Abstract
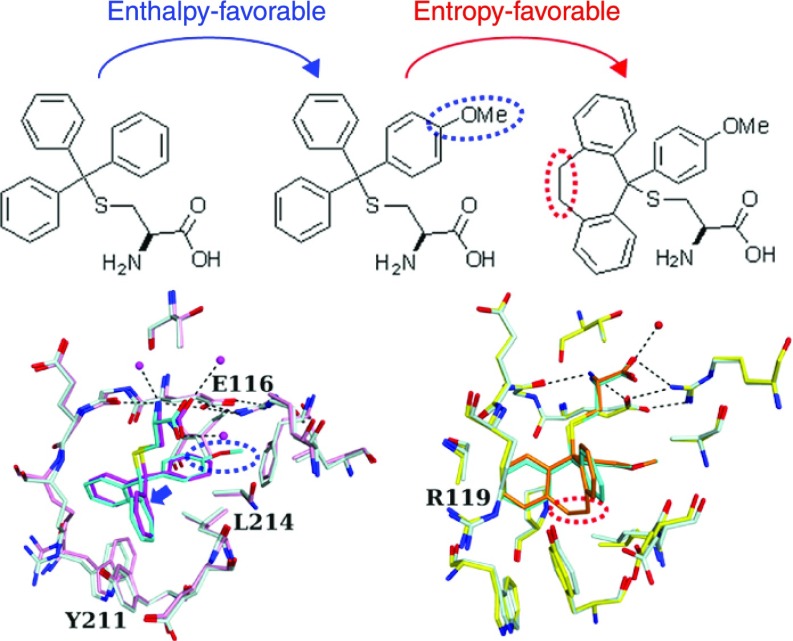
For a better understanding of protein–inhibitor interactions, we report structural, thermodynamic, and biological analyses of the interactions between S-trityl-l-cysteine (STLC) derivatives and the motor domain of kinesin spindle protein Eg5. Binding of STLC-type inhibitors to Eg5 was enthalpically driven and entropically unfavorable. The introduction of a para-methoxy substituent in one phenyl ring of STLC enhances its inhibitory activity resulting from a larger enthalpy gain possibly due to the increased shape complementarity. The substituent fits to a recess in the binding pocket. To avoid steric hindrance, the substituted STLC is nudged toward the side opposite to the recess, which enhances the interaction of Eg5 with the remaining part of the inhibitor. Further introduction of an ethylene linkage between two phenyl rings enhances Eg5 inhibitory activity by reducing the loss of entropy in forming the complex. This study provides valuable examples of enhancing protein–inhibitor interactions without forming additional hydrogen bonds.
Introduction
In recent years, the discovery and development of molecular-targeted agents have been carried out by high-throughput biological evaluation of chemical libraries consisting of small molecules. Hit compounds in the early stage of drug screening often have low affinity for their target biomolecules and show the expected biological activities only at high concentrations. Structure–activity relationship (SAR) studies using the structure-related molecules of initial hit compounds can lead to the identification of more potent compounds with high affinity for the target biomolecules.1,2 Crystal structure analysis is a powerful tool for the development of molecular-targeted agents. Detailed structural information on the interaction between a target protein and the hit compounds can help the rational design of small-molecule candidates with high affinity for the target in silico, which leads to structure-based drug design or fragment-based drug discovery.1,3−5 Then, the biological activity of individual designed compounds is confirmed by molecular biology-based compound evaluation. There are reports showing the crystal structures of target biomolecules in complex with inhibitors that are well developed to have high inhibitory activity toward the targets.6−9 However, few articles focus on the structural differences of protein–inhibitor complexes by using a series of structure-related derivatives with different affinities. For an efficient in silico design of compounds fitting more tightly into the binding pocket of a particular target protein, it is required to accumulate structural analyses of protein–inhibitor interactions between a target protein and a series of inhibitor derivatives. Thermodynamic analysis is also informative of the rational drug design in SAR research works. However, there are only a few reports on both of the crystal structures and thermodynamic analyses of protein–inhibitor complexes, such as HIV-1 protease10,11 and matrix metalloprotease.12 Studies are attempting to predict the binding affinities and thermodynamic parameters, such as enthalpic (ΔH) and entropic (−TΔS) contributions using high-resolution crystal structures, but it remains difficult to predict entropic components.13,14 More experimental data are needed to improve such prediction methods.
As the mitotic kinesin Eg5 is an attractive target for clinical cancer therapies,15 many Eg5 inhibitors, including S-trityl-l-cysteine (STLC), have been reported since the discovery of the first Eg5 inhibitor monastrol.16,17 Some of them, such as ispinesib and filanesib (ARRY-520), have been used in clinical trials as anticancer drugs.17 To date, there have been several structural studies of the Eg5 motor domain in complex with its inhibitors with different chemical scaffolds, such as monastrol,18 STLC,19 ispinesib,20,21 and others.22 These inhibitors bind to the same allosteric pocket of Eg5 formed by the L5 loop and α2 and α3 helices in the presence of adenosine 5′-triphosphate (ATP) or adenosine 5′-diphosphate (ADP) and inhibit the release of ADP from the protein.23,24 However, no comparative studies of crystal structure analysis and thermodynamic analysis using a series of the structure-related derivatives have been conducted, and sufficient lessons have not been obtained from the enormous efforts in the SAR studies.
STLC was found as a potent inhibitor for human Eg5 protein,25,26 and SAR studies using STLC derivatives have been reported by Kozielski’s and our groups.27,28 Here, we have performed crystal structure analyses and isothermal titration calorimetry (ITC) analyses using the Eg5 motor domain along with STLC and the two more potent derivatives: one with a para-methoxy substituent in one phenyl ring (PVEI0021) and the other with an ethylene linkage between two phenyl rings of PVEI0021 (PVEI0138). In this report, comparative studies of structural and thermodynamic analyses show how the substituents introduced into the initial hit STLC enhance the interaction of the STLC-type compounds with Eg5.
Results
STLC Derivatives Show Potent Inhibitory Effects against Eg5 ATPase Activity
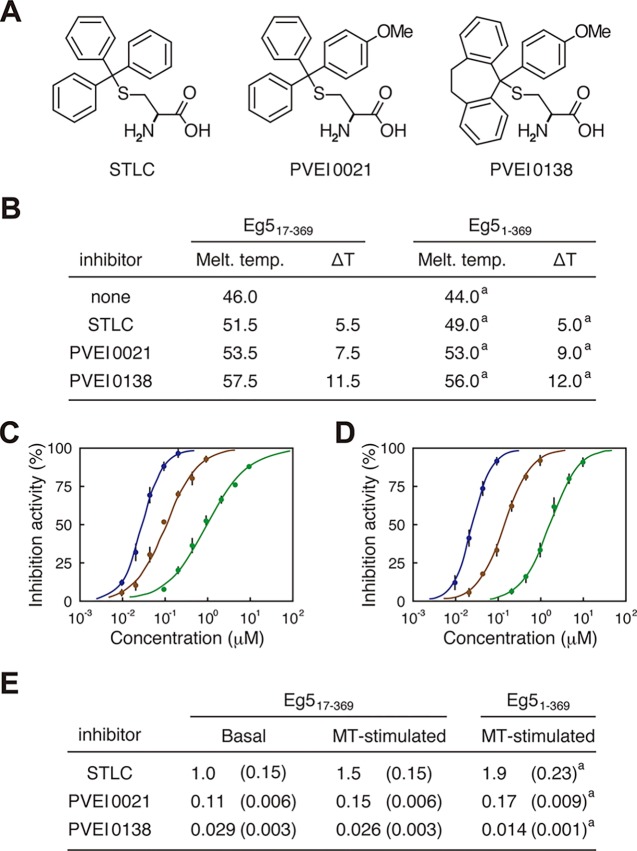
Two STLC derivatives, PVEI0021 and PVEI0138, were found as Eg5 inhibitors that worked in vitro and in cultured cells.25,28 PVEI0021 possesses a single para-methoxy substituent in one phenyl ring of the trityl group in STLC (Figure 1A), and was 10-fold more potent than STLC in inhibition of the ATPase activity of Eg5 motor domain consisting of the N-terminal 369 residues of Eg5 (Eg51–369).25 PVEI0138 is a cross-linked derivative with an ethylene linker between two phenyl rings of the trityl group in PVEI0021 (Figure 1A), and was 10-fold more potent than PVEI0021 in the inhibition of Eg5 enzymatic activity.28 In this study, we used an N-terminally truncated mutant lacking the first 16 residues, Eg517–369, to determine a high-resolution structure by producing high-quality crystals of Eg5 with the individual Eg5 inhibitors. Eg517–369 showed higher thermal stability than Eg51–369 (Figure 1B), although it showed about 65% ATPase activity compared to Eg51–369 in our ATPase assay conditions. Using Eg517–369, we evaluated the ability of the STLC derivatives to thermally stabilize the Eg5 motor domain (Figure 1B). The results of inhibitor-mediated changes in thermal denaturation temperatures were consistent with those using Eg51–369. Furthermore, the inhibitory effects of the STLC derivatives on the Eg517–369 ATPase activity in the presence and absence of taxol-stabilized microtubules (MTs) were also confirmed (Figure 1C,D). The half-maximal inhibitory concentration (IC50) values of the STLC derivatives are listed in Figure 1E, which were similar to those using Eg51–369.28 These results indicate that Eg517–369 is sensitive to the Eg5 inhibitors, similar to Eg51–369, and could be suitable for crystallization analyses. Therefore, we proceeded with structure determination of the Eg5 motor domain complexed with each STLC-type inhibitor using Eg517–369.
