Abstract.
Optical coherence tomography provides volumetric reconstruction of brain structure with micrometer resolution. Gray matter and white matter can be highlighted using conventional and polarization-based contrasts; however, vasculature in ex-vivo fixed brain has not been investigated at large scale due to lack of intrinsic contrast. We present contrast enhancement to visualize the vasculature by perfusing titanium dioxide particles transcardially into the mouse vascular system. The brain, after dissection and fixation, is imaged by a serial optical coherence scanner. Accumulation of particles in blood vessels generates distinguishable optical signals. Among these, the cross-polarization images reveal the vasculature organization remarkably well. The conventional and polarization-based contrasts are still available for probing the gray matter and white matter structures. The segmentation and reconstruction of the vasculature are presented by using a deep learning algorithm. Axonal fiber pathways in the mouse brain are delineated by utilizing the retardance and optic axis orientation contrasts. This is a low-cost method that can be further developed to study neurovascular diseases and brain injury in animal models.
Keywords: brain vasculature, polarization-sensitive optical coherence tomography, contrast enhancement, deep learning
1. Introduction
A comprehensive mouse brain vasculature atlas plays an important role in understanding the complicated brain functions and pathologies in mouse model, including neurodegenerative diseases, brain tumors, and strokes. Many imaging modalities have been implemented to visualize whole brain vasculature with different contrasts and at different scales. Micro-CT using the Microfil perfusion has been demonstrated to reveal complete mouse cerebrovasculature at resolution.1 Three-dimensional imaging of microvasculature in spinal cord has also been demonstrated using synchrotron radiation micro-CT with Microfil perfusion2 and x-ray synchrotron phase contrast tomography without contrast agent.3 For optical imaging, micro-optical sectioning tomography4 using Nissl staining5–7 and Indian ink perfusion,8 and serial two-photon tomography with fluorescence labeling9 have been reported to visualize large-scale mouse brain vasculature. Furthermore, tissue clearing methods have been combined with optical imaging to map brain vasculature. Aqueous-based optical clearing agents, such as SeeDB10 and Scale,11 have been utilized to clear fixed brains without quenching fluorescent signals. Microvasculature in thick slices of mouse brain has been visualized by using a fluorescent lipophilic dye and FocusClear, a water-soluble clearing agent, followed by confocal fluorescence microscopy.12 Light-sheet microscopy combined with hydrogel-based clearing method13,14 and solvent-based clearing agent15 have been demonstrated to reconstruct cerebral vasculature of the whole mouse brain at the single capillary level. These imaging techniques provide a brain vasculature atlas at micrometer resolution in ex-vivo configuration. Moreover, in-vivo imaging modalities including photoacoustic tomography with endogenous hemoglobin contrast have been applied to map cortical vasculature of the mouse brain.16
Optical coherence tomography (OCT) produces label-free and depth-resolved images of tissue structures at the micrometer-scale resolution,17 which has drawn attention in the medical imaging community. This includes the neuroimaging field that involves studying large-scale or whole brain microstructure and connectivity.18 In addition to conventional contrasts from OCT, polarization-sensitive optical coherence tomography (PS-OCT)19 provides birefringence information, which has been utilized to distinguish gray matter and white matter in the brain.20,21 Serial optical coherence scanner (SOCS), which integrates a spectral-domain PS-OCT and a tissue slicer, has been reported for reconstructing large-scale neuroanatomical circuitry of the brain at microscopic scale resolution18 and for revealing gray matter and white matter atrophy in spinocerebellar ataxia type 1 mouse models.22 In ex-vivo brain samples, however, the vasculature is not highlighted due to the lack of contrast. For in-vivo configuration, OCT-based angiography, which calculates the differences between sequential OCT signals at the same location to generate blood flow signals and visualize vascular networks,23 has been developed. OCT angiography techniques have been used to map cortical microvasculature through a cranial window,24 to transcranially map dynamic cerebrovascular perfusion,25 and to evaluate vasodynamics after stroke.26 These in-vivo OCT-based imaging techniques cannot provide volumetric neurovascular information of the whole brain due to limited imaging depth; thus, ex-vivo serial OCT imaging technique is necessary for mapping the vasculature network at large scales.
Deep learning algorithms have become increasingly powerful and able to perform tasks such as image segmentation,27 super-resolution imaging,28,29 and image denoising30 in biomedical research. This method requires large and annotated datasets and uses them to create a predictive tool to analyze other data, sometimes from different sources based on the trained model.31 Image segmentation is one of the applications wherein machine learning approaches outperform human and traditional parcellation methods, especially for dealing with large datasets.32 User-friendly tools have been developed and available to perform image segmentation tasks.27,33–35 Among those, U-Net is a robust and fast deep learning solution for image segmentation.35 U-Net-based algorithms have been applied and further developed to segment structure in OCT images such as vascular boundary.36
In this study, we present contrast enhancement of OCT signals by titanium dioxide () perfusion to visualize the brain vasculature. Serial imaging is performed on sagittal or coronal planes until the entire brain sample is imaged. Results show that accumulation of particles in blood vessels generates distinguished optical signals due to the strong scattering of particles. We find the cross-polarization images remarkably reveal the vasculature organization. Deep-learning-based segmentation and reconstruction of the vasculature is obtained from the cross-polarization images. In addition to the vascular network, we present the white matter organization of the same mouse brain.
