Abstract
The GAIT (gamma-interferon-activated inhibitor of translation) complex or miR-297-RISC (RNA-induced silencing complex), together with hnRNP L or hnRNP L-bearing complex, operates an RNA switch in myeloid cells that regulates stress-dependent expression of vascular endothelial growth factor-A (VEGFA). Here, we have shown that hnRNP L directs multiple hypoxia-inducible RNA switches simultaneously and regulates expression of these oncogenic genes in addition to VEGFA. Bioinformatic and polysome profiling-microarray screens have identified DNM1L (Dynamin 1-like) and PHF21A (PHD finger protein 21A) mRNAs as regulated at the translational level by GAIT-dependent, hnRNP L-directed RNA switches. We have also uncovered CDK6 (Cyclin dependent kinase 6), MKLN1 (Muskelin 1) and EIF5 (Eukaryotic initiation factor 5) as novel miR-297-dependent, hnRNP L-directed RNA switch transcripts. Src Kinase is required for the phosphorylation of hnRNP L and activation of the RNA switch pathway. Knockdown of hnRNP L sensitizes the human U937 monocytic cells under hypoxia stress but not in normoxia via inducing cell apoptosis partially due to the reduced translation of hnRNP L target mRNAs. Collectively, our findings suggest that commonly controlled genes by the hnRNP L-directed RNA switches form a translational regulon that promotes hypoxia resistance and cell survival.
Keywords: EPRS, hnRNP L, hypoxia, microRNA, RNA switch, translation
1. Introduction
Human protein-directed RNA switch features mutually exclusive interactions of two RNA binding proteins (RBPs) or ribonucleoprotein (RNP) complexes with two adjacent RNA elements, resulting in altered gene expression [1]. Our previous studies have shown that vascular endothelial growth factor-A (VEGFA) RNA switch is a founding member of stress-responsive RNA switches that respond to inflammation and hypoxia in human myeloid cells [1, 2]. However, it remains unclear whether the RNA switch pathway is more broadly present in human transcriptome and contributes to tumor cell survival.
Within the avascular cores of tumors, monocytes/Mϕ are simultaneously exposed to both inflammatory cytokines (such as IFN-γ) and hypoxia. We have shown that VEGFA expression in myeloid cells is translationally repressed by the GAIT (Gamma-Interferon Activated Inhibitor of Translation) pathway [3] (Fig. 1A, top). Treatment of human monocytic cells with IFN-γ induces the synthesis of VEGFA mRNA and protein for ~12-16 hr. However, the VEGFA mRNA translation is suppressed about 16 hr after IFN-γ treatment despite the presence of abundant VEGFA mRNA [3]. Translational silencing of VEGFA and other GAIT targets requires binding of the GAIT complex to its cognate GAIT element in the target mRNA 3'-untranslated region (UTR) [3]. The GAIT element is a defined 29-nt stem-loop with an internal bulge and specific sequence and structural features [4]. The GAIT complex is heterotetrameric bearing glutamyl-prolyl-tRNA synthetase (EPRS), ribosomal protein L13a, NS1-associated protein-1, and glyceraldehyde 3-phosphate dehydrogenase [3, 4].
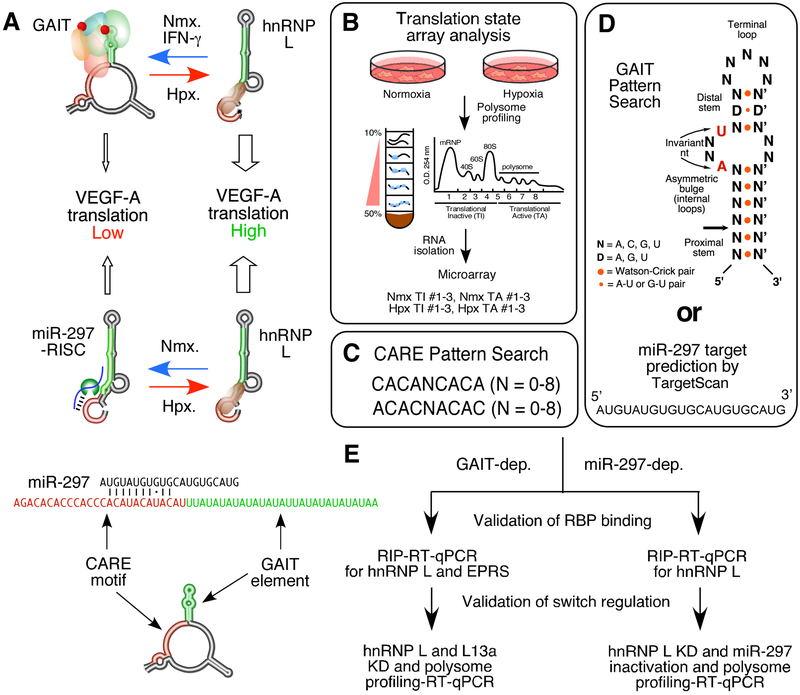
Fig. 1.
