Abstract
The Ca2+-sensing protein S100A11 of the S100 family is an important mediator of numerous biological functions and pathological conditions including cancer. The receptor for advanced glycation end products (RAGE) has been well accepted as the major receptor for several S100 family members. Here, we take the S100B protein as an antagonist to interfere with the interaction flanked by S100A11 and the RAGE V domain. We employed NMR spectroscopy to describe the interactions between the S100A11 and S100B proteins. 1H–15N heteronuclear single-quantum correlation-NMR titrations showed the potential binding dynamics of S100A11 and S100B interactions. In the HADDOCK program, we constructed the S100A11–S100B heterodimer complex that was then superimposed with the S100A11–S100B complex structure in the same orientation as the S100A11–RAGE V domain complex. This overlay analysis showed that S100B could interfere in the binding section of S100A11 and the RAGE V domain. Additionally, water-soluble tetrazolium-1 assay provided a functional read-out of the effects of these proteins in an in vitro cancer model. Our study establishes that the development of an S100B antagonist could perform a vital part in the treatment of S100- and RAGE-dependent human diseases.
Introduction
The name “S100” was given by Moore to the brain-specific proteins later identified as a mixture of S100A1 and S100B protein.1−3 These small, acidic proteins were first referred to as S100 owing to their ability to dissolve completely in 100% saturated ammonium sulfate at pH 7.1 The human S100 family is comprised of 21 similar proteins that are exclusive to vertebrates and includes two discrete EF-hand motifs with a helix–loop–helix structure. S100 family members range from 10 to 12 kDa, exhibit site-specific expression patterns, and efficiently form homo and heterodimers.4−6 These proteins have known interactions with calcium, zinc, and copper ions, and Ca2+ binding enables the numerous intracellular and extracellular regulatory effects of these proteins.7,8 All S100 family members are considered Ca2+-sensing proteins except for S100G, which acts as a Ca2+-modulating protein by buffering cytosolic Ca2+.9 S100 proteins are effective regulators of a variety of diverse task involving Ca2+ homeostasis, transcription, cell proliferation and differentiation, tissue development, and regeneration/repair. They are also involved in the origin and development of chronic inflammation, autoimmunity, contagious ailments, tumors, and their metastases.10
S100 proteins have recently attracted attention due to their involvement in various tumorigenic processes (e.g., tumor progression, metastasis) and altered expression patterns in multiple human cancers (e.g., pancreatic, lung).11−23 S100A11, first identified as Calgizzarin or S100C, was initially extracted from the smooth muscle of chicken gizzards and the cardiac muscle of pigs.24,25 It is expressed in the kidney, heart, lung, and most muscle tissues, with peak levels in the placenta and lowest in the brain.26 S100A11 mediates multiple biological processes by binding with target proteins such as receptor for advanced glycation end products (RAGE), Annexin A1, Annexin A2, p53, and Rad54B.27 However, its oncogenic role in many cancers has drawn the maximum interest.28 It appears to be associated with tumor progression in adenocarcinomas and is expressed excessively in various cancers such as lung,29 colorectal,30 breast,31 pancreatic,32,33 colon,34 laryngeal,35 and papillary thyroid carcinoma.36 Additionally, S100A11 is down-regulated in the bladder37 and ovarian cancer.38
The receptor for advanced glycation end products (RAGE) is a 35 kDa cell surface protein which is the member of the immunoglobulin superfamily. It is comprised of V, C1, and C2 domains reminiscent of 3 Ig-like regions, a transmembrane helix, and a cytoplasmic end respectively that mediates intracellular signaling.39−43 RAGE works as a pattern recognition receptor and thereby associates with a range of signaling molecules such as advanced glycation end products (AGEs), amphoterin, C3a, S100/calgranulin polypeptides, phosphatidylserine, amyloid β, and advanced oxidation protein products.44−50
RAGE binds multiple ligands and has been involved in an array of ailments (such as diabetic complications, cardiovascular disorders, chronic inflammation disorders, Alzheimer disease, and neurodegeneration).51 Furthermore, RAGE activation is linked to multiple cancers including lung,52 colorectal,53 pancreatic,54 kidney,55 prostate,56 ovarian,57 breast,58 and melanoma.59 In recent years, RAGE has emerged as a major receptor for the S100 family proteins, and this S100–RAGE interaction mediates cell migration, invasion, tumor growth, angiogenesis, and metastasis but the mechanism of action remains largely unknown.60−62 Therefore, interaction studies on RAGE and its associated ligands are desired to elucidate more insights into the development of different diseases and design the treatment modalities thereof.
The S100B protein is associated with a multigenic family of small proteins that binds calcium via EF-hand motifs.63,64 It is one of the earliest associates of the S100 protein family to be recognized and is the most active in the head region.3 S100B consists of S100 ββ homodimers.65 Many different cells including astrocytes,66 myoblasts,67 photoreceptor cells,68 skeletal muscle tissue,69 certain neuronal populations,70 oligodendroglial progenitor cells,71 and oligodendrocytes72 express this protein in varying amounts. Calcium-induced S100B exerts its intracellular and extracellular functions by interacting with other proteins. Previous studies have provided evidence of S100B’s involvement in several regulatory pathways concerning cell proliferation, differentiation, and morphology, calcium homeostasis, protein phosphorylation, transcription, microtubule and type III intermediate filament dynamics, enzyme activity, and metabolism.8,9,73,74 Interaction studies pertaining to S100B and RAGE has been performed earlier in detail.41,75,76 Depending on the microenvironment, secreted S100B has been shown to bind to RAGE in neurons, macrophages, myoblasts, astrocytes, and microglia toward carrying out a diversity of biological functions with variable effects (i.e., favorable or deleterious).77
An elevated amount of S100B has been noticed in a range of clinical circumstances such as brain injury and ischemia, neurodegeneration, inflammation, and mental disorders.78 S100B is secreted by glioblastoma cancer cells in vitro.79 Moreover, S100B is a firmly developed predictive indicator for malignant melanoma, and increased serum levels associated with negative patient outcomes.80,81
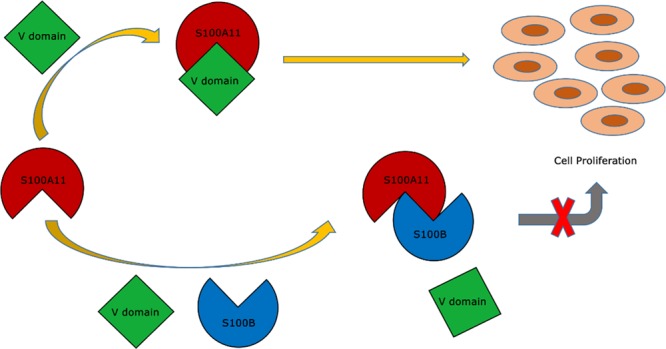
It was previously reported that S100A11 interacts with the S100B protein.82,83 In this study, we used the S100B protein as a probable candidate for an antagonist to hinder interactions flanked by S100A11 and the RAGE V domain, thereby inhibiting additional signal transduction. By employing the HADDOCK (High Ambiguity Driven protein–protein Docking) program, we constructed an S100A11–S100B heterodimer complex by compiling the data on interacting sites between S100A11 and S100B proteins from heteronuclear single-quantum correlation-NMR (HSQC-NMR) experiments. Then we superimposed the two complex structures (S100A11 and S100B complexes with the S100A11 and RAGE V domain complexes), which illustrated that S100B could interfere at the binding region of S100A11 and the RAGE V domain. Moreover, a water-soluble tetrazolium-1 (WST-1) assay confirmed that the S100B protein can hinder cell proliferation in vitro. This collaborative study provides structural insights into the S100A11–S100B heterodimer complex formation, which will facilitate the discovery of new therapies for S100- and RAGE-mediated diseases.