2. Materials and Methods
2.1. Sample Preparation
Ten-week-old wild-type FVB mice were used in this study. All animal treatments and experiments are in accordance with Institutional Animal Care and Use Committee at the University of Minnesota approved protocols. Animals were euthanatized and then perfused transcardially with 2 mg/ml (Spectrum Chemical Mfg. Corp., Gardena, California) suspension in deionized water. Dissected brains were kept in 10% buffered formalin for 24 h prior to imaging. This study presents results from three brains. Two of those brains were serially sectioned in coronal planes and the other brain was serially sectioned in sagittal planes.
2.2. System Setup and Imaging Procedure
The polarization maintaining fiber (PMF)-based spectral-domain PS-OCT system utilizes a 25-mW superluminescent diode with 840-nm center wavelength and 50-nm FWHM bandwidth, which yields an axial resolution of in tissue (refractive index ). A pair of galvanometer-based mirror scanners (Cambridge Technology, Bedford, Massachusetts) provides two-axis lateral (raster) scan of the light beam over the sample. A scan lens (LSM03-BB, Thorlabs, Inc., Newton, New Jersey) in the sample arm allows a large field of view () with a lateral resolution of . Details of the optical setup including the design, operation, and performance can be found in our previous publications.21,37 The brain samples were immersed in phosphate-buffered saline during imaging. After imaging the superficial region of the tissue block, a thick slice is removed to expose the deeper regions. The procedure is repeated until the entire brain sample is imaged. Details of serial scanning can be found in previous publications.18,21
2.3. Image Formation
Spectra from two orthogonal polarization channels are acquired by a single line-scan camera in a customized spectrometer. Inverse Fourier transform of interference-related spectral oscillations yields complex depth profiles for each polarization channel that are denoted as , where and indicate the amplitude and phase as a function of depth , and the subscripts represent the cross-polarization channel and copolarization channel. Multicontrasts of ex-vivo brain imaging are generated from the complex depth profiles. The contrasts include reflectivity , cross-polarization , retardance , and optic axis orientation . The relative axis orientation from PMF-based system is environment dependent due to a phase offset which can be dynamically removed by adding a calibration path.38
A cross-sectional image is generated by stacking a particular contrast of 1000 A-lines (depth profiles) that are acquired during a lateral scan. Eight hundred of these cross-sectional images are combined to form a volumetric dataset. En-face images are formed by two-dimensional (2-D) projections of the three-dimensional (3-D) datasets, and the pixel values are calculated by projecting the average or maximal value along the depth direction. Samples larger than the field of view require tile scanning to cover the entire area. For instance, we used two tiles, each of which was in size and had 13.6% overlap, to image the whole coronal plane. For each hemisphere, 55 en-face images were stacked. The 3-D stitching plugin in ImageJ was used to combine image stacks of the left and right hemispheres. The overlap was fused by linear blending. Afterward, Vaa3D software39 was used to visualize the whole brain volume.
2.4. Vasculature Identification
Due to the high scattering of particles accumulated in blood vessels, the brain vascular organization was highlighted in the OCT images. We utilized the U-Net architecture, which is a fully convolutional network, to perform deep-learning-based vasculature segmentation. The U-Net architecture was first developed by Ronneberger, Fischer, and Brox.35
For vessel segmentation at OCT natural resolution, we parcellated the large SOCS dataset recorded from the sagittally sectioned brain into 810 sagittal images (), each of which corresponds to a pixel depth (). Among these, we selected 30 images containing blood vessels in different regions to avoid biased annotation and manually labeled the vessels as the training set. The segmentation software, implemented with Python (v3.5.5) based on Keras (v2.2.2) and Tensorflow (v1.10.0), were revised from Ref. 40. After the model was trained, all images were fed into the model to perform vessel segmentation. The vessel segmentation on the coronal plane at OCT natural resolution also utilized the aforementioned model, and there was no separate training for it as general morphology of the vessels (long and short traces or dots) in two planes were similar.
For vessel segmentation on the en-face images, we used the SOCS dataset recorded from a coronally sectioned cerebrum. Each en-face coronal image () is derived from 30 pixels in depth (). The images were resized into and cropped into 32 tiles (). Each tile corresponds to area. We selected 30 tiles (out of 1760 tiles) with vessels in different regions to avoid biased annotation and manually labeled the blood vessels as a new training set for the en-face images. All cropped tiles were later segmented by the trained model. The segmented images were later stitched back to form the vessel map at the whole en-face coronal plane.
Segmentation accuracy typically reaches above 96% after five epochs. The Sørensen-Dice similarity coefficient between manual annotation and deep learning segmentation in the training dataset was calculated. The coefficient was (mean ± standard deviation) for vessel segmentation at OCT natural resolution and for segmentation on en-face images.
