Abstract
The micronutrient iron (Fe) is not only essential for plant survival and proliferation but also crucial for healthy human growth and development. Rice and wheat are the two leading staples globally; unfortunately, popular rice and wheat cultivars only have a minuscule amount of Fe content and mainly present in the outer bran layers. Unavailability of considerable Fe-rich rice and wheat germplasms limits the potential of conventional breeding to develop this micronutrient trait in both staples. Agronomic biofortification, defined as soil and foliar fertilizer application, has potential but remains quite challenging to improve grain Fe to the significant level. In contrast, recent accomplishments in genetic biofortification can help to develop Fe-enriched cereal grains to sustainably address the problem of “hidden hunger” when the roadmap from proof of concept to product and adoption can be achieved. Here, we highlight the different genetic biofortification strategies for rice and wheat and path to develop a product.
Keywords: biofortification, iron, rice, wheat, genes
Introduction
“Hidden hunger,” the cause of inadequate intake of key micronutrients, is a major problem globally affecting around 2 billion people worldwide, and 30–40% (GBD 2015 Disease and Injury Incidence and Prevalence Collaborators, 2016) of it is caused by iron (Fe) deficiency anemia (IDA). The people most vulnerable to IDA are women and children. IDA can hamper cognitive and physical development, reduce immunity, and enhance the risk of maternal and perinatal mortality. The breeding target to fulfill the 30% estimated average requirement (EAR) of woman and children recommended by the HarvestPlus program for Fe is 13 μg/g in polished rice or around five to sixfold increase of grain Fe in popular rice. While in wheat, it is 59 μg/g (dry weight) of Fe or around twofold (Bouis et al., 2011).
In developing and less developed countries, it is challenging to provide access to a more diverse diet that can ameliorate the micronutrient deficiency. Biofortification, the enhancement of bioavailable micronutrient in the edible parts of staple food by either conventional plant breeding, biotechnology techniques, or agronomic approaches can help to alleviate malnutrition in the regions where the main source of calories and micronutrients come from staples (Bouis and Saltzman, 2017).
Plant Fe Uptake and Translocation
Iron is an essential micronutrient in plants and is required frequently in various processes such as photosynthesis, respiration, or chlorophyll biosynthesis. Different strategies are known for the uptake of low soluble Fe(III) oxyhydrate from the rhizosphere in higher plants: (a) Strategy I (non-Graminaceae) implicating reduction of ferric Fe(III) chelate reduction at the root surface to allow absorption of ferrous Fe(II) at plasma membrane, (b) Strategy II (Graminaceae) is the chelation strategy involving mugineic acid (MA) biosynthesis and secretion, and (c) a combination of both (Connorton et al., 2017a).
Major key genes of uptake in Strategy I for dicots and non-grass species were identified from Arabidopsis, namely ferric-chelate reductase oxidase 2 (FRO2) (Robinson et al., 1999), Fe-regulated transporter 1 (IRT1) (Eide et al., 1996), and a large number of H+-ATPase (HA) genes that are responsible for proton and phenolic compound excretion into the rhizosphere to enhance the solubility of ferric ions (Kobayashi and Nishizawa, 2012).
Strategy II plants such as rice, wheat, barley, and maize secrete the high Fe affinity organic molecule phytosiderophore (PS), a MA family into the rhizosphere (Kobayashi et al., 2005; Suzuki et al., 2006; Borrill et al., 2014; Bashir et al., 2017). The basic scheme for genes involved in Fe homeostasis in rice as the model system is presented in Figure 1. The uptake and translocation process in wheat is similar to rice and has been comprehensively reviewed (Borrill et al., 2014; Connorton et al., 2017a).
FIGURE 1.
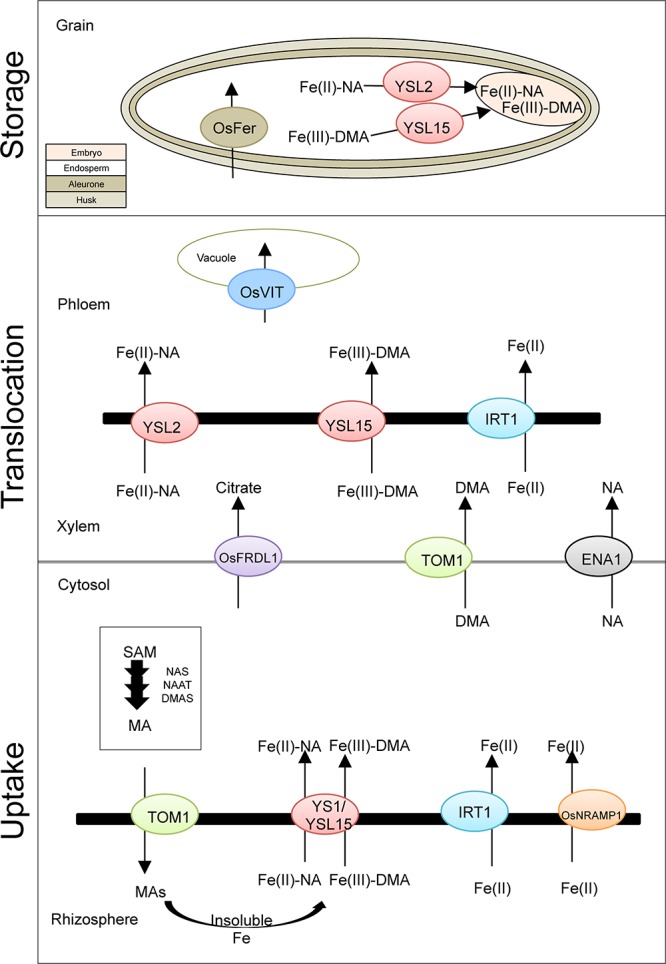
Basic scheme of iron uptake, translocation, and storage in rice. Presented are known genes involved in iron uptake and transport within the rice plant. TOM1, transporter of mugineic acid family phytosiderophores 1; YS1, yellow strip1; YSL15, yellow stripe 1-like; IRT1, iron-regulated transporter; OsNRAMP1, natural resistance-associated macrophage protein 1; OsFRDL1, ferric reductase defective like1; ENA, efflux transporter of nicotianamine; OsVIT, vacuolar iron transporter; OsFER, ferritin; MA, mugineic acid; DMA, 2′-deoxymugineic acid; NA, nicotianamine; SAM, S-adenosyl-L-methionine; NAS, nicotianamine synthase; NAAT, nicotianamine aminotransferase; DMAS, deoxymugineic acid synthase.
