Abstract
Molecular phylogenetic analyses have greatly advanced our understanding of phylogenetic relationships in Orobanchaceae, a model system to study parasitism in angiosperms. As members of this group may lack some genes widely used for phylogenetic analysis and exhibit varying degrees of accelerated base substitution in other genes, relationships among major clades identified previously remain contentious. To improve inferences of phylogenetic relationships in Orobanchaceae, we used two pentatricopeptide repeat (PPR) and three low-copy nuclear (LCN) genes, two of which have been developed for this study. Resolving power and level of support strongly differed among markers. Despite considerable incongruence among newly and previously sequenced markers, monophyly of major clades identified in previous studies was confirmed and, especially in analyses of concatenated data, strongly supported after the exclusion of a small group of East Asian genera (Pterygiella and Phtheirospermum) from the Euphrasia-Rhinanthus clade. The position of the Orobanche clade sister to all other parasitic Orobanchaceae may indicate that the shift to holoparasitism occurred early in the evolution of the family. Although well supported in analyses of concatenated data comprising ten loci (five newly and five previously sequenced), relationships among major clades, most prominently the Striga-Alectra clade, the Euphrasia-Rhinanthus clade, and the Castilleja-Pedicularis clade, were uncertain because of strongly supported incongruence also among well-resolving loci. Despite the limitations of using a few selected loci, congruence among markers with respect to circumscription of major clades of Orobanchaceae renders those frameworks for detailed, species-level, phylogenetic studies.
Keywords: low-copy nuclear genes, Orobanchaceae, parasitic plants, phylogeny, PPR genes
Introduction
Parasitic plants attach to other plants via a specialized organ, the haustorium, to obtain nutrients and water from their hosts (Kuijt, 1969). This renders parasitic plants of interest not only for plant scientists, who investigate structural, physiological, and molecular adaptations of parasitism (dePamphilis and Palmer, 1990; Cubero and Moreno, 1996; Joel et al., 2013) but also for farmers and applied scientists, because some parasitic plants are serious agricultural pests that can cause major yield losses (Parker and Riches, 1993). Within angiosperms, parasitism has evolved at least twelve times independently (Schneeweiss, 2013) and around 1% of all angiosperm species are parasitic plants, i.e., c. 4,500 species in about 20–30 families (Nickrent et al., 1998; Nickrent, 2019).
An excellent model system for studying the evolution of parasitism in plants is the family Orobanchaceae. Orobanchaceae is the largest parasitic family, comprising more than 2,000 species in about 90–115 genera (McNeal et al., 2013; Schneeweiss, 2013), and includes the full range of nutritional dependency from non-parasitic via photosynthetic parasitic (hemiparasitic) to non-photosynthetic parasitic (holoparasitic). Whereas parasitism has evolved only once in Orobanchaceae, the transition from hemi- to holoparasitism has occurred multiple times (Schneeweiss, 2013).
Molecular phylogenetic analyses have greatly advanced our understanding of phylogenetic relationships of Orobanchaceae. These have led to a greatly expanded circumscription of the family from the traditional Orobanchaceae, which previously comprised the exclusively holoparasitic Orobanche and a few related genera only (Beck-Mannagetta, 1930), to include all hemiparasites and the few holoparasites formerly placed in Scrophulariaceae (Young et al., 1999; Wolfe et al., 2005; Bennett and Mathews, 2006; McNeal et al., 2013). The sister group to parasitic Orobanchaceae is the Asian non-parasitic genus Lindenbergia, now commonly included in the thus-broadened Orobanchaceae (Young et al., 1999; Wolfe et al., 2005; Bennett and Mathews, 2006; Park et al., 2008; McNeal et al., 2013; but see Fischer, 2004). Only recently Rehmanniaceae (including two non-parasitic genera, Rehmannia and Triaenophora), the sister to Orobanchaceae (Albach et al., 2009; Xia et al., 2009), has been merged with Orobanchaceae as well (Angiosperm Phylogeny Group, 2016). The second major impact of molecular phylogenetic data concerns the identification of several major lineages within Orobanchaceae (dePamphilis et al., 1997; Wolfe and dePamphilis, 1998; Young et al., 1999; Bennett and Mathews, 2006; McNeal et al., 2013). These are the non-parasitic Lindenbergia clade; the small, hemiparasitic Cymbaria-Siphonostegia clade; the exclusively holoparasitic Orobanche clade; the exclusively hemiparasitic Castilleja-Pedicularis clade; the nearly exclusively hemiparasitic Euphrasia-Rhinanthus clade; the mainly tropical, mostly hemiparasitic Striga-Alectra clade; and the single genus Brandisia (Schneeweiss, 2013).
Despite these advances, our understanding of phylogenetic relationships within Orobanchaceae is hampered by two major shortcomings. The first is that about one third of the genera, especially those with tropical distributions, have not been studied yet using molecular phylogenetic tools. The second, which is the focus of this study, is that relationships among major clades were either poorly resolved (e.g., the position of Brandisia: Bennett and Mathews, 2006; McNeal et al., 2013) or suffered from, partly well supported, incongruent results from different markers (e.g., the position of Lindenbergia differed between two phytochrome genes: McNeal et al., 2013) or even from different data sets of the same marker (e.g., relationships among the Castilleja-Pedicularis clade, the Euphrasia-Rhinanthus clade, and the Striga-Alectra clade inferred from phytochrome A data were swapped in the study of McNeal et al., 2013, compared to that of Bennett and Mathews, 2006). These issues may be due to insufficient phylogenetic signal and/or marker-specific problems, such as substitution rate variation of plastid genes evolving under relaxed functional constraints (dePamphilis et al., 1997; Wolfe and dePamphilis, 1998; Wicke et al., 2013, 2016), paralogy issues in multi-copy genes such as ITS (Álvarez and Wendel, 2003) or in low-copy genes (Zimmer and Wen, 2012) such as phytochrome genes (Bennett and Mathews, 2006; McNeal et al., 2013). Evidently, additional nuclear low-copy markers, although no panacea for resolving all relationships, are needed to obtain a robust phylogenetic framework of Orobanchaceae.
A number of nuclear genes have recently been used to improve molecular phylogenetic analyses in plants. These include low-copy nuclear (LCN) Conserved Ortholog Set (COS) genes (Sang, 2002; Li M. et al., 2008; Duarte et al., 2010; Zhang et al., 2012; Zimmer and Wen, 2012, 2015; Babineau et al., 2013; Latvis et al., 2017) as well as multi-gene families, most notably pentatricopeptide repeat (PPR) genes (Yuan et al., 2009, 2010; Crowl et al., 2014). Members of the PPR protein family are sequence-specific RNA-binding proteins functioning in gene expression of chloroplasts and mitochondria (O’Toole et al., 2008; Barkan and Small, 2014), with over 400 members in the genomes of most plants sampled thus far (Yuan et al., 2010). Screening the model plants rice (Oryza sativa) and Arabidopsis thaliana, Yuan et al. (2010) found 127 PPR genes to be single copy, of which five were used to resolve phylogenetic relationships in selected Verbenaceae (Yuan et al., 2010). The applicability of LCN genes may decrease at deeper phylogenetic depth (e.g., of 274 LCN loci screened in Fabaceae by Choi et al., 2006, only ten markers were suitable at the family level), which may explain why beyond phytochrome genes (PHYA and PHYB) no LCN locus has been applied across the entire Orobanchaceae (but see Latvis et al., 2017, for a list of primers from a number of single-copy nuclear loci).
In this study, we analyze two PPR genes successfully applied in other angiosperms (Yuan et al., 2010) as well as three LCN loci, two newly established here, to infer phylogenetic relationships of major lineages within Orobanchaceae. We analyze these PPR and LCN loci both individually and jointly with previously used markers (plastid DNA, nuclear ITS, PHYA and PHYB). Specifically, we want to solve remaining uncertainties concerning (i) the unclear positions of Brandisia and the Cymbaria-Siphonostegia clade, (ii) the ambiguous support for monophyly of the Orobanche clade, and (iii) the contradicting relationships among the Castilleja-Pedicularis clade, the Euphrasia-Rhinanthus clade, and the Striga-Alectra clade inferred previously (Bennett and Mathews, 2006; McNeal et al., 2013). Additionally, we also want to assess the suitability of these markers at lower taxonomic levels using Odontites (from the Euphrasia-Rhinanthus clade), where recent phylogenetic work has revealed strong discrepancies among markers (Pinto-Carrasco et al., 2017; Gaudeul et al., 2018).
Materials and Methods
Plant Material
We included 56 species of 31 genera of Orobanchaceae (Table 1 and Supplementary Table S1). These taxa covered all major clades identified in previous studies (Bennett and Mathews, 2006; McNeal et al., 2013). Compared to McNeal et al. (2013), the most comprehensive phylogenetic study of Orobanchaceae to date, we have overall sparser taxon sampling, especially in the tropical Striga-Alectra clade and the Euphrasia-Pedicularis clade, but we include the following previously unsampled genera: Macrosyringion, Nothobartsia, Odontitella, Phtheirospermum (except Phtheirospermum japonicum), Rehmannia, and Triaenophora.
TABLE 1.
