Abstract
Individual differences in educational attainment are linked to differences in intelligence, and predict important social, economic, and health outcomes. Previous studies have found common genetic factors that influence educational achievement, cognitive performance and total brain volume (i.e., brain size). Here, in a large sample of participants from the UK Biobank, we investigate the shared genetic basis between educational attainment and fine-grained cerebral cortical morphological features, and associate this genetic variation with a related aspect of cognitive ability. Importantly, we execute novel statistical methods that enable high-dimensional genetic correlation analysis, and compute high-resolution surface maps for the genetic correlations between educational attainment and vertex-wise morphological measurements. We conduct secondary analyses, using the UK Biobank verbal–numerical reasoning score, to confirm that variation in educational attainment that is genetically correlated with cortical morphology is related to differences in cognitive performance. Our analyses relate the genetic overlap between cognitive ability and cortical thickness measurements to bilateral primary motor cortex as well as predominantly left superior temporal cortex and proximal regions. These findings extend our understanding of the neurobiology that connects genetic variation to individual differences in educational attainment and cognitive performance.
Keywords: cognitive performance, cortical thickness, educational attainment, genetic correlation, heritability
Introduction
Educational attainment as a heritable trait (Krapohl et al. 2014; Polderman et al. 2015) is predictive of many social, economic and health outcomes. Although linked to a range of diverse factors, it is highly phenotypically and genetically correlated with intelligence (Rietveld et al. 2014; Okbay et al. 2016; Savage et al. 2017; Sniekers et al. 2017), and has been successfully used as a proxy phenotype to facilitate the discovery of genetic variants associated with cognitive ability (Rietveld et al. 2014). In addition, polygenic scores of educational attainment are predictive of cognitive performance in adolescents and adults (Belsky et al. 2016; Selzam et al. 2017; Plomin and von Stumm 2018). Dissecting the biological bases of educational achievement may thus contribute to our understanding of cognition and adult functional outcomes.
Large-scale genome-wide association studies (GWAS) have found substantial genetic overlap between educational attainment and total brain volume (TBV) (i.e., brain size) (Adams et al. 2016; Okbay et al. 2016). In particular, a recent GWAS of educational attainment has identified genomic loci regulating brain-specific gene expression, and biological pathways involved in neural development (Okbay et al. 2016). Twin studies have implicated common genetic factors that influence both brain size and intelligence (Thompson et al. 2001; Posthuma et al. 2002, 2003; Toga and Thompson 2005; Hulshoff pol et al., 2006), a major contributor to the heritability of academic achievement (Krapohl et al. 2014). Our recent investigation further suggested that genetic influences on individual differences in educational attainment might be mediated by brain development (Elliott et al. 2018). However, to the best of our knowledge, no prior work has mapped the genetic correlations between educational attainment and fine-grained brain morphological measurements, likely due to methodological challenges and sample size (statistical power) limitations. Filling this knowledge gap represents an important next step in identifying the specific brain regions that lie in the pathway connecting genetics to educational outcomes.
In this study, we leverage the structural brain magnetic resonance imaging (MRI) scans and genomic data from a large sample of the UK Biobank participants (http://www.ukbiobank.ac.uk) (Sudlow et al. 2015) to investigate the shared genetic basis between educational attainment (years of schooling completed) and vertex-wise cortical thickness and surface area measurements. We also conduct secondary analyses, using the UK Biobank verbal–numerical reasoning score, to confirm that variation in years of education that is genetically correlated with cortical morphology is related to individual differences in cognitive ability. The verbal–numerical reasoning score assesses general cognitive ability and is heavily weighted towards math reasoning and vocabulary. Thus, it emphasizes learned knowledge, which has a strong relationship to educational attainment (Kaufman et al. 2009).
Well-established genetic correlation estimation methods such as genome-wide complex trait analysis (GCTA; also known as the GREML method) (Yang et al. 2011) and LD (linkage disequilibrium) score regression (Bulik-Sullivan et al., 2015) require either individual genotypes or GWAS summary statistics for the 2 traits of interest. However, to examine the genetic overlap between educational attainment/cognitive performance (verbal–numerical reasoning) and massive numbers of brain morphological measurements in a large sample, both GCTA and LD score regression can be computationally intractable. For example, one would have to run thousands of GWAS in order to use LD score regression, and multiple testing correction would be challenging in the presence of complex spatial correlation structures. Here we develop a computationally efficient method that enables high-dimensional genetic correlation estimation, and the empirical identification of brain regions that are genetically correlated with our constructs of interest above and beyond global brain volumetric measurements. The power requirements for this method as well as our interest in relating genetic variation, cognition and real world functional outcomes dictate our approach. We first conduct genetic correlation analyses based on our larger sample with data on educational attainment, and then relate the variation in educational attainment that is genetically correlated with cortical morphology to differences in cognitive performance. These analyses expand the literature on the genetic underpinnings and brain morphological correlates of educational attainment, and may contribute to our understanding of the neurobiology of cognitive ability.
Materials and Methods
The UK Biobank
UK Biobank is a prospective cohort study of approximately 500 000 individuals recruited across Great Britain during 2006–2010 (Sudlow et al. 2015). The protocol and consent were approved by the UK Biobank’s Research Ethics Committee. Details about the UK Biobank project are provided at http://www.ukbiobank.ac.uk. Data for the current analyses were obtained under an approved data request (ref: 32 568; previously 13 905).
Genetic Data
The genetic data for the UK Biobank comprised 488 377 samples. Two closely related Affymetrix arrays were used to genotype ∼800 000 markers spanning the genome. In addition, the dataset was phased and imputed to ∼96 million variants with the Haplotype Reference Consortium (HRC) (McCarthy et al. 2016) and the UK10K haplotype resource. We constrained all analyses to the HRC panel in the present study, which combines whole-genome sequence data from multiple cohorts of predominantly European ancestry, and thus covers a large majority of the common genetic variants in the European population.
The genetic data was quality controlled (QC) by the UK Biobank. Important information such as population structure and relatedness has been released. Details about the QC procedures can be found in Bycroft et al. (2017). We leveraged the QC metrics provided by the UK Biobank and removed samples that had mismatch between genetically inferred sex and self-reported sex, high genotype missingness or extreme heterozygosity, sex chromosome aneuploidy, and samples that were excluded from kinship inference and autosomal phasing. We removed one individual from each pair of the samples that were third degree or more closely related relatives, and restricted our analysis to participants that were estimated to have white British ancestry using principal component analysis (PCA).
Brain Imaging
We used the T1 structural brain MRI scans from 10 102 participants released by the UK Biobank in February 2017. FreeSurfer (Fischl 2012) version 6.0 was used to process the MRI scans. All processed images were manually inspected and those with processing errors, motion artifacts, poor resolution, pathologies (e.g., tumors) and other abnormalities were removed. Among the 9 229 participants that passed imaging QC, a subset of 7 818 unrelated white British participants (age, 45–79 y; female, 52.24%; see Table 1) additionally passed the genetic QC described above and were included in the analysis. We resampled subject-specific morphological measurements (cortical thickness and surface area) onto FreeSurfer’s fsaverage representation, which consists of 163,842 vertices per hemisphere with an inter-vertex distance of approximately 1-mm. We further smoothed the co-registered surface maps using a surface-based Gaussian kernel with 20-mm full width at half maximum (FWHM).
Table 1.
