Abstract
The adhesive and mechanical properties of a modular fusion protein consisting of two different types of binding units linked together via a flexible resilin-like-polypeptide domain are quantified. The adhesive domains have been constructed from fungal cellulose-binding modules (CBMs) and an amphiphilic hydrophobin HFBI. This study is carried out by single-molecule force spectroscopy, which enables stretching of single molecules. The fusion proteins are designed to self-assemble on the cellulose surface, leading into the submonolayer of proteins having the HFBI pointing away from the surface. A hydrophobic atomic force microscopy (AFM) tip can be employed for contacting and lifting the single fusion protein from the HFBI-functionalized terminus by the hydrophobic interaction between the tip surface and the hydrophobic patch of the HFBI. The work of rupture, contour length at rupture and the adhesion forces of the amphiphilic end domains are evaluated under aqueous environment at different pHs.
Introduction
In nature, many solutions are hierarchical and based on modular design that allows the flexibility to serve for multiple purposes.1 To understand how nature works and to be able to learn from it, engineering of materials and their quantitative understanding on a molecular level is essential. Genetic engineering is a powerful tool that can be employed for creating new modular designs of molecules.
Proteins that are both adhesive and elastic are attractive as molecular building blocks because of their ability to create elastic and energy dissipative interfaces between the components of hybrid materials. Examples of natural hybrid materials where the adhesive proteins are present at the interfaces are composite structures such as nacre2 and squid beak.3 In this study, a modular protein construct built from an elastic domain (resilin) and two adhesive functionalities (two cellulose-binding domains, dCBMs, and hydrophobin, HFBI) is investigated on the molecular level. Cellulose-binding modules (CBMs) are produced by different organisms and occur for instance in various types of cellulase enzymes.4−6 CBMs have a specific affinity to cellulose surfaces, but for increased affinity, they can be combined into double CBMs.7 Fusion proteins containing the dCBM have been investigated, for example, for stabilizing emulsions and as building blocks in biomimetic nanocomposites.8,9
Hydrophobins are small amphiphilic surfactant-like molecules produced by filamentous fungi.10 Class II hydrophobins have a diameter of 3 nm, and they have the tendency to form monolayer films at the air/water interface with a remarkably high shear elastic modulus, about 0.5 N m–1.10,11 Because of their properties, hydrophobins have been used to enhance the biocompatibility of surfaces for potential applications in biodevices. For example, HFBI-/collagen-modified poly- dimethylsiloxane surfaces have been designed to promote cell adhesion and growth.12 Similarly, the growth of neural stem cells was promoted on layers of serum proteins that are immobilized on hydrophobin-modified polylactic-co-glycolic acid surfaces: the better adhesion of serum proteins on the HFBI-modified surfaces and the consequent enhancement of cell immobilization improved the biocompatibility of implants after screening.13 HFBI can even be used to produce lipid-free membranes and vesicles.14
Resilin is an elastomeric protein present in the specialized regions of the insect cuticle and plays a key role in insect flight, jumping mechanism of fleas, and vocalization of cicadas.15 It is a rubberlike protein, according to the criteria of having high resilience and low stiffness, being able to deform reversibly without loss of energy.16,17 Furthermore, resilin is able to be deformed to large elongation under little force.18,19 When it is completely hydrated, resilin behaves close to a perfect rubber,20−22 has a rather low stiffness, and can be stretched to more than three times and compressed to one-third of its original length. Furthermore, when stressed and released again, it goes back to its initial state without having any residual deformations.20 For resilin in the elastic tendons of dragonflies and locust ligaments, Young’s moduli of 0.6–0.7 and 0.9 MPa were found, respectively.16 In the form of resilin-like polypeptide (RLP), resilin has been applied for tissue engineering and in the medical field.23−25
Single-molecule force spectroscopy (SMFS) is revealed as a great technique to study biological systems from cells26 to biomacromolecules.27,28 Concerning elastomeric proteins, previous studies have been carried out, for example, on tropoelastin29 and on REC (R = resilin, E = elastin, and C = collagen like molecules) polypeptides30 but not on resilin.
In this study, the mechanical and adhesive properties of two modular fusion proteins (Figure 1) were studied: the first protein was constructed from hydrophobin HFBI and a double CBM separated by a linker of 24 amino acids (HFBI–dCBM). For the second, the same building blocks were used but the linker region was replaced by a RLP (dCBM–RLP–HFBI).
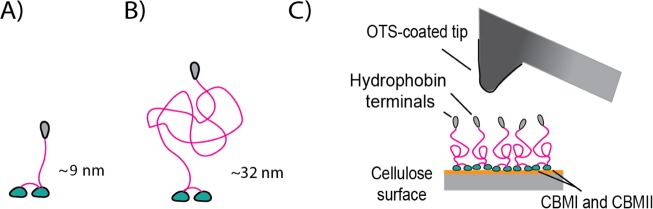
Figure 1.
Schematic images and sizes of the studied modular proteins. (A) HFBI–dCBM consisting of the terminal hydrophobin HFBI (gray globular domain) and the CBMs (green globular domain) combined with polypeptide linkers. (B) dCBM–RLP–HFBI protein where the binding modules are combined with the resilin-like peptide (red random coil domain). (C) Cartoon of the force spectroscopy setup made up of an OTS-functionalized tip approaching on a cellulose-coated surface with the proteins casted on it. The sizes are estimated from the amino acid sequences (see Figure S1 and Tables S1 and S2).
These proteins were used to study how adhesive proteins act as building blocks in hybrid nanomaterials. Therefore, the force response of both fusion proteins under tensile force using SMFS was recorded. The attachment of the molecule to the chosen surfaces occurred via self-assembly of the terminal units, dCBM being able to bind to cellulose and HFBI to a hydrophobic surface, allowing the linker region unbound and available for stretching. Similar approaches have been previously taken for studies of modular proteins such as titin-mimicking protein constructs.31−34 The importance of the single-molecule perspective on the development of nanocomposites with enhanced mechanical properties is evident because only by quantifying the smallest scale events, it is possible to progress toward higher levels of hierarchy.
Results and Discussion
SMFS Experiments
The SMFS experiment was carried out by approaching the surface-immobilized fusion proteins with a hydrophobic octadecyltrichlorosilane (OTS)-coated tip and then retracting the tip, as shown schematically in Figure 2A,B.
Figure 2.
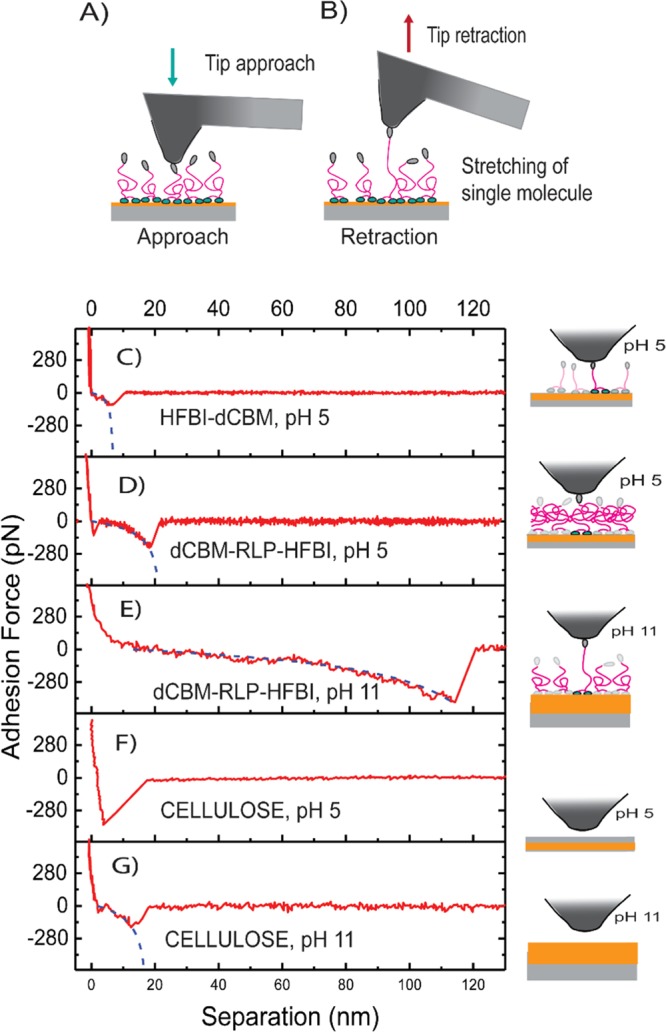
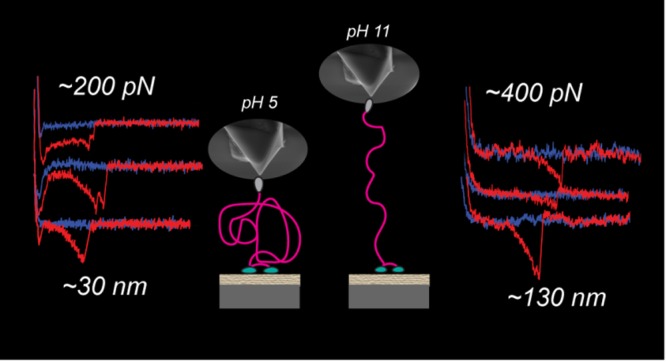
(A) Schematic representation of the SMFS experiment: as the hydrophobic tip is approaching the surface, the hydrophobins will attach to the tip. (B) Tip retraction leads to elongation of the attached protein and eventual detachment of the protein from the tip. Typical force/distance curves, with fit included, measured for (C) HFBI–dCBM molecule, (D) dCBM–RLP–HFBI molecule at pH 5, and (E) at pH 11 and for the (F) cellulose surface at pH 5 and (G) at pH 11. On the right column, a schematic of the molecules/surfaces involved in the stretching is reported.
