Abstract
Human Aichivirus A (AiV-A) is classified as a Kobuvirus, group IV positive sense single strand RNA viruses. The first outbreak of AiV-A was reported from Aichi Prefecture, Japan in 1989. AiV-A exists not only among clinical patients, such as diarrhea, but also in a variety of water environments, as its occurrence is reported across a wide geographical range, from developing to advanced countries. For diagnose of AiV-A from water samples, mostly polymerase chain reaction (PCR) system have been developed. However, loop-mediated isothermal amplification (LAMP) assay has not been applied. In this study, developed a LAMP method to achieve a rapid, specific and highly sensitive detection of AiV-A. The method developed in this study is aimed specifically at AiV-A. Through a specific and non-specific selection and sensitivity test process for the five prepared LAMP primer sets, one primer set and optimum reaction temperature were selected. A newly developed method was more rapid (approximately 2–8 h), specific and equivalent detection of AiV-A than with the conventional PCRs. In addition, confirm system of positive LAMP reaction was developed by using the restriction enzyme Aci I and Hae III. For evaluation and verification of developing LAMP assay, a was applied to twenty cDNA from groundwater samples. This study proved rapid and specific diagnosis of AiV-A from water samples, and it is also demanded to be applicable to other environmental, clinical and food samples.
Electronic supplementary material
The online version of this article (10.1007/s12088-019-00803-3) contains supplementary material, which is available to authorized users.
Keywords: AiV-A, Human Aichivirus A, LAMP, Loop-mediated isothermal amplification, Water sample
Aichivirus A (AiV-A) is classified as Kobuvirus, Picornaviridae and group IV (+) ssRNA virus [1]. AiV-A was first reported in 1989 in a stool specimen of non-bacterial gastroenteritis in the Aichi Prefecture, Japan [2]. AiV-A has been believed to be a local, endemic disease until the virus was detected among travelers to Japan and Southeast Asia [3]. However, it is now reported worldwide in Asia, Europe, North America, and South America [4–7]. Although the AiV-A can be detected in patients with diarrhea, detection frequency is very low at 0–4.1% [8]. In addition, mixed infection with other waterborne viruses can occur (i.e. Norovirus), and AiV-A characterized as being present in a wide range of geographical locations, from developing countries to advanced countries [9, 10]. It has also been reported that the AiV-A may exist in an aquatic environment under natural conditions [11]. According to a survey of water sources, effluents, rivers, and streams, the AiV-A gene was detected at a high rate in Japan, in the United States, and in Europe [12–14]. A method for detecting AiV-A specific genes by using reverse transcription nested polymerase chain reaction (RT-PCR) assay, have been reported [15–17]. It was also developed on the basis of RT-nested PCR for detection of AiV-A from national project in 2016–2018, Korea [18]. Since the conventional PCR method entails a long time after DNA extraction [19], it required extensive laboratory effort and labor costs. Thus, the loop-mediated isothermal amplification (LAMP) has been proposed as one of the methods for the rapid monitoring and mass treatment of specimens [18]. A number of PCR-based AiV-A detecting system has been developed, however, LAMP assay has not been applied. In this study, developed a rapid, specific and highly sensitive detection of AiV-A by using a LAMP assay.
In the design of a LAMP primers to detect AiV-A, the following sequences were collected from the National Center for Biotechnology Information (NCBI); (1) target viruses (AiV-A strains), (2) taxonomically similar viruses (AiV-B and C), and (3) eighteen enteric waterborne viruses. Five LAMP primer sets were designed by using PrimerExplorer that targets the P2A-B region of Human AiV-A (Table S1).
It was genetically synthesized to human AiV-A (NC_001918; location 4400-5199 nucleotide), and nucleic acid or plasmid of sixteen reference viruses (Rotavirus A, Echovirus 5, Norovirus GII, Coronavirus 229E, Enterovirus 68, Coxsackievirus A6, Coxsackievirus B1, Coxsackievirus B5, Enterovirus 71, Adenovirus 41, non-enteric adenovirus, Parvovirus B19, AiV-B, AiV-C, Sapovirus, and Hepatitisvirus A) were collected.