Flow chart of identification of hnRNP L-directed RNA switches. A. Schematic of GAIT- and miR-297-dependent, hnRNP L-directed VEGFA RNA switch. B. Translation state array analysis. C. CARE pattern search. D. GAIT element pattern search and miR-297 target prediction. E. Validation procedures of RNA switch targets.
Heterogeneous nuclear ribonucleoprotein L (hnRNP L) is a key post-transcriptional regulator of VEGFA expression. hnRNP L has three consensus RNA recognition motifs (RRM) and binds CA-rich element (CARE) sequences in noncoding regions of multiple transcripts [2, 5]. We have reported that hnRNP L operates a hypoxia-stimulated, binary conformation RNA switch that overrides IFN-g-induced GAIT-mediated translational silencing of VEGFA mRNA in human monocytic cells [1, 2]. Hypoxia activates VEGFA mRNA translation via binding of hnRNP L and double-stranded RNA binding protein 76 (DRBP76) to a 21-nt CARE and a long AU-rich stem-loop region of the VEGFA mRNA 3'-UTR, respectively [2]. The switch permits high-level VEGFA expression under combined inflammatory and hypoxic condition [1] (Fig. 1A). In the absence of IFN-γ stimulus, hnRNP L and miRNA-297-mediated RNA-induced silencing complex (miR-297-RISC) can bind to two overlapping RNA elements within the VEGFA mRNA 3'-UTR, namely, hnRNP L-binding 21-nt CARE and miR-297-targeting sequence [6] (Fig. 1A, bottom). In normoxia, miR-297-RISC binds to its target sequence within VEGFA 3'-UTR and silences VEGFA mRNA translation; however, in hypoxia, hnRNP L binds to CARE and blocks miR-297-RISC binding via spatial hindrance, elevates VEGFA mRNA translation, and thus promotes tumor growth [6, 7].
In our study, we found that the hnRNP L-directed RNA switch mechanism regulates the translation of a novel family of mRNA targets in hypoxia using a genome-wide screen. Src kinase is the proximal kinase responsible for phosphorylation of hnRNP L and the activation of the RNA switch pathway. The cytoplasmic hnRNP L and its directed RNA switch pathway are required for cell survival under hypoxic stress.
2. Materials and methods
2.1. Reagents
Phospho-safe extraction buffer was from Novagen. Reagents for protein purification were from Pierce. Primers, dNTP mix, TRIzol reagent, and one-step RT-PCR system were from Invitrogen. Protein A/G beads and siRNAs against L13a, Fyn, Src and Yes kinases were from Santa Cruz. hnRNP L siRNAs was from OriGene (SR302174). Src, JAK and FAK kinases were from Thermo Fisher. Herbimycin A, PF-573228, BAY 61-3606, and Cucurbitacin I were from Sigma.
2.2. Cell culture and transfection
Human U937 monocytic cells were cultured in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum, 2 mM glutamine, and 100 U/ml of penicillin and streptomycin at 37°C and 5% CO2. Cells were treated in normoxia (21% O2) or hypoxia (1% O2) for 8, 24 or 48 h. Cell lysates were prepared in extraction buffer containing protease inhibitors. To knockdown endogenous genes, U937 cells (5×106 cells) were transfected with gene-specific or scrambled siRNA (100-200 nM) using human monocyte nucleofector kit (Lonza). U937 cells were transfected with anti-miR or control inhibitors (200-400 nM, Ambion).
2.3. Analysis of RNA by RT-qPCR
To determine the expression of various mRNAs by real-time PCR analysis, total RNA (1 mg) was extracted by Trizol and subjected to reverse transcription (TaqMan RT kit, Applied Biosystems). qPCR amplification was performed using Taqman expression assay (Invitrogen) and an ABI Thermo Cycler (ABI Prism 7000 SDS).
2.4. RIP-RT-qPCR
Protein A/G beads (50 μl) were incubated with 500 μl of cell lysate (4 mg) for 1 h at 4°C with rotation to pre-clear. The cell lysate was centrifuged and the supernatant collected. Mouse anti-hnRNP L or anti-EPRS antibody (2 μg) was added (normal mouse or rabbit IgG was used as negative control) and the mixture incubated at 4°C for 1 h with rotation. Protein A/G beads (50 μl) were added and incubated at 4°C for 4 h. The beads were washed five times with 1 ml of wash buffer. Total IPed RNA (5 ml) and total RNA (200 ng) from 10% of lysates were extracted with Trizol for RT-qPCR.