List of taxa and source of sequence information (for details see Supplementary Table S1).
| Taxon | AT1G04780 | AT1G14610 | Agt1 | AT2G37230 | AT1G09680 | PHYA | PHYB | ITS | matK | rps2 |
| Rehmannia-Triaenophora Clade | ||||||||||
| Rehmannia piasezkii | + | + | + | + | + | + | + | +GB | + | +GB |
| Triaenophora shennongjiaensis | + | + | + | + | + | + | +GB | + | +GB | |
| Lindenbergia Clade | ||||||||||
| Lindenbergia muraria | + | + | + | +GB | +GB | +GB | +GB | +GB | ||
| Lindenbergia philippensis | + | + | + | + | + | +GB | +GB | +GB | +GB | +GB |
| Cymbaria-Siphonostegia Clade | ||||||||||
| Bungea trifida | + | + | + | + | + | +GB | + | +GB | +GB | +GB |
| Schwalbea americana | + | + | + | + | + | +GB | +GB | +GB | +GB | +GB |
| Orobanche Clade | ||||||||||
| Boschniakia himalaica | + | + | + | +GB | +GB | +GB | +GB | +GB | ||
| Cistanche phelypaea | + | + | + | + | +GB | +GB | +GB | |||
| Cistanche tubulosa | + | + | + | + | +GB | +GB | +GB | |||
| Epifagus virginiana | + | + | + | + | +GB | +GB | +GB | +GB | +GB | |
| Orobanche caryophyllacea | + | + | + | + | + | +GB | +GB | +GB | ||
| Orobanche flava | + | + | + | + | + | + | + | +GB | + | +GB |
| Orobanche gracilis | + | + | + | + | +GB | + | +GB | +GB | +GB | |
| Orobanche lycoctoni | + | + | + | + | + | + | +GB | + | ||
| Phelipanche aegyptiaca | +PPGP | +PPGP | +PPGP | +PPGP | +PPGP | +GB | +GB | +GB | ||
| Phelipanche arenaria | + | + | +GB | +GB | ||||||
| Incertae sedis | ||||||||||
| Brandisia hancei | + | + | + | + | + | +GB | +GB | +GB | +GB | +GB |
| Pterygiella Clade | ||||||||||
| Phtheirospermum tenuisectum | + | + | + | + | +GB | +GB | + | |||
| Pterygiella cylindrica | + | + | + | + | + | + | + | +GB | +GB | + |
| Pterygiella duclouxii | + | + | + | + | + | + | + | +GB | +GB | + |
| Castilleja-Pedicularis Clade | ||||||||||
| Pedicularis aspleniifolia | + | + | + | + | + | +GB | ||||
| Pedicularis decora | + | + | + | + | +GB | +GB | ||||
| Pedicularis densispica | + | + | + | + | + | +GB | +GB | +GB | +GB | +GB |
| Pedicularis elwesii | + | + | + | + | + | + | +GB | +GB | +GB | +GB |
| Pedicularis lachnoglossa | + | + | + | + | + | +GB | +GB | |||
| Pedicularis rex | + | + | +GB | +GB | ||||||
| Pedicularis rostrato spicata | + | + | + | + | + | |||||
| Pedicularis verticillata | + | + | + | + | + | +GB | +GB | |||
| Triphysaria pusilla | +PPGP | +PPGP | +PPGP | +GB | +GB | +GB | ||||
| Triphysaria versicolor | +PPGP | +PPGP | +PPGP | +PPGP | +PPGP | +GB | +GB | +GB | ||
| Euphrasia-Rhinanthus Clade | ||||||||||
| Bellardia trixago | + | + | + | +GB | +GB | +GB | +GB | +GB | ||
| Euphrasia frigida | + | + | + | + | +GB | +GB | ||||
| Euphrasia sinuata | + | + | + | + | ||||||
| Euphrasia stricta | + | + | +GB | + | +GB | +GB | +GB | |||
| Lathraea squamaria | + | + | +GB | + | + | +GB | +GB | +GB | +GB | |
| Macrosyringion longiflorum | + | + | + | + | +GB | +GB | ||||
| Melampyrum sylvaticum | + | +GB | + | + | +GB | +GB | +GB | +GB | ||
| Nothobartsia asperrima | + | + | + | + | + | +GB | +GB | |||
| Odontitella virgata | + | + | + | + | + | +GB | +GB | |||
| Odontites bolligeri | + | + | + | + | + | +GB | +GB | |||
| Odontites cebennensis | + | + | + | + | +GB | +GB | ||||
| Odontites luteus | + | + | + | + | +GB | +GB | ||||
| Odontites vernus | + | + | + | + | +GB | +GB | ||||
| Odontites viscosus | + | + | + | + | +GB | +GB | ||||
| Parentucellia latifolia | + | + | + | + | +GB | +GB | +GB | +GB | +GB | |
| Parentucellia viscosa | + | +GB | +GB | +GB | +GB | +GB | ||||
| Rhinanthus alectorolophus | + | + | +GB | + | + | +GB | +GB | +GB | +GB | +GB |
| Striga-Alectra Clade | ||||||||||
| Aeginetia indica | + | +4 | +GB | +GB | +GB | |||||
| Buchnera americana | + | + | + | + | +GB | +GB | +GB | +GB | ||
| Buchnera hispida | + | + | +GB | +GB | + | |||||
| Radamaea montana | + | + | +4 | + | +GB | +GB | +GB | +GB | +GB | |
| Striga bilabiata | + | +1 | + | +3 | +GB | +GB | +GB | +GB | +GB | |
| Striga gesnerioides | + | +3 | + | + | +GB | +GB | +GB | +GB | +GB | |
| Striga hermonthica | +PPGP | +PPGP | +PPGP | +PPGP | +PPGP | +GB | +GB | +GB | ||
| Out-groups | ||||||||||
| Paulownia sp.1 | +1KP | +1KP | +1KP | +1KP | +1KP | +GB | +GB | +GB | +GB | |
| Mimulus guttatus | +PZ | +PZ | +PZ | +PZ | +PZ | +GB | +GB | +GB | ||
| Total number | 50 | 47 | 39 | 51 | 43 | 31 | 30 | 34 | 34 | 31 |
+Indicate sequences newly obtained in this study (superscript numbers indicate number of clones, where applicable); +GB Indicate sequences from previous studies obtained from GenBank; +PPGP indicate sequences (EST libraries or combined builds) obtained from the Parasitic Plant Genome Project (PPGP) database (available from: http://ppgp.huck.psu.edu/; assessed on Feb 27th 2017); +1KP indicate sequences from the 1KP database (http://www.onekp.com/public_data.html; assessed on Feb 27th 2017); +PZ sequences from Phytozome 12.1 database (https://phytozome.jgi.doe.gov/pz/portal.html#; assessed on Feb 27th 2017). 1Sequences from the 1KP project (+1KP) are from P. fargesii, the remaining ones (+GB) are from P. tomentosa.
Marker Development
Our goal was to establish several low-copy markers that amplify well (ideally without requiring any cloning) across the entire family Orobanchaceae. To this end, we tested both already published and newly developed markers. To retrieve homologous LCN genes from Orobanchaceae, we conducted a BLASTN search (as implemented on the Parasitic Plant Genome Project1) on genes from Arabidopsis that have been shown to be low-copy in Arabidopsis, Populus, Vitis, and Oryza (Duarte et al., 2010) against unigenes from four Orobanchaceae species [Lindenbergia philippensis, Phelipanche (Orobanche) aegyptiaca, Striga hermonthica, Triphysaria versicolor] available from the Parasitic Plant Genome Project2 (PPGP, Yang et al., 2015) using an e-value of e–10. Of the thus retrieved loci, the 200+ longest ones were retained and aligned separately using Muscle 3.8.31 (Edgar, 2004) available as web-service from EMBL-EBI (McWilliam et al., 2013). We chose two species, for which genomic data are available, as outgroups: Paulownia fargesii (Paulowniaceae, the sister-group to Orobanchaceae), whose transcriptome data are available from the 1000 Plants (1KP) project3 (see Matasci et al., 2014, for details on this project), and Erythranthe guttata (syn. Mimulus guttatus, Phrymaceae, sister-group to the clade of Orobanchaceae plus Paulowniaceae), whose genome is available from Phytozome 12.14 (see Hellsten et al., 2013, for details on an earlier version genome annotation). Alignments were edited manually in BioEdit 7.2.1 (Hall, 1999). Primers were designed in conserved regions using Primer Premier 5.0 (Premier Biosoft International, Palo Alto, CA, United States) requiring primer lengths of 15–30 bp, GC contents of 40–60%, melting temperatures of 55–75°C, and avoiding repetitive motifs, hairpins, and the potential for dimer formation.
DNA Extraction, PCR, and Sequencing
Total genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. We amplified five PPR genes and 90 LCN genes. Most of those, however, could only be amplified and sequenced with limited success. Specifically, 16 of the 90 LCN genes (14.4%) could be PCR amplified from three to 27 species across the family (Supplementary Table S2), but failed to amplify across the entire family. Five loci gave reliable PCR amplification from at least 30 species of Orobanchaceae. These were the LCN gene Agt1 using modified forward and reverse primers from Li M. et al. (2008), two LCN genes (AT1G04780 and AT1G14610) identified here and two PPR genes (AT1G09680 and AT2G37230) using primers from Yuan et al. (2010); the primers used (including internal ones, where necessary) are listed in Table 2.
TABLE 2.
