Abstract
Metagenomic and metatranscriptomic time-series data covering a 52-day period in the fall of 2016 provide an inventory of bacterial and archaeal community genes, transcripts, and taxonomy during an intense dinoflagellate bloom in Monterey Bay, CA, USA. The dataset comprises 84 metagenomes (0.8 terabases), 82 metatranscriptomes (1.1 terabases), and 88 16S rRNA amplicon libraries from samples collected on 41 dates. The dataset also includes 88 18S rRNA amplicon libraries, characterizing the taxonomy of the eukaryotic community during the bloom. Accompanying the sequence data are chemical and biological measurements associated with each sample. These datasets will facilitate studies of the structure and function of marine bacterial communities during episodic phytoplankton blooms.
Subject terms: Element cycles, Metagenomics, Sequencing, Microbial ecology
| Design Type(s) | transcription profiling design • sequence assembly objective • biodiversity assessment objective |
| Measurement Type(s) | transcription profiling assay • marine metagenome • microbial community |
| Technology Type(s) | RNA sequencing • DNA sequencing • amplicon sequencing |
| Factor Type(s) | assay protocol • temporal_instant |
| Sample Characteristic(s) | marine metagenome • Monterey Bay • ocean biome |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Background & Summary
In pelagic marine ecosystems, a major proportion of primary production is transformed by heterotrophic microbes on the scale of hours to days1–3. Much of this rapidly-processed primary production is made available in the form of dissolved organic carbon (DOC), released from phytoplankton by direct excretion or through trophic interactions. Bacterial uptake of DOC produces living biomass and regenerates inorganic nutrients1.
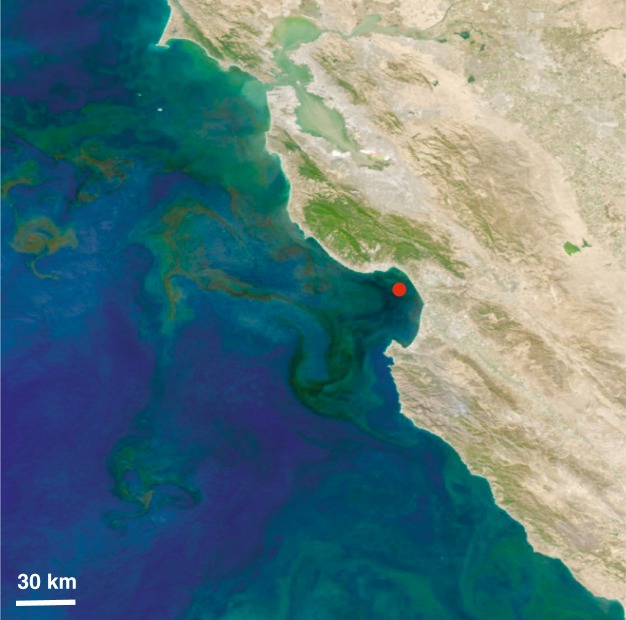
Monterey Bay is a coastal ecosystem with high primary production driven by frequent upwelling of nutrient-rich waters4,5. Intense phytoplankton blooms can develop6, and these vary dynamically in terms of taxonomic composition. In 2016, the fall phytoplankton bloom (Fig. 1) was dominated by an unusually intense bloom of the dinoflagellate Akashiwo sanguinea7. A. sanguinea cell abundances reached 4.9 × 106 cells L−1, and chlorophyll a concentrations reached 57 µg L−1 (at ~6 m depth) over the period spanning mid-September to mid-November. Here we present metagenomic, metatranscriptomic, and iTag data on the bacterial and archaeal communities during a 52-day period spanning this unusual plankton bloom in Monterey Bay (Table 1). iTag data on the eukaryotic microbial communities provides contextual information on community dynamics of the bloom-forming phytoplankton and grazer communities.
Fig. 1.
MODIS satellite image on September 26, 2016 of the phytoplankton bloom occurring in Monterey Bay and extending into the Pacific. The red dot represents the sampling station M0, located at 36.835 N, 121.901 W.
Table 1.
Sequence datasets from the fall bloom in Monterey Bay, CA, 2016.
| Source Name | Sampling Dates | Geographical Location | Sampling Method | Sequence Type | Sample Identifiers from GOLD (Gaxxx) or the JGI Portal (Project ID xxx) | BioProject Accession IDs from the NCBI SRA |
|---|---|---|---|---|---|---|
| Monterey Bay Station M0 | September 26 - November 16, 2016 | Monterey Bay, CA, USA, 36.835 N, 121.901 W | Autonomous collection by the Environmental Sample Processor and Niskin bottle sampling | All | Umbrella project PRJNA533622 | |
| Monterey Bay Station M0 | September 26 - November 16, 2016 | Monterey Bay, CA, USA, 36.835 N, 121.901 W | Autonomous collection by the Environmental Sample Processor and Niskin bottle sampling | Metagenomics | Ga0228601 - Ga0228678; GA0233393 - Ga0233402 | PRJNA467720 - PRJNA467773, PRJNA468208 - PRJNA468214, PRJNA502407 - PRJNA502427, PRJNA502440 - PRJNA502442 |
| Monterey Bay Station M0 | September 26 - November 16, 2016 | Monterey Bay, CA, USA, 36.835 N, 121.901 W | Autonomous collection by the Environmental Sample Processor and Niskin bottle sampling | Metatranscriptomics | Ga0228679 - Ga0232167; Ga0247556 - Ga0247607; Ga0256411 - Ga0256417 | PRJNA467774 - PRJNA467774, PRJNA468143 - PRJNA468143, PRJNA468299 - PRJNA468332, PRJNA502451 - PRJNA502468, PRJNA502608 - PRJNA502612 |
| Monterey Bay Station M0 | September 26 - November 16, 2016 | Monterey Bay, CA, USA, 36.835 N, 121.901 W | Autonomous collection by the Environmental Sample Processor and Niskin bottle sampling | 16S rRNA iTags | JGI Project ID 1190879 | PRJNA511156 - PRJNA511206, PRJNA511216 - PRJNA511252 |
| Monterey Bay Station M0 | September 26 - November 16, 2016 | Monterey Bay, CA, USA, 36.835 N, 121.901 W | Autonomous collection by the Environmental Sample Processor and Niskin bottle sampling | 18S rRNA iTags | JGI Project ID 1190880 | PRJNA511207 - PRJNA511215, PRJNA511253 - PRJNA511331 |
Methods
Sampling protocol
From September 26 through November 16, 2016, microbial cells were collected at Monterey Bay station M0 for sequence analysis. A moored autonomous robotic instrument, the Environmental Sample Processor (ESP)8, filtered up to 1 L of seawater sequentially through a 5.0 µm pore-size polyvinylidene fluoride filter to capture primarily eukaryotic microbes, which was stacked on top of a 0.22 µm pore-size polyvinylidene fluoride filter to capture primarily bacteria and archaea (Table 1). The samples were collected between 5 and 7 m depth at approximately 10 a.m. PST. Samples were collected daily except during October 7 – November 1 when the ESP was offline for repair. ESP filters were preserved with RNAlater at the completion of sample collection and stored in the instrument until retrieval. While the ESP was offline, grab samples were collected by Niskin bottle at the M0 mooring site 2–3 times per week, with time of sampling, depth of sampling, and filters the same as for the ESP samples except that filters were flash frozen in liquid nitrogen.
Environmental data (temperature, salinity, chlorophyll a fluorescence, light transmission, and dissolved O2 concentrations) were collected by a CTD instrument mounted with the ESP9. Additional environmental data were obtained from grab samples collected at the M0 mooring 2–3 times per week [total dimethylsulfoniopropionate concentration (DMSPt), dissolved DMSP concentration (DMSPd), DMSPd consumption rate, chlorophyll a, and cell counts by flow cytometry and microscopy]10,11 (Online-only Table 1).
DNA/RNA extraction
Total community nucleic acids for metagenome, metatranscriptome, and 16S iTag sequencing were obtained from the same 0.22 µm filter (0.22–5.0 μm size fraction) using the ZymoBIOMICS DNA/RNA Miniprep Kit (Zymo Research, Irvine CA). At extraction start, internal standards were added to the lysis buffer tube (see Usage Notes), and the filter was cut into small pieces under sterile conditions to facilitate extraction. RNA was treated according to the manufacturer’s instructions with in-column DNase I treatment. After elution, RNA was treated with Turbo DNase (Invitrogen, Carlsbad CA) and concentrated using Zymo RNA Clean and Concentrator (Zymo Research). Except for a few cases of low nucleic acid yields, duplicate filters were sequenced for each sample date.
DNA for 18S rRNA gene sequencing was extracted from the 5.0 μm filters using the DNeasy Plant Mini Kit (Qiagen, Venlo NL) with modifications. Filters were cut into pieces and added into a prepared lysis tube containing ~200 µl of 1:1 mixed 0.1 and 0.5 mm zirconia/silica beads (Biospec Products, Bartlesville, OK) and 400 μl Buffer AP1. Internal standards (see Usage Notes) were added just prior to extraction. Three freeze-thaw cycles were performed using liquid nitrogen and a 65 °C water bath. Following freeze-thaw, bead beating was performed for 10 min, followed by centrifugation at 8,000 rpm for 10 min to remove foam. Following centrifugation, 45 μl of proteinase K (>600 mAU/ml, Qiagen) was added to each tube and incubated at 55 °C for 90 min with gentle rotation. Filters were then removed and the tubes incubated at 55 °C for 1 h. The DNeasy kit protocol was resumed at the RNase A addition step. Final DNA was eluted in 75 μl of diluted (1:10) TE buffer.
Metagenome sequencing and analysis
Sequence data were generated at the Department of Energy (DOE) Joint Genome Institute (JGI) using Illumina technology. Libraries were constructed and sequenced using the HiSeq-2000 1TB platform (2 × 151 bp). For assembly, reads were trimmed and screened, and those with no mate pair were removed using BFC (v r181)12. Remaining reads were assembled using SPAdes (v 3.11.1)13. The read set was mapped to the final assembly and coverage information generated using BBMap (v 37.78)14 with default parameters. Assembled metagenomes were processed through the DOE JGI Metagenome Annotation Pipeline (MAP) and loaded into the Integrated Microbial Genomes and Microbiomes (IMG/M) platform15,16.
Metatranscriptome sequencing and analysis
Sequence data were generated at the DOE JGI using Illumina technology. Libraries were constructed and sequenced using the HiSeq-2500 1TB platform (2 × 151 bp). Metatranscriptome reads were assembled using MEGAHIT (v 1.1.2)17. Cleaned reads were mapped to the assembly using BBMap.
16S and 18S iTag sequencing and analysis
Sequence data were generated at the DOE JGI using Illumina technology. Primers 515FB18 (5′-GTGYCAGCMGCCGCGGTAA) and 806RB19 (5′-GGACTACNVGGGTWTCTAAT) were used for 16S rRNA gene amplification, and primers 565F (5′-CCAGCASCYGCGGTAATTCC) and 948R (5′-ACTTTCGTTCTTGATYRA) were used for 18S rRNA gene amplification20. Libraries were constructed and sequenced using the Illumina MiSeq platform (2 × 301 bp). Contaminant reads were removed using the kmer filter in BBDuk, and filtered reads were processed by the JGI iTagger (v 2.2) pipeline (https://bitbucket.org/berkeleylab/jgi_itagger).
To generate an overview of microbial community composition during the bloom (Figs 2 and 3), the 16S and 18S rRNA amplicon libraries (raw reads) were primer-trimmed using Cutadapt (v 1.18)21 and analyzed using QIIME2 (v 2018.6)22. The DADA223 plugin in QIIME2 was used to generate exact sequence variants (ESVs), which were classified using the QIIME2 naive Bayes classifier trained on 99% Operational Taxonomic Units (OTUs) from the SILVA rRNA database (v 132)24 after trimming to the primer region. Taxonomic bar plots were generated using QIIME2.
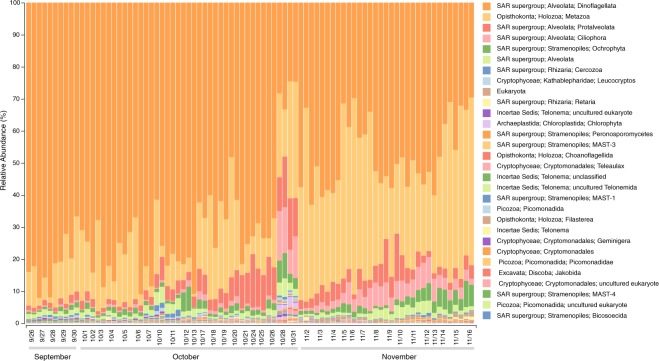
Fig. 2.
Relative abundance of bacterial and archaeal taxa at Monterey Bay station M0 during the fall of 2016. Samples were collected at ~6 m, and 16S rRNA genes were amplified from community DNA in the 0.22 to 5.0 µm size range. Taxonomic groups were defined based on exact sequence variants using DADA2 in QIIME 2 (https://qiime2.org) and assigned taxonomy with the naive Bayes q2-feature-classifier trained using the 515F/806R region from 99% operational taxonomic units from the SILVA 132 16S rRNA database. Assignments of the 30 most abundant taxa are given at the family level.
Fig. 3.
Relative abundance of eukaryotic taxa at Monterey Bay station M0 during the fall of 2016. Samples were collected at ~6 m, and 18S rRNA genes were amplified from community DNA in the >5.0 µm size range. Taxonomic groups were defined based on exact sequence variants using DADA2 in QIIME 2 (https://qiime2.org) and assigned taxonomy with the naive Bayes q2-feature-classifier trained using the 565F/948R region from 99% operational taxonomic units from the SILVA 132 18S rRNA database.
Data Records
The raw Illumina sequencing reads for metagenomes, metatranscriptomes, and 16S rRNA and 18S rRNA iTags are available from the NCBI Sequence Read Archive under 342 separate project IDs (summarised in Online-only Table 2) which we have gathered under a single BioProject umbrella ID25.
Contigs assembled within each individual metagenome and metatranscriptome are available from the JGI Integrated Microbial Genomes portal (Online-only Table 2).
Chemical and biological data associated with each sample are available at the Biological and Chemical Oceanography Data Management Office (BCO-DMO)9,10. Measured parameters include temperature, salinity, depth, light transmission, concentrations of dissolved oxygen and chlorophyll, concentration and consumption rates of DMSP, and cell counts for heterotrophic bacteria, Synechococcus, Akashiwo, and photosynthetic eukaryotes.
Technical Validation
For metagenomic and metatranscriptomic Illumina data, BBDuk (version 37.95; https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/bbduk-guide/) was used to remove contaminants, trim reads that contained adapter sequence, and trim reads where quality dropped to zero. BBDuk was used to remove reads that contained four or more ‘N’ bases, had an average quality score across the read <3, or had a minimum length ≤51 bp or 33% of the full read length. Reads mapped with BBMap to masked human, cat, dog and mouse references at >93% identity were separated into a chaff file. Reads aligned to common microbial contaminants were also separated into a chaff file. For metatranscriptomic data, reads containing ribosomal RNA and known JGI spike-in sequences were removed and placed into separate fastq files. The internal DNA and mRNA standards added for quantification purposes at the nucleic acid extraction step (see Usage Notes) were recovered at 0.5–5.0% of sequences as expected.
For 16S rRNA and 18S rRNA, BBDuk was used to remove contaminants and trim reads that contained adapter sequence. This program was also used to remove reads that contained one or more ‘N’ bases, had an average quality score across the read of <10, or had a minimum length ≤51 bp or 33% of the full read length. Reads mapped with BBMap to masked human, cat, dog and mouse references at >93% identity or aligned to common microbial contaminants were separated into a chaff file. The 16S and 18S rRNA reads amplified from the internal DNA standards added for quantification purposes (see Usage Notes) were recovered at their expected level (0.5–5.0% of sequences).
Sequence datasets were checked for consistency with the expected composition of coastal marine microbial communities. Taxonomic assignments of 16S and 18S rRNA ESVs matched those of marine microbes common in coastal areas in general26,27 and in Monterey Bay seawater in particular11 (Figs 2 and 3). Taxonomic assignments of protein-encoding genes from metagenomic datasets were likewise representative of coastal and Monterey Bay microbial communities, and had taxonomic assignments consistent with the iTag datasets.
Usage Notes
Sample processing included the addition of internal standards to allow for calculation of volume-based absolute copy numbers for each gene or transcript type (i.e., counts L−1 rather than % of sequence library)28,29. The DNA standards consisted of genomic DNA from Thermus thermophilus DSM7039 HB829 and Blautia producta strain VPI 4299 (American Type Culture Collection, Manassas, VA). mRNA standards consisted of custom-designed 1006 nt artificial transcripts29. Artificial transcript sequences are available at Addgene Plasmid Repository (https://www.addgene.org; products MTST5 and MTST6). All four standards (two DNA and two mRNA) were added to the 0.22 μm pore size samples at the initiation of nucleic acid extraction. In the case of 18S iTag samples, genomic DNA from Arabidopsis (BioChain Institute, Inc., Newark, CA) and Mus musculus (Millipore Sigma, Burlington MA) was similarly added to the 5.0 μm pore size samples at initiation of extraction. Added amounts of internal standards were estimated at ~1% of final yields of DNA or mRNA based on prior recoveries from similar filters. Actual yields averaged ~2% of reads. The internal standards should be removed from the raw data prior to analysis. Information on how internal standards can be used for volume-based quantification is available elsewhere29,30.
Environmental data collected in association with the nucleic acid samples are given in Online-only Table 1. Available data differ between sampling dates depending on whether sampling was done by the ESP, from Niskin grab samples, or both.
ISA-Tab metadata file
Acknowledgements
We thank B. Roman, B. Ussler, J. Figurski, C. Wahl, B. Kieft, S. Gifford, T. Pennington for sampling and protocol assistance, advice, and technical expertise; K. Selph for flow cytometric analysis, L. Ziccarelli for A. sanguinea microscopy analysis, J. Christmann and the crew of the R/V Shana Rae; the Moss Landing Marine Laboratories Small Boat Facility; J. Ryan for processing MODIS satellite images; and S. Sharma and C. Edwardson for bioinformatic assistance. This work was funded by NSF grants OCE-1342694, OCE-1342699, OCE-1342734, the DOE Community Science Program, and partial support from the David and Lucile Packard Foundation through funds allocated to the Monterey Bay Aquarium Research Institute (MBARI). The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Online-only Tables
Online-only Table 1.
Environmental measurements associated with nucleic acid samples. An “x” indicates available data at the Biological and Chemical Data Management Office (BCO-DMO), https://www.bco-dmo.org/project/541255.