The synthesis of MA is a conserved pathway starting from S-adenosyl-L-methionine and covers consecutive enzymatic reactions of nicotianamine synthase (NAS), nicotianamine aminotransferase (NAAT), and deoxymugineic acid synthase (DMAS) genes, producing the precursor of nine known types of MAs: 2′-deoxymugineic acid (DMA) (Higuchi et al., 1999; Takahashi et al., 1999; Bashir et al., 2006; Kobayashi and Nishizawa, 2012). In rice, the expression of these genes is highly affected by the level of Fe availability in the soil; most genes were identified in response to Fe deficiency. NAS is localized on the membrane of vesicles in root cells, while NAAT is within these vesicles, proposing the location of the MA biosynthesis. Interestingly, a diurnal pattern is known for the secretion of MA, with its peak in the morning hours (Takagi et al., 1984; Suzuki et al., 2006). The secretion of MA into the rhizosphere is facilitated by the expression of TOM1 (transporter of MA family PSs). TOM1 was first identified in rice and barley and belongs to the major facilitator superfamily (MFS), which is one of the largest group of membrane transport proteins that promote the transit of substrates across cell membranes in response to chemiosmotic ion gradient (Nozoye et al., 2011). Fe–MA complexes are then built and taken up by YELLOW STRIPE 1 (YS1) and YELLOW STRIPE 1-like (YSL1) transporter into root cells (Curie et al., 2001; Inoue et al., 2009). The Fe(III)–DMA complex absorbed in root cell cytosols is likely reduced by ascorbate and altered to be Fe(II)–NA. The cytosolic Fe(II)–NA then excreted to the xylem and created complexes predominantly with citrate or with DMA (Fe–DMA) and transport further (Yoneyama et al., 2015).
In addition to the strategy II Fe(III)–DMA complex and OsYSL15 transporter, rice carries a ferrous transporter (OsIRT1) in its genome, which allows the direct uptake of Fe(II). On the root surface, rice shows a low ferric-chelate reductase activity, indicating an adaptation to submerge and anaerobic conditions (Ishimaru et al., 2006). Other metal transporters were identified for both strategies like ZIP (zinc-regulated transporter, IRT-like protein) family or natural resistance-associated macrophage protein (NRAMP) (Guerinot, 2000; Lanquar et al., 2005; Calliatte et al., 2010).
A complex cascade involves in the Fe translocation in higher plants, including xylem and phloem loading/unloading, transport, and retranslocation within the plant from source to sink organ (Kim and Guerinot, 2007). To facilitate this translocation, different chelators such as citrate, MAs, and nicotianamine (NA) play a crucial role in symplast heavy metal homeostasis (Garcia-Oliveira et al., 2018). The cytosolic Fe(II)–NA then excreted to the xylem and created complexes predominantly with citrate or with DMA (Fe–DMA) and transport further in rice phloem while remaining to bind to DMA, citrate, and proteins (Yoneyama et al., 2015). FERRIC REDUCTASE DEFECTIVE LIKE 1 (OsFRDL1) in rice (Figure 1) encodes a citrate efflux transporter required for Fe translocation. This gene is primarily expressed in root pericycle cells. Knockout plants of OsFRDL1 display a mild defect in Fe homeostasis, which suggests alternative chelators for Fe xylem transport (Inoue et al., 2004; Yokosho et al., 2009).
The YSL family members (influx transporter) are involved in metal–NA chelate translocation (Curie et al., 2009). The rice YSL family consists of 18 members. The OsYSL2 transporter is responsible for the long-distance transfer into sink tissues like leaves and grains (Koike et al., 2004). It is a carrier of Fe(II)–NA, but not Fe(III)–MAs. The transport of Fe(III)–DMA is performed by OsYSL15, which is responsible for Fe root absorption and internal translocation for long distance as well as seedling growth. The Fe transporter OsYSL18 is specifically expressed in reproductive organ, such as the pollen and pollen tube, which suggests a specific role in fertilization. It also can be found in the phloem of laminar joints, indicating a part in phloem Fe transport (Kobayashi and Nishizawa, 2012). The ferrous transporter OsIRT1 and DMA effluxer TOM1 seem not only to be involved in Fe uptake but also in Fe translocation within the plant, due to its expression in vascular tissue in rice (Ishimaru et al., 2006; Nozoye et al., 2011). While the two efflux transporter of NA 1 (ENA1) and ENA2 were involved in NA transport (Nozoye et al., 2011).
Biofortification: Agronomic Vs./and Genetic in Rice and Wheat
Agronomic Biofortification
Cereal grain micronutrient content can be enriched through agronomic biofortification, which is a fertilizer-based application method to soil or to plant foliar (Cakmak and Kutman, 2017). Soil agronomic biofortification is easy and can be quite cost-effective (Garg et al., 2018). It is a short time solution, important to complement the genetic biofortification, particularly when the soil in the target region is limited to a readily available pool of micronutrient (Cakmak and Kutman, 2017). For rice, the main challenge is the translocation of the mineral from the vegetative part to the grain (Mabesa et al., 2013; Slamet-Loedin et al., 2015), since it is mostly grown in lowland irrigated areas where Fe is highly available.
Foliar feeding is established via plant tissue test or visual foliar. It is affected by endogenous (leaf anatomy), exogenous (pH, soil), as well as environmental factors. After foliar Fe application, it takes the plants 10–20 days to absorb 50% of the micronutrient (Alshall and El-Ramady, 2017). In contrast with the promising results of foliar fertilizer application to improve grain Zn in rice (Cakmak, 2008; Cakmak and Kutman, 2017), the increase of grain Fe using foliar fortification was modest. A similar phenomenon was observed in wheat, neither soil nor foliar applications in inorganic form (e.g., FeSO4) or chelated form (e.g., Fe–EDTA, Fe–EDDHA, or Fe–citrate) were reported to be effective for increasing grain Fe concentrations (Cakmak, 2008). A minor increase in grain Fe was observed with Fe-EDTA application and also nitrogen application in wheat (Cakmak and Kutman, 2017). Another study showed a foliar application of Fe–amino acid (Fe AA) modestly increased grain Fe concentration by 14.5% on average in rice and by 32.5% when 1% (w/v) NA was added (Yuan et al., 2012). A foliar application to reach a significant increase in grain Fe for biofortification remains challenging.
Genetic Biofortification
Efforts to enrich Fe in the rice grain by conventional breeding are constrained by the limited natural variation of polished grain Fe in rice germplasm. Over 20,000 different accessions of rice germplasm were tested and displayed a maximum of only 5–8 mg kg–1 in polished rice grains (Gregorio et al., 2000; Graham, 2003). To date none of the conventional breeding and molecular marker approaches reached grain Fe breeding target in rice and wheat to fulfill 30% EAR in women and children; therefore, this review focuses on the genetic biofortification through transgenic approaches.
