BBB damage induced by sera from patients with secondary progressive multiple sclerosis. Int. J. Mol. Med 2009, 24, 743–747. [DOI] [PubMed] [Google Scholar]
- [40].Blecharz KG, Haghikia A, Stasiolek M, Kruse N et al. , Glucocorticoid effects on endothelial barrier function in the murine brain endothelial cell line cEND incubated with sera from patients with multiple sclerosis. Mult. Scler 2010, 16, 293–302. [DOI] [PubMed] [Google Scholar]
- [41].Kidd BA, Ho PP, Sharpe O, Zhao X et al. , Epitope spreading to citrullinated antigens in mouse models of autoimmune arthritis and demyelination. Arthritis Res. Ther 2008, 10, R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kim JK, Mastronardi FG, Wood DD, Lubman DM et al. , Multiple sclerosis: an important role for posttranslational modifications of myelin basicprotein in pathogenesis. Mol. Cell Proteomics 2003, 2, 453–462. [DOI] [PubMed] [Google Scholar]
- [43].Moscarello MA, Mastronardi FG, Wood DD, The role of citrullinated proteins suggests a novel mechanism in the pathogenesis of multiple sclerosis. Neurochem. Res 2007, 32, 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Qi X, Lewin AS, Sun L, Hauswirth WW, Guy J, Mitochondrial protein nitration primes neurodegeneration in experimental autoimmune encephalomyelitis. J. Biol. Chem 2006, 281, 31950–31962. [DOI] [PubMed] [Google Scholar]
- [45].Kolln J, Ren HM, Da RR, Zhang Y et al. , Triosephosphate isomerase- and glyceraldehyde-3-phosphate dehydrogenase-reactive autoantibodies in the cerebrospinal fluid of patients with multiple sclerosis. J. Immunol 2006, 177, 5652–5658. [DOI] [PubMed] [Google Scholar]
- [46].Witte ME, Bo L, Rodenburg RJ, Belien JA et al. , Enhanced number and activity of mitochondria in multiple sclerosis lesions. J. Pathol 2009, 219, 193–204. [DOI] [PubMed] [Google Scholar]
- [47].LeBeau AM, Sevillano N, Markham K, Winter MB et al. , Imaging active urokinase plasminogen activator in prostate cancer. Cancer Res. 2015, 75, 1225–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Viola KL, Sbarboro J, Sureka R, De M et al. , Towards non-invasive diagnostic imaging of early-stage Alzheimer’s disease. Nat. Nanotechnol 2015, 10, 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Braunwald E, Shattuck lecture–cardiovascular medicine at the turn of the millennium: triumphs, concerns, and opportunities. N. Engl. J. Med 1997, 337, 1360–1369. [DOI] [PubMed] [Google Scholar]
- [50].Zimmer S, Grebe A, Latz E, Danger signaling in atherosclerosis. Circ. Res 2015, 116, 323–340. [DOI] [PubMed] [Google Scholar]
- [51].Narula J, Strauss HW, The popcorn plaques. Nat. Med 2007, 13, 532–534. [DOI] [PubMed] [Google Scholar]
- [52].Fernandez-Friera L, Ibanez B, Fuster V, Imaging subclinical atherosclerosis: is it ready for prime time? A review. J. Cardiovasc. Transl. Res 2014, 7, 623–634. [DOI] [PubMed] [Google Scholar]
- [53].Wang TJ, Gona P, Larson MG, Tofler GH et al. , Multiple biomarkers for the prediction of first major cardiovascular events and death. N. Engl. J. Med 2006, 355, 2631–2639. [DOI] [PubMed] [Google Scholar]
- [54].Musunuru K, Blumenthal RS, Biomarkers for prediction of cardiovascular events. N. Engl. J. Med 2007, 356, 1472; author reply 1474–1475. [DOI] [PubMed] [Google Scholar]
- [55].Napoli C, Zullo A, Picascia A, Infante T, Mancini FP, Recent advances in proteomic technologies applied to cardiovascular disease. J. Cell Biochem 2012, 114, 7–20. [DOI] [PubMed] [Google Scholar]
- [56].Shalhoub J, Sikkel MB, Davies KJ, Vorkas P A. et al. , Systems biology of human atherosclerosis. Vasc. Endovasc. Surg 2014, 48, 5–17. [DOI] [PubMed] [Google Scholar]
- [57].Eberini I, Wait R, Calabresi L, Sensi C et al. , A proteomic portrait of atherosclerosis. J. Proteomics 2013, 82, 92–112. [DOI] [PubMed] [Google Scholar]
- [58].Gordon SM, Deng J, Lu LJ, Davidson WS, Proteomic characterization of human plasma high density lipoprotein fractionated by gel filtration chromatography. J. Proteome Res 2010, 9, 5239–5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Vaisar T, Mayer P, Nilsson E, Zhao XQ et al. , HDL in humans with cardiovascular disease exhibits a proteomic signature. Clin. Chim. Acta 2010, 411, 972–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lepedda AJ, Cigliano A, Cherchi GM, Spirito R et al. , A proteomic approach to differentiate histologically classified stable and unstable plaques from human carotid arteries. Atherosclerosis 2009, 203, 112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].de la Cuesta F, Barderas MG, Calvo E, Zubiri I et al. , Secretome analysis of atherosclerotic and non-atherosclerotic arteries reveals dynamic extracellular remodeling during pathogenesis. J. Proteomics 2011, 75, 2960–2971. [DOI] [PubMed] [Google Scholar]
- [62].Malaud E, Piquer D, Merle D, Molina L et al. , Carotid atherosclerotic plaques: proteomics study after a low-abundance protein enrichment step. Electrophoresis 2012, 33, 470–482. [DOI] [PubMed] [Google Scholar]