| Sample Name | NCBI BioProject Accession | GOLD Project ID | Collection Date | Temperature (oC) | Salinity (psu) | Depth (m) | Dissolved Oxygen (ml/L) | Chlorophyll a (CTD) (ug/L) | Chlorophyll a (Niskin) (ug/L) | Light Transmission (%) | DMSPt (nM) | DMSPd (in situ) (nM) | DMSPd (lab) (nM) | DMSPd consumption rate (nM/d) | Photosynthetic Eukaryotes (cells /mL) | Heterotrophic Bacteria (cells/mL) | Synechococcus (cells/mL) | Akashiwo (cells/mL) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10D | PRJNA502409 | Gp0266599 | 11/10/2016 | x | x | x | x | x | x | |||||||||
| 11D | PRJNA467725 | Gp0266600 | 10/6/2016 | x | x | x | x | x | x | |||||||||
| 125D_r | PRJNA467773 | Gp0266668 | 11/15/2016 | x | x | x | x | x | x | |||||||||
| 14D | PRJNA502410 | Gp0266603 | 10/28/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 15D | PRJNA502411 | Gp0266604 | 10/13/2016 | x | x | x | x | x | x | x | x | x | ||||||
| 16D | PRJNA502412 | Gp0266605 | 10/17/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 18D | PRJNA502413 | Gp0266607 | 10/20/2016 | x | x | x | x | x | x | x | x | |||||||
| 1D | PRJNA467720 | Gp0266591 | 10/4/2016 | x | x | x | x | x | x | |||||||||
| 1D_r | PRJNA502427 | Gp0266664 | 9/26/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 20D | PRJNA467726 | Gp0266608 | 9/27/2016 | x | x | x | x | x | x | |||||||||
| 21D | PRJNA468208 | Gp0272196 | 10/24/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 22D | PRJNA502414 | Gp0266609 | 10/12/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 23D | PRJNA467727 | Gp0266610 | 10/6/2016 | x | x | x | x | x | x | |||||||||
| 24D | PRJNA502440 | Gp0272197 | 11/2/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| 25D | PRJNA502415 | Gp0266611 | 11/6/2016 | x | x | x | x | x | x | |||||||||
| 26D | PRJNA467728 | Gp0266612 | 10/1/2016 | x | x | x | x | x | x | |||||||||
| 27D | PRJNA468209 | Gp0272198 | 10/12/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 28D | PRJNA467729 | Gp0266613 | 10/4/2016 | x | x | x | x | x | x | |||||||||
| 29D | PRJNA467730 | Gp0266614 | 10/10/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 2D | PRJNA467721 | Gp0266592 | 11/16/2016 | x | x | x | x | x | x | |||||||||
| 30D | PRJNA502416 | Gp0266615 | 10/5/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 31D | PRJNA467731 | Gp0266616 | 10/26/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 32D | PRJNA468210 | Gp0272199 | 10/19/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 33D | PRJNA502417 | Gp0266617 | 9/28/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 34D | PRJNA467732 | Gp0266618 | 9/30/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 35D | PRJNA502441 | Gp0272200 | 10/20/2016 | x | x | x | x | x | x | x | x | |||||||
| 36D | PRJNA467733 | Gp0266619 | 11/14/2016 | x | x | x | x | x | x | |||||||||
| 36D_r | PRJNA467770 | Gp0266665 | 10/5/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 37D | PRJNA467734 | Gp0266620 | 10/25/2016 | x | x | x | x | x | x | x | ||||||||
| 38D | PRJNA468211 | Gp0272201 | 10/2/2016 | x | x | x | x | x | x | |||||||||
| 39D | PRJNA467735 | Gp0266621 | 9/28/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 3D | PRJNA467722 | Gp0266593 | 11/9/2016 | x | x | x | x | x | x | |||||||||
| 40D | PRJNA467736 | Gp0266622 | 11/4/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 41D | PRJNA467737 | Gp0266623 | 11/13/2016 | x | x | x | x | x | x | |||||||||
| 42D | PRJNA502418 | Gp0266624 | 9/27/2016 | x | x | x | x | x | x | |||||||||
| 43D | PRJNA468212 | Gp0272202 | 11/8/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 44D | PRJNA467738 | Gp0266625 | 9/30/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 45D | PRJNA467739 | Gp0266626 | 11/3/2016 | x | x | x | x | x | x | |||||||||
| 46D | PRJNA468213 | Gp0272203 | 10/21/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 47D | PRJNA467740 | Gp0266627 | 10/25/2016 | x | x | x | x | x | x | x | ||||||||
| 48D | PRJNA467741 | Gp0266628 | 10/31/2016 | x | x | x | x | x | x | x | x | x | ||||||
| 48D_r | PRJNA467771 | Gp0266666 | 11/2/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| 49D | PRJNA502442 | Gp0272204 | 10/3/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 4D | PRJNA467723 | Gp0266594 | 11/3/2016 | x | x | x | x | x | x | |||||||||
| 50D | PRJNA502419 | Gp0266629 | 10/18/2016 | x | x | x | x | x | x | x | x | |||||||
| 51D | PRJNA502420 | Gp0266630 | 11/13/2016 | x | x | x | x | x | x | |||||||||
| 52D | PRJNA467742 | Gp0266631 | 10/17/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 53D | PRJNA502421 | Gp0266632 | 9/29/2016 | x | x | x | x | x | x | |||||||||
| 54D | PRJNA468214 | Gp0272205 | 11/11/2016 | x | x | x | x | x | x | |||||||||
| 55D | PRJNA502422 | Gp0266633 | 11/8/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 56D | PRJNA467743 | Gp0266634 | 11/9/2016 | x | x | x | x | x | x | |||||||||
| 57D | PRJNA502423 | Gp0266635 | 11/6/2016 | x | x | x | x | x | x | |||||||||
| 58D | PRJNA502424 | Gp0266636 | 11/4/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 58D_r | PRJNA467772 | Gp0266667 | 11/4/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 59D | PRJNA467744 | Gp0266637 | 10/21/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 60D | PRJNA502425 | Gp0266638 | 11/12/2016 | x | x | x | x | x | x | |||||||||
| 61D | PRJNA502426 | Gp0266639 | 10/31/2016 | x | x | x | x | x | x | x | x | x | ||||||
| 62D | PRJNA467745 | Gp0266640 | 10/28/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 63D | PRJNA467746 | Gp0266641 | 10/24/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 64D | PRJNA467747 | Gp0266642 | 10/5/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 65D | PRJNA467748 | Gp0266643 | 11/2/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| 66D | PRJNA467749 | Gp0266644 | 10/3/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 67D | PRJNA467750 | Gp0266645 | 11/15/2016 | x | x | x | x | x | x | |||||||||
| 68D | PRJNA467751 | Gp0266646 | 11/7/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| 70D | PRJNA467752 | Gp0266647 | 10/7/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 71D | PRJNA467753 | Gp0266648 | 10/18/2016 | x | x | x | x | x | x | x | x | |||||||
| 73D | PRJNA467754 | Gp0266649 | 11/16/2016 | x | x | x | x | x | x | |||||||||
| 74D | PRJNA467755 | Gp0266650 | 11/11/2016 | x | x | x | x | x | x | |||||||||
| 75D | PRJNA467756 | Gp0266651 | 11/14/2016 | x | x | x | x | x | x | |||||||||
| 76D | PRJNA467757 | Gp0266652 | 11/10/2016 | x | x | x | x | x | x | |||||||||
| 77D | PRJNA467758 | Gp0266653 | 10/7/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 78D | PRJNA467759 | Gp0266654 | 9/26/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 79D | PRJNA467760 | Gp0266655 | 11/7/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| 7D | PRJNA502407 | Gp0266596 | 10/26/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 80D | PRJNA467761 | Gp0266656 | 10/11/2016 | x | x | x | x | x | x | x | ||||||||
| 81D | PRJNA467762 | Gp0266657 | 10/11/2016 | x | x | x | x | x | x | x | ||||||||
| 82D | PRJNA467763 | Gp0266658 | 10/10/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 84D | PRJNA467764 | Gp0266659 | 10/11/2016 | x | x | x | x | x | x | x | ||||||||
| 85D | PRJNA467765 | Gp0266660 | 10/13/2016 | x | x | x | x | x | x | x | x | x | ||||||
| 89D | PRJNA467766 | Gp0266661 | 11/5/2016 | x | x | x | x | x | x | |||||||||
| 8D | PRJNA467724 | Gp0266597 | 11/5/2016 | x | x | x | x | x | x | |||||||||
| 90D | PRJNA467768 | Gp0266662 | 11/8/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 91D | PRJNA467769 | Gp0266663 | 11/11/2016 | x | x | x | x | x | x | |||||||||
| 9D | PRJNA502408 | Gp0266598 | 11/15/2016 | x | x | x | x | x | x | |||||||||
| 26R | PRJNA468309 | Gp0290791 | 10/1/2016 | x | x | x | x | x | x | |||||||||
| 58R_r | PRJNA468331 | Gp0290824 | 11/4/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 125R_r | PRJNA468332 | Gp0290825 | 11/15/2016 | x | x | x | x | x | x | |||||||||
| 13R | PRJNA468304 | Gp0290783 | 10/1/2016 | x | x | x | x | x | x | |||||||||
| 12R | PRJNA468303 | Gp0290782 | 9/29/2016 | x | x | x | x | x | x | |||||||||
| 8R | PRJNA502451 | Gp0290778 | 11/5/2016 | x | x | x | x | x | x | |||||||||
| 85R | PRJNA468329 | Gp0290822 | 10/13/2016 | x | x | x | x | x | x | x | x | x | ||||||
| 76R | PRJNA502468 | Gp0290819 | 11/10/2016 | x | x | x | x | x | x | |||||||||
| 40R | PRJNA468314 | Gp0290799 | 11/4/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 53R | PRJNA468320 | Gp0290809 | 9/29/2016 | x | x | x | x | x | x | |||||||||
| 73R | PRJNA468325 | Gp0290817 | 11/16/2016 | x | x | x | x | x | x | |||||||||
| 23R | PRJNA468307 | Gp0290788 | 10/6/2016 | x | x | x | x | x | x | |||||||||
| 49R | PRJNA468318 | Gp0290807 | 10/3/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 17R | PRJNA468305 | Gp0290786 | 11/12/2016 | x | x | x | x | x | x | |||||||||
| 14R | PRJNA502455 | Gp0290784 | 10/28/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 44R | PRJNA468315 | Gp0290803 | 9/30/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 29R | PRJNA468143 | Gp0267189 | 10/10/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 18R | PRJNA467774 | Gp0266669 | 10/20/2016 | x | x | x | x | x | x | x | x | |||||||
| 24R | PRJNA468308 | Gp0290789 | 11/2/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| 79R | PRJNA468328 | Gp0290821 | 11/7/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| 22R | PRJNA467776 | Gp0266671 | 10/12/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 21R | PRJNA467775 | Gp0266670 | 10/24/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 60R | PRJNA502465 | Gp0290813 | 11/12/2016 | x | x | x | x | x | x | |||||||||
| 15R | PRJNA502456 | Gp0290785 | 10/13/2016 | x | x | x | x | x | x | x | x | x | ||||||
| 75R | PRJNA468326 | Gp0290818 | 11/14/2016 | x | x | x | x | x | x | |||||||||
| 3R | PRJNA468301 | Gp0290776 | 11/9/2016 | x | x | x | x | x | x | |||||||||
| 20R | PRJNA468306 | Gp0290787 | 9/27/2016 | x | x | x | x | x | x | |||||||||
| 91R | PRJNA468330 | Gp0290823 | 11/11/2016 | x | x | x | x | x | x | |||||||||
| 56R | PRJNA468322 | Gp0290811 | 11/9/2016 | x | x | x | x | x | x | |||||||||
| 46R | PRJNA468316 | Gp0290805 | 10/21/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 2R | PRJNA468300 | Gp0290775 | 11/16/2016 | x | x | x | x | x | x | |||||||||
| 38R | PRJNA502460 | Gp0290797 | 10/2/2016 | x | x | x | x | x | x | |||||||||
| 45R | PRJNA502464 | Gp0290804 | 11/3/2016 | x | x | x | x | x | x | |||||||||
| 36R | PRJNA468312 | Gp0290796 | 11/14/2016 | x | x | x | x | x | x | |||||||||
| 39R | PRJNA468313 | Gp0290798 | 9/28/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 68R | PRJNA502467 | Gp0290815 | 11/7/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| 30R | PRJNA468310 | Gp0290793 | 10/5/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 48R | PRJNA468317 | Gp0290806 | 10/31/2016 | x | x | x | x | x | x | x | x | x | ||||||
| 42R | PRJNA502462 | Gp0290801 | 9/27/2016 | x | x | x | x | x | x | |||||||||
| 25R | PRJNA502457 | Gp0290790 | 11/6/2016 | x | x | x | x | x | x | |||||||||
| 78R | PRJNA468149 | Gp0267195 | 9/26/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 63R | PRJNA468147 | Gp0267193 | 10/24/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 43R | PRJNA502463 | Gp0290802 | 11/8/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 66R | PRJNA502466 | Gp0290814 | 10/3/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 54R | PRJNA468321 | Gp0290810 | 11/11/2016 | x | x | x | x | x | x | |||||||||
| 41R | PRJNA502461 | Gp0290800 | 11/13/2016 | x | x | x | x | x | x | |||||||||
| 28R | PRJNA502458 | Gp0290792 | 10/4/2016 | x | x | x | x | x | x | |||||||||
| 32R | PRJNA468144 | Gp0267190 | 10/19/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 81R | PRJNA467786 | Gp0266685 | 10/11/2016 | x | x | x | x | x | x | x | ||||||||
| 70R | PRJNA468324 | Gp0290816 | 10/7/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 67R | PRJNA468148 | Gp0267194 | 11/15/2016 | x | x | x | x | x | x | |||||||||
| 33R | PRJNA502459 | Gp0290794 | 9/28/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 27R | PRJNA467777 | Gp0266672 | 10/12/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 34R | PRJNA468311 | Gp0290795 | 9/30/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 4R | PRJNA468302 | Gp0290777 | 11/3/2016 | x | x | x | x | x | x | |||||||||
| 57R | PRJNA468323 | Gp0290812 | 11/6/2016 | x | x | x | x | x | x | |||||||||
| 1R | PRJNA468299 | Gp0290774 | 10/4/2016 | x | x | x | x | x | x | |||||||||
| 47R | PRJNA467780 | Gp0266675 | 10/25/2016 | x | x | x | x | x | x | x | ||||||||
| 9R | PRJNA502452 | Gp0290779 | 11/15/2016 | x | x | x | x | x | x | |||||||||
| 77R | PRJNA468327 | Gp0290820 | 10/7/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 55R | PRJNA468145 | Gp0267191 | 11/8/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 10R | PRJNA502453 | Gp0290780 | 11/10/2016 | x | x | x | x | x | x | |||||||||
| 11R | PRJNA502454 | Gp0290781 | 10/6/2016 | x | x | x | x | x | x | |||||||||
| 58R | PRJNA467783 | Gp0266678 | 11/4/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 52R | PRJNA467782 | Gp0266677 | 10/17/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 51R | PRJNA468319 | Gp0290808 | 11/13/2016 | x | x | x | x | x | x | |||||||||
| 50R | PRJNA467781 | Gp0266676 | 10/18/2016 | x | x | x | x | x | x | x | x | |||||||
| 90R | PRJNA467788 | Gp0266687 | 11/8/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 80R | PRJNA467785 | Gp0266684 | 10/11/2016 | x | x | x | x | x | x | x | ||||||||
| 89R | PRJNA468151 | Gp0267197 | 11/5/2016 | x | x | x | x | x | x | |||||||||
| 35R | PRJNA467779 | Gp0266674 | 10/20/2016 | x | x | x | x | x | x | x | x | |||||||
| 71R | PRJNA467784 | Gp0266682 | 10/18/2016 | x | x | x | x | x | x | x | x | |||||||
| 31R | PRJNA467778 | Gp0266673 | 10/26/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 84R | PRJNA467787 | Gp0266686 | 10/11/2016 | x | x | x | x | x | x | x | ||||||||
| 61R | PRJNA468146 | Gp0267192 | 10/31/2016 | x | x | x | x | x | x | x | x | x | ||||||
| 36R_r | PRJNA468152 | Gp0267198 | 10/5/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 82R | PRJNA468150 | Gp0267196 | 10/10/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| MB_1025D_MT | PRJNA502611 | Gp0294703 | 10/25/2016 | x | x | x | x | x | x | x | ||||||||
| WCR_12_MT | PRJNA502608 | Gp0294698 | 9/26/2016 | x | x | x | x | x | x | |||||||||
| WCR_74_MT | PRJNA502609 | Gp0294699 | 11/6/2016 | x | x | x | x | x | x | |||||||||
| WCR_77_MT | PRJNA502610 | Gp0294700 | 11/7/2016 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| MB_1026D_MT | PRJNA502612 | Gp0294704 | 10/26/2016 | x | x | x | x | x | x | x | x | x | x | |||||
| 1_DF | #N/A | #N/A | 10/4/16 | x | x | x | x | x | x | |||||||||
| 3_DF | #N/A | #N/A | 11/9/16 | x | x | x | x | x | x | |||||||||
| 13_DF | #N/A | #N/A | 10/1/16 | x | x | x | x | x | x | |||||||||
| 26_DF | #N/A | #N/A | 10/1/16 | x | x | x | x | x | x | |||||||||
| 13_S | #N/A | #N/A | 10/1/16 | x | x | x | x | x | x | |||||||||
| 26_S | #N/A | #N/A | 10/1/16 | x | x | x | x | x | x | |||||||||
| 38_DF | #N/A | #N/A | 10/2/16 | x | x | x | x | x | x | |||||||||
| 38_S | #N/A | #N/A | 10/2/16 | x | x | x | x | x | x | |||||||||
| 24_DF | #N/A | #N/A | 11/2/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| 65_DF | #N/A | #N/A | 11/2/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| 48r_DF | #N/A | #N/A | 11/2/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| 24_S | #N/A | #N/A | 11/2/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| 65_S | #N/A | #N/A | 11/2/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| 48r_S | #N/A | #N/A | 11/2/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| 49_DF | #N/A | #N/A | 10/3/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 66_DF | #N/A | #N/A | 10/3/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 49_S | #N/A | #N/A | 10/3/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 66_S | #N/A | #N/A | 10/3/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 4_DF | #N/A | #N/A | 11/3/16 | x | x | x | x | x | x | |||||||||
| 45_DF | #N/A | #N/A | 11/3/16 | x | x | x | x | x | x | |||||||||
| 4_S | #N/A | #N/A | 11/3/16 | x | x | x | x | x | x | |||||||||
| 45_S | #N/A | #N/A | 11/3/16 | x | x | x | x | x | x | |||||||||
| 28_DF | #N/A | #N/A | 10/4/16 | x | x | x | x | x | x | |||||||||
| 1_S | #N/A | #N/A | 10/4/16 | x | x | x | x | x | x | |||||||||
| 28_S | #N/A | #N/A | 10/4/16 | x | x | x | x | x | x | |||||||||
| 40_DF | #N/A | #N/A | 11/4/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 58_DF | #N/A | #N/A | 11/4/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 58r_DF | #N/A | #N/A | 11/4/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 40_S | #N/A | #N/A | 11/4/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 58_S | #N/A | #N/A | 11/4/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 58r_S | #N/A | #N/A | 11/4/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 30_DF | #N/A | #N/A | 10/5/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 64_DF | #N/A | #N/A | 10/5/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 36r_DF | #N/A | #N/A | 10/5/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 30_S | #N/A | #N/A | 10/5/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 64_S | #N/A | #N/A | 10/5/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 36r_S | #N/A | #N/A | 10/5/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 8_DF | #N/A | #N/A | 11/5/16 | x | x | x | x | x | x | |||||||||
| 89_DF | #N/A | #N/A | 11/5/16 | x | x | x | x | x | x | |||||||||
| 8_S | #N/A | #N/A | 11/5/16 | x | x | x | x | x | x | |||||||||
| 89_S | #N/A | #N/A | 11/5/16 | x | x | x | x | x | x | |||||||||
| 11_DF | #N/A | #N/A | 10/6/16 | x | x | x | x | x | x | |||||||||
| 23_DF | #N/A | #N/A | 10/6/16 | x | x | x | x | x | x | |||||||||
| 11_S | #N/A | #N/A | 10/6/16 | x | x | x | x | x | x | |||||||||
| 23_S | #N/A | #N/A | 10/6/16 | x | x | x | x | x | x | |||||||||
| 25_DF | #N/A | #N/A | 11/6/16 | x | x | x | x | x | x | |||||||||
| 57_DF | #N/A | #N/A | 11/6/16 | x | x | x | x | x | x | |||||||||
| 25_S | #N/A | #N/A | 11/6/16 | x | x | x | x | x | x | |||||||||
| 57_S | #N/A | #N/A | 11/6/16 | x | x | x | x | x | x | |||||||||
| 70_DF | #N/A | #N/A | 10/7/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 77_DF | #N/A | #N/A | 10/7/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 70_S | #N/A | #N/A | 10/7/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 77_S | #N/A | #N/A | 10/7/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 68_DF | #N/A | #N/A | 11/7/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| 79_DF | #N/A | #N/A | 11/7/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| 68_S | #N/A | #N/A | 11/7/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| 79_S | #N/A | #N/A | 11/7/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| 43_DF | #N/A | #N/A | 11/8/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 55_DF | #N/A | #N/A | 11/8/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 90_DF | #N/A | #N/A | 11/8/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 43_S | #N/A | #N/A | 11/8/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 55_S | #N/A | #N/A | 11/8/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 90_S | #N/A | #N/A | 11/8/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 56_DF | #N/A | #N/A | 11/9/16 | x | x | x | x | x | x | |||||||||
| 3_S | #N/A | #N/A | 11/9/16 | x | x | x | x | x | x | |||||||||
| 56_S | #N/A | #N/A | 11/9/16 | x | x | x | x | x | x | |||||||||
| 29_DF | #N/A | #N/A | 10/10/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 82_DF | #N/A | #N/A | 10/10/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 29_S | #N/A | #N/A | 10/10/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 82_S | #N/A | #N/A | 10/10/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 10_DF | #N/A | #N/A | 11/10/16 | x | x | x | x | x | x | |||||||||
| 76_DF | #N/A | #N/A | 11/10/16 | x | x | x | x | x | x | |||||||||
| 10_S | #N/A | #N/A | 11/10/16 | x | x | x | x | x | x | |||||||||
| 76_S | #N/A | #N/A | 11/10/16 | x | x | x | x | x | x | |||||||||
| 80_DF | #N/A | #N/A | 10/11/16 | x | x | x | x | x | x | x | ||||||||
| 81_DF | #N/A | #N/A | 10/11/16 | x | x | x | x | x | x | x | ||||||||
| 84_DF | #N/A | #N/A | 10/11/16 | x | x | x | x | x | x | x | ||||||||
| 80_S | #N/A | #N/A | 10/11/16 | x | x | x | x | x | x | x | ||||||||
| 81_S | #N/A | #N/A | 10/11/16 | x | x | x | x | x | x | x | ||||||||
| 84_S | #N/A | #N/A | 10/11/16 | x | x | x | x | x | x | x | ||||||||
| 54_DF | #N/A | #N/A | 11/11/16 | x | x | x | x | x | x | |||||||||
| 74_DF | #N/A | #N/A | 11/11/16 | x | x | x | x | x | x | |||||||||
| 91_DF | #N/A | #N/A | 11/11/16 | x | x | x | x | x | x | |||||||||
| 54_S | #N/A | #N/A | 11/11/16 | x | x | x | x | x | x | |||||||||
| 74_S | #N/A | #N/A | 11/11/16 | x | x | x | x | x | x | |||||||||
| 91_S | #N/A | #N/A | 11/11/16 | x | x | x | x | x | x | |||||||||
| 22_DF | #N/A | #N/A | 10/12/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 27_DF | #N/A | #N/A | 10/12/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 22_S | #N/A | #N/A | 10/12/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 27_S | #N/A | #N/A | 10/12/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 17_DF | #N/A | #N/A | 11/12/16 | x | x | x | x | x | x | |||||||||
| 60_DF | #N/A | #N/A | 11/12/16 | x | x | x | x | x | x | |||||||||
| 17_S | #N/A | #N/A | 11/12/16 | x | x | x | x | x | x | |||||||||
| 60_S | #N/A | #N/A | 11/12/16 | x | x | x | x | x | x | |||||||||
| 15_DF | #N/A | #N/A | 10/13/16 | x | x | x | x | x | x | x | x | x | ||||||
| 85_DF | #N/A | #N/A | 10/13/16 | x | x | x | x | x | x | x | x | x | ||||||
| 15_S | #N/A | #N/A | 10/13/16 | x | x | x | x | x | x | x | x | x | ||||||
| 85_S | #N/A | #N/A | 10/13/16 | x | x | x | x | x | x | x | x | x | ||||||
| 41_DF | #N/A | #N/A | 11/13/16 | x | x | x | x | x | x | |||||||||
| 51_DF | #N/A | #N/A | 11/13/16 | x | x | x | x | x | x | |||||||||
| 41_S | #N/A | #N/A | 11/13/16 | x | x | x | x | x | x | |||||||||
| 51_S | #N/A | #N/A | 11/13/16 | x | x | x | x | x | x | |||||||||
| 36_DF | #N/A | #N/A | 11/14/16 | x | x | x | x | x | x | |||||||||
| 75_DF | #N/A | #N/A | 11/14/16 | x | x | x | x | x | x | |||||||||
| 36_S | #N/A | #N/A | 11/14/16 | x | x | x | x | x | x | |||||||||
| 75_S | #N/A | #N/A | 11/14/16 | x | x | x | x | x | x | |||||||||
| 9_DF | #N/A | #N/A | 11/15/16 | x | x | x | x | x | x | |||||||||
| 67_DF | #N/A | #N/A | 11/15/16 | x | x | x | x | x | x | |||||||||
| 125r_DF | #N/A | #N/A | 11/15/16 | x | x | x | x | x | x | |||||||||
| 9_S | #N/A | #N/A | 11/15/16 | x | x | x | x | x | x | |||||||||
| 67_S | #N/A | #N/A | 11/15/16 | x | x | x | x | x | x | |||||||||
| 125r_S | #N/A | #N/A | 11/15/16 | x | x | x | x | x | x | |||||||||
| 73_DF | #N/A | #N/A | 11/16/16 | x | x | x | x | x | x | |||||||||
| 2_S | #N/A | #N/A | 11/16/16 | x | x | x | x | x | x | |||||||||
| 73_S | #N/A | #N/A | 11/16/16 | x | x | x | x | x | x | |||||||||
| 2_DF | #N/A | #N/A | 11/16/16 | x | x | x | x | x | x | |||||||||
| 16_DF | #N/A | #N/A | 10/17/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 52_DF | #N/A | #N/A | 10/17/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 16_S | #N/A | #N/A | 10/17/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 52_S | #N/A | #N/A | 10/17/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 50_DF | #N/A | #N/A | 10/18/16 | x | x | x | x | x | x | x | x | |||||||
| 71_DF | #N/A | #N/A | 10/18/16 | x | x | x | x | x | x | x | x | |||||||
| 50_S | #N/A | #N/A | 10/18/16 | x | x | x | x | x | x | x | x | |||||||
| 71_S | #N/A | #N/A | 10/18/16 | x | x | x | x | x | x | x | x | |||||||
| 5_DF | #N/A | #N/A | 10/19/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 32_DF | #N/A | #N/A | 10/19/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 5_S | #N/A | #N/A | 10/19/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 32_S | #N/A | #N/A | 10/19/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 18_DF | #N/A | #N/A | 10/20/16 | x | x | x | x | x | x | x | x | |||||||
| 35_DF | #N/A | #N/A | 10/20/16 | x | x | x | x | x | x | x | x | |||||||
| 18_S | #N/A | #N/A | 10/20/16 | x | x | x | x | x | x | x | x | |||||||
| 35_S | #N/A | #N/A | 10/20/16 | x | x | x | x | x | x | x | x | |||||||
| 46_DF | #N/A | #N/A | 10/21/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 59_DF | #N/A | #N/A | 10/21/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 46_S | #N/A | #N/A | 10/21/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 59_S | #N/A | #N/A | 10/21/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 21_DF | #N/A | #N/A | 10/24/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 63_DF | #N/A | #N/A | 10/24/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 21_S | #N/A | #N/A | 10/24/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 63_S | #N/A | #N/A | 10/24/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 37_DF | #N/A | #N/A | 10/25/16 | x | x | x | x | x | x | x | ||||||||
| 47_DF | #N/A | #N/A | 10/25/16 | x | x | x | x | x | x | x | ||||||||
| 37_S | #N/A | #N/A | 10/25/16 | x | x | x | x | x | x | x | ||||||||
| 47_S | #N/A | #N/A | 10/25/16 | x | x | x | x | x | x | x | ||||||||
| 78_DF | #N/A | #N/A | 9/26/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 1r_DF | #N/A | #N/A | 9/26/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 78_S | #N/A | #N/A | 9/26/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 1r_S | #N/A | #N/A | 9/26/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 7_DF | #N/A | #N/A | 10/26/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 31_DF | #N/A | #N/A | 10/26/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 7_S | #N/A | #N/A | 10/26/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 31_S | #N/A | #N/A | 10/26/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 20_DF | #N/A | #N/A | 9/27/16 | x | x | x | x | x | x | |||||||||
| 42_DF | #N/A | #N/A | 9/27/16 | x | x | x | x | x | x | |||||||||
| 20_S | #N/A | #N/A | 9/27/16 | x | x | x | x | x | x | |||||||||
| 42_S | #N/A | #N/A | 9/27/16 | x | x | x | x | x | x | |||||||||
| 33_DF | #N/A | #N/A | 9/28/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 39_DF | #N/A | #N/A | 9/28/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 33_S | #N/A | #N/A | 9/28/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 39_S | #N/A | #N/A | 9/28/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 14_DF | #N/A | #N/A | 10/28/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 62_DF | #N/A | #N/A | 10/28/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 14_S | #N/A | #N/A | 10/28/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 62_S | #N/A | #N/A | 10/28/16 | x | x | x | x | x | x | x | x | x | x | |||||
| 12_DF | #N/A | #N/A | 9/29/16 | x | x | x | x | x | x | |||||||||
| 53_DF | #N/A | #N/A | 9/29/16 | x | x | x | x | x | x | |||||||||
| 12_S | #N/A | #N/A | 9/29/16 | x | x | x | x | x | x | |||||||||
| 53_S | #N/A | #N/A | 9/29/16 | x | x | x | x | x | x | |||||||||
| 34_DF | #N/A | #N/A | 9/30/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 44_DF | #N/A | #N/A | 9/30/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 34_S | #N/A | #N/A | 9/30/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 44_S | #N/A | #N/A | 9/30/16 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| 48_DF | #N/A | #N/A | 10/31/16 | x | x | x | x | x | x | x | x | x | ||||||
| 61_DF | #N/A | #N/A | 10/31/16 | x | x | x | x | x | x | x | x | x | ||||||
| 48_S | #N/A | #N/A | 10/31/16 | x | x | x | x | x | x | x | x | x | ||||||
| 61_S | #N/A | #N/A | 10/31/16 | x | x | x | x | x | x | x | x | x |
Online-only Table 2.
