Abstract
Purpose
To determine plasma metabolite and metabolic pathway differences between patients with type 2 diabetes with diabetic retinopathy (DR) and without retinopathy (diabetic controls), and between patients with proliferative DR (PDR) and nonproliferative DR (NPDR).
Methods
Using high-resolution mass spectrometry with liquid chromatography, untargeted metabolomics was performed on plasma samples from 83 DR patients and 90 diabetic controls. Discriminatory metabolic features were identified through partial least squares discriminant analysis, and linear regression was used to adjust for age, sex, diabetes duration, and hemoglobin A1c. Pathway analysis was performed using Mummichog 2.0.
Results
In the adjusted analysis, 126 metabolic features differed significantly between DR patients and diabetic controls. Pathway analysis revealed alterations in the metabolism of amino acids, leukotrienes, niacin, pyrimidine, and purine. Arginine, citrulline, glutamic γ-semialdehyde, and dehydroxycarnitine were key contributors to these pathway differences. A total of 151 features distinguished PDR patients from NPDR patients, and pathway analysis revealed alterations in the β-oxidation of saturated fatty acids, fatty acid metabolism, and vitamin D3 metabolism. Carnitine was a major contributor to the pathway differences.
Conclusions
This study demonstrates that arginine and citrulline-related pathways are dysregulated in DR, and fatty acid metabolism is altered in PDR patients compared with NPDR patients.
Keywords: diabetic retinopathy, arginine, citrulline, carnitine, metabolomics
Diabetic retinopathy (DR) is the leading cause of blindness in working-age adults, affecting approximately 93 million people worldwide.1 Its prevalence will likely increase as the global population with diabetes is projected to rise by 55% from 2013 to 2035.2 Vision impairment resulting from DR is a debilitating disease associated with lost independence and reduced quality of life.3 Current treatment options for DR, including laser photocoagulation and intravitreal injections, are both costly and invasive.
Clinically, DR is classified into two major stages, nonproliferative (NPDR) and proliferative diabetic retinopathy (PDR). NPDR is characterized by retinal microaneurysms and hemorrhages and may progress to PDR, which is defined by retinal neovascularization with the potential for severe vision loss.4 The strongest risk factors for DR include poor glycemic control and longer duration of diabetes. Some studies have suggested that hypertension and hyperlipidemia may also play a role.5 Maintaining adequate glycemic control is difficult, requiring rigorous dietary restrictions and precise medication management. Furthermore, strict glycemic control does not prevent the development of DR in all patients,6 and the causes of this clinical heterogeneity are not well understood.
Given the limited treatment options and incomplete understanding of DR pathogenesis, further studies are essential to optimize patient care. Metabolomics, the study of small molecules present in a biological sample, has the potential to uncover metabolic changes that distinguish healthy and disease states. Liquid chromatography coupled with high-resolution mass spectrometry (LC-MS) is a technique that can measure thousands of metabolites in biological fluids or tissues.7 We have previously used this approach to identify metabolites and metabolic pathways altered in neovascular AMD (NVAMD)8,9 and POAG.10 Metabolomics studies have identified changes in the pentose phosphate, arginine to proline, and ascorbic acid pathways, as well as altered plasma levels of glutamine and glutamate in patients with DR.11–14 However, many of these studies were limited by sample size, and none investigated differences between NPDR and PDR patients.
In this study, we performed untargeted metabolomics via LC-MS in plasma of patients with type 2 diabetes mellitus with and without DR. The goal was to identify metabolites or metabolic pathways altered in DR, as well as metabolic differences between patients with NPDR and PDR. Determining differences in these metabolic profiles could reveal molecular mechanisms of DR and PDR, facilitating the identification of new therapeutic targets.
Methods
Ethics Statement
This clinical case-control study was approved by the Vanderbilt University Human Research Protection Program. Research adhered to the tenets of the Declaration of Helsinki and was conducted in accordance with Health Insurance Portability and Accountability Act regulations. Written informed consent was obtained from all participants prior to study enrollment.
Study Participants
A total of 173 patients were recruited from the Retina Service of the Vanderbilt Eye Institute. All patients enrolled had a diagnosis of type 2 diabetes made by their primary care provider or endocrinologist and had been prescribed at least one diabetes medication. The diabetic controls (n = 90) included patients with a diagnosis of type 2 diabetes for at least 5 years and no clinical signs of DR as determined by dilated fundus examination by a retina specialist. DR cases (n = 83) were patients with type 2 diabetes and DR diagnosed on dilated fundus examination by a retina specialist. Presence of DR was confirmed and documented by color fundus photography (Zeiss 450+ fundus camera; Carl Zeiss Meditec, Dublin, CA, USA), and classified as NPDR or PDR. A diagnosis of NPDR was based on the presence of blot hemorrhages, microaneurysms, cotton-wool spots, or intraretinal microvascular abnormalities and no evidence of active PDR or history of treatment for PDR. A PDR diagnosis was based on the presence of neovascularization of the iris or retina or clinical evidence or documentation of treatment for PDR. Exclusion criteria for both cases and controls included nondiabetic retinopathy or retinal degeneration, glaucoma, and active ocular inflammation.
After obtaining written, informed consent, blood was drawn from each participant using a 21- or 23-G butterfly needle into 3-mL blood collection tubes containing 56 USP units of lithium heparin. These tubes were centrifuged for 10 minutes at 4°C for blood fractionation, and plasma was subsequently aliquoted in 1.5-mL conical tubes and immediately stored at −80°C.
