Abstract
Visuospatial working memory (WM), which is impaired in schizophrenia, depends on a distributed network including visual, posterior parietal, and dorsolateral prefrontal cortical regions. Within each region, information processing is differentially regulated by subsets of γ-aminobutyric acid (GABA) neurons that express parvalbumin (PV), somatostatin (SST), or vasoactive intestinal peptide (VIP). In schizophrenia, WM impairments have been associated with alterations of PV and SST neurons in the dorsolateral prefrontal cortex. Here, we quantified transcripts selectively expressed in GABA neuron subsets across four cortical regions in the WM network from comparison and schizophrenia subjects. In comparison subjects, PV mRNA levels declined and SST mRNA levels increased from posterior to anterior regions, whereas VIP mRNA levels were comparable across regions except for the primary visual cortex (V1). In schizophrenia subjects, each transcript in PV and SST neurons exhibited similar alterations across all regions, whereas transcripts in VIP neurons were unaltered in any region except for V1. These findings suggest that the contribution of each GABA neuron subset to inhibitory regulation of local circuitry normally differs across cortical regions of the visuospatial WM network and that in schizophrenia alterations of PV and SST neurons are a shared feature across these regions, whereas VIP neurons are affected only in V1.
Keywords: calretinin, parvalbumin, postmortem, somatostatin, vasoactive intestinal peptide
Introduction
Working memory (WM), the ability to transiently maintain and manipulate information to guide thought or behavior, is mediated by neural networks distributed across multiple cortical regions (Christophel et al. 2017). For example, the visuospatial WM network includes primary (V1) and association (V2) visual regions in the occipital cortex that encode low-level details of visual inputs, as well as higher order association regions, such as the posterior parietal cortex (PPC) and the dorsolateral prefrontal cortex (DLPFC) that process complex information (Eriksson et al. 2015; Christophel et al. 2017). Excitatory pyramidal neurons furnish the connections linking the cortical regions of the WM network, and inhibitory neurons that utilize γ-aminobutyric acid (GABA) shape pyramidal neuron firing during WM tasks (Rao et al. 2000; Constantinidis et al. 2002; Kim et al. 2016).
Cortical GABA neurons are classified into three major nonoverlapping subsets based on the expression of parvalbumin (PV), somatostatin (SST), or vasoactive intestinal peptide (VIP) (Pfeffer et al. 2013; Lake et al. 2016; Tremblay et al. 2016). These subsets appear to have distinct roles in cortical information processing. For example, PV neurons, via their perisomatic innervation of pyramidal neurons, regulate coincident input detection, response gain, and synchronization of nearby pyramidal neurons (Tremblay et al. 2016). In contrast, SST neurons, via their inputs to pyramidal neuron distal dendrites, modulate dendritic integration and contribute to processing and encoding of specific information by pyramidal neurons (Kim et al. 2016; Tremblay et al. 2016; Kamigaki and Dan 2017). Finally, VIP neurons, by virtue of their inhibitory synapses on SST neurons and to a lesser extent on PV neurons, provide disinhibition of pyramidal neurons (Pfeffer et al. 2013; Pi et al. 2013; Tremblay et al. 2016). In nonhuman primates, the neuronal density of each GABA neuron subset differs among cortical regions and these differences are thought to reflect regionally specific requirements of each subset for local information processing (Hayes et al. 1991; Kondo et al. 1994; Conde et al. 1996; Defelipe et al. 1999).
In subjects with schizophrenia, WM impairments, a core cognitive deficit in the illness (Silver et al. 2003; Barch and Ceaser 2012), have been attributed, at least in part, to altered inhibition in the DLPFC (Marin 2012), which appears to involve PV and SST neurons. First, multiple postmortem studies reported reduced levels of PV and SST mRNAs in the DLPFC of subjects with schizophrenia (Hashimoto et al. 2003; Morris et al. 2008; Mellios et al. 2009; Fung et al. 2010, 2014; Guillozet-Bongaarts et al. 2014; Chung et al. 2016b). Second, reduced transcript levels were also reported for KCNS3, a potassium channel subunit that is highly selective to PV neurons in the human neocortex (Georgiev et al. 2014), and LIM homeobox 6 (LHX6) transcription factor (Volk et al. 2012b), which is selectively expressed in both PV and SST neurons in the human neocortex (Georgiev et al. 2012). Third, μ-opioid receptor (MOR), which was primarily detected in most PV neurons and some SST neurons in the rodent hippocampus (Drake and Milner 2002; Stumm et al. 2004), exhibited elevated transcript levels in the DLPFC of schizophrenia subjects (Volk et al. 2012b). Finally, levels of GAD67, an enzyme for GABA synthesis, was reduced in PV neurons (Hashimoto et al. 2003; Curley et al. 2011). In contrast, transcripts selective for VIP neurons, including VIP and calretinin (CR) (Kubota et al. 1994; Gabbott and Bacon 1997; Lake et al. 2016), are relatively unaltered in the DLPFC of schizophrenia subjects (Hashimoto et al. 2003; Fung et al. 2010, 2014; Guillozet-Bongaarts et al. 2014; Chung et al. 2016b).
Given the functional differences among cortical regions of the visuospatial WM network, the distinct roles of the GABA neuron subsets in cortical processing, and the regionally different densities of each GABA neuron subset, we asked how the contribution of each GABA neuron subset to local circuitry differs across cortical regions of the WM network in unaffected comparison subjects. Furthermore, in schizophrenia, the selective alterations in PV and SST neurons in the DLPFC raise the question of whether the neural substrate for visuospatial WM dysfunction involves shared or distinct regional alterations in GABA neuron subsets. To answer these questions, we quantified transcript levels for markers of each GABA neuron subset across four cortical regions (V1, V2, PPC, and DLPFC) of the visuospatial WM network from 20 matched pairs of unaffected comparison and schizophrenia subjects.
Materials and Methods
Human Subjects
Brain specimens from unaffected comparison (n = 20) and schizophrenia subjects (n = 20) were obtained during autopsies conducted at the Allegheny County Medical Examiner’s Office, Pittsburgh, PA after consent was obtained from the next of kin. An independent committee of experienced research clinicians verified the absence of any psychiatric diagnosis, or made consensus DSM-IV diagnoses, for each subject on the basis of structured diagnostic interviews with family members and medical records. To reduce biological and control for experimental variances, each schizophrenia subject was matched with an unaffected comparison subject for sex and, as closely as possible for age and postmortem interval, and tissue samples from both members of each pair were always processed together. The mean age, postmortem interval, RNA integrity number (Bioanalyzer, Agilent Technologies), brain pH, and tissue storage time did not differ between subject groups (Table 1, Supplementary Table 1). All procedures were approved by the University of Pittsburgh’s Committee for the Oversight of Research and Clinical Training Involving the Dead and Institutional Review Board for Biomedical Research, as well as by the Ethics Committee of Kanazawa University Graduate School of Medical Sciences.
Table 1.
Summary of demographic and postmortem characteristics of human subjects
| Parameter | Unaffected comparison subjects | Subjects with schizophrenia | Statistics |
|---|---|---|---|
| Number | 20 | 20 | N/A |
| Sex, male/female | 14/6 | 14/6 | N/A |
| Race, white/black | 15/5 | 14/6 | χ 2 1 = 0.1; P = 0.72 |
| Age, years | 45.4 ± 11.6 | 44.3 ± 10.4 | t 38 = –0.3; P = 0.74 |
| PMI, hours | 15.4 ± 5.8 | 14.3 ± 6.4 | t 38 = –0.6; P = 0.57 |
| RIN | 8.3 ± 0.5 | 8.3 ± 0.6 | t 38 = –0.2; P = 0.87 |
| Brain pH | 6.7 ± 0.3 | 6.5 ± 0.3 | t 38 = –1.7; P = 0.10 |
| Storage time at –80 °C, Months | 111.9 ± 41.3 | 113.0 ± 48.8 | t 38 = 0.1; P = 0.94 |
Values are n or mean ± SD.
