Abstract
Studies have investigated CCR5 haplotypes (HHA, HHB, HHC, HHD, HHE, HHF*1, HHF*2, HHG*1, HHG*2), defined by seven 5’UTR single nucleotide polymorphisms (SNPs), CCR2-V64I and CCR5Δ32, in HIV-1 disease. CCR5 cis-regulatory regions were sequenced, CCR2-V64I and CCR5Δ32 genotyped, and compared in HIV-1-infected black South Africans: 71 HIV-1 controllers (23 elite controllers, 37 viraemic controllers (VCs), 11 high viral load long-term non-progressors) and 74 progressors. The HHE haplotype and 3’UTR +2919T>G SNP heterozygosity were underrepresented in total controllers and VCs vs. progressors (p=0.004; p=0.007 and p=0.002, pbonferroni=0.032; p=0.004, respectively). Possession of the +2919T>G SNP (dominant mode) was associated with HIV-1 progression (controllers vs. progressors: p=0.001, pbonferroni=0.016). The +2919T>G SNP is in linkage disequilibrium (LD; r2=0.73) with two 5’UTR SNPs (−2459G>A and −2135T>C;r2=1: 5’UTR-2SNP-hap). The 5’UTR-2SNP-hap was lower in total controllers and VCs vs. progressors (p=0.003, pbonferroni=0.048; p=0.01, respectively). Results suggest −2459G>A, −2135T>C, and +2919T>G as key CCR5 variants in HIV-1 control.
Keywords: CCR5, HIV-1, genetic variants, cis-regulatory regions, natural control, South Africa
1. Introduction
Unique individuals who are able to either naturally suppress HIV-1 viral load (VL) or maintain healthy CD4+ T cell counts with higher viral loads, and who exhibit slow progression of the disease without antiretroviral therapy (ART), are broadly termed HIV-1 controllers. Within this group there are rare individuals termed elite controllers (ECs) who suppress VL to less than 50 RNA copies/ml plasma, viraemic controllers (VCs) who tend to have low viral load set-points (generally <2000 RNA copies/ml) with sustained high CD4+ T cell counts, and high viral load long-term non-progressors (HVL LTNPs) who maintain high CD4+ T cell counts for prolonged periods without ARV treatment despite high viral loads (generally >10 000 RNA copies/ml), a similar phenotype to SIV-infected sooty mangabeys [1]. A number of factors, including viral, immunological and environmental, have been proposed to influence the ability of an individual to naturally control HIV-1 [2], however it is likely that different mechanisms may be responsible for the different modes of control.
The chemokine receptor CCR5, together with the CD4 receptor, is responsible for allowing HIV-1 entry into the target cell and is well studied with regards to HIV-1 disease. Lower CCR5 expression levels have been associated with slower HIV-1 disease progression [3]. The differential expression of CCR5 may be explained by polymorphisms in the cis-regulatory regions of CCR5 [4]. Although the 5’UTR has been extensively studied, surprisingly, little is known about the genetic variation in the CCR5 3’UTR, a region increasingly shown to be crucial for mRNA regulation.
One of the most studied genetic variations in CCR5 is the Δ32 mutation (CCR5Δ32), a 32 base pair deletion that results in non-functional CCR5 receptors. This variant is predominantly found in European populations and is virtually absent in African, East Asian, and American Indian populations [5]. The rs553615728 −4223C>T single nucleotide polymorphism (SNP) has been shown to disrupt a cytidine phosphate guanidine (CpG) dinucleotide in the c/s-region of CCR5, namely CpG-41, a binding site where DNA methylation occurs, and is uniquely found in individuals from southern Africa [6]. Additionally, there are nine previously described CCR5 haplotypes [7–9] that have been studied with respect to HIV-1 disease progression (HHA, HHB, HHC, HHD, HHE, HHF*1, HHF*2, HHG*1 and HHG*2). These haplotypes are defined by seven CCR5 5’UTR SNPs and the presence/absence of CCR5Δ32and a SNP in the coding region of chemokine receptor CCR2 (CCR2-V64I) [8,9].
This study was conducted to explore the genetic variation in the cis-regulatory regions of CCR5 and to explore the role of these variants on HIV-1 control in a population of HIV-1 infected black South African controllers and progressors. This information will help to formulate a protective and deleterious CCR5 genetic signature for black South African individuals infected with HIV-1 and will be useful in informing cure strategies.
2. Materials and Methods
2.1. Sample population
This study included black South African ART-naive HIV-1 infected HIV-1 controllers comprised of 23 elite controllers (ECs), 37 viraemic controllers (VCs) and 11 high viral load long-term non-progressors (HVL LTNPs). A shared defining feature of HIV-1 controllers is a CD4+ T cell count greater than 500 cells/μl of blood. In this study, VCs are defined as having low detectable viral loads (<2000 RNA copies/ml plasma), while rare individuals (less than 1% of HIV-1 infected individuals in previously studied cohorts [10]) who have undetectable viral loads (<50 RNA copies/ml) after infection with HIV-1 are termed ECs [11]. Individuals who maintain CD4+ T cell counts greater than 500 cells/μl for more than 7 years without the use of ART, but who do not suppress HIV-1 viral load (generally >10 000 RNA copies/ml), are termed high viral load long-term non-progressors (HVL LTNPs) in this study. This study also included 74 ART-naïve HIV-1 infected progressors who required initiation of ART upon enrolment and were not selected on basis of progression rate, i.e. not rapid progressors. Detailed cohort characteristics are described in Table 1.
Table 1.
Characteristics of study cohort
| Group | Number of patients | Age (years) | Gender | CD4+ T cell count* (cells/μl) | Viral load* (HIV RNA copies/ml) | Years since diagnosis |
|---|---|---|---|---|---|---|
| (n) | (Mean and range) | (% female) | (Median and IQR) | (Median and IQR) | (Median and IQR) | |
| ECs | 23 | 48 (27–66) | 78.2 | 693 (588 – 969) | <20 | 15 (8–16) |
| VCs | 37 | 42 (27–59) | 91.9 | 704 (587 – 910) | 598 (135 – 1180) | 8 (8–15) |
| HVL LTNPs | 11 | 43 (33–53) | 81.8 | 660 (606 – 749) | 22 410 (14 262 – 77 820) | 8 (8–11) |
| Progressors | 74 | 45 (30–73) | 83.8 | 177 (145 – 210) | 38 444 (19 790 – 103 314) | 6 (1–7) |
CD4+ T cell counts and viral loads of controllers used were from time of enrolment whereas for the progressors the last CD4 T cell count and viral load prior to ART initiation was used
ECs: elite controllers, VCs: viraemic controllers, HVL LTNPs: high viral load long-term non-progressors
Viral loads (RNA copies/ml plasma) were quantified using the COBAS®AmpliPrep/COBAS®Taqman® HIV-1 Test, v2.0 ultrasensitive tests (<20 RNA copies/ml) (Roche Diagnostic Systems, Inc, New Jersey, USA) and CD4+ T cell counts (cells/μl whole blood) were determined using the FACSCount System (Becton Dickinson, San Jose, California, USA). Written informed consent was obtained from all individuals participating in this study and ethics approval has been obtained from the Human Research Ethics Committee at the University of the Witwatersrand, Johannesburg.