At the time of the blood draw, patients were asked a standardized set of questions about disease history. Past medical history, comorbidity data, creatinine values, and hemoglobin A1c (HbA1c) measurements were obtained from each participant's electronic medical record (taken from the date closest to blood draw). Diagnoses of hypertension, coronary artery disease, and dyslipidemia were based on clinical notes from the primary care physician or cardiologist.
High-Resolution Untargeted Metabolomics Analysis
Plasma samples were thawed and analyzed by LC-MS at Emory University as previously described.8–10,15,16 Samples were randomized into batches of 20 prior to analysis. Pooled reference plasma was run prior to and after each batch for quality control and assurance. Plasma sample aliquots (65 μL) were treated with 130 μL acetonitrile (2:1 vol/vol) containing 3.5 μL of an internal isotopic standard mix,10,15,17 placed on ice for 30 minutes, and centrifuged for 10 minutes (16,100g at 4°C) to remove protein. The supernatants were loaded onto a Shimadzu autosampler maintained at 4°C and analyzed in triplicate using a Thermo LTQ Velos Orbitrap (Thermo Scientific, San Jose, CA, USA) and C18 column chromatography. Elution was obtained with a formic acid/acetonitrile gradient at a flow rate of 0.35 mL/min for the initial 6 minutes and 0.5 mL/min for the remaining 4 minutes. The first 2-minute period consisted of 5% solution A (2% [vol/vol] formic acid in water), 60% water, 35% acetonitrile, and the final 4-minute period was maintained at 5% solution A, 95% acetonitrile. The mass spectrometer was set to collect mass-to-charge ratio (m/z) from 85 to 2000 over 10 minutes at 60,000 mass resolution. Electrospray ionization was used in positive mode for detection.9,10,15,17
Peak Detection and Annotation
An adaptive processing software package, apLCMS v5.9.8 (in the public domain, http://web1.sph.emory.edu/apLCMS/), designed for use with high-resolution mass spectrometry data, was used for noise removal as well as for feature extraction, alignment, and quantification.18 With apLCMS, each metabolic feature is defined by a unique combination of m/z and retention time. To enhance the feature detection process and perform quality evaluation, systematic data re-extraction and statistical filtering were performed using xMSanalyzer v2.0.8 (in the public domain, http://sourceforge.net/projects/xmsanalyzer/).19 Each sample was analyzed in triplicate, and coefficient of variation (CV) was used to evaluate the quality of all features. Pearson correlation within technical replicates was used to evaluate the quality of samples.
Batch-effect correction was performed using ComBat.20 Computational annotation of the features to find database matches in The Human Metabolome Database (HMDB) version 3.521 was performed using the xMSannotator v1.3.2 package in R (R Project for Statistical Computing, Vienna Austria). This package employs a multilevel clustering procedure based on intensity across all samples, retention time, mass defect, and isotope/adduct patterns.22 Additionally, xMSannotator uses metabolic pathway associations to assign confidence levels for database matches. Confidence levels range from zero to three, designating annotations from no confidence to high confidence. This reduces the risk of false annotations and allows prioritization of computationally derived annotations for further experimental evaluation and confirmation.19 Additionally, an in-house database of previously confirmed metabolites based on comparison of adduct, m/z, retention time, and liquid chromatography-tandem mass spectrometry (LC-MS/MS) spectra to authentic standards or database spectra was used for metabolite identity confirmation.23–25 Metabolite identification levels were assigned based on the Metabolomics Standards Initiative26 criteria as follows: (1) confirmation using tandem mass spectrometry (MS/MS) and co-elution with authentic standards (level 1); and (2) confirmation by comparing experimental MS/MS spectra with MS/MS spectra in mzCloud or in silico predicted spectra retrieved from MetFrag (level 2).
Liquid Chromatography/Electrospray Ionization Tandem Mass Spectrometry
Metabolites identified in the pathway analysis and Wilcoxon rank-sum test, P < 0.05, were fragmented under collision-induced dissociation (CID) for further identification. Samples were analyzed using a Thermo LTQ Velos Orbitrap high-resolution (60,000 mass resolution) mass spectrometer (Thermo Fisher Scientific, San Diego, CA, USA) operated in positive ion mode with 10-minute C18 reversed-phase column chromatography and standard source conditions used for the untargeted metabolic profiling. Prior to analysis, plasma proteins were precipitated using acetonitrile:water (2:1 vol/vol) and allowed to sit on ice for 30 minutes. The supernatant was then carefully pipetted for MS/MS analysis. CID was accomplished using high purity N2 at a normalized collision energy of 35%. The MS/MS data were processed using the xcmsSet and xcmsFragments functions in XCMS to extract the MS/MS fragments associated with each parent mass,27–29 and the experimental spectra were compared with in silico fragmentation using MetFrag30 or the spectra available from mzCloud (in the public domain, https://www.mzcloud.org/).
Statistical and Bioinformatics Analysis
Descriptive statistics for demographic and clinical variables were calculated for the study population. Comparisons between DR patients and diabetic controls, as well as between PDR and NPDR patients, were made using a two-tailed t-test for continuous data (e.g., age, HbA1c, and diabetes duration) and the X2 test for categoric data (e.g., sex and presence of comorbidities). A Wilcoxon rank sum test was performed to compare creatinine values between the respective groups as these values were not normally distributed within the groups.