Sequences of primers used in this study.
| Primer | Sequence | References |
| AT1G09680 | ||
| AT1G09680_180f | ACCRCCCTWTCTCAAGCCATCCAAA | Yuan et al. (2010) |
| AT1G09680_1760r | TARTCAAGAACAAGCCCTTTCGCAC | Yuan et al. (2010) |
| AT1G09680_850f | GTTAGTTTCAATACTTTGATGAA | Yuan et al. (2010) |
| AT1G09680_850r | TTCATCAAAGTATTGAAACTAAC | Yuan et al. (2010) |
| AT2G37230 | ||
| AT2G37230_320f | GCCTGGACDACMCGTTTRCAGAA | Yuan et al. (2010) |
| AT2G37230_1770r | TCRAACAAGCTCTCCATCAC | Yuan et al. (2010) |
| AT2G37230_1800r | GCYGTCTGAACWCSYCCATCYTC | Yuan et al. (2010) |
| AT2G37230_512f | GGCAACAARGTYGAGTAAG | This study |
| AT2G37230_1066r | GATGAGGATTTGTGGGT | This study |
| AT1G14610 | ||
| AT1G14610f | RAGGCTAGARGAKGGDAACT | This study |
| AT1G14610r | AAACTGCCACCAYGARTA | This study |
| AT1G04780 | ||
| AT1G04780f | CMCTTCATYTGGCTGTTA | This study |
| AT1G04780r | TCYGDCGAGTCCATYTTA | This study |
| AT1G04780r511 | GGAGMACCWGCACCATCCAA | This study |
| Agt1 | ||
| Agt1f_oro | GATTTCCGCATGGAYGARTGGGG | Modified from Li M. et al. (2008) |
| Agt1r_oro | CCAYTCCTCCTTCTGASTGCAGTT | Modified from Li M. et al. (2008) |
Primers yielding the longest amplicon are underlined, the remaining primers are internal primers.
Amplification was done in a volume of 15.8 μL containing 0.3 U of KAPA3G Plant DNA Polymerase (Peqlab, Vienna, Austria), 7 μL of 2× PCR buffer, 0.5 μL of 10 μM primers, 0.7 μL DNA, and 7 μL water. PCR conditions for LCN loci amplification were: denaturation for 4 min at 94°C; 35 cycles each with 30 s at 94°C, 30 s at 48°C, 1 min at 72°C; and final elongation for 10 min at 72°C. For the PPR loci we used the protocol of Yuan et al. (2010). For species not included in previous studies (Bennett and Mathews, 2006; McNeal et al., 2013), we also generated PHYA, PHYB, matk, and rps2 sequences using primers and PCR conditions described by Li et al. (2016). PCR products were purified using 0.5 μL Exonuclease I and 1 μL FastAP thermo sensitive alkaline phosphatase (Thermo Fisher Scientific, St. Leon-Rot, Germany) following the manufacturer’s protocol. A mixture of 5 μL of purified template, 2 μL trehalose, 1.5 μL sequencing buffer, 0.5 μL of primer (10 μM), and 1 μL BigDye Terminator (Applied Biosystems, Foster City, CA, United States) was used in cycle sequencing. Reactions were purified on Sephadex G-50 Fine (GE Healthcare Bio-Sciences, Uppsala, Sweden) and sequenced on an ABI 3730 DNA Analyzer capillary sequencer (Applied Biosystems). For a few species from the Striga-Alectra clade direct sequencing of AT1G14610 and AT2G37230 did not result in clean reads (these samples are indicated in Table 1), and these sequences were cloned. To this end, purified PCR products were run on an agarose gel and target bands were isolated using the Quick Gel Extraction Kit (Invitrogen, Vienna, Austria). All PCR products were ligated to vector pGEM-T (Zoman, Beijing, China) and then were transformed into DH5alpha competent E. coli. After blue white screening on LB medium, eight white colonies were checked by colony PCR, and at least three positive colonies were sequenced with primers M13F and M13R.
Phylogenetic Analyses
Sequences were assembled and edited using SeqMan II 5.05 (DNAStar Inc., Madison, United States). Initial alignments of individual loci were made with Muscle 3.8.31 (Edgar, 2004) using the web-service available from EMBL-EBI (McWilliam et al., 2013) and manually adjusted using BioEdit 7.2.1 (Hall, 1999). Parsimony-informative sites were calculated using PAUP* 4.0a163 (Swofford, 2002). These five loci were analyzed separately as well as concatenated into a matrix containing 56 species. Furthermore, we generated a concatenated alignment of 56 species by combining five loci in this study with five loci used by McNeal et al. (2013), i.e., PHYA, PHYB, ITS, matK, and rps2. For all analyses (single markers and concatenated data sets), the best-fit substitution models as well as partitioning schemes for DNA sequence alignments (considering codon positions and introns, where applicable, for each marker) were identified via the Akaike Information Criterion (AIC; Akaike, 1974) using PartitionFinder 1.1.0 (Lanfear et al., 2012), employing the greedy algorithm. We tested those 24 models that are implemented in MrBayes. Maximum likelihood analyses were conducted using RAxML 8.1 (Stamatakis, 2014), employing the fast bootstrap approach (Stamatakis et al., 2008) with 1,000 bootstrap replicates and the GTRGAMMA model. Bayesian inference was done using MrBayes 3.2.3 (Ronquist and Huelsenbeck, 2003) using the partitioning schemes and substitution models identified before (see data matrices available in dryad under doi: 10.5061/dryad.31cf160). Values for all parameters, such as the shape of the gamma distribution or the substitution rates, were estimated during the analysis. Partitions were allowed to evolve under different rates (ratepr = variable). We ran four cold Monte Carlo Markov (MCMC) chains simultaneously starting from different random starting trees for 10 million generations, and sampled trees every 5,000th generation. We used Tracer 1.4 (Rambaut et al., 2018) to check the stability of output parameters from Bayesian analyses (i.e., ESS values of at least 200). After combining 1,800 trees from each run (i.e., after discarding 10% trees as burn-in, when the MCMC chain had reached stationarity, evident from standard deviations of split variances being below 0.01), posterior probabilities were estimated.
Possible discrepancies among phylogenetic relationships inferred from different markers (five newly sequenced here, five taken from McNeal et al., 2013) were visualized using super networks (Huson et al., 2004) as implemented in SplitsTree 4 (Huson and Bryant, 2006). To this end, phylogenetic super networks were obtained from the five newly sequenced loci and from all ten loci, i.e., the five newly sequenced ones plus those used by McNeal et al. (2013), with default parameter settings.
The evolution of parasitism was reconstructed on the maximum likelihood tree from the combined 10 loci using maximum parsimony as implemented in Mesquite 3.51 (Maddison and Maddison, 2018). Under the assumption that holoparasitism (i.e., non-photosynthetic parasitism) can only evolve via hemiparasitism (i.e., photosynthetic parasitism), as suggested by the sequence of genome reduction and gene loss in plastomes of parasitic plants (Wicke et al., 2016), we used ordered parsimony for these reconstructions.
Results
Maximum likelihood and Bayesian analyses resulted in topologically identical trees, with exceptions concerning only weakly supported nodes [bootstrap support (BS) < 0.8 and posterior probabilities (PP) < 0.95]; hence only maximum likelihood trees are shown (Figures 1, 2). All trees (maximum likelihood trees, consensus trees from the Bayesian analyses) are available in the nexus files (available in dryad under doi: 10.5061/dryad.31cf160).
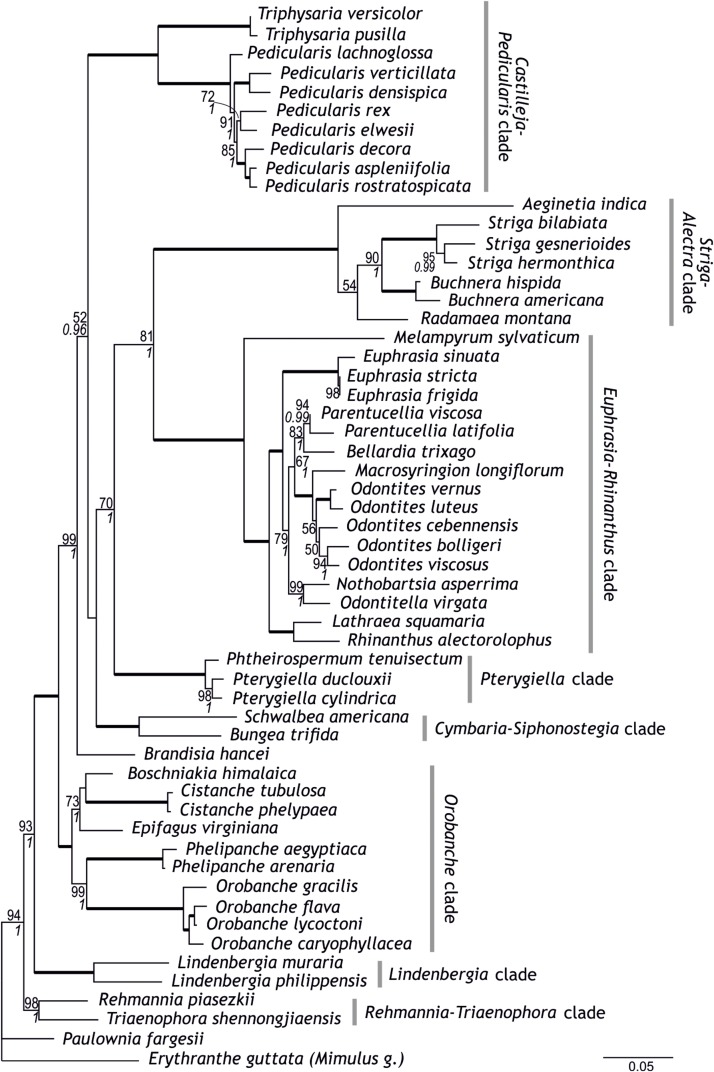
FIGURE 1.
Phylogenetic relationships within Orobanchaceae inferred using maximum likelihood on a combined data set of five loci newly sequenced for this study. Numbers at branches are maximum likelihood bootstrap support values of at least 50 and, in italics, posterior probabilities of at least 0.95; branches with maximum support are indicated by thick lines. Circumscription of major clades within Orobanchaceae is indicated.