The proteins were adhered on a thin cellulose layer via adsorption through the dCBM. Typical force/distance curves are represented in Figure 2C–G, where the typical retraction curve had only one peak that most probably shows the detachment event of a single molecule. More examples of force/distance curves are reported in Figure S2. For both the fusion proteins and the plain cellulose surface, the experiments were performed in 10 mM NaAc buffer solution at pH 5. dCBM–RLP–HFBI and the plain cellulose surface were also studied at pH 11 in 10 mM phosphate buffer.
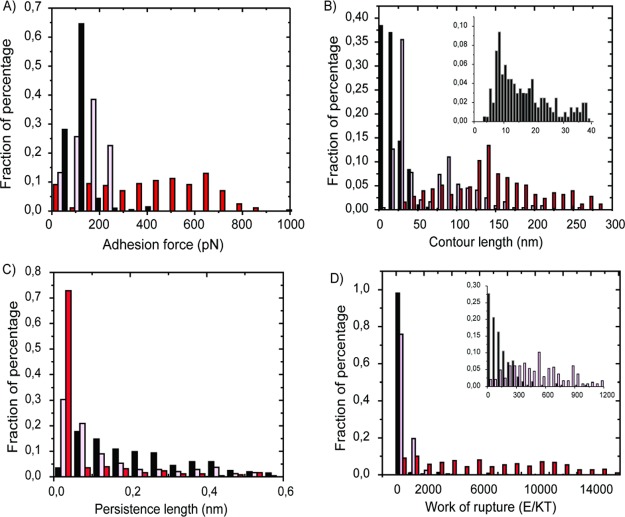
The force values corresponding to the last detachment of the tip from the surface were collected from the force/distance curves and are presented as a histogram in Figure 3A.
Figure 3.
Histograms summarizing the (A) adhesion force, (B) contour length, (C) persistence length, and (D) work of rupture for HFBI–dCBM at pH 5 (black bars) and dCBM–RLP–HFBI at pH 5 (pink bars) and at pH 11 (red bars). The graphs (B,D) show the insets respectively for the contour length of HFBI–dCBM and the work of rupture of HFBI–dCBM and dCBM–RLP–dCBM at pH 5 to point out the distribution of the values in the lower range of x-axis.
The distribution of the adhesive forces follows approximately a normal distribution. For HFBI–dCBM and dCBM–RLP–HFBI proteins at pH 5, the values vary between 50 and 200 pN, whereas at pH 11, the distribution is wider and the values vary between 0 and 800 pN. The contour length, the persistence length, and the work of rupture were extracted, as explained in the Materials and Methods section, and are presented in Figure 3B–D. The mean values and standard deviation of the data are presented in Table 1.
Table 1. Mean Values of the Adhesion Force, the Contour Length, the Persistence Length, and the Work of Rupture of the Studied Molecules.
| surface | adhesion force ± standard error (pN) | contour length ± standard error (nm) | persistence length ± standard error (nm) | work of rupture ± standard error (E/kT) |
|---|---|---|---|---|
| dCBM–RLP–HFBI on cellulose at pH 5 | 162 ± 4 | 29 ± 3 | 0.04 ± 0.001 | 485.3 ± 9 |
| 93 ± 6 | ||||
| HFBI–dCBM on cellulose at pH 5 | 86 ± 1 | 12 ± 1 | 0.2 ± 0.01 | 84.7 ± 5 |
| dCBM–RLP–HFBI on cellulose at pH 11 | 438 ± 36 | 64 ± 5 | 0.04 ± 2 | 6725 ± 29 |
| 133 ± 2 | ||||
| 149 ± 5 | ||||
| 243 ± 9 | ||||
| cellulose at pH 5 | 483 ± 3 | 1004 ± 6 | ||
| cellulose at pH 11 | 159.95 ± 5 | 18 ± 1 | 0.12 ± 0.1 | 270 ± 48 |
The reversed setup of the experiment, where the proteins were attached to the tip, was also studied and is discussed in the Supporting Information (Figures S3 and S4).
Thickness and Elasticity of the Cellulose and Proteins
To have a correct interpretation of the molecular response during the force spectroscopy experiments, it was important to know the consistence and the organization of the molecules and the cellulose on the top of the silicon surface.
The adsorption of the fusion proteins on the spin-coated cellulose was studied by measuring the adsorbed mass and the change in the dissipation of the quartz crystal microbalance (QCM-D) sensor simultaneously (Figure 4).
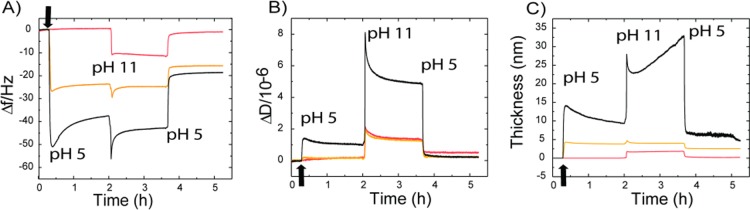
Figure 4.
QCM-D experiment on protein adsorption on the cellulose surface and consequent conformational changes. (A) Frequency change at pHs 5 and 11 on a cellulose-coated gold sensor (orange) and after injection (black arrow) of 0.1 mg mL−1 HFBI−dCBM (red) and dCBM−RLP−HFBI (black) on the cellulose surface. (B) Dissipation and (C) thickness change during the same experiment.
The data show a clear decrease of the frequency, indicating an adsorption of the proteins in NaAc buffer. Switching the pH to 11, a further decrease in Δf was observed for the RLP probably because of a conformational change and only a slight Δf change for HFBI–dCBM. The dissipation signal in the NaAc buffer rapidly increased during the adsorption of dCBM–RLP–HFBI but stabilized after the protein film was formed in a rather modest value near 10–6. At pH 11, the dissipation signal drove up to 8 × 10–6 meaning an increase of viscoelasticity because of the swelling. This swelling effect of the resilin is reflected by a change in the thickness from 10 nm at pH 5 to 30 nm at pH 11. The increase of the dissipation during the adsorption of HFBI–dCBM was less pronounced. A control measurement on cellulose was carried out to investigate the pH effect on the cellulose layer. Switching from pH 5 to 11, only modest changes in frequency and dissipation were observed.
The atomic force microscopy (AFM) images of Figure 5 visualize the different morphologies of the cellulose surface and the proteins at different environments. Comparing the images of the pure cellulose layer (Figure 5A,D) reveals that changing the pH from 5 to 11 leads to an increased roughness, although the cellulose layer does not swell significantly, as shown by QCM-D graphs. The pH in fact affects the consistence of the film that appears in a less ordered structure.
