Abstract
The purpose of this review is to highlight recent developments in small molecules and peptides that block the binding of coactivators to steroid receptors. These coactivator binding inhibitors bind at the coregulator binding groove, also known as Activation Function-2, rather than at the ligand-binding site of steroid receptors. Steroid receptors that have been targeted with coactivator binding inhibitors include the androgen receptor, estrogen receptor and progesterone receptor. Coactivator binding inhibitors may be useful in some case of resistance to currently prescribed therapeutics. The scope of the review includes small-molecule and peptide coactivator binding inhibitors for steroid receptors, with a particular focus on recent compounds that have been assayed in cell-based models.
Keywords: steroid receptor coactivator, androgen receptor, estrogen receptor, progesterone receptor, protein-protein interactions, coactivator binding inhibitors
Introduction
One of the most physiologically and pharmacologically important families of the nuclear receptor superfamily of transcription factors is the steroid receptor family (Type NR3), which comprises the androgen receptor (AR), estrogen receptors (ERs), progesterone receptor (PR), mineral corticoid receptor (MR), and glucocorticoid receptor (GR) (Nuclear Receptors Nomenclature Committee et al., 1999). Given their ability to bind small molecules, their importance in physiology, and their misregulation in many disease states, steroid receptors are an important class of drug targets. Approximately 16% of drugs bind to nuclear receptors, and steroid receptors comprise a major class of drug targets within the nuclear receptor superfamily (Santos et al., 2016).
Although there are many endogenous, natural, and synthetic small molecules that modulate the function of steroid receptors by binding to the ligand-binding pocket, there are problems that arise with the use of some of these ligands in the clinic, most notably resistance to such endocrine therapies. For instance, despite undeniable successes, one of the major unsolved problems in ER+ breast cancer therapy is resistance to endocrine therapy (Musgrove and Sutherland, 2009; Osborne and Schiff, 2010). One-third of breast cancer patients who are given the selective estrogen receptor modulator (SERM) tamoxifen will develop recurrent cancer within 15 years. (Musgrove and Sutherland, 2009) Likewise, even with the development of third-generation antiandrogens like enzalutamide (XTANDI®), the development of resistance to AR-directed therapies remains a significant medical problem in prostate cancer (Crawford et al., 2018; Watson et al., 2015).
In such cases, it would be advantageous to target alternate regions on the surface of steroid receptors. One of the first ideas proposed for alternative pharmacological regulation of nuclear receptors was through inhibition of the interaction of coactivators with steroid receptors, the details of which are described below. This approach was first described by McDonnell and coworkers in 1999 (Norris et al., 1999). Throughout the years since these initial reports, a great deal of work has been carried out to develop small-molecule and peptide-based coactivator binding inhibitors. This area has been reviewed previously, (Caboni and Lloyd, 2013; Moore et al., 2010; Shapiro et al., 2011; Tice and Zheng, 2016) and the goal of this review is to provide an update on work that has been carried out since 2008, with a particular emphasis on those coactivator binding inhibitors that have shown responses in cell-based assays of steroid receptor activation and function. Although the focus of this review is on the steroid receptors, coactivator binding inhibitors have been described for other nuclear receptors, including the vitamin D receptor (Mita et al., 2010; Nandhikonda et al., 2012), pregnane × receptor (Kholodovych et al., 2008; Welsh et al., 2007), thyroid hormone receptor, (Arnold et al., 2007; Hwang et al., 2009) peroxisome proliferator-activated receptors (Mettu et al., 2007) and retinoid × receptors (Chen et al., 2014; Hu et al., 2014). Many of these molecules have been reviewed in the current Special Issue. (See contributions by Arnold and coworkers (Mutchie et al., 2019), and Staudinger (Staudinger, 2019).)
The steroid receptors bind to coregulator proteins, including the Steroid Receptor Coactivators (SRCs, also known as Nuclear Receptor Coactivators, or NCOAs) (Horowitz et al., 1996). Many of the coactivators have an LXXLL motif displayed over two turns of an α-helix. This motif, originally discovered for binding to the estrogen receptor, was initially thought to be both necessary and sufficient for binding of coactivators to nuclear receptors, but more recent data suggest that non-LXXLL motifs may also bind to the coactivator binding groove, a region in the N-terminal F-domain of steroid receptors (Heery et al., 1997; Savkur and Burris, 2004). Many of the coactivator binding grooves are hydrophobic in nature, complementary to the hydrophobic LXXLL motif. Additionally, androgen receptor is capable of binding larger motifs, including FXXLF and WXXVW (Hur et al., 2004).
This coactivator binding groove is also known as Activation Function 2 (AF-2). Many steroid receptors also have an N-terminal Activation Function-1, which may also bind to coactivators and mediate gene transcription (Tora et al., 1989). Although the structural details of AF-2 binding to coactivators are well-understood, including through numerous x-ray crystal structures, the molecular level interaction of AF-1 with coactivators remains shrouded. Relatively little work has been carried out to discover inhibitors of AF-1 activity, although there are some papers that have described small-molecule inhibitors of AF-1 activity (De Mol et al., 2016; Sadar, 2011). In this review, we will focus on molecules that inhibit binding of coactivators to steroid receptors and block AF-2 activity.
Strategies for discovering coactivator binding inhibitors generally fall into two categories: design and screening. When designing inhibitors of a steroid receptor/coactivator interaction, there are two options for designing the molecule: 1) mimic the features of the coactivator and bind to the nuclear receptor, or 2) mimic the nuclear receptor and bind to the coactivator. Because the interaction occurs when amino acid sidechains from the coactivator bind to a hydrophobic groove on the surface of the nuclear receptor, it is much easier to design coactivator mimics that bind to the nuclear receptor, rather than to design a nuclear-receptor-mimicking hydrophobic surface. All known coactivator binding inhibitors function by binding to nuclear receptors rather than coactivators.
Designed inhibitors often use computational approaches, including docking or molecular dynamics simulations, to design molecules that may bind to the AF-2 binding cleft. For instance, native LXXLL-containing peptides have been used as a starting point to design constrained peptides and peptide mimetics that bind to the surface of steroid receptors (see below).
The other major strategy to discover coactivator binding inhibitors is screening. Blocking the interaction of steroid receptors with coactivators is inherently mechanism-based; thus, many of the assays that are used to screen and/or characterize these types of inhibitors are in vitro biochemical and biophysical assays where the readout is directly tied to inhibiting the protein-protein interaction between steroid receptor and coactivator. An important consideration in developing coactivator binding inhibitors is that the desired protein-protein interaction inhibition leads to antagonism of the receptor; however, indirectly blocking the receptor-coactivator interaction with a small molecule that binds in the steroid-binding pocket also leads to receptor antagonism, but this indirect antagonism is mechanistically undesirable. Receptor agonists that bind at the steroid-binding pocket are necessary to bias toward receptor conformations that are favorable for coactivator binding. To address this dilemma, coactivator binding inhibitor assays are often set up so that a high concentration of receptor agonist is used, often orders of magnitude greater than the agonist dissociation constant. Such high concentrations decrease the likelihood that a small molecule could displace an agonist to induce unfavorable coactivator-binding conformations.
In this review, we will review recent literature that has described small molecules and peptides that block AF-2 activity of steroid receptors, including the estrogen, androgen, and progesterone receptors. Particular emphasis has been placed on reviewing publications that have shown activity in cell culture and/or animal models.
Estrogen Receptor
Estrogen receptor (ER) plays important roles in reproductive, brain, bone, liver and cardiovascular tissues (Katzenellenbogen et al., 2000). Nuclear ER exists as two subtypes, ERα and ERβ, and the proportion and levels of each of these subtypes varies by tissue type. The domain architecture of ER comprises domains A-F. This review will focus on the C-terminal E/F domains, which contain the ligand binding domain and ligand-dependent activation function-2 (AF-2), also described as the coactivator binding groove (see above) (Kumar et al., 1987). Transcriptional activation in ER is dependent on the binding of an agonist, which induces a conformational change, exposing the DNA-binding domain and AF-2 leading to coactivator recruitment. Some reports have shown that the identity of the agonist can have effects on the recruitment of coactivators to the ligand-binding domain. (Aarts et al., 2013; Jeyakumar et al., 2011). Estrogen receptor agonists, selective estrogen receptor modulators, and selective estrogen receptor degraders are currently used in the clinic and form a mainstay of endocrine therapy. Selective estrogen receptor modulators (e.g., tamoxifen, raloxifene and toremifene) and a selective estrogen receptor degrader (fulvestrant) are currently used for the treatment of breast cancer (Howell et al., 2004). Selective estrogen receptor modulators are also used for osteoporosis (lasofoxifene and raloxifene) and/or hot flashes (bazedoxifene) (Pinkerton and Thomas, 2014). Although the use of selective estrogen modulators (SERMs) has been highly successful for the treatment of estrogen receptor-positive breast cancer, resistance to endocrine therapy is still a major unmet medical need. Additionally, occurrence of mutations in the estrogen receptor after treatment with endocrine therapy renders some endocrine therapies less effective (Fanning et al., 2016; Lipson et al., 2014; Merenbakh-Lamin et al., 2013; Robinson et al., 2013; Toy et al., 2013).
Coactivator binding inhibitors (CBIs) represent a potentially useful mechanism of ER antagonism. The first CBIs reported for ER were peptides containing the NR box LXXLL motif, common to most steroid-receptor coactivator proteins (Chang et al., 1999; Norris et al., 1999). The sequences flanking the LXXLL motif were modified to target either the ER-a or ER-b isoforms confirming that peptides can be specific and distinguish between isoforms. The first reported small-molecule CBIs for inhibiting the ER-SRC interaction were pyrimidine-based CBIs that mimicked the LXXLL motif of the SRC (Rodriguez et al., 2004). A variety of peptide, peptidomimetic and small-molecule CBIs have been developed since the first ones were reported in 1999 (for reviews see (Caboni and Lloyd, 2013; Moore et al., 2010; Shapiro et al., 2011) This review will describe more recent molecules which have been tested in cells.
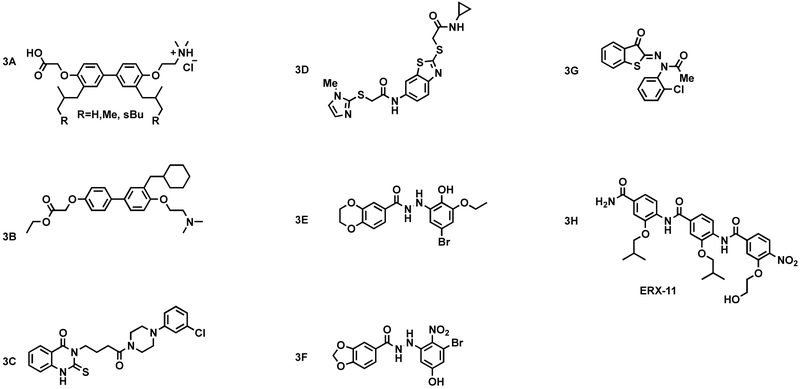
Williams et al. synthesized and characterized a set of compounds based on a bis-4,4’-oxyphenyl scaffold (Williams et al., 2009). These compounds have a tertiary amine and a carboxyl group on opposite ends to engage the ER “charge clamp” that surrounds the hydrophobic groove to which coactivators bind. A time-resolved fluorescence resonance energy transfer (TR-FRET) assay shows that one of the compounds (3A, Fig. 3) with R=Me blocks binding of an SRC fragment to ER with a Ki of 33 μM in presence of ligand. A radiometric competitive ligand-binding assay was used to ensure that the inhibitors were competing with coactivators instead of binding at the estrogen-binding site. A reporter gene assay was used to test these compounds in HEC-1 cells (which express nuclear receptor coactivators but not endogenous ERα). Only compound 3A (R=sBu) exhibited inhibitory activity, with an IC50 of ~2 μM and limited efficacy.