2.2. Standard polymerase chain reaction (PCR) amplification and sequencing of CCR5 5’UTR and 3’UTR
Genomic DNA was extracted from either whole blood or buffy coats of patients using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). Each region of interest was PCR amplified with primers designed using PrimerW software (Supplementary Table 1) using the EXPAND High Fidelity PCR System (Roche, Mannheim, Germany) with cycling conditions carried out according to manufacturer’s instructions. The resulting PCR products were purified and electrophoresed on an automated 3100 Genetic Analyser (Applied Biosystems, Foster City, California, USA), according to manufacturer’s instructions.
2.3. Sequence analysis
Resulting sequence chromatograms were analyzed using Sequencher software version 5.1 (Gene Codes Corporation, Ann Arbor, Michigan, USA). Imported sequences were aligned to a reference sequence obtained from the NCBI database (NCBI ref seqNC_000006.12). The numbering of CCR5 SNPs used in this study is as described by Mummidi et al. [7], with the first nucleotide of the translational start site designated as +1 and the nucleotide immediately upstream from that designated as −1.
2.4. CCR5 haplotype assignment
Individuals were assigned previously described CCR5 haplotypes (HHA, HHB, HHC, HHD, HHE, HHF*1, HHF*2, HHG*1, HHG*2) based on the presence or absence of seven 5’UTR SNPs at positions −2733 (rs2856758), −2554 (rs2734648), −2459 (rs1799987), −2135 (rs1799988), −2132 (rs41469351), −2086 (rs1800023) and −1835 (rs1800024), and the presence/absence of CCR2-V64I (rs1799864) and CCR5Δ32 (rs333) [12]. Individuals were genotyped for CCR2-V64I, the presence/absence of CCR5Δ32 and the CCR5 −4223C>T SNP (rs553615728) using allelic discrimination assays as previously described [6,13,14]. Some individuals from our cohort were previously genotyped for the CCR5 −4223C>T SNP [6]. The remaining individuals were genotyped using an allelic discrimination assay designed using available heterozygotes from the prior genotyping. The sequences of the primers and probes used for all applications are listed in Supplementary Table 1.
2.5. Linkage disequilibrium (LD) and Hardy-Weinberg equilibrium
The genotypic data generated for polymorphic loci were tested for linkage disequilibrium as well as deviation from Hardy-Weinberg equilibrium using the Haploview version 4.2 software [15].
2.6. Data analysis and statistics
To compare SNP and haplotype frequencies between respective groups, we used Fisher exact tests, which were conducted using VassarStats: website for statistical computation [16] (http://vassarstats.net/odds2x2.html), to calculate statistical significances and exact 95% confidence intervals (CI) of odds ratios (OR). Two-sided tests were used and statistical significance for all analyses was set at p<0.05. Correction for multiple comparisons was conducted on all p values generated using Bonferroni correction - p values significant post correction are shown as pbonferroni (the absence of Pbonferroni indicates that relationship was not significant post correction). The frequency of each haplotype was calculated by counting the number of haplotypic alleles and dividing by the total number of alleles. Visual analysis was used for determining haplotypes across gene regions.
3. Results
3.1. CCR5 5’UTR: variability and HIV-1 control
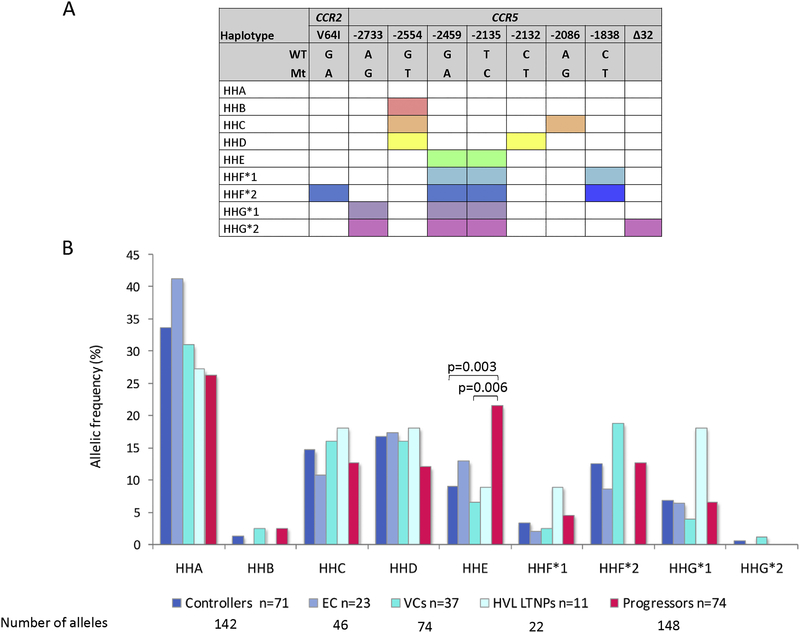
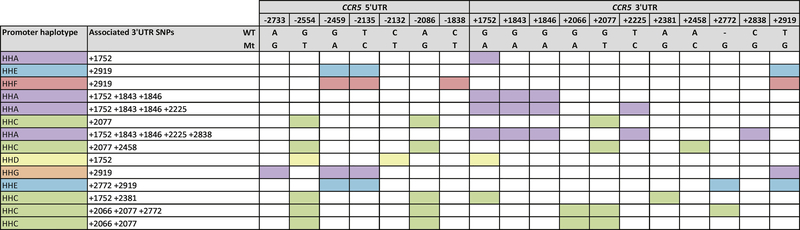
We amplified and sequenced the CCR5 promoter region and individuals were genotyped for the CCR5Δ32 deletion and the CCR2-V64I polymorphism. Previously defined haplotypes [9,12] were assigned to the 71 HIV-1 controllers and 74 HIV-1 progressors. The defining polymorphisms for the CCR5 haplotypes and allelic representation of these haplotypes in controllers, controller subgroups and progressors are shown in Figures 1A and 1B, respectively. Interestingly, one individual in our study, a viraemic controller, was heterozygous for the CCR5Δ32 deletion (haplotype HHG*2).
Figure 1.
A. Schematic showing polymorphisms forming each haplotype. Colour shaded boxes show the respective SNPs or indels that form each haplotype B. Bar graph showing the allelic frequencies (%) of previously defined CCR5 promoter haplotypes in black South African controllers, controller subgroups (ECs, VCs and HVL LTNPs) and progressors. ECs: elite controllers, VCs: viraemic controllers, HVL LTNPs: high viral load long-term non-progressors. WT: wild type allele, Mt: mutant allele
With regards to CCR5 promoter haplotypes, the most prominent relationship was seen with respect to the HHE haplotype. All controller subgroups showed low levels of HHE representation compared to progressors (21.6%; Figure 1), however this was only significant in the VCs (6.8%; p=0.006; OR=3.81; CI=1.42–10.23) and total controllers (9.2%; p=0.003; OR=2.74; CI=1.37–5.47, pbonferroni=0.048). Comparison of allelic representation of either HHE, HHF*1, HHF*2 or HHG*1 revealed significant underrepresentation in total controllers compared to progressors (p=0.02; OR=1.77; CI=1.1–2.86), however comparison of the genotypic representation of these same haplotypes (i.e. combining individuals with at least one copy of these haplotypes) revealed more significant underrepresentation in total controllers compared to progressors (p=0.003; OR=2.9; CI=1.41–5.47, pbonferroni=0.048). The combination of select CCR5 haplotypes (i.e. the genotypes) and the presence or absence of a particular allele (haplotype) can collectively influence CCR5 expression. HHA/HHC was significantly overrepresented in the total group of controllers compared to the progressors (p=0.028; OR=0.19; CI=0.04–0.92) and this was more significant in the VCs compared to the progressors (p=0.016; OR=0.14; CI=0.03–0.75). The large confidence intervals of these findings suggest that further study of HHA/HHC in larger cohorts would be helpful to determine true significance. The frequencies of all CCR5 haplotype genotypes detected in the controllers, controller subgroups and progressors are shown in Supplementary Table 2.