N/A, not applicable; PMI, postmortem interval; RIN, RNA integrity number.
Tissue Preparation
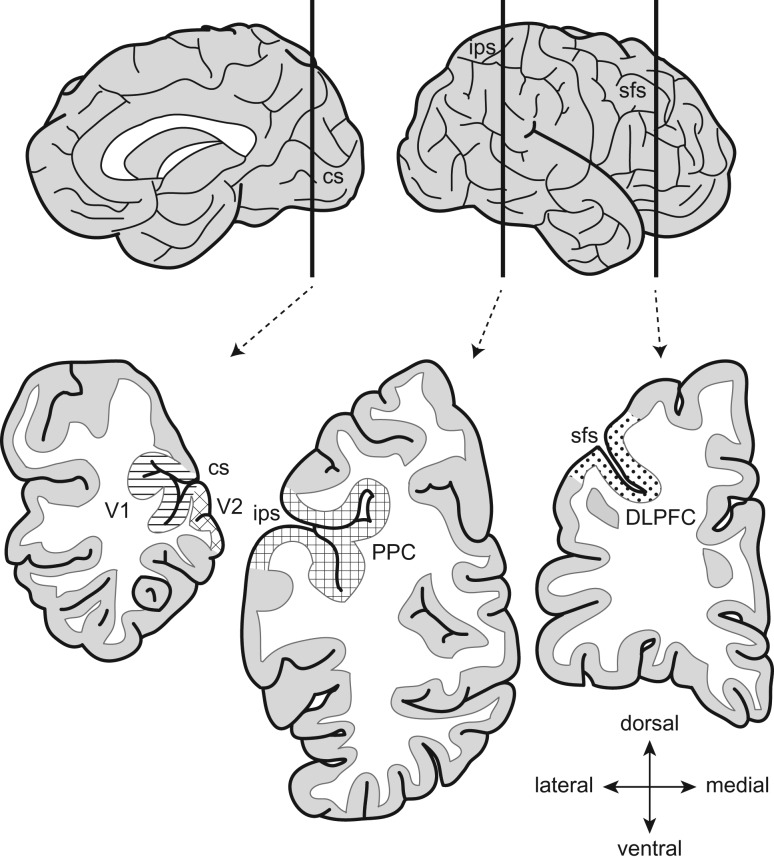
The right hemisphere of each brain was blocked coronally, immediately frozen, and stored at −80 °C. Four cortical regions (V1, V2, PPC, and DLPFC) were identified in Nissl-stained sections from the immersion-fixed left hemisphere based on cytoarchitectonic features and then sampled from the homotopic region in the fresh-frozen right hemisphere (Fig. 1); the identity of each sampled region was confirmed by Nissl staining of the adjacent sections (Hoftman et al. 2018). The gray matter (GM) of each region was dissected from 40-μm-thick cryostat sections in areas where the tissue was cut perpendicular to the pial surface and immediately homogenized in TRIzol reagent (Invitrogen).
Figure 1.
Cortical regions for RNA extraction. Schematic medial (left) and lateral (right) views of the right hemisphere of the human brain (top). Vertical bars indicate the approximate position of coronal blocks (bottom). The locations of sampled regions are represented by different patterns. V1, primary visual cortex; V2, association visual cortex; PPC, posterior parietal cortex; DLPFC, dorsolateral prefrontal cortex; cs, calcarine sulcus; ips, intraparietal sulcus; sfs, superior frontal sulcus.
Real-Time PCR Analyses
Total RNA was isolated from the homogenate samples of each cortical region using RNeasy lipid tissue mini kit (Qiagen) and converted into cDNA with High Capacity RNA-to-cDNA Kit (Applied Biosystems). The fragments of each transcript were amplified using Power SYBR Green PCR Master Mix and StepOnePlus real-time PCR system (Applied Biosystems). All primer sets (Supplementary Table 2) amplified specific, single products in dissociation curve analyses and had amplification efficiency ≥96% in standard curve analyses. All amplified fragments had expected sizes. For each subject pair and each cortical region, eight target transcripts selectively expressed in GABA neuron subsets and two internal control transcripts (beta-actin and cyclophilin-A) were amplified in one qPCR run as quadruplicates. Cycle threshold (CT) of each transcript was determined as the average across quadruplicates.
As the difference in cycle threshold (dCT) between the CT of each target transcript and the mean CT of beta-actin and cyclophilin-A represents log2-transformed expression ratio of the target transcript to the geometric mean of the two control transcripts, the expression levels of each target transcript were determined as 2−dCT.
For each target transcript in each region, the difference in expression levels between comparison and schizophrenia subjects in each pair was determined as −ddCt [−(dCT for the schizophrenia subject −dCT for the comparison subject)], and the within-pair expression ratio of schizophrenia and comparison subjects was determined as 2−ddCT.
Statistical Analyses
Mixed models treating observations from the four regions for each subject as repeated measurements were used to account for the within-subject correlation among four regions, and characterize the within- and between-subject variability. To compare the target transcript levels across the four regions in comparison subjects, the mixed model included transcript level as the dependent variable, region as the fixed effect, and sex, age, PMI, storage time, RIN, and brain pH as covariates. F-tests were used to assess the overall regional effect, followed by post hoc pairwise comparisons. To assess if the expression of these transcripts was altered in schizophrenia, the mixed model included transcript level as the dependent variable, diagnosis, region, and diagnosis-by-region interaction as the fixed effects, and the same set of covariates. The diagnosis effect and diagnosis-by-region interaction effect were tested using F-tests, followed by post hoc comparisons in each region. The denominator degrees of freedom (57 for the evaluation of regional effects on transcript levels in comparison subjects and 114 for the assessment of the effect of schizophrenia on transcript levels), computed using SAS PROC MIXED (“containment”), correspond to effective sample sizes that are larger than the numbers of subjects (20 and 40, respectively) but smaller than the numbers of samples (80 and 160, respectively) (Jiang 2007). The Holm simultaneous inference procedure (Holm 1979) was used to correct for multiple comparisons across eight transcripts (Volk et al. 2000; Morris et al. 2008) and reported P values for each transcript have been adjusted to correspond to the family-wise error rate of 0.05.
Results
Expression of GABA Neuron Transcripts in Unaffected Subjects
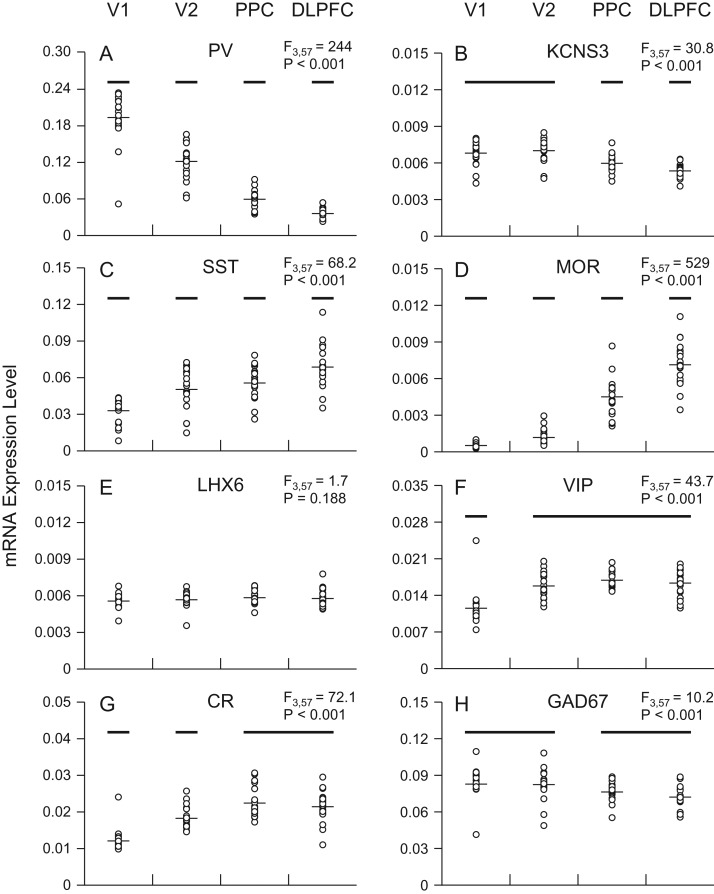
Transcripts selectively expressed in the three GABA neuron subsets exhibited distinctive expression patterns across the four regions of the visuospatial WM network. PV mRNA levels declined steeply from the posterior to anterior regions (F3,57 = 244, P < 0.001) and differed significantly among all four regions (Fig. 2A). Similarly, KCNS3 mRNA, which is selectively expressed in PV neurons in the human cortex (Georgiev et al. 2012), showed a posterior-to-anterior decline (F3,57 = 30.8, P < 0.001), although the differences in KCNS3 mRNA levels across regions were smaller and not significant between V1 and V2. KCNS3 mRNA levels were significantly higher in V1 and V2 than in PPC and significantly lower in DLPFC than in PPC (Fig. 2B).