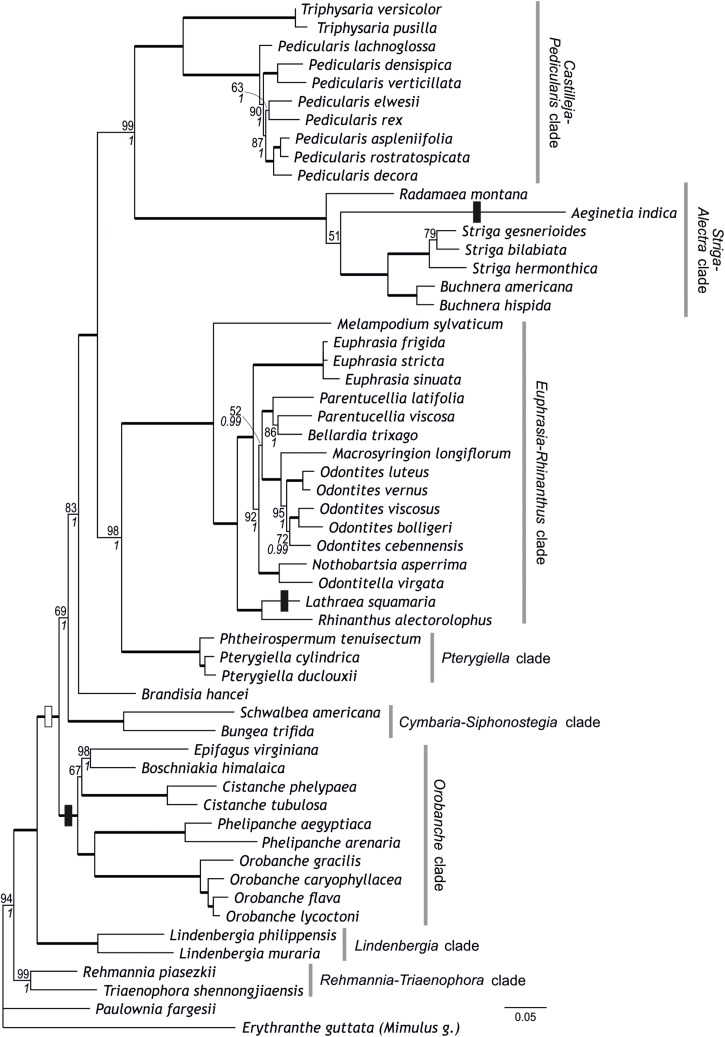
FIGURE 2.
Phylogenetic relationships within Orobanchaceae inferred using maximum likelihood on a combined data set of ten loci. Numbers at branches are maximum likelihood bootstrap support values of at least 50 and, in italics, posterior probabilities of at least 0.95; branches with maximum support are indicated by thick lines. Circumscription of major clades within Orobanchaceae is indicated. The transition to parasitism is indicated by a white box, transitions to holoparasitic (from hemiparasitic ancestors) are indicated by black boxes.
Single Markers
The five markers were successfully amplified from at least 30 of the 56 taxa (Table 1), thus after adding sequences from other sources (e.g., GenBank) each marker was available for at least 39 of the 56 taxa (Supplementary Table S1). Cloned sequences of a marker from the same species always formed well supported clades (data not shown), and only a single randomly chosen clone per marker and sample was used for final analyses. Alignment lengths of the markers used ranged from 289 bp in Agt1 to 1508 bp in AT1G09680, the two PPR genes (AT1G09680, AT2G37230) being the longest sequences (Table 3). Introns were present in AT1G14610 and Agt1. A few regions, most prominently the intron from Agt1, were excluded from phylogenetic analysis because they were not universally alignable across all taxa of the family (Table 3).
TABLE 3.
Sequences characteristics.
| Locus | Sequence length (bp) exon (intron) | Alignment length (bp) exon (intron) | Number of parsimony-informative sites |
| AT1G09680 | 786–1505 (0) | 15081 (0) | 734 |
| AT2G37230 | 391–1380 (0) | 13592 (0) | 533 |
| AT1G14610 | 250–362 (55–150) | 365 (1053) | 197 |
| AT1G04780 | 435–808 (0) | 811 (0) | 259 |
| Agt1 | 206–289 (220–818) | 289 (04) | 93 |
1A sample-specific insertion of 14 bps in Orobanche flava has been removed. 2An invariant motif of 21 bp at the 3′ terminus sequenced only for three Pedicularis species (P. aspleniifolia, P. rostratospicata, and P. verticillata) has been removed. 3Sample-specific insertions removed: motifs of 18 and 5 bps, respectively, in Melampyrum sylvaticum, a motif of 52 bps in Striga gesnerioides. 4Intron not universally alignable across all taxa and, therefore, removed.
The best markers with respect to level of resolution and support were the two PPR genes, AT1G09680, and AT2G37230. AT1G09680, the locus yielding the longest alignment (Table 3), provided good and often well-supported resolution across the entire phylogeny, including the backbone (Supplementary Figure S1). The second PPR gene, AT2G37230, yielding the second longest alignment (Table 3), showed reduced support (especially from maximum likelihood analysis) at the backbone, but usually high support among genera and species, except the Euphrasia-Rhinanthus clade (Supplementary Figure S2). Conflicts between the PPR genes concerned, for instance, the placement of the Cymbaria-Siphonostegia clade and of Brandisia, which received moderate to (especially in Bayesian analysis, if taking posterior probabilities of at least 0.95 into account) high support. The LCN loci AT1G14610 (Supplementary Figure S3) and AT1G04780 (Supplementary Figure S4), which have never been used in any phylogenetic study before, showed poor resolution at the backbone, but better and usually well-supported resolution among genera and species at least in some clades, such as the Castilleja-Pedicularis clade, the Euphrasia-Rhinanthus clade, or the Striga-Alectra clade (Supplementary Figures S3, S4). The locus yielding the shortest alignment (Table 3), Agt1, provided poor resolution at all levels in all clades except the Striga-Alectra clade (Supplementary Figure S5).
Single markers usually recovered the major clades identified previously (McNeal et al., 2013). Exceptions were the Orobanche clade inferred as polyphyletic, though not supported (BS < 50, PP < 0.5), by AT1G14610 data (Supplementary Figure S3), the Cymbaria-Siphonostegia clade inferred as polyphyletic, though not supported (BS < 50, PP < 0.5), by AT1G04780 (Supplementary Figure S4), and the Castilleja-Pedicularis clade inferred as paraphyletic, though not supported (BS < 50, PP < 0.5), by Agt1 (Supplementary Figure S5). The clade comprising Rehmannia and Triaenophora, henceforth referred to as Rehmannia-Triaenophora clade, was inferred as non-monophyletic not only by the two short markers AT1G04780 (Supplementary Figure S4) and Agt1 (Supplementary Figure S5), but also by one of the PPR genes (AT2G37230, Supplementary Figure S2), but in none of these cases did the lack of monophyly receive sufficient support. Congruently, a clade of several Pterygiella species and Phtheirospermum tenuisectum, the Pterygiella clade, was identified to be distinct from (all markers: Supplementary Figures S1–S5) and not sister to the Euphrasia-Rhinanthus clade (all markers except AT1G04780: Supplementary Figure S4).
Odontites (including Macrosyringion, where available) was inferred as monophyletic by three markers (the PPR gene AT1G09680, AT1G14610, and Atg1) with high support (BS 97–100, PP 1; Supplementary Figures S1, S3, S5), but not by the other two markers. Here, Odontites was either inferred as paraphyletic due to the, yet unsupported, inclusion of Melampyrum (the second PPR gene AT2G37230; Supplementary Figure S2) or as polyphyletic due to the, yet unsupported, placements of Macrosyringion, Nothobartsia, Odontitella, and Parentucellia (AT1G04780; Supplementary Figure S4). With the exception of the first PPR gene AT1G09680 (Supplementary Figure S1), relationships among Odontites species were poorly resolved and usually insufficiently supported (Supplementary Figures S2–S5). Nothobartsia and Odontitella were inferred as sister groups (BS 65–100, PP 0.97–1) in all but two of the shorter markers (AT1G14610 and Atg1; Supplementary Figures S3, S5).
Concatenated Markers
Following a supermatrix approach, we combined the five markers newly generated here. The thus combined data set comprised 4,437 nucleotide sites in 56 species. Whereas all previously identified major clades (including the Orobanche clade), the Rehmannia-Triaenophora clade, and the Pterygiella clade were recovered with high support (BS 98–100, PP 1), relationships among some of these clades were less certain (Figure 1). Possibly, this is due to conflicts among the genes, e.g., between the two PPR genes mentioned in the previous section, which is reflected in the network connecting major lineages of Orobanchaceae in the super network (Figure 3A). Major uncertainty was reflected by low support for a clade comprising the Cymbaria-Siphonostegia clade, the Pterygiella clade, the Euphrasia-Rhinanthus clade, and the Striga-Alectra clade (BS < 50, PP < 0.95) and for the node joining this clade with the Castilleja-Pedicularis clade (BP = 52, PP = 0.96; Figure 1). The Orobanche clade was well supported as sister to the remaining parasitic taxa (BS 100, PP 1), as were the remaining nodes uniting Lindenbergia and the parasitic taxa, and the node uniting these with Rehmannia and Triaenophora (BS 99–100, PP 1, Figure 1). Nothobartsia and Odontitella were inferred as sister taxa (BS 99, PP 1) well separated from Odontites (Figure 1). Odontites was inferred as monophyletic, but only from maximum likelihood and without support (BS 56), with Macrosyringion as sister (BS 100, PP 1).
