Abstract
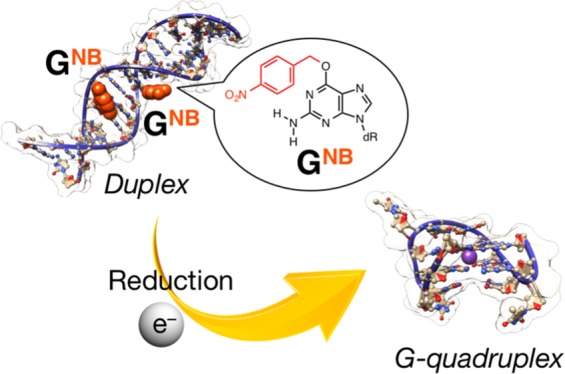
Stimuli-controlled structural transitions of nucleic acids have received growing attentions owing to their potential applications in the fields of chemical and synthetic biology. Here, we describe the development of reduction-responsive deoxyribonucleic acid (DNA) duplexes, in which guanine rings bearing a reduction-responsive cleavable nitrobenzyl (NB) group at the O6 position (GNB) are introduced at defined positions. We demonstrate that the artificial NB group can be removed in response to reduction stimulus without the dissociation of the intermolecular duplex structure, which comprises a G-quadruplex forming nucleic acid strand with one GNB and its complementary sequence with one mismatch pair. Meanwhile, another duplex that comprised a G-quadruplex forming nucleic acid strand with two GNB and its complementary sequence with three mismatch pairs exhibited reduction-responsive structural transitions from intermolecular duplex to intramolecular quadruplex. These findings might be useful for the development of DNA architectures endowed with reduction-responsive functions.
Introduction
Stimuli-responsive biomolecules, including nucleic acids, have attracted growing attention because of the possibility to control their functions and processes in biological environments. To this end, photocaged biomolecules have been explored for decades.1 When introducing photoremovable chemical groups into a target biomolecule, the activities of that biomolecule can be temporarily masked; then, they can be restored by an external stimulus, i.e., photoirradiation, to remove the chemical groups introduced. Moreover, recent progress and deeper understanding of bio-orthogonal chemical reactions have extended the scope of the stimuli: stimuli-triggered release of active biomolecules (decaging) is no longer limited to photostimuli.2 Some recent interesting examples are the inverse-demand Diels–Alder reaction-triggered decaging to release active proteins, in which tetrazine derivatives can be used as a trigger for the decaging, as described by Chen et al.3 In addition, Deiters et al. reported a phosphine-mediated Staudinger reduction capable of activating proteins, which are caged via an azido-functionalized amino acid.4 In those works, xenobiotic, but not seriously toxic, small molecules can trigger the selective decaging to restore active proteins even in living cells. In addition, endogenous compounds, such as reactive oxygen species (e.g., hydrogen peroxide), can be employed to induce the decaging of proteins with phenylboronic acid derivatives.5 Considering these seminal works on proteins and their progress, chemically caged nucleic acids are not yet sufficiently explored.
Nucleic acids hold inherently important biological activities, for example, even short strands produce RNA interference (RNAi).6 Moreover, artificially designed or explored functional nucleic acids, such as decoy deoxyribonucleic acid (DNA),7 aptamers,8 ribozymes,9 and DNAzymes,10 have been actively investigated. Stimuli-responsive control over the structure of nucleic acids would thus set the basis for several bioapplications. Moreover, photocaged nucleic acids have been explored and utilized as tools for biological research, such as light control of transcription, aptamers, antisense activity, and RNAi.1,11 However, examples of chemically caged nucleic acids and their applications as well as the understanding of their behavior are still rather limited compared to the recent substantial progress on stimuli-responsive proteins. The pioneering work of Micura et al. developed RNAs that contain one or two O6-trichloroethyl-modified guanines (GTCE) in their sequences. They are capable of exhibiting zinc-responsive transition in their folding structures accompanied with the recovery of guanine by the removal of the trichloroethyl groups.12 Recently, Saneyoshi and Ono et al. developed a O4-(4-nitrobenzyl)-modified thymine (TNB) and investigated the hypoxia(bioreduction)-responsive properties of oligodeoxynucleotides (ODNs) bearing it.13a Also, very recently, Morihiro and Obika et al. developed nucleobases bearing 4-boronobenzyl group (NBB) and introduced O4-(4-boronobenzyl)-modified thymine into antisense ODNs for hydrogen peroxide-triggered gene silencing.13b Not only nucleobases but also other moieties of the nucleic acids, e.g., phosphate diester14 and ribose,15 have been target sites for modification with chemically caged groups.16
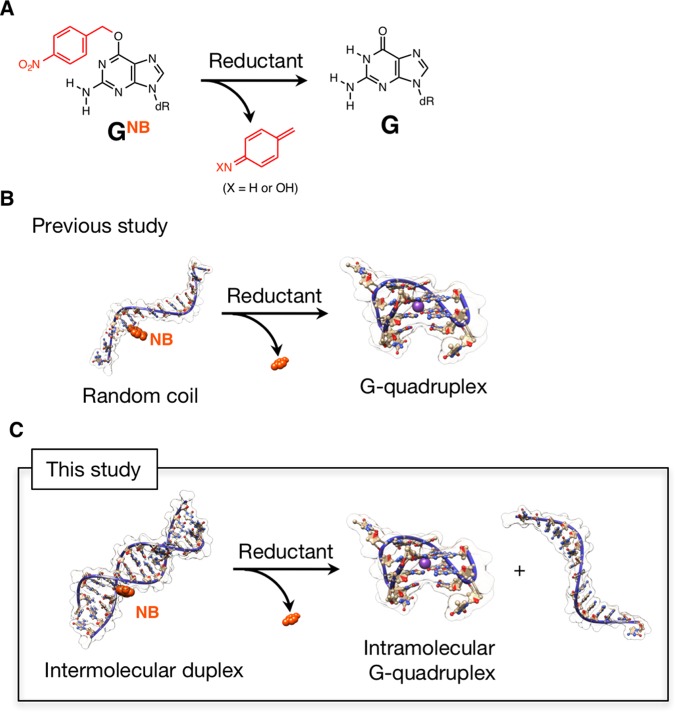
In view of the fact that the guanine exhibits unique structural polymorphs, such as G-quartet, compared with other nucleobases, we previously developed a reduction-responsive guanine (GNB, Figure 1A) bearing a nitrobenzyl (NB) group at the O6 position17 and introduced it into the 5′-end of a G-quadruplex forming DNA (15-mer, 5′-GGTTGGTGTGGTTGG-3′), which is known as thrombin-binding aptamer (TBA).8b In our previous study,17 we demonstrated that tertiary structural transition from random coil to intramolecular G-quadruplex (both are single-stranded structures) can be triggered by chemical and enzymatic reductions to convert GNB to G (Figure 1B). Such a stimuli-controlled transition of oligonucleotide tertiary structures might be useful for OFF–ON switching of their functions. Recently, reduction-responsive biomolecules as well as self-assembled nanomaterials have been attracting increasing attention mostly for cancer-related hypoxia investigations.18 Conversely, the experience on the selection of aptamers revealed that a significant proportion of DNA aptamers adopt G-quadruplex structures.19 In addition, polymorphic G-quadruplex structures are involved in telomere DNA structures and their corresponding biological activity.20 Therefore, stimuli-controlled formation of the G-quadruplex structures might provide potential opportunity to explore OFF–ON switching of target-binding functions or transcription regulation. In this study, we describe the reduction-responsive behavior of intermolecular duplexes, comprising TBAs with one or two GNB and their complementary sequences with a specific number of mismatches. TBA(8b) was selected as typical and functional G-quadruplex. Also, the relatively short length of TBA might be suitable for inducing duplex dissociation in response to a desired stimulus. In particular, we present a sequence design to enable the reduction stimuli-triggered dissociation of such intermolecular duplex to simultaneously release intramolecular TBA G-quadruplex (Figure 1C).