Figure 2.
Expression levels of transcripts selective to GABA neuron subsets across the four cortical regions of the visuospatial WM network in unaffected comparison subjects. In each graph, transcript name is at the top center and the mixed model result at the top right. Across regions, transcript levels of individual subjects are plotted as open circles and group means as horizontal lines. For each transcript, regions not sharing the same horizontal bar are statistically different on post hoc comparisons. V1, primary visual cortex; V2, association visual cortex; PPC, posterior parietal cortex; DLPFC, dorsolateral prefrontal cortex.
In contrast to PV neuron markers, SST mRNA levels increased from the posterior to anterior regions (F3,57 = 68.2, P < 0.001) and differed significantly among all four regions (Fig. 2C). In a manner remarkably similar to SST mRNA, levels of MOR mRNA, reported to be present in both PV and SST neurons (Drake and Milner 2002; Stumm et al. 2004), increased from the posterior to anterior regions (F3,57 = 529, P < 0.001) (Fig. 2D). In contrast, levels of LHX6 mRNA, which is also expressed by both PV and SST neurons (Georgiev et al. 2012), did not differ across the cortical regions (F3,57 = 1.7, P = 0.19) (Fig. 2E).
The expression of VIP mRNA was comparable across V2, PPC, and DLPFC, but significantly lower in V1 (F3,57 = 43.7, P < 0.001) (Fig. 2F). Similarly, CR mRNA, which is selectively expressed in most VIP neurons in the primate cortex (Gabbott and Bacon 1997; Lake et al. 2016), showed comparable expression levels in PPC and DLPFC, slightly but significantly lower levels in V2, and lowest levels in V1 (F3,57 = 72.1, P < 0.001) (Fig. 2G).
The levels of GAD67 mRNA, which is expressed in all three GABA neuron subsets, were significantly greater in visual (V1 and V2) than association (PPC and DLPFC) regions (F3,57 = 10.2, P < 0.001) (Fig. 2H).
Effect of Schizophrenia on GABA Neuron Transcript Levels in the Visuospatial WM Network
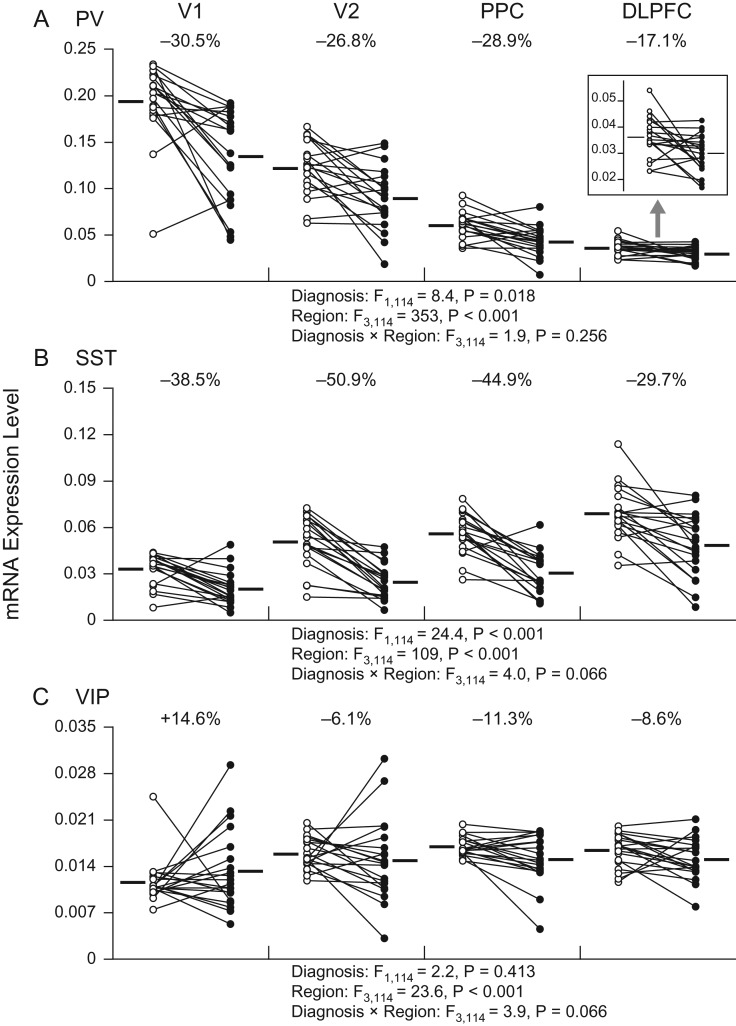
Relative to comparison subjects, schizophrenia subjects exhibited lower PV (−17% to −31%) and SST (−30% to −51%) mRNA levels in all four cortical regions of the visuospatial WM network (Fig. 3A,B). Our mixed model detected a significant effect of diagnosis for PV (F1,114 = 8.4, P = 0.02) and SST (F1,114 = 24.4, P < 0.001) mRNAs, but without significant diagnosis-by-region interaction effects (F3,114 = 1.9, P = 0.26 and F3,114 = 4.0, P = 0.07 for PV and SST mRNAs, respectively). In contrast, VIP mRNA expression levels did not show a significant effect of diagnosis (F1,114 = 2.2, P = 0.41) nor a diagnosis-by-region interaction (F3,114 = 3.9, P = 0.07) (Fig. 3C). These findings indicate that the effect of schizophrenia on transcript levels of the GABA neuron subset-specific markers is generally similar across the four cortical regions; however, given the marginal diagnosis-by-region interactions for SST and VIP mRNAs, we cannot exclude the presence of modest differences in expression levels across regions for these transcripts.
Figure 3.
Transcript levels in three GABA neuron subsets across the four cortical regions of the visuospatial WM network in subjects with schizophrenia and matched comparison subjects. Individual comparison and schizophrenia subjects are represented by open and filled circles, respectively, and the members of each pair are connected by lines. Horizontal bars indicate group means. The inset in panel A shows PV mRNA levels in the DLPFC with the expanded Y-axis for a better illustration of the effect of schizophrenia. V1, primary visual cortex; V2, association visual cortex; PPC, posterior parietal cortex; DLPFC, dorsolateral prefrontal cortex.
