Abstract
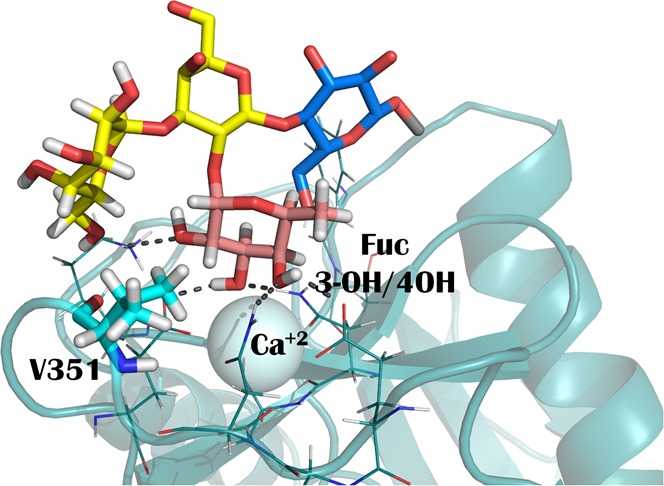
The dendritic cell-specific intracellular adhesion molecule-3-grabbing nonintegrin (DC-SIGN) is an important receptor of the immune system. Besides its role as pathogen recognition receptor (PRR), it also interacts with endogenous glycoproteins through the specific recognition of self-glycan epitopes, like LeX. However, this lectin represents a paradigmatic case of glycan binding promiscuity, and it also has been shown to recognize antigens with α1−α2 linked fucose, such as the histo blood group antigens, with similar affinities to LeX. Herein, we have studied the interaction in solution between DC-SIGN and the blood group A and B antigens, to get insights into the atomic details of such interaction. With a combination of different NMR experiments, we demonstrate that the Fuc coordinates the primary Ca2+ ion with a single binding mode through 3-OH and 4-OH. The terminal αGal/αGalNAc affords marginal direct polar contacts with the protein, but provides a hydrophobic hook in which V351 of the lectin perfectly fits. Moreover, we have found that αGal, but not αGalNAc, is a weak binder itself for DC-SIGN, which could endow an additional binding mode for the blood group B antigen, but not for blood group A.
One of the most prominent and studied members of the C-type lectin receptor (CLR) family is DC-SIGN. These carbohydrate binding proteins are profusely distributed on the surface of antigen-presenting cells, where they act as pathogen recognition receptor (PRRs) through the specific recognition of glycans on the pathogen surface.1,2 Infections by M. tuberculosis,3,4C. Albicans,5Leishmania,6 dengue,7 ebola,8 or HIV,3 for instance, have been reported to involve the participation of DC-SIGN. Interestingly, at the same time, this lectin plays important roles in the immune modulation and homeostasis through its interaction with endogenous glycoproteins where it recognizes self-glycans.9
The glycan binding specificity of this lectin is remarkably broad. Besides, interestingly, different glycan array data have reached contradictory results.10−13 They mostly agree in the facts that ligands include certain fucose-containing antigens, as well as high mannose and complex type N-glycans, although very subtle structural differences may influence the binding event.13 The presentation of glycan epitopes is extremely important for their interactions with receptors14 and this seems to be also a key element in DC-SIGN recognition events. For instance, the histo blood group A/B (BGA, BGB) antigens have been described to be recognized by DC-SIGN when presented on a long linker. However, only BGB was recognized when presented with a short one, while BGA was not.10
The interaction of DC-SIGN with endogenous partners remains highly unexplored. The LewisX (LeX) antigen has been shown to be recognized on cell surfaces,11 and the epitope recognized on the ICAM-3 glycoprotein,15 one of the known endogenous ligands of DC-SIGN. Obviously, the broad glycan specificity of the lectin raises the question of the biological significance of the interaction with other fucosylated self-epitopes. Fucose is commonly found in mammalian glycans as a terminal modification as part a of the blood group antigens: Lewis (α1−α3 and α1−α4 linked) and ABH types (α1−α2 linked). Recently, the detection of terminal αGal residues and higher fucosylation patterns in oral cancer cells has suggested a role of the blood group B (BGB) antigen, which could be related to its enhanced recognition by DC-SIGN and the escape of the cancer cells to the immune systems mediated by DC-SIGN.16
Herein, we have studied the structural details of the molecular recognition in solution of the histo blood group A and B antigens by the carbohydrate recognition domain of DC-SIGN (CRD DC-SIGN) by using nuclear magnetic resonance (NMR) and molecular modeling methods.17 The interaction between LeX and DC-SIGN, characterized by X-ray crystallography and NMR,10,18 occurs through the Fuc residue, which binds at the primary calcium binding site by coordinating the Ca2+ ion with hydroxyls 3-OH and 4-OH. Interestingly, the homologous C-type lectin langerin,19 with a similar glycan recognition profile with respect to fucosylated oligosaccharides,12 has been shown to also bind to the BGB antigen.20 In this case, however, the Fuc residue is attached to the Ca2+ ion through 2-OH and 3-OH.
In this scenario, and given the promiscuity and plasticity of this particular system, we aimed at shedding light onto the molecular details of the recognition of the histo blood group A and B antigens by CRD DC-SIGN in solution. Our data show that both BGA and BGB type VI tetrasaccharide antigens (compounds 1 and 2, respectively; see Chart 1) are recognized with a similar presentation and with comparable affinities, and disclose the relevant structural requirements for the interaction. In addition, our data suggest that, for 2, and not for 1, a second binding mode also may occur, in which the terminal αGal residue is bound at the primary Ca2+ binding site. Indeed, a simple αGal epitope is also a ligand for DC-SIGN through a weak, but specific interaction.
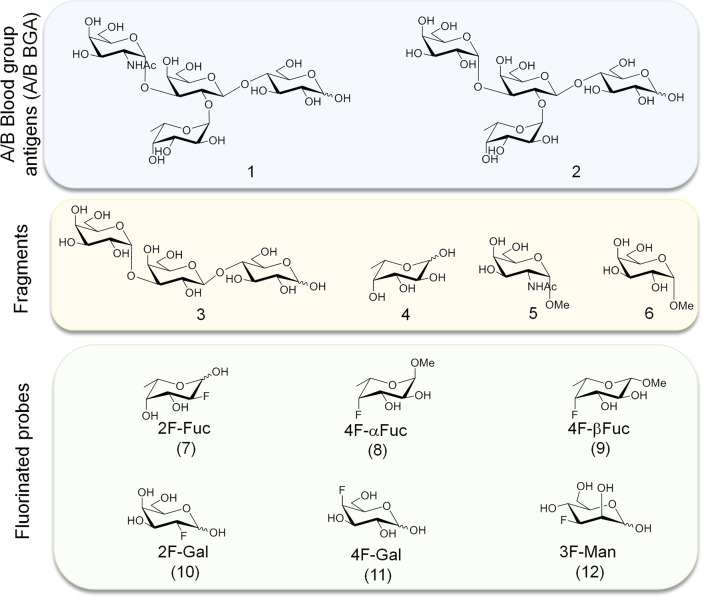
Chart 1. Glycan Structures Used in This Study: Histo Blood Group Antigens A and B Type VI Tetrasaccharides (1 and 2, Respectively); the Fragments: Galili-Type VI Trisaccharide (3), and the Monosaccharides Fuc (4), GalNAc-α-OMe (5) and Gal-α-OMe (6); and Monofluorinated Probes of Fuc (7, 8, 9), Gal (10, 11), and Man (12).