The metabolomics data were preprocessed and filtered prior to statistical analysis. A log2 transformation was applied to reduce heteroscedasticity and normalize results. Quantile normalization was performed to reduce between-sample variability.31 To increase confidence for selection of discriminating metabolites, data were filtered to include only those features present in at least 50% of all samples and present in at least 90% of either DR cases or diabetic controls. Similar criteria were used in comparing NPDR patients and PDR patients such that only features with nonmissing values present in at least 50% of all samples and in at least 90% of either NPDR or PDR samples were included for further analysis. Metabolic features that discriminate between the comparison groups were identified using the variable importance for projection (VIP) greater than or equal to 2 criterion based on partial least squares discriminant analysis (PLS-DA) implemented in the R package mixOmics.32 Features with VIP greater than one are generally considered important.33,34 In this study, a more stringent threshold of VIP ≥ 2 was used to prioritize selection of highly discriminatory features between DR patients and diabetic controls, and between PDR and NPDR patients. Ten-fold cross-validation method was used for evaluation of the selected features using a support vector machine classifier implemented in R package e1071.35 One-way hierarchical clustering analysis was performed to visualize the clustering pattern of discriminatory features using a Spearman rank correlation-based dissimilarity measure and the complete agglomeration method implemented in hclust() function in R.
Following feature selection, the fold change between the comparison groups was calculated for each of the discriminating features. In addition, multiple linear regressions were used to test the association between selected discriminatory features and disease status (DR versus diabetic control; PDR versus NPDR) using metabolite intensity as the dependent variable and adjusting for potential confounding variables including age, sex, HbA1c, and diabetes duration. All statistical analyses were performed in R.
Pathway Analysis
Discriminating features with VIP ≥ 1.5 were used to perform pathway analysis via Mummichog 2.0 (in the public domain, http://mummichog.org/), a program designed for untargeted metabolomics that combines metabolite annotation and metabolic pathway/network analysis.36,37 As Mummichog evaluates pathway level enrichment, a less stringent VIP cutoff was used to generate the input list of discriminatory features to prevent information loss and enhance the coverage of metabolites within individual pathways.33,38 Only significant pathways with an overlap size of three or greater are reported in this study. Individual features with VIP ≥ 1.5 and matches to [M+H]+ forms of metabolites from the top enriched pathways were further evaluated using Wilcoxon rank sum tests and MS/MS.
Results
Study Population
The study population consisted of 173 patients with type 2 diabetes, including 83 DR patients and 90 diabetic controls. The patient groups did not significantly differ in age or sex. Compared with diabetic controls, DR patients had a longer mean diabetes duration (P = 6.8 × 10−10) and a higher mean HbA1c (P = 0.012) (Table 1). The two groups did not differ significantly in rates of coronary artery disease, hypertension, dyslipidemia, or creatinine levels. When comparing PDR patients (n = 34) with NPDR patients (n = 49), no differences were observed in demographic or clinical characteristics (Table 1).
Table 1.
Study Population Characteristics
|
Characteristic |
Diabetic Controls (n
= 90) |
DR Patients (n
= 83) |
P
Value |
NPDR (n
= 49) |
PDR (n
= 34) |
P
Value |
| Age, y | 59.7 ± 10.0 | 57.9 ± 11.1 | 0.37 | 59.4 ± 11.3 | 55.7 ± 10.9 | 0.10 |
| Males, % | 52.3 | 63.2 | 0.21 | 61.4 | 65.6 | 0.89 |
| Diabetes duration, y | 11.0 ± 5.1 | 19.0 ± 9.0 | 6.8 × 10−10 | 17.7 ± 7.6 | 20.9 ± 10.6 | 0.28 |
| HbA1c, % | 7.9 ± 1.6 | 8.5 ± 1.9 | 0.012 | 8.6 ± 1.8 | 8.5 ± 2.1 | 0.97 |
| Creatinine, mg/dL | 0.86 ± 0.29 | 0.92 ± 0.46 | 0.13 | 0.92 ± 0.22 | 0.92 ± 0.64 | 0.88 |
| CAD, % | 25.6 | 22.9 | 0.82 | 22.4 | 23.5 | 1.0 |
| HTN, % | 76.7 | 73.5 | 0.76 | 79.6 | 64.7 | 0.21 |
| DLD, % | 74.4 | 61.5 | 0.10 | 67.4 | 52.9 | 0.27 |
Study groups were compared in terms of demographics and comorbidities. For age, diabetes duration, and HbA1c, the mean and standard deviations are presented and comparisons were made by t-test. For creatinine levels, the median and interquartile range are presented, and comparisons were made by Wilcoxon rank sum test. Sex and rates of comorbidities were compared by X2 test. HbA1c and creatinine levels taken from date closest to date of blood draw. CAD, coronary artery disease; HTN, hypertension; DLD, dyslipidemia.
DM Versus DR
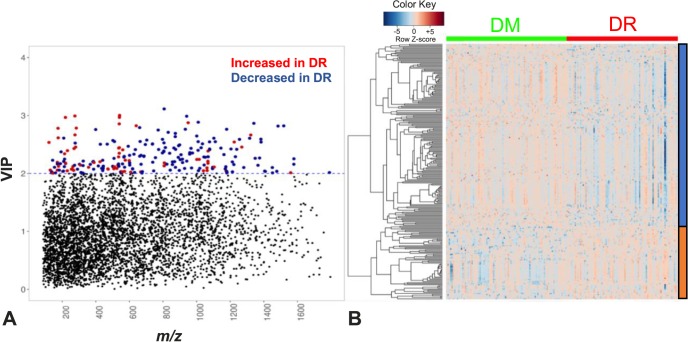
Mass spectral data generated via LC-MS were extracted and filtered to yield 10,306 features defined by m/z, retention time, and ion intensity. A metabolome-wide association study (MWAS) was performed to determine which metabolic features differed between DR patients and diabetic controls. Based on PLS-DA, 236 differed with VIP ≥ 2 (Fig. 1A). A 10-fold, cross-validation classification accuracy of 88.5% was achieved using the 236 discriminatory features. Of these, 167 were increased and 69 were decreased in the plasma of DR patients compared with diabetic controls. These features clustered into 33 subclusters identified using hierarchical clustering analysis (Fig. 1B).