3. Results
3.1. Cross-Polarization Intensity Reveals Contrast-Enhanced Vascular Network
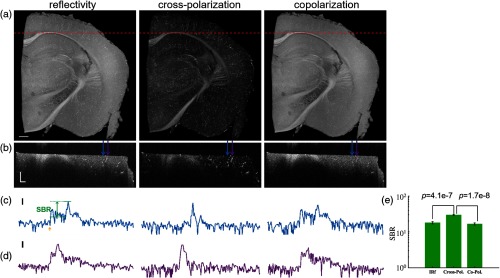
We first investigated which contrast provided by PS-OCT can better highlight the accumulation in blood vessels. Figure 1(a) shows the en-face coronal images of mouse brain at Bregma (right hemisphere). From left to right column, reflectivity, cross-polarization intensity and copolarization intensity are presented. The pixel value indicates the mean value of the corresponding contrast over a depth of . The red dashed lines on Fig. 1(a) mark the position of cross-sectional images that are presented in Fig. 1(b). The blue and purple arrows on Fig. 1(b) indicate the locations of two depth profiles that exhibit blood vessels in the deeper and shallower regions, respectively. These depth-profiles are shown in Figs. 1(c) and 1(d). The small yellow arrow indicates the tissue surface, and the range given in green represents the signal-to-background ratio (SBR), which is defined as the difference between the maximum value and the background value within a depth profile.
Fig. 1.
PS-OCT contrasts of a mouse brain with particles in the vasculature. (a) En-face coronal images are with mean values of corresponding contrasts over a depth of . (b) Cross-sectional images; red dashed lines in (a) indicate the location. (c), (d) Representative depth profiles; blue and purple arrows in (b) mark the locations; yellow arrow indicates the surface, and SBR is signal-to-background ratio. (e) Mean and s.e.m. of SBR. Scale bars: in (a), in (b), and 10 dB in (c) and (d).
For blood vessels in gray matter region, we calculated SBR of reflectivity, cross-polarization intensity, and copolarization intensity contrasts. Twenty-five A-lines were chosen for the analysis. Mean and standard error of the mean (s.e.m.) of the SBR values, as shown in Fig. 1(e), are for reflectivity, for cross-polarization intensity, and for copolarization intensity. One-way analysis of variance (ANOVA) with subsequent Dunn’s test for multiple comparisons was performed and SBR of cross-polarization intensity was proven significantly higher than that of reflectivity and copolarization intensity ( and , respectively). The cross-polarization intensity was found to be the most useful contrast to identify the blood vessels with content. Depolarization of light by scattering particles can yield cross-polarization backscatters in OCT,41 and accumulation of particles ( at 840 nm) in blood vessels can serve for the same purpose.
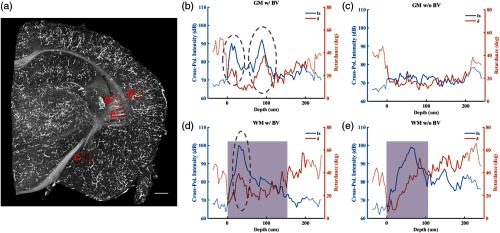
Blood vessels with content can be potentially segmented by simply applying a threshold to cross-polarization intensity in gray matter. In white matter, however, due to the high scattering and retardance of myelinated nerve fibers, the vessels may not be easily segmented. Figure 2 compares depth profiles of cross-polarization intensity and retardance contrasts in gray matter and white matter regions while with and without presence of blood vessels. Figure 2(a) shows en-face image of the same brain slice as Fig. 1 by projecting the maximum of cross-polarization intensity along the depth. Evidently, the maximum value projection highlights the blood vessels better than the mean value projection. This is especially true for visualizing the smaller blood vessels, which can determine the maximum value but provide limited contribution to the mean value. As labeled (b) to (e), the red dashed boxes on Fig. 2(a) indicate locations of brain regions whose depth-profiles are, respectively, shown in Figs. 2(b)–2(e). For each of these regions, we chose 12 adjacent A-lines and averaged cross-polarization intensity and retardance of the selected A-lines to present the general trends. Blue and orange traces in Figs. 2(b)–2(e) show averaged cross-polarization intensity and retardance, respectively. Solid lines represent signals within (top surface) and depth of the brain sample, and dotted lines represent background or low signal-to-noise ratio (SNR) regions. Shaded regions in Figs. 2(d) and 2(e) indicate white matter region. Gray dashed ovals mark the depth location of blood vessels in gray matter [Fig. 2(b)] and white matter [Fig. 2(d)]. Sudden increase and decrease of cross-polarization intensity are due to the existence of high scattering particles in blood vessels (dashed ovals). The particles also resulted in temporary increase of the retardance. On the other hand, both contrasts remained low with minimal fluctuation in the gray matter region where blood vessels are not exhibited [Fig. 2(c)].
Fig. 2.
Depth profiles of cross-polarization intensity and retardance in white matter (WM) and gray matter (GM) regions with and without blood vessels (BV). (a) En-face coronal image by projecting the maximum value of cross-polarization intensity. Twelve adjacent A-lines are chosen in each region indicated by red dashed boxes. (b)–(e) Averaged cross-polarization intensity (Ix, blue trace) and retardance (, orange trace) of selected A-lines. Dotted lines represent background or low SNR regions. Dashed ovals mark location of blood vessels. Shaded regions indicate white matter areas. Scale bar: .