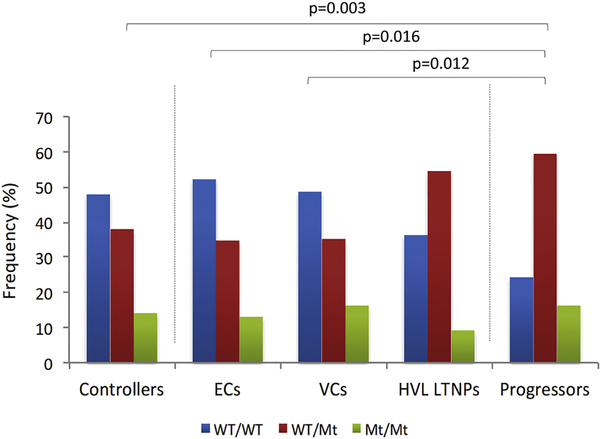
Two 5’UTR SNPs, common to the HHE, HHF and HHG haplotypes (−2459A and −2135C alleles), were in complete linkage disequilibrium in our study population (LD; r2=1). We hence analysed the effect of this 2-SNP haplotype (termed 5’UTR-2SNP-hap). Table 2 shows results of the comparison of representation of 5’UTR-2SNP-hap between controller and progressor groups and Figure 2 depicts its genotypic frequency in the respective groups. The minor allele of 5’UTR-2SNP-hap was significantly underrepresented in the total controller group compared to progressors (p=0.031; OR=1.72; 0=1.07–2.77). Heterozygosity for 5’UTR-2SNP-hap was significantly less prevalent in ECs compared to progressors (p=0.016; OR=3.67: 0=1.28–10.47), VCs compared to progressors (p=0.012; OR=3.38; 0=1.38–8.32) and most significantly in the total controller group compared to progressors (p=0.003; OR=3.08; 0=1.46–6.49, pbonferroni=0.048). Interestingly, heterozygosity for 5’UTR-2SNP-hap was very similar between HVL LTNPs and progressors (55% vs. 59%). Comparison of the 5’UTR-2SNP-hap in the dominant mode [i.e. wild type (WT)/mutant (Mt) + Mt/Mt] revealed similar results (Table 2). The genotypic frequencies of all identified 5’UTR SNPs in the respective groups are shown in Supplementary Table 3.
Table 2.
Comparison of the genotypic and allelic representation of CCR5 5’UTR-2SNP-hap (−2459G>A and −2135C>T) and CCR5 3’UTR +2919T>G SNP in controllers, controller subgroups and progressors
| Controllers vs. Progressors | ECs vs. Progressors | VCs vs. Progressors | HVL LTNPs vs. Progressors | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | CI | P | OR | CI | P | OR | CI | P | OR | CI | P | |
| 5’UTR-2SNP-hap | ||||||||||||
| 5’UTR-2SNP-hap – allelic | 1.72 | 1.07–2.77 | 0.03 | 1.94 | 0.96–3.94 | 0.087 | 1.67 | 0.93–2.98 | 0.11 | 1.49 | 0.59–3.76 | 0.49 |
| 5’UTR-2SNP-hap – heterozygosity (WT/Mt) | 3.08 | 1.46–6.49 | 0.003 (0.048*) | 3.67 | 1.28–10.47 | 0.017 | 3.38 | 1.38–8.32 | 0.012 | 1.63 | 0.41–6.47 | 0.71 |
| 5’UTR-2SNP-hap – dominant mode | 2.86 | 1.41–5.79 | 0.003 (0.048*) | 3.39 | 1.28–9.00 | 0.019 | 2.95 | 1.28–6.79 | 0.017 | 1.78 | 0.47–6.78 | 0.46 |
| 3’UTR +2919 SNP | ||||||||||||
| +2919T>G SNP – allelic | 1.9 | 1.18–3.09 | 0.01 | 1.89 | 0.93–3.83 | 0.08 | 1.83 | 1.02–3.31 | 0.06 | 2.21 | 0.82–5.95 | 0.17 |
| +2919T>G SNP – heterozygosity (WT/Mt) | 3.33 | 1.59–7.00 | 0.002 (0.032*) | 2.75 | 1.00–7.60 | 0.06 | 3.7 | 1.5–8.92 | 0.004 | 3.75 | 0.95–14.88 | 0.07 |
| +2919T>G SNP – dominant mode | 3.2 | 1.58–6.48 | 0.001 (0.016*) | 2.85 | 1.08–7.56 | 0.04 | 3.28 | 1.43–7.57 | 0.006 | 3.73 | 1.02–13.70 | 0.07 |
p value after Bonferroni correction for multiple comparisons. Other comparisons did not maintain significance after Bonferroni correction
Shaded blocks indicate significant comparisons
ECs: elite controllers, VCs: viraemic controllers, HVL LTNPs: high vi ral load long term non-progressors OR: odds ratio, CI: 95% confidence i nterval
Figure 2.
Bar graph showing the genotypic frequency of 5’UTR-2SNP-hap (−2459G>A and −2135T>C) in black South African controllers, controller subgroups (ECs, VCs and HVL LTNPs) and progressors. ECs: elite controllers, VCs: viraemic controllers, HVL LTNPs: high viral load long-term non-progressors. WT: wild type allele, Mt: mutant allele
The CCR5 −4223C>T SNP had previously been genotyped for a subset of these individuals (52 controllers and 66 progressors) [6]. We developed an allelic discrimination assay and genotyped the remaining 19 controllers and 8 progressors. Although there is very strong LD between the −4223C>T SNP and the HHA haplotype in our study, one individual out of the twelve possessing the −4223 T allele in the complete cohort (controllers and progressors) did not possess an HHA haplotype (a progressor). Among controllers, the −4223C>T SNP was exclusively found in the non-EC controllers. Furthermore, we noted that three out of five progressors that harboured the −4223C>T SNP also possessed the HHE haplotype. Although this SNP was not significantly overrepresented in controllers compared to progressors, taking into account that the HHE haplotype has reported high transcriptional promoter activity [18] and could therefore potentially negate the effect of the −4223C>T SNP, we considered removing the progressors with an HHE haplotype to be a more informative comparison. Viraemic controllers had significantly higher representation of the −4223C>T SNP in the absence of HHE compared to progressors (p=0.04; OR=0.18; 0 =0.03–0.97), and when we compared the non-EC controllers to progressors in the absence of HHE, this relationship was strengthened (p=0.03; OR=0.16; 0=0.03–0.82).