Figure 1.
(A) Reduction-triggered conversion of O6-nitrobenzyldeoxyguanosine (GNB) to deoxyguanosine (G). dR denotes deoxyribose. The typical reductant used in this study is sodium dithionite (Na2S2O4). (B, C) Schematic representations of reduction-triggered (B) tertiary structural transition from random coil to intramolecular chair-type antiparallel TBA G-quadruplex, reported in the previous study17 and (C) dissociation of intermolecular duplex to simultaneously form intramolecular TBA G-quadruplex with the release of a nearly complementary strand (one GNB is highlighted, see text for the details). The molecular models were constructed using the HyperChem software and visualized with the UCSF Chimera package.21
Results and Discussion
Sequence Design of Reduction-Responsive G-Quadruplex Forming DNAs Containing One or Two GNB
In this study, we prepared two TBAs (15-mer) containing GNB: TBA5NB (5′-GGTTGNBGTGTGGTTGG-3′, superscript before NB in “TBA5NB” is the position from the 5′-end) and TBA(5,8)NB2 (5′-GGTTGNBGTGNBTGGTTGG-3′), according to the previously reported method (see Supporting Information).17 According to the pioneer studies by Mayer et al., where a photocaged G is incorporated into TBA to destabilize or inhibit the formation of the G-quadruplex structure, at least one G from the eight involved in the quadruplex formation has to be replaced with the photocaged G; that is a G at positions 1, 2, 5, 6, 10, 11, 14, or 15 in the sequence shown above.22 In our previous study, we incorporated a GNB at position 1 (i.e., 5′-end), which enabled almost complete inhibition of the G-quadruplex formation and thus led to the development of a reduction-responsive G-quadruplex forming DNA (TBA1NB), as shown in Figure 1B. In this study, G was replaced with GNB at position 5 (near the center of the sequence), which is expected to have a substantial effect on the thermodynamic stability of duplexes comprising TBAs with GNB and their nearly complementary strands. The incapability of both TBA5NB and TBA(5,8)NB2 to form a G-quadruplex structure and the reduction stimulus-induced formation of a TBA G-quadruplex were evident from circular dichroism (CD) spectroscopic measurements, which are similar to those of TBA1NB (Figure S6).17
Duplex Formation: TBA with One GNB + Its Nearly Complementary Sequences
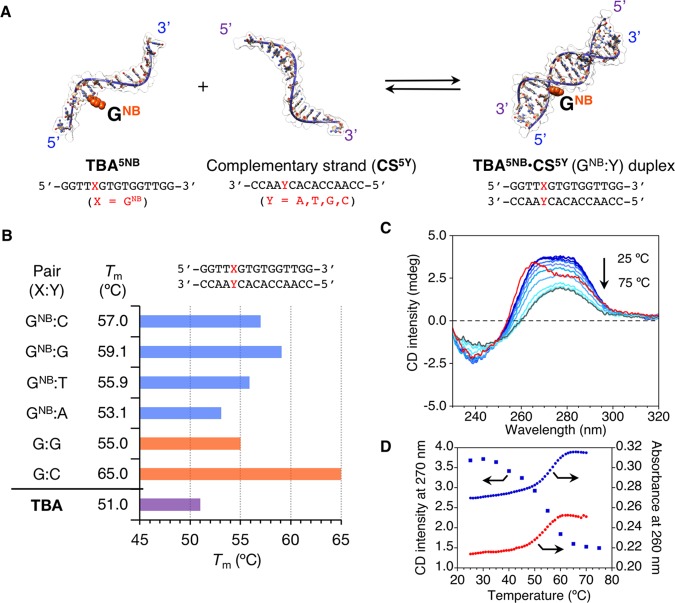
To understand the influence of GNB on the thermodynamic stability of the duplex (Figure 2A), melting temperatures (Tm) were measured by means of an ultraviolet (UV) absorption spectroscopy for the nearly complementary sequences 5′-GGTTXGTGTGGTTGG-3′ (X = GNB, TBA5NB) and 3′-CCAAYCACACCAACC-5′ (CS5Y, superscript is the position from the 3′-end). Different bases were incorporated at position Y (A or T or G or C) to give a series of TBA5NB·CS5Y (GNB:Y) duplexes. As summarized in Figure 2B, the following Tm hierarchy was observed in the order of decreasing stability: GNB:G > GNB:C > GNB:T > GNB:A.23 The four TBA5NB·CS5Y (GNB:Y = A or T or G or C) duplexes evidently showed lower thermodynamic stability compared to the perfectly matched native TBA·CS5C (G:C) duplex (Tm = 65.0 °C). Nonetheless, TBA5NB·CS5G duplex (GNB:G, Tm = 59.1 °C), which was the most stable among the four duplexes investigated, was thermodynamically more stable than TBA·CS5G duplex (G:G, Tm = 55.0 °C) by 4.1 °C (ΔTm: difference in thermodynamic stability between duplexes with or without NB group).24 This implies that duplexes bearing a GNB:G pair become unstable after the removal of the NB group, which might be promising to induce duplex dissociation in response to a reduction stimulus. The study was thus focused on the GNB:G pair.
Figure 2.
(A) Schematic representation of TBA5NB·CS5Y duplexes formation. Duplexes comprise 5′-GGTTGNBGTGTGGTTGG-3′ (TBA5NB) and 3′-CCAAYCACACCAACC-5′ (CS5Y) with different bases incorporated at position Y (A or T or G or C). (B) Thermal stabilities (Tm) of TBA5NB·CS5Y duplex and TBA G-quadruplex (see Figures S3 and S5). (C) CD spectral changes during thermal melting for TBA5NB·CS5G (blue lines: from 25 to 70 °C at a 5 °C interval, gray line: 75 °C) and CD spectrum of TBA·CS5C duplex (red line). (D) Representative thermal CD (270 nm, blue squares for TBA5NB·CS5G duplex) and UV (260 nm, blue circles for TBA5NB·CS5G, red circles for TBA·CS5G duplex) melting curves. Conditions: TBA5NB·CS5Y duplex (10 μM) and TBA (10 μM), 50 mM phosphate buffer (pH 7.6) with 100 mM KCl for (B) and 100 mM N-(2-hydroxyethyl)piperazine-N′-ethanesulfonic acid (HEPES) (pH 7.0) buffer with 0.1 mM ethylenediaminetetraacetate (EDTA) and 105 mM KCl for (C, D), and 0.1 cm cell length.
