Abstract
People who score higher on intelligence tests tend to have larger brains. Twin studies suggest the same genetic factors influence both brain size and intelligence. This has led to the hypothesis that genetics influence intelligence partly by contributing to the development of larger brains. We tested this hypothesis using four large imaging genetics studies (combined N = 7965) with polygenic scores derived from a genome-wide association study (GWAS) of educational attainment, a correlate of intelligence. We conducted meta-analysis to test associations among participants’ genetics, total brain volume (i.e., brain size), and cognitive test performance. Consistent with previous findings, participants with higher polygenic scores achieved higher scores on cognitive tests, as did participants with larger brains. Participants with higher polygenic scores also had larger brains. We found some evidence that brain size partly mediated associations between participants’ education polygenic scores and their cognitive test performance. Effect sizes were larger in the population-based samples than in the convenience-based samples. Recruitment and retention of population-representative samples should be a priority for neuroscience research. Findings suggest promise for studies integrating GWAS discoveries with brain imaging to understand neurobiology linking genetics with cognitive performance.
Keywords: brain volume, intelligence, MRI, polygenic scores
Introduction
People who score higher on tests of intelligence tend to have larger brains, as measured by ex vivo brain weight and in vivo magnetic resonance imaging (MRI) (van Valen 1974; Haier et al. 2004; McDaniel 2005; Pietschnig et al. 2015). Twin studies indicate this relationship partly reflects genetic factors that influence both brain size (i.e., volume) and intelligence (Posthuma et al. 2002, 2003; Toga and Thompson 2005; Deary et al. 2010). These findings suggest the hypothesis that one path through which genetic differences between people influence individual differences in intelligence is by contributing to the development of larger brains. This hypothesis can now be tested using molecular genetic data.
A recent genome-wide association study (GWAS) of educational attainment identified dozens of genetic variants that showed substantial enrichment for genes expressed during brain development (Okbay et al. 2016). Follow-up studies further identified associations between an aggregate measure of GWAS-discovered influences on education, called a polygenic score, and intelligence, including in young children who had not yet entered school (Belsky et al. 2016; Selzam et al. 2017). These findings implicate brain development and intelligence in the pathway connecting people’s genetics to their educational outcomes. Further, GWAS research has discovered polygenic variants associated with brain size (inferred through intracranial volume) (Adams et al. 2016) that also overlaps with variants associated educational attainment (Okbay et al. 2016). Now, studies are needed to test if genetics discovered in GWAS of education are associated with in vivo intermediate phenotypes, like brain size, that could constitute a biological pathway linking genetic variation to differences in intelligence and educational attainment.
We analyzed data from four imaging genetics studies from the United Kingdom (UK Biobank), New Zealand (Dunedin Study), and the United States (Brain Genomics Superstruct Project (GSP) and Duke Neurogenetics Study (DNS)), including 7965 participants, to test associations among a polygenic score for educational attainment, cognitive test performance, and brain size. We hypothesized that, consistent with previous findings, 1) participants with higher education polygenic scores would have higher cognitive test scores and 2) that participants with larger brains as measured by total brain volume would have higher cognitive test scores. We further posed the novel hypotheses that participants with higher education polygenic scores would have larger brains and that brain size would mediate the association between the education polygenic score and cognitive test performance. We combined results across our four imaging genetics datasets using random effects meta-analysis. We also examined heterogeneity between the datasets under the hypothesis that effect sizes might differ between the population-based UK Biobank and Dunedin Study samples and the GSP and DNS samples, for which range in cognitive performance is more restricted.
Methods
Participants
We analyzed data from European-descent participants in the United Kingdom-based UK Biobank (Sudlow et al. 2015; Miller et al. 2016), a population-based volunteer sample (N = 5691), the New Zealand-based Dunedin Study, a population-representative birth cohort (N = 596) (Poulton et al. 2015), and two studies in the United States consisting primarily of university students, the GSP (Holmes et al. 2015) (N = 1163), and the DNS (Elliott et al. 2018) (N = 515). Sample sizes reflect participants with available structural MRI, cognitive testing, and genetic data (Table 1). Samples are described in detail in the Supplementary Material and Table 1.
Table 1.
Samples and measures included in analysis
| Sample | Cognitive test | Total brain volume (cm3) |
|---|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
Note: Polygenic scores for all samples were computed based on the most recent GWAS of educational attainment (Okbay et al. 2016) following established methods.
