Abstract
Background
The blood flow rate of brain arteriovenous malformations (bAVMs) is an important clinical characteristic closely associated with the hemorrhage risk and radiosurgery obliteration rate of bAVMs. However, the underlying molecular properties remain unclear. To identify potential key molecules, signaling pathways, and vascular cell types involved, we compared gene expression profiles between bAVMs with high flow rates and low flow rates (LFR) and validated the functions of selected key molecules in vitro.
Methods and Results
We performed RNA‐sequencing analysis on 51 samples, including 14 high flow rate bAVMs and 37 LFR bAVMs. Functional pathway analysis was performed to identify potential signals influencing the flow rate phenotype of bAVMs. Candidate genes were investigated in bAVM specimens by immunohistochemical staining. Migration, tube formation, and proliferation assays were used to test the effects of candidate genes on the phenotypic properties of cultured human umbilical vein endothelial cells and human brain vascular smooth muscle cells. We identified 250 upregulated and 118 downregulated genes in LFR bAVMs compared with high flow rate bAVMs. Wnt signaling was activated in the LFR group via upregulation of FZD10 and MYOC. Immunohistochemical staining showed that vascular endothelial and smooth muscle cells of LFR bAVMs exhibited increased FZD10 and MYOC expression. Experimentally elevating these genes promoted human umbilical vein endothelial cells and migration and tube formation by activating canonical Wnt signaling in vitro.
Conclusions
Our results suggest that canonical Wnt signaling mediated by FZD10 and MYOC is activated in vascular endothelial and smooth muscle cells in LFR bAVMs.
Keywords: intracranial arteriovenous malformations, gene expression, hemodynamics, Wnt signaling pathway
Subject Categories: Cerebrovascular Malformations, Gene Expression & Regulation, Hemodynamics, Angiogenesis
Clinical Perspective
What Is New?
RNA‐sequencing of brain arteriovenous malformation (bAVM) tissue revealed differences in gene expression profiles of bAVMs with different blood flow rate phenotypes.
FZD10 and MYOC were upregulated and Wnt signaling was activated in the low flow rate bAVMs.
We found that overexpression of FZD10 and MYOC could activate canonical Wnt signaling and promote angiogenesis responses in endothelial and smooth muscle cells, which might induce high resistance and lead to low blood flow rate phenotypes.
What Are the Clinical Implications?
The blood flow rate of bAVMs is closely associated with the hemorrhage risk, obliteration rate after radiosurgery, and occurrence of normal perfusion pressure breakthrough after surgical resection.
The current study may help identify the potential causes of different blood flow phenotypes and provide a possibility for medical treatment to prevent bAVM rupture.
Introduction
Brain arteriovenous malformations (bAVMs) consist of abnormal tangles of dilated vascular structures, called a nidus, which connect arteries and veins directly without the intervening capillary beds.1 They are one of the major causes of intracranial hemorrhage and/or subarachnoid hemorrhage, leading to substantial morbidity and mortality, especially in children and young adults.2, 3 Current intervention options include neurosurgery, embolization, and stereotactic radiotherapy. The primary goal of these interventions is to prevent new or recurrent hemorrhage, but all these procedures have considerable risks and complications.4, 5
The blood flow rate is regarded as an important clinical characteristic that is closely associated with hemorrhage risk, obliteration rate after radiosurgery, and the occurrence of normal perfusion pressure breakthrough after surgical resection.6, 7, 8 Generally, the blood flow rate reflects the hemodynamic and angioarchitectural features of bAVMs, including an imbalance between inflow and outflow, vascular resistance, and morphological patterns.9, 10 Although the blood flow rate of bAVMs is important, the underlying molecular mechanisms remain unclear.
Studies investigating differences in gene expression in bAVMs with different flow rates may identify genes and pathways involved in this phenotype. Previous studies on bAVM transcriptomes were small scale and used microarray techniques.11, 12 RNA‐sequencing (RNA‐Seq) is a more advanced gene expression analysis method that can identify biomarkers across the broadest range of mRNAs with high efficiency and sensitivity.13 In our study, we divided bAVM patients into a high flow rate (HFR) group and a low flow rate (LFR) group according to the blood flow rate obtained from radiological information. RNA‐Seq was performed on the bAVM surgical samples, and we investigated the differentially expressed genes and related pathways between the 2 groups. The functions of differentially expressed genes of interest were further investigated in vitro. Our findings may help identify the mechanisms behind the different blood flow phenotypes and provide a possibility for medical treatment to prevent progression or bleeding of bAVMs.
Methods
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Patients and Samples
All study participants were Chinese and recruited at Beijing Tiantan Hospital, Capital Medical University. Samples were collected from consecutive patients who underwent surgical treatment for bAVMs in our department. Hereditary hemorrhagic telangiectasia was clinically excluded according to the Curaçao Criteria.14 Patients whose samples passed quality control for RNA‐Seq were enrolled in our study. Finally, 51 patients were enrolled from September 2016 to November 2017. All subjects provided informed consent, and the study was approved by the institutional review board of Beijing Tiantan Hospital, Capital Medical University. Clinical diagnoses were confirmed by digital subtraction angiography and histologic evaluation in the hospital's pathology department. For sample preparation, once the bAVM was resected, the brain tissue was removed in the operating room, and 2 to 3 g bAVM tissue was collected for RNA‐Seq. Specimens were stored in liquid nitrogen within 5 minutes.