Because the white matter is birefringent, it yields changes in cross-polarization intensity and retardance. For instance, a bundle of myelinated axonal fibers changes the polarization state of light resulting in a cumulative increase of retardance as seen in Fig. 2(e). This also yields an increase in cross-polarization intensity, which eventually decays to zero due to attenuation of light in tissue. Although one would expect the characteristics of these contrasts to be different in blood vessel containing [Fig. 2(d)] and noncontaining [Fig. 2(e)] white matter, for instance, faster and temporary changes by particles, complications could arise for segmentation and identification of vascular and white matter networks as simple thresholding would not be sufficient.
3.2. Deep-Learning-Based Vasculature Segmentation at OCT Natural Resolution
To identify blood vessels in different brain regions, we applied the deep learning segmentation algorithm on cross-polarization images at OCT natural resolution. Vessel segmentation was performed on large-scale imaging data, which was acquired from the right hemisphere of a mouse brain by serial imaging and sectioning (sagittal) procedure. From the imaging data, a volume was selected for the analysis. Deep learning-based vessel segmentation was performed on 2-D images. Sagittal and coronal images were separately used for segmentation.
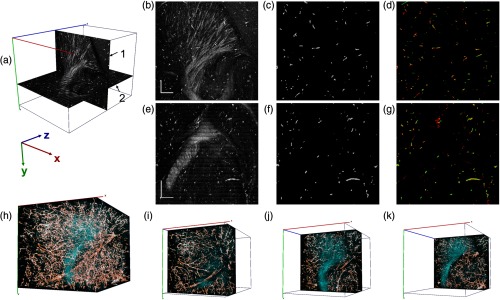
Figure 3 shows details of the vasculature segmentation. The coordinate system in Fig. 3(a) includes (1) one sagittal view and (2) one coronal view from the 3-D dataset. These views represent optical slices with a thickness of one pixel, which corresponds to 3.3 and for the sagittal and coronal images, respectively. Figures 3(b) and 3(e) show representative cross-polarization images on the sagittal and coronal planes. In addition to the blood vessels highlighted by contrast enhancement of particles, abundant fiber tracts are visible in the striatum region. Figure 3(c) shows the result of blood vessel segmentation from the sagittal image presented in Fig. 3(b). Similarly, Fig. 3(f) illustrates the result of blood vessel segmentation from the coronal image presented in Fig. 3(e). Apparently, some segments of the blood vessel network were not captured by applying the deep learning algorithm on a single 2-D image. The imaging data are 3-D; therefore, a number of 2-D imaging stacks (sagittal, coronal, and horizontal) can be obtained, and segmentation of the images from different views can allow whether or not a particular pixel belongs to a blood vessel. When the cylindrical shape is considered, the minimal representation of the blood vessel in a particular view would be a small circle (cross-section). On the other hand, other orthogonal views will show the blood vessel as a long line (long axis). Therefore, one would think that application of the algorithm on imaging stacks of two orthogonal views could be sufficient for identifying the blood vessels.
Fig. 3.
Deep-learning-based vessel segmentation at OCT natural resolution. (a) Sagittal (1) and coronal (2) planes of 3-D dataset. (b) Sagittal and (e) coronal views of cross-polarization images. Vessel segmentation from (c) the sagittal and (f) coronal images. Composite images in (d) and (g) show two separate segmentation results from sagittal (red) and coronal (green) views. (h) 3-D reconstruction of vasculature in a brain volume of . In addition, cross-polarization intensity is given in cyan. (i)–(k) Three composite thin layers () of the same dataset. The -stack of the whole volume is shown in Video 1 (MP4, 11.1 MB [URL: https://doi.org/10.1117/1.NPh.6.3.035004.1]). Scale bar: .
We applied the segmentation algorithm on the sagittal and coronal images of the 3-D dataset. This yields two separate segmentation results for the same brain block. Coronal segmentation results were reconstructed into 3-D dataset, and then volumetric segmentation results were read from sagittal view. This was done to compare the two separate segmentation results on sagittal plane. Figure 3(d) shows a composite image with red channel representing the segmentation of the sagittal image [same as Fig. 3(c)] and green channel representing the result reconstructed from the segmentations of all coronal images. The pixels shown in yellow are segmented by both methods. Similarly, sagittal segmentation results were reconstructed into 3-D dataset, and then volumetric segmentation results were read from coronal view. Figure 3(g) shows the composite image with green channel representing the segmentation of the coronal image [same as Fig. 3(f)] and red channel representing the results obtained from segmentations of all sagittal images. It is clear that segmentation performed on a single plane does not completely identify the blood vessels as the composite images show forming longer segments from shorter fragments in different colors. Therefore, it is better to reconstruct the vascular network by combining the segmentation from the two planes.
Figure 3(h) shows a volumetric reconstruction of the brain block. Deep learning-based vasculature is obtained by combining the segmentation results from the sagittal and coronal images. The segmented vasculature is presented with orange channel. In addition, Fig. 3(h) presents the cross-polarization intensity with cyan channel. For more descriptive views of vessels tracts, Figs. 3(i)–3(k) display three composite thin layers () that start at position of 244, 914, and . The -stack of the whole volume is shown in Video 1.