3.2. CCR5 3’UTR: variability and HIV-1 control
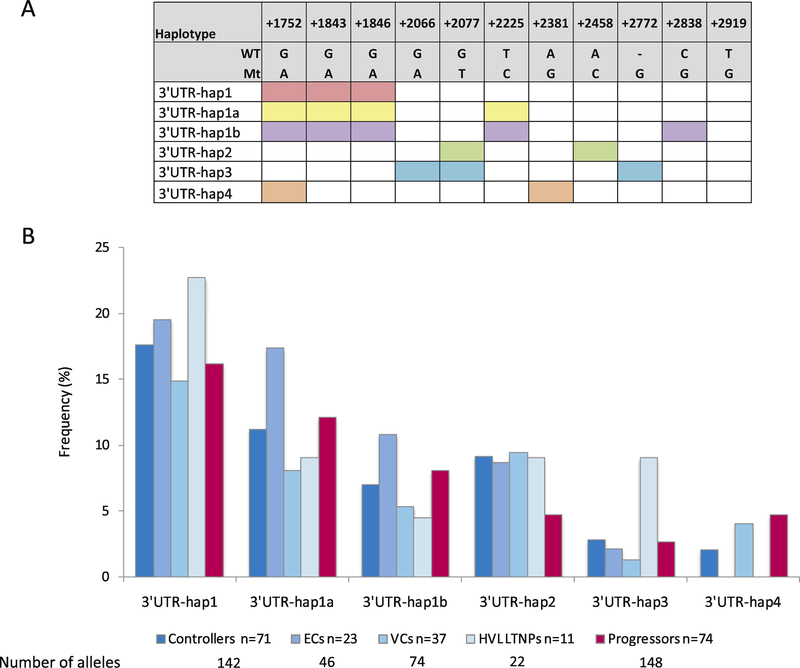
Sequencing the CCR5 3’UTR and analyzing the resulting chromatograms revealed eleven previously reported variants at positions +1752 (rs41495153), +1843 (rs41418945), +1846 (rs41466044), +2066 [17], +2077 (rs1800874), +2225 (rs41535253), +2381 (rs550958125), +2458 (rs3188094), +2772 [17], +2838 (rs41512547) and +2919 (rs746492). The genotypic frequencies of all identified 3’UTR variants in the respective groups are shown in Supplementary Table 3.
A CCR5 3’UTR indel (G insertion) at position +2772 was identified in black and Caucasian South African HIV-1-uninfected populations in a study by Picton et al. [17]. In our study, this indel was found to be underrepresented in the total controllers compared to progressors (p=0.08; OR=2.8; CI=0.95–8.36). Additionally, the 3’UTR +2919T>G SNP was differentially represented between controllers and progressors. Table 2 shows results of the comparison of representation of the 3’UTR +2919T>G SNP between controller and progressor groups. The minor allele (G) showed trends of lower representation in both ECs and VCs compared to progressors (Table 2) and was significantly underrepresented in the total controllers compared to progressors (p=0.01; OR=1.90; 0=1.18–3.09). Comparison of +2919T>G genotypes showed heterozygosity (TG) to be underrepresented in all controller groups compared to progressors (Table 2), and was significant in the VCs (p=0.004; OR=3.65; CI=1.50–8.92) and the total group comparison (p=0.002; OR=3.33; CI=1.59–6.99, pbonferroni=0.032). Comparison of the +2919T>G SNP in the dominant mode (i.e. TG+GG) revealed the most significant result however, with lower representation in all controller subgroups compared to progressors (Table 2), that were significant in ECs (p=0.04; OR=2.85; CI=1.08–7.56), VCs (p=0.006; OR=3.28; CI=1.42–7.57), but most significant in the total controller group comparison with 49.3% representation in the controllers compared to 75.7% in progressors (p=0.001; OR=3.2; CI=1.58–6.48, pbonferroni,=0.016).
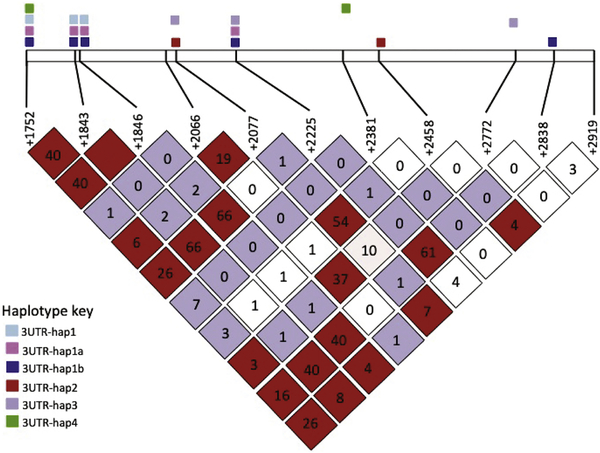
Visual examination of the data revealed obvious linkage between select SNPs in the CCR5 3’UTR and were thus analysed as haplotypes. The linkage disequilibrium (LD) plot showing linkage between the 3’UTR SNPs is shown in Figure 3. In total, six novel haplotypes were identified. The polymorphisms making up each haplotype and the allelic frequency of these haplotypes in controllers, controller subgroups and progressors are shown in Figures 4A and 4B, respectively. No 3’UTR haplotypes were found to differ significantly in representation between the controllers or controller subgroups compared to progressors. Linkage patterns between the SNPs in the CCR5 3’UTR were complex and interesting. Single nucleotide polymorphisms +1843G>A and +1846G>A were in complete LD (r2=1) and had directional LD with the +1752G>A SNP (r2=0.4); i.e. they always occurred with the upstream +1752G>A SNP (3’UTR-hap1), however the +1752G>A SNP was found in the absence of the +1843G>A/+1846G>A haplotype. The +2225T>C SNP also had directional LD with 3’UTR-hap1 (r2=0.66; 3’UTR Hap1a) and the +2838C>G SNP had directional LD with 3’UTR-hap1a. The +2458A>C SNP had directional linkage with the +2077G>T SNP (r2=0.54; 3’UTR-hap2). Except for 7 individuals (6 controllers and 1 progressor), the +2077G>T SNP never occurred with 3’UTR-hap1a (43/50). When occurring together, SNPs +2066G>A and +2077G>T were in complete directional LD with the +2772 indel (r2=1; 3’UTR-hap3). The +2381A>G SNP had directional linkage with the +1752 SNPG>A (r2=1; 3’UTR-hap4).
Figure 3.
Linkage disequilibrium (LD) plot depicting the linkage between CCR5 3’UTR variants. The numbers in the triangles indicate the r2 values, with 0 indicating no linkage and blank red triangles (100) indicating complete linkage between the respective variants. Polymorphisms making up the various 3’UTR haplotypes are marked with coloured boxes at the top of the figure; SNPs not part of the haplotypes are not marked with coloured boxes
Figure 4.