Similar to PV and SST mRNAs, KCNS3 and GAD67 mRNA levels were significantly lower by −10% to −22% (F1,114 = 21.6, P < 0.001) and by −8% to −19% (F1,114 = 9.3, P = 0.02), respectively, in schizophrenia, with similar effects of illness across the four regions (diagnosis-by-region interaction effect: F3,114 = 3.1, P = 0.12 and F3,114 = 2.3, P = 0.25 for KCNS3 and GAD67 mRNAs, respectively) (Fig. 4). MOR mRNA levels were significantly higher by +25% to +66% in schizophrenia across the four regions (diagnosis effect: F1,114 = 9.0, P = 0.02), without a significant regional difference in the magnitudes of upregulation (diagnosis-by-region interaction effect: F3,114 = 1.9, P = 0.26) (Fig. 4). For CR mRNA, although the effect of diagnosis was not significant (F1,114 = 1.8, P = 0.41), we detected a significant diagnosis-by-region interaction (F3,114 = 5.8, P < 0.01), indicating a region-specific effect of schizophrenia on CR expression (Fig. 4). Indeed, post hoc analysis revealed that CR levels were unaltered in V2, PPC, and DLPFC but were significantly upregulated by +41% in V1 (t114 = 3.4, P < 0.001) of subjects with schizophrenia. For LHX6 mRNA, we failed to detect a significant effect of diagnosis (F1,114 = 0.4, P = 0.56) or diagnosis-by-region interaction (F3,114 = 4.0, P = 0.07) (Fig. 4).
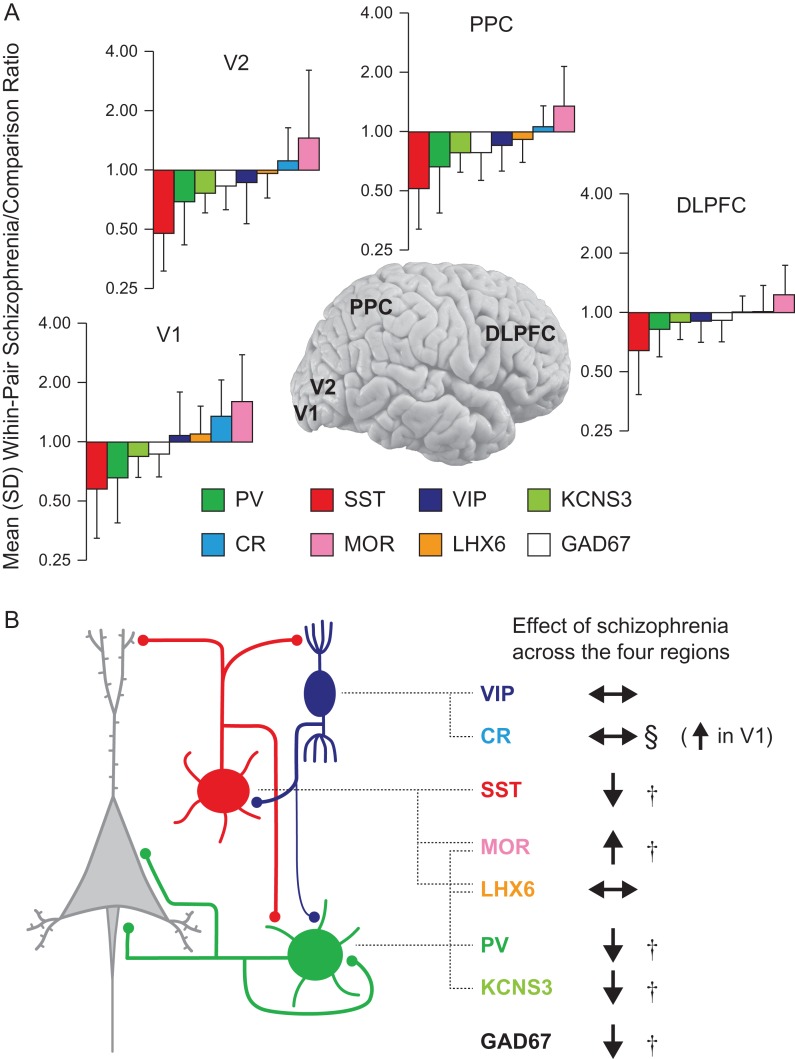
Figure 4.
Summary of findings. (A) Eight GABA neuron transcripts were rank-ordered by the magnitude of change in schizophrenia of their expression levels in each of the four cortical regions of the visuospatial WM network. Note that the rank orders are remarkably similar across the four regions; the only exception is the reversal of GAD67 and VIP mRNAs in DLPFC. V1, primary visual cortex; V2, association visual cortex; PPC, posterior parietal cortex; DLPFC, dorsolateral prefrontal cortex. (B) PV-, SST- and VIP-positive subsets of GABA neurons are schematically presented in green, red, and dark blue, respectively (left). Broken lines connect each GABA neuron subset and selectively expressed transcripts (middle). For each transcript, the effect of schizophrenia on its expression level across the WM network is indicated by arrows (right). † indicates significant diagnosis effect, § indicates significant diagnosis-by-region interaction effect.
Magnitudes of the Effect of Schizophrenia Across Cortical Regions
For each transcript in each region, the magnitude of the effect of schizophrenia was calculated as the mean within-pair, schizophrenia-to-comparison subject expression ratio across the 20 pairs. Then, for each region, the eight transcripts were rank-ordered by the magnitude of the differences (Fig. 4A). In all four regions, SST mRNA exhibited the largest decrease in schizophrenia, followed by PV, KCNS3, and GAD67 mRNAs. MOR mRNA showed the greatest increase in schizophrenia across all four regions. The increases of CR mRNA were smaller than those of MOR mRNA and were not significant across the regions except for V1. These findings indicate that in the visuospatial WM network the effect of schizophrenia on transcript levels in PV and SST neurons is conserved across the four cortical regions, whereas the effect on CR mRNA levels is selective to V1 (Fig. 4).
Discussion
Here, we quantified expression levels of eight transcripts selectively expressed in the three separate subsets of cortical GABA neurons (Fig. 4B) (Pfeffer et al. 2013; Lake et al. 2016; Tremblay et al. 2016) across four cortical regions of the visuospatial WM network from 20 matched pairs of comparison and schizophrenia subjects. Our findings are unique in that the examination of multiple regions within a well-described network revealed that 1) unaffected comparison subjects exhibit regional differences in the expression levels of these transcripts, 2) regional expression patterns are distinct across different classes of transcripts defined by their expression in each GABA neuron subset, and 3) the relative effect of schizophrenia on each transcript (except for CR) is conserved across nodes of the visuospatial WM network (Fig. 4).
In this study, transcript levels were determined in total GM of specific cortical regions from the right hemisphere. Although it is not possible to completely exclude a minor contribution of glial cells to the measured transcript levels, these cells should not represent a major confound, as previous in situ hybridization studies demonstrated that all transcripts studied here are predominantly expressed in neurons, but not in glial cells, in the primate neocortex (Benson et al. 1991; Akbarian et al. 1995; Peckys and Landwehrmeyer 1999; Volk et al. 2000; Hashimoto et al. 2003; Morris et al. 2008; Georgiev et al. 2012; Joshi et al. 2015). Furthermore, the current observations in the right DLPFC are consistent with previous analyses of the left DLPFC. For example, lower PV, SST, and GAD67 mRNA levels and unaltered CR mRNA levels in schizophrenia have been replicated in the studies of the right or left DLPFC (Akbarian et al. 1995; Mellios et al. 2009; Fung et al. 2010). In addition, recent studies using quantitative immunohistochemistry revealed reduced GAD67 and PV protein levels and unaltered CR protein levels in the left DLPFC of schizophrenia subjects (Curley et al. 2011; Enwright et al. 2016; Rocco et al. 2016; Chung et al. 2016a).