The CD spectrum of TBA5NB·CS5G duplex before thermal melting exhibited positive signals centered at 270 nm and negative signals at 240 nm, which can be compared to that of perfectly matched native TBA·CS5C duplex (Figure 2C). The observed small deviation (i.e., enhanced positive CD intensity at ∼280 nm for TBA5NB·CS5G duplex compared to TBA·CS duplex) suggests a contribution of the NB group under the chiral environment of a double-stranded helical structure because NB has an absorption band at ∼280 nm.17 CD spectral changes during the thermal melting of TBA5NB·CS5G duplex were characteristic for the double-stranded helical structure of ODNs (a red shift of the zero-crossing wavelength from 257 to 260 nm and a decrease in the intensity of CD signals, Figure 2C). Moreover, both the UV and CD thermal melting experiments of TBA5NB·CS5G duplex gave similar melting curves, as shown in Figure 2D. Also, almost no hysteresis was observed (Figure S4), indicating fast hybridization kinetics of the duplex. Overall, although the exact structure of the TBA5NB·CS5G duplex, i.e., the location or dynamics of NB in the double-stranded helical structure, remains elusive and requires further studies, these results suggest that TBA5NB·CS5G duplex can adopt the standard double-stranded helical structure (most probably a B-form conformation) without significant structural perturbations.
Reduction Stimuli-Responsive Properties of TBA5NB·CS5G Duplex with One GNB:G Pair: Reduction-Responsive Duplex Exhibiting No Dissociation
From thermal denaturation experiments with CD spectroscopy under the Tm measurement conditions similar to the duplexes (Figure S5), the Tm of intramolecular G-quadruplex structure of TBA was evaluated as 51.0 °C (Figure 2B, similar to the reported value25). Therefore, the thermodynamic stability of the related intermolecular duplexes bearing one GNB:G pair and the intramolecular TBA G-quadruplex is TBA5NB·CS5G duplex (Tm = 59.1 °C) > TBA·CS5G duplex (Tm = 55.0 °C) > TBA G-quadruplex (Tm = 51.0 °C). This indicates that TBA·CS5G, which was obtained after the reduction-triggered removal of NB from TBA5NB·CS5G, is more stable than the intramolecular TBA G-quadruplex structure.
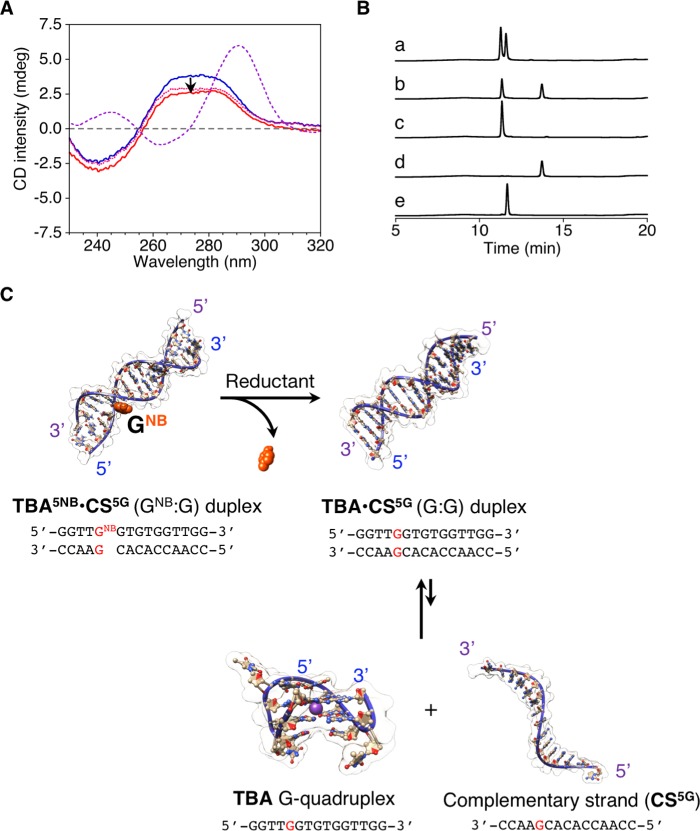
To investigate the reduction-responsive conformational change of TBA5NB·CS5G duplex, Na2S2O4 was employed as reductant to induce the removal of NB. As shown in Figure 3A, only a small change was observed in the CD spectra of the TBA5NB·CS5G duplex, even after addition of a sufficient amount of Na2S2O4 that was determined in our previous study to allow for almost quantitative removal of the NB group. Furthermore, as shown in Figure 3B, the conversion from TBA5NB (tR = 13.7 min) to TBA (tR = 11.6 min) after the addition of Na2S2O4 was confirmed by ion-pair reversed-phase high performance liquid chromatography (IP RP HPLC) analyses as well as mass spectra (MS, data not shown). The peaks in the chromatogram were assigned using authentic samples as references or were based on the mass spectra (tR = 11.3 min for CS5G). Conversely, in the absence of CS5G, single-stranded TBA5NB showed a sharp increase in the CD signal at 290 nm after the addition of Na2S2O4 under similar conditions (vide supra, Figure S6), which indicates the reduction-responsive folding of TBA5NB to form the TBA G-quadruplex. These results suggest that the reduction-triggered removal of NB from the TBA5NB·CS5G duplex yielded mostly the TBA·CS5G duplex with neither duplex dissociation nor TBA G-quadruplex formation (Figure 3C).
Figure 3.
(A) CD spectra of the TBA5NB·CS5G duplex before (blue line) and after (red line) the addition of Na2S2O4. CD spectrum of a solution containing TBA and CS5G (pink dotted line) and the theoretical sum of the CD spectra of TBA and CS5G (purple dashed line) are also shown. (B) IP RP HPLC chromatograms of (a) TBA5NB·CS5G duplex after the addition of Na2S2O4, (b) TBA5NB·CS5G duplex before the addition of Na2S2O4, (c) CS5G, (d) TBA5NB, and (e) TBA (detection at 260 nm). Conditions: TBA5NB·CS5G (10 μM), Na2S2O4/TBA5NB·CS5G = 1050 ([Na2S2O4] = 10.5 mM), 100 mM HEPES (pH 7.0) buffer with 0.1 mM EDTA, and 105 mM KCl, 30 °C for (A) CD spectrum and 0.1 cm cell length. (C) Schematic representations showing the reduction-responsive properties of the intermolecular duplex (TBA5NB·CS5G duplex).