Results
Lectin Chemical Shift Perturbation Analysis
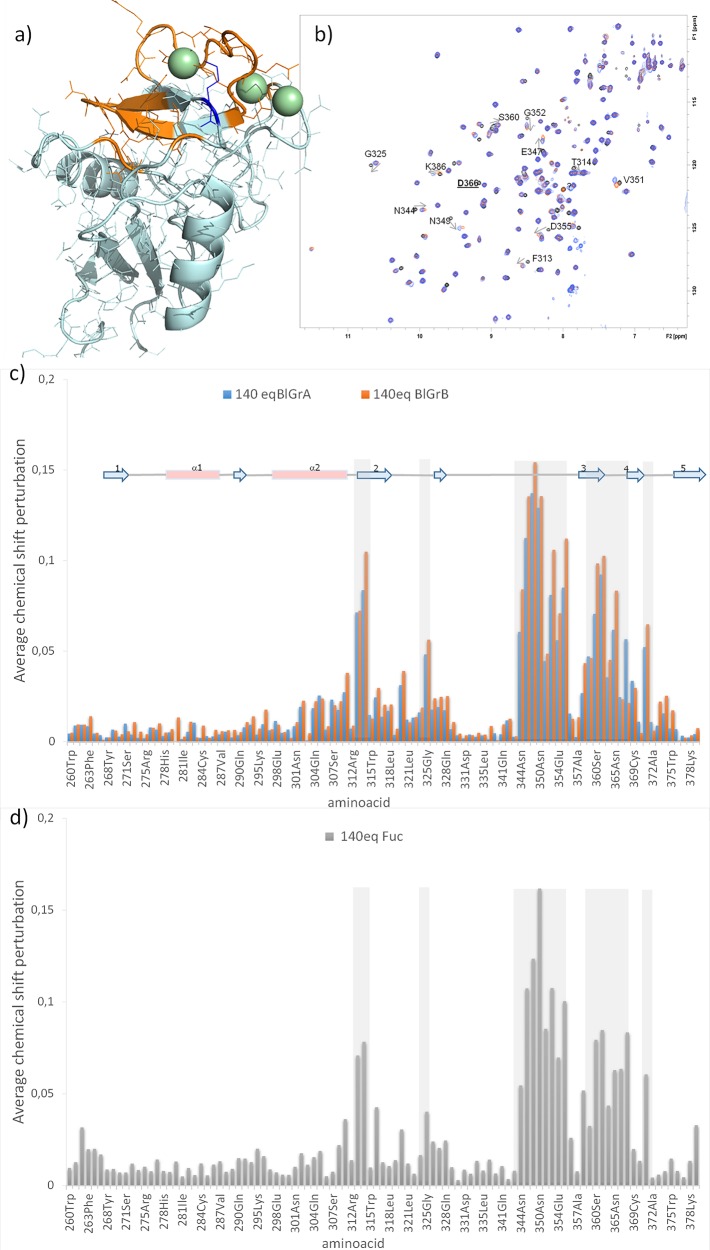
1H–15N HSQC titration experiments on 15N-labeled CRD DC-SIGN were performed to deduce the protein region involved in ligand binding and to estimate binding affinities. Data were acquired for the blood group tetrasaccharides (1 and 2), as well as for the monosaccharide Fuc (4). The addition of the tetrasaccharides produced very similar chemical shift perturbation profiles on the protein 1H–15N backbone resonances for the same number of equivalents (Figures 1a–c). As expected, the residues around the primary Ca2+ binding site—namely, the residues in the long loop and β-strands 3 and 4—were the ones affected the most. In addition, residues F313, T314, and L371, in the nearby loops were also perturbed (Figure 1a). Generally, the observed chemical shift perturbations were more pronounced for the B tetrasaccharide 2, with the only exception of K368, which was more affected in the presence of 1, the A antigen. The affected protein region is large enough to think about the existence of an extended interaction surface on the protein, where, besides the monosaccharide attached to the primary Ca2+ binding site, additional glycan residue(s) would establish further contacts with the protein, as reported for LeX. However, the chemical shift perturbation produced by the simple monosaccharide Fuc (Figure 1d) is strikingly similar to that for the tetrasaccharides (Figure 1c). In particular, no further residues are affected by the tetrasaccharides, compared to Fuc. This result suggests that the molecular recognition of the tetrasaccharides primarily involves the interaction with the Fuc moiety.
Figure 1.
Chemical shift perturbations for the interaction of CRD DC-SIGN with tetrasaccharides (1 and 2) and with Fuc (4). (a) In orange amino acids with chemical shifts most perturbed upon the addition of BGB and BGA. In blue K368, affected more with BGA than with BGB. (b) Superimposition of 1H–15N HSQC spectra (black, apo DC-SIGN; orange, in the presence of 140 equiv of BGB; blue, in the presence of 140 equiv of BGA). Some affected crosspeaks are annotated. Residue D366 that disappear in the middle points of the titration is underlined. (c, d) Average chemical shift perturbation upon the addition of BGA, BGB and Fuc. [D366 is not included in the plot; average chemical shift perturbations were calculated using the formula {1/2[δH2 + (0.2δN)2]}1/2, where δH and δN are the chemical shift change in 1H and 15N, respectively (in ppm), between the apo and bound forms.]
The titration curves allowed estimating the binding affinities. The best binder is 2 (KD = 2.3 ± 0.6 mM), followed by Fuc (KD = 4.1 ± 0.7 mM) and 1, which displays the weakest affinity (KD = 7.6 ± 1.4 mM). A detailed inspection of the titration curves highlighted some remarkable issues. In the case of Fuc, a nonlinear chemical shift perturbation was clear for certain residues (Figure S1 in the Supporting Information). For 2, although the nonlinear trajectories were not that obvious, however, the curve fitting (average chemical shift perturbation vs protein:ligand ratio) showed a step in the experimental data at ca. 1:50 lectin:ligand equivalents, which is an effect that is not observed for 1 (Figure S2 in the Supporting Information). At the same time, for both tetrasaccharides, some cross peaks diminish their intensity or disappear in the intermediate points of the titration, while others increase their intensity in the presence of the ligands (see Figure S1). Thus, although the recognition site on the lectin is basically the same for the three ligands, different dynamic processes occur, depending on the particular sugars.
Saturation-Transfer Difference (STD-NMR): The Ligand Point of View
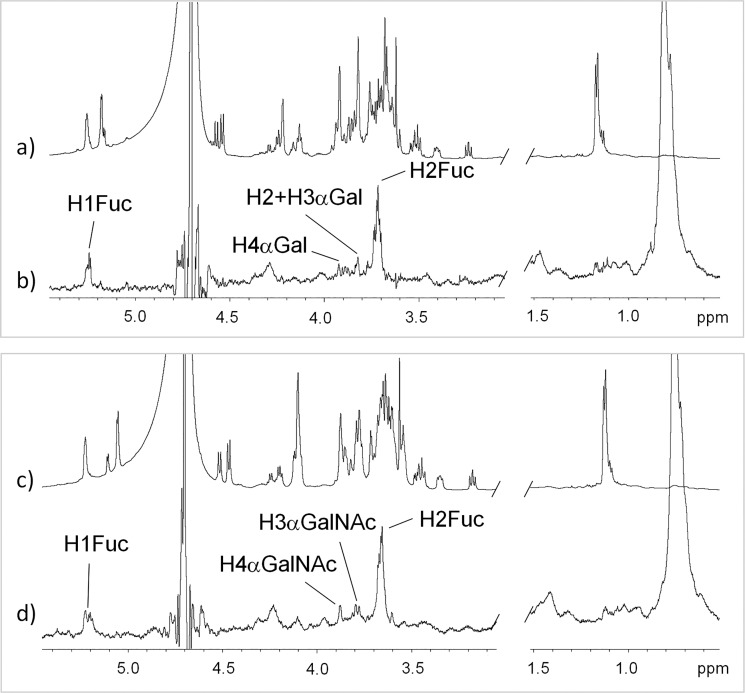
1H NMR STD experiments21 were performed to get information on the ligand epitopes for the different analogues (Figure 2). Higher STD intensities were obtained for protein irradiation in the aliphatic region, while temperature did not significantly affect the STD response. Because of the mutarrotation at the reducing-end residue (Glc), two separate resonances (for α and β anomers) are observed for some protons. The strongest STD was observed for Fuc H2, for both ligands 1 and 2. Weaker STD intensities were observed for Fuc H1, αGalNAc H4, and αGalNAc H3, for 1, and for Fuc H1, αGal H4, and αGal H3/H2 (overlapped) for 2. No STDs were observed neither for the Fuc methyl group, nor for any of the protons of the Glc reducing-end residue. These data indicate that both tetrasaccharides are presented in a similar way to interact with DC-SIGN, displaying Fuc H2 close to the protein aliphatic side chain(s).22
Figure 2.