Sample sizes and demographics of the nonimaging samples for the genome-wide association analyses of educational attainment and the verbal–numerical reasoning score, and the neuroimaging sample. There is no overlap between the nonimaging samples and the neuroimaging sample
| Trait | Sample size | Age range (y) | Female (%) |
|---|---|---|---|
| Educational attainment | 332,613 | 39–72 | 53.77 |
| Verbal–numerical reasoning score | 108,147 | 40–70 | 53.51 |
| Neuroimaging measurements | 7 818 | 45–79 | 52.24 |
Educational Attainment
Following Okbay et al. (2016), we mapped each of the educational qualifications collected from the UK Biobank participants (UK Biobank field ID: 6138) to 1 of the 7 categories defined in the 1997 International Standard Classification of Education (ISCED) of the United Nations Educational, Scientific and Cultural Organization, and imputed the number of years of schooling completed for each ISCED category. The mapping is shown in Supplementary Tables S1 and S2. Of all the participants that passed genetic QC, 332 613 (age, 39–72 y; female, 53.77%; years of education, 14.8 ± 5.1 y) had years of schooling imputed at the baseline assessment visit (2006–2010) and were used in the GWAS of educational attainment. There was no overlap between the GWAS sample and the neuroimaging sample.
Test of Verbal–Numerical Reasoning
The verbal–numerical reasoning score (UK Biobank field ID: 20 016; labeled as fluid intelligence score) used in the present study is an unweighted sum of the number of correct answers given to the 13 higher-order reasoning questions (Lyall et al. 2016) in the UK Biobank touchscreen questionnaire. Participants who did not answer all of the 13 questions within the allotted 2-min limit were scored as 0 for each of the unattempted questions. Of all the participants that passed genetic QC, 108 147 (age, 40–70 y; female, 53.51%; verbal–numerical reasoning score, 6.2 ± 2.1) had verbal–numerical reasoning scores at the baseline assessment visit (2006–2010) and were used in the GWAS. There was no overlap between the GWAS sample of the verbal–numerical reasoning score and the neuroimaging sample.
GWAS of Educational Attainment and the Verbal–Numerical Reasoning Score
We performed GWAS of educational attainment (years of schooling completed) and the verbal–numerical reasoning score in 332 613 and 108 147 UK Biobank participants, respectively. In addition to the sample QC described above, we filtered out genetic markers with minor allele frequency <1% and imputation quality score <0.8. A total of 7 656 609 and 7 658 275 imputed SNPs on the HRC panel were included in the 2 GWAS, respectively. Association tests were conducted using SNPTEST v2.5.2 (Marchini and Howie 2010). For each genetic marker, a linear regression model was fitted, adjusting for age (at the baseline assessment visit), sex, age2, age × sex, age2 × sex, genotype array, UK Biobank assessment center, and top 10 principal components (PC) of the genotype data as covariates. GWAS results were visualized using FUMA (Watanabe et al. 2017) and the R package qqman (Turner 2014).
Estimator for SNP Heritability
Consider the linear model , where is an vector of covariate-adjusted and standardized phenotypes, is an matrix of genotypes with each column normalized to mean zero and variance one, is an vector of (random) SNP effect sizes, and is an vector of residuals. In Supplementary Material, we show that under a polygenic model, the following moment-matching estimators for SNP heritability are asymptotically equivalent:
| (1) |
where, is a constant, is the empirical genetic relationship matrix, is the average LD score across the genome, is the marginal effect size estimate of the -th variant with being the corresponding statistic, is the average statistic across the genome, is a weighted average of the genotype, that is, an vector of individual-specific polygenic scores, and denotes inner product, that is, .
We note that is the LD score regression estimator based on GWAS summary statistics, with the intercept constrained to one and the reciprocal of the LD score as the regression weight (Bulik-Sullivan et al., 2015). is the Haseman–Elston regression estimator based on individual genotypes (Haseman and Elston 1972; Elston et al. 2000; Golan et al. 2014; Ge, Chen, et al. 2017). formulates SNP heritability estimation as a polygenic score analysis. The equivalence between and has been established both theoretically and empirically in prior work (Bulik-Sullivan 2015; Ge, Chen, et al. 2017; Zhou 2017).
Estimator for SNP Coheritability
Consider the bivariate model and , where and are and vectors of covariate-adjusted and standardized phenotypes, and are and matrices of standardized genotypes, and are vectors of SNP effect sizes, and are and vectors of residuals, respectively. Without loss of generality, we assume that the first samples are identical for the 2 phenotypes. In Supplementary Material, we show that under a polygenic model, the following moment-matching estimators for SNP coheritability are asymptotically equivalent:
| (2) |
where, is a constant, , , is the phenotypic correlation between the 2 traits, and are marginal effect size estimates of the -th variant, with and being the corresponding statistics, respectively, is the average product of statistics across the genome, and are individual-specific polygenic scores.
We note that is the LD score regression estimator based on GWAS summary statistics, with a constrained intercept and the reciprocal of the LD score as the regression weight (Bulik-Sullivan et al., 2015). is the Haseman–Elston regression estimator based on individual genotypes. formulates SNP coheritability estimation as a polygenic score analysis, and thus enables coheritability analysis when GWAS summary statistics are available for one trait and individual genotypes are available for the other trait. The equivalence between and has been established in prior work (Bulik-Sullivan 2015).
Statistical Genetic Analysis
For all heritability and genetic correlation analyses, we used SNPs in the HapMap3 panel whose LD scores have been computed and released as part of the LD score regression software. We further filtered out genetic markers with imputation quality score <0.9, missing rate >1%, minor allele frequency <1%, and significant deviation from Hardy–Weinberg equilibrium (P < 1 × 10−10) in the UK Biobank. A total of 870 962 SNPs were used in the heritability and genetic correlation analyses.
The SNP heritability of educational attainment, denoted as , was computed using the LD score regression estimator in Eq. (1) and the summary statistics of the education GWAS in the UK Biobank. The SNP heritability of the verbal–numerical reasoning score was computed similarly. The SNP heritability of the cortical thickness measurement at vertex , denoted as , was computed using the Haseman–Elston regression estimator in Eq. (1) and individual genotypes of the imaging sample (N = 7818). We adjusted for age (at the imaging visit), sex, age2, age × sex, age2×sex, handedness, genotype array, and top 10 PCs of the genotype data as covariates. We also controlled for the TBV to remove global genetic influences on brain size. Vertex-wise estimates , , where, V is the total number of vertices, form a surface map for the heritability of cortical thickness measurements. The surface map for the heritability of surface area measurements was constructed similarly.
The SNP coheritability between educational attainment and the cortical thickness measurement at vertex , denoted as , were computed using the estimator in Eq. (2). More specifically, the summary statistics of the education GWAS (N = 332 613) were used to calculate an individual-specific polygenic score in the imaging sample (N = 7818) where individual genotypes were available. The polygenic score was then correlated with the cortical thickness measurement at each cortical location and properly scaled to produce the coheritability estimate . Since there was no overlap between the education GWAS sample and the neuroimaging sample, the bias term in the estimator, that is, , was set to zero. We adjusted for age (at the imaging visit), sex, age2, age × sex, age2 × sex, handedness, TBV, genotype array, and top 10 PCs of the genotype data as covariates in the coheritability (polygenic score) analysis. The genetic correlation between educational attainment and the cortical thickness measurement at each vertex was then computed as follows:
| (3) |
The genetic correlations between the verbal–numerical reasoning score and cortical thickness measurements were computed similarly.
Vertex-wise estimates , , form a surface map for the genetic correlations between educational attainment and cortical thickness measurements. Clusters on the surface map can be defined by spatially contiguous vertices with P-values below a threshold. To assess the significance of the size (number of vertices) of a cluster while accounting for the spatial correlation of cortical thickness measurements, we employed the following permutation procedure. We recomputed and thresholded the P-value map using a permuted polygenic score, and recorded the maximal cluster size across the 2 hemispheres for each permutation . For an observed cluster with size , the family-wise error (FWE) corrected P-value was then computed as follows (Westfall and Young 1993):
| (4) |
10 000 permutations were used in this study. We repeated the genetic correlation analyses using vertex-wise surface area measurements.