Figure 1.
Sensitivity of Eg517–369 to STLC, PVEI0021, and PVEI0138. (A) Chemical structures of STLC, PVEI0021, and PVEI0138. (B) Table of the ability of each inhibitor to thermally stabilize Eg517–369 in the presence of ATP. The melting temperatures (°C) of Eg517–369 in the absence and presence of inhibitors were determined by differential scanning fluorimetry (DSF). The inhibitor-mediated thermal shifts (°C) are presented as ΔT. The melting temperatures are the averages of at least four independent experiments at 0.5 °C intervals. (C, D) Concentration–response curves of the inhibitors on the basal (C) and MT-stimulated (D) ATPase activities of Eg517–369. The experimental data and calculated curves of STLC, PVEI0021, and PVEI0138 are presented in green, brown, and blue, respectively. Each data point represents the means of at least three independent experiments with standard deviations. (E) Table for the IC50 values (μM) of each inhibitor in the ATPase assay using Eg517–369. The values were calculated using the same data shown in (C) and (D) with standard deviations shown in parentheses. In (B) and (E), values of IC50, melting temperature, and ΔT when using Eg51–369 are also presented for reference. a Values were previously reported by our group.28
Overall Structures
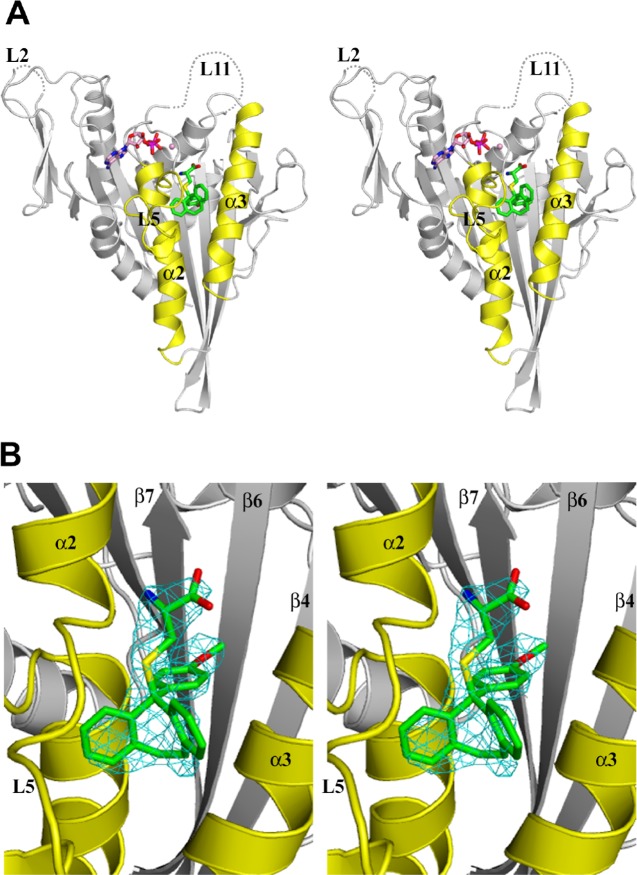
Crystals of Eg517–369 in complex with PVEI0138 or PVEI0021 were obtained in two crystal forms: one belonged to space group P21, and the other to C2. The structures of the Eg517–369–PVEI0138 complex (C2 type) and the Eg517–369–PVEI0021 complex (P21 and C2 types) were determined at resolutions of 2.6, 2.2, and 2.7 Å (Table 1). All of these structures contain two molecules (subsequently named A and B) in an asymmetric unit. The final model of Eg517–369–PVEI0138 contains residues 16–366, one Mg2+ ADP, and one PVEI0138 for molecules A and B. Residue 16 is a methionine derived from the expression vector (see Supporting Information). Residues 55–58 (L2) and 272–286 (L11) are missing for molecules A and B (Figure 2A). These loop regions are also missing in other Eg5 structures.21,29,30 Both the final models of Eg517–369–PVEI0021 (P21 and C2 types) contain residues 16–367, one Mg2+ ADP, and one PVEI0021 for molecules A and B. Residues 55–60 (L2) and 272–286 (L11) are missing for molecules A and B. PVEI0138 and PVEI0021 have well-defined electron densities in the structures and are located in the inhibitor-binding pocket bordered by helix α2, loop L5, and helix α3 (Figures 2B and S1), as observed in the Eg51–368–STLC complex.19 ADP also has well-defined electron densities in all of the structures, and each ADP is located in the ADP-binding pocket in the same orientation observed in the Eg51–368–STLC complex19 (Figure S2).
Table 1. Data Collection and Refinement Statistics.
| Eg517–369–PVEI0138 | Eg517–369–PVEI0021 (P21) | Eg517–369–PVEI0021 (C2) | |
|---|---|---|---|
| Data Collection | |||
| X-ray source | SPring-8 BL26B1 | SPring-8 BL26B1 | SPring-8 BL26B1 |
| detector | CCD MSC Saturn A200 | CCD Rayonix MX225 | CCD Rayonix MX225 |
| space group | C2 | P21 | C2 |
| cell dimensions | |||
| a, b, c (Å) | 158.35, 50.58, 93.69 | 95.29, 50.72, 83.36 | 155.16, 50.73, 94.31 |
| β (deg) | 102.20 | 112.99 | 102.19 |
| wavelength (Å) | 1.0000 | 1.0000 | 1.0000 |
| resolution range (Å) | 20.00–2.60 (2.74–2.60)a | 20.00–2.20 (2.32–2.20)a | 20.00–2.70 (2.75–2.70)a |
| no. of observed reflections | 98 040 | 127 609 | 69 714 |
| no. of unique reflections | 22 457 (3209) | 36 080 (4216) | 20 540 (953) |
| Rmerge (I)b | 0.083 (0.377) | 0.060 (0.290) | 0.104 (0.565) |
| completeness | 0.992 (0.988) | 0.959 (0.775) | 0.988 (0.945) |
| average I/σ | 13.1 (3.8) | 15.2 (3.4) | 18.6 (2.2) |
| Wilson B-factor (Å2) | 33.2 | 24.6 | 48.6 |
| Refinement | |||
| resolution range (Å) | 19.52–2.60 | 19.83–2.20 | 19.74–2.70 |
| no. of reflections used | 20 191 | 32 407 | 17 799 |
| Rc/Rfreed | 0.225/0.283 | 0.205/0.244 | 0.227/0.283 |
| no. of nonhydrogen atoms | |||
| protein | 5248 | 5226 | 5226 |
| ligand | 116 | 112 | 112 |
| solvent | 83 | 129 | 40 |
| average B factors (Å2) | |||
| protein | 50.2 | 35.5 | 65.3 |
| ligand | 40.0 | 25.4 | 43.2 |
| solvent | 34.3 | 24.8 | 48.0 |
| rms deviations from ideality | |||
| bond lengths (Å) | 0.011 | 0.009 | 0.011 |
| bond angles (deg) | 1.543 | 1.420 | 1.578 |
| Ramachandran plote (%) | |||
| favored region | 97.1 | 99.1 | 99.2 |
| allowed region | 2.9 | 0.9 | 0.8 |
| outlier region | 0.0 | 0.0 | 0.0 |
Values in parentheses are for the highest-resolution shell.