To facilitate an efficient and targeted genetic biofortification in rice five key steps can be addressed: (a) enhanced uptake, (b) increase translocation to grain, (c) specialization of Fe storage toward endosperm, (d) decrease of anti-nutritions, and (e) increase of bioavailability (Mulualem, 2015). Both single approach or combination of multiple approaches have been applied in genetic biofortification. The first attempt to obtain an Fe-enriched rice was reported by Goto et al. (1999, Table 1), focusing on improving the Fe storage protein. An increase of grain Fe concentration was attained in brown rice (up to 38 μg/g) compared to the wildtype Japonica cv. Kitaake (∼14.3 μg/g) through ectopic overexpression (OE) of SoyFerH1 in the endosperm. Several studies followed using a similar approach of OE lines of SoyferH1, differing in the genetic background (Swarna, IR68144, BR29, IR64, M12) and usage of promoters (OsGluB1, OsGtbl, CluB1, GluB4, OsG1b) (Slamet-Loedin et al., 2015). In two attempts, an elevated Fe level in the polished rice (up to ∼9.2 or ∼7.6 μg/g) was obtained and stable over several generations (Khalekuzzaman et al., 2006; Oliva et al., 2014); however, in particular case, there was no significant increase observed (Drakakaki et al., 2000). Not only was the soybean storage gene utilized to improve the Fe levels within the grain, but also the effect of overexpressing the rice ferritin gene (OsFer2) was analyzed. Higher Fe concentrations were found in T3 rice seeds (up to ∼15.9 μg/g) compared to the control Indica cv. Pusa-Sugandh II with ∼7 μg/g (Paul et al., 2012). Other studies focus on the genes for Fe uptake and translocation in the plant such as the development of OE lines of OsYSL15 (Lee et al., 2009a), responsible for the uptake of Fe(III)–DMA, and OsYSL2 (Ishimaru et al., 2010) for the uptake of Fe(II)–NA from the rhizosphere. The OsYSL2 OE study indicated a higher Fe content of ∼7.5 μg/g in polished rice compared to its counterpart (∼1.8 μg/g) in the T1 generation (Table 1). Only minimal elevated Fe concentrations were detected for OsYSL15 OE lines in T1 brown rice (Lee et al., 2009a), similar to the OE of OsYSL9 (Senoura et al., 2017). The alternative approach is to increase the expression of the NAS genes in rice, either by adding a 35S promoter enhancer in OsNAS3 and OsNAS2 gene through T-DNA activation tagging (Lee et al., 2009b, 2012), or by an endosperm OE of OsNAS1 (Zheng et al., 2010), or constitutive OE of OsNAS2 gene (Johnson et al., 2011). The endosperm OE of OsNAS1 resulted in 19 μg/g Fe level in brown rice compared to its wildtype (12 μ/g) and reduced to 5 mg/g after polishing (Zheng et al., 2010). Better results were achieved in the OE OsNAS2 plants with a Fe level-up to 19 μg/g in polished rice (Johnson et al., 2011) in comparison with the Nipponbare wildtype (4.5 μg/g), or in the OsNAS3 and OsNAS2 activation tag plants with 12 and 10 μg/g Fe in milled rice respectively compared to the wildtype (4 μg/g) (Lee et al., 2009b, 2012). A high Fe increase up to 55 μg/g was reported in the endosperm of japonica rice by OE of OsNAS1 and HvNAAT genes (Diaz-Benito et al., 2018). The Fe level is exceptionally high for the starchy endosperm suggesting either Fe contamination or the presence aleurone layer. The Fe level in the earlier generation was 18 μg/g (Banakar et al., 2017b) that already fulfill the nutritive target.
TABLE 1.
Review on transgenic approaches to develop iron-rich rice and wheat grains.
|
Iron ([c] in μg/g) |
Generation | Seed status | Growth condition | Cultivar | References | ||
| TG | WT | ||||||
| RICE | Genes involved in iron uptake and translocation | ||||||
| Rice gene overexpression | |||||||
| OsIRT1 | ∼12 | ∼10 | T3 | Brown | Paddy field | Japonica cv. Dongjin | Lee and An, 2009 |
| TOM1 | ∼18 | ∼15 | T1 | Brown | Hydroponic | Japonica cv. Tsukinohikari | Nozoye et al., 2011 |
| OsYSL15 | ∼14 | ∼12 | T1 | Brown | Paddy field | Japonica cv. Dongjin | Lee et al., 2009a |
| OsNAS1 | up to ∼19 | ∼12 | Brown | Field | Japonica cv. Xiushui 110 | Zheng et al., 2010 | |
| OsNAS2 | ∼10 | ∼4 | Milled | Greenhouse | Japonica cv. Dongjin | Lee et al., 2012 | |
| OsNAS3 | ∼12 | ∼4 | T1 | Milled | Greenhouse | Oryza sativa L. (cv EYI 105) | Lee et al., 2009b |
| OsYSL2 | ∼7.5 | ∼1.8 | T1 | Polished | Glasshouse | Japonica cv. Tsukinohikari | Ishimaru et al., 2010 |
| OsNAS1, OsNAS2, OSNAS3 | up to ∼19 | ∼4.5 | T1 | Polished | Glasshouse | Japonica cv. Nipponbare | Johnson et al., 2011 |
| OsNAS1 | up to ∼40 | ∼20 | T4 | Endosperm | Greenhouse | Oryza sativa L. (cv EYI 105) | Diaz-Benito et al., 2018 |
| OsYSL13 | ∼15 | ∼11 | Brown | Greenhouse | Japonica cv. Zhonghua 11 | Zhang et al., 2018 | |
| Overexpression of gene of different species | |||||||
| HvNAS1 | ∼8.5 | ∼4 | T2 | Polished | Greenhouse | Japonica cv. Tsukinohikari | Masuda et al., 2009 |
| HvYS1 | up to ∼9 | ∼4 | T2 | Polished | Oryza sativa L. (cv EYI 105) | Banakar et al., 2017a | |
| AtIRT1 | up to ∼4.86 | ∼2.28 | T3 | Polished | Greenhouse | Japonica cv. Taipei 309 | Boonyaves et al., 2016 |
| Genes involved in storage | |||||||
| Rice gene overexpression | |||||||
| OsFer2 | up to ∼15.9 | ∼7 | T3 | Milled | Greenhouse | Indica cv. Pusa-Sugandh II | Paul et al., 2012 |
| Soybean gene overexpression | |||||||
| SoyferH1 | up to ∼38 | ∼14.3 | T1 | Brown | Greenhouse | Japonica cv. Kitaake | Goto et al., 1999 |
| SoyferH1 | up to ∼25 | ∼17 | T3–T6 | Brown | Greenhouse | Japonica cv. Kitaake | Qu et al., 2005 |
| SoyferH1 | ∼18 | ∼18 | T2 | Brown | Greenhouse | Indica cv. M12 | Drakakaki et al., 2000 |
| SoyferH1 | up to ∼37 | ∼10 | T2 | Milled | Screenhouse | Indica cv. IR68144 | Vasconcelos et al., 2003 |
| SoyferH1 | up to ∼16 | ∼6.75 | BC2F5 | Milled | Greenhouse | Indica cv. Swarna | Paul et al., 2014 |
| SoyferH1 | up to ∼9.2 | ∼3.8 | T3 | Polished | Greenhouse | Indica cv. BR29 | Khalekuzzaman et al., 2006 |
| SoyferH1 | up to ∼7.6 | ∼3.3 | T4 | Polished | Greenhouse | Indica cv. IR64 | Oliva et al., 2014 |
| Genes involved in iron deficiency response | |||||||
| Rice gene overexpression | |||||||
| OsIRO2 | up to ∼15.5 | ∼6 | T1 | Brown | Greenhouse | Japonica cv. Tsukinohikari | Ogo et al., 2011 |
| Genes involved in inter-cellular/intra-cellular transport and storage | |||||||
| Rice gene silencing/knock-down mutant | |||||||
| OsVIT1 | ∼26 | ∼20 | Brown | Paddy field | Japonica cv. Zhonghua 11 | Zhang et al., 2012 | |
| OsVIT2 | ∼28 | ∼20 | Brown | Paddy field | Japonica cv. Dongjin | Zhang et al., 2012 | |