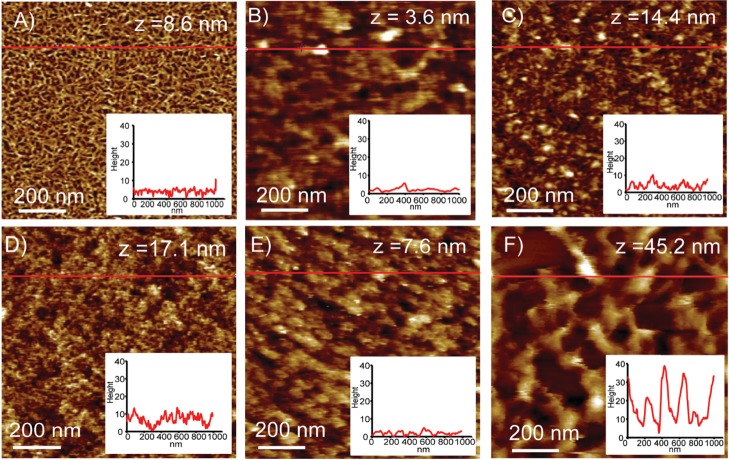
Figure 5.
AFM images of the (A) cellulose surface at pH 5; (B) HFBI–dCBM and (C) dCBM–RLP–HFBI drop-casted on the cellulose at pH 5; (D) cellulose surface at pH 11; and (E) HFBI–dCBM and (F) dCBM–RLP–HFBI drop-casted on the cellulose at pH 11. Insets represent a section analysis of the position indicated by the red line. The images were recorded on wet conditions under the respective buffer. Height range z from black to white is given in each image.
The adsorption of dCBM–RLP–HFBI on the cellulose at pH 5 increases the surface roughness, and the proteins are visible as small, randomly organized features having a height of 5–7 nm (Figure 5C). At pH 11, the protein film was strongly swollen, which was observed as increased roughness and smeared image of the surface having up to 40 nm height variation (Figure 5F). For HFBI–dCBM, it was possible to observe a change in the morphological structure at different conditions, but no swelling could be observed at pH 11. Additional AFM images at pH 5 are reported in Figure S5.
Extension of the Molecules
For comparing the experimental values obtained for the dimensions of the molecules, a theoretical size of the proteins was estimated based on the amino acid sequence and the conformation of different regions, assuming alternative random coils and extended linker regions (Figure S1). Because the binding modules are globular and cross-linked, it may be assumed that they are stable under the tensile stress applied in the AFM experiment and thus will not extend in the experiment. The RLP is an unstructured protein, which in the relaxed form assumes a random coil conformation stabilized by weak interactions between the hydrophobic amino acids.35 Thus, it was assumed that the resilin can have either a random coil or an extended conformation, especially under the tensile stress, and thus the length of the folded RLP was estimated by calculating both the radius of gyration and the extended length. All of the values calculated in this manner for both proteins are given in Tables S1 and S2.
The estimated dimensions of HFBI–dCBM match well with the measured contour length. The experimental contour length of dCBM–RLP–HFBI at pH 5, however, followed a bimodal distribution having two probable values, a shorter one, 29 nm, indicating a coiled structure and a longer one, 93 nm indicating a more extended conformation (Figure 6A,B).
Figure 6.
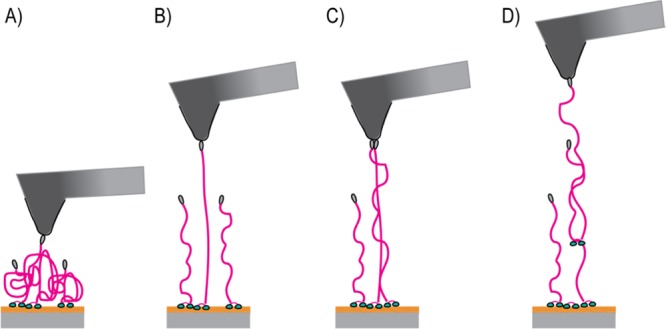
Schematic of different conformations of the resilin-like peptide at the detachment of the molecule. (A) At pH 5, resilin assumes a random coil conformation. (B) At pH 11, resilin assumes an extended conformation. (C,D) Attachment of multiple entangled resilin molecules at pH 11.
At pH 11, four Gaussian curves were fitted to the data to obtain a good overall fit. The mean values of the contour lengths at pH 11 were 64, 133, 149, and 243 nm. When comparing the estimated lengths (32 nm folded and 136 nm extended) with the measured contour lengths, it appears that the shorter observed detachment length at pH 5, matches well with the conformation where RLP is assumed to have the random coil structure. The second mode of detachment at pH 5 has a contour length of ca. 90 nm which is between the values of the partially and fully extended proteins. This indicates that dCBM–RLP–HFBI is partly stretched before detachment at pH 5. At pH 11, the experimental contour length had more quantized values, indicating more possible conformations. The shortest value represents a swollen coil, and the mid values that represent the majority of the measurements, 133 and 149, are close to the fully extended chain. At elevated pH, the cellulose also slightly swells to some extent as presented in the discussion of the AFM images and becomes softer (see section Elasticity of the Protein Layer). This could well explain the third and fourth contour length that are above the theoretical extended chain value, possibly because of the stretching of the cellulose substrate before the detachment of the protein. The control measurements on the plain cellulose surface showed values of the contour length of 10–30 nm that may refer to the “loosening of some cellulose chains”. In Figure S6, the histograms of the rupture force, the work of rupture, and the contour and persistence length for plain cellulose are presented. The values covered a range between 0 and 200 pN of force, 0–600E/kT of energy, and 10–25 nm of contour length. Entanglement of the proteins at elevated pH may also lead to the contour lengths exceeding 200 nm (Figure 6D).
For the persistence length of the proteins, the values were typically considerably smaller than 0.36 nm, which would be expected for a polypeptide.36 Only HFBI–dCBM had a significant number of measured values on the typical range, whereas most of the values of the RLP proteins were much lower than expected. In the literature, low persistence length values have been reported for situations where more filaments than only one are stretched simultaneously during the tip–protein detachment.36 Entanglement of the resilins and their mutual interactions would explain such behavior in the system studied here (Figure 6D). To explicit this statement, a closer analysis of the distributions of the measured data for dCBM–RLP–HFBI at pH 11 is reported in Figure S7.
The event of stretching a single molecule was observed in almost 2% for HFBI–dCBM and almost 6 and 18% for dCBM–RLP–HFBI at pH 5 and 11, respectively.
Adhesion of the Molecules
The QCM-D experiment (Figure 4) indicates that both the fusion proteins adhered to the cellulose surface as a monolayer. The estimated molecular densities imply a slightly lower density for the larger RLP-containing fusion protein, which can be explained with the larger dimensions of this molecule. In the SMFS experiments, a much lower protein concentration of 0.1 μM was used to obtain a very low molecular density on the surface and hence to increase the probability of single-molecule pickup events.