1H STD-NMR experiments for the interaction of CRD DC-SIGN with (a, b) 2 and (c, d) 1. Spectra (a) and (c) are reference (off-resonance) spectra, whereas spectra (b) and (d) are STD spectra with on-resonance irradiation in the aliphatic region, using a 50 ms Gaussian pulse.
The Bound Ligand Conformation: Transferred NOESY
trNOESY experiments23 were acquired for 1/5 ratio samples of CRD DC-SIGN/tetrasaccharide and compared to the NOESY spectra of the free ligands. At 298 K and 600 MHz, the free ligands showed NOE effects close to zero, with both positive and negative NOEs and crosspeaks dominated by a double-quantum contribution, even at long mixing times. At 800 MHz, both tetrasaccharides exhibited weak negative NOE effect. In contrast, in the presence of the protein, all NOEs became strong and negative at either field, indicating that they are trNOEs.
The comparison of the interglycosidic ROEs/NOEs between the free and bound ligands indicated that the bound conformations remains the same as that existing in solution for both 1 and 2 tetrasaccharides. These branched glycans are known to be fairly rigid and they display a single very major conformation in solution, in which Fuc H1 and H2 are packed against the α-face of the αGal/αGalNAc residue. This is also the bound conformation in the complex with DC-SIGN, as confirmed by the key trNOEs between αGal/αGalNAc H1 with βGal H4 and H3, Fuc H1 with βGal H2 and H3 and αGal/αGalNAc H3 (Figure S3). However, the analysis of the trNOESY spectra revealed a foremost issue: intermolecular NOE crosspeaks between protein and ligand protons. As shown in Figures 3a and 3b, H1 Fuc (δ 5.26 ppm) has a clear NOE with protein signals at δ 0.77 ppm. Indeed, no ligand protons are present at that chemical shift and this cross peak is not present in the NOESY spectrum of the free protein (spectrum superimposed in blue in Figures 3a and 3b). Moreover, additional NOE crosspeaks between Gal H3α and Fuc H2 and the protein signals at δ 0.77 ppm were also observed, which again are not present in the NOESY spectrum of the free protein. This high field region (<1 ppm) is exclusive of the methyl protons of Val, Leu, and Ile residues. The herein employed CRD construct of DC-SIGN contains 3 Ile, 11 Leu, and 6 Val residues. Among those, only V351 is located at the primary Ca2+ binding site and expected to be close to the ligand. Thus, these protein signals should correspond to the Hγ of V351, which display intermolecular NOEs with Fuc H1 and H2 and with αGal H3. This assignment was confirmed by 1H–15N HSQC-TOCSY (Figure S4 in the Supporting Information). Interestingly, V351 has been reported to be essential for binding to one of the known endogenous glycoprotein receptors, ICAM-3. Fittingly, the V351G DC-SIGN mutant was unable to bind ICAM-3.24 The X-ray structure of the complex with LeX also highlights the key van der Waals contact.20
Figure 3.
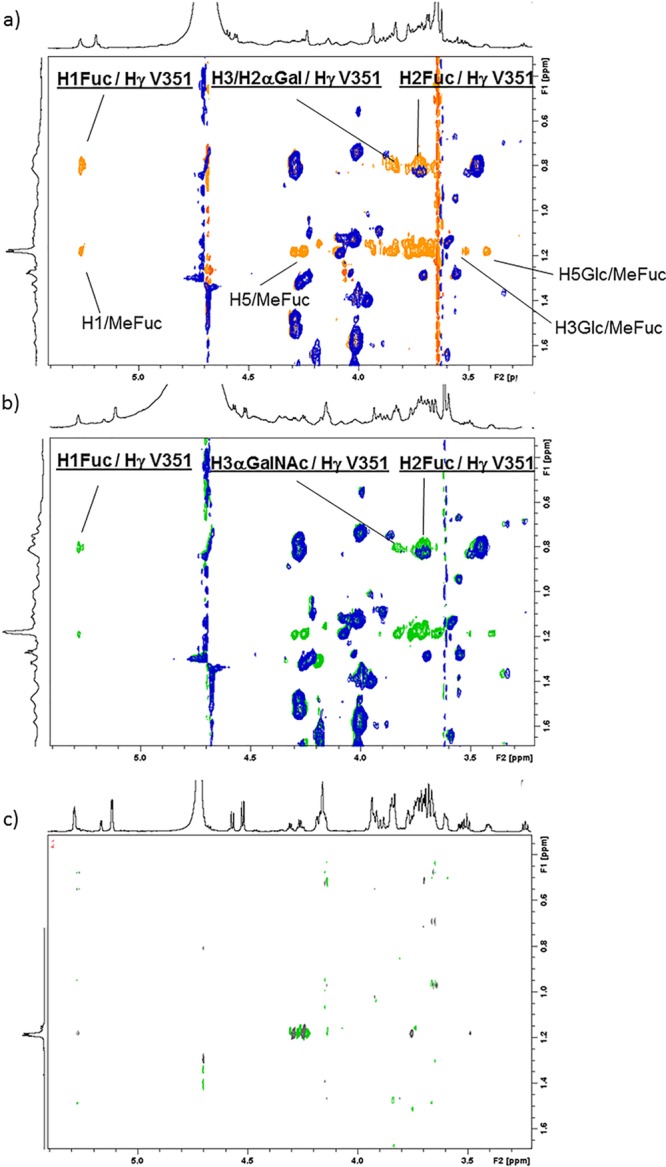
Quest of protein/ligand intermolecular NOEs. (a) Superimposition of: the NOESY of the CRD of DC-SIGN (blue), and the trNOESY of the complex of the CRD DC-SIGN with 2 (lectin/ligand molar ratio = 1/5, orange). The new crosspeaks in the orange spectrum correspond to intramolecular NOE of the ligand (all those from MeFuc, some of which are annotated) and intermolecular NOE (underlined). (b) Superimposition of the NOESY of the CRD of DC-SIGN (blue), and the trNOESY of the complex of the CRD DC-SIGN with 1 (lectin/ligand molar ratio = 1/5, green). (c) NOESY spectrum of tetrasaccharide 1. All spectra were acquired at 298 K at 800 MHz and with 400 ms of mixing time.