- [63].de la Cuesta F, Alvarez-Llamas G, Maroto AS, Donado A et al. , A proteomic focus on the alterations occurring at the human atherosclerotic coronary intima. Mol. Cell Proteomics 2011, 10, M110 003517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Seibert TA, Hibbert B, Chen YX, Rayner K et al. , Serum heat shock protein 27 levels represent a potential therapeutic target for atherosclerosis: observations from a human cohort and treatment of female mice. J. Am. Coll. Cardiol 2013, 62, 1446–1454. [DOI] [PubMed] [Google Scholar]
- [65].Martin-Ventura JL, Duran MC, Blanco-Colio LM, Meilhac O et al. , Identification by a differential proteomic approach of heat shock protein 27 as a potential marker of atherosclerosis. Circulation 2004, 110,2216–2219. [DOI] [PubMed] [Google Scholar]
- [66].Jozefowicz-Okonkwo G, Wierzbowska-Drabik K, Kasielski M, Trzos E et al. , Is Hsp27 a marker of myocardial ischaemia? Kardiol. Pol 2009, 67, 947–952. [PubMed] [Google Scholar]
- [67].Kardys I, Rifai N, Meilhac O, Michel JB et al. , Plasma concentration of heat shock protein 27 and risk of cardiovascular disease: a prospective, nested case-control study. Clin. Chem 2008, 54, 139–146. [DOI] [PubMed] [Google Scholar]
- [68].Ghayour-Mobarhan M, Saber H, Ferns GA, The potential role of heat shock protein 27 in cardiovascular disease. Clin. Chim. Acta 2012, 413, 15–24. [DOI] [PubMed] [Google Scholar]
- [69].Ghayour-Mobarhan M, Sahebkar A, Parizadeh SM, Moohebati M et al. , Antibody titres to heat shock protein 27 are elevated in patients with acute coronary syndrome. Int. J. Exp. Pathol 2008, 89, 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Azarpazhooh MR, Mobarra N, Parizadeh SM, Tavallaie S et al. , Serum high-sensitivity C-reactive protein and heat shock protein 27 antibody titers in patients with stroke and 6-month prognosis. Angiology 2010, 61, 607–612. [DOI] [PubMed] [Google Scholar]
- [71].Blanco-Colio LM, Martin-Ventura JL, Carrero JJ, Yilmaz MI et al. , Vascular proteomics and the discovery process of clinical biomarkers: the case of TWEAK. Proteomics Clin. Appl 2011, 5, 281–288. [DOI] [PubMed] [Google Scholar]
- [72].Blanco-Colio LM, Martin-Ventura JL, Munoz-Garcia B, Orbe J et al. , Identification of soluble tumor necrosis factor-like weak inducer of apoptosis (sTWEAK) as a possible biomarker of subclinical atherosclerosis. Arterioscler. Thromb. Vasc. Biol 2007, 27, 916–922. [DOI] [PubMed] [Google Scholar]
- [73].Sastre C, Fernandez-Laso V, Madrigal-Matute J, Munoz-Garcia B et al. , Genetic deletion or TWEAK blocking antibody administration reduce atherosclerosis and enhance plaque stability in mice. J. Cell Mol. Med 2014, 18, 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Jelic-Ivanovic Z, Bujisic N, Spasic S, Bogavac-Stanojevic N et al. , Circulating sTWEAK improves the prediction of coronary artery disease. Clin. Biochem 2009, 42, 1381–1386. [DOI] [PubMed] [Google Scholar]
- [75].Chorianopoulos E, Rosenberg M, Zugck C, Wolf J et al. , Decreased soluble TWEAK levels predict an adverse prognosis in patients with chronic stable heart failure. Eur. J. Heart Fail 2009, 11, 1050–1056. [DOI] [PubMed] [Google Scholar]
- [76].Martin-Ventura JL, Lindholt JS, Moreno JA, Vega de Ceniga M et al. , Soluble TWEAK plasma levels predict expansion of human abdominal aortic aneurysms. Atherosclerosis 2010, 214, 486–489. [DOI] [PubMed] [Google Scholar]
- [77].Turkmen K, Tonbul HZ, Erdur FM, Toker A et al. , Soluble TWEAK independently predicts atherosclerosis in renal transplant patients. BMC Nephrol. 2013, 14, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Sircar G, Saha B, Bhattacharya SG, Saha S, Allergic asthma biomarkers using systems approaches. Front Genet 2014, 4, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Lam E, dos Santos CC, Advances in molecular acute lung injury/acute respiratory distress syndrome and ventilator-induced lung injury: the role of genomics, proteomics, bioinformatics and translational biology. Curr. Opin. Crit. Care 2008, 14, 3–10. [DOI] [PubMed] [Google Scholar]
- [80].Barnett N, Ware LB, Biomarkers in acute lung injury–marking forward progress. Crit. Care Clin 2011, 27, 661–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Tunceroglu H, Shah A, Porhomayon J, Nader ND, Biomarkers of lung injury in critical care medicine: past, present, and future. Immunol. Invest 2013, 42, 247–261. [DOI] [PubMed] [Google Scholar]
- [82].Terracciano R, Pelaia G, Preiano M, Savino R, Asthma and COPD proteomics: current approaches and future directions. Proteomics Clin. Appl 2014, 9, 203–220. [DOI] [PubMed] [Google Scholar]
- [83].Gharib SA, Nguyen EV, Lai Y, Plampin JD et al. , Induced sputum proteome in healthy subjects and asthmatic patients. J. Allergy Clin. Immunol 2011, 128, 1176–1184, e1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Bloemen K, Van Den Heuvel R, Govarts E, Hooyberghs J et al. , A new approach to study exhaled proteins as potential biomarkers for asthma. Clin. Exp. Allergy 2010, 41, 346–356. [DOI] [PubMed] [Google Scholar]
- [85].Lee TH, Jang AS, Park JS, Kim TH et al. , Elevation of S100 calcium binding protein A9 in sputum of neutrophilic inflammation in severe uncontrolled asthma. Ann. Allergy Asthma Immunol 2013, 111, 268–275, e261. [DOI] [PubMed] [Google Scholar]
