Abstract
The emergence of bacterial resistance and hesitance in approving new drugs has bolstered research on membrane-active agents such as antimicrobial peptides and their synthetic derivatives as therapeutic alternatives against bacterial infections. Herein, we document the action of aryl-alkyl-lysines on liposomes mimicking bacterial membranes using solid-state nuclear magnetic resonance spectroscopy. A significant perturbation of the lipid thickness and order parameter of the lipid membrane was observed upon treatment with this class of compounds. Encouraged by these results, the ability of the most active compound (NCK-10) to interact with aggregates of lipopolysaccharides (LPSs) was studied. In vitro experiments showed that NCK-10 was able to prevent the LPS-induced stimulation of proinflammatory cytokines such as tumor necrosis factor-α and interleukin-6. The compound could also disrupt the biofilms of Pseudomonas aeruginosa in vitro and bring down the bacterial burden by more than 99% in a mice model of burn infections caused by the biofilms of P. aeruginosa.
Introduction
Antimicrobial resistance (AMR) has been highlighted as an extremely serious challenge facing world health today. The O’Neill commission has estimated that by 2050, AMR will claim 10 million lives and cost the global economy $100 trillion.1 The global scientific community has been working collectively to solve this problem using various strategies and several new antibacterial compounds are undergoing clinical trials.2 In one such strategy, antimicrobial peptides which work by targeting the bacterial membrane or modulating the host immune system are being tried for possible antibacterial agents.3−5 In fact, membrane-active compounds that do not suffer from inherent problems of antimicrobial peptides such as stability, toxicity, and expenditure offer more advantages.6,7 Several of such compounds were designed based on the premise that antimicrobial peptides partition into the membrane interface rather than adopting transmembrane alignments.8,9 The important role of electrostatic forces in the selective interactions of cationic antimicrobial peptides with negatively charged bacterial membranes has been emphasized in various designs, which include α-peptides,10 β-peptides,11,12 oligoacyl lysines,13 oligoureas,14 α-AApeptides,15 arylamide foldamers,16 antimicrobial polymers,17−23 cationic amphiphiles,24 β2,2-amino acid derivatives,25 and designs based on the amino acid conjugates of lipids.26−28 Although almost all the compounds described above are known to act on the membranes, few studies have looked into the atomistic details of the perturbation caused to the membranes.
Nuclear magnetic resonance (NMR) spectroscopy is a powerful tool to attain atomistic information on the structure and dynamics of molecules as well as their interactions. Although multidimensional solution NMR spectroscopy has been traditionally used to provide high-resolution structures of globular biomolecules,29,30 solid-state NMR methods have been developed for the structural investigation of fibrils, supramolecular aggregates, and membrane components.31−35 These techniques have also been developed to investigate the interactions of membrane-active compounds including antimicrobial peptides with phospholipid bilayers.36 Therefore, solid-state NMR spectroscopy offers valuable insights into the macroscopic phase properties, curvature strain, and fatty acyl chain order of membrane constituents.37
Aryl-alkyl-lysines are some of the simplest examples of membrane-active agents. These small molecules involve easy synthesis and are not substrates for proteolysis. Interestingly, they have been found to inhibit the growth of bacteria, fungi, viruses, and parasites.38−43 Their antimicrobial activity could be mostly attributed to their ability to compromise microbial membranes. In order to investigate their interactions with bacterial membranes, in this report, we have used solid-state NMR spectroscopy as a tool. First, solid-state NMR spectroscopy provided information about the macroscopic behavior of model membranes made from phosphatidylethanolamine (POPE) and phosphatidylglycerol (POPG) lipid bilayers, in the presence of varying concentrations of different aryl-alkyl-lysines. Upon identification of the compound which perturbs the membrane lipids most effectively, we extrapolated the application of the compounds to other systems, which possess negative charge and lipid tails, for example, the lipopolysaccharides (LPSs) of Gram-negative bacteria. We investigated the ability of the best compound to inhibit the LPS-mediated stimulation of cytokines such as tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6). Further, we also tested the ability of the compound to disrupt the biofilms of Pseudomonas aeruginosa. Finally, we have validated the efficacy of the compound in a murine model of burn infections caused by the biofilms of P. aeruginosa.
Results
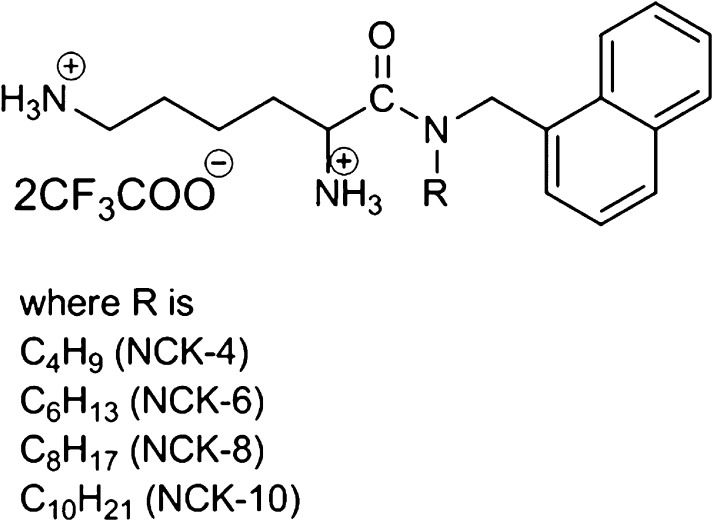
The compounds used in this study consisted of a naphthalene ring, an l-lysine moiety, and a variable alkyl chain. Depending on the length of the chain, the compounds (Figure 1) were NCK-4 (butyl chain), NCK-6 (hexyl chain), NCK-8 (octyl chain), and NCK-10 (decyl chain).
Figure 1.
Chemical structures of the compounds used in the study.
Vesicle Deformation, Inhomogeneity in Order Parameters, and Membrane Thickness by Solid-State NMR Spectroscopy
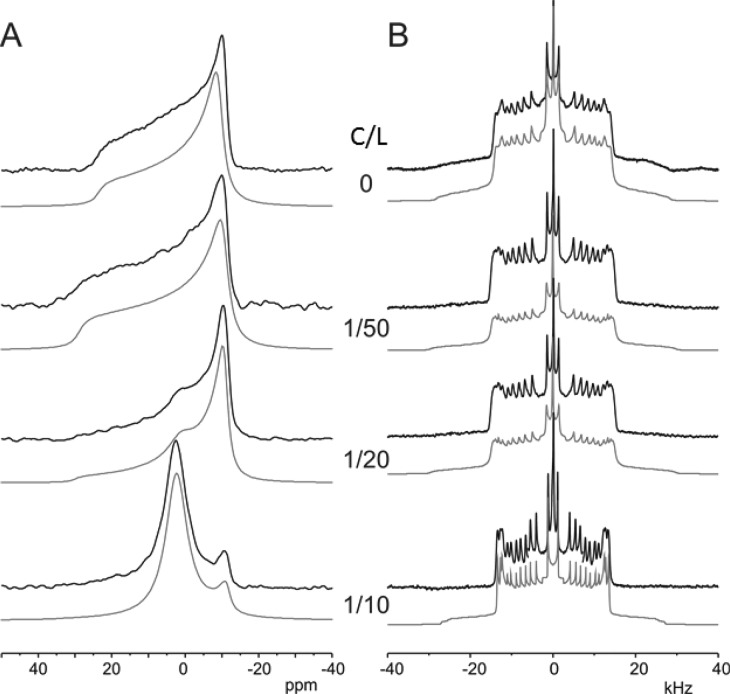
To investigate the macroscopic behavior of model POPE/POPG (3:1) lipid bilayers as a function of compound-to-lipid (C/L) ratio of proton-decoupled 31P, the solid-state NMR spectra for all the four compounds (NCK-4, NCK-6, NCK-8, and NCK-10) were acquired. The data have been furnished in Table 1, and the NMR spectra that have been obtained for NCK-10 and are shown in Figure 2. In the absence of the compounds and at 310 K, the spectra exhibited a chemical shift anisotropy around 45 ppm (Figure 2) in agreement with the membranes in the liquid-crystalline state.44 At 7.1 T, the phosphorus spectrum is typical of quasi-spheroidal liposomes with no preferential orientation in the magnetic field: an intense peak at ca. −15 ppm and a marked left shoulder at +30 ppm. Upon compound addition at a ratio of 1/50, the 31P solid-state NMR spectrum represents a spherical shape of the liposomes. In contrast, at a C/L ratio of 1/20, the shoulder at ca. +30 ppm loses its intensity and becomes barely detectable, indicative of a prolate distribution of phospholipid alignments. In addition, the intensities close to the isotropic chemical shift position become visible. At a C/L ratio of 1/10, the 31P solid-state NMR spectrum is characterized by an isotropic resonance, indicating a fast tumbling of smaller lipid aggregates. Spectral simulation (gray lines) can be performed where the deformation of vesicles is illustrated by the ratio c/a, which represents the ellipsoid long-to-short axis ratio (Table 1). The values of c/a close to 1 represent spherical vesicles, which is the case for the lipids POPE/POPG (3:1) in the absence of the compounds. Upon addition of all the compounds at a C/L ratio of 1/50, no change in the shape was observed (c/a = 1.1). At a C/L ratio of 1/20, a significant change was observed only in the case of NCK-10 (c/a = 1.5). However, at a C/L ratio of 1/10, changes were observed for all compounds, with the c/a values of 1.5, 1.6, and 1.7 for NCK-4, NCK-6, and NCK-8, respectively (Table 1). For NCK-10, the damage was such that no c/a value could be determined (c/a = ∞). Thus, it was concluded that NCK-10 was able to perturb the liposomes most efficiently.