The rupture forces observed for HFBI–dCBM and dCBM–RLP–HFBI at pH 5 were in the range between 50 and 200 pN and can be attributed to the hydrophobic interaction between the hydrophobic side of the fusion protein and the OTS tip. These values are in accordance with the previous measurements where a single molecule has been in contact between the tip and the surface.37−39 Moreover, theoretical simulations predict that HFBI adsorbs to CH3-SAM preferably with its hydrophobic patch toward the surface.40 Previous studies have shown for polymers41 and DNA strands42 as well as for proteins37 and peptides43 binding toward hydrophobic surfaces a range of force values between 50 and 200 pN. Adhesin proteins expressed by bacteria showed a similar behavior toward the hydrophobic surfaces.44
The rupture forces obtained for dCBM–RLP–HFBI at pH 11, however, are much larger than those commonly recorded for single molecules. The change in the pH should mainly affect the conformation of the resilin module. This was proven by previous QCM-D studies that have shown a conformational change from a globular structure at pH 5 to an extended coiled structure at pH 11 where a stronger protein–water interaction is observed, forming a relatively soft layer of the extended coil or brushlike conformations.45 However, this is not directly related to the measured force, namely, the rupture force of the hydrophobins. One possibility for higher rupture forces is the simultaneous attachment of several hydrophobins on the tip surface (Figure 6C), possibly because of the entanglement of the RLP linkers. Earlier studies show similar observations where the higher forces were considered to be related to the detachment of two or few more molecules on the tip–surface contact.44 The AFM topography image of the dCBM–RLP–HFBI layer at pH 11 shows a softer surface and increased roughness that indicates that the molecules are protruding in more extended conformation which may lead to entangled brushlike structures (Figure 5D), which makes it less probable to obtain attachment of the individual molecule to the AFM tip. The shape of the distribution of the rupture force values for dCBM–RLP–HFBI at pH 11 (red bars in Figure 3A) also hints at an overlay of at least two or three distributions (with mean values as approximately on time, two times and three times the mean rupture force of dCBM–RLP–HFBI at pH 5). It is also known that the resilin and other structure-forming proteins go through a precursor conformation as a part of self-assembly, leading to the formation of filaments or other structures with higher order.35 The tendency toward such structural transformation may explain the strong mutual interactions and entanglement.
On the basis of the force curves alone, it is very difficult to draw a definite conclusion which of the attachments are breaking in the experiment but there are factors that point toward certain events: first of all, on the basis of the rupture forces measured, it can be safely assumed that the polypeptide backbone of the protein does not break but the molecule detaches from one of the interfaces, either dCBM/cellulose or HFBI/OTS-Si. Second, the fact that the binding forces at pH 5 are on a similar level for both the proteins indicates that the linker region does not play a significant role in the attachment to the surface. Third, the reason for assuming that the dCBM has stronger adherence is found in the success of the repeated approach/retraction cycles. If the dCBM had been detached, this would have led to continuous picking up of molecules on the tip in each cycle. Moreover, the attachment of the tip-bound molecule on the cellulose surface may have been sterically hindered by the other molecules occupying the surface. Because neither accumulation of the molecules on the probe nor steric hindrance was observed, but the cycling of single-molecule attachment/detachment could be repeated, it is very likely that really the detachment of the HFBI was measured.
The energy needed for the rupture of the molecules was quantified as the work of rupture. The differences between the studied fusion proteins and the resilin conformation are clearly observed in the work of rupture histograms in Figure 3D. Resilin at pH 11 has a very wide distribution of the work of rupture that extends to 10 000kT range, whereas at pH 5, the values are in 1000kT range. It is obvious that the resilin linker requires larger work to be extended because of its larger extension and also the higher rupture forces compared to the protein where only the simple linker can be extended in prior detachment. Even though all the data do not precisely represent stretching of a single molecule, resilin’s ability to dissipate energy upon tensile stress is obvious. The resilin exon I is a very elastic domain having repetitive hydrophilic regions and a few hydrophobic regions. At pH 5, it appears that the uncoiling force of the resilin was higher than the rupture force of the hydrophobin. However, at pH 11, the rupture force exceeded the uncoiling force and the molecule could be extended to its full length. In earlier work, it has been observed that at elevated pH, the unordered secondary structure of the resilin goes through a change toward a more ordered β turn structure, which could explain the strong bundling.46 The bundling of the extended molecules, on the other hand, can be the reason for simultaneous attachment of several hydrophobins to the tip, which is observed as the increased rupture force.
Elasticity of the Protein Layer
By analyzing the force curve recorded during the approach, the stiffness of the layer formed from the cellulose and the proteins can be estimated. The approach curve represents the indentation of the AFM probe to the soft layer of the molecules, and the slope at the close proximity to the surface is a measure of the surface elasticity (Figure 7).47−49
Figure 7.
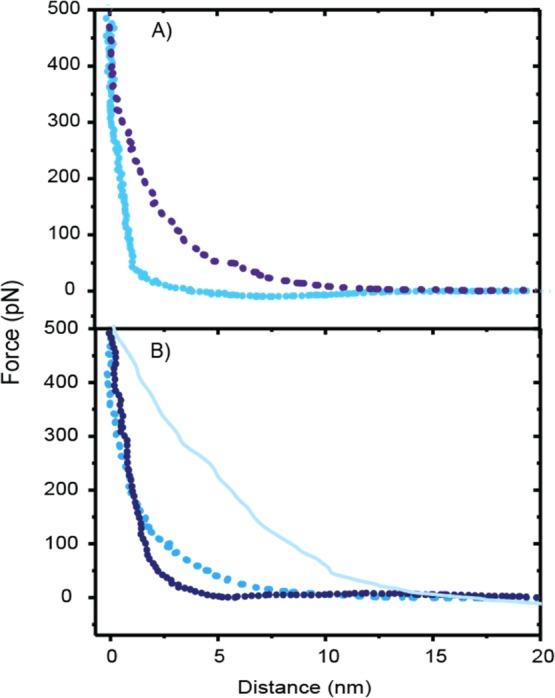
(A) Examples of approach curves on the cellulose surface at pH 5 (light blue dots) and pH 11 (dark blue small dots). (B) Examples of approach curves on the HFBI–dCBM layer at pH 5 (blue dots), dCBM–RLP–HFBI layer at pH 5 (violet small dots), and at pH 11 (light blue line).
When the protein layer is indented, the slope of the measured force curve will change according to the sample stiffness; the steeper is the slope, the stiffer is the layer. Although there is no remarkable difference in the slope between the two fusion proteins at pH 5, on the other hand, the resilin fusion protein layer appears stiffer at pH 5 compared to that at pH 11, which means that the layer becomes very soft once the resilin in swollen. As a comparison, the approach curves on the plain cellulose surface are presented in Figure 7A. The stiffness of the spin-coated cellulose layer was clearly lower at pH 11 compared to pH 5.
Conclusions
By employing SMFS, modular proteins containing adhesive domains and elastic linker region were characterized. The focus was on the mechanical properties of the elastic exon I resilin-like peptide by having it as a linker domain in between the adhesive units, hydrophobin HFBI, and double CBM, readily able to self-assemble in between a hydrophobic AFM tip and a cellulose surface. We could quantify the rupture force of a single hydrophobin, the molecular dimensions of the studied fusion proteins, and the work of rupture of the molecules and make observations on the elasticity of the studied molecular layers. The attachment of the double CBM on a cellulose model surface appeared to be stable under the applied tensile stress.
The effect of the pH on the resilin conformation and its consequences on the mechanical properties of the protein layers were studied. At pH 5, the resilin-like peptide was in a random coil conformation and could not be fully extended by pulling the molecule, whereas at pH 11, full extension of the RLP was obtained. The difference in the elastic moduli of the RLP layer at random and swollen conformations was qualitatively confirmed by the AFM indentation. On the basis of the AFM topography imaging and the SMFS data, at pH 11, the entanglement of the resilin chains appeared to be strong, which indicates the strong tendency toward networklike structures. By defining the binding and elastic properties of these fusion proteins, it has been carried out as a step forward in the design of hybrid materials.
Materials and Methods
Proteins
HFBI–dCBM is a recombinant chimeric protein consisting of two CBMs from Trichoderma reesei, whose production, preparation, and purification is described elsewere.7,9,32 The second construct, dCBM–RLP–HFBI, contains similar domains and hydrophobins (HFBI) connected together with a RLP.23,35 The sequences of the proteins are presented in the Supporting Information (Figure S8). The proteins were dissolved in sodium acetate and phosphate buffers (Sigma-Aldrich).
Production, Expression, and Purification of the dCBM–RLP–HFBI Fusion Protein
Synthetic genes encoding Drosophila melanogaster Rec1-resilin (RLP) and the T. reesei HFBI and CBMs were codon-optimized for T. reesei and synthesized. The T. reesei CBHII (cellobiohydrolase II; Cel6A) and CBHI (cellobiohydrolase I; Cel7a) CBMs were separated by a linker (Figure S8). A C-terminal Strep-tag (WSHPQFEK) was added after the HFBI to the protein to enable detection of the protein and affinity purification. The synthetic gene fragments were cloned into an expression plasmid containing cbh1 promoter, secretion carrier and terminator, hygromycin selection marker, and targeting sequence for the cbh1 locus. dCBM–RLP–HFBI was expressed as a CBHI carrier protein fusion with a KEX2 protease cleavage site, NVISKR, between the carrier and the dCBM–RLP–HFBI protein50 (Figure S9). The ligation mixtures were transformed into Escherichia coli by electroporation, and colonies were selected on the kanamycin agar plates.