Transcript Expression Patterns Across WM Network in the Healthy State
In comparison subjects, PV and SST mRNAs exhibited decreasing and increasing posterior-to-anterior expression gradients, respectively, across the cortical regions, whereas VIP mRNA levels were comparable across the anterior three regions but significantly lower in V1 (Fig. 2). The regional expression patterns of PV and KCNS3 mRNAs and of VIP and CR mRNAs were similar, consistent with their coexpression in different subsets of GABA neurons (Kubota et al. 1994; Gabbott and Bacon 1997; Georgiev et al. 2012; Lake et al. 2016). Interestingly, prior studies in nonhuman primates demonstrated that the densities of each GABA neuron subset differ among cortical regions (Hayes et al. 1991; Kondo et al. 1994; Conde et al. 1996; Defelipe et al. 1999). For example, the density of PV neurons was reported to be greater in early visual regions compared with DLPFC (Conde et al. 1996), consistent with our findings of decreasing PV mRNA levels from posterior to anterior regions. In addition, both the densities of SST neurons and fibers were lower in V1 than in the superior frontal cortex (Hayes et al. 1991), consistent with our observation of an increasing posterior-to-anterior gradient of SST mRNA levels. Therefore, the cross-regional patterns of transcripts appear to represent the neuronal densities of GABA neuron subsets that express those transcripts across the regions of the WM network. Consequently, the absence of regional difference in LHX6 mRNA levels across cortical regions likely reflects its expression in both PV and SST neurons (Georgiev et al. 2012). Although MOR was detected in most PV neurons and some SST neurons in the rodent hippocampus (Drake and Milner 2002; Stumm et al. 2004), the selectivity of MOR expression to any GABA neuron subsets has not been assessed in the human cortex. However, the regional pattern of MOR mRNA expression that was strikingly similar to that of SST mRNA might suggest more predominant expression of MOR in SST neurons than in other GABA neurons in human neocortex.
The different regional patterns of PV, SST, and VIP mRNA levels might reflect regional differences in the relative number of synaptic targets for each GABA neuron subset. Although both PV and SST neurons promiscuously innervate most of neighboring pyramidal neurons within their axonal arbors (Karnani et al. 2014), they furnish inhibitory synapses on distinctive subcellular domains of these pyramidal neurons, perisomatic regions versus distal dendrites, respectively (Fig. 4B) (Tremblay et al. 2016). On the other hand, VIP neurons specifically target SST neurons and, to a lesser extent, PV neurons within their horizontally narrow, vertically elongated axon arbors (Fig. 4B) (Karnani et al. 2016; Tremblay et al. 2016). In primates, the densities of neurons in V1 and V2 are approximately three times and approximately two times higher, respectively, than in DLPFC, with the density in PPC intermediate (Haug 1987; Collins et al. 2010). Given that pyramidal neurons represent 75–80% of all neurons across the neocortex (Hendry et al. 1987), the densities of pyramidal neuron somata would be expected to decline from posterior to anterior regions. In our analysis, PV mRNA levels were 5.4 times, 3.4 times, and 1.7 times higher in V1, V2, and PPC, respectively, than in DLPFC (Fig. 2A). Given that PV neurons principally innervate the perisomatic region of pyramidal neurons, the regional pattern of PV mRNA levels suggests that the number of PV neurons varies directly with the number of pyramidal neuron somata across regions. In contrast, the posterior-to-anterior increase in SST mRNA levels (Fig. 2C) suggests that the ratio of SST neurons to pyramidal neurons is greater in anterior regions relative to posterior regions. This apparent mismatch might reflect regional differences in the size of pyramidal neuron dendritic arbors that are innervated by SST neurons. For example, in monkeys, the mean dendritic field areas of pyramidal neurons were 1.3 times, 2.5–3.5 times, and >5 times larger in V2, PPC, and DLPFC, respectively, than in V1 (Elston and Rosa 1997; Elston 2000). The total number of dendritic branches per pyramidal neuron showed a similar posterior-to-anterior increase (Elston and Rosa 1997; Elston 2000), indicating a substantial increase in the complexity of dendritic integration by pyramidal neurons from posterior to anterior regions. Thus, more SST neurons appear to provide inhibitory inputs to a relatively greater amount of pyramidal neuron dendrites in anterior than in posterior regions. Although VIP neurons innervate almost exclusively SST neurons in the mouse V1 (Pfeffer et al. 2013), several studies provided evidence for their direct inhibition of both SST and PV neurons in multiple regions, including the auditory, motor and prefrontal cortices in rodents (Donato et al. 2013; Pi et al. 2013; Lagler et al. 2016). Therefore, the relatively similar VIP mRNA levels across V2, PPC, and DLPFC (Fig. 2F) might reflect a similar number of targets for VIP neurons among these regions, as the increase in SST neurons from posterior to anterior regions would be balanced by the decrease in PV neurons across these regions (Hayes et al. 1991; Kondo et al. 1994; Conde et al. 1996). On the other hand, the lower VIP mRNA levels in V1 (Fig. 2F) might correspond to a smaller number of VIP neuron targets compared with other regions, if VIP neurons innervate mainly SST neurons (Pfeffer et al. 2013) whose density was lowest in V1 (Hayes et al. 1991).
Transcript Changes Across WM Network in Schizophrenia
For all transcripts expressed in PV and SST neurons, the effect of schizophrenia on expression levels was markedly conserved across the four cortical regions of the WM network (Fig. 4A). Although a recent study of layer 3 transcript levels in the same four cortical regions detected a lower GAD67 mRNA levels only in DLPFC of schizophrenia subjects (Hoftman et al. 2018), the change in GAD67 mRNA levels in layer 3 might not represent alterations in PV and SST neurons in total GM. Consistent with this idea, deficits in GAD67 mRNA expression in total GM have been reported in multiple cortical regions, including DLPFC, anterior cingulate cortex (ACC), orbitofrontal cortex (OFC), and superior temporal gyrus (STG), of schizophrenia subjects (Akbarian et al. 1995; Woo et al. 2004; Thompson et al. 2009; Joshi et al. 2012). The lower GAD67 mRNA levels in ACC, OFC, and STG indicate that alterations in GABA neurons are present in neocortical areas outside of the visuospatial WM network in schizophrenia. Indeed, we previously found that PV, SST, and GAD67 mRNA levels in total GM were lower across four cortical regions (i.e., DLPFC, ACC, primary motor cortex, and V1) that belong to different functional domains in schizophrenia, using 9–12 matched subject pairs, which do not overlap with the current 20 pairs except for one pair (Hashimoto et al. 2008b). Other groups have also reported that PV and SST mRNA levels were lower in total GM of neocortical areas outside of the visuospatial WM network, including ACC and OFC (Fung et al. 2014; Joshi et al. 2015; McMeekin et al. 2016). Together, these findings indicate that the alteration of both PV and SST neurons is shared by a wide range of cortical regions in schizophrenia.
Several lines of evidence suggest that the conserved alterations of transcript levels in PV and SST neurons across cortical regions are unlikely to be the consequence of other factors commonly associated with schizophrenia. First, in our previous analyses, none of comorbid factors, such as the presence of substance abuse/dependence at the time of death, the use of antidepressants, benzodiazepine/anticonvulsant or antipsychotics at the time of death, and suicide as the cause of death, had a significant effect on the expression of PV, SST, KCNS3, MOR, or GAD67 mRNAs in the DLPFC of schizophrenia subjects (Morris et al. 2008; Hashimoto et al. 2008a,b; Curley et al. 2011; Volk et al. 2012b; Georgiev et al. 2014). Second, none of these transcripts exhibited altered expression in the DLPFC of monkeys chronically exposed to antipsychotics at clinically relevant doses (Volk et al. 2000; Hashimoto et al. 2003; Morris et al. 2008; Hashimoto et al. 2008a; Volk et al. 2012b; Georgiev et al. 2014). Third, the comparable magnitudes of expression changes of each transcript across the four cortical regions argue against a potential influence of antipsychotics on these transcripts, as any effects of antipsychotics would be likely to differ across these regions, which markedly differ in the densities of both dopamine axons (Lewis et al. 1987; Gaspar et al. 1989) and dopamine receptors (Lidow et al. 1989). Finally, using the current data, we confirmed that none of the comorbid factors had a significant effect on the transcript levels in any cortical region of schizophrenia subjects (Supplementary Table 3).