Table 1. Deformation (c/a) Ratios of POPE/POPG (3:1 Molar Ratio) Liposomes in the Presence of NCK-4, NCK-6, NCK-8, and NCK-10 at Increasing C/L Ratios.
| NCK-4 | NCK-6 | NCK-8 | NCK-10 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C/L | 0 | +1/50 | +1/20 | +1/10 | +1/50 | +1/20 | +1/10 | +1/50 | +1/20 | +1/10 | +1/50 | +1/20 | +1/10 |
| c/a | 1.1 | 1.1 | 1.1 | 1.5 | 1.1 | 1.2 | 1.6 | 1.1 | 1.2 | 1.7 | 1.1 | 1.5 | ∞ |
Figure 2.
Experimental (black) and simulated (gray) proton-decoupled (A) 31P solid-state NMR spectra of POPE/POPG liposomes and (B) 2H solid-state NMR spectra of POPE/POPE-2H31/POPG (2:1:1) in the absence and presence of NCK-10 (C/L = 1/50, 1/20, 1/10) at 37 °C.
In order to shed more light on the interactions between NCK-10 and lipid fatty acyl chains, the 2H solid-state NMR spectra were recorded with deuterated palmitoyl chains [POPE/POPE-2H31/POPG (2:1:1)] in the presence of increasing concentrations of NCK-10. As can be seen from the spectra shown in Figure 2B, each of the different CD2 and CD3 segments contributed a quadrupolar powder pattern. The characteristic quadrupolar splitting for each site is defined by the distance between the two main peaks from each segment.45−47 In a static state, the splitting would amount to 125 kHz; however, motions and cis–trans isomerizations in a liquid-crystalline bilayer contribute to a reduced value.
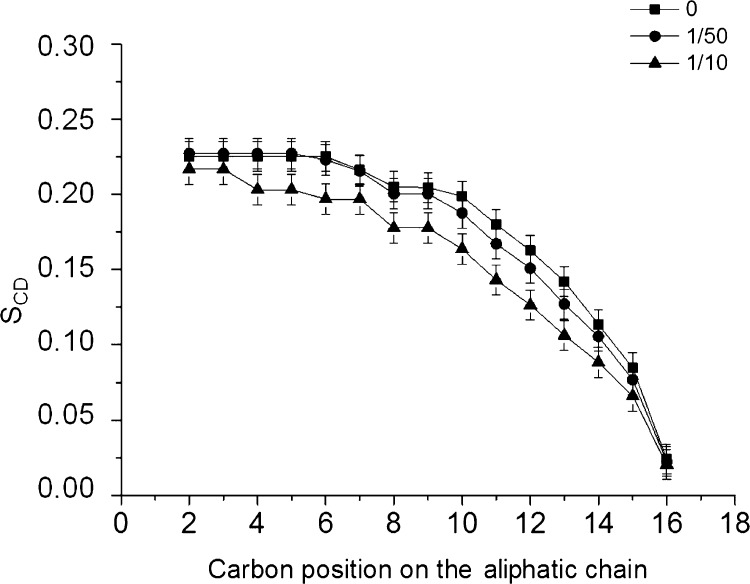
This phenomenon can also be described by the ratio of the measured and maximal value or the deuterium order parameter SCD. Understandably enough, more freedom is displayed by the segments in the hydrophobic interior in comparison to the interfacial region. Consequently, a decrease in the order parameters is noted with an increasing distance from the carbonyls. The values for the quadrupolar splittings were obtained from the 2H solid-state NMR spectra. Subsequently, the order parameters SCD were calculated and plotted against the carbon position of the labeled lipids (Figure 3). Upon addition of 2 and 10 mol % of NCK-10 (a C/L of 1/50 and 1/10), the 2H solid-state NMR spectra of the quadrupolar splittings decrease as a function of NCK-10 concentration (Figure 3). This decrease in the quadrupolar splitting for all positions on the aliphatic chain (from the glycerol backbone to the center of the bilayer) is associated with a decrease in the order parameter (SCD), from the so-called plateau region representing the first CH2 segments to the terminal methyl group (Table S1). This change in lipid configuration is related to a reduction of the hydrophobic chain length of the bilayers from 2 × 24.8 Å to 2 × 21.8 Å for lipids associated with 1/10 NCK-10.
Figure 3.
Order parameters of liposomes made of POPE/POPE-2H31/POPG (2:1:1) at 90% hydration as a function of carbon position in the presence of NCK-10 at different C/L ratios.
Ability To Interact with LPS Aggregates
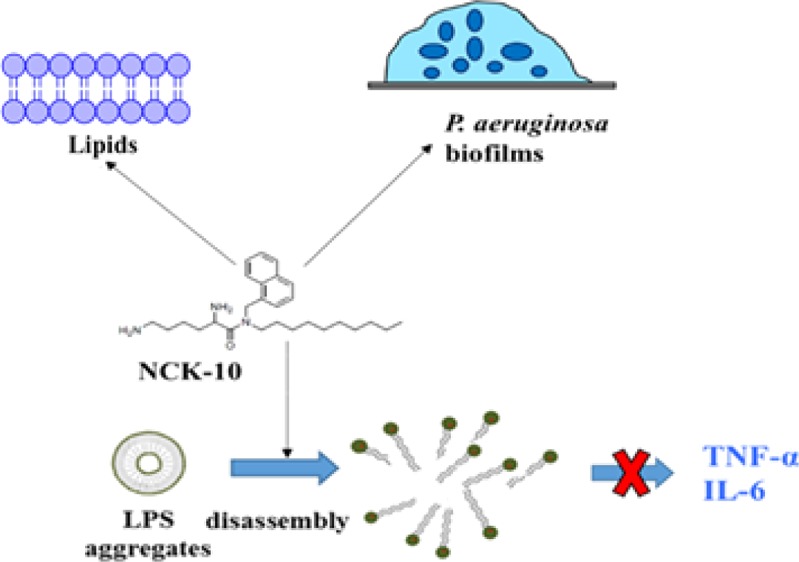
From the experimental studies described above, it is clear that NCK-10 perturbs the bacterial membranes. Because the compound contains two positive charges and one decyl chain, we reasoned that it might interact with other negatively charged amphipathic molecules such as LPSs. LPS is a component of Gram-negative bacteria and forms aggregates in an aqueous solution. These aggregates are recognized by host cells initiating a cascade of reactions upon recognition, which ultimately leads to sepsis.48,49 Sepsis, a global problem, contributes to around 8 million deaths per year.50 Unfortunately, there is no dedicated treatment for sepsis, and antibiotics, which themselves can trigger sepsis, remain the drugs of choice for treatment.50 LPSs when released are recognized by the host immune systems and prompt them to over-react. Uncontrolled inflammation is often a result of this over-reaction, leading to multiorgan failure or death.4 This uncontrolled inflammation, in turn, is mediated by an increased production of TNF-α and IL-6 from macrophages/monocytes.4,50 Several antimicrobial peptides and their mimics have been reported to interact with LPS aggregates and subsequently prevent inflammation caused by the release of cytokines such as TNF-α and IL-6.4,5,51−53 We surmised that if NCK-10 interacts with LPSs, it would also prevent inflammatory responses and ultimately sepsis.