The final expression construct pMIs124 was digested with KpnI-XhoI, and the 10 kb integration fragment was isolated and transformed into protoplasts of T. reesei strain M658 carrying deletions for extracellular proteases.51,52 Transformed colonies were selected based on the hygromycin resistance (125 μg/mL) and carried out as described previously.53 Replacement of the cbh1 locus by the transformed expression construct DNA was confirmed by polymerase chain reaction (PCR) (Table S3). The final strain was designated M1230.
A T. reesei strain M1230 expressing dCBM–RLP–HFBI was grown in the 24-well plates in TrMM plus 40 g/L lactose, 20 g/L spent grain extract, 8.6 g/L diammonium citrate, 5.4 g/L Na2SO4, and 100 mM 1,4-piperazinedipropanesulfonic acid at pH 4.5, shaking at 28 °C at 800 rpm (Infors AG). TrMM contains 15.0 g/L KH2PO4, 2.4 mM MgSO4·7H2O, 4.1 mM CaCI2·H2O, 3.7 mg/L CoCI2, 5 mg/L FeSO4·7H2O, 1.4 mg/L ZnSO4·7H2O, and 1.6 mg/L MnSO4·7H2O. The culture was started by inoculating 1 × 107 spores into a 50 mL growth medium. The T. reesei fungus secreted the dCBM–RLP–HFBI fusion protein into the culture medium. One liter of culture was grown for 5 days in the 24-well plates (4 mL per well in multiple plates). The growth medium was filtered through a glass microfiber filter to remove the mycelium, and 0.5 mM phenylmethylsulfonyl fluoride was added to the clarified culture supernatant for stabilization.
The fusion protein was purified by aqueous two-phase separation (ATPS).9 Briefly, to the centrifuged supernatant, a Triton X-114 surfactant (Sigma-Aldrich) was added to a final concentration of 4%. After gentle mixing at 22–24 °C, the solution was allowed to settle in a separation funnel, where the Triton phase was separated. Acetate buffer (50 mM NaAc and 40 mM ethylenediaminetetraacetic acid, pH 5.0) was added to the detergent phase to a final concentration of 5% and mixed gently first. After this, isobutanol was added to the detergent phase, 10 times of the detergent phase volume, and again mixed gently. The samples from ATPS purification were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, Gel Code Blue Stain, Pierce) and after blotting to nitrocellulose filters by western blotting. The membrane was blocked with 5% nonfat milk in a TBST buffer (50 mM Tris, 150 mM NaCl, 0.05% Tween, pH 7.4) and then probed with a strep-Tactin AP 35 conjugate (1:2000 in TBST, IBA Gmbh) followed by chromogenic detection with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Promega). The lower phase from isobutanol extraction was purified further by preparative reversed-phase chromatography using a Vydac C4 (20 cm) column and a gradient elution from 0.1% trifluoroacetic acid (TFA) to 100% acetonitrile (ACN) containing 0.1% TFA acid. After evaporation of ACN and TFA, the peak fractions were pooled and lyophilized.
The samples in the growth medium were collected from the two culture wells on days 3, 4, 5, and 6, and the mycelium was spun down. The supernatant was diluted so that 0.7 μL could be loaded in 20 μL volume per well into a 4–20% Criterion SDS-PAGE gel (Bio-Rad). After separating the proteins in the SDS-PAGE gel, they were transferred to a nitrocellulose membrane using a turbo blotter (Bio-Rad). Immunodetection was done with a mouse anti-Strep tag antibody (IBA #2-1507-001) diluted to 0.5 μg/mL in TBST. The secondary antibody was goat antimouse IgG 1:30 000 diluted in TBST (IRDye 680RD goat antimouse IgG; Li-cor #926-68070). The protein standards (human VEGF receptor 2 protein fragment) were loaded into the gel corresponding to 400, 200, 100, 50, and 25 ng (Abcam #ab182692). The membranes were scanned with the LI-COR Odyssey CLx Infrared Imaging System at 700 nm.
The dCBM–RLP–HFBI expression level in the culture supernatant was determined to be 260 mg/L on day 5 of the culture (Figure S10A). The culture supernatant containing the fusion protein was processed and purified via ATPS and reversed-phase chromatography and was around >97% pure (Figure S10B). From 1 L of culture supernatant, 60 mg of dCBM–RLP–HFBI was recovered after freeze-drying.
Cellulose Functionalization of the Surface
The cellulose model surfaces were prepared on the silicon oxide surface by converting trimethylsilylcellulose (TMSC) into cellulose.54,55 In detail, a 0.5 × 0.5 cm2 piece of silicon oxide surface was spin-coated with 50 μL of 1 mg mL–1 TMSC56 (purity > 98%, friendly gift by Prof. Dr. Thomas Heinze, Friedrich Schiller University Jena) dissolved in hexane at a spinning speed of 5000 rpm for 60 s. After that, the TMSC-coated surface was carefully placed under 12 M hydrochloric acid (Sigma-Aldrich) for 2 min.
Characterization of the Cellulose Films
Finally, to investigate the presence of the cellulose layer, both the films of TMSC and cellulose were characterized by contact angle measurements (SCA 20, Data Physics), ellipsometry (EP3 Imaging Ellipsometer Nanofilm Technology, Accurion, Germany), AFM (Bruker, BioFastScan), and X-ray photoelectron spectroscopy (XPS, ESCA Lab Mk II photoelectronspectrometer, by Vacuum Generators, England). The values reported in Table S4 and Figure S11A–D are in agreement with the values given in the literature.57 The cellulose layer had a thickness of 5 nm in comparison with the thickness of 7.8 nm of the TMSC film. The cellulose contact angle, however, reveals a higher hydrophilicity for the cellulose rather than the TMSC (see Table S4). To choose the most suitable cellulose coating for the experiments, the cellulose surfaces having concentrations of 10, 5, and 1 mg mL–1 have been prepared and characterized. The thinnest and most homogeneous cellulose surface was the one prepared from the 1 mg mL–1 solution (see Table S4).
The conversion from TMSC to cellulose upon HCl treatment is also supported by XPS experiments. In Figure S11A, the C 1s spectrum (Al Kα radiation, h̵ω = 1486.6 eV, take-off angle = 45°, Shirley background correction) of the TMSC sample displays three prominent peaks that can be assigned to the photoemission from the C 1s orbital with the main peak at the lowest binding energy representing an overlap of carbon bond as C–Si and C–C. The two minor peaks at higher binding energy can be assigned to the C–O–C and C–O bonds. Despite a small shift of binding energies (probably caused by a slight charging of the sample), the spectral shape is very close to that observed.57 After HCl treatment, the C 1s spectrum in Figure S11B excels by a strong reduction of the C–C- and C–Si-related peak intensities, giving evidence that the silyl groups have been eliminated from the TMSC and that a conversion to cellulose has taken place. This observation is also in accordance with the previous results reported in the literature.57
Silanization of the AFM Probes
Probes based on the silicon nitride cantilevers with mounted tips have been functionalized via the silanization reaction. Briefly, the AFM probes were plasma-cleaned for 3 min. Then, OTS (Sigma-Aldrich) was added to a solution of bicyclohexyl (Acros Organics) and CCl4 (Sigma Aldrich) in a molar ratio of 479:1, and the probes have been left immersed for 15 min. After that, the probes were rinsed with chloroform and dried in air. All of the functionalized probes have been used in the week of the preparation.