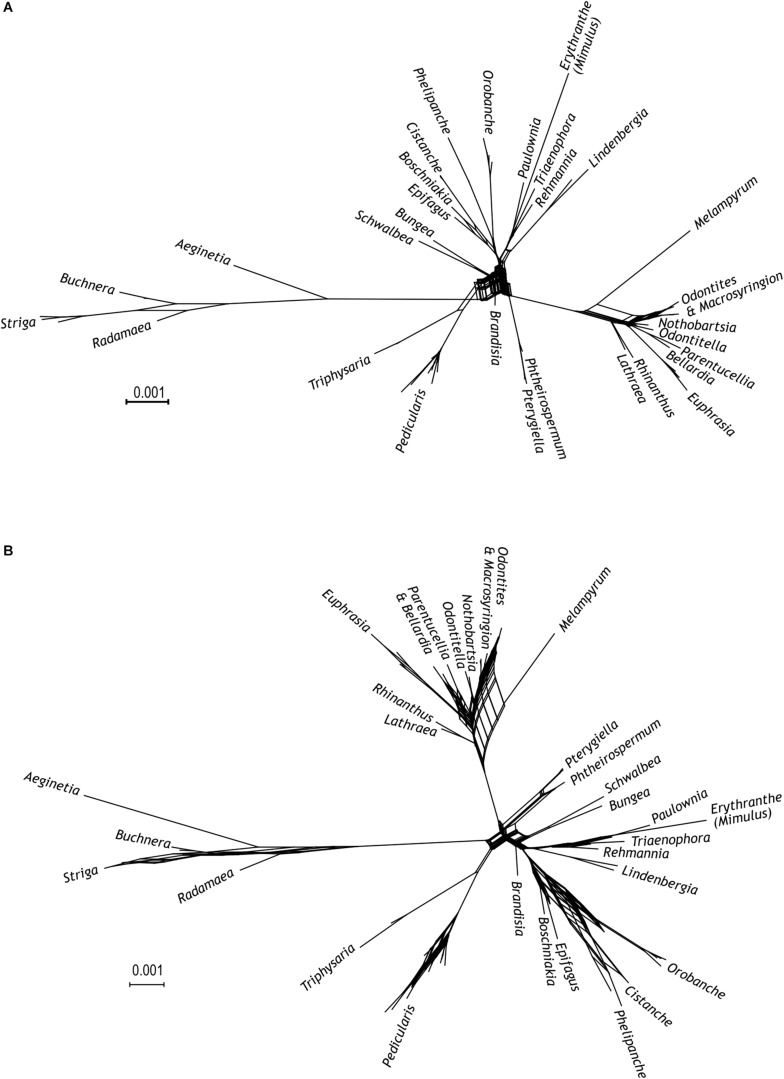
FIGURE 3.
Super network from the maximum likelihood trees. Input trees are from (A) each of the five newly sequenced markers (AT1G09680, AT2G37230, AT1G14610, AT1G04780, and Agt1) and (B) from each of the ten used markers (five newly sequenced markers and PHYA, PHYB, ITS, matK, and rps2).
Combining the newly developed loci with the five loci of McNeal et al. (2013) resulted in a matrix comprising 11,093 nucleotide sites from 56 species. All previously identified major clades (including the Orobanche clade), the Rehmannia-Triaenophora clade, and the Pterygiella clade were recovered with (nearly) maximum support (BS 99–100, PP 1; Figure 2). Relationships among major clades tended to reflect those recovered by McNeal et al. (2013), rather than relationships inferred by analyses of the five newly developed markers. Specifically, the Striga-Alectra clade was inferred as well-supported sister (BS 99, PP 1) to the Castilleja-Pedicularis clade (Figure 2) instead of to the Euphrasia-Rhinanthus clade (BS 81, PP 1; Figure 1) and the Pterygiella clade was inferred as sister to the Euphrasia-Rhinanthus clade (BS 98, PP 1; Figure 2) instead of to the clade comprising the Euphrasia-Rhinanthus clade and the Striga-Alectra clade (BS 70, PP 1; Figure 1). Brandisia was placed as sister to the clade comprising the Striga-Alectra clade, the Castilleja-Pedicularis clade, the Euphrasia-Rhinanthus clade, and the Pterygiella clade (BS 83, PP 1; Figure 2), whereas from the five marker analyses the relationship of Brandisia to any of these clades remained unclear (Figure 1). The Cymbaria-Siphonostegia clade was inferred as sister to all other hemiparasitic Orobanchaceae, yet this did not receive strong support (BS 69, PP 1; Figure 2). With respect to the phylogenetic positions of the Orobanche clade, the Lindenbergia clade, and the Rehmannia-Triaenophora clade as subsequent sisters to the hemiparasitic clades, the ten marker analyses agreed with the five marker analyses, yet with higher support (BS 94–100, PP 1; Figure 2). Despite the often-high bootstrap support values, there was considerable incongruence among markers with respect to phylogenetic relationships, as is reflected in reticulate relationships among major lineages in the super network including all ten markers (Figure 3B).
Ancestral character state reconstruction suggested that parasitism (i.e., hemiparasitism) evolved only once in the sister of the Lindenbergia clade (Figure 2). The ancestor of the clade including all parasitic taxa was inferred to be hemiparasitic (Figure 2).
Discussion
Phylogenetic Utility of PPR Genes and Three LCN Loci in Orobanchaceae
Analyses of two PPR genes, AT1G09680 and AT2G37230, indicated resolved, though not necessarily well-supported, relationships among major clades of Orobanchaceae and among Odontites species (Supplementary Figures S1, S2). This confirms the high potential of PPR genes for molecular phylogenetic studies from the family to the species level (Yuan et al., 2010; Barkan and Small, 2014; Crowl et al., 2014), notwithstanding issues of incongruence among markers from the level of major clades to the infrageneric level, as in Odontites (Supplementary Figures S1, S2). In line with decreasing length and concomitantly decreasing number of informative sites (measured here as number of parsimony-informative sites: Table 3), phylogenetic resolution and support were lower, especially at the backbone, in inferences from the LCN loci AT1G14610 and AT1G04780 (Supplementary Figures S3, S4), coding for an aminoacyl-tRNA ligase and an ankyrin repeat family protein, respectively (Isner et al., 2012; Ometto et al., 2012), and especially from the LCN locus Agt1 (Supplementary Figure S5), encoding a peroxisomal photorespiratory enzyme (Liepman and Olsen, 2003). Whereas the readily amplifiable and alignable AT1G14610 and AT1G04780, to our knowledge, have not previously been used for phylogenetic purposes, Agt1 has been suggested as phylogenetically useful locus (Li M. et al., 2008; López-Pujol et al., 2012; Gonzalez, 2014), an assessment that is not supported by our analyses of Orobanchaceae.
Practical limitations of LCN loci as used here include the difficulty in designing primers and in obtaining reliable amplification. Here, screening of more than 200 loci resulted in identification of only a few that could be used over the desired broad taxonomic range. Even some PPR genes successfully used by Yuan et al. (2010) failed to work in Orobanchaceae. Reasons for this are unclear, but may include poor primer match due to the phylogenetic distance between Verbenaceae and Orobanchaceae, evolutionary rate variation, or pseudogene formation in Orobanchaceae. It can, however, be expected that enrichment procedures, such as target-capture (Lemmon and Lemmon, 2013; Zimmer and Wen, 2015; Johnson et al., 2019) will essentially eliminate the need for the time-consuming search for suitable loci (a pipeline for identifying loci amenable to target-capture in Orobanchaceae has been suggested recently: Li et al., 2017). Using phylogenomic approaches with hundreds of loci is also expected to help resolve phylogenetic relationships in the presence of incongruence among loci (Buddenhagen et al., 2016; Crowl et al., 2017; Léveillé-Bourret et al., 2018), as long as heterogeneous population-genetic processes are taken into account (Bravo et al., 2019).
Phylogenetic Relationships Among Major Clades
Although there are conflicts among phylogenies generated using different markers and their combinations (Figures 1–3 and Supplementary Figures S1–S5; McNeal et al., 2013), circumscription of major clades as identified previously (Bennett and Mathews, 2006; McNeal et al., 2013) is mostly confirmed from single marker analyses (Supplementary Figures S1–S5) and is well-supported from the combined data (Figures 1, 2). This is also the case for Brandisia, which is corroborated as a distinct lineage. The only modification to the circumscription of major clades concerns the Pterygiella clade [represented by P. tenuisectum (Pterygiella tenuisecta), Pterygiella cylindrica, and Pt. duclouxii], comprising Pterygiella, Phtheirospermum (except Ph. japonicum), and Xizangia (Yu et al., 2018). This small group was inferred as sister to the Euphrasia-Rhinanthus clade by McNeal et al. (2013), a relationship also supported by the combined 10-marker data set (Figure 2). This position is, however, found neither by the newly sequenced markers (except AT1G04780; Supplementary Figure S4), analyzed individually (Supplementary Figures S1–S3, S5) or jointly (Figure 1), nor by ITS and plastid data used by Yu et al. (2018). A closer relationship of the Pterygiella clade to Lindenbergia and Brandisia, as suggested by fruit and seed characters (Dong et al., 2015), is not supported by the nuclear data, but, with respect to Brandisia, by plastid data (Yu et al., 2018). Given these uncertainties and the deep divergence of Pterygiella and relatives from the Euphrasia-Rhinanthus clade, even if inferred as sister taxa (Figure 2), we consider it prudent to recognize this small group of East Asian genera as the Pterygiella clade distinct from the Euphrasia-Rhinanthus clade until its precise position within the family has been ascertained.