A. Schematic showing polymorphisms that form each haplotype. Colour shaded boxes show the respective SNPs or indels that form each haplotype B. Bar graph showing the allelic frequencies (%) of CCR5 3’UTR haplotypes in black South African controllers, controller subgroups (ECs, VCs and HVL LTNPs) and progressors. ECs: elite controllers, VCs: viraemic controllers, HVL LTNPs: high viral load long-term non-progressors. WT: wild type allele, Mt: mutant allele
3.3. Haplotypes spanning the CCR5 5’UTR and 3’UTR
Linkage disequilibrium between SNPs in the CCR5 5’UTR and the CCR5 3’UTR regions was investigated. Visual analysis and Haploview software revealed fourteen haplotypes spanning the CCR5 5’UTR and 3’UTR regions (Figure 5). Picton et al. (2010) [17] previously identified four of the fourteen haplotypes: HHA and +1752G>A SNP, HHE and +2919T>G SNP, HHF and +2919T>G SNP, HHC and +2077G>T SNP, in order of decreasing frequency.
Figure 5.
Schematic showing polymorphisms that form haplotypes spanning the CCR5 5’UTR and 3’UTR in black South African controllers, controller subgroups (ECs, VCs and HVL LTNPs) and progressors. Colour shaded boxes show the respective SNPs or indels that form each haplotype. 3’UTR SNPs that have linkage with the same root 5’UTR haplotype are shown in the same colour. ECs: elite controllers, VCs: viraemic controllers, HVL LTNPs: high viral load long-term non-progressors. WT: wild type allele, Mt: mutant allele
Possession of both 5’UTR-2SNP-hap and the +2919T>G SNP (TG+GG) was significantly underrepresented in total controllers compared to progressors (p=0.009; OR=1.97; CI=1.2–3.24) and this relationship was strengthened when looking at possession of both 5’UTR-2SNP-hap and +2919T>G SNP heterozygosity (p=0.004; OR=2.84; CI=1.43–5.66). Elite controllers also had significantly less 5’UTR-2SNP-hap and the +2919T>G SNP (TG+GG) and 5’UTR-2SNP-hap heterozygosity and +2919T>G SNP heterozygosity compared to progressors (p=0.04; OR=2.29; CI=1.08–4.87 and p=0.031; OR=3.16; CI=1.12–8.90, respectively). The +2919T>G SNP showed strong LD with the HHE, HHF*1, HHF*2 and HHG*1 promoter haplotypes (r2=0.73, D’=0.89). Comparison of individuals having both the HHE haplotype and the +2919T>G SNP (TG+GG) showed a significant association with HIV-1 progression when comparing progressors to VCs (p=0.010; OR=3.51; CI=1.30–9.46) and the total group of controllers (p=0.008; OR=2.61; CI=1.30–5.22). More significantly, progressors were more likely to have both the HHE haplotype and +2919T>G SNP heterozygosity (p=0.003; OR=3.5; CI=1.5–8.2, pbonferroni=0.048). Since the +2919T>G SNP is in LD with HHE, HHF and HHG, we also looked at the combinatorial effect of having these haplotypes together with the +2919T>G SNP. The representation of individuals with either HHE and/or HHF and/or HHG*1 together with the 3’UTR +2919T>G SNP (TG+GG) was again underrepresented in the total group of controllers when compared to progressors (p=0.027; OR=1.80; CI=1.09–2.89) and in the ECs compared to progressors (p=0.04; OR=2.29; CI=1.08–4.87).
When comparing controllers and progressors who possessed HHE or HHF*1 or HHF*2 or HHG*1 and the +2772−/G indel, the total controllers had significantly lower instances of this occurring (p=0.009; OR=6.67; CI=1.44–31.02). In addition, when comparing ECs + VCs to progressors as well as the total controller group to progressors, progressors were more likely to possess an HHE promoter haplotype, the 3’UTR +2772 insertion and the 3’UTR +2919T>G SNP (p=0.042; OR=8.17; 0=1.00–66.43 and p=0.056; OR=4.77; CI= 0.99–22.94, respectively). However, it is necessary to again note the large confidence intervals of these findings, suggesting that further study in larger cohorts is needed to determine true significance.
3.4. Potential markers for HIV-1 progression
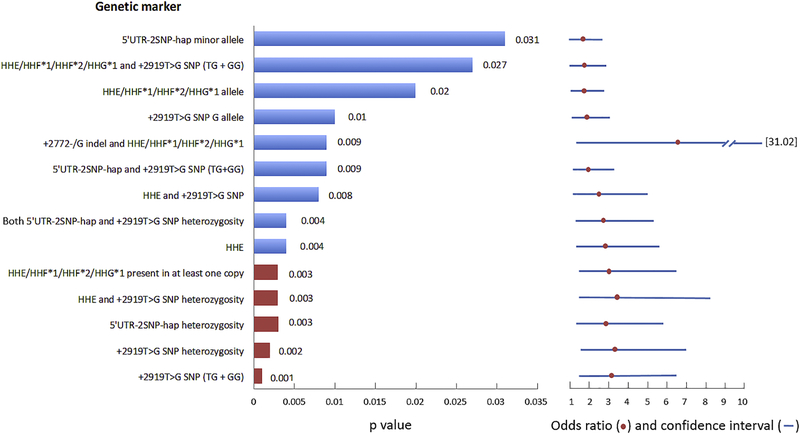
The variants that were the strongest indicators of HIV-1 progression in our study were the 3’UTR +2919T>G SNP in dominant mode (controllers vs. progressors: p=0.001, pbonferroni=0.016), heterozygosity for the 3’UTR +2919T>G SNP (controllers vs. progressors: p=0.002, pbonferroni=0.032) and heterozygosity for 5’UTR-2SNP-hap (controllers vs. progressors: p=0.003, pbonferroni=0.048). A summary of all deleterious associations (total controllers vs. progressors) is shown in Figure 6.
Figure 6.
Schematic showing a summary of the p values of all significant CCR5 genetic markers predictive of HIV-1 progression in black South Africans (HIV-1 total controllers vs. progressors). The smaller the p value is, the more predictive the biomarker. Red bars indicate p values that maintain significance post correction for multiple comparisons (Bonferroni). Blue horizontal lines indicate the 95% confidence intervals and the red circles indicate the odds ratios for the respective comparisons
4. Discussion
In this study, we characterized polymorphisms (SNPs and indels) and intragenic haplotypes found within the cis-regulatory regions of CCR5 in a group of subtype C infected black HIV-1 controllers and progressors from South Africa. Previously unreported polymorphisms and haplotypes have been identified and previously defined haplotypes within the CCR5 gene have been expanded upon. Two promoter variants and one 3’UTR variant were found to significantly differentiate progressors from controllers.
The −4223C>T SNP, shown to disrupt the CpG-41 site [6], is found to be more prevalent in individuals with the CCR5 HHA haplotype and although the −4223C>T SNP was associated with protection from HIV-1 acquisition in black South Africans in a previous study [6], associations did not reach statistical significance in that study or in ours. In our study, all seven controllers with this SNP had at least one HHA haplotype. In the progressors, four of the five individuals possessing the −4223 T allele had at least one HHA haplotype, however, three of the five progressors possessing the −4223 T allele also had an HHE haplotype. It is also interesting to note that no ECs possessed the −4223 T allele, indicating that the −4223C>T polymorphism may only be helpful in the presence of higher viraemia. Exclusion of non-viraemic controllers (i.e. ECs) and progressors harbouring the HHE haplotype from the comparison revealed a significant protective effect of the −4223C>T SNP, with higher representation in controllers with higher viraemia (VCs + HVL LTNPs) compared to progressors.