Results and Discussion
Binding Interface (S100A11/S100B) Region in S100A11
The binding regions among proteins or amongst proteins and their associated ligands are generally recognized by employing 1H–15N HSQC-NMR experiments.84,85 The residues near the S100A11 and S100B interfaces were recognized by analyzing the resonances on the HSQC-NMR spectra of free S100A11 and compared to those of the S100A11–S100B complex.
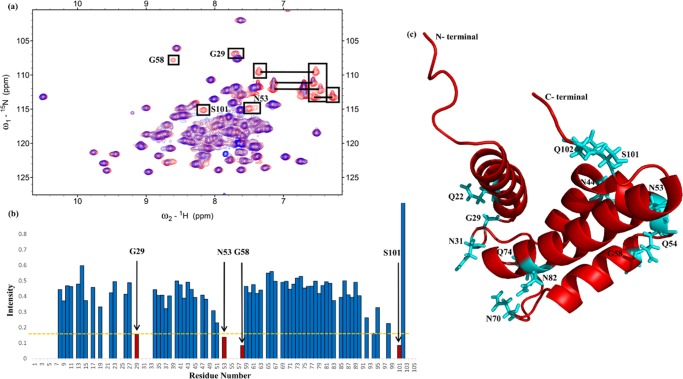
Figure 1a displays the overlaid HSQC-NMR spectra of only nitrogen-15-labeled S100A11 (red color) and nitrogen-15-labeled S100A11 complexed with unlabeled S100B (blue color). Some of the NMR signals decreased upon the addition of free S100B to nitrogen-15-labeled S100A11. The observed HSQC-NMR indications of the S100A11–S100B complex were significantly inferior to those obtained with free S100A11. This occurrence was owing to 15N nuclei near the interaction spots of the proteins among S100A11 and S100B.
Figure 1.
Analysis of the binding interface (S100A11/S100B) region in S100A11. (a) 1H–15N HSQC spectra overlay of free 15N S100A11 (red) and 15N S100A11 binding to unlabeled S100B (blue). Cross-peaks illustrating changes in intensities are shown in boxes. Horizontal lines connect the NH2 side chains. (b) The cross-peak intensity plot (I/I0) where (I) denotes the cross-peak intensity of S100A11–S100B complex and (I0) indicates the early intensity of free S100A11 versus a number of amino-acid residues (1–105) illustrated as a bar diagram. (c) A picture illustration of the S100A11 monomer with residues exhibiting decreases in cross-peak signals distinguished by the cyan color.
The 15N nuclei surrounding the interface region were affected by the presence of other proteins in close vicinity, which resulted in lowering the cross-peaks on the HSQC-NMR spectrum. The S100 protein residues were influenced due to the formation of a complex with their target proteins. The 1H–15N HSQC-NMR spectra from the free S100A11 and the S100A11–S100B complex were matched to detect the perturbation or reduction in the intensity of the S100A11 residues near the interface region.
A bar graph that compares the residues from both spectra is shown in Figure 1b. Some residues from the HSQC cross-peaks had considerably inferior intensities (Figure 1b). These S100A11 residues (red color) were G29, N53, G58, and S101. Residues along with the NH2 side chains of asparagines and glutamines (namely Q22, N31, N44, Q54, N70, Q74, N82, and Q102) were also included (as labeled in boxes that are connected by horizontal lines in Figure 1a, however, their specific side-chain assignments were unavailable) for plotting on the three-dimensional (3D) construction of the S100A11 monomer (Figure 1c).
Binding Interface (S100A11/S100B) in S100B
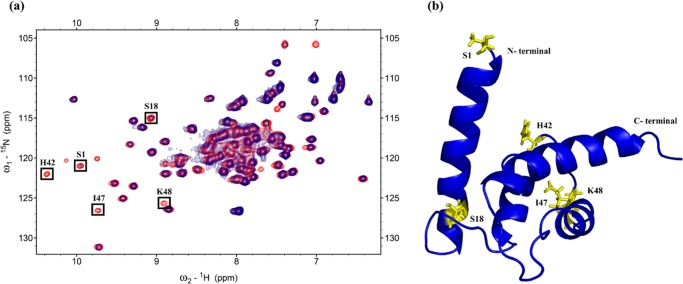
From HSQC-NMR experiments, the spectra of the free nitrogen-15-labeled S100B area were overlaid on the spectra of the nitrogen-15-labeled S100B in the complex with unlabeled S100A11 (Figure 2a).
Figure 2.
Analysis of the binding interface (S100A11/S100B) region in S100B. (a) 1H–15N HSQC spectra overlay of free 15N S100B (red) and 15N S100B binding to unlabeled S100A11 (blue). Cross-peaks showing changes in intensities are shown in boxes. (b) Cartoon representation of the S100B monomer with residues exhibiting a decrease in cross-peak signals are shown in yellow.
Cross-peaks with reduced intensities (line broadening) were identified and marked including S1, S18, H42, I47, and K48. The S1, S18, H42, I47, and K48 residues (red color) were mapped onto the 3D construction of the S100B monomer protein (Figure 2b).
Fundamental Depiction of the S100A11–S100B Complex
To compute the protein–protein interactions, the basic model of the S100A11–S100B complex was built via the HADDOCK program. The difference in the intensities and the HSQC-NMR spectra information analysis from S100A11 and S100B provided the data for Ambiguous Interaction Constraints (AIRs). As input parameters in HADDOCK, the residues with a difference in the intensities (HSQC-NMR) indicated the constraints for computation. The HADDOCK outcomes produced models for the heterodimeric S100A11–S100B complex. The structure of Ca2+-bound S100A11 was attained from PDB ID: 2LUC and the model of S100B were assimilated from PDB ID: 1UWO. Almost 2000 complex models were created in HADDOCK by rigid-body minimization. The final 200 models using the lowermost energy outcomes following water refinement were engaged for additional examination. The S100A11–S100B complex model is depicted in Figure 3.
Figure 3.

Cartoon representing S100A11–S100B complex created with the HADDOCK program. S100A11 is depicted in red and S100B in blue color. Residues near the interaction sites are illustrated in cyan (from the S100A11 side) and yellow (from S100B side) respectively.
In total, two binding sites were identified involving residues S101 and Q102 from S100A11 with S1 from S100B at the first binding site; and N53 and Q54 from S100A11 with H42 from S100B at the second binding site. The structural stereochemistry of the complex was checked through PROCHECK analysis provided online by the European Bioinformatics Institute.86 The Ramachandran plot revealed the presence of 82.1% residues in the maximum favored areas, 13.4% in furthermore permitted areas, 2.8% in allowed regions, and only 1.7% in disallowed regions. The all-inclusive average score of G-factors was found to be 0.19, which is in the expected range (data not included).
Dissociation Constant
In solution, S100A11 and S100B proteins usually occur in homodimeric forms. We first got the HSQC spectrum of 15N S100A11 (Figure 4) in the homodimer form alone.
Figure 4.
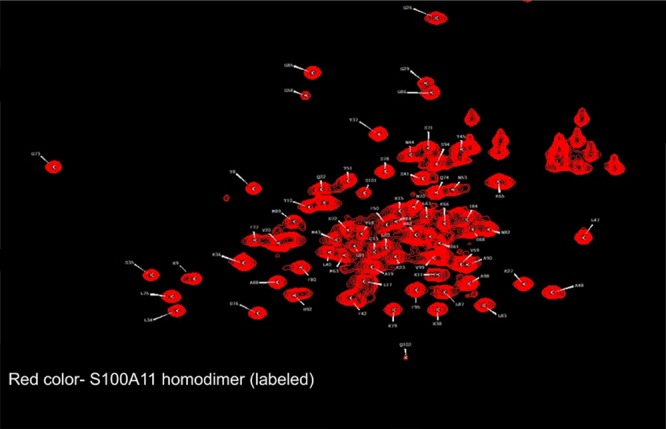
1H–15N HSQC spectra of free 15N S100A11 (red).