Monofluorinated Probes
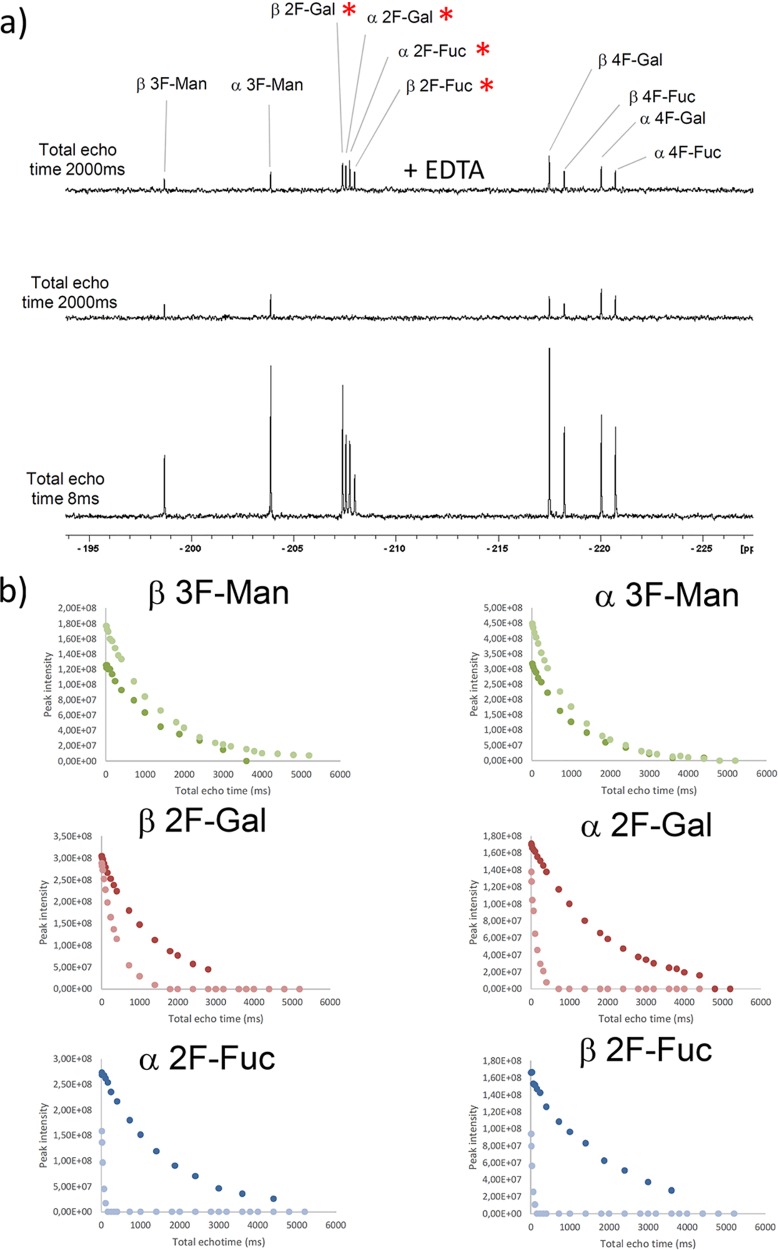
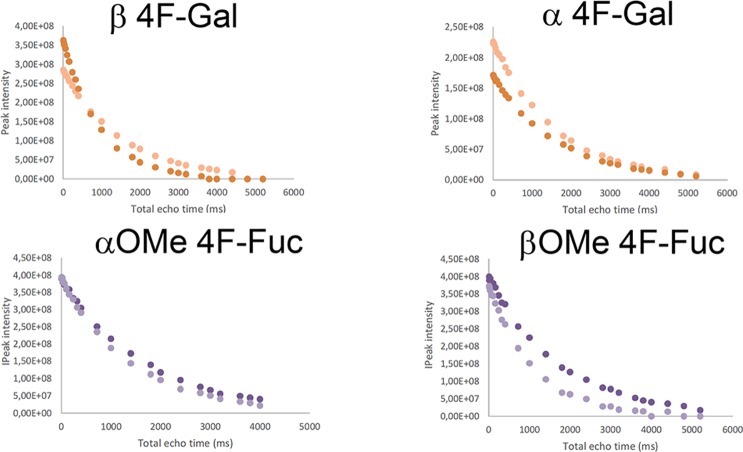
The substitution of a pyranose hydroxyl group by a fluorine atom has long been used to explore its participation in direct contacts with receptors.25,26 Since 19F NMR experiments are extremely useful to monitor glycan–lectin interactions,27 we have herein used a small library of simple monofluorinated monosaccharides and 19F CPMG experiments28 to define the hydroxyl pair coordination to the Ca2+ ion at the primary DC-SIGN binding site. In particular, we have employed 2-F-Fuc (7) and the α and β methyl glycosides of 4-F-Fuc (8 and 9), hypothesizing that the presence of fluorine at key positions of these molecules would abrogate the binding event through the 2-OH/3-OH and 3-OH/4-OH pairs, respectively. In addition, we have tested the corresponding Gal probes, 2-F-Gal and 4-F-Gal (10 and 11, respectively), and 3-F-Man (12) as negative control, since this molecule is completely unable to coordinate calcium in either way.29 As described, the variations in 19F transverse relaxation rates (R2), in the absence and presence of the lectin, can be used as a probe for detecting binding through simple 19F CPMG experiments. Figure 4a shows three NMR spectra (at the same scale) of the corresponding 19F CMPG experiments performed for a mixture of the monofluorinated compounds in the presence of CRD DC-SIGN. In the spectrum below, acquired with a total echo time of 8 ms, all the 19F NMR signals corresponding to every monosaccharide in the library are observed. In the spectrum in the middle (total echo time of 2000 ms), the 19F NMR signals for β2-F-Gal, α2-F-Gal, α2-F-Fuc, and β2-F-Fuc (highlighted with an asterisk) have disappeared, while those for 3F-Man, 4F-Gal, and 4F-Fuc remain. All the signals were recovered after the addition of EDTA (also see Figure S6 in the Supporting Information). Figure 4b gathers the complete 19F T2 decay for each species, in the absence and the presence of the lectin.
Figure 4.
19F CPMG experiments for the fluorinated monosaccharide small library. (a) The employed concentrations were 40 μM CRD DC-SIGN and 0.8 mM of each monosaccharide. [(Bottom) The first spectrum was acquired with 8 ms total echo time; (middle) the spectrum was acquired with 2000 ms echo time; (top) the spectrum with 2000 ms echo time after the addition of 20 mM EDTA-d12. The red asterisks highlight those 19F signals whose T2 values show a faster decay.] (b) Representation of the 19F T2 decay curves obtained for every species in the mixture, in the absence (dark color) and the presence of DC-SIGN (light color).
From these data, it becomes clear that 2-F-Fuc is a DC-SIGN binder (both α and β anomers), because of the pronounced change in the T2 curves (free and bound). Remarkably, neither α- nor β-OMe-4-F-Fuc bind DC-SIGN.
This result unambiguously indicates that Fuc binds DC-SIGN exclusively through 3-OH and 4-OH. Strikingly, the 19F T2 of 2-F-Gal, especially for the α-anomer, was importantly affected by the presence of the protein, indicating that αGal is also recognized by DC-SIGN. Thus, in this case, Ca2+ coordination also occur through 3-OH and 4-OH hydroxyls.
The Interaction with the Antigen Fragments
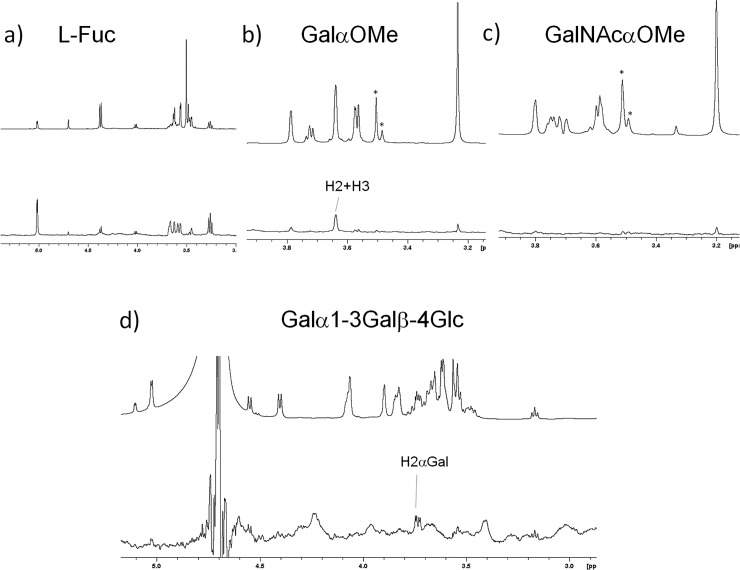
In order to further corroborate whether α-Gal is a ligand for DC-SIGN, we performed STD experiments with Galα-OMe (6), the Galα1–3Galβ1–4Glc trisaccharide (3) present in the B antigen, with GalNAcα-OMe (5) (present only in the A antigen), and l-Fuc (4) (Figure 5). In addition to the expected STD for Fuc, both GalαOMe and Galα1–3Galb1–4Glc showed an STD effect, which disappeared upon the addition of EDTA to the sample (see Figure S7 in the Supporting Information). In contrast, GalNAcαOMe showed no STD signals. For both αGal-containing moieties, exclusive STDs were observed for Gal H2. Although the Galβ anomer also showed some perturbation in the T2 19F relaxation, STD experiments with lactose (Galβ1–4Gc) (data not shown) showed no STD signal, in agreement with previous reports.
