Abstract
BACKGROUND:
Clinical practice guidelines recommend estimation of glomerular filtration rate (eGFR) using validated equations based on serum creatinine (eGFRcr), cystatin C (eGFRcys), or both (eGFRcr-cys). However, when compared with the measured GFR (mGFR), only eGFRcr-cys meets recommended performance standards. Our goal was to develop a more accurate eGFR method using a panel of metabolites without creatinine, cystatin C, or demographic variables.
METHODS:
An ultra-performance liquid chromatography-tandem mass spectrometry assay for acetylthreonine, phenylacetylglutamine, pseudouridine, and tryptophan was developed, and a 20-day, multiinstrument analytical validation was conducted. The assay was tested in 2424 participants with mGFR data from 4 independent research studies. A new GFR equation (eGFRmet) was developed in a random subset (n = 1615) and evaluated in the remaining participants (n = 809). Performance was assessed as the frequency of large errors [estimates that differed from mGFR by at least 30% (1 — P30); goal <10%].
RESULTS:
The assay had a mean imprecision (≤10% intraassay, ≤6.9% interassay), linearity over the quantitative range (r2 > 0.98), and analyte recovery (98.5%– 113%). There was no carryover, no interferences observed, and analyte stability was established. In addition, 1 — P30 in the validation set for eGFRmet (10.0%) was more accurate than eGFRcr (13.1%) and eGFRcys (12.0%) but not eGFRcr-cys (8.7%). Combining metabolites, creatinine, cystatin C, and demographics led to the most accurate equation (7.0%). Neither equation had substantial variation among population subgroups.
CONCLUSIONS:
The new eGFRmet equation could serve as a confirmatory test for GFR estimation.
Glomerular filtration rate (GFR)7 is generally accepted as the best overall index of kidney function, and a decrease in GFR has diagnostic, prognostic, and patient treatment implications (1,2). GFR can be measured (mGFR) based on the clearance of an exogenous filtration marker, but this method is impractical for routine practice. Instead, an estimated GFR (eGFR) is calculated using the serum concentration of an endogenous filtration marker [creatinine (eGFRcr), cystatin C (eGFRcys), or both (eGFRcr-cys)] and demographic variables (age, sex, and race) (3).
The 2012 Kidney Disease Improving Global Outcomes guidelines recommend using eGFRcr as an initial test, followed by confirmatory tests based on eGFRcys or eGFRcr-cys when more accurate GFR assessment would affect clinical decision-making (1). However, performance of eGFR can be undermined in clinical settings in which either creatinine or cystatin C is unreliable as a filtration marker (4). Additionally, inclusion of demographic characteristics may be a disadvantage in GFR estimation because their relationships with filtration markers may vary across populations and race/ethnicity coefficients may reflect nonbiological factors and may not be readily available. For these reasons, further advances in GFR estimation will likely require new filtration markers.
The discovery of several promising metabolites that correlate highly with mGFR or eGFR (5–8) was followed by a report suggesting that a panel of metabolites more accurately estimates GFR than eGFRcr or eGFRcys, and is as accurate as eGFRcr-cys, without including creatinine, cys-tatin C, or demographic characteristics (9). Here, we describe the development and analytical validation of an ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS), laboratory-developed test that quantitatively measures 4 serum metabolites (N-acetylthreonine, pseudouridine, phenyla-cetylglutamine, and tryptophan), as well as the development and clinical validation of an equation based on these 4 metabolites alone (eGFRmet) in 4 independent research studies, totaling 2424 participants with mGFR data.
Materials and Methods
ANALYTICAL VALIDATION
Reagents and instrumentation.
Mass spectrometric grade formicacid (98%), ammonium formate (≥98%), bilirubin, L-tryptophan, and Intralipid® were obtained from Sigma- Aldrich, along with HPLC-grade acetonitrile and methanol (VWR), BSA (IgG, fatty acid, and protease-free) (GenDEPOT), PBS (Fisher Scientific), β-pseudouridine and acetyl-L-threonine (Santa Cruz Biotechnology), phenylacetyl-L-glutamine, β-pseudouridine-13C,15N2, and L-tryptophan-d5 (Toronto Research Chemicals), Nα-(phenyl-d5-acetyl)-L-glutamine and N-acetyl-d3-L- threonine-2,3-day2 (C/D/N Isotopes), and human serum (Bioreclamation). Deionized water (18 mol/LΏ) was purified using a Hydro water purification system. Centrifugation was done with a Sorvall ST-40R centrifuge (Thermo Scientific).
Calibrators, internal standards, and quality control (QC) samples.
The assay quantitative range for each analyte was established to encompass the 95% reference interval from the analysis of 2199 samples during assay development (see Fig. 1 in the Data Supplement that accompanies the online version of this article at http://www.clinchem.org/content/vol65/issue3). Eight calibrators, prepared by spiking aqueous analyte solutions into surrogate matrix (7.5% BSA in PBS), were used to cover the quantification ranges. The working internal standard solution was made by adding internal standards into acetonitrile/ methanol/water/formic acid extraction solution (88: 10:2:0.2) at 0.300, 0.400, 0.600, and 1.50 μg/mL for N-acetyl-d3-L-threonine-2,3-d2, Nα-(phenyl-d5-acetyl)-L-glutamine, β-pseudouridine-13C,15N2, and L-tryptophan-d5, respectively. Calibration standards were stored at −80 °C; stock solutions, calibration spiking solutions, and internal standard solutions were stored at 4 °C.
Low-, mid-, and high-level QC samples were prepared from lots of human serum of appropriate analyte concentrations with fortification of analytes as necessary. Lower limit of quantification (LLOQ) QC samples were prepared independently from the calibration standards by diluting purified analytes into a BSA solution at the concentration of the lowest standard for all analytes. QC samples were stored at −80 °C.
Sample preparation.
Sample preparation was carried out in a polypropylene 96-well plate. Each well received 175 μL of working internal standard, except for blanks that received 175 μL of blank extraction solvent. For standards, QCs, and study samples, 25 μL of appropriate matrix (thawed on ice) was transferred to corresponding wells; for blanks and blanks with internal standard, 25 μL of BSA solution was transferred. The plate was capped, vortex-mixed for 2 min at ambient temperature, and centrifuged for 10 min at 3488g at 4 °C. An aliquot (150 μL) ofsupernatant was transferred to a new plate for LC-MS/MS analysis.
Chromatography.
An Agilent 1290 Infinity ultra-high performance liquid chromatography system, equipped with a binary solvent pump, a refrigerated autosampler (4 °C), and a column heater (60 °C), was used with a hydrophilic interaction liquid chromatography column (Waters ACQUITY UPLC® BEH Amide, 1.7 μm, 2.1 × 150 mm). Mobile phases were 20 mmol/L ammonium formate in water with 1% formic acid and acetonitrile. Acetonitrile/water (50:50) was used for needle wash. A linear gradient elution was carried out with a gradient of 12% to 42% mobile phase A. The flow rate was 550 μL/min, and the total run time was 3.7 min. A fixed aliquot of 1.0 μL of the final supernatant was injected for each sample. The eluent was directly introduced into the electrospray source of a mass spectrometer.
Mass spectrometry.