Results
GWAS of Educational Attainment
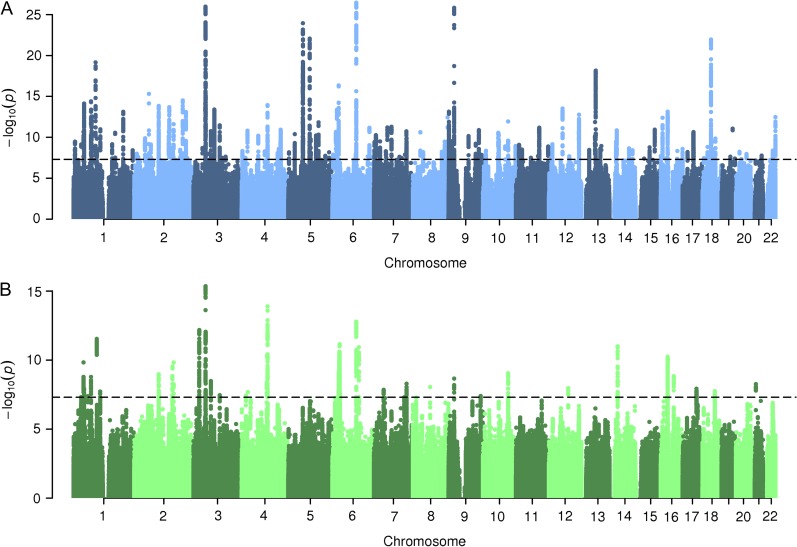
Genome-wide association analysis of educational attainment (years of schooling completed; N = 332 613) in the UK Biobank identified 158 independent genome-wide significant loci. Figure 1A shows the Manhattan plot for the GWAS. Supplementary Figure S1 provides additional information on each of the genome-wide significant regions. The heritability of educational attainment was estimated to be 0.156 (s.e. 0.004).
Figure 1.
(A) Manhattan plot for the genome-wide association analysis of educational attainment (years of schooling completed) in the UK Biobank (N = 332 613). (B) Manhattan plot for the genome-wide association analysis of the verbal–numerical reasoning score in the UK Biobank (N = 108 147). In both panels, the dash line indicates the genome-wide significant threshold P < 5 × 10–8.
Heritability of Cortical Thickness
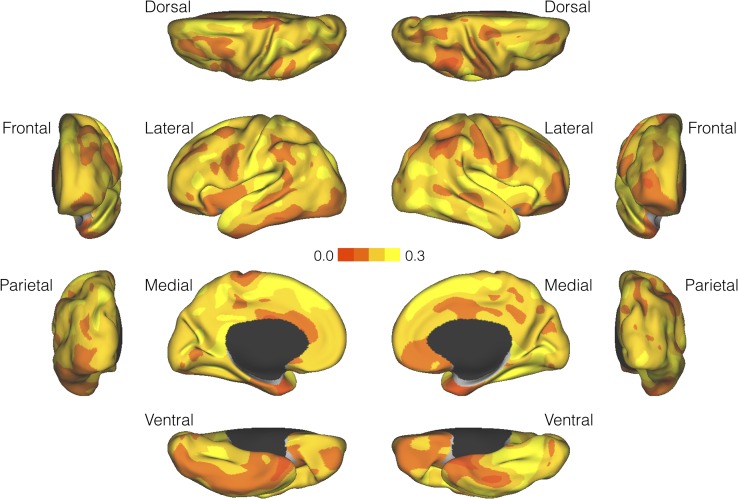
We estimated the SNP heritability of vertex-wise cortical thickness measurements using an unbiased and computationally efficient moment-matching method. As an empirical justification, our method produced virtually identical heritability estimates to LD score regression when applied to the average cortical thickness measurements in 68 regions of interest (ROIs; 34 ROIs per hemisphere) defined by the Desikan-Killiany atlas (Desikan et al. 2006) (Supplementary Fig. S2, left; also see Supplementary Material for a theoretical treatment). As shown in Figure 2, fine-grained cortical thickness measurements were moderately heritable across the cortical mantle.
Figure 2.
Surface maps for the SNP heritability of cortical thickness measurements (N = 7818).
Genetic Correlation Between Educational Attainment and Cortical Thickness
Given that educational attainment and cortical thickness measurements were both heritable, we sought to examine whether they have a shared genetic basis. An empirical comparison of the genetic correlation between educational attainment and the average cortical thickness measurement in each of the 68 Desikan-Killiany ROIs estimated by the proposed polygenic score analysis and LD score regression showed that the 2 methods produced almost identical estimates (Supplementary Fig. S2, right). Theoretical equivalence between the 2 methods is established in Supplementary Material.
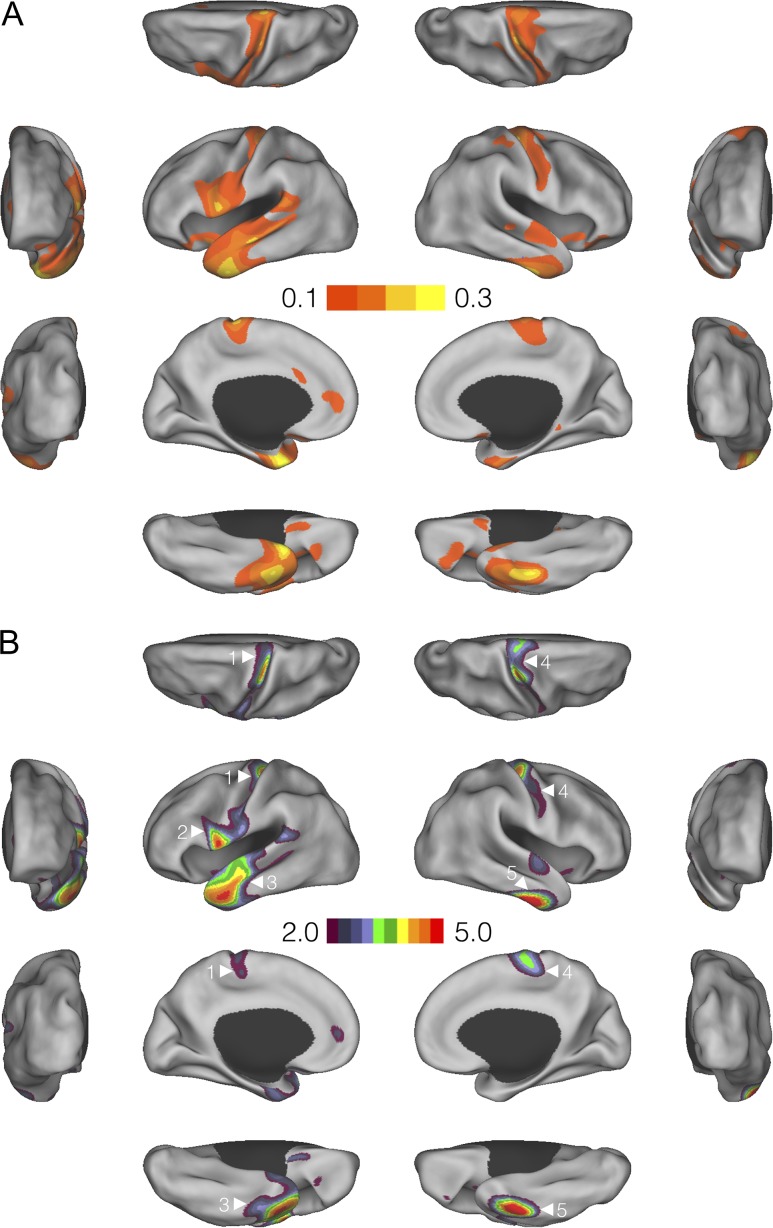
Figure 3A and B shows surface maps for the genetic correlation and its statistical significance between educational attainment and cortical thickness measurements, respectively. Moderate and positive genetic correlations were observed in bilateral motor cortex and predominantly left superior temporal cortex and proximal regions. We thresholded the significance map using P = 0.01 as the threshold (Fig. 3B), and assessed the significance of the size of each identified cluster (spatially contiguous vertices) and computed their FWE corrected P-values using a permutation procedure. Statistically significant clusters were observed in bilateral primary motor cortex (cluster 1, PFWE = 0.033; cluster 2, PFWE = 0.042; and cluster 4, PFWE = 0.012). Cluster 2 on the left hemisphere also extended into the pars opercularis (also known as Brodmann area 44 or BA44), which is part of the Broca’s speech area. In addition, cluster 3 (PFWE = 0.005) spanned the left temporal pole and superior temporal cortex, and overlapped with the auditory cortex and the Wernicke’s language area. The right inferiortemporal cortex (cluster 5) also showed a strong genetic correlation with educational attainment but did not survive multiple testing correction (PFWE = 0.110). Adjusting for Townsend deprivation index, a proxy for socioeconomic status (SES), as a covariate in the analyses produced highly similar results (Supplementary Fig. S3).