Radiological Review
All patients underwent simultaneous biplanar digital subtraction angiography as the diagnostic method or part of a treatment plan with an image frame rate of 4 frames per second and injector‐controlled contrast injection rates (4 cm3/s for a total of 8 cm3). Angiographic, magnetic resonance, and computed tomography images available for each patient were evaluated by consensus between 2 researchers who were blinded to the clinical information. The blood flow rate of AVMs was estimated as previously described by determining the number of digital subtraction angiography frames between the first depiction of the nidus and the first visualization of a vein (HFR: venous drainage seen in <2 frames after nidal visualization; LFR: venous drainage seen in 2 or more frames after nidal visualization).9
RNA Isolation, Library Preparation, and Sequencing
Total RNA was isolated using the TRIzol method. Then, RNA degradation and contamination were monitored on 1% agarose gels. We checked RNA purity by using a NanoPhotometer spectrophotometer (IMPLEN, CA), and RNA concentration was measured using a Qubit RNA Assay Kit with a Qubit 2.0 Fluorometer (Life Technologies, CA). RNA integrity was assessed using an RNA Nano 6000 Assay Kit with a Bioanalyzer 2100 system (Agilent Technologies, CA).
A total amount of 4 μg RNA per sample was used as input material for the RNA sample preparations. Ribosomal RNA was removed by using an Epicentre Ribo‐zero rRNA Removal Kit (Epicentre, WI), and the rRNA‐free residue was cleaned by ethanol precipitation. Sequencing libraries were generated using rRNA‐depleted RNA with an NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, MA) following the manufacturer's recommendations. Fragmentation was carried out using divalent cations at an elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). First strand cDNA was synthesized using a random hexamer primer and M‐MuLV Reverse Transcriptase (RNaseH‐). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. In the reaction buffer, dTTP was replaced by dUTP. The remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation, the 3′ ends of DNA fragments were ligated with NEBNext Adaptors with a hairpin loop structure to prepare for hybridization. The library fragments were purified with an AMPure XP system (Beckman Coulter, Beverly, MA). Then, 3 μL USER Enzyme (NEB, MA) was used with cDNA at 37°C for 15 minutes followed by 5 minutes at 95°C before polymerase chain reaction. Polymerase chain reaction was then performed with Phusion High‐Fidelity DNA polymerase, Universal polymerase chain reaction primers, and Index (X) Primer. Finally, the products were purified (AMPure XP system), and the library quality was assessed on an Agilent Bioanalyzer 2100. The clustering of the index‐coded samples was performed on a cBot Cluster Generation System using a TruSeq PE Cluster Kit v3‐cBot‐HS (Illumina) according to the manufacturer's instructions. After cluster generation, the libraries were sequenced on the Illumina HiSeq platform, and 150‐bp paired‐end reads were generated.
Quality Control and Data Analysis
Raw data in fastq format were first processed through in‐house Perl scripts. In this step, clean data were obtained by removing reads containing adaptors, reads containing poly‐N, and low‐quality reads from the raw data. At the same time, Q20, Q30, and GC content of the clean data were calculated. All downstream analyses were based on high‐quality clean data. Reference genome and gene model annotation files were downloaded directly from the genome website. The index of the reference genome was built using bowtie2 v2.2.8, and paired‐end clean reads were aligned to the reference genome using Hisat2 v2.0.5.
Hg19 RefSeq (RNA sequences, GRCh37) was downloaded from the UCSC Genome Browser (http://genome.ucsc.edu). The clean reads were aligned with both genome hg19 and transcript reference using STAR v2.2.1, and gene expression was calculated by RSEM v1.3.0 using FPKM (fragments per kilobase of exon per million fragments mapped). Transcripts with a P<0.05 were considered differentially expressed. Functional enrichment analysis of upregulated genes (fold change ≥2, P<0.01) in LFR bAVMs was implemented by the WebGestalt website (http://webgestalt.org/option.php), including Gene Ontology and Kyoto Encyclopedia of Genes and Genomes. Terms with P values <0.05 were considered significantly enriched in differentially expressed genes. R v3.5.1 was used for analysis of the gene expression data.
Immunohistochemistry
The tissue sections were incubated with primary antibody, FZD10 (1:500, ab150564, Abcam) or MYOC (1:500, ab41552, Abcam), overnight at 4°C and then incubated with a biotinylated secondary antibody at room temperature for 1 hour, followed by incubation with horseradish peroxidase‐labeled streptavidin for 30 minutes. After washing with Tris‐buffer, the sections were stained with diamino benzidine, and nuclei were counterstained with hematoxylin. Specimens were observed, and images were captured with an EVOS FL Auto 2 Imaging System (Invitrogen). We measured the expression semiquantitatively as previously described.15 Scoring of immunoreactivity was as follows: 0, no staining; 1, mild staining; 2, moderate staining; and 3, intense staining. Two researchers who were blinded to the clinical information performed the measurements, and data were collected as the average of the 2 observations.
Cell Culture and Treatment
Human umbilical vein endothelial cells (HUVECs) were isolated from umbilical veins (ScienCell, Carlsbad, CA). Cells were cultured in endothelial cell medium (ScienCell) supplemented with endothelial cell growth supplement and 5% fetal bovine serum (ScienCell). Human brain vascular smooth muscle cells (HBVSMCs) were isolated from arteries and arterioles of the human brain (ScienCell) and were cultured in smooth muscle cell basal medium (ScienCell) supplemented with smooth muscle cell growth supplement and 2.5% fetal bovine serum (ScienCell). All cells were cultured according to the recommended protocols. For plasmid DNA transfection, cells were transfected with a GV219 vector expressing FZD10 or MYOC using Lipofectamine 3000 (Invitrogen) according to the manufacturer's instructions. As controls, cells were transfected in parallel with a GV219 empty vector without the insert.
Reverse Transcription Quantitative Polymerase Chain Reaction
Total RNA was isolated from transfected cells using the TRIzol reagent (Invitrogen). RNA was cleaned with gDNA Eraser (Takara) to remove DNA contamination. One microgram of purified RNA was reverse transcribed using a PrimeScript RT reagent Kit (Takara). Quantitative polymerase chain reaction was performed using TB Green Premix Ex Taq (Takara) with a QuantStudio 3 System (Applied Biosystems) with specific primers designed for amplicons of 75 to 150 bp. GAPDH was used as a reference gene. The primer sequences are listed in Table S1.
