Abstract
Microvascular proliferation (MVP), an aberrant vascular structure containing multilayered mitotically active endothelial- and smooth-muscle cells/pericytes, is a histopathological hallmark of glioblastoma multiforme (GBM). Although MVP tends to be associated with high-grade glioma, it has also been detected in WHO grade I pilocytic astrocytoma (PA). However, little is known about the mechanism underlying its formation. Using TP53 point mutations as a marker for tumor-derived cells, we earlier reported that MVP was partially converted from tumor cells via mesenchymal transition. In the current study we used the KIAA1549-BRAF fusion gene as a marker to assess whether MVPs in PA contained tumor-derived cells and/or phenotypically distinct tumor cells expressing vascular markers. cDNA synthesized from frozen tissue of six PA patients operated at our institute was analyzed to detect the KIAA1549-BRAF fusion gene by reverse transcription polymerase chain reaction (RT-PCR) assay. The breakpoint in the fusion gene was identified by long and accurate PCR (LA-PCR) and Sanger sequencing of genomic DNA. Distinct tumor cells and cellular components of MVP were obtained by laser microdissection. For the qualitative and quantitative detection of the KIAA1549-BRAF fusion gene we performed genomic and digital PCR assays. Fluorescence in situ hybridization (FISH) was used to assess gene fusion in cellular components of MVP. Samples from three PA patients harbored the KIAA1549 exon 15, BRAF exon 9 fusion gene. In two patient samples with abundant MVP, RT-PCR assay detected strong bands arising from the KIAA1549-BRAF fusion gene in both tumor cells and cellular components of MVP. Digital PCR showed that vis-à-vis tumor tissue, its relative expression in cellular components of MVP was 42% in one- and 76% in another sample. FISH revealed amplified signals in both tumor cells and cellular components of MVP indicative of tandem duplication. Our findings suggest that in patients with PA, some cellular components of MVP contained tumor derived cell and/or phenotypically distinct tumor cells expressing vascular markers.
Introduction
Gliomas, the most common type of brain tumors, are characterized by their infiltrative growth and high vascularity. Among them, glioblastoma multiforme (GBM) is the most aggressive and lethal. It is most highly neovascularized and contains vascular structures, i.e. areas of microvascular proliferation (MVP), that are complex structures composed of both proliferating endothelial and smooth-muscle cells [1–4]. MVP is abundant in areas adjacent to necrosis and at the edge of tumor invasion where rapidly proliferating tumor cells increase the demand for oxygen [5]. Consequently, vascular endothelial growth factor A (VEGF-A), induced by HIF1α and hypoxia, is thought to be the most important mediator for assembling these special vascular structures [6, 7]. In randomized phase III trials of bevacizumab, a monoclonal antibody against human VEGF-A, the overall survival time of patients with newly-diagnosed glioblastoma was not significantly increased [8, 9]. Recent work indicated that vascular endothelial cells derived from tumor-initiating cells play an important role in resistance to anti-VEGF therapy [10].
Glioma stem-like cells (GSCs) can transdifferentiate into vascular endothelial cells [10–12]. It was shown that endothelial cells in GBM surgical samples carried the same genomic alteration as tumor cells, and GSCs characterized as CD133-positive could develop into endothelial cells. We also showed that cellular components of MVP in GBM harbored the same p53 mutation as surrounding tumor cells as analyzed by immunostaining and Sanger sequencing with the laser microdissection method [13]. These findings suggested that tumor neovascularization in GBM was partially achieved via endothelial differentiation of GSCs. Additionally, it has been reported that tumor derived endothelial cells showed resistance to anti-VEGF therapies [10–12]. As aberrant vascular structures and chemoresistant phenotypes may be strongly involved in gliomas, treatments that target tumor-derived endothelial cells may overcome their resistance to anti-VEGF therapies.
Although MVP is generally a hallmark of GBM and is characteristic in high-grade glioma, it was observed in pilocytic astrocytoma (PA), a grade I astrocytoma. PA is a circumscribed, slow-growing tumor that includes abundant MVP comprised of so-called ‘glomeruloid’ vessels that form complex structures composed of both proliferating endothelial- and smooth-muscle cells [4, 14, 15]. Although some studies showed that the expression of VEGF was similar in PA and GBM, and was considered to elicit MVP in PA, very little is known [16]. And same as GBM, MVP in PA is also considered as the candidate therapeutic target. Anti-VEGF therapies have been used to treat PA patients [17, 18]. In some adults with recurrent PA unresponsive to conventional therapy, bevacizumab was efficacious, however, in pediatric glioma patients, bevacizumab and irinotecan showed no effect [17, 18]. Resistance to anti-VEGF therapies must be overcome in PA patients. To develop anti-angiogenic strategies for their treatment, the mechanisms underlying neovascularization in PA must be identified.
In the present study, we investigated the possibility that MVPs in PA, as in GBM, harbor tumor derived endothelial cells and/or phenotypically distinct tumor cells expressing vascular markers. The KIAA1549-BRAF fusion gene, resulting from duplication of the BRAF gene at 7q34, is used as a marker of tumor derived cells and phenotypically distinct tumor cells expressing vascular markers; it is the most frequent genetic alteration in PA (>70%) [19]. To evaluate whether cellular components of MVP in PA harbor the KIAA1549-BRAF fusion, tumor samples from patients treated at our institution were analyzed by Sanger sequencing for breakpoints. With primers designed around the breakpoint, the presence of the KIAA1549-BRAF fusion in cellular components of MVP, strictly collected by laser microdissection methodology, were qualitatively and quantitatively evaluated by genomic and digital PCR, respectively. FISH analysis was also employed to detect KIAA1549-BRAF fusion.