Rmerge(I) = ∑hkl∑j|Ij(hkl) – ⟨I(hkl)⟩|/∑hkl∑jIj(hkl), where Ij(hkl) is the intensity of an individual reflection and ⟨I(hkl)⟩ is the mean intensity of that reflection.
R = ∑hkl||Fobs| – |Fcalc||/∑hkl|Fobs|, where |Fobs| and |Fcalc| are the observed and calculated structure factor amplitudes, respectively.
Rfree is calculated for 10% of the reflections randomly excluded from refinement.
Values were calculated with RAMPAGE.31
Figure 2.
Crystal structure of the Eg517–369–PVEI0138 complex (stereo view). Molecule A was used to generate these figures. (A) Front view shown with a ribbon representation. PVEI0138 (green) and Mg2+ ADP (pink) are shown as ball-and-stick models (N atom, blue; O atom, red; S atom, yellow; P atom, magenta). The dotted lines indicate the disordered regions. Helix α2, loop L5, and helix α3 are colored yellow. (B) Close-up view of PVEI0138 and its binding pocket. The Fo–Fc omit map of PVEI0138 was calculated with phases from the model without PVEI0138, contoured at 3σ, and colored cyan. The values of the real space correlation coefficient of PVEI0138 are 0.899 in chain A and 0.888 in chain B. This view is almost the same as that in (A).
The structures of the Eg5 motor domain in complex with STLC-type inhibitors were superposed well with each other using PDBeFold32 (Table S2 and Figures S2 and S3). As for the two structures of Eg517–369–PVEI0021 (P21 and C2 types), the patterns of crystal packing can be regarded as being nearly the same between the P21- and C2-type PVEI0021 complexes (see Figure S4 for details). Therefore, in the following description, the structure of the P21-type PVEI0021 complex is used as a PVEI0021-bound structure to compare with other STLC-type inhibitor-complexed structures. Because molecules A of the Eg5 motor domain in complex with PVEI0138, PVEI0021 (P21 type), and STLC are in similar structures to root-mean-square deviation (rmsd) values less than 0.8 Å (Table S2), these three structures are used to discuss the difference in the structures of the Eg5 motor domain bound to each inhibitor in the following description (see Table S2 and Figures S2 and S3 for details).
Differences in Inhibitor Binding among STLC-type Inhibitor Complexes
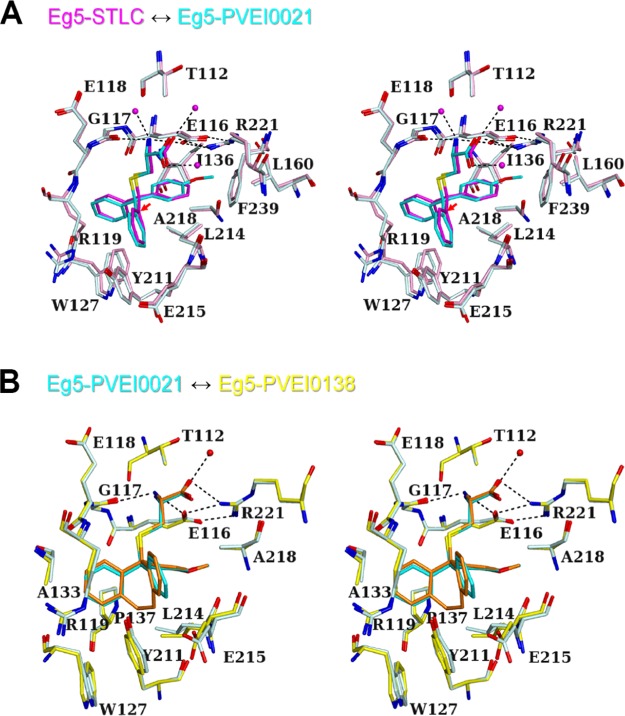
The structures of inhibitor-binding sites of the Eg5 motor domain in complex with STLC, PVEI0021, and PVEI0138 were compared. The amino N atom of the cysteine moiety of STLC is hydrogen-bonded to Glu116 Oε1 and Gly117 O atoms. The carboxy O atom of the cysteine moiety of STLC is hydrogen-bonded to Arg221 Nη1.19 These hydrogen bonds are commonly observed in three structures of STLC, PVEI0021, and PVEI0138 complexes (Figure 3). The salt bridges between the side chains of Glu116 and Arg221 are also commonly observed in the three structures, although the salt bridges are not observed in the inhibitor-free structure of Eg5–ADP·Mg2+.33 The surface area of Eg5 interfacing with inhibitor was calculated as shown in Table S3A. Glu116, Arg119, Leu214, and Ala218 are common residues with large interface areas. We name each portion of the inhibitor-binding pockets as follows: “Top”, “Bottom”, “Left-side”, “Right-side”, “Front”, and “Back”, as shown in Figure 4B. Both methoxy moieties of PVEI0021 and PVEI0138 are surrounded by the side chains of Top Glu116 and Arg221, Bottom Leu214, Right-side Ile136, Leu160, and Phe239 and form van der Waals interactions with them. In previously determined crystal structure of the Eg5 motor domain in complex with an STLC derivative having a para-substituted chlorine in one phenyl ring, the para-chlorophenyl ring is located in a similar pocket to the para-methoxyphenyl group of PVEI0021.34 The surface area of Top Glu116 interfacing with PVEI0021 (41 Å2) is larger than that with STLC (38 Å2, Table S3A). The surface area of Bottom Leu214 interfacing with PVEI0021 (40 Å2) is also larger than that with STLC (36 Å2, Table S3A). These two differences are ascribable to the addition of van der Waals interactions of the para-methoxy substituent with the side chains of Top Glu116 and Bottom Leu214. The other residues of Top Arg221, and Right-side Ile136, Leu160, and Phe239 show no increased interface area (Table S3A). For the STLC and PVEI0021 complexes, the side chains of Top and Bottom residues are nearly in the same locations, but those of Right-side residues shift to a small extent (Figure 3A). Differences in the locations are as follows: Top Glu116 Cδ, 0.3 Å; Arg221 Cζ, 0.2 Å; Bottom Leu214 Cγ, 0.3 Å; Right-side Ile136 Cβ, 0.4 Å; Leu160 Cγ, 0.3 Å; and Phe239 Cγ, 0.6 Å. The shift of the Right-side wall broadens the pocket volume around the para-substituted phenyl (3 in Figure 4B) to a small extent (Figure 4A,B).
Figure 3.
Comparison of the inhibitor-binding sites of the Eg5 motor domain in complex with STLC-type inhibitors (stereo view). On the basis of the structure of the Eg517–369–PVEI0138 complex (yellow), the structures of PVEI0021 (P21 type, cyan) and STLC (Protein Data Bank (PDB) code 2WOG, pink) complexes were superposed. Molecule A was used to generate these figures. PVEI0138 (orange), PVEI0021 (cyan), and STLC (magenta) are shown as ball-and-stick models. (A) Superposition of STLC (pink) and PVEI0021 (P21 type, cyan) complexes. Water molecules of the STLC complex are also shown as spheres. Hydrogen bonds of the STLC complex are shown by the dotted lines. The red arrow indicates the molecular displacement from STLC to PVEI0021. (B) Superposition of PVEI0021 (P21 type, cyan) and PVEI0138 (yellow) complexes. A water molecule of the PVEI0138 complex is also shown as a sphere. Hydrogen bonds of the PVEI0138 complex are shown by the dotted lines.