Figure 1.
Metabolic features different between DR patients and diabetic controls. (A) Manhattan plot of the VIP and m/z of 10,306 features shows a total of 236 features were significantly different between DR patients (n = 83) and diabetic controls (n = 90) by PLS-DA using a VIP ≥ 2.0 (threshold indicated by blue dashed horizontal line). Significant metabolic features increased (red dots) or decreased (blue dots) in DR patients compared with diabetic controls are indicated. (B) One-way hierarchical clustering based on the intensity of significant metabolite features selected by PLS-DA (VIP ≥ 2.0, 236 m/z features) identified clusters of features that were increased (orange) or decreased (blue) in DR patients.
To account for potential confounding effects of demographic or clinical differences between DR patients and diabetic controls, we performed linear regression analyses for each of the 236 discriminatory features with metabolite intensity as the dependent variable and DR status, age, sex, diabetes duration, and HbA1c levels as independent variables in the model. Regressions were performed in the 162 patients (93.6%) with all available clinical data (76 DR patients, 86 diabetic controls). A total of 126 metabolic features differed between DR patients and diabetic controls by both PLS-DA (VIP ≥ 2) and adjusted linear regression analysis (P < 0.05). Eighteen of 126 discriminating features were assigned tentative annotations at a medium or high confidence level using xMSannotator22 (Supplemental Table S1). These features include multiple adducts of arginine, citrulline, and acylcarnitines.
We performed pathway analysis using the 832 features discriminating between DR patients and diabetic controls with VIP ≥ 1.5. We used VIP ≥ 1.5 in order to enhance the coverage of metabolites in individual pathways and reduce information loss.38 This demonstrated enrichment of nine metabolic pathways (Table 2), five of which (metabolism of alanine and aspartate, arginine and proline, aspartate and asparagine, niacin, and urea cycle/amino group) are related to arginine metabolism. The other four pathways included pyrimidine, leukotriene, purine, and lysine metabolism.
Table 2.
Pathways Altered in DR Patients Compared With Diabetic Controls
|
Pathway |
Overlapping Features |
Pathway Size |
P
Value |
| Niacin metabolism | 4 | 5 | 2.3 × 10−4 |
| Alanine and aspartate metabolism | 4 | 6 | 9.1 × 10−4 |
| Arginine and proline metabolism | 6 | 14 | 1.4 × 10−3 |
| Aspartate and asparagine metabolism | 6 | 16 | 2.9 × 10−3 |
| Pyrimidine metabolism | 4 | 8 | 3.3 × 10−3 |
| Leukotriene metabolism | 4 | 9 | 5.8 × 10−3 |
| Purine metabolism | 3 | 8 | 0.029 |
| Urea cycle/amino group metabolism | 5 | 20 | 0.042 |
| Lysine metabolism | 3 | 10 | 0.047 |
Pathway analysis was performed using Mummichog 2.0 on the 832 features discriminating between DR patients and diabetic controls identified by PLS-DA with a VIP ≥ 1.5. Overlapping features represents the number of metabolites enriched in the pathway, while pathway size describes the total number of metabolites in each pathway.
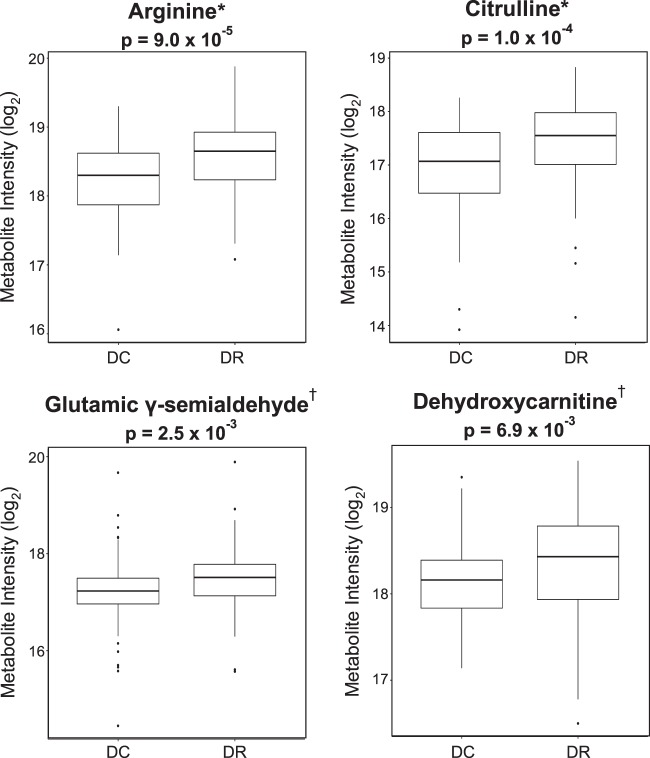
To prioritize features for targeted evaluation using MS/MS and the in-house metabolite library,23,24 we focused on features with VIP ≥ 1.5 in the top pathways that both corresponded to [M+H]+ forms of metabolites and were significant (P < 0.05) in a Wilcoxon rank sum test comparing the plasma levels of these metabolites in DR patients and diabetic controls. Those metabolites confirmed with Metabolomics Standards Initiative (MSI)26 level 1 or 2 identification included arginine, citrulline, glutamic γ-semialdehyde, and dehydroxycarnitine (Fig. 2).
Figure 2.