Statistical Analysis
Statistical analyses were accomplished by using PRISM (GraphPad version 7.0) and SPSS (version 25.0). For clinical data, patients with HFR or LFR bAVMs were compared using descriptive statistics. For age and Spetzler‐Martin grade, Mann–Whitney U tests were performed. Sex and hemorrhage were compared using Pearson χ2 tests. Fisher exact tests were used to compare seizure, deep venous drainage, perforating artery, and location. The Mann–Whitney U tests were used to investigate the differential expression of FZD10 and MYOC in tissue sections, and Student t tests were used to analyze the results of in vitro experiments. All results are expressed as the mean±SD. P<0.05 was considered to indicate statistical significance in all cases.
Additional methodologies are described in Data S1.
Results
bAVM Flow Rate Types and Other Characteristics of the Study Population
A total of 51 patients with bAVMs were included in our study. None of the patients had familial bAVM or hereditary hemorrhagic telangiectasia. The bAVMs were further classified as HFR (n=14, 27.45%) or LFR (n=37, 72.55%) according to the flow types indicated by digital subtraction angiography (Figure 1). No significant differences were found between the 2 groups regarding sex, history of seizure, hemorrhagic presentation, or angioarchitectural features. The baseline characteristics of all patients are summarized in Table.
Figure 1.
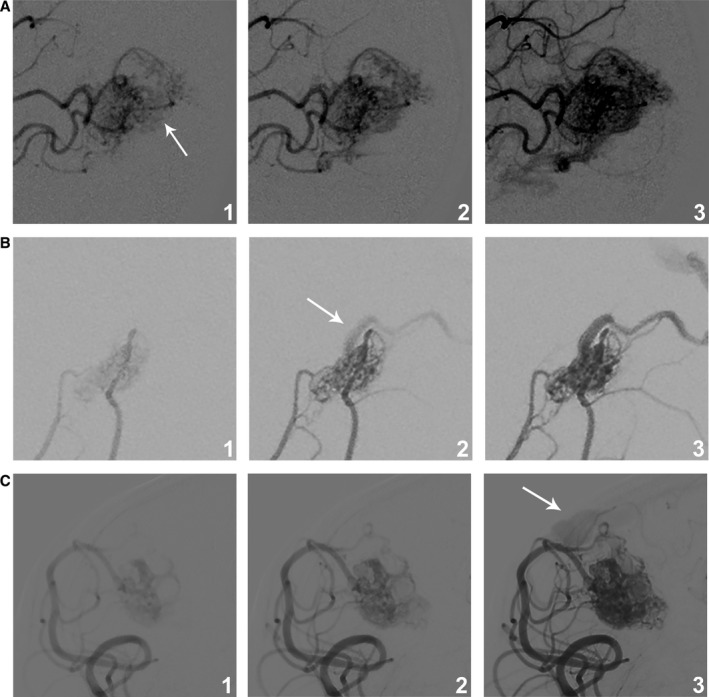
bAVM blood flow rate subtypes. The blood flow rate was classified as high if the vein was visible at the same time as the nidus (A) or at 1 frame after visualization of the nidus (B), and the blood flow rate was classified as low if the vein was visualized later than 2 frames after depiction of the nidus (C). The same frame rate of 4 images per second was used in all instances, and machine‐assisted injections with a constant rate increase were performed. The arrows denote the first visualization of the vein. bAVM indicates brain arteriovenous malformations.
Table 1.
Baseline Characteristics of the Study Participants
| Characteristic | High Flow Rate (n=14) | Low Flow Rate (n=37) | P Value |
|---|---|---|---|
| Sex, female | 3 (21%) | 18 (49%) | 0.078 |
| Age, y | 33±19 | 27±14 | 0.347 |
| Clinical presentation | |||
| Seizure | 4 (29%) | 14 (38%) | 0.744 |
| Hemorrhage | 7 (50%) | 18 (49%) | 0.931 |
| Angioarchitectural features | |||
| Deep venous drainage | 1 (7%) | 13 (35%) | 0.077 |
| Perforating artery | 0 (0%) | 4 (11%) | 0.565 |
| Location | |||
| Frontal | 6 (43%) | 15 (41%) | 0.835 |
| Temporal | 4 (29%) | 10 (27%) | |
| Parietal | 2 (14%) | 8 (22%) | |
| Occipital | 2 (14%) | 2 (5%) | |
| Cerebellar | 0 (0%) | 2 (5%) | |
| Spetzler‐Martin | |||
| 1 | 5 (36%) | 5 (14%) | 0.030a |
| 2 | 6 (43%) | 12 (32%) | |
| 3 | 2 (14%) | 14 (38%) | |
| 4 | 1 (7%) | 6 (16%) | |
| 5 | 0 (0%) | 0 (0%) | |
P value indicates statistical significance (P≤0.05).
Differential Expression Profile Between HFR and LFR bAVMs
The analysis compared gene expression profiles between 14 HFR bAVMs and 37 LFR bAVMs to identify genes showing consistent differences in expression. A total of 368 genes were identified as differentially expressed (Figure 2A), and 250 genes presented more than 2‐fold upregulated expression in LFR bAVMs (Table S2). A marked upregulation of SBK3 expression was seen in LFR bAVMs with a 9.5‐fold change. In addition, the expression of 118 genes was downregulated by more than 2‐fold in LFR bAVMs (Table S2), and the most obvious decrease in expression of LFR bAVMs was observed for IGHV3‐9, with an 8.3‐fold change.
Figure 2.