- [86].Roth MJ, Parks BA, Ferguson JT, Boyne MT 2nd, Kelleher NL, “Proteotyping”: population proteomics of human leukocytes using top down mass spectrometry. Anal. Chem 2008, 80, 2857–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Markowitz J, Carson WE 3rd, Review of S100A9 biology and its role in cancer. Biochim. Biophys. Acta 2012, 1835, 100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Halayko AJ, Ghavami S, S100A8/A9: a mediator of severe asthma pathogenesis and morbidity? Can. J. Physiol. Pharmacol 2009, 87, 743–755. [DOI] [PubMed] [Google Scholar]
- [89].Goyette J, Geczy CL, Inflammation-associated S100 proteins: new mechanisms that regulate function. Amino Acids 2010, 41,821–842. [DOI] [PubMed] [Google Scholar]
- [90].Lim SY, Raftery MJ, Goyette J, Geczy CL, S-glutathionylation regulates inflammatory activities of S100A9. J. Biol. Chem 2010, 285, 14377–14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Schnapp LM, Donohoe S, Chen J, Sunde DA et al. , Mining the acute respiratory distress syndrome proteome: identification of the insulin-like growth factor (IGF)/IGF-binding protein-3 pathway in acute lung injury. Am. J. Pathol 2006, 169, 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Ahasic AM, Zhai R, Su L, Zhao Y et al. , IGF1 and IGFBP3 in acute respiratory distress syndrome. Eur. J. Endocrinol 2011, 166, 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Lutmer J, Watkins D, Chen CL, Velten M, Besner G, Heparin-binding epidermal growth factor-like growth factor attenuates acute lung injury and multiorgan dysfunction after scald burn. J. Surg. Res 2013, 185, 329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Liu D, Mao P, Huang Y, Liu Y et al. , Proteomic analysis of lung tissue in a rat acute lung injury model: identification of PRDX1 as a promoter of inflammation. Mediators Inflamm. 2014, 2014,469358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Menoret A, Kumar S, Vella AT, Cytochrome b5 and cytokeratin 17 are biomarkers in bronchoalveolar fluid signifying onset of acute lung injury. PLoS One 2012, 7, e40184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Chen X, Shan Q, Jiang L, Zhu B, Xi X, Quantitative proteomic analysis by iTRAQfor identification of candidate biomarkers in plasma from acute respiratory distress syndrome patients. Biochem. Biophys. Res. Commun 2013, 441, 1–6. [DOI] [PubMed] [Google Scholar]
- [97].Chang DW, Hayashi S, Gharib SA, Vaisar T et al. , Proteomic and computational analysis of bronchoalveolar proteins during the course of the acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med 2008, 178, 701–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Ware LB, Koyama T, Zhao Z, Janz DR et al. , Biomarkers of lung epithelial injury and inflammation distinguish severe sepsis patients with acute respiratory distress syndrome. Crit. Care 2013, 17, R253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Agrawal A, Zhuo H, Brady S, Levitt J et al. , Pathogenetic and predictive value of biomarkers in patients with ALI and lower severity of illness: results from two clinical trials. Am. J. Physiol. Lung Cell Mol. Physiol 2012, 303, L634–L639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ware LB, Koyama T, Billheimer DD, Wu W et al. , Prognostic and pathogenetic value of combining clinical and biochemical indices in patients with acute lung injury. Chest 2010, 137, 288–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Liu B, Yin Q, Chen YX,Zhao YZ, Li CS, Role of Presepsin (sCD14-ST) and the CURB65 scoring system in predicting severity and outcome of community-acquired pneumonia in an emergency department. Respir. Med 2014, 108, 1204–1213. [DOI] [PubMed] [Google Scholar]
- [102].Aschner Y, Zemans RL, Yamashita CM, Downey GP, Matrix metalloproteinases and protein tyrosine kinases: potential noveltargets in acute lung injury and ARDS. Chest 2014, 146, 1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Hemmer B, Nessler S, Zhou D, Kieseier B, Hartung HP, Immunopathogenesis and immunotherapy of multiple sclerosis. Nat. Clin. Pract. Neurol 2006, 2, 201–211. [DOI] [PubMed] [Google Scholar]
- [104].Trapp BD, Nave KA, Multiple sclerosis: an immune or neurodegenerative disorder? Annu. Rev. Neurosci 2008, 31, 247–269. [DOI] [PubMed] [Google Scholar]
- [105].Fakhoury M, Role of immunity and inflammation in the pathophysiology of neurodegenerative diseases. Neurodegener. Dis 2015, 15, 63–69. [DOI] [PubMed] [Google Scholar]
- [106].Tsushima K, King LS, Aggarwal NR, De Gorordo A et al. , Acute lung injury review. Intern. Med 2009, 48, 621–630. [DOI] [PubMed] [Google Scholar]
- [107].Aggarwal NR, King LS, D’Alessio F R., Diverse macrophage populations mediate acute lung inflammation and resolution. Am. J. Physiol. Lung Cell Mol. Physiol 2014, 306, L709–L725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Han S, Mallampalli RK, The acute respiratory distress syndrome: from mechanism to translation. J. Immunol 2015, 194, 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Lambrecht BN, Hammad H,The immunology of asthma. Nat. Immunol 2014, 16, 45–56. [DOI] [PubMed] [Google Scholar]