| OsVIT2 | ∼8 | ∼5 | Polished | Japonica cv. Dongjin | Bashir et al., 2013 | ||
| OsYSL9 | up to ∼2.5 | 1 | Polished | Japonica cv. Tsukinohikari | Senoura et al., 2017 | ||
| OsDMAS1 | ∼5 | ∼5 | Polished | Japonica cv. Dongjin | Bashir et al., 2017 | ||
| Combined strategies | |||||||
| Multigene overexpression | |||||||
| PyFerritin+rgMT+phyA | ∼22 | ∼10 | T1 | Brown | Greenhouse | Japonica cv. Taipei 309 | Lucca et al., 2002 |
| OsYSL2+, SoyFerH2+, HvNAS1 | up to ∼4 | ∼1 | T3 | Polished | Paddy field | Japonica cv. Tsukinohikari | Masuda et al., 2012 |
| HvNAS1, HvNAS1+, HvNAAT, IDS3 | up to ∼7.3 | ∼5.8 | T1 | Polished | Paddy field | Japonica cv. Tsukinohikari | Suzuki et al., 2008 |
| HvNAS1+, OsYSL2, SoyFerH2+ | ∼6.3 (∼5.02) | ∼3.2 (∼1.46) | T1(T2) | Polished | Greenhouse | Tropical Japonica cv. Paw San Yin | Aung et al., 2013 |
| AtNAS1+, Pvferritin+, Afphytase | up to ∼7 | ∼1 | T1 | Polished | Hydroponic | Japonica cv. Taipei 309 | Wirth et al., 2009 |
| AtIRT1, PvFERRITIN, AtNAS1 | up to ∼10.46 | ∼2.7 | T2 | Polished | Greenhouse | Japonica cv. Nipponbare | Boonyaves et al., 2017 |
| GmFERRITIN, OsNAS2 | ∼15 | ∼2.5 | T3 | Polished | Field | Indica cv. IR64 | Trijatmiko et al., 2016 |
| AtNAS1, AtFRD3, PvFer | up to ∼11.08 | ∼2.05 | T3 | Polished | Greenhouse | Japonica cv. Nipponbare | Wu et al., 2018 |
| AtNAS1, PvFer, AtNRAMP3 | up to ∼13.65 | ∼2.72 | T2 | Polished | Greenhouse | Indica cv. IR64 | Wu et al., 2019 |
| AtNAS1, PvFER, ZmPSY, PaCRT1 | up to ∼6.02 | ∼1.82 | T3 | Polished | Greenhouse | Japonica cv. Nipponbare | Singh et al., 2017a |
| OsNAS1, HvNAATb | up to ∼55 | ∼20 | T4 | Endosperm | Greenhouse | Oryza sativa L. (cv EYI 105) | Diaz-Benito et al., 2018 |
| OsNAS1, HvHAATb | ∼up to 18 | ∼4 | T3 | Endosperm | Hydroponic | Oryza sativa L. (cv EYI 105) | Banakar et al., 2017b |
| WHEAT | |||||||
| Genes involved in iron uptake and translocation | |||||||
| Single gene over expression | |||||||
| OsNAS2 | up to ∼80 | 40 | Whole grain | Field | Triticum aestivum (cv Bob White) | Beasley et al., 2019 | |
| OsNAS2 | up to ∼22 | 14 | Flour | Field | Triticum aestivum (cv Bob White) | Beasley et al., 2019 | |
| Genes involved in storage | |||||||
| Ta FERRITIN | Up to ∼130 | 72 | T2 | Whole grain | Greenhouse | Triticum aestivum (cv Bob White) | Borg et al., 2012 |
| Ta FERRITIN | Average 88.5 | 70 | T2 | Flour | Greenhouse | Triticum aestivum (cv Bob White) | Borg et al., 2012 |
| Genes involved in inter-cellular/intra-cellular transport and storage | |||||||
| Ta VIT | Up to 20 | 10 | T1 | Flour | Connorton et al., 2017b | ||
| Combined strategy | |||||||
| Multigene overexpression | |||||||
| OsNAS2 +/or PvFERRITIN | up to ∼93.1 | 42.7 | T4 | Whole grain | Greenhouse | Triticum aestivum (cv Bob White) | Singh et al., 2017b |
| OsNAS2, +/or PvFERRITIN | 53.3 | 21.4 | Flour | Greenhouse | Triticum aestivum (cv Bob White) | Singh et al., 2017b | |
A few studies use the silencing approach to reduce the Fe content in rice grain. Zhang et al. (2012) developed vascular Fe transport (OsVIT) silencing lines, aiming to interrupt the transport of Fe into the flag leaves. A comparable approach was used by Bashir et al. (2013), achieving a Fe level of ∼8 μg/g in polished rice. Only one gene involved in the Fe deficiency response was used to increase the Fe concentration till now. The constitutive expression of OsIRO2 resulted in ∼15.5 μg/g Fe in T1 brown rice seeds in comparison with the wildtype (∼6 μg/g) (Ogo et al., 2011).
The most promising results for Fe-enriched rice grains in tropical Indica rice were developed by Trijatmiko et al. (2016) and Wu et al. (2019) by multigene OE. By expressing the endosperm storage gene PvFER, the chelator AtNAS1 gene and an intracellular iron stores AtNRAMP3 in one cassette, the level of 13.65 μg/g iron was reached in the greenhouse condition (Wu et al., 2019). A slightly higher level of Fe of 15 μg/g concentration in polished grain Fe coupled with high Zn was shown in the results of Trijatmiko et al. (2016) in two field trials (Table 1; Trijatmiko et al., 2016). Generally, the field condition resulted in a lower grain Fe compared to the glasshouse setting (Masuda et al., 2012). These reported studies serve as a proof of concept for the potential product of Fe-biofortified rice.
In wheat, using the marker-assisted breeding a modest increase in Fe by 18% was observed in near-isogenic lines of NAM-B1 (Distelfeld et al., 2007). The first transgenic approach in wheat reported a significant increase in grain Fe achieved by the OE of the wheat FERRITIN gene (Borg et al., 2012; Table 1). This OE of TaFer1-A gene increased to 50–85% higher Fe content in wheat grains; however, the TaFer1-A genes presence was not stable over the generations (Borg, personal communication). The recent report (Singh et al., 2017b) showed significant progress that OE of OsNAS2 produced up to 93.1 μg/g of Fe in the wheat grain in the greenhouse, it surpasses the recommended nutritive breeding target. While using VACUOLAR IRON TRANSPORTER (TaVIT), a double of Fe concentration was obtained in flour but not in whole grain (Connorton et al., 2017b). Another recent study OE of OsNAS2 in wheat (Beasley et al., 2019) reported up to 80 μg/g of Fe concentration in wheat grain under field condition.
Potential of Genome Editing Approaches
A recent development in the genome editing tool, Clustered Regulatory Interspaced Palindromic Repeats (CRISPR) for precise modification within the genome, gives researchers a possibility for accurate targeting of genes or genomic regions. This technology has been used in rice to improve yield and stress resistance (Jaganathan et al., 2018; Mishra et al., 2018). The potential example to use CRISPR-based approach is to knockdown OsVIT2 to achieve the increase of grain Fe, similar to the published T-DNA insertion silencing of this gene (Bashir et al., 2013) in different rice cultivars. The development of Fe-enriched rice and wheat grains can also benefit from this method by tweaking the expression of genes involved in Fe homeostasis by editing the regulatory element of Fe homeostasis genes.
Trait Selection and Post-Harvest Effect on Iron and Bioavailability
The accuracy of grain Fe measurement requires consistent preparation, contamination free, and a standardized processing method to reach a consistent milling degree to select the product. The most accurate technique for Fe measurement is ICP-OES (Elzain et al., 2016).