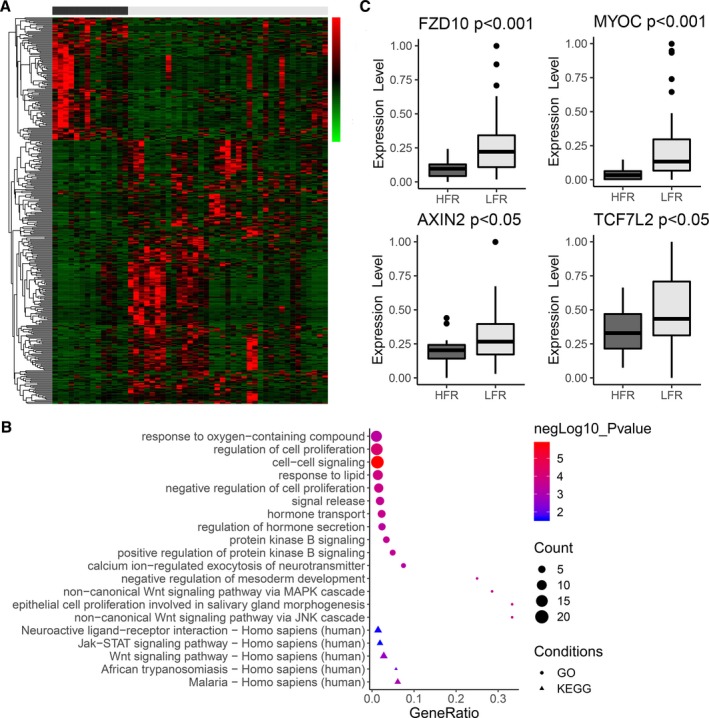
mRNA expression profiling in bAVMs. A, Gene expression heatmap of differentially expressed mRNAs (P≤0.05 and fold change ≥2 or ≤0.5) in HFR vs LFR bAVM tissues. The x‐axis shows each bAVM patient (black=HFR; gray=LFR), and the y‐axis shows individual genes. In the heatmap cells, red indicates high gene expression (ie, upregulated expression) relative to the median expression; green indicates low expression (ie, downregulated expression); black indicates that expression is similar to the median. B, Top 15 terms of GO analysis (ranked by P value) and KEGG analysis enriched by upregulated genes in LFR bAVMs. C, The expression levels of FZD10, MYOC, and downstream molecules in canonical Wnt signaling. bAVM indicates brain arteriovenous malformations; HFR, high flow rate; LFR, low flow rate.
Wnt Signaling Is Activated in LFR bAVMs
To identify potential functional pathways that influence the flow rate phenotype of bAVMs, we performed Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analysis of differentially expressed genes. Gene Ontology analysis identified that Wnt signaling, an important signaling pathway associated with embryonic angiogenesis and the development of bAVMs,15, 16 was activated in the LFR group with the upregulation of FZD10 and MYOC (Figure 2B; Table S3). FZD10 is a member of the family of Frizzled proteins, which are 7‐transmembrane domain proteins, and is known as a receptor of WNT7b, which may influence downstream molecules through Wnt signaling.17, 18 MYOC belongs to a family of glycosylated proteins containing a C‐terminal olfactomedin domain and can interact with secreted inhibitors of Wnt signaling, such as sFRP1 and sFRP3, as a modulator of the Wnt signaling pathway.19, 20
FZD10 and MYOC were found in the noncanonical Wnt signaling pathway via Jun N‐terminal kinase and MAPK in Gene Ontology analysis, and Kyoto Encyclopedia of Genes and Genomes pathway analysis did not identify which type of Wnt signaling, canonical or noncanonical, was affected by the 2 molecules (Figure 2B; Table S3). Further investigation of differentially expressed genes (including a fold change <2) found that AXIN2 and TCF7L2, the target genes of the canonical Wnt pathway, showed 1.2‐ (P<0.05) and 1.3‐fold (P<0.05) increases in expression in the LFR group, respectively. The expression of FZD10, MYOC, and downstream molecules of the canonical Wnt signaling pathway in bAVMs with different blood flow rates are shown in Figure 2C. The specific functions of FZD10 and MYOC in canonical or noncanonical Wnt signaling need to be verified in vascular cells.
FZD10 and MYOC Are Relatively Highly Expressed in Vascular Endothelial Cells and Smooth Muscle Cells of LFR bAVMs
To validate the differential expression of FZD10 and MYOC at the protein level and investigate the cell types expressing them, we performed immunohistochemistry on bAVM specimens. We detected the expression of FZD10 and MYOC in 44 specimens (LFR, n=37; HFR, n=14). Immunohistochemistry analysis revealed the expression of FZD10 in 26 samples (23/30 LFR and 3/14 HFR) and MYOC in 38 samples (30/30 LFR and 8/14 HFR). FZD10 was observed in vascular endothelial cells (ECs), smooth muscle cells (SMCs) (Figure 3A), and brain tissues far from the nidus. MYOC was also detected in ECs and SMCs of bAVMs (Figure 3B). We observed that the expression levels of FZD10 and MYOC in ECs and SMCs showed the same trend.
Figure 3.
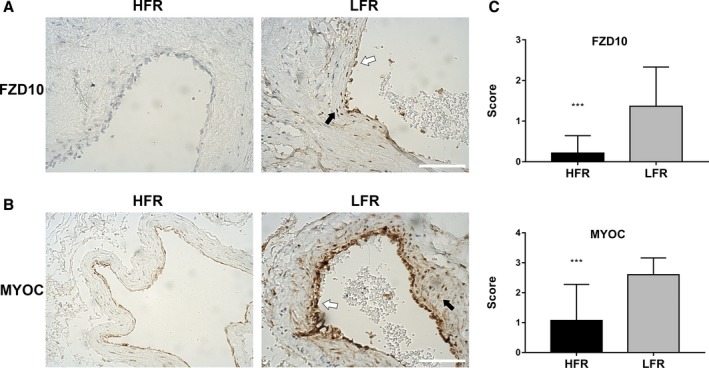
Detection of FZD10 and MYOC in bAVM tissue samples. Immunohistochemical staining of bAVM tissue samples with differential flow rate subtypes show strong staining for FZD10 (A) and MYOC (B) in LFR bAVM tissue. Endothelial cells lining the vascular lumen (white arrows) and vascular smooth muscle cells in the vessel wall (black arrows) both show staining for FZD10 and MYOC. The scale bar corresponds to 200 μm. C, Semiquantitative grading of FZD10 and MYOC expression levels in the vascular structure of bAVMs. ***P<0.001. bAVM indicates brain arteriovenous malformations; HFR, high flow rate; LFR, low flow rate.