Materials and methods
Patients and tumor samples
The Institutional Review Board of Miyazaki University Hospital, Japan, approved our study (approval number 0–0175). All patients provided written informed consent. We enrolled six newly diagnosed PA patients who had undergone surgery at our institution between November 2010 and February 2015. The general patient characteristics, including the age, sex, and tumor location, were obtained from medical records and are shown in Table 1. Tissue specimens (n = 6) obtained at surgery either underwent histological examination or were frozen in liquid nitrogen and stored at -80°C until processing for genomic DNA and total RNA extraction.
Table 1. Patient characteristics.
| Patient | Age (years) | Sex | Tumor location |
|---|---|---|---|
| S1 | 3 | Female | Cerebellar vermis |
| S2 | 43 | Male | Temporal |
| S3 | 11 | Male | Occipital |
| S4 | 17 | Female | Cerebellar hemisphere |
| S5 | 14 | Male | Frontal |
| S6 | 5 | Male | Basal ganglia~Temporal |
cDNA synthesis and genomic DNA extraction
Total RNA was extracted from frozen tumor tissues using the Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. Reverse transcription was with SuperScript VILO cDNA Synthesis Master Mix (Invitrogen, Grand Island, NY, USA) according to the manufacturer’s instructions in a thermal cycler at 25°C for 10 min, followed by 42°C for 60 min, 85°C for 5 min, and cooling to 4°C.
Genomic DNA was isolated from frozen tissues using the QIAamp DNA Mini Kit (Qiagen). To obtain micro-dissected samples we used paraffin-embedded sections and the QIAamp DNA Micro Kit (Qiagen) according to the manufacturer’s instructions. cDNA and genomic DNA samples were stored at -20°C until analysis.
Collection of tumor and MVP constituent cells
Formalin-fixed, paraffin-embedded tissue was cut into 2-μm-thick sections, mounted on membrane slides (1.0 PEN NF, Carl Zeiss, Oberkochen, Germany), and stained with hematoxylin and eosin for the detection of MVP. We identified glomeruloid vessels as MVP. Tumor samples were micro-dissected with a laser under a PALM Micro Beam Laser Capture Microdissection microscope (Carl Zeiss); dissected samples were collected with Adhesive Cap 500 Clear (Carl Zeiss). Approximately 1,000–5,000 tumor cells and cellular components of MVP (30–40 MVPs) were collected for analysis.
Polymerase chain reaction (PCR)
Reverse transcription PCR (RT-PCR) assays were performed in a total reaction volume of 25 μl containing 12.5 μl Go Taq HS (Promega, Madison, WI, USA), 9.5 μl distilled water, 1 μl of 10 μM of each primer, and 1 μl of cDNA- or genomic DNA samples. PCR amplification was in a GeneAmp PCR System 9700 (Applied Biosystems, Waltham, MA, USA). The thermal cycling conditions were: initial denaturation (95°C, 2 min), 35 cycles of denaturation (95°C, 30 sec), annealing (58°C, 30 sec), extension (72°C, 20 sec), final extension (72°C, 5 min). The PCR products were separated by electrophoresis on 2% agarose gels; the target bands were cut from the gels.
Long and accurate PCR (LA-PCR) assays were performed in a total reaction volume of 50 μl containing 10 x LA-PCR Buffer II (5 μl, Takara Bio Inc., Shiga, Japan), a dNTP mixture (8 μl), distilled water (35.5 μl), 1 μl of 10 μM of each primer, and 1 μl of a genomic DNA sample. PCR amplification was in a GeneAmp PCR System 9700 (Applied Biosystems). The thermal cycling conditions were: initial denaturation (94°C, 1 min), 30 cycles of denaturation (98°C, 10 sec), annealing (68°C, 20 min), final extension (72°C, 5 min). The PCR products were separated by electrophoresis on 0.9% agarose gels; the target bands were cut from the gels.
Digital PCR assays were on a QuantStudio 3D Digital PCR System (Thermo Fisher Scientific, Waltham, MA, USA). To 6 μl of genomic DNA we added 1.525 μl nuclease-free water, 7.25 μl QuantStudio 3D Digital PCR Master Mix, and 0.725 μl TaqMan Copy Number Reference Assay for detecting RNase P. To detect the KIAA-BRAF fusion gene we combined 5 μl of genomic DNA with 7.25 μl QuantStudio 3D Digital PCR Master Mix, 1.35 μl of each primer, and 0.375 μl of a TaqMan probe. Sample mixes were loaded onto chips and placed in the ProFlex 2x Flat PCR System (Thermo Fisher Scientific). The program was: stage 1 (96°C, 10 min), 40 cycles of stage 2a (54°C, 2 min), stage 2b (98°C, 30 sec), stage 3 (60°C, 2 min). The end-point fluorescence of the partitions on the chips was analyzed with the QuantStudio 3D Analysis Suite Cloud Software (Thermo Fisher Scientific). Table 2 shows the primer- and the TaqMan probe sequences.