Figure 3.
Small-molecule inhibitors of the estrogen receptor - steroid receptor coactivator interaction.
Based on these promising data, Weiser et al. designed a set of similar analogs. These compounds were tested for their ability to block the activity of estrogen receptor and androgen receptor using luciferase reporter genes. Inhibition of an ER reporter gene was measured in HepG2 cells transfected with VP-ERα pM-GRIP1-LxxLL2, reporter gene 5xGal4-Luc3 and a normalization control pCMV-β-gal. (Weiser et al., 2012) Compound 3A (Fig. 3; R=sBu), bearing the largest substituent, was the best antagonist and exhibited no activity in the absence of 17β-estradiol (E2). A second assay employing MCF-7 (ER+) breast cancer cells was utilized to evaluate the ability of these compounds to inhibit native genes regulated by ER. RNA was harvested after a 14 h incubation period with compound (20 μM) and either vehicle or estradiol and analyzed by quantitative-PCR. This group analyzed the expression of known ERα genes SDF1, PR, and PS2, in addition to negative control IDH3A. Three compounds exhibited a modest decrease in the expression of ERα target genes, and no decrease in gene transcription was observed in the absence of estradiol.
In more advanced work, Weiser et al. synthesized a series of biphenyl peptidomimetics bearing different substitutions on each phenyl ring. These compounds were tested for their ability to inhibit ERa activity in a cotransfection reporter gene assay. 3xERE-TATA-luciferase and estrogen receptor (RST7-ERα)) were transfected into ER-negative SKBR3 breast cancer cells with a normalization control pCMV-β -gal. Cells were then treated with serial two-fold dilutions of the synthesized biphenyl compounds with and without 1 nM of E2 and incubated for 40 h prior to assaying. Three compounds were found to decrease reporter gene activity at concentrations <20 μM, both in the presence and absence of E2, suggesting that these compounds bind to the ligand-binding pocket and not the coactivator binding-groove (Weiser et al., 2014). To confirm these results, compounds were subjected to a mammalian two-hybrid competitive inhibition assay employing the estrogen receptor (VP16-ERα)) and the coactivator peptide pM-GRIP1 LXXLL. Again, it was observed that compounds that provided inhibition of reporter gene activity were active in the absence of E2. To determine whether these compounds were binding in the ligand-binding pocket, the reporter gene assay was repeated with increasing concentrations of E2. If these compounds were true CBIs, then the EC50 of E2 should not be affected by the concentration of inhibitor present. Although no compound was identified as solely a coactivator binding inhibitor, compound 3B (Fig. 3) appeared to have mixed inhibition of the ligand-binding pocket and the coactivator-binding groove. Further synthesis of analogous compounds would be required to produce a true coactivator binding inhibitor for ER±.
Using a TR-FRET assay, Sun et al. carried out a high-throughput screen to identify novel coactivator binding inhibitors (CBIs) capable of disrupting the estrogen receptor/coactivator interaction in presence of estradiol. Two scaffolds—3C and 3D (Fig. 3)—exhibited desirable inhibition activity (Sun et al., 2011). Follow-up medicinal chemistry, reporter gene assays, and in silico structural analysis led to the optimization of two new series of ER coactivator binding inhibitors which were active in the low micromolar range. The authors noted, however, that these CBIs did not seem to span the entirety of the surface area of the SRC-binding region. The authors concluded that CBI potency was thus entropically limited due to reduced displacement of surface-bound water molecules. Based on this information, they inferred that screening compound libraries containing larger molecules may lead to the discovery of CBIs with greater potency, even if these molecules may have a higher-than-normal molecular weight.
Singh et al. used a virtual screening approach to identify new small molecules that could bind to the AF-2 site of ERα. Two compounds that were identified in this work were 3E and 3F (Fig. 3), which both belonged to the carbohydrazide class of small molecules (Singh et al., 2015). Using the ERα-positive cell line, T47D-KBluc, the authors showed that the in silico hits reduced transcriptional activity of ERα at the luciferase reporter. Upon confirmation of the small molecules’ binding to AF-2, and not the ligand-binding pocket, the authors showed that the compounds could inhibit the growth of MCF-7 cells, as well as tamoxifen-resistant TamR3 and TamR6 cells (3F and 3E had IC50 values of 6.0 and 7.8 μM, respectively). To confirm the slowing of cell growth was through inhibition of ERα, TamR3 and TamR6 cells were transfected with a luciferase reporter gene. Using MCF7 cells and subsequent qRT-PCR analysis, the authors showed that 2E was able to reduce the expression of genes controlled by ERα.
Singh et al. used molecular dynamics simulations on a series of benzothiophenone derivatives to enhance target affinity and improve drug-like properties of a previously reported coactivator binding inhibitor (3E, Fig. 3). The addition of a methyl group to the benzothiophenone core of a lead compound led to a significantly more potent compound, 3G (Singh et al., 2018). A two-hybrid assay showed that this compound inhibits ERα via the inhibition of ERα-coactivator interaction. Luciferase assays highlighted the selectivity of the compound for ERα. No activity was observed for AR, GR and PR. The authors also demonstrated the anti-proliferative effects of this non-cytotoxic compound in ERα+ cell lines, along with a downregulation of ERE-driven genes by the compound. A major advantage of this AF-2 inhibitor is its similar effectiveness against clinically important mutant forms of ERα.
Using a peptidomimetic approach, Raj et al. have designed and optimized a well-characterized ER-coregulator interaction inhibitor, ERX-11 (3H, Fig. 3). This compound has been observed to be a potential therapeutic for wild-type and mutant ER-positive breast cancers as it blocks proliferation and induces apoptosis (Raj et al., 2017). This molecule contains a serine-mimicking hydroxyethyl group and has been shown to block estradiol-induced proliferation in ER-positive breast cancer cell lines. Biotinylated 3H was used to pull down endogenous ER in ZR-75 nuclear extracts, indicating that the molecule interacts with purified ER in vitro and within a cell. Using a NanoBiT assay, the authors showed that the ER–PELP1 interaction could be blocked by treatment with ERX-11. RNA-seq analyses indicated that ERX-11 altered the expression of 880 estradiol-regulated genes. Tumors treated with ERX-11 were observed to be significantly smaller, exhibited less proliferation and increased apoptosis, as compared to the vehicle control.
In addition to small molecules, peptides have also been designed to block the estrogen receptor/coactivator interaction. It has been argued in the past that peptides make poor drugs because they can be conformationally flexible, impermeable to cells, and unstable to proteases, but several recent advances in peptide chemistry have addressed these shortcomings (Henninot et al., 2018). One of the most successful approaches uses “stapled” peptides. Briefly, hydrocarbon “stapled” peptides are unnatural peptides that are linked into an α-helical conformation by linking two sidechains together with a robust, accessible reaction called ring-closing metathesis. (Blackwell and Grubbs, 1998)(Schafmeister et al., 2000) Stapled peptides specifically have been found to have improvements in pharmacology, target affinity, proteolytic resistance, serum half-life, and cell penetration as compared to non-stapled peptides. (Verdine and Hilinski, 2012). The advantages of developing a peptide coactivator binding inhibitor has led to the development of steroid receptor coactivator mimics, which show inhibitory activity in the presence of estradiol.
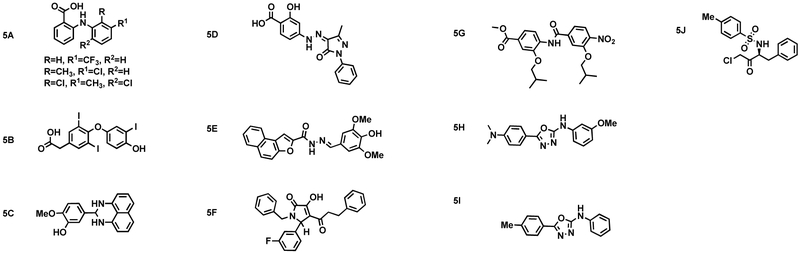
Phillips et al. designed and tested stapled peptide inhibitors of the ER-SRC interaction. Because the interaction between ER and SRC is mediated by an α-helical LXXLL motif, the authors used this motif as a starting point for development. Phillips et al. used the strategy of hydrocarbon stapling to stabilize the peptides into a helical conformation. They determined that an optimal inhibitor of the protein-protein interaction was 4A, (Fig. 4) with a KD of 75 and 155 nM for ERα and ERβ, respectively (Phillips et al., 2011).. Although 4A was not tested in more advanced assays, this work provided the basis for further developments of stapled peptide inhibitors of the ER-SRC interaction.
Figure 4.
Peptide inhibitors of the estrogen receptor - steroid receptor coactivator interaction.
Building on this work, Speltz et al. showed that the stapled peptides produced in the Phillips work were not active in cell culture. To address this, the authors made an alteration to one of the previously reported stapled peptides. Based on molecular dynamics simulations, four arginine residues were added to the N-terminus to create stapled peptide 4B, (Fig. 4) (Speltz et al., 2018a). This positively charged peptide showed an even better IC50 value of 5.0 nM. Using MCF-7 cells and the fluorescein isothiocyanate (FITC)-labeled versions of the peptides, the authors showed that there was greater cell permeation with 4B, compared to the original stapled peptide. 4B reduced the transcription of several genes regulated by the ER, and it reduced proliferation of estradiol-stimulated MCF-7 cells. An RNA-seq experiment showed that the peptide modestly reversed the expression of ER-regulated genes, although it was not as efficacious as 4-hydroxytamoxifen.
Demizu et al. designed and synthesized several LXXLL motif-containing stabilized helical peptides. Peptidomimetic estrogen receptor modulators (PERMS) 4C and 4D (Fig. 4), which contain disulfide bridges that stabilize their α-helical structures, were first described by Spatola et al (Galande et al., 2005; Leduc et al., 2003). Demizu et al. found that 4C and 4D exhibited strong inhibitory activity, with IC50 values of 35 and 13 nM, respectively, suggesting that α-helicity was important for peptides’ inhibitory activity. To observe ER-mediated transcriptional inhibition by PERM at the cellular level, the authors also synthesized R7-conjugated peptides. Heptaarginine fragments have been shown to be cell-penetrant, so their inclusion in peptide sequences may improve peptide membrane permeability (Schmidt et al., 2010; Yang and Hinner, 2015). The conjugation of PERM to a heptaarginine (R7) fragment was effective at promoting cellular uptake of PERM and inhibiting ER transcription (Demizu et al., 2015). These peptides were able to efficiently penetrate ER-positive T47D cells, and the inhibitory activity of R7 fragment-containing PERM molecules was shown to be higher with the R7 fragment in the C-terminus rather than the N-terminus.