A Sciex 5500 QTRAP was used in this study and operated in positive multiple reaction monitoring mode. Detailed mass spectrometry settings for each analyte are listed in Table 1 of the online Data Supplement. Data were acquired and processed using Analyst® 1.6.2 software. For quantification, peak area ratios of analyte to internal standard were fitted against the concentrations of the calibration standards by weighted (1/x2) linear least-squares regression analysis. The resulting slope and intercept of the calibration curve were used to calculate the concentrations in unknown samples.
CLINICAL VALIDATION
Overall approach.
Our goal was to develop and validate a GFR-estimating equation (eGFRmet) using a panel of novel metabolites only (not including creatinine, cystatin C, or age, sex, and race). A cross-sectional design was used to compare the performance of eGFRmet and GFR- estimating equations to measured GFR.
Study populations and measurement methods.
Four populations from research studies were selected such that the total study population was diverse across the range of GFR and racial/ethnic composition (10–13) (see Table 2 in the online Data Supplement). Given the known variation across geographical areas, we restricted the studies to North America and Europe, as previously done (4, 14). Participants were included from the African American Study of Kidney Disease and Hypertension (AASK) who had a GFR measurement performed during the screening visit (n = 1609) (10), Modification of Diet in Renal Disease (MDRD) Study participants at the 12-month follow-up visit (n = 678) (11), Age, Gene/Environment Susceptibility (AGES)-Kidney Study (n = 764) (12),and Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease (CRISP) Study (n = 197) (13).
The total participant population was 3,248, which was divided randomly into data sets for development (50%; n = 1624, n = 1618 with metabolites, n = 1615 with complete measurements) and validation (25%; n = 812, n = 811 with metabolites, n = 809 for complete measurements), with the remaining 25% reserved for future validation. The eGFRmet equation was developed in the development data set, and its coefficients were fixed for subsequent analyses. In a sensitivity analysis, we developed eGFRmet in 3 studies (MDRD, AGES-Kidney, and CRISP studies) and validated it in 1 study (AASK). Table 2 in the online Data Supplement shows the GFR and laboratory cystatin C measurements used in these studies (4). For consistency with the approach of developing a panel assayed by mass spectrometry, we used an analytically validated UPLC-MS/MS creatinine assay (CV, 3.4%).
Analytical methods.
Metabolite concentrations, creatinine, cystatin C, and GFR were log-transformed. Multiple linear regression was used to develop GFR-estimating equations with the metabolites, with and without age, sex, and race, or creatinine and cystatin C. The reference equations were based on the Chronic Kidney Disease Epidemiology (CKD-EPI) equations recommended for clinical use (1, 4, 14). To optimize the CKD-EPI equations’ performance in these populations, we refit the equations without changing the knots for the 2-slope linear splines for creatinine and cystatin C. Additionally, the performance of other GFR-estimating equations using creatinine and cystatin C was evaluated (15–19).
Statistics.
mGFR and eGFR were expressed per 1.73 m2 body surface area. The performance of eGFR equations in estimating mGFR was evaluated in the development and validation data sets. Performance was quantified using established metrics (14). The 2002 Kidney Disease Outcomes Quality Initiative guidelines considered an eGFR within 30% of mGFR as generally satisfactory for clinical practice, and as a performance measure, recommended that >90% of the eGFRs determined in a validation population be within 30% of the mGFR (P30 > 90%) (19), equivalent to <10% of eGFRs with large errors (quantified as 1 — P30 <10%). The root mean square error (RMSE) was used on the log scale to approximate the SD of the percent error in estimation. Bias was expressed as the mGFR - eGFR. To calculate P values, we used a signed rank test for RMSE, McNemar κ2 for 1 — P30, and bootstrapping for bias implemented in Stata version 14.2 (StataCorp LP).
Classification of all equations was assessed by evaluating the concordance for eGFR and mGFR by GFR categories and by area under the ROC curve for detecting mGFR threshold of 60 mL/min/1.73 m2. Improvement in participant classification to mGFR ≥60 mL/min/1.73 m2 by eGFRmet and other equations compared with eGFRcr was evaluated using net reclassification index statistic.
Subgroups were defined by eGFR (≥90, 60–89, 45–59, 30–44, <30 mL/min/1.73 m2), age (<40, 40–64, ≥65 years), sex, race, body mass index (BMI; <20, 20–25, 26–30, >30 kg/m2), and study. For evaluation of bias, 1 — P30, and reclassification in subgroups, the development and validation data sets were combined to avoid small sample size. P < 0.05 was considered significant for all comparisons, without adjustment for multiple comparisons.
Results
ANALYTICAL VALIDATION
Method development.
All analytes ionized well in positive mode using electrospray, and product ions were selected for quantification using individual tuning. Because of the analytes’ low molecular mass, suitable alternative product ions were not found to use transition ratios in this method. The corresponding multiple reaction monitoring transitions of isotopically labeled internal standards were selected to match those of the analytes. Hydrophilic interaction liquid chromatographic conditions were developed to retain and separate the analytes (see Fig. 2 in the online Data Supplement).
Imprecision and linearity.
Intraassay and interassay imprecision of analyte values in serum was evaluated at 3 QC levels (low, mid, and high). During the validation, 3 replicates per QC level were analyzed in each run, in 2 runs per day over 20 days (20). Interassay imprecision for every analyte at each QC level is shown in Table 1 (see also Table 3 in the online Data Supplement for intraassay imprecision). For each QC level, 120 replicates were included in the interassay calculations with resulting CVs ≤6.4%. Multiinstrument validation on 3 identical LC- MS/MS systems used 5 replicates of QCs analyzed in each run, with 5 runs per instrument (20). Twenty-five replicates per QC level were used to calculate the interassay imprecision per instrument (see Table 4 in the online Data Supplement), with 75 replicates used to calculate the interinstrument variability across systems (CV ≤6.9%) (Table 1).
Table 1.
Interassay imprecision forserum QCs.
| Validation (n = 120) |
Multiinstrument validationa (n = 75) |
||||
|---|---|---|---|---|---|
| Analyte | Level | Mean concentration, μg/mL | CV, % | Mean concentration, μg/mL | CV, % |
| Acetylthreonine | Low | 0.0774 | 5.2 | 0.0775 | 5.8 |
| Mid | 0.892 | 5.1 | 0.867 | 5.5 | |
| High | 1.78 | 4.8 | 1.69 | 4.8 | |
| Phenylacetylglutamine | Low | 0.329 | 5.2 | 0.337 | 4.2 |
| Mid | 8.17 | 4.0 | 8.30 | 4.5 | |
| High | 16.0 | 3.8 | 16.1 | 4.8 | |
| Pseudouridine | Low | 1.27 | 5.2 | 1.30 | 6.9 |
| Mid | 16.4 | 4.2 | 16.5 | 5.7 | |
| High | 32.1 | 3.7 | 31.7 | 5.6 | |
| Tryptophan | Low | 4.08 | 6.4 | 4.13 | 4.6 |
| Mid | 44.7 | 4.3 | 44.6 | 4.0 | |
| High | 88.0 | 4.3 | 87.3 | 4.7 | |
Data reported are from all 3 LC-MS/MSsystemstested.
Table 4.