Education Polygenic Score
We computed our polygenic score based on GWAS of educational attainment rather than GWAS of cognitive performance because educational attainment is a proxy phenotype for cognitive performance (Rietveld et al. 2014) and the polygenic score for educational attainment is more predictive of cognitive performance than polygenic scores from GWAS of cognitive performance (Plomin and von Stumm 2018). Education polygenic scores were computed from genome-wide single-nucleotide polymorphism (SNP) data based on GWAS results published by the Social Science Genetics Association Consortium (Okbay et al. 2016) following the methods described by Dudbridge (2013) according to the procedure used in our previous work (Belsky et al. 2016). Genetic data from the Dunedin Study were imputed to 1000 genomes (Abecasis et al. 2012), data from all other studies were not imputed. Following established practice (Wray et al. 2007; Dudbridge 2013; Okbay et al. 2016), we computed polygenic scores using data from all SNPs included in the EA2 GWAS. SNPs were not clumped or pruned for LD prior to analysis (Ware et al. 2017). Briefly, for each study, we matched SNPs in the study’s genetic database with published educational attainment GWAS results (Okbay et al. 2016). We then multiplied the education-associated allele of each SNP by the GWAS-estimated effect size and computed the average of these products across all SNPs. Polygenic scores were standardized within each study to have M = 0, SD = 1 for analysis.
Cognitive Performance
Cognitive performance was measured in the UK Biobank using 13 reason and logic puzzles (Lyall et al. 2016). Cognitive performance was measured in the Dunedin, GSP, and DNS studies using intelligence tests (the Wechsler Adult Intelligence Scale-IV [WAIS-IV] (Wechsler 1997) in the Dunedin Study, the Shipley Institute of Living Scale (Zachary 1986) in GSP, and the Wechsler Abbreviated Scale of Intelligence [WASI] (Wechsler 2013) in the DNS).
Total Brain Volume
Total brain volume was measured from high resolution, T1-weighted MRI images. In the UK Biobank, total brain volume was estimated using SIENAX (Smith et al. 2002). In the Dunedin, GSP, and DNS studies, images were processed using the FreeSurfer processing pipeline.
Statistical Analyses
We tested associations using linear regression models. Models were adjusted for sex. Models including the polygenic score were adjusted for the first 10 principal components estimated from the genome-wide SNP data to account for any residual population stratification within the European-descent samples analyzed (Price et al. 2006). Models of UK biobank and GSP data were adjusted for age. (The Dunedin Study is a single-year birth cohort and DNS participants vary in age by only by 1–2 years.). In addition to age, models in the GSP were also adjusted for scanner, console version, and head coil (12 vs. 32 channel) because the GSP was collected across multiple sites. Analyses of individual studies were conducted in R (version 3.4.0). Linear regressions were performed using the lm function. Mediation analyses were performed using a system of equations approach (Preacher and Hayes 2008) implemented with the “mediation” package (Tingley et al. 2014) in R, using nonparametric bootstrapping with 1000 iterations. The system of equations includes three regressions; the first regression tests’ association between the predictor (polygenic score [PGS]) and outcome (intelligence quotient [IQ]), the second regression tests’ association between the predictor (PGS) and the mediator (TBV) and the third regression tests’ multivariate association between the predictor (PGS) and outcome (IQ) with covariate adjustment for the mediator (total brain volume [TBV]). If the regression coefficient between predictor and outcome is significantly smaller in the third model than the first, the inference of mediation is made. Coefficients from these regressions are combined using the formula originally proposed by Sobel (2007). Standard errors are computed using the bootstrap method described by Preacher and Hayes (2008). We combined estimates across studies using random effects meta-analysis (DerSimonian and Laird 1986) implemented using STATA (version 15).
Results
Participants with Higher Polygenic Scores Performed Better on Cognitive Tests
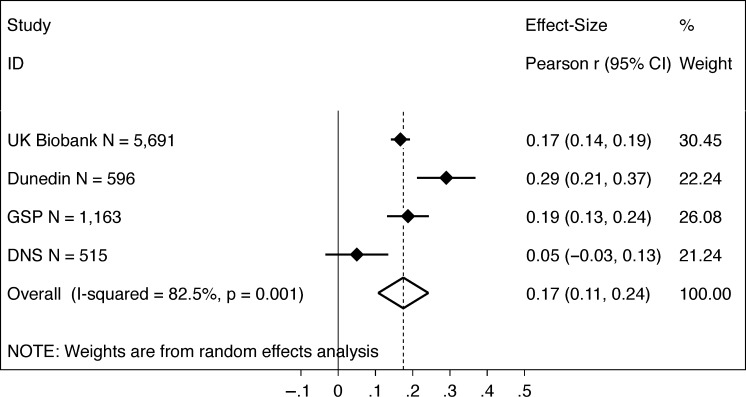
As anticipated, participants with higher polygenic scores performed better on cognitive tests (Fig. 1). Meta-analysis estimated the cross-study effect size as r = 0.18 (P < 0.001; 95% CI [0.11, 0.24]) with evidence of heterogeneity in effect sizes across studies (I2 83%, P = 0.001; τ2= 0.004). Effect sizes were statistically significant in UK Biobank (r = 0.17, P < 0.001), Dunedin Study (r = 0.28, P < 0.001) and GSP (r = 0.19, P < 0.001) but not in the DNS (r = 0.05, P = 0.220).
Figure 1.