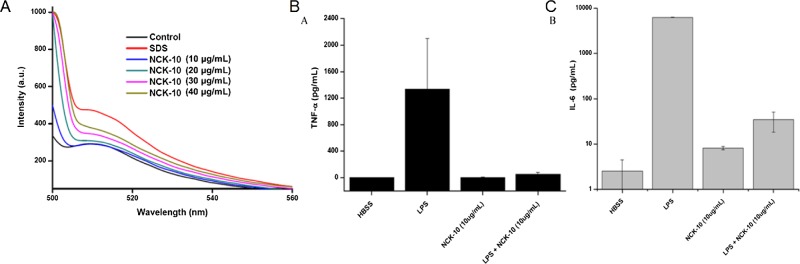
At first, we checked the ability of NCK-10 to interact with a BODIPY dye conjugate of LPSs. In water, LPSs usually self-assemble to form aggregates, and because of aggregation, the fluorescence of BODIPY in BODIPY-LPS is quenched. However, dissociation of the aggregates leads to an increase in the fluorescence of BODIPY. Such disaggregation is often due to the interaction with an amphiphilic molecule, such as sodium dodecyl sulfate (SDS).51,54,55Figure 4A also shows that SDS (2%) was able to increase the intensity of the fluorescence by several orders of magnitude in comparison to control measurements (blank). NCK-10, which was amphiphilic as well, was able to interact with LPS aggregates at concentrations of 10 μg/mL. With an increasing concentration, the response was enhanced, proving that NCK-10 was able to dissociate the LPS aggregates.
Figure 4.
Anti-inflammatory properties of NCK-10. (A) Ability of NCK-10 to interact with LPS-BODIPY. A dose-dependent response can be observed. SDS (2%) was used as a positive control. The ability of NCK-10 to suppress the LPS-induced stimulation of (B) TNF-α and (C) IL-6.
Ability To Prevent the LPS-Induced Stimulation of Proinflammatory Cytokines TNF-α and IL-6
We surmised that if the compound can dissociate aggregates of LPS, it can prevent proinflammatory responses that ultimately lead to sepsis. Thus, we proceeded to check the response of LPSs on immune cells in the presence and absence of NCK-10. The experiment was performed with human peripheral blood mononuclear cells (hPBMCs), which are known to circulate in blood and produce inflammatory responses upon interaction with bacterial endotoxins such as LPSs. Ficoll-Hypaque density centrifugation was used to isolate hPBMCs from fresh human blood. Then, the hPBMCs were stimulated with LPS (the concentration used was 20 ng/mL) in the presence and absence of 10 μg/mL of NCK-10. The secretion of proinflammatory cytokines, TNF-α and IL-6, was quantified using ELISA. As can be seen from Figure 4B,C, NCK-10 on its own did not elicit any response. However, LPS successfully stimulated the expression of both TNF-α and IL-6. However, in the presence of NCK-10, the proinflammatory response of LPS was suppressed. This experiment proved that NCK-10 could not only treat bacterial infection but also prevent sepsis.
Ability To Disrupt the Preformed Biofilms of P. aeruginosa
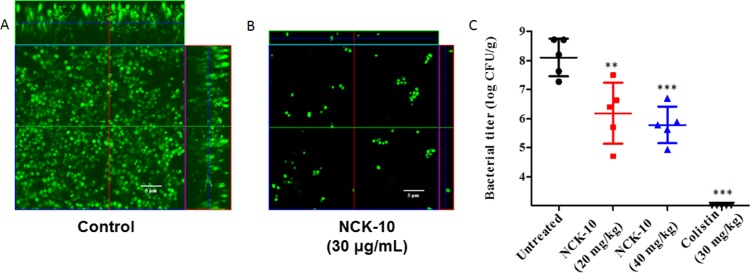
P. aeruginosa is notorious for forming bacterial biofilms.56 These biofilms are stable communities within which bacteria survive with limited resources. Biofilms are recalcitrant to antibiotic treatment and pose an important problem for health care.56,57 The biofilm matrix of Pseudomonas sp. has several negatively charged components such as alginates, LPS, DNA, and so forth.58−61 Outer membrane vesicles also contribute to the biofilm matrix.62−64 We had earlier seen that aryl-alkyl-lysines infiltrate the outer membrane of Gram-negative bacteria.38 In this report, we demonstrated that NCK-10 also interacts with LPS. On the basis of these observations, we surmised that NCK-10 would be able to disrupt the biofilm matrix of P. aeruginosa. NCK-10 was reported to inhibit various strains of P. aeruginosa at minimum inhibitory concentrations ranging from 3 to 7 μg/mL.38,39 In order to visualize the effect of the compound on the biofilms of P. aeruginosa, we treated 72 h old biofilms (grown on a glass cover slip) of a clinical isolate of P. aeruginosa R590 (described earlier) at a concentration of 30 μg/mL for 24 h and subsequently stained the biofilm with SYTO-9. As can be seen from Figure 5A, in the untreated case, the biofilm was thick (12 μm) and uniform, whereas upon treatment with the compound (Figure 5B), only a monolayer of cells remained (thickness 3.6 μm). Furthermore, the cells were distributed unevenly. This proved that the compound indeed exhibited an antibiofilm activity against P. aeruginosa.
Figure 5.
Antibiofilm activity of NCK-10 against P. aeruginosa. (A) Untreated biofilms and (B) NCK-10-treated biofilms. (C) In vivo activity of NCK-10 against burn infections caused by the biofilms of P. aeruginosa. Bacterial count after treatment with NCK-10 or colistin in comparison to control. Student’s t-test was used to establish the statistical significance between the values (p-value <0.05 was considered to be significant; ***p < 0.001).
Next, we tested if the bacterial cells within the biofilms of P. aeruginosa were lysed upon treatment with the compound. When 72 h preformed biofilms of P. aeruginosa were incubated with the compound at concentrations of 15 and 30 μg/mL for 24 h, a 3.6 logs and 4.8 logs reduction (compared to the control) in bacterial count was observed.