We overlapped the HSQC spectrum of the S100A11 homodimer alone (as shown in red color, Figure 5) with the HSQC spectrum of the S100A11 homodimer (15N-labeled) mixed with the S100B homodimer (unlabeled) in a 1:1 ratio (as illustrated in blue color, Figure 5). We found that the two HSQC spectra were exactly the same. This showed that the homodimer of S100A11 does not interact with the homodimer of S100B.
Figure 5.
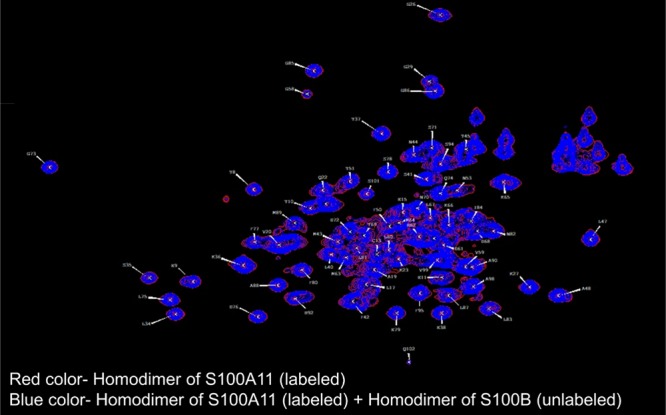
1H–15N HSQC spectra overlay of free 15N S100A11 (red) and 15N S100A11 homodimer + S100B homodimer mixture (blue).
Similar results were observed when interaction studies were performed through the isothermal titration calorimetry experiment where titrations were performed by injecting 10 μL aliquots of the S100B homodimer (1 mM, 28 injections) into 0.1 mM of the S100A11 homodimer in the cell (and vice-versa). No binding was observed (data not included). The supposed results were due to the fact that the homodimer of S100A11 did not interact with the homodimer of S100B. However, the monomer of S100A11 will interact with the monomer of S100B to form an S100A11–S100B heterodimer. This conclusion was supported by the following HSQC experiments:
S100A11 (15N-labeled) and S100B (unlabeled) proteins were initially in their homodimeric forms. First, both the proteins were added together (1:1 ratio) followed by denaturation (using 8 M urea to ensure that the monomer was formed for both S100A11 and S100B proteins) and renaturation (removal of 8 M urea by exchanging it with excess deionized water) promoting heterodimer formation. The HSQC spectrum was acquired for this heterodimer. Then this spectrum was overlapped with the HSQC spectrum of the free S100A11 homodimer. The resulting NMR spectra showed a decrease in peak intensities which may be due to the formation of heterodimers (Figure 6). This confirmed the possibility of S100A11–S100B heterodimer formation. So, to get the idea about the dissociation constant, we used the data from the chemical shift perturbations to calculate Kd.
Figure 6.
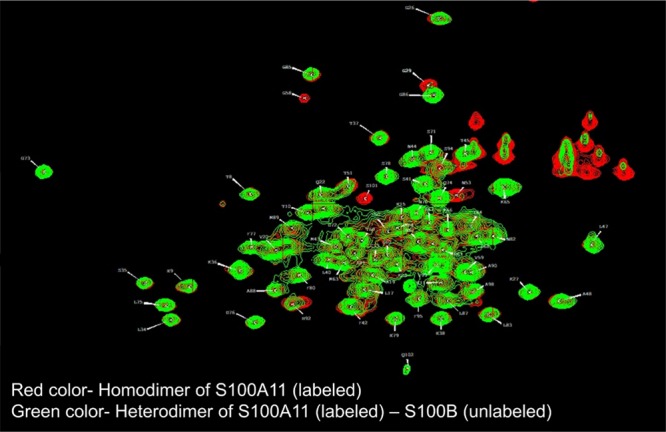
1H–15N HSQC spectra overlay of free 15N S100A11 (red) and 15N S100A11–S100B heterodimers (green).
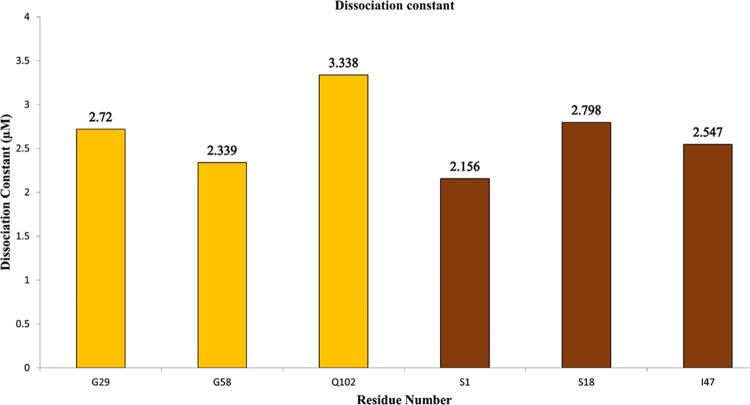
On the basis of the alterations from the chemical shift perturbation, HSQC-NMR titration spectrum analysis can be employed to compute the dissociation constant (Kd).85 The HSQC-NMR spectra of nitrogen-15-labeled S100A11 titrated with unlabeled S100B (and vice-versa) designated the cross-peaks that altered considerably. Residues G29, G58, and Q102 were selected when 15N S100A11 was titrated with unlabeled S100B and residues S1, S18, and I47 were selected with inverse titration for our best model with the succeeding equation
In the above equation, Δδobs represents the variation detected in the shift among the free and bound-form (1:1 molar ratio). Δδmax represents the maximal shift change at the 1:3 molar ratio, and [P]t and [L]t are the net concentrations of the protein (S100A11) and the ligand (S100B). These equations compute Kd values from the shifts found at diverse concentrations of proteins. At a molar ratio of 1:3 of 15N S100A11 to unlabeled S100B, the mean Kd was ∼2.8 μM. Similarly, inverse calculations (i.e., 15N S100B to unlabeled S100A11) resulted in a mean Kd value of about ∼2.5 μM. Taken together, the average dissociation constant was around ∼2.65 μM (Figure 7). These results indicated that the dissociation constant (Kd) of S100A11 was in the micromolar range.
Figure 7.
Dissociation constant. Kd calculated by the designated residues detected in 15N S100A11 HSQC titrations with S100B (yellow) and vice-versa (brown). The overall average Kd is about 2.65 μM.
WST-1 Assay
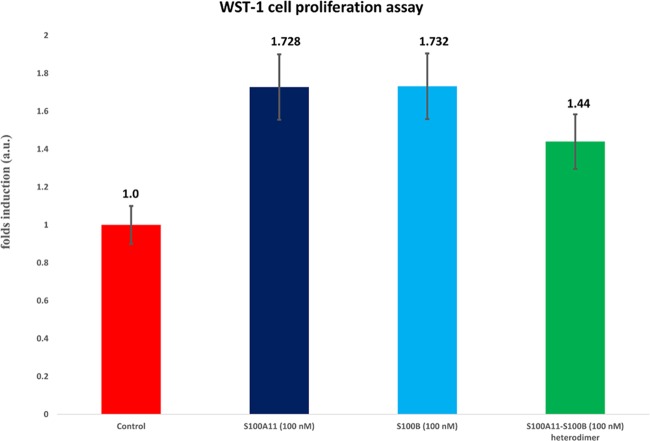
A WST-1-based cell proliferation assay was employed to describe the downstream stimulation of roles transmitted by the RAGE V domain through S100A11. We chose SW-480 cells for our functional assays since they have epithelial-like morphology and are often used to study cancer in vitro. SW-480 cell is a cell line with high expression of RAGE and low expression of the S100A11 protein. Therefore, the role of endogenous S100A11 is not influential and changes in the cell activity can be viewed clearly. The SW-480 cells were treated for 48 h with S100A11 and S100B prior to analysis for viability using the WST-1 assay.