Plasma levels of arginine, citrulline, dehydroxycarnitine, and glutamic γ-semialdehyde are elevated in DR patients. Metabolites enriched in the pathway analyses were further analyzed with a Wilcoxon rank sum test and using LC-MS/MS, revealing arginine, citrulline, dehydroxycarnitine, and glutamic γ-semialdehyde levels are significantly increased in DR patients compared with diabetic controls. DC, diabetic controls. *Confidence level 1 by MSI standards, identities verified via LC-MS/MS with authentic standards. †Confidence level 2 by MSI standards, putative annotation based on MS/MS spectral matching using MetFrag and mzCloud.
NPDR Versus PDR
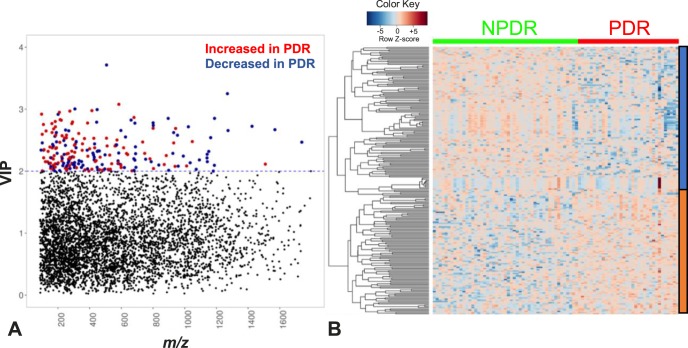
To determine metabolic features that differ between PDR and NPDR, we performed an MWAS comparing PDR patients (n = 34) with NPDR patients (n = 49). Of 10,306 metabolic features that met quality control and filtering criteria, 219 features that distinguish NPDR and PDR groups were identified based on PLS-DA with a VIP ≥ 2.0 (Fig. 3A). A 10-fold cross-validation classification accuracy of 91.7% was obtained using these 219 discriminatory features. Of these discriminating features, 108 were increased and 111 were decreased in the plasma of PDR patients compared with NPDR patients. These features clustered into 81 subclusters identified using hierarchic clustering analysis (Fig. 3B).
Figure 3.
Metabolic features that differ between NPDR and PDR patients. (A) Manhattan plot of the VIP and m/z of 10,306 features showing a total of 219 features were significantly different between PDR patients (n = 34) and NPDR patients (n = 49) using a VIP ≥ 2.0 (threshold indicated by the blue dashed horizontal line). Significant metabolic features increased (red dots) or decreased (blue dots) in PDR compared with NPDR patients are indicated. (B) One-way hierarchical clustering based on the intensity of significant metabolite features selected by PLS-DA (VIP ≥ 2.0, 219 m/z features) identified clusters of features that were increased (orange) or decreased (blue) in PDR patients.
To account for potential confounding effects of demographic or clinical differences between NPDR and PDR patients, we performed linear regression analyses for each of the 219 discriminatory features with metabolite intensity as the dependent variable and PDR status, age, sex, diabetes duration, and HbA1c as independent variables in the model. Regressions were performed in the 76 DR patients (91.6%) with complete clinical data (44 NPDR, 32 PDR). A total of 151 metabolic features differed between NPDR patients and PDR patients by both PLS-DA (VIP ≥ 2) and adjusted linear regression analysis (P < 0.05). Using xMSannotator, 46 of 151 discriminating features were matched to known metabolites with medium or high confidence (Supplemental Table S2). These features included matches to multiple carnitine adducts, acylcarnitines, and fatty acids.
Pathway analysis using the 766 features with VIP ≥ 1.5 identified enrichment in β-oxidation of saturated fatty acids, fatty acid metabolism, and vitamin D3 metabolism (Table 3). A Wilcoxon rank sum test was performed on the metabolites that contributed to the enrichment of these pathways to prioritize metabolites for further mass spectral analysis. The identity of carnitine, elevated in PDR patients compared with NPDR patients, was confirmed via LC-MS/MS (MSI level 1) (Fig. 4).
Table 3.
Pathways Altered in PDR Patients Compared With NPDR Patients
|
Pathway |
Overlapping Features |
Pathway Size |
P
Value |
| Saturated fatty acids β-oxidation | 3 | 6 | 0.032 |
| Fatty acid metabolism | 4 | 10 | 0.038 |
| Vitamin D3 metabolism | 3 | 7 | 0.048 |
Pathway analysis was performed using Mummichog 2.0 on the 766 features identified by PLS-DA with a VIP ≥ 1.5. Overlapping features represents the number of metabolites enriched in the pathway, while pathway size describes the total number of metabolites in each pathway.
Figure 4.
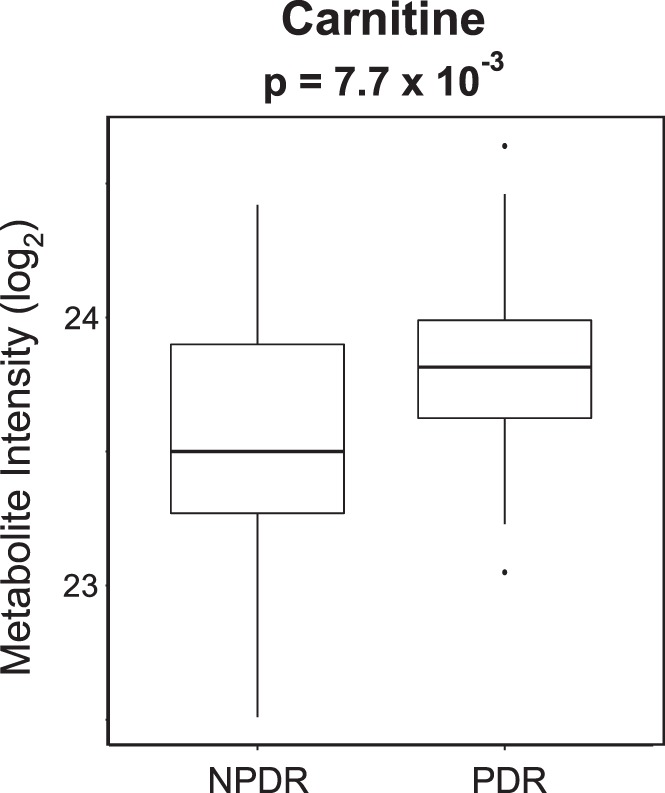
Plasma levels of carnitine are elevated in PDR patients. Plasma levels of carnitine are significantly increased in PDR compared with NPDR patients. Comparison was made using Wilcoxon rank sum test with P value shown. The identity of carnitine was verified via LC-MS/MS with authentic standards (MSI level 1).