Figure 3.
Vertex-wise genetic correlations between educational attainment and cortical thickness measurements. (A) Surface maps for the genetic correlation estimates. (B) Surface maps for the –log10P-values of the genetic correlations. Clusters identified by a cluster-forming threshold of P = 0.01 are shown. Family-wise error corrected significant (or marginally significant) clusters are annotated.
Analyses of the Verbal–Numerical Reasoning Score
Analyses of educational attainment indicated that cortical thickness in several regions, including the primary motor cortex and Broca’s speech and Wernicke’s language areas, may have shared genetic origins with cognitive ability. We thus conducted a secondary analysis using the verbal–numerical reasoning score in the UK Biobank, which captures general cognitive ability but particularly knowledge of learned material, to investigate whether the variation in years of schooling that is genetically correlated with cortical thickness is related to individual differences in cognitive performance.
Genome-wide analysis of the verbal–numerical reasoning score in the UK Biobank (N = 108 147) identified 35 genome-wide significant loci, among which 17 overlapped with the genomic loci identified in the educational attainment GWAS. Figure 1B shows the Manhattan plot for the GWAS. Supplementary Figure S4 provides additional information on each of the genome-wide significant regions. The heritability of the verbal–numerical reasoning score was 0.247 (s.e. 0.008). Educational attainment and the verbal–numerical reasoning score were both phenotypically (Pearson correlation r = 0.353; N = 110 233; P < 1e-323) and genetically (genetic correlation rg = 0.710; s.e. 0.016; P < 1e-323) correlated.
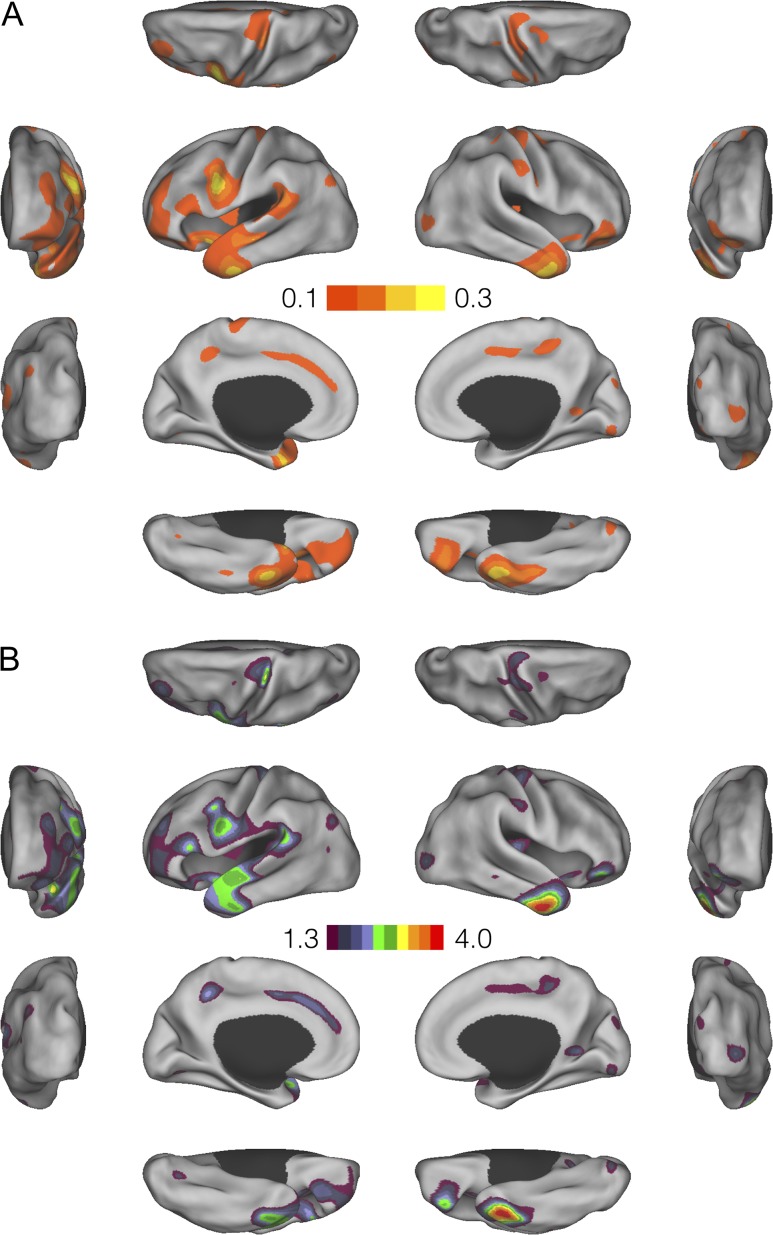
Figure 4A and B shows surface maps for the genetic correlation and its statistical significance (thresholded at the nominal P = 0.05) between the verbal–numerical reasoning score and cortical thickness measurements, respectively. Both maps showed similar patterns to the surface maps for educational attainment, with positive genetic correlations observed in the predominantly left inferior precentral gyrus (including Broca’s speech area), superior temporal cortex (including auditory cortex), supramarginal gyrus (including Wernicke’s language area), and proximal regions. The genetic correlation estimates showed a similar range to those between educational attainment and cortical thickness measurements, but were less statistically significant due to the reduced power afforded by the smaller number of subjects in the verbal–numerical reasoning GWAS relative to the education GWAS.
Figure 4.
Vertex-wise genetic correlations between the verbal–numerical score and cortical thickness measurements. (A) Surface maps for the genetic correlation estimates. (B) Surface maps for the −log10P-values of the genetic correlations, thresholded at uncorrected P = 0.05.
Surface area Analyses
We repeated all analyses using surface area measurements. Vertex-wise surface area measurements were substantially less heritable than cortical thickness measurements (Supplementary Fig. S5). In contrast to cortical thickness, no significant cluster of genetic correlations between educational attainment and surface area measurements was identified (all PFWE > 0.10; Supplementary Fig. S6).