DNA and RNA sample information.
| Sample Name | env_biome | env_feature | env_material | geo_loc_name | Collection Date | Latitude and Longitude | Volume Seawater Filtered (ml) | Type | JGI Project ID | JGI Sequencing Project ID | JGI Sequencing Project Name | JGI Sample ID | JGI Contigs Link | NCBI Project ID | NCBI BioProject Accession | NCBI SRA Accession ID | NCBI BioSample Accession | GOLD Project ID | Instrument | Sequencing Run Mode | Total Bases | Assembly Method |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/10/2016 | 36.835 N, 121.901 W | 350 | Metagenome | 1174520 | 1174688 | 10D metaG | 166744 | https://genome.jgi.doe.gov/portal/10DmetaG_FD/10DmetaG_FD.download.html | 502409 | PRJNA502409 | SRP174084 | SAMN10350899 | Gp0266599 | Illumina HiSeq-2000 | 2 × 151 | 10,341,052,894 | SPAdes assembler 3.11.1 |
| 10R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/10/2016 | 36.835 N, 121.901 W | 350 | Metatranscriptome | 1174786 | 1174930 | 10R metaT | 166830 | https://genome.jgi.doe.gov/portal/10RmetaT_FD/10RmetaT_FD.download.html | 502453 | PRJNA502453 | SRP174124 | SAMN10350900 | Gp0290780 | Illumina HiSeq-2000 | 2 × 151 | 11,461,972,100 | MEGAHIT v1.1.2 |
| 11D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/6/2016 | 36.835 N, 121.901 W | 250 | Metagenome | 1174522 | 1174689 | 11D metaG | 166745 | https://genome.jgi.doe.gov/portal/11DmetaG_FD/11DmetaG_FD.download.html | 467725 | PRJNA467725 | SRP155249 | SAMN09201852 | Gp0266600 | Illumina HiSeq-2000 | 2 × 151 | 19,576,899,642 | SPAdes assembler 3.11.1 |
| 11R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/6/2016 | 36.835 N, 121.901 W | 250 | Metatranscriptome | 1174789 | 1174931 | 11R metaT | 166831 | https://genome.jgi.doe.gov/portal/11RmetaT_FD/11RmetaT_FD.download.html | 502454 | PRJNA502454 | SRP174126 | SAMN10351123 | Gp0290781 | Illumina HiSeq-2000 | 2 × 151 | 11,561,507,374 | MEGAHIT v1.1.2 |
| 125D_r | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/15/2016 | 36.835 N, 121.901 W | 950 | Metagenome | 1174678 | 1174767 | 125D_r metaG | 166823 | https://genome.jgi.doe.gov/portal/125D_rmetaG_FD/125D_rmetaG_FD.download.html | 467773 | PRJNA467773 | SRP155430 | SAMN09202115 | Gp0266668 | Illumina HiSeq-2000 | 2 × 151 | 6,328,886,556 | SPAdes assembler 3.11.1 |
| 125R_r | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/15/2016 | 36.835 N, 121.901 W | 950 | Metatranscriptome | 1174921 | 1174975 | 125R_r metaT | 166875 | https://genome.jgi.doe.gov/portal/125R_rmetaT_FD/125R_rmetaT_FD.download.html | 468332 | PRJNA468332 | SRP163296 | SAMN09200099 | Gp0290825 | Illumina HiSeq-2000 | 2 × 151 | 7,601,810,818 | MEGAHIT v1.1.2 |
| 12R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/29/2016 | 36.835 N, 121.901 W | 125 | Metatranscriptome | 1174792 | 1174932 | 12R metaT | 166832 | https://genome.jgi.doe.gov/portal/12RmetaT_FD/12RmetaT_FD.download.html | 468303 | PRJNA468303 | SRP163181 | SAMN09201820 | Gp0290782 | Illumina HiSeq-2000 | 2 × 151 | 7,804,474,562 | MEGAHIT v1.1.2 |
| 13R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/1/2016 | 36.835 N, 121.901 W | 125 | Metatranscriptome | 1174795 | 1174933 | 13R metaT | 166833 | https://genome.jgi.doe.gov/portal/13RmetaT_FD/13RmetaT_FD.download.html | 468304 | PRJNA468304 | SRP163185 | SAMN09202081 | Gp0290783 | Illumina HiSeq-2000 | 2 × 151 | 7,778,654,770 | MEGAHIT v1.1.2 |
| 14D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/28/2016 | 36.835 N, 121.901 W | 250 | Metagenome | 1174528 | 1174692 | 14D metaG | 166748 | https://genome.jgi.doe.gov/portal/14DmetaG_FD/14DmetaG_FD.download.html | 502410 | PRJNA502410 | SRP174083 | SAMN10350898 | Gp0266603 | Illumina HiSeq-2000 | 2 × 151 | 8,488,758,242 | SPAdes assembler 3.11.1 |
| 14R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/28/2016 | 36.835 N, 121.901 W | 250 | Metatranscriptome | 1174798 | 1174934 | 14R metaT | 166834 | https://genome.jgi.doe.gov/portal/14RmetaT_FD/14RmetaT_FD.download.html | 502455 | PRJNA502455 | SRP173017 | SAMN10350972 | Gp0290784 | Illumina HiSeq-2000 | 2 × 151 | 8,659,605,984 | MEGAHIT v1.1.2 |
| 15D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/13/2016 | 36.835 N, 121.901 W | 100 | Metagenome | 1174530 | 1174693 | 15D metaG | 166749 | https://genome.jgi.doe.gov/portal/15DmetaG_FD/15DmetaG_FD.download.html | 502411 | PRJNA502411 | SRP174080 | SAMN10351355 | Gp0266604 | Illumina HiSeq-2000 | 2 × 151 | 7,691,258,084 | SPAdes assembler 3.11.1 |
| 15R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/13/2016 | 36.835 N, 121.901 W | 100 | Metatranscriptome | 1174801 | 1174935 | 15R metaT | 166835 | https://genome.jgi.doe.gov/portal/15RmetaT_FD/15RmetaT_FD.download.html | 502456 | PRJNA502456 | SRP173008 | SAMN10350971 | Gp0290785 | Illumina HiSeq-2000 | 2 × 151 | 9,108,232,722 | MEGAHIT v1.1.2 |
| 16D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/17/2016 | 36.835 N, 121.901 W | 200 | Metagenome | 1174532 | 1174694 | 16D metaG | 166750 | https://genome.jgi.doe.gov/portal/16DmetaG_FD/16DmetaG_FD.download.html | 502412 | PRJNA502412 | SRP174089 | SAMN10350897 | Gp0266605 | Illumina HiSeq-2000 | 2 × 151 | 9,245,335,890 | SPAdes assembler 3.11.1 |
| 17R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/12/2016 | 36.835 N, 121.901 W | 400 | Metatranscriptome | 1174804 | 1174936 | 17R metaT | 166836 | https://genome.jgi.doe.gov/portal/17RmetaT_FD/17RmetaT_FD.download.html | 468305 | PRJNA468305 | SRP163182 | SAMN09201819 | Gp0290786 | Illumina HiSeq-2000 | 2 × 151 | 8,651,241,792 | MEGAHIT v1.1.2 |
| 18D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/20/2016 | 36.835 N, 121.901 W | 200 | Metagenome | 1174536 | 1174696 | 18D metaG | 166752 | https://genome.jgi.doe.gov/portal/18DmetaG_FD/18DmetaG_FD.download.html | 502413 | PRJNA502413 | SRP174085 | SAMN10351120 | Gp0266607 | Illumina HiSeq-2000 | 2 × 151 | 8,004,892,332 | SPAdes assembler 3.11.1 |
| 18R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/20/2016 | 36.835 N, 121.901 W | 200 | Metatranscriptome | 1174985 | 1175087 | 18R metaT | 166879 | https://genome.jgi.doe.gov/portal/18RmetaT_FD/18RmetaT_FD.download.html | 467774 | PRJNA467774 | SRP155442 | SAMN09201439 | Gp0266669 | Illumina HiSeq-2000 | 2 × 151 | 8,874,292,952 | MEGAHIT v1.1.2 |
| 1D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/4/2016 | 36.835 N, 121.901 W | 200 | Metagenome | 1174504 | 1174680 | 1D metaG | 166736 | https://genome.jgi.doe.gov/portal/1D_rmetaG_FD/1D_rmetaG_FD.download.html | 467720 | PRJNA467720 | SRP155234 | SAMN09201680 | Gp0266591 | Illumina HiSeq-2000 | 2 × 151 | 6,639,302,994 | SPAdes assembler 3.11.1 |
| 1D_r | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/26/2016 | 36.835 N, 121.901 W | 95.1 | Metagenome | 1174670 | 1174763 | 1D_r metaG | 166819 | https://genome.jgi.doe.gov/portal/1DmetaG_FD/1DmetaG_FD.download.html | 502427 | PRJNA502427 | SRP174125 | SAMN10351067 | Gp0266664 | Illumina HiSeq-2000 | 2 × 151 | 38,554,088,892 | SPAdes assembler 3.11.1 |
| 1R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/4/2016 | 36.835 N, 121.901 W | 200 | Metatranscriptome | 1174768 | 1174924 | 1R metaT | 166824 | https://genome.jgi.doe.gov/portal/1RmetaT_FD/1RmetaT_FD.download.html | 468299 | PRJNA468299 | SRP155435 | SAMN09201822 | Gp0290774 | Illumina HiSeq-2000 | 2 × 151 | 10,940,653,962 | MEGAHIT v1.1.2 |
| 20D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/27/2016 | 36.835 N, 121.901 W | 73.5 | Metagenome | 1174538 | 1174697 | 20D metaG | 166753 | https://genome.jgi.doe.gov/portal/20DmetaG_FD/20DmetaG_FD.download.html | 467726 | PRJNA467726 | SRP155254 | SAMN09202066 | Gp0266608 | Illumina HiSeq-2000 | 2 × 151 | 13,033,898,710 | SPAdes assembler 3.11.1 |
| 20R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/27/2016 | 36.835 N, 121.901 W | 73.5 | Metatranscriptome | 1174807 | 1174937 | 20R metaT | 166837 | https://genome.jgi.doe.gov/portal/20RmetaT_FD/20RmetaT_FD.download.html | 468306 | PRJNA468306 | SRP163187 | SAMN09202082 | Gp0290787 | Illumina HiSeq-2000 | 2 × 151 | 9,121,070,440 | MEGAHIT v1.1.2 |
| 21D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/24/2016 | 36.835 N, 121.901 W | 250 | Metagenome | 1174540 | 1174698 | 21D metaG | 166754 | https://genome.jgi.doe.gov/portal/21DmetaG_FD/21DmetaG_FD.download.html | 468208 | PRJNA468208 | SRP155253 | SAMN09201605 | Gp0272196 | Illumina HiSeq-2000 | 2 × 151 | 7,000,627,270 | SPAdes assembler 3.11.1 |
| 21R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/24/2016 | 36.835 N, 121.901 W | 250 | Metatranscriptome | 1174988 | 1175088 | 21R metaT | 166880 | https://genome.jgi.doe.gov/portal/21RmetaT_FD/21RmetaT_FD.download.html | 467775 | PRJNA467775 | SRP155445 | SAMN09201531 | Gp0266670 | Illumina HiSeq-2000 | 2 × 151 | 9,023,160,530 | MEGAHIT v1.1.2 |
| 22D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/12/2016 | 36.835 N, 121.901 W | 100 | Metagenome | 1174542 | 1174699 | 22D metaG | 166755 | https://genome.jgi.doe.gov/portal/22DmetaG_FD/22DmetaG_FD.download.html | 502414 | PRJNA502414 | SRP174086 | SAMN10350896 | Gp0266609 | Illumina HiSeq-2000 | 2 × 151 | 7,011,736,038 | SPAdes assembler 3.11.1 |
| 22R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/12/2016 | 36.835 N, 121.901 W | 100 | Metatranscriptome | 1174991 | 1175089 | 22R metaT | 166881 | https://genome.jgi.doe.gov/portal/22RmetaT_FD/22RmetaT_FD.download.html | 467776 | PRJNA467776 | SRP155447 | SAMN09202116 | Gp0266671 | Illumina HiSeq-2000 | 2 × 151 | 8,949,209,488 | MEGAHIT v1.1.2 |
| 23D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/6/2016 | 36.835 N, 121.901 W | 150 | Metagenome | 1174544 | 1174700 | 23D metaG | 166756 | https://genome.jgi.doe.gov/portal/23DmetaG_FD/23DmetaG_FD.download.html | 467727 | PRJNA467727 | SRP155978 | SAMN09201453 | Gp0266610 | Illumina HiSeq-2000 | 2 × 151 | 7,254,602,022 | SPAdes assembler 3.11.1 |
| 23R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/6/2016 | 36.835 N, 121.901 W | 150 | Metatranscriptome | 1174810 | 1174938 | 23R metaT | 166838 | https://genome.jgi.doe.gov/portal/23RmetaT_FD/23RmetaT_FD.download.html | 468307 | PRJNA468307 | SRP163179 | SAMN09201818 | Gp0290788 | Illumina HiSeq-2000 | 2 × 151 | 8,584,843,770 | MEGAHIT v1.1.2 |
| 24D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/2/2016 | 36.835 N, 121.901 W | 425 | Metagenome | 1174546 | 1174701 | 24D metaG | 166757 | https://genome.jgi.doe.gov/portal/24DmetaG_FD/24DmetaG_FD.download.html | 502440 | PRJNA502440 | SRP174090 | SAMN10351047 | Gp0272197 | Illumina HiSeq-2000 | 2 × 151 | 6,136,433,734 | SPAdes assembler 3.11.1 |
| 24R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/2/2016 | 36.835 N, 121.901 W | 425 | Metatranscriptome | 1174813 | 1174939 | 24R metaT | 166839 | https://genome.jgi.doe.gov/portal/24RmetaT_FD/24RmetaT_FD.download.html | 468308 | PRJNA468308 | SRP163186 | SAMN09201817 | Gp0290789 | Illumina HiSeq-2000 | 2 × 151 | 8,896,620,114 | MEGAHIT v1.1.2 |
| 25D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/6/2016 | 36.835 N, 121.901 W | 575 | Metagenome | 1174548 | 1174702 | 25D metaG | 166758 | https://genome.jgi.doe.gov/portal/25DmetaG_FD/25DmetaG_FD.download.html | 502415 | PRJNA502415 | SRP174108 | SAMN10351090 | Gp0266611 | Illumina HiSeq-2000 | 2 × 151 | 7,480,494,700 | SPAdes assembler 3.11.1 |
| 25R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/6/2016 | 36.835 N, 121.901 W | 575 | Metatranscriptome | 1174816 | 1174940 | 25R metaT | 166840 | https://genome.jgi.doe.gov/portal/25RmetaT_FD/25RmetaT_FD.download.html | 502457 | PRJNA502457 | SRP174129 | SAMN10351332 | Gp0290790 | Illumina HiSeq-2000 | 2 × 151 | 10,195,628,720 | MEGAHIT v1.1.2 |
| 26D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/1/2016 | 36.835 N, 121.901 W | 125 | Metagenome | 1174550 | 1174703 | 26D metaG | 166759 | https://genome.jgi.doe.gov/portal/26DmetaG_FD/26DmetaG_FD.download.html | 467728 | PRJNA467728 | SRP155258 | SAMN09201452 | Gp0266612 | Illumina HiSeq-2000 | 2 × 151 | 7,294,852,280 | SPAdes assembler 3.11.1 |
| 26R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/1/2016 | 36.835 N, 121.901 W | 125 | Metatranscriptome | 1174819 | 1174941 | 26R metaT | 166841 | https://genome.jgi.doe.gov/portal/26RmetaT_FD/26RmetaT_FD.download.html | 468309 | PRJNA468309 | SRP163184 | SAMN09202122 | Gp0290791 | Illumina HiSeq-2000 | 2 × 151 | 6,828,158,996 | MEGAHIT v1.1.2 |
| 27D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/12/2016 | 36.835 N, 121.901 W | 100 | Metagenome | 1174552 | 1174704 | 27D metaG | 166760 | https://genome.jgi.doe.gov/portal/27DmetaG_FD/27DmetaG_FD.download.html | 468209 | PRJNA468209 | SRP155263 | SAMN09202468 | Gp0272198 | Illumina HiSeq-2000 | 2 × 151 | 8,451,209,978 | SPAdes assembler 3.11.1 |
| 27R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/12/2016 | 36.835 N, 121.901 W | 100 | Metatranscriptome | 1174994 | 1175090 | 27R metaT | 166882 | https://genome.jgi.doe.gov/portal/27RmetaT_FD/27RmetaT_FD.download.html | 467777 | PRJNA467777 | SRP155449 | SAMN09202117 | Gp0266672 | Illumina HiSeq-2000 | 2 × 151 | 10,817,948,644 | MEGAHIT v1.1.2 |
| 28D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/4/2016 | 36.835 N, 121.901 W | 149.5 | Metagenome | 1174554 | 1174705 | 28D metaG | 166761 | https://genome.jgi.doe.gov/portal/28DmetaG_FD/28DmetaG_FD.download.html | 467729 | PRJNA467729 | SRP155261 | SAMN09201851 | Gp0266613 | Illumina HiSeq-2000 | 2 × 151 | 4,854,715,534 | SPAdes assembler 3.11.1 |
| 28R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/4/2016 | 36.835 N, 121.901 W | 149.5 | Metatranscriptome | 1174822 | 1174942 | 28R metaT | 166842 | https://genome.jgi.doe.gov/portal/28RmetaT_FD/28RmetaT_FD.download.html | 502458 | PRJNA502458 | SRP174134 | SAMN10350970 | Gp0290792 | Illumina HiSeq-2000 | 2 × 151 | 10,456,791,978 | MEGAHIT v1.1.2 |
| 29D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/10/2016 | 36.835 N, 121.901 W | 100 | Metagenome | 1174556 | 1174706 | 29D metaG | 166762 | https://genome.jgi.doe.gov/portal/29DmetaG_FD/29DmetaG_FD.download.html | 467730 | PRJNA467730 | SRP155264 | SAMN09202067 | Gp0266614 | Illumina HiSeq-2000 | 2 × 151 | 5,913,326,704 | SPAdes assembler 3.11.1 |
| 29R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/10/2016 | 36.835 N, 121.901 W | 100 | Metatranscriptome | 1174997 | 1175091 | 29R metaT | 166883 | https://genome.jgi.doe.gov/portal/29RmetaT_FD/29RmetaT_FD.download.html | 468143 | PRJNA468143 | SRP155448 | SAMN09201429 | Gp0267189 | Illumina HiSeq-2000 | 2 × 151 | 8,853,622,562 | MEGAHIT v1.1.2 |
| 2D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/16/2016 | 36.835 N, 121.901 W | 1000 | Metagenome | 1174506 | 1174681 | 2D metaG | 166737 | https://genome.jgi.doe.gov/portal/2DmetaG_FD/2DmetaG_FD.download.html | 467721 | PRJNA467721 | SRP155237 | SAMN09201419 | Gp0266592 | Illumina HiSeq-2000 | 2 × 151 | 7,983,910,278 | SPAdes assembler 3.11.1 |
| 2R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/16/2016 | 36.835 N, 121.901 W | 1000 | Metatranscriptome | 1174771 | 1174925 | 2R metaT | 166825 | https://genome.jgi.doe.gov/portal/2RmetaT_FD/2RmetaT_FD.download.html | 468300 | PRJNA468300 | SRP155438 | SAMN09202080 | Gp0290775 | Illumina HiSeq-2000 | 2 × 151 | 9,198,635,516 | MEGAHIT v1.1.2 |
| 30D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/5/2016 | 36.835 N, 121.901 W | 200 | Metagenome | 1174558 | 1174707 | 30D metaG | 166763 | https://genome.jgi.doe.gov/portal/30DmetaG_FD/30DmetaG_FD.download.html | 502416 | PRJNA502416 | SRP174104 | SAMN10350895 | Gp0266615 | Illumina HiSeq-2000 | 2 × 151 | 6,613,959,758 | SPAdes assembler 3.11.1 |
| 30R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/5/2016 | 36.835 N, 121.901 W | 200 | Metatranscriptome | 1174825 | 1174943 | 30R metaT | 166843 | https://genome.jgi.doe.gov/portal/30RmetaT_FD/30RmetaT_FD.download.html | 468310 | PRJNA468310 | SRP163180 | SAMN09201816 | Gp0290793 | Illumina HiSeq-2000 | 2 × 151 | 9,853,104,246 | MEGAHIT v1.1.2 |
| 31D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/26/2016 | 36.835 N, 121.901 W | 200 | Metagenome | 1174560 | 1174708 | 31D metaG | 166764 | https://genome.jgi.doe.gov/portal/31DmetaG_FD/31DmetaG_FD.download.html | 467731 | PRJNA467731 | SRP155266 | SAMN09201850 | Gp0266616 | Illumina HiSeq-2000 | 2 × 151 | 6,324,142,740 | SPAdes assembler 3.11.1 |
| 31R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/26/2016 | 36.835 N, 121.901 W | 200 | Metatranscriptome | 1175000 | 1175092 | 31R metaT | 166884 | https://genome.jgi.doe.gov/portal/31RmetaT_FD/31RmetaT_FD.download.html | 467778 | PRJNA467778 | SRP155456 | SAMN09201438 | Gp0266673 | Illumina HiSeq-2000 | 2 × 151 | 12,414,596,672 | MEGAHIT v1.1.2 |
| 32D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/19/2016 | 36.835 N, 121.901 W | 200 | Metagenome | 1174562 | 1174709 | 32D metaG | 166765 | https://genome.jgi.doe.gov/portal/32DmetaG_FD/32DmetaG_FD.download.html | 468210 | PRJNA468210 | SRP155268 | SAMN09201604 | Gp0272199 | Illumina HiSeq-2000 | 2 × 151 | 7,234,605,696 | SPAdes assembler 3.11.1 |
| 32R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/19/2016 | 36.835 N, 121.901 W | 200 | Metatranscriptome | 1175003 | 1175093 | 32R metaT | 166885 | https://genome.jgi.doe.gov/portal/32RmetaT_FD/32RmetaT_FD.download.html | 468144 | PRJNA468144 | SRP155455 | SAMN09202305 | Gp0267190 | Illumina HiSeq-2000 | 2 × 151 | 10,512,598,256 | MEGAHIT v1.1.2 |
| 33D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/28/2016 | 36.835 N, 121.901 W | 75 | Metagenome | 1174564 | 1174710 | 33D metaG | 166766 | https://genome.jgi.doe.gov/portal/33DmetaG_FD/33DmetaG_FD.download.html | 502417 | PRJNA502417 | SRP174107 | SAMN10351036 | Gp0266617 | Illumina HiSeq-2000 | 2 × 151 | 8,943,864,994 | SPAdes assembler 3.11.1 |
| 33R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/28/2016 | 36.835 N, 121.901 W | 75 | Metatranscriptome | 1174828 | 1174944 | 33R metaT | 166844 | https://genome.jgi.doe.gov/portal/33RmetaT_FD/33RmetaT_FD.download.html | 502459 | PRJNA502459 | SRP174133 | SAMN10350969 | Gp0290794 | Illumina HiSeq-2000 | 2 × 151 | 10,664,122,830 | MEGAHIT v1.1.2 |
| 34D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/30/2016 | 36.835 N, 121.901 W | 94 | Metagenome | 1174566 | 1174711 | 34D metaG | 166767 | https://genome.jgi.doe.gov/portal/34DmetaG_FD/34DmetaG_FD.download.html | 467732 | PRJNA467732 | SRP155270 | SAMN09201849 | Gp0266618 | Illumina HiSeq-2000 | 2 × 151 | 5,589,222,116 | SPAdes assembler 3.11.1 |
| 34R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/30/2016 | 36.835 N, 121.901 W | 94 | Metatranscriptome | 1174831 | 1174945 | 34R metaT | 166845 | https://genome.jgi.doe.gov/portal/34RmetaT_FD/34RmetaT_FD.download.html | 468311 | PRJNA468311 | SRP163189 | SAMN09202123 | Gp0290795 | Illumina HiSeq-2000 | 2 × 151 | 10,817,986,998 | MEGAHIT v1.1.2 |
| 35D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/20/2016 | 36.835 N, 121.901 W | 200 | Metagenome | 1174568 | 1174712 | 35D metaG | 166768 | https://genome.jgi.doe.gov/portal/35DmetaG_FD/35DmetaG_FD.download.html | 502441 | PRJNA502441 | SRP174106 | SAMN10351042 | Gp0272200 | Illumina HiSeq-2000 | 2 × 151 | 8,308,168,886 | SPAdes assembler 3.11.1 |