Reduction Stimuli-Responsive Properties of the TBA(5,8)NB2·CS(5,8)G7T Duplex with Two GNB:G Pairs: Transition from Intermolecular Duplex to Intramolecular Quadruplex
To induce the intermolecular duplex dissociation after reduction-triggered removal of NB and simultaneous intramolecular quadruplex folding (reduction-induced transition from duplex to quadruplex), the minimum requirements would be as follows: (i) the initial sequences of a duplex containing GNB should possess enough thermodynamic stability to maintain intermolecular duplex structure at a given temperature and (ii) thermodynamic stability of the duplex after the removal of NB should become at least lower than that of the intramolecular TBA G-quadruplex, hopefully, below the designated temperature. To satisfy these criteria and based on the results mentioned above, the number of GNB:G pairs was increased in the initial sequence of the intermolecular duplex. In particular, the G at position 8 in the TBA sequence in addition to G at position 5 was replaced with GNB to obtain TBA(5,8)NB2. Furthermore, based on calculations with the DINAMelt software,27 one additional mismatch T:T pair was carefully introduced in the middle of the sequence (position 7 from the 5′-end of TBA) to modulate the thermodynamic stability of the duplex; i.e., to decrease the stability of the duplex after removal of NB while maintaining the stability of the initial duplex at a given temperature. Thereby, the TBA(5,8)NB2·CS(5,8)G7T duplex was constructed (TBA(5,8)NB2: 5′-GGTTGNBGTGNBTGGTTGG-3′; CS(5,8)G7T: 3′-CCAAGCTGACCAACC-5′).
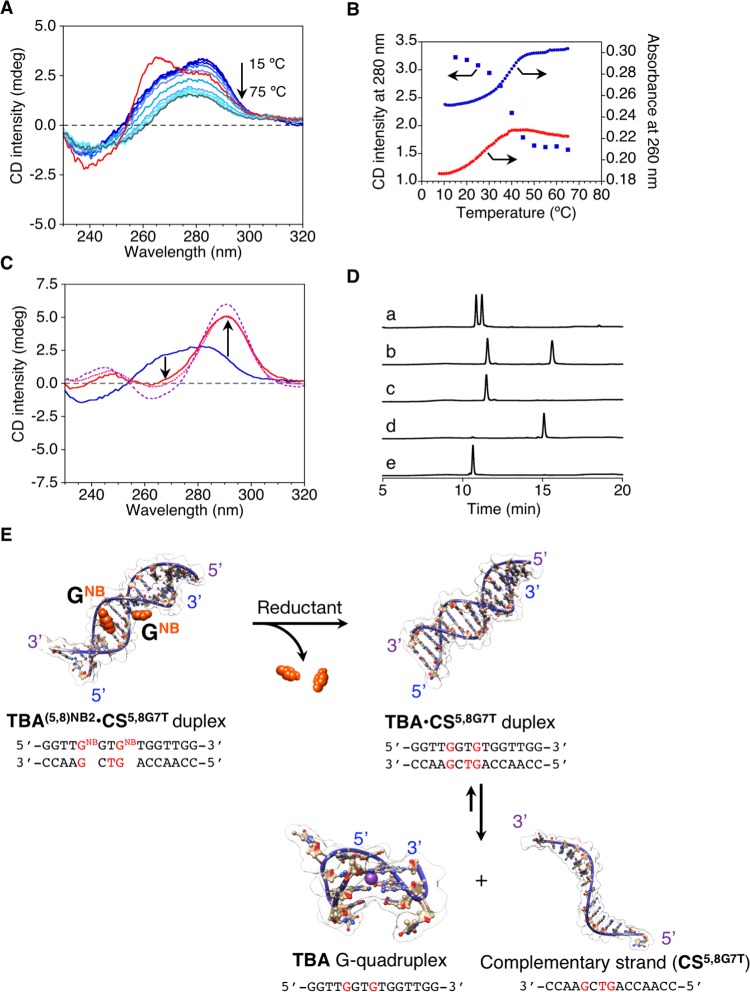
From thermal UV denaturation experiments, Tm of the TBA(5,8)NB2·CS(5,8)G7T duplex was evaluated to be 40.8 °C (Figure 4A). In addition, TBA(5,8)NB2·CS(5,8)G7T duplex showed a CD spectrum similar to that of the perfectly matched native TBA·CS duplex, and the CD melting curve was comparable to the UV melting curve (Figure 4B). Moreover, almost no hysteresis was observed as in the case of TBA5NB·CS5G duplex (Figure S4). These results suggest that the TBA(5,8)NB2·CS(5,8)G7T duplex can also adopt the standard double-stranded helical structure. Notably, Tm of TBA·CS(5,8)G7T duplex, which should be obtained after the removal of NB from the TBA(5,8)NB2·CS(5,8)G7T duplex, was evaluated to be 27.9 °C according to the UV melting curve (Figure 4B). The large difference in the thermal stability of TBA(5,8)NB2·CS(5,8)G7T duplex before and after the removal of the NB groups compared to that of TBA5NB·CS5G duplex (12.9 °C for TBA(5,8)NB2·CS(5,8)G7T duplex against 4.1 °C for TBA5NB·CS5G duplex as ΔTm) encouraged us to investigate its reduction-responsive structural transition. On the basis of the Tm values and the melting curves, temperature was set at 30 °C, where TBA(5,8)NB2·CS(5,8)G7T duplex was thermodynamically stable but TBA·CS(5,8)G7T duplex became unstable.
Figure 4.
(A) CD spectral changes during thermal melting for TBA(5,8)NB2·CS(5,8)G7T (blue lines: from 15 to 70 °C at a 5 °C interval, gray line: 75 °C) and CD spectrum of TBA·CS5C (red line). (B) Representative thermal CD (280 nm, blue squares for TBA(5,8)NB2·CS(5,8)G7T) and UV (260 nm, blue circles for TBA(5,8)NB2·CS(5,8)G7T, red circles for TBA·CS(5,8)G7T) melting curves. (C) CD spectra of the TBA(5,8)NB2·CS(5,8)G7T duplex before (blue line) and after (red line) addition of Na2S2O4. CD spectrum of a solution containing TBA and CS(5,8)G7T (pink dotted line) and the theoretical sum of the CD spectra of TBA and CS(5,8)G7T (purple dashed line) are also shown. The experimental temperature was set at 30 °C. (D) IP RP HPLC chromatograms of (a) TBA(5,8)NB2·CS(5,8)G7T duplex after addition of Na2S2O4, (b) TBA(5,8)NB2·CS(5,8)G7T duplex before addition of Na2S2O4, (c) CS(5,8)G7T, (d) TBA(5,8)NB2, and (e) TBA (detection at 260 nm). Conditions: TBA(5,8)NB2·CS(5,8)G7T (10 μM), Na2S2O4/TBA(5,8)NB2·CS(5,8)G7T = 1050 ([Na2S2O4] = 10.5 mM), 100 mM HEPES (pH 7.0) buffer with 0.1 mM EDTA and 105 mM KCl, and 0.1 cm cell length. (E) Schematic representations showing reduction-triggered dissociation of double-stranded helix to simultaneously form the G-quadruplex with the release of a nearly complementary strand.