- [110].Shoenfeld Y, Sherer Y, Harats D, Artherosclerosis as an infectious, inflammatory and autoimmune disease. Trends Immunol. 2001, 22, 293–295. [DOI] [PubMed] [Google Scholar]
- [111].Hulsmans M, Van Dooren E, Holvoet P, Mitochondrial reactive oxygen species and risk of atherosclerosis. Curr. Atheroscler. Rep 2012, 14, 264–276. [DOI] [PubMed] [Google Scholar]
- [112].Catherman AD, Skinner OS, Kelleher NL, Top down proteomics: facts and perspectives. Biochem. Biophys. Res. Commun 2014, 445, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Komatsu S, Edman sequencing of proteins from 2D gels. Methods Mol. Biol 2007, 355, 211–217. [DOI] [PubMed] [Google Scholar]
- [114].Anderson L, Six decades searching for meaning in the proteome. J. Proteomics 2014, 107, 24–30. [DOI] [PubMed] [Google Scholar]
- [115].Vandermarliere E, Mueller M, Martens L, Getting intimate with trypsin, the leading protease in proteomics. Mass Spectrom. Rev 2013, 32, 453–465. [DOI] [PubMed] [Google Scholar]
- [116].Hildonen S, Halvorsen TG, Reubsaet L, Why less is more when generating tryptic peptides in bottom-up proteomics. Proteomics 2014, 14,2031–2041. [DOI] [PubMed] [Google Scholar]
- [117].Soldi M, Cuomo A, Bonaldi T, Improved bottom-up strategy to efficiently separate hypermodified histone peptides through ultra-HPLC separation on a bench top Orbitrap instrument. Proteomics 2014, 14, 2212–2225. [DOI] [PubMed] [Google Scholar]
- [118].Stastna M, Van Eyk JE, Post-translational modifications of lysine and evolving role in heart pathologies–recent developments. Proteomics 2014, 15, 1164–1180. [DOI] [PubMed] [Google Scholar]
- [119].Burska AN, Hunt L, Boissinot M, Strollo R et al. , Autoantibodies to posttranslational modifications in rheumatoid arthritis. Mediators Inflamm. 2014, 2014, 492873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Copeland RJ, Han G, Hart GW, O-GlcNAcomics-revealing roles of O-GlcNAcylation in disease mechanisms and development of potential diagnostics. Proteomics Clin. Appl 2013, 7, 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Anbalagan M, Huderson B, Murphy L, Rowan BG, Post-translational modifications of nuclear receptors and human disease. Nucl. Recept Signal 2012, 10, e001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Cargile BJ, Bundy JL, Stephenson JL Jr., Potential for false positive identifications from large databases through tandem mass spectrometry. J. Proteome Res 2004, 3, 1082–1085. [DOI] [PubMed] [Google Scholar]
- [123].Vuckovic D, Dagley LF, Purcell AW, Emili A, Membrane proteomics by high performance liquid chromatography-tandem mass spectrometry: analytical approaches and challenges. Proteomics 2012, 13, 404–423. [DOI] [PubMed] [Google Scholar]
- [124].Chahrour O, Cobice D, Malone J, Stable isotope labelling methods in mass spectrometry-based quantitative proteomics. J. Pharm. Biomed. Anal 2015. [DOI] [PubMed] [Google Scholar]
- [125].Calderon-Gonzalez KG, Valero Rustarazo ML, Labra-Barrios ML, Bazan-Mendez CI et al. , Determination of the protein expression profiles of breast cancer cell lines by quantitative proteomics using iTRAQ labelling and tandem mass spectrometry. J. Proteomics 2015, 124, 50–78. [DOI] [PubMed] [Google Scholar]
- [126].Sajic T, Liu Y, Aebersold R, Using data-independent, high-resolution mass spectrometry in protein biomarker research: perspectives and clinical applications. Proteomics Clin. Appl 2015, 9, 307–321. [DOI] [PubMed] [Google Scholar]
- [127].Brun V, Masselon C, Garin J, Dupuis A, Isotope dilution strategies for absolute quantitative proteomics. J. Proteomics 2009, 72, 740–749. [DOI] [PubMed] [Google Scholar]
- [128].Craft GE, Chen A, Nairn AC, Recent advances in quantitative neuroproteomics. Methods 2013, 61, 186–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Lanucara F, Eyers CE, Top-down mass spectrometry for the analysis of combinatorial post-translational modifications. Mass Spectrom. Rev 2012, 32, 27–42. [DOI] [PubMed] [Google Scholar]
- [130].Gregorich ZR, Ge Y, Top-down proteomics in health and disease: challenges and opportunities. Proteomics 2014, 14, 1195–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Fonslow BR, Yates JR 3rd, Capillary electrophoresis applied to proteomic analysis. J. Sep. Sci 2009, 32, 1175–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Mazur MT, Cardasis HL, Spellman DS, Liaw A et al. , Quantitative analysis of intact apolipoproteins in human HDL by top-down differential mass spectrometry. Proc. Natl. Acad. Sci. USA 2010, 107, 7728–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Edwards RL, Griffiths P, Bunch J, Cooper HJ, Top-down proteomics and direct surface sampling of neonatal dried blood spots: diagnosis of unknown hemoglobin variants. J. Am. Soc. Mass Spectrom 2012, 23, 1921–1930. [DOI] [PubMed] [Google Scholar]