Figure 4.
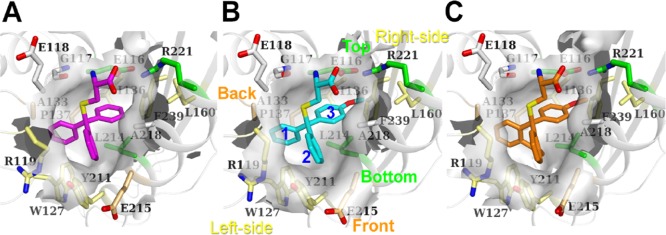
Surface representation of the inhibitor-binding pockets calculated with CASTp.35 (A) Eg51–368–STLC complex; (B) Eg517–369–PVEI0021 complex (P21 type); (C) Eg517–369–PVEI0138 complex. Molecule A was used to generate these figures. STLC (magenta), PVEI0021 (cyan), and PVEI0138 (orange) are shown as stick models. The views are almost the same as those in Figure 3A. In (B), the names of the walls forming the binding pocket are also labeled with the same color as the side chains shown as stick models: Top (Glu116 and Arg221) and Bottom (Leu214), green; Front (Glu215) and Back (Ala133 and Pro137), orange; Left-side (Arg119, Trp127, and Tyr211); and Right-side (Ile136, Leu160, and Phe239); yellow. Phenyl or methoxyphenyl groups of PVEI0021 are numbered.
Due to the presence of the para-methoxy substituent, the C atom in the center of the trityl group of PVEI0021 was displaced 0.5 Å from that of STLC (Figure 3A). Compared to STLC, PVEI0138 and para-chloro STLC, as a whole molecule, also shift 0.6 Å to the Left-side (Figures 3A and S5A,D). For reference, the locations of ADP are nearly identical in the four STLC-type inhibitor complexes (Figure S2). The differences in the positions of ADP molecules are less than 0.1 Å in the four structures (Figure S5C,D). Because of the shift, the two phenyl rings (1 and 2 in Figure 4B) easily interact with neighboring residues of Left-side Arg119, Trp127, and Tyr211, and Front Glu215. Comparing the STLC and PVEI0021 complexes, the side chains of the Left-side residues shift to some extent (Figure 3A). Differences in the locations are as follows: Left-side Arg119 Cδ, 0.9 Å; Trp127 Cβ, 0.6 Å; Tyr211 Cε1, 0.3 Å; and Front Glu215 Cβ, 0.3 Å. These residues move away from the inhibitor and thus broaden the pocket volume around two phenyl rings 1 and 2 (Figure 4A,B). The surface area of Left-side Tyr211 interfacing with PVEI0021 (25 Å2) is larger than that with STLC (22 Å2, Table S3A). These differences may contribute to the higher affinity of PVEI0021 with Eg5 than that of STLC.
In addition to a methoxy substituent to the phenyl group, PVEI0138 possesses an ethylene linker between the other two phenyl rings 1 and 2 of the trityl group (Figure 1A). There are no additional hydrogen-bonding interactions between the protein and PVEI0138 compared to PVEI0021. Residues of Left-side Arg119, Trp127, and Tyr211, Back Pro137, Bottom Leu214, and Front Glu215 are located around and in van der Waals interactions with the ethylene linker of PVEI0138 (Figure 3B). Comparing PVEI0021 and PVEI0138 complexes, the side chains of Bottom and Front residues shift to some extent (Figure 3B). Differences in the locations are as follows: Left-side Arg119 Cδ, 0.2 Å; Trp127 Cβ, 0.2 Å; Tyr211 Cε1, 0.1 Å; Back Pro137 Cβ, 0.2 Å; Bottom Leu214 Cβ, 0.6 Å; and Front Glu215 Cβ, 0.5 Å. Because the ethylene linker connects two phenyl rings 1 and 2, the distances between the two C atoms attaching the ethylene linker of PVEI0138 (2.9 Å) is shorter than those of PVEI0021 (3.3 Å), STLC (3.1 Å), and para-chloro STLC (3.2 Å, Figure S5B). As a result, the two phenyl ring moieties of PVEI0138 approach the side chain of Left-side Arg119, and the surface area of Arg119 interfacing with PVEI0138 (34 Å2) is larger than that with PVEI0021 (31 Å2, Table S3A), although the location of the Left-side wall is nearly the same between the PVEI0021 and PVEI0138 complexes. The side chain of Front Glu215 also comes close to the ethylene linker of PVEI0138 in comparison to PVEI0021, and the surface area of Glu215 interfacing with PVEI0138 (19 Å2) is larger than that with PVEI0021 (15 Å2, Table S3A). The increase in the surface area interfacing with PVEI0138 compared to PVEI0021 contributes to the increased van der Waals interactions. In contrast, the side chain of Bottom Leu214 moves away from the ethylene linker of PVEI0138 in comparison to PVEI0021, and the surface area of Leu214 interfacing with PVEI0138 (35 Å2) is smaller than that with PVEI0021 (40 Å2, Table S3A). According to the comparison between PVEI0021 and PVEI0138 complexes, the addition of an ethylene linker broadens the Bottom space and shrinks the Front space, although the Left-side and Back walls show no remarkable change (Figure 4B,C). These differences may result in a more stable interaction of PVEI0138 with Eg5 than that of PVEI0021.
The volumes of the binding pocket of STLC, PVEI0021, and PVEI0138 complexes are 474, 507, and 558 Å3, respectively (Table S3B). In addition, the molecular surface areas of the binding pocket of STLC, PVEI0021, and PVEI0138 complexes are 346, 394, and 426 Å2, respectively (Table S3B). The volume and surface area of the pocket become larger with the introduction of a para-methoxy substituent and an ethylene linker, and accordingly, the inhibitory effects become larger. To investigate how well the STLC-type inhibitors fit into the binding pocket, we calculated the shape complementarity (Sc) values of the interfaces between Eg5 motor domain and STLC-type inhibitors with the program Sc36 (Table S3C), which is often used to evaluate the interfaces of protein–protein interactions37,38 or protein–ligand interactions.39,40 Sc values can range from 0 to 1. Interfaces with Sc = 1 fit perfectly, whereas interfaces with Sc = 0 mean topologically uncorrelated surfaces. All of the STLC-type inhibitors show excellent shape complementarity with the binding pocket. The Sc values of STLC, PVEI0021, and PVEI0138 complexes are 0.785, 0.811, and 0.814, respectively (Table S3C). These values are much higher than the values for antibody–antigen interfaces (0.64–0.68).36 The results suggests that the introduction of a para-methoxy substituent and an ethylene linker leads to higher shape complementarity.