Table 2. Primer sequences.
| Primers for screening with cDNA and LA-PCR | |
| primer | Sequence |
| KIAA ex15 Fwd | ATAAAACGTTCTCCCAAGCCTCGCCGGAAACACCA |
| KIAA ex16 Fwd | AGCCGATGTGCAGACACCATCCTCGGTGGAACTGG |
| BRAF ex9 Rev | CCATCACCACGAAATCCTTGGTCTCTAATCAAGTC |
| Primers for detecting Breakpoint | |
| primer | Sequence |
| FP0 | GAAACACCAGGTCAACGGCT |
| FP1 | TAGGGGAGAGAGAACCAGCC |
| FP2 | GTCCCTTCACTTCTCCAGCC |
| FP3 | CAGCCAGCTGCTTGTTCTTG |
| FP4 | GTTATAGCCGCTGGGGACTC |
| FP5 | TCCTCACAGAGGGTATGGGG |
| FP6 | TGGCTGTTTAGGGGCTTTCA |
| RP0 | GACCAAGGATTTCGTGGTGA |
| RP1 | TGGATGGATAAGGGCACACT |
| RP2 | GAGGGATTGAATGGCCCCAA |
| RP3 | GGCAAACACTACTAGCCCCA |
| RP4 | GACTTAGGAAGAGCCAGCCC |
| RP5 | CACCATTCCCCAAGCCTTCT |
| RP6 | TGGACAGACTGAGAACAACCC |
| Primers for Sanger sequencing of PCR amplicons | |
| primer | Sequence |
| KIAA ex15 Fwd | GAAACACCAGGTCAACGGCT |
| BRAF ex9 Rev | GACCAAGGATTTCGTGGTGA |
| Primers for LCM samples | |
| primer | Sequence |
| S1 Fwd | CCTTATGCAACCAGCCATTT |
| S1 Rev | AACACAATGCATAGCCCACA |
| S6 Fwd | TGCCAAGAGAACCCAGAAAT |
| S6 Rev | CAAACCACTATGGAAGGCAAA |
| Primers and Taqman probes for digital PCR | |
| primer and Taqman | Sequence |
| S1B Fwd | GATCGCTTTAGATAGACCTTATGCAA |
| S1B Rev | ACAGTAAGTCACTAAAAGTTTTGGAACAA |
| S1B Taqman | ATTTCTTAGTTCATCTGTTCTC |
| S6B Fwd | TCTGCCAAGAGAACCCAGAAA |
| S6B Rev | CAAACACTACTAGCCCCAGGATTAAA |
| S6B Taqman | CTATCAATTAAAACCAGCTCTGT |
Sanger sequencing
To extract DNA fragments from gels for sequencing, the QIAquick Gel Extraction Kit (Qiagen) was used. Each product was sequenced on an ABI PRISM 310- or 3130xl Genetic Analyzer (Applied Biosystems). Sequencing reactions were performed using a Big Dye Terminator version 1.1 or version 3.1 Cycle Sequencing Kit (Applied Biosystems). The conditions were: initial denaturation (96°C, 1 min), 25 cycles of denaturation (30 sec), annealing (50°C, 5 sec), extension (60°C, 4 min). Sequence Scanner Software version 2 (Applied Biosystems) was used for sequence analysis.
Detection of fusion breakpoints in genomic DNA
To detect fusion breakpoints, LA-PCR products were analyzed by Sanger sequencing. KIAA intron 15 and BRAF intron 8 that included breakpoints were approximately 7 kb in length. Using our seven sequencing primers located in each intron at 1,000-bp intervals, the area including breakpoints was identified by state-of-base calling; breakpoints were detected by detailed analysis of the base sequences (S1 Fig).
Fluorescence in situ hybridization (FISH)
Dual-color interphase FISH was performed for KIAA-BRAF fusion gene detection. Probes that annealed the duplication site of BRAF was labeled with Texas red, and the centromere of chromosome 7q was labeled with fluorescein isothiocyanate. Both probes were purchased from GSP Laboratory Inc. (BRAF1/CEN7q Dual Color FISH Probe, product No. GC115, Kobe, Japan).
Sections were cut at a 2-μm thickness. After having been deparaffinized and boiled in pretreatment solution for 30 minutes, the slides were washed twice in 2 x saline-sodium citrate buffer (SSC) for 5 minutes. The slides were digested in protease solution at 37°C for 10–20 minutes, were rinsed twice with 2 x SSC, and were dehydrated with a 70%/100% ethanol series. The probes (10 μl) were applied, and the sections were covered with cover glasses and rubber cement. After having been denatured on a hot plate at 75°C for 5 minutes, the slides were incubated at 37°C for 26–50 hr in a humid chamber. After hybridization, the slides were washed with 2 x SSC/0.3% NP-40 for 5 minutes, 2 x SSC/0.3% NP-40 at 72°C for 1–2 minutes, and then 2 x SSC at room temperature for 5 minutes. The nuclei were counterstained with 4ʹ,6-diamidino-2-phenylindole, and signals were visualized by fluorescence microscopy.