Educational attainment polygenic score associations with cognitive test scores. The figure shows a graph of effect sizes for analyses of the UK Biobank, Dunedin Study (Dunedin), GSP, and DNS samples (solid blue diamonds) and the cross-study effect size estimated from random effects meta-analysis (open blue diamond). Gray boxes around the solid blue diamonds show the weighting of study-specific estimates in the meta-analysis (larger gray boxes indicate higher weights). 95% CIs for estimates are shown as error bars for the study-specific estimates and as the left and right extremes of the diamond for the meta-analysis effect size. The meta-analysis estimate of between-study heterogeneity (I2) is listed to the left of the open blue diamond showing the meta-analysis effect size. The table to the right of the effect size graph reports values for effect sizes, 95% CIs, and meta-analysis weights.
Participants with Larger Brains had Higher Cognitive Test Scores
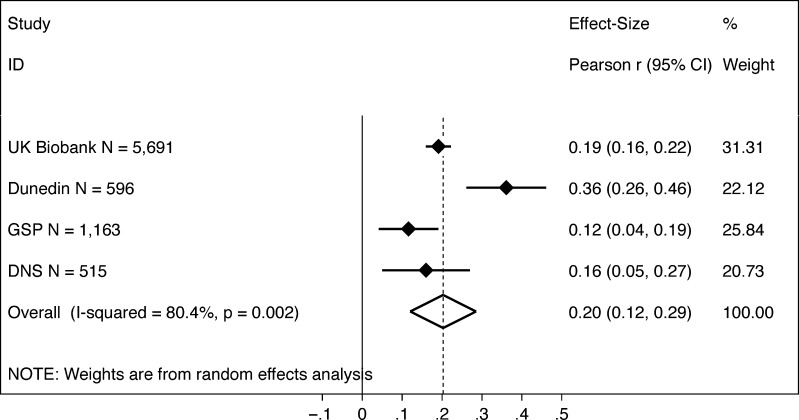
We next tested if participants with larger brains performed better on cognitive tests. As anticipated, participants with larger brains (i.e., those with higher total brain volume) performed better on cognitive tests (Fig. 2). Meta-analysis estimated the cross-study effect size as r = 0.20 (P < 0.001; 95% CI [0.12, 0.29]) with evidence of heterogeneity in effect sizes across studies (I2 = 75.8%, P = 0.002; τ2 = 0.005). Effect sizes were statistically significant in all studies (UK Biobank r = 0.19, P < 0.001; Dunedin Study r = 0.35, P < 0.001; GSP r = 0.12, P = 0.002; DNS r = 0.16, P = 0.004).
Figure 2.
Associations between brain size and cognitive test scores. The figure shows a graph of effect sizes for analyses of the UK Biobank, Dunedin Study (Dunedin), GSP, and DNS samples (solid blue diamonds) and the cross-study effect size estimated from random effects meta-analysis (open blue diamond). Gray boxes around the solid blue diamonds show the weighting of study-specific estimates in the meta-analysis (larger gray boxes indicate higher weights). 95% CIs for estimates are shown as error bars for the study-specific estimates and as the left and right extremes of the diamond for the meta-analysis effect size. The meta-analysis estimate of between-study heterogeneity (I2) is listed to the left of the open blue diamond showing the meta-analysis effect size. The table to the right of the effect size graph reports values for effect sizes, 95% CIs, and meta-analysis weights.
Participants with Higher Polygenic Scores for Educational Attainment had Larger Brains in Two Samples
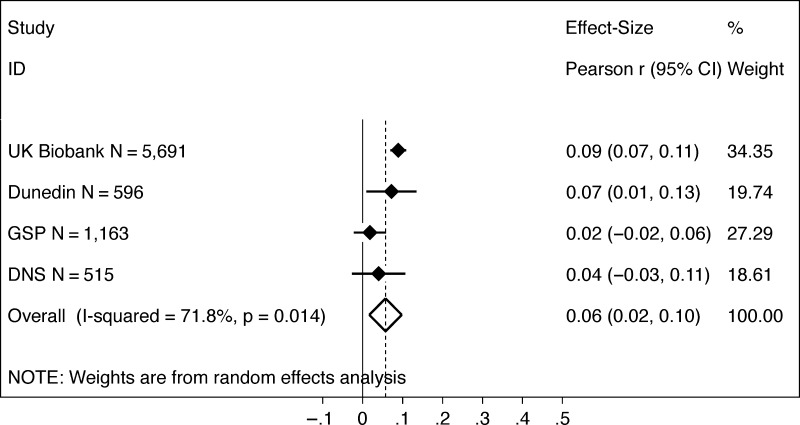
Finally, we tested if participants with higher polygenic scores tended to have larger brains. Meta-analysis estimated the cross-study effect size as r = 0.06 (P = 0.006; 95% CI [0.02, 0.10]) (Fig. 3). The test for evidence of heterogeneity in effect sizes across studies was statistically significant at the α = 0.05 level (I2 = 71.8%, P = 0.014; τ2 = 0.001). Participants with higher polygenic scores had larger brains in the UK Biobank (r = 0.09, P < 0.001) and the Dunedin Study (r = 0.07, P = 0.024). Effect sizes were smaller and not statistically significant in the GSP r = 0.02, P = 0.380 and DNS r = 0.04, P = 0.288.