3.3. Reconstruction of White Matter on En-Face Images
We investigated identification and reconstruction of white matter from our datasets. Figure 4(a) shows an en-face image created by maximally projecting the cross-polarization signal of a depth of 0.1 mm for a lateral area at lateral 1.2 mm (sagittal view). Not only the white matter but also the vasculature is present in the image. In order to separate these structures, we first performed vessel segmentation on the brain block at OCT natural resolution as described in Sec. 2.4. After finding the vessel pixels in the dataset, the pixels within the volume of interest were projected on the en-face plane. The en-face image shown in Fig. 4(b) represents the blood vessels which can be perceived as artifacts for white matter analysis. By excluding blood vessel pixels in the 3-D dataset and projecting the remaining pixels on the plane, white matter structures can be highlighted in the en-face images. Figures 4(c) and 4(d) show the en-face image of cross-polarization intensity that are calculated this way by taking maximum value and mean value along the depth, respectively. Suppression of the blood vessels is evident in these images. Compared with the maximum projection method [Fig. 4(c)], averaging the cross-polarization intensity along the depth [Fig. 4(d)] is more effective in removing the blood vessel-related artifacts, and therefore, it reveals the white matter architecture better. This is an expected outcome, since the maximum projection method can still emphasize the high intensity pixels of the blood vessels when it is not fully segmented.
Fig. 4.
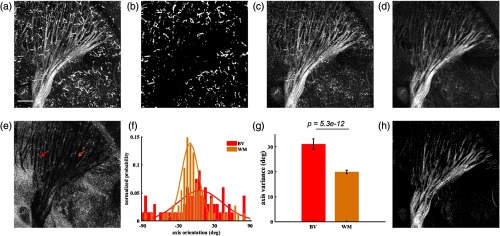
White matter reconstruction after vascular segmentation. (a) En-face cross-polarization image with maximum projection. (b) En-face projection of blood vessel segmentation at OCT natural resolution. After removing segmented vessel pixels, en-face cross-polarization images are created with (c) maximum and (d) mean value projection. (e) Variation of axis orientation along depth. (f) Histogram of axis orientation values along single A-lines in blood vessel (BV, red) and white matter (WM, orange) regions, whose locations are indicated by color-coded arrows in (e). (g) Mean and s.e.m. of variation of axis orientation (18 A-lines in BV and WM areas). (h) White matter structure map by removing pixels that corresponds to high variation of axis orientation in (e) and low cross-polarization intensity in (d). Scale bar: .
Optic axis orientation is an additional contrast of our optical imaging system. It indicates the direction of optical anisotropy (birefringence) in the plane. Since myelinated axons exhibit birefringence, the axis orientation contrast has been found useful for optical tractography of the brain.20 Due to directional alignment of axons in fiber bundles, variance of the axis orientation in white matter is expected to be smaller than that of a nonbirefringent tissue. This is due to the calculation of axis orientation from the phase of the co- and cross-polarization channels, and phase is not reliable when the cross-polarization channel remains low for nonbirefringent tissue. However, the particles yield high co- and cross-polarization signals that complicates differentiation of white matter and blood vessel networks. On the other hand, randomly scattering particles yields high variation in optic axis orientation. This is not surprising since accumulation in blood vessels is not structurally organized. We calculated the axis variation value of each A-line by histogram analysis that relies on binning the orientation values along a depth profile into 5 deg intervals and formed an en-face axis variation map as shown in Fig. 4(e). Dark color of the white matter network is due to low variation of axis orientation. Figure 4(f) shows distributions of axis orientation values for two separate A-lines. Red plot is the histogram of an A-line from a blood vessel region, and orange plot is from a white matter region without vessels. Correspondingly, these regions are indicated by red and orange arrows on Fig. 4(e). Gaussian fit was performed on the normalized probability values for each A-line. Standard deviation of the Gaussian fit was used to compare the variations of the axis orientation values. Standard deviations for the distributions shown in Fig. 4(f) are 14.3 deg for the white matter curve and 34.0 deg for the blood vessel curve. For a better comparison, eighteen A-lines were chosen for each region. Standard deviation of the Gaussian fit was calculated for each A-line, and the mean and s.e.m. of the deviations were analyzed. As Fig. 4(g) shows, optic axis values in white matter A-lines varies significantly less than that of blood vessels (). Mean and s.e.m. of axis variation of selected blood vessel region and white matter are and , respectively.
We utilized variation of the axis orientation contrast [low values in Fig. 4(e)] to obtain a binary mask for preserving the white matter and removing the blood vessels on the en-face image. In addition, a certain value threshold was applied to cross-polarization en-face image to remove low scattering regions. The resulting en-face image shows white matter structures in Fig. 4(h).