Activity against a Biofilm Model of Burn Infections Caused by P. aeruginosa R590 Inflicted on Mice
In order to investigate the efficacy of the compound in animal models of infection, we first created burn wounds in mice and then infected the wounds with 107 cfu/mL of P. aeruginosa R590. In order to facilitate biofilm formation, the wounds were not treated for 24 h. After 24 h, the wounds were subsequently treated for 6 days with NCK-10 at 20 or 40 mg/kg once a day. Colistin was used as a positive control at a concentration of 30 mg/kg. As can be seen from Figure 5C, the bacterial burden was reduced by 1.9 logs and 2.3 logs (>99% reduction) upon treatment with 20 and 40 mg/kg of NCK-10, respectively. Although colistin was more efficacious, it has to be noted that bacteria find it easier to develop resistance to colistin in comparison to NCK-10.39
Discussion
In the fight against bacterial resistance, membrane-active agents represent promising alternatives to conventional antibiotics. Both natural antimicrobial peptides and their synthetic mimics have entered clinical trials for treatment against infections caused by microbes.6 Although synthetic membrane-active agents offer more advantages over natural peptides, more studies need to be performed delineating their interactions with membranes at an atomistic level. Aryl-alkyl-lysines and related compounds are microbicidal compounds with activities against bacteria, fungi, parasites, and viruses.26,38−43,65 A representative compound, NCK-10, was found to be a potent compound, which was nontoxic to mammalian cells at its microbicidal concentrations.38,40,43 In this study, we have documented the interaction of aryl-alkyl-lysines with lipids simulating bacterial membranes using solid-state NMR spectroscopy. Additionally, we have probed the ability of NCK-10 to prevent inflammatory responses caused by interactions with other negatively charged lipophilic moieties such as LPS. The compound could also eradicate the biofilms of P. aeruginosa both in vitro and in vivo.
Investigations using 31P and 2H NMR revealed that the interaction with NCK-10 induces supramolecular rearrangements within the vesicles that lead to deformation (observable in the 7.1 T magnetic field of the NMR spectrometer). In Figure 6, a schematic of the interactions between NCK-10 and the liposomes, based on the observations from NMR studies, has been furnished. Furthermore, upon increasing the NCK-10-to-lipid ratio, isotropic resonance intensities appeared. This effect was observed by the 31P solid-state NMR spectroscopy of the phospholipid head groups and the 2H solid-state NMR spectra of the deuterium-labeled fatty acyl chains (Figure 2A,B), indicating that the changes in the supramolecular features of the membranes, rather than local conformational properties, contribute to spectral alterations. These spectral changes resemble those observed in the presence of magnetically oriented bicelles,66−69 which with the addition of high amounts of NCK-10 becomes small enough to tumble freely in all directions (isotropic bicelles or micelles).8,70 The deuterium NMR spectra indicate a decrease in the C–2H order parameter concomitant with the thinning of the membrane in the presence of NCK-10. This decrease in thickness agrees well with the observations made when amphiphilic peptides partition into the head group region.71
Figure 6.
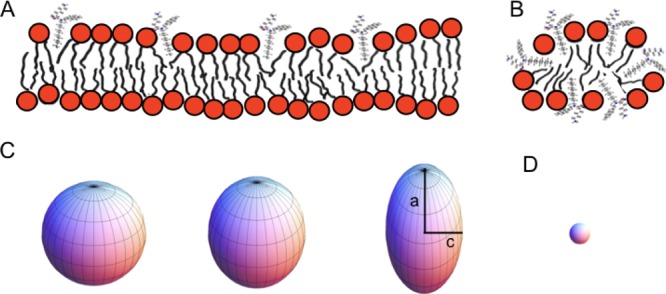
Schematic model of the partitioning of NCK-10. It is assumed that the charged and polar functional groups remain in contact with water, and at the membrane interface, the aliphatic side chains partition into the hydrophobic region of the membrane. (A) At low concentrations of membrane-interacting NCK-10, this causes a decrease in the order parameter of the phospholipid-deuterated fatty acyl chains. (B) In the presence of high NCK-10 concentrations, membranes of high curvature are formed, possibly micelles. (C) Liposomes have been shown to deform in the strong magnetic fields of modern NMR spectrometers. Spherical liposomes and magnetically deformed structures are shown. From left to right, the a/c values are 1, ≈1.1, and ≈1.5. D. In comparison, small vesicles or micelles tumble fast in the magnetic field giving isotropic line shapes. The sphere was created using the Mathematica program.
Our hypothesis that NCK-10 would be able to interact with LPS aggregates, which are also negatively charged and contain lipid tails, is proved in Figure 4. Because of its ability to disassemble the aggregates of LPSs, we further hypothesized that NCK-10 would prevent LPSs from invoking a cytokine response. Indeed, the stimulation of TNF-α and IL-6 production was hindered by the addition of the compound. This opens up the possibility to use NCK-10 as an antisepsis therapy.
As mentioned earlier, the biofilms formed by P. aeruginosa are composed of several anionic lipophilic components and are also recalcitrant to antibiotic therapy. The ability of NCK-10 to act on P. aeruginosa biofilms is indeed an important finding. Drugs against any infection caused by multidrug-resistant P. aeruginosa are scarce, and therapy against biofilms caused by P. aeruginosa is nonexistent. Therefore, the most important finding here is that the compound was active in murine models of burn infections caused by the biofilms of P. aeruginosa. This opens up the possibility of NCK-10 to be used in the therapy against topical infections caused by P. aeruginosa biofilms.
Conclusions
In summary, this study gives a comprehensive understanding of the interactions aryl-alkyl-lysines have with bacterial membranes. Solid-state NMR spectra prove that the density, thickness, and order parameters of the lipid bilayer are significantly perturbed, which ultimately lead to the disordering and lysis of bacterial membranes. Further, the compound was able to interact with other medically relevant anionic lipophilic moieties such as LPS and biofilms of P. aeruginosa, extending its possible use as anti-inflammatory and antibiofilm formulations. The activity of the compound against P. aeruginosa biofilms in a murine model of burn infections further opens up its possibility as an anti-infective topical agent.
Materials and Methods
The synthetic protocols used for the preparation of the compounds have been described in previous publications.38 Colistin was bought from Sigma-Aldrich (Bangalore, India). All lipids were purchased from Avanti Polar Lipids (Alabaster, AL, USA). Escherichia coli BODIPY-LPS [BODIPYFL (503/513)] and E. coli 0111:B4 LPS were obtained from Molecular Probes, Life Technologies, USA. P. aeruginosa strain PA R590 was acquired from the Department of Neuromicrobiology, National Institute of Mental Health and Neuro Sciences, Hosur Road, Bangalore 560029, India. Culture media were from HiMedia and Sigma-Aldrich (India). SYTO-9 was bought from Thermo Fischer, Bangalore, India.
Preparation of Liposomes
Multilamellar vesicles were prepared by co-dissolving of lipids [POPE/POPG (3:1) or POPE/POPE-2H31/POPG (2:1:1)] in the absence and presence of compounds at the aforementioned compound-to-lipid ratio in a mixture of chloroform/methanol (2:1, v/v). Complete mixing of the components was ensured, and then the solvent was evaporated using nitrogen gas. The samples were exposed to high vacuum using a lyophilizer further to remove any traces of solvent and then rehydrated at h = 0.9 [h = mass of water/the total mass of the system (phospholipids and water)]. Samples were made homogeneous by repeatedly vortexing the liposomes for 40 s, freezing in liquid nitrogen for 30 s, and heating in a water bath at 40 °C for 10 min. After one to three freeze–thaw–vortex cycles, the compounds embedded in the liposomes were obtained and transferred into MAS rotors, which are used as containers for static solid-state NMR measurements.
Solid-State NMR Spectroscopy
The solid-state NMR spectra were recorded and analyzed following previously reported protocols.72,73 A dedicated Bruker AVANCE NMR spectrometer (7.4 T) was used to run the solid-state NMR experiments. The proton-decoupled 31P NMR spectra were acquired at 121.5 MHz using a phase-cycled Hahn-echo pulse sequence. The following acquisition parameters were typically used: a spectral window of 32 kHz and a π/2 pulse width of 4.30 μs. An interpulse delay of 30 μs and a recycle delay of 2 s were used. A line broadening of 150 Hz was usually applied prior to Fourier transformation. H3PO4 (85% in H2O, 0 ppm) was used as a standard for the calibration of phosphorus chemical shifts. Samples were pre-equilibrated (15 min at 40 ± 1 °C) before the NMR signal was acquired.