The distribution of CCR5 gene promoter haplotypes is highly variable amongst ethnic groups. The various haplotypes have different promoter activity, correlating with differential cell surface CCR5 expression [6,7,18–20]. Joshi et al. [18] calculated the relative promoter activity (RPA) of each CCR5 5’UTR haplotype in vitro using a luciferase-based assay and normalizing to the ancestral HHA haplotype. The combined relative promoter activity based on both alleles was also calculated. HHA, HHB, HHC and HHD had a low RPA (i.e. low expected CCR5 expression) whereas HHE, HHF and HHG had a high RPA (i.e. high expected CCR5 expression) [18].
The HHE haplotype has been described as deleterious in the context of HIV-1 control across multiple ethnicities and studies [12,21–23]. In agreement, in this study, HHE was significantly underrepresented in the total group of controllers compared to progressors as well as in the VCs compared to the progressors. Possession of either HHE, HHF*1, HHF*2 or HHG*1 associated significantly with progression, although comparisons of just HHE possession gave a more significant result. While the HHE haplotype has been identified as deleterious, homozygosity for HHE (HHE/HHE) was not shown to associate with disease-modifying effects in African Americans whereas in Caucasians, HHE homozygosity, but not HHE heterozygosity, associated with disease acceleration [12]. Therefore, it is not only an individual CCR5 haplotype that has consequences on HIV-1 disease, but also the combinatorial (additive and/or interactive) effect of an individual’s CCR5 genotype.
The CCR5 haplotype pair (genotype) in an individual and its relationship to the CCR5 levels on CD4+ T cells is partly related to whether one or both haplotypes have increased or decreased sensitivity to activation-associated demethylation [6]. In our study, only one individual, a progressor, had the HHE/HHE genotype, indicating that it is a rare genotype in black South Africans and may not be contributing to HIV-1 disease progression or acquisition. Genotypes with at least one HHE haplotype, compared with genotypes lacking an HHE haplotype, have been associated with higher CCR5 levels [6].
In our study, HHA/HHC was significantly overrepresented in the total group of controllers compared to the progressors as well as in the VCs compared to the progressors. In African Americans, possession of an HHC haplotype has been associated with disease acceleration [12]. However, if an HHC haplotype was paired with one of the haplotypes that was associated with protection in African Americans (HHA or HHF*2), the disease-accelerating effects of the HHC haplotype were negated, as can be seen in our black South African population, where the pairing of HHA (protective in African Americans [12]) and HHC (deleterious in African Americans [12]) i.e. the HHA/HHC genotype, was found to be protective. The HHA and HHC haplotypes have been reported to have reduced sensitivity to activation-associated demethylation [6], possibly revealing a functional cause as to why HHA/HHC may be protective in black South Africans.
It is interesting to note that, while not significant, there was a complete absence of HHF*2, a relatively prevalent haplotype, in the HVL LTNP group. Also, the HHG*1 haplotype was found at a comparatively high frequency (18.2%) compared to the other two controller subgroups (ECs=6.5%; VCs=4.1%) and the progressors (6.8%), suggesting a reciprocal relationship of these two haplotypes in the HVL LTNPs compared to the other groups. Since HHF*2 is defined by the presence of the CCR2-V64I variant, CCR2 may be playing a role in control in this group. HVL LTNPs are a very unique group in adults [24–27] and to our knowledge, we have the largest cohort of such individuals worldwide (n=11). This makes it very difficult to verify the role of HHF*2 in this method of control of HIV-1 disease in different cohorts.
Several individual SNPs located in the CCR5 promoter have been reported to affect the expression of CCR5. The −2459G>A SNP (rs1799987) is in complete LD with the −2135T>C SNP (rs1799988).The −2459G>A SNP (and therefore the −2135T>C SNP by association) has been linked to differences in CCR5 expression levels on CD14+ monocytes [28] and has been associated with the rate of progression to AIDS [29–31]. In our study, the −2459G>A and −2135T>C SNPs (5’UTR-2SNP-hap) were significantly underrepresented in controllers compared to progressors. 5’UTR-2SNP-hap is common to the HHE, HHF and HHG haplotypes (which have been reported to be high promoter activity haplotypes [7,18]). Individuals heterozygous for 5’UTR-2SNP-hap were significantly less likely to control HIV-1 in total group comparisons as well as in ECs and VCs compared to progressors and possession of the minor allele of 5’UTR-2SNP-hap was significantly underrepresented in controllers when compared to progressors. In a cohort of self-identified white and black patients, a study observed that the −2459G>A SNP had a strong association with the time taken to achieve virologic success of highly active anti-retroviral therapy (HAART) in black but not in white patients (p=0.04), and that this association increased with stronger African ancestry [32].
The 3’UTR plays a major role in gene expression and regulation by influencing the localization, stability, export, and translation efficiency of an mRNA [33].The linkage patterns in the 3’UTR were complex, suggesting interesting selection pressures that are likely to have consequential functional implications that have not yet been elucidated. In the CCR5 3’UTR, the +2919T>G SNP G allele was significantly underrepresented in controllers compared to progressors. Possession of the +2919T>G SNP in the dominant mode was significantly associated with HIV-1 control. In addition, the +2919T>G TG genotype was significantly underrepresented in controllers when compared to progressors. Interestingly, the allelic and genotypic representation of the +2919T>G SNP in progressors in our study was more similar to a European population than other African populations, suggesting that the represe ntation in controllers is more reflective of the background population and that the representation in progressors is skewed (see Supplementary Figure 1). The +2919T>G SNP was in strong LD with the CCR5 promoter HHE, HHF*1, HHF*2 and HHG*1 haplotypes and subsequently 5’UTR-2SNP-hap (common to these haplotypes), thus begging the question as to which polymorphism is functionally driving the deleterious effect on HIV-1 control in black South Africans. While possession of the HHE haplotype alone, the HHE haploty pe with the +2919T>G SNP, or any of the remaining three deleterious haplotypes (HHF*1, HHF*2, HHG*1) with the +2919T>G SNP all significantly associated with HIV-1 progression (p=0.004; p=0.003, pbonferroni=0.048; and p=0.027 respectively), there was a stronger association with progression when looking at possession of the +2919T>G SNP in the dominant mode alone (p=0.001. pbonferroni=0.016). To our knowledge, no other studies have associated this SNP with increased risk of HIV-1 disease acquisition or more rapid progression of HIV-1 in any other population.