In one of the stapled peptides designed to fit the coactivator binding pocket, reported by Phillips et al., it was noted that the crosslinked sidechains could replace interacting residues Ile689 and Leu 693. Speltz et al. synthesized a stapled peptide, designed to fit the coactivator binding pocket, that replaced Ile689 with an S-γ-methylated stapling amino acid. This molecule, 4E (Fig. 4), had an IC50 of 89 nM in vitro vs. 390 nM for the peptide that had no methyl group (Speltz et al., 2016). 4E also had high helicity. X-ray crystallography and molecular dynamics studies indicated that minimization of syn-pentane strain between two methyl groups at the α- and γ-positions was responsible for increased helicity.
Zhao et al. have developed a thioether-tethered helical peptide. This 11-mer peptide mimics the native SRC protein with a cysteine in the i position and an unnatural amino acid (mS5) in the i+4 position (Zhao et al., 2016). The binding affinity of peptide 4F was tested using a fluorescence polarization assay with the stapled peptide having a binding affinity of 91 nM, and the unstapled an affinity of 188 nM. In comparison to the binding affinity of the native SRC sequence (2500 nM), these peptides bound to the receptor with much higher affinity.
Further developments by Zhao et al. have included the development and synthesis of a thiol-yne tethered helical peptide (Tian et al., 2016). A fluorescence polarization assay indicated a Kd of 100 nM for ERγ and 177 nM for ERβ. In an effort to determine the permeability and cellular uptake of peptide 4G, the authors used flow cytometry analysis using HEK293T cells. The constrained peptide had five-fold higher uptake as compared to the unconstrained peptide. Serum stability was enhanced significantly, almost 18-fold, as compared to the unconstrained peptide, with the authors concluding that the presence of the tether enhanced protease resistance. The authors completed an MTT assay to determine the effect on cell viability that these peptides may have. The stapled peptide lacked cytotoxicity towards ER-negative MDA-MB-231 cells and exhibited cytotoxicity towards ER-positive MCF-7 cells. Hydrocarbon-stapled peptides may cause non-optimal levels of membrane lysis; therefore, Zhao et al. used a lactate dehydrogenase (LDH) release assay to determine the membrane lysis capability of these thiol-yne peptides in HeLa cells. The thiol-yne peptides had a smaller effect on the disruption of membranes as compared to the all-hydrocarbon peptides, providing these thiol-yne peptides with a potential advantage.
Xie et al. developed N-terminal isoaspartic acid-tethered helical peptides 4H that target the ER-SRC interaction (Fig. 4). The authors determined the effect of the tethered helical peptide in MCF-7 cells after exposure to 20 μM peptide for 24 hours with the following results: the constrained peptide showed 64.5% cell viability after exposure, while the linear peptide showed 98.5% cell viability after exposure. The stabilized peptide had dose-dependent inhibition of MCF7 cell viability (12 μM). A fluorescence polarization assay indicated specificity for ERα as compared to ERβ (4.4:1), progesterone receptor (3.7:1) and vitamin D receptor (30:1)
Speltz et al. prepared orthogonally stapled peptides and studied their effects on helicity, stability, and affinity (Speltz et al., 2018b). Computational analysis of a previously described peptide, (Fig. 4I) bearing an olefin-staple, demonstrated that an Arg692-Asp696 salt-bridge increased its helicity. Using this orthogonal stapling approach, the authors designed and prepared the bicyclic peptide 4J using solid phase peptide synthesis. 4J was “cross-stitched”, replacing the Arg692-Asp696 salt-bridge in 4I with an amide-based covalent linkage to provide an additional lactam staple and potentially increase α-helicity. Using a TR-FRET assay to measure the peptides’ ability to inhibit coactivator recruitment to estrogen receptor, the authors concluded that, while the added hydrocarbon staple enhanced proteolytic and helical stability, affinity was lowered, possibly due to less optimal hydrophobic interactions between the staple and estrogen receptor surface. Thus, the non-interacting lactam in 4K best enhanced binding affinity.
Fuchs et al. have used ribosome display to discover and optimize the binding motif of LXXLL and discovered a PXLXXLLXXP binding motif (Fuchs et al., 2013). Molecular modeling and x-ray crystallography were used to analyze this optimized binding motif. The prolines were determined to be helix primers, optimizing the helix dipole and allowing for better recognition of the peptide. The presence of prolines also aids in inducing conformational constraints, and stabilizing hydrogen bonding. X-ray crystallography confirms the presence of the prolines above the charge clamp residues, confirming the initial hypothesis. Circular dichroism measurements in the presence of trifluoroethanol were observed to have strong helicity. A fluorescence polarization assay provided Ki values of 0.025-75 μM, with the proline-containing peptides being more selective or equally selective for ERβ.
Androgen Receptor
Androgen receptor (AR) plays important roles in reproductive, neural, bone, immune, hemopoietic and cardiovascular tissues (Davey, 2016; Zhao et al., 2016). Unlike the ER, there is only one AR isoform (Narayanan et al., 2018). The domain architecture of AR comprises domains A-F. This review will focus on the C-terminal E/F domains, which contain the ligand binding domain and ligand-dependent activation function-2 (AF-2), also described as the coactivator binding groove (see above) (Brinkmann et al., 1989). Particular to the androgen receptor is binding function (BF)-3, a regulatory surface located near AF-2. Recently, during the screening of compounds for inhibition of coactivator binding, compounds have been discovered which bind to BF-3 and allosterically modulate AR function (Estebanez-Perpina et al., 2007). Other examples of BF-3 inhibitors (Ban et al., 2014; Lack et al., 2011) will not be discussed in this review, but allosteric inhibitors have been reviewed in this Special Issue by Brunsveld and coworkers (Meijer et al., 2019).
A unique feature of the androgen receptor is the so-called “N/C interaction.” In its N-terminal region, AR contains a phenyalanine-rich motif (23FQNLF27) which seems to interact with the C-terminal AF-2 (He et al., 2000). This N/C interaction may compete with coregulators for binding to the AR AF-2 region (Chang and McDonnell, 2005). An outcome of this observation is that AR is able to accommodate larger hydrophobic motifs than other members of the nuclear receptor family. Indeed, several groups have shown that peptides that contains (F/W)XXLF motifs bind to the androgen receptor (He et al., 2004; Hur et al., 2004), and some of these motifs are expressed in coregulators that are unique to AR (He et al., 2002). AR can bind larger motifs selectively, and this size-exclusion principle may be used to develop AR-selective CBIs (Chang and McDonnell, 2005).
Similarly to SERMs, selective androgen receptor modulators (SARMs) were developed in an effort to exhibit tissue-selectivity. The initial development of SARMs was for a treatment for muscle-wasting conditions (Crawford et al., 2016; Dalton et al., 2011; Gao et al., 2005). Further potential clinical uses of SARMs include the treatment of osteoporosis (Gao et al., 2005; Kearbey et al., 2007; Mohler et al., 2005) and breast cancer (Coss et al., 2014; Schwartzberg et al., 2017). AR has been successfully targeted for the development of treatments for prostate cancer, with the development of anti-androgens like flutamide, bicalutamide, and, most recently, enzalutamide (TANDI®); however, as with ER-directed therapies, the development of resistance to therapies targeting AR is a significant problem in the effective treatment of prostate cancer (Nakazawa et al., 2017). Coactivator binding inhibitors (CBIs) represent a potentially useful mechanism of AR antagonism. The first CBIs reported for AR were peptides containing a modified NR box motif (F/W)XXL(F/W) mimicking the LXXLL motif, common to most steroid-receptor coactivator proteins (He et al., 2002; Hsu et al., 2003).The first reported small-molecule CBIs for inhibiting the AR-SRC interaction were peptidomimetic pyrimidine-based CBIs that mimicked the modified NR box motif (F/W)XXL(F/W) (Gunther et al., 2009). This review will describe more recent molecules which have been assayed in cell culture.
Compounds prepared by Weiser et al. to block the ER/coactivator interaction (see above) were also analyzed for their ability to inhibit the interaction of androgen receptor and coactivator in a MMTV-luc reporter gene assay in CV-1 cell line transfected with exogenous AR (Weiser et al., 2014). Bicalutamide, a nonsteroidal antiandrogen was used as an antagonist control, and R1881, a potent anabolic steroid was used to activate AR. Only amino-ester analogs exhibited inhibitory action in the luciferase reporter gene assay; however, toxicity at higher doses was observed in all but one active compound. This study shows that these peptidomimetics are capable of inhibiting nuclear receptor-coactivator and can be tuned for selective inhibition.
Axerio-Cilies et al. virtually screened ~4 million compounds and discovered six potential candidates that were hypothesized to inhibit AR AF-2 (Axerio-Cilies et al., 2011). These compounds were tested using FP and cell-based eGFP AR transcription assays, and all showed IC50 values in the 4-36 μM range in both assays. Biolayer interferometry showed that 2,3-dihydro-1H-sperimidine 5C and 1H-pyrazol-5-(4H)-one 5D (Fig. 5) directly and reversibly interact with AF-2 in AR. An androgen displacement assay ensured that compounds did not bind to the hormone-binding site of AR.
Figure 5.
Small-molecule inhibitors of the androgen receptor - steroid receptor coactivator interaction.
Crystallography of 2,3-dihydro-1H-spermidine 5C showed a very similar binding arrangement to the one predicted by the Glide program. Structural changes occurred at AF-2 when compound 5C was bound: repositioning of Lys720 to engage in a cation-π interaction and Met734 pushed away from the site, likely caused by repulsion with compound 5C.
Caboni, et al. discovered and characterized a series of diarylhydrazides that blocked binding of coactivators to AR, which may be useful in developing non-ligandbinding pocket interventions for prostate cancer (Caboni et al., 2012). Compound 5E, identified using a virtual screen, was found to have antagonistic activity in a TR-FRET coactivator displacement assay. Fluorescence polarization assays showed no androgen displacement at the ligand-binding pocket. Further, TR-FRET assays suggested that the diarylhydrazides are AR antagonists and PR partial agonists, but that they do not displace coactivators from glucocorticoid receptor (GR), ERα, or ERβ. Compound 5C also showed low cytotoxicity, reduction in DHT-dependent cell proliferation and DHT- and CPA-stimulated PSA expression in prostate cancer cell lines. Caboni et al. further investigated the structure-activity relationship of the diarylhydrazide antiandrogens and discovered eight novel compounds which interact with AF-2 (Caboni et al., 2013).