After the addition of Na2S2O4 to the TBA(5,8)NB2·CS(5,8)G7T duplex at 30 °C, a remarkable increase in the CD intensity at 292 nm, which is characteristic of the TBA G-quadruplex, was observed within 3 min (Figure 4C). Also, the spectrum after addition of Na2S2O4 was almost identical to the mixture of the TBA G-quadruplex and CS(5,8)G7T and was comparable to a simple theoretical sum of each spectrum (Figure 4C). The difference between the experimental spectrum of the mixture containing TBA G-quadruplex and CS(5,8)G7T and the theoretical sum of each spectrum implies that the system might not reach to the equilibrium state to provide the complete formation of TBA G-quadruplex under these conditions; in other words, TBA·CS(5,8)G7T duplex and the TBA G-quadruplex would be in equilibrium, as depicted in Figure 4E.26 IP RP HPLC analyses (Figure 4D) as well as mass spectra clearly indicate an almost quantitative conversion from TBA(5,8)NB2 (tR = 15.1 min) to TBA after addition of Na2S2O4. These results support that the dissociation of the TBA(5,8)NB2·CS(5,8)G7T duplex and the simultaneous intramolecular G-quadruplex folding of TBA were essentially triggered after addition of the reducing reagent Na2S2O4.
Stimuli-controlled transition at high-order structures of nucleic acids is useful for various applications.28 Nevertheless, single-stranded oligonucleotides are mostly utilized as input signals, which can induce structural transitions following strand exchange reactions via taking advantage of complementary duplex formation, the so called strand displacement, in particular, using the single-stranded toehold motif.29 In contrast, examples of tertiary structural transition triggered by stimuli other than such single-stranded oligonucleotides are still rather limited. For example, Miyoshi et al. describe that molecular crowding conditions can induce the transition from duplex to quadruplex, which is highly valuable to gain deep understanding of biological events.30 Very recently, Manderville et al. developed a TBA with a fluorescent guanine derivative that allows monitoring structural transitions from duplex to G-quadruplex by fluorescence spectral changes in response to the binding of the modified TBA to thrombin.31 Zhou et al. explored a photoresponsive G-quadruplex binding molecule to modulate G-quadruplex formation in response to light irradiation as external stimulus.32 In addition, Liu et al. reported pH-driven reversible conformational changes between four- and double-stranded structures on a thin gold surface.33 This is, to the best of our knowledge, the first example wherein DNA structural transition was induced from duplex to quadruplex in response to reduction stimuli. Considering our previous study, it is reasonably expected that this NB group-based reduction responsiveness can be extended to biomolecule responsiveness after coupling with enzymatic reactions.34
Conclusions
We successfully constructed reduction-responsive DNA duplexes, in which guanine rings bearing a reduction-responsive cleavable NB group, were introduced at discrete positions and sequences. Moreover, we found that the duplexes exhibit reduction-responsive structural transitions, depending on their sequences. Not only structural transition from intermolecular duplex to intramolecular quadruplex was observed but also the removal of artificial NB groups from intermolecular duplex without its dissociation was demonstrated. Both phenomena are useful for exploring switch functions of biological events, involving DNA duplex and/or quadruplex structures. To explain the behavior of tertiary structures of DNA that contain guanine rings with NB, comprehensive structural studies, such as NMR spectroscopy and X-ray crystallography, may be necessary. Further research along these lines is currently in progress in our laboratory.
Materials and Methods
Unless stated otherwise, all commercial reagents were used as received. Thrombin-binding aptamer (TBA, d(GGTTGGTGTGGTTGG)) and other oligonucleotides, purified by ion-pair reversed-phase, high-performance liquid chromatography (IP RP HPLC), were synthesized by Fasmac Corp. (Kanagawa, Japan). The concentration of TBA was determined at 260 nm and 75 °C using a molar absorption coefficient (1.46 × 105 M–1 cm–1).25 All water used in the experiments refers to ultrapure water obtained from a Millipore system having a specific resistance of 18 MΩ cm. IP RP HPLC was conducted with a Shimadzu Prominence instrument LC-20AD and SPD-20A equipped with a GL Science Inertsil ODS-3 column (150 mm × 4.6 mm I.D., 5 μm) for analysis and GL Science Inertsil ODS-3 column (150 mm × 10 mm I.D., 5 μm) for purifications in a column oven CTO-20A. Matrix-assisted laser desorption/ionization (MALDI) coupled with time-of-flight (TOF) mass spectra were recorded using a Shimadzu AXIMA-CFR plus mass spectrometer. UV–vis spectra were obtained using a Jasco V-630 or a Shimadzu UV-2450 spectrophotometer.
Solid-Phase Oligonucleotide Synthesis
Oligonucleotide synthesis for TBA5NB and TBA(5,8)NB2 was carried out on an NTS H-6 DNA/RNA synthesizer (Nihon Techno Service) using the phosphoramidite method (commercially available phosphoramidite for dG and dC with synthetized dGNB reported previously17) at a 1 μmol scale. After cleavage from the solid support, deprotection of bases and phosphates was performed in aqueous ammonia (1.0 mL, 28%) at 55 °C for 12 h. The purification of the “trityl-on” oligonucleotide was carried out on IP RP HPLC ((GL Science Inertsil ODS-3 column (150 mm × 10 mm I.D.)), linear gradient from A/B = 100:0 (A: 0.1 M triethylamine acetate (TEAA) containing 5% acetonitrile, B: acetonitrile) to 50:50 over 30 min was used with a flow rate of 3.0 mL/min, detection wavelength = 260 nm). The purified “trityl-on” oligonucleotide was treated with 80% CH3COOH for 15 min at 37 °C to remove the dimethoxytrityl residues. The detritylated oligomer was purified by IP RP HPLC ((GL Science Inertsil ODS-3 column (150 mm × 10 mm I.D.)), linear gradient from A/B = 100:0 (A: 0.1 M TEAA containing 5% acetonitrile, B: acetonitrile) to 50:50 over 30 min was used with a flow rate of 3.0 mL/min, detection wavelength = 260 nm). IP RP HPLC charts are shown in Figure S1. The identity of the oligonucleotides TBA5NB and TBA(5,8)NB2 has been established by MALDI-TOF MS (matrix: 3-hydroxypicolinic acid (3-HPA), negative, Figure S2), and the observed molecular weights were in good agreement with their structures: TBA5NB, calcd for [M – H]−: 4874.1, found: 4876.1 and TBA(5,8)NB2, calcd. for [M – H]−: 5030.1, found: 5031.3. The oligonucleotide concentrations were determined by UV spectroscopy. The molar extinction coefficient (at 260 nm) ratio of dGNB/dG (=1.15) was used to determine the concentrations of TBANB and TBA(5,8)NB2 by measurements of absorbance (260 nm) at 75 °C.