Unpolished brown rice and wheat have a higher concentration and variation of Fe in the aleurone layers, wherein most of the Fe in rice and wheat is stored. Fe in cereal is stored in vacuoles in complex with phytate (Borg et al., 2012), and localized in the aleurone layer and embryo parts. Minimal phytate amount has been detected in the endosperm (Saenchai et al., 2016). Depending on the genotype, the rice bran can have one to five aleurone layers (Del Rosario et al., 1968). Unfortunately, dietary factors that inhibit cereal Fe absorption in human include phytic acid (myoinositol hexaphosphate) together with polyphenols (Gibson et al., 2010; Moretti et al., 2014), tannins, fiber, or hemagglutinins (Welch and Graham, 2004; Holme et al., 2012). Fe in cereal bran is chelated by phytic acid and forms an insoluble complex in the gastrointestinal tract (Iqbal et al., 1994). Rice is predominantly consumed in polished form, while wheat consumption is mixed between brown and white flour. To effectively reduce the “hidden hunger,” it is crucial that the added biofortified grain Fe is bioavailable.
Iron bound to ferritin is most likely to be highly available (Lönnerdal et al., 2006). NA has also been suggested as a Fe bioavailability enhancer in rice (Zheng et al., 2010). The correlation of the bioavailability of grain Fe with the presence of NA and ferritin was shown in rice (Trijatmiko et al., 2016). Recently in wheat, the OE lines of OsNAS2 show that the endosperm Fe is not co-localized with phosphorous but likely with NA (Beasley et al., 2019).
Pathway of a Genetic Biofortification: Proof of Concept to a Product
The path from proof of concept to the product started from the development of commercial event amenable for deregulation, the trait and agronomic testing phase in multiple confined fields, the regulatory science phase, and varietal registration phase (McDougall, 2011). To pass the regulatory requirement, GM rice needs to be screened for single locus insertion containing one or more copies and characterized at the molecular level of the actual insert, the absence of vector backbone or other unintended change, and heritability and followed by comprehensive food and environmental safety studies to prove product safety (Heck et al., 2005). In the biofortification study by Trijatmiko et al. (2016), the highest concentrations were obtained in the lines having two copies of transgenes in one insertion locus. Insertion of more than one copy of a transgene does not impede event deregulation as has been shown in some commercially released crops events (Canola-23-18-17, Soya-GU262) with multiple copies (Health Canada, 1999; ISAAA, 2019).
Collection of a comprehensive safety data of food and feed use of Fe-biofortified rice to develop a robust safety regulatory dossier is crucial. In most countries, the assessment follows the Codex Alimentarius guideline, which requires the description of the donor organism; genetic modification and its characterization; and assessment of the possible toxicity, allergenicity, and compositional analysis (FAO, 2003). For the environmental evaluation of nutrition trait, the necessary assessment could be to monitor the relevant insect pest population and change in seed vigor to assess the weediness potential.
Development and submission of a robust biosafety dossier, an excellent agronomic performance, enhancement of Fe bioavailability, and bioefficacy (Bouis and Saltzman, 2017) within the local context of the staple food processing, cooking, and eating habit, and a strong support of the seed sector and health sector in target countries will lay a path for deployment and adoption of Fe-enriched rice. Significant progress in genetic biofortification in recent years can provide a sustainable food-based solution to complement other interventions to reduce iron deficiency anemia in the target communities.
Author Contributions
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Russell Reinke, the IRRI healthier rice project leader, Joe Tohme, The HarvestPlus biotech project coordinator, and Lawrence Kent, the BMGF program officer for Healthier rice for their support.
Footnotes
Funding. This work was supported by the HarvestPlus – title: Developing high-iron and high-zinc transgenic rice to alleviate iron deficiency in Bangladesh and Southeast Asia Agreement No. 2015H5315 and the BMGF Healthier Rice Project.
References
- Alshall T., El-Ramady H. (2017). Foliar application: from plant nutrition to biofortification. Environ. Biodiver. Soil Secur. 1 71–83. 10.21608/jenvbs.2017.1089.1006 [DOI] [Google Scholar]
- Aung M. S., Masuda H., Kobayashi T., Nakanishi H., Yamakawa T., Nishizawa N. K. (2013). Iron biofortification of myanmar rice. Front. Plant Sci. 4:158. 10.3389/fpls.2013.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banakar R., Alvarez-Fernandez A., Abadia J., Capell T., Christou P. (2017a). The expression of heterologous Fe (III) phytosiderophore transporter HvYS1 in rice increases Fe uptake, translocation and seed loading and excludes heavy metals by selective Fe transport. Plant Biotechnol. J. 15 423–432. 10.1111/pbi.12637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banakar R., Alvarez-Fernandez A., Diaz-Benito P., Abadia J., Capell T., Christou P. (2017b). Phytosiderophores determine thresholds for iron and zinc accumulation in biofortified rice endosperm while inhibiting the accumulation of cadmium. J. Exp. Bot. 68 4983–4995. 10.1093/jxb/erx304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir K., Inoue H., Nagasaka S., Takahashi M., Nakanishi H., Mori S., et al. (2006). Cloning and characterization of deoxymugineic acid synthase genes from graminaceous plants. J. Biol. Chem. 281 32395–32402. 10.1074/jbc.m604133200 [DOI] [PubMed] [Google Scholar]
- Bashir K., Nozoye T., Nagasaka S., Rasheed S., Miyauchi N., Seki M., et al. (2017). Paralogs and mutants show that one DMA synthase functions in iron homeostasis in rice. J. Exp. Bot. 68 1785–1795. 10.1093/jxb/erx065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir K., Takahashi R., Akhtar S., Ishimaru Y., Nakanishi H., Nishizawa N. K. (2013). The knockdown of OsVIT2 and MIT affects iron localization in rice seed. Rice 6:31. 10.1186/1939-8433-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley J. T., Bonneau J. P., Sanchez-Palacios J. T., Moreno-Moyano L. T., Callahan D. L., Tako E., et al. (2019). Metabolic engineering of bread wheat improves grain iron concentration and bioavailability. Plant Biotechnol. J. 10.1111/pbi.13074 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyaves K., Gruissem W., Bhullar N. K. (2016). NOD promoter-controlled AtIRT1 expression functions synergistically with NAS and FERRITIN genes to increase iron in rice grains. Plant Mol. Biol. 90 207–215. 10.1007/s11103-015-0404-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyaves K., Wu T.-Y., Gruissem W., Bhullar N. K. (2017). Enhanced Grain iron levels in rice expressing an iron-regulated metal transporter, nicotianamine synthase, and ferritin gene cassette. Front. Plant Sci. 8:130. 10.3389/fpls.2017.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg S., Brinch-Pedersen H., Tauris B., Madsen L. H., Darbani B., Noeparvar S., et al. (2012). Wheat ferritins: improving the iron content of the wheat grain. J. Cereal Sci. 56 204–213. 10.1016/J.Jcs.2012.03.005 [DOI] [Google Scholar]