Baseline patient characteristics of the development and validation data sets.
| Development (n = 1615) |
Validation (n = 809) |
|||
|---|---|---|---|---|
| Characteristic | Mean (SD) 5th-95th percentile | Mean (SD) 5th-95th percentile | ||
| Age, years | 59(16) | 32–82 | 58(16) | 30–82 |
| Male, % | 57 | 57 | ||
| Black, % | 52 | 51 | ||
| Diabetes, % | 3.2 | 4.0 | ||
| BMI, kg/m2 | 29(6) | 21–41 | 29 (6) | 21–40 |
| mGFR, mL/min/1.73 m2 | 55 (26) | 16–98 | 56 (25) | 16–98 |
| mGFR category, n | ||||
| <15 | 62 | 33 | ||
| 15–29 | 240 | 112 | ||
| 30–44 | 300 | 143 | ||
| 45–59 | 330 | 147 | ||
| 60–89 | 557 | 306 | ||
| ≥90 | 126 | 68 | ||
| Creatinine, mg/dL | 1.9 (1.2) | 0.8–4.1 | 1.8 (1.1) | 0.8–4.2 |
| Cystatin C, mg/L | 1.6 (0.8) | 0.8–3.3 | 1.5 (0.8) | 0.8–3.2 |
| Pseudouridine, μg/mL | 1.6 (1.1) | 0.7–3.8 | 1.6 (1.1) | 0.7–3.8 |
| Phenylacetylglutamine, μg/mL | 1.7 (1.9) | 0.3–4.8 | 1.6 (1.6) | 0.3–4.7 |
| Acetylthreonine, μg/mL | 0.16(0.1) | 0.08–0.37 | 0.16(0.1) | 0.07–0.37 |
| Tryptophan, μg/mL | 12(3) | 7–17 | 12(3) | 7–17 |
Linear responses (r2 > 0.98) were observed over a 100-fold range for acetylthreonine, pseudouridine, and tryptophan and over a 200-fold range for phenylacetyl- glutamine on all instruments (see Table 2 here and Tables 5 and 6 in the online Data Supplement).
Table 2.
Analytical ranges and interassay linearity data.
| Range | Validation (n = 40) | Multiinstrument validationa (n =15) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Analyte | LLOQ | ULOQb | Mean slope (SD) | Mean intercept (SD) | Mean r2 | Mean S | Mean slope (SD) | Mean intercept (SD) | Mean r2 | Mean S |
| Acetylthreonine | 0.0200 | 2.00 | 0.521 (0.0284) | 0.000320(0.000559) | 0.9989 | 0.0212 | 0.663(0.0326) | 0.0000905 (0.000623) | 0.9987 | 0.0282 |
| Phenylacetylglutamine | 0.100 | 20.0 | 0.453 (0.0186) | −0.00127(0.00258) | 0.9986 | 0.224 | 0.594 (0.0463) | 0.00300 (0.00306) | 0.9980 | 0.237 |
| Pseudouridine | 0.400 | 40.0 | 0.236 (0.00852) | 0.00119 (0.00491) | 0.9981 | 0.639 | 0.268 (0.00856) | 0.00113 (0.00644) | 0.9971 | 0.737 |
| Tryptophan | 1.00 | 100 | 0.0306 (0.00189) | −0.00210 (0.00294) | 0.9960 | 1.37 | 0.0348 (0.00375) | −0.00390 (0.00287) | 0.9956 | 1.51 |
Data reported are from all 3 LC-MS/MS systems tested.
ULOQ, upper limit of quantitation; S, standard error of the regression.
LLOQ. Recovery and imprecision for each analyte at the LLOQ were evaluated by analyzing the LLOQ QC in triplicate in 2 runs per day for 15 days. Interassay recovery and imprecision were calculated using 90 replicates of the LLOQ QC with resulting recoveries between 95.5% and 102% with CVs ≤9.3% (see Table 3 here and Table 7 in the online Data Supplement for intraassay data). In the multiinstrument validation, 5 replicates of the LLOQ QC were analyzed per run, with 5 runs per instrument. Twenty-five replicates were included in the interassay calculations per instrument (see Table 8 in the online Data Supplement), with 75 replicates included in the combined instrument data. Interinstrument recoveries were between 95.4% and 104% with CVs ≤8.8% (Table 3). The signal-to-noise ratio for every analyte at the LLOQ exceeded the 5:1 specification.
Table 3.
Interassay imprecision and recovery of analyte values at the LLOQ.
| Analyte | LLOQ QC Actual concentration, μg/mL |
Validation (n = 90) | Multiinstrument validationa (n = 75) |
||||
|---|---|---|---|---|---|---|---|
| Mean measured concentration, μg/mL |
CV, % |
Recoveryb, % | Mean measured concentration, μg/mL |
CV, % |
CV, Recoveryb, % |
||
| Acetylthreonine | 0.0200 | 0.0203 | 6.5 | 102 | 0.0199 | 6.7 | 99.5 |
| Phenylacetylglutamine | 0.100 | 0.0955 | 8.6 | 95.5 | 0.0954 | 6.7 | 95.4 |
| Pseudouridine | 0.400 | 0.393 | 7.0 | 98.3 | 0.394 | 8.8 | 98.5 |
| Tryptophan | 1.00 | 1.02 | 9.3 | 102 | 1.04 | 7.9 | 104 |
Data reported are from all 3 LC-MS/MS systems tested.
Recovery of the LLOQ QC measurement is the comparison of the quantification of the independently prepared LLOQ QC sample against the nominal concentration.
Extraction recovery.
To assess the extraction recovery of analytes, mid QC samples were fortified with concentrations of analytes equivalent to standard E (0.600, 3.00, 12.0, and 30.0 μg/mL for acetylthreonine, phenylacetyl- glutamine, pseudouridine, and tryptophan, respectively). Six replicates of the recovery samples were extracted and analyzed alongside triplicate control mid QC samples in the corresponding run. Extraction recoveries of the added analytes ranged from 98.5% to 113% (see Table 9 in the online Data Supplement).
Interference and selectivity.
Icteric, lipemic, and hemolytic interferences on analyte quantification were assessed using 4 concentrations ofbilirubin (34.7–139 μg/mL), Intralipid (0.625–2.50 μL/mL), or hemolyzed red blood cells (0.25%–4%) spiked into low and high QC samples. The target interference concentrations spanned the range of interference indices established on an in-house Abbott Architect ci4100 analyzer. Each sample was extracted and analyzed in triplicate and compared with solvent-spiked controls at each QC level. Analyte quantification values showed biases of ≤9.8% (see Table 10 in the online Data Supplement).
Representative pharmaceuticals from commonly prescribed classes of drugs (e.g., statins, pain relievers, nonsteroidal antiinflammatory drugs) were spiked into low and high QC samples at approximately twice their therapeutic concentrations to investigate quantitative interference. Spiked samples were extracted and analyzed in triplicate and compared with solvent-spiked controls at each QC level. Bias ranged from –4.9% to 4.1%, indicating that those pharmaceuticals did not interfere with quantification (see Table 11 in the online Data Supplement).
Collection tube interference was evaluated using blood collected from 10 individual donors into serum nonseparator and separator tubes. The matrix was handled per standard processing procedures for each tube type, and the resulting serum from each donor was extracted in triplicate and compared for differences in over-all analyte quantification. The differences ranged from –7.8% to 7.2% (data not shown). Transfer tube interference (Nalgene cryovial) was also tested by storage with water refrigerated for 3 days or with matrix at the low and high QC levels on ice for 30 min. The transfer tube interference showed analyte bias between −4.8% and 1.5% (data not shown).
Solutions of commercially available isomers of the 4 analytes were tested to assess the selectivity of the method. These solutions were extracted and evaluated for interfering peaks at the retention times of the corresponding analytes. No interfering peaks were identified (see Table 12 in the online Data Supplement).