Thermodynamics of STLC-type Inhibitors Binding to Eg5
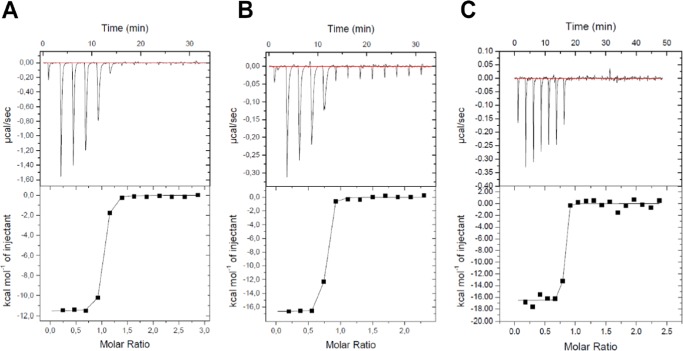
To analyze the thermodynamics of STLC-type inhibitors binding to Eg5, ITC experiments were performed using Eg517–369 (Figure 5). The resulting thermodynamic parameters were determined and calculated as shown in Table 2. A binding stoichiometry for STLC was 0.94 ± 0.003, close to 1. However, each binding stoichiometry for PVEI0021 and PVEI0138 was relatively low, probably due to the low solubility of PVEI0021 and PVEI0138 in this study. Each KD value was 67 ± 8.8 nM for STLC, 6.3 ± 1.8 nM for PVEI0021, and <1 nM for PVEI0138. The approximate KD value for PVEI0138 was 0.9 nM, although the value was under a detection limit. PVEI0021 showed 1 order of magnitude higher affinity to Eg517–369 than STLC, and PVEI0138 showed approximately 1 order of magnitude higher affinity to Eg517–369 than PVEI0021. The binding of STLC to Eg517–369 was enthalpically driven (ΔH = −11.6 ± 0.1 kcal/mol) and entropically neutral or a bit unfavorable (−TΔS = 1.8 kcal/mol). In comparison to STLC, the binding of PVEI0021 to Eg517–369 was enthalpically more favorable by 5.0 kcal/mol (ΔH = −16.6 ± 0.1 kcal/mol), but was entropically more unfavorable by 3.7 kcal/mol (−TΔS = 5.5 kcal/mol). The para-methoxy substituent contributed to an enthalpically more stable interaction with Eg517–369. In the binding of PVEI0138, a change in enthalpy (ΔH = −16.5 ± 0.3 kcal/mol) was similar to that of PVEI0021. However, a change in entropy (−TΔS = 4.2 kcal/mol) was more favorable by 1.3 kcal/mol than that of PVEI0021. In total, the values of Gibbs energy ΔG of STLC, PVEI0021, and PVEI0138 are −9.8, −11.2, and −12.3 kcal/mol, respectively, and Eg517–369–STLC-type inhibitor complexes become more stable in this order. These results were consistent with those obtained by our biochemical analyses, including Eg5 ATPase assays and differential scanning fluorimetry (Figure 1).28
Figure 5.
Isothermal titration calorimetry of Eg517–369 with STLC-type inhibitors. Titration results of STLC (A), PVEI0021 (B), and PVEI0138 (C) into Eg517–369 are shown. Raw thermograms of ITC measurements after baseline correction (top) and integrated heats of injection (bottom) are shown for each interaction.
Table 2. Thermodynamic Parameters of ITC Experiments between Eg517–369- and STLC-type Inhibitors.
| ligand | N (sites) | KDa (nM) | ΔH (kcal/mol) | –TΔS (kcal/mol) | ΔG (kcal/mol) |
|---|---|---|---|---|---|
| STLC | 0.94 ± 0.003 | 67 ± 8.8 | –11.6 ± 0.1 | 1.8 | –9.8 |
| PVEI0021 | 0.70 ± 0.003 | 6.3 ± 1.8 | –16.6 ± 0.1 | 5.5 | –11.2 |
| PVEI0138 | 0.77 ± 0.008 | <1 | –16.5 ± 0.3 | 4.2 | –12.3 |
KD values were calculated from ITC-derived KA.
Discussion
Here, we report the structural and thermodynamic analyses of kinesin spindle protein Eg5 in complex with STLC-type inhibitors using X-ray crystallography and ITC. This study provides two interesting examples of the effects of the substituent introduction to enhance the protein–inhibitor interactions without forming additional new hydrogen bonds between the protein and the inhibitor. One example suggests that the shape complementarity of inhibitors in the binding pockets of the target protein can be used as a valuable indicator for designing more potent inhibitors in SAR studies. The other example provides scientific evidence for the well-known but empirical strategy of introducing cyclization cross-linkers to obtain stronger inhibitors.
In SAR research, effective methods to design more potent bioactive compounds based on the chemical structures of the initial hit compounds are desired. In computer-aided designs of more potent inhibitors, valid calculated values are required as useful indicators for predicting which compounds with similar chemical structures are superior in the inhibitory activity. The surface area of proteins interfacing with inhibitors is often described in scientific reports of structural analysis of protein–inhibitor complexes because it is believed to correlate with the van der Waals interaction. The shape complementarity, originally devised as an indicator of protein–protein interactions, has been previously proposed to be used as an indicator for the evaluation of the inhibitor structures.36−40 According to the conventional lock-and-key model about the relationship between enzymes and substrates, the shape complementarity is considered useful for the optimization of protein–inhibitor interactions. However, the values of shape complementarity are not often mentioned in the papers reporting SAR studies of inhibitors or structural analysis of protein–inhibitor complexes. In the case of Eg5 and PVEI0021 shown in this study, the increase in the Eg5 inhibitory activity is accompanied by the improvement in the shape complementarity (Table S3C). The para-methoxy substituent of PVEI0021, the substituent added onto STLC, is surrounded by the Top, Bottom, and Right-side residues of the pocket (Figure 4B), which contributes to a higher shape complementarity with PVEI0021 than STLC (Figures 3A and 4A,B). In addition, the para-methoxy substituent causes a steric hindrance with the neighboring Right-side residues and thus the inhibitor molecule is totally shifted by 0.5 Å to the Left-side wall compared to STLC (Figures 3A and S5A). By approaching closer to the Left-side wall than STLC, PVEI0021 also shows a higher shape complementarity at the site opposite to the para-methoxy substituent. This is the first report showing that a placement shift of the inhibitor within the binding pocket caused by the introduction of an additional substituent can result in a significant improvement in the shape complementarity between the protein and the inhibitor. In the conversion from STLC to PVEI0021, the shape complementarity becomes higher from 0.785 to 0.811 (Table S3C). At the same time, the surface area of the binding pocket interfacing with the inhibitor increases from 288 to 294 Å2 (Table S3A). The shape complementarity of the protein–inhibitor interaction, rather than the interface area, seems to correlate with the van der Waals interaction between the protein and the inhibitor, and also with the large gain of enthalpy in forming the protein–inhibitor complex (Table 2). Therefore, this study suggests that, in the computer-aided modeling of a protein complex with newly designed candidate derivatives, the shape complementarity of protein–inhibitor complexes can work as one of the effective indicators for predicting the inhibitory potency of the candidate derivatives. The idea of quantifying spatial complementarity in the binding interface between a protein and an inhibitor is quite significant. It is desirable to develop better methods for evaluating spatial complementarity of protein–inhibitor interactions.
When designing more potent inhibitors based on the initial hit compounds, introduction of cyclization cross-linkers is known as a plausible strategy. PVEI0138 is a cyclized derivative of PVEI0021 with an ethylene cross-linkage between two phenyl rings, and is a successful example of the strategy. It is a more potent Eg5 inhibitor in the Eg5 ATPase assay and the DSF than PVEI0021 (Figure 1). Our studies provide scientific evidence for the effectiveness of the cyclization strategy. In the conversion from PVEI0021 to PVEI0138, the structural analysis shows the same state in the hydrogen bonds of the protein–inhibitor complexes. The striking differences between PVEI0138 and PVEI0021 complexes were observed only in a change in entropy in forming the protein–inhibitor complex. Although both of the complexes were entropically unfavorable, the complex formation of the Eg5 motor domain with PVEI0138 caused more reduced loss of entropy (4.2 kcal/mol) than with PVEI0021 (5.5 kcal/mol). The main reason for this is thought to be that PVEI0138, due to the presence of the cyclization cross-linkage between two phenyl rings, is a more rigid molecule with a lower degree of freedom compared to PVEI0021. The presence of the cross-linkage increases the volume of the binding pocket for PVEI0138 compared to PVEI0021. This cramped accommodation of PVEI0138 in the binding pocket may also contribute to the reduction of the loss of entropy in the degree of freedom of the PVEI0138 complex. In the structural analysis, even though the two phenyl rings are cross-linked with an ethylene linker, the spatial position of the two phenyl groups of PVEI0138 is almost the same as that of PVEI0021 (Figure 3B). There are no significant differences in the shape complementarity and the interface area between the protein and each inhibitor, and the gain level of enthalpy in forming the protein–inhibitor complex is quite similar in the two complexes. Therefore, it is considered that the reduced loss of entropy in forming the PVEI0138 complex contributes directly to the binding affinity of the inhibitor to the protein. Our previous SAR study showed that the Eg5 inhibitory activity drastically decreases if the length of the cross-linker is longer or shorter than that of the ethylene cross-linker.28 In the case of such cyclized derivatives, the gain in enthalpy in protein–inhibitor complex formation is thought to be much smaller than the reduced loss of entropy produced by conversion of candidate compounds to more rigid molecules.