Immunohistochemistry
Serial sections (2 μm) were mounted on MAS-coated slides (Matsunami Glass Ind.,Ltd, Osaka, Japan) and processed for heat-induced antigen retrieval by autoclave with citrate buffer (pH 6.0) for OLIG2 or EDTA buffer (pH 9.0) for glial fibrillary acidic protein, CD34, and α-SMA, followed by treatment with 3% H2O2 in PBS for 10 minutes and were washed thrice in PBS. After blocking in 10% normal goat serum in PBS for 1 hour, the sections were incubated in moist chambers overnight at 4°C with antibodies to glial fibrillary acidic protein (rabbit polyclonal, prediluted; Dako Cytomation, Glostrup, Denmark), OLIG2 (rabbit polyclonal, dilution 1:100; IBL, Fujioka, Japan), CD34 (mouse monoclonal, dilution 1:50; Dako Cytomation), and α-SMA (mouse monoclonal, dilution 1:100; Dako Cytomation). After washing with PBS, the corresponding secondary antibody reaction was conducted. 3,3′-diaminobenzidine (DAB) solution was used for visualization. The slides were counterstained with hematoxylin, were dehydrated, and were cover-slipped.
Statistics
The relative expression of the KIAA1549-BRAF fusion gene/RNase-P gene on digital PCR images was calculated using triplicate data from each sample (S1 and S6) analyzed with QuantStudio 3D Analysis Suite Cloud Software. Comparison between the samples (S1 and S6) was performed by Student’s t-test. Differences were considered significant at p<0.05. Statistical analyses were performed with Stat Mate III software (ATMS, Tokyo, Japan).
Results
Detection of the KIAA-BRAF fusion gene in patients with Pilocytic astrocytoma (PA)
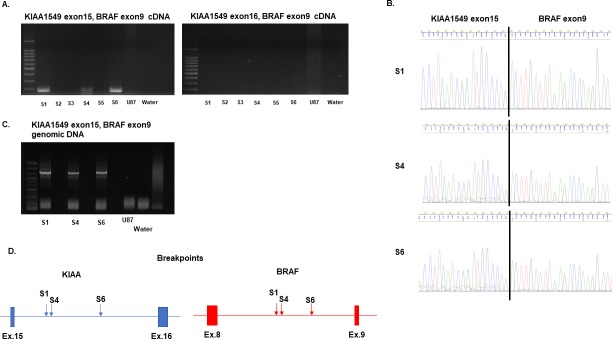
To identify the KIAA-BRAF fusion gene in six patients with PA, we performed RT-PCR with synthesized cDNA from frozen tumor tissues. Table 1 shows the clinical characteristics of the 6 patients. PCR primers were used to assess frequent fusion patterns such as KIAA1549 exon 15, BRAF exon 9 and KIAA1549 exon 16, BRAF exon 9. KIAA1549 exon 15, BRAF exon 9 fusions were found in three samples (Fig 1A). Sanger sequencing verified KIAA1549 exon15, BRAF exon 9 fusion junctions in these samples (Fig 1B). In our series, no sample exhibited the KIAA1549 exon 16, BRAF exon 9 fusion pattern (Fig 1A).
Fig 1. Screening for patient samples harboring the KIAA-BRAF fusion gene and breakpoint detection.
(A) RT-PCR was performed to detect samples harboring the KIAA-BRAF fusion gene. cDNA of U87 and water were the negative controls. Among the six patient samples, S1, S4, and S6 expressed the KIAA1549 exon15, BRAF exon 9 fusion gene (left). None of the six samples manifested the KIAA1549 exon16, BRAF exon 9 fusion gene (right). (B) Sanger sequencing of S1, S4, and S6 revealed the KIAA1549 exon15, BRAF exon 9 fusion junctions. (C) LA-PCR assay of genomic DNA performed to obtain long amplified products including breakpoints. The genome of U87 and water were the negative controls. S1, S4, and S6 contained amplified products around 7 kb in length. (D) Breakpoints in S1, S4, and S6 detected by Sanger sequencing. Although the fusion pattern was the same, the breakpoints were different. For details, see S8 Fig.
Determination of the breakpoints in the KIAA-BRAF fusion gene
To obtain the cellular components of MVP in PA, we used laser microdissection to collect cells from formalin-fixed, paraffin-embedded (FFPE) slides. However, as genomic DNA derived from FFPE slides is usually fragmented, we identified the accurate breakpoints of each KIAA1549-BRAF fusion gene.
In the three samples harboring the KIAA1549-BRAF fusion gene (S1, S4, S6), amplified products including breakpoints were obtained by long and accurate PCR (LA-PCR) with genomic DNA purified from frozen tissues (Fig 1C). Sanger sequencing using 7 sequencing primers at 1,000-bp intervals for each intron detected breakpoints in KIAA1549 intron 15 and BRAF intron 8 (Fig 1D and S2–S8 Figs). As in another report, each sample featured a distinct breakpoint despite exhibiting the same fusion pattern [20].
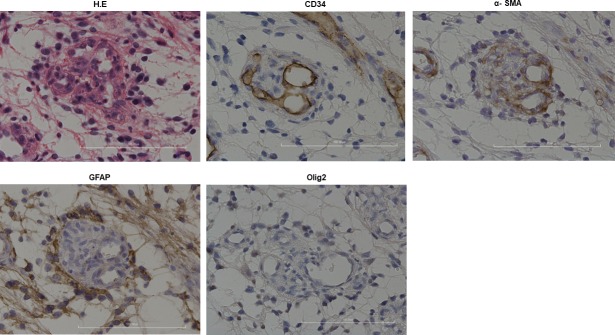
Histological characteristics of MVP in PA
To evaluate MVP in PA, we performed immunohistochemical studies. Neither of the tumor cell markers glial fibrillary acidic protein (GFAP) and Olig2 was detected. However, the vessel markers CD34 and α-smooth muscle actin (α-SMA) were observed (Fig 2). This means that none of MVP was directly invaded by surrounding original tumor cells.