Patient specimen linearity.
Varying proportions of the low and high QC samples, prepared from separate serum lots, were mixed to form 8 different concentrations to assess whether matrix components would interfere with analyte measurement. Each concentration was analyzed in triplicate, and bias was calculated as the deviation of the mean measured value from the calculated theoretical value. These values were plotted against each other, and a linear regression was calculated for each analyte. Biases ranged from −9.4% to 0.5%, and linear responses were observed for all analytes with r2 > 0.9980 (see Table 13 in the online Data Supplement).
Matrix effect.
Matrix effect was evaluated by performing a postcolumn infusion experiment using an internal standard solution during injection of 10 individual lots of serum collected in 2 different tube types (separator vs nonseparator) and extracted without internal standards. By monitoring the internal standard transitions, any suppression or enhancement at the retention time of the analytes can be observed. Only tryptophan was affected by significant matrix effect in the retention area. Tryptophan elutes just before an area of suppression that exceeds 25% of the unaffected signal. Because the coeluting internal standards in this assay are isotopically labeled, and peak area ratios of the analyte to internal standard were used for the quantification, any minor matrix effect should have occurred similarly for both and be compensated for in the final concentration calculation.
Carryover.
Analyte carryover was determined by injecting a blank sample (7.5% BSA solution) immediately after the highest standard. Peak areas in the blank sample were compared with the corresponding peak areas in the lowest standard. There was no detectable carryover for acetylthreonine, pseudouridine, or tryptophan. Carryover was observed only once for phenylacetylglu-tamine at 4% of the concentration of standard A (data not shown).
Stability.
QC samples at 3 concentration levels were used to investigate the stability of analytes in human serum. Short-term stability experiments were conducted in triplicate, and bias was determined by comparing mean values of stressed samples against those of nonstressed samples. For room temperature stability, QC samples were stored on the benchtop for 8, 24, and 48 h. The biases from the control QC values were between −7.5% and 14.7% for all analytes across all timepoints (see Table 14 in the online Data Supplement). For refrigerator stability, QC samples were stored at 4 °C for 3 and 7 days. Additionally, serum from 10 individual donors collected in 2 different tube types (separator vs nonseparator) was stored at 4 °C for 2, 3, 4, and 6 days and compared with fresh matrix that was immediately processed. The biases for the QC samples ranged from −8.1% to 10.0% (see Table 14 in the online Data Supplement), and the individual donors were between −15.5% and 13.1% from the fresh sample values (see Table 15 in the online Data Supplement). Freeze—thaw stability was evaluated on the QCs after 5 freeze—thaw cycles at −80 °C with biases ranging from −2.4% and 4.3% (see Table 14 in the online Data Supplement). Fresh vs frozen stability was conducted using serum from 10 individual donors with biases ranging between −9.6% and 13.1% from their corresponding control samples (see Table 15 in the online Data Supplement). Long-term stability of the QCs was examined at 1 year. All QC levels showed <5% bias for all analytes compared with their initial values (see Table 14 in the online Data Supplement).
Blood collected in serum separator tubes was drawn from 3 individual donors to test analyte stability in whole blood. One tube was processed immediately for each donor (control); a second tube of blood was left at room temperature for 1 h; the third and fourth tubes were stored at 4 °C for 4 and 8 h before analysis. Analytes were stable in whole blood at room temperature for 1 h with biases ranging from −2.5% to 12.0%, and stability was established at 4 °C for 8 h with biases between −2.6% and 13.3% (see Table 16 in the online Data Supplement).
Extract stability was tested by reanalyzing QC extracts after 3 days at 4 °C and quantifying against the original standard curve. The mean percent biases from the initial QC values were between −5.0% and 11.3% (data not shown). Run injection stability was also established for 3 days by reinjecting a complete plate after storage at 4 °C and comparing analyte values with the initial injection.
Algorithm stability was conducted by assessing the total variability of the eGFRmet equation in the 3 QC levels over the 20-day validation, taking into account the combined contribution of the 4 analytes’ imprecision. The data showed that the eGFRmet score variability in QCs was not more than ±5 points (mL/min/1.73 m2) (with 1 outlier) and CVs <5.1% (see Fig. 3 in the online Data Supplement).
CLINICAL VALIDATION
Study populations.
Clinical characteristics of the participants in the development and validation data sets are shown in Table 4. In the development data set, mean (SD) age was 59 (16) years, 57% were men, 52% were black, and 3.2% had diabetes. Mean (SD) BMI was 29 (6) kg/m2. Mean (SD) mGFRwas 55 (26) mL/min/1.73 m2. The clinical characteristics were similar in the validation data set.
Correlations.
Correlations of the 4 novel metabolites with mGFRin the development dataset were −0.93, −0.90, –0.63, and +0.62 for pseudouridine, acetylthreonine, phenylacetylglutamine, and tryptophan, respectively, vs –0.84 and −0.92 for creatinine and cystatin C, respectively (see Table 17 in the online Data Supplement). After adjusting for mGFR, partial correlations of pseudouridine, acetylthreonine, creatinine, and cystatin C with each other were ≥0.4, whereas the other metabolites were minimally correlated (≤0.1).
Equations in development data set.
In the 4 novel metabolites’ equation (eGFRmet), pseudouridine had the largest (negative) coefficient, whereas the coefficient for tryptophan is positive (Table 5, upper panel). The coefficients and standardized coefficients for this and all other equations developed in the development data set are shown in Table 18 of the online Data Supplement.
Table 5.
eGFRmet equation and performance of GFR-estimating equations in the development and validation data sets.
| eGFRmet equationa | |||||
|---|---|---|---|---|---|
| In(eGFRmet) = 3.336 – 0.805 * In(pseudouridine) − 0.047 * In(phenylacetylglutamine) − 0.075 * ln(acetylthreonine) + 0.266 * In(tryptophan) | |||||
| eGFRmet = 28.106 * pseudouridine−0 805 * phenylacetylglutamine 0047 * acetylthreonine−0.075 * tryptophan0.266 | |||||
| Development (n = 1615) |
Validation (n = 809) |
||||
| Estimating equationsb | 1 - P30, % | RMSE | 1 -P30,% | RMSE | Median bias, mL/min/1.73 m2 |
| Reference equations | |||||
| 1. eGFRcr | 15.734567 | 0.225234567 | 13.134567 | 0.21934567 | 0.378 |
| 2. eGFRcys | 12.134567 | 0.203134567 | 12.0367 | 0.20934567 | 0.74247 |
| 3. eGFRcr-cys | 8.41267 | 0.1791247 | 8.7127 | 0.182127 | 0.403 |
| Equations using novel metabolites | |||||
| 4. Metabolites only (eGFRmet) | 9.41267 | 0.186123567 | 10.017 | 0.1881267 | −0.0102 |
| 5. Metabolites + demographics | 8.81267 | 0.1801247 | 9.817 | 0.1851267 | 0.181 |
| 6. Metabolites + cr-cys | 7.112345 | 0.1741247 | 8.312 | 0.17712457 | −0.100 |
| 7. Metabolites + demographics + cr-cys | 6.512345 | 0.164123456 | 7.012345 | 0.169123456 | 0.1142 |
Equations were modeled on the natural logarithmic scale. Units for coefficients are mL/min/1.73m2 per 1.0 μg/mL. Eq. 4 was developed in the full data set (n = 1618) and chosen as the primary equation before running the validation samples. All other equations were developed in the data set with complete cystatin C (n=1615). Eq. 4 developed in sample with n = 1615 rather than n = 1618; all coefficients are the same except for pseudouridine, which is −0.804 instead of −0.805.