| 35R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/20/2016 | 36.835 N, 121.901 W | 200 | Metatranscriptome | 1175006 | 1175094 | 35R metaT | 166886 | https://genome.jgi.doe.gov/portal/35RmetaT_FD/35RmetaT_FD.download.html | 467779 | PRJNA467779 | SRP155457 | SAMN09201437 | Gp0266674 | Illumina HiSeq-2000 | 2 × 151 | 12,257,738,778 | MEGAHIT v1.1.2 |
| 36D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/14/2016 | 36.835 N, 121.901 W | 825 | Metagenome | 1174570 | 1174713 | 36D metaG | 166769 | https://genome.jgi.doe.gov/portal/36D_rmetaG_FD/36D_rmetaG_FD.download.html | 467733 | PRJNA467733 | SRP155275 | SAMN09202068 | Gp0266619 | Illumina HiSeq-2000 | 2 × 151 | 9,178,069,618 | SPAdes assembler 3.11.1 |
| 36D_r | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/5/2016 | 36.835 N, 121.901 W | 200 | Metagenome | 1174672 | 1174764 | 36D_r metaG | 166820 | https://genome.jgi.doe.gov/portal/36DmetaG_FD/36DmetaG_FD.download.html | 467770 | PRJNA467770 | SRP155426 | SAMN09201533 | Gp0266665 | Illumina HiSeq-2000 | 2 × 151 | 5,877,069,792 | SPAdes assembler 3.11.1 |
| 36R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/14/2016 | 36.835 N, 121.901 W | 825 | Metatranscriptome | 1174834 | 1174946 | 36R metaT | 166846 | https://genome.jgi.doe.gov/portal/36R_rmetaT_FD/36R_rmetaT_FD.download.html | 468312 | PRJNA468312 | SRP163183 | SAMN09201815 | Gp0290796 | Illumina HiSeq-2000 | 2 × 151 | 9,511,697,776 | MEGAHIT v1.1.2 |
| 36R_r | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/5/2016 | 36.835 N, 121.901 W | 200 | Metatranscriptome | 1175078 | 1175118 | 36R_r metaT | 166910 | https://genome.jgi.doe.gov/portal/36RmetaT_FD/36RmetaT_FD.download.html | 468152 | PRJNA468152 | SRP155603 | SAMN09201303 | Gp0267198 | Illumina HiSeq-2000 | 2 × 151 | 14,183,895,080 | MEGAHIT v1.1.2 |
| 37D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/25/2016 | 36.835 N, 121.901 W | 200 | Metagenome | 1174572 | 1174714 | 37D metaG | 166770 | https://genome.jgi.doe.gov/portal/37DmetaG_FD/37DmetaG_FD.download.html | 467734 | PRJNA467734 | SRP155273 | SAMN09202069 | Gp0266620 | Illumina HiSeq-2000 | 2 × 151 | 5,904,284,220 | SPAdes assembler 3.11.1 |
| 38D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/2/2016 | 36.835 N, 121.901 W | 175 | Metagenome | 1174574 | 1174715 | 38D metaG | 166771 | https://genome.jgi.doe.gov/portal/38DmetaG_FD/38DmetaG_FD.download.html | 468211 | PRJNA468211 | SRP155280 | SAMN09202469 | Gp0272201 | Illumina HiSeq-2000 | 2 × 151 | 7,854,968,358 | SPAdes assembler 3.11.1 |
| 38R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/2/2016 | 36.835 N, 121.901 W | 175 | Metatranscriptome | 1174837 | 1174947 | 38R metaT | 166847 | https://genome.jgi.doe.gov/portal/38RmetaT_FD/38RmetaT_FD.download.html | 502460 | PRJNA502460 | SRP173009 | SAMN10351182 | Gp0290797 | Illumina HiSeq-2000 | 2 × 151 | 9,291,317,504 | MEGAHIT v1.1.2 |
| 39D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/28/2016 | 36.835 N, 121.901 W | 75 | Metagenome | 1174576 | 1174716 | 39D metaG | 166772 | https://genome.jgi.doe.gov/portal/39DmetaG_FD/39DmetaG_FD.download.html | 467735 | PRJNA467735 | SRP155282 | SAMN09201451 | Gp0266621 | Illumina HiSeq-2000 | 2 × 151 | 9,772,488,366 | SPAdes assembler 3.11.1 |
| 39R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/28/2016 | 36.835 N, 121.901 W | 75 | Metatranscriptome | 1174840 | 1174948 | 39R metaT | 166848 | https://genome.jgi.doe.gov/portal/39RmetaT_FD/39RmetaT_FD.download.html | 468313 | PRJNA468313 | SRP155441 | SAMN09202124 | Gp0290798 | Illumina HiSeq-2000 | 2 × 151 | 9,537,885,102 | MEGAHIT v1.1.2 |
| 3D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/9/2016 | 36.835 N, 121.901 W | 175 | Metagenome | 1174508 | 1174682 | 3D metaG | 166738 | https://genome.jgi.doe.gov/portal/3DmetaG_FD/3DmetaG_FD.download.html | 467722 | PRJNA467722 | SRP155238 | SAMN09201454 | Gp0266593 | Illumina HiSeq-2000 | 2 × 151 | 6,667,059,814 | SPAdes assembler 3.11.1 |
| 3R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/9/2016 | 36.835 N, 121.901 W | 175 | Metatranscriptome | 1174774 | 1174926 | 3R metaT | 166826 | https://genome.jgi.doe.gov/portal/3RmetaT_FD/3RmetaT_FD.download.html | 468301 | PRJNA468301 | SRP155439 | SAMN09201821 | Gp0290776 | Illumina HiSeq-2000 | 2 × 151 | 9,116,905,256 | MEGAHIT v1.1.2 |
| 40D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/4/2016 | 36.835 N, 121.901 W | 400 | Metagenome | 1174578 | 1174717 | 40D metaG | 166773 | https://genome.jgi.doe.gov/portal/40DmetaG_FD/40DmetaG_FD.download.html | 467736 | PRJNA467736 | SRP155287 | SAMN09201848 | Gp0266622 | Illumina HiSeq-2000 | 2 × 151 | 8,607,890,598 | SPAdes assembler 3.11.1 |
| 40R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/4/2016 | 36.835 N, 121.901 W | 400 | Metatranscriptome | 1174843 | 1174949 | 40R metaT | 166849 | https://genome.jgi.doe.gov/portal/40RmetaT_FD/40RmetaT_FD.download.html | 468314 | PRJNA468314 | SRP163274 | SAMN09201262 | Gp0290799 | Illumina HiSeq-2000 | 2 × 151 | 8,161,434,334 | MEGAHIT v1.1.2 |
| 41D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/13/2016 | 36.835 N, 121.901 W | 350 | Metagenome | 1174580 | 1174718 | 41D metaG | 166774 | https://genome.jgi.doe.gov/portal/41DmetaG_FD/41DmetaG_FD.download.html | 467737 | PRJNA467737 | SRP155292 | SAMN09201681 | Gp0266623 | Illumina HiSeq-2000 | 2 × 151 | 7,636,600,916 | SPAdes assembler 3.11.1 |
| 41R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/13/2016 | 36.835 N, 121.901 W | 350 | Metatranscriptome | 1174846 | 1174950 | 41R metaT | 166850 | https://genome.jgi.doe.gov/portal/41RmetaT_FD/41RmetaT_FD.download.html | 502461 | PRJNA502461 | SRP174136 | SAMN10351183 | Gp0290800 | Illumina HiSeq-2000 | 2 × 151 | 10,445,115,752 | MEGAHIT v1.1.2 |
| 42D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/27/2016 | 36.835 N, 121.901 W | 75 | Metagenome | 1174582 | 1174719 | 42D metaG | 166775 | https://genome.jgi.doe.gov/portal/42DmetaG_FD/42DmetaG_FD.download.html | 502418 | PRJNA502418 | SRP174105 | SAMN10350894 | Gp0266624 | Illumina HiSeq-2000 | 2 × 151 | 9,179,809,138 | SPAdes assembler 3.11.1 |
| 42R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/27/2016 | 36.835 N, 121.901 W | 75 | Metatranscriptome | 1174849 | 1174951 | 42R metaT | 166851 | https://genome.jgi.doe.gov/portal/42RmetaT_FD/42RmetaT_FD.download.html | 502462 | PRJNA502462 | SRP173010 | SAMN10350968 | Gp0290801 | Illumina HiSeq-2000 | 2 × 151 | 10,001,448,458 | MEGAHIT v1.1.2 |
| 43D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/8/2016 | 36.835 N, 121.901 W | 225 | Metagenome | 1174584 | 1174720 | 43D metaG | 166776 | https://genome.jgi.doe.gov/portal/43DmetaG_FD/43DmetaG_FD.download.html | 468212 | PRJNA468212 | SRP155294 | SAMN09201603 | Gp0272202 | Illumina HiSeq-2000 | 2 × 151 | 7,671,333,030 | SPAdes assembler 3.11.1 |
| 43R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/8/2016 | 36.835 N, 121.901 W | 225 | Metatranscriptome | 1174852 | 1174952 | 43R metaT | 166852 | https://genome.jgi.doe.gov/portal/43RmetaT_FD/43RmetaT_FD.download.html | 502463 | PRJNA502463 | SRP174135 | SAMN10350967 | Gp0290802 | Illumina HiSeq-2000 | 2 × 151 | 10,303,982,092 | MEGAHIT v1.1.2 |
| 44D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/30/2016 | 36.835 N, 121.901 W | 94.1 | Metagenome | 1174586 | 1174721 | 44D metaG | 166777 | https://genome.jgi.doe.gov/portal/44DmetaG_FD/44DmetaG_FD.download.html | 467738 | PRJNA467738 | SRP155297 | SAMN09201847 | Gp0266625 | Illumina HiSeq-2000 | 2 × 151 | 8,361,881,096 | SPAdes assembler 3.11.1 |
| 44R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/30/2016 | 36.835 N, 121.901 W | 94.1 | Metatranscriptome | 1174855 | 1174953 | 44R metaT | 166853 | https://genome.jgi.doe.gov/portal/44RmetaT_FD/44RmetaT_FD.download.html | 468315 | PRJNA468315 | SRP163282 | SAMN09202125 | Gp0290803 | Illumina HiSeq-2000 | 2 × 151 | 8,829,029,192 | MEGAHIT v1.1.2 |
| 45D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/3/2016 | 36.835 N, 121.901 W | 300 | Metagenome | 1174588 | 1174722 | 45D metaG | 166778 | https://genome.jgi.doe.gov/portal/45DmetaG_FD/45DmetaG_FD.download.html | 467739 | PRJNA467739 | SRP155298 | SAMN09201682 | Gp0266626 | Illumina HiSeq-2000 | 2 × 151 | 6,945,518,612 | SPAdes assembler 3.11.1 |
| 45R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/3/2016 | 36.835 N, 121.901 W | 300 | Metatranscriptome | 1174858 | 1174954 | 45R metaT | 166854 | https://genome.jgi.doe.gov/portal/45RmetaT_FD/45RmetaT_FD.download.html | 502464 | PRJNA502464 | SRP174137 | SAMN10351124 | Gp0290804 | Illumina HiSeq-2000 | 2 × 151 | 9,459,974,538 | MEGAHIT v1.1.2 |
| 46D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/21/2016 | 36.835 N, 121.901 W | 200 | Metagenome | 1174590 | 1174723 | 46D metaG | 166779 | https://genome.jgi.doe.gov/portal/46DmetaG_FD/46DmetaG_FD.download.html | 468213 | PRJNA468213 | SRP155299 | SAMN09201602 | Gp0272203 | Illumina HiSeq-2000 | 2 × 151 | 7,398,835,108 | SPAdes assembler 3.11.1 |
| 46R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/21/2016 | 36.835 N, 121.901 W | 200 | Metatranscriptome | 1174861 | 1174955 | 46R metaT | 166855 | https://genome.jgi.doe.gov/portal/46RmetaT_FD/46RmetaT_FD.download.html | 468316 | PRJNA468316 | SRP163285 | SAMN09202126 | Gp0290805 | Illumina HiSeq-2000 | 2 × 151 | 9,167,903,090 | MEGAHIT v1.1.2 |
| 47D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/25/2016 | 36.835 N, 121.901 W | 200 | Metagenome | 1174592 | 1174724 | 47D metaG | 166780 | https://genome.jgi.doe.gov/portal/47DmetaG_FD/47DmetaG_FD.download.html | 467740 | PRJNA467740 | SRP155307 | SAMN09201846 | Gp0266627 | Illumina HiSeq-2000 | 2 × 151 | 7,252,908,708 | SPAdes assembler 3.11.1 |
| 47R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/25/2016 | 36.835 N, 121.901 W | 200 | Metatranscriptome | 1175012 | 1175096 | 47R metaT | 166888 | https://genome.jgi.doe.gov/portal/47RmetaT_FD/47RmetaT_FD.download.html | 467780 | PRJNA467780 | SRP155461 | SAMN09201301 | Gp0266675 | Illumina HiSeq-2000 | 2 × 151 | 11,208,036,608 | MEGAHIT v1.1.2 |
| 48D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/31/2016 | 36.835 N, 121.901 W | 300 | Metagenome | 1174594 | 1174725 | 48D metaG | 166781 | https://genome.jgi.doe.gov/portal/48D_rmetaG_FD/48D_rmetaG_FD.download.html | 467741 | PRJNA467741 | SRP155306 | SAMN09201450 | Gp0266628 | Illumina HiSeq-2000 | 2 × 151 | 7,977,827,394 | SPAdes assembler 3.11.1 |
| 48D_r | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/2/2016 | 36.835 N, 121.901 W | 550 | Metagenome | 1174674 | 1174765 | 48D_r metaG | 166821 | https://genome.jgi.doe.gov/portal/48DmetaG_FD/48DmetaG_FD.download.html | 467771 | PRJNA467771 | SRP155431 | SAMN09201532 | Gp0266666 | Illumina HiSeq-2000 | 2 × 151 | 8,114,732,752 | SPAdes assembler 3.11.1 |
| 48R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/31/2016 | 36.835 N, 121.901 W | 300 | Metatranscriptome | 1174864 | 1174956 | 48R metaT | 166856 | https://genome.jgi.doe.gov/portal/48RmetaT_FD/48RmetaT_FD.download.html | 468317 | PRJNA468317 | SRP163284 | SAMN09201261 | Gp0290806 | Illumina HiSeq-2000 | 2 × 151 | 9,928,027,426 | MEGAHIT v1.1.2 |
| 49D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/3/2016 | 36.835 N, 121.901 W | 325 | Metagenome | 1174596 | 1174726 | 49D metaG | 166782 | https://genome.jgi.doe.gov/portal/49DmetaG_FD/49DmetaG_FD.download.html | 502442 | PRJNA502442 | SRP174109 | SAMN10351356 | Gp0272204 | Illumina HiSeq-2000 | 2 × 151 | 8,952,643,228 | SPAdes assembler 3.11.1 |
| 49R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/3/2016 | 36.835 N, 121.901 W | 325 | Metatranscriptome | 1174867 | 1174957 | 49R metaT | 166857 | https://genome.jgi.doe.gov/portal/49RmetaT_FD/49RmetaT_FD.download.html | 468318 | PRJNA468318 | SRP163276 | SAMN09201260 | Gp0290807 | Illumina HiSeq-2000 | 2 × 151 | 8,642,543,890 | MEGAHIT v1.1.2 |
| 4D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/3/2016 | 36.835 N, 121.901 W | 325 | Metagenome | 1174510 | 1174683 | 4D metaG | 166739 | https://genome.jgi.doe.gov/portal/4DmetaG_FD/4DmetaG_FD.download.html | 467723 | PRJNA467723 | SRP155240 | SAMN09201857 | Gp0266594 | Illumina HiSeq-2000 | 2 × 151 | 7,717,664,360 | SPAdes assembler 3.11.1 |
| 4R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/3/2016 | 36.835 N, 121.901 W | 325 | Metatranscriptome | 1174777 | 1174927 | 4R metaT | 166827 | https://genome.jgi.doe.gov/portal/4RmetaT_FD/4RmetaT_FD.download.html | 468302 | PRJNA468302 | SRP161170 | SAMN09201263 | Gp0290777 | Illumina HiSeq-2000 | 2 × 151 | 10,907,101,460 | MEGAHIT v1.1.2 |
| 50D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/18/2016 | 36.835 N, 121.901 W | 200 | Metagenome | 1174598 | 1174727 | 50D metaG | 166783 | https://genome.jgi.doe.gov/portal/50DmetaG_FD/50DmetaG_FD.download.html | 502419 | PRJNA502419 | SRP174111 | SAMN10351174 | Gp0266629 | Illumina HiSeq-2000 | 2 × 151 | 7,899,520,304 | SPAdes assembler 3.11.1 |
| 50R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/18/2016 | 36.835 N, 121.901 W | 200 | Metatranscriptome | 1175015 | 1175097 | 50R metaT | 166889 | https://genome.jgi.doe.gov/portal/50RmetaT_FD/50RmetaT_FD.download.html | 467781 | PRJNA467781 | SRP155462 | SAMN09201714 | Gp0266676 | Illumina HiSeq-2000 | 2 × 151 | 11,745,765,426 | MEGAHIT v1.1.2 |
| 51D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/13/2016 | 36.835 N, 121.901 W | 400 | Metagenome | 1174600 | 1174728 | 51D metaG | 166784 | https://genome.jgi.doe.gov/portal/51DmetaG_FD/51DmetaG_FD.download.html | 502420 | PRJNA502420 | SRP174112 | SAMN10351052 | Gp0266630 | Illumina HiSeq-2000 | 2 × 151 | 6,003,670,004 | SPAdes assembler 3.11.1 |
| 51R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/13/2016 | 36.835 N, 121.901 W | 400 | Metatranscriptome | 1174870 | 1174958 | 51R metaT | 166858 | https://genome.jgi.doe.gov/portal/51RmetaT_FD/51RmetaT_FD.download.html | 468319 | PRJNA468319 | SRP163283 | SAMN09202127 | Gp0290808 | Illumina HiSeq-2000 | 2 × 151 | 11,738,448,570 | MEGAHIT v1.1.2 |
| 52D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/17/2016 | 36.835 N, 121.901 W | 200 | Metagenome | 1174602 | 1174729 | 52D metaG | 166785 | https://genome.jgi.doe.gov/portal/52DmetaG_FD/52DmetaG_FD.download.html | 467742 | PRJNA467742 | SRP155308 | SAMN09201449 | Gp0266631 | Illumina HiSeq-2000 | 2 × 151 | 5,968,743,100 | SPAdes assembler 3.11.1 |
| 52R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/17/2016 | 36.835 N, 121.901 W | 200 | Metatranscriptome | 1175018 | 1175098 | 52R metaT | 166890 | https://genome.jgi.doe.gov/portal/52RmetaT_FD/52RmetaT_FD.download.html | 467782 | PRJNA467782 | SRP155463 | SAMN09202118 | Gp0266677 | Illumina HiSeq-2000 | 2 × 151 | 11,715,640,926 | MEGAHIT v1.1.2 |
| 53D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/29/2016 | 36.835 N, 121.901 W | 125 | Metagenome | 1174604 | 1174730 | 53D metaG | 166786 | https://genome.jgi.doe.gov/portal/53DmetaG_FD/53DmetaG_FD.download.html | 502421 | PRJNA502421 | SRP174114 | SAMN10351091 | Gp0266632 | Illumina HiSeq-2000 | 2 × 151 | 10,237,618,498 | SPAdes assembler 3.11.1 |
| 53R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/29/2016 | 36.835 N, 121.901 W | 125 | Metatranscriptome | 1174873 | 1174959 | 53R metaT | 166859 | https://genome.jgi.doe.gov/portal/53RmetaT_FD/53RmetaT_FD.download.html | 468320 | PRJNA468320 | SRP163278 | SAMN09201259 | Gp0290809 | Illumina HiSeq-2000 | 2 × 151 | 8,487,681,914 | MEGAHIT v1.1.2 |
| 54D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/11/2016 | 36.835 N, 121.901 W | 425 | Metagenome | 1174606 | 1174731 | 54D metaG | 166787 | https://genome.jgi.doe.gov/portal/54DmetaG_FD/54DmetaG_FD.download.html | 468214 | PRJNA468214 | SRP155314 | SAMN09202493 | Gp0272205 | Illumina HiSeq-2000 | 2 × 151 | 8,189,482,886 | SPAdes assembler 3.11.1 |
| 54R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/11/2016 | 36.835 N, 121.901 W | 425 | Metatranscriptome | 1174876 | 1174960 | 54R metaT | 166860 | https://genome.jgi.doe.gov/portal/54RmetaT_FD/54RmetaT_FD.download.html | 468321 | PRJNA468321 | SRP163277 | SAMN09201279 | Gp0290810 | Illumina HiSeq-2000 | 2 × 151 | 10,432,362,896 | MEGAHIT v1.1.2 |
| 55D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/8/2016 | 36.835 N, 121.901 W | 175 | Metagenome | 1174608 | 1174732 | 55D metaG | 166788 | https://genome.jgi.doe.gov/portal/55DmetaG_FD/55DmetaG_FD.download.html | 502422 | PRJNA502422 | SRP174115 | SAMN10351092 | Gp0266633 | Illumina HiSeq-2000 | 2 × 151 | 9,803,128,682 | SPAdes assembler 3.11.1 |
| 55R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/8/2016 | 36.835 N, 121.901 W | 175 | Metatranscriptome | 1175021 | 1175099 | 55R metaT | 166891 | https://genome.jgi.doe.gov/portal/55RmetaT_FD/55RmetaT_FD.download.html | 468145 | PRJNA468145 | SRP155489 | SAMN09201664 | Gp0267191 | Illumina HiSeq-2000 | 2 × 151 | 11,401,846,316 | MEGAHIT v1.1.2 |
| 56D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/9/2016 | 36.835 N, 121.901 W | 174.4 | Metagenome | 1174610 | 1174733 | 56D metaG | 166789 | https://genome.jgi.doe.gov/portal/56DmetaG_FD/56DmetaG_FD.download.html | 467743 | PRJNA467743 | SRP155320 | SAMN09201683 | Gp0266634 | Illumina HiSeq-2000 | 2 × 151 | 4,883,065,180 | SPAdes assembler 3.11.1 |
| 56R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/9/2016 | 36.835 N, 121.901 W | 174.4 | Metatranscriptome | 1174879 | 1174961 | 56R metaT | 166861 | https://genome.jgi.doe.gov/portal/56RmetaT_FD/56RmetaT_FD.download.html | 468322 | PRJNA468322 | SRP163275 | SAMN09202128 | Gp0290811 | Illumina HiSeq-2000 | 2 × 151 | 9,136,023,970 | MEGAHIT v1.1.2 |
| 57D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/6/2016 | 36.835 N, 121.901 W | 550 | Metagenome | 1174612 | 1174734 | 57D metaG | 166790 | https://genome.jgi.doe.gov/portal/57DmetaG_FD/57DmetaG_FD.download.html | 502423 | PRJNA502423 | SRP174120 | SAMN10351051 | Gp0266635 | Illumina HiSeq-2000 | 2 × 151 | 8,240,164,526 | SPAdes assembler 3.11.1 |
| 57R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/6/2016 | 36.835 N, 121.901 W | 550 | Metatranscriptome | 1174882 | 1174962 | 57R metaT | 166862 | https://genome.jgi.doe.gov/portal/57RmetaT_FD/57RmetaT_FD.download.html | 468323 | PRJNA468323 | SRP163281 | SAMN09201278 | Gp0290812 | Illumina HiSeq-2000 | 2 × 151 | 10,912,252,070 | MEGAHIT v1.1.2 |
| 58D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/4/2016 | 36.835 N, 121.901 W | 400 | Metagenome | 1174614 | 1174735 | 58D metaG | 166791 | https://genome.jgi.doe.gov/portal/58D_rmetaG_FD/58D_rmetaG_FD.download.html | 502424 | PRJNA502424 | SRP174121 | SAMN10351093 | Gp0266636 | Illumina HiSeq-2000 | 2 × 151 | 10,007,475,472 | SPAdes assembler 3.11.1 |
| 58D_r | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/4/2016 | 36.835 N, 121.901 W | 425 | Metagenome | 1174676 | 1174766 | 58D_r metaG | 166822 | https://genome.jgi.doe.gov/portal/58DmetaG_FD/58DmetaG_FD.download.html | 467772 | PRJNA467772 | SRP155434 | SAMN09202114 | Gp0266667 | Illumina HiSeq-2000 | 2 × 151 | 7,642,683,196 | SPAdes assembler 3.11.1 |