- Borrill P., Connorton J. M., Balk J., Miller A. J., Sanders D., Uauy C. (2014). Biofortification of wheat grain with iron and zinc: integrating novel genomic resources and knowledge from model crops. Front. Plant Sci. 5:53. 10.3389/fpls.2014.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouis H. E., Hotz C., McClafferty B., Meenakshi J. V., Pfeiffer W. H. (2011). Biofortification: a new tool to reduce micronutrient malnutrient. Food Nutr. Bull. 32 S31–S40. 10.1021/es010549d [DOI] [PubMed] [Google Scholar]
- Bouis H. E., Saltzman A. (2017). Improving nutrition through biofortification: a review of evidence from HarvestPlus, 2003 through 2016. Glob. Food Secur. 12 49–58. 10.1016/j.gfs.2017.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmak I. (2008). Enrichment of cereal grains with zinc: Agronomic or genetic biofortification? Plant Soil 302 1–17. 10.1007/s11104-007-9466-3 [DOI] [Google Scholar]
- Cakmak I., Kutman U. B. (2017). Agronomic biofortification of cereals with zinc: a review. Eur. J. Soil Sci. 69 172–180. 10.1111/ejss.12437 [DOI] [Google Scholar]
- Calliatte R., Schikora A., Briat J. F., Mari S., Curie C. (2010). High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell 22 904–917. 10.1105/tpc.109.073023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connorton J. M., Balk J., Rodriguez-Celma J. (2017a). Iron homeostasis in plants – a brief overview. Metallomics 9 813–823. 10.1039/c7mt00136c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connorton J. M., Jones E. R., Rodríguez-Ramiro I., Fairweather-Tait S., Uauy C., Balk J. (2017b). Wheat vacuolar iron transporter TaVIT2 transports Fe and Mn and is effective for biofortification. Plant Physiol. 174 2434–2444. 10.1104/pp.17.00672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C., Cassin G., Couch D., Divol F., Higuchi K., Le Jean M, et al. (2009). Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporter. Ann. Bot. 103 1–11. 10.1093/aob/mcn207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C., Panaviene Z., Loulergue C., Dellaporta S. L., Briat J. F., Walker E. L. (2001). Maize yellow stripe 1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409 346–349. 10.1038/35053080 [DOI] [PubMed] [Google Scholar]
- Del Rosario A. R., Briones V. P., Vidal A. J., Juliano B. O. (1968). Composition and endosperm structure of developing and mature rice kernel. Cereal Chem. 45 225–235. [Google Scholar]
- Diaz-Benito P., Banakar R., Rodriguez-Menendez S., Capell T., Pereiro R., Christou P., et al. (2018). Iron and zinc in the embryo and endosperm of rice (Oryza sativa L.) seeds in contrasting 2’-deoxymugineic acid/ nicotianamine scenarios. Front. Plant Sci. 9:1190. 10.3389/fpls.2018.01190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A., Cakmak I., Peleg Z., Ozturk L., Yazici A. M., Budak H., et al. (2007). Multiple QTL-effects of wheat Gpc-B1 locus on grain protein and micronutrient concentrations. Physiol. Plant 129 635–643. 10.1111/j.1399-3054.2006.00841.x [DOI] [Google Scholar]
- Drakakaki G., Christou P., Stöger E. (2000). Constitutive expression of soybean ferritin cDNA in transgenic wheat and rice results in increased iron levels in vegetative tissues but not in seeds. Transgenic Res. 9 445–452. 10.1023/A:1026534009483 [DOI] [PubMed] [Google Scholar]
- Eide D., Broderius M., Fett J., Guerinot M. L. (1996). A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. U.S.A. 93 5624–5628. 10.1073/pnas.93.11.5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzain A. H., Ebrahim A. M., Ali Eltoum M. S. (2016). Comparison between XRF, PIXE and ICP-OES Techniques Applied For Analysis of Some Medicinal Plants. IOSR-JAC 9 6–12. 10.9790/5736-0904010612 [DOI] [Google Scholar]
- FAO (2003). Guideline for the Conduct of Food Safety Assessment of Foods Derived From Recombinant-DNA Plants. Available at: http://www.fao.org/fileadmin/user_upload/gmfp/docs/CAC.GL_45_2003.pdf (accessed June 12, 2019). [Google Scholar]
- Garcia-Oliveira A. L., Chander S., Ortiz R., Menkir A., Gedil M. (2018). Genetic basis and breeding perspectives of grain iron and zinc enrichment in cereals. Front. Plant Sci. 9:937. 10.3389/fpls.2018.00937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg M., Sharma N., Sharma S., Kapoor P., Kumar A., Chunduri V., et al. (2018). Biofortified crops generated by breeding, agronomy, and transgenic approaches are improving lives of millions of people around the world. Front. Nutr. 5:12. 10.3389/fnut.2018.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD 2015 Disease and Injury Incidence and Prevalence Collaborators (2016). Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388 1545–1602. 10.1016/S0140-6736(16)31678-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson R. S., Bailey K. B., Gibbs M., Ferguson E. L. (2010). A review of phytate, iron, zinc, and calcium concentrations in plant-based complementary foods used in low-income countries and implications for bioavailability. Food Nutr. Bull. 31(2 Suppl.), S134–S146. 10.1177/15648265100312S206 [DOI] [PubMed] [Google Scholar]
- Goto F., Yoshihara T., Shigemoto N., Toki S., Takaiwa F. (1999). Iron fortification of rice seed by the soybean ferritin gene. Nat. Biotechnol. 17 282–286. 10.1038/7029 [DOI] [PubMed] [Google Scholar]
- Graham R. D. (2003). Biofortification: a global challenge. Int. Rice Res. Notes 28 4–8. [Google Scholar]
- Gregorio G. B., Senadhira D., Htut H., Graham R. D. (2000). Breeding for trace mineral density in rice. Food Nutr. Bull. 21 382–386. 10.1177/156482650002100407 [DOI] [Google Scholar]
- Guerinot M. L. (2000). The ZIP family of metal transporters. Biochim. Biophys. Acta 1465 190–198. 10.1016/s0005-2736(00)00138-3 [DOI] [PubMed] [Google Scholar]
- Health Canada (1999). ARCHIVED - High Lauric Acid Canola Lines 23-198, 23-18-17. Available at: https://www.canada.ca/en/health-canada/services/food-nutrition/genetically-modified-foods-other-novel-foods/approved-products/high-lauric-acid-canola-lines-23-198-23-18-17.html (accessed October, 1999). [Google Scholar]
- Heck G. R., Armstrong C. L., Astwood J. D., Behr C. F., Bookout J. T., Brown S. M., et al. (2005). Development and characterization of a CP4 EPSPS-based, glyphosate-tolerant corn event. Crop Sci. 44 329–339. 10.2135/cropsci2005.0329 [DOI] [Google Scholar]