Serum-starved SW-480 cells were matured with S100A11 (100 nM). A 1.728-fold increase was witnessed in the living cell count over the serum-free control (Figure 8, lane 2). This result can be elucidated by the point that S100A11 binds to the RAGE V domain and, thus, induced cell proliferation.87 Similarly, S100B showed a 1.732-fold increase compared to the serum-free control (Figure 8, lane 3) indicating that S100B does bind to RAGE and the interaction of S100B (similar to S100A11) with endogenous RAGE contributes to cell proliferation. However, a 1.44-fold decrease was observed in cells treated with the S100A11–S100B (100 nM) heterodimer protein (Figure 8, lane 4). These results suggest that the S100B protein potentially interferes with the contact between S100A11 and the RAGE V domain, in addition to inhibiting cell proliferation activity.
Figure 8.
WST-1 assay analysis. The SW-480 cells were treated with 100 nM of S100A11 (dark blue), 100 nM of S100B (light blue), and 100 nM of S100A11–S100B heterodimers (green). The proportional cell counts after subsequent treatment with S100A11 and S100B are depicted as fold inductions with serum-free media alone as the control (red).
S100B Inhibits the S100A11–RAGE V Domain
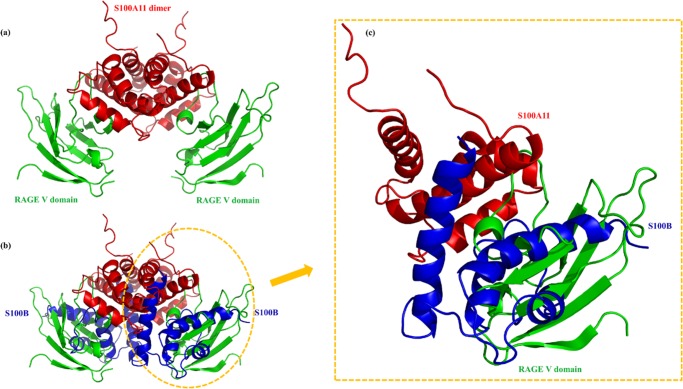
S100A11 is well-known to exist in a noncovalent homodimeric form having an antiparallel conformation. Molecular interactions between S100A11 and the RAGE V domain have shown that S100A11 binding to the RAGE V domain could eventually trigger an autophosphorylation process leading to numerous signal transduction cascades that regulate cell proliferation and migration.87,88 Here, we used a similar S100A11–RAGE V domain complex model for our studies (Figure 9a).87
Figure 9.
S100B interfering S100A11–RAGE V domain. (a) The complex model of the S100A11–RAGE V domain constructed via HADDOCK. (b) Superimposition of the complex between S100A11 (red) and the RAGE V domain (green), with the complex amid S100A11 (red) and S100B (blue). (c) The magnified depiction of the clear and significant hindrance to S100A11–RAGE V domain associated with the S100B protein.
Analysis of the S100A11 HSQC spectra with and without unlabeled S100B revealed the S100A11 residues involved in the interaction with S100B. These participating residues near the interaction sites from the S100A11 side were N53, Q54, S101, and Q102. Likewise, S100A11 residues involved in the interaction with the RAGE V domain were N53, Q54, and A90 respectively (Figure 5b).87 Interaction studies related to S100A11 and the RAGE V domain have been performed previously in detail.87 Binding affinity values indicated a stronger interaction between S100A11 and the RAGE V domain when compared to the S100A11–S100B interaction. However, it may be noted that the interaction of S100A11 either with the RAGE V domain or S100B involved a common interaction site (s). Therefore, comparing the S100A11–S100B complex model (obtained through the HADDOCK program) with the S100A11–RAGE V domain complex (constructed in the similar manner87), S100B showed considerable interference. This indicates successful inhibition of the formation of S100A11–RAGE V domain complex (Figure 9b).
Furthermore, through the WST-1 assay it can be understood well that the treatment of exogenous S100A11 and S100B, separately, to SW-480 cells resulted in cell proliferation compared to the control cells. This indicated the interaction of both S100A11 and S100B with the endogenous RAGE V domain giving rise to higher cell counts. However, treatment of the SW-480 cells with the S100A11–S100B heterodimer protein led to a decrease in cell proliferation significantly which indicated that S100B has obstructed the greatly desired interface position of S100A11 and the RAGE V domain needed for higher cell count. In conclusion, an association of S100A11 with S100B has blocked significantly the RAGE signaling causing cell proliferation, and we hypothesize that S100B (or similar antagonist) will disrupt the S100A11–RAGE-mediated signaling significantly in cancerous cells when introduced in higher concentrations.
Conclusions
The biological importance of S100 family proteins associated with RAGE is well-known and S100–RAGE-mediated signaling pathways have been investigated for their role in the progress of many human cancers.89,90 For instance, S100A11 and RAGE signaling have been shown to modulate osteoarthritis pathogenesis.91,92 Additionally, the association of RAGE was found to enhance S100A11 secretion in human squamous cancer cells.28 S100–RAGE-mediated signaling is likely a relevant therapeutic target for the treatment of S100–RAGE-related disorders. In an experimental mouse model, blocking S100–RAGE interactions resulted in the reduction of colonic inflammation and decreased infiltration of inflammatory cells.46,93
Here, we report a study that details the interactions between S100A11 and S100B proteins, proving the interference of S100B in the binding of the S100A11–RAGE V domain in vitro. These data propose that an improvement in the S100B antagonist could greatly impact S100 protein- and RAGE-dependent human diseases. The results from the complex structure analysis confirmed that S100B interferes at the interface region of S100A11 and the RAGE V domain, further highlighting the therapeutic potential of this pathway. In addition, treatment with the S100A11–S100B heterodimer protein in vitro resulted in lower cell numbers in a cancer model.
Overall, our analysis provides prominent structural insights into S100A11–S100B heterodimer complex formation and suggests that an antagonist similar to S100B could be promising for the discovery of new therapies pertaining to S100- and RAGE-mediated diseases.
Materials and Methods
Materials
All the HSQC-NMR experiments were performed utilizing 99% pure isotope-tagged 15N-ammonium chloride (15NH4Cl) and deuterium oxide (D2O). Solutions were formulated using Milli-Q water and sample buffers for the said experiments were purified with a 0.22 μm aseptic device. SW-480 cancer cells were acquired from the American Type Culture Collection (CCL-288).
Expression, Labeling, and Purification
Human S100A1188 and S100B94,95 proteins were expressed and purified as per the protocols mentioned earlier. The purity and identity of the acquired proteins were determined by the sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE) gel method and confirmed by electrospray ionization mass spectrometry (ESI-MS) analysis (Supporting Information, Figures S1 and S2).