3.4. Blood Vessel and White Matter Segmentation of the Whole Mouse Cerebrum
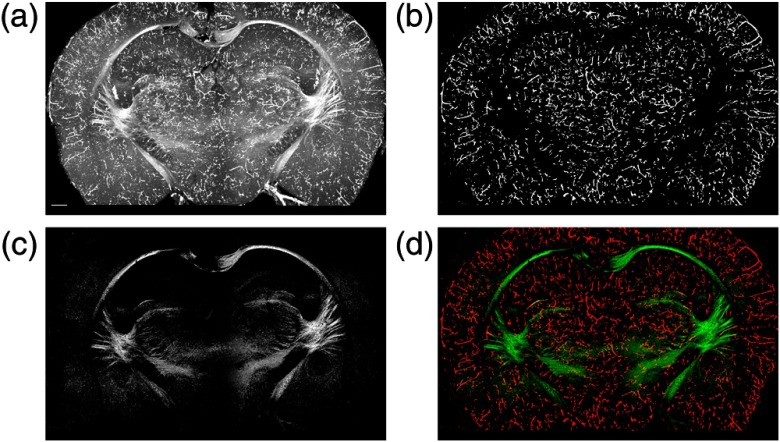
Figure 5(a) shows an en-face cross-polarization image on coronal plane at Bregma . Pixel intensity corresponds to the maximum value along depth. As described in Sec. 2.4, a whole coronal en-face image was resized and cropped into 32 tiles. Deep learning segmentation was applied on these tiles to identify blood vessels. Results from the tiles were stitched back to reconstruct the vascular network. The en-face image in Fig. 5(b) shows blood vessels on the coronal plane. Alternatively, blood vessels could have been segmented at OCT resolution and projected onto the en-face plane as in Fig. 4(b); however, this is time-consuming if only the blood vessels on the en-face view is needed. In addition, we found that segmentation of en-face images showed more continuous segments.
Fig. 5.
Blood vessel segmentation and white matter reconstruction. (a) En-face coronal image of cross-polarization with maximum projection. (b) Vessel segmentation on the image. (c) Reconstruction of white matter. (d) Composite image of blood vessel (red) and white matter (green) networks. The -stack reconstruction for the whole cerebrum is shown in Video 2 (MP4, 10.9 MB [URL: https://doi.org/10.1117/1.NPh.6.3.035004.2]). Scale bar: .
As described in Sec. 3.3, we created white matter maps on en-face images by (i) segmenting blood vessels at OCT natural resolution, (ii) excluding these vessel pixels in 3-D dataset, (iii) calculating en-face images, and (iv) masking out unwanted pixels that correspond to high variation of axis orientation and low crosspolarized intensity. Figure 5(c) shows the resulting en-face image of white matter. Figure 5(d) shows the composite image with red and green channels representing blood vessel and white matter structures. With the composite images, Video 2 shows the -stack reconstruction of the vascular and white matter networks of the whole cerebrum.
4. Discussion
One of the limitations of the study is that the vessel segmentation relying on U-net architecture would not perfectly pick up all blood vessels such as some smaller dots or discontinued fragments. One reason could be that the nonrepresentative and inconsistent annotations cause the failure of U-net to perform the actual task.27 In this study, vasculature segmentation was realized on 2-D images, and the results were improved by combining the segmentation from two different views (sagittal and coronal). However, an alternative and possibly a better approach would be to develop a segmentation architecture that works on 3-D data.
Lateral resolution of the current imaging configuration () is larger than the diameter of arterial and venous terminal branches, reported as and , respectively.7 Thus, some small branches might not be picked up by the imaging system. Microscope objectives can be incorporated into the SOCS to improve lateral resolution to reveal more detailed vessel branches. At last, an imperfect accumulation may induce some fragments of the vasculature. The diameter of the particles we used is expected to be 300 nm as other commonly used commercial products, which is much smaller than vessels diameter.
Corpus callosum was shown with high crosspolarized intensity in the coronal images. However, the sagittal view, which captures this structure in part, shows it with low crosspolarized intensity [Figs. 3(b) and 4)]. For the white matter, it is known that large inclination angle of fibers with respect to the imaging plane can reduce the PS-OCT signals such as reflectivity and apparent birefringence.20 Imaging at different illumination angles can be implemented to avoid misinterpretations, which also allows calculation of 3-D optic axis orientation and quantification of actual birefringence.38
Mapping the brain vasculature in detail is important to understand the brain anatomy and functions. However, contrast enhancement by particles does not allow us to distinguish arteries and veins, which are known to be structurally different. One interesting direction to map the vasculature and white matter could be combining contrast enhanced SOCS and fluorescence microscopy with an artery-specific dye Alexa 633.42
5. Conclusions
We transcardially perfused particles into the vascular system to enhance PS-OCT contrasts for visualization of vasculature in ex-vivo brain at a low cost. A deep learning algorithm, U-net architecture, was used to identify blood vessels at OCT natural resolution. The algorithm was also applied to en-face images. We found that axis orientation contrast within A-lines in white matter varies significantly less than that of the accumulation in blood vessels, which is not structurally organized. Thus, the finding helps better identification of white matter structures in this study. Both brain vasculature and white matter network were reconstructed. This work realized large-scale visualization and segmentation of brain vasculature in ex-vivo imaging configuration at high resolution. The future advances of this work involve intriguing applications in studying vascular disorders in animal disease models, such as stroke and cancer, and brain injury.
Supplementary Material
Acknowledgments
The work was supported by a research grant from the US National Science Foundation (NSF, CBET-1510674) and by a Graduate School Doctoral Dissertation Fellowship at the University of Minnesota (to C.J.L.). High-performance computing resources were provided by Minnesota Supercomputing Institute. The authors also thank Orion Rainwater for his assistance with sample preparation.