- Higuchi K., Suzuki K., Nakanishi H., Yamaguchi H., Nishizawa N. K., Mori S. (1999). Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol. 119 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holme I. B., Dioisio G., Brinch-Pedersen H., Wendt T., Madsen C. K., Vincze E., et al. (2012). Cisgenic barley with improved phytase activity. Plant Biotechnol. J. 10 237–247. 10.1111/j.1467-7652.2011.00660.x [DOI] [PubMed] [Google Scholar]
- Inoue H., Kobayashi T., Nozoye T., Takahashi M., Kakei Y., Suzuki K., et al. (2009). Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J. Biol. Chem. 284 3470–3479. 10.1074/jbc.M806042200 [DOI] [PubMed] [Google Scholar]
- Inoue H., Suzuji M., Takahashi M., Nakanishi H., Mori S., Nishizawa N. K. (2004). A rice FRD3-like (OsFRDL1) gene is expressed in the cells involved in long-distance transport. Soil Sci. Plant Nutr. 50 1133–1140. 10.1080/00380768.2004.10408586 [DOI] [Google Scholar]
- Iqbal T. H., Lewis K. O., Copper B. T. (1994). Phytase activity in the human and rat small intestine. Gut 35 1233–1236. 10.1136/gut.35.9.1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISAAA (2019). International Service for the Acquisition of Agri-Biotech Applications. http://www.isaaa.org/gmapprovaldatabase/event/default.asp?EventID=164 [Google Scholar]
- Ishimaru Y., Masuda H., Bashir K., Inoue H., Tsukamoto T., Takahashi M., et al. (2010). Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 62 379–390. 10.1111/j.1365-313X.2010.04158.x [DOI] [PubMed] [Google Scholar]
- Ishimaru Y., Suzuki M., Tsukamoto T., Suzuki K., Nakazono M., Kobayashi T., et al. (2006). Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 45 335–346. 10.1111/j.1365-313x.2005.02624.x [DOI] [PubMed] [Google Scholar]
- Jaganathan D., Ramasamy K., Sellamuthu G., Jayabalan S., Venkataraman G. (2018). CRISPR for crop improvement: an update review. Front. Plant Sci. 9:985. 10.3389/fpls.2018.00985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. A. T., Kyriacou B., Callahan D. L., Carruthers L., Stangoulis J., Lombi E., et al. (2011). Constitutive overexpression of OsNAS gene family reveals single-gene strategies for effective iron- and zinc- biofortification of rice endosperm. PLoS One 6:e24476. 10.1371/journal.pone.0024476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalekuzzaman M., Datta K., Oliva N., Alam M. F., Joarder I., Datta S. K. (2006). Stable integration, expression and inheritance of the ferritin gene in transgenic elite indica rice cultivar BR29 with enhanced iron level in the endosperm. Indian J. 5 26–31. [Google Scholar]
- Kim S. A., Guerinot M. L. (2007). Mining iron: iron uptake and transport in plants. FEBS Lett. 581 2273–2280. 10.1016/j.febslet.2007.04.043 [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Nishizawa N. K. (2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63 131–152. 10.1146/annurev-arplant-042811-105522 [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Suzuki M., Inoue H., Itai R. N., Takahashi M., Nakanishi H., et al. (2005). Expression of iron-acquisition-related genes in iron-deficient rice is co-ordinately induced by partially conserved iron-deficiency-responsive elements. J. Exp. Bot. 56 1305–1316. 10.1093/jxb/eri131 [DOI] [PubMed] [Google Scholar]
- Koike S., Inoue H., Mizuno D., Takahashi M., Nakanishi H., Mori S., et al. (2004). OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in phloem. Plant J. 39 415–424. 10.1111/j.1365-313x.2004.02146.x [DOI] [PubMed] [Google Scholar]
- Lanquar V., Lelièvre F., Bolte S., Hamès C., Alcon C., Neumann D., et al. (2005). Mobilization of vacuolar iron by AtNRAMP3 and At NRAMP4 is essential for seed germination on low iron. EMBO J. 24 4041–4051. 10.1038/sj.emboj.7600864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., An G. (2009). Over-expression of OsIRT leads to increased iron and zinc accumulations in rice. Plant. Cell Environ. 32 408–416. 10.1111/j.1365-3040.2009.01935.x [DOI] [PubMed] [Google Scholar]
- Lee S., Chiecko J. C., Kim S. A., Walker E. L., Lee Y., Guerinot M. L., et al. (2009a). Disruption of OsYSL15 leads to iron inefficiency in rice plants. Plant Physiol. 150 786–800. 10.1104/pp.109.135418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Jeon U. S., Lee S. J., Kim Y.-K., Persson D. P., Husted S., et al. (2009b). Iron fortification of rice seeds through activation of the nicotianamine synthase gene. Proc. Natl. Acad. Sci. U.S.A. 106 22014–22019. 10.1073/pnas.0910950106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Kim Y.-S., Jeon U.-S., Kim Y.-K., Schjoerring J. K., An G. (2012). Activation of rice nicotianamine synthase 2 (OsNAS2) enhances iron availability for biofortification. Mol. Cells 33 269–275. 10.1007/s10059-012-2231-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lönnerdal B., Bryant A., Liu X., Theil E. C. (2006). Iron absorption from soybean ferritin in nonanemic women. Am. J. Clin. Nutr. 83 103–107. 10.1093/ajcn/83.1.103 [DOI] [PubMed] [Google Scholar]
- Lucca P., Hurrell R., Potrykus I. (2002). Fighting iron deficiency anemia with iron-rich rice. J. Am. Coll. Nutr. 21 184–190. 10.1080/07315724.2002.10719264 [DOI] [PubMed] [Google Scholar]
- Mabesa R. L., Impa S. M., Grewal D., Johnson-Beebout S. E. (2013). Contrasting grain-Zn response of biofortification rice (Oryza sativa L.) breeding lines to foliar Zn application. Field Crops Res. 149 223–233. 10.1016/j.fcr.2013.05.012 [DOI] [Google Scholar]
- Masuda H., Ishimaru Y., Aung M. S., Kobayashi T., Kakei Y., Takahashi M., et al. (2012). Iron biofortification in rice by the introduction of multiple genes involved in iron nutrition. Sci. Rep. 2 1–7. 10.1038/srep00543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H., Usuda K., Kobayashi T., Ishimaru Y., Kakei Y., Takahashi M., et al. (2009). Overexpression of the barley nicotianamine synthase gene HvNAS1 increases iron and zinc concentrations in rice grains. Rice 2 155–166. 10.1007/s12284-009-9031-9031 [DOI] [Google Scholar]
- McDougall A. (2011). The Cost and Time Involved in the Discovery, Development and Authorisation of a New Plant Biotechnology Derived Trait, a Consultancy Study for Crop Life International. Available at: https://croplife.org/wp-content/uploads/2014/04/Getting-a-Biotech-Crop-to-Market-Phillips-McDougall-Study.pdf (accessed April 14, 2015). [Google Scholar]
- Mishra R., Joshi R. K., Zhao K. (2018). Genome editing in rice: recent advances, challenges, and future implications. Front. Plant Sci. 9:1361. 10.33389/fpls.2018.01361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti D., Biebinger R., Bruins M. J., Hoeft B., Kraemer K. (2014). Bioavailability of iron, zinc, folic acid, and vitamin A from fortified maize. Ann. N. Y. Acad. Sci. 1312 54–65. 10.1111/nyas.12297 [DOI] [PubMed] [Google Scholar]
- Mulualem T. (2015). Application of bio-fortification through plant breeding to improve the value of staple crops. Biomed. Biotechnol. 3 11–19. 10.12691/bb-3-1-3 [DOI] [Google Scholar]