The AiV-A was subjected to a specific reaction for five LAMP primer sets. The LAMP reaction mixture amounted to a total of 20 μL, containing 1 μL of the template (AiV-A plasmid), 1 μL of the LAMP primers [F3 and B3 (10 pmol)], 2 μL of FIP and BIP (10 pmol), 1.5 μL of Bst-polymerase (8 unit/μL), 2 μL of dNTP (10 mM), 2 μL of 10× buffer, and 7.5 μL of nucleic acid free water. Regarding the LAMP condition, it reacted for 10 min at 95 °C before reacting for 1 h under four different conditions [60–63 °C (1 °C interval)], thereby completing the reaction of all five LAMP primer sets. Among the five LAMP primer sets, a non-specific reaction occurred in the C and E sets. The AiV-A specific reaction was found in the LAMP primers A and B sets, while no reaction was observed under the four conditions in set D. In particular, LAMP primer set A exhibited specific reactions under all four conditions. Among the varying temperatures in which a reaction occurred in sets A and B, 62 °C was selected (Fig. S1). A check of the non-specific reaction of sixteen reference viruses in the two LAMP primer sets with AiV-A specific reaction showed no response in the references of both primer sets (Fig. S2). Furthermore, the AiV-A gene was diluted tenfold to evaluate the relative detection sensitivity of the two LAMP primer sets. The detection sensitivity of set A was about 10 times higher than that of set B (Fig. S3A). For the comparison of LAMP and conventional PCR, 1 conventional PCR primer set was selected according to recently reported with high sensitivity [18]. A comparison of the detection sensitivity of the human AiV-A with 1 conventional PCR sets showed that the highest detection sensitivity was at 10−6, which was about 1 fg, and was equivalent to that of LAMP primer set A (Fig. S3B).
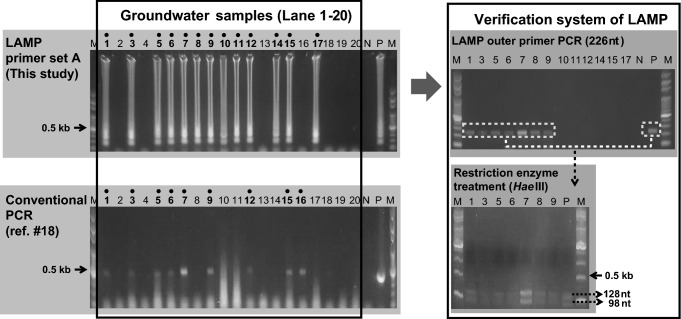
The specific response to LAMP primer set A was re-verified, and PCR amplification was performed on the LAMP outer primer set (F3 and B3) with its composition shown below: a total of 25 μL was added with 1 μL of the template (AiV-A plasmid) 1 μL each of F3 and B3 (10 pmol), 0.1 μL of HS Taq polymerase (2.5 units/μl, GeneAll, Korea), 2 μL of dNTP (10 mM), 2.5 μL of 10× buffer, and 17.4 μL of nucleic acid free water. The conditions were 5 min at 94 °C, and 3 min at 72 °C after repeating 35 cycles (94 °C for 30 s, 62 °C for 30 s, and 72 °C for 45 s). The PCR product (226 bp) reacted using two restriction enzyme, Aci I (C/CGC) and Hae III (GG/CC) (10 units) with 37 °C for 2 h. Restriction enzyme treatment resulted with gel electrophoresis, PCR products were digested with 152 + 74 bp (Aci I) and 128 + 98 bp (Hae III) (Fig. S4 and S5).
