Abstract
Rice is staple food of nearly half the world’s population. Rice yields must therefore increase to feed ever larger populations. By colonising rice and other plants, Herbaspirillum spp. stimulate plant growth and productivity. However the molecular factors involved are largely unknown. To further explore this interaction, the transcription profiles of Nipponbare rice roots inoculated with Herbaspirillum seropedicae were determined by RNA-seq. Mapping the 104 million reads against the Oryza sativa cv. Nipponbare genome produced 65 million unique mapped reads that represented 13,840 transcripts each with at least two-times coverage. About 7.4% (1,014) genes were differentially regulated and of these 255 changed expression levels more than two times. Several of the repressed genes encoded proteins related to plant defence (e.g. a putative probenazole inducible protein), plant disease resistance as well as enzymes involved in flavonoid and isoprenoid synthesis. Genes related to the synthesis and efflux of phytosiderophores (PS) and transport of PS-iron complexes were induced by the bacteria. These data suggest that the bacterium represses the rice defence system while concomitantly activating iron uptake. Transcripts of H. seropedicae were also detected amongst which transcripts of genes involved in nitrogen fixation, cell motility and cell wall synthesis were the most expressed.
Subject terms: Gene expression analysis, Transcriptomics
Introduction
To answer the ever increasing demand for cereals, genetic improvement of rice and the concomitant development of bio-fertilisers are promising, low environmental-impact solutions. After water, the most limiting nutrient in plant development is nitrogen and nitrogenous fertilisers have been heavily used in rice cultivation1. Heavy use of nitrogen fertilisers causes environmental damage including contamination of ground-water and the release of nitrogen oxides.
An alternative to the use of nitrogenous fertilisers is to employ plant-associated micro-organisms that fix nitrogen. Herbaspirillum seropedicae is an endophytic diazotroph that can colonise many plants and improve their productivity (reviewed by Monteiro et al. and Chubatsu et al.2,3). Inoculation of rice with H. seropedicae increased root and shoot biomass by 38 to 54% and 22 to 50% respectively4 part of which was attributable to biological nitrogen fixation4–7. Pankievicz et al.8 showed that the nitrogen fixed by H. seropedicae and Azospirillum brasilense was rapidly incorporated into Setaria viridis. Other factors, including production of phyto-hormones by the bacteria stimulate plant growth and several authors have observed that the increase in biomass of inoculated plants is dependent on the plant genotype4,9.
Transcriptome based studies are powerful tools to detect differentially expressed genes (DEG) and discover novel molecular processes10–16. Expression analyses (using EST sequencing and RT-qPCR) of rice roots inoculated with H. seropedicae suggested that genes related to auxin and ethylene syntheses as well as defence are modulated by the microorganism in a cultivar dependent manner17. Therefore, the purpose of the present study was to determine the effect of H. seropedicae on the gene expression of rice roots by RNA-seq transcriptome analyses. The data showed a genome wide repression of plant defence genes and activation of iron uptake systems, which seems an important feature for successful root colonization.
Results and Discussion
Transcriptional analyses
H. seropedicae enters rice roots via cracks at the points of lateral root emergence and later (three to 15 days) colonises the intercellular spaces, aerenchyma, cortical cells and vascular tissue4,6,7,18. 14 days after inoculation we observed an increase of weight of roots and leaf but this increase was not statistically significant. The number of endophytic H. seropedicae reached approximately 105 to 106 CFU per gram of fresh root weight one to two days after inoculation (DAI), with a peak at three DAI (Supplementary Fig. 1). For this reason roots were collected for RNA-seq analyses three DAI when the population of H. seropedicae had stabilised in the intercellular spaces and xylem7. Rice plants inoculated in parallel with the samples used for RNA extraction contained 4.2 × 105 endophytic CFU.g−1 of fresh roots and 4.4 × 108 epiphytic CFU.g−1 of root at three DAI.
Sixty-four percent of the reads (103,563,118) were finally used for mapping to the reference rice and H. seropedicae genomes. Illustration of RNA-seq analyses is shown in Supplementary Fig. 2 and the numbers of reads mapped to each reference genome are listed in Supplementary Table 1. Mapping on the rice genome database (http://rice.plantbiology.msu.edu/) produced 22 million unique mapped reads representing 13,837 expressed transcripts.
Differentially expressed genes
Statistical analyses were performed using DESeq software19 and comparison of non-inoculated with inoculated samples revealed 1,014 differentially expressed transcripts (P < 0.05) (Supplementary Table 2), with 255 having fold-changes higher than two. A heat map constructed using the expression profiles of these transcripts showed the inoculated sample clustering clearly separated from the control samples (Supplementary Fig. 3). In addition, functional categorisation of these set of genes was performed using MapMan (http://mapman.gabipd.org/) (Fig. 1B) and Blast2GO20 followed by GO enrichment with Gene Set Enrichment Analysis (GSEA)21. In the Biological Process category the following GOs terms were enriched: homeostatic process, drug metabolic process, carboxylic acid metabolic process, transmembrane transport, ion transport, response to chemical, regulation of transcription, DNA-templated, oxidation-reduction process and small molecule biosynthetic process (Supplementary Table 3).
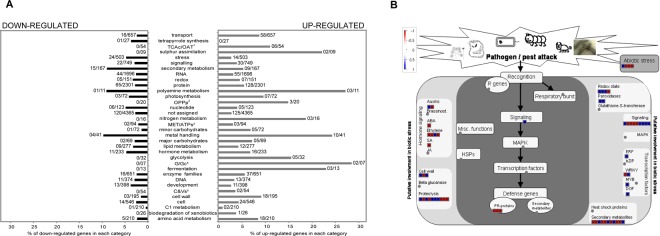
Figure 1.
Transcripts differentially expressed in rice roots colonised by H. seropedicae were grouped according to metabolic categories according to MapMan. Overview metabolism is shown in Panel A and biotic and abiotic stress in Panel B. (A) Up-regulated genes are shown in the right-hand column (in gray) and down-regulated genes in the left column (in black). Numbers of regulated genes and total numbers of expressed genes are shown for each category. 1TCA cycle/organic acids: 2oxidative pentose phosphate pathway; 3mitochondrial electron transport/ATP synthesis; 4gluconeogenesis/glyoxylate cycle; 5cofactor and vitamin synthesis. (B) The scheme was constructed using only genes with fold change ≥2; ≤2. Small squares represent up-regulated (blue) or down-regulated (red) genes.
MapMan analyses also revealed several rice roots pathways regulated by H. seropedicae colonisation. Considering the number of regulated genes in relation to the number of expressed genes in each category, the main MapMan categories down-regulated by H. seropedicae were: metal handling (9.0%; 4/41), polyamine metabolism (9.1%; 1/11), secondary metabolism (9%; 15/167), hormone metabolism (4.7%; 11/233); stress (4.8%; 24/503) and nucleotide metabolism (4.9%; 6/123). Among genes up-regulated by bacteria were those involved in gluconeogenesis/glyoxylate cycles (28.6%; 02/07), polyamine metabolism (27.3%; 03/11), metal handling (24.4%; 10/41), fermentation (23.1%; 03/13) and nitrogen metabolism (18.8%; 03/16) (Fig. 1A). Since MapMan was specifically developed to cover plant-specific pathways and is widely used by plant researchers, some of genes of these regulated categories are analysed in more detail below.
Biotic and abiotic stresses
Among the 255 differentially expressed genes (>2fold), 59 were stress-related (30 repressed and 29 induced) in the following categories: secondary metabolites, hormone signalling, cell-wall, proteolysis, PR-proteins, signalling, transcription factors, redox-state, abiotic stress and peroxidases (Fig. 1B and Supplementary Table 4).
Secondary metabolism
Amongst the secondary metabolic pathways with higher numbers of regulated genes were those involved in phenylpropanoid and isoprenoid synthesis (Supplementary Table 4). Phenylpropanoids synthesised by deamination of L-phenylalanine are eventually converted to p-coumaric acid, a precursor of flavonoids and lignin22,23. Additionally, H. seropedicae modulated expression of four genes involved in flavonoid synthesis. Amongst them, the gene encoding chalcone isomerase (CHI), which catalyses the synthesis of naringenin from tetrahydroxy-chalcone24, was repressed 2.1-fold. Naringenin is a key intermediate in the synthesis of other compounds including flavonol [flavonol synthase (FS), repressed 8.3-fold by H. seropedicae], and anthocyanins [dihydroxiflavonol 4-reductase (DRF), repressed 2.5-fold]. In addition, isoflavone reductase (IFR) transcript, involved in isoflavone synthesis, was repressed 1.3-fold (P = 0.01). RT-qPCR of FS confirmed repression by H. seropedicae (Fig. 2) but to a much lower extent (1.8-fold). Previously, Balsanelli et al.25 showed that narigenin has antimicrobial activity against H. seropedicae. Naoumkina el al.24 reported that flavonone-3-β-hydroxylase was induced in rice following infection with Xanthomonas oryzae. In addition, nematode cysts stimulate isoflavone synthesis (including chalcone reductases, chalcone isomerase, isoflavon 2′-hydroxylase, isoflavones and isoflavone reductase synthase) in soybeans. Flavonols such as quercetin23 exhibit antimicrobial activity possibly by binding and inhibiting DNA gyrase26. The data thus indicate that down-regulation of flavonoid/isoflavone synthesis by H. seropedicae is part of the attenuation of the defence system in rice roots that is necessary to host an endophyte.
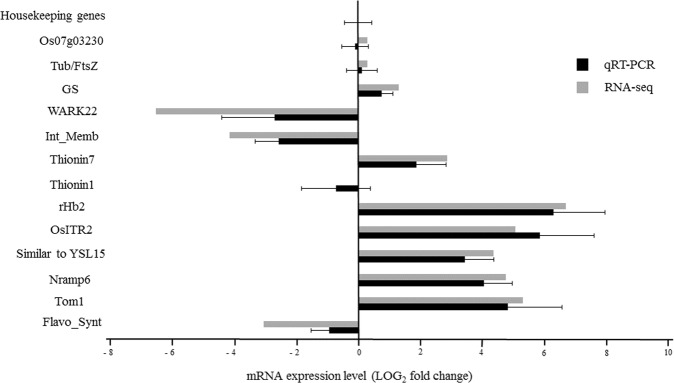
Figure 2.
Confirmation of differential expression of rice genes by qRT-PCR and RNA-Seq. The results are average of three independent samples and error bars represent, the standard deviation. The reference genes used for the analysis were actin 1, tubulin beta-2 chain and conserved hypothetical protein.
Isoprenoid (or terpenoid) synthesis was also modulated by H. seropedicae (Supplementary Table 4 and Fig. 3). Isoprenoids have diverse biological functions and are derived from isomeric compounds including isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP)27 via two pathways: the mevalonate pathway (MEV) and the methyl-erythritol phosphate pathway (MEP) (Fig. 3).