Figure 5.
1H NMR STD experiments with fragments of the blood group antigens: STD (below) and reference NMR spectra (top) recorded for the interaction of the CRD of DC-SIGN with (a) l-Fuc (4), (b) Galα-OMe (6), (c) GalNAcα-OMe (5), and (d) the Galα1–3Galβ1–4Glc trisaccharide (3).
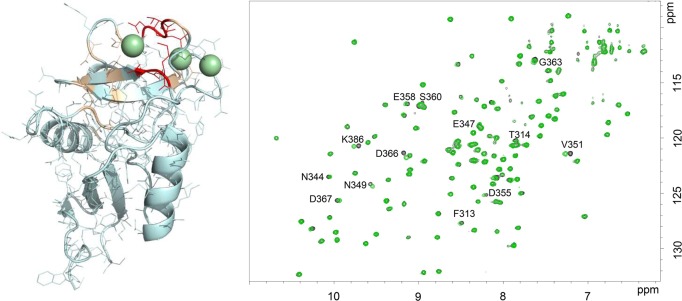
One additional proof of the interaction of trisaccharide 3 with DC-SIGN was obtained by monitoring the chemical shift perturbations of the lectin signals in the 1H–15N HSQC spectrum of the CRD DC-SIGN upon the addition of an excess of the trisaccharide. Figure 6 shows the superimposition of the spectra in the absence and in the presence of 40 equiv of 3. Although the chemical shift perturbations are smaller in magnitude, with respect to those observed for the interaction with simple Fuc and with the Fuc-containing glycans 1 and 2, specific crosspeaks were affected by the presence of the trisaccharide. Without being exhaustive, the affected region is essentially the same as for the interaction with ligands 1, 2, and 4, with a slightly different profile (see Figure S5 in the Supporting Information). In particular, N349, N350, V351, D366, and K368 were the most affected residues (also affected with ligands 1, 2, and 4). Thus, in agreement with the 19F-T2 relaxation and the 1H-STD experiments, the Galα moiety is indeed a ligand for DC-SIGN, although the interaction is weaker than that for the blood group A and B antigens, and for LeX.
Figure 6.
1H–15N HSQC spectra recorded for the CRD of DC-SIGN without ligand (black) and upon the addition of 40 equiv of compound 3 (green). On the left-hand side, the three-dimensional (3D) structure of the lectin where residues affected by the addition of 3 have been highlighted in wheat, and the most affected ones are shown in red.
Modeling
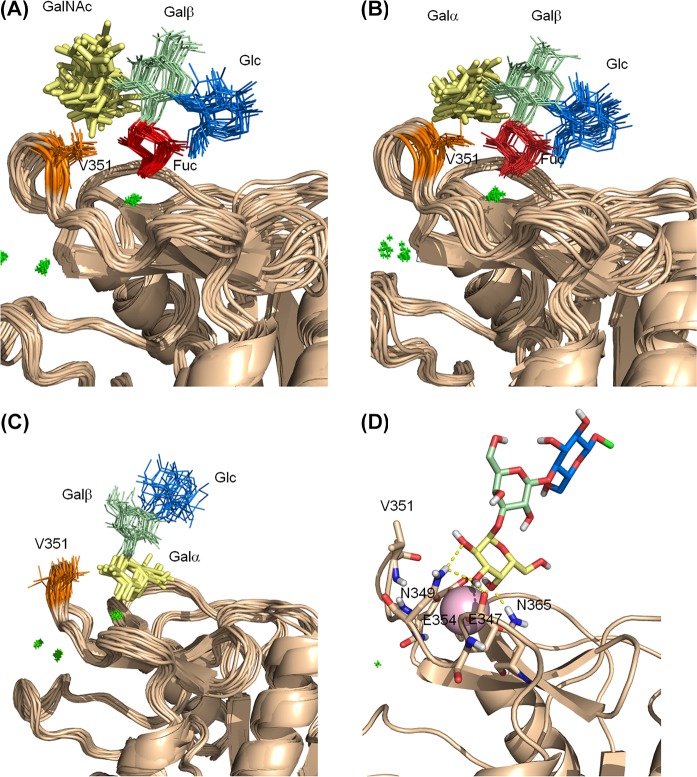
The analysis of the different experimental NMR data permitted us to gather key information for building structural models for the complexes of 1 and 2 with DC-SIGN. The trNOESY data strongly suggest that Fuc H2 is oriented toward Val351. The observed STD fully support this scenario, in which Fuc H2 shows, by far, the strongest STD response when the on-resonance irradiation frequency is set on the protein aliphatic region. At the same time, the 19F T2 experiments indicate that the Ca2+ coordination occurs through 3-OH and 4-OH of Fuc, and not using the 2-OH/3-OH pair. The Fuc binding mode found in the X-ray crystallographic structure of the complex DC-SIGN/LeX fully comply with these observations. In addition, the chemical shift perturbations on the protein backbone resonances are essentially the same for the simple monosaccharide Fuc and for the tetrasaccharides. Thus, a manual docking protocol was employed for fitting the Fucose residues of 1 and 2 in the binding site of DC-SIGN. The obtained starting geometries were minimized and subjected to MD simulations. Both complexes were stable along the entire 100 ns MD run. The analysis of the structural parameters for both complexes was essentially the same. (See Figure 7.)
Figure 7.
Structural models for the complexes of CRD DC-SIGN with tetrasaccharides 1 and 2, and trisaccharide 3 obtained from MD simulations: (A, B) superimposition of 20 frames extracted from the 100 ns MD simulations performed for the complexes with 1 (panel (A)) and 2 (panel (B)), where the binding to the calcium ion occurs through the Fuc residue; (C, D) the complex for the Galα1–3Galβ1–4Glc trisaccharide 3, as deduced from the corresponding MD simulations.
For the weak interaction with the αGal moiety, the NMR data indicate that Gal H2 is positioned close to V351, while the Ca2+ coordination occurs through the 3-OH/4-OH pair. Thus, the starting geometry of the Galα1–3Galβ1–4Glc trisaccharide bound to the DC-SIGN binding site was generated by compiling with these structural prerequisites. The complex was stable along a 200 ns MD simulation. The same geometry was then used for docking 2 into DC-SIGN, but with the terminal αGal in the primary binding site. Again, the complex was stable throughout the complete MD simulation (see Figure S8 in the Supporting Information).
Discussion
DC-SIGN can be considered as a paradigmatic case of glycan binding promiscuity. This behavior has been related to its different biological roles, which include both pathogen recognition and interaction with self-glycoproteins. In both cases, DC-SIGN is involved in the recognition of either pathogenic or self-glycans. How these different recognition processes are regulated at a molecular level is not fully understood. The understanding of the structural differences at the molecular level is an important piece of information. DC-SIGN is known to recognize certain fucosylated as well as high-mannose glycans, which implies a large number of structures present both in host and pathogenic cells. The atomic level structural information provided by X-ray crystallography has shown that Man or Fuc residues bind at the primary Ca2+ recognition site. Interestingly, Man has been shown to be recognized in two alternative binding modes,30 which would be in dynamic equilibrium in solution,31 and would contribute to enhance the affinity for high mannose N-glycans. For Fuc, a single binding mode has been reported, involving Ca2+ coordination through 3-OH and 4-OH, although langerin, which is a homologous lectin, recognizes Fuc through 2-OH and 3-OH. Avidity and multivalence have been reported to be important elements in the recognition events by DC-SIGN, where these phenomena highly increase the low affinities observed for the monosaccharides. These effects are presumably amplified for the full length form of DC-SIGN, which occurs when the neck domain is present, promoting DC-SIGN arrangement into tetramers.32
Herein, we have used the CRD of DC-SIGN for studying the molecular recognition in solution of the blood group A and B antigens. We show that BGB is recognized with an affinity similar to that for the LeX antigen, while the BGA binds slightly weakly. In both cases, the interaction is based on the attachment of Fuc, which coordinates the primary calcium exclusively with 3-OH and 4-OH. MD simulations show a stable hydrogen bonding network in which 4-OH act as an H-bond donor to E347 and acceptor from N350, 3-OH is donor to E376 and acceptor to N365, and 2-OH is donor to E354, in agreement with previous structures. In addition, occasional hydrogen bonds involve the terminal Galα residue, in particular 6-OH with NH of V351, and 6-OH or 2-OH with K368. Importantly, our experimental data highlights the significant role of V351, whose side chain perfectly packs against the hydrophobic cleft formed by the α-face of αGal and H1/H2 Fuc, confirming this interaction as a key contact for branched fucosylated oligosaccharides.