| 58R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/4/2016 | 36.835 N, 121.901 W | 400 | Metatranscriptome | 1175024 | 1175100 | 58R metaT | 166892 | https://genome.jgi.doe.gov/portal/58R_rmetaT_FD/58R_rmetaT_FD.download.html | 467783 | PRJNA467783 | SRP155488 | SAMN09201436 | Gp0266678 | Illumina HiSeq-2000 | 2 × 151 | 11,675,230,004 | MEGAHIT v1.1.2 |
| 58R_r | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/4/2016 | 36.835 N, 121.901 W | 425 | Metatranscriptome | 1174918 | 1174974 | 58R_r metaT | 166874 | https://genome.jgi.doe.gov/portal/58RmetaT_FD/58RmetaT_FD.download.html | 468331 | PRJNA468331 | SRP163295 | SAMN09200098 | Gp0290824 | Illumina HiSeq-2000 | 2 × 151 | 7,024,482,250 | MEGAHIT v1.1.2 |
| 59D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/21/2016 | 36.835 N, 121.901 W | 200 | Metagenome | 1174616 | 1174736 | 59D metaG | 166792 | https://genome.jgi.doe.gov/portal/59DmetaG_FD/59DmetaG_FD.download.html | 467744 | PRJNA467744 | SRP155321 | SAMN09201444 | Gp0266637 | Illumina HiSeq-2000 | 2 × 151 | 8,505,201,538 | SPAdes assembler 3.11.1 |
| 60D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/12/2016 | 36.835 N, 121.901 W | 475 | Metagenome | 1174618 | 1174737 | 60D metaG | 166793 | https://genome.jgi.doe.gov/portal/60DmetaG_FD/60DmetaG_FD.download.html | 502425 | PRJNA502425 | SRP174119 | SAMN10351050 | Gp0266638 | Illumina HiSeq-2000 | 2 × 151 | 7,531,245,498 | SPAdes assembler 3.11.1 |
| 60R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/12/2016 | 36.835 N, 121.901 W | 475 | Metatranscriptome | 1174885 | 1174963 | 60R metaT | 166863 | https://genome.jgi.doe.gov/portal/60RmetaT_FD/60RmetaT_FD.download.html | 502465 | PRJNA502465 | SRP173012 | SAMN10351125 | Gp0290813 | Illumina HiSeq-2000 | 2 × 151 | 9,036,901,228 | MEGAHIT v1.1.2 |
| 61D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/31/2016 | 36.835 N, 121.901 W | 290 | Metagenome | 1174620 | 1174738 | 61D metaG | 166794 | https://genome.jgi.doe.gov/portal/61DmetaG_FD/61DmetaG_FD.download.html | 502426 | PRJNA502426 | SRP174118 | SAMN10351049 | Gp0266639 | Illumina HiSeq-2000 | 2 × 151 | 8,327,099,756 | SPAdes assembler 3.11.1 |
| 61R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/31/2016 | 36.835 N, 121.901 W | 290 | Metatranscriptome | 1175030 | 1175102 | 61R metaT | 166894 | https://genome.jgi.doe.gov/portal/61RmetaT_FD/61RmetaT_FD.download.html | 468146 | PRJNA468146 | SRP155487 | SAMN09201304 | Gp0267192 | Illumina HiSeq-2000 | 2 × 151 | 12,760,280,972 | MEGAHIT v1.1.2 |
| 62D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/28/2016 | 36.835 N, 121.901 W | 250 | Metagenome | 1174622 | 1174739 | 62D metaG | 166795 | https://genome.jgi.doe.gov/portal/62DmetaG_FD/62DmetaG_FD.download.html | 467745 | PRJNA467745 | SRP155334 | SAMN09201274 | Gp0266640 | Illumina HiSeq-2000 | 2 × 151 | 7,880,205,894 | SPAdes assembler 3.11.1 |
| 63D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/24/2016 | 36.835 N, 121.901 W | 250 | Metagenome | 1174624 | 1174740 | 63D metaG | 166796 | https://genome.jgi.doe.gov/portal/63DmetaG_FD/63DmetaG_FD.download.html | 467746 | PRJNA467746 | SRP155339 | SAMN09202070 | Gp0266641 | Illumina HiSeq-2000 | 2 × 151 | 8,133,342,596 | SPAdes assembler 3.11.1 |
| 63R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/24/2016 | 36.835 N, 121.901 W | 250 | Metatranscriptome | 1175036 | 1175104 | 63R metaT | 166896 | https://genome.jgi.doe.gov/portal/63RmetaT_FD/63RmetaT_FD.download.html | 468147 | PRJNA468147 | SRP155506 | SAMN09202403 | Gp0267193 | Illumina HiSeq-2000 | 2 × 151 | 10,302,220,526 | MEGAHIT v1.1.2 |
| 64D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/5/2016 | 36.835 N, 121.901 W | 199.2 | Metagenome | 1174626 | 1174741 | 64D metaG | 166797 | https://genome.jgi.doe.gov/portal/64DmetaG_FD/64DmetaG_FD.download.html | 467747 | PRJNA467747 | SRP155340 | SAMN09202071 | Gp0266642 | Illumina HiSeq-2000 | 2 × 151 | 8,079,854,168 | SPAdes assembler 3.11.1 |
| 65D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/2/2016 | 36.835 N, 121.901 W | 400 | Metagenome | 1174628 | 1174742 | 65D metaG | 166798 | https://genome.jgi.doe.gov/portal/65DmetaG_FD/65DmetaG_FD.download.html | 467748 | PRJNA467748 | SRP155355 | SAMN09201541 | Gp0266643 | Illumina HiSeq-2000 | 2 × 151 | 6,290,782,310 | SPAdes assembler 3.11.1 |
| 66D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/3/2016 | 36.835 N, 121.901 W | 250 | Metagenome | 1174630 | 1174743 | 66D metaG | 166799 | https://genome.jgi.doe.gov/portal/66DmetaG_FD/66DmetaG_FD.download.html | 467749 | PRJNA467749 | SRP155353 | SAMN09202072 | Gp0266644 | Illumina HiSeq-2000 | 2 × 151 | 4,930,952,112 | SPAdes assembler 3.11.1 |
| 66R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/3/2016 | 36.835 N, 121.901 W | 250 | Metatranscriptome | 1174888 | 1174964 | 66R metaT | 166864 | https://genome.jgi.doe.gov/portal/66RmetaT_FD/66RmetaT_FD.download.html | 502466 | PRJNA502466 | SRP174138 | SAMN10350916 | Gp0290814 | Illumina HiSeq-2000 | 2 × 151 | 10,330,806,336 | MEGAHIT v1.1.2 |
| 67D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/15/2016 | 36.835 N, 121.901 W | 1000 | Metagenome | 1174632 | 1174744 | 67D metaG | 166800 | https://genome.jgi.doe.gov/portal/67DmetaG_FD/67DmetaG_FD.download.html | 467750 | PRJNA467750 | SRP155356 | SAMN09201540 | Gp0266645 | Illumina HiSeq-2000 | 2 × 151 | 6,280,811,478 | SPAdes assembler 3.11.1 |
| 67R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/15/2016 | 36.835 N, 121.901 W | 1000 | Metatranscriptome | 1175045 | 1175107 | 67R metaT | 166899 | https://genome.jgi.doe.gov/portal/67RmetaT_FD/67RmetaT_FD.download.html | 468148 | PRJNA468148 | SRP155500 | SAMN09201529 | Gp0267194 | Illumina HiSeq-2000 | 2 × 151 | 10,617,839,518 | MEGAHIT v1.1.2 |
| 68D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/7/2016 | 36.835 N, 121.901 W | 575 | Metagenome | 1174634 | 1174745 | 68D metaG | 166801 | https://genome.jgi.doe.gov/portal/68DmetaG_FD/68DmetaG_FD.download.html | 467751 | PRJNA467751 | SRP155364 | SAMN09202073 | Gp0266646 | Illumina HiSeq-2000 | 2 × 151 | 5,858,217,744 | SPAdes assembler 3.11.1 |
| 68R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/7/2016 | 36.835 N, 121.901 W | 575 | Metatranscriptome | 1174891 | 1174965 | 68R metaT | 166865 | https://genome.jgi.doe.gov/portal/68RmetaT_FD/68RmetaT_FD.download.html | 502467 | PRJNA502467 | SRP173013 | SAMN10350915 | Gp0290815 | Illumina HiSeq-2000 | 2 × 151 | 9,554,211,222 | MEGAHIT v1.1.2 |
| 70D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/7/2016 | 36.835 N, 121.901 W | 207.5 | Metagenome | 1174636 | 1174746 | 70D metaG | 166802 | https://genome.jgi.doe.gov/portal/70DmetaG_FD/70DmetaG_FD.download.html | 467752 | PRJNA467752 | SRP155365 | SAMN09201539 | Gp0266647 | Illumina HiSeq-2000 | 2 × 151 | 6,470,832,294 | SPAdes assembler 3.11.1 |
| 70R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/7/2016 | 36.835 N, 121.901 W | 207.5 | Metatranscriptome | 1174894 | 1174966 | 70R metaT | 166866 | https://genome.jgi.doe.gov/portal/70RmetaT_FD/70RmetaT_FD.download.html | 468324 | PRJNA468324 | SRP163290 | SAMN09202129 | Gp0290816 | Illumina HiSeq-2000 | 2 × 151 | 10,602,724,418 | MEGAHIT v1.1.2 |
| 71D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/18/2016 | 36.835 N, 121.901 W | 200 | Metagenome | 1174638 | 1174747 | 71D metaG | 166803 | https://genome.jgi.doe.gov/portal/71DmetaG_FD/71DmetaG_FD.download.html | 467753 | PRJNA467753 | SRP155366 | SAMN09201443 | Gp0266648 | Illumina HiSeq-2000 | 2 × 151 | 8,687,854,460 | SPAdes assembler 3.11.1 |
| 71R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/18/2016 | 36.835 N, 121.901 W | 200 | Metatranscriptome | 1175048 | 1175108 | 71R metaT | 166900 | https://genome.jgi.doe.gov/portal/71RmetaT_FD/71RmetaT_FD.download.html | 467784 | PRJNA467784 | SRP155502 | SAMN09202119 | Gp0266682 | Illumina HiSeq-2000 | 2 × 151 | 12,304,237,416 | MEGAHIT v1.1.2 |
| 73D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/16/2016 | 36.835 N, 121.901 W | 1000 | Metagenome | 1174640 | 1174748 | 73D metaG | 166804 | https://genome.jgi.doe.gov/portal/73DmetaG_FD/73DmetaG_FD.download.html | 467754 | PRJNA467754 | SRP155376 | SAMN09202074 | Gp0266649 | Illumina HiSeq-2000 | 2 × 151 | 6,269,792,102 | SPAdes assembler 3.11.1 |
| 73R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/16/2016 | 36.835 N, 121.901 W | 1000 | Metatranscriptome | 1174897 | 1174967 | 73R metaT | 166867 | https://genome.jgi.doe.gov/portal/73RmetaT_FD/73RmetaT_FD.download.html | 468325 | PRJNA468325 | SRP163289 | SAMN09201277 | Gp0290817 | Illumina HiSeq-2000 | 2 × 151 | 8,528,379,736 | MEGAHIT v1.1.2 |
| 74D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/11/2016 | 36.835 N, 121.901 W | 425 | Metagenome | 1174642 | 1174749 | 74D metaG | 166805 | https://genome.jgi.doe.gov/portal/74DmetaG_FD/74DmetaG_FD.download.html | 467755 | PRJNA467755 | SRP155377 | SAMN09201442 | Gp0266650 | Illumina HiSeq-2000 | 2 × 151 | 7,965,995,034 | SPAdes assembler 3.11.1 |
| 75D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/14/2016 | 36.835 N, 121.901 W | 825 | Metagenome | 1174644 | 1174750 | 75D metaG | 166806 | https://genome.jgi.doe.gov/portal/75DmetaG_FD/75DmetaG_FD.download.html | 467756 | PRJNA467756 | SRP155385 | SAMN09201538 | Gp0266651 | Illumina HiSeq-2000 | 2 × 151 | 5,308,517,646 | SPAdes assembler 3.11.1 |
| 75R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/14/2016 | 36.835 N, 121.901 W | 825 | Metatranscriptome | 1174900 | 1174968 | 75R metaT | 166868 | https://genome.jgi.doe.gov/portal/75RmetaT_FD/75RmetaT_FD.download.html | 468326 | PRJNA468326 | SRP163291 | SAMN09201276 | Gp0290818 | Illumina HiSeq-2000 | 2 × 151 | 9,110,384,170 | MEGAHIT v1.1.2 |
| 76D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/10/2016 | 36.835 N, 121.901 W | 350 | Metagenome | 1174646 | 1174751 | 76D metaG | 166807 | https://genome.jgi.doe.gov/portal/76DmetaG_FD/76DmetaG_FD.download.html | 467757 | PRJNA467757 | SRP155391 | SAMN09201302 | Gp0266652 | Illumina HiSeq-2000 | 2 × 151 | 7,179,999,868 | SPAdes assembler 3.11.1 |
| 76R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/10/2016 | 36.835 N, 121.901 W | 350 | Metatranscriptome | 1174903 | 1174969 | 76R metaT | 166869 | https://genome.jgi.doe.gov/portal/76RmetaT_FD/76RmetaT_FD.download.html | 502468 | PRJNA502468 | SRP173016 | SAMN10351184 | Gp0290819 | Illumina HiSeq-2000 | 2 × 151 | 8,145,636,412 | MEGAHIT v1.1.2 |
| 77D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/7/2016 | 36.835 N, 121.901 W | 201.5 | Metagenome | 1174648 | 1174752 | 77D metaG | 166808 | https://genome.jgi.doe.gov/portal/77DmetaG_FD/77DmetaG_FD.download.html | 467758 | PRJNA467758 | SRP155398 | SAMN09201684 | Gp0266653 | Illumina HiSeq-2000 | 2 × 151 | 6,424,234,902 | SPAdes assembler 3.11.1 |
| 77R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/7/2016 | 36.835 N, 121.901 W | 201.5 | Metatranscriptome | 1174906 | 1174970 | 77R metaT | 166870 | https://genome.jgi.doe.gov/portal/77RmetaT_FD/77RmetaT_FD.download.html | 468327 | PRJNA468327 | SRP163294 | SAMN09202166 | Gp0290820 | Illumina HiSeq-2000 | 2 × 151 | 11,267,917,772 | MEGAHIT v1.1.2 |
| 78D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/26/2016 | 36.835 N, 121.901 W | 92.5 | Metagenome | 1174650 | 1174753 | 78D metaG | 166809 | https://genome.jgi.doe.gov/portal/78DmetaG_FD/78DmetaG_FD.download.html | 467759 | PRJNA467759 | SRP155399 | SAMN09201441 | Gp0266654 | Illumina HiSeq-2000 | 2 × 151 | 7,805,644,812 | SPAdes assembler 3.11.1 |
| 78R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/26/2016 | 36.835 N, 121.901 W | 92.5 | Metatranscriptome | 1175054 | 1175110 | 78R metaT | 166902 | https://genome.jgi.doe.gov/portal/78RmetaT_FD/78RmetaT_FD.download.html | 468149 | PRJNA468149 | SRP155516 | SAMN09202306 | Gp0267195 | Illumina HiSeq-2000 | 2 × 151 | 10,213,209,650 | MEGAHIT v1.1.2 |
| 79D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/7/2016 | 36.835 N, 121.901 W | 550 | Metagenome | 1174652 | 1174754 | 79D metaG | 166810 | https://genome.jgi.doe.gov/portal/79DmetaG_FD/79DmetaG_FD.download.html | 467760 | PRJNA467760 | SRP155406 | SAMN09201685 | Gp0266655 | Illumina HiSeq-2000 | 2 × 151 | 5,990,380,192 | SPAdes assembler 3.11.1 |
| 79R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/7/2016 | 36.835 N, 121.901 W | 550 | Metatranscriptome | 1174909 | 1174971 | 79R metaT | 166871 | https://genome.jgi.doe.gov/portal/79RmetaT_FD/79RmetaT_FD.download.html | 468328 | PRJNA468328 | SRP163293 | SAMN09201275 | Gp0290821 | Illumina HiSeq-2000 | 2 × 151 | 8,943,879,490 | MEGAHIT v1.1.2 |
| 7D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/26/2016 | 36.835 N, 121.901 W | 200 | Metagenome | 1174514 | 1174685 | 7D metaG | 166741 | https://genome.jgi.doe.gov/portal/7DmetaG_FD/7DmetaG_FD.download.html | 502407 | PRJNA502407 | SRP174087 | SAMN10351037 | Gp0266596 | Illumina HiSeq-2000 | 2 × 151 | 10,931,783,316 | SPAdes assembler 3.11.1 |
| 80D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/11/2016 | 36.835 N, 121.901 W | 100 | Metagenome | 1174654 | 1174755 | 80D metaG | 166811 | https://genome.jgi.doe.gov/portal/80DmetaG_FD/80DmetaG_FD.download.html | 467761 | PRJNA467761 | SRP155408 | SAMN09201537 | Gp0266656 | Illumina HiSeq-2000 | 2 × 151 | 6,844,779,566 | SPAdes assembler 3.11.1 |
| 80R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/11/2016 | 36.835 N, 121.901 W | 100 | Metatranscriptome | 1175057 | 1175111 | 80R metaT | 166903 | https://genome.jgi.doe.gov/portal/80RmetaT_FD/80RmetaT_FD.download.html | 467785 | PRJNA467785 | SRP155515 | SAMN09201524 | Gp0266684 | Illumina HiSeq-2000 | 2 × 151 | 12,009,368,844 | MEGAHIT v1.1.2 |
| 81D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/11/2016 | 36.835 N, 121.901 W | 100 | Metagenome | 1174656 | 1174756 | 81D metaG | 166812 | https://genome.jgi.doe.gov/portal/81DmetaG_FD/81DmetaG_FD.download.html | 467762 | PRJNA467762 | SRP155407 | SAMN09202075 | Gp0266657 | Illumina HiSeq-2000 | 2 × 151 | 8,013,377,022 | SPAdes assembler 3.11.1 |
| 81R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/11/2016 | 36.835 N, 121.901 W | 100 | Metatranscriptome | 1175060 | 1175112 | 81R metaT | 166904 | https://genome.jgi.doe.gov/portal/81RmetaT_FD/81RmetaT_FD.download.html | 467786 | PRJNA467786 | SRP155517 | SAMN09201523 | Gp0266685 | Illumina HiSeq-2000 | 2 × 151 | 10,557,788,932 | MEGAHIT v1.1.2 |
| 82D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/10/2016 | 36.835 N, 121.901 W | 100 | Metagenome | 1174658 | 1174757 | 82D metaG | 166813 | https://genome.jgi.doe.gov/portal/82DmetaG_FD/82DmetaG_FD.download.html | 467763 | PRJNA467763 | SRP155413 | SAMN09201536 | Gp0266658 | Illumina HiSeq-2000 | 2 × 151 | 5,810,730,358 | SPAdes assembler 3.11.1 |
| 82R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/10/2016 | 36.835 N, 121.901 W | 100 | Metatranscriptome | 1175063 | 1175113 | 82R metaT | 166905 | https://genome.jgi.doe.gov/portal/82RmetaT_FD/82RmetaT_FD.download.html | 468150 | PRJNA468150 | SRP155535 | SAMN09201528 | Gp0267196 | Illumina HiSeq-2000 | 2 × 151 | 16,445,750,354 | MEGAHIT v1.1.2 |
| 84D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/11/2016 | 36.835 N, 121.901 W | 100 | Metagenome | 1174660 | 1174758 | 84D metaG | 166814 | https://genome.jgi.doe.gov/portal/84DmetaG_FD/84DmetaG_FD.download.html | 467764 | PRJNA467764 | SRP155419 | SAMN09201535 | Gp0266659 | Illumina HiSeq-2000 | 2 × 151 | 7,118,809,232 | SPAdes assembler 3.11.1 |
| 84R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/11/2016 | 36.835 N, 121.901 W | 100 | Metatranscriptome | 1175066 | 1175114 | 84R metaT | 166906 | https://genome.jgi.doe.gov/portal/84RmetaT_FD/84RmetaT_FD.download.html | 467787 | PRJNA467787 | SRP155539 | SAMN09202120 | Gp0266686 | Illumina HiSeq-2000 | 2 × 151 | 12,527,235,424 | MEGAHIT v1.1.2 |
| 85D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/13/2016 | 36.835 N, 121.901 W | 100 | Metagenome | 1174662 | 1174759 | 85D metaG | 166815 | https://genome.jgi.doe.gov/portal/85DmetaG_FD/85DmetaG_FD.download.html | 467765 | PRJNA467765 | SRP155420 | SAMN09201686 | Gp0266660 | Illumina HiSeq-2000 | 2 × 151 | 7,178,048,646 | SPAdes assembler 3.11.1 |
| 85R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/13/2016 | 36.835 N, 121.901 W | 100 | Metatranscriptome | 1174912 | 1174972 | 85R metaT | 166872 | https://genome.jgi.doe.gov/portal/85RmetaT_FD/85RmetaT_FD.download.html | 468329 | PRJNA468329 | SRP163292 | SAMN09199668 | Gp0290822 | Illumina HiSeq-2000 | 2 × 151 | 8,012,925,230 | MEGAHIT v1.1.2 |
| 89D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/5/2016 | 36.835 N, 121.901 W | 575 | Metagenome | 1174664 | 1174760 | 89D metaG | 166816 | https://genome.jgi.doe.gov/portal/89DmetaG_FD/89DmetaG_FD.download.html | 467766 | PRJNA467766 | SRP155421 | SAMN09201534 | Gp0266661 | Illumina HiSeq-2000 | 2 × 151 | 7,163,560,498 | SPAdes assembler 3.11.1 |
| 89R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/5/2016 | 36.835 N, 121.901 W | 575 | Metatranscriptome | 1175069 | 1175115 | 89R metaT | 166907 | https://genome.jgi.doe.gov/portal/89RmetaT_FD/89RmetaT_FD.download.html | 468151 | PRJNA468151 | SRP155534 | SAMN09201527 | Gp0267197 | Illumina HiSeq-2000 | 2 × 151 | 12,255,826,816 | MEGAHIT v1.1.2 |
| 8D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/5/2016 | 36.835 N, 121.901 W | 775 | Metagenome | 1174516 | 1174686 | 8D metaG | 166742 | https://genome.jgi.doe.gov/portal/8DmetaG_FD/8DmetaG_FD.download.html | 467724 | PRJNA467724 | SRP155246 | SAMN09201853 | Gp0266597 | Illumina HiSeq-2000 | 2 × 151 | 9,439,829,628 | SPAdes assembler 3.11.1 |
| 8R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/5/2016 | 36.835 N, 121.901 W | 775 | Metatranscriptome | 1174780 | 1174928 | 8R metaT | 166828 | https://genome.jgi.doe.gov/portal/8RmetaT_FD/8RmetaT_FD.download.html | 502451 | PRJNA502451 | SRP175188 | SAMN10351181 | Gp0290778 | Illumina HiSeq-2000 | 2 × 151 | 7,929,114,492 | MEGAHIT v1.1.2 |
| 90D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/8/2016 | 36.835 N, 121.901 W | 175 | Metagenome | 1174666 | 1174761 | 90D metaG | 166817 | https://genome.jgi.doe.gov/portal/90DmetaG_FD/90DmetaG_FD.download.html | 467768 | PRJNA467768 | SRP155424 | SAMN09201440 | Gp0266662 | Illumina HiSeq-2000 | 2 × 151 | 10,726,324,864 | SPAdes assembler 3.11.1 |
| 90R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/8/2016 | 36.835 N, 121.901 W | 175 | Metatranscriptome | 1175072 | 1175116 | 90R metaT | 166908 | https://genome.jgi.doe.gov/portal/90RmetaT_FD/90RmetaT_FD.download.html | 467788 | PRJNA467788 | SRP155602 | SAMN09201435 | Gp0266687 | Illumina HiSeq-2000 | 2 × 151 | 11,924,015,188 | MEGAHIT v1.1.2 |
| 91D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/11/2016 | 36.835 N, 121.901 W | 475 | Metagenome | 1174668 | 1174762 | 91D metaG | 166818 | https://genome.jgi.doe.gov/portal/91DmetaG_FD/91DmetaG_FD.download.html | 467769 | PRJNA467769 | SRP155425 | SAMN09202113 | Gp0266663 | Illumina HiSeq-2000 | 2 × 151 | 6,496,286,666 | SPAdes assembler 3.11.1 |