Figure 3.
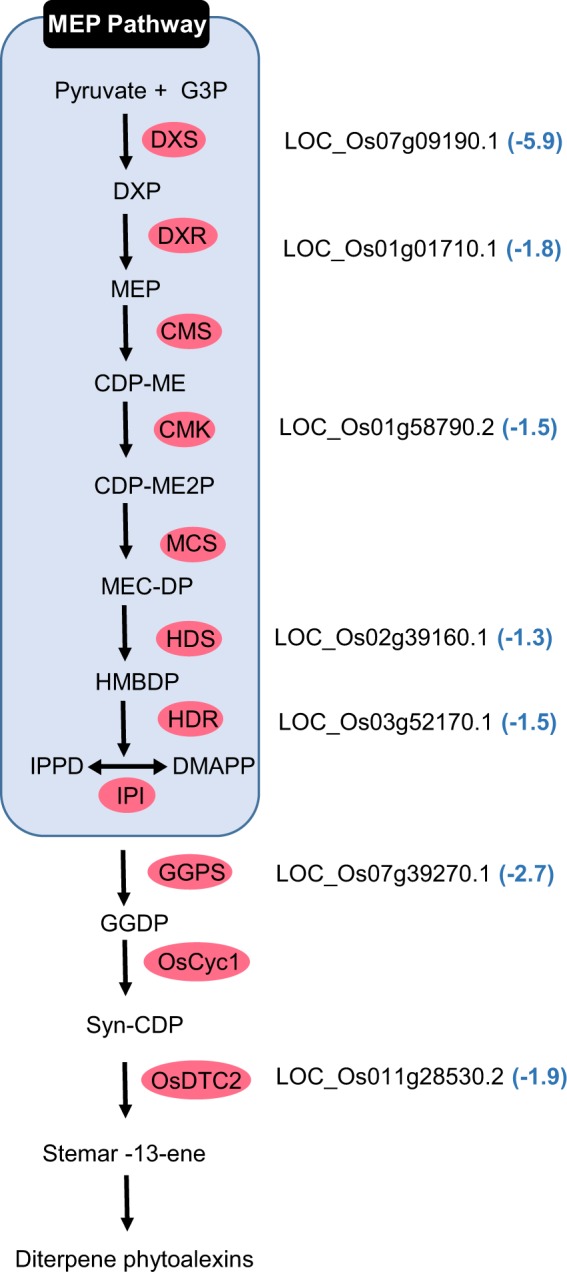
Isoprenoid synthesis genes down-regulated in rice roots colonised by H. seropedicae. The names of the genes differentially expressed are shown and the numbers in parentheses represent the fold change. The components of the MEP pathway leading to geranylgeranyl diphosphate synthesis and the diterpenoid- phytoalexin pathway are: G3P, glyceraldehyde-3-phosphate; DXP, 1-deoxy-D-xylulose 5-phosphate; MEP, 2-C-methyl-D-erythritol 4-phosphate; CDP-ME, 4-(cytidine 5-diphospho)-2-C-methyl-D-erythritol; CDP-ME2P, 2-phospho-4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol; MEC-DP, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate; HMBDP, 1-hydroxy-2-methyl-2-butenyl 4-diphosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GGDP, geranylgeranyl diphosphate; and CDP, copalyl diphosphate. Enzymes are indicated in rose coloured circles: DXS, 1-deoxy-D-xylulose 5-phosphate synthase; DXR, DXP reductoisomerase; CMS, CDP-ME synthase; CMK, CDP-ME kinase; MCS, MECDP synthase; HDS, HMBDP synthase; HDR, HMBDP reductase; IPI, IPP isomerase; GGPS, GGDP synthase; OsCyc1, syn-CDP synthase; OsCyc2, ent-CDP synthase; OsDTC2, stemar-13-ene synthase.
In rice, H. seropedicae repressed genes of the MEP pathway (Supplementary Table 4 and Fig. 3). The most repressed transcript (5.9 times) encodes 1-deoxy-D-xylulose 5-phosphate synthase (DXS) the first enzyme of the pathway that synthesises 1-deoxy-D-xylulose 5-phosphate (DXP) from pyruvate and D-glyceraldehyde 3-phosphate. DXP is a precursor of antimicrobial compounds including phytoalexins. Chitin induces the MEP pathway in cultured rice cells and as a result phytoalexins accumulate28. Furthermore, treatment of the cells with inhibitors of the enzymes DXS and 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) impeded chitin-dependent accumulation of phytoalexin. The authors suggested that activation of the MEP pathway is required to meet the demand for isoprenoid and phytoalexin synthesis in infected cells. In agreement with our results Drogue et al.29 showed the repression of a gene encoding stemar-13-ene synthase (OsDTC2), an enzyme involved in synthesis of phytoalexins in rice, which was repressed 1.9-fold in H. seropedicae treated rice roots (Fig. 3) and two-fold when infected with Azospirillum spp.29. It thus seems as if H. seropedicae represses production of defence-related isoprenoids in rice roots perhaps to allow the bacteria to enter and rapidly colonise the intercellular spaces and xylem.
Oxidative stress responses
In this study H. seropedicae modulated expression of five redox state genes including those for non-symbiotic hemoglobin 2 and peroxiredoxin. One peroxiredoxin transcript (LOC_Os07g44440.1) induced 3.7 and the other LOC_Os01g16152.1 was repressed 2.1 fold. Peroxiredoxins are a group of H2O2-decomposing antioxidant enzymes related to the redox state. In addition to the reduction of H2O2, peroxiredoxin proteins also detoxify alkyl hydroperoxides and peroxinitrite30,31.
Plants respond to attacks by pathogens with rapid increases in reactive oxygen species (ROS) such as superoxide and H2O232. Peroxidases produce ROS that could cause oxidative damage to proteins, DNA, and lipids. Many defects in the immune system of mature A. thaliana plants with reduced expression of two key peroxidase genes, PRX33 or PRX34, were observed33. Silencing the French-bean class III peroxidase (FBP1) in A. thaliana impaired the oxidative burst and rendered plants more susceptible to bacterial and fungal pathogens34. Proteomic studies of rice roots seven days after inoculation with H. seropedicae showed induction of ascorbate peroxidases35, though the genes encoding these enzymes were not affected in our RNA-seq analyses. Moreover, seven days after inoculation, ROS levels in Herbaspirillum rubrisubalbicans attached to rice roots had increased suggesting that the bacteria were subject to oxidative stresses36. In this work, two peroxidase genes were induced by H. seropedicae in inoculated roots.
Cell wall
A variety of diazotrophic microorganisms such as H. seropedicae Z67, H. rubrisubalbicans and A. brasilense produce cell-wall degrading enzymes6,18. Interestingly, the H. seropedicae SmR1 genome did not reveal genes coding for known cellulases, pectinases or any other cell-wall degrading enzymes37. Nevertheless, rice roots inoculated with H. seropedicae induced a gene (2.0-fold) encoding a β-D-xylosidase and repressed 2.4-fold a gene coding for a polygalacturonase suggesting re-modeling of plant cell wall. In A. thaliana, β-D-xylosidase has been shown to be involved in secondary cell-wall hemi-cellulose metabolism and plant development38, but little is known about the function of this enzyme.
Other differentially expressed genes involved in cell-wall metabolism code for proteins similar to an expansin11 (induced 2.6-fold). Expansins have a loosening effect on plant cell-walls and function in cell enlargement as well as in diverse developmental processes in which cell-wall modification occurs including elongation. In addition, they promote elongation of root-hairs39,40 and root-hair initiation41.
Expansins have been correlated with plant-bacteria interactions. In tobacco, Bacillus subtilis G1, a plant growth promoting bacterium, induced the expression of two expansins NtEXP2 and NtEXP642. Also, inoculation of Melilotus alba with Sinorhizobium meliloti lead to enhanced MaEXP1 mRNA levels in roots and nodules43. Together, these results suggest that inoculation with H. seropedicae also led to modification of plant cell wall which may facilitate bacterial colonization of inner tissue by loosening cell wall.
Plant immune responses
The plant immune system can sense and respond to pathogen attacks in two manners: the first involves recognition of pathogen/microbe associated molecular patterns (PAMPs) or damage associated molecular patterns (DAMPs) by surface pattern-recognition receptors (PRRs) resulting in pattern-triggered immunity (PTI). Second, resistance proteins (R) that recognise pathogen effectors are expressed leading to effector-triggered immunity (ETI)44–47. The receptors that perceive PAMPs or DAMPs have an ectodomain potentially involved in ligand binding, a single transmembrane domain and, most of times, a intracellular kinase domain47. Amongst the genes with the highest expression differences seen in RNA-seq data was a wall-associated receptor kinase-like (WAK) 22 precursor (LOC_Os10g07556.1), repressed 93-fold (6.5-fold by RT-qPCR) in H. seropedicae inoculated roots. Cell wall-associated receptor kinases (WAKs) contain an extracellular domain composed of one or more epidermal growth factor (EGF) repeats. Animal proteins containing these repeats are known to bind small peptides48. In A. thaliana WAKs bind to cross-linked pectin cell wall, pathogen- or damage-induced pectin fragments and oligogalacturonides, thus regulating cell expansion or stress response depending on the state of the pectin49–51. Several studies have described the role of WAK genes in rice resistance to pathogens52–55. Plant proteins domain architecture consist of cell-wall pectin binding extracellular region, the EGF-like domain, and a kinase domain. Analyses of rice loss-of-function mutants of WAK genes showed that individual genes are important for resistance against M. oryzae. OsWAK14, OsWAK91 and OsWAK92 positively regulate resistance while OsWAK112d is a negative regulator of blast resistance55. Cayrol et al. (2016) demonstrated that OsWAK14, OsWAK91 and OsWAK92 can form homo- and hetero-complexes and hypothesized that the loss of function of any of these proteins may destabilize the complex and affect their functioning. In A. thaliana the WAK22 orthologous gene (AtWAKL10) encodes a functional guanylyl cyclase which is co-expressed with pathogen defence related genes56. Thus, the wall-associated receptor kinase-like 22 gene of rice could be a candidate for surface pattern-recognition receptor and its repression may allow H. seropedicae to evade activation of the plant-defence system.
A cysteine-rich receptor-like protein kinase (CRK) (LOC_Os04g56430.1) and a serine/threonine-protein kinase At1g18390 precursor (LOC_Os05g47770.1) were induced 3.2-fold and 2-fold, respectively, while a lectin-like receptor kinase 7 (LOC_Os07g03790.1) and SHR5-receptor-like kinase (LOC_Os08g10320.1) were repressed 2.7 and 2.5-fold respectively in the presence of H. seropedicae (Supplementary Table 4). The latter protein has 75% identity with sugarcane SHR5 receptor kinase repressed by colonisation with diazotrophic endophytes57. The authors suggested that the expression levels of this gene were inversely related to the efficiency of beneficial plant-bacterial interactions57. Besides in Arabidopsis cysteine rich receptor-like kinase 5 protein is involved in regulation of growth, development, and acclimatory responses58.