Caboni et al. used molecular topology in the discovery of a novel coactivatorbinding inhibitor, 5F (Fig.5) (Caboni et al., 2014). A TR-FRET assay was conducted to confirm inhibition of coactivator binding, and a fluorescence polarization assay confirmed that the compound did not bind in the ligand binding pocket. The IC50 for this compound was 24 μM, similar to that of other known coactivator binding inhibitors. Molecular docking studies suggested that this compound binds to AF-2 with higher affinity than to BF-3.
Raj and coworkers designed a peptidomimetic compound mimicking the LXXLL motif common to the interaction of nuclear receptors and coactivators (Ravindranathan et al., 2013). Using the rigid structure of a bis-benzamide scaffold, Raj and coworkers designed molecule 5G (Fig.5). To determine whether 5G successfully blocked the interaction of androgen receptor with coregulators, co-immunoprecipitation studies were completed. LNCaP cells preincubated with 5G were shown to block the AR-coactivator interaction with an IC50 of 40 nM, even in the presence of dihydrotestosterone. The peptidomimetic was also selective for the androgen receptor as it had no effect on the interaction of estrogen receptor β and coactivator. An ARE-luciferase model was used to determine the effect on transcriptional activity, with the compound decreasing dihydrotestosterone-induced activity to basal levels. An MTT assay illustrated that 5G inhibits DHT-induced proliferation. 5G was also observed to inhibit growth of xenograft tumors in vivo and to inhibit AR expression of human tumors ex vivo.
Hsu et al. used phage display to develop small-molecule peptidomimetic inhibitors of androgen receptor. Phage display indicated enrichment of the FXXFY motif in mutated androgen receptor (bicalutamide-induced W741L point mutation) (Hsu et al., 2017). The use of peptidomimetic molecules has had some success for other nuclear receptors; therefore, Hsu et al. developed oxadiazole-containing peptidomimetics containing two hydrophobic rings to mimic the FXXFY motif discovered by phage display. AR transcriptional reporter assay was completed in PC-3 cells with most of the molecules having IC50 values of about 1 μM. 5H and 5I, the most effective inhibitors, were further tested in in vitro prostate cancer cell line growth assay with IC50 values of 110 μM. These two molecules were also observed to suppress prostate cancer cell xenografts in mouse models. The molecules were confirmed to target the AF-2 site of androgen receptor by determining the effect on AR-cofactor interactions within 0.1-1 μM.
Using an AR/TIF2 biosensor assay in a PC3 cell line, Fancher et al. identified nonsteroidal hit 5J that could inhibit the interaction between AR/TIF2 (Fancher et al., 2016). The IC50 values of the hit compounds were in the range of 0.9 to 2.4 μM. The authors showed that these hits did not compete with binding for the hormone site with dihydrotestosterone. Using LnCaP, C4-2, and 22-Rv1 (all AR-positive cell lines), the authors showed general growth inhibition of these cells lines with the non-steroidal hits.
To address the knowledge gap surrounding the specific requirements for AR N-terminal domain (NTD)/coactivator interactions, Dubbink et al. performed computer modelling, cell-based studies, and mutation-based analysis to investigate the interaction of the AR ligand binding domain (LBD) with the AR FXXLF motif and the transcriptional intermediary factor 2 (TIF2) LXXLL motif (Dubbink et al., 2004). The authors reported that AR-LBD did not interact with FXXLF and LXXLL motifs according to the classic charge clamp model of binding, with hydrogen bonding on one side of the charge clamp and significant hydrophobic interaction within the deep coactivator binding site being seemingly required for high peptide-binding affinity. They found that optimal peptide binding requires either two L residues for TIF2, or two F residues for TIF2 or AR at the +1 and +5 positions. The FXXLF motif was observed to bind preferentially to AR due to the presence of the phenylalanine residues, which interact with the deep hydrophobic coactivator groove, with which the LXXLL motif will also interact. Additionally, through comparison of binding experiments involving peptide interaction with AR and ERα LBDs, the authors determined that the shape of a NR coactivator binding groove can be critical to selective peptide interaction, with the FXXLF motif being specific for AR rather than ERα due to the AR’s deeper hydrophobic coactivator binding groove.
In order to study the specificity of androgen receptor ligand-binding domain (AR LBD) interactions with the known FXXLF motif, the authors used pairwise systematic analysis, swapping F and L residues in three FXXLF and nine LXXLL motifs. At positions +1 and +5, bulky F residues were found to be major contributors to strong interaction with AR LBD and to block binding to most other nuclear receptor (NR) LBDs (Dubbink et al., 2006). These residues dominated over flanking sequences in determining AR LBD specificity. For F and L substitutions at +1 and +5, yeast two-hybrid assays were carried out to directly compare peptide binding to different NR LBDs and to screen for AR LBD-interacting peptides. Four charged residues, K720, K717, R726, and E897, were found to modulate peptide binding independent of whether F or L was at the +1 or +5 positions. Structural studies of AR LBD demonstrated that FXXLF motifs hydrogen-bond with both K720 and E897, while LXXLL motifs only hydrogen-bond with K720. The authors carried out a mammalian one-hybrid assay using F23L/F27L-AR with alanine substitutions at K717, K720, R726, and E897. Binding capacity of AR-interacting peptides was reduced with E897A substitution, and although individual K720A, R726A, or K717A substitutions had no effect, double mutations strongly reduced AR LBD interaction and triple mutations completely abolished AR LBD peptide interaction. Using random mutagenesis at positions +2 and +3 of the FXXLF motif, the authors found that many different amino acid combinations gave strong interactions; all amino acid residues were detected, apart from M at the +2 position and F and Y at the +3 position. The authors carried out a yeast two-hybrid screen using a randomized AR FXXLF peptide expression library for the solvent-exposed positions 2 and 3 and assessed AR LBD interaction with a liquid β-galactosidase assay. A preference was shown for E at +2 and K or R at +3, although the reason for this preference was not clear. The authors presumed that charged residues at these positions increase stability of the peptide conformation.
van de Wijngaart surveyed the amino-acid requirements at the +4 (Leu) position of FXXLF motifs in either partner (AR vs. cofactors ARA54, ARA70) using a yeast two-hybrid read-out (van de Wijngaart et al., 2006. They found Leu → Phe or Met substitutions were the only ones that conserved interaction specificity and function. These substitutions served to enhance AR selectivity over progesterone receptor (PR) in ARA70. The subsequent search for FXXFF and FXXMF motif-bearing peptides turned up PAK6 (FXXMF) and gelsolin (FXXFF) as strong AR interactors. The interactions were confirmed by in vivo FRET assay using Hep3B cultures (normalized FRET efficiency > 0.1), site-directed mutagenesis (which abolished these PPIs) and a mammalian one-hybrid read out. Two additional FXXFF/FXXMF-bearing peptides (supervillin and cdc37) did, however, show weak AR LBD interaction. While this work did not develop an inhibitor directly, the approach potentially functions to uncover new foundations on which to develop peptide inhibitors.
To develop AR/coactivator binding inhibitors, Nakka et al. utilized two SRC-1 peptides, P100 and P200, that interact with the AR and not the PR (Nakka et al., 2013). In a mammalian two-hybrid assays, these peptides inhibited the SRC-1/AR interaction (a reduction in SRC-2/AR interaction was also noted). Even though the two peptides disrupted the transcriptional activity of AR (and its variant ARV7) and repressed all the AR-induced genes tested, they showed no similar activity for the VDR. The peptides were also tested for PSA inhibition using a double immunofluorescence technique and were found to reduce PSA expression in LNCaP cells. The peptides did not alter PC-3 cell proliferation. Unsurprisingly, the peptides did not affect the proliferation of PC-3 cells as these cells lack the AR).
To study the loss of specificity of androgen receptors (AR) in prostate cancer patients who develop Anti-androgen Withdrawal Syndrome (AWS), the authors compared the interactions of mutated AR and wild-type AR with certain peptides. Because anti-androgens/mutant AR complex has a similar binary structure as 5α-dihydrotestosterone (DHT)-bound wild-type-AR, the authors hypothesized that when bound to a ligand (i.e. DHT/wt-AR, hydroxyflutamide/T877A-AR, and bicalutamide (CDX)/W741L-AR), AR may recognize common AR-associated peptide motifs. The authors used bacterially expressed DHT/wt-AR DBD-LBD and hydroxyflutamide/T877A-AR DBD-LBD proteins as baits to screen potential peptides using phage display (Hsu et al., 2014). AR bound to similar peptides that contained an FXX(F/H/L/W/Y)Y motif cluster, with an extra hydrogen bond between the Tyr residue in the +5 position and AR greatly increasing binding affinity. BUD31 peptide motif was demonstrated to be the best inhibitor of LNCaP cell growth, suppressing both DHT-mediated AR and hydroxyflutamide-mediated T877A-AR transactivation. Cell work was completed with BUD31-transfected LNCaP cells in co-immunoprecipitation (Co-IP) and chromatin immunoprecipitation (ChIP) assays. Additionally, the PC-3 cell line was used in mammalian two-hybrid assays and reporter gene assays to confirm the screened peptides did in fact interact with ARs.
Li et al. explored the interaction of semenogelin I (SgI) as a coactivator of androgen receptor (Li et al., 2018). SgI contains multiple iterations of the LXXLL motif, so the authors attempted to elucidate their role in the interactions with AR. In the presence of zinc, SgI induced proliferation. A mutant SgI, with a modified LXXLL motif (to LXXAA), did not induce proliferation or migration as illustrated by the results of a MTT assay. Luciferase and co-immunoprecipitation assays confirmed that the mutated SgI is unable to interact with AR. The mutant SgI also failed to induce AR transactivation, confirming that the LXXLL motif is essential for interaction of SgI with AR. Finally, the effect of a SgI peptide with or without the LXXLL motif on prostate cancer cell lines LNCaP and C4-2 was observed using an MTT assay. Proliferation of both cell lines in the presence of zinc was inhibited by the wild-type peptide but not the peptide lacking the LXXLL motif; however, in the absence of zinc, neither peptide had an inhibitory effect on proliferation. Li et al speculate that the inhibition of the LXXLL motif interaction with AR could be a potential target for decreasing AR activity and tumor growth.
Progesterone Receptor
Progesterone receptor (PR) plays an important role in the development and differentiation of female reproductive tissue (Li and O’Malley, 2003). PR exists as two subtypes, PR-A and PR-B, which have different transactivation properties and the proportion and levels of each of these subtypes varies by tissue type (Kastner et al., 1990). Similarly to ER and AR, the domain architecture of PR comprises domains A-F. This review will focus on the C-terminal E/F domains, which contain the ligand binding domain and ligand-dependent activation function-2 (AF-2), also described as the coactivator binding groove (see above) (Hill et al., 2012). Progesterone receptor agonists have been used for menopausal hormone therapy, birth control and other gynecological disorders (Erkkola and Landgren, 2005). Antiprogestins, including Mifeprestone (Mifeprex®), are used clinically for termination of pregnancy. Selective nonsteroidal progesterone receptor modulators have been developed and have been shown to have mixed agonist-antagonist activity in in vitro transcription assays (Tabata et al., 2003). Coactivator binding inhibitors (CBIs) represent a potentially useful mechanism of PR antagonism. Similarly to ER and AR, the progesterone receptor interacts with steroid-receptor coactivators through the NR box motif LXXLL common to most steroid-receptor coactivator proteins (Rowan and O’Malley, 2000). The first reported CBIs for inhibiting the PR-SRC interaction were recently developed and are described below.