This study further emphasizes the need for population specific studies with regards to potential genetic markers and HIV-1 control. Individuals in sub-Saharan Africa remain severely understudied when compared with other HIV-1 infected populations around the world. Overall, our results reproduce other studies with regards to the CCR5 HHE haplotype being deleterious for HIV-1 disease progression. We found that the HHA haplotype and HHA/HHC genotype associated with protection from HIV-1 disease progression, while the HHE haplotype associated with deleterious disease outcomes in our population. We have characterized novel haplotypes in the 3’UTR as well as haplotypes spanning the CCR5 5’UT R and 3’UTR. Our results suggest that two CCR5 promoter SNPs (−2459G>A and −2135T>C) and one CCR53’UTR SNP (+2919T>G) may be key functional variants with regards to HIV-1 control in black South Africans. Limitations of our study include the small number of individuals in our cohorts, however the extreme phenotypes of these groups often negate the need to test extremely large numbers. It is important to note that only select comparisons remained significant after Bonferroni correction for multiple comparisons, thus future work including functional studies will help to reinforce our findings.
In conclusion, we propose that possession of the 3’UTR +2919T>G SNP in the dominant mode, the strongest predictor for HIV-1 progression in this study (p=0.001, pbonferroni,=0.016), can be used as a marker for accelerated disease progression in black South Africans.
Supplementary Material
Acknowledgements
CT, MP, AP – conception and design of the study
NM, RC, OE – recruitment of patients
GK, MP, AP, FK – acquisition of data and analysis
GK, MP – interpretation of data
SL, MP – design of an assay
GK, MP – drafting the article
MP, CT, WH, SA – revising it critically for important intellectual content
MP, CT – final approval of the version to be submitted.
Funding sources
This work is based on the research supported by grants awards from the Strategic Health Innovation Partnerships (SHIP) Unit of the South African Medical Research Council, a grantee of the Bill & Melinda Gates Foundation, and the South African Research Chairs Initiative of the Department of Science and Technology and National Research Foundation of South Africa.
Footnotes
None of the authors have any potential financial conflicts of interest related to this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Silvestri G, AIDS pathogenesis: a tale of two monkeys., J. Med. Primatol 37 Suppl 2 (2008) 6–12. doi : 10.1111/j.1600-0684.2008.00328.x. [DOI] [PubMed] [Google Scholar]
- [2].Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, Cirulli ET, Urban TJ, Zhang K, Gumbs CE, Smith JP, Castagna A, Cozzi-Lepri A, De Luca A, Easterbrook P, Günthard HF, Mallal S, Mussini C, Dalmau J, Martinez-Picado J, Miro JM, Obel N, Wolinsky SM, Martinson JJ, Detels R, Margolick JB, Jacobson LP, Descombes P, Antonarakis SE, Beckmann JS, O’Brien SJ, Letvin NL, McMichael AJ, Haynes BF, Carrington M, Feng S, Telenti A, Goldstein DB, N.C. for H.V.I. (CHAVI), Common Genetic Variation and the Control of HIV-1 in Humans, PLOS Genet. 5 (2009) e1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].de Roda Husman A M K, M C, et al. , Association between CCR5 Genotype and the Clinical Course of HIV-1 Infection, Ann. Intern. Med 127 (1997) 882–890. doi: 10.7326/0003-4819-127-10-199711150-00004. [DOI] [PubMed] [Google Scholar]
- [4].Menten P, Wuyts A, Van Damme J, Macrophage inflammatory protein-1, Cytokine Growth Factor Rev. 13 (2002) 455–481. doi: 10.1016/S1359-6101(02)00045-X. [DOI] [PubMed] [Google Scholar]
- [5].Sabeti PC, Walsh E, Schaffner SF, Varilly P, Fry B, Hutcheson HB, Cullen M, Mikkelsen TS, Roy J, Patterson N, Cooper R, Reich D, Altshuler D, O’Brien S, Lander ES, The Case for Selection at CCR5-Δ32, PLOS Biol. 3 (2005) e378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gornalusse GG, Mummidi S, Gaitan AA, Jimenez F, Ramsuran V, Picton A, Rogers K, Manoharan MS, Avadhanam N, Murthy KK, Martinez H, Murillo A. Molano, Chykarenko ZA, Hutt R, Daskalakis D, Shostakovich-Koretskaya L, Karim S. Abdool, Martin JN, Deeks SG, Hecht F, Sinclair E, Clark RA, Okulicz J, Valentine FT, Martinson N, Tiemessen CT, Ndung’u T, Hunt PW, He W, Ahuja SK, Epigenetic mechanisms, T-cell activation, and CCR5 genetics interact to regulate T-cell expression of CCR5, the major HIV-1 coreceptor, Proc. Natl. Acad. Sci. U. S. A 112 (2015) E4762–E4771. doi: 10.1073/pnas.1423228112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mummidi S, Bamshad M, Ahuja SS, Gonzalez E, Feuillet PM, Begum K, Galvis MC, Kostecki V, Valente AJ, Murthy KK, Haro L, Dolan MJ, Allan JS, Ahuja SK, Evolution of Human and Non-human Primate CC Chemokine Receptor 5 Gene and mRNA: Potential Roles for Haplotype and mRNA Diversity, Differential Haplotype-Specific Transcriptional Activity, and Altered Transcription Factor Binding to Polymorphis Nucleotides, J. Biol. Chem 275 (2000) 18946–18961. doi: 10.1074/jbc.M000169200. [DOI] [PubMed] [Google Scholar]
- [8].Mangano A, Gonzalez E, Dhanda R, Catano G, Bamshad M, Bock A, Duggirala R, Williams K, Mummidi S, Clark RA, Ahuja SS, Dolan MJ, Bologna R, Sen L, Ahuja SK, Concordance between the CC Chemokine Receptor 5 Genetic Determinants That Alter Risks of Transmission and Disease Progression in Children Exposed Perinatally to Human Immunodeficiency Virus, J. Infect. Dis 183 (2001) 1574–1585. [DOI] [PubMed] [Google Scholar]
- [9].Smith MW, Dean M, Carrington M, Winkler C, Huttley GA, Lomb DA, Goedert JJ, O'Brien TR, Jacobson LP, Kaslow R, Buchbinder S, Vittinghoff E, Vlahov D, Hoots K, Hilgartner MW, M.A.C.S. (MACS) Study Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE, O'Brien SJ, Contrasting Genetic Influence of CCR2 and CCR5 Variants on HIV-1 Infection and Disease Progression, Science (80−. ). 277 (1997) 959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- [10].Okulicz JF, Marconi VC, Landrum ML, Wegner S, Weintrob A, Ganesan A, Hale B, Crum-Cianflone N, Delmar J, Barthel V, Quinnan G, Agan BK, Dolan MJ, Clinical outcomes of elite controllers, viremic controllers, and long-term nonprogressors in the US Department of Defense HIV natural history study., J. Infect. Dis 200 (2009) 1714–1723. doi : 10.1086/646609. [DOI] [PubMed] [Google Scholar]
- [11].Saez-Cirion A, Pancino G, Sinet M, Venet A, Lambotte O, HIV controllers: how do they tame the virus?, Trends Immunol. 28 (2007) 532–540. doi: 10.1016/j.it.2007.09.002. [DOI] [PubMed] [Google Scholar]
- [12].Gonzalez E, Bamshad M, Sato N, Mummidi S, Dhanda R, Catano G, Cabrera S, McBride M, Cao XH, Merrill G, O’Connell P, Bowden DW, Freedman BI, Anderson SA, Walter EA, Evans JS, Stephan KT, Clark RA, Tyagi S, Ahuja SS, Dolan MJ, Ahuja SK, Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes., Proc. Natl. Acad. Sci. U. S. A 96 (1999) 12004–12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Picton AC, HIV-1 Coreceptor CCR5: Gene Characterization and Expression, University of the Witwatersrand, 2013. [Google Scholar]
- [14].Loubser SA, The Multiple Roles of HLA in HIV Immunity and Treatment, University of the Witwatersrand, 2015. [Google Scholar]
- [15].Barrett JC, Fry B, Maller J, Daly MJ, Haploview: analysis and visualization of LD and haplotype maps., Bioinformatics. 21 (2005) 263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- [16].VassarStats: Statistical Computation Web Site., (n.d.). doi:citeulike-article-id:11901322.