Among the regulated genes that have functions involved in defence (in dark gray squares in Fig. 1B), as determined by MapMan, three genes encoding PR-proteins were repressed (LOC_Os10g25870.1, LOC_Os11g07680.1, LOC_Os02g38392.1, 2.5, 3.4 and 2.7-fold respectively) while one transcript that encoded a lipase called EDS1 (enhanced disease susceptibility 1) was induced 2-fold. The role of EDS1 in defence is well described in A. thaliana and is required for resistance conditioned by R genes which encode proteins that contain nucleotide-binding sites and leucine-rich repeats (NBS-LLR)45,59–62. Mutant eds1 seedlings exhibited enhanced susceptibility to the biotrophic oomycete Peronospora parasifica63. Analysis of EDS1 and PR-gene expression showed induction of both genes after inoculation with Pseudomonas syringae or treatment with salicylic acid (SA)64. In addition, in the A. thaliana eds1 mutant, the expression of PR-proteins was undetectable. When the eds1 mutant was treated with SA however, expression of PR was detected. The authors suggest that EDS1 functions upstream of PR1-mRNA accumulation. Among the 3 transcript for PR-proteins repressed, one (LOC_Os02g38392.1) is a NBS-LRR disease resistance protein. Therefore, in H. seropedicae- rice interaction an inverse correlation between PR-protein and EDS1 expression was observed, perhaps suggesting a fine regulation of these defence systems by H. seropedicae.
In previous work down-regulation of genes associated with defence was observed in rice plants inoculated with H. seropedicae such as a putative probenazole inducible protein (PBZ1, LOC_Os12g36840.1) that was repressed 3.6-fold as shown by RT-qPCR analysis17. The RNA-seq data showed that this transcript (LOC_Os12g36840.1) was repressed 2.9-fold. Another two transcript coding proteins similar to PBZ1 (LOC_Os12g36830.1 and LOC_Os12g36850.1) were also repressed (7.2 and 4.1, respectively). Kawahara et al.65 using an RNA-seq approach to study the transcriptome of rice inoculated with the blast fungus M. oryzae observed that the same PBZ1 genes detected in our study were induced 273 and 233-fold upon inoculation with the pathogen. The induction of PBZ1 gene by pathogens has been considered as a molecular marker for rice defence response66.
Another gene regulated by H. seropedicae that is related to defence codes for a thionin, a small cysteine-rich protein that occurs in a broad range of plant species67. Thionins are known for their toxicity to plant pathogens and several studies showed that their over-expression is related to increased resistance to diseases68–71. Brusamarello-Santos et al.17 showed 5-fold repression of thionin genes from chromosome 6 seven days after inoculation with H. seropedicae but these thionins from chromosome 6 were not regulated in roots three days after inoculation with H. seropedicae. Since there are 15 thionin genes (according to the Rice Genome Annotation Project RGAP7) sharing high identity in chromosome 6, the unmapped reads were mapped to the rice genome as well as to chromosome 6 separately. An average of 480 reads of control libraries and 136 of inoculated libraries mapped on thionin genes, a result that is in accordance with the repression pattern observed in rice roots seven days after inoculation with H. seropedicae17. Interestingly, a thionin transcript from chromosome 7 (LOC_Os07g24830.1) was induced 7.2-fold in the presence of H. seropedicae three DAI. Time-dependent regulation of thionin was also observed by Ji et al.71 in rice roots infected with Meloidogyne graminicola. Straub et al.72 observed only a few defence-related genes induced in the transcriptome of Miscanthus sinensis inoculated with Herbaspirillum frisingense helping to explain why this bacterium can effectively invade and colonise plants72.
The repression of genes related to defence is necessary perhaps to allow the bacteria to enter and rapidly colonise the intercellular space and xylem. However, it seems as if a balance between induction and repression of defence-related genes allows H. seropedicae to survive inside rice tissues while concomittantly activating some defence responses to protect the plant from pathogens.
Phytohormones
Auxins regulate diverse physiological processes such as vascular tissue differentiation, lateral root initiation and have also been linked to defence in plant-pathogen interactions73–76. Auxin response elements (AuxREs), when bound to auxin response factors (ARFs), control auxin-dependent gene expression. The Aux/IAA protein family members that inhibit ARFs mediate this regulation77.
Four differentially expressed transcript related to auxin signalling were identified in rice roots inoculated with H. seropedicae. The transcript LOC_Os08g24790.1 encoding an auxin-responsive protein was repressed 2.1-fold (Table 1) whereas the transcript coding for auxin-induced proteins, LOC_Os09g25770.1 and LOC_Os05g01570.1, were repressed 1.8 and 2.5-fold, respectively. Repression of genes related to auxin signalling was reported in rice roots inoculated with H. seropedicae17. Brusamarello-Santos et al.17 found that ARF2-like, IAA 11 and IAA18 were repressed 1.4, 1.5 and 2.8-fold, respectively, in the presence of H. seropedicae 7 days after inoculation. These genes were not regulated in our data probably due to the time difference of the cDNA library construction and expression levels too low.
Table 1.
Rice genes modulated by colonisation with H. seropedicae that are involved in phyto-hormone signalling and the MTA cycle.
| Locus name | Gene product/description* | Fold-change | Pvalue |
|---|---|---|---|
| LOC_Os01g67030.1 | Auxin-responsive protein, putative, expressed | 2.5 | 4.99E-06 |
| LOC_Os08g24790.1 | AIR12, putative, expressed | 0.49 | 0.04 |
| LOC_Os09g25770.1 | Auxin-induced protein 5NG4, putative, expressed | 0.55 | 0.01 |
| LOC_Os05g01570.1 | Auxin-induced protein 5NG4, putative, expressed | 0.40 | 0.05 |
| LOC_Os09g28050.1 |
1-aminocyclopropane-1-carboxylate synthase family protein (ACC synthase) (RAPDB) Asparate aminotransferase (MSU) |
6.1 | 2.11E-54 |
| LOC_Os03g63900.1 | 1-aminocyclopropane-1-carboxylate oxidase 2 (ACC oxidase) | 0.50 | 7.88E-04 |
| LOC_Os01g04800.1 |
B3 DNA binding domain containing protein (MSU) APETALA2/ethylene-responsive element binding protein 129 (RAPDB) |
2.2 | 3.67E-03 |
| LOC_Os06g02220.1 | MTA/SAH nucleosidase | 2.6 | 1.83E-11 |
| LOC_Os04g57400.1 | Methylthioribose kinase | 6.1 | 3.20E-18 |
| LOC_Os04g57410.1 | Methylthioribose kinase | 10.2 | 1.16E-12 |
| LOC_Os06g04510.1 | Similar to enolase 1 | 1.9 | 0.03 |
| LOC_Os03g06620.1 | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase | 7.5 | 5.94E-65 |
| LOC_Os10g28360.1 | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase | 0.47 | 6.14E-03 |
| LOC_Os01g22010.3 | S-adenosylmethionine synthetase (SAM) | 1.9 | 1.72E-05 |
*Gene product name/description of the both rice database were used: MSU (Rice Genome Annotation Project) and RAPDB (The Rice Annotation Project).
H. seropedicae can produce IAA in the presence of tryptophan, thus suggesting the plant growth promotion effect may be due to bacterial derived auxin78. In addition exogenous application of auxin lead to increase in lateral root numbers79. In the absence of clear regulation of auxin-dependent genes and phenotype, the results suggest that auxin does not play an important role in Nipponbare rice roots colonization by H. seropedicae. On the other hand the repression of auxin signalling has been correlated to defence responses. Auxin levels have been correlated with susceptibility to pathogens74 that are able to produce high levels of auxin80. It has also been shown that pathogen-associated molecular patterns (PAMPs) induce the expression of a miRNA that negatively regulates mRNAs for F-box auxin receptors leading to resistance to P. syringae in Arabidopsis81. The observed repression in several genes involved with defence in the rice-H. seropedicae interaction opens the question whether auxin could be important for bacteria survival inside the plant by down-regulating defence system. Further studies are needed to elucidate if and how auxin signalling participates in plant-bacterial interactions.
Ethylene is also involved in several biological processes that activate defence responses and adventitious root-growth in rice and other plants82–84. Ethylene can be synthesised by oxidation of 1-aminocyclopropane-1-carboxylate (ACC) by ACC oxidase. ACC is synthesised from adenosylmethionine (AdoMet) by ACC synthase (ACS). ACS is up regulated 6-fold and ACC oxidase is repressed 2-fold by inoculation with H. seropedicae. An ethylene response factor (ERF) (LOC_Os01g04800.1) transcript was also induced 2.2-fold in inoculated roots. These data indicate that ethylene synthesis is attenuated in the presence of bacteria. Moreover Alberton et al.35 measured the level of ethylene in inoculated rice roots (seven days after inoculation) and found a decrease of ethylene levels. Furthermore, Valdameri et al.36 detected induction of ACC oxidase in rice plants inoculated with the pathogen H. rubrisubalbicans. These results suggest that the ethylene pathway is differentially modulated in the presence of pathogens and beneficial endophytic bacteria that promote plant growth.
Salicylic acid is derived from phenolic compounds and is involved in response to attack by pathogens85,86. In rice roots inoculated with H. seropedicae, a SA-dependent carboxyl methyltransferase family protein gene (LOC_Os11g15340.2) was repressed 40-fold. A member of this family is salicylic acid carboxyl methyltransferase (SAMT) that catalyses the formation of methyl salicylate (MeSA) from SA87. MeSA is an essential signal for systemic acquired resistance (SAR) in tobacco plants. In addition mutations in SAMT showed that this gene is required for SAR88.
SA signalling is differentially regulated by members of the WRKY transcription factor family89. In inoculated rice roots, two WRKY transcription factors were regulated, one repressed (2.3-fold) and one induced (2.0-fold). The induced gene encodes a WRKY51 similar to WRKY11 of Arabidopsis, whereas the repressed gene encodes WRKY46, which was shown to be induced by SA in Arabidopsis. WRKY11 is a negative regulator of resistance90 and Arabidopsis plants in which WRKY46 was over-expressed were more resistant to P. syringae91. These results are in agreement with the hypothesis that the SA signalling and defence system are attenuated in the presence of the H. seropedicae.