Discussion
This MWAS used untargeted high-resolution LC-MS to identify the metabolites and metabolic pathways altered in DR. After adjusting for potential cofounders, we discovered 126 metabolic features that distinguish DR patients and diabetic controls. Pathway analysis using discriminating features revealed alterations in multiple pathways, including the metabolism of multiple amino acids, leukotrienes, niacin, pyrimidine, and purine. Arginine, citrulline, glutamic γ-semialdehyde, and dehydroxycarnitine were key contributors to the differences in the metabolic pathways identified, and their identities were confirmed by LC-MS/MS. We also identified 151 features that distinguished PDR from NPDR patients after adjusting for age, sex, diabetes duration, and HbA1c. Pathway analysis using discriminating features revealed alterations in the β-oxidation of saturated fatty acids, fatty acid metabolism, and vitamin D3 metabolism. Carnitine, a key molecule in oxidation and metabolism of fatty acids, was elevated in PDR patients compared with NPDR patients.
The relationship between arginine metabolism and DR is complex, as arginine is a key molecule in several overlapping pathways with numerous metabolic fates in the body. In the urea cycle, arginine is cleaved by arginase to form ornithine and urea. Ornithine is then converted back to arginine through the intermediate citrulline, completing the cycle. Alternatively, ornithine may also be metabolized to proline through the intermediate glutamic γ-semialdehyde.39 Increased levels of proline, ornithine, citrulline, and arginine have been found in the vitreous of patients with PDR,11 and elevations in serum arginine were discovered in patients with severe DR.40 Our study identified increased plasma levels of arginine, citrulline, and glutamic γ-semialdehyde in patients with DR, consistent with these prior studies. Studies in mouse models of DR and in bovine retinal endothelial cells cultured in high glucose found elevated arginase activity, also implicating derangements in arginine metabolism as a mediator of DR.41,42 Our results provide further evidence that alterations in urea cycle metabolites, particularly arginine and citrulline, are associated with DR.
Arginine also serves as a substrate for the enzyme nitric oxide synthase, which catalyzes a reaction that produces citrulline and nitric oxide (NO), a vasodilator that plays a critical role in maintaining the health of the vascular endothelium. NO synthase is inhibited by asymmetric dimethylarginine (ADMA), which has been reported to be elevated in plasma,43 serum,40 and aqueous humor44 of patients with DR. Given our current findings and these previous studies, it is possible that dysregulated arginine metabolism may be linked not only through urea cycle metabolites, but also through ADMA and NO.
This study also identified increased levels of an acylcarnitine, dehydroxycarnitine, in DR patients compared with diabetic controls, and increased plasma carnitine in patients with PDR compared with NPDR. Carnitine is essential for the transport of long chain fatty acids into mitochondria via acylcarnitine intermediates prior to β-oxidation. Carnitine has been shown in some studies to be decreased in type 2 diabetes and has even been suggested as a potential adjuvant in the treatment of diabetes.45 A recent review by Bene et al.45 describes studies that report decreased carnitine levels in type 2 diabetes and in various diabetic complications, but also cites other studies in which no relationship between carnitine and diabetic complications was seen.46–48 Our results are consistent with a previous study, which compared PDR patients with nondiabetic controls and found elevated acylcarnitine levels in vitreous samples.11 It is likely that differences in study design and clinical phenotyping, as well as small sample sizes in some studies, contribute to the inconsistencies thus far in the literature regarding carnitine metabolites and DR. Of note, our lab recently demonstrated that the carnitine shuttle pathway is altered in NVAMD.9 Like PDR, NVAMD is characterized by new vascular growth. Given that evidence demonstrates carnitine metabolites are elevated in both PDR and NVAMD, it is possible that changes in fatty acid metabolism are related to ischemia or neovascularization. A more detailed investigation is necessary to further characterize the relationship between carnitine metabolites and PDR.
The large cohort of DR patients and diabetic controls enrolled from the same institution with standardized sample collection and processing are strengths of the study. However, because most patients were Caucasian and all from the same geographic region, the generalizability of these results is not yet certain. Untargeted high-resolution LC-MS is a sensitive technique providing broad coverage of endogenous, environmental, and dietary metabolites. This sensitivity increases the chances of identifying metabolites that are important in DR pathology, but it is important in untargeted studies to account for potential confounding factors and to provide confirmation of metabolite identity. For this analysis, we confirmed that cases and controls were similar in rates of critical systemic conditions. However, these comorbidity assignments were based on clinical notes and treatment history, and it is possible that there may have been undiagnosed conditions in patients included in the study. We performed linear regressions to ensure that discriminating metabolites differed between cases and controls when adjusting for potential confounding variables. We also used LC-MS/MS to confirm the molecular identity of the most critical discriminating features. Additionally, this metabolomics study is the first to specifically compare the metabolic profiles of PDR and NPDR patients.