Understanding protein–inhibitor interactions at the molecular and atomic levels is a major issue in the field of medicinal chemistry. Understanding them in terms of thermodynamics is also important. Due to the thermal motion of solvent molecules, proteins and inhibitors in solution under physiological conditions are always fluctuating during their association, complex formation, and dissociation. By structural and thermodynamic analyses in addition to biochemical analysis, this study shows an importance of the shape complementarity as an indicator of the stability of the protein–inhibitor binding state. It also supports the effectiveness of introducing a cyclization cross-linkage as a strategy for designing more potent inhibitors. Performing both crystal structure analysis and thermodynamic analysis is beneficial for finding small but important differences in protein−inhibitor interactions. A deep understanding of protein–inhibitor interactions by several different analytical methods will lead to the establishment of better ways to design more potent inhibitors in SAR studies. There may be points of view to which we did not pay enough attention in this study, even though they are important. More structural and thermodynamic studies on protein–inhibitor interactions are required to validate existing strategies for designing more potent inhibitors and to develop various inhibitor design strategies. In conclusion, this study not only provides valuable information for rational drug design, but also shows the importance of executing biochemical analysis, structural analysis, and thermodynamic analysis in parallel to establish highly versatile and rational drug design methodology.
Materials and Methods
Eg5 Inhibitors
STLC, PVEI0021, and PVEI0138 (Figure 1A) were synthesized as described previously (compounds 1 and 4f in ref (25); compounds 3, 3b, and 5b in ref (28)).25,28
Preparation and Biochemical Assay of Eg5
Eg517–369 was expressed in Escherichia coli BL21(DE3) CodonPlus RIL as a C-terminal His6 fusion protein. The expression plasmid and protein purification from bacterial extracts were described in the Supporting Information. Enzymatic assays to evaluate the basal and MT-stimulated ATPase activities of Eg517–369 and differential scanning fluorimetry analyses to examine the thermal stability of Eg517–369 were performed using Eg517–369 instead of Eg51–369, along with ATP, as described previously.25,28,29
Protein Crystallization
The purified protein was mixed with each inhibitor at a molar ratio of 1:5. Crystallization was performed using the sitting-drop vapor diffusion method at 20 °C. Crystallization drops were prepared by mixing 0.5 μL of the protein–inhibitor solution and 0.5 μL of the reservoir solution. In the case of the Eg517–369–PVEI0138 complex, the protein–inhibitor solution contained 17.4 mg/mL (0.41 mM) Eg517–369, 2.0 mM PVEI0138 in buffer A, and 5% (w/v) sucrose. Buffer A contained 50 mM piperazine-1,4-bis(2-ethanesulfonic acid) (PIPES)–NaOH (pH 6.8), 0.4 M NaCl, 1 mM ADP, 2 mM MgCl2, 1 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA)–NaOH, and 1 mM tris(2-carboxyethyl)phosphine (TCEP)–HCl. The reservoir solution contained 30% (w/v) poly(ethylene glycol) (PEG) 3350, 0.1 M 2-morpholinoethanesulfonic acid (MES)–NaOH (pH 6.5), and 0.2 M ammonium sulfate. Rod-shaped crystals grew to an approximate size of 0.1 × 0.05 × 0.05 mm3.
In the case of the Eg517–369–PVEI0021 complex, crystallization conditions were almost the same as in the case of Eg517–369–PVEI0138. The protein–inhibitor solution contained 15.5 mg/mL (0.37 mM) Eg517–369, 2.0 mM PVEI0021 in buffer A, and 5% (w/v) sucrose. The reservoir solution contained 24–34% (w/v) PEG3350, 0.1 M MES–NaOH (pH 6.5), and 0.2 M ammonium sulfate. Two types of crystals appeared from almost the same crystallization conditions: one belonged to space group P21 and the other to C2.
X-ray Data Collection and Structure Determination
A crystal of the Eg517–369–PVEI0138 complex was cryoprotected in a solution containing 30% (w/v) sucrose, 34% (w/v) PEG3350, 0.1 M MES–NaOH (pH 6.5), 0.2 M ammonium sulfate, and buffer A and flash-frozen at 100 K. Each crystal of the Eg517–369–PVEI0021 complex belonging to P21 and C2 types was cryoprotected in a solution containing 20% (w/v) sucrose, 34% (w/v) PEG3350, 0.1 M MES–NaOH (pH 6.5), 0.2 M ammonium sulfate, and buffer A and flash-frozen at 100 K. All X-ray diffraction data were collected at SPring-8 (Harima, Japan). Data from the Eg517–369–PVEI0138 and Eg517–369–PVEI0021 (P21 type) complexes were processed and scaled with XDS41 and SCALA,42 and those from the Eg517–369–PVEI0021 complex (C2 type) were processed and scaled with HKL2000.43
The structure of the Eg517–369–PVEI0138 complex was determined using a molecular replacement method with the program MOLREP44 in the CCP4 suite.45 The structure of the Eg51–368–STLC complex (PDB code, 2WOG; chain A)19 was used as an initial model. Structural refinement was performed with REFMAC546 and PHENIX,47 and manual model fitting was achieved with Coot.48 The structures of the Eg517–369–PVEI0021 complex (P21 and C2 types) were determined using almost the same procedure as in the Eg517–369–PVEI0138 complex. Data collection and refinement statistics are summarized in Table 1.
The least-squares fitting between the two structures was performed with PDBeFold32 using all of the residues. Accessible surface areas were calculated with AREAIMOL in the CCP4 suite.45 The area and volume of surface pockets were calculated with CASTp.35 The shape complementarities of the interface between the Eg5 motor domain and STLC-type inhibitors were calculated with Sc.36 All molecular figures were produced with PyMOL (http://www.pymol.org/).
Isothermal Titration Calorimetry
Binding affinities and thermal parameters of three inhibitors, STLC, PVEI0021, and PVEI0138, to Eg517–369 were measured using ITC (MicroCal iTC200, Malvern). The measurements were performed at 25 °C with a reference power of 5 μcal/s. STLC (3 μL, 750 μM) in the syringe was injected into 50 μM Eg517–369 in the cell, and a total of 13 injections were performed. Similarly, 3 μL of 120 μM PVEI0021 in the syringe was injected into 10 μM Eg517–369 in the cell, and a total of 13 injections were performed. Similarly, 2 μL (or 1.5 μL) of 120 μM PVEI0138 in the syringe was injected into 10 μM Eg517–369 in the cell, and a total of 19 injections (or 25 injections) were performed. Cell and syringe solutions were in the ITC buffer containing 50 mM PIPES–NaOH (pH 6.8), 0.4 M NaCl, 1 mM ADP, 2 mM MgCl2, 1 mM EGTA–NaOH, 1 mM TCEP–HCl, 5% (w/v) sucrose, and 0.75% (v/v) dimethyl sulfoxide. Parallel experiments were performed by injecting each inhibitor into the ITC buffer, and then the heat of dilution was controlled prior to fitting the data. Experiments using the same conditions were all run in duplicate. ITC data were analyzed using Origin software (MicroCal) and were fitted using a one-site binding model.
Acknowledgments
The authors thank the staff of SPring-8 for their assistance in X-ray data collection (Proposal Nos. 2014B1991, 2015A1059, and 2015B2059). They also thank M. Hirose (Malvern Instruments, Japan) for her advice in measuring ITC data, and S. Makita and H. Nagao (Institute for Molecular Science, Okazaki, Japan) for their assistance in measuring ITC data. They extend their thanks to Y. Hashizume and K. Terai for their help in these experiments. This study was supported by JSPS KAKENHI Grant Numbers 26440034 and 17K07316 to H.Y. from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), and a grant entitled “Drug Discovery Program” from Pharma Valley Center to A.A.