We demonstrate that, opposite to mannose, fucose binds DC-SIGN through a single binding mode.33 However, we found that the αGal epitope, which is present in the BGB, is also able to weakly bind DC-SIGN. Indeed, 1H-STD and 19F-R2 experiments indicate that αGal and the trisaccharide Galα1–3Galβ1–4Glc interact with the lectin. In contrast, αGalNAc, which is present in the BGA, does not bind DC-SIGN, probably due to the steric clash of the NHAc group on C2 precisely with the V351 side chain. Even though the affinity for this interaction is very low, it could be relevant in the biological context, where glycan density on cell surface can be very high. Surface glycan density, in fact, has been proposed to be one of the regulatory mechanisms of immunological lectins34 that could act as another level of regulation besides the loose preferences of DC-SIGN for individual glycan epitopes. In fact, this additional weak interaction for αGal may be at the origin of the differences observed in glycan array data, where the BGB was shown to bind DC-SIGN, regardless the chemical nature and length of the linker, while the BGA was shown to be binder only when presented with a long linker and not with a shorter one. In this way, longer linkers would make the main epitope, Fuc, fully accessible, and thus both BGB and BGA would bind. However, with shorter linkers, this key epitope could be hidden, and thus BGB could still bind through the αGal epitope, while BGA could not.
This work discloses fine atomic structural details for the interaction between the fucosylated A/B blood group self-antigens with DC-SIGN. Moreover, our findings highlight, once more, the promiscuity of this lectin at the molecular level, revealing a weak interaction also with the αGal, but not the αGalNAc epitope, which may have key implications in different immunological events from cancer16 to allergy.35
Materials and Methods
Glycans
Compounds 1–4 were purchased from Elycityl, compound 5 from Toronto Research Chemicals and compound 6 from Sigma. Fluorinated monosaccharides were purchased from Carbosynth, except for compounds 8 and 9 that were synthesized as described below.
Synthesis of the Methyl 4-Deoxy-4-fluorofucoside Anomers 8 and 9
The synthesis of 8 and 9 (Scheme 1) started from the partially protected methyl fucoside3613 by Lattrell-Dax inversion of the 4-OH group. Surprisingly, fluorination of 14 was unsuccessful, but after benzoate removal to methyl-α-l-quinovoside 15, selective fluorination proceeded in low yield on gram scale to give the desired methyl 4-deoxy-4-fluoro-α-l-fucoside 8 next to unreacted 15 (37%). The β-anomer was subsequently obtained as a mixture of anomers (α:β 2:1) by acid-catalyzed equilibration.
Scheme 1. Synthesis of 8 and 9.
Protein Expression and Purification
The DNA fragment coding for the CRD DC-SIGN fragment (H254-A404 with an additional His-Tag at the N-terminus) was inserted into a pET15b plasmid (Thermo Fischer Scientific), amplified in E. coli DH5α cells, and subsequently transformed into E. coli BL21/DE3 cells. The culture was grown overnight at 37 °C in an LB medium in the presence of ampicillin. When optical density reached 0.6 (600 nm), protein expression was induced with isopropyl-1-thio-β-d-galactopyranoside (IPTG) and the culture was allowed to grow for 3 h. For 13C,15N-labeled and 15N-labeled samples, a small preculture in LB grown overnight at 37 °C was transferred to M9 minimal medium containing 1 g/L 15NH4Cl and 20% w/v U–13C-glucose, or 1 g/L 15NH4Cl, respectively. The culture was harvested by centrifugation at 4500 rpm, and the final pellet was resuspended in lysis buffer (10 mM Tris-HCl pH 8) and sonicated at 4 °C. The inclusion bodies were recovered by ultracentrifugation at 30 000 rpm for 1 h. The pellet was then dissolved in Tris-HCl 100 mM (pH 8) with 6 M urea and 0.01% v/v β-mercaptoethanol. The residual insoluble fraction was sedimented by ultracentrifugation at 40 000 rpm × 2.5 h, and the supernatant was decanted. The protein was then refolded through subsequent dialysis against 4 M, 2 M, and no urea (in 100 mM Tris-HCl pH 8, 100 mM NaCl, 10 mM CaCl2, 0.01% (v/v) β-mercaptoethanol). Residual unfolded proteins were removed by ultracentrifugation, and the soluble folded protein was then purified through mannose-sepharose affinity chromatography and further purified by size exclusion chromatography in a HiLoad 16/60 Superdex 75 column eluting with 25 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH 8. Finally, the fractions containing the pure monomeric CRD were washed with 25 mM Tris-HCl, 150 mM NaCl, 4 mM CaCl2, 2 mM DTT, pH 8 to remove the EDTA, and concentrated using 10 MWCO membrane filters. For NMR experiments, the buffer was eventually changed to 25 mM Tris-d11, 150 mM NaCl, 4 mM CaCl2, 2 mM DTT-d10, in H2O/D2O 9:1 or D2O. The presence of the lectin was monitored by 4%–12% SDS-PAGE (band at 18–19 kDa) and the protein concentrations were quantified by the absorbance at 280 nm (ελ = 52 855 M–1 cm–1, estimated from ProtParam).
NMR Experiments
The backbone resonance assignment of the CRD DC-SIGN was performed at 37 °C on a 800 MHz Bruker spectrometer equipped with a cryoprobe, on a sample containing 200 μM of double-labeled protein in the presence of 30 mM d-mannose (133 equiv). 3D HNCO, HN(CA)CO, HN(CO)CA and HNCA, and HN(CO)CACB and HNCACB experiments were acquired and assigned. The entire analysis provided the unambiguous identification of 86% of the expected NH signals for the CRD. The spectra were processed with Bruker TopSpin 3.5 and analyzed via CARANMR 1.9.1. Most of the assignments agreed with those18 deposited in the Biological Magnetic Resonance Data Bank (BMRB 19931), with minor discrepancies. The assignment have been deposited in the BMRB with the accession code 27854.
1H–15N-HSQC-based were performed on 120 μM 15N-labeled CRD samples, at 37 °C. Six or ten points were recorded for each ligand. Averaged chemical shift perturbation (CSP) and dissociation constants (KD) were calculated using the CcpNmr Analysis 2.4.2 software.37
STD experiments were performed as described38 at 600 MHz, using standard Bruker pulse sequences without water suppression nor protein filter. Protein saturation was achieved with a Gaussian-shaped pulse of 50 ms at both the aliphatic and aromatic regions. Saturation build-up curves were obtained with saturation times from 0.5 to 3.5 s in steps of 0.5 s. Samples contained 60 μM of protein and 4.2 mM glycan (1:70 lectin/ligand) in buffered D2O (pD 8).
Transferred NOESY spectra were acquired at 600 and 800 MHz with 1/5 protein/tetrasaccharide ratio samples (183 μM of protein), with mixing times of 200, 400, 600, and 800 ms, at 298 K.
19F CPMG spectra were acquired in a 600 MHz spectrometer equipped with a Bruker selective 19F–1H decoupling (SEF) probe at 298 K, on samples containing 40 uM of protein and 0.8 mM of the fluorinated probes. The standard CPMG Bruker pulse sequence was modified as described.39 Twenty four (24) points were acquired with total echo times from 8 to 5200 ms, with τ = 2 ms. Data were analyzed with the T1T2 relaxation module of Topspin3.5.