Colucci and Ortlund presented a high-resolution crystal structure of the ancestral 3-ketosteroid receptor liganded with progesterone that shows the weak agonist mifepristone bound at the coactivator binding groove (Colucci and Ortlund, 2013). This surprising observation may lead to the development of a possible scaffold for coactivator binding inhibitors. The authors originally set out to observe ligand exchange at significantly high excess of mifepristone in vitro for ancSR2. Because ligand exchange is possible with other ancestral SRs, this lack of ligand exchangeability was unique to ancSR2.
Kobayashi et al. aimed understand how agonist and/or antagonist-activated progesterone receptor interferes with NF-kB signaling in breast cancer cells (Kobayashi et al., 2010). Two types of responsive genes were identified that reflect at least two different mechanisms by which PR can interface with NF-kB. The authors hypothesized that a functional AF-2 distinguishes the mechanism of inhibition of the type I and type II responsive genes (Kobayashi et al., 2010). To determine the role of AF-2 in PR gene expression, a coactivator binding inhibitor was required. A screen of CBIs from a library of LXXLL-containing peptides identified peptide 6 (Fig. 6) which interacted well with agonist-activated hPR-A and hPR-B. LX23 also inhibited progesterone-dependent activation of transfected 2XPRE-tk-Luc reporter. To determine the effect AF-2 had in PR-mediated inhibition of inflammatory cytokines, Gal4-DBD-LX23 or control Gal4-DBD-LXAA was overexpressed in T47D-A18 cells using an adenovirus-based expression vector. The impact of these peptides on endogenous target genes was analyzed, and it was found that R5020-mediated repression of both CCL2 and CCL20 expression was reversed by peptide 7, but no effect was observed on IL-8 and CCL4 expression. This indicated that the two types of gene responses from the PR were AF-2 dependent (Type I) or AF-2 independent (Type II). This work is interesting and important due to the finding of two distinct mechanisms the PR employs to inhibit NF-kB target gene transcription.
Figure 6.
Peptide inhibitor of the progesterone receptor-steroid receptor coactivator
Conclusion
Since McDonnell and coworkers developed the first coactivator binding inhibitors, (Norris et al., 1999) there have been a number of advances, and in this review we have cataloged recent advances in small-molecule and peptide coactivator binding inhibitors that target steroid receptors. The most highly developed CBIs are those that target the ER and AR. Indeed, two of the most well-characterized CBIs, ERX-11 and 5G, are targeted to the ER and AR, respectively (Raj et al., 2017) (Ravindranathan et al., 2013). ERX-11 and 5G are some of the only CBIs that have robust activity in animal models.
A number of opportunities remain for the development of CBIs as both chemical probes and as viable therapeutic strategies. Several groups have defined the effects of CBIs on the expression of a small number of genes, but few groups have compared CBIs to appropriate selective nuclear receptor modulators (SNRMs) using genome- and transcriptome-wide approaches (Raj et al., 2017; Speltz et al., 2018a).
Similarly, the development of CBIs has been somewhat hampered by the lack of available crystal structures. Although crystal structures of AF-2-targeting peptides are rather common, there are few crystal structures of small molecules that bind at the AF-2 site (Wang et al., 2006), and there is a possibility that these structures are artifacts of the crystallization conditions.
Because coactivators are shared among other transcription factors, selectivity for one particular steroid receptor/coactivator pair has not been demonstrated. Selectivity for a particular nuclear receptor seems achievable, given slight differences in the coactivator binding grooves (Geistlinger and Guy, 2003), but it is unclear whether selectivity for one particular receptor/coactivator pair will ever be achievable or whether such activity is necessary or desirable. This point is important because a lack of selectivity may manifest itself as toxicity, and, as the field of coactivator binding inhibitors progresses, care must be taken to monitor for off-target toxicity.
Lastly, while this review has focused on molecules that directly block the protein-protein interaction between coactivators and receptors, there is a growing literature on molecules that block the enzymatic activity of coactivators (Song et al., 2016; Wang et al., 2014, 2011). Although these molecules require further optimization, they provide a useful starting point for creating new chemical probes of coactivator function and activity, and they may one day lead to exciting new therapeutics.
Overall, the future for coactivator binding inhibitors is bright. The history of CBIs, outlined here, coupled with opportunities mentioned in the previous paragraphs will continue to create a fertile ground of exploration for molecular endocrinologists, chemical biologists, and drug discovery scientists alike.
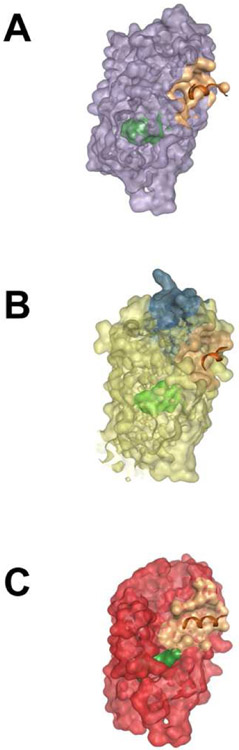
Figure 1.
A surface representation of a nuclear receptor. Shown are the ligand-binding domains of three steroid receptors, ligand binding pocket (green), binding function-3 (teal), activation function 2 (beige), and coactivator peptide (brown). (A) The estrogen receptor ligand binding domain (PDB: 5WGD; lilac). (B) The androgen receptor ligand binding domain (PDB: 5JJM; yellow). (C) The progesterone receptor ligand binding domain (PDB: 4OAR; red).
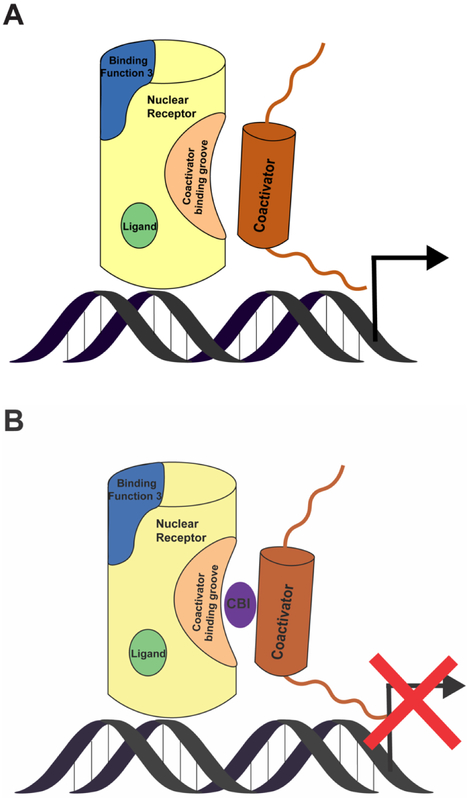
Figure 2.
A schematic representation of a nuclear receptor. Shown are nuclear receptor ligand binding domain (yellow), ligand binding pocket (green), binding function-3 (blue), activation function 2 (beige), coactivator peptide (brown), and coactivator binding inhibitor (CBI-purple). (A) The coactivator interacts with the nuclear receptor and leads to transcription. (B) The coactivator binding inhibitor prevents coactivator from interacting with nuclear receptor and leads to inhibition of transcription.
Highlights.
Small molecules and peptides block binding of coactivators to steroid receptors.
Coactivator binding inhibitors have been discovered through screening and design.
The most advanced coactivator binding inhibitors have been tested in animal models.
Further optimization and characterization are needed to enable clinical trials.