Semiquantitative grading of FZD10 and MYOC expression levels was performed in the vascular structures of bAVMs (Figure 3C). FZD10 had a relatively low expression level of 0.21±0.11 in HFR bAVMs, while the expression level reached 1.37±0.18 in LFR bAVMs (P<0.001). The expression level of MYOC was 1.07±0.32 in HFR bAVMs and 2.60±0.10 in LFR bAVMs (P<0.001). The results were consistent with RNA‐Seq analysis.
FZD10 and MYOC Improve Cell Migration and Tube Formation of HUVECs and HBVSMCs
To investigate the functional significance of FZD10 and MYOC in the ECs and SMCs of bAVMs, we transfected HUVECs and HBVSMCs with FZD10‐ or MYOC‐overexpressing plasmids and GV219 empty vector. The results of reverse transcription polymerase chain reaction suggested that HUVECs and HBVSMCs expressed low levels of FZD10 and MYOC mRNA (Figure S1); therefore, we did not try to downregulate their expression by siRNA. We determined optimal transfection concentrations and confirmed that the plasmids were taken up by cells and expressed (Figure 4A). Several assays were performed to test the effects of FZD10 or MYOC upregulation on tube formation, cell migration, and cell proliferation. After transfection with FZD10‐overexpressing plasmids, increased mobility of transfected cells was observed in wound healing assays (Figure 4B), and the microvasculature formed by HUVECs and HBVSMCs was significantly increased compared with that of the control cells (Figure 4C). Similar findings were also apparent in tube formation and wound healing assays of MYOC‐transfected HUVECs and HBVSMCs (Figure 4B and 4C). However, FZD10 or MYOC did not influence the proliferation of HUVECs and HBVSMCs (data not shown). These results suggest that FZD10 and MYOC can promote the angiogenesis responses of HUVECs and HBVSMCs by changing the biological behavior of cells.
Figure 4.
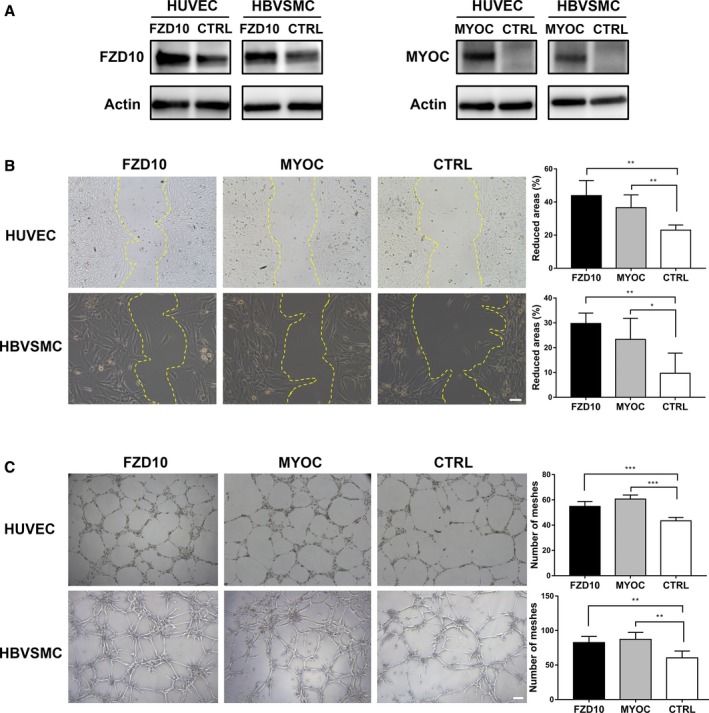
Effects of FZD10 and MYOC on HUVECs and HBVSMCs. A, Western blotting analysis of HUVECs and HBVSMCs transfected with a pcDNA3 vector overexpressing FZD10/MYOC or negative control (CTRL). B, Effects of FZD10/MYOC on migration of HUVECs and HBVSMCs. The scale bar corresponds to 100 μm. C, Effects of FZD10/MYOC on tube formation of HUVECs and HBVSMCs. The scale bar corresponds to 200 μm. One representative experiment out of 5 is shown. *P<0.05; **P<0.025; ***P<0.001. HBVSMCs indicates human brain vascular smooth muscle cells; HUVECs, human umbilical vein endothelial cells.
FZD10 and MYOC Activate Canonical β‐Catenin/Wnt Signaling in HUVECs and HBVSMCs
Previous studies found that FZD10 and MYOC could activate both canonical and noncanonical Wnt signaling.18, 20, 21, 22 To investigate the specific pathway modulated by the 2 genes, we detected the downstream molecules of canonical and noncanonical Wnt signaling in HUVECs and HBVSMCs. Overexpression of FZD10 in HUVECs and HBVSMCs increased the mRNA levels of AXIN2 and TCF‐1 (Figure 5D), the target genes of the canonical Wnt pathway. In contrast, FZD10 did not influence Jun N‐terminal kinase activation (Figure 5E), a hallmark of noncanonical Wnt activation. After transfection with FZD10, the total level of β‐catenin in HUVECs was elevated slightly, which might mean that the degradation of β‐catenin was inhibited (Figure 5E). However, the total level of β‐catenin in HBVSMCs remained unchanged. Although β‐catenin did not show an obvious increase, its nuclear translocation in both HUVECs and HBVSMCs was confirmed by immunofluorescence (Figure 5A, 5B, and 5C), which is direct evidence of canonical Wnt pathway activation.
Figure 5.