Thermal Denaturation Experiments
Duplex Sample Preparation
Hybridization to construct duplexes was accomplished by mixing equal amounts of purified TBAs with or without GNB (TBA, TBANB, or TBA(5,8)NB2) and their complemented strands with designated number of mismatches to a final concentration of 100 μM in (A) 10 mM phosphate buffer (pH 7.6) containing 100 mM KCl or (B) 100 mM HEPES buffer (pH 7.0) containing 105 mM KCl. The samples were heated at 95 °C for 5 min followed by cooling to room temperature over a period of approximately 1 h. The samples were then stored at 4 °C until experiments were accomplished.
Ultraviolet (UV) Melting Experiments
UV melting experiments were carried out on a Jasco V-630 spectrometer equipped with a programmable temperature-control unit (Jasco ETCS-761) using a 0.1 cm quartz cell unless otherwise noted. The melting temperatures (Tms) were determined from the maximum in the first derivatives of the melting curves (absorbance at 260 nm against temperature, 0.5 °C/min). Oligonucleotides were diluted at a final concentration of 10 μM in aqueous buffers.
Circular Dichroism (CD) Spectral Measurements and Melting Experiments
CD experiments were carried out on a Jasco J-820 spectropolarimeter equipped with a programmable temperature-control unit (Julabo HP-4) using a 0.1 cm quartz cell unless otherwise noted. CD spectra were obtained by using a 1 nm slit width and a scanning step of 0.1 nm from 320 to 230 nm. Each spectrum was an average of eight scans with the buffer background subtracted. The melting temperatures (Tms) were determined from the maximum in the first derivatives of the melting curves (CD intensity at 292 nm against temperature, 1.0 °C/min).
Monitoring Reduction-Responsive Structural Change by CD and IP RP HPLC
Stock solution of oligonucleotide duplexes was diluted with an aqueous buffer to afford solutions containing oligonucleotide duplexes (10 μM). The reductant (Na2S2O4) was then added to adjust ca. 10 mM as the final concentrations in the mixture. After incubation at designated temperature (25, 30, or 35 °C), CD spectral changes were measured under the conditions mentioned above. Also, the reaction mixture was subjected to IP RP HPLC analysis (GL Science Inertsil ODS-3 column (150 mm × 4.6 mm I.D., 5 μm), linear gradient from A/B = 100:0 (A: 0.1 M TEAA containing 5% acetonitrile, B: acetonitrile) to 50:50 over 30 min was used with a flow rate of 1.0 mL/min, detection wavelength = 260 nm, column oven temperature = 40 °C). The identity of the starting oligonucleotides (TBA5NB, TBA(5,8)NB2) and the desired reduction product (TBA) were established by comparing the retention time of authentic standard as well as MALDI-TOF MS spectra. Experiment was repeated in at least duplicates.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research (B) (M.I., No. 15H03836) of the Japan Society for the Promotion of Science. Financial supports from Mochida Memorial Foundation for Medical and Pharmaceutical Research and Daiichi Sankyo Foundation of Life Science to M.I. are also acknowledged. We acknowledge Division of Instrumental Analysis, Life Science Research Centre, Gifu University for the maintenance of the instruments and kind support (especially, Prof. Yasunori Oumi, Prof. Shigeo Takashima, Dr. Yoshiharu Sawada, Yoshiko Wakihara and Kayoko Toyoshi). The authors would like to thank Enago (www.enago.jp) for the English language review.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b01177.
Figures showing characterization of oligonucleotides; thermal denaturation experiments; and reduction-responsive property (Figures S1–S8) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- For recent reviews; a Klán P.; Šolomek T.; Bochet C. G.; Blanc A.; Givens R.; Rubina M.; Popik V.; Kostiko A.; Wirz J. Photoremovable protecting groups in chemistry and biology: reaction mechanisms and efficacy. Chem. Rev. 2013, 113, 119–191. 10.1021/cr300177k. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Brieke C.; Rohrbach F.; Gottschalk A.; Mayer G.; Heckel A. Light-controlled tools. Angew. Chem., Int. Ed. 2012, 51, 8446–8476. 10.1002/anie.201202134. [DOI] [PubMed] [Google Scholar]; c Yu H.; Li J.; Wu D.; Qiua Z.; Zhang Y. Chemistry and biological applications of photo-labile organic molecules. Chem. Soc. Rev. 2010, 39, 464–473. 10.1039/B901255A. [DOI] [PubMed] [Google Scholar]; d Lee H.-M.; Larson D. R.; Lawrence D. S. Illuminating the chemistry of life: design, synthesis, and applications of “caged” and related photoresponsive compounds. ACS Chem. Biol. 2009, 4, 409–427. 10.1021/cb900036s. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Ellis-Davies G. C. R. Caged compounds: photorelease technology for control of cellular chemistry and physiology. Nat. Methods 2007, 4, 619–628. 10.1038/nmeth1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- For recent reviews; a Li J.; Chem P. R. Development and application of bond cleavage reactions in bioorthogonal chemistry. Nat. Chem. Biol. 2016, 12, 129–137. 10.1038/nchembio.2024. [DOI] [PubMed] [Google Scholar]; b Völker T.; Meggers E. Transition-metal-mediated uncaging in living human cells—an emerging alternative to photolabile protecting groups. Curr. Opin. Chem. Biol. 2015, 25, 48–54. 10.1016/j.cbpa.2014.12.021. [DOI] [PubMed] [Google Scholar]
- a Li J.; Jia S.; Chen P. R. Diels-Alder reaction-triggered bioorthogonal protein decaging in living cells. Nat. Chem. Biol. 2014, 10, 1003–1005. 10.1038/nchembio.1656. [DOI] [PubMed] [Google Scholar]; b Fan X.; Ge Y.; Lin F.; Yang Y.; Zhang G.; Ngai W. S. C.; Lin Z.; Zheng S.; Wang J.; Zhao J.; Li J.; Chen P. R. Optimized tetrazine derivatives for rapid bioorthogonal decaging in living cells. Angew. Chem., Int. Ed. 2016, 55, 14046–14050. 10.1002/anie.201608009. [DOI] [PubMed] [Google Scholar]