Acknowledgments/Funding:
This study was funded by the University of Illinois Cancer Center (to T.W.M.). T.E.S. was funded by training grant T32AT007533, Office of the Director, National Institutes of Health (OD) and National Center for Complementary and Integrative Health (NCCIH). We thank Dr. Sean Fanning (University of Chicago) for helpful discussions during the revision of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aarts JMMJG, Wang S, Houtman R, Van Beuningen RMGJ, Westerink WMA, Van De Waart BJ, Rietjens IMCM, Bovee TFH, 2013. Robust array-based coregulator binding assay predicting ERα-agonist potency and generating binding profiles reflecting ligand structure. Chem. Res. Toxicol 26, 336–346. 10.1021/tx300463b [DOI] [PubMed] [Google Scholar]
- Arnold LA, Kosinski A, Estébanez-Perpiñá E, Fletterick RJ, Guy RK, 2007. Inhibitors of the interaction of a thyroid hormone receptor and coactivators: Preliminary structure-activity relationships. J. Med. Chem 50, 5269–5280. 10.1021/jm070556y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axerio-Cilies P, Lack NA, Nayana MR, Chan KH, Yeung A, Leblanc E, Guns ES, Rennie PS, Cherkasov A, 2011. Inhibitors of androgen receptor activation function-2 (AF2) site identified through virtual screening. J Med Chem 54, 6197–6205. 10.1021/jm200532b [DOI] [PubMed] [Google Scholar]
- Ban F, Munuganti RSN, Cherkasov A, Li H, Leblanc E, Frewin K, Rennie PS, 2014. Discovery of 1 H -Indole-2-carboxamides as Novel Inhibitors of the Androgen Receptor Binding Function 3 (BF3). J. Med. Chem 57, 6867–6872. 10.1021/jm500684r [DOI] [PubMed] [Google Scholar]
- Blackwell HE, Grubbs RH, 1998. Highly {Efficient} {Synthesis} of {Covalently} {Cross}-{Linked} {Peptide} {Helices} by {Ring}-{Closing} {Metathesis}. Angew. Chemie Int. Ed. 37, 3281–3284. [DOI] [PubMed] [Google Scholar]
- Brinkmann AO, Klaasen P, Kuiper GGJM, van der Korput JAGM, Bolt J, de Boer W, Smit A, Faber PW, van Rooij HCJ, Geurts van Kessel A, Voorhorst MM, Mulder E, Trapman J, 1989. Structure and function of the androgen receptor. Urol. Res 17, 87–93. 10.1007/BF00262026 [DOI] [PubMed] [Google Scholar]
- Caboni L, Egan B, Kelly B, Blanco F, Fayne D, Meegan MJ, Lloyd DG, 2013. Structure-activity relationships in non-ligand binding pocket (Non-LBP) diarylhydrazide antiandrogens. J. Chem. Inf. Model 53, 2116–2130. 10.1021/ci400189m [DOI] [PubMed] [Google Scholar]
- Caboni L, Gálvez-Llompart M, Gálvez J, Blanco F, Rubio-Martinez J, Fayne D, Lloyd DG, 2014. Molecular topology applied to the discovery of 1-benzyl-2-(3-fluorophenyl)-4-hydroxy-3-(3-phenylpropanoyl)-2 h -pyrrole-5-one as a non-ligand-binding-pocket antiandrogen. J. Chem. Inf. Model. 54, 2953–2966. 10.1021/ci500324f [DOI] [PubMed] [Google Scholar]
- Caboni L, Kinsella GK, Blanco F, Fayne D, Jagoe WN, Carr M, Williams DC, Meegan MJ, Lloyd DG, 2012. “True” antiandrogens-selective non-ligand-binding pocket disruptors of androgen receptor-coactivator interactions: novel tools for prostate cancer. J Med Chem 55, 1635–1644. 10.1021/jm201438f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caboni L, Lloyd DG, 2013. Beyond the Ligand-Binding Pocket: Targeting Alternate Sites in Nuclear Receptors Laura. Med. Res. Rev 33, 1081–1118. 10.1002/med [DOI] [PubMed] [Google Scholar]
- Chang C, Norris JD, Grøn H, Paige LA, Hamilton PT, Kenan DJ, Fowlkes D, McDonnell DP, 1999. Dissection of the LXXLL Nuclear Receptor-Coactivator Interaction Motif Using Combinatorial Peptide Libraries: Discovery of Peptide Antagonists of Estrogen Receptors α and β. Mol. Cell. Biol. 19, 8226–8239. https://doi.Org/10.1128/mcb.19.12.8226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CY, McDonnell DP, 2005. Androgen receptor-cofactor interactions as targets for new drug discovery. Trends Pharmacol. Sci 26, 225–228. 10.1016/j.tips.2005.03.002 [DOI] [PubMed] [Google Scholar]
- Chen F, Zhou H, Liu J, Chen L, Su Y, Zhang X, 2014. Regulation of the nongenomic actions of retinoid X receptor-α by targeting the coregulator-binding sites. Acta Pharmacol. Sin 36, 102–112. 10.1038/aps.2014.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colucci JK, Ortlund EA, 2013. X-ray crystal structure of the ancestral 3-ketosteroid receptor-progesterone-mifepristone complex shows mifepristone bound at the coactivator binding interface. PLoS One 8, e80761 10.1371/journal.pone.0080761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coss CC, Jones A, Dalton JT, 2014. Selective androgen receptor modulators as improved androgen therapy for advanced breast cancer. Steroids. 10.1016/j.steroids.2014.06.010 [DOI] [PubMed] [Google Scholar]
- Crawford ED, Schellhammer PF, McLeod DG, Moul JW, Higano CS, Shore N, Denis L, Iversen P, Eisenberger MA, Labrie F, 2018. Androgen Receptor Targeted Treatments of Prostate Cancer: 35 Years of Progress with Antiandrogens. J. Urol 10.1016/j.juro.2018.04.083 [DOI] [PubMed] [Google Scholar]
- Crawford J, Prado CMM, Johnston MA, Gralla RJ, Taylor RP, Hancock ML, Dalton JT, 2016. Study Design and Rationale for the Phase 3 Clinical Development Program of Enobosarm, a Selective Androgen Receptor Modulator, for the Prevention and Treatment of Muscle Wasting in Cancer Patients (POWER Trials). Curr. Oncol. Rep 10.1007/s11912-016-0522-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton JT, Barnette KG, Bohl CE, Hancock ML, Rodriguez D, Dodson ST, Morton RA, Steiner MS, 2011. The selective androgen receptor modulator GTx-024 (enobosarm) improves lean body mass and physical function in healthy elderly men and postmenopausal women: Results of a double-blind, placebo-controlled phase II trial. J. Cachexia. Sarcopenia Muscle. 10.1007/s13539-011-0034-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey RAMG, 2016. Androgen Receptor Structure, Function and Biology: From Bench to Bedside. Clin Biochem Rev 37, 3–15. 10.1038/sc.1992.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mol E, Fenwick RB, Phang CTW, Buzón V, Szulc E, De La Fuente A, Escobedo A, García J, Bertoncini CW, Estébanez-Perpiñá E, McEwan IJ, Riera A, Salvatella X, 2016. EPI-001, A Compound Active against Castration-Resistant Prostate Cancer, Targets Transactivation Unit 5 of the Androgen Receptor. ACS Chem. Biol 11, 2499–2505. 10.1021/acschembio.6b00182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demizu Y, Misawa T, Nagakubo T, Kanda Y, Okuhira K, Sekino Y, Naito M, Kurihara M, 2015. Structural development of stabilized helical peptides as inhibitors of estrogen receptor (ER)-mediated transcription. Bioorg Med Chem 23, 4132–4138. 10.1016/j.bmc.2015.06.067 [DOI] [PubMed] [Google Scholar]
- Dubbink HJ, Hersmus R, Pike AC, Molier M, Brinkmann AO, Jenster G, Trapman J, 2006. Androgen receptor ligand-binding domain interaction and nuclear receptor specificity of FXXLF and LXXLL motifs as determined by L/F swapping. Mol Endocrinol 20, 1742–1755. 10.1210/me.2005-0348 [DOI] [PubMed] [Google Scholar]
- Dubbink HJ, Hersmus R, Verma CS, van der Korput HA, Berrevoets CA, van Tol J, Ziel-van der Made AC, Brinkmann AO, Pike AC, Trapman J, 2004. Distinct recognition modes of FXXLF and LXXLL motifs by the androgen receptor. Mol Endocrinol 18, 2132–2150. 10.1210/me.2003-0375 [DOI] [PubMed] [Google Scholar]
- Erkkola R, Landgren BM, 2005. Role of progestins in contraception. Acta Obstet. Gynecol. Scand 84, 207–216. 10.1111/j.0001-6349.2005.00759.x [DOI] [PubMed] [Google Scholar]
- Estebanez-Perpina E, Arnold LA, Nguyen P, Rodrigues ED, Mar E, Bateman R, Pallai P, Shokat KM, Baxter JD, Guy RK, Webb P, Fletterick RJ, 2007. A surface on the androgen receptor that allosterically regulates coactivator binding. Proc Natl Acad Sci U S A 104, 16074–16079. 10.1073/pnas.0708036104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancher AT, Hua Y, Camarco DP, Close DA, Strock CJ, Johnston PA, 2016. Reconfiguring the AR-TIF2 Protein-Protein Interaction HCS Assay in Prostate Cancer Cells and Characterizing the Hits from a LOPAC Screen. Assay Drug Dev Technol 14, 453–477. 10.1089/adt.2016.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning SW, Mayne CG, Dharmarajan V, Carlson KE, Martin TA, Novick SJ, Toy W, Green B, Panchamukhi S, Katzenellenbigen BS, Tajkhorshid E, Griffin PR, Shen Y, Chandarlapaty S, Katzenellenbogen JA, Greene GL, 2016. Estrogen receptor alpha somatic mutations Y537S and D538G confer breast cancer endocrine resistance by stabilizing the activating function-2 binding conformation. Elife 5, 1–25. 10.7554/elife.12792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S, Nguyen HD, Phan TTP, Burton MF, Nieto L, De Vries-Van Leeuwen IJ, Schmidt A, Goodarzifard M, Agten SM, Rose R, Ottmann C, Milroy LG, Brunsveld L, 2013. Proline primed helix length as a modulator of the nuclear receptor-coactivator interaction. J. Am. Chem. Soc 135, 4364–4371. https://doi.Org/10.1021/ja311748r [DOI] [PubMed] [Google Scholar]
- Galande AK, Bramlett KS, Trent JO, Burris TP, Wittliff JL, Spatola AF, 2005. Potent Inhibitors of LXXLL-Based Protein-Protein Interactions. ChemBioChem 6, 1991–1998. 10.1002/cbic.200500083 [DOI] [PubMed] [Google Scholar]
- Gao W, Reiser PJ, Coss CC, Phelps MA, Kearbey JD, Miller DD, Dalton JT, 2005. Selective androgen receptor modulator treatment improves muscle strength and body composition and prevents bone loss in orchidectomized rats. Endocrinology 10.1210/en.2005-0572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geistlinger TR, Guy RK, 2003. Novel selective inhibitors of the interaction of individual nuclear hormone receptors with a mutually shared steroid receptor coactivator 2. J. Am. Chem. Soc 125, 6852–6853. 10.1021/ja0348391 [DOI] [PubMed] [Google Scholar]
- Gunther JR, Parent AA, Katzenellenbogen JA, 2009. Alternative inhibition of androgen receptor signaling: Peptidomimetic pyrimidines as direct androgen receptor/coactivator disruptors. ACS Chem. Biol 4, 435–440. 10.1021/cb900043e [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Gampe RT, Kole AJ, Hnat AT, Stanley TB, An G, Stewart EL, Kalman RI, Minges JT, Wilson EM, 2004. Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance. Mol. Cell 16, 425–438. 10.1016/j.molcel.2004.09.036 [DOI] [PubMed] [Google Scholar]
- He B, Kemppainen JA, Wilson EM, 2000. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J. Biol. Chem 275, 22986–22994. 10.1074/jbc.M002807200 [DOI] [PubMed] [Google Scholar]