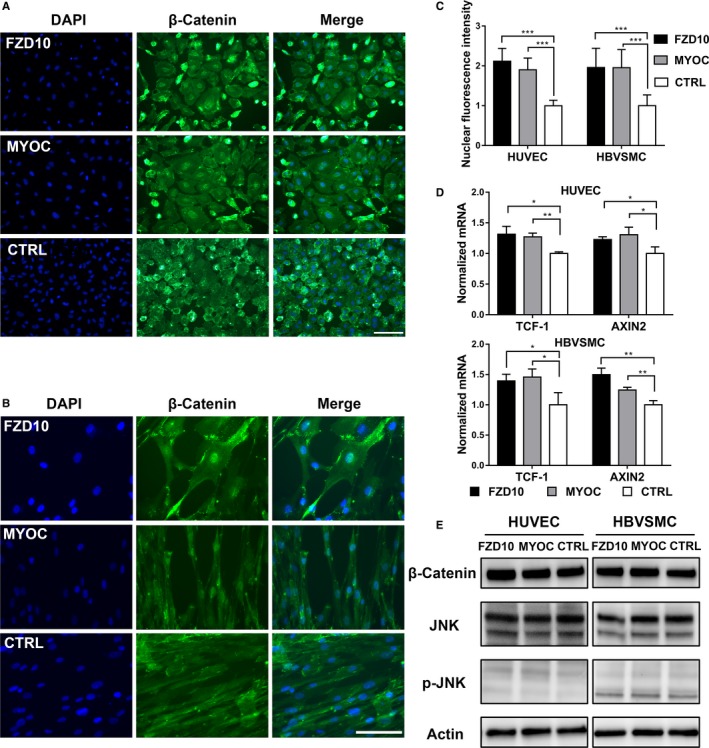
Wnt signaling is influenced by FZD10 or MYOC in HUVECs and HBVSMCs. Immunofluorescent staining for β‐catenin in HUVECs (A) and HBVSMCs (B) transfected with a pcDNA3 vector overexpressing FZD10/MYOC or negative control (CTRL). C, Nuclear β‐catenin level was quantified by measuring fluorescence intensity in the cell nucleus. Data were derived from 3 randomly selected fields. D, RT‐qPCR for AXIN2 and TCF‐1 expression in HUVECs and HBVSMCs transfected as in (A, B). E, Western blotting analysis for total JNK (t‐JNK), p‐JNK (Thr183/Tyr185), and β‐catenin in HUVECs and HBVSMCs transfected as in (A, B). *P<0.05; **P<0.025; ***P<0.001. The scale bar corresponds to 200 μm. These data are representative of 3 independent experiments. DAPI indicates 4′,6‐diamidino‐2‐phenylindole; HBVSMCs, human brain vascular smooth muscle cells; RT‐qPCR, reverse transcription quantitative polymerase chain reaction; HUVECs, human umbilical vein endothelial cells.
Similarly, transfection of HUVECs and HBVSMCs with the MYOC‐overexpressing plasmid led to the upregulation of AXIN2 and TCF‐1 and did not influence the phosphorylation of Jun N‐terminal kinase (Figure 5D and 5E). In agreement with canonical Wnt pathway activation, nuclear β‐catenin staining was also enhanced in HUVECs and HBVSMCs (Figure 5A, 5B, and 5C), even if the total level of β‐catenin did not change noticeably.
Discussion
The blood flow rate of bAVMs is an important hemodynamic parameter that can influence the clinical decision and efficacy of treatments.7, 9 However, the underlying molecular properties remain unclear. In this research, we performed RNA‐Seq to identify differences in the gene expression between surgical bAVM tissues from the HFR group and the LFR group and to facilitate the understanding of the molecular mechanisms in bAVMs with different flow rates. We identified FZD10 and MYOC in vascular ECs and SMCs as key molecules, and Wnt signaling was activated in the LFR group patients. The function of FZD10 and MYOC was investigated in cultured ECs and SMCs. We found that overexpression of the 2 molecules could activate canonical Wnt signaling to promote angiogenesis responses, such as cell migration and tube formation, which might induce high resistance and lead to phenotypes of low blood flow rate.
A previous study based on microarray analysis identified several neuron‐related genes, including NPY, SYT1, NeuroD, and EFNB3, which were downregulated in the HFR bAVMs.12 This result explained only the neuronal network injury caused by the lack of perfusion in the perinidal area, which might be the outcome rather than the cause of high blood flow rate.23 In our study, RNA‐Seq data and functional pathway analysis suggested that FZD10 and MYOC had relatively high expression levels in LFR bAVMs. They were involved in the activation of Wnt signaling, which plays important roles in central nervous system angiogenesis and regulates vessel density during embryonic development.16, 24, 25, 26 In cultured ECs and SMCs, we found that overexpressed FZD10 and MYOC could activate canonical rather than noncanonical Wnt signaling, which was consistent with a previous study.17, 20 We postulated that the upregulation of canonical Wnt signaling might increase the blood vessel density of bAVMs, eventually resulting in an increase in vascular resistance and a reduction in blood flow rate. Our study implies that targeting FZD10, MYOC, and canonical Wnt signaling might modulate the blood flow rate in bAVMs.
The morphologic and functional changes in vascular ECs and SMCs are involved in the pathophysiological process of bAVMs.27, 28 Whole‐exome sequencing of bAVMs showed that some vascular ECs contained KRAS mutations, and further investigation revealed that cultured ECs that had the same mutation were phenotypically larger and elongated, demonstrated faster migration, and had more cytoskeletal actin projections.29 SMCs derived from bAVMs formed tubes in culture, which were longer than those formed by normal brain vascular SMCs. The migration and proliferation of bAVM SMCs also exceeded those of normal brain vascular SMCs.30 These findings suggest that the pathogenesis of bAVMs is likely because of abnormal vascular ECs and SMCs. Whether the function of vascular ECs and SMCs affects the phenotype development of bAVMs is still unknown. In this study, we observed that overexpressed FZD10 and MYOC were mainly concentrated in vascular ECs and SMCs of LFR bAVMs. In vitro experiments further verified that overexpression of FZD10 and MYOC improved the migration and tube formation of ECs and SMCs. These results suggested that ECs and SMCs might play essential roles in the regulation of different flow rate phenotype development in bAVMs as well.