In contrast to the generally well-supported circumscription of major clades, phylogenetic relationships among these clades are not consolidated yet (Figure 3). One such area of uncertainty concerns relationships among the Castilleja-Pedicularis clade, the Euphrasia-Rhinanthus clade, and the Striga-Alectra clade. Bennett and Mathews (2006), using PHYA including obvious paralogs, inferred the Striga-Alectra clade as sister to the Euphrasia-Rhinanthus clade (with bootstrap support of at least 80), together being sister-group to the Castilleja-Pedicularis clade (with maximum bootstrap support). In contrast, McNeal et al. (2013), using, among others, PHYA excluding obvious paralogs, found the Striga-Alectra clade to be sister to the Castilleja-Pedicularis clade (with bootstrap support of at least 99 in analyses of PHYA and PHYB separately as well as combined in their 5-marker dataset), jointly being sister to the Euphrasia-Rhinanthus clade (including the Pterygiella clade). While PPR genes (Supplementary Figures S1, S2) and our 5-marker combined dataset (Figure 1) support the hypothesis of Bennett and Mathews (2006), i.e., the sister-group relationship of the Striga-Alectra clade and the Euphrasia-Rhinanthus clade, the 10-marker combined data set (Figure 2) agrees with the hypothesis of McNeal et al. (2013), i.e., a sister-group relationship of the Striga-Alectra clade and the Castilleja-Pedicularis clade. The reasons for these conflicts are unknown, but potentially include sampling of paralogs as is evident from the large effect their inclusion has on the inferred relationships (compare the PHYA trees inferred by Bennett and Mathews, 2006, with those inferred by McNeal et al., 2013). Paralogs have also been reported from PPR genes (AT2G37230 has experienced a recent gene duplication in Glandularia and Verbena of Verbenaceae: Yuan et al., 2010), although copies recovered in Orobanchaceae appear to be orthologs (Supplementary Figure S2). It has been shown that already tiny subsets of large phylogenomic data sets may drive contentious relationships (Shen et al., 2017), and this might also be the case here, but additional data will be needed to test this.
The position of Brandisia as sister to the mostly hemiparasitic clades excluding the Cymbaria-Siphonostegia clade is well supported by the 10-marker combined data (Figure 2). However, the uncertain position of Brandisia in previous studies (Bennett and Mathews, 2006; McNeal et al., 2013) and in the newly sequenced genes, whether analyzed individually or jointly (Figure 1 and Supplementary Figures S1–S5), warrants caution with respect to its phylogenetic position.
The Cymbaria-Siphonostegia clade, comprising five hemiparasitic genera (ca. 20 species) distributed mainly in Eurasia (Schneeweiss, 2013), has been inferred as sister-group to all other parasitic Orobanchaceae (Bennett and Mathews, 2006; McNeal et al., 2013). Whereas its precise position remains ambiguous, PPR genes (Supplementary Figures S1, S2), the 5-marker combined (Figure 1), and the 10-marker combined analyses (Figure 2) suggest that the Cymbaria-Siphonostegia clade is sister to, or even nested among, the mostly hemiparasitic clades (Brandisia, Castilleja-Pedicularis clade, Euphrasia-Rhinanthus clade, Pterygiella clade, Striga-Alectra clade). A consequence of the altered position of the Cymbaria-Siphonostegia clade is that the exclusively holoparasitic and, except for the shortest markers used (Supplementary Figures S4, S5), well-supported Orobanche clade is sister to all other parasitic clades (Figures 1, 2). Although this may suggest that holoparasitism evolved early in parasitic Orobanchaceae, conservation of the chlorophyll synthesis in holoparasitic Phelipanche (Wickett et al., 2011) despite loss of photosynthesis and the concomitant reductions in the plastid genome (Wicke et al., 2013, 2016) may be interpreted as evidence for a comparatively recent loss of photosynthetic functionality, i.e., a transition to holoparasitism, only in the stem lineage of the Orobanche clade, in line with results from ancestral character state reconstruction (Figure 2).
Lindenbergia is sister to the parasitic Orobanchaceae, although high support for this position is only achieved from the concatenated data sets (Figures 1, 2). The close relationship of Lindenbergia to parasitic lineages is supported not only by molecular-phylogenetic evidence (Young et al., 1999; Olmstead et al., 2001; McNeal et al., 2013), but also by palynological and leaf stomatal closure data (Hjertson, 1995; Bennett and Mathews, 2006). Sister to Lindenbergia and other Orobanchaceae is the Rehmannia-Triaenophora clade (here represented by the newly sampled Triaenophora shennongjiaensis and Rehmannia piasezkii, Table 1) endemic to China (Chin, 1979; Li et al., 2005; Li X.D. et al., 2008). A close relationship of Rehmannia and/or Triaenophora to Orobanchaceae has been suggested before (Albach et al., 2009; Xia et al., 2009), which eventually has led to the extension of Orobanchaceae to include both genera (Angiosperm Phylogeny Group, 2016).
Conclusion
We analyzed the potential of five nuclear genes (two PPR genes and three LCN genes) to address phylogenetic relationships within Orobanchaceae focusing on major clades identified previously. Of those, the longer markers (the two PPR genes, AT1G09680 and AT2G37230, and the LCN locus AT1G04780) consistently performed better in inferring relationships within and among major clades than the two short markers (LCN loci AT1G14610 and Agt1). Whereas extension of the data set (increasing sequence length by adding more loci) clearly improves resolving power, at least when concatenating loci, and corroborates and refines circumscription of major clades, this study also highlights the limits of sequencing hand-picked loci for phylogenetic purposes. These are, among others, the large effort to establish suitable nuclear loci and the inability to deal with incongruence among loci through species tree estimation methods as these methods cannot be applied because of the too low number of sequenced loci. We expect that already available phylogenomic approaches, once applied to Orobanchaceae, will help to resolve relationships among major clades. This notwithstanding, congruence among markers in inference of major clades of Orobanchaceae allows these major clades to be taken as frameworks for detailed, species-level, phylogenetic studies in this family, a model for studying plant parasitism.
Data Availability
Newly generated sequences are available in GenBank under accession numbers MK588398–MK588632. Data matrices are available at Dryad under doi: 10.5061/dryad.31cf160.
Author Contributions
GS and XL designed the study. XL and TF generated and analyzed the data. XL and GS drafted the manuscript. XL, CR, and GS finalized the manuscript. All authors read and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Da Pan for sampling and assistance in executing software. We are grateful to Hong Wang, Wenbin Yu, María Montserrat Martínez Ortega, Daniel Pinto Carrasco, Clemens Pachschwöll, Xiaodong Li, Susann Wicke, Yanxia Sun and all other friends who helped us with sampling or providing material. We also thank Sarah Mathews (Australian National University) and Elfriede Grasserbauer and Michael H. J. Barfuss (University of Vienna) for their help in lab work.
Funding. This study was funded by the China Scholarship Council Grant No. 201206100007 (to XL).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00902/full#supplementary-material
Phylogenetic relationships within Orobanchaceae inferred using maximum likelihood on an AT1G09680 data set. Numbers at branches are maximum likelihood bootstrap support values of at least 50 and, in italics, posterior probabilities of at least 0.95. Circumscription of major clades within Orobanchaceae is indicated.
Phylogenetic relationships within Orobanchaceae inferred using maximum likelihood on an AT2G37230 data set. Numbers at branches are maximum likelihood bootstrap support values of at least 50 and, in italics, posterior probabilities of at least 0.95. Circumscription of major clades within Orobanchaceae is indicated.
Phylogenetic relationships within Orobanchaceae inferred using maximum likelihood on an AT1G14610 data set. Numbers at branches are maximum likelihood bootstrap support values of at least 50 and, in italics, posterior probabilities of at least 0.95. Circumscription of major clades within Orobanchaceae is indicated.
Phylogenetic relationships within Orobanchaceae inferred using maximum likelihood on an AT1G04780 data set. Numbers at branches are maximum likelihood bootstrap support values of at least 50 and, in italics, posterior probabilities of at least 0.95. Circumscription of major clades within Orobanchaceae is indicated.
Phylogenetic relationships within Orobanchaceae inferred using maximum likelihood on an Agt1 data set. Numbers at branches are maximum likelihood bootstrap support values of at least 50 and, in italics, posterior probabilities of at least 0.95. Circumscription of major clades within Orobanchaceae is indicated.
List of taxa, locality and voucher information, and sequence source (GenBank accession numbers or other databases).
List of low-copy nuclear genes successfully PCR-amplified in up to 27 taxa.