2H NMR spectra were recorded at 46 MHz by means of a quadrupolar echo pulse sequence, with a π/2 pulse width of 3.4 μs and an interpulse delay of 50 μs. An exponential apodization resulting in 200 Hz line broadening was applied prior to Fourier transformation starting on the top of the echo signal. The samples were allowed to equilibrate for at least 15 min at 40 ± 1 °C before the NMR signal was acquired.
Spectral Simulation and Determination of c/a Deformation Ratios
The spectral intensity distribution upon deformation of vesicles was analyzed using a Fortran program home written for the purpose. The algorithm first calculates the free induction decay and then performs its Fourier transformation.73 Parameters such as chemical shift anisotropy (estimated from the experimental spectrum), line widths, weight (w), isotropic chemical shift, and c/a ratio provided inputs for the calculation using a trial and error process. Once the simulated spectrum matched with the experimental one, the only adjustable variable remaining was the ellipsoid c/a ratio. By considering an ellipsoidal distribution of bilayer orientations, the probability of an alignment at angle Θ with respect to the magnetic field is74
where Θ is the angle between a directional vector and B0 and c/a represent the ellipsoid long-to-short axis ratio.69,75,76
Determination of Experimental Order Parameters
The classical deconvolution method of Bloom was applied first, followed by spectral simulation to the refinement of attributions, which allowed a good overlap of the experimental and calculated spectra. The experimental spectrum was simulated by entering six to eight measured quadrupolar splittings; any nonresolved splittings were matched with the best powder pattern intensities thereafter by varying the signals that represent 16 labeled positions of the aliphatic chain. The equation linking the NMR-measured quadrupolar splittings in the lipid bilayers, ΔνQ, to SCD order parameters is45,46
where AQ is the static electric quadrupolar coupling constant (167 kHz) for methyl and methylene groups. Often, the plateau is measured where the quadrupolar splittings correspond to positions k = 2 to 8–10. In addition, the measurements of the terminal C14-2H3 order parameter report on the dynamics at the center of the bilayer.
Ability To Interact with LPS-BODIPY
Binding and dissociation of LPSs were performed using a previously published protocol.51 Briefly, a stock solution of 100 μg/mL of E. coli BODIPY-LPS [BODIPYFL (503/513), Molecular Probes, Life Technologies, USA] was diluted to 10 μg/mL in 1× phosphate-buffered saline (PBS; pH 7.4) and sonicated for 2 min every time prior to use. Two milliliters of this sonicated solution of BODIPY-LPS was transferred to a quartz cuvette at a concentration of 500 ng/mL. Subsequently, the respective concentrations of NCK-10 in PBS were added to it, and the fluorescence was measured using a λS55 fluorescence spectrophotometer (PerkinElmer). The parameters used were as follows: excitation wavelength: 485 nm; emission wavelengths: 500–700 nm; excitation slit width = 5 nm; emission slit width = 15 nm; and temperature: 25 °C. Three independent experiments were performed, and the data furnished are a representative image of one experiment.
Ability of NCK-10 To Inhibit the LPS-Induced Stimulation of Proinflammatory Cytokines
This experiment was performed following a previously published protocol.51 Briefly, hPBMCs were freshly isolated and seeded in 24-well plates for 3 h [1 × 106 cells in 1 mL of Roswell Park Memorial Institute medium (Gibco)]. To these wells, 20 ng/mL of E. coli 0111:B4 LPS (Molecular Probes, Life Technologies, USA) was added in the presence or absence of NCK-10 (10 μg/mL). Hanks’ balanced salt solution and only NCK-10 were also added as controls. After incubation for 18–24 h, ELISA was performed on the cell culture supernatants for cytokines TNF-α and IL-6. ELISA kit was bought from BD Biosciences, and manufacturer’s instructions were followed for the experiment.
Biofilm Disruption Assay
These experiments were performed using slightly modified versions of previously published protocols.39,40
Confocal Imaging
Sterilized glass cover slips (diameter 13 mm) were positioned in six-well plates. To the wells containing the cover slips, 2 mL of mid-log phase culture of P. aeruginosa (R590), at a concentration of 105 CFU/mL in a nutrient broth containing 0.1% NaCl and 0.1% glucose (complete medium), was added. The plate was incubated at 33 °C for 72 h under a stationary condition. Then, the medium was aspirated, and unbound bacteria were carefully removed by washing off the cover slips with 1× PBS (pH = 7.4). Biofilms containing cover slips were then placed into another six-well plate, which were supplemented with 2 mL of complete medium without (untreated control) or with NCK-10 (30 μg/mL). The plates were incubated further at 33 °C for 24 h. After 24 h, the supernatant was aspirated, and unbound bacterial cells were removed by washing with PBS. The cover slips were carefully removed from the wells and placed on the glass slides, stained with 10 μL SYTO-9 (3 μM), and observed under a Zeiss 510 Meta confocal laser scanning microscope. An LSM 5 Image examiner was used to prepare the final images.39,40
Ability To Lyse Bacteria Embedded within the Biofilms40
Biofilms were prepared on sterilized cover slips exactly as described above. However, instead of imaging, the bacteria embedded within were counted. Both the NCK-10-treated and -untreated biofilms (on glass cover slips) were incubated in 100 μL of trypsin–ethylenediaminetetraacetic acid (0.25%) (GIBCO) for 5 min. Then, the surfaces of the cover slips containing the biofilms were scraped off with a pipette tip. The resulting cell suspension was then vortexed at a high speed for 1 min to break up the clumps of bacteria and homogenize the solution. This vortexed solution was serially diluted, plated on nutrient agar plates, and incubated for 24 h at 37 °C to obtain bacterial counts. After 24 h of incubation, the plates were counted for bacterial colonies, and cell viability was expressed as log (CFU/mL) and compared with the untreated control.40 The experiment was performed twice.
In Vivo Infection Studies
These experiments were performed using slightly modified versions of previously published protocols.39,40 The mice experiments were approved by the Institutional Animal Ethics Committee. Female Balb/c mice (6–8 weeks old) weighing 22–25 g were used for the experiment. At first, the mice were anesthetized by injecting a ketamine–xylazine cocktail intraperitoneally. Once anesthetized, around 1 cm2 of their dorsal skin surface was shaved and cleansed. Around 6 mm burn wounds were created in the shaved area by applying a 120 s heated brass bar for 10 s.39 Subsequently, the burn wounds were infected with a mid-log phase bacterial inoculum of 107 CFU of P. aeruginosa (PA R590) prepared in PBS. The treatment was started 24 h post-infection, to allow sufficient time for biofilm formation. Burn wounds were treated every 24 h for 6 days with 40 μL of NCK-10 (at concentrations of 20 and 40 mg/kg) solution and colistin (30 mg/kg) solution in saline. Only saline was applied to mice in the untreated control group. Mice were sacrificed 7 days post-injury; the infected muscle tissue was excised, weighed, and homogenized in 10 mL of PBS. These homogenized solutions were serially diluted and then plated on MacConkey agar (Himedia, India). Bacteria were counted after 24 h, and the results were analyzed and reported as a log (CFU/g) of tissue.39,40p value was calculated using unpaired Student’s t-test (two-tailed two samples assuming equal variances) between the control group and the treatment group (a value of p < 0.05 was considered significant).39,40
Acknowledgments
J.H. thanks Prof. C. N. R. Rao for his constant support and encouragement. B.B. is grateful for financial contributions from the Agence Nationale de la Recherche (projects TRANSPEP 07-PCV-0018, MemPepSyn 14-CE34-0001-01, InMembrane 15-CE11-0017-01, and the LabEx Chemistry of Complex Systems 10-LABX-0026_CSC), the University of Strasbourg, the CNRS, the Région Alsace and the RTRA International Center of Frontier Research in Chemistry as well as to the Institut Universitaire de France for providing additional time to be dedicated to research. The authors are grateful to Arnaud Marquette for providing the Mathematica illustration of a sphere.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b01052.