HSQC-NMR Titration Experiments
All HSQC-NMR experiments were executed inside a Varian 700 MHz spectrometer at 25 °C by utilizing a cryogenic probe. Using an NMR buffer (100 mM NaCl, 5 mM CaCl2, 25 mM Tris/HCl, 1 mM dithiothreitol, 10% D2O, 7.5 pH), an unlabeled S100B protein was supplemented to 15N-labeled-S100A11 at molar ratios of 0:1, 1:1, and 3:1. As these proteins occur in homodimeric forms, they were initially denatured in the presence of 8 M urea before associating as heterodimers.83,96,97 The 8 M urea was completely exchanged with high amounts of water (dialysis will facilitate and ensure the heterodimer formation) and finally replaced with NMR buffer. The converse titration experiments (15N-labeled S100B and unlabeled S100A11) with the similar protocol and molar ratios were carried out as well. The resulting NMR spectra were handled with the VNMRJ 2.3 program and studied further by Sparky 3.198 toward identifying any chemical perturbations, shifts in the cross peaks, or variations in the peak intensities of the protein.
Molecular Docking
The S100A11–S100B complex model was obtained by using the HADDOCK program (version 2.2).99−101 For docking studies, NMR coordinates for S100A11 and S100B were recovered from the Protein Data Bank (PDB ID: 2LUC and PDB ID: 1UWO, respectively).88,94 The residues with detectable chemical shift perturbations or decreased intensity based on HSQC titrations were assimilated for future interaction studies. These residues were included in the Ambiguous Interaction Constraints (AIR) parameters and the residues were assigned as active and passive residues based on the NACCESS program.102 Residues with a comparative accessible surface area >30%, plus side chains and backbone, were considered as active while those <30% were considered as passive residues. A total of 2000 structures were considered for rigid-body docking by using the usual HADDOCK procedure including the optimized potential for liquid simulation parameters.103 Five hundred structures were then considered for semi-flexible refinement, and the next 200 lowest energy structures were examined. All possible structures in clusters were studied and analyzed by the PyMOL program.104
Cell Proliferation Assay
The WST-1 [4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate] cell proliferation assay was executed as per the manufacturer’s (Roche) guidelines.
Acknowledgments
The authors humbly express their gratitude to the associates of the Varian 700 MHz NMR facility at the Instrumentation Center of the Chemistry Department of National Tsing Hua University, and electrospray ionization-MS facility at the Instrumentation Center of National Chiao Tung University, Hsinchu, Taiwan.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b00922.
SDS–PAGE and ESI-MS (PDF)
Accession Codes
UniProtKB—P31949 (S10AB_HUMAN), UniProtKB—P04271 (S100B_HUMAN).
This research was sponsored by the Ministry of Science and Technology (MOST) of Taiwan (Grant number MOST 104-2113-M-007-019-MY3) and in part by China Medical University (Grant number CMU104-S-02, Taiwan).
The authors declare no competing financial interest.
Supplementary Material
References
- Moore B. W. A soluble protein characteristic of the nervous system. Biochem. Biophys. Res. Commun. 1965, 19, 739–744. 10.1016/0006-291X(65)90320-7. [DOI] [PubMed] [Google Scholar]
- Moore B. W.; McGregor D. Chromatographic and electrophoretic fractionation of soluble proteins of brain and liver. J. Biol. Chem. 1965, 240, 53. [PubMed] [Google Scholar]
- Isobe T.; Okuyama T. The Amino-Acid Sequence of the α Subunit in Bovine Brain S-100a Protein. FEBS J. 1981, 116, 79–86. 10.1111/j.1432-1033.1981.tb05303.x. [DOI] [PubMed] [Google Scholar]
- Zimmer D. B.; Eubanks J. O.; Ramakrishnan D.; Criscitiello M. F. Evolution of the S100 family of calcium sensor proteins. Cell Calcium 2013, 53, 170–179. 10.1016/j.ceca.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Marenholz I.; Heizmann C. W.; Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature). Biochem. Biophys. Res. Commun. 2004, 322, 1111–1122. 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- Schäfer B. W.; Heizmann C. W. The S100 family of EF-hand calcium-binding proteins: functions and pathology. Trends Biochem. Sci. 1996, 21, 134–140. 10.1016/S0968-0004(96)80167-8. [DOI] [PubMed] [Google Scholar]
- Heizmann C. W.; Cox J. A. New perspectives on S100 proteins: a multi-functional Ca 2+-, Zn 2+-and Cu 2+-binding protein family. Biometals 1998, 11, 383–397. 10.1023/A:1009212521172. [DOI] [PubMed] [Google Scholar]
- Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int. J. Biochem. Cell Biol. 2001, 33, 637–668. 10.1016/S1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- Santamaria-Kisiel L.; Rintala-Dempsey A. C. Shaw GS. Calcium-dependent and -independent interactions of the S100 protein family. Biochem. J. 2006, 396, 201–214. 10.1042/BJ20060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato R.; R Cannon B.; Sorci G.; et al. Functions of S100 proteins. Curr. Mol. Med. 2013, 13, 24–57. 10.2174/156652413804486214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.; Zhang S.; Fernig D. G.; Martin-Fernandez M.; Rudland P. S.; Barraclough R. Mutually antagonistic actions of S100A4 and S100A1 on normal and metastatic phenotypes. Oncogene 2005, 24, 1445–1454. 10.1038/sj.onc.1208291. [DOI] [PubMed] [Google Scholar]
- Bulk E.; Sargin B.; Krug U.; et al. S100A2 induces metastasis in non-small cell lung cancer. Clin. Cancer Res. 2009, 15, 22–29. 10.1158/1078-0432.CCR-08-0953. [DOI] [PubMed] [Google Scholar]
- Kang M.; Lee H. S.; Lee Y. J.; et al. S100A3 suppression inhibits in vitro and in vivo tumor growth and invasion of human castration-resistant prostate cancer cells. Urology 2015, 85, 273.e9–273.e15. 10.1016/j.urology.2014.09.018. [DOI] [PubMed] [Google Scholar]
- Ochiya T.; Takenaga K.; Endo H. Silencing of S100A4, a metastasis-associated protein, in endothelial cells inhibits tumor angiogenesis and growth. Angiogenesis 2014, 17, 17–26. 10.1007/s10456-013-9372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Zhang K.; Jiang X.; Zhang J. S100A6 as a potential serum prognostic biomarker and therapeutic target in gastric cancer. Dig. Dis. Sci. 2014, 59, 2136–2144. 10.1007/s10620-014-3137-z. [DOI] [PubMed] [Google Scholar]
- Nasser M. W.; Qamri Z.; Deol Y. S.; et al. S100A7 enhances mammary tumorigenesis through upregulation of inflammatory pathways. Cancer Res. 2012, 72, 604–615. 10.1158/0008-5472.CAN-11-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa M.; Williams R.; Wang L.; Vogl T.; Srikrishna G. S100A8/A9 activate key genes and pathways in colon tumor progression. Mol. Cancer Res. 2011, 9, 133–148. 10.1158/1541-7786.MCR-10-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebhardt S.; Muller-Decker K.; Bestvater F.; Hershfinkel M.; Mayer D. Impact of S100A8/A9 expression on prostate cancer progression in vitro and in vivo. J. Cell Physiol. 2014, 229, 661–671. 10.1002/jcp.24489. [DOI] [PubMed] [Google Scholar]