2.5. Polysome profiling
Cycloheximide (CHX, 100 μg/ml, Sigma) was added to cells for 15 min and then collected. 107 cells were suspended in 350 μl TMK lysis buffer. The lysates were centrifuged at 12,000 rpm for 10 min and the supernatants collected. RNase inhibitor (2 μl, 40 U/μl) and CHX (50 μl, 100 μg/μl) were added in 50 ml each of 10% and 50% sucrose gradient solutions. Cytosolic lysates were loaded on the sucrose gradient and centrifuged at 29,000 rpm for 4 h, and 12 fractions of ~0.5-1 ml were collected, including free RNP, 40S, 60S, 80S, and polysome fractions. Total RNA was extracted from individual fractions, purified by RNeasy minikit (Qiagen), and used for RT-qPCR.
2.6. In vitro phosphorylation assay
Cell lysates (0.5 mg) and purified recombinant GST-hnRNP L (1 μg) [2] was pre-incubated for 10 min in kinase assay buffer (50 mM Tris-HCl, pH 7.6, 1 mM dithiothreitol, 10 mM MgCl2, and 1 mM CaCl2) containing phosphatase inhibitor cocktail. The reaction was initiated by incubation with 5 μCi [γ-32P]ATP (PerkinElmer) for 20 min at 37°C and terminated by adding 5×SDS gel-loading dye followed by heat-denaturation. Phosphorylation was determined by SDS-PAGE and autoradiography. To determine responsible kinase for hnRNP L phosphorylation, recombinant wild-type or Y359F mutant His-tagged hnRNP L (1 μg) [7] was incubated with recombinant Src, JAK or FAK (25 ng) and [γ-32P]ATP (1 μCi) for 20 min.
2.7. Translation state array analysis using Illumina GeneChip and data analysis
RNA (1 μg) from pooled non-translational and translational fractions was subjected to labeling reaction using a TargetAmp™-Nano labeling kit (Illumina) combining first and second strand cDNA synthesis by reverse transcription, purification of cDNA, and synthesis of biotin-labeled antisense cRNAs using in vitro transcription with biotin-NTP mix. Biotinylated cRNAs were purified and quantitated and subjected to hybridization with a Human HT-12 v4.0 Expression Beadchip (Illumina). The chips were washed and scanned using a BeadArray reader. Microarray data from Illumina Genome Studio were exported to Excel and Access to determine translationally up- and down-regulated genes. Average signal intensities and p-value (“avg” and “pv_count” in Table S1) were calculated for each probe id from three separate experiments and for each of four samples (NmxTI, normoxia, translationally inactive pool; NmxTA, normoxia, translationally active pool; HpxTI, hypoxia, translationally inactive pool; HpxTA, hypoxia, translationally active pool). A “pv_count” of 3 indicates a p-value < 0.05 in all three experiments. We defined NmxTA/NmxTI and HpxTA/HpxTI are the relative translation efficiencies in normoxia and hypoxia, respectively. A “final_ratio” defined as (HpxTA/HpxTI)/(NmxTA/NmxTI) was calculated to compare the relative translation efficiency in hypoxia versus normoxia and determine transcripts translationally activated in hypoxia. Translationally up- and down-regulated genes were determined by final_ratio > 1.5 and < 0.67 (pv_counti ≥ 2, i = HpxTI, NmxTA), respectively. An average value was calculated for a given gene in case of multiple probe ids for a given gene, to estimate an average final ratio for that gene.
2.8. CA-rich element pattern search
We used an in-house shell script to search two human 3'-UTR databases (PACdb and UTRef) using the search pattern “CACANCACA OR ACACNACAC; N=0…8” [5]. CARE sequences and their positions in the five mRNA candidates and VEGFA (Table S2) were validated using PatSearch [8].
2.9. Statistical analysis
All quantitative data were presented as mean±SD and analyzed using Prism 7 software. For a comparison between 2 groups, a Student t test was performed. Statistical significance was assumed at a value of P≤0.05.
3. Results
The cytoplasmic hnRNP L can bind to VEGFA mRNA and other transcripts in several cell lines [1, 9, 10], implying that hnRNP L may direct RNA switches for various genes in multiple cell types. To determine novel transcripts regulated by hnRNP L-directed switching mechanisms, we applied three independent analyses to the human transcriptome, identifying (i) mRNAs translationally activated by hypoxia (Fig. 1A-B), (ii) CARE-bearing, potential hnRNP L target mRNAs (Fig. 1A, C), and (iii) Potential mRNA targets negatively regulated by the GAIT complex or miR-297-RISC (Fig. 1A, D). We overlapped the three datasets and retrieved the common hits for further confirmation (Fig. 1E). RIP (RNA binding protein immunoprecipitation) and RT-qPCR were performed to determine the binding of target transcripts by the trans-acting proteins. siRNA-directed knockdown of critical RNA switch mediators followed by polysome profiling and RT-qPCR was performed to show the switch regulation.