Experimental order parameters and bilayer thickness calculations for POPE/POPE-2H31/POPG (2:1:1) systems and order parameters plots for NCK-4, NCK-6, NCK-8, and NCK-10 embedded in POPE/POPE-2H31/POPG (2:1:1) (PDF)
Author Contributions
This manuscript was written through contributions from all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf
- Butler M. S.; Blaskovich M. A.; Cooper M. A. Antibiotics in the clinical pipeline at the end of 2015. J. Antibiot. 2017, 70, 3–24. 10.1038/ja.2016.72. [DOI] [PubMed] [Google Scholar]
- Fjell C. D.; Hiss J. A.; Hancock R. E. W.; Schneider G. Designing antimicrobial peptides: form follows function. Nat. Rev. Drug Discovery 2012, 11, 37–51. 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- Hancock R. E. W.; Nijnik A.; Philpott D. J. Modulating immunity as a therapy for bacterial infections. Nat. Rev. Microbiol. 2012, 10, 243–254. 10.1038/nrmicro2745. [DOI] [PubMed] [Google Scholar]
- Hancock R. E. W.; Sahl H.-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- Ghosh C.; Haldar J. Membrane-Active Small Molecules: Designs Inspired by Antimicrobial Peptides. ChemMedChem 2015, 10, 1606–1624. 10.1002/cmdc.201500299. [DOI] [PubMed] [Google Scholar]
- Scorciapino M. A.; Rinaldi A. C. Antimicrobial peptidomimetics: reinterpreting nature to deliver innovative therapeutics. Front. Immunol. 2012, 3, 171. 10.3389/fimmu.2012.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechinger B. Detergent-like properties of magainin antibiotic peptides: a 31P solid-state NMR spectroscopy study. Biochim. Biophys. Acta 2005, 1712, 101–108. 10.1016/j.bbamem.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Moulay G.; Leborgne C.; Mason A. J.; Aisenbrey C.; Kichler A.; Bechinger B. Histidine-rich designer peptides of the LAH4 family promote cell delivery of a multitude of cargo. J. Pept. Sci. 2017, 23, 320–328. 10.1002/psc.2955. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Mant C. T.; Farmer S. W.; Hancock R. E. W.; Vasil M. L.; Hodges R. S. Rational Design of α-Helical Antimicrobial Peptides with Enhanced Activities and Specificity/Therapeutic Index. J. Biol. Chem. 2005, 280, 12316–12329. 10.1074/jbc.m413406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter E. A.; Wang X.; Lee H.-S.; Weisblum B.; Gellman S. H. Erratum: Non-haemolytic β-amino-acid oligomers. Nature 2000, 405, 298. 10.1038/35012704. [DOI] [PubMed] [Google Scholar]
- Hamuro Y.; Schneider J. P.; DeGrado W. F. De Novo Design of Antibacterial β-Peptides. J. Am. Chem. Soc. 1999, 121, 12200–12201. 10.1021/ja992728p. [DOI] [Google Scholar]
- Radzishevsky I. S.; Rotem S.; Bourdetsky D.; Navon-Venezia S.; Carmeli Y.; Mor A. Improved antimicrobial peptides based on acyl-lysine oligomers. Nat. Biotechnol. 2007, 25, 657–659. 10.1038/nbt1309. [DOI] [PubMed] [Google Scholar]
- Violette A.; Averlant-Petit M. C.; Semetey V.; Hemmerlin C.; Casimir R.; Graff R.; Marraud M.; Briand J.-P.; Rognan D.; Guichard G. N,N’-Linked Oligoureas as Foldamers: Chain Length Requirements for Helix Formation in Protic Solvent Investigated by Circular Dichroism, NMR Spectroscopy, and Molecular Dynamics. J. Am. Chem. Soc. 2005, 127, 2156–2164. 10.1021/ja044392b. [DOI] [PubMed] [Google Scholar]
- Padhee S.; Hu Y.; Niu Y.; Bai G.; Wu H.; Costanza F.; West L.; Harrington L.; Shaw L. N.; Cao C.; Cai J. Non-hemolytic α-AApeptides as antimicrobial peptidomimetics. Chem. Commun. 2011, 47, 9729–9731. 10.1039/c1cc13684d. [DOI] [PubMed] [Google Scholar]
- Choi S.; Isaacs A.; Clements D.; Liu D.; Kim H.; Scott R. W.; Winkler J. D.; DeGrado W. F. De novo design and in vivo activity of conformationally restrained antimicrobial arylamide foldamers. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 6968–6973. 10.1073/pnas.0811818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilker M. F.; Nüsslein K.; Tew G. N.; Coughlin E. B. Tuning the hemolytic and antibacterial activities of amphiphilic polynorbornene derivatives. J. Am. Chem. Soc. 2004, 126, 15870–15875. 10.1021/ja045664d. [DOI] [PubMed] [Google Scholar]
- Kuroda K.; DeGrado W. F. Amphiphilic polymethacrylate derivatives as antimicrobial agents. J. Am. Chem. Soc. 2005, 127, 4128–4129. 10.1021/ja044205+. [DOI] [PubMed] [Google Scholar]
- Gelman M. A.; Weisblum B.; Lynn D. M.; Gellman S. H. Biocidal activity of polystyrenes that are cationic by virtue of protonation. Org. Lett. 2004, 6, 557–560. 10.1021/ol036341+. [DOI] [PubMed] [Google Scholar]
- Nederberg F.; Zhang Y.; Tan J. P. K.; Xu K.; Wang H.; Yang C.; Gao S.; Guo X. D.; Fukushima K.; Li L.; Hedrick J. L.; Yang Y.-Y. Biodegradable nanostructures with selective lysis of microbial membranes. Nat. Chem. 2011, 3, 409–414. 10.1038/nchem.1012. [DOI] [PubMed] [Google Scholar]
- Haldar J.; An D.; Alvarez de Cienfuegos L.; Chen J.; Klibanov A. M. Polymeric coatings that inactivate both influenza virus and pathogenic bacteria. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 17667–17671. 10.1073/pnas.0608803103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppu D. S. S. M.; Akkapeddi P.; Manjunath G. B.; Yarlagadda V.; Hoque J.; Haldar J. Polymers with tunable side-chain amphiphilicity as non-hemolytic antibacterial agents. Chem. Commun. 2013, 49, 9389–9391. 10.1039/c3cc43751e. [DOI] [PubMed] [Google Scholar]
- Sambhy V.; Peterson B. R.; Sen A. Antibacterial and hemolytic activities of pyridinium polymers as a function of the spatial relationship between the positive charge and the pendant alkyl tail. Angew. Chem. 2008, 47, 1250–1254. 10.1002/anie.200702287. [DOI] [PubMed] [Google Scholar]
- Hoque J.; Akkapeddi P.; Yarlagadda V.; Uppu D. S. S. M.; Kumar P.; Haldar J. Cleavable cationic antibacterial amphiphiles: synthesis, mechanism of action, and cytotoxicities. Langmuir 2012, 28, 12225–12234. 10.1021/la302303d. [DOI] [PubMed] [Google Scholar]
- Haug B. E.; Stensen W.; Kalaaji M.; Rekdal Ø.; Svendsen J. S. Synthetic antimicrobial peptidomimetics with therapeutic potential. J. Med. Chem. 2008, 51, 4306–4314. 10.1021/jm701600a. [DOI] [PubMed] [Google Scholar]
- Ghosh C.; Konai M. M.; Sarkar P.; Samaddar S.; Haldar J. Designing Simple Lipidated Lysines: Bifurcation Imparts Selective Antibacterial Activity. ChemMedChem 2016, 11, 2367–2371. 10.1002/cmdc.201600400. [DOI] [PubMed] [Google Scholar]