Protein Attachment
CBMs have a strong binding affinity to cellulose so that simply by allowing the proteins spontaneously adsorb on the cellulose-coated silicon surfaces, it was possible to create a stable protein layer on the sample. HFBI–dCBM was dissolved in a buffer solution of 10 mM NaAc at pH 5 for a final concentration of 0.1 μM, and a 50 μL droplet of the solution was placed on top of the sample. After 10 min, the samples were carefully rinsed with 10 mM NaAc to remove the protein that was not adsorbed on the cellulose. The same procedure was adopted for dCBM–RLP–HFBI. The cantilever was calibrated in liquid by using the thermal tune technique.58,59
AFM Imaging
Topographical images have been recorded by AFM in the tapping mode with a Bruker Dimension FastScan Bio instrument. For measurement in air, the images were scanned using silicon cantilevers (Olympus) with a resonance frequency of 300 kHz and a force constant of 26 N m–1. The scanned image size was 1 × 1 μm2. For the measurements in liquid environment, FastScan-D and SNL cantilevers having resonance frequencies of 110 and 65 kHz, and force constants of 0.25 and 0.35 N m–1, respectively, were used.
Atomic Force Microscopy–Single-Molecule Force Spectroscopy
Briefly, the SMFS technique is based on measuring the force between a cantilever of an AFM tip functionalized with the molecule of interest and a surface to which the studied molecule has an affinity. The interacting forces between the protein and the surface cause deflection of the cantilever, which is measured by the position of a laser beam reflected by the cantilever on a photodiode. This signal will go to a detector that will convert it to an electrical response, which is analyzed in terms of a force/distance curve. Experiments were conducted in two ways: (1) between the silanized AFM tip and the protein-functionalized cellulose surface, and (2) silanized surface and proteins on the cellulose-functionalized AFM tip (see Supporting Information, Figure S3). The experiments were carried out in buffer at room temperature with a Bruker Dimension FastScan Bio instrument. Each experiment consists of at least 500–600 single force/distance curves. The force measurements were carried out using a z-range of 500 nm, a scan rate of 1 Hz, a spring constant of 0.03 N m–1, and a relative force trigger of 0.1 V. The rupture forces were evaluated with NanoScope software (Bruker, Santa Barbara, CA) by calculating the difference between the force at rupture in the last stretching event and the baseline for every single curve.59 Baseline correction was carried out using MATLAB software. The histogram values have been extracted from around 200–300 curves for each experiment. The Gaussian curves were fitted to the data to obtain mean values and their standard error. When applicable, multiple peaks were included to obtain a representative overall fit.
Data Analysis
The energy related to the detachment process as well as the contour length and persistence length of the molecules was evaluated by comparing the force/distance curves with the wormlike chain model. This model describes the molecules as semiflexible polymer chains having a contour length L consisting of rigid parts having a length P (persistence length). For the tensile force curve, the model yields eq 1, where T is the temperature, F is the adhesion force, l is the persistence length, L is the contour length, and z is the extension length. The data in Figure 3B–D are obtained from the curves described by this equation, which were fitted to the measured data. The work of rupture was then calculated by integrating the fitted model force/distance curve until the point of rupture.
| 1 |
QCM-D Measurements
Before functionalization, the QCM-D gold sensors (Q-Sense; Gothenburg, Sweden) were exposed to UV light and ozone in an ozonator (BioForce Nanosciences Inc., California USA). By this treatment, the surface was cleaned from the traces of organic contaminants and rendered hydrophilic.60 After that, the QCM-sensors were functionalized with TMSC as described elsewhere.54,55 The sensors were spin-coated twice with toluene (4000 rpm for 15 s) as a cleaning procedure after which TMSC was spin-coated at 4000 rpm and an acceleration of 2200 rpm s–1 for 45 s. The TMSC spin-coated films were then converted into cellulose by exposure to 2 M HCl vapor under vacuum of −0.1 MPa. Before the experiments, the cellulose-coated QCM sensors were left to stabilize overnight in water.
The QCM-D measurements were performed in flow mode using a Q-Sense E4 instrument (Q-Sense, Sweden). AT-cut quartz crystals, with a fundamental frequency of 5 MHz, were purchased from Q-Sense. The measurements were carried out at 22 °C using a flow rate of 100 μL min–1. To obtain information about the stability of the cellulose coatings and the binding of the HFBI–dCBM and dCBM–RLP–HFBI proteins on them, the frequency and dissipation changes of the cellulose-coated sensors during exposure to 0.1 mg mL–1 protein solutions in 10 mM sodium acetate at pH 5 and phosphate buffer at pH 11 were measured. Also, the response of a cellulose-coated sensor was measured.
Acknowledgments
The authors acknowledge Riitta Suihkonen for the technical assistance in the protein purification, Markku Saloheimo and Nina Aro for help in planning the Trichoderma expression, Jussi Joensuu for help with cloning strategies and protein purification, and Marika Vitikainen for help with protein purification. We would also like to thank Thomas Heinze for providing the cellulose, Gerhard Wenz for advice with the cellulose film preparation, Nicolas Thewes for valuable discussion about the force measurements, and finally Ann Westerholm-Parvinen for help with protein purification and editing of the manuscript.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b01133.
Sizes of the molecules, protein amino acidic sequences and PCR primers, XPS, AFM, ellipsometry data describing the conversion from TMSC to cellulose, additional force distance curves, presentation of the reversed setup, description of Voigt model and Sauerbrey equation for the QCM-D experiments, AFM images of HFBI–dCBM and dCBM–RLP–HFBI, and histograms of plain cellulose at pH 11 (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Funding for A.G. and P.L. has been granted from the Academy of Finland Center of Excellence Program via the “Center of Excellence in Molecular Engineering of Biosynthetic Hybrid Materials” (“HYBER”) and for A.G., H.H., S.G., F.M., and K.J. from the German Research Foundation (DFG) via the Collaborative Research Center SFB 1027 “Physical modeling of nonequilibrium processes in biological Systems”.
The authors declare no competing financial interest.