3.1. Identification of translationally upregulated mRNAs by genome-wide translation state array analysis in hypoxia-stressed monocytes
mRNAs subject to hypoxia-inducible translational activation were identified by translation state microarray analysis (Fig. 1B). Human U937 monocytic cells were treated with IFN-γ for 24 h under normoxic or hypoxic conditions. Cell lysates were fractionated by sucrose gradient centrifugation to separate translating and non-translating RNA pools which were subjected to microarray analysis. Candidate transcripts (2731) were translationally activated by greater than 1.5-fold (P < 0.05) during hypoxia compared to normoxia (Table S1). As positive controls, the translation efficiency of two established hnRNP L-binding mRNAs VEGFA and BCL2 was increased by 1.88-fold and 2.97-fold, respectively. As negative controls, GAIT component genes EPRS, NSAP1, L13a were not translationally activated by hypoxia. To identify candidate mRNAs regulated by hnRNP L, we applied an in-house search algorithm (PERL script) to query two human 3'-UTR databases (PACdb and UTRef) [11, 12] for mRNAs with CARE motifs, with a relaxed pattern containing a minimum of two CACA or ACAC high-score motifs separated by at most 8 nt [5] (Fig. 1C, S1A). 3'-UTR CARE sequences were predicted in 6036 human mRNAs, including VEGFA and BCL2, both experimentally validated hnRNP L-binding targets [9, 13] (Table S2, Fig. S1A).
3.2. Identification of novel GAIT-dependent, hnRNP L-directed RNA switch transcripts
To determine novel mRNAs regulated by hnRNP L- and GAIT-dependent switching mechanisms (Fig. 2A), we applied a pattern-matching algorithm, PatSearch [8] developed in-house to query a 3'-UTR database [12] for mRNAs with sequence motifs and secondary structures matching the archetypal human Cp GAIT element [4] (Fig. 1D). Predicted GAIT elements in the 3'-UTR of 52 annotated human mRNAs were found, including Cp and VEGFA (Table S3).
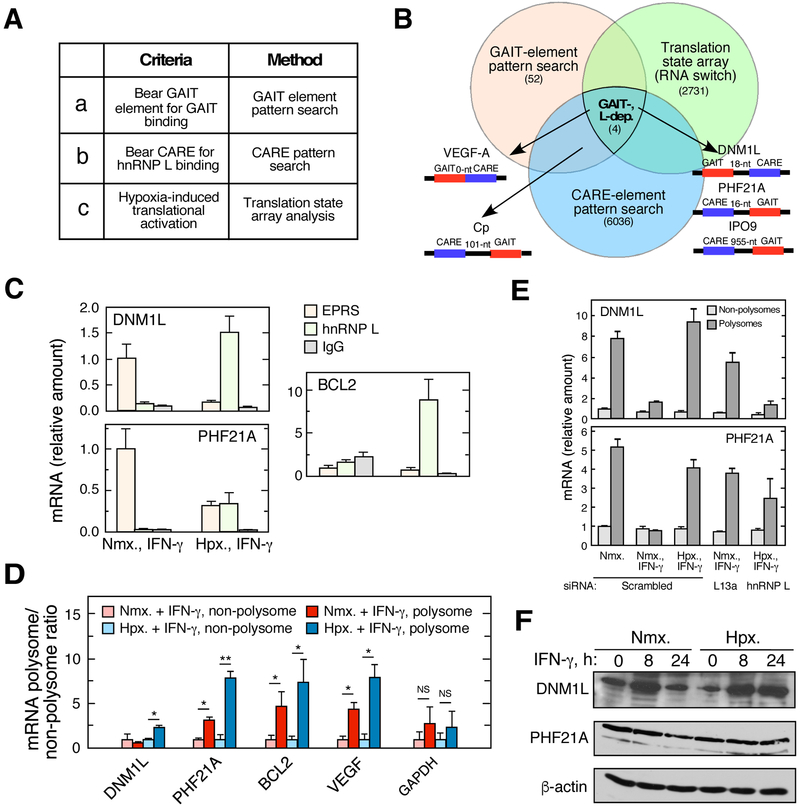
Fig. 2.
Identification of DNM1L and PHF21A as GAIT-dependent, hnRNP L-directed RNA switch transcript. A. Applied criteria and method. B. Boolean diagram of potential mRNAs regulated by GAIT-dependent, hnRNP L-directed RNA switches. Results based on bioinformatic pattern search for GAIT element and CARE, and translation state array analysis. Targets selected for experimental validation are shown. C. Condition-dependent binding of candidate switch mRNAs to GAIT and hnRNP L. U937 cells were treated with IFN-γ under normoxia or hypoxia. Lysates were subjected to IP with anti-hnRNP L or -EPRS antibodies (or IgG) coupled with RT-qPCR. D. Translation state analysis of selected mRNAs for hypoxia-driven switch activity. Polysome profiling of lysates from U937 cells treated with IFN-γ in normoxia or hypoxia, coupled with RT-qPCR. E. Translation state of RNA switch mRNAs following gene-specific knockdown. F. Hypoxia antagonized IFN-γ-activated inhibition of DNM1L and PHF21A protein expression in monocytic cells.