Metal ion metabolism
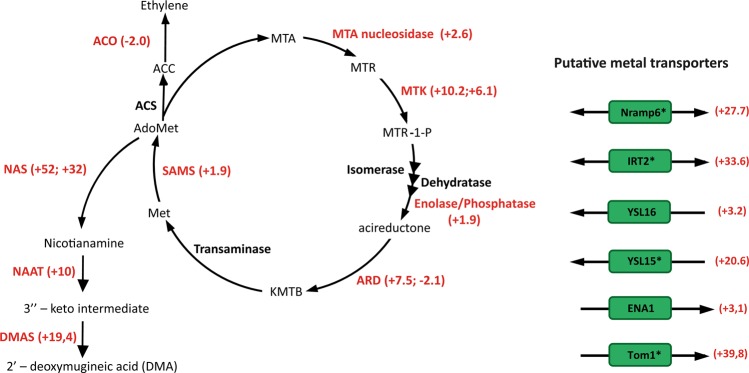
Several genes related to metal transport were differentially expressed, most of them up regulated in the presence of H. seropedicae. Amongst the 20 most highly regulated rice genes, eight were related to metal transport (Table 2 and 3). Phyto-siderophore synthesis starts with production of nicotianamine (NA) from S-adenosylmethionine, which in turn is derived from 5′-methylthioadenosine of the methionine salvage pathway (MTA cycle) (Fig. 4). Transcripts of enzymes of the MTA cycle encoding SAM, MTA nucleosidase, MTR kinase, E1 enolase/phosphatase and acireductone dioxygenase (ARD) were induced 1.9, 2.6, 6.0, 1.9 and 7.5-times, respectively, in roots colonised by H. seropedicae (Fig. 4). Using proteomic and RT-qPCR analyses, Alberton et al.35 observed similar expression patterns in rice roots inoculated with H. seropedicae.
Table 2.
Rice genes highly regulated (fold change > 10) by inoculation with H. seropedicae.
| Locus name | Gene product/description* | Fold change | P-value |
|---|---|---|---|
| Up-regulated rice genes | |||
| LOC_Os02g20360.1 | Ttyrosine aminotransferase, putative, expressed, Similar to Nicotianamine aminotransferase A (RAPDB) | 10 | 5.24E-30 |
| LOC_Os04g57410.1 | Methylthioribose kinase, putative. Expressed | 10.2 | 1.16E-12 |
| LOC_Os03g26210.1 | Helix-loop-helix DNA-binding domain containing protein. Expressed | 12.6 | 4.10E-09 |
| LOC_Os01g72370.1 | Helix-loop-helix DNA-binding domain containing protein. Expressed | 16.4 | 3.36E-74 |
| LOC_Os11g15624.1 | Expressed protein | 17.7 | 1.22E-25 |
| LOC_Os03g13390.2 | Oxidoreductase. aldo/keto reductase family protein, putative. Expressed | 19.4 | 1.61E-10 |
| LOC_Os02g43410.1 | Transposon protein, putative. Unclassified. Expressed | 21 | 3.97E-81 |
| LOC_Os10g11889.2 | Expressed protein | 23 | 3.29E-113 |
| LOC_Os01g65110.1 | POT family protein. Expressed | 26 | 6.63E-14 |
| LOC_Os07g15460.1 | Metal transporter Nramp6, putative. Expressed | 27 | 3.56E-61 |
| LOC_Os03g03724.1 | Expressed protein | 28 | 0.02 |
| LOC_Os03g19427.1 | Nicotianamine synthase, putative. expressed | 32 | 1.02E-22 |
| LOC_Os03g46454.1 | Metal cation transporter, putative. expressed | 34 | 4.02E-08 |
| LOC_Os06g19095.1 | Expressed protein | 40 | 1.51E-14 |
| LOC_Os11g04020.1 | Major facilitator superfamily antiporter, putative. expressed | 40 | 8.84E-51 |
| LOC_Os03g19420.2 | Nicotianamine synthase, putative. expressed | 52 | 5.57E-80 |
| LOC_Os12g18410.1 | Expressed protein | 70 | 2.07E-05 |
| LOC_Os03g12510.1 | Non-symbiotic hemoglobin 2, putative. expressed | 104 | 5.58E-24 |
| LOC_Os04g51660.1 | Transferase family protein, putative, expressed | 0.08 | 2.79E-03 |
| LOC_Os04g59020.1 | Integral membrane protein, putative, expressed | 0.08 | 3.41E-03 |
| LOC_Os09g23300.1 | Integral membrane protein, putative, expressed | 0.06 | 3.63E-05 |
| LOC_Os11g15340.2 |
SAM dependent carboxyl methyltransferase family protein, putative, expressed |
0.03 | 1.54E-03 |
| LOC_Os01g55690.1 | Glutelin, putative, expressed | 0.02 | 0.02 |
| LOC_Os12g38040.1 | Metallothionein family protein, expressed | 0.02 | 3.72E-22 |
| LOC_Os10g07556.1 | Wall-associated receptor kinase-like 22 precursor, putative, expressed | 0.01 | 5.22E-08 |
*Gene product name/description of the both rice database were used: MSU (Rice Genome Annotation Project) and RAPDB (The Rice Annotation Project).
Table 3.
Differentially expressed genes in rice roots colonised by H. seropedicae involved in uptake and transport of metals.
| Locus name | Gene product/description* | Fold change | p-value |
|---|---|---|---|
| LOC_Os01g22010.3 | S-adenosylmethionine synthetase, putative, expressed | 1.9 | 1.72E-05 |
| LOC_Os03g19427.1 | Nicotianamine synthase. putative. expressed | 32 | 1.02E-22 |
| LOC_Os03g19420.2 | Nicotianamine synthase. putative. expressed | 52 | 5.57E-80 |
| LOC_Os02g20360.1 | tyrosine aminotransferase, putative, expressed (MSU), Similar to Nicotianamine aminotransferase (RAPDB) | 10 | 5.24E-30 |
| LOC_Os03g13390.2 |
Oxidoreductase. aldo/keto reductase family protein, putative. Expressed (MSU) Similar to NADPH-dependent codeinone reductase, gene name: deoxymugineic acid synthase1 (RAPDB) |
19.4 | 1.61E-10 |
| LOC_Os11g04020.1 | Major facilitator superfamily antiporter, putative, expressed (TOM1) | 40 | 8.84E-51 |
| LOC_Os11g05390.1 | Transporter, major facilitator family, putative, expressed (ENA1) | 3.1 | 5.80E-03 |
| LOC_Os03g46470.1 | Metal cation transporter, putative, expressed (OsIRT1) | 4.3 | 4.09E-14 |
| LOC_Os03g46454.1 | Metal cation transporter, putative, expressed (OsIRT2) | 34 | 4,02E-08 |
| LOC_Os04g45900.1 |
Transposon protein, putative, unclassified, expressed (MSU) Similar to Metal-nicotianamine transporter YSL2, Gene symbol synonym: OsYSL16 (RAPDB) |
3.2 | 1.23E-07 |
| LOC_Os02g43410.1 |
Transposon protein, putative. Unclassified. Expressed (MSU) Iron-phytosiderophore transporter, Iron homeostasis (Os02t0650300-01); Similar to Iron-phytosiderophore transporter YSL15. (Os02t0650300-02) (RAP-DB) |
21 | 3.97E-81 |
| LOC_Os07g15460.1 | Metal transporter Nramp6, putative. Expressed | 27 | 3.56E-61 |
*Gene product name/description of the both rice database were used: MSU (Rice Genome Annotation Project) and RAPDB (The Rice Annotation Project).
Figure 4.
Differentially expressed genes in rice roots following colonisation by H. seropedicae. Genes involved in siderophore synthesis and transport, the methionine salvage pathway and ethylene synthesis are shown. Numbers in parentheses represent the fold change. H. seropedicae SmR1 induces methionine recycling and mugineic acid (MA) synthesis as well as the expression of transporters involved in iron metabolism. The expression of those genes marked with an asterisk was confirmed by RT-qPCR Abbreviations: AdoMet, S-adenosylmethionine; ACC, 1-aminocyclopropane-1-carboxylate; ACS, 1-aminocyclopropane-1-carboxylate synthase; ACO, 1-aminocyclopropane-1-carboxylate oxidase; MTA, 5′-methylthioadenosine; MTR, 5′-methylthioribose; MTK, methylthioribose kinase; MTR-1-P, 5′-methylthioribose-1-phosphate; KMTB, 2-keto-4-methylthiobutyrate; ARD, acireductone dioxygenase; SAMS, S-adenosylmethionine synthetase; NAS, nicotianamine synthase; NAAT nicotianamine aminotransferase; DMAS, deoxymugineic acid synthase; Tom1, transporter of mugineic acid 1; ENA1 (efflux transporters of nicotianamine 1); Nramp6, Natural Resistance-Associated Macrophage Protein; IRT2(iron-regulated transporter 2); YSL16 (yellow strip-like gene 16); YSL15 (yellow strip-like gene 15).
The synthesis of NA and MAs involves a set of enzymes including S-adenosylmethionine synthase (SAM) that catalyses the adenylation of L-methionine to S-adenosylmethionine, nicotianamine synthase (NAS) that converts S-adenosylmethionine to nicotianamine, and nicotianamine aminotransferase (NAAT) that catalyses the amino transfer of NA to produce the 3”-keto intermediate that is reduced by deoxymugineic acid synthase (DMAS) to produce 2′-deoxymugineic acid (DMA)92–95.
Two nicotianamine synthase (NAS) genes of rice - LOC_Os03g19420.2 and LOC_Os03g19427.1 were induced (52- and 32-fold, respectively) in inoculated rice roots. LOC_Os03g19427.1 was also induced in rice roots inoculated with Azospirillum spp.29. Increased levels of nicotianamine have been shown to increase Fe uptake in rice plants96. Furthermore, in Lotus japonicus inoculated with Mesorhizobium loti, nicotianamine synthase 2 was expressed only in nodules pointing to a role in symbiotic nitrogen fixation97. A tomato mutant defective in the synthesis of nicotianamine was affected in iron metabolism98. In addition, NAAT (LOC_Os02g20360.1) and DMAS (LOC_Os03g13390.2) were also induced 10-fold and 19-times respectively in colonised rice roots. Iron deficiency provoked induction of rice gene OsNAAT195.
Recently, members of a major facilitator super-family have been described as essential to the efflux of MA and NA in rice. TOM1 (transporter of mugineic acid 1) is involved in the efflux of MA, while ENA1 (efflux transporters of nicotianamine 1) and ENA2 in the efflux of NA99. TOM1 (LOC_Os11g04020.1) and ENA1 (LOC_Os11g05390.1) were induced 40 and 3-fold in colonised roots. Induction of TOM1 was confirmed by RT-qPCR (Fig. 2).