References
- Akaike H. (1974). A new look at the statistical model identification. IEEE Trans. Automatic Control 19 716–723. 10.1109/TAC.1974.1100705 [DOI] [Google Scholar]
- Albach D. C., Yan K., Jensen S. R., Li H.-Q. (2009). Phylogenetic placement of Triaenophora (formerly Scrophulariaceae) with some implications for the phylogeny of Lamiales. Taxon 58 749–756. 10.1002/tax.583005 [DOI] [Google Scholar]
- Álvarez I., Wendel J. F. (2003). Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogen. Evol. 29 417–434. 10.1016/S1055-7903(03)00208-2 [DOI] [PubMed] [Google Scholar]
- Angiosperm Phylogeny Group (2016). An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 181 1–20. 10.1111/boj.12385 26027755 [DOI] [Google Scholar]
- Babineau M., Gagnon E., Bruneau A. (2013). Phylogenetic utility of 19 low copy nuclear genes in closely related genera and species of caesalpinioid legumes. South Afr. J. Bot. 89 94–105. 10.1016/j.sajb.2013.06.018 [DOI] [Google Scholar]
- Barkan A., Small I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65 415–442. 10.1146/annurev-arplant-050213-040159 [DOI] [PubMed] [Google Scholar]
- Beck-Mannagetta G. (1930). “Orobanchaceae,” in Das Pflanzenreich IV 261, ed. Engler A. (Leipzig: Wilhelm Engelmann; ), 1–348. [Google Scholar]
- Bennett J., Mathews S. (2006). Phylogeny of the parasitic plant family Orobanchaceae inferred from phytochrome A. Am. J. Bot. 93 1039–1051. 10.3732/ajb.93.7.1039 [DOI] [PubMed] [Google Scholar]
- Bravo G. A., Antonelli A., Bacon C. D., Bartoszek K., Blom M. P. K., Huynh S., et al. (2019). Embracing heterogeneity: coalescing the Tree of Life and the future of phylogenomics. PeerJ 7:e6399. 10.7717/peerj.6399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddenhagen C., Lemmon A. R., Lemmon E. M., Bruhl J., Cappa J., Clement W. L., et al. (2016). Anchored phylogenomics of angiosperms I: assessing the robustness of phylogenetic estimates. bioRxiv [Preprint]. 10.1101/086298 [DOI] [Google Scholar]
- Chin T. L. (1979). “Rehmannia and Triaenophora,” in Flora Reipublicae Popularis Sinicae 67 Part 2 Scrophulariaceae, eds Tsoong P. C., Yang H. P. (Bejing: Science Press; ), 212–222. [Google Scholar]
- Choi H. K., Luckow M., Doyle J., Cook D. R. (2006). Development of nuclear gene-derived molecular markers linked to legume genetic maps. Mol. Genet. Genom. 276 56–70. 10.1007/s00438-006-0118-8 [DOI] [PubMed] [Google Scholar]
- Crowl A. A., Mavrodiev E., Mansion G., Haberle R., Pistarino A., Kamari G., et al. (2014). Phylogeny of Campanuloideae (Campanulaceae) with emphasis on the utility of nuclear pentatricopeptide repeat (PPR) genes. PLoS One 9:e94199. 10.1371/journal.pone.0094199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowl A. A., Myers C., Cellinese N. (2017). Embracing discordance: phylogenomic analyses provide evidence for allopolyploidy leading to cryptic diversity in a Mediterranean Campanula (Campanulaceae) clade. Evolution 71 913–922. 10.1111/evo.13203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubero J. I., Moreno M. T. (1996). “Parasitic plant science: a quarter century,” in Advances in Parasitic Plant Research, eds Moreno M. T., Cubero J. I., Berner D., Joel D., Musselman L. J., Parker C. (Sevilla: Consejería de Agricultura y Pesca, Junta de Andalucía; ), 15–21. [Google Scholar]
- dePamphilis C. W., Palmer J. D. (1990). Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature 348 337–339. 10.1038/348337a0 [DOI] [PubMed] [Google Scholar]
- dePamphilis C. W., Young N. D., Wolfe A. D. (1997). Evolution of plastid gene rps2 in a lineage of hemiparasitic and holoparasitic plants: many losses of photosynthesis and complex patterns of rate variation. Proc. Natl. Acad Sci. U.S.A. 94 7367–7372. 10.1073/pnas.94.14.7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L.-N., Wang H., Wortley A. H., Li D. Z., Lu L. (2015). Fruit and seed morphology in some representative genera of tribe Rhinantheae sensu lato (Orobanchaceae) and related taxa. Plant Syst. Evol. 301 479–500. 10.1007/s00606-014-1087-8 [DOI] [Google Scholar]
- Duarte J. M., Wall P. K., Edger P. P., Landherr L. L., Ma H., Pires J. C., et al. (2010). Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomical levels. BMC Evol. Biol. 10:61–79. 10.1186/1471-2148-10-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 32 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer E. (2004). “Scrophulariaceae,” in The Families and Genera of Vascular Plants VII: Flowering Plants, Dicotyledons, Lamiales (Excluding Acanthaceae, Including Avicenniaceae), ed. Kadereit J. W. (Berlin: Springer; ), 333–432. [Google Scholar]
- Gaudeul M., Siljak-Yakovlev S., Jang T. S., Rouhan G. (2018). Reconstructing species relationships within the recently diversified genus Odontites Ludw. (Orobanchaceae): evidence for extensive reticulate evolution. Int. J. Plant Sci. 179 1–20. 10.1086/694763 [DOI] [Google Scholar]
- Gonzalez L. A. (2014). Phylogenetics and Mating System Evolution in the Southern South American Valeriana (Valerianaceae). Master’s thesis,University of New Orleans: New Orleans, LO. [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41 95–98. [Google Scholar]
- Hellsten U., Wright K. M., Jenkins J., Shu S., Yuan Y., Wessler S. R., et al. (2013). Fine-scale variation in meiotic recombination in Mimulus inferred from population shotgun sequencing. Proc. Natl. Acad. Sci. U.S.A. 110 19478–19482. 10.1073/pnas.1319032110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjertson M. L. (1995). Taxonomy, phylogeny, and biogeography of Lindenbergia (Scrophulariaceae). Bot. J. Linn. Soc. 119 265–321. 10.1016/S0024-4074(95)80002-6 [DOI] [Google Scholar]
- Huson D. H., Bryant D. (2006). Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23 254–267. 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- Huson D. H., Dezulian T., Kloepper T., Steel M. A. (2004). Phylogenetic super-networks from partial trees. IEEE ACM Trans. Comp. Biol. Bioinform. 1 151–158. 10.1109/TCBB.2004.44 [DOI] [PubMed] [Google Scholar]
- Isner J. C., Nuhse T., Maathuis F. J. (2012). The cyclic nucleotide cGMP is involved in plant hormone signalling and alters phosphorylation of Arabidopsis thaliana root proteins. J. Exp. Bot. 63 3199–3205. 10.1093/jxb/ers045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joel D. M., Gressel J., Musselman L. J. (eds) (2013). Parasitic Orobanchaceae. Parasitic Mechanisms and Control Strategies. Berlin: Springer. [Google Scholar]
- Johnson M., Pokorny L., Dodsworth S., Botigue L. R., Cowan R. S., Devault A., et al. (2019). A universal probe set for targeted sequencing of 353 nuclear genes from any flowering plant designed using k-medoids clustering. Syst. Biol. 68 594–606. 10.1093/sysbio/syy086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijt J. (1969). The Biology of Parasitic Flowering Plants. Berkeley, CA: University of California Press. [Google Scholar]
- Lanfear R., Calcott B., Ho S. Y., Guindon S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29 1695–1701. 10.1093/molbev/mss020 [DOI] [PubMed] [Google Scholar]
- Latvis M., Jacobs S. J., Mortimer S. M. E., Richards M., Blischak P. D., Mathews S., et al. (2017). Primers for Castilleja and their utility across Orobanchaceae: II. single-copy nuclear loci. Appl. Plant Sci. 5:1700038. 10.3732/apps.1700038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon E. M., Lemmon A. R. (2013). High-throughput genomic data in systematics and phylogenetics. Annu. Rev. Ecol. Evol. Syst. 44 99–121. 10.1146/annurev-ecolsys-110512-135822 [DOI] [Google Scholar]
- Léveillé-Bourret É., Starr J. R., Ford B. A., Lemmon E. M., Lemmon A. R. (2018). Resolving rapid radiations within angiosperm families using anchored phylogenomics. Syst. Biol. 67 94–112. 10.1093/sysbio/syx050 [DOI] [PubMed] [Google Scholar]
- Li M., Wunder J., Bissoli G., Scarponi E., Gazzani S., Barbaro E., et al. (2008). Development of COS genes as universally amplifiable markers for phylogenetic reconstructions of closely related plant species. Cladistics 24 727–745. 10.1111/j.1096-0031.2008.00207.x [DOI] [Google Scholar]
- Li X., Hao B., Pan D., Schneeweiss G. M. (2017). Marker development for phylogenomics: the case of Orobanchaceae, a plant family with contrasting nutritional modes. Front. Plant Sci. 8:1973. 10.3389/fpls.2017.01973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Jang T.-S., Temsch E. M., Kato H., Takayama K., Schneeweiss G. M. (2016). Molecular and karyological data confirm that the enigmatic genus Platypholis from Bonin-Islands (SE Japan) is phylogenetically nested within Orobanche (Orobanchaceae). J. Plant Res. 130 273–280. 10.1007/s10265-016-0888-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. D., Li J. Q., Zan Y. Y. (2005). A new species of Triaenophora (Scrophulariaceae) from China. Novon 15 559–561. [Google Scholar]
- Li X. D., Zan Y. Y., Li J. Q., Yang S. Z. (2008). A numerical taxonomy of the genera Rehmannia and Triaenophora (Scrophulariaceae). J. Syst. Evol. 46 730–737. [Google Scholar]
- Liepman A. H., Olsen L. J. (2003). Alanine aminotransferase homologs catalyze the glutamate:glyoxylate aminotransferase reaction in peroxisomes of Arabidopsis. Plant Physiol. 131 215–227. 10.1104/pp.011460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Pujol J., Garcia-Jacas N., Susanna A., Vilatersana R. (2012). Should we conserve pure species or hybrid species? Delimiting hybridization and introgression in the Iberian endemic Centaurea podospermifolia. Biol. Conserv. 152 271–279. 10.1016/j.biocon.2012.03.032 [DOI] [Google Scholar]