- Phipps K. D.; Surette A. P.; O’Connell P. A.; Waisman D. M. Plasminogen receptor S100A10 is essential for the migration of tumor-promoting macrophages into tumor sites. Cancer Res. 2011, 71, 6676–6683. 10.1158/0008-5472.CAN-11-1748. [DOI] [PubMed] [Google Scholar]
- Hao J.; Wang K.; Yue Y.; et al. Selective expression of S100A11 in lung cancer and its role in regulating proliferation of adenocarcinomas cells. Mol. Cell. Biochem. 2012, 359, 323–332. 10.1007/s11010-011-1026-8. [DOI] [PubMed] [Google Scholar]
- Zhao F.-T.; Jia Z.-S.; Yang Q.; Song L.; Jiang X.-J. S100A14 Promotes the Growth and Metastasis of Hepatocellular Carcinoma. Asian Pac. J. Cancer Prev. 2013, 14, 3831–3836. 10.7314/APJCP.2013.14.6.3831. [DOI] [PubMed] [Google Scholar]
- Wang H.; Zhang L.; Zhang I. Y.; et al. S100B promotes glioma growth through chemoattraction of myeloid-derived macrophages. Clin. Cancer Res. 2013, 19, 3764–3775. 10.1158/1078-0432.CCR-12-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakhel S.; Padilla L.; Adan J.; et al. S100P antibody-mediated therapy as a new promising strategy for the treatment of pancreatic cancer. Oncogenesis 2014, 3, e92 10.1038/oncsis.2014.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todoroki H.; Kobayashi R.; Watanabe M.; Minami H.; Hidaka H. Purification, characterization, and partial sequence analysis of a newly identified EF-hand type 13-kDa Ca (2+)-binding protein from smooth muscle and non-muscle tissues. J. Biol. Chem. 1991, 266, 18668–18673. [PubMed] [Google Scholar]
- Ohta H.; Sasaki T.; Naka M.; et al. Molecular cloning and expression of the cDNA coding for a new member of the S100 protein family from porcine cardiac muscle. FEBS Lett. 1991, 295, 93–96. 10.1016/0014-5793(91)81393-M. [DOI] [PubMed] [Google Scholar]
- Inada H.; Naka M.; Tanaka T.; Davey G. E.; Heizmann C. W. Human S100A11 exhibits differential steady-state RNA levels in various tissues and a distinct subcellular localization. Biochem. Biophys. Res. Commun. 1999, 263, 135–138. 10.1006/bbrc.1999.1319. [DOI] [PubMed] [Google Scholar]
- Leclerc E.; Fritz G.; Vetter S. W.; Heizmann C. W. Binding of S100 proteins to RAGE: an update. Biochim. Biophys. Acta, Mol. Cell Res. 2009, 1793, 993–1007. 10.1016/j.bbamcr.2008.11.016. [DOI] [PubMed] [Google Scholar]
- Sakaguchi M.; Sonegawa H.; Murata H.; et al. S100A11, an dual mediator for growth regulation of human keratinocytes. Mol. Biol. Cell 2008, 19, 78–85. 10.1091/mbc.e07-07-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T.; Hao J.; Xu A.; et al. Determination of metastasis-associated proteins in non-small cell lung cancer by comparative proteomic analysis. Cancer Sci. 2007, 98, 1265–1274. 10.1111/j.1349-7006.2007.00514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.; Wang X.; Wang S.; et al. Colorectal cancer progression correlates with upregulation of S100A11 expression in tumor tissues. Int. J. Colorectal Dis. 2008, 23, 675–682. 10.1007/s00384-008-0464-6. [DOI] [PubMed] [Google Scholar]
- Fan Ca2+-binding protein S100A11: A novel diagnostic marker for breast carcinoma. Oncol. Rep. 2010, 23, 1301–1308. 10.3892/or_00000764. [DOI] [PubMed] [Google Scholar]
- Ohuchida K.; Mizumoto K.; Ohhashi S.; et al. S100A11, a putative tumor suppressor gene, is overexpressed in pancreatic carcinogenesis. Clin. Cancer Res. 2006, 12, 5417–5422. 10.1158/1078-0432.CCR-06-0222. [DOI] [PubMed] [Google Scholar]
- Xiao M. B.; Jiang F.; Ni W. K.; et al. High expression of S100A11 in pancreatic adenocarcinoma is an unfavorable prognostic marker. Med. Oncol. 2012, 29, 1886–1891. 10.1007/s12032-011-0058-y. [DOI] [PubMed] [Google Scholar]
- Meding S.; Balluff B.; Elsner M.; et al. Tissue-based proteomics reveals FXYD3, S100A11 and GSTM3 as novel markers for regional lymph node metastasis in colon cancer. J. Pathol. 2012, 228, 459–470. 10.1002/path.4021. [DOI] [PubMed] [Google Scholar]
- Wang C.; Zhang Z.; Li L.; et al. S100A11 is a migration-related protein in laryngeal squamous cell carcinoma. Int. J. Med. Sci. 2013, 10, 1552–1559. 10.7150/ijms.5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anania M. C.; Miranda C.; Vizioli M. G.; et al. S100A11 overexpression contributes to the malignant phenotype of papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2013, 98, E1591–E1600. 10.1210/jc.2013-1652. [DOI] [PubMed] [Google Scholar]
- Memon A. A.; Sorensen B. S.; Meldgaard P.; Fokdal L.; Thykjaer T.; Nexo E. Down-regulation of S100C is associated with bladder cancer progression and poor survival. Clin. Cancer Res. 2005, 11, 606–611. [PubMed] [Google Scholar]
- Liu Y.; Han X.; Gao B. Knockdown of S100A11 expression suppresses ovarian cancer cell growth and invasion. Exp. Ther. Med. 2015, 9, 1460–1464. 10.3892/etm.2015.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A. M.; Vianna M.; Gerlach M.; et al. Isolation and characterization of two binding proteins for advanced glycosylation end products from bovine lung which are present on the endothelial cell surface. J. Biol. Chem. 1992, 267, 14987–14997. [PubMed] [Google Scholar]
- Neeper M.; Schmidt A.; Brett J.; et al. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J. Biol. Chem. 1992, 267, 14998–5004. [PubMed] [Google Scholar]
- Dattilo B. M.; Fritz G.; Leclerc E.; Kooi C. W.; Heizmann C. W.; Chazin W. J. The extracellular region of the receptor for advanced glycation end products is composed of two independent structural units. Biochemistry 2007, 46, 6957–6970. 10.1021/bi7003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson B. I.; Carter A. M.; Harja E.; et al. Identification, classification, and expression of RAGE gene splice variants. FASEB J. 2008, 22, 1572–1580. 10.1096/fj.07-9909com. [DOI] [PubMed] [Google Scholar]
- Rauvala H.; Rouhiainen A. Physiological and pathophysiological outcomes of the interactions of HMGB1 with cell surface receptors. Biochim. Biophys. Acta, Gene Regul. Mech. 2010, 1799, 164–170. 10.1016/j.bbagrm.2009.11.012. [DOI] [PubMed] [Google Scholar]
- Xie J.; Mendez J. D.; Mendez-Valenzuela V.; Aguilar-Hernandez M. M. Cellular signalling of the receptor for advanced glycation end products (RAGE). Cell Signal 2013, 25, 2185–2197. 10.1016/j.cellsig.2013.06.013. [DOI] [PubMed] [Google Scholar]
- Hori O.; Brett J.; Slattery T.; et al. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin mediation of neurite outgrowth and co-expression of rage and amphoterin in the developing nervous system. J. Biol. Chem. 1995, 270, 25752–25761. 10.1074/jbc.270.43.25752. [DOI] [PubMed] [Google Scholar]