Supplementary Material
References
- Kushner A. M.; Guan Z. Modular Design in Natural and Biomimetic Soft Materials. Angew. Chem., Int. Ed. 2011, 50, 9026–9057. 10.1002/anie.201006496. [DOI] [PubMed] [Google Scholar]
- Suzuki M.; Saruwatari K.; Kogure T.; Yamamoto Y.; Nishimura T.; Kato T.; Nagasawa H. An Acidic Matrix Protein, Pif, Is a Key Macromolecule for Nacre Formation. Science 2009, 325, 1388–1390. 10.1126/science.1173793. [DOI] [PubMed] [Google Scholar]
- Linder M. B. Biomaterials: Recipe for Squid Beak. Nat. Chem. Biol. 2015, 11, 455–456. 10.1038/nchembio.1842. [DOI] [PubMed] [Google Scholar]
- Várnai A.; Mäkelä M. R.; Djajadi D. T.; Rahikainen J.; Hatakka A.; Viikari L.. Advances in Applied Microbiology; Sariaslani S., Gadd G. M., Eds.; Academic Press: Dundee, Scotland, 2014; pp 103–165. [DOI] [PubMed] [Google Scholar]
- Henrissat B. Cellulases and Their Interaction with Cellulose. Cellulose 1994, 1, 169–196. 10.1007/bf00813506. [DOI] [Google Scholar]
- Carvalho A. L.; Goyal A.; Prates J. A. M.; Bolam D. N.; Gilbert H. J.; Pires V. M. R.; Ferreira L. M. A.; Planas A.; Romão M. J.; Fontes C. M. G. A. The Family 11 Carbohydrate-Binding Module of Clostridium Thermocellum Lic26A-Cel5E Accommodates β-1,4- and β-1,3–1,4-Mixed Linked Glucans at a Single Binding Site. J. Biol. Chem. 2004, 279, 34785–34793. 10.1074/jbc.m405867200. [DOI] [PubMed] [Google Scholar]
- Linder M.; Salovuori I.; Ruohonen L.; Teeri T. T. Characterization of a Double Cellulose-Binding Domain. Synergistic High Affinity Binding to Crystalline Cellulose. J. Biol. Chem. 1996, 271, 21268–21272. 10.1074/jbc.271.35.21268. [DOI] [PubMed] [Google Scholar]
- Laaksonen P.; Walther A.; Malho J.-M.; Kainlauri M.; Ikkala O.; Linder M. B. Genetic Engineering of Biomimetic Nanocomposites: Diblock Proteins, Graphene, and Nanofibrillated Cellulose. Angew. Chem., Int. Ed. 2011, 50, 8688–8691. 10.1002/anie.201102973. [DOI] [PubMed] [Google Scholar]
- Linder M. B.; Qiao M.; Laumen F.; Selber K.; Hyytiä T.; Nakari-Setälä T.; Penttilä M. E. Efficient Purification of Recombinant Proteins Using Hydrophobins as Tags in Surfactant-Based Two-Phase Systems. Biochemistry 2004, 43, 11873–11882. 10.1021/bi0488202. [DOI] [PubMed] [Google Scholar]
- Linder M. B. Hydrophobins: Proteins That Self Assemble at Interfaces. Curr. Opin. Colloid Interface Sci. 2009, 14, 356–363. 10.1016/j.cocis.2009.04.001. [DOI] [Google Scholar]
- Lienemann M.; Grunér M. S.; Paananen A.; Siika-aho M.; Linder M. B. Charge-Based Engineering of Hydrophobin HFBI: Effect on Interfacial Assembly and Interactions. Biomacromolecules 2015, 16, 1283–1292. 10.1021/acs.biomac.5b00073. [DOI] [PubMed] [Google Scholar]
- Hou S.; Yang K.; Qin M.; Feng X.-Z.; Guan L.; Yang Y.; Wang C. Patterning of Cells on Functionalized Poly(Dimethylsiloxane) Surface Prepared by Hydrophobin and Collagen Modification. Biosens. Bioelectron. 2008, 24, 912–916. 10.1016/j.bios.2008.07.045. [DOI] [PubMed] [Google Scholar]
- Li X.; Hou S.; Feng X.; Yu Y.; Ma J.; Li L. Patterning of Neural Stem Cells on Poly(Lactic-Co-Glycolic Acid) Film Modified by Hydrophobin. Colloids Surf., B 2009, 74, 370–374. 10.1016/j.colsurfb.2009.07.039. [DOI] [PubMed] [Google Scholar]
- Hähl H.; Vargas J. N.; Griffo A.; Laaksonen P.; Szilvay G.; Lienemann M.; Jacobs K.; Seemann R.; Fleury J.-B. Pure Protein Bilayers and Vesicles from Native Fungal Hydrophobins. Adv. Mater. 2017, 29, 1602888. 10.1002/adma.201602888. [DOI] [PubMed] [Google Scholar]
- Bennet-Clark H. The First Description of Resilin. J. Exp. Biol. 2007, 210, 3879–3881. 10.1242/jeb.001339. [DOI] [PubMed] [Google Scholar]
- Elvin C. M.; Carr A. G.; Huson M. G.; Maxwell J. M.; Pearson R. D.; Vuocolo T.; Liyou N. E.; Wong D. C. C.; Merritt D. J.; Dixon N. E. Synthesis and Properties of Crosslinked Recombinant Pro-Resilin. Nature 2005, 437, 999–1002. 10.1038/nature04085. [DOI] [PubMed] [Google Scholar]
- Andersen S. O.; Weis-Fogh T.. Resilin. A Rubberlike Protein in Arthropod Cuticle. In Advances in Insect Physiology; Beament J. W. L., Treherne J. E., Wigglesworth V. B., Eds.; Academic Press, 1964; Vol. 2, pp 1–65. [Google Scholar]
- Gosline J.; Lillie M.; Carrington E.; Guerette P.; Ortlepp C.; Savage K. Elastic Proteins: Biological Roles and Mechanical Properties. Philos. Trans. R. Soc., B 2002, 357, 121–132. 10.1098/rstb.2001.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papi M. The L2 Motivational Self System, L2 Anxiety, and Motivated Behavior: A Structural Equation Modeling Approach. System 2010, 38, 467–479. 10.1016/j.system.2010.06.011. [DOI] [Google Scholar]
- Weis-Fogh T. Molecular Interpretation of the Elasticity of Resilin, a Rubber-like Protein. J. Mol. Biol. 1961, 3, 648–667. 10.1016/s0022-2836(61)80028-4. [DOI] [Google Scholar]
- Weis-Fogh T. Thermodynamic Properties of Resilin, a Rubber-like Protein. J. Mol. Biol. 1961, 3, 520–531. 10.1016/s0022-2836(61)80018-1. [DOI] [Google Scholar]
- Michels J.; Gorb S. N. Detailed Three-Dimensional Visualization of Resilin in the Exoskeleton of Arthropods Using Confocal Laser Scanning Microscopy. J. Microsc. 2012, 245, 1–16. 10.1111/j.1365-2818.2011.03523.x. [DOI] [PubMed] [Google Scholar]
- Balu R.; Whittaker J.; Dutta N. K.; Elvin C. M.; Choudhury N. R. Multi-Responsive Biomaterials and Nanobioconjugates from Resilin-like Protein Polymers. J. Mater. Chem. B 2014, 2, 5936–5947. 10.1039/c4tb00726c. [DOI] [PubMed] [Google Scholar]
- Li L.; Kiick K. L. Transient Dynamic Mechanical Properties of Resilin-Based Elastomeric Hydrogels. Front. Chem. 2014, 2, 21. 10.3389/fchem.2014.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Tong Z.; Jia X.; Kiick K. L. Resilin-like Polypeptide Hydrogels Engineered for Versatile Biological Function. Soft Matter 2013, 9, 665–673. 10.1039/c2sm26812d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D. J.; Dufrêne Y. F. Atomic Force Microscopy: A Nanoscopic Window on the Cell Surface. Trends Cell Biol. 2011, 21, 461–469. 10.1016/j.tcb.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Puchner E. M.; Gaub H. E. Force and Function: Probing Proteins with AFM-Based Force Spectroscopy. Curr. Opin. Struct. Biol. 2009, 19, 605–614. 10.1016/j.sbi.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Li Q.; Zhang T.; Pan Y.; Ciacchi L. C.; Xu B.; Wei G. AFM-Based Force Spectroscopy for Bioimaging and Biosensing. RSC Adv. 2016, 6, 12893–12912. 10.1039/c5ra22841g. [DOI] [Google Scholar]
- Baldock C.; Oberhauser A. F.; Ma L.; Lammie D.; Siegler V.; Mithieux S. M.; Tu Y.; Chow J. Y. H.; Suleman F.; Malfois M.; et al. Shape of Tropoelastin, the Highly Extensible Protein That Controls Human Tissue Elasticity. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 4322–4327. 10.1073/pnas.1014280108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbrana F.; Fotia C.; Bracalello A.; Baldini N.; Marletta G.; Ciapetti G.; Bochicchio B.; Vassalli M. Multiscale Characterization of a Chimeric Biomimetic Polypeptide for Stem Cell Culture. Bioinspir. Biomim. 2012, 7, 046007. 10.1088/1748-3182/7/4/046007. [DOI] [PubMed] [Google Scholar]