Fig 2. Histopathological findings of MVP in PA.
H & E staining: MVP exhibiting a glomerular structure. CD34: Inner layer of MVP was stained against this marker. α-SMA: Cellular components of MVP were stained diffusely against this marker. GFAP and Olig2: No cellular components of MVP were stained against these markers. Original magnification: 20x. Scale bar: 100 μm.
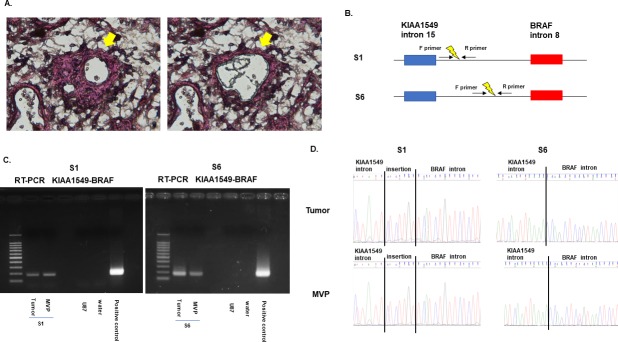
Detection of KIAA-BRAF fusion in MVP by PCR
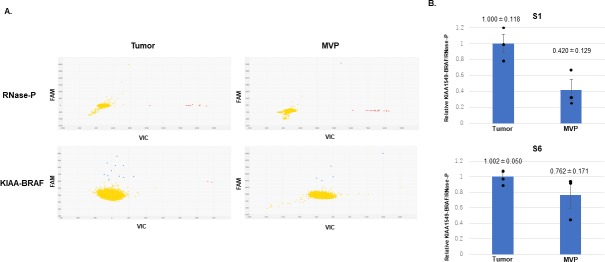
To determine whether cellular components of MVP and tumor cells in PA harbored the same KIAA1549-BRAF fusion gene, we evaluated cells collected from FFPE slides by laser microdissection (Fig 3A and S9 Fig). Two samples (S1 and S6) with abundant MVP were selected for analysis. In both samples, PCR yielded bands for the KIAA1549-BRAF fusion gene in tumor cells and cellular components of MVP (Fig 3B and 3C). PCR analysis was performed in triplicate to ascertain reproducibility (data not shown). Sanger sequencing confirmed the expected breakpoints in the amplified products (Fig 3D). These data suggest that some cellular components of MVP contained tumor derived cell and/or phenotypically distinct tumor cells expressing vascular markers. Additionally, to further analyze the ratio of cells harboring the KIAA1549-BRAF fusion gene in cellular components of MVP compared to tumor cells, digital PCR with customized primers and Taqman probes was carried out. The results showed that the ratio of cells harboring the KIAA1549-BRAF fusion was 42% in cellular components of MVP compared to tumor cells in S1 and 76% in S6 (Fig 4 and S10 Fig). There was no significant difference in the ratio of cells harboring the KIAA1549-BRAF fusion gene in cellular components of MVP compared to tumor cells between S1 and S6 (p = 0.096).
Fig 3. Evaluation of the KIAA-BRAF fusion gene in cellular components of MVP by genomic PCR.
(A) Cellular components of MVP were collected from FFPE slides with laser microdissection (arrows). (B) Special primers designed for around breakpoints. (C) PCR analysis with the special primers was performed for fusion gene detection. Samples S1 and S6 exhibited a positive band for tumor and for cellular components of MVP. (D) Sanger sequencing was performed to confirm the amplified products. In S1 and S6, tumor cells and cellular components of MVP exhibited the same base sequence, including breakpoints.
Fig 4. Digital PCR for quantitative fusion gene evaluation.
(A) Raw digital PCR data for S1. RNase-P was employed as an internal control. Red dots showed droplets with RNase-P DNA and blue dots with the KIAA-BRAF fusion gene. Yellow dots showed droplets with no DNA. (B) The relative expression of the KIAA-BRAF fusion gene in tumor cells and cellular components of MVP is shown. The ratio of cells harboring the KIAA-BRAF fusion gene (cellular components of MVP vs. tumor cells) was 42% in S1 and 76% in S6. The results are expressed as the mean +- S.E.M., n = 3.
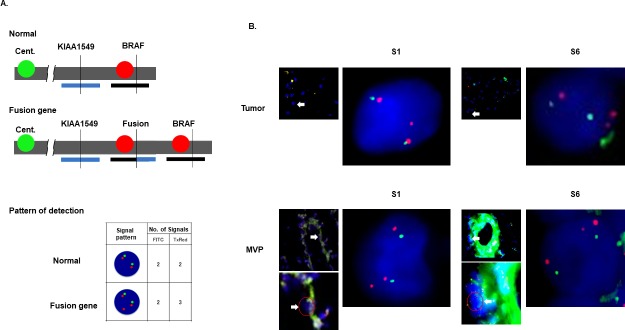
Detection of KIAA-BRAF fusion in MVP by FISH
FISH was used to visually evaluate the KIAA1549-BRAF fusion in MVP. In PA, tandem duplication at 7q34 results in such fusion. Therefore, the combination signal pattern of BRAF-specific probes placed on the duplicated site (red) and on the centromere of chromosome 7 (green) detected the KIAA1549-BRAF fusion gene (Fig 5A). Not only tumor cells but also some cellular components of MVP exhibited a positive signal pattern for the fusion gene (Fig 5B). These findings also demonstrated that MVP in PA contained tumor derived cell and/or phenotypically distinct tumor cells expressing vascular markers.