Modeling
MD simulations were run using Amber 12.40 The initial pdb coordinates for CRD DC-SIGN were derived from the crystal structure Protein Database (PDB) 1SL5. The magnesium ion was replaced by calcium, and the desired glycan manually docked into the binding site. The glycan structures were built in the Glycam web. The complexes were prepared in explicit water (TIP3P model) and minimized in two steps before starting the simulations. ff99SB and GLYCAM_06h force fields were used for treating the protein and the ligands, respectively. MD simulations of 100 or 200 ns were analyzed with cpptraj.
Acknowledgments
We thank Agencia Estatal de Investigacion of Spain and the European Research Council for financial support. B.L., J.-B.V., and J.M. thank the Industrial Biotechnology Catalyst (Innovate UK, BBSRC, EPSRC, BB/M028941/1) and the EPSRC (core capability EP/K039466/1) for funding. We also thank J. F. Espinosa (Lilly Research Laboratories, Alcobendas, Spain), for providing the modified NMR pulse sequence for the CPMG experiments.
Glossary
ABBREVIATIONS
- BGA
Blood group A type 6
- BGB
Blood group B type 6
- CLR
C-type lectin receptor
- CPMG
Carr–Purcell–Meiboom–Gill sequence experiment
- CRD
carbohydrate recognition domain
- DC
dendritic cell
- DC-SIGN
dendritic cell-specific ICAM-3 grabbing nonintegrin
- Fuc
l-fucopyranose
- Gal
d-galactopyranose
- Glc
d-glucopyranose
- GlcNAc
N-acetyl-2-d-glucosaminopyranose
- HIV
human immunodeficiency virus
- HSQC
heteronuclear single quantum spectroscopy
- ICAM-3
intracellular adhesion molecule 3
- LeX
Lewis X trisaccharide
- MD
molecular dynamics
- NOE
nuclear Overhauser effect
- NOESY
nuclear Overhauser effect spectroscopy
- PRR
pathogen recognition receptor
- ROE
rotating-frame overhauser enhancement
- STD
saturation transfer difference
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.9b00458.
Figures S1–S8, detailed synthetic procedures and characterization data (PDF)
Author Contributions
The manuscript was written through contributions of all authors.
Agencia Estatal de Investigacion of Spain: Grant Nos. CTQ2015-64597-C2-1P (to J.J.-B.), CTQ2015-64597-C2-2P (to F.J.C.), SEV-2016-0644 Severo Ochoa Excellence Accreditation (to CIC bioGUNE), Ramón y Cajal fellowship RYC-2014–15969 (to A.A.), and FPU14/10147 (to P.V.). European Research Council (RECGLYCANMR, Advanced Grant No. 788143, to J.J.-B.). Industrial Biotechnology Catalyst (Innovate UK, BBSRC, EPSRC, BB/M028941/1); EPSRC (No. EP/K039466/1).
The authors declare no competing financial interest.
Supplementary Material
References
- Rodriguez E.; Schetters S. T. T.; van Kooyk Y. (2018) The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat. Rev. Immunol. 18, 204–211. 10.1038/nri.2018.3. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek T. B.; den Dunnen J.; Gringhuis S. I. (2009) Pathogen recognition by DC-SIGN shapes adaptive immunity. Future Microbiol. 4, 879–890. 10.2217/fmb.09.51. [DOI] [PubMed] [Google Scholar]
- van Kooyk Y.; Appelmelk B.; Geijtenbeek T. B. H. (2003) A fatal attraction: Mycobacterium tuberculosis and HIV-1 target DC-SIGN to escape immune surveillance. Trends Mol. Med. 9, 153–159. 10.1016/S1471-4914(03)00027-3. [DOI] [PubMed] [Google Scholar]
- Pitarque S.; Herrmann J. L.; Duteyrat J. L.; Jackson M.; Stewart G. R.; Lecointe F.; Payre B.; Schwartz O.; Young D. B.; Marchal G.; Lagrange P. H.; Puzo G.; Gicquel B.; Nigou J.; Neyrolles O. (2005) Deciphering the molecular bases of Mycobacterium tuberculosis binding to the lectin DC-SIGN reveals an underestimated complexity. Biochem. J. 392, 615–624. 10.1042/BJ20050709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambi A.; Netea M. G.; Mora-Montes H. M.; Gow N. A.; Hato S. V.; Lowman D. W.; Kullberg B. J.; Torensma R.; Williams D. L.; Figdor C. G. (2008) Dendritic cell interaction with Candida albicans critically depends on N-linked mannan. J. Biol. Chem. 283, 20590–20599. 10.1074/jbc.M709334200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparrós E.; Serrano D.; Puig-Kröger A.; Riol L.; Lasala F.; Martinez I.; Vidal-Vanaclocha F.; Delgado R.; Rodríguez-Fernández J. L.; Rivas L.; Corbí A. L.; Colmenares M. (2005) Role of the C-type lectins DC-SIGN and L-SIGN in Leishmania interaction with host phagocytes. Immunobiology 210, 185–193. 10.1016/j.imbio.2005.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P.; Ridilla M.; Patel P.; Betts L.; Gallichotte E.; Shahidi L.; Thompson N. L.; Jacobson K. (2017) Beyond attachment: Roles of DC-SIGN in dengue virus infection. Traffic 18, 218–231. 10.1111/tra.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez C. P.; Lasala F.; Carrillo J.; Muñiz Ó.; Corbí A. L.; Delgado R. (2002) C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 76, 6841–6844. 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Vallejo J. J.; van Kooyk Y. (2013) The physiological role of DC-SIGN: a tale of mice and men. Trends Immunol. 34, 482–486. 10.1016/j.it.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Guo Y.; Feinberg H.; Conroy E.; Mitchell D. A.; Alvarez R.; Blixt O.; Taylor M. E.; Weis W. I.; Drickamer K. (2004) Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat. Struct. Mol. Biol. 11, 591–598. 10.1038/nsmb784. [DOI] [PubMed] [Google Scholar]
- van Liempt E.; Bank C. M.; Mehta P.; Garcia-Vallejo J. J.; Kawar Z. S.; Geyer R.; Alvarez R. A.; Cummings R. D.; Kooyk Y.; van Die I. (2006) Specificity of DC-SIGN for mannose- and fucose-containing glycans. FEBS Lett. 580, 6123–613. 10.1016/j.febslet.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Holla A.; Skerra A. (2011) Comparative analysis reveals selective recognition of glycans by the dendritic cell receptors DC-SIGN and Langerin. Protein Eng., Des. Sel. 24 (9), 659–669. 10.1093/protein/gzr016. [DOI] [PubMed] [Google Scholar]
- Brzezicka K.; Echeverria B.; Serna S.; van Diepen A.; Hokke C. H.; Reichardt N. C. (2015) Synthesis and microarray-assisted binding studies of core xylose and fucose containing N-glycans. ACS Chem. Biol. 10, 1290–302. 10.1021/cb501023u. [DOI] [PubMed] [Google Scholar]
- Gimeno A.; Reichardt N. C.; Cañada F. J.; Perkams L.; Unverzagt C.; Jiménez-Barbero J.; Ardá A. (2017) NMR and Molecular Recognition of N-Glycans: Remote Modifications of the Saccharide Chain Modulate Binding Features. ACS Chem. Biol. 12, 1104–1112. 10.1021/acschembio.6b01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoevska V.; Nollau P.; Lucka L.; Grunow D.; Klampe B.; Uotila L. M.; Samsen A.; Gahmberg C. G.; Wagener C. (2007) DC-SIGN binds ICAM-3 isolated from peripheral human leukocytes through Lewis x residues. Glycobiology 17, 324–333. 10.1093/glycob/cwl073. [DOI] [PubMed] [Google Scholar]