Fe+++ ions chelated by PS need to be transported inside the cell. In gramineous plants, two groups of Fe-MA transporters are present: ZmYS1100 and the YSL (yellow strip-like) transporter family101. Inoue et al.101 analysed the expression of 18 YSL genes in rice and observed induction of OsYSL15 and OSYSL16 genes under iron-deficiency. Other studies have shown that OsYSL2 (LOC_Os02g43370) takes up Fe2+-NA102 and OsYSL16 Fe3+-DMA (LOC_Os04g45900.1)103. Here we showed induction (3.2-fold) of OsYSL16 (LOC_Os04g45900.1) and 21-fold increase of transcripts encoding a gene similar to OSYSL15 (LOC_Os02g43410.1). Induction of OSYSL15 was confirmed by RT-qPCR (Fig. 2). Furthermore, OsIRT1 (iron-regulated transporter 1) and OsIRT2 were also induced under low Fe conditions104. We detected induction of OsIRT2 (LOC_Os03g46454.1) (34-fold in RNA-seq and 93-fold in RT-qPCR) (Fig. 2).
Other metal transporters such as Nramp6 (LOC_Os07g15460.1) were also up regulated (27-fold) in colonised rice roots, a result confirmed by RT-qPCR (Fig. 2). Members of the natural resistance-associated macrophage protein (NRAMP) family are transition metal cation/proton co-transporters or anti-porters of broad specificity. AtNRAMP6 of Arabidopsis is up-regulated in response to iron deficiency and is involved with metal mobilization from vacuoles to cytosol105. Induction of Nramp6 was observed in rice roots colonised by H. seropedicae. In a previous study in rice inoculated with Azospirillum spp the expression of this gene was also induced29. In addition a gene for an integral membrane protein (LOC_Os09g23300.1) named OsVIT2 was 17.7-fold repressed by H. seropedicae. This gene is involved in transport of Fe/Zn into vacuoles and is up-regulated in rice roots with excess Fe. Knockout/ knockdown of this gene led to Fe accumulation in seeds106,107. Together, these results suggest that colonised roots respond in such a manner as to accumulate Fe.
Interestingly the transcript with the highest fold change [104-times – a result confirmed using RT-qPCR (Fig. 2)] in colonised roots codes for the non-symbiotic hemoglobin 2 (LOC_Os03g12510.1). High levels of non-symbiotic hemoglobin 2 could help buffer free oxygen and protect bacterial nitrogenase. Arredondo-Peter et al.108 studied the repression of two hemoglobins, Hb1 and Hb2, in rice. The Hb2 induced by H. seropedicae is very similar to the one described by Arredondo-Peter et al. (1997) (coverage 82% with an identity of 97%). In addition, Lira-Ruan, Sarath and Arredondo-Peter109 studied the synthesis of hemoglobins in rice under normal and stress conditions, coming to the conclusion that Hbs are not part of a generalised stress response. They demonstrated that Hb1 is expressed in different rice organs (root and leaves) during plant development. In etiolated rice plants under 02 limiting conditions the Hb levels increase109. This increase suggest that Hb expression maybe due to reduced O2 levels in the presence of the bacteria which make the root environment microaerophilic. The higher requirement for Fe needed for incorporation into Hb may partially explain activation of siderophore synthesis and Fe accumulation. We also found a H. seropedicae bacterioferritin (Hsero_1195) gene induced (2.2-fold), but symptoms of Fe deficiency in colonised rice plants were not observed.
Iron homeostasis has been related to plant defence, ROS accumulation and immunity. Also, Fe deficiency triggers accumulation of antimicrobial phenolics compounds. It has been suggested recently that Fe sequestration by bacterial siderophore could be a signal for pathogen infection110. However, bacterial genes involved in siderophore biosynthesis were not observed among the H. seropedicae genes expressed in rice roots. Also, transcriptomic analysis of H. seropedicae attached to wheat and mayze roots did not show iron metabolism genes up-regulated111,112. These results suggest that the effect of bacteria on plant iron mebabolism is more complex than those caused by iron sequestration.
H. seropedicae transcripts detected in rice roots
The libraries from inoculated roots were mapped against the H. seropedicae genome (24,263 reads representing 0.06% of the total reads) (Supplementary Table 1). Amongst the 4,085 annotated genes of H. seropedicae, 287 were expressed in rice roots (at least one-fold coverage (this set of genes was called H. seropedicae expressed genes) (Supplementary Table 5). After ribosomal genes, the most abundant functional classes found were unknown, energy production and conversion, amino acid transport and metabolism, cell motility and cell wall (Table 4). Comparison of expressed genes of H. seropedicae detected in plants with bacterial genes expressed in culture revealed only 16 differences [p-value < 0.05 using the DESeq statistical package (Table 4)].
Table 4.
Genes of H. seropedicae regulated during interaction with rice roots.
| Locus_tag | Fold Change | p-value | ID Feature | Description | COG |
|---|---|---|---|---|---|
| HSERO_RS17470 | 0.4 | 7.0E-04 | qor |
qor NADPH:quinone oxidoreductase protein 4007887:4008915 forward |
C;R - Energy production and conversion;General function prediction only |
| HSERO_RS05670 | 30 | 6.20E-03 | Hsero_1130 |
Hsero_1130 ABC-type dipeptide transporter, periplasmic peptide-binding protein 1287409:1289031 forward |
E - Amino acid transport and metabolism |
| HSERO_RS23580 | 27 | 7.70E-03 | urtA |
UrtA ABC-type urea transport system, periplasmic component protein 5407999:5409252 forward |
E - Amino acid transport and metabolism |
| HSERO_RS00420 | 43 | 0.02 | glnK | GlnK nitrogen regulatory PII-like protein 99052:99390 forward | E - Amino acid transport and metabolism |
| HSERO_RS07390 | 0.2 | 2.0E-04 | ttuC | TtuC tartrate dehydrogenase protein 1690575:1691654 reverse | G - Carbohydrate transport and metabolism |
| HSERO_RS05635 | 84 | 0.02 | Hsero_1123 |
Hsero_1123 family II aminotransferase protein 1278563:1279939 reverse |
H - Coenzyme transport and metabolism |
| HSERO_RS07590 | 0.1 | 2.0E-03 | rplS | RplS 50S ribosomal subunit protein L19 1731532:1731915 forward | J - Translation, ribosomal structure and biogenesis |
| HSERO_RS03380 | 0.3 | 0.02 | ompW2 | OmpW2 outer membrane W protein 741190:741927 forward | M - Cell wall |
| HSERO_RS12905 | 0.4 | 5.0E-03 | lon | Lon ATP-dependent protease LA protein 2940893:2943301 reverse | O - Posttranslational modification, protein turnover, chaperones |
| HSERO_RS00425 | 20 | 0.04 | amtB |
AmtB ammonium transporter transmembrane protein 99406:100938 forward |
P - Inorganic ion transport and metabolism |
| HSERO_RS14575 | 0.1 | 1.10E-03 | Hsero_2905 |
Hsero_2905 conserved hypothetical protein 3301039:3301272 reverse |
S - Function unknown |
| HSERO_RS00415 | 15 | 0.03 | Hsero_0083 |
Hsero_0083 membrane protein 98251:99039 forward |
S - Function unknown |
| HSERO_RS13665 | 90 | 1.90E-03 | Hsero_2723 |
Hsero_2723 methyl-accepting chemotaxis transmembrane protein 3101951:3103597 reverse |
T;N - Signal transduction mechanisms;Cell motility |
| HSERO_RS06095 | 0.3 | 0.04 | trnK |
TrnK tRNA-Lys 1374570:1374645 reverse |
trnK |
| HSERO_RS07375 | 0.2 | 1.7E-03 | trnL |
TrnL tRNA-Leu 1689660:1689744 reverse |
trnL |
| HSERO_RS07775 | 0.1 | 0.03 | trnS |
TrnS tRNA-Ser 1781783:1781873 forward |
trnS |
Genes involved in nitrogen fixation (nif) were found amongst genes classified as “energy production and conversion” and “amino acid transport and metabolism”. Nif-genes encode proteins involved in the synthesis, maturation and assembly of the nitrogenase complex3. nifD and nifH (coverage 1.7 and 1.2 respectively) were highly expressed in H. seropedicae colonising wheat and maize roots111,112. Two ferredoxin genes (fdxA and fdxN, coverage 1.9 and 1.6 respectively) important for nitrogenase activity were also identified113. The promoters of nif genes are activated by NifA that, in turn, is regulated by the Ntr system. When comparisons were made between free-living H. seropedicae and those grown in association with rice, glnK and amtB of the Ntr system were induced 43 and 20-times respectively in the plant-bacterial interaction. Pankievicz et al.111 also found that amtB was induced in H. seropedicae attached to maize roots. Furthermore, fixN and fixP (both 1.6X coverage) were also detected in planta along with an urtA ABC-type urea transport system (Hsero_4713) (27-fold of induction in planta).
Twenty-three genes related to cell motility were found, four with >2X coverage. Amongst these were cheW (a positive regulator of CheA), flhD, fliC and pilZ (Hsero_2062) that encodes a type IV pilus assembly protein. A methyl-accepting chemotaxis trans-membrane protein (Hsero_2723) was induced 90-fold when compared with expression in culture114. This gene was also found to be up-regulated in epiphytic H. seropedicae colonising wheat and maize111,112. Methyl-accepting chemotaxis proteins interact with Che proteins to detect signals from the environment.
Among the 19 cell-wall related genes, seven were covered at least 2-fold and an outer-membrane porin (Hsero_4295) was induced 28-fold. Another membrane-protein (Hsero_0083) of unknown function was induced 14-fold. These could be proteins that H. seropedicae uses to recognise rice.