Biographies
Tianqi Li received her BS degree in electrical engineering from the University of Science and Technology of China in 2015. She is a PhD candidate in biomedical engineering at the University of Minnesota. Her research interests are optical imaging techniques development and structural and functional applications related to neurodegenerative diseases.
Chao J. Liu received his BS degree in optoelectronics engineering from the Huazhong University of Science and Technology, China, and his PhD from the University of Minnesota. He is now a postdoctoral associate at the Centre for Magnetic Resonance Research at the University of Minnesota. He is currently interested in understanding neurovascular coupling in mammalian brains with three-photon microscopy and functional MRI.
Taner Akkin received his BSc and MSc degrees in electrical and electronics engineering from Çukurova University, Turkey, in 1995 and 1997, and his PhD from the University of Texas at Austin in 2003. After postdoctoral studies at Harvard Medical School/Wellman Center for Photomedicine, Massachusetts General Hospital, he joined the Department of Biomedical Engineering at the University of Minnesota, in 2005, where he is an associate professor. He develops optical imaging systems to study neural structure and function.
Disclosures
The authors have no relevant financial interests in this article and no potential conflicts of interest to disclose.
References
- 1.Ghanavati S., et al. , “A perfusion procedure for imaging of the mouse cerebral vasculature by x-ray micro-CT,” J. Neurosci. Methods 221, 70–77 (2014). 10.1016/j.jneumeth.2013.09.002 [DOI] [PubMed] [Google Scholar]
- 2.Cao Y., et al. , “Three-dimensional imaging of microvasculature in the rat spinal cord following injury,” Sci. Rep. 5(1), 12643 (2015). 10.1038/srep12643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fratini M., et al. , “Simultaneous submicrometric 3D imaging of the micro-vascular network and the neuronal system in a mouse spinal cord,” Sci. Rep. 5(1), 8514 (2015). 10.1038/srep08514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Q., et al. , “Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain,” Science 330(6009), 1404–1408 (2010). 10.1126/science.1191776 [DOI] [PubMed] [Google Scholar]
- 5.Wu J., et al. , “Direct 3D analyses reveal barrel-specific vascular distribution and cross-barrel branching in the mouse barrel cortex,” Cereb. Cortex 26(1), 23–31 (2016). 10.1093/cercor/bhu166 [DOI] [PubMed] [Google Scholar]
- 6.Wu J., et al. , “3D BrainCV: simultaneous visualization and analysis of cells and capillaries in a whole mouse brain with one-micron voxel resolution,” Neuroimage 87, 199–208 (2014). 10.1016/j.neuroimage.2013.10.036 [DOI] [PubMed] [Google Scholar]
- 7.Xiong B., et al. , “Precise cerebral vascular atlas in stereotaxic coordinates of whole mouse brain,” Front. Neuroanat. 11, 1–16 (2017). 10.3389/fnana.2017.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang T., et al. , “Indian-ink perfusion based method for reconstructing continuous vascular networks in whole mouse brain,” PLoS One 9(1), e88067 (2014). 10.1371/journal.pone.0088067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amato S. P., et al. , “Whole brain imaging with serial two-photon tomography,” Front. Neuroanat. 10, 31 (2016). 10.3389/fnana.2016.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ke M.-T., Fujimoto S., Imai T., “SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction,” Nat. Neurosci. 16(8), 1154–1161 (2013). 10.1038/nn.3447 [DOI] [PubMed] [Google Scholar]
- 11.Hama H., et al. , “Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain,” Nat. Neurosci. 14(11), 1481–1488 (2011). 10.1038/nn.2928 [DOI] [PubMed] [Google Scholar]
- 12.Moy A. J., Wiersma M. P., Choi B., “Optical histology: a method to visualize microvasculature in thick tissue sections of mouse brain,” PLoS One 8(1), e53753 (2013). 10.1371/journal.pone.0053753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalyanasundaram S., et al. , “Structural and molecular interrogation of intact biological systems,” Nature 497(7449), 332–337 (2013). 10.1038/nature12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Giovanna A. P., et al. , “Whole-brain vasculature reconstruction at the single capillary level,” Sci. Rep. 8(1), 1–11 (2018). 10.1038/s41598-018-30533-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lugo-Hernandez E., et al. , “3D visualization and quantification of microvessels in the whole ischemic mouse brain using solvent-based clearing and light sheet microscopy,” J. Cereb. Blood Flow Metab. 37(10), 3355–3367 (2017). 10.1177/0271678X17698970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stein E. W., Maslov K., Wang L. V., “Noninvasive, in vivo imaging of the mouse brain using photoacoustic microscopy,” J. Appl. Phys. 105(10), 102027 (2009). 10.1063/1.3116134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang D., et al. , “Optical coherence tomography,” Science 254(5035), 1178–1181 (1991). 10.1126/science.1957169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H., Zhu J., Akkin T., “Serial optical coherence scanner for large-scale brain imaging at microscopic resolution,” Neuroimage 84, 1007–1017 (2014). 10.1016/j.neuroimage.2013.09.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Boer J. F., et al. , “Two-dimensional birefringence imaging in biological tissue by polarization-sensitive optical coherence tomography,” Opt. Lett. 22(12), 934–936 (1997). 10.1364/OL.22.000934 [DOI] [PubMed] [Google Scholar]