Glossary
Abbreviations
- SAR
structure–activity relationship
- STLC
S-trityl-l-cysteine
- ITC
isothermal titration calorimetry
- MT
microtubule
- IC50
half-maximal inhibitory concentration
- rmsd
root-mean-square deviation
- Sc
shape complementarity
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b00778.
Primers used; least-squares fitting between two structures; surface area of Eg5 interfacing with inhibitor (Tables S1–S3); close-up view of PVEI0021; superposition of three structures of the Eg5 motor domain; plots of Cα-atom distances between two structures; two types of crystal packing of Eg5–PVEI0021 complexes; comparison of the inhibitor and ADP structures (Figures S1–S5); and additional methods (PDF)
Accession Codes
The atomic coordinates and structure factors have been deposited in the Protein Data Bank Japan (PDBj) with the accession codes 5ZO7 (Eg517–369–PVEI0138 complex), 5ZO8 (Eg517–369–PVEI0021 complex, P21 type), and 5ZO9 (Eg517–369–PVEI0021 complex, C2 type).
Author Contributions
∥ H.Y. and J.-i.S. contributed equally to this work.
Author Contributions
H.Y. and K.S. performed the protein crystallization, structure analysis, and ITC analysis with assistance from N.K., Y.I., and K.H. J.-i.-S. performed the biochemical studies. N.O. synthesized the STLC-type Eg5 inhibitors. A.A. and H.H. supervised this study. H.Y. and J.-i.-S. wrote the manuscript. All authors commented on the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Wermuth C. G.; Villoutreix B.; Grisoni S.; Oliver A.; Rocher J. P.. Strategies in the Search for New Lead Compounds or Original Working Hypotheses. In The Practice of Medicinal Chemistry, 4th ed.; Wermuth C. G., Aldous D., Raboisson P., Rognan D., Eds.; Elsevier Academic Press: London, 2015; pp 73–99. [Google Scholar]
- Inglese J.; Shamu C. E.; Guy R. K. Reporting data from high-throughput screening of small-molecule libraries. Nat. Chem. Biol. 2007, 3, 438–441. 10.1038/nchembio0807-438. [DOI] [PubMed] [Google Scholar]
- Lounnas V.; Ritschel T.; Kelder J.; McGuire R.; Bywater R. P.; Foloppe N. Current progress in structure-based rational drug design marks a new mindset in drug discovery. Comput. Struct. Biotechnol. J. 2013, 5, e201302011 10.5936/csbj.201302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigglesworth M. J.; Murray D. C.; Blackett C. J.; Kossenjans M.; Nissink J. W. M. Increasing the delivery of next generation therapeutics from high throughput screening libraries. Curr. Opin. Chem. Biol. 2015, 26, 104–110. 10.1016/j.cbpa.2015.04.006. [DOI] [PubMed] [Google Scholar]
- Tawada M.; Suzuki S.; Imaeda Y.; Oki H.; Snell G.; Behnke C. A.; Kondo M.; Tarui N.; Tanaka T.; Kuroita T.; Tonimoto M. Novel approach of fragment-based lead discovery applied to rennin inhibitors. Bioorg. Med. Chem. 2016, 24, 6066–6074. 10.1016/j.bmc.2016.09.065. [DOI] [PubMed] [Google Scholar]
- Parlow J. J.; Case B. L.; Dice T. A.; Fenton R. L.; Hayes M. J.; Jones D. E.; Neumann W. L.; Wood R. S.; Lachance R. M.; Girard T. J.; Nicholson N. S.; Clare M.; Stegeman R. A.; Steven A. M.; Stallings W. C.; Kurumbail R. G.; South M. S. Design, parallel synthesis, and crystal structures of pyrazinone antithrombotics as selective inhibitors of the tissue factor VIIa complex. J. Med. Chem. 2003, 46, 4050–4062. 10.1021/jm030131l. [DOI] [PubMed] [Google Scholar]
- Dymock B.; Barril X.; Beswick M.; Collier A.; Davies N.; Drysdale M.; Fink A.; Fromont C.; Hubbard R. E.; Massey A.; Surgenor A.; Wright L. Adenine derived inhibitors of the molecular chaperone HSP90–SAR explained through multiple X-ray structures. Bioorg. Med. Chem. Lett. 2004, 14, 325–328. 10.1016/j.bmcl.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Deng H.; Bannister T. D.; Jin L.; Babine R. E.; Quinn J.; Nagafuji P.; Celatka C. A.; Lin J.; Lazarova T. I.; Rynkiewicz M. J.; Bibbins F.; Pandey P.; Gorga J.; Meyers H. V.; Abdel-Meguid S. S.; Strickler J. E. Synthesis, SAR exploration, and X-ray crystal structures of factor XIa inhibitors containing an α-ketothiazole arginine. Bioorg. Med. Chem. Lett. 2006, 16, 3049–3054. 10.1016/j.bmcl.2006.02.052. [DOI] [PubMed] [Google Scholar]
- Eidam O.; Ramagnoli C.; Caselli E.; Babaoglu K.; Pohlhaus D. T.; Karpiak J.; Bonnet R.; Shoichet B. K.; Prati F. Design, synthesis, crystal structures, and antimicrobial activity of sulfonamide boronic acids as β-lactamase inhibitors. J. Med. Chem. 2010, 53, 7852–7863. 10.1021/jm101015z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafont V.; Armstrong A. A.; Ohtaka H.; Kiso Y.; Amzel L. M.; Freire E. Compensating enthalpic and entropic changes hinder binding affinity optimization. Chem. Biol. Drug Des. 2007, 69, 413–422. 10.1111/j.1747-0285.2007.00519.x. [DOI] [PubMed] [Google Scholar]
- King N. M.; Prabu-Jeyabalan M.; Bandaranayake R. M.; Nalam M. N. L.; Nalivaika E. A.; Özen A.; Haliloğlu T.; Yılmaz N. K.; Schiffer C. A. Extreme entropy–enthalpy compensation in a drug-resistant variant of HIV-1 protease. ACS Chem. Biol. 2012, 7, 1536–1546. 10.1021/cb300191k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini I.; Calderone V.; Fragai M.; Giachetti A.; Loconte M.; Luchinat C.; Maletta M.; Nativi C.; Yeo K. J. Exploring the subtleties of drug–receptor interactions: the case of matrix metalloproteinases. J. Am. Chem. Soc. 2007, 129, 2466–2475. 10.1021/ja065156z. [DOI] [PubMed] [Google Scholar]
- Tang Y. T.; Marshall G. R. PHOENIX: a scoring function for affinity prediction derived using high-resolution crystal structures and calorimetry measurements. J. Chem. Inf. Model. 2011, 51, 214–228. 10.1021/ci100257s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S.; Litterman N. K.; Lipinski C. A.; Bunin B. A. Thermodynamic proxies to compensate for biases in drug discovery methods. Pharm. Res. 2016, 33, 194–205. 10.1007/s11095-015-1779-y. [DOI] [PubMed] [Google Scholar]
- Sakowicz R.; Finer J. T.; Beraud C.; Crompton A.; Lewis E.; Fritsch A.; Lee Y.; Mak J.; Moody R.; Turincio R.; Chabala J. C.; Gonzales P.; Roth S.; Weitman S.; Wood K. W. Antitumor activity of a kinesin inhibitor. Cancer Res. 2004, 64, 3276–3280. 10.1158/0008-5472.CAN-03-3839. [DOI] [PubMed] [Google Scholar]
- Mayer T. U.; Kapoor T. M.; Haggarty S. J.; King R. W.; Schreiber S. L.; Mitchison T. J. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 1999, 286, 971–974. 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- El-Nassan H. B. Advances in the discovery of kinesin spindle protein (Eg5) inhibitors as antitumor agents. Eur. J. Med. Chem. 2013, 62, 614–631. 10.1016/j.ejmech.2013.01.031. [DOI] [PubMed] [Google Scholar]
- Yan Y.; Sardana V.; Xu B.; Homnick C.; Halczenko W.; Buser C. A.; Schaber M.; Hartman G. D.; Huber H. E.; Kuo L. C. Inhibition of a mitotic motor protein: where, how, and conformational consequences. J. Mol. Biol. 2004, 335, 547–554. 10.1016/j.jmb.2003.10.074. [DOI] [PubMed] [Google Scholar]