Performance for all equations (1–7) is compared with each of the other equations. Reference equations are CKD-EPI equations refit for the development data set. Performance in the validation data set is for algorithms fixed in the development data set (coefficients are not refit to the validation data set). No underline marking indicates that the equation is better than the numbered (1–7) comparison equation at P < 0.05; an underline marking indicates that the equation is worse than the numbered comparison equation at P < 0.05. Bias is defined as mGFR−eGFR, so a positive value indicates an underestimate, whereas a negative value indicates an overestimate. Bias in the development sample is approximately zero because all equations including the reference equations were fit in the development data set.
Performance.
Performance of the equations developed in the development and validation data sets is compared in Table 5 (lower panel). In the development data set, 1 — P30 of eGFRmet (9.4%) was significantly lower than both eGFRcr and eGFRcys (15.7% and 12.1%, respectively) but not significantly different than eGFRcr-cys (8.4%). Addition of demographic characteristics, creatinine, and cystatin C improved 1 — P30 (6.5%), which was significantly better than for eGFRcr-cys. RMSE for eGFRmet was significantly lower than for eGFRcr and eGFRcys but not as low as eGFRcr-cys, although RMSE for the equation with all filtration markers and demographics was the best. In the validation data set, bias was minimal and 1 — P30 and RMSE were generally similar to values observed in the development data set. Performance of eGFRmet varied across studies (see Table 19 in the online Data Supplement), but sensitivity analysis showed generally similar findings for comparisons of GFR-estimating equations developed in 3 of the studies (MDRD, AGES, and CRISP) and validated in the remaining study (AASK) (see Table 2Q in the online Data Supplement).
Performance of the CKD-EPI equations without refitting and of other equations for eGFRcr, eGFRcys, and eGFRcr-cys is shown in Table 21 of the online Data Supplement. As expected, the CKD-EPI equations with-out refitting had variable bias in the development and validation data sets; thus, 1 — P30 and RMSE were slightly higher than the equations refit in the development data set.
Classification and reclassification.
Concordance of eGFR categories with mGFR and area under the ROC curve for mGFR threshold of 60 mL/min/1.73 m2 was better for eGFRmet than eGFRcr, but not better than eGFRcys or eGFRcr-cys, and was best for the equation with all filtration markers and demographics (see Table 22 in the online Data Supplement).
eGFRmet led to significant reclassification compared with eGFRcr (net reclassification index, 5.6; 95% CI, 2.4–8.7) (Table 6). Using an eGFR threshold of 60 mL/min/1.73 m2, we found that eGFRmet reclassified 349 of 2424 (14.4%) participants. Reclassification was more often correct than incorrect in participants with mGFR >60 mL/min/1.73 m2 but not in participants with mGFR <60 mL/min/1.73 m2. In the subgroup with eGFRcr of 45 to 59 mL/min/1.73 m2 (n = 577), eGFRmet correctly reclassified 127 of200 (63.5%) with mGFR ≥60 mL/min/1.73 m2, but incorrectly reclassified 68 of 377 (18%) with mGFR <60 mL/min/1.73 m2. The net reclassification index was also significant for eGFRcys (Eq. 2), eGFRcr-cys (Eq. 3), and eGFRmet + demographics + cr-cys (Eq. 7) (see Tables 23–25 in the online Data Supplement).
Table 6.
Reclassification of participants in the combined development and validation data sets using eGFRmet vs eGFRcr according to mGFR greater and lower than 60 mL/min/1.73 m2.
| Total group | Subgroup with mGFR <60 mL/min/1.73 m2 |
Subgroup with mGFR ≥60 mL/min/1.73 m2 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| eGFRcr range, mL/min/1.73 m2 |
n | Total reclassified n (%) |
n | Correctly reclassifieda n (%) |
Incorrectly reclassifiedb n (%) |
Net difference, % |
n | Correctly reclassifiedb n (%) |
Incorrectly reclassifieda n (%) |
Net difference, % |
Overall NRIc
(95% Cl) |
| Overall | 2424 | 349 (14.4) | 1,367 | 72 (5.3) | 72 (5.3) | 0 | 1057 | 132 (12.5) | 73 (6.9) | 5.6 | 5.6 (2.4–8.7) |
| 30–89 | 1864 | 347 (18.6) | 942 | 72(7.6) | 72 (7.6) | 0 | 922 | 132 (14.3) | 71 (7.7) | 6.6 | 6.6 (2.7–10.5) |
| 45–74 | 1123 | 322 (28.7) | 495 | 68(13.7) | 68 (13.7) | 0 | 628 | 127 (20.2) | 59 (9.4) | 10.8 | 10.8 (4.5–17.1) |
| 60–74 | 546 | 127 (23.3) | 118 | 68(57.6) | NAd | NA | 428 | NA | 59 (13.8) | NA | NA |
| 45–59 | 577 | 195 (33.8) | 377 | NA | 68 (18.0) | NA | 200 | 127 (63.5) | NA | NA | NA |
eGFRcr ≥ 60 and eGFRmet < 60.
eGFRcr < 60 and eGFRmet ≥ 60.
Values for NRI can be from −200 to 200. P < 0.001 for all.
NA, not applicable; NRI, net reclassification index.
Performance in subgroups.
The performance of equations in subgroups defined by eGFR, age, sex, race, and BMI in the combined development and validation data sets is shown in Fig. 1. Equations using novel metabolites generally had only minor variation in bias across subgroups. Improvement in 1 — P30 compared with eGFRcr and eGFRcys was observed in most subgroups. Results were similar in the separate development and validation data-bases (see Figs. 4 and 5 in the online Data Supplement).
Fig.1. Performance of GFR-estimating equations in subgroups in the combined development and validation data sets.