The cDNAs of a total of twenty samples from groundwater, were collected to verify AiV-A detection. The collected cDNAs reacted by using the most sensitive method among the LAMP primer set A developed in this study, and the conventional PCR method [18]. The LAMP results showed that AiV-A was positive in 13 of 20 samples (65%), but conventional PCR results showed 9 of 20 samples (45%). Among the AiV-A positive samples of conventional PCR and LAMP assay, 8 samples were same positive results, additionally 5 samples were positive results at LAMP assay (sample number 8, 10, 11, 14 and 17). One sample showed positive results at conventional PCR but result of LAMP assay was negative result (sample number 16). However, conventional PCR showed that non-specific reaction from sample tests (sample number 10, 11 and 17). For verification of LAMP reaction, PCR amplification with 13 positive samples was performed on the LAMP outer primer set. After amplification, 7 samples showed that 226 bp positive reaction from LAMP outer primer set. Outer primer positive samples were cut due to the restriction enzyme, Hae III (10 units) from PCR products, with 37 °C, 2 h. Digested from PCR products were confirmed with 128 + 98 bp (Fig. 1).
Fig. 1.
Comparison of LAMP and conventional PCR system for the detection of Aichivirus A from twenty groundwater samples. Lane M, 100 bp step DNA Ladder marker (Genepia, Korea); Lane 1–20, groundwater samples; N negative control, P positive control
In this study, developed a LAMP assay that could rapid, specific, and sensitive detection of AiV-A from water samples. There have been cases of SYBR or hydroxynaphthol blue application for more rapid monitoring of the LAMP assay, which may reduce the time and cost of electrophoresis [19]. Comparison between LAMP assay and conventional PCR, LAMP assay have advantages in high sensitivity, high specificity and reaction time. LAMP assay can be shortened the total duration time about 2–3 h as compared to conventional PCR methods, or by about 8 h when compared to the RT-nested PCR used in the environment. Also, despite of same sensitivity results from conventional PCR and LAMP assay (Fig. S3A), LAMP assay can detection lower concentration virus in environmental samples, almost 20% were highly specific and sensitive compare to conventional PCR methods (Fig. 1). Since the primers used in the LAMP reaction anneal specifically to the six locations, only a specific pathogen can be detected more accurately than through the conventional PCR. Furthermore, since LAMP amplification can be re-verified with the restriction enzyme Hae III. This study is expected to be useful as a diagnose AiV-A from water environment, and also applicable to other environmental, clinical and food samples.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) Grant funded by the Korea Government (MSIT) (No. 2017R1A2B1009303).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pringle CR. Virus taxonomy at the XIth international congress of virology, Sydney, Australia, 1999. Arch Virol. 1999;144:2065–2070. doi: 10.1007/s007050050728. [DOI] [PubMed] [Google Scholar]
- 2.Yang S, Zhang W, Shen Q, Yang Z, Zhu J, Cui L, Hua X. Aichi virus strains in children with gastroenteritis, China. Emerg Infect Dis. 2009;15:1703–1705. doi: 10.3201/eid1510.090522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamashita T, Sakae K, Kobayashi S, Ishihara Y, Miyake T, Mubina A, Isomura S. Isolation of cytopathic small round virus (Aichi virus) from Pakistani children and Japanese travelers from Southeast Asia. Microbiol Immunol. 1995;39:433–435. doi: 10.1111/j.1348-0421.1995.tb02225.x. [DOI] [PubMed] [Google Scholar]
- 4.Yamashita T, Kobayashi S, Sakae K, Nakata S, Chiba S, Ishihara Y, Isomura S. Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J Infect Dis. 1991;164:954–957. doi: 10.1093/infdis/164.5.954. [DOI] [PubMed] [Google Scholar]