Potential Pathogenic Mechanisms Underlying Transcript Changes
Instead, our findings are more likely to reflect pathogenic mechanisms affecting gene expression that are common to PV and SST neurons and conserved across cortical regions. For example, the expression of PV, SST, KCNS3, and GAD67 mRNAs are all activity dependent (Marty and Onteniente 1997; Lee et al. 2015; Cohen et al. 2016; Mardinly et al. 2016). Thus, their downregulation in schizophrenia likely reflects the reduced activity of PV and SST neurons, which might be secondary to lower activity in neighboring pyramidal neurons which provide the largest source of excitatory drive to PV and SST neurons (Tremblay et al. 2016). Consistent with this idea, the density of pyramidal neuron dendritic spines, the site of most excitatory synapses, was found to be decreased across various cortical regions, including the DLPFC, OFC, temporal association cortex, and hippocampus, in subjects with schizophrenia (Glausier and Lewis 2013). Although spine density was not significantly lower in V1 of schizophrenia subjects (Glantz and Lewis 2000), excitatory feedback from other cortical regions might be reduced in this region. Another, but not mutually exclusive, possibility is that alterations in PV and SST neurons are due to shared disruptions in their early development caused by genetic and/or environmental insults, as these neurons, but not VIP neurons, have a common site of origin and common regulators of their proliferation, migration and phenotypic specification (Volk and Lewis 2014). Interestingly, transcripts of some of these regulators, including transcription factors and chemokine receptors, were found to be upregulated in the DLPFC of subjects with schizophrenia (Volk et al. 2015, 2016). These changes might reflect compensatory responses to maintain normal development of these neurons in the face of insults.
Functional Implications
The differences in transcript levels observed across cortical nodes of the visuospatial WM network in schizophrenia (Fig. 4A,B) might contribute to the neuronal substrate for WM impairments in the illness. Both PV and SST neurons play important roles in generating neural oscillations in wide frequency ranges (Buzsaki and Wang 2012; Amilhon et al. 2015; Chen et al. 2017; Veit et al. 2017) and synchronization of these oscillations across multiple cortical regions regulates information processing in distributed neuronal networks (Bastos et al. 2015; Spellman et al. 2015), including the visuospatial WM network (Palva et al. 2010). In addition, a suppression of SST neuron activities in V1 was shown to reduce context-dependent visually induced oscillations (Chen et al. 2017; Veit et al. 2017). Therefore, alterations of PV and SST neurons across the cortical regions and of VIP neurons in V1, which primarily inhibit local SST neurons (Pfeffer et al. 2013), could have a profound impact on the generation and synchronization of cortical oscillations across the visuospatial WM network, contributing to the WM impairments in schizophrenia.
As both PV- and SST-positive GABA neuron subsets can be further divided into subtypes with distinctive morphological and functional properties and laminar distributions, the identification of affected subtypes will be important for further understanding the pathophysiology of cortical network dysfunction in schizophrenia. For example, a minority of SST neurons directly connects distant cortical regions via long-range projections and could contribute to the mechanism for synchronizing oscillations among multiple cortical regions (Tomioka and Rockland 2007; Tamamaki and Tomioka 2010). Consequently, the lower SST mRNA levels across the cortical regions might reflect alterations of direct inter-regional inhibition within the WM network in schizophrenia. However, this possibility remains speculative due to the absence of a specific marker of long-range projecting SST neurons in the human cortex. Interestingly, a recent single-cell RNA sequencing study of mouse cortical neurons reported multiple transcripts that were differentially expressed among different subtypes of each GABA neuron subset (Paul et al. 2017). These transcripts could be used in the future studies to identify affected subtypes of PV and SST neurons across cortical regions in schizophrenia.
Conclusions
In summary, our analyses of transcript levels across cortical regions in the visuospatial WM network revealed that the transcripts associated with each GABA neuron subset had distinct regional expression patterns in comparison subjects, suggesting that the contribution of each GABA neuron subset to the inhibitory regulation of local circuitry normally differs across cortical regions of the WM network. We also found that each transcript expressed in PV and SST neurons was affected in a similar fashion across these regions in subjects with schizophrenia (Fig. 4), suggesting that alterations of PV and SST neurons represent a shared feature across diverse cortical regions, including those for the visuospatial WM network.
Supplementary Material
Funding
This work was supported by Japan Society for the Promotion of Science (Grants-in-Aid 15H01280, 16H05372 to T.H.) and National Institutes of Health (P50 MH103204 to D.A.L.).
Notes
David A. Lewis currently receives investigator-initiated research support from Pfizer. In 2016–2018, he served as a consultant in the areas of target identification and validation and new compound development to Merck. The other authors declare no conflict of interest.
References
- Akbarian S, Kim JJ, Potkin SG, Hagman JO, Tafazzoli A, Bunney WE Jr., Jones EG. 1995. Gene expression for glutamic acid decarboxylase is reduced without loss of neurons in prefrontal cortex of schizophrenics. Arch Gen Psychiatry. 52:258–266. [DOI] [PubMed] [Google Scholar]
- Amilhon B, Huh CY, Manseau F, Ducharme G, Nichol H, Adamantidis A, Williams S. 2015. Parvalbumin interneurons of hippocampus tune population activity at theta frequency. Neuron. 86:1277–1289. [DOI] [PubMed] [Google Scholar]
- Barch DM, Ceaser A. 2012. Cognition in schizophrenia: core psychological and neural mechanisms. Trends Cogn Sci. 16:27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos AM, Vezoli J, Bosman CA, Schoffelen JM, Oostenveld R, Dowdall JR, De Weerd P, Kennedy H, Fries P. 2015. Visual areas exert feedforward and feedback influences through distinct frequency channels. Neuron. 85:390–401. [DOI] [PubMed] [Google Scholar]
- Benson DL, Isackson PJ, Jones EG. 1991. In situ hybridization reveals VIP precursor mRNA-containing neurons in monkey and rat neocortex. Brain Res Mol Brain Res. 9:169–174. [DOI] [PubMed] [Google Scholar]
- Buzsaki G, Wang XJ. 2012. Mechanisms of gamma oscillations. Annu Rev Neurosci. 35:203–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Zhang Y, Li X, Zhao X, Ye Q, Lin Y, Tao HW, Rasch MJ, Zhang X. 2017. Distinct inhibitory circuits orchestrate cortical beta and gamma band oscillations. Neuron. 96:1403–1418 e1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophel TB, Klink PC, Spitzer B, Roelfsema PR, Haynes JD. 2017. The distributed nature of working memory. Trends Cogn Sci. 21:111–124. [DOI] [PubMed] [Google Scholar]
- Chung DW, Fish KN, Lewis DA. 2016. a. Pathological basis for deficient excitatory drive to cortical parvalbumin interneurons in schizophrenia. Am J Psychiatry. 173:1131–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung DW, Volk DW, Arion D, Zhang Y, Sampson AR, Lewis DA. 2016. b. Dysregulated ErbB4 splicing in schizophrenia: selective effects on parvalbumin expression. Am J Psychiatry. 173:60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM, Ma H, Kuchibhotla KV, Watson BO, Buzsaki G, Froemke RC, Tsien RW. 2016. Excitation-transcription coupling in parvalbumin-positive interneurons employs a novel CaM kinase-dependent pathway distinct from excitatory neurons. Neuron. 90:292–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CE, Airey DC, Young NA, Leitch DB, Kaas JH. 2010. Neuron densities vary across and within cortical areas in primates. Proc Natl Acad Sci USA. 107:15927–15932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde F, Lund JS, Lewis DA. 1996. The hierarchical development of monkey visual cortical regions as revealed by the maturation of parvalbumin-immunoreactive neurons. Brain Res Dev Brain Res. 96:261–276. [DOI] [PubMed] [Google Scholar]