- Luo J.; Liu Q.; Morihiro K.; Deiters A. Small-molecule control of protein function through Staudinger reduction. Nat. Chem. 2016, 8, 1027–1034. 10.1038/nchem.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Chen Z.-j.; Tian Z.; Kallio K.; Oleson A. L.; Ji A.; Borchardt D.; Jian D.-e.; Remington S. J.; Ai H.-w. The N–B interaction through a water bridge: understanding the chemoselectivity of a fluorescent protein based probe for peroxynitrite. J. Am. Chem. Soc. 2016, 138, 4900–4907. 10.1021/jacs.6b01285. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hoang T. T.; Smith T. P.; Raines R. T. A. Boronic acid conjugate of angiogenin that shows ROS-responsive neuroprotective activity. Angew. Chem., Int. Ed. 2017, 56, 2619–2622. 10.1002/anie.201611446. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Wang M.; Sun S.; Neufeld C. I.; Perez-Ramirez B.; Xu Q. Reactive oxygen species-responsive protein modification and its intracellular delivery for targeted cancer therapy. Angew. Chem., Int. Ed. 2014, 53, 13444–13448. 10.1002/anie.201407234. [DOI] [PubMed] [Google Scholar]; d Brustad E.; Bushey M. L.; Lee J. W.; Groff D.; Liu W.; Schultz P. G. A genetically encoded boronate-containing amino acid. Angew. Chem., Int. Ed. 2008, 47, 8220–8223. 10.1002/anie.200803240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Elbashir S. M.; Harborth J.; Lendeckel W.; Yalcin A.; Weber K.; Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. 10.1038/35078107. [DOI] [PubMed] [Google Scholar]; b Hannon G. J. RNA interference. Nature 2002, 418, 244–251. 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- a Morishita R.; Sugimoto T.; Aoki M.; Kida I.; Tomita N.; Moriguchi A.; Maeda K.; Sawa Y.; Kaneda Y.; Higaki J.; Ogihara T. In vivo transfection of cis element “decoy” against nuclear factor-kappaB binding site prevents myocardial infarction. Nat. Med. 1997, 3, 894–899. 10.1038/nm0897-894. [DOI] [PubMed] [Google Scholar]; b Bielinska A.; Shivdasani R. A.; Zhang L. Q.; Nabel G. J. Regulation of gene expression with double-stranded phosphorothioate oligonucleotides. Science 1990, 250, 997–1000. 10.1126/science.2237444. [DOI] [PubMed] [Google Scholar]
- a Ellington A. D.; Szostak J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]; b Bock L. C.; Griffin L. C.; Latham J. A.; Vermaas E. H.; Toole J. J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]; For recent review; c Poolsup S.; Kim C.-Y. Therapeutic applications of synthetic nucleic acid aptamers. Curr. Opin. Biotechnol. 2017, 48, 180–186. 10.1016/j.copbio.2017.05.004. [DOI] [PubMed] [Google Scholar]
- a Kruger K.; Grabowski P. J.; Zaug A. J.; Sands J.; Gottschling D. E.; Cech T. R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982, 31, 147–157. 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]; b Guerrier-Takada C.; Gardiner K.; Marsh T.; Pace N.; Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983, 35, 849–857. 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]; c Robertson D. L.; Joyce G. F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990, 344, 467–468. 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]; For review.; d Hammann C.; Luptak A.; Perreault J.; de la Pena M. The ubiquitous hammerhead ribozyme. RNA 2012, 18, 871–885. 10.1261/rna.031401.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Ponce-Salvatierra A.; Wawrzyniak-Turek K.; Steuerwald U.; Höbartner C.; Pena V. Crystal structure of a DNA catalyst. Nature 2016, 529, 231–234. 10.1038/nature16471. [DOI] [PubMed] [Google Scholar]; b Breaker R. R.; Joyce G. F. A DNA enzyme that cleaves RNA. Chem. Biol. 1994, 1, 223–229. 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]; For review; c Silverman S. K. Catalytic DNA (deoxyribozymes) for synthetic applications—current abilities and future prospects. Chem. Commun. 2008, 3467–3485. 10.1039/b807292m. [DOI] [PubMed] [Google Scholar]
- For review; a Liu Q.; Deiters A. Optochemical control of deoxyoligonucleotide function via a nucleobase-caging approach. Acc. Chem. Res. 2014, 47, 45–55. 10.1021/ar400036a. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Tang X.; Dmochowski I. J. Regulating gene expression with light-activated oligonucleotides. Mol. BioSyst. 2007, 3, 100–110. 10.1039/B614349K. [DOI] [PubMed] [Google Scholar]
- Höbartner C.; Mittendorfer H.; Breuker K.; Micura R. Triggering of RNA Secondary structures by a functionalized nucleobase. Angew. Chem., Int. Ed. 2004, 43, 3922–3925. 10.1002/anie.200460068. [DOI] [PubMed] [Google Scholar]
- a Saneyoshi H.; Hiyoshi Y.; Iketani K.; Kondo K.; Ono A. Bioreductive deprotection of 4-nitrobenzyl group on thymine base in oligonucleotides for the activation of duplex formation. Bioorg. Med. Chem. Lett. 2015, 25, 5632–5635. 10.1016/j.bmcl.2015.10.025. [DOI] [PubMed] [Google Scholar]; b Mori S.; Morihiro K.; Okuda T.; Kasahara Y.; Obika S. Hydrogen peroxide-triggered gene silencing in mammalian cells through boronated antisense oligonucleotides. Chem. Sci. 2018, 9, 1112–1118. 10.1039/C7SC04318J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Meade B. R.; Gogoi K.; Hamil A. S.; Palm-Apergi C.; van den Berg A.; Hagopian J. C.; Springer A. D.; Eguchi A.; Kacsinta A. D.; Dowdy C. F.; Presente A.; Lönn P.; Kaulich M.; Yoshioka N.; Gros E.; Cui X.-S.; Dowdy S. F. Efficient delivery of RNAi prodrugs containing reversible charge-neutralizing phosphotriester backbone modifications. Nat. Biotechnol. 2014, 32, 1256–1261. 10.1038/nbt.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Saneyoshi H.; Ono A. Development of protecting groups for prodrug-type oligonucleotide medicines. Chem. Pharm. Bull. 2018, 66, 147–154. 10.1248/cpb.c17-00696. [DOI] [PubMed] [Google Scholar]
- Ochi Y.; Nakagawa O.; Sakaguchi K.; Wada S.-i.; Urata H. A post-synthetic approach for the synthesis of 2’-O-methyldithiomethyl-modified oligonucleotides responsive to a reducing environment. Chem. Commun. 2013, 49, 7620–7622. 10.1039/c3cc43725f. [DOI] [PubMed] [Google Scholar]
- For reviewIkeda M.; Kabumoto M. Chemically caged nucleic acids. Chem. Lett. 2017, 46, 634–640. 10.1246/cl.161063. [DOI] [Google Scholar]
- Ikeda M.; Kamimura M.; Hayakawa Y.; Shibata A.; Kitade Y. Reduction-responsive guanine incorporated into G-quadruplex-forming DNA. ChemBioChem 2016, 17, 1304–1307. 10.1002/cbic.201600164. [DOI] [PubMed] [Google Scholar]