- Nozoye T., Nagaska S., Kobayashi T., Takahashi M., Sato Y., Sato Y., et al. (2011). Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J. Biol. Chem. 286 5446–5454. 10.1074/jbc.M110.180026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogo Y., Itai R. N., Kobayashi T., Aung M. S., Nakanishi H., Nishizawa N. K. (2011). OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol. Biol. 75 593–605. 10.1007/s11103-011-9752-6 [DOI] [PubMed] [Google Scholar]
- Oliva N., Chadha-Mohanty P., Poletti S., Abrigo E., Atienza G., Torrizo L., et al. (2014). Large-scale production and evaluation of marker-free indica rice IR64 expressing phytoferritin genes. Mol. Breed. 33 23–37. 10.1007/s11032-013-9931-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Ali N., Datta S. K., Datta K. (2014). Development of an iron-enriched high-yieldings indica rice cultivar by introgression of a high-iron trait from transgenic iron-biofortified rice. Plant Foods Hum. Nutr. 69 203–208. 10.1007/s11130-014-0431-z [DOI] [PubMed] [Google Scholar]
- Paul S., Ali N., Gayen D., Datta S. K., Datta K. (2012). Molecular breeding of Osfer2 gene to increase iron nutrition in rice grain. GM Crops Food 3 310–316. 10.4161/gmcr.22104 [DOI] [PubMed] [Google Scholar]
- Qu L. Q., Yoshihara T., Ooyama A., Goto F., Takaiwa F. (2005). Iron accumulation does not parallel the high expression level of ferritin in transgenic rice seeds. Planta 222 225–233. 10.1007/s00425-005-1530-8 [DOI] [PubMed] [Google Scholar]
- Robinson N. J., Procter C. M., Connolly E. L., Guerinot M. L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397 694–697. 10.1038/17800 [DOI] [PubMed] [Google Scholar]
- Saenchai C., Prom-u-thai C., Lordkaew S., Rouached H., Rerkasem B. (2016). Distribution of iron and Zn in plant and grain of different rice genotypes grown under aerobic and wetland conditions. J. Cereal Sci. 71 108–115. 10.1016/j.jcs.2016.08.007 [DOI] [Google Scholar]
- Senoura T., Sakashita E., Kobayashi T., Takahashi M., Aung M. S., Masuda H., et al. (2017). The iron-chelate transporter OsYSL9 plays a role in iron distribution in developing rice grains. Plant Mol. Biol. 95 375–387. 10.1007/s11103-017-0656-y [DOI] [PubMed] [Google Scholar]
- Singh S. P., Gruissem W., Bhullar N. K. (2017a). Single genetic locus improvement if iron, zinc and β-carotene content in rice grains. Sci. Rep. 7:6883. 10.1038/s41598-017-07198-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. P., Keller B., Gruissem W., Bhullar N. K. (2017b). Rice nicotianamine synthase 2 expression improves dietary iron and zinc levels in wheat. Theor. Appl. Genet 130 283–292. 10.1007/s00122-016-2808-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamet-Loedin I. H., Johnson-Beebout S. E., Impa S., Tsakirpaloglou N. (2015). Enriching rice with Zn and Fe while minimizing Cd risk. Front. Plant Sci. 6:121. 10.3389/fpls.2015.00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Morikawa K. C., Nakanishi H., Takahashi M., Saigusa M., Mori S., et al. (2008). Transgenic rice lines that include barley genes have increased tolerance to low iron availability in a calcareous paddy soil. Soil Sci. Plant Nutr. 54 77–85. 10.1111/j.1747-0765.2007.00205.x [DOI] [Google Scholar]
- Suzuki M., Takahashi M., Tsukamoto T., Watanabe S., Matsuhashi S., Yazaki J., et al. (2006). Biosynthesis and secretion of mugineic acid family phytosiderophores in zinc-deficient barely. Plant J. 48 85–97. 10.1111/j.1365-313x.2006.02853.x [DOI] [PubMed] [Google Scholar]
- Takagi S., Nomoto K., Takemoto S. (1984). Physiological aspect of mugineic acid, a possible phytosiderophore of graminaceous plants. J. Plant Nutr. 7 469–477. 10.1080/01904168409363213 [DOI] [Google Scholar]
- Takahashi M., Yamaguchi H., Nakanishi H., Shioiri T., Nishizawa N. K., Mori S. (1999). Cloning two genes for nicotianamine aminotransferase, a critical enzyme in iron acquisition (StrategyII) in graminaceous plants. Plant Physiol. 121 947–956. 10.1104/pp.121.3.947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trijatmiko K. R., Dueñas C., Tsakirpaloglou N., Torrizo L., Arines F. M., Adeva C., et al. (2016). Biofortified indica rice attains iron and zinc nutrition dietary targets in the field. Sci. Rep. 6:19792. 10.1038/srep19792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos M., Datta K., Oliva N., Khalekuzzaman M., Torrizo L., Krishnan S., et al. (2003). Enhanced iron and zinc accumulation in transgenic rice with the ferritin gene. Plant Sci. 164 371–378. 10.1016/S0168-9452(02)00421-1 23750162 [DOI] [Google Scholar]
- Welch R. M., Graham R. D. (2004). Breeding for micronutrients in staple food crops from human nutrition perspective. J. Exp. Bot. 55 353–365. 10.1093/jxb/erh064 [DOI] [PubMed] [Google Scholar]
- Wirth J., Poletti S., Aeschlimann B., Yakandawala N., Drosse B., Osorio S., et al. (2009). Rice endosperm iron biofortification by targeted and synergistic action of nicotianamine synthase and ferritin. Plant Biotechnol. J. 7 631–644. 10.111/j.1467-7652.2009.00430.x [DOI] [PubMed] [Google Scholar]
- Wu T.-Y., Gruissem W., Bhullar N. K. (2018). Facilitated citrate-dependent iron translocation increases rice endosperm iron and zinc concentrations. Plant Sci. 270 13–22. 10.1016/.plantsci.2018.02.002 [DOI] [PubMed] [Google Scholar]
- Wu T.-Y., Gruissem W., Bhullar N. K. (2019). Targeting intracellular transport combined with efficient uptake and storage significantly increases grain iron and zinc levels in rice. Plant Biotechnol. J. 17 9–20. 10.111/pbi.12943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosho K., Yamaji N., Ueno D., Mitani N., Ma J. F. (2009). OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiol. 149 297–305. 10.1104/pp.108.128132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama T., Ishikawa S., Fujimaki S. (2015). Route and regulation of zinc, cadmium, and iron transport in rice plants (Oryza sativa L.) during vegetative growth and grain filling: Metal transporters, metal speciation, grain Cd reduction and Zn and Fe biofortification. Int. J. Mol. Sci. 16 19111–19129. 10.3390/ijms160819111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Wu L., Yang C., Lv Q. (2012). Effects of iron and zinc foliar applications on rice plants and their grain accumulation and grain nutritional quality. J. Sci. Food Agric. 93 254–261. 10.1002/jsfa.5749 [DOI] [PubMed] [Google Scholar]
- Zhang C., Shinwari K. I., Luo L., Zheng L. (2018). OsYSL13 is involved in iron distribution in rice. Int. J. Mol. Sci. 19:3537. 10.3390/ijms19113537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xu Y.-H., Yi H.-Y., Gong J.-M. (2012). Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J. 72 400–410. 10.1111/j.1365-313X.2012.05088.x [DOI] [PubMed] [Google Scholar]
- Zheng L., Cheng Z., Ai C., Jiang X., Bei X., Zheng Y., et al. (2010). Nicotianamine, a novel enhancer of rice iron bioavailability to humans. PLoS One 5:e10190. 10.1371/journal.pone.0010190 [DOI] [PMC free article] [PubMed] [Google Scholar]