Materials and Methods
Plant material and growth conditions
Testas were removed from seeds of rice (Oryza sativa ssp japonica cv. Nipponbare, kindly provided by the Instituto Riograndense do arroz, IRGA – Avenida Missões 342, Porto Alegre, RS, Brazil), then disinfected with 70% (v/v) ethanol for 5 min followed by 30 min soaking in 8% sodium hypochlorite (1 mL per seed) containing 0.1% v/v Triton-X100. After rinsing 20 times with sterile water, the seeds were treated with 0.025% (v/v) Vitavax-Thiram (Chentura, Avenida Nações Unidas 4777, Alto de Pinheiros, São Paulo, SP, Brazil) fungicide solution and stirred (120 rpm) for 24 h in the dark at 30 °C. The seeds were then transferred to 0.7% water-agar and left for two days to germinate after which the seedlings were inoculated with 1 mL of Herbaspirillum seropedicae strain SmR1 (108 cells/seedling) for 30 minutes while control seedlings were treated with 1 mL of N-free NFbHP-malate medium115 (controls). Seedlings were washed with sterile water and transferred to glass tubes (25 cm long, 2.5 cm diameter) containing propylene beads and 25 mL of modified Hoagland’s solution116 without nitrogen (1 mM KH2PO4, 1 mM K2HPO4, 2 mM MgSO4.7H2O, 2 mM CaCl2.2H2O, 1 mL/L micronutrient solution (H3BO3 2.86 g.L−1, MnCl2.4H2O 1.81 g.L−1, ZnSO4.7H2O 0.22 g.L−1, CuSO4.5H2O 0.08 g.L−1, Na2MoO4.2H2O 0.02 g.L−1) and 1 mL.L−1 Fe-EDTA solution (Na2H2EDTA.2H2O 13.4 g.L−1 and FeCl3.6H2O 6 g.L−1)), pH 6.5–7.0. Plants were cultivated at 24 °C under 14 h light and 10 h dark for 3 days. H. seropedicae was cultivated in NFbHP malate medium containing 5 mM glutamate as the nitrogen source. Cells were shaken (120 rpm) overnight at 30 °C, then centrifuged, washed once with N-free NFbHP-malate and suspended in the same medium to OD600 = 1 (corresponding to 108 cells.mL−1). Strain SmR1117 is a spontaneous streptomycin resistant mutant of strain Z78118. For colonisation assays roots from one plant were washed in 70% v/v ethanol for 1 min, 1% chloramine T for 1 min followed by three washes with sterile water119. At least 5 plants were used per data point. The roots were then crushed with a mortar and pestle in 1 mL of NFbHP-malate and serial dilutions (10−1 to 10−5) were plated onto solid NFbHP-malate containing 20 mM NH4Cl. Two days later the cells were counted to determine the colony forming units (CFU) per gram of fresh root.
RNA isolation and construction of libraries
Three days after inoculation with H. seropedicae (6 days after the disinfection procedure) the roots were separated from the aerial part and immediately stored in RNA later™ (Life Technologies, Foster City, CA, USA). Total RNA was extracted from roots of five rice plants for each biological replicates (using a RNAqueous kit (Ambion, Austin, TX, USA). Contaminating genomic DNA was eliminated with RNase-free DNase I (Ambion) for 30 min at 37 °C. Total RNA (7 to 10 µg) was depleted of ribosomal RNA by treatment with a RiboMinus™ Plant Kit for RNA-Seq (Invitrogen, Carlsbad, CA, USA). The integrity and quality of the total RNA was checked spectrophotometrically and by agarose gel electrophoresis. Whole Transcriptome Analysis RNA Kits™ (Life Technologies) were used on 500 ng purified RNA to construct the libraries and sequencing was performed in a SOLiD4 (Life Technologies) sequencer. Two independent libraries were constructed for each condition (control and inoculated). Three of these libraries (two from control sample and one from inoculated sample) were run twice to check for technical reproducibility.
Sequencing and data analysis
SOLiD sequencing produced 161 million 50 bp reads that were analysed by SAET software (Applied Biosystems – Foster City, CA, EUA) to improve base calling (command shown in Supplementary material 1), followed by quality trimming using the CLC Genomics Workbench (CLC bio, a QIAGEN Company, Silkeborgvej 2, DK-8000 Aarhus C, Denmark) (quality scores higher than 0.05 and reads with less than 20 bp were discarded). Then the reads were mapped to the rice genome database from the MSU Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) using CLC Genomics Workbench and the following parameters: a minimum length fraction of 95%, minimum similarity of 90% and only one hit. Differential expression was analysed using DESeq.19, genes covered at least twice were considered expressed and regulated when expression changed two times and the p-value was lower than 0.05. For Blast2GO analyses, protein sequences encoded by differentially expressed genes were used to search non-redundant (nr) database with BLASTp. Then, Gene Ontology (GO) terms were assigned based on the top BLAST hit using Blast2GO20 with default parameters. The results of GO annotation were combined for Gene Set enrichment analysis (GSEA)21 using Blast2GO (default parameters), which identifies GO terms enriched as a result of differential expression between samples. A heat map showing the expression profiles of the differentially expressed genes normalized by CPM (counts per million) was constructed using an in-house script (Supplementary material 2). MapMan analyses were performed with differentially expressed transcript table as input using default parameters.
Quantification of mRNA levels using RT-qPCR
Reverse transcription quantitative PCR (RT-qPCR) analyses were used to evaluate gene expression under the conditions described above. Total RNA was isolated from rice roots using the TRI Reagent (Sigma, St. Louis, MO, USA) and contamination with genomic DNA was removed with DNase I (Life Technologies). The integrity and quality of the total RNA was confirmed by spectrophotometric analyses and electrophoresis. cDNA was produced from 1 μg DNase-treated total RNA using high-capacity cDNA reverse transcription kits (Life Technologies). The cDNA reaction was diluted 60 times before quantitative PCR using Power SYBR-Green PCR Master Mix on a Step One Plus Real Time-PCR System (both from Life Technologies). Primer sequences are listed in Supplementary Table 6 and were designed with the Primer express 3.0 software (Applied Biosystems) and the NCBI primer designing tool using the genome sequence of O. sativa ssp. japonica cv. Nipponbare. Calibration curves for all primer sets were linear over four orders of magnitude (R2 = 0.98 to 0.99) and efficiencies were 90% or higher. mRNA expression levels were normalised using the expression levels of actin 1, tubulin beta-2 chain (beta-2 tubulin)120 and a hypothetical protein (protein kinase)121 using geNorm 3.4 software122. The relative expression level was calculated according to Pfaffl123. Three independent samples were analysed for each condition and each sample was assayed in triplicate.
Supplementary information
Acknowledgements
We are grateful M.G. Yates for critical reading of the manuscript, Leonardo M. Cruz and Rodrigo A. Cardoso for bioinformatics support. Instituto Riograndense do Arroz (IRGA) is thanked for providing seeds. We are also thankful to Roseli Prado, Marilza Doroti Lamour and Valter A. Baura for technical assistance. This work was supported by National Institute of Science and Technology of Biological Nitrogen Fixation (INCT – BNF), Council for Scientific and Technological Development (CNPq), Fundação Araucária and Coordination of Improvement of Higher-Education Personnel (CAPES).
Author Contributions
L.C.C.B.-S. did the plant growth experiments and RNA purification. L.C.C.B.-S., D.C.-N., M.Z.T.-S. and H.F. prepared the sequencing libraries and sequencing. L.C.C.B.-S., E.M.S., R.C., K.L. and A.B.-S. performed bioinformatics analyse. L.C.C.B.-S., D.A., R.A.M., W.J.B., F.O.P., R.W. and E.M.S. designed the project and wrote the main manuscript text. L.C.C.B.-S. and V.G. did the qRT-PCR experiments and analysis. All the authors reviewed the manuscript.
Data Availability
The data that support the findings of this study are openly available. RNA-Seq data from this study have been deposited at the NCBI under the BioProject accession No. PRJNA489273 and BioSample Nos. SAMN09942067, SAMN09942069, SAMN09942071 and SAMN09953899.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-45866-w.
References
- 1.Ladha JK, Reddy PM. Nitrogen fixation in rice systems: state of knowledge and future prospects. Plant and Soil. 2003;252:151–167. doi: 10.1023/A:1024175307238. [DOI] [Google Scholar]
- 2.Monteiro RA, et al. Herbaspirillum-plant interactions: microscopical, histological and molecular aspects. Plant and Soil. 2012;356:175–196. doi: 10.1007/s11104-012-1125-7. [DOI] [Google Scholar]
- 3.Chubatsu LS, et al. Nitrogen fixation control in Herbaspirillum seropedicae. Plant Soil. 2012;356:197–207. doi: 10.1007/s11104-011-0819-6. [DOI] [Google Scholar]
- 4.Gyaneshwar P, James EK, Reddy PM, Ladha JK. Blackwell Science Ltd Herbaspirillum colonization increases growth and nitrogen accumulation in aluminium-tolerant rice varieties. New Phytologist. 2002;154:131–145. doi: 10.1046/j.1469-8137.2002.00371.x. [DOI] [Google Scholar]
- 5.James EK. Nitrogen fixation in endophytic and associative symbiosis. Field Crops Research. 2000;65:197–209. doi: 10.1016/S0378-4290(99)00087-8. [DOI] [Google Scholar]
- 6.James EK, et al. Infection and colonization of rice seedlings by the plant growth-promoting bacterium Herbaspirillum seropedicae Z67. Mol. Plant Microbe Interact. 2002;15:894–906. doi: 10.1094/MPMI.2002.15.9.894. [DOI] [PubMed] [Google Scholar]
- 7.Roncato-Maccari LDB, et al. Endophytic Herbaspirillum seropedicae expresses nif genes in gramineous plants. FEMS Microbiol. Ecol. 2003;45:39–47. doi: 10.1016/S0168-6496(03)00108-9. [DOI] [PubMed] [Google Scholar]
- 8.Pankievicz VCS, et al. Robust biological nitrogen fixation in a model grass-bacterial association. Plant J. 2015;81:907–919. doi: 10.1111/tpj.12777. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki K, et al. Impact of plant genotype and nitrogen level on rice growth response to inoculation with Azospirillum sp. strain B510 under paddy field conditions. Soil Science and Plant Nutrition. 2010;56:636–644. doi: 10.1111/j.1747-0765.2010.00499.x. [DOI] [Google Scholar]
- 10.Nobuta K, et al. An expression atlas of rice mRNAs and small RNAs. Nat. Biotechnol. 2007;25:473–477. doi: 10.1038/nbt1291. [DOI] [PubMed] [Google Scholar]
- 11.Mizuno H, et al. Massive parallel sequencing of mRNA in identification of unannotated salinity stress-inducible transcripts in rice (Oryza sativa L.) BMC Genomics. 2010;11:683. doi: 10.1186/1471-2164-11-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu H, Gao Y, Wang J. Transcriptomic analysis of rice (Oryza sativa) developing embryos using the RNA-Seq technique. PLoS ONE. 2012;7:e30646. doi: 10.1371/journal.pone.0030646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu, X.-H. et al. Friend or foe: differential responses of rice to invasion by mutualistic or pathogenic fungi revealed by RNAseq and metabolite profiling. Sci Rep5 (2015). [DOI] [PMC free article] [PubMed]
- 14.Wakasa Y, et al. RNA sequencing-mediated transcriptome analysis of rice plants in endoplasmic reticulum stress conditions. BMC Plant Biology. 2014;14:101. doi: 10.1186/1471-2229-14-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magbanua Zenaida V, Arick Mark, Buza Teresia, Hsu Chuan-Yu, Showmaker Kurt C, Chouvarine Philippe, Deng Peng, Peterson Daniel G, Lu Shien. Transcriptomic dissection of the rice – Burkholderia glumae interaction. BMC Genomics. 2014;15(1):755. doi: 10.1186/1471-2164-15-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shankar R, Bhattacharjee A, Jain M. Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Scientific Reports. 2016;6:23719. doi: 10.1038/srep23719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brusamarello-Santos, L. C. C. et al. Differential gene expression of rice roots inoculated with the diazotroph Herbaspirillum seropedicae. Plant Soil (2011).