- [134].Zhang J, Guy MJ, Norman HS, Chen YC et al. , Top-down quantitative proteomics identified phosphorylation of cardiac troponin I as a candidate biomarker for chronic heart failure. J. Proteome Res 2011, 10, 4054–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Ansong C, Wu S, Meng D, Liu X et al. , Top-down proteomics reveals a unique protein S-thiolation switch in Salmonella typhimurium in response to infection-like conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 10153–10158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Auclair JR, Boggio KJ, Petsko GA, Ringe D, Agar JN, Strategies for stabilizing superoxide dismutase (SOD1), the protein destabilized in the most common form of familial amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA 2010, 107, 21394–21399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Borges CR, Oran PE, Buddi S, Jarvis JW et al. , Building multidimensional biomarker views of type 2 diabetes on the basis of protein microheterogeneity. Clin. Chem 2011, 57,719–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Desiderio C, D’Angelo L, Rossetti DV, lavarone F et al. , Cerebrospinal fluid top-down proteomics evidenced the potential biomarker role of LVV- and VV-hemorphin-7 in posterior cranial fossa pediatric brain tumors. Proteomics 2012, 12, 2158–2166. [DOI] [PubMed] [Google Scholar]
- [139].Genitori L, Lena G, [Cranial injuries in infants]. Soins Gynecol. Obstet. Pueric Pediatr 1990, 107, 8–11. [PubMed] [Google Scholar]
- [140].Savaryn JP, Catherman AD, Thomas RM, Abecassis MM, Kelleher NL, The emergence of top-down proteomics in clinical research. Genome Med. 2013, 5, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Messana I, Cabras T, lavarone F, Vincenzoni F et al. , Unraveling the different proteomic platforms. J. Sep. Sci 2013, 36, 128–139. [DOI] [PubMed] [Google Scholar]
- [142].Schlautman JD, Rozek W, Stetler R, Mosley RL et al. , Multidimensional protein fractionation using ProteomeLab PF 2D for profiling amyotrophic lateral sclerosis immunity: a preliminary report. Proteome Sci. 2008, 6, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Menoret A, McAleer JP, Ngoi SM, Ray S et al. ,The oxazolidinone derivative locostatin induces cytokine appeasement. J. Immunol 2009, 183, 7489–7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Ruelle V, Falisse-Poirrier N, Elmoualij B, Zorzi D et al. , An immuno-PF2D-MS/MS proteomic approach for bacterial antigenic characterization: to Bacillus and beyond. J. Proteome Res 2007, 6, 2168–2175. [DOI] [PubMed] [Google Scholar]
- [145].Lee HJ, Kwon MS, Lee EY, Cho SY, Paik Y K., Establishment of a PF2D-MS/MS platform for rapid profiling and semiquantitative analysis of membrane protein biomarkers. Proteomics 2008, 8, 2168–2177. [DOI] [PubMed] [Google Scholar]
- [146].McDonald T, Sheng S, Stanley B, Chen D et al. , Expanding the subproteome of the inner mitochondria using protein separation technologies: one- and two-dimensional liquid chromatography and two-dimensional gel electrophoresis. Mol. Cell Proteomics 2006, 5, 2392–2411. [DOI] [PubMed] [Google Scholar]
- [147].Hurst RE, Kyker KD, Dozmorov MG, Takemori N et al. , Proteome-level display by 2-dimensional chromatography of extracellular matrix-dependent modulation of the phenotype of bladder cancer cells. Proteome Sci. 2006, 4, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Ornstein DK, Tyson DR, Proteomics for the identification of new prostate cancer biomarkers. Urol. Oncol 2006, 24, 231–236. [DOI] [PubMed] [Google Scholar]
- [149].Sheng S, Chen D, Van Eyk JE, Multidimensional liquid chromatography separation of intact proteins by chromatographic focusing and reversed phase of the human serum proteome: optimization and protein database. Mol. Cell Proteomics 2006, 5, 26–34. [DOI] [PubMed] [Google Scholar]
- [150].Kim M, Lee SH, Min J, Kobayashi F et al. , The utilization of Triton X-100 for enhanced two-dimensional liquidphase proteomics. J. Biomed. Biotechnol 2011, 2011, 213643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Sanchez-Juanes F, Muniz MC, Raposo C, Rodriguez-Prieto S et al. , Unveiling the rat urinary proteome with three complementary proteomics approaches. Electrophoresis 2013, 34, 2473–2483. [DOI] [PubMed] [Google Scholar]
- [152].Menoret A, Drew DA, Miyamoto S, Nakanishi M et al. , Differential proteomics identifies PDIA3 as a novel chemoprevention target in human colon cancer cells. Mol. Carcinog 2012, 53 (Suppl 1), E11–E22. [DOI] [PubMed] [Google Scholar]
- [153].Soldi M, Sarto C, Valsecchi C, Magni F et al. , Proteome profile of human urine with two-dimensional liquid phase fractionation. Proteomics 2005, 5, 2641–2647. [DOI] [PubMed] [Google Scholar]
- [154].Lee HJ, Kang MJ, Lee EY, Cho SY et al. , Application of a peptide-based PF2D platform for quantitative proteomics in disease biomarker discovery. Proteomics 2008, 8, 3371–3381. [DOI] [PubMed] [Google Scholar]
- [155].Yano H, Kuroda S, Buchanan BB, Disulfide proteome in the analysis of protein function and structure. Proteomics 2002, 2, 1090–1096. [DOI] [PubMed] [Google Scholar]
- [156].Suberbielle E, Gonzalez-Dunia D, Pont F, High reproducibility of two-dimensional liquid chromatography using pH-driven fractionation with a pressure-resistant electrode. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci 2008, 871, 125–129. [DOI] [PubMed] [Google Scholar]