- Hofmann M. A.; Drury S.; Fu C.; et al. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell 1999, 97, 889–901. 10.1016/S0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- Du Yan S.; Chen X.; Fu J.; et al. RAGE and amyloid-β peptide neurotoxicity in Alzheimer’s disease. Nature 1996, 382, 685–691. 10.1038/382685a0. [DOI] [PubMed] [Google Scholar]
- He M.; Kubo H.; Morimoto K.; et al. Receptor for advanced glycation end products binds to phosphatidylserine and assists in the clearance of apoptotic cells. EMBO Rep. 2011, 12, 358–364. 10.1038/embor.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan B. H.; Li X.; Winkler A. R.; et al. Complement C3a, CpG oligos, and DNA/C3a complex stimulate IFN-alpha production in a receptor for advanced glycation end product-dependent manner. J. Immunol. 2010, 185, 4213–4222. 10.4049/jimmunol.1000863. [DOI] [PubMed] [Google Scholar]
- Zhou L. L.; Cao W.; Xie C.; et al. The receptor of advanced glycation end products plays a central role in advanced oxidation protein products-induced podocyte apoptosis. Kidney Int. 2012, 82, 759–770. 10.1038/ki.2012.184. [DOI] [PubMed] [Google Scholar]
- Tesarova P.; Cabinakova M.; Mikulova V.; Zima T.; Kalousova M. RAGE and its ligands in cancer–culprits, biomarkers, or therapeutic targets?. Neoplasma 2015, 62, 353–364. 10.4149/neo_2015_061. [DOI] [PubMed] [Google Scholar]
- Jing R.-R.; Cui M.; Sun B.-L.; Yu J.; Wang H.-M. Tissue-specific expression profiling of receptor for advanced glycation end products and its soluble forms in esophageal and lung cancer. Genet. Test. Mol. Biomarkers 2010, 14, 355–361. 10.1089/gtmb.2009.0064. [DOI] [PubMed] [Google Scholar]
- Dahlmann M.; Okhrimenko A.; Marcinkowski P.; et al. RAGE mediates S100A4-induced cell motility via MAPK/ERK and hypoxia signaling and is a prognostic biomarker for human colorectal cancer metastasis. Oncotarget 2014, 5, 3220 10.18632/oncotarget.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R.; Tang D.; Lotze M. T.; Zeh H. J. III AGER/RAGE-mediated autophagy promotes pancreatic tumorigenesis and bioenergetics through the IL6-pSTAT3 pathway. Autophagy 2012, 8, 989–991. 10.4161/auto.20258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L.; Zhong K.; Sun Z.; Wu G.; Ding G. Receptor for advanced glycation end products (RAGE) partially mediates HMGB1-ERKs activation in clear cell renal cell carcinoma. J. Cancer Res. Clin. Oncol. 2012, 138, 11–22. 10.1007/s00432-011-1067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C.-B.; Bao J.-M.; Lu Y.-J.; et al. Co-expression of RAGE and HMGB1 is associated with cancer progression and poor patient outcome of prostate cancer. Am. J. Cancer Res. 2014, 4, 369–377. [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Hou X.; Zi S.; Wang Y.; Chen L.; Kong B. Polymorphisms of receptor for advanced glycation end products and risk of epithelial ovarian cancer in Chinese patients. Cell Physiol. Biochem. 2013, 31, 525–531. 10.1159/000350073. [DOI] [PubMed] [Google Scholar]
- Tesařová P.; Kalousová M.; Jachymova M.; Mestek O.; Petruželka L.; Zima T. Receptor for advanced glycation end products (RAGE)—soluble form (sRAGE) and gene polymorphisms in patients with breast cancer. Cancer Invest. 2007, 25, 720–725. 10.1080/07357900701560521. [DOI] [PubMed] [Google Scholar]
- Popa I.; Ganea E.; Petrescu S. M. Expression and subcellular localization of RAGE in melanoma cells. Biochem. Cell Biol. 2014, 92, 127–136. 10.1139/bcb-2013-0064. [DOI] [PubMed] [Google Scholar]
- Kierdorf K.; Fritz G. RAGE regulation and signaling in inflammation and beyond. J. Leukocyte Biol. 2013, 94, 55–68. 10.1189/jlb.1012519. [DOI] [PubMed] [Google Scholar]
- Padilla L.; Dakhel S.; Hernandez J. L. S100 to receptor for advanced glycation end-products binding assay: looking for inhibitors. Biochem. Biophys. Res. Commun. 2014, 446, 404–409. 10.1016/j.bbrc.2014.02.143. [DOI] [PubMed] [Google Scholar]
- Leclerc E.; Heizmann C. The importance of Ca2+/Zn2+ signaling S100 proteins and RAGE in translational medicine. Front. Biosci., Scholar Ed. 2011, 3, 1232–1262. 10.2741/s223. [DOI] [PubMed] [Google Scholar]
- Baudier J.; Glasser N.; Gerard D. Ions binding to S100 proteins. I. Calcium-and zinc-binding properties of bovine brain S100 alpha alpha, S100a (alpha beta), and S100b (beta beta) protein: Zn2+ regulates Ca2+ binding on S100b protein. J. Biol. Chem. 1986, 261, 8192–8203. [PubMed] [Google Scholar]
- Amburgey J. C.; Abildgaard F.; Starich M. R.; Shah S.; Hilt D. C.; Weber D. J. 1H, 13C and 15N NMR assignments and solution secondary structure of rat Apo-S100β. J. Biomol. NMR 1995, 6, 171–179. 10.1007/BF00211781. [DOI] [PubMed] [Google Scholar]
- Drohat A. C.; Amburgey J. C.; Abildgaard F.; Starich M. R.; Baldisseri D.; Weber D. J. Solution structure of rat apo-S100B (ββ) as determined by NMR spectroscopy. Biochemistry 1996, 35, 11577–11588. 10.1021/bi9612226. [DOI] [PubMed] [Google Scholar]
- Rickmann M.; Wolff J. S100 protein expression in subpopulations of neurons of rat brain. Neuroscience 1995, 67, 977–991. 10.1016/0306-4522(94)00615-C. [DOI] [PubMed] [Google Scholar]
- Sorci G.; Bianchi R.; Giambanco I.; Rambotti M.; Donato R. Replicating myoblasts and fused myotubes express the calcium-regulated proteins S100A1 and S100B. Cell Calcium 1999, 25, 93–106. 10.1054/ceca.1998.0012. [DOI] [PubMed] [Google Scholar]
- Rambotti M. G.; Giambanco I.; Spreca A.; Donato R. S100B and S100A1 proteins in bovine retina: their calcium-dependent stimulation of a membrane-bound guanylate cyclase activity as investigated by ultracytochemistry. Neuroscience 1999, 92, 1089–1101. 10.1016/S0306-4522(99)00074-3. [DOI] [PubMed] [Google Scholar]
- Arcuri C.; Giambanco I.; Bianchi R.; Donato R. Annexin V, annexin VI, S100A1 and S100B in developing and adult avian skeletal muscles. Neuroscience 2002, 109, 371–388. 10.1016/S0306-4522(01)00330-X. [DOI] [PubMed] [Google Scholar]
- Vives V.; Alonso G.; Solal A. C.; Joubert D.; Legraverend C. Visualization of S100B-positive neurons and glia in the central nervous system of EGFP transgenic mice. J. Comp. Neurol. 2003, 457, 404–419. 10.1002/cne.10552. [DOI] [PubMed] [Google Scholar]
- Deloulme J. C.; Raponi E.; Gentil B. J.; et al. Nuclear expression of S100B in oligodendrocyte progenitor cells correlates with differentiation toward the oligodendroglial lineage and modulates oligodendrocytes maturation. Mol. Cell Neurosci. 2004, 27, 453–465. 10.1016/j.mcn.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Hachem S.; Aguirre A.; Vives V.; Marks A.; Gallo V.; Legraverend C. Spatial and temporal expression of S100B in cells of oligodendrocyte lineage. Glia 2005, 51, 81–97. 10.1002/glia.20184. [DOI] [PubMed] [Google Scholar]
- Heizmann C. W.; Fritz G.; Schafer B. S100 proteins: structure, functions and pathology. Front. Biosci., Landmark Ed. 2002, 7, 1356–1368. 10.2741/A846. [DOI] [PubMed] [Google Scholar]