In summary, this metabolomics analysis demonstrates that arginine metabolism is dysregulated in DR and that fatty acid metabolism is altered in PDR. Further studies are necessary to determine how these pathways are related to DR and the more advanced PDR. Characterizing these relationships may lead to the identification of novel therapeutic targets for this vision-threatening disease.
Supplementary Material
Acknowledgments
The authors thank Vi Linh Tran for her technical assistance.
Supported by National Institutes of Health (Bethesda, MD, USA) Grants R01 EY022618, P30 EY008126, P30 ES019776, U2C ES030163, and S10 OD018006, a grant from the International Retinal Research Foundation (Birmingham, AL, USA), and an unrestricted departmental grant to Vanderbilt University Medical Center from Research to Prevent Blindness (New York, NY, USA).
Disclosure: K. Sumarriva, None; K. Uppal, None; C. Ma, None; D.J. Herren, None; Y. Wang, None; I.M. Chocron, None; C. Warden, None; S.L. Mitchell, None; L.G. Burgess, None; M.P. Goodale, None; M.P. Osborn, None; A.J. Ferreira, None; J.C. Law, None; E.F. Cherney, None; D.P. Jones, None; M.A. Brantley Jr, None
References
- 1.Yau JWY, Rogers SL, Kawasaki R, et al. Global prevalence and major risk factors of diabetic retinopathy. Diabetes Care. 2012;35:556–564. doi: 10.2337/dc11-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guariguata L, Whiting DR, Hambleton I, Beagley J, Linnenkamp U, Shaw JE. Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res Clin Pract. 2014;103:137–149. doi: 10.1016/j.diabres.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Sinclair AJ, Bayer AJ, Girling AJ, Woodhouse KW. Older adults, diabetes mellitus and visual acuity: a community-based case-control study. Age Ageing. 2000;29:335–339. doi: 10.1093/ageing/29.4.335. [DOI] [PubMed] [Google Scholar]
- 4.Antonetti DA, Klein R, Gardner TW. Diabetic retinopathy. N Engl J Med. 2012;366:1227–1239. doi: 10.1056/NEJMra1005073. [DOI] [PubMed] [Google Scholar]
- 5.Cheung N, Mitchell P, Wong TY. Diabetic retinopathy. Lancet. 2010;376:124–136. doi: 10.1016/S0140-6736(09)62124-3. [DOI] [PubMed] [Google Scholar]
- 6.UK Prospective Diabetes Study (UKPDS) Group. Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33) Lancet. 1998;352:837–853. [PubMed] [Google Scholar]
- 7.Walker DI, Go Y-M, Liu K, Pennell KD, Jones DP. Population screening for biological and environmental properties of the human metabolic phenotype: implications for personalized medicine. In: Holmes E, Nicholson J, editors. Metabolic Phenotyping in Personalized and Public Healthcare. Boston, MA: Academic Press;; 2016. pp. 167–211. [Google Scholar]
- 8.Osborn MP, Park Y, Parks MB, et al. Metabolome-wide association study of neovascular age-related macular degeneration. PLoS One. 2013;8:e72737. doi: 10.1371/journal.pone.0072737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell SL, Uppal K, Williamson SM, et al. The carnitine shuttle pathway is altered in patients with neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2018;59:4978–4985. doi: 10.1167/iovs.18-25137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burgess LG, Uppal K, Walker DI, et al. Metabolome-wide association study of primary open angle glaucoma. Invest Ophthalmol Vis Sci. 2015;56:5020–5028. doi: 10.1167/iovs.15-16702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paris LP, Johnson CH, Aguilar E, et al. Global metabolomics reveals metabolic dysregulation in ischemic retinopathy. Metabolomics. 2016;12:15. doi: 10.1007/s11306-015-0877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barba I, Garcia-Ramírez M, Hernández C, et al. Metabolic fingerprints of proliferative diabetic retinopathy: an 1H-NMR-based metabonomic approach using vitreous humor. Invest Ophthalmol Vis Sci. 2010;51:4416–4421. doi: 10.1167/iovs.10-5348. [DOI] [PubMed] [Google Scholar]
- 13.Chen L, Cheng C-Y, Choi H, et al. Plasma metabonomic profiling of diabetic retinopathy. Diabetes. 2016;65:1099–1108. doi: 10.2337/db15-0661. [DOI] [PubMed] [Google Scholar]
- 14.Rhee SY, Jung ES, Park HM, et al. Plasma glutamine and glutamic acid are potential biomarkers for predicting diabetic retinopathy. Metabolomics. 2018;14:89. doi: 10.1007/s11306-018-1383-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soltow QA, Strobel FH, Mansfield KG, Wachtman L, Park Y, Jones DP. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics Off J Metabolomic Soc. 2013;9:S132–S143. doi: 10.1007/s11306-011-0332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson JM, Yu T, Strobel FH, Jones DP. A practical approach to detect unique metabolic patterns for personalized medicine. Analyst. 2010;135:2864–2870. doi: 10.1039/c0an00333f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frediani JK, Jones DP, Tukvadze N, et al. Plasma metabolomics in human pulmonary tuberculosis disease: a pilot study. PLoS One. 2014;9:e108854. doi: 10.1371/journal.pone.0108854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu T, Park Y, Johnson JM, Jones DP. apLCMS–adaptive processing of high-resolution LC/MS data. Bioinforma Oxf Engl. 2009;25:1930–1936. doi: 10.1093/bioinformatics/btp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uppal K, Soltow QA, Strobel FH, et al. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics. 2013;14:15. doi: 10.1186/1471-2105-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostat Oxf Engl. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 21.Wishart DS, Jewison T, Guo AC, et al. HMDB 3.0–The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uppal K, Walker DI, Jones DP. xMSannotator: an R package for network-based annotation of high-resolution metabolomics data. Anal Chem. 2017;89:1063–1067. doi: 10.1021/acs.analchem.6b01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Go Y-M, Walker DI, Liang Y, et al. Reference standardization for mass spectrometry and high-resolution metabolomics applications to exposome research. Toxicol Sci Off J Soc Toxicol. 2015;148:531–543. doi: 10.1093/toxsci/kfv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Go Y-M, Liang Y, Uppal K, et al. Metabolic characterization of the common marmoset (Callithrix jacchus) PLoS One. 2015;10:e0142916. doi: 10.1371/journal.pone.0142916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Accardi CJ, Walker DI, Uppal K, et al. High-resolution metabolomics for nutrition and health assessment of armed forces personnel. J Occup Environ Med. 2016;58:S80–S88. doi: 10.1097/JOM.0000000000000770. [DOI] [PubMed] [Google Scholar]
- 26.Sumner LW, Amberg A, Barrett D, et al. Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 28.Tautenhahn R, Böttcher C, Neumann S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics. 2008;9:504. doi: 10.1186/1471-2105-9-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benton HP, Want EJ, Ebbels TMD. Correction of mass calibration gaps in liquid chromatography-mass spectrometry metabolomics data. Bioinforma Oxf Engl. 2010;26:2488–2489. doi: 10.1093/bioinformatics/btq441. [DOI] [PubMed] [Google Scholar]
- 30.Ruttkies C, Schymanski EL, Wolf S, Hollender J, Neumann S. MetFrag relaunched: incorporating strategies beyond in silico fragmentation. J Cheminformatics. 2016;8:3. doi: 10.1186/s13321-016-0115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel RM, Roback JD, Uppal K, Yu T, Jones DP, Josephson CD. Metabolomics profile comparisons of irradiated and nonirradiated stored donor red blood cells. Transfusion (Paris) 2015;55:544–552. doi: 10.1111/trf.12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohart F, Gautier B, Singh A, Cao K-AL. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13:e1005752. doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uppal K, Salinas JL, Monteiro WM, et al. Plasma metabolomics reveals membrane lipids, aspartate/asparagine and nucleotide metabolism pathway differences associated with chloroquine resistance in Plasmodium vivax malaria. PLoS One. 2017;12:e0182819. doi: 10.1371/journal.pone.0182819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikström C, Wold S. Multi- and Megavariate Data Analysis: Part II: Advanced Applications and Method Extensions. Umea: Umetrics Inc;; 2006. [Google Scholar]
- 35.Vapnik V. The Nature of Statistical Learning Theory 2nd ed. New York: Springer-Verlag;; 2000. [Google Scholar]
- 36.Li S, Park Y, Duraisingham S, et al. Predicting network activity from high throughput metabolomics. PLoS Comput Biol. 2013;9:e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S, Sullivan NL, Rouphael N, et al. Metabolic phenotypes of response to vaccination in humans. Cell. 2017;169:862–877. doi: 10.1016/j.cell.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol. 2012;8:e1002375. doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris SM. Arginine metabolism: boundaries of our knowledge. J Nutr. 2007;137:1602S–1609S. doi: 10.1093/jn/137.6.1602S. [DOI] [PubMed] [Google Scholar]
- 40.Abhary S, Kasmeridis N, Burdon KP, et al. Diabetic retinopathy is associated with elevated serum asymmetric and symmetric dimethylarginines. Diabetes Care. 2009;32:2084–2086. doi: 10.2337/dc09-0816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narayanan SP, Rojas M, Suwanpradid J, Toque HA, Caldwell RW, Caldwell RB. Arginase in retinopathy. Prog Retin Eye Res. 2013;36:260–280. doi: 10.1016/j.preteyeres.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel C, Rojas M, Narayanan SP, et al. Arginase as a mediator of diabetic retinopathy. Front Immunol. 2013;4:173. doi: 10.3389/fimmu.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malecki MT, Undas A, Cyganek K, et al. Plasma asymmetric dimethylarginine (ADMA) is associated with retinopathy in type 2 diabetes. Diabetes Care. 2007;30:2899–2901. doi: 10.2337/dc07-1138. [DOI] [PubMed] [Google Scholar]
- 44.Sugai M, Ohta A, Ogata Y, et al. Asymmetric dimethylarginine (ADMA) in the aqueous humor of diabetic patients. Endocr J. 2007;54:303–309. doi: 10.1507/endocrj.k06-140. [DOI] [PubMed] [Google Scholar]
- 45.Bene J, Hadzsiev K, Melegh B. Role of carnitine and its derivatives in the development and management of type 2 diabetes. Nutr Diabetes. 2018;8:8. doi: 10.1038/s41387-018-0017-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poorabbas A, Fallah F, Bagdadchi J, et al. Determination of free L-carnitine levels in type II diabetic women with and without complications. Eur J Clin Nutr. 2007;61:892–895. doi: 10.1038/sj.ejcn.1602594. [DOI] [PubMed] [Google Scholar]
- 47.Tamamoğullari N, Siliğ Y, İçağasioğlu S, Atalay A. Carnitine deficiency in diabetes mellitus complications. J Diabetes Complications. 1999;13:251–253. doi: 10.1016/s1056-8727(99)00052-5. [DOI] [PubMed] [Google Scholar]
- 48.Liepinsh E, Skapare E, Vavers E, et al. High l-carnitine concentrations do not prevent late diabetic complications in type 1 and 2 diabetic patients. Nutr Res. 2012;32:320–327. doi: 10.1016/j.nutres.2012.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.