Figure 3.
Educational attainment polygenic score associations with brain size. The figure shows a graph of effect sizes for analyses of the UK Biobank, Dunedin Study (Dunedin), GSP, and DNS samples (solid blue diamonds) and the cross-study effect size estimated from random effects meta-analysis (open blue diamond). Gray boxes around the solid blue diamonds show the weighting of study-specific estimates in the meta-analysis (larger gray boxes indicate higher weights). 95% CIs for estimates are shown as error bars for the study-specific estimates and as the left and right extremes of the diamond for the meta-analysis effect size. The meta-analysis estimate of between-study heterogeneity (I2) is listed to the left of the open blue diamond showing the meta-analysis effect size. The table to the right of the effect size graph reports values for effect sizes, 95% CIs, and meta-analysis weights.
Brain Size was a Weak Mediator of the Polygenic Score Associations with Cognitive Test Scores in Two Study Samples
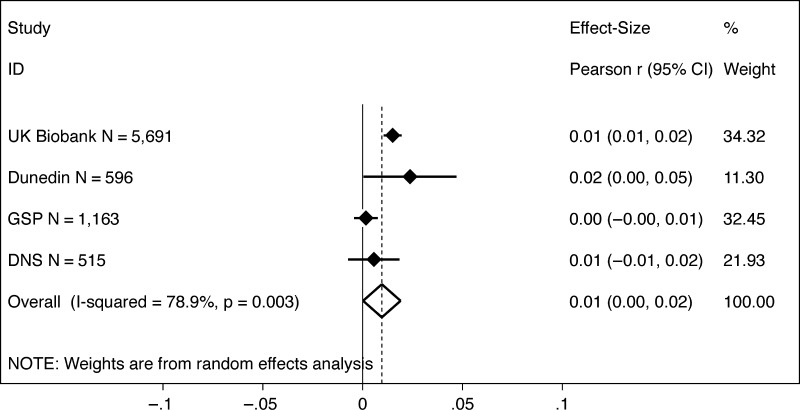
To test the hypothesis that larger brains mediated the polygenic score association with intelligence, we used the system of equations described by Baron and Kenny (1986) and the methods described by Preacher and Hayes (2008). Meta-analysis estimated the cross-study indirect effect to be b = 0.01, 95% CI [0.00, 0.02], P = 0.045, with evidence of heterogeneity in effect sizes across studies (I2 = 79.5%, P = 0.003; τ2 = 0.000) (Fig. 4). The mediation effect was statistically significant in the UK Biobank (b = 0.02, 95% CI [0.01, 0.02], P < 0.001) and the Dunedin Study (b = 0.02, 95% CI [0.00, 0.05], P = 0.028). We did not find evidence of a mediation effect in the GSP (b = 0.00, 95% CI [0.00, 0.00], P = 0.36) or DNS (b = 0.01, 95% CI [−0.00, 0.02], P = 0.24) (for details see Supplementary Table S2).
Figure 4.
Mediation effect of brain size on the association between the polygenic score for educational attainment and cognitive test scores. The figure shows a graph of effect sizes for analyses of the UK Biobank, Dunedin Study (Dunedin), GSP, and DNS samples (solid blue diamonds) and the cross-study effect size estimated from random effects meta-analysis (open blue diamond). Gray boxes around the solid blue diamonds show the weighting of study-specific estimates in the meta-analysis (larger gray boxes indicate higher weights). 95% CIs for estimates are shown as error bars for the study-specific estimates and as the left and right extremes of the diamond for the meta-analysis effect size. The meta-analysis estimate of between-study heterogeneity (I2) is listed to the left of the open blue diamond showing the meta-analysis effect size. The table to the right of the effect size graph reports values for effect sizes, 95% CIs, and meta-analysis weights.
Sensitivity Analysis: Associations Among Polygenic Scores, Brain Size, and Cognitive Test Scores were Partially Attenuated by Range Restriction
UK Biobank and Dunedin Study participants’ polygenic scores, brain size, and cognitive test performance were positively correlated, with similar effect sizes (Dunedin Study effect sizes for analyses including IQ were somewhat larger, possibly reflecting greater measurement precision of the WAIS as compared with the UK Biobank reason and logic puzzle test). By comparison, effect sizes for these associations were smaller among GSP and DNS participants. To test if this difference could reflect the relatively restricted range of cognitive test performance in the GSP and DNS samples relative to the population-based UK Biobank and Dunedin samples, we conducted sensitivity analysis. Cognitive test scores were on average, 1–1.5 SDs higher in the GSP and DNS samples as compared with the general population and 30–50% less variable, indicating restricted range (Table 1). Sensitivity analysis restricted the UK Biobank sample—the largest study in our analysis—to participants with cognitive test scores 1 SD above the mean (i.e., scores of 9–13; n = 1391) for which the variance was approximately 45% of the full-sample variance. In this restricted sample, associations among participants’ polygenic scores, brain size, and cognitive test performance were attenuated by roughly 1/3 to 1/2 relative to the full-sample estimates (Supplementary Table S3). Parallel analysis testing restriction at the other end of the cognitive test score distribution yielded similar results (Supplementary Table S4). Statistical correction of effect sizes for range restriction using Thorndike’s formula (Stauffer and Mendoza 2001) yielded similar results (Supplementary Table S7).