- [157].Wang L, Yang W, Ju W, Wang P et al. , A proteomic study of Hutchinson-Gilford progeria syndrome: application of 2D-chromotography in a premature aging disease. Biochem. Biophys. Res. Commun 2012, 417, 1119–1126. [DOI] [PubMed] [Google Scholar]
- [158].Wang Y, Wu R, Cho KR, Shedden KA et al. , Classification of cancer cell lines using an automated two-dimensional liquid mapping method with hierarchical clustering techniques. Mol. Cell Proteomics 2006, 5, 43–52. [DOI] [PubMed] [Google Scholar]
- [159].Deford JH, Nuss JE, Amaning J, English RD et al. , High-throughput liquid-liquid fractionation of multiple protein post-translational modifications. J. Proteome Res 2009, 8, 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Grebenova D, Roselova P, Pluskalova M, Halada P et al. , Proteins implicated in the increase of adhesivity induced by suberoylanilide hydroxamic acid in leukemic cells. J. Proteomics 2012, 77, 406–422. [DOI] [PubMed] [Google Scholar]
- [161].Nakanishi M, Menoret A, Belinsky GS, Giardina C et al. , Utilizing endoscopic technology to reveal real-time proteomic alterations in response to chemoprevention. Proteomics Clin. Appl 2007, 1, 1660–1666. [DOI] [PubMed] [Google Scholar]
- [162].Shin YK, Lee HJ, Lee JS, Paik YK, Proteomic analysis of mammalian basic proteins by liquid-based two-dimensional column chromatography. Proteomics 2006, 6, 1143–1150. [DOI] [PubMed] [Google Scholar]
- [163].Barre O, Solioz M, Improved protocol for chromatofocusing on the ProteomeLab PF2D. Proteomics 2006, 6, 5096–5098. [DOI] [PubMed] [Google Scholar]
- [164].Poste G, Biospecimens, biomarkers, and burgeoning data: the imperative for more rigorous research standards. Trends Mol. Med 2012, 18, 717–722. [DOI] [PubMed] [Google Scholar]
- [165].Fuzery AK, Levin J, Chan MM, Chan DW, Translation of proteomic biomarkers into FDA approved cancer diagnostics: issues and challenges. Clin. Proteomics 2013, 10, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Wright KT, Vella AT, RKIP contributes to IFN-gamma synthesis by CD8+ T cells after serial TCR triggering in systemic inflammatory response syndrome. J. Immunol 2013, 191, 708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Beckhove P, Warta R, Lemke B, Stoycheva D et al. , Rapid T cell-based identification of human tumor tissue antigens by automated two-dimensional protein fractionation. J. Clin. Invest 2010, 120, 2230–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Falisse-Poirrier N, Ruelle V, ElMoualij B, Zorzi D et al. , Advances in immunoproteomics for serological characterization of microbial antigens. J. Microbiol. Methods 2006, 67, 593–596. [DOI] [PubMed] [Google Scholar]
- [169].Kunnath-Velayudhan S, Davidow AL, Wang HY, Molina DM et al. , Proteome-scale antibody responses and outcome of Mycobacterium tuberculosis infection in nonhuman primates and in tuberculosis patients. J. Infect. Dis 2012, 206, 697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Pitarch A, Nombela C, Gil C, Serum antibody signature directed against Candida albicans Hsp90 and enolase detects invasive candidiasis in non-neutropenic patients. J. Proteome Res 2014, 13, 5165–5184. [DOI] [PubMed] [Google Scholar]
- [171].Koerber JT, Thomsen ND, Hannigan BT, Degrado WF, Wells JA, Nature-inspired design of motif-specific antibody scaffolds. Nat. Biotechnol 2013, 31, 916–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Tran JC, Zamdborg L, Ahlf DR, Lee JE et al. , Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature 2011, 480, 254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Catherman AD, Durbin KR, Ahlf DR, Early BP et al. , Large-scale top-down proteomics of the human proteome: membrane proteins, mitochondria, and senescence. Mol. Cell Proteomics 2013, 12, 3465–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Sherma ND, Borges CR, Trenchevska O, Jarvis JW et al. , Mass spectrometric immunoassay for the qualitative and quantitative analysis of the cytokine macrophage migration inhibitory factor (MIF). Proteome Sci. 2014, 12,52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Ye H, Mandal R, Catherman A, Thomas P M. et al. , Top-down proteomics with mass spectrometry imaging: a pilot study towards discovery of biomarkers for neurodevelopmental disorders. PLoS One 2014, 9, e92831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Wei B, Rogers BJ, Wirth MJ, Slip flow in colloidal crystals for ultraefficient chromatography. J. Am. Chem. Soc 2012, 134, 10780–10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Wu Z, Wei B, Zhang X, Wirth MJ, Efficient separations of intact proteins using slip-flow with nano-liquid chromatography-mass spectrometry. Anal. Chem 2014, 86, 1592–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Chamot-Rooke J, Mikaty G, Malosse C, Soyer M et al. , Posttranslational modification of pili upon cell contact triggers N. meningitidis dissemination. Science 2011,331,778–782. [DOI] [PubMed] [Google Scholar]