- 20.Wang H., et al. , “Reconstructing micrometer-scale fiber pathways in the brain: multi-contrast optical coherence tomography based tractography,” Neuroimage 58(4), 984–992 (2011). 10.1016/j.neuroimage.2011.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu C. J., et al. , “Visualizing and mapping the cerebellum with serial optical coherence scanner,” Neurophotonics 4(1), 011006 (2016). 10.1117/1.NPh.4.1.011006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C. J., et al. , “Polarization-sensitive optical coherence tomography reveals gray matter and white matter atrophy in SCA1 mouse models,” Neurobiol. Dis. 116(April), 69–77 (2018). 10.1016/j.nbd.2018.05.003 [DOI] [PubMed] [Google Scholar]
- 23.Chen C.-L., Wang R. K., “Optical coherence tomography based angiography [invited],” Biomed. Opt. Express 8(2), 1056–1082 (2017). 10.1364/BOE.8.001056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srinivasan V. J., et al. , “Rapid volumetric angiography of cortical microvasculature with optical coherence tomography,” Opt. Lett. 35(1), 43–45 (2010). 10.1364/OL.35.000043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia Y., Wang R. K., “Optical micro-angiography images structural and functional cerebral blood perfusion in mice with cranium left intact,” J. Biophotonics 4(1–2), 57–63 (2011). 10.1002/jbio.201000001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu L., et al. , “Spectral Doppler optical coherence tomography imaging of localized ischemic stroke in a mouse model,” J. Biomed. Opt. 15(6), 066006 (2010). 10.1117/1.3505016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falk T., et al. , “U-Net: deep learning for cell counting, detection, and morphometry,” Nat. Methods 16(1), 67–70 (2019). 10.1038/s41592-018-0261-2 [DOI] [PubMed] [Google Scholar]
- 28.Weigert M., et al. , “Content-aware image restoration: pushing the limits of fluorescence microscopy,” Nat. Methods 15(12), 1090–1097 (2018). 10.1038/s41592-018-0216-7 [DOI] [PubMed] [Google Scholar]
- 29.Wang H., et al. , “Deep learning enables cross-modality super-resolution in fluorescence microscopy,” Nat. Methods 16(1), 103–110 (2019). 10.1038/s41592-018-0239-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Batson J., Royer L., “Noise2Self: blind denoising by self-supervision,” arXiv:1901.11365 (2019).
- 31.Webb S., “Deep learning for biology,” Nature 554(7693), 555–557 (2018). 10.1038/d41586-018-02174-z [DOI] [PubMed] [Google Scholar]
- 32.Strack R., “Deep learning in imaging,” Nat. Methods 16(1), 17–17 (2019). 10.1038/s41592-018-0267-9 [DOI] [PubMed] [Google Scholar]
- 33.McQuin C., et al. , “CellProfiler 3.0: next-generation image processing for biology,” PLoS Biol. 16(7), e2005970 (2018). 10.1371/journal.pbio.2005970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haberl M. G., et al. , “CDeep3M—Plug-and-play cloud-based deep learning for image segmentation,” Nat. Methods 15(9), 677–680 (2018). 10.1038/s41592-018-0106-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronneberger O., Fischer P., Brox T., “U-net: convolutional networks for biomedical image segmentation,” Lect. Notes Comput. Sci. 9351, 234–241 (2015). 10.1007/978-3-319-24574-4 [DOI] [Google Scholar]
- 36.Wu C., et al. , “Automatic boundary segmentation of vascular Doppler optical coherence tomography images based on cascaded U-net architecture,” OSA Contin. 2(3), 677–689 (2019). 10.1364/OSAC.2.000677 [DOI] [Google Scholar]
- 37.Wang H., Al-Qaisi M. K., Akkin T., “Polarization-maintaining fiber based polarization-sensitive optical coherence tomography in spectral domain,” Opt. Lett. 35(2), 154–156 (2010). 10.1364/OL.35.000154 [DOI] [PubMed] [Google Scholar]
- 38.Liu C. J., et al. , “Quantifying three-dimensional optic axis using polarization-sensitive optical coherence tomography,” J. Biomed. Opt. 21(7), 070501 (2016). 10.1117/1.JBO.21.7.070501 [DOI] [PubMed] [Google Scholar]
- 39.Peng H., et al. , “V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets,” Nat. Biotechnol. 28(4), 348–353 (2010). 10.1038/nbt.1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhixuhao, “Implementation of deep learning framework—Unet, using Keras,” https://github.com/zhixuhao/unet (accessed 26 November 2018).
- 41.Schmitt J. M., Xiang S. H., “Cross-polarized backscatter in optical coherence tomography of biological tissue,” Opt. Lett. 23(13), 1060–1062 (1998). 10.1364/OL.23.001060 [DOI] [PubMed] [Google Scholar]
- 42.Shen Z., et al. , “An artery-specific fluorescent dye for studying neurovascular coupling,” Nat. Methods 9(3), 273–276 (2012). 10.1038/nmeth.1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.