Discussion
We analyzed data from four imaging genetics studies in the United Kingdom, New Zealand, and United States to test if genetic associations with cognitive performance were mediated by differences in brain size. As anticipated, we found that participants with higher educational attainment polygenic scores tended to score higher on tests of cognitive performance, as did those with larger brains. We also found new information that participants with higher education polygenic scores tended to have larger brains. In mediation analysis, brain size accounted for only a small fraction of the association between participants’ educational attainment polygenic scores and their cognitive performance, and this mediation effect was statistically significant in the population-based UK Biobank and Dunedin samples but not in the GSP and DNS samples.
Effect size variation across the samples we analyzed followed a consistent pattern; effect sizes were larger in the population-based UK Biobank and Dunedin Study samples than in the GSP and DNS samples (see Figs (1–4) and Supplementary Table S1). One reason for these differences may be the more restricted range of variation in cognitive performance in the GSP and DNS samples arising from, for example, overrepresentation of university-educated individuals. Such range restriction biases association estimates (Mendoza and Mumford 1987; Bland and Altman 2011) and has previously been shown to bias brain imaging research (Falk et al. 2013; Lewinn et al. 2017). In these relatively high-IQ and restricted-range samples, average cognitive performance was 1–1.5 SDs above the general population mean and the variance was reduced by 30–50%. We conducted sensitivity analysis in a UK Biobank subsample selected to have high cognitive performance similar to the GSP and DNS samples. In this sample with restricted range of cognitive test performance, effect sizes were attenuated by roughly 30–50%. We obtained similar estimates when we performed a statistical correction for range restriction using Thorndike’s formula (Stauffer and Mendoza 2001). Selective observation of high-cognitive performance individuals in the GSP and DNS samples may have contributed to the lower effect size estimates in these samples and to overall heterogeneity across samples in our meta-analysis.
We acknowledge limitations of our current analyses, which can be addressed in future research. First, analyses were restricted to European-descent participants. We focused on European-descent participants to match the population studied in the GWAS of educational attainment. Application of GWAS results from European-descent samples to compute polygenic scores for samples of different ancestry has uncertain validity (Martin et al. 2017). As GWAS of education and related phenotypes in non-European samples become available, replication in additional populations will be needed. Second, polygenic scores were measured with substantial error. Genetic effect sizes thus represent lower bound estimates. As larger sample GWAS become available, error in polygenic score measurement will decline and effect sizes can be expected to increase (Cesarini and Visscher 2017). A third education polygenic score is available, but we were unable to use EA3 to compare across cohorts because the discovery sample included all of UK Biobank (Supplementary Table S6 reports EA3 for the other samples). Measurement error may also affect the other variables in our analysis. For example, as noted by Gignac and Bates (2017), effect size estimates from more reliable cognitive tests, such as the WAIS administered in the Dunedin Study, tend to be larger compared with effect size estimates from briefer less reliable cognitive tests. We report effect sizes disattenuated for estimated measurement error and reliability using the approach proposed by Tucker-drob (2017) in Supplementary Table S8. Third, total brain volume is only one route through which the genetics linked with educational attainment could affect cognitive performance. We studied this specific phenotype because it is the best-replicated neural correlate of cognitive function (Pietschnig et al. 2015). As more refined neural phenotypes of cognitive function are developed, including measures of cortical thickness, surface area, gyrification, and brain function, it will be important to test their potential mediating role in linking genetics with cognitive performance. Importantly, the hunt for neural phenotypes mediating genetic associations with cognitive performance need not assume that education-linked genetics directly affect brain development. For example, there is evidence that exposure to education increases cognitive performance (Ritchie and Tucker-Drob 2018). It could be that higher education-linked genetics, and higher IQs, lead to more education, which in turn enhances brain size and other neural phenotypes.
We also cannot rule out age differences as a potential explanation for the difference in findings between the population-based UK Biobank and Dunedin Study samples as compared with the GSP and DNS samples. UK Biobank and Dunedin Study participants were measured in midlife, whereas GSP and DNS samples primarily included young adults. Among midlife UK Biobank participants, restricting the range of cognitive performance to be similar to the GSP and DNS samples reduced effect sizes for associations among polygenic scores, brain size, and cognitive test performance. Population-based samples including both young and midlife individuals with DNA, MRI, and cognitive testing are needed to evaluate whether genetic associations with brain volume and cognitive performance vary with age. A final concern is potential reverse causation between brain size and cognitive function. Higher cognitive ability and related educational and socioeconomic attainments may be protective of age-related decline in brain volume or they may promote brain development. As GWAS of these phenotypes become available, new and developing methods may help address this question (Burgess et al. 2015; Grotzinger et al. 2018). Ultimately, longitudinal studies with repeated measures of brain volume and cognition will be needed to further inform our understanding of the relationship between cognitive development and brain development.