Discussion
In this article, we examined the genetic overlap between educational attainment and fine-grained brain morphological measurements. Leveraging the large-scale brain imaging and genomic data in the UK Biobank, we found a shared genetic basis between years of schooling and cortical thickness measurements in bilateral primary motor cortex and predominantly left superior temporal cortex. A secondary analysis of the verbal–numerical reasoning score confirmed that the variation underlying education achievement that is genetically correlated with cortical thickness is related to individual differences in cognitive performance.
Although educational attainment is a complex behavioral trait that is linked to intelligence, personality, family environments, and many social factors, it is highly phenotypically and genetically correlated with intelligence (Okbay et al. 2016; Savage et al. 2017; Sniekers et al. 2017), and has been successfully used as a proxy phenotype to identify genetic variants associated with general cognitive ability (Rietveld et al. 2014). Recent studies have associated the polygenic scores of educational attainment with cognitive test scores and brain size (Belsky et al. 2016; Selzam et al. 2017; Plomin and von Stumm 2018), and further implicated that genetic influences on educational outcomes might be mediated through brain development and intelligence (Elliott et al. 2018). Within the UK biobank, tests of cognitive functioning were brief and only completed by a subsample of the participants. In contrast, educational qualifications, which can be mapped directly to years of schooling, were available for virtually all UK Biobank participants. Therefore, we leveraged educational attainment as a proxy for general cognitive ability in the identification of genetic overlap between cerebral cortical morphology and cognitive performance. We used this trait in our discovery analysis because its size and objective reliability would boost the statistical power relative to the brief cognitive screening tests with substantially smaller sample sizes in the UK Biobank.
Of the 13 questions that comprise the UK Biobank verbal–numerical reasoning score, 6 relate to numeric reasoning, 4 relate to vocab/verbal reasoning, and 3 are logic questions. Conventionally, the 10 verbal and numeric reasoning questions would fall within the domain of crystallized ability, which reflects learned knowledge, including material absorbed in the educational setting (Wilhelm 2004). Logic questions relate to novel problem-solving, and fall within the domain of fluid intelligence (Wilhelm 2004). Thus, our cognition measure, which is labeled as “fluid intelligence” in the UK Biobank directory, captures both crystallized and fluid intelligence (the major domains in a commonly accepted model of general ability) (Horn and Cattell 1966), but is more heavily loaded towards crystallized knowledge. Given that crystallized ability would be expected to increase over time whereas fluid ability would decrease, our conceptualization of this measurement is consistent with Lyall et al. (2016) that this score in the UK Biobank improved slightly over a roughly 4-year period. Measurements of crystallized ability are well-known to relate to educational attainment, and more recently measurements of fluid ability have been associated with this outcome (Kaufman et al. 2009). As such, educational attainment was considered well-suited to serve as a proxy index of this cognitive ability measurement in our analyses. This rationale was supported by the large genetic correlation between these measures in our sample. That said, future studies that map the genetic correlations between cortical morphology and comprehensive cognitive measures will be interesting.
Our analyses of educational attainment and the verbal–numerical reasoning score produced highly similar surface maps for genetic correlations and localized a common genetic basis between cognitive performance and cortical thickness measurements in bilateral primary motor cortex and predominately left inferior precentral gyrus, pars opercularis, superior temporal cortex, supramarginal gyrus, and their adjacent regions. Intriguingly, some of these regions overlap with the auditory cortex, Broca’s speech area, and Wernicke’s language area, suggesting that educational attainment and our cognitive measurement may have common genetic origins with auditory and language-related brain regions. The distinctiveness of language and cognition has been debated over the 20th century (Harris 2003). Our data provides evidence of potentially shared biological underpinnings that, if confirmed, could contribute to this line of inquiry. The primary motor cortex and its vicinity have been found to be consistently activated during reasoning tasks (Acuna et al. 2002; Prado et al. 2011), and implicated in lesion mapping of intelligence (Glascher et al. 2010; Woolgar et al. 2010). Therefore, our results also suggest that cognitive performance may have a shared genetic basis with brain regions involved in motor processes.
Previous studies have used neuroimaging to examine the relationships between intelligence, cognitive ability, education, and brain structure in vivo (Luders et al. 2009; Cox et al. 2016; Sabuncu et al. 2016). Higher intelligence has been associated with larger brains (Wickett et al. 2000; McDaniel 2005; Witelson et al. 2006; Rushton and Ankney 2009; Pietschnig et al. 2015), and greater total and regional gray matter volumes (Andreasen et al. 1993; Flashman et al. 1997; MacLullich et al. 2002; Colom et al. 2006; Haier et al. 2004, 2009). Beyond TBV, positive correlations have been reported between scores of cognitive tests and cortical thickness measurements in a number of cortical regions (Narr et al. 2007; Choi et al. 2008; Karama et al. 2009; Joshi et al. 2011; Menary et al. 2013). Our results expanded this literature by identifying brain regions in which cortical thickness measurements are genetically correlated with cognitive performance above and beyond global brain volumetric measurements. Cortical surface area has also been associated with general cognitive ability (Colom et al. 2013; Fjell et al. 2013; Schnack et al. 2014), and some recent studies have suggested that the phenotypic and genetic correlations between brain volume and cognitive performance are predominantly driven by surface area rather than cortical thickness (Vuoksimaa et al. 2014; Cox et al. 2018); however, we did not observe significant genetic correlations between educational attainment and surface area measurements in our analyses.
We note that we used different methods to estimate heritability for our imaging and nonimaging data due to computational considerations. As shown by prior studies (Bulik-Sullivan 2015; Ge, Chen, et al. 2017) and in this paper (see Supplementary Material and Supplementary Fig. S2), LD score regression and Haseman–Elston regression for heritability estimation are asymptotically equivalent under certain conditions. LD score regression is flexible and computationally efficient when summary statistics of large-scale GWAS are available. Haseman–Elston regression requires individual-level genotypes but is particularly useful when analyzing massive numbers of phenotypes and conducting GWAS for each phenotype is computationally infeasible. Given their equivalence, we selected the most appropriate method for each analysis based on joint consideration of data types available and computational cost.
A major contribution of this study is that we developed a novel statistical method to examine the shared genetic basis between educational attainment and cortical morphology at high spatial resolution. Prior studies along this line used the twin design and largely focused on global brain volumes. Existing methods for genetic correlation analyses in unrelated individuals either require individual-level genotypes (e.g., GCTA or GREML) (Yang et al. 2011) or GWAS summary statistics (e.g., LD score regression) (Bulik-Sullivan et al., 2015) for both traits, which become challenging to apply in high-dimensional settings. We filled this technical gap by formulating genetic correlation estimation as a polygenic score analysis. More specifically, the summary statistics of the educational attainment (or verbal–numerical reasoning) GWAS were used to weight individual genotypes of the imaging sample and calculate an individual-specific polygenic score. The polygenic score was then correlated with the morphological measurement at each cortical location and properly scaled and normalized to produce a genetic correlation estimate. Our method is thus highly computationally efficient and can be applied to estimate the genetic correlation between any trait (e.g., a cognitive, behavioral, or disease phenotype) whose GWAS summary statistics are available, and a high-dimensional phenotype, such as the MRI-derived vertex-wise cortical thickness or surface area measurements in the present study. Permutation procedures can also be devised to enable flexible statistical inferences, such as the cluster-wise analysis (Friston et al. 1994, 1996) on the surface map of genetic correlations. Although cluster-wise inference combined with the relatively large amount of smoothing applied in this study may reduce spatial resolution, this approach follows our prior expectation that genetic influences on cortical morphology are spatially distributed. Standard polygenic score analyses may achieve similar results and findings but our method avoids the selection of the GWAS P-value threshold (or screening multiple thresholds) used in the calculation of polygenic scores, and produce more interpretable estimates (i.e., genetic correlation) at no additional computational cost.