- Li H.; Oberhauser A. F.; Fowler S. B.; Clarke J.; Fernandez J. M. Atomic Force Microscopy Reveals the Mechanical Design of a Modular Protein. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 6527–6531. 10.1073/pnas.120048697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson B. R.; Bogomolovas J.; Labeit S.; Granzier H. Single Molecule Force Spectroscopy on Titin Implicates Immunoglobulin Domain Stability as a Cardiac Disease Mechanism. J. Biol. Chem. 2013, 288, 5303–5315. 10.1074/jbc.m112.401372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke W. A.; Kulke M.; Li H.; Fujita-Becker S.; Neagoe C.; Manstein D. J.; Gautel M.; Fernandez J. M. PEVK Domain of Titin: An Entropic Spring with Actin-Binding Properties. J. Struct. Biol. 2002, 137, 194–205. 10.1006/jsbi.2002.4468. [DOI] [PubMed] [Google Scholar]
- Reversible Unfolding of Individual Titin Immunoglobulin Domains by AFM|Science. http://science.sciencemag.org/content/276/5315/1109 (accessed Jun 14, 2017). [DOI] [PubMed]
- Qin G.; Hu X.; Cebe P.; Kaplan D. L. Mechanism of Resilin Elasticity. Nat. Commun. 2012, 3, 1003. 10.1038/ncomms2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbrana F.; Lorusso M.; Canale C.; Bochicchio B.; Vassalli M. Effect of Chemical Cross-Linking on the Mechanical Properties of Elastomeric Peptides Studied by Single Molecule Force Spectroscopy. J. Biomech. 2011, 44, 2118–2122. 10.1016/j.jbiomech.2011.05.028. [DOI] [PubMed] [Google Scholar]
- Fahs A.; Louarn G. Plant Protein Interactions Studied Using AFM Force Spectroscopy: Nanomechanical and Adhesion Properties. Phys. Chem. Chem. Phys. 2013, 15, 11339–11348. 10.1039/c3cp51007g. [DOI] [PubMed] [Google Scholar]
- Bizzarri A. R.; Cannistraro S. The Application of Atomic Force Spectroscopy to the Study of Biological Complexes Undergoing a Biorecognition Process. Chem. Soc. Rev. 2010, 39, 734–749. 10.1039/b811426a. [DOI] [PubMed] [Google Scholar]
- Yadavalli V. K.; Forbes J. G.; Wang K. Functionalized Self-Assembled Monolayers on Ultraflat Gold as Platforms for Single Molecule Force Spectroscopy and Imaging. Langmuir 2006, 22, 6969–6976. 10.1021/la060320h. [DOI] [PubMed] [Google Scholar]
- Peng C.; Liu J.; Zhao D.; Zhou J. Adsorption of Hydrophobin on Different Self-Assembled Monolayers: The Role of the Hydrophobic Dipole and the Electric Dipole. Langmuir 2014, 30, 11401–11411. 10.1021/la502595t. [DOI] [PubMed] [Google Scholar]
- Friedsam C.; Del Campo Bécares A.; Jonas U.; Seitz M.; Gaub H. E. Adsorption of Polyacrylic Acid on Self-Assembled Monolayers Investigated by Single-Molecule Force Spectroscopy. New J. Phys. 2004, 6, 9. 10.1088/1367-2630/6/1/009. [DOI] [PubMed] [Google Scholar]
- Shlyakhtenko L. S.; Dutta S.; Li M.; Harris R. S.; Lyubchenko Y. L. Single-Molecule Force Spectroscopy Studies of APOBEC3A–Single-Stranded DNA Complexes. Biochemistry 2016, 55, 3102–3106. 10.1021/acs.biochem.6b00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C. D.; Wang C.; Acevedo-Vélez C.; Gellman S. H.; Abbott N. L. Modulation of Hydrophobic Interactions by Proximally Immobilized Ions. Nature 2015, 517, 347–350. 10.1038/nature14018. [DOI] [PubMed] [Google Scholar]
- Alsteens D.; Beaussart A.; Derclaye S.; El-Kirat-Chatel S.; Park H. R.; Lipke P. N.; Dufrêne Y. F. Single-Cell Force Spectroscopy of Als-Mediated Fungal Adhesion. Anal. Methods 2013, 5, 3657–3662. 10.1039/c3ay40473k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong M. Y.; Dutta N. K.; Choudhury N. R.; Kim M.; Elvin C. M.; Hill A. J.; Thierry B.; Vasilev K. A PH-Responsive Interface Derived from Resilin-Mimetic Protein Rec1-Resilin. Biomaterials 2010, 31, 4434–4446. 10.1016/j.biomaterials.2010.02.019. [DOI] [PubMed] [Google Scholar]
- Mayavan S.; Dutta N. K.; Choudhury N. R.; Kim M.; Elvin C. M.; Hill A. J. Self-Organization, Interfacial Interaction and Photophysical Properties of Gold Nanoparticle Complexes Derived from Resilin-Mimetic Fluorescent Protein Rec1-Resilin. Biomaterials 2011, 32, 2786–2796. 10.1016/j.biomaterials.2010.12.030. [DOI] [PubMed] [Google Scholar]
- Variola F. Atomic Force Microscopy in Biomaterials Surface Science. Phys. Chem. Chem. Phys. 2015, 17, 2950–2959. 10.1039/c4cp04427d. [DOI] [PubMed] [Google Scholar]
- Roduit C.; Sekatski S.; Dietler G.; Catsicas S.; Lafont F.; Kasas S. Stiffness Tomography by Atomic Force Microscopy. Biophys. J. 2009, 97, 674–677. 10.1016/j.bpj.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigmatullin R.; Lovitt R.; Wright C.; Linder M.; Nakari-Setälä T.; Gama M. Atomic Force Microscopy Study of Cellulose Surface Interaction Controlled by Cellulose Binding Domains. Colloids Surf., B 2004, 35, 125–135. 10.1016/j.colsurfb.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Landowski C. P.; Mustalahti E.; Wahl R.; Croute L.; Sivasiddarthan D.; Westerholm-Parvinen A.; Sommer B.; Ostermeier C.; Helk B.; Saarinen J.; et al. Enabling Low Cost Biopharmaceuticals: High Level Interferon Alpha-2b Production in Trichoderma Reesei. Microb. Cell Fact. 2016, 15, 104. 10.1186/s12934-016-0508-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multiple Proteases Deficient Filamentous Fungal Cells and Methods of Use Thereof.
- Landowski C. P.; Huuskonen A.; Wahl R.; Westerholm-Parvinen A.; Kanerva A.; Hänninen A.-L.; Salovuori N.; Penttilä M.; Natunen J.; Ostermeier C.; et al. Enabling Low Cost Biopharmaceuticals: A Systematic Approach to Delete Proteases from a Well-Known Protein Production Host Trichoderma Reesei. PLoS One 2015, 10, e0134723 10.1371/journal.pone.0134723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penttilä M.; Nevalainen H.; Rättö M.; Salminen E.; Knowles J. A Versatile Transformation System for the Cellulolytic Filamentous Fungus Trichoderma Reesei. Gene 1987, 61, 155–164. 10.1016/0378-1119(87)90110-7. [DOI] [PubMed] [Google Scholar]
- Kontturi E.; Thüne P. C.; Niemantsverdriet J. W. Cellulose Model Surfaces Simplified Preparation by Spin Coating and Characterization by X-Ray Photoelectron Spectroscopy, Infrared Spectroscopy, and Atomic Force Microscopy. Langmuir 2003, 19, 5735–5741. 10.1021/la0340394. [DOI] [Google Scholar]
- Kontturi E.; Thüne P. C.; Niemantsverdriet J. W. Novel Method for Preparing Cellulose Model Surfaces by Spin Coating. Polymer 2003, 44, 3621–3625. 10.1016/s0032-3861(03)00283-0. [DOI] [Google Scholar]
- Schaub M.; Wenz G.; Wegner G.; Stein A.; Klemm D. Ultrathin Films of Cellulose on Silicon Wafers. Adv. Mater. 1993, 5, 919–922. 10.1002/adma.19930051209. [DOI] [Google Scholar]
- Mohan T.; Kargl R.; Doliška A.; Vesel A.; Köstler S.; Ribitsch V.; Stana-Kleinschek K. Wettability and Surface Composition of Partly and Fully Regenerated Cellulose Thin Films from Trimethylsilyl Cellulose. J. Colloid Interface Sci. 2011, 358, 604–610. 10.1016/j.jcis.2011.03.022. [DOI] [PubMed] [Google Scholar]
- Hutter J. L.; Bechhoefer J. Calibration of Atomic-force Microscope Tips. Rev. Sci. Instrum. 1993, 64, 1868–1873. 10.1063/1.1143970. [DOI] [Google Scholar]
- Carpick R.; Batteas J.; Boer M.. Scanning Probe Studies of Nanoscale Adhesion Between Solids in the Presence of Liquids and Monolayer Films. In Springer Handbook of Nanotechnology; Bhushan B., Ed.; Springer: Berlin, 2007; pp 951–980. [Google Scholar]
- Keller C. A.; Kasemo B. Surface Specific Kinetics of Lipid Vesicle Adsorption Measured with a Quartz Crystal Microbalance. Biophys. J. 1998, 75, 1397–1402. 10.1016/s0006-3495(98)74057-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.