Within the bounds of these limitations, our findings contribute to evidence that genetics discovered in GWAS of educational attainment influence brain development and cognitive function. Bioinformatic analysis of education GWAS results have identified enrichment of variants near genes expressed in brain development, specifically neural proliferation, neural development, and dendrite formation (Okbay et al. 2016). Epidemiologic analysis of an education GWAS-based polygenic score found that children who carried more education-associated genetic variants scored higher on cognitive tests as early as age 5 and that polygenic score-associated differences in cognitive test scores grew larger from middle childhood through adolescence (Belsky et al. 2016, 2018). Several studies have reported that an education GWAS-based polygenic score is predictive of cognitive test performance in adolescents and adults (Domingue et al. 2015; Selzam et al. 2017; Plomin and von Stumm 2018). Here, we show that adults with higher education GWAS-based polygenic scores have larger brains and score higher on cognitive tests as compared with peers with lower polygenic scores. Evidence for larger brains as a statistical mediator of polygenic score associations with cognitive performance was mixed in our analysis. But findings suggest promise for future neuroscientific investigation of education-linked genetics. One design to complement formal mediation analysis is gene–environment interaction analysis to test if exposures that slow brain growth or restrict brain size, for example, Zika virus (Calvet et al. 2016), diminish associations between genetics and cognitive performance.
Our finding that genetics associated with educational and socioeconomic attainments are also related to brain volume has implications for research on effects of poverty on the developing brain. Childhood poverty exposure is associated with smaller brain volumes (Luby et al. 2013; Hair et al. 2015). Education polygenic scores also tend to be lower in children growing up in poorer families, a gene–environment correlation that presumably reflects the effects of education-linked genetics on parents’ economic attainments, which children inherit along with their genotypes (Belsky et al. 2016). Studies that include controls for education genetics could complement intervention studies (Brody et al. 2017) to help rule out potential confounding in associations between poverty and brain development.
A challenge facing research on how genetics affect the brain is the lack of population-representative samples with available brain imaging data. Human brain imaging research has typically been conducted in samples similar to those in the GSP and DNS whose data we analyzed (Sears 1986; Peterson and Merunka 2014). Our findings illustrate how studies of samples preselected for high levels of cognitive functioning and related characteristics impose limitations on analysis of cognition-related neurobiology. Opportunities to understand the brain afforded by 21st-century measurement technologies must still reckon with 20th-century discoveries about selection bias (Berkson 1946; Heckman 1979). Efforts to recruit more representative samples that reflect the full range of cognitive functioning in the population are needed.
Individual differences in cognitive performance have a partial genetic etiology (Plomin and Deary 2015; Plomin and von Stumm 2018). This genetic etiology should be evident in individual differences in brain biology. As GWAS discoveries for intelligence and related traits clarify genetic etiology, follow-up in genetically informed brain imaging studies can shed light on the neurobiological correlates of this genetic variation. Our findings not only encourage enthusiasm for this research but also highlight the limitations of the existing data resources. Recruiting and retaining samples that are representative of the general population must be a priority in neuroscience research.
Supplementary Material
Funding
This research was conducted using the UK Biobank Resource (project ID #28 174). The Dunedin Multidisciplinary Health and Development Study is supported by the New Zealand Health Research Council (NZ HRC), New Zealand Ministry of Business Innovation and Employment (NZ MBIE), National Institute on Aging (grants R01AG032282 and R01AG049789), and UK Medical Research Council (grant MR/P005918/1). The Duke Neurogenetics Study received support from Duke University as well as US-National Institutes of Health (grants R01DA033369 and R01DA031579). National Science Foundation Graduate Research Fellowship (grant no. NSF DGE-1644868 to M.L.E.). D.W.B. is supported by an early-career fellowship from the Jacobs Foundation; National Institutes of Health (grant K99AG054573 to T.G.); National Institute of Mental Health (grant K01MH099232 to A.J.H.).
Notes
The authors thank to members of the Advisory Board for the Dunedin Neuroimaging Study. Conflict of Interest: None declared.