| 91R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/11/2016 | 36.835 N, 121.901 W | 475 | Metatranscriptome | 1174915 | 1174973 | 91R metaT | 166873 | https://genome.jgi.doe.gov/portal/91RmetaT_FD/91RmetaT_FD.download.html | 468330 | PRJNA468330 | SRP163297 | SAMN09199667 | Gp0290823 | Illumina HiSeq-2000 | 2 × 151 | 9,122,005,734 | MEGAHIT v1.1.2 |
| 9D | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/15/2016 | 36.835 N, 121.901 W | 975 | Metagenome | 1174518 | 1174687 | 9D metaG | 166743 | https://genome.jgi.doe.gov/portal/9DmetaG_FD/9DmetaG_FD.download.html | 502408 | PRJNA502408 | SRP174088 | SAMN10351267 | Gp0266598 | Illumina HiSeq-2000 | 2 × 151 | 10,053,837,002 | SPAdes assembler 3.11.1 |
| 9R | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/15/2016 | 36.835 N, 121.901 W | 975 | Metatranscriptome | 1174783 | 1174929 | 9R metaT | 166829 | https://genome.jgi.doe.gov/portal/9RmetaT_FD/9RmetaT_FD.download.html | 502452 | PRJNA502452 | SRP174123 | SAMN10350901 | Gp0290779 | Illumina HiSeq-2000 | 2 × 151 | 11,212,078,576 | MEGAHIT v1.1.2 |
| MB_1025D_MT | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/25/2016 | 36.835 N, 121.901 W | 200 | Metatranscriptome | 1188669 | 1188681 | MB_1025D metaT | 175198 | https://genome.jgi.doe.gov/portal/MB_1025DmetaT_FD/MB_1025DmetaT_FD.download.html | 502611 | PRJNA502611 | SRP174142 | SAMN10350584 | Gp0294703 | Illumina HiSeq-2000 | 2 × 151 | 30,614,719,084 | MEGAHIT v1.1.2 |
| MB_1026D_MT | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/26/2016 | 36.835 N, 121.901 W | 200 | Metatranscriptome | 1188672 | 1188682 | MB_1026D metaT | 175199 | https://genome.jgi.doe.gov/portal/MB_1026DmetaT_FD/MB_1026DmetaT_FD.download.html | 502612 | PRJNA502612 | SRP174143 | SAMN10350798 | Gp0294704 | Illumina HiSeq-2000 | 2 × 151 | 37,885,516,762 | MEGAHIT v1.1.2 |
| WCR_12_MT | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/26/2016 | 36.835 N, 121.901 W | 200 | Metatranscriptome | 1188654 | 1188676 | WCR_12 metaT | 175193 | https://genome.jgi.doe.gov/portal/WCR_12metaT_FD/WCR_12metaT_FD.download.html | 502608 | PRJNA502608 | SRP174139 | SAMN10350732 | Gp0294698 | Illumina HiSeq-2000 | 2 × 151 | 34,281,550,234 | MEGAHIT v1.1.2 |
| WCR_74_MT | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/6/2016 | 36.835 N, 121.901 W | 425 | Metatranscriptome | 1188657 | 1188677 | WCR_74 metaT | 175194 | https://genome.jgi.doe.gov/portal/WCR_74metaT_FD/WCR_74metaT_FD.download.html | 502609 | PRJNA502609 | SRP174140 | SAMN10350604 | Gp0294699 | Illumina HiSeq-2000 | 2 × 151 | 36,435,144,246 | MEGAHIT v1.1.2 |
| WCR_77_MT | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/7/2016 | 36.835 N, 121.901 W | 625 | Metatranscriptome | 1188660 | 1188678 | WCR_77 metaT | 175195 | https://genome.jgi.doe.gov/portal/WCR_77metaT_FD/WCR_77metaT_FD.download.html | 502610 | PRJNA502610 | SRP174141 | SAMN10350815 | Gp0294700 | Illumina HiSeq-2000 | 2 × 151 | 37,188,680,754 | MEGAHIT v1.1.2 |
| 1_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/4/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178203 | 511260 | PRJNA511260 | SRP174784 | SAMN10636594 | Gp0358229 | Illumina MiSeq | 2 × 301 | 92,259,510 | n/a | |
| 1_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/4/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178291 | 511216 | PRJNA511216 | SRP174685 | SAMN10636692 | Gp0358321 | Illumina MiSeq | 2 × 301 | 76,656,272 | n/a | |
| 10_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/10/16 | 36.835 N, 121.901 W | 350 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178211 | 511268 | PRJNA511268 | SRP174801 | SAMN10636691 | Gp0358237 | Illumina MiSeq | 2 × 301 | 104,195,966 | n/a | |
| 10_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/10/16 | 36.835 N, 121.901 W | 350 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178299 | 511224 | PRJNA511224 | SRP174679 | SAMN10636635 | Gp0358330 | Illumina MiSeq | 2 × 301 | 105,784,042 | n/a | |
| 11_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/6/16 | 36.835 N, 121.901 W | 250 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178212 | 511269 | PRJNA511269 | SRP174800 | SAMN10636604 | Gp0358238 | Illumina MiSeq | 2 × 301 | 86,498,370 | n/a | |
| 11_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/6/16 | 36.835 N, 121.901 W | 250 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178300 | 511225 | PRJNA511225 | SRP174672 | SAMN10636702 | Gp0358332 | Illumina MiSeq | 2 × 301 | 7,791,686 | n/a | |
| 12_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/29/16 | 36.835 N, 121.901 W | 125 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178213 | 511270 | PRJNA511270 | SRP174803 | SAMN10636591 | Gp0358239 | Illumina MiSeq | 2 × 301 | 92,700,174 | n/a | |
| 12_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/29/16 | 36.835 N, 121.901 W | 125 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178301 | 511226 | PRJNA511226 | SRP174673 | SAMN10636675 | Gp0358333 | Illumina MiSeq | 2 × 301 | 56,683,718 | n/a | |
| 125r_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/15/16 | 36.835 N, 121.901 W | 950 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178290 | 511215 | PRJNA511215 | SRP174796 | SAMN10636674 | Gp0358320 | Illumina MiSeq | 2 × 301 | 106,222,298 | n/a | |
| 125r_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/15/16 | 36.835 N, 121.901 W | 950 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178378 | 511162 | PRJNA511162 | SRP174734 | SAMN10637061 | Gp0358415 | Illumina MiSeq | 2 × 301 | 962,598 | n/a | |
| 13_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/1/16 | 36.835 N, 121.901 W | 125 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178214 | 511271 | PRJNA511271 | SRP174843 | SAMN10636660 | Gp0358240 | Illumina MiSeq | 2 × 301 | 102,382,140 | n/a | |
| 13_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/1/16 | 36.835 N, 121.901 W | 125 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178302 | 511227 | PRJNA511227 | SRP174671 | SAMN10636634 | Gp0358334 | Illumina MiSeq | 2 × 301 | 51,512,538 | n/a | |
| 14_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/28/16 | 36.835 N, 121.901 W | 250 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178215 | 511272 | PRJNA511272 | SRP174793 | SAMN10636590 | Gp0358241 | Illumina MiSeq | 2 × 301 | 105,082,110 | n/a | |
| 14_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/28/16 | 36.835 N, 121.901 W | 250 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178303 | 511228 | PRJNA511228 | SRP174676 | SAMN10636701 | Gp0358335 | Illumina MiSeq | 2 × 301 | 87,379,698 | n/a | |
| 15_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/13/16 | 36.835 N, 121.901 W | 100 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178216 | 511273 | PRJNA511273 | SRP174791 | SAMN10636605 | Gp0358243 | Illumina MiSeq | 2 × 301 | 101,095,666 | n/a | |
| 15_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/13/16 | 36.835 N, 121.901 W | 100 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178304 | 511229 | PRJNA511229 | SRP174674 | SAMN10636700 | Gp0358336 | Illumina MiSeq | 2 × 301 | 76,358,282 | n/a | |
| 16_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/17/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178217 | 511274 | PRJNA511274 | SRP174790 | SAMN10636960 | Gp0358244 | Illumina MiSeq | 2 × 301 | 125,671,714 | n/a | |
| 16_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/17/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178305 | 511230 | PRJNA511230 | SRP174675 | SAMN10636714 | Gp0358337 | Illumina MiSeq | 2 × 301 | 81,619,762 | n/a | |
| 17_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/12/16 | 36.835 N, 121.901 W | 400 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178218 | 511275 | PRJNA511275 | SRP174792 | SAMN10636958 | Gp0358245 | Illumina MiSeq | 2 × 301 | 98,466,732 | n/a | |
| 17_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/12/16 | 36.835 N, 121.901 W | 400 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178306 | 511231 | PRJNA511231 | SRP174677 | SAMN10636699 | Gp0358338 | Illumina MiSeq | 2 × 301 | 87,798,088 | n/a | |
| 18_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/20/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178219 | 511276 | PRJNA511276 | SRP174795 | SAMN10636606 | Gp0358246 | Illumina MiSeq | 2 × 301 | 119,638,470 | n/a | |
| 18_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/20/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178307 | 511232 | PRJNA511232 | SRP174701 | SAMN10636698 | Gp0358339 | Illumina MiSeq | 2 × 301 | 81,561,970 | n/a | |
| 1r_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/26/16 | 36.835 N, 121.901 W | 95.1 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178286 | 511211 | PRJNA511211 | SRP174819 | SAMN10636639 | Gp0358316 | Illumina MiSeq | 2 × 301 | 92,464,190 | n/a | |
| 1r_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/26/16 | 36.835 N, 121.901 W | 95.1 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178374 | 511158 | PRJNA511158 | SRP174753 | SAMN10636615 | Gp0358410 | Illumina MiSeq | 2 × 301 | 23,979,466 | n/a | |
| 2_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/16/16 | 36.835 N, 121.901 W | 1000 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178204 | 511261 | PRJNA511261 | SRP174785 | SAMN10636593 | Gp0358230 | Illumina MiSeq | 2 × 301 | 100,366,644 | n/a | |
| 2_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/16/16 | 36.835 N, 121.901 W | 1000 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178292 | 511217 | PRJNA511217 | SRP174686 | SAMN10636930 | Gp0358322 | Illumina MiSeq | 2 × 301 | 70,867,440 | n/a | |
| 20_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/27/16 | 36.835 N, 121.901 W | 73.5 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178220 | 511277 | PRJNA511277 | SRP174794 | SAMN10636665 | Gp0358247 | Illumina MiSeq | 2 × 301 | 76,705,636 | n/a | |
| 20_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/27/16 | 36.835 N, 121.901 W | 73.5 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178308 | 511233 | PRJNA511233 | SRP174733 | SAMN10636697 | Gp0358340 | Illumina MiSeq | 2 × 301 | 2,140,712 | n/a | |
| 21_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/24/16 | 36.835 N, 121.901 W | 250 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178221 | 511278 | PRJNA511278 | SRP174797 | SAMN10636659 | Gp0358248 | Illumina MiSeq | 2 × 301 | 132,824,076 | n/a | |
| 21_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/24/16 | 36.835 N, 121.901 W | 250 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178309 | 511234 | PRJNA511234 | SRP174732 | SAMN10636715 | Gp0358341 | Illumina MiSeq | 2 × 301 | 102,678,324 | n/a | |
| 22_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/12/16 | 36.835 N, 121.901 W | 100 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178222 | 511279 | PRJNA511279 | SRP174821 | SAMN10636709 | Gp0358249 | Illumina MiSeq | 2 × 301 | 87,594,010 | n/a | |
| 22_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/12/16 | 36.835 N, 121.901 W | 100 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178310 | 511235 | PRJNA511235 | SRP174738 | SAMN10636609 | Gp0358342 | Illumina MiSeq | 2 × 301 | 75,881,498 | n/a | |
| 23_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/6/16 | 36.835 N, 121.901 W | 150 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178223 | 511280 | PRJNA511280 | SRP174820 | SAMN10636658 | Gp0358250 | Illumina MiSeq | 2 × 301 | 119,028,644 | n/a | |
| 23_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/6/16 | 36.835 N, 121.901 W | 150 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178311 | 511236 | PRJNA511236 | SRP174740 | SAMN10636633 | Gp0358343 | Illumina MiSeq | 2 × 301 | 72,300,200 | n/a | |
| 24_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/2/16 | 36.835 N, 121.901 W | 425 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178224 | 511281 | PRJNA511281 | SRP174823 | SAMN10636666 | Gp0358251 | Illumina MiSeq | 2 × 301 | 106,448,650 | n/a | |
| 24_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/2/16 | 36.835 N, 121.901 W | 425 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178312 | 511237 | PRJNA511237 | SRP174735 | SAMN10636922 | Gp0358345 | Illumina MiSeq | 2 × 301 | 61,332,362 | n/a | |
| 25_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/6/16 | 36.835 N, 121.901 W | 575 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178225 | 511282 | PRJNA511282 | SRP174824 | SAMN10636657 | Gp0358252 | Illumina MiSeq | 2 × 301 | 117,012,546 | n/a | |
| 25_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/6/16 | 36.835 N, 121.901 W | 575 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178313 | 511238 | PRJNA511238 | SRP174700 | SAMN10636920 | Gp0358346 | Illumina MiSeq | 2 × 301 | 103,392,898 | n/a | |
| 26_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/1/16 | 36.835 N, 121.901 W | 125 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178226 | 511283 | PRJNA511283 | SRP174822 | SAMN10636667 | Gp0358253 | Illumina MiSeq | 2 × 301 | 129,192,210 | n/a | |
| 26_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/1/16 | 36.835 N, 121.901 W | 125 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178314 | 511239 | PRJNA511239 | SRP174702 | SAMN10636610 | Gp0358347 | Illumina MiSeq | 2 × 301 | 80,336,900 | n/a | |
| 27_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/12/16 | 36.835 N, 121.901 W | 100 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178227 | 511284 | PRJNA511284 | SRP174826 | SAMN10636957 | Gp0358254 | Illumina MiSeq | 2 × 301 | 98,704,522 | n/a | |
| 27_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/12/16 | 36.835 N, 121.901 W | 100 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178315 | 511240 | PRJNA511240 | SRP174737 | SAMN10636696 | Gp0358348 | Illumina MiSeq | 2 × 301 | 101,889,704 | n/a | |
| 28_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/4/16 | 36.835 N, 121.901 W | 149.5 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178228 | 511285 | PRJNA511285 | SRP174827 | SAMN10636595 | Gp0358255 | Illumina MiSeq | 2 × 301 | 116,352,152 | n/a | |
| 28_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/4/16 | 36.835 N, 121.901 W | 149.5 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178316 | 511241 | PRJNA511241 | SRP174736 | SAMN10636689 | Gp0358349 | Illumina MiSeq | 2 × 301 | 74,780,440 | n/a | |
| 29_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/10/16 | 36.835 N, 121.901 W | 100 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178229 | 511286 | PRJNA511286 | SRP174825 | SAMN10636956 | Gp0358256 | Illumina MiSeq | 2 × 301 | 100,751,322 | n/a | |
| 29_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/10/16 | 36.835 N, 121.901 W | 100 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178317 | 511163 | PRJNA511163 | SRP174699 | SAMN10636676 | Gp0358350 | Illumina MiSeq | 2 × 301 | 85,079,456 | n/a | |
| 3_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/9/16 | 36.835 N, 121.901 W | 175 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178205 | 511262 | PRJNA511262 | SRP174781 | SAMN10636603 | Gp0358231 | Illumina MiSeq | 2 × 301 | 57,795,612 | n/a | |
| 3_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/9/16 | 36.835 N, 121.901 W | 175 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178293 | 511218 | PRJNA511218 | SRP174680 | SAMN10637051 | Gp0358323 | Illumina MiSeq | 2 × 301 | 66,809,358 | n/a | |
| 30_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/5/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178230 | 511287 | PRJNA511287 | SRP174817 | SAMN10636954 | Gp0358257 | Illumina MiSeq | 2 × 301 | 108,967,418 | n/a | |
| 30_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/5/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178318 | 511164 | PRJNA511164 | SRP174720 | SAMN10636632 | Gp0358351 | Illumina MiSeq | 2 × 301 | 85,140,258 | n/a | |
| 31_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/26/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178231 | 511288 | PRJNA511288 | SRP174847 | SAMN10636596 | Gp0358258 | Illumina MiSeq | 2 × 301 | 98,156,100 | n/a | |
| 31_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/26/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178319 | 511165 | PRJNA511165 | SRP174722 | SAMN10636631 | Gp0358352 | Illumina MiSeq | 2 × 301 | 65,760,674 | n/a | |
| 32_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/19/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178232 | 511289 | PRJNA511289 | SRP174845 | SAMN10636656 | Gp0358259 | Illumina MiSeq | 2 × 301 | 108,968,622 | n/a | |
| 32_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/19/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178320 | 511166 | PRJNA511166 | SRP174714 | SAMN10636918 | Gp0358353 | Illumina MiSeq | 2 × 301 | 54,899,992 | n/a | |
| 33_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/28/16 | 36.835 N, 121.901 W | 75 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178233 | 511290 | PRJNA511290 | SRP174816 | SAMN10636655 | Gp0358260 | Illumina MiSeq | 2 × 301 | 130,152,400 | n/a | |
| 33_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/28/16 | 36.835 N, 121.901 W | 75 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178321 | 511167 | PRJNA511167 | SRP174684 | SAMN10636916 | Gp0358354 | Illumina MiSeq | 2 × 301 | 110,829,404 | n/a | |
| 34_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/30/16 | 36.835 N, 121.901 W | 94 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178234 | 511291 | PRJNA511291 | SRP174841 | SAMN10636668 | Gp0358261 | Illumina MiSeq | 2 × 301 | 150,864,812 | n/a | |
| 34_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/30/16 | 36.835 N, 121.901 W | 94 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178322 | 511168 | PRJNA511168 | SRP174713 | SAMN10637055 | Gp0358355 | Illumina MiSeq | 2 × 301 | 95,840,206 | n/a | |
| 35_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/20/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178235 | 511292 | PRJNA511292 | SRP174839 | SAMN10636654 | Gp0358262 | Illumina MiSeq | 2 × 301 | 111,955,746 | n/a | |
| 35_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/20/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178323 | 511169 | PRJNA511169 | SRP174715 | SAMN10636630 | Gp0358356 | Illumina MiSeq | 2 × 301 | 88,798,612 | n/a | |
| 36_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/14/16 | 36.835 N, 121.901 W | 825 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178236 | 511293 | PRJNA511293 | SRP174838 | SAMN10636669 | Gp0358264 | Illumina MiSeq | 2 × 301 | 102,925,746 | n/a | |
| 36_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/14/16 | 36.835 N, 121.901 W | 825 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178324 | 511170 | PRJNA511170 | SRP174712 | SAMN10636677 | Gp0358358 | Illumina MiSeq | 2 × 301 | 82,905,032 | n/a | |