To apply this new genetic correlation estimation method, we conducted genome-wide association analyses of educational attainment and the verbal–numerical reasoning score in the UK Biobank, and identified 158 and 35 independent genome-wide significant loci, respectively. Previous studies have conducted large-scale genome-wide meta-analyses of educational attainment (Okbay et al. 2016) and general intelligence (Savage et al. 2017; Sniekers et al. 2017), and GWAS with a similar or larger total sample size than the present study exists. Although we could have leveraged the summary statistics of existing GWAS in our analyses, we performed GWAS in the UK Biobank to exclude all participants that had neuroimaging data, and thus protected the genetic correlation estimates from potential bias induced by sample overlap.
Findings in this analysis should be generalized with caution to populations with different sample characteristics with respect to age range, sex composition, ancestry groups, SES, or other environmental exposures. Educational attainment in our analysis reflects life-time academic achievement, while both human intelligence and the cortex undergo rapid development in childhood and adolescence, and age-related decline and degeneration in late adulthood. More importantly, a number of studies have found that the relationship between cognitive ability and brain morphology is dynamic over time and sex-dependent (Reiss et al. 1996; Gur et al. 1999; Shaw et al. 2006; Witelson et al. 2006; Burgaleta et al. 2014). In this study, we controlled for age, sex, age2, age × sex, age2 × sex in all the analyses to remove the (potentially nonlinear) effect of age and sex on cognitive performance, brain structure, and their correlations. However, since the UK Biobank only recruited middle- and older-aged participants, our results may not be generalizable to other age ranges. In addition, genetic influences on cognitive performance may be moderated by educational opportunities and SES (Deary et al., 2010; Hanscombe et al. 2012; Von Stumm and Plomin 2015). Although adjusting for Townsend deprivation index, a measure of material deprivation that can serve as a proxy for SES, did not have a notable impact on the results, our findings should be interpreted in light of the fact that UK Biobank participants are on average more educated and have higher SES than the general population (Tyrrell et al. 2016; Ge et al., 2017).
Although we identified genetic overlap between educational attainment and cortical thickness measurements in several brain regions, these correlations do not necessarily indicate causal relationships. Future work is needed to shed light on whether genetic influences on cognitive performance are mediated through brain morphology. Also, genetic correlation is a genome-wide metric and does not provide any information about specific genes that might underlie both cognitive ability and brain structure. Further statistical and molecular genetic analyses are needed to dissect their genetic overlap. Lastly, in addition to gray matter volumes, cortical thickness and surface area measurements, white matter volumes, diffusion tensor imaging derived measurements, functional MRI task activations, and indices of complex brain networks have also been associated with cognitive performance (Luders et al. 2009; Deary et al. 2010). Given that a range of features derived from brain morphology, resting state networks, and the structural and functional connectomes are substantially heritable (Glascher et al., 2010; Thompson et al. 2013; Kochunov et al. 2015; Ge et al. 2015, 2016; Ge, Holmes, et al. 2017), integration of multimodal imaging data might provide further insights into possible neural mechanisms of educational attainment and cognitive ability.
Supplementary Material
Funding
This research was carried out in part at the Athinoula A. Martinos Center for Biomedical Imaging at the Massachusetts General Hospital (MGH), using resources provided by the Center for Functional Neuroimaging Technologies, P41EB015896, a P41 Biotechnology Resource Grant supported by the National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH). This work involved the use of instrumentation supported by the NIH Shared Instrumentation Grant Program (Grant numbers S10RR023043 and S10RR023401), and the Enterprise Research Infrastructure & Services (ERIS) at Partners Healthcare. This research was also funded in part by NIH Grants K99AG054573 (T.G.); R01MH095904 and R01MH109562 (D.J.H.); R01AG053949 and R01LM012719 (M.R.S.); and K24MH094614 (J.W.S.). J.W.S. is a Tepper Family MGH Research Scholar and was also supported in part by a gift from the Demarest Lloyd, Jr. Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article. This research has been conducted using the UK Biobank resource under an approved data request (ref: 32 568; previously 13 905).
Notes
Conflict of Interest: None declared.
References
- Acuna BD, Eliassen JC, Donoghue JP, Sanes JN. 2002. Frontal and parietal lobe activation during transitive inference in humans. Cereb Cortex. 12(12):1312–1321. [DOI] [PubMed] [Google Scholar]
- Adams HHH, Hibar DP, Chouraki V, Stein JL, Nyquist PA. 2016. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat Neurosci. 19(12):1569–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen NC, Flaum M, Swayze V 2nd, O’leary DS, Alliger R, Cohen G, Ehrhardt J, Yuh WT. 1993. Intelligence and brain structure in normal individuals. Am J Psychiatry. 150(1):130–134. [DOI] [PubMed] [Google Scholar]
- Belsky DW, Moffitt TE, Corcoran DL, Domingue B, Harrington H, Hogan S, Houts R, Ramrakha S, Sugden K, Williams BS, et al. . 2016. The genetics of success: how single-nucleotide polymorphisms associated with educational attainment relate to life-course development. Psychol Sci. 27(7):957–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B. Relationship between LD score and Haseman-Elston regression. bioRxiv, p. 018283, 2015.
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, ReproGen Consortium; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3, Duncan L, Perry JR, Patterson N, Robinson EB, et al. . 2015. An atlas of genetic correlations across human diseases and traits. Nat Genet. 47(11):1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics C, Patterson N, Daly MJ, Price AL, Neale BM. 2015. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 47(3):291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgaleta M, Johnson W, Waber DP, Colom R, Karama S. 2014. Cognitive ability changes and dynamics of cortical thickness development in healthy children and adolescents. Neuroimage. 84:810–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft C, Freeman C, Petkova D, Band G, Elliott LT Genome-wide genetic data on ∼500,000 UK Biobank participants. bioRxiv, p. 166298, 2017.