References
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature. 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams HHH, Hibar DP, Chouraki V, Stein JL, Nyquist PA, Rentería ME, Trompet S, Arias-Vasquez A, Seshadri S, Desrivières S, et al. 2016. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat Neurosci. 19:1569–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron RM, Kenny DA. 1986. The moderator-mediator variable distinction in social psychological-research: conceptual, strategic, and statistical considerations. J Pers Soc Psychol. 51:1173–1182. [DOI] [PubMed] [Google Scholar]
- Belsky DW, Domingue BW, Wedow R, Arseneault L, Boardman JD, Caspi A, Conley D, Fletcher JM, Freese J, Herd P, et al. 2018. Genetic analysis of social mobility in five longitudinal studies. Proc Natl Acad Sci. 115:E7275–E7284. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, Moffitt TE, Corcoran DL, Domingue B, Harrington H, Hogan S, Houts R, Ramrakha S, Sugden K, Williams BS, et al. 2016. The genetics of success: how single-nucleotide polymorphisms associated with educational attainment relate to life-course development. Psychol Sci. 27:957–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkson J. 1946. Limitations of the application of fourfold table analysis to hospital data. Biometrics. 2:27–53. [PubMed] [Google Scholar]
- Bland JM, Altman DG. 2011. Statistics notes: correlation in restricted ranges of data. BMJ. 343:577. [DOI] [PubMed] [Google Scholar]
- Brody GH, Gray JC, Yu T, Barton AW, Beach SRH, Galvan A, MacKillop J, Windle M, Chen E, Miller GE, et al. 2017. Protective prevention effects on the association of poverty with brain development. JAMA Pediatr. 171:46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Scott RA, Timpson NJ, Smith GD, Thompson SG. 2015. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 30:543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvet G, Aguiar RS, Melo ASO, Sampaio SA, de Filippis I, Fabri A, Araujo ESM, de Sequeira PC, de Mendonça MCL, de Oliveira L, et al. 2016. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect Dis. 16:653–660. [DOI] [PubMed] [Google Scholar]
- Cesarini D, Visscher PM. 2017. Genetics and educational attainment. NPJ Sci Learn. 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deary IJ, Penke L, Johnson W. 2010. The neuroscience of human intelligence differences. Nat Rev Neurosci. 11:201–211. [DOI] [PubMed] [Google Scholar]
- DerSimonian R, Laird N. 1986. Meta-analysis in clinical trials. Control Clin Trials. 7:177–188. [DOI] [PubMed] [Google Scholar]
- Domingue BW, Belsky DW, Conley D, Harris KM, Boardman JD. 2015. Polygenic influence on educational attainment. AERA Open. 1:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F. 2013. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 9:e1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott ML, Romer A, Knodt AR, Hariri AR. 2018. A connectome-wide functional signature of transdiagnostic risk for mental illness. Biol Psychiatry. 84:452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk EB, Hyde LW, Mitchell C, Faul J, Gonzalez R, Heitzeg MM, Keating DP, Langa KM, Martz ME, Maslowsky J, et al. 2013. What is a representative brain? Neuroscience meets population science. Proc Natl Acad Sci. 110:17615–17622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B. 2012. FreeSurfer. Neuroimage. 62:774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gignac GE, Bates TC. 2017. Brain volume and intelligence: the moderating role of intelligence measurement quality. Intelligence. 64:18–29. [Google Scholar]
- Grotzinger AD, Rhemtulla M, Vlaming R, de, Ritchie SJ, Mallard TT, Hill WD, Ip HF, McIntosh AM, Deary IJ, Koellinger PD, et al. 2018. Genomic SEM Provides Insights into the Multivariate Genetic Architecture of Complex Traits. bioRxiv. 305029. [DOI] [PMC free article] [PubMed]
- Haier RJ, Jung RE, Yeo RA, Head K, Alkire MT. 2004. Structural brain variation and general intelligence. Neuroimage. 23:425–433. [DOI] [PubMed] [Google Scholar]
- Hair NL, Hanson JL, Wolfe BL, Pollak SD. 2015. Association of child poverty, brain development, and academic achievement. JAMA Pediatr. 169:822–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman JJ. 1979. Sample selection bias as a specification error. Econometrica. 47:153. [Google Scholar]
- Holmes AJ, Hollinshead MO, O’Keefe TM, Petrov VI, Fariello GR, Wald LL, Fischl B, Rosen BR, Mair RW, Roffman JL, et al. 2015. Brain genomics superstruct project initial data release with structural, functional, and behavioral measures. Sci Data. 2:150031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinn KZ, Sheridan MA, Keyes KM, Hamilton A, McLaughlin KA. 2017. Sample composition alters associations between age and brain structure. Nat Commun. 8:874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby J, Belden A, Botteron K, Marrus N, Harms MP, Babb C, Nishino T, Barch D. 2013. The effects of poverty on childhood brain development. JAMA Pediatr. 167:1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyall DM, Cullen B, Allerhand M, Smith DJ, Mackay D, Evans J, Anderson J, Fawns-Ritchie C, McIntosh AM, Deary IJ, et al. 2016. Cognitive test scores in UK biobank: data reduction in 480,416 participants and longitudinal stability in 20,346 participants. PLoS One. 11:e0156366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, Daly MJ, Bustamante CD, Kenny EE. 2017. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 100:635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel MA. 2005. Big-brained people are smarter: a meta-analysis of the relationship between in vivo brain volume and intelligence. Intelligence. 33:337–346. [Google Scholar]