Fig 5. FISH analysis for the detection of the KIAA-BRAF fusion gene.
(A) FISH probes at the duplication site of the BRAF gene (7q34) are labeled with Texas red. The centromere of chromosome 7q (7q11.21) is labeled with fluorescein isothiocyanate (FITC, green). Due to tandem duplication at 7q34 leading to the expression of the KIAA-BRAF fusion gene, cells with the fusion gene demonstrated gain of a red signal (total: three signals) in comparison with the green signal (two signals). (B) In both S1 and S6, tumor cells and cellular components of MVP were positive for the KIAA-BRAF fusion gene.
Discussion
New insight of the mechanism of GBM neovascularization that GSCs could transdifferentiate into vascular endothelial cells was much impressive not only for one of the mechanisms of the resistance of anti- VEGF therapies but for the interaction of GSCs with vascular compartments took part in the microenvironment [10–12, 20]. By contrast, little is known about neovascularization, vascular niche, and the existence of GSCs in PA. In this study, we evaluated whether vascular endothelial cells of PA contained tumor derived cells and/or phenotypically distinct tumor cells expressing vascular markers. This is the first report considering the origin of MVP in PA, and our results might shed light on the microenvironment in low-grade glioma.
To analyze samples harvested by laser microdissection from FFPE slides, we first identified the accurate breakpoint in each sample. Although three fusion-positive samples exhibited the same fusion pattern (KIAA1549 exon 15, BRAF exon 9), the breakpoints were different. Lawson et al. also reported that the breakpoint of each samples was totally different in their analysis of 43 low-grade astrocytoma patients, and these fusions could be categorized into three groups: “seamless transition,” “presence of short inserted,” and “breakpoint microhomology.” This terminology is based on the sequence alignment of the fusion breakpoint [21]. In our fusion positive samples, S6 showed a continuous transition from KIAA1549 intron 15 to BRAF intron 8 and, thus, was categorized as “seamless transition” (S3 Fig), S1 showed the 7 bp inserted sequence at the breakpoint and categorized as “presence of short inserted” (S5 Fig), and S4 showed the shared 3 bp sequence between the end of KIAA1549 and the beginning of BRAF and categorized as “breakpoint microhomology” (S7 Fig). We think that these results document the accuracy of our method for detecting breakpoints.
Our result showed that the ratio of cells harboring the KIAA1549-BRAF fusion gene in cellular components of MVP to tumor cells was different from S1 (42%) to S6 (76%). Soda et al. also showed a difference among GBM samples in the rate of endothelial cells derived from tumor cells (2.0–37.8%) [10]. They suggested that the observed rate variation was attributable to the location of the analyzed endothelial cells in the tumor, or to the tumor size per se. At present, we cannot rule out sampling bias due to the heterogeneity of our tumor cells and of the cellular components of MVP. We are collecting more samples to identify factors responsible for the inter-sample difference in the rate of cellular components of MVP that harbor the KIAA1549-BRAF fusion gene in patients with PA.
In this study, we demonstrated that MVP in PA contained tumor derived cell and/or phenotypically distinct tumor cells expressing vascular markers by analyzing the presence of the KIAA-BRAF fusion gene via PCR and FISH methods. In GBM, Vitiani et al. demonstrated that GSCs cultured in endothelial condition medium could generate progeny with the phenotypic and functional features of endothelial cells, resulting in tube formation and CD31 expression. GSCs injected into mice produced tumor xenografts, the vessels of which were composed primarily of human endothelial cells [11]. However, in PA, it is difficult to perform such in vitro and in vivo experiments because there are currently no methods for selecting and culturing GSCs from these tumors. Additionally, we also have no commercially available cell lines of PA. Thus, it will be critical to establish such methods to handle GSCs in PA and perform such verification experiments in the future.
Conclusions
We provide evidence that MVP in PA might contain tumor derived cell and/or phenotypically distinct tumor cells expressing vascular markers. Our work sheds light on not only the resistance of PA to anti-VEGF therapies but also on the microenvironment in low-grade glioma.
Supporting information
Sequencing primers (7 each) to be located at KIAA and BRAF introns at 1,000-bp intervals were designed to detect breakpoints.
(TIF)
S6 amplicons were subjected to Sanger sequencing. Red circles indicate that the base sequence annealed with the sequence primer. State-of-base calling identified areas of interest in the intron that included the breakpoint. In S6 the breakpoint was located between primers FP4 and RP3.
(TIF)
The breakpoint in S6 was detected by Sanger sequencing using the RP3 primer. The fusion was categorized as a “seamless transition” from KIAA intron 15 to BRAF intron 8.
(TIF)
S1 amplicons were subjected to Sanger sequencing. The breakpoint was located between the FP1 and the RP5 primers.
(TIF)
The breakpoint in S1 was detected by Sanger sequencing using the FP1 primer. The fusion was categorized as “presence of short insert”. A 7-bp sequence was inserted at the breakpoint.
(TIF)
PCR analysis with FP1 and RP5 primers revealed bands in S1 and S4. As in S1 (S4 Fig.), the breakpoint in S4 was located between primers FP1 and RP5.