- Maddison W. P., Maddison D. R. (2018). Mesquite: a Modular System for Evolutionary Analysis. Version 3.51. Available at: http://www.mesquiteproject.org (accessed April 29, 2019). [Google Scholar]
- Matasci N., Hung L. H., Yan Z., Carpenter E. J., Wickett N. J., Mirarab S., et al. (2014). Data access for the 1,000 Plants (1KP) project. GigaScience 3:17. 10.1186/2047-217X-3-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal J., Bennett J. R., Wolfe A. D., Mathews S. (2013). Phylogeny and origins of holoparasitism in Orobanchaceae. Am. J. Bot. 100 971–983. 10.3732/ajb.1200448 [DOI] [PubMed] [Google Scholar]
- McWilliam H., Li W., Uludag M., Squizzato S., Park Y. M., Buso N., et al. (2013). Analysis tool web services from the EMBL-EBI. Nucl. Acids Res. 41 W597–W600. 10.1093/nar/gkt376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickrent D. L. (2019). The Parasitic Plant Connection. Available at: https://parasiticplants.siu.edu/ (accessed April 12, 2019). [Google Scholar]
- Nickrent D. L., Duff R. J., Colwell A. E., Wolfe A. D., Young N. D., Steiner K. E., et al. (1998). “Molecular phylogenetic and evolutionary studies of parasitic plants,” in Molecular Systematics of Plants II, eds Soltis D. E., Soltis P. S., Doyle J. J. (Boston, MA: Springer; ), 211–241. 10.1007/978-1-4615-5419-6_8 [DOI] [Google Scholar]
- Olmstead R. G., dePamphilis C. W., Wolfe A. D., Young N. D., Elison W. J., Reeves P. A. (2001). Disintegration of the Scrophulariaceae. Am. J. Bot. 88 348–362. 10.2307/2657024 [DOI] [PubMed] [Google Scholar]
- Ometto L., Li M., Bresadola L., Varotto C. (2012). Rates of evolution in stress-related genes are associated to habitat preference in two Cardamine lineages. BMC Evol. Biol. 12:7. 10.1186/1471-2148-12-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole N., Hattori M., Andres C., Iida K., Lurin C., Schmitz-Linneweber C., et al. (2008). On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 25 1120–1128. 10.1093/molbev/msn057 [DOI] [PubMed] [Google Scholar]
- Park J. M., Manen J. F., Colwell A. E., Schneeweiss G. M. (2008). A plastid gene phylogeny of the non-photosynthetic parasitic Orobanche (Orobanchaceae) and related genera. J. Plant Res. 121 365–376. 10.1007/s10265-008-0169-5 [DOI] [PubMed] [Google Scholar]
- Parker C., Riches C. R. (1993). Parasitic Weeds of the World: Biology and Control. Wallingford: CAB International. [Google Scholar]
- Pinto-Carrasco D., Scheunert A., Heubl G., Rico E., Martinez-Ortega M. M. (2017). Unravelling the phylogeny of the root-hemiparasitic genus Odontites (tribe Rhinantheae, Orobanchaceae): evidence for five main lineages. Taxon 66 886–908. 10.12705/664.6 [DOI] [Google Scholar]
- Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A. (2018). Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67 901–904. 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Sang T. (2002). Utility of low-copy nuclear gene sequence in plant phylogenetics. Crit. Rev. Biochem. Mol. Biol. 37 121–147. 10.1080/10409230290771474 [DOI] [PubMed] [Google Scholar]
- Schneeweiss G. M. (2013). “Phylogenetic relationships and evolutionary trends in Orobanchaceae,” in Parasitic Orobanchaceae: Parasitic Mechanisms and Control Strategies, eds Joel D. M., Gressel J., Musselman L. J. (Berlin: Springer; ), 243–265. 10.1007/978-3-642-38146-1_14 [DOI] [Google Scholar]
- Shen X. X., Hittinger C. T., Rokas A. (2017). Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nat. Ecol. Evol. 1:0126. 10.1038/s41559-017-0126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57 758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Swofford D. L. (2002). PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4b10. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Wicke S., Müller K. F., dePamphilis C. W., Quandt D., Bellot S., Schneeweiss G. M. (2016). Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc. Natl. Acad. Sci. U.S.A. 113 9045–9050. 10.1073/pnas.1607576113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S., Müller K. F., dePamphilis C. W., Quandt D., Wickett N. J., Zhang Y., et al. (2013). Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 25 3711–3725. 10.1105/tpc.113.113373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett N. J., Honaas L. A., Wafula E. K., Das M., Huang K., Wu B., et al. (2011). Transcriptomes of the parasitic plant family Orobanchaceae reveal surprising conservation of chlorophyll synthesis. Curr. Biol. 21 2098–2104. 10.1016/j.cub.2011.11.011 [DOI] [PubMed] [Google Scholar]
- Wolfe A. D., dePamphilis C. W. (1998). The effect of relaxed functional constraints on the photosynthetic gene rbcL in photosynthetic and nonphotosynthetic parasitic plants. Mol. Biol. Evol. 15 1243–1258. 10.1093/oxfordjournals.molbev.a025853 [DOI] [PubMed] [Google Scholar]
- Wolfe A. D., Randle C. P., Liu L., Steiner K. E. (2005). Phylogeny and biogeography of Orobanchaceae. Folia Geobot. 40 115–134. 10.1007/BF02803229 [DOI] [Google Scholar]
- Xia Z., Wang Y. Z., Smith J. F. (2009). Familial placement and relations of Rehmannia and Triaenophora (Scrophulariaceae s.l.) inferred from five gene regions. Am. J. Bot. 96 519–530. 10.3732/ajb.0800195 [DOI] [PubMed] [Google Scholar]
- Yang Z., Wafula E. K., Honaas L. A., Zhang H., Das M., Fernandez-Aparicio M., et al. (2015). Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Mol. Biol. Evol. 32 767–790. 10.1093/molbev/msu343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N. D., Steiner K. E., dePamphilis C. W. (1999). The evolution of parasitism in Scrophulariaceae/Orobanchaceae: plastid gene sequences refute an evolutionary transition series. Ann. Miss. Bot. Garden 86 876–893. 10.2307/2666173 [DOI] [Google Scholar]
- Yu W. B., Randle C. P., Lu L., Wang H., Yang J. B., dePamphilis C. W., et al. (2018). The hemiparasitic plant Phtheirospermum (Orobanchaceae) is polyphyletic and contains cryptic species in the Hengduan Mountains of southwest China. Front. Plant Sci. 9:142. 10.3389/fpls.2018.00142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y. W., Liu C., Marx H. E., Olmstead R. G. (2009). The pentatricopeptide repeat (PPR) gene family, a tremendous resource for plant phylogenetic studies. New Phytol. 182 272–283. 10.1111/j.1469-8137.2008.02739.x [DOI] [PubMed] [Google Scholar]
- Yuan Y. W., Liu C., Marx H. E., Olmstead R. G. (2010). An empirical demonstration of using pentatricopeptide repeat (PPR) genes as plant phylogenetic tools: phylogeny of Verbenaceae and the Verbena complex. Mol. Phylogen. Evol. 54 23–35. 10.1016/j.ympev.2009.08.029 [DOI] [PubMed] [Google Scholar]
- Zhang N., Zeng L., Shan H., Ma H. (2012). Highly conserved low-copy nuclear genes as effective markers for phylogenetic analyses in angiosperms. New Phytol. 195 923–937. 10.1111/j.1469-8137.2012.04212.x [DOI] [PubMed] [Google Scholar]
- Zimmer E. A., Wen J. (2012). Using nuclear gene data for plant phylogenetics: progress and prospects. Mol. Phylogen. Evol. 65 774–785. 10.1016/j.ympev.2012.07.015 [DOI] [PubMed] [Google Scholar]
- Zimmer E. A., Wen J. (2015). Using nuclear gene data for plant phylogenetics: progress and prospects II. next-gen approaches. J. Syst. Evol. 53 371–379. 10.1111/jse.12174 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic relationships within Orobanchaceae inferred using maximum likelihood on an AT1G09680 data set. Numbers at branches are maximum likelihood bootstrap support values of at least 50 and, in italics, posterior probabilities of at least 0.95. Circumscription of major clades within Orobanchaceae is indicated.
Phylogenetic relationships within Orobanchaceae inferred using maximum likelihood on an AT2G37230 data set. Numbers at branches are maximum likelihood bootstrap support values of at least 50 and, in italics, posterior probabilities of at least 0.95. Circumscription of major clades within Orobanchaceae is indicated.
Phylogenetic relationships within Orobanchaceae inferred using maximum likelihood on an AT1G14610 data set. Numbers at branches are maximum likelihood bootstrap support values of at least 50 and, in italics, posterior probabilities of at least 0.95. Circumscription of major clades within Orobanchaceae is indicated.
Phylogenetic relationships within Orobanchaceae inferred using maximum likelihood on an AT1G04780 data set. Numbers at branches are maximum likelihood bootstrap support values of at least 50 and, in italics, posterior probabilities of at least 0.95. Circumscription of major clades within Orobanchaceae is indicated.
Phylogenetic relationships within Orobanchaceae inferred using maximum likelihood on an Agt1 data set. Numbers at branches are maximum likelihood bootstrap support values of at least 50 and, in italics, posterior probabilities of at least 0.95. Circumscription of major clades within Orobanchaceae is indicated.
List of taxa, locality and voucher information, and sequence source (GenBank accession numbers or other databases).
List of low-copy nuclear genes successfully PCR-amplified in up to 27 taxa.
Data Availability Statement
Newly generated sequences are available in GenBank under accession numbers MK588398–MK588632. Data matrices are available at Dryad under doi: 10.5061/dryad.31cf160.