- [17].Picton ACP, Paximadis M, Tiemessen CT, Genetic variation within the gene encodi ng the HIV-1 CCR5 coreceptor in two South African populations, Infect. Genet. Evol 10 (2010) 487–494. doi : 10.1016/j.meegid.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Joshi A, Punke EB, Sedano M, Beauchamp B, Patel R, Hossenlopp C, Alozie OK, Gupta J, Mukherjee D, Garg H, CCR5 promoter activity correlates with HIV disease progression by regulating CCR5 cell surface expression and CD4 T cell apoptosis, Sci. Rep 7 (2017) 232. doi: 10.1038/s41598-017-00192-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jiang D, Mummidi S, Ahuja SK, Jarrett HW, CCR5 promoter haplotype transcription complex characterization, J. Health Care Poor Underserved. 22 (2011) 73–90. doi: 10.1353/hpu.2011.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liu H, Nakayama EE, Theodorou I, Nagai Y, Likanonsakul S, Wasi C, Debre P, Iwamoto A, Shioda T, Polymorphisms in CCR5 chemokine receptor gene in Japan, Int. J. Immunogenet 34 (2007) 325–335. doi : 10.1111/j.1744-313X.2007.00694.x. [DOI] [PubMed] [Google Scholar]
- [21].Martin MP, Dean M, Smith MW, Winkler C, Gerrard B, Michael NL, Lee B, Doms RW, Margolick J, Buchbinder S, Goedert JJ, O’Brien TR, Hilgartner MW, Vlahov D, O’Brien SJ, Carrington M, Genetic acceleration of AIDS progression by a promoter variant of CCR5., Science. 282 (1998) 1907–1911. [DOI] [PubMed] [Google Scholar]
- [22].Tang J, Shelton B, Makhatadze NJ, Zhang Y, Schaen M, Louie LG, Goedert JJ, Seaberg EC, Margolick JB, Mellors J, Kaslow RA, Distribution of Chemokine Receptor <em>CCR2</em> and <em>CCR5</em> Genotypes and Their Relative Contribution to Human Immunodeficiency Virus Type 1 (HIV-1) Seroconversion, Early HIV-1 RNA Concentration in Plasma, and Later Disease, J. Virol 76 (2002) 662–672. doi: 10.1128/JVI.76.2.662-672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hladik F, Liu H, Speelmon E, Livingston-Rosanoff D, Wilson S, Sakchalathorn P, Hwangbo Y, Greene B, Zhu T, McElrath MJ, Combined effect of CCR5-Delta32 heterozygosity and the CCR5 promoter polymorphism −2459 A/G on CCR5 expression and resistance to human immunodeficiency virus type 1 transmission, J. Virol 79 (2005) 11677–11684. doi: 10.1128/JVI.79.18.11677-11684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Muenchhoff M, Adland E, Karimanzira O, Crowther C, Pace M, Csala A, Leitman E, Moonsamy A, McGregor C, Hurst J, Groll A, Mori M, Sinmyee S, Thobakgale C, Tudor-Williams G, Prendergast AJ, Kloverpris H, Roider J, Leslie A, Shingadia D, Brits T, Daniels S, Frater J, Willberg CB, Walker BD, Ndung’u T, Jooste P, Moore PL, Morris L, Goulder P, Nonprogressing HIV-infected children share fundamental immunological features of nonpathogenic SIV infection., Sci. Transl. Med 8 (2016) 358ra125. doi : 10.1126/scitranslmed.aag1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ssewanyana I, Elrefaei M, Dorsey G, Ruel T, Jones NG, Gasasira A, Kamya M, Nakiwala J, Achan J, Charlebois E, Haviir D, Cao H, Profile of T Cell Immune Responses in HIV-infected Children from Uganda, J. Infect. Dis 196 (2007) 1667–1670. doi: 10.1086/522013. [DOI] [PubMed] [Google Scholar]
- [26].Klatt NR, Bosinger SE, Peck M, Richert-Spuhler LE, Heigele A, Gile JP, Patel N, Taaffe J, Julg B, Camerini D, Torti C, Martin JN, Deeks SG, Sinclair E, Hecht FM, Lederman MM, Paiardini M, Kirchhoff F, Brenchley JM, Hunt PW, Silvestri G, Limited HIV Infection of Central Memory and Stem Cell Memory CD4+ T Cells Is Associated with Lack of Progression in Viremic Individuals, PLOS Pathog. 10 (2014) e1004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Goulder P, Deeks SG, HIV control: Is getting there the same as staying there?, PLOS Pathog. 14 (2018) e1007222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Salkowitz JR, Bruse S, Meyerson H, Valdez H, Mosier D, Harding C, Zimmerman P, Lederman MM, CCR5 promoter polymorphism determines macrophage CCR5 density and magnitude of HIV-1 propagation in vitro, 2003. doi: 10.1016/S1521-6616(03)00147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].McDermott DH, Zimmerman PA, Guignard F, Kleeberger CA, Leitman SF, Murphy PM, CCR5 promoter polymorphism and HIV-1 disease progression. Multicenter AIDS Cohort Study (MACS)., Lancet (London, England). 352 (1998) 866–870. [DOI] [PubMed] [Google Scholar]
- [30].Clegg AO, Ashton LJ, Biti RA, Badhwar P, Williamson P, Kaldor JM, Stewart GJ, CCR5 promoter polymorphisms, CCR5 59029A and CCR5 59353C, are under represented in HIV-1-infected long-term non- progressors. The Australian Long-Term Non-Progressor Study Group., AIDS. 14 (2000) 103–108. [DOI] [PubMed] [Google Scholar]
- [31].Knudsen TB, Kristiansen TB, Katzenstein TL, Eugen-Olsen J, Adverse effect of the CCR5 promoter −2459A allele on HIV-1 disease progression., J. Med. Virol 65 (2001) 441–444. [PubMed] [Google Scholar]
- [32].Cheruvu VK, Igo RP Jr, Jurevic RJ, Serre D, Zimmerman PA, Rodriguez B, Mehlotra RK, African ancestry influences CCR5 −2459G>A genotype-associated virologic success of highly active antiretroviral therapy, J. Acquir. Immune Defic. Syndr. 66 (2014) 102 −107. doi: 10.1097/QAI.0000000000000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mayr C, Regulation by 3’-Untranslated Regions, Annu. Rev. Genet 51 (2017) 171–194. doi: 10.1146/annurev-genet-120116-024704. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.