(TIF)
The RP5 primer was used. The breakpoint in S4 was detected by sequencing. This fusion was categorized as “breakpoint microhomology”. We observed a shared 3-bp sequence between the end of KIAA and the start of BRAF.
(TIF)
The location of breakpoints between KIAA intron 15 and BRAF intron 8 in S1, S4, and S6 is shown. Note the inter-sample difference in the breakpoint. S1: Magenta: KIAA intron 15, Purple: BRAF intron 8. S4: Aqua: KIAA intron 15, Green: BRAF intron 8. S6: Red: KIAA intron 15, Blue: BRAF intron 8.
(TIF)
Tumor samples were collected from tumor tissue without MVP. Tumor samples and the cellular components of MVP were collected from the same tissue.
(TIF)
Raw digital PCR data for S6 are shown. Samples from tumor cells and cellular MVP components were independently analyzed. Top: Red and yellow dots indicate droplets with RNase-P DNA and droplets with no DNA, respectively. Bottom: Green and red dots identify droplets with the KIAA-BRAF fusion gene and droplets with no DNA, respectively.
(TIF)
Raw data of Figs 1B, 3D, 4A, 4B, 5 and S3, S5, S7, S8 and S9 Figs are shown by power point or excel files. Original sequencing data and digital PCR data could be seen with adequate software (Sequence scanner version 2 and QuantStudio 3D Analysis Suite Cloud). These raw data are also available at Dryad digital repository (DOI: https://doi.org/10.5061/dryad.bv44rk5).
(ZIP)
Acknowledgments
We thank Ayumi Nagatomo, Itsuki Morinaga, and members of the Chromocenter of the Kobe Research Institute for their technical assistance and U. Petralia for editorial assistance.
Data Availability
All relevant data are within the paper, figures, its Supporting Information files, and the Dryad digital repository (Detection of the KIAA1549-BRAF fusion gene in cells forming microvascular proliferation of pilocytic astrocytoma, DOI: https://doi.org/10.5061/dryad.bv44rk5).
Funding Statement
This work was supported by JSPS KAKENHI Grant Number JP16K10763 (Hideo Takeshima).
References
- 1.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–20. Epub 2016/05/10. 10.1007/s00401-016-1545-1 . [DOI] [PubMed] [Google Scholar]
- 2.Haddad SF, Moore SA, Schelper RL, Goeken JA. Vascular smooth muscle hyperplasia underlies the formation of glomeruloid vascular structures of glioblastoma multiforme. Journal of neuropathology and experimental neurology. 1992;51(5):488–92. Epub 1992/09/01. 10.1097/00005072-199209000-00002 . [DOI] [PubMed] [Google Scholar]
- 3.Nagashima T, Hoshino T, Cho KG. Proliferative potential of vascular components in human glioblastoma multiforme. Acta Neuropathol. 1987;73(3):301–5. Epub 1987/01/01. . [DOI] [PubMed] [Google Scholar]
- 4.NEUROPATHOLOGY., BLACKWOOD W, GREENFIELD JG. Greenfield's Neuropathology. By W. Blackwood [and Others], Etc: Edward Arnold; 1963.
- 5.Brat DJ, Van Meir EG. Glomeruloid microvascular proliferation orchestrated by VPF/VEGF: a new world of angiogenesis research. Am J Pathol. 2001;158(3):789–96. Epub 2001/03/10. 10.1016/S0002-9440(10)64025-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaur B, Khwaja FW, Severson EA, Matheny SL, Brat DJ, Van Meir EG. Hypoxia and the hypoxia-inducible-factor pathway in glioma growth and angiogenesis. Neuro Oncol. 2005;7(2):134–53. Epub 2005/04/16. 10.1215/S1152851704001115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plate KH, Breier G, Weich HA, Risau W. Vascular endothelial growth factor is a potential tumour angiogenesis factor in human gliomas in vivo. Nature. 1992;359(6398):845–8. Epub 1992/10/29. 10.1038/359845a0 . [DOI] [PubMed] [Google Scholar]
- 8.Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R, et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):709–22. Epub 2014/02/21. 10.1056/NEJMoa1308345 . [DOI] [PubMed] [Google Scholar]
- 9.Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):699–708. Epub 2014/02/21. 10.1056/NEJMoa1308573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soda Y, Marumoto T, Friedmann-Morvinski D, Soda M, Liu F, Michiue H, et al. Transdifferentiation of glioblastoma cells into vascular endothelial cells. Proc Natl Acad Sci U S A. 2011;108(11):4274–80. Epub 2011/01/26. 10.1073/pnas.1016030108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ricci-Vitiani L, Pallini R, Biffoni M, Todaro M, Invernici G, Cenci T, et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468(7325):824–8. Epub 2010/11/26. 10.1038/nature09557 . [DOI] [PubMed] [Google Scholar]
- 12.Wang R, Chadalavada K, Wilshire J, Kowalik U, Hovinga KE, Geber A, et al. Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010;468(7325):829–33. Epub 2010/11/26. 10.1038/nature09624 . [DOI] [PubMed] [Google Scholar]
- 13.Kawasoe T, Takeshima H, Yamashita S, Mizuguchi S, Fukushima T, Yokogami K, et al. Detection of p53 mutations in proliferating vascular cells in glioblastoma multiforme. J Neurosurg. 2015;122(2):317–23. Epub 2014/11/22. 10.3171/2014.10.JNS132159 . [DOI] [PubMed] [Google Scholar]