The intersection of the three screens yielded four potential target mRNAs translationally regulated by hnRNP L- and GAIT-dependent RNA switches (Fig. 2B). VEGFA mRNA, a validated RNA switch, was shown up as a positive control in the triple screen. However, despite having both GAIT (nt 78-106 in 3'-UTR) and CARE (nt 208-219) elements, Cp was not translationally upregulated indicated by the translation state analysis, consistent with the fact that Cp mRNA is not subject to hypoxia-induced RNA switching [1]. Possibly, the distance between the elements, 101 nt, is greater than the limit for RNA switch function (<20-nt), which does not permit the competition between the two RNA elements and their associated proteins [2]. IPO9 mRNA contains CARE and GAIT elements, but the distance is 955 nt. Therefore, IPO9 is unlikely to possess a functional GAIT-dependent RNA switch. Two candidate mRNAs DNM1L (dynamin 1-like) and PHF21A (PHD finger protein 21A) were chosen for validation since the distance between the two RNA elements is within 18-nt and 15-nt, respectively (Fig. 2B, S1B-C). Besides, VEGFA was used as a positive control and BCL2 (contain no GAIT element) was chosen as a negative control (Fig. S1B). DNM1L mRNA was exclusively bound by EPRS under IFN-γ treatment in normoxia and almost completely shifted to be associated with hnRNP L in hypoxia (Fig. 2C). PHF21A mRNA was bound by EPRS in normoxia and partially shifted to interact with hnRNP L in hypoxia. In contrast, EPRS did not bind to BCL2 mRNA in both conditions while hnRNP L strongly bound it in hypoxia (Fig. 2C). Transcripts that interacted with EPRS in normoxia and with hnRNP L in hypoxia were further evaluated for a hypoxia-dependent translational shift as determined by polysome profiling coupled with RT-qPCR (Fig. 2D). DNM1L and PHF21A mRNAs were associated with translationally silent non-polysome fractions in normoxia under IFN-γ stimulation. These two transcripts were shifted to be associated with translationally active polysome fractions in hypoxia (Fig. 2D). Following siRNA-mediated knockdown of the GAIT component RPL13a, polysome association of DNM1L and PHF21A mRNAs was restored in cells treated with IFN-γ under normoxia (Fig. 2E). Likewise, knockdown of hnRNP L (Fig. S2A) reduced the polysome association of the transcripts in cells treated with IFN-γ under hypoxia (Fig. 2E). Protein expression of DNM1L and PHF21A was reduced in normoxia with IFN-γ stimulation and restored in hypoxia without any significant change in mRNA expression (Fig. 2F, S2B). Taken together, DNM1L and PHF21A were validated experimentally as functioning as GAIT-dependent hnRNP L-directed RNA switches.
3.3. Identification of novel miR-297-dependent, hnRNP L-directed RNA switch transcripts
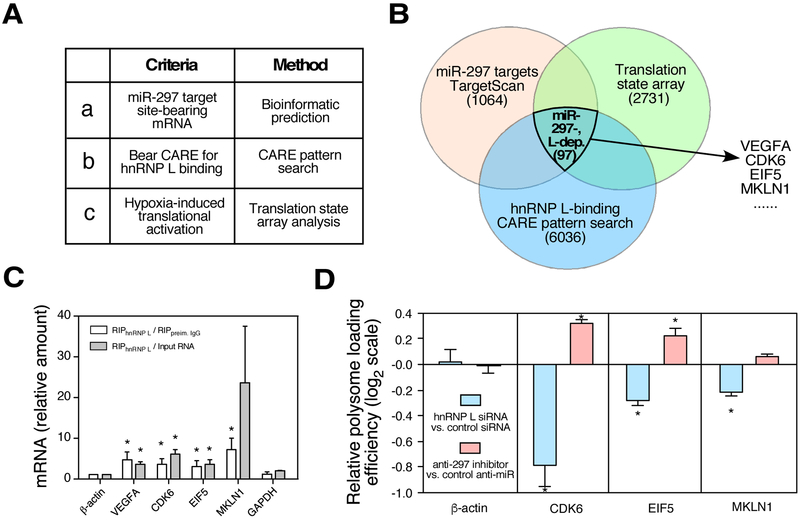
To identify additional candidate oncogenic mRNAs regulated by miR-297-RISC-dependent hnRNP L-directed RNA switches besides VEGFA in human monocytic cells (Fig. 3A), we obtained 1064 potential common miR-297 target mRNAs in both human and mouse using the TargetScan web tool (Fig. 1D, Table S4). From the intersection of predicted miR-297 and hnRNP L targets screening and translation state array analysis, we identified 97 potential mRNAs regulated by miR-297 and hnRNP L (Table S5). We chose three candidate RNA switch genes among oncogenic genes for validation, including CDK6, MKLN1, and EIF5. hnRNP L interacted with all three transcripts in hypoxia (Fig. 3C). Knockdown of hnRNP L or inactivation of miR-297 reduced and increased the translation efficiency of the three mRNAs, respectively (Fig. 3D). These data suggest that the three mRNAs are regulated by miR-297-dependent hnRNP L-directed RNA switch pathway.