- Kaan H. Y. K.; Ulaganathan V.; Hackney D. D.; Kozielski F. An allosteric transition trapped in an intermediate state of a new kinesin-inhibitor complex. Biochem. J. 2010, 425, 55–60. 10.1042/BJ20091207. [DOI] [PubMed] [Google Scholar]
- Zhang B.; Liu J. F.; Xu Y.; Ng S. C. Crystal structure of HsEg5 in complex with clinical candidate CK0238273 provides insight into inhibitory mechanism, potency, and specificity. Biochem. Biophys. Res. Commun. 2008, 372, 565–570. 10.1016/j.bbrc.2008.05.074. [DOI] [PubMed] [Google Scholar]
- Talapatra S. K.; Schüttelkopf A. W.; Kozielski F. The structure of the ternary Eg5–ADP–ispinesib complex. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2012, 68, 1311–1319. 10.1107/S0907444912027965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S. M.; Collins I. Recent findings and future directions for interpolar mitotic kinesin inhibitors in cancer therapy. Future Med. Chem. 2016, 8, 463–489. 10.4155/fmc.16.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga Z.; Kapoor T. M.; Mitchison T. J. Evidence that monastrol is an allosteric inhibitor of the mitotic kinesin Eg5. Chem. Biol. 2002, 9, 989–996. 10.1016/S1074-5521(02)00212-0. [DOI] [PubMed] [Google Scholar]
- Lad L.; Luo L.; Carson J. D.; Wood K. W.; Hartman J. J.; Copeland R. A.; Sakowicz R. Mechanism of inhibition of human KSP by ispinesib. Biochemistry 2008, 47, 3576–3585. 10.1021/bi702061g. [DOI] [PubMed] [Google Scholar]
- Ogo N.; Oishi S.; Matsuno K.; Sawada J.; Fujii N.; Asai A. Synthesis and biological evaluation of l-cysteine derivatives as mitotic kinesin Eg5 inhibitors. Bioorg. Med. Chem. Lett. 2007, 17, 3921–3924. 10.1016/j.bmcl.2007.04.101. [DOI] [PubMed] [Google Scholar]
- DeBonis S.; Skoufias D. A.; Indorato R. L.; Liger F.; Marquet B.; Laggner C.; Joseph B.; Kozielski F. Structure-activity relationship of S-trityl-l-cysteine analogues as inhibitors of the human mitotic kinesin Eg5. J. Med. Chem. 2008, 51, 1115–1125. 10.1021/jm070606z. [DOI] [PubMed] [Google Scholar]
- Good J. A. D.; Wang F.; Rath O.; Kaan H. Y. K.; Talapatra S. K.; Podgórski D.; MacKay S. P.; Kozielski F. Optimized S-trityl-l-cysteine-based inhibitors of kinesin spindle protein with potent in vivo antitumor activity in lung cancer xenograft models. J. Med. Chem. 2013, 56, 1878–1893. 10.1021/jm3014597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogo N.; Ishikawa Y.; Sawada J.; Matsuno K.; Hashimoto A.; Asai A. Structure-guided design of novel l-cysteine derivatives as potent KSP inhibitors. ACS Med. Chem. Lett. 2015, 6, 1004–1009. 10.1021/acsmedchemlett.5b00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama H.; Sawada J.; Katoh S.; Matsuno K.; Ogo N.; Ishikawa Y.; Hashimoto H.; Fujii S.; Asai A. Structural basis of new allosteric inhibition in kinesin spindle protein Eg5. ACS Chem. Biol. 2015, 10, 1128–1136. 10.1021/cb500939x. [DOI] [PubMed] [Google Scholar]
- Ulaganathan V.; Talapatra S. K.; Rath O.; Pannifer A.; Hackney D. D.; Kozielski F. Structural insights into a unique inhibitor binding pocket in kinesin spindle protein. J. Am. Chem. Soc. 2013, 135, 2263–2272. 10.1021/ja310377d. [DOI] [PubMed] [Google Scholar]
- Lovell S. C.; Davis I. W.; Arendall W. B. III; de Bakker P. I. W.; Word J. M.; Prisant M. G.; Richardson J. S.; Richardson D. C. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins: Struct., Funct., Bioinf. 2003, 50, 437–450. 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Krissinel E.; Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004, 60, 2256–2268. 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Turner J.; Anderson R.; Guo J.; Beraud C.; Fletterick R.; Sakowicz R. Crystal structure of the mitotic spindle kinesin Eg5 reveals a novel conformation of the neck-linker. J. Biol. Chem. 2001, 276, 25496–25502. 10.1074/jbc.M100395200. [DOI] [PubMed] [Google Scholar]
- Kaan H. Y. K.; Weiss J.; Menger D.; Ulaganathan V.; Tkocz K.; Laggner C.; Popowycz F.; Joseph B.; Kozielski F. Structure–activity relationship and multidrug resistance study of new S-trityl-L-cysteine derivatives as inhibitors of Eg5. J. Med. Chem. 2011, 54, 1576–1586. 10.1021/jm100991m. [DOI] [PubMed] [Google Scholar]
- Dundas J.; Ouyang Z.; Tseng J.; Binkowski A.; Turpaz Y.; Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006, 34, W116–W118. 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M. C.; Colman P. M. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993, 234, 946–950. 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- Stamper C. C.; Zhang Y.; Tobin J. F.; Erbe D. V.; Ikemizu S.; Davis S. J.; Stahl M. L.; Seehra J.; Somers W. S.; Mosyak L. Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses. Nature 2001, 410, 608–611. 10.1038/35069118. [DOI] [PubMed] [Google Scholar]
- Cohen A.; Rosenthal E.; Shifman J. M. Analysis of structural features contributing to weak affinities of ubiquitin/protein interactions. J. Mol. Biol. 2017, 429, 3353–3362. 10.1016/j.jmb.2017.09.003. [DOI] [PubMed] [Google Scholar]
- Reinherz E. L.; Tan K.; Tang L.; Kern P.; Liu J.; Xiong Y.; Hussey R. E.; Smolyar A.; Hare B.; Zhang R.; Joachimiak A.; Chang H. C.; Wagner G.; Wang J. The crystal structure of a T cell receptor in complex with peptide and MHC class II. Science 1999, 286, 1913–1921. 10.1126/science.286.5446.1913. [DOI] [PubMed] [Google Scholar]
- Sami M.; Rizkallah P. J.; Dunn S.; Molloy P.; Moysey R.; Vuidepot A.; Baston E.; Todorov P.; Li Y.; Gao F.; Boulter J. M.; Jakobsen B. K. Crystal structures of high affinity human T-cell receptors bound to peptide major histocompatibility complex reveal native diagonal binding geometry. Protein Eng., Des. Sel. 2007, 20, 397–403. 10.1093/protein/gzm033. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006, 62, 72–82. 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z.; Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. 10.1016/s0076-6879(97)76066-x. [DOI] [PubMed] [Google Scholar]
- Vagin A.; Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 22–25. 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- Winn M. D.; Ballard C. C.; Cowtan K. D.; Dodson E. J.; Emsley P.; Evans P. R.; Keegan R. M.; Krissinel E. B.; Leslie A. G. W.; McCoy A.; McNicholas S. J.; Murshudov G. N.; Pannu N. S.; Potterton E. A.; Powell H. R.; Read R. J.; Vagin A.; Wilson K. S. Overview of the CCP4 suite and current developments. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011, 67, 235–242. 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N.; Skubák P.; Lebedev A. A.; Pannu N. S.; Steiner R. A.; Nicholls R. A.; Winn M. D.; Long F.; Vagin A. A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011, 67, 355–367. 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkóczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L. W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004, 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.