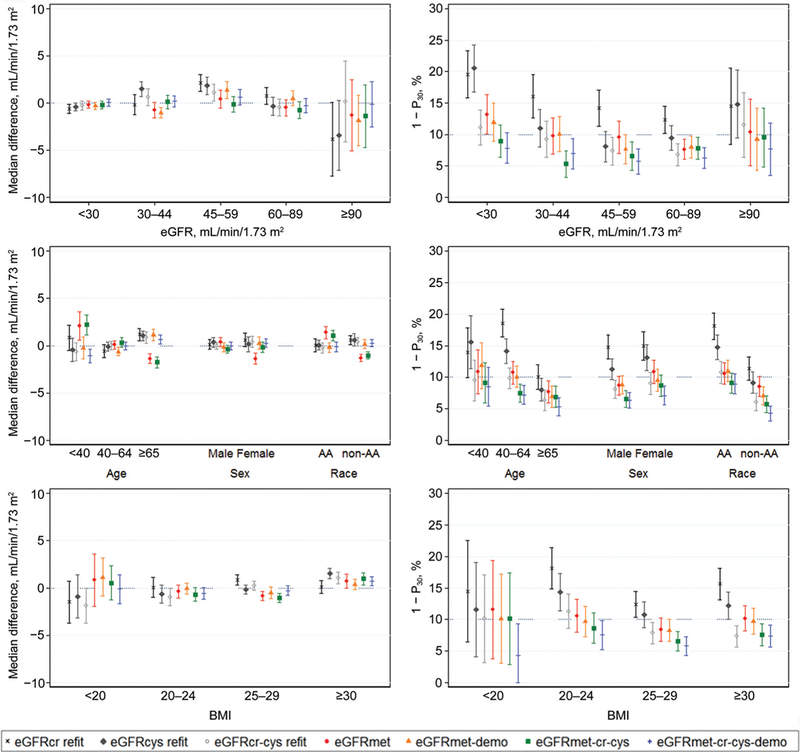
The left 3 panels show median difference (mGFR-eGFR) in mL/min/1.73 m2 (a positive value indicates an underestimate, whereas a negative value indicatesan overestimate). Theright3 panels show theproportionof large errors (1 - P30). Equations: eGFRcr refit, eGFRfrom creatinine (includes demographics) refit in the combined database; eGFRcys refit, eGFR from cystatin C (includes demographics) refit in the combined database; eGFRcr-cys refit, eGFR from creatinine and cystatin C (includes demographics) refit in the combined database; eGFRmet (eGFR from metabolites only) developed in the combined database; eGFRmet-demo, eGFR from metabolites and demographic characteristics developed in the combined database; eGFRmet-cr-cys, eGFR from metabolites, creatinine, and cystatin Cdeveloped in the combined database;eGFRmet- cr-cys-demo, eGFR from metabolites, creatinine, cystatin C, and demographic characteristics developed in the combined database. Sample size for each eGFR subgroup varies among equations. Sample size for subgroups-age (years): <40,295; 40–64,1232; ≥65,897; sex: male, 1385; female, 1039; race: African American (AA), 1255; other, 1169; BMI (kg/m2): <20, 69; 20–24,557; 25–29,931; ≥30, 867.
Discussion
Despite progress made in developing more precise and accurate GFR equations, their performance is limited by non-GFR determinants that affect the serum concentration of the endogenous filtration markers used to estimate GFR. Only eGFRcr-cys meets the recommended performance standard that <10% of estimates differ by >30% from measured GFR (1 — P30 < 10%). Here, we sought to develop anew GFR-estimating equation based on a panel of metabolites that was more accurate than eGFRcr and eGFRcys and independent of creatinine, cystatin C, or demographic characteristics. We now report the development and analytical validation of a UPLC-MS/MS method for the quantitative measurement of 4 novel filtration markers in human serum (acetylthreonine, phenylacetylglutamine, pseudouridine, and tryptophan) and the development and clinical validation of an algorithm (eGFRmet) to provide a more accurate assessment ofGFR. The analytical performance of this assay was validated in a clinical laboratory using Clinical and Laboratory Standards Institute guidelines and on 3 identical LC-MS/MS systems. All analytes passed acceptance criteria for each test conducted and were stable within the timeframes established. The accuracy of eGFRmet (1 — P30 of 9.4% in development and 10.0% in validation), even without the use of demographic variables, was greater than eGFRcr and eGFRcys and consistent among subgroups defined by eGFR, age, sex, race, and BMI, thereby offering an alternative and independent means for accurate GFR estimation. eGFRmet was not more accurate than eGFRcr-cys (1 — P30 of 8.4%), but combining the 4 eGFRmet metabolites with creatinine, cystatin C, and demographic characteristics provided the most accurate GFR estimate (1 — P30 of 6.5% in development and 7.0% in validation). These results have implications for further research and patient treatment in clinical practice.
The serum concentrations of all endogenous filtration markers are influenced by their non-GFR determinants, including generation, tubular reabsorption and secretion, and extrarenal elimination. In principle, markers that are less influenced by non-GFR determinants may provide a more accurate estimate of GFR, and using a panel offiltration markers whose non-GFR determinants are not strongly correlated with each other would reduce error in GFR estimates (21). Our results support these principles: eGFRmet, based on metabolites with varying correlation with each other, is more accurate than eGFRcr and eGFRcys. Although it does not require creatinine, cystatin C, or demographics characteristics, combining those with eGFRmet markers led to a panel more accurate than eGFRcr-cys.
Serum creatinine is affected by muscle mass, diet, and drugs inhibiting its tubular secretion. Serum cystatin C is affected by obesity, inflammation, smoking, and alterations in thyroid and adrenal hormones (22, 23). These conditions are commonly encountered in patients with kidney diseases and other illnesses limiting the accuracy and clinical utility of eGFRcr and eGFRcys. Although demographic characteristics (age, sex, and race) can serve as surrogates for some non-GFR determinants, the coefficients for the demographic characteristics represent average values for the relationships between the marker and its non-GFR determinants and do not account for the variation in other non-GFR determinants that occur in individuals with disease. The 4 markers in eGFRmet were among 15 targeted assays developed on metabolites correlated with mGFR (9). The 4 eGFRmet metabolites were selected based on a combination of factors: representation of distinct metabolic pathways, highest negative or positive correlations with mGFR, low correlations with each other after accounting for correlation with mGFR in most cases, and availability of reference materials.
Pseudouridine, phenylacetylglutamine, and acetyl- threonine are metabolic end-products eliminated in the urine and, thus, may have value as filtration markers. Pseudouridine, an abundant modified nucleoside (24), is the product of pseudouridine synthases’ action on RNA uridine residues. Although its function is not fully understood, pseudouridine is considered a tRNA stabilizing agent and potentially important for RNA translation. The perception of a steady pseudouridine production and its urinary excretion supports its potential as a filtration marker, although it appears to be reabsorbed and filtered, and some clinical situations alter its production (25–29). Phenylacetylglutamine, a microbiome-derived uremic solute (30), is an end-product of phenylalanine metabolism that is filtered, secreted, and normally found in human urine (31, 32). Association of phenylacetylglutamine with GFR and kidney disease was reported (5, 6). Acetylthreonine, the acetylated form of threonine, is the product of a normal enzymatic posttranslational amino acid acetylation process for which multiple roles have been suggested (33). Presence of acetyl-amino acids in blood matrices was documented, and the correlation between acetyl-amino acid concentrations and GFR was reported (5, 6). Tryptophan, an essential amino acid, was positively rather than negatively correlated with GFR. Kynurenine pathway upregulation, along with the increased production of indoxyl sulfate, a gut-derived tryptophan metabolite, was documented in chronic kidney disease and may explain, in part, the lower serum tryptophan concentrations observed with chronic kidney disease progression (34). Preliminary internal data show no significant change in tryptophan levels in fasted and nonfasted participants (data not shown). Further testing in diverse experimental and clinical settings as the development of the assay continues will help assess the potential non-GFR determinants of these markers and the gen- eralizability of our results. Because neither reference methods nor certified reference materials exist to evaluate accuracy of analyte measurement, it was necessary to approximate trueness using recovery experiments. How-ever, as the measurands are combined in an algorithm that was developed and validated against large patient cohorts to estimate GFR, then accuracy of the assay and resulting score (eGFRmet) is dependent on maintaining the integrity and reproducibility of the assay conditions and performance.
Strengths ofthis work include the extensive analyses demonstrating the validity and reproducibility of the assay and the assessment of eGFRmet performance in large retrospective and prospective cohorts offering a diverse data set with a wide range of GFR, ages, and high percentage of African Americans (5–7, 9, 35). Additionally, we compared performance with “best fit” eGFRcr, eGFRcys, and eGFRcr-cys, rather than other known equations, which optimizes their performance and avoids possible bias because ofvariation in assays for serum creatinine and cystatin C that may persist even after assay standardization (36–38). We evaluated the performance ofcreatinine, cystatin C, and demographic characteristics plus novel metabolites, which suggests that further improvement is possible.