- Donato R. Intracellular and extracellular roles of S100 proteins. Microsc. Res. Tech. 2003, 60, 540–551. 10.1002/jemt.10296. [DOI] [PubMed] [Google Scholar]
- Ostendorp T.; Leclerc E.; Galichet A.; et al. Structural and functional insights into RAGE activation by multimeric S100B. EMBO J. 2007, 26, 3868–3878. 10.1038/sj.emboj.7601805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.; Boyington J. C. The 1.5 Å crystal structure of human receptor for advanced glycation endproducts (RAGE) ectodomains reveals unique features determining ligand binding. J. Biol. Chem. 2010, 285, 40762–40770. 10.1074/jbc.M110.169276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato R.; Sorci G.; Riuzzi F.; et al. S100B’s double life: intracellular regulator and extracellular signal. Biochim. Biophys. Acta, Mol. Cell Res. 2009, 1793, 1008–1022. 10.1016/j.bbamcr.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Nash D. L.; Bellolio M. F.; Stead L. G. S100 as a marker of acute brain ischemia: a systematic review. Neurocrit. Care 2008, 8, 301–307. 10.1007/s12028-007-9019-x. [DOI] [PubMed] [Google Scholar]
- Dagdan E.; Morris D. W.; Campbell M.; et al. Functional assessment of a promoter polymorphism in S100B, a putative risk variant for bipolar disorder. Am. J. Med. Genet., Part B 2011, 156B, 691–699. 10.1002/ajmg.b.31211. [DOI] [PubMed] [Google Scholar]
- Egberts F.; Pollex A.; Egberts J.-H.; Kaehler K. C.; Weichenthal M.; Hauschild A. Long-term survival analysis in metastatic melanoma: serum S100B is an independent prognostic marker and superior to LDH. Onkologie 2008, 31, 380–384. 10.1159/000135492. [DOI] [PubMed] [Google Scholar]
- Weide B.; Richter S.; Buttner P.; et al. Serum S100B, lactate dehydrogenase and brain metastasis are prognostic factors in patients with distant melanoma metastasis and systemic therapy. PLoS One 2013, 8, e81624 10.1371/journal.pone.0081624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi R.; Giambanco I.; Arcuri C.; Donato R. Subcellular localization of S100A11 (S100C) in LLC-PK1 renal cells: Calcium-and protein kinase c–dependent association of S100A11 with S100B and vimentin intermediate filaments. Microsc. Res. Tech. 2003, 60, 639–651. 10.1002/jemt.10305. [DOI] [PubMed] [Google Scholar]
- Deloulme J. C.; Assard N.; Mbele G. O.; Mangin C.; Kuwano R.; Baudier J. S100A6 and S100A11 are specific targets of the calcium- and zinc-binding S100B protein in vivo. J. Biol. Chem. 2000, 275, 35302–35310. 10.1074/jbc.M003943200. [DOI] [PubMed] [Google Scholar]
- Takeuchi K.; Wagner G. NMR studies of protein interactions. Curr. Opin. Struct. Biol. 2006, 16, 109–117. 10.1016/j.sbi.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Williamson M. P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 73, 1–16. 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Laskowski R. A.; Rullmann J. A. C.; MacArthur M. W.; Kaptein R.; Thornton J. M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Huang Y. K.; Chou R. H.; Yu C. Tranilast Blocks the Interaction between the Protein S100A11 and Receptor for Advanced Glycation End Products (RAGE) V Domain and Inhibits Cell Proliferation. J. Biol. Chem. 2016, 291, 14300–14310. 10.1074/jbc.M116.722215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung K. W.; Chang Y. M.; Yu C. NMR structure note: the structure of human calcium-bound S100A11. J. Biomol. NMR 2012, 54, 211–215. 10.1007/s10858-012-9661-2. [DOI] [PubMed] [Google Scholar]
- Sparvero L. J.; Asafu-Adjei D.; Kang R.; et al. RAGE (Receptor for Advanced Glycation Endproducts), RAGE ligands, and their role in cancer and inflammation. J. Transl. Med. 2009, 7, 17 10.1186/1479-5876-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato R. RAGE: a single receptor for several ligands and different cellular responses: the case of certain S100 proteins. Curr. Mol. Med. 2007, 7, 711–724. 10.2174/156652407783220688. [DOI] [PubMed] [Google Scholar]
- Cecil D. L.; Johnson K.; Rediske J.; Lotz M.; Schmidt A. M.; Terkeltaub R. Inflammation-Induced Chondrocyte Hypertrophy Is Driven by Receptor for Advanced Glycation End Products. J. Immunol. 2005, 175, 8296–8302. 10.4049/jimmunol.175.12.8296. [DOI] [PubMed] [Google Scholar]
- Cecil D. L.; Terkeltaub R. Transamidation by transglutaminase 2 transforms S100A11 calgranulin into a procatabolic cytokine for chondrocytes. J. Immunol. 2008, 180, 8378–8385. 10.4049/jimmunol.180.12.8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S. S.; Wu Z.-Y.; Zhang H. P.; et al. Suppression of experimental autoimmune encephalomyelitis by selective blockade of encephalitogenic T-cell infiltration of the central nervous system. Nat. Med. 2003, 9, 287–293. 10.1038/nm831. [DOI] [PubMed] [Google Scholar]
- Smith S. P.; Shaw G. S. A novel calcium-sensitive switch revealed by the structure of human S100B in the calcium-bound form. Structure 1998, 6, 211–222. 10.1016/S0969-2126(98)00022-7. [DOI] [PubMed] [Google Scholar]
- Gupta A. A.; Chou R. H.; Li H.; Yang L. W.; Yu C. Structural insights into the interaction of human S100B and basic fibroblast growth factor (FGF2): Effects on FGFR1 receptor signaling. Biochim. Biophys. Acta, Proteins Proteomics 2013, 1834, 2606–2619. 10.1016/j.bbapap.2013.09.012. [DOI] [PubMed] [Google Scholar]
- Baudier J.; Gerard D. Ions binding to S100 proteins. II. Conformational studies and calcium-induced conformational changes in S100 alpha alpha protein: the effect of acidic pH and calcium incubation on subunit exchange in S100a (alpha beta) protein. J. Biol. Chem. 1986, 261, 8204–8212. [PubMed] [Google Scholar]
- Pröpper C.; Huang X.; Roth J.; Sorg C.; Nacken W. Analysis of the MRP8-MRP14 protein-protein interaction by the two-hybrid system suggests a prominent role of the C-terminal domain of S100 proteins in dimer formation. J. Biol. Chem. 1999, 274, 183–188. 10.1074/jbc.274.1.183. [DOI] [PubMed] [Google Scholar]
- Goddard T.; Kneller D.. SPARKY 3; University of California: San Francisco, 2008.
- van Zundert G. C. P.; Rodrigues J.; Trellet M.; et al. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- de Vries S. J.; van Dijk M.; Bonvin A. M. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010, 5, 883–897. 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- Dominguez C.; Boelens R.; Bonvin A. M. HADDOCK: a protein–protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003, 125, 1731–1737. 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Hubbard S. J.; Thornton J. M.. NACCESS, Computer Program, Department of Biochemistry and Molecular Biology; University College London: London, 1993.
- Linge J. P.; Williams M. A.; Spronk C. A.; Bonvin A. M.; Nilges M. Refinement of protein structures in explicit solvent. Proteins 2003, 50, 496–506. 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- Schrödinger, LLC, The PyMOL Molecular Graphics System, Version 1.7.4.5, 2015
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.