Fig. 3.
Identification of multiple oncogenic genes as miR-297-dependent, hnRNP L-directed RNA switch transcripts. A. Applied criteria and method. B. Boolean diagram of potential mRNAs regulated by miR-297-dependent, hnRNP L-directed RNA switches. Results based on bioinformatic search for miR-297 target and CARE-bearing mRNAs, and translation state array analysis. Targets selected for further experimental validation are indicated. C. Hypoxia-induced binding of candidate switch mRNAs to hnRNP L. Lysates from hypoxia-treated U937 cells were subjected to IP with anti-hnRNP L antibody (or IgG) coupled with RT-qPCR. D. Translation-state analysis of selected mRNAs for hypoxia-driven switch activity. Polysome profiling of lysates from U937 cells cultured in normoxia or hypoxia, coupled with RT-qPCR. Translation efficiency is expressed as log2 ratio of polysome-derived mRNA to non-polysome-derived mRNA.
3.4. Src kinase is responsible for hypoxia-activated phosphorylation of hnRNP L
Since hnRNP L drives the active translation of CARE-bearing mRNAs [2, 10], we ought to examine the location of hnRNP L in polysome profiling. We found that hnRNP L was primarily localized in non-translating fractions in normoxia and translocated partially to translating polysome fractions upon hypoxic treatment (Fig. 4A). Our previous studies have shown that hypoxia induces Tyr359 phosphorylation of hnRNP L by an unidentified kinase, resulting in marked cytoplasmic accumulation [2]. Induction of Tyr phosphorylation by hypoxia suggests the activity of a non-receptor Tyr kinase such as a member of the Src, Syk, Fak, or Jak kinase families. To identify the kinase family, U937 cells were treated with a spectrum of pharmacological inhibitors, and cell lysates incubated in the presence of 32P-labeled ATP and recombinant GST-hnRNP L. In vitro phosphorylation of GST-hnRNP L was dramatically reduced by herbimycin A, an Src-family kinase inhibitor, but not by inhibitors of Syk, Fak, and Jak kinases, i.e., BAY 61-3606, PF-573228, and cucurbitacin I, respectively (Fig. 4B). Of the 11 SH3 domain-containing Src family kinases, Src kinase itself is a prime candidate because, unlike Fyn and Yes, it is known to be hypoxia-activated [14], and also phosphorylates multiple hnRNP targets (including hnRNP K, AB, A3, and DL) [15]. Gene-specific, siRNA-mediated knockdown of Src kinase in hypoxia-treated U937 cells confirmed its important role in hnRNP L phosphorylation (Figure 4C). Finally, recombinant Src kinase drives in vitro phosphorylation of wild-type hnRNP L, but not the phosphorylation site mutant Tyr359-to-Phe (Y359F), establishing it as the proximal kinase (Figure 4D). Inhibition of Src kinase by Herbimycin A reduced hnRNP L binding to VEGFA mRNA (Figure S2C). These data are consistent with a mechanism in which hypoxic stress induces Src kinase-mediated phosphorylation of hnRNP L, increasing its cytoplasmic level, and promoting translation of VEGFA and other mRNAs [2].
Fig. 4.
Src kinase directs hnRNP L phosphorylation for translational activation of RNA switch mRNAs. A. Localization of hnRNP L in polysome profiling fractions of U937 cells treated in normoxia and hypoxia. B. Src-family kinase is required for hnRNP L phosphorylation. Lysates from 4-h, Hpx.-treated U937 cells were used as kinase source for in vitro phosphorylation of purified GST-hnRNP L. Kinase inhibitors: BAY 61-3606 (1 μM), herbimycin A (1 μM), PF-573228 (500 nM), and cucurbitacin I (1 μM). C. Src kinase phosphorylates hnRNP L in vivo. U937 cells were transfected with Src-family kinase-specific siRNAs for 24 h and then cultured in Hpx. for 8 h. Lysates were subjected to IP with anti-hnRNP L antibody and immunoblot with anti-phospho-Tyr antibody. D. In vitro phosphorylation of wild-type and mutant hnRNP L. Wild-type or Y359F mutant hnRNP L was incubated with Src, FAK, or JAK kinase, and phosphorylation determined by immunoblot with anti-phospho-Tyr antibody. E. Caspase 3 and PARP cleavage and expression of DNM1L and CDK6 proteins after knockdown of hnRNP L. F. Schematic model of hnRNP L-directed RNA switches in promoting tumor cell survival.