- Ghosh C.; Sarkar P.; Samaddar S.; Uppu D. S. S. M.; Haldar J. L-Lysine based lipidated biphenyls as agents with anti-biofilm and anti-inflammatory properties that also inhibit intracellular bacteria. Chem. Commun. 2017, 53, 8427–8430. 10.1039/C7CC04206J. [DOI] [PubMed] [Google Scholar]
- Konai M. M.; Haldar J. Fatty Acid Comprising Lysine Conjugates: Anti-MRSA Agents That Display In Vivo Efficacy by Disrupting Biofilms with No Resistance Development. Bioconjugate Chem. 2017, 28, 1194–1204. 10.1021/acs.bioconjchem.7b00055. [DOI] [PubMed] [Google Scholar]
- Protein NMR Spectroscopy: Principles and Practices, 2nd ed.; Cavanagh J., Fairbrother W. J., Palmer A. P. III, Rance M., Skelton N. J., Eds.; Elsevier: Oxford, 2006. [Google Scholar]
- NMR in Biological Research: Peptides and Proteins; Wüthrich K., Ed.; North Holland/American Elesevier, 1976. [Google Scholar]
- Barnes A. B.; De Paëpe G.; van der Wel P. C. A.; Hu K.-N.; Joo C.-G.; Bajaj V. S.; Mak-Jurkauskas M. L.; Sirigiri J. R.; Herzfeld J.; Temkin R. J.; Griffin R. G. High-Field Dynamic Nuclear Polarization for Solid and Solution Biological NMR. Appl. Magn. Reson. 2008, 34, 237–263. 10.1007/s00723-008-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Q. Z.; Daviso E.; Can T. V.; Markhasin E.; Jawla S. K.; Swager T. M.; Temkin R. J.; Herzfeld J.; Griffin R. G. High frequency dynamic nuclear polarization. Acc. Chem. Res. 2013, 46, 1933–1941. 10.1021/ar300348n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker L. A.; Baldus M. Characterization of membrane protein function by solid-state NMR spectroscopy. Curr. Opin. Struct. Biol. 2014, 27, 48–55. 10.1016/j.sbi.2014.03.009. [DOI] [PubMed] [Google Scholar]
- Meier B. H.; Böckmann A. The structure of fibrils from ’misfolded’ proteins. Curr. Opin. Struct. Biol. 2015, 30, 43–49. 10.1016/j.sbi.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Kaplan M.; Pinto C.; Houben K.; Baldus M. Nuclear magnetic resonance (NMR) applied to membrane–protein complexes. Q. Rev. Biophys. 2016, 49, e15 10.1017/s003358351600010x. [DOI] [PubMed] [Google Scholar]
- Bechinger B.; Resende J. M.; Aisenbrey C. The structural and topological analysis of membrane-associated polypeptides by oriented solid-state NMR spectroscopy: Established concepts and novel developments. Biophys. Chem. 2011, 153, 115–125. 10.1016/j.bpc.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Bechinger B.; Salnikov E. S. The membrane interactions of antimicrobial peptides revealed by solid-state NMR spectroscopy. Chem. Phys. Lipids 2012, 165, 282–301. 10.1016/j.chemphyslip.2012.01.009. [DOI] [PubMed] [Google Scholar]
- Ghosh C.; Manjunath G. B.; Akkapeddi P.; Yarlagadda V.; Hoque J.; Uppu D. S. S. M.; Konai M. M.; Haldar J. Small molecular antibacterial peptoid mimics: the simpler the better!. J. Med. Chem. 2014, 57, 1428–1436. 10.1021/jm401680a. [DOI] [PubMed] [Google Scholar]
- Ghosh C.; Manjunath G. B.; Konai M. M.; Uppu D. S. S. M.; Paramanandham K.; Shome B. R.; Ravikumar R.; Haldar J. Aryl-alkyl-lysines: Membrane-Active Small Molecules Active against Murine Model of Burn Infection. ACS Infect. Dis. 2016, 2, 111–122. 10.1021/acsinfecdis.5b00092. [DOI] [PubMed] [Google Scholar]
- Ghosh C.; Manjunath G. B.; Konai M. M.; Uppu D. S. S. M.; Hoque J.; Paramanandham K.; Shome B. R.; Haldar J. Aryl-Alkyl-Lysines: Agents That Kill Planktonic Cells, Persister Cells, Biofilms of MRSA and Protect Mice from Skin-Infection. PLoS One 2015, 10, e0144094 10.1371/journal.pone.0144094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowall S.; Bewley K.; Watson R.; Vasan S.; Ghosh C.; Konai M.; Gausdal G.; Lorens J.; Long J.; Barclay W.; Garcia-Dorival I.; Hiscox J.; Bosworth A.; Taylor I.; Easterbrook L.; Pitman J.; Summers S.; Chan-Pensley J.; Funnell S.; Vipond J.; Charlton S.; Haldar J.; Hewson R.; Carroll M. Antiviral Screening of Multiple Compounds against Ebola Virus. Viruses 2016, 8, 277. 10.3390/v8110277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh C.; Chaubey S.; Tatu U.; Haldar J. Aryl-alkyl-lysines: small molecular membrane-active antiplasmodial agents. MedChemComm 2017, 8, 434–439. 10.1039/c6md00589f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh C.; Yadav V.; Younis W.; Mohammad H.; Hegazy Y. A.; Seleem M. N.; Sanyal K.; Haldar J. Aryl-alkyl-lysines: Membrane-Active Fungicides That Act against Biofilms of Candida albicans. ACS Infect. Dis. 2017, 3, 293–301. 10.1021/acsinfecdis.6b00192. [DOI] [PubMed] [Google Scholar]
- Seelig J.; Seelig A. Lipid conformation in model membranes and biological membranes. Q. Rev. Biophys. 1980, 13, 19–61. 10.1017/s0033583500000305. [DOI] [PubMed] [Google Scholar]
- Davis J. H. The description of membrane lipid conformation, order and dynamics by 2H-NMR. Biochim. Biophys. Acta 1983, 737, 117–171. 10.1016/0304-4157(83)90015-1. [DOI] [PubMed] [Google Scholar]
- Seelig J. Deuterium magnetic resonance: theory and application to lipid membranes. Q. Rev. Biophys. 1977, 10, 353–418. 10.1017/s0033583500002948. [DOI] [PubMed] [Google Scholar]
- Wolf J.; Aisenbrey C.; Harmouche N.; Raya J.; Bertani P.; Voievoda N.; Süss R.; Bechinger B. pH-Dependent Membrane Interactions of the Histidine-Rich Cell-Penetrating Peptide LAH4-L1. Biophys. J. 2017, 113, 1290–1300. 10.1016/j.bpj.2017.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. The immunopathogenesis of sepsis. Nature 2002, 420, 885–891. 10.1038/nature01326. [DOI] [PubMed] [Google Scholar]
- Aderem A.; Ulevitch R. J. Toll-like receptors in the induction of the innate immune response. Nature 2000, 406, 782–787. 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- Uppu D. S. S. M.; Ghosh C.; Haldar J. Surviving sepsis in the era of antibiotic resistance: are there any alternative approaches to antibiotic therapy?. Microb. Pathog. 2015, 80, 7–13. 10.1016/j.micpath.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Uppu D. S. S. M.; Haldar J. Lipopolysaccharide Neutralization by Cationic-Amphiphilic Polymers through Pseudoaggregate Formation. Biomacromolecules 2016, 17, 862–873. 10.1021/acs.biomac.5b01567. [DOI] [PubMed] [Google Scholar]
- Bowdish D. M. E.; Davidson D. J.; Lau Y. E.; Lee K.; Scott M. G.; Hancock R. E. W. Impact of LL-37 on anti-infective immunity. J. Leukocyte Biol. 2005, 77, 451–459. 10.1189/jlb.0704380. [DOI] [PubMed] [Google Scholar]