There are several limitations of the analyses. First, sample handling in the MDRD and AASK studies did not follow a standardized protocol, and the storage period was many years. However, the creatinine data collected by LC-MS/MS in this study and the original clinical chemistry data were virtually equivalent. The results from the MDRD and AASK studies were consistent with the AGES and CRISP studies, suggesting sampling integrity. Second, acetylthreonine is highly correlated with pseudouridine, and tryptophan is not a retained solute, which may limit their usefulness in GFR estimation. Third, GFR measurement is known to be imprecise, which inflates the reported 1 — P30 and, therefore, would bias the results to the null (21). Fourth, participants with comorbid conditions were underrepresented, which might also bias toward the null, as eGFRcr and eGFRcys perform well in the absence ofcomorbid conditions, and, thus, the full advantages of eGFRmet might be underestimated in patients for whom creatinine and cystatin C are expected to perform well. Finally, validation in other data sets, especially in clinical settings in which creatinine or cystatin C are less reliable filtration markers, is desirable.
There is an unmet clinical need for a GFR-estimating equation that is more accurate than the eGFRcr or eGFRcys, particularly in clinical settings with large variation in the non-GFR determinants of serum concentrations of creatinine and cystatin C and in which clinical decision-making requires precise knowledge of the GFR. There is also an unmet clinical need for a GFR estimation equation that does not require demographic characteristics. eGFRmet may satisfy both these needs; it does not require age, sex, or race while being more accurate than the “best fit” eGFRcr and eGFRcys using the CKD-EPI equations. As with eGFRcr-cys, it could be used as a confirmatory and independent test for decreased eGFRcr. eGFRmet is not more accurate than eGFRcr- cys in this cohort; however, the “panel eGFR” approach can enable even more accurate GFR estimation. For example, future strategies would be to consider additional filtration markers or algorithms that optimize GFR estimates based on agreement among a subset of the markers.
In conclusion, we have demonstrated the successful development and analytical validation of a robust and reproducible UPLC-MS/MS method quantitating 4 human serum metabolites and described the development and clinical validation ofalgorithms for the estimation of GFR using said metabolites. Our results show that eGFRmet, which does not include creatinine, cystatin C, or demographic characteristics, is more accurate than either eGFRcr or eGFRcys. Like eGFRcr-cys, it may be useful as a confirmatory test for GFR estimation, but unlike eGFRcr-cys, it is independent of creatinine and cystatin C. GFR estimation can likely be improved further by use ofadditional filtration markers. Evaluation in additional study populations will be necessary to confirm the generalizability of these observations but also high-light the clinical settings in which eGFRmet may offer a significantly superior clinical value over current methods.
Supplementary Material
Acknowledgments:
The authors thank Sara Couture for assistance in manuscript preparation.
Research Funding: J. Coresh, peGFR R01 grant #5007386-SERV, funding from National Kidney Foundation and National Institute of Health to institution; L.A. Inker, peGFR R01 grant#5007386-SERV; V. Gudnason, Age, Gene/Environment Susceptibility—Reykjavik Study (AGES-Reykjavik) was funded by NIH contracts N01-AG-1– 2100 and HHSN27120120022C the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Alth-ingi (the Icelandic Parliament); V.E. Torres, NIH R01DK097020 Estimating GFR from a Panel of Endogenous Filtration Markers to institution; A.S. Levey, peGFR R01 grant #5007386-SERV, NI- DDK to institution.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, preparation of manuscript, or final approval of manuscript.
7 Nonstandard abbreviations:
- GFR
glomerular filtration rate
- mGFR
measured glomerular filtration rate
- eGFR
estimated glomerular filtration rate
- eGFRcr
estimated glomerular filtration rate using serum creatinine
- eGFRcys
estimated glomerular filtration rate using serum cystatin C
- eGFRcr-cys
estimate glomerular filtration rate using serum creatinine and cystatin C
- UPLC-MS/MS
ultra-performance liquid chromatography-tandem massspectrometry
- eGFRmet
estimated glomerular filtration rate using metabolite panel
- QC
quality control
- LLOQ
lower limit of quantification
- AASK
African American Study of Kidney Disease and Hypertension
- MDRD
Modification of Diet in Renal Disease Study
- AGES
Age, Gene/Environment Susceptibility Study
- CRISP
Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease
- CKD-EPI
Chronic Kidney Disease Epidemiology Collaboration; RMSE, root mean square error
- BMI
body mass index
Footnotes
Employment orLeadership: T.A. Freed, Metabolon, Inc.; D.R. Toal, Metabolon, Inc.; K.E.T.Vroom, Metabolon, Inc.; M.L. Oyaski, Metabolon, Inc.; L.A. Ford, Metabolon, Inc.; R. Perichon, Metabolon, Inc.
Consultant or Advisory Role: L.A. Inker, Tricida Inc, Omeros Corp.
Stock Ownership: R. Perichon, Metabolon, Inc.; K.E.T. Vroom, Metabolon, Inc.;J.E. Wulff,Metabolon, Inc.; L. Ford, Metabolon Inc.
Honoraria: None declared.
Expert Testimony: None declared.
Patents: T.A. Freed, US 62/435,967; J. Coresh, PCT/US2015/ 044567 (provisional patent); L.A. Inker, PCT/US2015/044567 (provisional patent); R. Perichon, US2014/037762; K.D. Goodman,US 62/435,967; D.M. Hauser, US 62/435,967; K.E.T. Vroom, US 62/ 435,967; L.A. Ford, US 62/435,967; A.S. Levey, PCT/US2015/ 044567 (provisional patent).
Other Remuneration: L.A. Ford, AACC Mass Spectrometry and Separation Sciences Division abstract award. Tufts Medical Center, John Hopkins University and Metabolon Inc haveacollaboration agreement to develop a product to estimate GFR from a panel of markers.