3.5. Dysfunctional hnRNP L-directed RNA switch reduces target mRNA translation and promotes cell apoptosis
hnRNP L-mediated RNA switch is a conserved mechanism in multiple cell types, including monocytes, Hela, and glioma cells [2, 10, 16]. siRNA-medicated knockdown of hnRNP L under hypoxic but not normoxic condition sensitized the U937 cells to undergo robust apoptosis indicated by activated caspase 3 and PARP cleavage (Fig. 4E). Translation of multiple RNA switch target mRNAs was reduced, such as DNM1L and CDK6 (Fig. 4E).
4. Discussion
Our previous studies have shown that VEGFA is regulated by hypoxia-activated hnRNP L-directed RNA switch [1, 2, 7]. By bioinformatic analyses and experimental validation, we uncovered two GAIT-dependent, hnRNP L-directed RNA switch transcripts, DNM1L, and PHF21A (Fig. 4F). DNM1L controls mitochondria fission and dynamics and promotes cancer progression [17]. PHF21A is involved in chromatin remodeling, and its dysregulation is associated with glioma tumorigenesis [18]. We found that protein components and mechanisms utilized by the VEGFA mRNA switch are applied to distinct mRNA switches. HnRNP L recognizes DNM1L and PHF21A mRNAs through sequence and structure analogous to the VEGFA switch region, i.e., CARE and GAIT elements (Fig. S1B-C). The proximity of the elements is consistent with a steric hindrance mechanism. Therefore, our work suggests that stress-inducible, protein-directed RNA switches based on combinatorial use of nearby RNA elements are widespread regulatory mechanisms of gene expression in mammalian cells.
Besides, we have found several miR-297-dependent, hnRNP L-directed RNA switch transcripts, including CDK6, EIF5, and MKLN1 (Fig. 4F). CDK6 is a cyclin-dependent kinase required for cell cycle progression and considered as an anti-cancer pharmacological target [19]. EIF5 is a translation initiation factor for protein synthesis and promotes oncogenesis [20]. MKLN1 is a cytoskeleton-associated protein which is critical for cytoskeleton organization and cell adhesion [21]. The six hnRNP L-directed RNA switch genes may contribute to cell survival and tumorigenesis via various pathways (Fig. 4F). Therefore, reducing cytoplasmic hnRNP L under hypoxia decreases the translation of multiple target oncogenic mRNAs, thereby impairing cell survival under hypoxic stress. Taken together, our results suggest that hnRNP L-directed RNA switches contribute to hypoxia resistance and cell survival.
Supplementary Material
HIGHLIGHTS.
Identification of hnRNP L-directed, GAIT-dependent RNA switches; experimental validation of DNM1L and PHF21A
Identification of hnRNP L-directed, miR-297-RISC-dependent RNA switches including CDK6, MKLN1 and EIF5
Src kinase is a proximal kinase that phosphorylates Tyr359 residue of hnRNP L and promotes RNA switch in hypoxia
hnRNP L-mediated RNA switch is an oncogenic translational regulon that contributes to cell survival
Acknowledgments
We are grateful to Paul Fox for helpful discussion, LRI Genomics Core for microarray experiment, and Qiuqing Wang for bioinformatic consulting.
Sources of support
P.Y. was supported by NIH R01 (HL132899) and AHA SDG (13SDG15970006).