- He B, Minges JT, Lee LW, Wilson EM, 2002a. The FXXLF motif mediates androgen receptor-specific interactions with coregulators. J. Biol. Chem 277, 10226–10235. 10.1074/jbc.M111975200 [DOI] [PubMed] [Google Scholar]
- He B, Minges JT, Lee LW, Wilson EM, 2002b. The FXXLF motif mediates androgen receptor-specific interactions with coregulators. J. Biol. Chem 277, 10226–10235. 10.1074/jbc.M111975200 [DOI] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG, 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 10.1038/42750 [DOI] [PubMed] [Google Scholar]
- Henninot A, Collins JC, Nuss JM, 2018. The Current State of Peptide Drug Discovery: Back to the Future? J. Med. Chem 61, 1382–1414. 10.1021/acs.jmedchem.7b00318 [DOI] [PubMed] [Google Scholar]
- Hill KK, Roemer SC, Churchill MEA, Edwards DP, 2012. Structural and functional analysis of domains of the progesterone receptor. Mol. Cell. Endocrinol 348, 418–429. 10.1016/j.mce.2011.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz KB, Jackson TA, Bain DL, Richer JK, Takimoto GS, Tung L, 1996. Nuclear receptor coactivators and corepressors. Mol. Endocrinol 10, 1167–1177. 10.1621/nrs.01001 [DOI] [PubMed] [Google Scholar]
- Howell SJ, Johnston SRD, Howell A, 2004. The use of selective estrogen receptor modulators and selective estrogen receptor down-regulators in breast cancer. Best Pract. Res. Clin. Endocrinol. Metab 18, 47–66. 10.1016/j.beem.2003.08.002 [DOI] [PubMed] [Google Scholar]
- Hsu CL, Chen YL, Yeh S, Ting HJ, Hu YC, Lin H, Wang X, Chang C, 2003. The use of phage display technique for the isolation of androgen receptor interacting peptides with (F/W)XXL(F/W) and FXXLY new signature motifs. J. Biol. Chem 278, 23691–23698. 10.1074/jbc.M211908200 [DOI] [PubMed] [Google Scholar]
- Hsu CL, Liu JS, Lin TW, Chang YH, Kuo YC, Lin AC, Ting HJ, Pang ST, Lee LY, Ma WL, Lin CC, Wu WG, 2017. Characterization of a novel androgen receptor (AR) coregulator RIPK1 and related chemicals that suppress AR-mediated prostate cancer growth via peptide and chemical screening. Oncotarget 8, 69508–69519. 10.18632/oncotarget.17843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CL, Liu JS, Wu PL, Guan HH, Chen YL, Lin AC, Ting HJ, Pang ST, Yeh SD, Ma WL, Chen CJ, Wu WG, Chang C, 2014. Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis. Mol Oncol 8, 1575–1587. 10.1016/j.molonc.2014.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Su Y, Zhang X, Chen F, Liu J, Huang M, 2014. Identification of a New RXRα Antagonist Targeting the Coregulator-Binding Site. ACS Med. Chem. Lett 5, 736–741. 10.1021/ml5000405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur E, Pfaff SJ, Sturgis Payne E, Grøn H, Buehrer BM, Fletterick RJ, 2004. Recognition and accommodation at the androgen receptor coactivator binding interface. PLoS Biol. 10.1371/journal.pbio.0020274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JY, Arnold LA, Zhu F, Kosinski A, Mangano TJ, Setola V, Roth BL, Guy RK, 2009. Improvement of pharmacological properties of irreversible thyroid receptor coactivator binding inhibitors. J Med Chem 52, 3892–3901. 10.1021/jm9002704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyakumar M, Carlson KE, Gunther JR, Katzenellenbogen JA, 2011. Exploration of dimensions of estrogen potency: Parsing ligand binding and coactivator binding affinities. J. Biol. Chem 286, 12971–12982. 10.1074/jbc.M110.205112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner P, Krust A, Turcotte B, Stropp U, Tora L, Gronemeyer H, Chambon P, 1990. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 9, 1603–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzenellenbogen BS, Choi I, Delage-Mourroux R, Ediger TR, Martini PGV, Montano M, Sun J, Weis K, Katzenellenbogen JA, 2000. Molecular mechanisms of estrogen action: selective ligands and receptor pharmacology. J. Steroid Biochem. Mol. Biol 74, 279–285. [DOI] [PubMed] [Google Scholar]
- Kearbey JD, Gao W, Narayanan R, Fisher SJ, Wu D, Miller DD, Dalton JT, 2007. Selective androgen receptor modulator (SARM) treatment prevents bone loss and reduces body fat in ovariectomized rats. Pharm. Res 10.1007/s11095-006-9152-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholodovych V, Gera L, Welsh WJ, Bachmann K, Ekins S, Sinz M, Gal J, Ai N, Mani S, 2008. Computational Discovery of Novel Low Micromolar Human Pregnane X Receptor Antagonists. Mol. Pharmacol 74, 662–672. 10.1124/mol.108.049437 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Stice JP, Kazmin D, Wittmann BM, Kimbrel EA, Edwards DP, Chang CY, McDonnell DP, 2010. Mechanisms of progesterone receptor inhibition of inflammatory responses in cellular models of breast cancer. Mol Endocrinol 24, 2292–2302. 10.1210/me.2010-0289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Green S, Stack G, Berry M, Jin J., Chambon P, 1987. Functional domains of the human estrogen receptor. Cell 51, 941–951. 10.1016/0960-0760(92)90402-5 [DOI] [PubMed] [Google Scholar]
- Lack NA, Axerio-Cilies P, Tavassoli P, Han FQ, Chan KH, Feau C, LeBlanc E, Guns ET, Guy RK, Rennie PS, Cherkasov A, 2011. Targeting the binding function 3 (BF3) site of the human androgen receptor through virtual screening. J Med Chem 54, 8563–8573. 10.1021/jm201098n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc AM, Trent JO, Wittliff JL, Bramlett KS, Briggs SL, Chirgadze NY, Wang Y, Burris TP, Spatola AF, 2003. Helix-stabilized cyclic peptides as selective inhibitors of steroid receptor-coactivator interactions. Proc Natl Acad Sci U S A 100, 11273–11278. 10.1073/pnas.1934759100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Chen J, Kashiwagi E, Mizushima T, Han B, Inoue S, Ide H, Izumi K, Miyamoto H, 2018. The interaction between androgen receptor and semenogelin I: A synthetic LxxLL peptide antagonist inhibits the growth of prostate cancer cells. Br. J. Cancer 118, 416–420. 10.1038/bjc.2017.404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, O’Malley BW, 2003. Unfolding the Action of Progesterone Receptors. J. Biol. Chem 278, 39261–39264. 10.1074/jbc.r300024200 [DOI] [PubMed] [Google Scholar]
- Lipson D, Dvir A, Meric-Bernstam F, Miller VA, Hawryluk M, Ferrer-Lozano J, Otto G, Brown M, Ross JS, Sun J, Jarosz M, Wolf I, Balko JM, Frampton G, Perez-Fidalgo JA, Pusztai L, Arteaga CL, Gomez H, Giltnane J, Cristofanilli M, Schnitt S, Stephens P, Rubinek T, Yelensky R, Buchwalter G, Come SE, Gilmore L, Soussan-Gutman L, Gonzalez-Angulo AM, Jeselsohn R, Cronin MT, 2014. Emergence of Constitutively Active Estrogen Receptor- Mutations in Pretreated Advanced Estrogen Receptor-Positive Breast Cancer. Clin. Cancer Res 1757–1768. 10.1158/1078-0432.ccr-13-2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer FA, Leijten-van de Gevel IA, de Vries RMJM, Brunsveld L, 2019. Allosteric small molecule modulators of nuclear receptors. Mol. Cell. Endocrinol 485, 20–34. 10.1016/j.mce.2019.01.022 [DOI] [PubMed] [Google Scholar]
- Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, Dvir A, Soussan-Gutman L, Jeselsohn R, Yelensky R, Brown M, Miller VA, Sarid D, Rizel S, Klein B, Rubinek T, Wolf I, 2013. D538G mutation in estrogen receptor-α: A novel mechanism for acquired endocrine resistance in breast cancer. Cancer Res. 73, 6856–6864. 10.1158/0008-5472.CAN-13-1197 [DOI] [PubMed] [Google Scholar]
- Mettu NB, Stanley TB, Dwyer MA, Jansen MS, Allen JE, Hall JM, McDonnell DP, 2007. The nuclear receptor-coactivator interaction surface as a target for peptide antagonists of the peroxisome proliferator-activated receptors. Mol Endocrinol 21, 2361–2377. 10.1210/me.2007-0201 [DOI] [PubMed] [Google Scholar]
- Mita Y, Dodo K, Noguchi-Yachide T, Miyachi H, Makishima M, Hashimoto Y, Ishikawa M, 2010. LXXLL peptide mimetics as inhibitors of the interaction of vitamin D receptor with coactivators. Bioorg Med Chem Lett 20, 1712–1717. 10.1016/j.bmcl.2010.01.079 [DOI] [PubMed] [Google Scholar]
- Mohler ML, Nair VA, Hwang DJ, Rakov IM, Patil R, Miller DD, 2005. Nonsteroidal tissue selective androgen receptor modulators: a promising class of clinical candidates. Expert Opin. Ther. Pat 10.1517/13543776.15.11.1565 [DOI] [Google Scholar]
- Moore TW, Mayne CG, Katzenellenbogen JA, 2010. Minireview: Not picking pockets: nuclear receptor alternate-site modulators (NRAMs). Mol Endocrinol 24, 683–695. 10.1210/me.2009-0362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrove EA, Sutherland RL, 2009. Biological determinants of endocrine resistance in breast cancer. Nat. Rev. Cancer 9, 631–643. 10.1038/nrc2713 [DOI] [PubMed] [Google Scholar]
- Mutchie TR, Yu OB, Di Milo ES, Arnold LA, 2019. Alternative binding sites at the vitamin D receptor and their ligands. Mol. Cell. Endocrinol 485, 1–8. 10.1016/j.mce.2019.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa M, Paller C, Kyprianou N, 2017. Mechanisms of Therapeutic Resistance in Prostate Cancer. Curr. Oncol. Rep 19 10.1007/s11912-017-0568-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakka M, Agoulnik IU, Weigel NL, 2013. Targeted disruption of the p160 coactivator interface of androgen receptor (AR) selectively inhibits AR activity in both androgen-dependent and castration-resistant AR-expressing prostate cancer cells. Int J Biochem Cell Biol 45, 763–772. 10.1016/j.biocel.2012.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandhikonda P, Lynt WZ, McCallum MM, Ara T, Baranowski AM, Yuan NY, Pearson D, Bikle DD, Guy RK, Arnold LA, 2012. Discovery of the first irreversible small molecule inhibitors of the interaction between the vitamin D receptor and coactivators. J Med Chem 55, 4640–4651. 10.1021/jm300460c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan R, Coss CC, Dalton JT, 2018. Development of Selective Androgen Receptor Modulators (SARMs). Mol. Cell. Endocrinol 465, 134–142. 10.1016/j.trsl.2014.0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris JD, Paige LA, Christensen DJ, Chang CY, Huacani MR, Fan D, Hamilton PT, Fowlkes DM, McDonnell DP, 1999. Peptide antagonists of the human estrogen receptor. Science (80-. ). 285, 744–746. 10.1126/science.285.5428.744 [DOI] [PubMed] [Google Scholar]