- Mendoza JL, Mumford M. 1987. Corrections for Attenuation and Range Restriction on the Predictor. J Educ Stat. 12:282–293. [Google Scholar]
- Miller KL, Alfaro-Almagro F, Bangerter NK, Thomas DL, Yacoub E, Xu J, Bartsch AJ, Jbabdi S, Sotiropoulos SN, Andersson JLR, et al. 2016. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat Neurosci. 19:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA, Turley P, Chen G-B, Emilsson V, Meddens SFW, et al. 2016. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 533:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RA, Merunka DR. 2014. Convenience samples of college students and research reproducibility. J Bus Res. 67:1035–1041. [Google Scholar]
- Pietschnig J, Penke L, Wicherts JM, Zeiler M, Voracek M. 2015. Meta-analysis of associations between human brain volume and intelligence differences: How strong are they and what do they mean? Neurosci Biobehav Rev. 57:411–432. [DOI] [PubMed] [Google Scholar]
- Plomin R, Deary IJ. 2015. Genetics and intelligence differences: five special findings. Mol Psychiatry. 20:98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R, von Stumm S. 2018. The new genetics of intelligence. Nat Rev Genet. 19:148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posthuma D, Baaré WFC, Pol HEH, Kahn RS, Boomsma DI, De Geus EJC. 2003. Genetic correlations between brain volumes and the WAIS-III dimensions of verbal comprehension, working memory, perceptual organization, and processing speed. Twin Res. 6:131–139. [DOI] [PubMed] [Google Scholar]
- Posthuma D, De Geus EJC, Baaré WFC, Hulshoff Pol HE, Kahn RS, Boomsma DI. 2002. The association between brain volume and intelligence is of genetic origin. Nat Neurosci. 5:83–84. [DOI] [PubMed] [Google Scholar]
- Poulton R, Moffitt TE, Silva PA. 2015. The Dunedin multidisciplinary health and development study: overview of the first 40 years, with an eye to the future. Soc Psychiatry Psychiatr Epidemiol. 50:679–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preacher KJ, Hayes AF. 2008. Asymptotic and resampling strategies for assessing and comparing indirect effects in multiple mediator models. Behav Res Methods. 40:879–891. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 38:904–909. [DOI] [PubMed] [Google Scholar]
- Rietveld CA, Esko T, Davies G, Pers TH, Turley P, Benyamin B, Chabris CF, Emilsson V, Johnson AD, Lee JJ, Leeuw CD. 2014. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc Natl Acad Sci. 111:13790–13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie SJ, Tucker-Drob EM. 2018. How much does education improve intelligence? A meta-analysis. Psychol Sci. 29:1358–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears DO. 1986. College sophomores in the laboratory. influences of a narrow data base on social psychology’s view of human nature. J Pers Soc Psychol. 51:515–530. [Google Scholar]
- Selzam S, Krapohl E, von Stumm S, O’Reilly PF, Rimfeld K, Kovas Y, Dale PS, Lee JJ, Plomin R. 2017. Predicting educational achievement from DNA. Mol Psychiatry. 22:267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Zhang Y, Jenkinson M, Chen J, Matthews PM, Federico A, De Stefano N. 2002. Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. Neuroimage. 17:479–489. [DOI] [PubMed] [Google Scholar]
- Sobel ME. 2007. Identification of causal parameters in randomized studies with mediating variables. J Educ Behav Stat. 33:230–251. [Google Scholar]
- Stauffer JM, Mendoza JL. 2001. The proper sequence for correcting correlation coefficients for range restriction and unreliability. Psychometrika. 66:63–68. [Google Scholar]
- Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, et al. 2015. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingley D, Yamamoto T, Hirose K, Keele L, Imai K. 2014. Mediation: R package for causal mediation analysis. J Stat Softw. 59:1–38.26917999 [Google Scholar]
- Toga AW, Thompson PM. 2005. Genetics of brain structure and intelligence. Annu Rev Neurosci. 28:1–23. [DOI] [PubMed] [Google Scholar]
- Tucker-drob EM. 2017. Measurement Error Correction of Genome-Wide Polygenic Scores in Prediction Samples. bioRxiv.
- van Valen L. 1974. Brain size and intelligence in man. Am J Phys Anthropol. 40:417–423. [DOI] [PubMed] [Google Scholar]
- Ware EB, Schmitz LL, Faul JD, Gard A, Mitchell C, Smith JA, Zhao W, Weir D, Kardia SL. 2017. Heterogeneity in polygenic scores for common human traits. bioRxiv. 106062.
- Wechsler D. 1997. WAIS‐III administration and scoring manual. San Antonio, TX: The Psychological Corporation. [Google Scholar]
- Wechsler D. 2013. WASI -II: Wechsler abbreviated scale of intelligence—second edition. J Psychoeduc Assess. 31:337–341. [Google Scholar]
- Wray NR, Goddard ME, Visscher PM. 2007. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 17:1520–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachary RA. 1986. Shipley Institute of Living Scale: Revised Manual. Los Angeles East Psychol Serv.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.