Currently, clinical decisions for bAVMs are still in a dilemma, particularly when dealing with certain difficult cases, such as large bAVMs located near or in the functional areas.31, 32 Alternative novel therapeutic strategies with higher safety and efficacy need to be explored. The blood flow rate of bAVMs, which is an important factor that has a relationship with prognosis, could be a potential target for intervention. By performing RNA‐Seq on bAVM tissues dissected from patients, we found differential gene expression profiling between bAVMs with different flow rates, providing a potential therapeutic target to regulate the blood flow rate, which might provide a promising perspective for bAVM treatment.
Sources of Funding
This study was supported by the “National Key Research and Development Program of China during the 13th Five‐Year Plan Period” (Grant No. 2016YFC1301803, Principle Investigator: Professor Yong Cao and Grant No. 2016YFC1301801, Principle Investigator: Professor Shuo Wang) and the “Key Project of Beijing Municipal Science & Technology Commission” (Grant No. D161100003816006, Principal Investigator: Professor Shuo Wang and Grant No. D161100003816005, Principle Investigator: Professor Jizong Zhao).
Disclosures
None.
Supporting information
Data S1. Supplemental Methods.
Table S1. Primers Used for RT‐qPCR
Table S2. Differential Expression Profile
Table S3. GO and KEGG Analysis Enriched by Upregulated Genes in Low Flow Rate bAVMs
Figure S1. Expression level of FZD10 (A) and MYOC (B) in HUVECs and HBVSMCs after transfection.
Acknowledgments
We thank Zhicen Li and Ji Ma for their contributions to the data collection. We also thank Yang Liu, Jiayu Li, and Haoyu Li for their assistance in manuscript preparation.
(J Am Heart Assoc. 2019;8:e012746 DOI: 10.1161/JAHA.119.012746.)
References
- 1. Solomon RA, Connolly ES. Arteriovenous malformations of the brain. N Engl J Med. 2017;376:1859–1866. [DOI] [PubMed] [Google Scholar]
- 2. Fullerton HJ, Achrol AS, Johnston SC, McCulloch CE, Higashida RT, Lawton MT, Sidney S, Young WL. Long‐term hemorrhage risk in children versus adults with brain arteriovenous malformations. Stroke. 2005;36:2099–2104. [DOI] [PubMed] [Google Scholar]
- 3. Meyer‐Heim AD, Boltshauser E. Spontaneous intracranial haemorrhage in children: aetiology, presentation and outcome. Brain Dev. 2003;25:416–421. [DOI] [PubMed] [Google Scholar]
- 4. Derdeyn CP, Zipfel GJ, Albuquerque FC, Cooke DL, Feldmann E, Sheehan JP, Torner JC. Management of brain arteriovenous malformations: a scientific statement for healthcare professionals from the American Heart Association/American Stroke Association. Stroke. 2017;48:e200–e224. [DOI] [PubMed] [Google Scholar]
- 5. Mohr JP, Parides MK, Stapf C, Moquete E, Moy CS, Overbey JR, Al‐Shahi SR, Vicaut E, Young WL, Houdart E, Cordonnier C, Stefani MA, Hartmann A, von Kummer R, Biondi A, Berkefeld J, Klijn CJ, Harkness K, Libman R, Barreau X, Moskowitz AJ. Medical management with or without interventional therapy for unruptured brain arteriovenous malformations (Aruba): a multicentre, non‐blinded, randomised trial. Lancet. 2014;383:614–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen X, Cooke DL, Saloner D, Nelson J, Su H, Lawton MT, Hess C, Tihan T, Zhao Y, Kim H. Higher flow is present in unruptured arteriovenous malformations with silent intralesional microhemorrhages. Stroke. 2017;48:2881–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Taeshineetanakul P, Krings T, Geibprasert S, Menezes R, Agid R, Terbrugge KG, Schwartz ML. Angioarchitecture determines obliteration rate after radiosurgery in brain arteriovenous malformations. Neurosurgery. 2012;71:1071–1078; discussion 1079. [DOI] [PubMed] [Google Scholar]
- 8. Rangel‐Castilla L, Spetzler RF, Nakaji P. Normal perfusion pressure breakthrough theory: a reappraisal after 35 years. Neurosurg Rev. 2015;38:399–404:discussion 404–5. [DOI] [PubMed] [Google Scholar]
- 9. Ma L, Chen XL, Chen Y, Wu CX, Ma J, Zhao YL. Subsequent haemorrhage in children with untreated brain arteriovenous malformation: higher risk with unbalanced inflow and outflow angioarchitecture. Eur Radiol. 2017;27:2868–2876. [DOI] [PubMed] [Google Scholar]
- 10. Fukuoka S, Takanashi M, Seo Y, Suematsu K, Nakamura J. Radiosurgery for arteriovenous malformations with gamma‐knife: a multivariate analysis of factors influencing the complete obliteration rate. J Clin Neurosci. 1998;5(suppl):68–71. [DOI] [PubMed] [Google Scholar]
- 11. Weinsheimer SM, Xu H, Achrol AS, Stamova B, McCulloch CE, Pawlikowska L, Tian Y, Ko NU, Lawton MT, Steinberg GK, Chang SD, Jickling G, Ander BP, Kim H, Sharp FR, Young WL. Gene expression profiling of blood in brain arteriovenous malformation patients. Transl Stroke Res. 2011;2:575–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Takagi Y, Aoki T, Takahashi JC, Yoshida K, Ishii A, Arakawa Y, Kikuchi T, Funaki T, Miyamoto S. Differential gene expression in relation to the clinical characteristics of human brain arteriovenous malformations. Neurol Med Chir. 2014;54:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. ‘t Hoen PA, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RH, de Menezes RX, Boer JM, van Ommen GJ, den Dunnen JT. Deep sequencing‐based expression analysis shows major advances in robustness, resolution and inter‐lab portability over five microarray platforms. Nucleic Acids Res. 2008;36:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shovlin CL, Guttmacher AE, Buscarini E, Faughnan ME, Hyland RH, Westermann CJ, Kjeldsen AD, Plauchu H. Diagnostic criteria for hereditary hemorrhagic telangiectasia (rendu‐osler‐weber syndrome). Am J Med Genet. 2000;91:66–67. [DOI] [PubMed] [Google Scholar]