- a Wilson W. R.; Hay M. P. Targeting hypoxia in cancer therapy. Nat. Rev. Cancer 2011, 11, 393–410. 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]; b Thambi T.; Park J. H.; Lee D. S. Hypoxia-responsive nanocarriers for cancer imaging and therapy: recent approaches and future perspectives. Chem. Commun. 2016, 52, 8492–8500. 10.1039/C6CC02972H. [DOI] [PubMed] [Google Scholar]
- Platella C.; Riccardi C.; Montesarchio D.; Roviello G. N.; Musumeci D. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biochim. Biophys. Acta, Gen. Subj. 2017, 1861, 1429–1447. 10.1016/j.bbagen.2016.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Siddiqui-Jain A.; Grand C. L.; Bearss D. J.; Hurley L. H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 11593–11598. 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Biffi G.; Tannahill D.; McCafferty J.; Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Bochman M. L.; Paeschke K.; Zakian V. A. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012, 13, 770–780. 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Mayer G.; Kröck L.; Mikat V.; Engeser M.; Heckel A. Light-induced formation of G-quadruplex DNA secondary structures. ChemBioChem 2005, 6, 1966–1970. 10.1002/cbic.200500198. [DOI] [PubMed] [Google Scholar]
- This is the similar order of decreasing thermal stability reported previously for O6-benzylguanine (GB) paired with the canonical bases in double-stranded helical structures of ODNs. GB as well as O6-methylguanine (GMe) is frequently found as O6-alkylguanine lesions.; a Gong J.; Sturla S. J. A synthetic nucleoside probe that discerns a DNA adduct from unmodified DNA. J. Am. Chem. Soc. 2007, 129, 4882–4883. 10.1021/ja070688g. [DOI] [PubMed] [Google Scholar]; b Gaffney B. L.; Jones R. A. Thermodynamic comparison of the base pairs formed by the carcinogenic lesion O6-methylguanine with reference both to Watson-Crick pairs and to mismatched pairs. Biochemistry 1989, 28, 5881–5889. 10.1021/bi00440a026. [DOI] [PubMed] [Google Scholar]
- The phosphate buffer was used for the evaluation of Tm due to temperature dependent pKa of HEPES. The HEPES buffer was used for the other experiments in relationship to our previous study. Nevertheless, there was no significant difference in thermal melting behaviors in this study as far as we tested.
- Kankia B. I.; Marky L. A. Folding of the thrombin aptamer into a G-quadruplex with Sr2+: stability, heat, and hydration. J. Am. Chem. Soc. 2001, 123, 10799–10804. 10.1021/ja010008o. [DOI] [PubMed] [Google Scholar]
- CD spectrum of the mixture containing TBA G-quadruplex and CS(5,8)G7T became almost identical to the theoretical sum of each spectrum at higher temperature such as 35 °C (Figure S7). However, at 35 °C, the considerable portion (ca. 40%) of the initial TBA(5,8)NB2·CS(5,8)G7T duplex would dissociate into two unfolded single-stranded structures judging from its UV thermal melting curve. The CD spectral change at 25 °C is also presented in Figure S8, which shows the smaller change compared with that at 30 °C. Thereby, temperature near 30 °C would be optimal conditions to induce structural transition from duplex to quadruplex as large as possible in this system.
- Markham N. R.; Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005, 33, W577–W581. 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Alberti P.; Mergny J.-L. DNA duplex–quadruplex exchange as the basis for a nanomolecular machine. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 1569–1573. 10.1073/pnas.0335459100. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Uno S.-n.; Dohno C.; Bittermann H.; Malinovskii V. L.; Häner R.; Nakatani K. A light-driven supramolecular optical switch. Angew. Chem., Int. Ed. 2009, 48, 7362–7365. 10.1002/anie.200903251. [DOI] [PubMed] [Google Scholar]; For reviews; c Beissenhirtz M. K.; Willner I. DNA-based machines. Org. Biomol. Chem. 2006, 4, 3392–3401. 10.1039/b607033g. [DOI] [PubMed] [Google Scholar]; d Seeman N. C. From genes to machines: DNA nanomechanical devices. Trends Biochem. Sci. 2005, 30, 119–125. 10.1016/j.tibs.2005.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Krishnan Y.; Simmel F. C. Nucleic acid based molecular devices. Angew. Chem., Int. Ed. 2011, 50, 3124–3156. 10.1002/anie.200907223. [DOI] [PubMed] [Google Scholar]
- a Yurke B.; Turberfield A. J.; Mills A. P. Jr.; Simmel F. C.; Neumann J. L. A DNA-fuelled molecular machine made of DNA. Nature 2000, 406, 605–608. 10.1038/35020524. [DOI] [PubMed] [Google Scholar]; For review; b Zhang D. Y.; Seelig G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 2011, 3, 103–113. 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- Miyoshi D.; Matsumura S.; Nakano S.-i.; Sugimoto N. Duplex dissociation of telomere DNAs induced by molecular crowding. J. Am. Chem. Soc. 2004, 126, 165–169. 10.1021/ja036721q. [DOI] [PubMed] [Google Scholar]
- Fadock K. L.; Manderville R. A.; Sharma P.; Wetmore S. D. Optimization of fluorescent 8-heteroaryl-guanine probes for monitoring protein-mediated duplex→G-quadruplex exchange. Org. Biomol. Chem. 2016, 14, 4409–4419. 10.1039/C6OB00474A. [DOI] [PubMed] [Google Scholar]
- Wang X.; Huang J.; Zhou Y.; Yan S.; Weng X.; Wu X.; Deng M.; Zhou X. Conformational switching of G-quadruplex DNA by photoregulation. Angew. Chem., Int. Ed. 2010, 49, 5305–5309. 10.1002/anie.201002290. [DOI] [PubMed] [Google Scholar]
- Liu D.; Bruckbauer A.; Abell C.; Balasubramanian S.; Kang D.-J.; Klenerman D.; Zhou D. A reversible pH-driven DNA nanoswitch array. J. Am. Chem. Soc. 2006, 128, 2067–2071. 10.1021/ja0568300. [DOI] [PubMed] [Google Scholar]
- a Ikeda M.; Tanida T.; Yoshii T.; Kurotani K.; Onogi S.; Urayama K.; Hamachi I. Installing logic-gate responses to a variety of biological substances in supramolecular hydrogel–enzyme hybrids. Nat. Chem. 2014, 6, 511–518. 10.1038/nchem.1937. [DOI] [PubMed] [Google Scholar]; b Shigemitsu H.; Fujisaku T.; Onogi S.; Yoshii T.; Ikeda M.; Hamachi I. Preparation of supramolecular hydrogel–enzyme hybrids exhibiting biomolecule-responsive gel degradation. Nat. Protoc. 2016, 11, 1744–1756. 10.1038/nprot.2016.099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.