- 18.Elbeltagy A, et al. Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Appl. Environ. Microbiol. 2001;67:5285–5293. doi: 10.1128/AEM.67.11.5285-5293.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Götz S, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrer J-L, Austin MB, Stewart C, Jr, Noel JP. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 2008;46:356–370. doi: 10.1016/j.plaphy.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassan S., Mathesius U. The role of flavonoids in root-rhizosphere signalling: opportunities and challenges for improving plant-microbe interactions. Journal of Experimental Botany. 2012;63(9):3429–3444. doi: 10.1093/jxb/err430. [DOI] [PubMed] [Google Scholar]
- 24.Naoumkina MA, et al. Genome-wide analysis of phenylpropanoid defence pathways. Mol. Plant Pathol. 2010;11:829–846. doi: 10.1111/j.1364-3703.2010.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balsanelli E, et al. Herbaspirillum seropedicae rfbB and rfbC genes are required for maize colonization. Environ. Microbiol. 2010;12:2233–2244. doi: 10.1111/j.1462-2920.2010.02187.x. [DOI] [PubMed] [Google Scholar]
- 26.Plaper A, et al. Characterization of quercetin binding site on DNA gyrase. Biochem. Biophys. Res. Commun. 2003;306:530–536. doi: 10.1016/S0006-291X(03)01006-4. [DOI] [PubMed] [Google Scholar]
- 27.Chen F, Tholl D, Bohlmann J, Pichersky E. The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011;66:212–229. doi: 10.1111/j.1365-313X.2011.04520.x. [DOI] [PubMed] [Google Scholar]
- 28.Okada A, et al. Elicitor induced activation of the methylerythritol phosphate pathway toward phytoalexins biosynthesis in rice. Plant Mol. Biol. 2007;65:177–187. doi: 10.1007/s11103-007-9207-2. [DOI] [PubMed] [Google Scholar]
- 29.Drogue B, et al. Plant root transcriptome profiling reveals a strain-dependent response during Azospirillum-rice cooperation. Front Plant Sci. 2014;5:607. doi: 10.3389/fpls.2014.00607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhee SG, Chae HZ, Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic. Biol. Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 31.Dietz K-J, et al. The function of peroxiredoxins in plant organelle redox metabolism. J. Exp. Bot. 2006;57:1697–1709. doi: 10.1093/jxb/erj160. [DOI] [PubMed] [Google Scholar]
- 32.Apostol I, Heinstein PF, Low PS. Rapid Stimulation of an Oxidative Burst during Elicitation of Cultured Plant Cells: Role in Defense and Signal Transduction. Plant Physiol. 1989;90:109–116. doi: 10.1104/pp.90.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daudi A, et al. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell. 2012;24:275–287. doi: 10.1105/tpc.111.093039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bindschedler LV, et al. Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J. 2006;47:851–863. doi: 10.1111/j.1365-313X.2006.02837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alberton D, et al. Comparative proteomics analysis of the rice roots colonized by Herbaspirillum seropedicae strain SmR1 reveals induction of the methionine recycling in the plant host. J. Proteome Res. 2013;12:4757–4768. doi: 10.1021/pr400425f. [DOI] [PubMed] [Google Scholar]
- 36.Valdameri G, et al. Herbaspirillum rubrisubalbicans, a mild pathogen impairs growth of rice by augmenting ethylene levels. Plant Mol. Biol. 2017;94:625–640. doi: 10.1007/s11103-017-0629-1. [DOI] [PubMed] [Google Scholar]
- 37.Pedrosa FO, et al. Genome of Herbaspirillum seropedicae strain SmR1, a specialized diazotrophic endophyte of tropical grasses. PLoS Genet. 2011;7:e1002064. doi: 10.1371/journal.pgen.1002064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goujon T, et al. AtBXL1, a novel higher plant (Arabidopsis thaliana) putative beta-xylosidase gene, is involved in secondary cell wall metabolism and plant development. Plant J. 2003;33:677–690. doi: 10.1046/j.1365-313X.2003.01654.x. [DOI] [PubMed] [Google Scholar]
- 39.Lin C, Choi H-S, Cho H-T. Root hair-specific EXPANSIN A7 is required for root hair elongation in Arabidopsis. Mol. Cells. 2011;31:393–397. doi: 10.1007/s10059-011-0046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.ZhiMing Y, et al. Root hair-specific expansins modulate root hair elongation in rice. Plant J. 2011;66:725–734. doi: 10.1111/j.1365-313X.2011.04533.x. [DOI] [PubMed] [Google Scholar]
- 41.Kwasniewski M, Szarejko I. Molecular cloning and characterization of beta-expansin gene related to root hair formation in barley. Plant Physiol. 2006;141:1149–1158. doi: 10.1104/pp.106.078626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S, et al. Molecular mechanism of plant growth promotion and induced systemic resistance to tobacco mosaic virus by Bacillus spp. J. Microbiol. Biotechnol. 2009;19:1250–1258. doi: 10.4014/jmb.0901.008. [DOI] [PubMed] [Google Scholar]
- 43.Giordano W, Hirsch AM. The expression of MaEXP1, a Melilotus alba expansin gene, is upregulated during the sweetclover-Sinorhizobium meliloti interaction. Mol. Plant Microbe Interact. 2004;17:613–622. doi: 10.1094/MPMI.2004.17.6.613. [DOI] [PubMed] [Google Scholar]
- 44.Abramovitch RB, Anderson JC, Martin GB. Bacterial elicitation and evasion of plant innate immunity. Nat. Rev. Mol. Cell Biol. 2006;7:601–611. doi: 10.1038/nrm1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 46.McDowell JM, Simon SA. Molecular diversity at the plant-pathogen interface. Dev. Comp. Immunol. 2008;32:736–744. doi: 10.1016/j.dci.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 47.Couto D, Zipfel C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016;16:537–552. doi: 10.1038/nri.2016.77. [DOI] [PubMed] [Google Scholar]
- 48.Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 2009;21:177–184. doi: 10.1016/j.ceb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 49.Kohorn, B. D. & Kohorn, S. L. The cell wall-associated kinases, WAKs, as pectin receptors. Front. Plant Sci. 3 (2012). [DOI] [PMC free article] [PubMed]
- 50.Kohorn BD. The state of cell wall pectin monitored by wall associated kinases: A model. Plant Signal Behav. 2015;10:e1035854. doi: 10.1080/15592324.2015.1035854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kohorn BD. Cell wall-associated kinases and pectin perception. J. Exp. Bot. 2016;67:489–494. doi: 10.1093/jxb/erv467. [DOI] [PubMed] [Google Scholar]
- 52.Li H, Zhou S-Y, Zhao W-S, Su S-C, Peng Y-L. A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol. Biol. 2009;69:337–346. doi: 10.1007/s11103-008-9430-5. [DOI] [PubMed] [Google Scholar]
- 53.Cayrol B, Delteil A, Gobbato E, Kroj T, Morel J-B. Three wall-associated kinases required for rice basal immunity form protein complexes in the plasma membrane. Plant Signal Behav. 2016;11:e1149676. doi: 10.1080/15592324.2016.1149676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harkenrider M, et al. Overexpression of Rice Wall-Associated Kinase 25 (OsWAK25) Alters Resistance to Bacterial and Fungal Pathogens. PLoS ONE. 2016;11:e0147310. doi: 10.1371/journal.pone.0147310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delteil A, et al. Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol. 2016;16:17. doi: 10.1186/s12870-016-0711-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meier S, et al. The Arabidopsis wall associated kinase-like 10 gene encodes a functional guanylyl cyclase and is co-expressed with pathogen defense related genes. PLoS ONE. 2010;5:e8904. doi: 10.1371/journal.pone.0008904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vinagre F, et al. SHR5: a novel plant receptor kinase involved in plant-N2-fixing endophytic bacteria association. J. Exp. Bot. 2006;57:559–569. doi: 10.1093/jxb/erj041. [DOI] [PubMed] [Google Scholar]
- 58.Burdiak P, Rusaczonek A, Witoń D, Głów D, Karpiński S. Cysteine-rich receptor-like kinase CRK5 as a regulator of growth, development, and ultraviolet radiation responses in Arabidopsis thaliana. J Exp Bot. 2015;66:3325–3337. doi: 10.1093/jxb/erv143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aarts N, et al. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA. 1998;95:10306–10311. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McHale L, Tan X, Koehl P, Michelmore RW. Plant NBS-LRR proteins: adaptable guards. Genome Biol. 2006;7:212. doi: 10.1186/gb-2006-7-4-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bartsch M, et al. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell. 2006;18:1038–1051. doi: 10.1105/tpc.105.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Venugopal SC, et al. Enhanced disease susceptibility 1 and salicylic acid act redundantly to regulate resistance gene-mediated signaling. PLoS Genet. 2009;5:e1000545. doi: 10.1371/journal.pgen.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parker JE, et al. Plant Cell. 1996. Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes; pp. 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Falk A, et al. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci USA. 1999;96:3292–3297. doi: 10.1073/pnas.96.6.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kawahara Y, et al. Simultaneous RNA-Seq Analysis of a Mixed Transcriptome of Rice and Blast Fungus Interaction. PLoS ONE. 2012;7:e49423. doi: 10.1371/journal.pone.0049423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim ST, et al. Proteomics analysis of rice lesion mimic mutant (spl1) reveals tightly localized probenazole-induced protein (PBZ1) in cells undergoing programmed cell death. J. Proteome Res. 2008;7:1750–1760. doi: 10.1021/pr700878t. [DOI] [PubMed] [Google Scholar]
- 67.Florack DEA, Stiekema WJ. Thionins: properties, possible biological roles and mechanisms of action. Plant Molecular Biology. 1994;26:25–37. doi: 10.1007/BF00039517. [DOI] [PubMed] [Google Scholar]
- 68.Iwai T, et al. Enhanced resistance to seed-transmitted bacterial diseases in transgenic rice plants overproducing an oat cell-wall-bound thionin. Mol. Plant Microbe Interact. 2002;15:515–521. doi: 10.1094/MPMI.2002.15.6.515. [DOI] [PubMed] [Google Scholar]
- 69.Choi Y, Choi YD, Lee JS. Antimicrobial activity of gamma-thionin-like soybean SE60 in E. coli and tobacco plants. Biochem. Biophys. Res. Commun. 2008;375:230–234. doi: 10.1016/j.bbrc.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 70.Muramoto N, et al. Transgenic sweet potato expressing thionin from barley gives resistance to black rot disease caused by Ceratocystis fimbriata in leaves and storage roots. Plant Cell Rep. 2012;31:987–997. doi: 10.1007/s00299-011-1217-5. [DOI] [PubMed] [Google Scholar]