Footnotes
Conflict of interest
The authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ray PS, Jia J, Yao P, Majumder M, Hatzoglou M, Fox PL, A stress-responsive RNA switch regulates VEGFA expression, Nature, 457 (2009) 915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Yao P, Potdar AA, Ray PS, Eswarappa SM, Flagg AC, Willard B, Fox PL, The HILDA complex coordinates a conditional switch in the 3'-untranslated region of the VEGFA mRNA, PLoS Biol, 11 (2013) e1001635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yao P, Potdar AA, Arif A, Ray PS, Mukhopadhyay R, Willard B, Xu Y, Yan J, Saidel GM, Fox PL, Coding Region Polyadenylation Generates a Truncated tRNA Synthetase that Counters Translation Repression, Cell, 149 (2012) 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sampath P, Mazumder B, Seshadri V, Gerber CA, Chavatte L, Kinter M, Ting SM, Dignam JD, Kim S, Driscoll DM, Fox PL, Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation, Cell, 119 (2004) 195–208. [DOI] [PubMed] [Google Scholar]
- [5].Hui J, Hung LH, Heiner M, Schreiner S, Neumuller N, Reither G, Haas SA, Bindereif A, Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing, EMBO J, 24 (2005) 1988–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jafarifar F, Yao P, Eswarappa SM, Fox PL, Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L, EMBO J, 30 (2011) 1324–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yao P, Wu J, Lindner D, Fox PL, Interplay between miR-574-3p and hnRNP L regulates VEGFA mRNA translation and tumorigenesis, Nucleic Acids Res, 45 (2017) 7950–7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Grillo G, Licciulli F, Liuni S, Sbisa E, Pesole G, PatSearch: A program for the detection of patterns and structural motifs in nucleotide sequences, Nucleic Acids Res, 31 (2003) 3608–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lee DH, Lim MH, Youn DY, Jung SE, Ahn YS, Tsujimoto Y, Lee JH, hnRNP L binds to CA repeats in the 3'UTR of bcl-2 mRNA, Biochem Biophys Res Commun, 382 (2009) 583–587. [DOI] [PubMed] [Google Scholar]
- [10].Kefas B, Floyd DH, Comeau L, Frisbee A, Dominguez C, Dipierro CG, Guessous F, Abounader R, Purow B, A miR-297/hypoxia/DGK-alpha axis regulating glioblastoma survival, Neuro Oncol, 15 (2013) 1652–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Brockman JM, Singh P, Liu D, Quinlan S, Salisbury J, Graber JH, PACdb: PolyA Cleavage Site and 3'-UTR Database, Bioinformatics, 21 (2005) 3691–3693. [DOI] [PubMed] [Google Scholar]
- [12].Grillo G, Turi A, Licciulli F, Mignone F, Liuni S, Banfi S, Gennarino VA, Horner DS, Pavesi G, Picardi E, Pesole G, UTRdb and UTRsite (RELEASE 2010): a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs, Nucleic Acids Res, 38 (2010) D75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shih SC, Claffey KP, Regulation of human vascular endothelial growth factor mRNA stability in hypoxia by heterogeneous nuclear ribonucleoprotein L, J Biol Chem, 274 (1999) 1359–1365. [DOI] [PubMed] [Google Scholar]
- [14].Mukhopadhyay D, Tsiokas L, Zhou XM, Foster D, Brugge JS, Sukhatme VP, Hypoxic induction of human vascular endothelial growth factor expression through c-Src activation, Nature, 375 (1995) 577–581. [DOI] [PubMed] [Google Scholar]
- [15].Amanchy R, Zhong J, Molina H, Chaerkady R, Iwahori A, Kalume DE, Gronborg M, Joore J, Cope L, Pandey A, Identification of c-Src tyrosine kinase substrates using mass spectrometry and peptide microarrays, Journal of proteome research, 7 (2008) 3900–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rossbach O, Hung LH, Khrameeva E, Schreiner S, Konig J, Curk T, Zupan B, Ule J, Gelfand MS, Bindereif A, Crosslinking-immunoprecipitation (iCLIP) analysis reveals global regulatory roles of hnRNP L, RNA Biol, 11 (2014) 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xie LL, Shi F, Tan Z, Li Y, Bode AM, Cao Y, Mitochondrial network structure homeostasis and cell death, Cancer Sci, 109 (2018) 3686–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liu M, Xu Z, Du Z, Wu B, Jin T, Xu K, Xu L, Li E, Xu H, The Identification of Key Genes and Pathways in Glioma by Bioinformatics Analysis, J Immunol Res, 2017 (2017) 1278081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fogli S, Del Re M, Curigliano G, van Schaik RH, Lancellotti P, Danesi R, Drug-drug interactions in breast cancer patients treated with CDK4/6 inhibitors, Cancer Treat Rev, 74 (2019) 21–28. [DOI] [PubMed] [Google Scholar]
- [20].Kozel C, Thompson B, Hustak S, Moore C, Nakashima A, Singh CR, Reid M, Cox C, Papadopoulos E, Luna RE, Anderson A, Tagami H, Hiraishi H, Slone EA, Yoshino KI, Asano M, Gillaspie S, Nietfeld J, Perchellet JP, Rothenburg S, Masai H, Wagner G, Beeser A, Kikkawa U, Fleming SD, Asano K, Overexpression of eIF5 or its protein mimic 5MP perturbs eIF2 function and induces ATF4 translation through delayed re-initiation, Nucleic Acids Res, 44 (2016) 8704–8713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Adams JC, Seed B, Lawler J, Muskelin, a novel intracellular mediator of cell adhesive and cytoskeletal responses to thrombospondin-1, EMBO J, 17 (1998) 4964–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.