- 15. Hermanto Y, Takagi Y, Ishii A, Yoshida K, Kikuchi T, Funaki T, Mineharu Y, Miyamoto S. Immunohistochemical analysis Sox17 associated pathway in brain arteriovenous malformations. World Neurosurg. 2015;87:573–583. [DOI] [PubMed] [Google Scholar]
- 16. Corada M, Nyqvist D, Orsenigo F, Caprini A, Giampietro C, Taketo MM, Iruela‐Arispe ML, Adams RH, Dejana E. The Wnt/beta‐catenin pathway modulates vascular remodeling and specification by upregulating D114/notch signaling. Dev Cell. 2010;18:938–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gong C, Qu S, Lv XB, Liu B, Tan W, Nie Y, Su F, Liu Q, Yao H, Song E. BRMS1L suppresses breast cancer metastasis by inducing epigenetic silence of FZD10. Nat Commun. 2014;5:5406. [DOI] [PubMed] [Google Scholar]
- 18. Fukukawa C, Nagayama S, Tsunoda T, Toguchida J, Nakamura Y, Katagiri T. Activation of the non‐canonical Dvl‐Rac1‐JNK pathway by frizzled homologue 10 in human synovial sarcoma. Oncogene. 2009;28:1110–1120. [DOI] [PubMed] [Google Scholar]
- 19. Kwon HS, Lee HS, Ji Y, Rubin JS, Tomarev SI. Myocilin is a modulator of Wnt signaling. Mol Cell Biol. 2009;29:2139–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shen X, Ying H, Yue BY. Wnt activation by wild type and mutant myocilin in cultured human trabecular meshwork cells. PLoS One. 2012;7:e44902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hot B, Valnohova J, Arthofer E, Simon K, Shin J, Uhlen M, Kostenis E, Mulder J, Schulte G. FZD10‐Gα13 signalling axis points to a role of FZD10 in CNS angiogenesis. Cell Signal. 2017;32:93–103. [DOI] [PubMed] [Google Scholar]
- 22. Kwon HS, Tomarev SI. Myocilin, a glaucoma‐associated protein, promotes cell migration through activation of integrin‐focal adhesion kinase‐serine/threonine kinase signaling pathway. J Cell Physiol. 2011;226:3392–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takagi Y, Kikuta K, Nozaki K, Fujimoto M, Hayashi J, Hashimoto N. Neuronal expression of Fas‐associated death domain protein and caspase‐8 in the perinidal parenchyma of cerebral arteriovenous malformations. J Neurosurg. 2007;106:275–282. [DOI] [PubMed] [Google Scholar]
- 24. Tata M, Ruhrberg C, Fantin A. Vascularisation of the central nervous system. Mech Dev. 2015;138:26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Corada M, Morini MF, Dejana E. Signaling pathways in the specification of arteries and veins. Arterioscler Thromb Vasc Biol. 2014;34:2372–2377. [DOI] [PubMed] [Google Scholar]
- 26. Phng LK, Potente M, Leslie JD, Babbage J, Nyqvist D, Lobov I, Ondr JK, Rao S, Lang RA, Thurston G, Gerhardt H. Nrarp coordinates endothelial Notch and Wnt signaling to control vessel density in angiogenesis. Dev Cell. 2009;16:70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Crist AM, Lee AR, Patel NR, Westhoff DE, Meadows SM. Vascular deficiency of Smad4 causes arteriovenous malformations: a mouse model of hereditary hemorrhagic telangiectasia. Angiogenesis. 2018;21:363–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Frösen J, Joutel A. Smooth muscle cells of intracranial vessels: from development to disease. Cardiovasc Res. 2018;114:501–512. [DOI] [PubMed] [Google Scholar]
- 29. Nikolaev SI, Vetiska S, Bonilla X, Boudreau E, Jauhiainen S, Jahromi BR, Khyzha N, DiStefano PV, Suutarinen S, Kiehl TR, Pereira VM, Herman AM, Krings T, Andrade‐Barazarte H, Tung T, Valiante T, Zadeh G, Tymianski M, Rauramaa T, Ylä‐Herttuala S, Wythe JD, Antonarakis SE, Frösen J, Fish JE, Radovanovic I. Somatic activating kRAS mutations in arteriovenous malformations of the brain. N Engl J Med. 2018;378:250–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang J, Song J, Qu M, Wang Y, An Q, Song Y, Yan W, Wang B, Wang X, Zhang S, Chen X, Zhao B, Liu P, Xu T, Zhang Z, Greenberg DA, Wang Y, Gao P, Zhu W, Yang GY. MicrorRNA‐137 and microrNA‐195* inhibit vasculogenesis in brain arteriovenous malformations. Ann Neurol. 2017;82:371–384. [DOI] [PubMed] [Google Scholar]
- 31. Lawton MT; UCSF Brain Arteriovenous Malformation Study Project . Spetzler‐Martin Grade III arteriovenous malformations: surgical results and a modification of the grading scale. Neurosurgery. 2003;52:740–748: discussion 748–749. [DOI] [PubMed] [Google Scholar]
- 32. Reinard KA, Pabaney AH, Basheer A, Phillips SB, Kole MK, Malik GM. Surgical management of giant intracranial arteriovenous malformations: a single center experience over 32 years. World Neurosurg. 2015;84:1765–1778. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supplemental Methods.
Table S1. Primers Used for RT‐qPCR
Table S2. Differential Expression Profile
Table S3. GO and KEGG Analysis Enriched by Upregulated Genes in Low Flow Rate bAVMs
Figure S1. Expression level of FZD10 (A) and MYOC (B) in HUVECs and HBVSMCs after transfection.