| 36r_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/5/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178287 | 511212 | PRJNA511212 | SRP174787 | SAMN10636638 | Gp0358317 | Illumina MiSeq | 2 × 301 | 146,827,800 | n/a | |
| 36r_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/5/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178375 | 511159 | PRJNA511159 | SRP174747 | SAMN10636685 | Gp0358412 | Illumina MiSeq | 2 × 301 | 29,935,654 | n/a | |
| 37_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/25/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178237 | 511294 | PRJNA511294 | SRP174811 | SAMN10636708 | Gp0358265 | Illumina MiSeq | 2 × 301 | 116,812,682 | n/a | |
| 37_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/25/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178325 | 511171 | PRJNA511171 | SRP174717 | SAMN10636914 | Gp0358359 | Illumina MiSeq | 2 × 301 | 93,110,136 | n/a | |
| 38_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/2/16 | 36.835 N, 121.901 W | 175 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178238 | 511295 | PRJNA511295 | SRP174810 | SAMN10636953 | Gp0358266 | Illumina MiSeq | 2 × 301 | 115,647,812 | n/a | |
| 38_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/2/16 | 36.835 N, 121.901 W | 175 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178326 | 511172 | PRJNA511172 | SRP174730 | SAMN10636716 | Gp0358360 | Illumina MiSeq | 2 × 301 | 61,039,188 | n/a | |
| 39_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/28/16 | 36.835 N, 121.901 W | 75 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178239 | 511296 | PRJNA511296 | SRP174818 | SAMN10636597 | Gp0358267 | Illumina MiSeq | 2 × 301 | 140,738,570 | n/a | |
| 39_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/28/16 | 36.835 N, 121.901 W | 75 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178327 | 511173 | PRJNA511173 | SRP174719 | SAMN10636695 | Gp0358361 | Illumina MiSeq | 2 × 301 | 46,600,218 | n/a | |
| 4_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/3/16 | 36.835 N, 121.901 W | 325 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178206 | 511263 | PRJNA511263 | SRP174799 | SAMN10636592 | Gp0358232 | Illumina MiSeq | 2 × 301 | 84,373,912 | n/a | |
| 4_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/3/16 | 36.835 N, 121.901 W | 325 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178294 | 511219 | PRJNA511219 | SRP174687 | SAMN10636928 | Gp0358325 | Illumina MiSeq | 2 × 301 | 61,727,274 | n/a | |
| 40_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/4/16 | 36.835 N, 121.901 W | 400 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178240 | 511297 | PRJNA511297 | SRP174808 | SAMN10636710 | Gp0358268 | Illumina MiSeq | 2 × 301 | 120,755,782 | n/a | |
| 40_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/4/16 | 36.835 N, 121.901 W | 400 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178328 | 511174 | PRJNA511174 | SRP174718 | SAMN10636694 | Gp0358362 | Illumina MiSeq | 2 × 301 | 69,728,456 | n/a | |
| 41_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/13/16 | 36.835 N, 121.901 W | 350 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178241 | 511298 | PRJNA511298 | SRP174809 | SAMN10636653 | Gp0358269 | Illumina MiSeq | 2 × 301 | 153,685,784 | n/a | |
| 41_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/13/16 | 36.835 N, 121.901 W | 350 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178329 | 511175 | PRJNA511175 | SRP174716 | SAMN10636693 | Gp0358363 | Illumina MiSeq | 2 × 301 | 105,190,470 | n/a | |
| 42_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/27/16 | 36.835 N, 121.901 W | 75 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178242 | 511299 | PRJNA511299 | SRP174813 | SAMN10636652 | Gp0358270 | Illumina MiSeq | 2 × 301 | 120,916,516 | n/a | |
| 42_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/27/16 | 36.835 N, 121.901 W | 75 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178330 | 511176 | PRJNA511176 | SRP174731 | SAMN10636601 | Gp0358364 | Illumina MiSeq | 2 × 301 | 66,539,060 | n/a | |
| 43_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/8/16 | 36.835 N, 121.901 W | 225 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178243 | 511300 | PRJNA511300 | SRP174814 | SAMN10636951 | Gp0358271 | Illumina MiSeq | 2 × 301 | 161,805,560 | n/a | |
| 43_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/8/16 | 36.835 N, 121.901 W | 225 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178331 | 511177 | PRJNA511177 | SRP174724 | SAMN10636912 | Gp0358365 | Illumina MiSeq | 2 × 301 | 64,745,702 | n/a | |
| 44_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/30/16 | 36.835 N, 121.901 W | 94.1 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178244 | 511301 | PRJNA511301 | SRP174815 | SAMN10636949 | Gp0358272 | Illumina MiSeq | 2 × 301 | 107,242,688 | n/a | |
| 44_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/30/16 | 36.835 N, 121.901 W | 94.1 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178332 | 511178 | PRJNA511178 | SRP174721 | SAMN10636911 | Gp0358366 | Illumina MiSeq | 2 × 301 | 81,440,366 | n/a | |
| 45_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/3/16 | 36.835 N, 121.901 W | 300 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178245 | 511302 | PRJNA511302 | SRP174831 | SAMN10636598 | Gp0358273 | Illumina MiSeq | 2 × 301 | 90,206,690 | n/a | |
| 45_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/3/16 | 36.835 N, 121.901 W | 300 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178333 | 511179 | PRJNA511179 | SRP174726 | SAMN10637056 | Gp0358367 | Illumina MiSeq | 2 × 301 | 100,148,720 | n/a | |
| 46_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/21/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178246 | 511303 | PRJNA511303 | SRP174830 | SAMN10636651 | Gp0358274 | Illumina MiSeq | 2 × 301 | 60,635,848 | n/a | |
| 46_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/21/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178334 | 511180 | PRJNA511180 | SRP174723 | SAMN10636629 | Gp0358368 | Illumina MiSeq | 2 × 301 | 87,755,346 | n/a | |
| 47_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/25/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178247 | 511304 | PRJNA511304 | SRP174837 | SAMN10636707 | Gp0358275 | Illumina MiSeq | 2 × 301 | 115,867,542 | n/a | |
| 47_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/25/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178335 | 511181 | PRJNA511181 | SRP174725 | SAMN10636628 | Gp0358369 | Illumina MiSeq | 2 × 301 | 66,638,390 | n/a | |
| 48_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/31/16 | 36.835 N, 121.901 W | 300 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178248 | 511305 | PRJNA511305 | SRP174828 | SAMN10636711 | Gp0358276 | Illumina MiSeq | 2 × 301 | 65,209,242 | n/a | |
| 48_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/31/16 | 36.835 N, 121.901 W | 300 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178336 | 511182 | PRJNA511182 | SRP174728 | SAMN10636678 | Gp0358371 | Illumina MiSeq | 2 × 301 | 74,671,478 | n/a | |
| 48r_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/2/16 | 36.835 N, 121.901 W | 550 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178288 | 511213 | PRJNA511213 | SRP174782 | SAMN10636673 | Gp0358318 | Illumina MiSeq | 2 × 301 | 127,629,418 | n/a | |
| 48r_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/2/16 | 36.835 N, 121.901 W | 550 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178376 | 511160 | PRJNA511160 | SRP174748 | SAMN10636614 | Gp0358413 | Illumina MiSeq | 2 × 301 | 58,653,462 | n/a | |
| 49_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/3/16 | 36.835 N, 121.901 W | 325 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178249 | 511306 | PRJNA511306 | SRP174846 | SAMN10636706 | Gp0358277 | Illumina MiSeq | 2 × 301 | 95,586,764 | n/a | |
| 49_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/3/16 | 36.835 N, 121.901 W | 325 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178337 | 511183 | PRJNA511183 | SRP174729 | SAMN10636627 | Gp0358372 | Illumina MiSeq | 2 × 301 | 83,797,798 | n/a | |
| 5_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/19/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178207 | 511264 | PRJNA511264 | SRP174798 | SAMN10636664 | Gp0358233 | Illumina MiSeq | 2 × 301 | 44,540,776 | n/a | |
| 5_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/19/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178295 | 511220 | PRJNA511220 | SRP174681 | SAMN10636926 | Gp0358326 | Illumina MiSeq | 2 × 301 | 30,100,602 | n/a | |
| 50_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/18/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178250 | 511307 | PRJNA511307 | SRP174836 | SAMN10636705 | Gp0358278 | Illumina MiSeq | 2 × 301 | 84,935,578 | n/a | |
| 50_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/18/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178338 | 511184 | PRJNA511184 | SRP174727 | SAMN10636679 | Gp0358373 | Illumina MiSeq | 2 × 301 | 93,846,382 | n/a | |
| 51_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/13/16 | 36.835 N, 121.901 W | 400 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178251 | 511308 | PRJNA511308 | SRP174844 | SAMN10636650 | Gp0358279 | Illumina MiSeq | 2 × 301 | 108,360 | n/a | |
| 51_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/13/16 | 36.835 N, 121.901 W | 400 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178339 | 511185 | PRJNA511185 | SRP174710 | SAMN10636626 | Gp0358374 | Illumina MiSeq | 2 × 301 | 103,485,606 | n/a | |
| 52_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/17/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178252 | 511309 | PRJNA511309 | SRP174833 | SAMN10636670 | Gp0358280 | Illumina MiSeq | 2 × 301 | 98,717,766 | n/a | |
| 52_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/17/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178340 | 511186 | PRJNA511186 | SRP174706 | SAMN10636625 | Gp0358375 | Illumina MiSeq | 2 × 301 | 84,900,662 | n/a | |
| 53_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/29/16 | 36.835 N, 121.901 W | 125 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178253 | 511310 | PRJNA511310 | SRP174832 | SAMN10636947 | Gp0358281 | Illumina MiSeq | 2 × 301 | 110,774,622 | n/a | |
| 53_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/29/16 | 36.835 N, 121.901 W | 125 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178341 | 511187 | PRJNA511187 | SRP174711 | SAMN10636680 | Gp0358376 | Illumina MiSeq | 2 × 301 | 86,629,606 | n/a | |
| 54_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/11/16 | 36.835 N, 121.901 W | 425 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178254 | 511311 | PRJNA511311 | SRP174773 | SAMN10636712 | Gp0358282 | Illumina MiSeq | 2 × 301 | 62,542,984 | n/a | |
| 54_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/11/16 | 36.835 N, 121.901 W | 425 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178342 | 511188 | PRJNA511188 | SRP174708 | SAMN10636624 | Gp0358377 | Illumina MiSeq | 2 × 301 | 80,089,478 | n/a | |
| 55_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/8/16 | 36.835 N, 121.901 W | 175 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178255 | 511312 | PRJNA511312 | SRP174768 | SAMN10636704 | Gp0358284 | Illumina MiSeq | 2 × 301 | 62,783,182 | n/a | |
| 55_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/8/16 | 36.835 N, 121.901 W | 175 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178343 | 511189 | PRJNA511189 | SRP174709 | SAMN10636623 | Gp0358378 | Illumina MiSeq | 2 × 301 | 48,110,636 | n/a | |
| 56_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/9/16 | 36.835 N, 121.901 W | 174.4 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178256 | 511313 | PRJNA511313 | SRP174769 | SAMN10636945 | Gp0358285 | Illumina MiSeq | 2 × 301 | 68,732,146 | n/a | |
| 56_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/9/16 | 36.835 N, 121.901 W | 174.4 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178344 | 511190 | PRJNA511190 | SRP174707 | SAMN10636909 | Gp0358379 | Illumina MiSeq | 2 × 301 | 62,297,368 | n/a | |
| 57_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/6/16 | 36.835 N, 121.901 W | 550 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178257 | 511314 | PRJNA511314 | SRP174835 | SAMN10636607 | Gp0358286 | Illumina MiSeq | 2 × 301 | 123,612,874 | n/a | |
| 57_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/6/16 | 36.835 N, 121.901 W | 550 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178345 | 511191 | PRJNA511191 | SRP174704 | SAMN10636907 | Gp0358380 | Illumina MiSeq | 2 × 301 | 97,596,240 | n/a | |
| 58_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/4/16 | 36.835 N, 121.901 W | 400 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178258 | 511315 | PRJNA511315 | SRP174834 | SAMN10636943 | Gp0358287 | Illumina MiSeq | 2 × 301 | 130,885,636 | n/a | |
| 58_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/4/16 | 36.835 N, 121.901 W | 400 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178346 | 511192 | PRJNA511192 | SRP174705 | SAMN10636602 | Gp0358381 | Illumina MiSeq | 2 × 301 | 84,473,242 | n/a | |
| 58r_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/4/16 | 36.835 N, 121.901 W | 425 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178289 | 511214 | PRJNA511214 | SRP174783 | SAMN10636637 | Gp0358319 | Illumina MiSeq | 2 × 301 | 118,631,926 | n/a | |
| 58r_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/4/16 | 36.835 N, 121.901 W | 425 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178377 | 511161 | PRJNA511161 | SRP174758 | SAMN10636613 | Gp0358414 | Illumina MiSeq | 2 × 301 | 371,434 | n/a | |
| 59_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/21/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178259 | 511316 | PRJNA511316 | SRP174829 | SAMN10636649 | Gp0358288 | Illumina MiSeq | 2 × 301 | 70,527,912 | n/a | |
| 59_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/21/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178347 | 511193 | PRJNA511193 | SRP174703 | SAMN10636681 | Gp0358382 | Illumina MiSeq | 2 × 301 | 80,643,318 | n/a | |
| 60_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/12/16 | 36.835 N, 121.901 W | 475 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178260 | 511317 | PRJNA511317 | SRP174761 | SAMN10636648 | Gp0358289 | Illumina MiSeq | 2 × 301 | 94,567,578 | n/a | |
| 60_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/12/16 | 36.835 N, 121.901 W | 475 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178348 | 511194 | PRJNA511194 | SRP174695 | SAMN10636622 | Gp0358383 | Illumina MiSeq | 2 × 301 | 69,084,316 | n/a | |
| 61_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/31/16 | 36.835 N, 121.901 W | 290 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178261 | 511318 | PRJNA511318 | SRP174765 | SAMN10636941 | Gp0358290 | Illumina MiSeq | 2 × 301 | 68,405,862 | n/a | |
| 61_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/31/16 | 36.835 N, 121.901 W | 290 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178349 | 511195 | PRJNA511195 | SRP174689 | SAMN10636905 | Gp0358384 | Illumina MiSeq | 2 × 301 | 84,761,600 | n/a | |
| 62_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/28/16 | 36.835 N, 121.901 W | 250 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178262 | 511319 | PRJNA511319 | SRP174764 | SAMN10636608 | Gp0358291 | Illumina MiSeq | 2 × 301 | 53,843,482 | n/a | |
| 62_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/28/16 | 36.835 N, 121.901 W | 250 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178350 | 511196 | PRJNA511196 | SRP174688 | SAMN10636621 | Gp0358385 | Illumina MiSeq | 2 × 301 | 71,817,998 | n/a | |
| 63_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/24/16 | 36.835 N, 121.901 W | 250 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178263 | 511320 | PRJNA511320 | SRP174766 | SAMN10636684 | Gp0358292 | Illumina MiSeq | 2 × 301 | 246,218 | n/a | |
| 63_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/24/16 | 36.835 N, 121.901 W | 250 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178351 | 511197 | PRJNA511197 | SRP174749 | SAMN10636903 | Gp0358387 | Illumina MiSeq | 2 × 301 | 54,614,042 | n/a | |
| 64_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/5/16 | 36.835 N, 121.901 W | 199.2 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178264 | 511321 | PRJNA511321 | SRP174767 | SAMN10636647 | Gp0358293 | Illumina MiSeq | 2 × 301 | 88,243,568 | n/a | |
| 64_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/5/16 | 36.835 N, 121.901 W | 199.2 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178352 | 511198 | PRJNA511198 | SRP174750 | SAMN10637058 | Gp0358388 | Illumina MiSeq | 2 × 301 | 57,253,812 | n/a | |
| 65_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/2/16 | 36.835 N, 121.901 W | 400 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178265 | 511322 | PRJNA511322 | SRP174763 | SAMN10636686 | Gp0358294 | Illumina MiSeq | 2 × 301 | 122,270,414 | n/a | |
| 65_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/2/16 | 36.835 N, 121.901 W | 400 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178353 | 511199 | PRJNA511199 | SRP174751 | SAMN10636682 | Gp0358389 | Illumina MiSeq | 2 × 301 | 91,420,924 | n/a | |
| 66_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/3/16 | 36.835 N, 121.901 W | 250 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178266 | 511323 | PRJNA511323 | SRP174762 | SAMN10636646 | Gp0358295 | Illumina MiSeq | 2 × 301 | 123,413,010 | n/a | |
| 66_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/3/16 | 36.835 N, 121.901 W | 250 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178354 | 511200 | PRJNA511200 | SRP174752 | SAMN10636901 | Gp0358390 | Illumina MiSeq | 2 × 301 | 53,405,226 | n/a | |
| 67_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/15/16 | 36.835 N, 121.901 W | 1000 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178267 | 511324 | PRJNA511324 | SRP174772 | SAMN10636713 | Gp0358296 | Illumina MiSeq | 2 × 301 | 101,911,978 | n/a | |
| 67_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/15/16 | 36.835 N, 121.901 W | 1000 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178355 | 511201 | PRJNA511201 | SRP174698 | SAMN10637059 | Gp0358391 | Illumina MiSeq | 2 × 301 | 85,830,150 | n/a | |
| 68_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/7/16 | 36.835 N, 121.901 W | 575 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178268 | 511325 | PRJNA511325 | SRP174771 | SAMN10636703 | Gp0358297 | Illumina MiSeq | 2 × 301 | 118,754,734 | n/a | |
| 68_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/7/16 | 36.835 N, 121.901 W | 575 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178356 | 511202 | PRJNA511202 | SRP174692 | SAMN10636620 | Gp0358392 | Illumina MiSeq | 2 × 301 | 69,421,436 | n/a | |