- Easton D. M.; Nijnik A.; Mayer M. L.; Hancock R. E. W. Potential of immunomodulatory host defense peptides as novel anti-infectives. Trends Biotechnol. 2009, 27, 582–590. 10.1016/j.tibtech.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld Y.; Lev N.; Shai Y. Effect of the Hydrophobicity to Net Positive Charge Ratio on Antibacterial and Anti-Endotoxin Activities of Structurally Similar Antimicrobial Peptides. Biochemistry 2010, 49, 853–861. 10.1021/bi900724x. [DOI] [PubMed] [Google Scholar]
- Ong Z. Y.; Gao S. J.; Yang Y. Y. Short Syntheticβ-Sheet Forming Peptide Amphiphiles as Broad Spectrum Antimicrobials with Antibiofilm and Endotoxin Neutralizing Capabilities. Adv. Funct. Mater. 2013, 23, 3682–3692. 10.1002/adfm.201202850. [DOI] [Google Scholar]
- Flemming H.-C.; Wingender J.; Szewzyk U.; Steinberg P.; Rice S. A.; Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L.; Costerton J. W.; Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Jennings L. K.; Storek K. M.; Ledvina H. E.; Coulon C.; Marmont L. S.; Sadovskaya I.; Secor P. R.; Tseng B. S.; Scian M.; Filloux A.; Wozniak D. J.; Howell P. L.; Parsek M. R. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in thePseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. U.S.A. 2015, 112, 11353–11358. 10.1073/pnas.1503058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong J. N. C.; Yildiz F. H. Biofilm Matrix Proteins. Microbiol. Spectrum 2015, 3, MB-0004-2014. 10.1128/microbiolspec.MB-0004-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.; Sun J.; Ding W.; Lin J.; Tian R.; Lu L.; Liu X.; Shen X.; Qian P. Y. Extracellular matrix-associated proteins form an integral and dynamic system during Pseudomonas aeruginosa biofilm development. Front. Cell. Infect. Microbiol. 2015, 5, 40. 10.3389/fcimb.2015.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku M.; Roschitzki B.; Riedel K.; Eberl L. Identification of proteins associated with the Pseudomonas aeruginosa biofilm extracellular matrix. J. Proteome Res. 2012, 11, 4906–4915. 10.1021/pr300395j. [DOI] [PubMed] [Google Scholar]
- Couto N.; Schooling S. R.; Dutcher J. R.; Barber J. Proteome Profiles of Outer Membrane Vesicles and Extracellular Matrix of Pseudomonas aeruginosa Biofilms. J. Proteome Res. 2015, 14, 4207–4222. 10.1021/acs.jproteome.5b00312. [DOI] [PubMed] [Google Scholar]
- Murphy K.; Park A. J.; Hao Y.; Brewer D.; Lam J. S.; Khursigara C. M. Influence of O polysaccharides on biofilm development and outer membrane vesicle biogenesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2014, 196, 1306–1317. 10.1128/jb.01463-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C.; Kuehn M. J. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat. Rev. Microbiol. 2015, 13, 605–619. 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh C.; Sarkar P.; Samaddar S.; Uppu D. S. S. M.; Haldar J. l-Lysine based lipidated biphenyls as agents with anti-biofilm and anti-inflammatory properties that also inhibit intracellular bacteria. Chem. Commun. 2017, 53, 8427–8430. 10.1039/c7cc04206j. [DOI] [PubMed] [Google Scholar]
- Sanders C. R.; Prosser R. S. Bicelles: a model membrane system for all seasons?. Structure 1998, 6, 1227–1234. 10.1016/s0969-2126(98)00123-3. [DOI] [PubMed] [Google Scholar]
- Loudet C.; Diller A.; Grélard A.; Oda R.; Dufourc E. J. Biphenyl phosphatidylcholine: a promoter of liposome deformation and bicelle collective orientation by magnetic fields. Prog. Lipid Res. 2010, 49, 288–297. 10.1016/j.plipres.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Picard F.; Paquet M.-J.; Levesque J.; Bélanger A.; Auger M. 31 P NMR First Spectral Moment Study of the Partial Magnetic Orientation of Phospholipid Membranes. Biophys. J. 1999, 77, 888–902. 10.1016/s0006-3495(99)76940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sani M.; Separovic F.. Solid-State NMR Studies of Antimicrobial Peptide Interactions with Specific Lipid Environments. In Advances in Biological Solid-State NMR: Proteins and Membrane-Active Peptides; Separovic F., Naito A., Eds.; Royal Society of Chemistry Publishing: Cambridge, United Kingdom, 2014; pp 287–303. [Google Scholar]
- Laguerre A.; Löhr F.; Henrich E.; Hoffmann B.; Abdul-Manan N.; Connolly P. J.; Perozo E.; Moore J. M.; Bernhard F.; Dötsch V. From Nanodiscs to Isotropic Bicelles: A Procedure for Solution Nuclear Magnetic Resonance Studies of Detergent-Sensitive Integral Membrane Proteins. Structure 2016, 24, 1830–1841. 10.1016/j.str.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toke O.; Maloy W. L.; Kim S. J.; Blazyk J.; Schaefer J. Secondary Structure and Lipid Contact of A Peptide Antibiotic In Phospholipid Bilayers By REDOR. Biophys. J. 2004, 87, 662–674. 10.1529/biophysj.103.032706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo M. R.; Ghosh A.; Harmouche N.; Casciaro B.; Luca V.; Bortolotti A.; Cappiello F.; Stella L.; Bhunia A.; Bechinger B.; Mangoni M. L. Membrane perturbing activities and structural properties of the frog-skin derived peptide Esculentin-1a(1-21)NH 2 and its Diastereomer Esc(1-21)-1c: Correlation with their antipseudomonal and cytotoxic activity. Biochim. Biophys. Acta 2017, 1859, 2327–2339. 10.1016/j.bbamem.2017.09.009. [DOI] [PubMed] [Google Scholar]
- Harmouche N.; Aisenbrey C.; Porcelli F.; Xia Y.; Nelson S. E. D.; Chen X.; Raya J.; Vermeer L.; Aparicio C.; Veglia G.; Gorr S.-U.; Bechinger B. Solution and Solid-State Nuclear Magnetic Resonance Structural Investigations of the Antimicrobial Designer Peptide GL13K in Membranes. Biochemistry 2017, 56, 4269–4278. 10.1021/acs.biochem.7b00526. [DOI] [PubMed] [Google Scholar]
- Harmouche N.Les liposomes biphényles: un nouveau modèle de biomembrane magnétique fluorescent. Caractérisations par RMN des solides, microscopies optiques et électroniques et SAXS, Bordeaux 1. Ph.D. Thesis, University of Bordeux, France, 2013. [Google Scholar]
- Pott T.; Dufourc E. J. Action of melittin on the DPPC-cholesterol liquid-ordered phase: a solid state 2H-and 31P-NMR study. Biophys. J. 1995, 68, 965–977. 10.1016/s0006-3495(95)80272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet C.; Diller A.; Grélard A.; Oda R.; Dufourc E. J. Biphenyl phosphatidylcholine: a promoter of liposome deformation and bicelle collective orientation by magnetic fields. Prog. Lipid Res. 2010, 49, 289–297. 10.1016/j.plipres.2010.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.