- Constantinidis C, Williams GV, Goldman-Rakic PS. 2002. A role for inhibition in shaping the temporal flow of information in prefrontal cortex. Nat Neurosci. 5:175–180. [DOI] [PubMed] [Google Scholar]
- Curley AA, Arion D, Volk DW, Asafu-Adjei JK, Sampson AR, Fish KN, Lewis DA. 2011. Cortical deficits of glutamic acid decarboxylase 67 expression in schizophrenia: clinical, protein, and cell type-specific features. Am J Psychiatry. 168:921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defelipe J, Gonzalez-Albo MC, Del Rio MR, Elston GN. 1999. Distribution and patterns of connectivity of interneurons containing calbindin, calretinin, and parvalbumin in visual areas of the occipital and temporal lobes of the macaque monkey. J Comp Neurol. 412:515–526. [DOI] [PubMed] [Google Scholar]
- Donato F, Rompani SB, Caroni P. 2013. Parvalbumin-expressing basket-cell network plasticity induced by experience regulates adult learning. Nature. 504:272–276. [DOI] [PubMed] [Google Scholar]
- Drake CT, Milner TA. 2002. Mu opioid receptors are in discrete hippocampal interneuron subpopulations. Hippocampus. 12:119–136. [DOI] [PubMed] [Google Scholar]
- Elston GN. 2000. Pyramidal cells of the frontal lobe: all the more spinous to think with. J Neurosci. 20:RC95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston GN, Rosa MG. 1997. The occipitoparietal pathway of the macaque monkey: comparison of pyramidal cell morphology in layer III of functionally related cortical visual areas. Cereb Cortex. 7:432–452. [DOI] [PubMed] [Google Scholar]
- Enwright JF, Sanapala S, Foglio A, Berry R, Fish KN, Lewis DA. 2016. Reduced labeling of parvalbumin neurons and perineuronal nets in the dorsolateral prefrontal cortex of subjects with schizophrenia. Neuropsychopharmacology. 41:2206–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson J, Vogel EK, Lansner A, Bergstrom F, Nyberg L. 2015. Neurocognitive architecture of working memory. Neuron. 88:33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung SJ, Fillman SG, Webster MJ, Shannon Weickert C. 2014. Schizophrenia and bipolar disorder show both common and distinct changes in cortical interneuron markers. Schizophr Res. 155:26–30. [DOI] [PubMed] [Google Scholar]
- Fung SJ, Webster MJ, Sivagnanasundaram S, Duncan C, Elashoff M, Weickert CS. 2010. Expression of interneuron markers in the dorsolateral prefrontal cortex of the developing human and in schizophrenia. Am J Psychiatry. 167:1479–1488. [DOI] [PubMed] [Google Scholar]
- Gabbott PL, Bacon SJ. 1997. Vasoactive intestinal polypeptide containing neurones in monkey medial prefrontal cortex (mPFC): colocalisation with calretinin. Brain Res. 744:179–184. [DOI] [PubMed] [Google Scholar]
- Gaspar P, Berger B, Febvret A, Vigny A, Henry JP. 1989. Catecholamine innervation of the human cerebral cortex as revealed by comparative immunohistochemistry of tyrosine hydroxylase and dopamine-beta-hydroxylase. J Comp Neurol. 279:249–271. [DOI] [PubMed] [Google Scholar]
- Georgiev D, Arion D, Enwright JF, Kikuchi M, Minabe Y, Corradi JP, Lewis DA, Hashimoto T. 2014. Lower gene expression for KCNS3 potassium channel subunit in parvalbumin-containing neurons in the prefrontal cortex in schizophrenia. Am J Psychiatry. 171:62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev D, Gonzalez-Burgos G, Kikuchi M, Minabe Y, Lewis DA, Hashimoto T. 2012. Selective expression of KCNS3 potassium channel alpha-subunit in parvalbumin-containing GABA neurons in the human prefrontal cortex. PLoS One. 7:e43904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glantz LA, Lewis DA. 2000. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 57:65–73. [DOI] [PubMed] [Google Scholar]
- Glausier JR, Lewis DA. 2013. Dendritic spine pathology in schizophrenia. Neuroscience. 251:90–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillozet-Bongaarts AL, Hyde TM, Dalley RA, Hawrylycz MJ, Henry A, Hof PR, Hohmann J, Jones AR, Kuan CL, Royall J, et al. . 2014. Altered gene expression in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol Psychiatry. 19:478–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Arion D, Unger T, Maldonado-Aviles JG, Morris HM, Volk DW, Mirnics K, Lewis DA. 2008. a. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 13:147–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Bazmi HH, Mirnics K, Wu Q, Sampson AR, Lewis DA. 2008. b. Conserved regional patterns of GABA-related transcript expression in the neocortex of subjects with schizophrenia. Am J Psychiatry. 165:479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Volk DW, Eggan SM, Mirnics K, Pierri JN, Sun Z, Sampson AR, Lewis DA. 2003. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J Neurosci. 23:6315–6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug H. 1987. Brain sizes, surfaces, and neuronal sizes of the cortex cerebri: a stereological investigation of man and his variability and a comparison with some mammals (primates, whales, marsupials, insectivores, and one elephant). Am J Anat. 180:126–142. [DOI] [PubMed] [Google Scholar]
- Hayes TL, Cameron JL, Fernstrom JD, Lewis DA. 1991. A comparative analysis of the distribution of prosomatostatin-derived peptides in human and monkey neocortex. J Comp Neurol. 303:584–599. [DOI] [PubMed] [Google Scholar]
- Hendry SH, Schwark HD, Jones EG, Yan J. 1987. Numbers and proportions of GABA-immunoreactive neurons in different areas of monkey cerebral cortex. J Neurosci. 7:1503–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoftman GD, Dienel SJ, Bazmi HH, Zhang Y, Chen K, Lewis DA. 2018. Altered gradients of glutamate and gamma-aminobutyric acid transcripts in the cortical visuospatial working memory network in schizophrenia. Biol Psychiatry. 83:670–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm S. 1979. A simple sequentially rejective multiple test procedure. Scand J Stat. 6:65–70. [Google Scholar]
- Jiang J. 2007. Linear and Generalized Linear Mixed Models and Their Applications. New York: Springer-Verlag. [Google Scholar]
- Joshi D, Catts VS, Olaya JC, Shannon Weickert C. 2015. Relationship between somatostatin and death receptor expression in the orbital frontal cortex in schizophrenia: a postmortem brain mRNA study. NPJ Schizophr. 1:14004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi D, Fung SJ, Rothwell A, Weickert CS. 2012. Higher gamma-aminobutyric acid neuron density in the white matter of orbital frontal cortex in schizophrenia. Biol Psychiatry. 72:725–733. [DOI] [PubMed] [Google Scholar]
- Kamigaki T, Dan Y. 2017. Delay activity of specific prefrontal interneuron subtypes modulates memory-guided behavior. Nat Neurosci. 20:854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani MM, Agetsuma M, Yuste R. 2014. A blanket of inhibition: functional inferences from dense inhibitory connectivity. Curr Opin Neurobiol. 26:96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani MM, Jackson J, Ayzenshtat I, Hamzehei Sichani A, Manoocheri K, Kim S, Yuste R. 2016. Opening holes in the blanket of inhibition: localized lateral disinhibition by VIP interneurons. J Neurosci. 36:3471–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Jeong H, Lee J, Ghim JW, Her ES, Lee SH, Jung MW. 2016. Distinct roles of parvalbumin- and somatostatin-expressing interneurons in working memory. Neuron. 92:902–915. [DOI] [PubMed] [Google Scholar]