- 5.Yip CC, Lo KL, Que TL, Lee RA, Chan KH, Yuen KY, Woo PC, Lau SK. Epidemiology of human parechovirus, Aichi virus and salivirus in fecal samples from hospitalized children with gastroenteritis in Hong Kong. Virol J. 2014;11:182. doi: 10.1186/1743-422X-11-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drexler JF, Baumgarte S, Luna LKS, Bludau ME, Lukashev AN, Drosten C. Aichi virus shedding in high concentrations in patients with acute diarrhea. Emerg Infect Dis. 2011;17:1544–1548. doi: 10.3201/eid1708.101556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh DY, Silva PA, Hauroeder B, Diedrich S, Cardoso DD, Schreier E. Molecular characterization of the first Aichi viruses isolated in Europe and in South America. Arch Virol. 2006;151:1199–1206. doi: 10.1007/s00705-005-0706-7. [DOI] [PubMed] [Google Scholar]
- 8.Sdiri-Loulizi K, Hassine M, Gharbi-Khelifi H, Sakly N, Chouchane S, Guediche MN, Pothier P, Aouni M, AmbertBalay K. Detection and genomic characterization of Aichi viruses in stool samples from children in Monastir, Tunisia. J Clin Microbiol. 2009;47:2275–2278. doi: 10.1128/JCM.00913-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Svraka S, Duizer E, Vennema H, de Bruin E, van der Veer B, Dorresteijn B, Koopmans M. Etiological role of viruses in outbreaks of acute gastroenteritis in The Netherlands from 1994 through 2005. J Clin Microbiol. 2007;45:1389–1394. doi: 10.1128/JCM.02305-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Guyader FS, Le Saux JC, Ambert-Balay K, Krol J, Serais O, Parnaudeau S, Giraudon H, Delmas G, Pommepuy M, Pothier P, Atmar RL. Aichi virus, norovirus, astrovirus, enterovirus, and rotavirus involved in clinical cases from a french oyster-related gastroenteritis outbreak. J Clin Microbiol. 2008;46:4011–4017. doi: 10.1128/JCM.01044-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pham NTK, Khamrin P, Nguyen TA, Kanti DS, Phan TG, Okitsu S, Ushijima H. Isolation and molecular characterization of Aichi viruses from fecal specimens collected in Japan, Bangladesh, Thailand, and Vietnam. J Clin Microbiol. 2007;45:2287–2288. doi: 10.1128/JCM.00525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamashita T, Adachi H, Hirose E, Nakamura N, Ito M, Yasui Y, Kobayashi S, Minagawa H. Molecular detection and nucleotide sequence analysis of a new Aichi virus closely related to canine kobuvirus in sewage samples. J Med Microbiol. 2014;63:715–720. doi: 10.1099/jmm.0.070987-0. [DOI] [PubMed] [Google Scholar]
- 13.Lodder WJ, Rutjes SA, Takumi K, Husman MR. Aichi virus in sewage and surface water, the Netherlands. Emerg Infect Dis. 2013;19:1222–1230. doi: 10.3201/eid1908.130312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alcala A, Vizzi E, Rodriguez-Diaz J, Zambrano J, Betacourt W, Liparandi F. Molecular detection and characterization of Aichi viruses in sewage-polluted waters of venezuela. Appl Environ Microbiol. 2010;76:4113–4115. doi: 10.1128/AEM.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitajima M, Haramoto E, Phanuwam C, Katayama H. Prevalance and genetic diversity of Aichi viruses in wastewater and river water in japan. Appl Environ Microbiol. 2011;77:2184–2187. doi: 10.1128/AEM.02328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitajima M, Gerba CP. Aichi virus 1: Enviromental Occurrence and Behavior. Pathogens. 2015;4:256–268. doi: 10.3390/pathogens4020256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park S, Lee S, Chung H, Park J, Park S, Kim J, Choi B, Joo Y, Kim C, Lee B. Development and verification of genetically diagnostic method for the detection of non-regulated viruses from water environment (I) Incheon: Water Supply and Sewerage Research Division Environmental Infrastructure Research Department National Institute of Environmental Research; 2016. [Google Scholar]
- 18.Lee S, Kim JH, Choi JY, Jang WC. Loop-mediated isothermal amplification assay to rapidly detect Wheat streak mosaic virus in quarantined plants. Plant Pathol J. 2015;31:438–440. doi: 10.5423/PPJ.NT.06.2015.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S, Kim H, Lee JY, Rho JY. Development of rapid and highly sensitive detection of Bean common mosaic necrosis virus in leguminous crops using loop-mediated isothermal amplification assay. J Virol Methods. 2017;249:117–120. doi: 10.1016/j.jviromet.2017.08.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.