- [179].Dong X, Sumandea CA, Chen YC, Garcia-Cazarin ML et al. , Augmented phosphorylation of cardiac troponin I in hypertensive heart failure. J. Biol. Chem 2011,287,848–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Bendall SC, Simonds EF, Qiu P, Amir el AD et al. , Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011, 332, 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Han L, Qiu P, Zeng Z, Jorgensen JL et al. , Single-cell mass cytometry reveals intracellular survival/proliferative signaling in FLT3-ITD-mutated AML stem/progenitor cells. Cytometry A 2015, 87, 346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Angelo M, Bendall SC, Finck R, Hale MB et al. , Multiplexed ion beam imaging of human breast tumors. Nat. Med 2014, 20,436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Giesen C, Wang HA, Schapiro D, Zivanovic N et al. , Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 2014, 11, 417–422. [DOI] [PubMed] [Google Scholar]
- [184].Yang P, Liu K, Activity-based protein profiling: recent advances in probe development and applications. Chembiochem 2015, 16,712–714. [DOI] [PubMed] [Google Scholar]
- [185].Sadler NC, Wright AT, Activity-based protein profiling of microbes. Curr. Opin. Chem. Biol 2015, 24C, 139–144. [DOI] [PubMed] [Google Scholar]
- [186].Speers AE, Cravatt BF, Profiling enzyme activities in vivo using click chemistry methods. Chem. Biol 2004, 11, 535–546. [DOI] [PubMed] [Google Scholar]
- [187].Niphakis MJ, Cravatt BF, Enzyme inhibitor discovery by activity-based protein profiling. Annu. Rev. Biochem 2014, 83, 341–377. [DOI] [PubMed] [Google Scholar]
- [188].Bachovchin DA, Ji T, Li W, Simon GM et al. , Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc. Natl. Acad. Sci. USA 2010, 107, 20941–20946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Hunerdosse DM, Morris PJ, Miyamoto DK, Fisher KJ et al. , Chemical genetics screening reveals KIAA1363 as a cytokine-lowering target. ACS Chem. Biol 2014, 9, 2905–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Maaty WS, Steffens JD, Heinemann J, Ortmann AC et al. , Global analysis of viral infection in an archaeal model system. Front Microbiol. 2012, 3, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Willems LI, Overkleeft HS, van Kasteren SI, Current developments in activity-based protein profiling. Bioconjug. Chem 2014, 25, 1181–1191. [DOI] [PubMed] [Google Scholar]
- [192].Krysiak J, Breinbauer R, Activity-based protein profiling for natural product target discovery. Top Curr. Chem 2011, 324, 43–84. [DOI] [PubMed] [Google Scholar]
- [193].Gao G, Xuan C, Yang Q, Liu XC et al. , Identification of altered plasma proteins by proteomic study in valvular heart diseases and the potential clinical significance. PLoS One 2013, 8, e72111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Rosenling T, Stoop MP, Attali A, van Aken H et al. , Profiling and identification of cerebrospinal fluid proteins in a rat EAE model of multiple sclerosis. J. Proteome Res 2012, 11, 2048–2060. [DOI] [PubMed] [Google Scholar]
- [195].Guergova-Kuras M, Kurucz I, Hempel W, Tardieu N et al. , Discovery of lung cancer biomarkers by profiling the plasma proteome with monoclonal antibody libraries. Mol. Cell Proteomics 2011, 10, M111 010298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Fung ET, Wilson AM, Zhang F, Harris N et al. , A biomarker panel for peripheral arterial disease. Vasc. Med 2008, 13, 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Silva E, Bourin S, Sabounchi-Schutt F, Laurin Y et al. , A quantitative proteomic analysis of soluble bronchoalveolar fluid proteins from patients with sarcoidosis and chronic beryllium disease. Sarcoidosis Vasc. Diffuse Lung Dis 2007, 24, 24–32. [PubMed] [Google Scholar]
- [198].Urbonavicius S, Lindholt JS, Delbosc S, Urbonaviciene G et al. , Proteins associated with the size and expansion rate of the abdominal aortic aneurysm wall as identified by proteomic analysis. Interact. Cardiovasc. Thorac. Surg 2010, 11, 433–441. [DOI] [PubMed] [Google Scholar]
- [199].Lessner G, Schmitt O, Haas SJ, Mikkat S et al. , Differential proteome of the striatum from hemiparkinsonian rats displays vivid structural remodeling processes. J. Proteome Res 2010, 9, 4671–4687. [DOI] [PubMed] [Google Scholar]
- [200].Kelsen SG, Duan X, Ji R, Perez O et al. , Cigarette smoke induces an unfolded protein response in the human lung: a proteomic approach. Am. J. Respir. Cell Mol. Biol 2008, 38, 541–550. [DOI] [PubMed] [Google Scholar]
- [201].Rozek W, Horning J, Anderson J, Ciborowski P, Sera proteomic biomarker profiling in HIV-1 infected subjects with cognitive impairment. Proteomics Clin. Appl 2008, 2, 1498–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Chang CC, Chen SH, Ho SH, Yang CY et al. , Proteomic analysis of proteins from bronchoalveolar lavage fluid reveals the action mechanism of ultrafine carbon black-induced lung injury in mice. Proteomics 2007, 7, 4388–4397. [DOI] [PubMed] [Google Scholar]
- [203].Howes JM, Richardson VR, Smith KA, Schroeder V et al. , Complement C3 is a novel plasma clot component with anti-fibrinolytic properties. Diab. Vasc. Dis. Res. 2012, 9, 216–225. [DOI] [PubMed] [Google Scholar]