- Choi YY, Shamosh NA, Cho SH, DeYoung CG, Lee MJ, Lee JM, Kim SI, Cho ZH, Kim K, Gray JR, et al. . 2008. Multiple bases of human intelligence revealed by cortical thickness and neural activation. J Neurosci. 28(41):10323–10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colom R, Burgaleta M, Román FJ, Karama S, Álvarez-Linera J, Abad FJ, Martínez K, Quiroga MÁ, Haier RJ. 2013. Neuroanatomic overlap between intelligence and cognitive factors: morphometry methods provide support for the key role of the frontal lobes. Neuroimage. 72:143–152. [DOI] [PubMed] [Google Scholar]
- Colom R, Jung RE, Haier RJ. 2006. Distributed brain sites for the g-factor of intelligence. Neuroimage. 31(3):1359–1365. [DOI] [PubMed] [Google Scholar]
- Cox SR, Bastin ME, Ritchie SJ, Dickie DA, Liewald DC, Muñoz Maniega S, Redmond P, Royle NA, Pattie A, Valdés Hernández M, et al. . 2018. Brain cortical characteristics of lifetime cognitive ageing. Brain Struct Funct. 223(1):509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox SR, Dickie DA, Ritchie SJ, Karama S, Pattie A, Royle NA, Corley J, Aribisala BS, Valdés Hernández M, Muñoz Maniega S, et al. . 2016. Associations between education and brain structure at age 73 years, adjusted for age 11 IQ. Neurology. 87(17):1820–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deary IJ, Johnson W. 2010. Intelligence and education: causal perceptions drive analytic processes and therefore conclusions. Int J Epidemiol. 39(5):1362–1369. [DOI] [PubMed] [Google Scholar]
- Deary IJ, Penke L, Johnson W. 2010. The neuroscience of human intelligence differences. Nat Rev Neurosci. 11(3):201–211. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, et al. . 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 31(3):968–980. [DOI] [PubMed] [Google Scholar]
- Elliott ML, Belsky DW, Anderson K, Corcoran DT, Ge T A polygenic score for higher educational attainment is associated with larger brains. bioRxiv, p. 287490, 2018. [DOI] [PMC free article] [PubMed]
- Elston RC, Buxbaum S, Jacobs KB, Olson JM. 2000. Haseman and Elston revisited. Genet Epidemiol. 19(1):1–17. [DOI] [PubMed] [Google Scholar]
- Fischl B. 2012. Freesurfer. Neuroimage. 62(2):774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell AM, Westlye LT, Amlien I, Tamnes CK, Grydeland H, Engvig A, Espeseth T, Reinvang I, Lundervold AJ, Lundervold A, et al. . 2015. High-expanding cortical regions in human development and evolution are related to higher intellectual abilities. Cereb Cortex. 25(1):26–34. [DOI] [PubMed] [Google Scholar]
- Flashman LA, Andreasen NC, Flaum M, Swayze VW. 1997. Intelligence and regional brain volumes in normal controls. Intelligence. 25(3):149–160. [Google Scholar]
- Friston KJ, Holmes A, Poline JB, Price CJ, Frith CD. 1996. Detecting activations in PET and fMRI: levels of inference and power. Neuroimage. 4(3):223–235. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Worsley KJ, Frackowiak RSJ, Mazziotta JC, Evans AC. 1994. Assessing the significance of focal activations using their spatial extent. Hum Brain Mapp. 1(3):210–220. [DOI] [PubMed] [Google Scholar]
- Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, Sprosen T, Collins R, Allen NE. 2017. Comparison of sociodemographic and health-related characteristics of UK biobank participants with those of the general population. Am J Epidemiol. 186(9):1026–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T, Chen CY, Neale BM, Sabuncu MR, Smoller JW. 2017. Phenome-wide heritability analysis of the UK Biobank. PLoS Genet. 13(4):e1006711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T, Holmes AJ, Buckner RL, Smoller JW, Sabuncu MR. 2017. Heritability analysis with repeat measurements and its application to resting-state functional connectivity. Proc Natl Acad Sci USA. 114(21):5521–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T, Nichols TE, Lee PH, Holmes AJ, Roffman JL, Buckner RL, Sabuncu MR, Smoller JW. 2015. Massively expedited genome-wide heritability analysis (MEGHA). Proc Natl Acad Sci USA. 112(8):2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T, Reuter M, Winkler AM, Holmes AJ, Lee PH, Tirrell LS, Roffman JL, Buckner RL, Smoller JW, Sabuncu MR. 2016. Multidimensional heritability analysis of neuroanatomical shape. Nature Commun. 7:13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Winkler AM, Kochunov P, Almasy L, Duggirala R, Carless MA, Curran JC, Olvera RL, Laird AR, Smith SM, et al. . 2010. Genetic control over the resting brain. Proc Natl Acad Sci USA. 107(3):1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glascher J, Rudrauf D, Colom R, Paul LK, Tranel D, Damasio H, Adolphs R. 2010. Distributed neural system for general intelligence revealed by lesion mapping. Proc Natl Acad Sci USA. 107(10):4705–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan D, Lander ES, Rosset S. 2014. Measuring missing heritability: inferring the contribution of common variants. Proc Natl Acad Sci USA. 111(49):E5272–E5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur RC, Turetsky BI, Matsui M, Yan M, Bilker W, Hughett P, Gur RE. 1999. Sex differences in brain gray and white matter in healthy young adults: correlations with cognitive performance. J Neurosci. 19(10):4065–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haier RJ, Colom R, Schroeder DH, Condon CA, Tang C, Eaves E, Head K. 2009. Gray matter and intelligence factors: is there a neuro-g. Intelligence. 37(2):136–144. [Google Scholar]
- Haier RJ, Jung RE, Yeo RA, Head K, Alkire MT. 2004. Structural brain variation and general intelligence. Neuroimage. 23(1):425–433. [DOI] [PubMed] [Google Scholar]
- Hanscombe KB, Trzaskowski M, Haworth CMA, Davis OSP, Dale PS, Plomin R. 2012. Socioeconomic status (SES) and children’s intelligence (IQ): in a UK-representative sample SES moderates the environmental, not genetic, effect on IQ. PLoS One. 7(2):e30320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CL. 2003. Language and cognition In: Encyclopedia of cognitive science. London: MacMillan. [Google Scholar]
- Haseman JK, Elston RC. 1972. The investigation of linkage between a quantitative trait and a marker locus. Behav Genet. 2(1):3–19. [DOI] [PubMed] [Google Scholar]
- Horn JL, Cattell RB. 1966. Refinement and test of the theory of fluid and crystallized general intelligences. J Educ Psychol. 57(5):253–270. [DOI] [PubMed] [Google Scholar]
- Hulshoff pol HE, Schnack HG, Posthuma D, Mandl RCW, Baare WF, van Oel C, van Haren NE, Collins DL, Evans AC, Amunts K, et al. . 2006. Genetic contributions to human brain morphology and intelligence. J Neurosci. 26(40):10235–10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Leporé N, Joshi SH, Lee AD, Barysheva M, Stein JL, McMahon KL, Johnson K, de Zubicaray GI, Martin NG, et al. . 2011. The contribution of genes to cortical thickness and volume. Neuroreport. 22(3):101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karama S, Ad-Dab’bagh Y, Haier RJ, Deary IJ, Lyttelton OC, Lepage C, Evans AC. 2009. Positive association between cognitive ability and cortical thickness in a representative US sample of healthy 6 to 18 year-olds. Intelligence. 37:145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman AS, Kaufman JC, Liu X, Johnson CK. 2009. How do educational attainment and gender relate to fluid intelligence, crystallized intelligence, and academic skills at ages 22–90 years? Arch Clin Neuropsychol. 24(2):153–163. [DOI] [PubMed] [Google Scholar]
- Kochunov P, Jahanshad N, Marcus D, Winkler A, Sprooten E, Nichols TE, Wright SN, Hong LE, Patel B, Behrens T, et al. . 2015. Heritability of fractional anisotropy in human white matter: a comparison of Human Connectome Project and ENIGMA-DTI data. Neuroimage. 111:300–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapohl E, Rimfeld K, Shakeshaft NG, Trzaskowski M, McMillan A, Pingault JB, Asbury K, Harlaar N, Kovas Y, Dale PS, et al. . 2014. The high heritability of educational achievement reflects many genetically influenced traits, not just intelligence. Proc Natl Acad Sci USA. 111(42):15273–15278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luders E, Narr KL, Thompson PM, Toga AW. 2009. Neuroanatomical correlates of intelligence. Intelligence. 37(2):156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyall DM, Cullen B, Allerhand M, Smith DJ, Mackay D, Evans J, Anderson J, Fawns-Ritchie C, McIntosh AM, Deary IJ, et al. . 2016. Cognitive test scores in UK Biobank: data reduction in 480,416 participants and longitudinal stability in 20,346 participants. PLoS One. 11(4):e0154222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLullich AMJ, Ferguson KJ, Deary IJ, Seckl JR, Starr JM, Wardlaw JM. 2002. Intracranial capacity and brain volumes are associated with cognition in healthy elderly men. Neurology. 59(2):169–174. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B. 2010. Genotype imputation for genome-wide association studies. Nat Rev Genet. 11(7):499–511. [DOI] [PubMed] [Google Scholar]
- McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, et al. , Haplotype Reference Consortium . 2016. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 48(10):1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel M. 2005. Big-brained people are smarter: a meta-analysis of the relationship between in vivo brain volume and intelligence. Intelligence. 33(4):337–346. [Google Scholar]
- Menary K, Collins PF, Porter JN, Muetzel R, Olson EA, Kumar V, Steinbach M, Lim KO, Luciana M. 2013. Associations between cortical thickness and general intelligence in children, adolescents and young adults. Intelligence. 41(5):597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narr KL, Woods RP, Thompson PM, Szeszko P, Robinson D, Dimtcheva T, Gurbani M, Toga AW, Bilder RM. 2007. Relationships between IQ and regional cortical gray matter thickness in healthy adults. Cereb Cortex. 17(9):2163–2171. [DOI] [PubMed] [Google Scholar]
- Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA, Turley P, Chen GB, Emilsson V, Meddens SFW, et al. . 2016. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 533(7604):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietschnig J, Penke L, Wicherts JM, Zeiler M, Voracek M. 2015. Meta-analysis of associations between human brain volume and intelligence differences: how strong are they and what do they mean? Neurosci Biobehav Rev. 57:411–432. [DOI] [PubMed] [Google Scholar]
- Plomin R, von Stumm S. 2018. The new genetics of intelligence. Nat Rev Genet. 19:148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polderman TJC, Benyamin B, De Leeuw CA, Sullivan PF, Van Bochoven A, Visscher PM, Posthuma D. 2015. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet. 47(7):702–709. [DOI] [PubMed] [Google Scholar]
- Posthuma D, Baaré WFC, Hulshoff pol HE, Kahn RS, Boomsma DI, De Geus EJC. 2003. Genetic correlations between brain volumes and the WAIS-III dimensions of verbal comprehension, working memory, perceptual organization, and processing speed. Twin Res. 6(2):131–139. [DOI] [PubMed] [Google Scholar]
- Posthuma D, De Geus EJC, Baaré WFC, Pol HEH, Kahn RS, Boomsma DI. 2002. The association between brain volume and intelligence is of genetic origin. Nat Neurosci. 5(2):83–84. [DOI] [PubMed] [Google Scholar]
- Prado J, Chadha A, Booth JR. 2011. The brain network for deductive reasoning: a quantitative meta-analysis of 28 neuroimaging studies. J Cogn Neurosci. 23(11):3483–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss AL, Abrams MT, Singer HS, Ross JL, Denckla MB. 1996. Brain development, gender and IQ in children: a volumetric imaging study. Brain. 119(5):1763–1774. [DOI] [PubMed] [Google Scholar]
- Rietveld CA, Esko T, Davies G, Pers TH, Turley P, Benyamin B, Chabris CF, Emilsson V, Johnson AD, Lee JJ, et al. . 2014. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc Natl Acad Sci USA. 111(38):13790–13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton JP, Ankney CD. 2009. Whole brain size and general mental ability: a review. Int J Neurosci. 119(5):692–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabuncu MR, Ge T, Holmes AJ, Smoller JW, Buckner RL, Fischl B, Alzheimer’s Disease Neuroimaging I. 2016. Morphometricity as a measure of the neuroanatomical signature of a trait. Proc Natl Acad Sci USA. 113(39):E5749–E5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage JE, Jansen PR, Stringer S, Watanabe K, Bryois J GWAS meta-analysis (N=279,930) identifies new genes and functional links to intelligence. bioRxiv, p. 184853, 2017.
- Schnack HG, Van Haren NEM, Brouwer RM, Evans A, Durston S, Boomsma DI, Kahn RS, Hulshoff Pol HE. 2015. Changes in thickness and surface area of the human cortex and their relationship with intelligence. Cereb Cortex. 25(6):1608–1617. [DOI] [PubMed] [Google Scholar]
- Selzam S, Krapohl E, von Stumm S, O’reilly PF, Rimfeld K, Kovas Y, Dale PS, Lee JJ, Plomin R. 2017. Predicting educational achievement from DNA. Mol Psychiatry. 22(2):267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Greenstein D, Lerch J, Clasen L, Lenroot R, Gogtay N, Evans A, Rapoport J, Giedd J. 2006. Intellectual ability and cortical development in children and adolescents. Nature. 440(7084):676–679. [DOI] [PubMed] [Google Scholar]
- Sniekers S, Stringer S, Watanabe K, Jansen PR, Coleman JRI, Krapohl E, Taskesen E, Hammerschlag AR, Okbay A, Zabaneh D, et al. . 2017. Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence. Nat Genet. 49(7):1107–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, et al. . 2015. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12(3):e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Cannon TD, Narr KL, Van Erp T, Poutanen VP, Huttunen M, Lönnqvist J, Standertskjöld-Nordenstam CG, Kaprio J, Khaledy M, et al. . 2001. Genetic influences on brain structure. Nat Neurosci. 4(12):1253–1258. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Ge T, Glahn DC, Jahanshad N, Nichols TE. 2013. Genetics of the connectome. Neuroimage. 80:475–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga AW, Thompson PM. 2005. Genetics of brain structure and intelligence. Annu Rev Neurosci. 28:1–23. [DOI] [PubMed] [Google Scholar]
- Turner SD. qqman: an R package for visualizing GWAS results using QQ and manhattan plots. bioRxiv, 005165, 2014.
- Tyrrell J, Jones SE, Beaumont R, Astley CM, Lovell R, Yaghootkar H, Tuke M, Ruth KS, Freathy RM, Hirschhorn JN, et al. . 2016. Height, body mass index, and socioeconomic status: Mendelian randomisation study in UK biobank. BMJ. 352:i582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Stumm S, Plomin R. 2015. Socioeconomic status and the growth of intelligence from infancy through adolescence. Intelligence. 48:30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuoksimaa E, Panizzon MS, Chen CH, Fiecas M, Eyler LT, Fennema-Notestine C, Hagler DJ, Fischl B, Franz CE, Jak A, et al. . 2015. The genetic association between neocortical volume and general cognitive ability is driven by global surface area rather than thickness. Cereb Cortex. 25(8):2127–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Taskesen E, van Bochoven A, Posthuma D. 2017. Functional mapping and annotation of genetic associations with FUMA. Nature Commun. 8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PH, Young SS. 1993. Resampling-based multiple testing: examples and methods for p-value adjustment. New York: John Wiley & Sons. [Google Scholar]
- Wickett JC, Vernon PA, Lee DH. 2000. Relationships between factors of intelligence and brain volume. Pers Individ Dif. 29(6):1095–1122. [Google Scholar]
- Wilhelm O. 2004. Measuring reasoning ability In: Wilhelm O, Engle RW, editors. Handbook of understanding and measuring intelligence. London: Sage Publications; p. 373–392. [Google Scholar]
- Witelson SF, Beresh H, Kigar DL. 2006. Intelligence and brain size in 100 postmortem brains: sex, lateralization and age factors. Brain. 129(2):386–398. [DOI] [PubMed] [Google Scholar]
- Woolgar A, Parr A, Cusack R, Thompson R, Nimmo-Smith I, Torralva T, Roca M, Antoun N, Manes F, Duncan J. 2010. Fluid intelligence loss linked to restricted regions of damage within frontal and parietal cortex. Proc Natl Acad Sci USA. 107(33):14899–14902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. 2011. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 88(1):76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. 2017. A unified framework for variance component estimation with summary statistics in genome-wide association studies. Ann Appl Stat. 11(4):2027–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.