| 7_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/26/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178208 | 511265 | PRJNA511265 | SRP174805 | SAMN10636663 | Gp0358234 | Illumina MiSeq | 2 × 301 | 101,529,708 | n/a | |
| 7_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/26/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178296 | 511221 | PRJNA511221 | SRP174682 | SAMN10636924 | Gp0358327 | Illumina MiSeq | 2 × 301 | 70,919,814 | n/a | |
| 70_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/7/16 | 36.835 N, 121.901 W | 207.5 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178269 | 511326 | PRJNA511326 | SRP174777 | SAMN10636645 | Gp0358298 | Illumina MiSeq | 2 × 301 | 118,522,964 | n/a | |
| 70_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/7/16 | 36.835 N, 121.901 W | 207.5 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178357 | 511203 | PRJNA511203 | SRP174690 | SAMN10636683 | Gp0358393 | Illumina MiSeq | 2 × 301 | 77,110,180 | n/a | |
| 71_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/18/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178270 | 511327 | PRJNA511327 | SRP174770 | SAMN10636687 | Gp0358299 | Illumina MiSeq | 2 × 301 | 75,922,434 | n/a | |
| 71_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/18/16 | 36.835 N, 121.901 W | 200 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178358 | 511204 | PRJNA511204 | SRP174693 | SAMN10636899 | Gp0358394 | Illumina MiSeq | 2 × 301 | 72,771,566 | n/a | |
| 73_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/16/16 | 36.835 N, 121.901 W | 1000 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178271 | 511328 | PRJNA511328 | SRP174776 | SAMN10636644 | Gp0358300 | Illumina MiSeq | 2 × 301 | 43,210,958 | n/a | |
| 73_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/16/16 | 36.835 N, 121.901 W | 1000 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178359 | 511205 | PRJNA511205 | SRP174691 | SAMN10636897 | Gp0358395 | Illumina MiSeq | 2 × 301 | 58,997,806 | n/a | |
| 74_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/11/16 | 36.835 N, 121.901 W | 425 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178272 | 511329 | PRJNA511329 | SRP174774 | SAMN10636688 | Gp0358301 | Illumina MiSeq | 2 × 301 | 96,386,822 | n/a | |
| 74_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/11/16 | 36.835 N, 121.901 W | 425 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178360 | 511206 | PRJNA511206 | SRP174696 | SAMN10636895 | Gp0358396 | Illumina MiSeq | 2 × 301 | 61,698,378 | n/a | |
| 75_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/14/16 | 36.835 N, 121.901 W | 825 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178273 | 511330 | PRJNA511330 | SRP174775 | SAMN10636643 | Gp0358302 | Illumina MiSeq | 2 × 301 | 112,452,998 | n/a | |
| 75_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/14/16 | 36.835 N, 121.901 W | 825 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178361 | 511242 | PRJNA511242 | SRP174694 | SAMN10636671 | Gp0358397 | Illumina MiSeq | 2 × 301 | 94,110,660 | n/a | |
| 76_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/10/16 | 36.835 N, 121.901 W | 350 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178274 | 511331 | PRJNA511331 | SRP174760 | SAMN10636939 | Gp0358304 | Illumina MiSeq | 2 × 301 | 105,368,662 | n/a | |
| 76_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/10/16 | 36.835 N, 121.901 W | 350 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178362 | 511243 | PRJNA511243 | SRP174697 | SAMN10636893 | Gp0358398 | Illumina MiSeq | 2 × 301 | 81,985,176 | n/a | |
| 77_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/7/16 | 36.835 N, 121.901 W | 201.5 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178275 | 511253 | PRJNA511253 | SRP174842 | SAMN10636599 | Gp0358305 | Illumina MiSeq | 2 × 301 | 59,957,394 | n/a | |
| 77_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/7/16 | 36.835 N, 121.901 W | 201.5 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178363 | 511244 | PRJNA511244 | SRP174743 | SAMN10636611 | Gp0358399 | Illumina MiSeq | 2 × 301 | 95,181,618 | n/a | |
| 78_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/26/16 | 36.835 N, 121.901 W | 92.5 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178276 | 511254 | PRJNA511254 | SRP174802 | SAMN10636642 | Gp0358306 | Illumina MiSeq | 2 × 301 | 54,360,600 | n/a | |
| 78_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 9/26/16 | 36.835 N, 121.901 W | 92.5 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178364 | 511245 | PRJNA511245 | SRP174744 | SAMN10636619 | Gp0358400 | Illumina MiSeq | 2 × 301 | 61,418,448 | n/a | |
| 79_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/7/16 | 36.835 N, 121.901 W | 550 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178277 | 511255 | PRJNA511255 | SRP174788 | SAMN10636937 | Gp0358307 | Illumina MiSeq | 2 × 301 | 131,771,780 | n/a | |
| 79_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/7/16 | 36.835 N, 121.901 W | 550 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178365 | 511246 | PRJNA511246 | SRP174746 | SAMN10636891 | Gp0358401 | Illumina MiSeq | 2 × 301 | 100,075,276 | n/a | |
| 8_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/5/16 | 36.835 N, 121.901 W | 775 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178209 | 511266 | PRJNA511266 | SRP174804 | SAMN10636662 | Gp0358235 | Illumina MiSeq | 2 × 301 | 123,461,772 | n/a | |
| 8_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/5/16 | 36.835 N, 121.901 W | 775 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178297 | 511222 | PRJNA511222 | SRP174678 | SAMN10637053 | Gp0358328 | Illumina MiSeq | 2 × 301 | 80,934,084 | n/a | |
| 80_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/11/16 | 36.835 N, 121.901 W | 100 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178278 | 511256 | PRJNA511256 | SRP174789 | SAMN10636935 | Gp0358308 | Illumina MiSeq | 2 × 301 | 95,350,780 | n/a | |
| 80_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/11/16 | 36.835 N, 121.901 W | 100 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178366 | 511247 | PRJNA511247 | SRP174745 | SAMN10636890 | Gp0358402 | Illumina MiSeq | 2 × 301 | 57,104,516 | n/a | |
| 81_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/11/16 | 36.835 N, 121.901 W | 100 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178279 | 511257 | PRJNA511257 | SRP174786 | SAMN10636600 | Gp0358309 | Illumina MiSeq | 2 × 301 | 79,994,362 | n/a | |
| 81_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/11/16 | 36.835 N, 121.901 W | 100 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178367 | 511248 | PRJNA511248 | SRP174739 | SAMN10636612 | Gp0358403 | Illumina MiSeq | 2 × 301 | 62,038,508 | n/a | |
| 82_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/10/16 | 36.835 N, 121.901 W | 100 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178280 | 511258 | PRJNA511258 | SRP174806 | SAMN10636933 | Gp0358310 | Illumina MiSeq | 2 × 301 | 107,646,028 | n/a | |
| 82_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/10/16 | 36.835 N, 121.901 W | 100 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178368 | 511249 | PRJNA511249 | SRP174741 | SAMN10636888 | Gp0358404 | Illumina MiSeq | 2 × 301 | 34,461,490 | n/a | |
| 84_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/11/16 | 36.835 N, 121.901 W | 100 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178281 | 511259 | PRJNA511259 | SRP174840 | SAMN10636641 | Gp0358311 | Illumina MiSeq | 2 × 301 | 87,418,226 | n/a | |
| 84_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/11/16 | 36.835 N, 121.901 W | 100 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178369 | 511250 | PRJNA511250 | SRP174742 | SAMN10636690 | Gp0358405 | Illumina MiSeq | 2 × 301 | 92,202,922 | n/a | |
| 85_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/13/16 | 36.835 N, 121.901 W | 100 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178282 | 511207 | PRJNA511207 | SRP174812 | SAMN10637049 | Gp0358312 | Illumina MiSeq | 2 × 301 | 394,310 | n/a | |
| 85_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 10/13/16 | 36.835 N, 121.901 W | 100 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178370 | 511251 | PRJNA511251 | SRP174757 | SAMN10636618 | Gp0358406 | Illumina MiSeq | 2 × 301 | 84,782,670 | n/a | |
| 89_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/5/16 | 36.835 N, 121.901 W | 575 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178283 | 511208 | PRJNA511208 | SRP174778 | SAMN10636932 | Gp0358313 | Illumina MiSeq | 2 × 301 | 17,458 | n/a | |
| 89_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/5/16 | 36.835 N, 121.901 W | 575 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178371 | 511252 | PRJNA511252 | SRP174755 | SAMN10636617 | Gp0358407 | Illumina MiSeq | 2 × 301 | 62,152,286 | n/a | |
| 9_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/15/16 | 36.835 N, 121.901 W | 975 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178210 | 511267 | PRJNA511267 | SRP174807 | SAMN10636661 | Gp0358236 | Illumina MiSeq | 2 × 301 | 122,328,808 | n/a | |
| 9_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/15/16 | 36.835 N, 121.901 W | 975 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178298 | 511223 | PRJNA511223 | SRP174683 | SAMN10636636 | Gp0358329 | Illumina MiSeq | 2 × 301 | 81,732,336 | n/a | |
| 90_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/8/16 | 36.835 N, 121.901 W | 175 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178284 | 511209 | PRJNA511209 | SRP174779 | SAMN10636640 | Gp0358314 | Illumina MiSeq | 2 × 301 | 94,883,026 | n/a | |
| 90_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/8/16 | 36.835 N, 121.901 W | 175 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178372 | 511156 | PRJNA511156 | SRP174756 | SAMN10637063 | Gp0358408 | Illumina MiSeq | 2 × 301 | 37,095,842 | n/a | |
| 91_DF | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/11/16 | 36.835 N, 121.901 W | 475 | Ribosomal iTag | 1190879 | 1190881 | Phytoplankton community itags pl1 | 178285 | 511210 | PRJNA511210 | SRP174780 | SAMN10636672 | Gp0358315 | Illumina MiSeq | 2 × 301 | 104,717,900 | n/a | |
| 91_S | ocean_biome | ocean | water | Monterey Bay, CA, USA | 11/11/16 | 36.835 N, 121.901 W | 475 | Ribosomal iTag | 1190880 | 1190883 | Bacterial community itags | 178373 | 511157 | PRJNA511157 | SRP174754 | SAMN10636616 | Gp0358409 | Illumina MiSeq | 2 × 301 | 76,699,014 | n/a |
Author Contributions
B.N. managed the project, conducted field sampling, prepared RNA and DNA samples, analyzed data, and wrote the manuscript. C.B.S. prepared RNA and DNA samples. C.M.T. conducted field sampling. K.E. conducted field sampling and chemical data analysis. R.M. managed field sampling and instruments and contributed to manuscript writing. C.M.P. managed field sampling and instruments and contributed to manuscript writing. C.A.S. supervised field sampling and contributed to manuscript writing. J.M.B. supervised field sampling and contributed to manuscript writing. M.H. participated in sequencing and analysis. A.C. participated in sequencing and analysis. B.F. participated in sequencing and analysis. B.F. participated in sequencing and analysis. S.R. participated in sequencing and analysis. K.P. participated in sequencing and analysis. N.V. participated in sequencing and analysis. S.M. participated in sequencing and analysis. T.B.K.R. participated in sequencing and analysis. C.D. participated in sequencing and analysis. A.C. participated in sequencing and analysis. N.N.I. participated in sequencing and analysis. N.C.K. supervised sequencing and analysis. T.G.R. coordinated sequencing and analysis. W.B.W. designed the project and contributed to manuscript writing. R.P.K. designed the project, conducted field sampling, supervised chemical data analysis, and contributed to manuscript writing. E.A.E.F. supervised sequencing and analysis. M.A.M. designed and supervised the project and wrote the manuscript with contributions from all authors.
Code Availability
Software versions and parameters used are as follows:
BFC v r181
MEGAHIT v 1.1.2: –k-list 23, 43, 63, 83, 103, 123
SPAdes v 3.11.1: -m 2000, -k 33, 55, 77, 99, 127 –meta
BBDuk v 38.08 for 16S, v 38.06 for 18S
BBMap v 37.78
iTagger v 2.2
For 16S iTags:
Cutadapt v 1.18: –interleaved -g GTGYCAGCMGCCGCGGTAA -G
GGACTACNVGGGTWTCTAAT -m 275 –discard-untrimmed
QIIME2 v 2018.6:
qiime dada2 denoise-paired\
–p-trunc-len-f 210\
–p-trunc-len-r 181
For 18S itags:
Cutadapt v 1.18: –interleaved -g CCAGCASCYGCGGTAATTCC -G
ACTTTCGTTCTTGATYRA -m 275 –discard-untrimmed
QIIME2 v2018.6:
qiime dada2 denoise-paired\
–p-trunc-len-f 259\
–p-trunc-len-r 200
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
ISA-Tab metadata
is available for this paper at 10.1038/s41597-019-0132-4.
References
- 1.Azam F, et al. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–263. doi: 10.3354/meps010257. [DOI] [Google Scholar]
- 2.Moran MA. The global ocean microbiome. Science. 2015;350:aac8455. doi: 10.1126/science.aac8455. [DOI] [PubMed] [Google Scholar]
- 3.Williams P. Microbial contribution to overall marine plankton metabolism-direct measurements of respiration. Ocean Acta. 1981;4:359–364. [Google Scholar]
- 4.Pennington JT, Chavez FP. Seasonal fluctuations of temperature, salinity, nitrate, chlorophyll and primary production at station H3/M1 over 1989–1996 in Monterey Bay, California. Deep Sea Res Part II: Top Stud Oceanogr. 2000;47:947–973. doi: 10.1016/S0967-0645(99)00132-0. [DOI] [Google Scholar]
- 5.Ryan JP, et al. Influences of upwelling and downwelling winds on red tide bloom dynamics in Monterey Bay, California. Cont Shelf Res. 2009;29:785–795. doi: 10.1016/j.csr.2008.11.006. [DOI] [Google Scholar]
- 6.Schulien JA, Peacock MB, Hayashi K, Raimondi P, Kudela RM. Phytoplankton and microbial abundance and bloom dynamics in the upwelling shadow of Monterey Bay, California, from 2006 to 2013. Mar Ecol Prog Ser. 2017;572:43–56. doi: 10.3354/meps12142. [DOI] [Google Scholar]
- 7.Wells BK, et al. State of the California Current 2016–17: Still anything but normal in the north. CalCOFI. Rep. 2017;58:1–55. [Google Scholar]
- 8.Scholin Christopher, Birch James, Jensen Scott, Marin III Roman, Massion Eugene, Pargett Douglas, Preston Christina, Roman Brent, Ussler III William. The Quest to Develop Ecogenomic Sensors: A 25-Year History of the Environmental Sample Processor (ESP) as a Case Study. Oceanography. 2017;30(4):100–113. doi: 10.5670/oceanog.2017.427. [DOI] [Google Scholar]
- 9.Moran MA. 2019. Environmental data from CTD during the Fall 2016 ESP deployment in Monterey Bay, CA. Biological and Chemical Oceanography Data Management Office (BCO-DMO) [DOI]
- 10.Moran MA, Kiene RP. 2019. Environmental data from Niskin bottle sampling during the Fall 2016 ESP deployment in Monterey Bay, CA. Biological and Chemical Oceanography Data Management Office (BCO-DMO) [DOI]
- 11.Nowinski B, et al. Microdiversity and temporal dynamics of marine bacterial dimethylsulfoniopropionate genes. Environ Microbiol. 2019;12:1687–1701. doi: 10.1111/1462-2920.14560. [DOI] [PubMed] [Google Scholar]
- 12.Li H. BFC: correcting Illumina sequencing errors. Bioinformatics. 2015;31:2885–2887. doi: 10.1093/bioinformatics/btv290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comp Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bushnell, B. BBMap: a fast, accurate, splice-aware aligner. Lawrence Berkeley National Laboratory, DOE Joint Genome Institute, https://jgi.doe.gov/news-publications/scientific-posters/bb_user-meeting-2014-poster-final/ (2014).
- 15.Chen I-MA, et al. IMG/M v. 5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2018;47:D666–D677. doi: 10.1093/nar/gky901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huntemann M, et al. The standard operating procedure of the DOE-JGI Metagenome Annotation Pipeline (MAP v. 4) Stand Genomic Sci. 2016;11:17. doi: 10.1186/s40793-016-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li D, et al. MEGAHITv1. 0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 18.Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 19.Apprill A, McNally S, Parsons R, Weber L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol. 2015;75:129–137. doi: 10.3354/ame01753. [DOI] [Google Scholar]
- 20.Stoeck T, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol. 2010;19:21–31. doi: 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- 21.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 22.Bolyen, E. et al. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. Preprint at, 10.7287/peerj.preprints.27295v2 (2018).
- 23.Callahan BJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nature Meth. 2016;13:581. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.2018. BioProject. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA533622
- 26.Gifford SM, Sharma S, Booth M, Moran MA. Expression patterns reveal niche diversification in a marine microbial assemblage. ISME J. 2013;7:281. doi: 10.1038/ismej.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Satinsky BM, et al. Microspatial gene expression patterns in the Amazon River Plume. Proc Nat Acad Sci USA. 2014;111:11085–11090. doi: 10.1073/pnas.1402782111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moran MA, et al. Sizing up metatranscriptomics. ISME J. 2013;7:237. doi: 10.1038/ismej.2012.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satinsky, B. M., Gifford, S. M., Crump, B. C. & Moran, M. A. In Meth Enzymol Vol. 531 (ed. DeLong, E. F.) 237–250 (Elsevier, 2013). [DOI] [PubMed]
- 30.Lin Y, Gifford S, Ducklow H, Schofield O, Cassar N. Towards quantitative microbiome community profiling using internal standards. Appl Environ Microbiol. 2019;85:e02634–02618. doi: 10.1128/AEM.02634-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Moran MA. 2019. Environmental data from CTD during the Fall 2016 ESP deployment in Monterey Bay, CA. Biological and Chemical Oceanography Data Management Office (BCO-DMO) [DOI]
- Moran MA, Kiene RP. 2019. Environmental data from Niskin bottle sampling during the Fall 2016 ESP deployment in Monterey Bay, CA. Biological and Chemical Oceanography Data Management Office (BCO-DMO) [DOI]
- 2018. BioProject. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA533622
Supplementary Materials
Data Availability Statement
Software versions and parameters used are as follows:
BFC v r181
MEGAHIT v 1.1.2: –k-list 23, 43, 63, 83, 103, 123
SPAdes v 3.11.1: -m 2000, -k 33, 55, 77, 99, 127 –meta
BBDuk v 38.08 for 16S, v 38.06 for 18S
BBMap v 37.78
iTagger v 2.2
For 16S iTags:
Cutadapt v 1.18: –interleaved -g GTGYCAGCMGCCGCGGTAA -G
GGACTACNVGGGTWTCTAAT -m 275 –discard-untrimmed
QIIME2 v 2018.6:
qiime dada2 denoise-paired\
–p-trunc-len-f 210\
–p-trunc-len-r 181
For 18S itags:
Cutadapt v 1.18: –interleaved -g CCAGCASCYGCGGTAATTCC -G
ACTTTCGTTCTTGATYRA -m 275 –discard-untrimmed
QIIME2 v2018.6:
qiime dada2 denoise-paired\
–p-trunc-len-f 259\
–p-trunc-len-r 200