- 14.Giannini C, Scheithauer BW, Burger PC, Christensen MR, Wollan PC, Sebo TJ, et al. Cellular proliferation in pilocytic and diffuse astrocytomas. Journal of neuropathology and experimental neurology. 1999;58(1):46–53. Epub 1999/02/12. 10.1097/00005072-199901000-00006 . [DOI] [PubMed] [Google Scholar]
- 15.Collins VP, Jones DT, Giannini C. Pilocytic astrocytoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129(6):775–88. Epub 2015/03/21. 10.1007/s00401-015-1410-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sie M, de Bont ES, Scherpen FJ, Hoving EW, den Dunnen WF. Tumour vasculature and angiogenic profile of paediatric pilocytic astrocytoma; is it much different from glioblastoma? Neuropathol Appl Neurobiol. 2010;36(7):636–47. Epub 2010/08/14. 10.1111/j.1365-2990.2010.01113.x . [DOI] [PubMed] [Google Scholar]
- 17.Wasilewski A, Mohile N. Durable response to bevacizumab in adults with recurrent pilocytic astrocytoma. CNS Oncol. 2018;7(3):Cns26 Epub 2018/04/10. 10.2217/cns-2017-0039 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gururangan S, Chi SN, Young Poussaint T, Onar-Thomas A, Gilbertson RJ, Vajapeyam S, et al. Lack of efficacy of bevacizumab plus irinotecan in children with recurrent malignant glioma and diffuse brainstem glioma: a Pediatric Brain Tumor Consortium study. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28(18):3069–75. Epub 2010/05/19. 10.1200/jco.2009.26.8789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones DT, Kocialkowski S, Liu L, Pearson DM, Backlund LM, Ichimura K, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68(21):8673–7. Epub 2008/11/01. 10.1158/0008-5472.CAN-08-2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Audia A, Conroy S, Glass R, Bhat KPL. The Impact of the Tumor Microenvironment on the Properties of Glioma Stem-Like Cells. Front Oncol. 2017;7:143 Epub 2017/07/26. 10.3389/fonc.2017.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawson AR, Hindley GF, Forshew T, Tatevossian RG, Jamie GA, Kelly GP, et al. RAF gene fusion breakpoints in pediatric brain tumors are characterized by significant enrichment of sequence microhomology. Genome Res. 2011;21(4):505–14. Epub 2011/03/12. 10.1101/gr.115782.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequencing primers (7 each) to be located at KIAA and BRAF introns at 1,000-bp intervals were designed to detect breakpoints.
(TIF)
S6 amplicons were subjected to Sanger sequencing. Red circles indicate that the base sequence annealed with the sequence primer. State-of-base calling identified areas of interest in the intron that included the breakpoint. In S6 the breakpoint was located between primers FP4 and RP3.
(TIF)
The breakpoint in S6 was detected by Sanger sequencing using the RP3 primer. The fusion was categorized as a “seamless transition” from KIAA intron 15 to BRAF intron 8.
(TIF)
S1 amplicons were subjected to Sanger sequencing. The breakpoint was located between the FP1 and the RP5 primers.
(TIF)
The breakpoint in S1 was detected by Sanger sequencing using the FP1 primer. The fusion was categorized as “presence of short insert”. A 7-bp sequence was inserted at the breakpoint.
(TIF)
PCR analysis with FP1 and RP5 primers revealed bands in S1 and S4. As in S1 (S4 Fig.), the breakpoint in S4 was located between primers FP1 and RP5.
(TIF)
The RP5 primer was used. The breakpoint in S4 was detected by sequencing. This fusion was categorized as “breakpoint microhomology”. We observed a shared 3-bp sequence between the end of KIAA and the start of BRAF.
(TIF)
The location of breakpoints between KIAA intron 15 and BRAF intron 8 in S1, S4, and S6 is shown. Note the inter-sample difference in the breakpoint. S1: Magenta: KIAA intron 15, Purple: BRAF intron 8. S4: Aqua: KIAA intron 15, Green: BRAF intron 8. S6: Red: KIAA intron 15, Blue: BRAF intron 8.
(TIF)
Tumor samples were collected from tumor tissue without MVP. Tumor samples and the cellular components of MVP were collected from the same tissue.
(TIF)
Raw digital PCR data for S6 are shown. Samples from tumor cells and cellular MVP components were independently analyzed. Top: Red and yellow dots indicate droplets with RNase-P DNA and droplets with no DNA, respectively. Bottom: Green and red dots identify droplets with the KIAA-BRAF fusion gene and droplets with no DNA, respectively.
(TIF)
Raw data of Figs 1B, 3D, 4A, 4B, 5 and S3, S5, S7, S8 and S9 Figs are shown by power point or excel files. Original sequencing data and digital PCR data could be seen with adequate software (Sequence scanner version 2 and QuantStudio 3D Analysis Suite Cloud). These raw data are also available at Dryad digital repository (DOI: https://doi.org/10.5061/dryad.bv44rk5).
(ZIP)
Data Availability Statement
All relevant data are within the paper, figures, its Supporting Information files, and the Dryad digital repository (Detection of the KIAA1549-BRAF fusion gene in cells forming microvascular proliferation of pilocytic astrocytoma, DOI: https://doi.org/10.5061/dryad.bv44rk5).