- Nuclear Receptors Nomenclature Committee, Auwerx J, Baulieu E, Beato M, Backer-Andre M, Burbach H, Camerino G, Chambon P, Cooney A, Dejean A, Dreyer C, Evans RM, Gannon V, Giguere H, Gronemeyer H, Gustafson JA, Laude V, Lazar MA, Mangelsdorf DJ, Milbrandt J, Milgrom E, Moore DD, O’Malley B, Parker M, Parker K, Perlmann T, Pfahl M, Rosenfeld MG, Samuels H, Schutz G, Sladek FM, Stunnenberg HG, Spedding M, Thummel C, Tasi MJ, Umesono K, Vennstrom B, Wahli W, Weinberger C, Willson TM, Yamamoto K, 1999. A Unified Nomenclature System for the Nuclear Receptor Superfamily. Cell 97, 161–163. 10.1016/S0092-8674(00)80726-6 [DOI] [PubMed] [Google Scholar]
- Osborne CK, Schiff R, 2010. Mechanisms of Endocrine Resistance in Breast Cancer. Annu. Rev. Med 62, 233–247. 10.1146/annurev-med-070909-182917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C, Roberts LR, Schade M, Bazin R, Bent A, Davies NL, Moore R, Pannifer AD, Pickford AR, Prior SH, Read CM, Scott A, Brown DG, Xu B, Irving SL, 2011. Design and structure of stapled peptides binding to estrogen receptors. J Am Chem Soc 133, 9696–9699. 10.1021/ja202946k [DOI] [PubMed] [Google Scholar]
- Pinkerton JV, Thomas S, 2014. Use of SERMs for treatment in postmenopausal women. J. Steroid Biochem. Mol. Biol 142, 142–154. 10.1016/j.jsbmb.2013.12.011 [DOI] [PubMed] [Google Scholar]
- Raj GV, Sareddy GR, Ma S, Lee TK, Viswanadhapalli S, Li R, Liu X, Murakami S, Chen CC, Lee WR, Mann M, Krishnan SR, Manandhar B, Gonugunta VK, Strand D, Tekmal RR, Ahn JM, Vadlamudi RK, 2017. Estrogen receptor coregulator binding modulators (ERXs) effectively target estrogen receptor positive human breast cancers. Elife 6 10.7554/eLife.26857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindranathan P, Lee TK, Yang L, Centenera MM, Butler L, Tilley WD, Hsieh JT, Ahn JM, Raj GV, 2013. Peptidomimetic targeting of critical androgen receptor-coregulator interactions in prostate cancer. Nat. Commun 4, 1911–1923. 10.1038/ncomms2912 [DOI] [PubMed] [Google Scholar]
- Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, Kalyana-Sundaram S, Wang R, Ning Y, Hodges L, Gursky A, Siddiqui J, Tomlins SA, Roychowdhury S, Pienta KJ, Kim SY, Roberts JS, Rae JM, Van Poznak CH, Hayes DF, Chugh R, Kunju LP, Talpaz M, Schott AF, Chinnaiyan AM, 2013. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat. Genet 45, 1446–1451. 10.1038/ng.2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AL, Tamrazi A, Collins ML, Katzenellenbogen JA, 2004. Design, synthesis, and in vitro biological evaluation of small molecule inhibitors of estrogen receptor alpha coactivator binding. J Med Chem 47, 600–611. 10.1021/jm030404c [DOI] [PubMed] [Google Scholar]
- Rowan BG, O’Malley BW, 2000. Progesterone receptor coactivators. Steroids 65, 545–549. 10.1016/S0039-128X(00)00112-4 [DOI] [PubMed] [Google Scholar]
- Sadar MD, 2011. Small Molecule Inhibitors Targeting the "Achilles' Heel" of Androgen Receptor Activity. Cancer Res. 71, 1208–1213. 10.1158/0008-5472.CAN_10-3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, Karlsson A, Al-Lazikani B, Hersey A, Oprea TI, Overington JP, 2016. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov 16, 19–34. 10.1038/nrd.2016.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur RS, Burris TP, 2004. The coactivator LXXLL nuclear receptor recognition motif. J. Pept. Res 10.1111/j.1399-3011.2004.00126.x [DOI] [PubMed] [Google Scholar]
- Schafmeister CE, Po J, Verdine GL, 2000. An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides [8]. J. Am. Chem. Soc 122, 5891–5892. 10.1021/ja000563a [DOI] [Google Scholar]
- Schmidt N, Mishra A, Lai GH, Wong GCL, 2010. Arginine-rich cell-penetrating peptides. FEBS Lett. 584, 1806–1813. 10.1016/j.febslet.2009.11.046 [DOI] [PubMed] [Google Scholar]
- Schwartzberg LS, Yardley DA, Elias AD, Patel M, Lorusso P, Burris HA, Gucalp A, Peterson AC, Blaney ME, Steinberg JL, Gibbons JA, Traina TA, 2017. A phase I/Ib study of enzalutamide alone and in combination with endocrine therapies in women with advanced breast cancer. Clin. Cancer Res 10.1158/1078-0432.CCR-16-2339 [DOI] [PubMed] [Google Scholar]
- Shapiro DJ, Mao C, Cherian MT, 2011. Small molecule inhibitors as probes for estrogen and androgen receptor action. J. Biol. Chem 286, 4043–4048. 10.1074/jbc.R110.203026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Munuganti RS, Leblanc E, Lin YL, Leung E, Lallous N, Butler M, Cherkasov A, Rennie PS, 2015. In silico discovery and validation of potent small-molecule inhibitors targeting the activation function 2 site of human oestrogen receptor alpha. Breast Cancer Res 17, 27 10.1186/s13058-015-0529-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Munuganti RSN, Lallous N, Dalal K, Yoon JS, Sharma A, Yamazaki T, Cherkasov A, Rennie PS, 2018. Benzothiophenone Derivatives Targeting Mutant Forms of Estrogen Receptor-alpha in Hormone-Resistant Breast Cancers. Int J Mol Sci 19 10.3390/ijms19020579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Chen J, Zhao M, Zhang C, Yu Y, Lonard DM, Chow D-C, Palzkill T, Xu J, O’Malley BW, Wang J, 2016. Development of potent small-molecule inhibitors to drug the undruggable steroid receptor coactivator-3. Proc. Natl. Acad. Sci 113, 4970–4975. 10.1073/pnas.1604274113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speltz TE, Danes JM, Stender JD, Frasor J, Moore TW, 2018a. A Cell-Permeable Stapled Peptide Inhibitor of the Estrogen Receptor/Coactivator Interaction. ACS Chem Biol 13, 676–684. 10.1021/acschembio.7b01016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speltz TE, Fanning SW, Mayne CG, Fowler C, Tajkhorshid E, Greene GL, Moore TW, 2016. Stapled Peptides with gamma-Methylated Hydrocarbon Chains for the Estrogen Receptor/Coactivator Interaction. Angew Chem Int Ed Engl 55, 4252–4255. 10.1002/anie.201510557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speltz TE, Mayne CG, Fanning SW, Siddiqui Z, Tajkhorshid E, Greene GL, Moore TW, 2018b. A “cross-stitched” peptide with improved helicity and proteolytic stability. Org Biomol Chem 16, 3702–3706. 10.1039/c8ob00790j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger JL, 2019. Clinical applications of small molecule inhibitors of Pregnane X receptor. Mol. Cell. Endocrinol 485, 61–71. 10.1016/j.mce.2019.02.002 [DOI] [PubMed] [Google Scholar]
- Sun A, Moore TW, Gunther JR, Kim MS, Rhoden E, Du Y, Fu H, Snyder JP, Katzenellenbogen JA, 2011. Discovering small-molecule estrogen receptor alpha/coactivator binding inhibitors: high-throughput screening, ligand development, and models for enhanced potency. ChemMedChem 6, 654–666. 10.1002/cmdc.201000507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata Y, Iizuka Y, Shinei R, Kurihara KI, Okonogi T, Hoshiko S, Kurata Y, 2003. CP8668, a novel orally active nonsteroidal progesterone receptor modulator with tetrahydrobenzindolone skeleton. Eur. J. Pharmacol 461, 73–78. 10.1016/S0014-2999(02)02958-8 [DOI] [PubMed] [Google Scholar]
- Tian Y, Li J, Zhao H, Zeng X, Wang D, Liu Q, Niu X, Huang X, Xu N, Li Z, 2016. Stapling of unprotected helical peptides via photo-induced intramolecular thiol-yne hydrothiolation. Chem Sci 7, 3325–3330. 10.1039/c6sc00106h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice CM, Zheng YJ, 2016. Non-canonical modulators of nuclear receptors. Bioorganic Med. Chem. Lett 26, 4157–4164. 10.1016/j.bmcl.2016.07.067 [DOI] [PubMed] [Google Scholar]
- Tora L, White J, Brou C, Diane T, Webster N, Scheer E, Chambon P, 1989. The Human Estrogen Receptor Has Two Independent I–59. [DOI] [PubMed] [Google Scholar]
- Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, Li Z, Gala K, Fanning S, King TA, Hudis C, Chen D, Taran T, Hortobagyi G, Greene G, Berger M, Baselga J, Chandarlapaty S, 2013. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet 45, 1439–1445. 10.1038/ng.2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdine GL, Hilinski GJ, 2012. Stapled peptides for intracellular drug targets, 1st ed, Methods in Enzymology. Elsevier Inc; 10.1016/B978-0-12-396962-0.00001-X [DOI] [PubMed] [Google Scholar]
- Wang Y, Chirgadze NY, Briggs SL, Khan S, Jensen EV, Burris TP, 2006. A second binding site for hydroxytamoxifen within the coactivator-binding groove of estrogen receptor beta. Proc. Natl. Acad. Sci 103, 9908–9911. 10.1073/pnas.0510596103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lonard DM, Yu Y, Chow D-C, Palzkill TG, O’Malley BW, 2011. Small Molecule Inhibition of the Steroid Receptor Coactivators, SRC-3 and SRC-1. Mol. Endocrinol. 25, 2041–2053. 10.1210/me.2011-1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lonard DM, Yu Y, Chow DC, Palzkill TG, Wang J, Qi R, Matzuk AJ, Song X, Madoux F, Hodder P, Chase P, Griffin PR, Zhou S, Liao L, Xu J, O’malley BW, 2014. Bufalin is a potent small-molecule inhibitor of the steroid receptor coactivators SRC-3 and SRC-1. Cancer Res. 74, 1506–1517. 10.1158/0008-5472.CAN-13-2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PA, Arora VK, Sawyers CL, 2015. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer 15, 701–711. 10.1038/nrc4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser PT, Chang CY, McDonnell DP, Hanson RN, 2014. 4,4’-Unsymmetrically substituted 3,3’-biphenyl alpha helical proteomimetics as potential coactivator binding inhibitors. Bioorg Med Chem 22, 917–926. 10.1016/j.bmc.2013.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser PT, Williams AB, Chang CY, McDonnell DP, Hanson RN, 2012. 3,3’-Disubstituted bipolar biphenyls as inhibitors of nuclear receptor coactivator binding. Bioorganic Med. Chem. Lett 22, 6587–6590. 10.1016/j.bmcl.2012.09.007 [DOI] [PubMed] [Google Scholar]
- Welsh WJ, Iyer M, Bachmann K, Swaan PW, Ai N, Reschly EJ, Patel R, Chang C, Kholodovych V, Ekins S, Sinz M, Krasowski MD, Mani S, 2007. Human Pregnane X Receptor Antagonists and Agonists Define Molecular Requirements for Different Binding Sites. Mol. Pharmacol 72, 592–603. 10.1124/mol.107.038398 [DOI] [PubMed] [Google Scholar]
- Williams AB, Weiser PT, Hanson RN, Gunther JR, Katzenellenbogen JA, 2009. Synthesis of biphenyl proteomimetics as estrogen receptor-alpha coactivator binding inhibitors. Org Lett 11, 5370–5373. 10.1021/ol901999f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang NJ, Hinner MJ, 2015. Getting Across the Cell Membrane: An Overview for Small Molecules, Peptides, and Proteins. Methods Mol. Biol 1266, 29–53. https://doi.Org/10.1007/978-1-4939-2272-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Zhang Q, Li Z, 2016. Constructing thioether-tethered cyclic peptides via on-resin intra-molecular thiol-ene reaction. J Pept Sci 22, 540–544. 10.1002/psc.2902 [DOI] [PubMed] [Google Scholar]