- Kondo H, Hashikawa T, Tanaka K, Jones EG. 1994. Neurochemical gradient along the monkey occipito-temporal cortical pathway. Neuroreport. 5:613–616. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Hattori R, Yui Y. 1994. Three distinct subpopulations of GABAergic neurons in rat frontal agranular cortex. Brain Res. 649:159–173. [DOI] [PubMed] [Google Scholar]
- Lagler M, Ozdemir AT, Lagoun S, Malagon-Vina H, Borhegyi Z, Hauer R, Jelem A, Klausberger T. 2016. Divisions of identified parvalbumin-expressing basket cells during working memory-guided decision making. Neuron. 91:1390–1401. [DOI] [PubMed] [Google Scholar]
- Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung HL, Chen S, et al. . 2016. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 352:1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Royston SE, Vest MO, Ley DJ, Lee S, Bolton EC, Chung HJ. 2015. N-methyl-D-aspartate receptors mediate activity-dependent down-regulation of potassium channel genes during the expression of homeostatic intrinsic plasticity. Mol Brain. 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Campbell MJ, Foote SL, Goldstein M, Morrison JH. 1987. The distribution of tyrosine hydroxylase-immunoreactive fibers in primate neocortex is widespread but regionally specific. J Neurosci. 7:279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidow MS, Goldman-Rakic PS, Rakic P, Innis RB. 1989. Dopamine D2 receptors in the cerebral cortex: distribution and pharmacological characterization with [3H]raclopride. Proc Natl Acad Sci USA. 86:6412–6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardinly AR, Spiegel I, Patrizi A, Centofante E, Bazinet JE, Tzeng CP, Mandel-Brehm C, Harmin DA, Adesnik H, Fagiolini M, et al. . 2016. Sensory experience regulates cortical inhibition by inducing IGF1 in VIP neurons. Nature. 531:371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O. 2012. Interneuron dysfunction in psychiatric disorders. Nat Rev Neurosci. 13:107–120. [DOI] [PubMed] [Google Scholar]
- Marty S, Onteniente B. 1997. The expression pattern of somatostatin and calretinin by postnatal hippocampal interneurons is regulated by activity-dependent and -independent determinants. Neuroscience. 80:79–88. [DOI] [PubMed] [Google Scholar]
- McMeekin LJ, Lucas EK, Meador-Woodruff JH, McCullumsmith RE, Hendrickson RC, Gamble KL, Cowell RM. 2016. Cortical PGC-1alpha-dependent transcripts are reduced in postmortem tissue from patients with schizophrenia. Schizophr Bull. 42:1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N, Huang HS, Baker SP, Galdzicka M, Ginns E, Akbarian S. 2009. Molecular determinants of dysregulated GABAergic gene expression in the prefrontal cortex of subjects with schizophrenia. Biol Psychiatry. 65:1006–1014. [DOI] [PubMed] [Google Scholar]
- Morris HM, Hashimoto T, Lewis DA. 2008. Alterations in somatostatin mRNA expression in the dorsolateral prefrontal cortex of subjects with schizophrenia or schizoaffective disorder. Cereb Cortex. 18:1575–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palva JM, Monto S, Kulashekhar S, Palva S. 2010. Neuronal synchrony reveals working memory networks and predicts individual memory capacity. Proc Natl Acad Sci USA. 107:7580–7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A, Crow M, Raudales R, He M, Gillis J, Huang ZJ. 2017. Transcriptional architecture of synaptic communication delineates GABAergic neuron identity. Cell. 171:522–539 e520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckys D, Landwehrmeyer GB. 1999. Expression of mu, kappa, and delta opioid receptor messenger RNA in the human CNS: a 33P in situ hybridization study. Neuroscience. 88:1093–1135. [DOI] [PubMed] [Google Scholar]
- Pfeffer CK, Xue M, He M, Huang ZJ, Scanziani M. 2013. Inhibition of inhibition in visual cortex: the logic of connections between molecularly distinct interneurons. Nat Neurosci. 16:1068–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi HJ, Hangya B, Kvitsiani D, Sanders JI, Huang ZJ, Kepecs A. 2013. Cortical interneurons that specialize in disinhibitory control. Nature. 503:521–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SG, Williams GV, Goldman-Rakic PS. 2000. Destruction and creation of spatial tuning by disinhibition: GABA(A) blockade of prefrontal cortical neurons engaged by working memory. J Neurosci. 20:485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco BR, Lewis DA, Fish KN. 2016. Markedly lower glutamic acid decarboxylase 67 protein levels in a subset of boutons in schizophrenia. Biol Psychiatry. 79:1006–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver H, Feldman P, Bilker W, Gur RC. 2003. Working memory deficit as a core neuropsychological dysfunction in schizophrenia. Am J Psychiatry. 160:1809–1816. [DOI] [PubMed] [Google Scholar]
- Spellman T, Rigotti M, Ahmari SE, Fusi S, Gogos JA, Gordon JA. 2015. Hippocampal-prefrontal input supports spatial encoding in working memory. Nature. 522:309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumm RK, Zhou C, Schulz S, Hollt V. 2004. Neuronal types expressing mu- and delta-opioid receptor mRNA in the rat hippocampal formation. J Comp Neurol. 469:107–118. [DOI] [PubMed] [Google Scholar]
- Tamamaki N, Tomioka R. 2010. Long-range GABAergic connections distributed throughout the neocortex and their possible function. Front Neurosci. 4:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M, Weickert CS, Wyatt E, Webster MJ. 2009. Decreased glutamic acid decarboxylase(67) mRNA expression in multiple brain areas of patients with schizophrenia and mood disorders. J Psychiatr Res. 43:970–977. [DOI] [PubMed] [Google Scholar]
- Tomioka R, Rockland KS. 2007. Long-distance corticocortical GABAergic neurons in the adult monkey white and gray matter. J Comp Neurol. 505:526–538. [DOI] [PubMed] [Google Scholar]
- Tremblay R, Lee S, Rudy B. 2016. GABAergic Interneurons in the neocortex: from cellular properties to circuits. Neuron. 91:260–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit J, Hakim R, Jadi MP, Sejnowski TJ, Adesnik H. 2017. Cortical gamma band synchronization through somatostatin interneurons. Nat Neurosci. 20:951–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Austin MC, Pierri JN, Sampson AR, Lewis DA. 2000. Decreased glutamic acid decarboxylase 67 messenger RNA expression in a subset of prefrontal cortical gamma-aminobutyric acid neurons in subjects with schizophrenia. Arch Gen Psychiatry. 57:237–245. [DOI] [PubMed] [Google Scholar]
- Volk DW, Chitrapu A, Edelson JR, Lewis DA. 2015. Chemokine receptors and cortical interneuron dysfunction in schizophrenia. Schizophr Res. 167:12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Edelson JR, Lewis DA. 2016. Altered expression of developmental regulators of parvalbumin and somatostatin neurons in the prefrontal cortex in schizophrenia. Schizophr Res. 177:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Lewis DA. 2014. Early developmental disturbances of cortical inhibitory neurons: contribution to cognitive deficits in schizophrenia. Schizophr Bull. 40:952–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Matsubara T, Li S, Sengupta EJ, Georgiev D, Minabe Y, Sampson A, Hashimoto T, Lewis DA. 2012. a. Deficits in transcriptional regulators of cortical parvalbumin neurons in schizophrenia. Am J Psychiatry. 169:1082–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Radchenkova PV, Walker EM, Sengupta EJ, Lewis DA. 2012. b. Cortical opioid markers in schizophrenia and across postnatal development. Cereb Cortex. 22:1215–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo TU, Walsh JP, Benes FM. 2004. Density of glutamic acid decarboxylase 67 messenger RNA-containing neurons that express the N-methyl-D-aspartate receptor subunit NR2A in the anterior cingulate cortex in schizophrenia and bipolar disorder. Arch Gen Psychiatry. 61:649–657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.