- 71.Ji H, et al. The role of thionins in rice defence against root pathogens. Mol. Plant Pathol. 2015;16:870–881. doi: 10.1111/mpp.12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Straub D, Yang H, Liu Y, Tsap T, Ludewig U. Root ethylene signalling is involved in Miscanthus sinensis growth promotion by the bacterial endophyte Herbaspirillum frisingense GSF30(T) J. Exp. Bot. 2013;64:4603–4615. doi: 10.1093/jxb/ert276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rogg LE, Bartel B. Auxin signaling: derepression through regulated proteolysis. Dev. Cell. 2001;1:595–604. doi: 10.1016/S1534-5807(01)00077-6. [DOI] [PubMed] [Google Scholar]
- 74.Kazan K, Manners JM. Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci. 2009;14:373–382. doi: 10.1016/j.tplants.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 75.McSteen P. Auxin and Monocot Development. Cold Spring Harbor Perspectives in Biology. 2010;2(3):a001479–a001479. doi: 10.1101/cshperspect.a001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Adamowski M, Friml J. PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell. 2015;27:20–32. doi: 10.1105/tpc.114.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guilfoyle TJ. The PB1 domain in auxin response factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell. 2015;27:33–43. doi: 10.1105/tpc.114.132753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bastian F, et al. Production of indole-3-acetic acid and gibberellins A1 and A3 by Acetobacter diazotrophicus and Herbaspirillum seropedicae in chemically-defined culture media. Plant Growth Regulation. 1998;24:7–11. doi: 10.1023/A:1005964031159. [DOI] [Google Scholar]
- 79.Inukai Y, et al. Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell. 2005;17:1387–1396. doi: 10.1105/tpc.105.030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jameson P. Cytokinins and auxins in plant-pathogen interactions – An overview. Plant Growth Regulation. 2000;32:369–380. doi: 10.1023/A:1010733617543. [DOI] [Google Scholar]
- 81.Navarro L, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 82.Lorbiecke R, Sauter M. Adventitious Root Growth and Cell-Cycle Induction in Deepwater Rice. Plant Physiol. 1999;119:21–30. doi: 10.1104/pp.119.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 84.Robert-Seilaniantz A, Grant M, Jones JDG. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- 85.Vlot AC, Dempsey DA, Klessig DF. Salicylic Acid, a Multifaceted Hormone to Combat Disease. Annual Review of Phytopathology. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- 86.An C, Mou Z. Salicylic acid and its function in plant immunity. J Integr Plant Biol. 2011;53:412–428. doi: 10.1111/j.1744-7909.2011.01043.x. [DOI] [PubMed] [Google Scholar]
- 87.Ross JR, Nam KH, D’Auria JC, Pichersky E. S-Adenosyl-L-methionine:salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Arch. Biochem. Biophys. 1999;367:9–16. doi: 10.1006/abbi.1999.1255. [DOI] [PubMed] [Google Scholar]
- 88.Park S-W, Kaimoyo E, Kumar D, Mosher S, Klessig DF. Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science. 2007;318:113–116. doi: 10.1126/science.1147113. [DOI] [PubMed] [Google Scholar]
- 89.Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007;10:366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 90.Journot-Catalino N, Somssich IE, Roby D, Kroj T. The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell. 2006;18:3289–3302. doi: 10.1105/tpc.106.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hu Y, Dong Q, Yu D. Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 2012;185–186:288–297. doi: 10.1016/j.plantsci.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 92.Shojima S, et al. Biosynthesis of Phytosiderophores: In Vitro Biosynthesis of 2′-Deoxymugineic Acid from l-Methionine and Nicotianamine. Plant Physiol. 1990;93:1497–1503. doi: 10.1104/pp.93.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Higuchi K, et al. Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol. 1999;119:471–480. doi: 10.1104/pp.119.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bashir K, et al. Cloning and characterization of deoxymugineic acid synthase genes from graminaceous plants. J. Biol. Chem. 2006;281:32395–32402. doi: 10.1074/jbc.M604133200. [DOI] [PubMed] [Google Scholar]
- 95.Inoue H, et al. Identification and localisation of the rice nicotianamine aminotransferase gene OsNAAT1 expression suggests the site of phytosiderophore synthesis in rice. Plant Mol. Biol. 2008;66:193–203. doi: 10.1007/s11103-007-9262-8. [DOI] [PubMed] [Google Scholar]
- 96.Lee S, et al. Iron fortification of rice seeds through activation of the nicotianamine synthase gene. PNAS. 2009;106:22014–22019. doi: 10.1073/pnas.0910950106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hakoyama T, et al. Nicotianamine synthase specifically expressed in root nodules of Lotus japonicus. Planta. 2009;230:309–317. doi: 10.1007/s00425-009-0944-0. [DOI] [PubMed] [Google Scholar]
- 98.Ling HQ, Pich A, Scholz G, Ganal MW. Genetic analysis of two tomato mutants affected in the regulation of iron metabolism. Mol. Gen. Genet. 1996;252:87–92. doi: 10.1007/BF02173208. [DOI] [PubMed] [Google Scholar]
- 99.Nozoye T, et al. Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J. Biol. Chem. 2011;286:5446–5454. doi: 10.1074/jbc.M110.180026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Curie C, et al. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature. 2001;409:346–349. doi: 10.1038/35053080. [DOI] [PubMed] [Google Scholar]
- 101.Inoue H, et al. Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J. Biol. Chem. 2009;284:3470–3479. doi: 10.1074/jbc.M806042200. [DOI] [PubMed] [Google Scholar]
- 102.Ishimaru Y, et al. Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 2010;62:379–390. doi: 10.1111/j.1365-313X.2010.04158.x. [DOI] [PubMed] [Google Scholar]
- 103.Kakei Y, et al. OsYSL16 plays a role in the allocation of iron. Plant Mol. Biol. 2012;79:583–594. doi: 10.1007/s11103-012-9930-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ishimaru Y, et al. Rice plants take up iron as an Fe3+ -phytosiderophore and as Fe2+ Plant J. 2006;45:335–346. doi: 10.1111/j.1365-313X.2005.02624.x. [DOI] [PubMed] [Google Scholar]
- 105.Thomine S, Lelièvre F, Debarbieux E, Schroeder JI, Barbier-Brygoo H. AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. The Plant Journal. 2003;34:685–695. doi: 10.1046/j.1365-313X.2003.01760.x. [DOI] [PubMed] [Google Scholar]
- 106.Zhang Y, Xu Y-H, Yi H-Y, Gong J-M. Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J. 2012;72:400–410. doi: 10.1111/j.1365-313X.2012.05088.x. [DOI] [PubMed] [Google Scholar]
- 107.Bashir K, et al. The knockdown of OsVIT2 and MIT affects iron localization in rice seed. Rice (N Y) 2013;6:31. doi: 10.1186/1939-8433-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Arredondo-Peter R, et al. Rice hemoglobins. Gene cloning, analysis, and O2-binding kinetics of a recombinant protein synthesized in Escherichia coli. Plant Physiol. 1997;115:1259–1266. doi: 10.1104/pp.115.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lira-Ruan V, Sarath G, Klucas RV, Arredondo-Peter R. Synthesis of hemoglobins in rice (Oryza sativa var. Jackson) plants growing in normal and stress conditions. Plant Science. 2001;161:279–287. doi: 10.1016/S0168-9452(01)00411-3. [DOI] [PubMed] [Google Scholar]
- 110.Aznar A, Chen NWG, Thomine S, Dellagi A. Immunity to plant pathogens and iron homeostasis. Plant Sci. 2015;240:90–97. doi: 10.1016/j.plantsci.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 111.Pankievicz VCS, et al. RNA-seq transcriptional profiling of Herbaspirillum seropedicae colonizing wheat (Triticum aestivum) roots. Plant Mol. Biol. 2016;90:589–603. doi: 10.1007/s11103-016-0430-6. [DOI] [PubMed] [Google Scholar]
- 112.Balsanelli E, et al. Molecular adaptations of Herbaspirillum seropedicae during colonization of the maize rhizosphere. Environ. Microbiol. 2016;18:2343–2356. doi: 10.1111/1462-2920.12887. [DOI] [PubMed] [Google Scholar]
- 113.Souza ALF, et al. The involvement of the nif-associated ferredoxin-like genes fdxA and fdxN of Herbaspirillum seropedicae in nitrogen fixation. J. Microbiol. 2010;48:77–83. doi: 10.1007/s12275-009-0077-y. [DOI] [PubMed] [Google Scholar]
- 114.Tadra-Sfeir MZ, et al. Genome wide transcriptional profiling of Herbaspirillum seropedicae SmR1 grown in the presence of naringenin. Front Microbiol. 2015;6:491. doi: 10.3389/fmicb.2015.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Klassen G, Pedrosa FO, Souza EM, Funayama S, Rigo L. Effect of nitrogen compounds on nitrogenase activity in Herbaspirillum seropedicae. Canadian Journal of Microbiology. 1997;43:887–891. doi: 10.1139/m97-129. [DOI] [Google Scholar]
- 116.Hoagland, D. & Arnon, D. The water-culture method for growing plants without soil (1950).
- 117.Souza EM, Pedrosa FO, Rigo LU, Machado HB, Yates MG. Expression of the nifA gene of Herbaspirillum seropedicae: role of the NtrC and NifA binding sites and of the −24/−12 promoter element. Microbiology (Reading, Engl.) 2000;146(Pt 6):1407–1418. doi: 10.1099/00221287-146-6-1407. [DOI] [PubMed] [Google Scholar]
- 118.Baldani JI, Baldani VLD, Seldin L, Döbereiner J. Characterization of Herbaspirillum seropedicae gen. nov., sp. nov., a Root-Associated Nitrogen-Fixing Bacterium. Int J Syst Bacteriol. 1986;36:86–93. doi: 10.1099/00207713-36-1-86. [DOI] [Google Scholar]
- 119.Dobereiner, J., Baldani, V. L. D. & Baldani, J. I. Como isolar e identificar bactérias diazotróficas de plantas não-leguminosas. (EMBRAPA, 1995).
- 120.Jain M, Nijhawan A, Tyagi AK, Khurana JP. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006;345:646–651. doi: 10.1016/j.bbrc.2006.04.140. [DOI] [PubMed] [Google Scholar]
- 121.Narsai R, Ivanova A, Ng S, Whelan J. Defining reference genes in Oryza sativa using organ, development, biotic and abiotic transcriptome datasets. BMC Plant Biol. 2010;10:56. doi: 10.1186/1471-2229-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034.1–research0034.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available. RNA-Seq data from this study have been deposited at the NCBI under the BioProject accession No. PRJNA489273 and BioSample Nos. SAMN09942067, SAMN09942069, SAMN09942071 and SAMN09953899.