- Chen J.-T.; Chen C.-H.; Ku K.-L.; Hsiao M.; Chiang C.-P.; Hsu T.-L.; Chen M.-H.; Wong C.-H. (2015) Glycoprotein B7-H3 overexpression and aberrant glycosylation in oral cancer and immune response. Proc. Natl. Acad. Sci. U. S. A. 112, 13057–62. 10.1073/pnas.1516991112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda A.; Jimenez-Barbero J. (2018) The recognition of glycans by protein receptors. Insights from NMR spectroscopy. Chem. Commun. 54, 4761–476. 10.1039/C8CC01444B. [DOI] [PubMed] [Google Scholar]
- Pederson K.; Mitchell D. A.; Prestegard J. H. (2014) Structural characterization of the DC-SIGN-Lewis(X) complex. Biochemistry 53, 5700–5709. 10.1021/bi5005014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatwell L.; Holla A.; Kaufer B. B.; Skerra A. (2008) The carbohydrate recognition domain of Langerin reveals high structural similarity with the one of DC-SIGN but an additional, calcium-independent sugar-binding site. Mol. Immunol. 45, 1981–94. 10.1016/j.molimm.2007.10.030. [DOI] [PubMed] [Google Scholar]
- Feinberg H.; Taylor M. E.; Razi N.; McBride R.; Knirel Y. A.; Graham S. A.; Drickamer K.; Weis W. I. (2011) Structural basis for langerin recognition of diverse pathogen and mammalian glycans through a single binding site. J. Mol. Biol. 405, 1027–39. 10.1016/j.jmb.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.; Meyer B. (1999) Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew. Chem., Int. Ed. 38, 1784–1787. . [DOI] [PubMed] [Google Scholar]
- Monaco S.; Tailford L. E.; Juge N.; Angulo J. (2017) Angulo, Differential epitope mapping by STD NMR spectroscopy to reveal the nature of protein-ligand contacts. Angew. Chem., Int. Ed. 56, 15289–15293. 10.1002/anie.201707682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti R.; Perez S.; Arda A.; Imberty A.; Jimenez-Barbero J.; Silipo A.; Molinaro A. (2016) ″Rules of Engagement″ of protein-glycoconjugate interactions: a molecular view achievable by using NMR spectroscopy and molecular modeling. ChemistryOpen 5, 274–279. 10.1002/open.201600024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geijtenbeek T. B.; van Duijnhoven G. C.; van Vliet S. J.; Krieger E.; Vriend G.; Figdor C. G.; van Kooyk Y. (2002) Identification of different binding sites in the dendritic cell-specific receptor DC-SIGN for intercellular adhesion molecule 3 and HIV-1. J. Biol. Chem. 277, 11314–11320. 10.1074/jbc.M111532200. [DOI] [PubMed] [Google Scholar]
- Lemieux R. U. (1989) Rhône-Poulenc Lecture. The origin of the specificity in the recognition of oligosaccharides by proteins. Chem. Soc. Rev. 18, 347–374. 10.1039/CS9891800347. [DOI] [Google Scholar]
- Solís D.; Fernández P.; Díaz-Mauriño T.; Jiménez-Barbero J.; Martín-Lomas M. (1993) Hydrogen-bonding pattern of methyl beta-lactoside binding to the Ricinus communis lectins. Eur. J. Biochem. 214, 677–683. 10.1111/j.1432-1033.1993.tb17968.x. [DOI] [PubMed] [Google Scholar]
- Unione L.; Alcalá M.; Echeverria B.; Serna S.; Ardá A.; Franconetti A.; Cañada F. J.; Diercks T.; Reichardt N.; Jiménez-Barbero J. (2017) Fluoroacetamide moieties as NMR spectroscopy probes for the molecular recognition of GlcNAc-containing sugars: modulation of the CH-π stacking interactions by different fluorination patterns. Chem. - Eur. J. 23, 3957–3965. 10.1002/chem.201605573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalvit C.; Piotto M. (2017) 19F NMR transverse and longitudinal relaxation filter experiments for screening: a theoretical and experimental analysis. Magn. Reson. Chem. 55, 106–114. 10.1002/mrc.4500. [DOI] [PubMed] [Google Scholar]
- Note that, except for compounds 9 and 10, all of the other monosaccharides undergo mutarrotation in solution, and will thus exist as α- and β-anomers, therefore providing two fluorine signals in the 19F NMR spectrum.
- Feinberg H.; Castelli R.; Drickamer K.; Seeberger P. H.; Weis W. I. (2006) Multiple modes of binding enhance the affinity of DC-SIGN for high mannose N-linked glycans found on viral glycoproteins. J. Biol. Chem. 282, 4202–4209. 10.1074/jbc.M609689200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi C.; Alfarano P.; Sutkeviciute I.; Sattin S.; Ribeiro-Viana R.; Fieschi F.; Bernardi A.; Weiser J.; Rojo J.; Angulo J.; Nieto P. M. (2016) Detection and quantitative analysis of two independent binding modes of a small ligand responsible for DC-SIGN clustering. Org. Biomol. Chem. 14, 335–344. 10.1039/C5OB02025E. [DOI] [PubMed] [Google Scholar]
- Mitchell D. A.; Fadden A. J.; Drickamer K. (2001) Subunit organization and binding to multivalent ligands. J. Biol. Chem. 276, 28939–28945. 10.1074/jbc.M104565200. [DOI] [PubMed] [Google Scholar]
- This is true for glycosylated fucose, where OH1 is not available for calcium coordination. However, our experiments suggest that free Fuc can also weakly bind DC-SIGN through OH1 and OH2, and this is probably the reason for the nonlinear trajectory of the chemical shift perturbations in the DC-SIGN titration with Fuc.
- Dam T. K.; Brewer F. C. (2010) Lectins as pattern recognition molecules: the effects of epitope density in innate immunity. Glycobiology 20 (9), 1061–1064. 10.1093/glycob/cwq084. [DOI] [PubMed] [Google Scholar]
- Wolver S. E.; Sun D. R.; Commins S. P.; Schwartz L. B. (2013) A peculiar cause of anaphylaxis: no more steak? The journey to discovery of a newly recognized allergy to galactose-alpha-1,3-galactose found in mammalian meat. J. Gen. Int. Med. 28, 322–325. 10.1007/s11606-012-2144-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindhorst T. K.; Thiem J. (1991) Synthesis of 4-deoxy and 4-deoxy-4-halogeno derivatives of L-fucose as potential enzyme inhibitors. Carbohydr. Res. 209, 119–129. 10.1016/0008-6215(91)80150-L. [DOI] [Google Scholar]
- Vranken W. F.; Boucher W.; Stevens T. J.; Fogh R. H.; Pajon A.; Llinas M.; Ulrich E. L.; Markley J. L.; Ionides J.; Laue E. D. (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins: Struct., Funct., Genet. 59, 687–696. 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Santarsia S.; Grosso A. S.; Trovão F.; Jiménez-Barbero J.; Carvalho A. L.; Nativi C.; Marcelo F. (2018) Molecular recognition of a Thomsen-Friedenreich antigen mimetic targeting human galectin-3. ChemMedChem 13, 2030–2036. 10.1002/cmdc.201800525. [DOI] [PubMed] [Google Scholar]
- Urick A. K.; Calle L. P.; Espinosa J. F.; Hu H.; Pomerantz W. C. (2016) Protein-observed fluorine NMR is a complementary ligand discovery method to 1H CPMG ligand-observed NMR. ACS Chem. Biol. 11, 3154–3164. 10.1021/acschembio.6b00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D. A., Darden T. A., Cheatham T. E. III, Simmerling C. L., Wang J., Duke R. E., Luo R., Walker R. C., Zhang W., Merz K. M., Roberts B., Hayik S., Roitberg A., Seabra G., Swails J., Götz A. W., Kolossváry I., Wong K. F., Paesani F., Vanicek J., Wolf R. M., Liu J., Wu X., Brozell S. R., Steinbrecher T., Gohlke H., Cai Q., Ye X., Wang J., Hsieh M.-J., Cui G., Roe D. R., Mathews D. H., Seetin M. G., Salomon-Ferrer R., Sagui C., Babin V., Luchko T., Gusarov S., Kovalenko A., and Kollman P. A. (2012), AMBER 12; University of California: San Francisco, CA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.