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
References
- 1.Kidney Disease: Improving Global Outcomes (KDIGO) CKD Workgroup. KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int 2013;3 Suppl:S1–150. [DOI] [PubMed] [Google Scholar]
- 2.Stevens LA, Coresh J, Greene T, Levey AS. Assessing kidney function-measured and estimated glomerular filtration rate. N Engl J Med 2006;354:2473–83. [DOI] [PubMed] [Google Scholar]
- 3.Levey AS, Inker LA, Coresh J. GFR estimation: from physiology to public health. Am J Kidney Dis 2014;63:820–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inker LA, Schmid CH, Tighiouart H, Eckfeldt JH, Feldman HI, Greene T, et al. Estimating glomerular filtration rate from serum creatinine and cystatin C. N Engl J Med 2012;367:20–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niewczas MA, Sirich TL, Mathew AV, Skupien J, Mohney RP, Warram JH, et al. Uremic solutes and risk of endstage renal disease in type 2 diabetes: metabolomic study.KidneyInt 2014;85:1214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solini A, Manca ML, Penno G, Pugliese G, Cobb JE, Fer- rannini E. Prediction ofdeclining renal function and al¬buminuria in patients with type 2 diabetes by metabo-lomics. J Clin Endocrinol Metab 2016;101:696–704. [DOI] [PubMed] [Google Scholar]
- 7.Niewczas MA, Mathew AV, Croall S, Byun J, Major M, Sabisetti VS,et al. Circulatingmodifiedmetabolitesandarisk ofESRDinpatientswithtype1diabetesandchronickidney disease. Diabetes Care 2017;40:383–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coresh J, Inker LA, Levey AS. Precise estimation of glo¬merular filtration rate from multiple blood biomarkers [Abstract]. J Am Soc Nephrol 2014;25:52A. [Google Scholar]
- 9. Coresh J, Inker LA, Sang Y, Chen J, Shafi T, Post WS, et al. Metabolomic profiling to improve glomerular filtration rate estimation: a proof of conceptstudy. [Epub ahead of print] Nephrol Dial Transplant April 30,2018. as doi: 10.1093/ndt/gfy094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis J, Agodoa L, Cheek D, Greene T, Middleton J, O’Connor D, et al. Comparison of cross-sectional renal function measurements in African Americans with hypertensive nephrosclerosis and of primary formulas to estimate glomerular filtration rate. Am J Kidney Dis 2001;38:744–53. [DOI] [PubMed] [Google Scholar]
- 11.Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med 1999;130:461–70. [DOI] [PubMed] [Google Scholar]
- 12.Fan L, Levey AS, Gudnason V, Eiriksdottir G, Andresdot- tir MB, Gudmundsdottir H, et al. Comparing GFR estimating equations using cystatin C and creatinine in elderly individuals. J Am Soc Nephrol 2015;26:1982–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman AB, Guay-Woodford LM, Grantham JJ, Torres VE, Bae KT, Baumgarten DA, et al. Renal structure in early autosomal-dominant polycystic kidney disease (ADPKD): the Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease (CRISP) cohort. Kidney Int 2003;64:1035–45. [DOI] [PubMed] [Google Scholar]
- 14.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, et al. A new equation to estimate glomerular filtrationrate.AnnInternMed 2009;150:604–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaeffner ES, Ebert N, Delanaye P, Frei U, Gaedeke J, Jakob O,et al. Two novel equations to estimate kidney function in persons aged 70 years or older. Ann Intern Med 2012;157:471–81. [DOI] [PubMed] [Google Scholar]
- 16.Nyman U, Grubb A, Larsson A, Hansson LO, Flodin M, Nordin G, et al. The revised Lund-Malmö GFR estimating equation outperforms MDRD and CKD-EPI across GFR, age and BMI intervals in a large Swedish population. Clin Chem Lab Med 2014;52:815–24. [DOI] [PubMed] [Google Scholar]
- 17.Grubb A, Horio M, Hansson LO, Björk J, Nyman U, Flodin M, et al. Generation of a new cystatin C-based estimating equationforglomerularfiltrationratebyuseof 7 assays standardized to the international calibrator. Clin Chem 2014;60:974–86. [DOI] [PubMed] [Google Scholar]
- 18.Pottel H, Hoste L, Dubourg L, Ebert N, Schaeffner E, Eriksen BO, et al. An estimated glomerular filtration rate equation for the full age spectrum. Nephrol Dial Transplant 2016;31:798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pottel H, Delanaye P, Schaeffner E, Dubourg L, Erik-sen BO, Melsom T, et al. Estimating glomerular filtration rate for the full age spectrum from serum creatinine and cystatin C. Nephrol Dial Transplant 2017;32:497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.CLSI. Evaluation of precision of quantitative measurement procedures; approved guideline 3rd Ed. CLSI document EP05-A3. Wayne (PA): Clinical and Laboratory Standards Institute; 2014. [Google Scholar]
- 21.Kwong YT, Stevens LA, Selvin E, Zhang YL, Greene T, Van Lente F, et al. Imprecision of urinary iothalamate clearance as a gold-standard measure of GFR decreases the diagnostic accuracy of kidney function estimating equations. Am J Kidney Dis 2010;56:39 −49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Foster MC,Tighiouart H, Anderson AH, Beck GJ, Contreras G, et al. Non-GFR determinants of low-molecular-weight serum protein filtration markers in CKD. Am J Kidney Dis 2016;68:892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foster MC, Levey AS, Inker LA, Shafi T, Fan L, Gudnason V, et al. Non-GFR determinants of low-molecular-weightserum protein filtration markers in the elderly: AGES-Kidney and MESA-Kidney. Am J Kidney Dis 2017;70:406–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, et al. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acid Res 2013;41:D262–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spenkuch F,Motorin Y, Helm M.Pseudouridine:still mysterious, but never a fake (uridine)! RNA Biol 2014;11:1540–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drahovsky D, Winkler A, Skoda J. Increased urinary pseudouridine excretion in rats following irradiation. Nature 1964;201:411–2. [DOI] [PubMed] [Google Scholar]
- 27.Rasmuson T, Bjork GR. Urinary excretion of pseudouridine and prognosis of patients with malignant lymphoma. Acta Oncol 1995;34:61–7. [DOI] [PubMed] [Google Scholar]
- 28.Weissman S, Eisen AZ, Lewis M, Karon M. Pseudouridine metabolism. III. Studies with isotopically labeled pseudouridine. J Lab Clin Med 1962;60:40 −7. [PubMed] [Google Scholar]
- 29.Dlugajczyk A, Eiler J. Lack of catabolism of 5-ribosyuridine in man. Nature 1966;212:611–2. [DOI] [PubMed] [Google Scholar]
- 30.Barrios C,Beaumont M,Pallister T,Villar J,Goodrich JK, Clark A, et al. Gut-microbiota-metabolite axis in early renal function decline. PLoS One 2015;10:e0134311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moldave K, Meister A. Synthesis of phenylacetylglu- tamine by human tissue. J Biol Chem 1957;229:463–76. [PubMed] [Google Scholar]
- 32.Leong S,Sao J, Taussig A, Plummer N, Meyer T, Sirich T. Residual function effectively controls plasma concentrations of secreted solutes in patients on twice weekly hemodialysis. J Am Soc Nephrol 2018;29:1992–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drazic A, Myklebust LM, Ree R, Arnesen T. The world of protein acetylation. Biochim Biophys Acta 2016;1864:1372–401. [DOI] [PubMed] [Google Scholar]
- 34.Saito K, Fujigaki S, Heyes MP, Shibata K, Takemura M, Fujii H, et al. Mechanism of increases in L-kynurenine and quinolinicacid in renal insufficiency. Am J Physiol Renal Physiol 2000;279:F565–72. [DOI] [PubMed] [Google Scholar]
- 35.Sekula P, Goek ON, Quaye L, Barrios C, Levey AS, Romisch-Margl W, et al. A metabolome-wide association study of kidney function and disease in the general population. J Am Soc Nephrol 2016;27:1175–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Killeen AA, Ashwood ER, Ventura CB, Styer P. Recent trends in performance and current state of creatinine assays. Arch Pathol Lab Med 2013;137:496–502. [DOI] [PubMed] [Google Scholar]
- 37.Eckfeldt JH, Karger AB, Miller WG, Rynders GP, Inker LA. Performance in measurement of serum cystatin C by laboratories participating in the College of American Pathologists 2014 CYS survey. Arch Pathol Lab Med 2015;139:888–93. [DOI] [PubMed] [Google Scholar]
- 38.Bargnoux AS, Pieroni L, Cristol JP, Kuster N, Delanaye P, Carlier MC, et al. Multicenter evaluation of cystatin C measurement after assay standardization. Clin Chem 2017;63:833–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