- [204].Cai XW, Shedden K, Ao X, Davis M et al. , Plasma proteomic analysis may identify new markers for radiation-induced lung toxicity in patients with non-small-cell lung cancer. Int. J. Radiat. Oncol. Biol. Phys 2010, 77, 867–876. [DOI] [PubMed] [Google Scholar]
- [205].De Souza AI, Cardin S, Wait R, Chung YL et al. , Proteomic and metabolomic analysis of atrial profibrillatory remodelling in congestive heart failure. J. Mol. Cell Cardiol 2010, 49,851–863. [DOI] [PubMed] [Google Scholar]
- [206].He P, Naka T, Serada S, Fujimoto M et al. , Proteomics-based identification of alpha-enolase as a tumor antigen in non-small lung cancer. Cancer Sci. 2007, 98, 1234–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Haas B, Serchi T, Wagner DR, Gilson G et al. , Proteomic analysis of plasma samples from patients with acute myocardial infarction identifies haptoglobin as a potential prognostic biomarker. J. Proteomics 2011, 75, 229–236. [DOI] [PubMed] [Google Scholar]
- [208].Chen R, Vendrell I, Chen CP, Cash D et al. , Proteomic analysis of rat plasma following transient focal cerebral ischemia. Biomark. Med 2011, 5, 837–846. [DOI] [PubMed] [Google Scholar]
- [209].Kim N, Lee Y, Kim H, Joo H et al. , Potential biomarkers for ischemic heart damage identified in mitochondrial proteins by comparative proteomics. Proteomics 2006, 6, 1237–1249. [DOI] [PubMed] [Google Scholar]
- [210].Noben JP, Dumont D, Kwasnikowska N, Verhaert P et al. , Lumbarcerebrospinal fluid proteome in multiple sclerosis: characterization by ultrafiltration, liquid chromatography, and mass spectrometry. J. Proteome Res 2006, 5, 1647–1657. [DOI] [PubMed] [Google Scholar]
- [211].Cheng JM, Akkerhuis KM, Meilhac O, Oemrawsingh RM et al. , Circulating osteoglycin and NGAL/MMP9 complex concentrations predict 1-year major adverse cardiovascular events after coronary angiography. Arterioscler. Thromb. Vasc. Biol 2014, 34, 1078–1084. [DOI] [PubMed] [Google Scholar]
- [212].Cuadrado E, Rosell A, Penalba A, Slevin M et al. , Vascular MMP-9/TIMP-2 and neuronal MMP-10 up-regulation in human brain after stroke: a combined laser microdissection and protein array study. J. Proteome Res 2009, 8, 3191–3197. [DOI] [PubMed] [Google Scholar]
- [213].Xu D, Li Y, Li X, Wei LL et al. , Serum protein S100A9, SOD3, and MMP9 as new diagnostic biomarkers for pulmonary tuberculosis by iTRAQ-coupled two-dimensional LC-MS/MS. Proteomics 2014, 15, 58–67. [DOI] [PubMed] [Google Scholar]
- [214].Yin X, Subramanian S, Hwang SJ, O’Donnell CJ et al. , Protein biomarkers of new-onset cardiovascular disease: prospective study from the systems approach to biomarker research in cardiovascular disease initiative. Arterioscler. Thromb. Vasc. Biol 2014, 34, 939–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Ahn JM, Sung HJ, Yoon YH, Kim BG et al. , Integrated glycoproteomics demonstrates fucosylated serum paraoxonase 1 alterations in small cell lung cancer. Mol. Cell Proteomics 2014, 13, 30–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Cubedo J, Ramaiola I, Padro T, Martin-Yuste V et al. , High-molecular-weight kininogen and the intrinsic coagulation pathway in patients with de novo acute myocardial infarction. Thromb. Haemost 2013, 110, 1121–1134. [DOI] [PubMed] [Google Scholar]
- [217].Signor L, Tigani B, Beckmann N, Falchetto R, Stoeckli M, Two-dimensional electrophoresis protein profiling and identification in rat bronchoalveolar lavage fluid following allergen and endotoxin challenge. Proteomics 2004, 4, 2101–2110. [DOI] [PubMed] [Google Scholar]
- [218].Zamilpa R, Lopez EF, Chiao YA, Dai Q et al. , Proteomic analysis identifies in vivo candidate matrix metalloproteinase-9 substrates in the left ventricle post-myocardial infarction. Proteomics 2010, 10, 2214–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Ning M, Sarracino DA, Kho AT, Guo S et al. , Proteomic temporal profile of human brain endothelium after oxidative stress. Stroke 2010, 42, 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Mundt F, Johansson HJ, Forshed J, Arslan S et al. , Proteome screening of pleural effusions identifies galectin 1 as a diagnostic biomarker and highlights several prognostic biomarkers for malignant mesothelioma. Mol. Cell Proteomics 2013, 13,701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Martin-Rojas T, Gil-Dones F, Lopez-Almodovar LF, Padial LR et al. , Proteomic profile of human aortic stenosis: insights into the degenerative process. J. Proteome Res 2012, 11, 1537–1550. [DOI] [PubMed] [Google Scholar]
- [222].Passadore I, Iadarola P, Di Poto C, Giuliano S et al. , 2-DE and LC-MS/MS for a comparative proteomic analysis of BALf from subjects with different subsets of inflammatory myopathies. J. Proteome Res 2009, 8, 2331–2340. [DOI] [PubMed] [Google Scholar]
- [223].Giusti L, Da Valle Y, Bonotti A, Donadio E et al. , Comparative proteomic analysis of malignant pleural mesothelioma evidences an altered expression of nuclear lamin and filament-related proteins. Proteomics Clin. Appl 2014, 8, 258–268. [DOI] [PubMed] [Google Scholar]